Abstract
Background
Hepatocellular carcinoma (HCC) is known to be the second leading cause of cancer-related mortality worldwide. For improving the prognosis as well as reducing the rate of mortality, early diagnosis of HCC is a must.
Aims
This study was conducted to assess the ability of the serum expression of exosomal miR-18a and miR-222 to differentiate and diagnose patients with HCC, patients with liver cirrhosis, and healthy controls.
Methods
This study included 51 patients with liver cirrhosis, 51 patients with HCC on top of hepatitis C virus (HCV) infection, and 50 healthy controls.
Results
miR-18a and miR-222 were assessed using reverse transcription-polymerase chain reaction. MiR-18a and miR-222 levels were significantly higher in the liver cirrhosis and HCC groups than the control group (p ˂ 0.001). However, no statistically significant difference was found between patients with HCC and liver cirrhosis (p = 0.4 for miR-18a and p = 0.1 for miR-222). ROC curve analyses to evaluate the diagnostic performances of the two miRNAs as important noninvasive diagnostic markers revealed a best cutoff value of 2 for miR-18a to differentiate between liver cirrhosis, HCC, and healthy controls. And for mir-222, a cutoff value of 1.7 and 1.9 showed the highest specificity for discrimination between liver cirrhosis, HCC, and healthy controls, respectively. Moreover, logistic regression model revealed that miR-18a expression was independent predictive factor in HCC patients (p = 0.004), while miR-222 expression was independent predictive factor in liver cirrhosis patients (p < 0.001).
Conclusion
miR-18a and miR-222 were significantly discriminative markers between patients with liver cirrhosis and HCC and healthy individuals. Therefore, they have a prognostic rather than a diagnostic value. Moreover, miR-18a and miR-222 could be useful in identifying liver injuries, including fibrosis and cirrhosis.
Keywords: Hepatitis C virus, hepatocellular carcinoma, reverse transcription-polymerase chain reaction, miRNAs
Introduction
Hepatitis C virus (HCV) infection has the highest prevalence worldwide and is one of the major public health problems in Egypt. 1 HCV infection and its long-term complications are responsible for more than 67% of liver-related morbidities in Egypt. 2 Cirrhosis and subsequent hepatocellular carcinoma (HCC) are common sequelae of chronic HCV infection. El Ghoroury et al. reported that HCC accounts for 13% of Egyptian cancers. Moreover, 94% of HCC cases were preceded by chronic HCV infection. 3
HCC is the sixth most common cancer and the second cause of cancer-related mortality worldwide. 4 Studies have reported that the prevalence of HCC, which is mainly virally mediated, is increasing; however, currently available antiviral therapies for hepatitis B virus and HCV infections might help in controlling its increasing prevalence. 5 The heterogeneous nature of the tumor and late diagnosis have triggered the need for novel biomarkers for early diagnosis and new therapeutic targets to improve the prognosis of HCC. 6 Moreover, nearly 60% of patients develop recurrence or even distant metastasis after treatment. Therefore, the establishment of an effective method to predict the prognosis of HCC is warranted. 7 Nowadays, new genetic biomarkers, such as circulating microRNAs in peripheral blood, have been developed and become the focus of early diagnosis of HCC. 8
Exosomes are membrane-bound vesicles ranging in size from 40 nm to 150 nm. They transport DNA, RNA, extracellular matrix proteins, lipids, receptors, transcription factors, and enzymes to various sites in the body for performing various functions. Tumor cell-derived exosomes play an important role in the pathogenesis of cancer cells by supporting their invasion and metastasis. They transmit the tumor signals that determine the direction of metastasis and promote epithelial–mesenchymal transformation and angiogenesis. 9 Moreover, they are involved in the progression of liver fibrosis, viral hepatitis, alcoholic liver disease, and various precancerous liver diseases. 10
Previous studies have revealed that variable excess amounts of exosomes are released by different cancer cells.11,12 Thus, exosomes seem to be useful and specific biomarkers that can be used for the early diagnosis of tumors and/or the detection of tumor metastasis. In addition, exosomes are protected by cell membranes, are non-degradable in nature, and carry large amounts of tumor cell information, making them the best biomarkers for noninvasive diagnosis. 12
MicroRNAs (miRNAs) are short non-coding RNAs that inhibit target genes by regulating post-transcriptional gene expression within the genome. The binding of the miRNAs to the 3′ untranslated region within the messenger RNA (mRNA) results in either mRNA degradation or protein translation inhibition. miRNAs target many mRNAs involved in several biological processes, such as the cell cycle, apoptosis, metabolism, development, and differentiation. Circulatory miRNAs exist in body fluids and are highly resistant to degradation by RNases. 13
Several miRNAs are associated with impaired inflammatory signaling pathways that stimulate hepatic stellate cells and result in liver fibrosis. Dysregulation of miRNAs is observed in various cancer types, such as HCC, wherein tumor suppressor miRNAs that target oncogenes are downregulated and onco-miRNA that target tumor suppressor genes are upregulated. Established carcinogenesis-related transcription factors suppress some miRNAs, such as MYC, whereas other miRNA changes occur via epigenetic regulation owing to DNA methylation and histone modifications. In addition, some miRNA processing machinery genes, such as DROSHA, TRBP, and AGO2, are lost, resulting in a reduction of mature miRNA synthesis and HCC development. 14
Accordingly, miRNAs are reported as a novel serum biomarker that can be used to diagnose HCC. 15 Moreover, they are tissue-specific, reflecting a reliable correlation with liver cirrhosis and HCV-mediated HCC pathogenesis, and can be used as noninvasive biomarkers for the disgnosis of HCC. 16 Many circulatory miRNA serum levels, including those of miR-320c, miR-198, miR-134, miR-483-5p, miR-92a, and miR-20a, were reportedly increased in HCV and HCV-mediated fibrosis. miR-16 serum levels have been proposed as conventional biomarkers in the diagnosis of HCC. 17
miR-18a is strongly associated with oncogenesis and plays an important role in cancer development, that is, it either promotes oncogenesis or inhibits the progression of malignancy by overexpressing or targeting the factors involved in different pathways. Considering this observation, miR-18a should be extensively studied as a potential biomarker for cancer diagnosis. 18 miR-18a was found to promote the progression of mesothelioma as well as lung, cervical, prostate, and gastric cancers; inhibit the progression of colorectal and breast cancers; and reduce pancreatic progenitor cell proliferation. These effects are attributable to the regulation of downstream targets involved in cancer development, such as interferon regulatory factor 2 in non-small-cell lung carcinoma. In cervical carcinoma, miR-18a was associated with the upregulation of PD-L1 expression, which is involved in cancer progression. 18
The mechanism of miR-18a in HCC progression is still unclear. A study by Liu et al. suggested that miR-18a promotes the proliferation and progression of HCC cells by targeting KLF4, which has a tumor-suppressive effect, and its downstream P21, which is a negative regulator of the cell cycle. 19
MiR-222 plays an important role in inflammation and fibrosis; its upregulation denotes fibrotic progression in animal studies. 20 High levels of miR-222 were detected in activated mouse hepatic stellate cells, which indicated fibrotic progression in animal models. 20 A homologous miRNA to miR-222 is miR-221 (miR-221/222), which is significantly overexpressed in several diseases. 21 They were found to be highly deregulated in HCC tissues. 14 miR-221 targets many key tumor suppressors, such as CDKN1B/p27, CDKN1C/p57, PTEN, TIMP3, and DDIT4.
Upregulated serum miR-222 and miR-18a levels have been described in patients with liver cirrhosis following HCV infection and liver cancer compared with healthy individuals.22,23 Many studies highlight the expression levels of miR-222 and their correlation with disease progression in Egyptian patients. Abo ElAtta et al. reported a significant increase in miR-222 expression levels in rheumatoid arthritis. 24 Ahmed et al. found that miR-222 was significantly upregulated in inflammatory breast cancer. 25 Among liver diseases, El-Guendy et al. found that miR-222 expression levels were increased in severe chronic HCV infection. 26 Further, Motawi et al. found that miR-222 serum levels were significantly elevated in patients with HCV but not in patients with liver cancer, compared with controls. 20 In contrast, Ali et al. revealed that the median serum levels of miR-222 were significantly reduced in HCV compared with those in controls and significantly elevated in HCC samples compared with those in controls. 23 However, Atef et al. found that miR-222 was significantly upregulated in the sera of HCC cases and HCV-infected patients compared with that in the sera of controls. 27
In terms of miR-18a testing in the Egyptian population, Ibrahim et al. found that overexpressed miR-18a could be a noninvasive biomarker for prostate cancer (PC) screening and help in the differentiation of PC from benign prostatic hyperplasia. 28 Moreover, plasma expressions of miR-18a significantly correlated with positive nodal metastasis. 29 Zekri et al. stated that miR-18a was significantly upregulated in patients with colonic polyps but not in those with inflammatory bowel disease. 30 Khalifa et al. also found that miR-18a was overexpressed in patients with chronic lymphocytic leukemia compared to controls. 31 To the best of our knowledge, ours was the first study that evaluated the expression level of miR-18a in HCC.
This study aimed to assess the usefulness of exosomal serum miR-18a and miR-222 expression in differentiating between and diagnosing patients with hepatic cirrhosis and HCC following HCV infection compared to healthy individuals and to correlate this expression with other laboratory findings.
Material and methods
This case-control study included 152 subjects from the outpatient clinics of the National Research Centre and the National Cancer Institute in Cairo from June 2017 to March 2019. They were divided into three groups:
Group I
included 51 patients newly diagnosed with early-stage HCC following HCV infection.
Group II
included 51 patients with liver cirrhosis following HCV infection.
Group III
(control group) included 50 healthy subjects.
The mean size of the lesions in patients with HCC was approximately 2 cm or smaller and was detected on magnetic resonance imaging and four-phase computed tomography (CT). HCC was diagnosed based on the presence of typical histological findings or imaging characteristics according to the guidelines of the Korean Liver Cancer Study Group and the American Association for the Study of Liver Diseases. 32 HCC staging was determined using the Barcelona Clinic Liver Cancer staging system. 33 Liver cirrhosis was diagnosed based on histological examination and/or clinical findings of portal hypertension. 34 Confirmed cases underwent repeated imaging studies within 6 months of enrollment to exclude HCC.
Data from all the subjects were obtained from personal interviews, medical records, and pathology reports. The exclusion criteria were presence of a positive hepatitis B surface antigen, extrahepatic dissemination, associated malignancies other than HCC, or any other systemic or chronic disease; a history of causes for congenital or chronic liver diseases other than HCV; and previous treatment for HCC or antiviral therapy for HCV.
The 50 healthy controls enrolled in this study sought a routine health checkup in the National Research Centre clinics. They met the following requirements: being seronegative for alpha-fetoprotein (AFP) with no evidence of any liver disease, which was confirmed based on a qualitative anti-HCV antibody test, nor any associated malignancy or systemic or chronic disease.
The study protocol was approved by the Ethics Committee of the National Research Centre (ethical clearance number is 16,291), and the study was conducted according to the guidelines of the Declaration of Helsinki. Each participant provided written informed consent.
Methodology
All patients and controls were subjected to complete clinical assessment and laboratory investigations including the following:
Clinical examination
A detailed history was obtained, including history of smoking, alcohol intake, occupational exposure to chemicals, drug intake, and previous diseases. A thorough clinical assessment and total body examination with abdominal ultrasound and/or CT were performed when indicated to detect tumor size and extension.
Laboratory investigations
A blood sample (5 mL) was drawn from each subject then divided, processed, and stored according to the requirements and precautions of each test. All study participants underwent the following investigations: Conventional liver evaluation tests that include assessment of the levels of aspartate aminotransferase (AST), alanine aminotransferase (ALT), total bilirubin, and serum albumin. In addition, serum urea and creatinine levels were examined using a clinical chemistry analyzer (AU400; Olympus America, Pennsylvania, USA). AFP levels were assessed using enzyme-linked immunosorbent assay following instructions on kits purchased from Immunospec Co. (California, USA).
Exosomal extraction and detection
The exosomes were isolated from serum using Total Exosome Isolation Reagent (Invitrogen Corp., Waltham, MA, USA; catalog number 4,478,360) according to the manufacturer’s instructions. We added 100 µl of the serum to 20 µl of the reagent, mixed well, and incubated the mixture at a temperature of 2°C–8°C. The mixture was then centrifuged at 10,000 g for 10 minutes. Next, the supernatant was removed, and the pellet formed at the bottom of the tube was resuspended in 30 µl of phosphate buffer solution; this suspension contained exosomes ready for analysis. The characteristics of the isolated exosomes were verified using transmission electron microscopy.
MiRNAs were extracted from the exosomes according to the serum/plasma miReasy kit protocol, catalog number: 217,184 (Qiagen, Hilden, Germany). All isolated miRNA were quantified using NanoDrop 1000 (Nanodrop, Wilmington, Delaware, USA). 35 After the extraction of miRNA, cDNA synthesis was performed using TaqMan microRNA Reverse Transcription Kit (Applied Biosystems Corp., Carlsbad, CA, USA; catalog number: 4,366,596) by adding 7 µl of the master mix with 3 µl of the primer used and 5 µl of isolated miRNA and microRNA-specific stem-loop primers (part of the TaqMan microRNA Assay Kit; Applied Biosystems). The resulting double-stranded (ds) cDNA was a template for an in vitro transcription (IVT) reaction. 36
Real-time quantitative polymerase chain reaction (qRT-PCR) was performed using the Stratagene Mx3000P Real-Time PCR System (Applied Biosystems) for the detection of the miR-18a (5′ UAAGGUGCAUCUAGUGCAGAUAG 3′) and miR-222 (5′CUCAGUAGCCAGUGUAGAUCCU 3′) primers (ThermoFisher Scientific, catalog number 4,427,975). The following 2−ΔΔCt formula 37 was used to calculate miRNA levels in the serum: ΔCT = (CT of internal reference—CT of target miRNA). The relative expression levels of miR-222 and miR-18a were calculated and normalized to those of endogenous miR-16-5p (5′ UAGCAGCACGUAAAUAUUGGCG 3′) (Applied Biosystems).
Statistical analysis
The data were analyzed using Microsoft Excel 2010 and Statistical Package for the Social Sciences (version 24.0; IBM Corp., Armonk, NY, USA). Continuous normally distributed variables were represented as mean ± standard deviation with 95% confidence interval, whereas non-normally distributed variables were represented as median with 25 and 75 percentiles. Categorical variables were represented as frequencies and percentages, and p-values < 0.05 were used to denote statistical significance. Student’s t-test was used to compare the means of normally distributed variables between the groups, and the Mann–Whitney U-test was used to compare non-normally distributed variables between the groups. The χ2 test or Fisher’s exact test was used to determine the distribution of categorical variables between the groups. The diagnostic performance of miR-18a and miR-222, evaluated based on their sensitivity and specificity, was assessed using receiver operating characteristic (ROC) curves, which were created using IBM SPSS advanced statistics version 24 (IBM Corp., Armonk, NY, USA). Spearman’s rank correlation coefficient (r) was used to show the correlation between different parameters in this study. Effect modifications were evaluated via stratification. Statistical interaction was assessed by including the main effect variables and their product terms in the logistic regression model. The difference between the groups was considered statistically significant when the p-value was <0.05.
Results
Demographic and biochemical profiles of the patients and controls are presented in Table 1. The results of the control group were within the expected range/concentrations for all the biochemical parameters tested. The ALT, AST, bilirubin, and AFP levels were significantly higher in the HCC and liver cirrhosis groups than those in the control group (p ˂ 0.001). Albumin levels were significantly lower in the HCC and liver cirrhosis groups than those in the control group (p ˂ 0.001). Urea and creatinine levels were elevated only in the HCC group (Table 1). The HCC group had significantly different results for all examined biochemical parameters, except albumin, compared to the liver cirrhosis group (Table 1).
Table 1.
Demographic, clinical, and laboratory data of the studied groups.
| Variable | Control | Cirrhosis | HCC | p-value | ||
|---|---|---|---|---|---|---|
| N = 50 | N = 51 | N = 51 | P1 | P2 | P3 | |
| Age (years) | 56.0 ± 6.3 | 54.5 ± 4.6 | 54.5 ± 4.6 | 0.1 | 0.1 | 0.9 |
| Gender F/M | 24 26 | 26/25 | 26/25 | 0.9 | 0.9 | 0.99 |
| AST (U/L) | 28.0 (25.0–32.0) | 52.0 (45.0–90.0) | 160.0 (80.0–2970.0) | 0.001** | 0.001** | 0.001** |
| ALT (U/L) | 29.5 (27.0–33.0) | 56.0 (41.3–75.0) | 180.0 (89.0–2700.0) | 0.001** | 0.001** | 0.001** |
| Albumin (g/L) | 4.3 ± 0.4 | 3.2 ± 0.5 | 3.1 ± 0.6 | 0.001** | 0.001** | 0.4 |
| Bilirubin (μm/L) | 0.6 ± 0.1 | 1.4 ± 0.5 | 1.7 ± 0.7 | 0.001** | 0.001** | 0.01* |
| Urea(mg/dL) | 33.7 ± 5.7 | 31.5 ± 9.6 | 149.2 ± 68.5 | 0.1 | 0.001** | 0.001** |
| Creatinine (mg/dL) | 1.0 ± 0.2 | 1.1 ± 0.3 | 2.4 ± 0.8 | 0.1 | 0.001** | 0.001** |
| AFP (μg/L) | 2.7 (2.5–3.3) | 5.9 (4.4–16.2) | 310.0 (100.0–2000.0) | 0.001** | 0.001** | 0.001** |
| miRNA 18a | 0.7 (0.4–1.5) | 2.1 (1.1–3.9) | 1.4 (0.6–5.9) | 0.001** | 0.001** | 0.4 |
| miRNA 222 | 0.9 (0.5–1.2) | 2.4 (1.8–5.0) | 2.8 (0.5–4.2) | 0.001** | 0.01* | 0.1 |
Age as well as albumin, bilirubin, urea, and creatinine levels are represented as mean ± standard deviation and were analyzed using the Student’s t and χ2 tests. However, AST, ALT, and AFP are represented as medians with interquartile ranges (25%–75%) and were analyzed using the Mann–Whitney U-test.
P1 = Cirrhosis and control, P2 = HCC and control, P3 = HCC and cirrhosis. *p-values < 0.05 indicated statistical significance; **p-values < 0.01 indicated high levels of statistical significance.
Serum miRNA signature in patients with HCC and liver cirrhosis
The results of this study showed that the serum levels of miR-18a and miR-222 were different in patients with HCC and liver cirrhosis compared with those in the healthy control group. In addition, statistically significant differences were observed between patients with HCC, patients with liver cirrhosis, and healthy controls (p ˂ 0.001). However, no statistically significant difference was found between patients with HCC and those with liver cirrhosis (p = 0.4 for miR-18a, and p = 0.1 for miR-222) (Table 1).
Correlation between miR-18a, miR-222, and laboratory investigations
Our results revealed a positive correlation between miR-18a and AST, ALT, bilirubin, AFP, urea, and creatinine levels (r = 0.286, 0.372, 273, 0.357, 0.343, and 0.399, respectively; p ˂ 0.001), and a negative correlation between miR-18a and albumin levels (r = −0.253; p = 0.002). In addition, miR-222 levels were positively correlated with AST, ALT, AFP, urea, and creatinine levels (r = 0.469, 0.487, 0.122, 0.571, 0.413, 0.406, and 0.571, respectively; p ˂ 0.001) and negatively correlated with albumin levels (r = −0.231; p = 0.004). Moreover, miR-18a and miR-222 levels were positively correlated (r = 0.556; p ˂ 0.001) (Table 2).
Table 2.
Correlation of serum miR-18a and miR-222 levels in healthy controls, patients with liver cirrhosis, and patients with Hepatocellular carcinoma.
| miRNA 18a | miRNA222 | |||
|---|---|---|---|---|
| R | P | r | P | |
| Age | −0.109 | 0.181 | −0.011 | 0.892 |
| AST | 0.286** | 0.001 | 0.469** | 0.001 |
| ALT | 0.372** | 0.001 | 0.487** | 0.001 |
| Albumin | −0.253** | 0.002 | −0.231** | 0.004 |
| Bilirubin | 0.273** | 0.001 | 0.122 | 0.134 |
| Urea | 0.343** | 0.001 | 0.413** | 0.001 |
| Creatinine | 0.399** | 0.001 | 0.406** | 0.001 |
| AFP | 0.357** | 0.001 | 0.571** | 0.001 |
| miRNA 18a | 0.556** | 0.001 | ||
| miRNA 222 | 0.556** | 0.001 | ||
*p-values < 0.05 indicated statistical significance; **p-values < 0.01 indicated high levels of statistical significance.
Evaluation of the miR-18a and miR-222 distributions in patients with HCC and those with liver cirrhosis
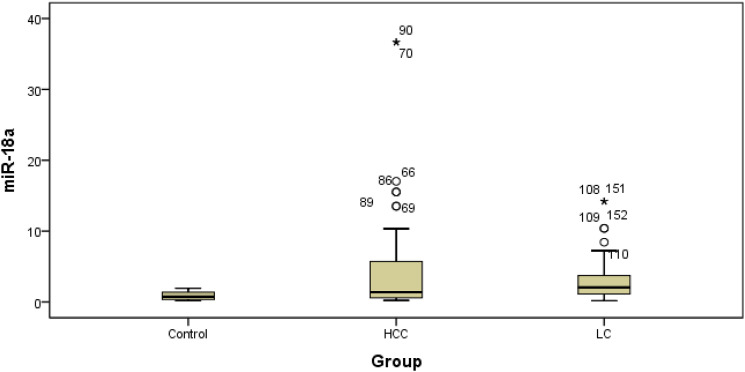
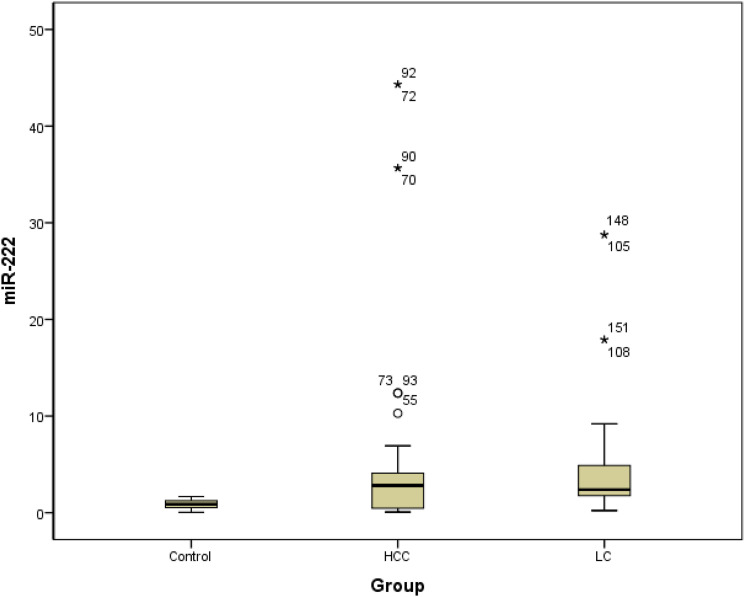
Figures 1 and 2 show the differential expression of miR-18a and miR-222 in different groups. Box plots of ΔCT values of the tested serum miRNAs in patients with liver cirrhosis and those with HCC compared with healthy controls are also depicted. The relative serum range normalized to the internal control miR-16-5p was represented on the Y-axis (ΔCT). The marker was not normally distributed among the studied individuals. The mean values for miR-18a were 3.35 ± 3.4, 4.84 ± 7.89, and 0.89 ± 0.17, and the median values were 2.06, 1.38, and 0.74 for patients with liver cirrhosis, patients with HCC, and healthy controls, respectively. The mean values for miR-222 were 4.73 ± 6.07, 5.64 ± 10.59, and 0.85 ± 0.48, and the median values were 2.38, 2.80, and 0.87 for patients with liver cirrhosis, patients with HCC, and healthy controls, respectively. When we compared the median range of miRNAs in liver cirrhosis or HCC to that of healthy subjects, it was considered significant (p < 0.01).
Figure 1.
Box plot of miR-18a in the studied groups. LC: Liver cirrhosis, HCC: Hepatocellular carcinoma.
Figure 2.
Box plot of miR-222 in the studied groups. LC: Liver cirrhosis, HCC: Hepatocellular carcinoma.
ROC analysis to determine the best discriminative value between patients with HCC, patients with liver cirrhosis, and healthy controls
We tried to find an optimal cutoff value using ROC analysis for both miR-18a and miR-222. When patients with liver cirrhosis were compared with the control group, the AUC was 0.8 for miR-18a, and a cutoff value of 2 was the best discriminative cutoff with 100% specificity and 52.9% sensitivity (p < 0.001). The AUC for miR-222 in patients with liver cirrhosis compared with the control group was 0.9, and a cutoff value of 1.7 was the best discriminative cutoff with 100% specificity and 76.5% sensitivity (p = 0.001). When patients with hepatocellular carcinoma were compared with the control group, the AUC was 0.72 for miR-18a, and a cutoff value of 2 was the best discriminative cutoff with 100% specificity and 45.1% sensitivity (p < 0.001). The AUC for miR-222 patients with hepatocellular carcinoma compared with the control group was 0.68, and a cutoff value of 1.9 was the best discriminative cutoff with 100% specificity and 57% sensitivity (p = 0.002) (Figure 3). However, this discriminative power lost significance when comparing the liver cirrhosis and HCC groups.
Figure 3.
ROC (Receiver Operating Characteristic) curve for the diagnostic potential of the differentially expressed individual and combined serum miRNAs. (a)miR-18a and (b) miR-222; in discrimination between liver cirrhosis and healthy controls. (c)miR-18a and (d) miR-222; in discrimination between HCC and healthy controls.
Logistic regression to estimate risk and calculate the odds ratio
Multivariate analysis revealed that miR-18a expression was independent predictive factor in HCC patients, as for every unit increase in miR-18a there is 2 times risk increase to be in HCC group (p = 0.004) (Table 3). While miR-222 expression was independent predictive factor in liver cirrhosis patients as for every unit increase in miR-222 there is 8 times risk increase to be in liver cirrhosis group (p < 0.001) (Table 4).
Table 3.
Logistic regression model to estimate risk in Hepatocellular carcinoma group.
| Beta coefficient | Standard error | p value | Odds ratio | 95% C.I. for OR | ||
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| miR-18a | 0.746 | 0.260 | 0.004 | 2.108 | 1.266 | 3.508 |
Table 4.
Logistic regression model to estimate risk between liver cirrhosis group.
| Beta coefficient | Standard error | p value | Odds ratio | 95% C.I. for OR | ||
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| miR-222 | 2.136 | 0.481 | <0.001 | 8.464 | 3.296 | 21.731 |
Discussion
The incidence of HCC varies widely worldwide and is increasing in Egypt. HCC mostly develops following HCV infection, which is prevalent in Africa and the Middle East. 38 Moreover, more than one-third of patients with HCC are diagnosed at the terminal stage or when liver failure occurs and die within few months due to the silent asymptomatic development of liver pathologies. Therefore, early diagnosis is necessary for the prevention and treatment of patients with liver cirrhosis and HCC. 39 MiRNAs have attracted great attention in the cancer research field. The dysregulation of circulating and tumor-derived miRNAs has been observed in patients with HCC. 40
Although the development of HCC following HCV infection remains to be elucidated, autoimmune and genetic associations may play a role. CTLA4, a regulatory glycoprotein, is a suppressor of the T-cell-mediated immune response. In genotype 4 (G4) HCV infection, CTLA4 function is impaired owing to either genetic mechanisms or the stimulation of co-stimulatory molecules. Signal transducer and activator of transcription 4 (STAT4) are reported to regulate type I interferon (IFN), interleukin (IL)-23, and IL-12 involved in the inflammation process. Dysregulation of STAT4 activity leads to suppression of the immune response and development of autoimmune disorders. 41 Many miRNAs target some proteins, such as SOCS1, CTLA4, and STAT, which are involved in the inflammation and tumor pathways. 42
MiRNA detection can be performed using conventional techniques, such as quantitative polymerase chain reaction (qPCR), northern blotting, or microarray, or new technology-based techniques, which involve signal amplification, such as hybridization chain reaction, nanoparticle-based amplification, rolling circle amplification, and isothermal exponential amplification. A combination of conventional and new technology-based techniques can increase the sensitivity of detection. 43 Further, miRNA-Seq is a comprehensive pipeline analysis designed to identify and quantify miRNAs by determining their differential expression while making miRNA target predictions. 44 Here, we used qRT-PCR to detect exosomal miR-18a and miR-222 as potential noninvasive diagnostic markers in patients with liver cirrhosis and hepatocellular carcinoma following HCV infection, as upregulated serum levels have been described in these patients compared with healthy individuals.
The levels of the liver enzymes in the liver function tests in the HCC and liver cirrhosis groups were elevated; however, AFP levels were only elevated in the HCC group. Similar to our results, Motawi et al. found a significant elevation in ALT, AST, and AFP in liver cancer compared to the control group. 20 This finding was concordant with previous studies in which liver function tests were specific to liver diseases and the levels of the liver enzymes tested were always elevated following any liver injury. Many studies reported elevated liver enzymes more frequently in HCC than in liver cirrhosis. 45 However, a study in Nigeria reported elevated liver enzyme levels in only 30% of HCC cases. There was no statistically significant difference between cirrhosis and HCC in terms of the level of elevation. The authors concluded that there was no specific diagnostic pattern for liver function tests in HCC. 46 This difference may be attributable to ethnic variations.
An important difference in our study is that all cases were preceded with HCV infection, which causes severe inflammation and injury to liver cells and may have augmented the findings. 47 AFP levels may increase in nearly 40% of liver cirrhosis cases; however, this increase fluctuates parallel to the inflammatory activity. Very high levels are present in 70% of HCC cases at diagnosis, and levels higher than 400–500 ng/mL are considered diagnostic for HCC. 47
There was a significant positive correlation of miR-18a and miR-222 levels with AST, ALT, bilirubin, AFP, urea, and creatinine (p ˂ 0.001) levels and a negative correlation of miR-18a and miR-222 levels with albumin levels (p = 0.002 and 0.004, respectively). Importantly, among the studied patients in both liver cirrhosis and HCC groups, multivariate analysis revealed that high expression of miR-18a and miR-222 to be an independent prognostic factor for elevated enzymes and decreased albumin production. These findings correlate with the liver pathogenicity and reflect that higher marker levels are associated with disease progression in elevated enzymes and decreased liver function of albumin formation. 48 Yasser et al. reported similar results in terms of the correlation of miR-222 levels with ALT, AST, and albumin levels following disease progression from chronic HCV infection to cirrhosis; higher values were found in the HCC group. 13 Moreover, Motawi et al. reported a significant correlation between HCV and low albumin levels. 20
The serum expression level of miR-18a was significantly higher in the liver cirrhosis and HCC groups than that in the control group. Similar to our results, miR-18a was overexpressed in Egyptian patients with colonic polyps, PC, and CLL.28-31 To the best of our knowledge, miR-18a did not correlate before to liver cirrhosis, and only its tumorigenic activity was known; however, the mechanism remains unclear. Murakmi et al. proposed a panel of miRNAs to predict treatment response (IFNs) in chronic HCV infection and found that miR-18a was upregulated in the liver tissue of IFN non-responders. 49 This finding may also be associated with the aggressive nature of miR-18a as an oncomiR; however, the follow-up of patients, whether they developed HCC or not later, was not the aim of their study. 49
In agreement with our results, Morita et al. found an increase in miR-18a expression in primary and recurrent HCC cases. Increased expression levels correlate with higher associated tumor marker levels, a large tumor size, and a high recurrence rate. The researchers also found that in HCC cells, miR-18a regulated tumor necrosis factor-alpha-induced protein 3 (TNFAIP3) expression. 50 MiR-18a was shown to enhance the malignant progression of many cancers, such as those of the cervix, lung, and breast. This situation occurs through the interaction of many pathways. For example, lung carcinoma was found to downregulate IRF2, which has apoptotic and antiproliferative activity. A positive correlation was found between miR-18a and NFκB; however, it was not subjected to further investigation.
On the other hand, miR-18a is known for its dual activity as it inhibits the progression of breast, colorectal, as well as lung cancers. 18 In nasopharyngeal carcinoma, Mai et al. proposed a novel oncogenic activity of miR-18a: downregulation of the SMG1, resulting in mTOR pathway activation. They suggested that the activation of NF-κB might lead to its binding to the promoter regions of miR-18a and the induction of miR-18a expression and that the introduction of miR-18a inhibitors can help control the disease. 51
Liu et al. found that miR-18a was highly expressed in HCC and suggested that miR-18a enhances the motility and progression of cancer cells by downregulating KLF4, a known tumor suppressor, in HCC. 19 Sanchez-Mejias et al. observed an increased expression of miR-18a in HCC and proposed a novel mechanism targeting the SOCS5, a negative regulator of the JAK/STAT cytokine signaling pathway. 52 Moreover, previous studies have proved that miR-18a may target ERα, suggesting its role in the progression of HCC in women.53,54
In contrast to our study results, Zhang et al. stated that miR-18a is downregulated in patients with HCC, demonstrating a protective effect and good overall survival. 55 These findings conflict with the results of this study and the results of those published in the literature. This concept might refer to several elements that affect the expression pattern of miRNAs in other studies using different specimens and methods and housekeeping transcripts for miRNA expression normalization, resulting in experimental bias and may lead to different miRNA expression profiles. 56 For example, Zhang et al. examined HCC in different populations, which may be related to ethnic differences, including all stages of the disease, and may have another associated factor related to survival. Moreover, they worked on a fresh tumor tissue sample, which can raise the question of whether a lower level exists in the tumor tissue due to its shedding into the peripheral blood. Therefore, normalization should be an essential step in removing almost all nonbiological variations, consecutively ensuring accurate miRNA expression profiles. 55
ROC curve analysis for determining the diagnostic ability of the miR-18a marker was performed and showed that a cutoff value of 2 was the best discriminative value with 100% specificity and 52.9% and 45.1% sensitivity for both liver cirrhosis and HCC, respectively, (p < 0.001) when compared to healthy controls. However, this discriminative power lost significance when comparing the cirrhotic and HCC groups. We chose this cutoff value with the best specificity based on our aim as a marker of initial diagnosis; however, these results suggest the introduction of the marker as an indicator of disease progression rather than being disease-specific and should be combined with other clinical, imaging, and laboratory criteria for differentiation.
The serum expression level of miR-222 was higher in the liver cirrhosis and HCC groups than in healthy controls. Previous studies on Egyptian patients reported high miR-222 expression levels in patients with rheumatoid arthritis and inflammatory breast cancer.24,25 In contrast, in terms of liver diseases, a previous study reported increased miR-222 expression levels in patients with severe HCV infection 26 and cirrhosis but not in those with liver cancer compared with controls. 20 In contrast, Ali et al. reported significantly elevated serum levels of miR-222 in patients with HCC and reduced levels in patients with HCV compared to controls. 23 However, Atef et al. found that miR-222 was significantly upregulated in the sera of HCC cases and HCV-infected patients compared to controls. 27
Similar to our results, Ogawa et al. found an increased expression of miR-221/222 in fibrotic livers, and this expression was increased 1.4-fold (p < 0.01) compared to that in healthy controls. 57 In a review by Wang et al., the upregulation of miR-222 was mentioned as a driver of hepatic fibrosis progression to liver cirrhosis and then HCC; thus, it may be elevated in the precancerous stages. 58 Wang et al. demonstrated a significant reduction in miR-222 in patients with renal cell carcinomas. 59 Many studies stated that miR-221 could promote tumor progression in liver cancer by inhibiting apoptosis and increasing cell growth. 21 MiR-221 and miR-222 are a homologous pair of miRNAs encoded in the same region. They have overlapping functions, act as oncomiR or oncosuppressor-miRs, and are overexpressed in many tumors, such as thyroid, breast, epithelial, and hepatic cancer.14,60 They achieve their oncogenic roles by downregulating the expression of some tumor suppressors or costimulators of pathways, such as p57, PTEN, and especially CDKN1B (p27, Kip1). 60
ROC curve analysis for the determination of the diagnostic ability of the miR-222 marker was performed and showed that a cutoff value of 1.7 and 1.9 are the best discriminative values with 100% specificity and 76.5% and 57% sensitivity for liver cirrhosis and HCC, respectively, (p = 0.001 and 0.002, respectively) when comparing the cirrhotic and the HCC groups with the controls. Similar to miR-18a, choosing this cutoff was based on the best specificity in our aim as a marker of initial diagnosis; however, similar to miR-18a, these results can suggest the introduction of the marker as an indicator of disease progression rather than being disease-specific and should be combined with other clinical, imaging, and laboratory criteria for differentiation.
Similar to miR-18a, many studies have suggested that miR-222 is associated with nuclear factor-kappa B (NF-κB). It is upregulated by nuclear factor-kappa B (NF-κB), NF-κB activator, TNF-α, and transforming growth factor-α (TGF-α), where the NF-κB p65 subunit binds to two separate regions of the miR-222 promoter, resulting in the upregulation of miR-222 expression. 51 The fact that miR-18a and miR-222 are related to the NF-κB pathway could explain their parallel levels, patterns of increase, and significance in liver diseases.
In summary, like Chen et al., we believe that miRNAs are stable, reproducible, and consistent. 61 We provided a suggestion for introducing new biomarkers for the detection of liver disease progression as miR-18a and miR-222 were significantly altered in the serum of patients with HCC and liver cirrhosis compared to those in healthy controls. This finding might offer a novel alternative therapeutic target for HCC and associated liver diseases.
Conclusion
miR-18a and miR-222 were significantly discriminative markers in patients with HCV-induced liver cirrhosis and HCC and healthy individuals. Therefore, they have predictive rather than diagnostic value. Moreover, miR-18a and miR-222 might be useful in diagnosing liver injuries or progression, including fibrosis and cirrhosis, and HCC.
Limitation
The main limitation of this study was the unavailability of a large number of patients. Further, we did not perform a power analysis to calculate the sample size selected for this study.
Acknowledgements
We thank all colleagues in National Research Centre and National Cancer Institute, Cairo University.
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by National Research Centre.
Ethical approval: The protocol for this study was approved by the Ethics Committee of the National Research Centre (number: 16291) and according to Declaration of Helsinki guidelines.
Informed consent: Each participant of this study provided informed written consent before the study.
ORCID iDs
Mahmoud M Kamel https://orcid.org/0000-0003-0264-3096
Lamiaa A Fathalla https://orcid.org/0000-0002-2918-9922
References
- 1.Elgharably A, Gomaa AI, Crossey MM, et al. (2017) Hepatitis C in Egypt-past, present, and future. Int J Gen Med 10: 1–6. DOI: 10.2147/ijgm.s119301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atta MM, Atta HM, Gad MA, et al. (2016) Clinical significance of vascular endothelial growth factor in hepatitis C related hepatocellular carcinoma in Egyptian patients. J Hepatocell Carcinoma 3: 19–24. DOI: 10.2147/jhc.s86708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elghoroury EA, Maksoud SAA, Kandil D, et al. (2017) Expression of microRNAs-21 and-223 in hepatocellular carcinoma in hepatitis C virus infected Egyptian population. Journal of applied pharmaceutical sciences 7: 052–057. [Google Scholar]
- 4.Kim DW, Talati C, Kim R. (2017) Hepatocellular carcinoma (HCC): beyond sorafenib-chemotherapy. J Gastrointest Oncol 8: 256–265. DOI: 10.21037/jgo.2016.09.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mak LY, Cruz-Ramón V, Chinchilla-López P, et al. (2018) Global epidemiology, prevention, and management of hepatocellular carcinoma. American Society of Clinical Oncology educational book American Society of Clinical Oncology annual meeting 38: 262–279. DOI: 10.1200/edbk_200939. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X-D, Zhao M. (2019) Molecular diagnosis and therapy of hepatocellular carcinoma: achievements and challenges. Hepatoma research 56: 267–275. [Google Scholar]
- 7.Pan Q, Qin F, Yuan H, et al. (2021) Normal tissue adjacent to tumor expression profile analysis developed and validated a prognostic model based on Hippo‐related genes in hepatocellular carcinoma. Cancer Medicine 10: 3139–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song T, Li L, Wu S, et al. (2021) Peripheral blood genetic biomarkers for the early diagnosis of hepatocellular carcinoma. Frontiers in oncology: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Shi K, Chen Y, et al. (2021) Exosomes and their role in cancer progression. Frontiers in oncology 11: 639159. DOI: 10.3389/fonc.2021.639159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Lu Z, Zhao X. (2019) Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J Hematol Oncol 12: 133. DOI: 10.1186/s13045-019-0806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soung YH, Nguyen T, Cao H, et al. (2016) Emerging roles of exosomes in cancer invasion and metastasis. BMB reports 49: 18–25. DOI: 10.5483/BMBRep.2016.49.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, Yang Y, Sun L, et al. (2020) Exosomal microRNAs as liquid biopsy biomarkers in hepatocellular carcinoma. OncoTargets and therapy 13: 2021–2030. DOI: 10.2147/ott.s232453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasser MB, Abdellatif M, Emad E, et al. (2021) Circulatory miR-221 & miR-542 expression profiles as potential molecular biomarkers in hepatitis C virus mediated liver cirrhosis and hepatocellular carcinoma. Virus Research 296: 198341. DOI: 10.1016/j.virusres.2021.198341. [DOI] [PubMed] [Google Scholar]
- 14.Morishita A, Oura K, Tadokoro T, et al. (2021) MicroRNAs in the pathogenesis of hepatocellular carcinoma: A review. Cancers 13: 514. DOI: 10.3390/cancers13030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang N, Zhou J, Li Q, et al. (2019) miR-96 exerts carcinogenic effect by activating AKT/GSK-3β/β-catenin signaling pathway through targeting inhibition of FOXO1 in hepatocellular carcinoma. Cancer Cell International 19: 38. DOI: 10.1186/s12935-019-0756-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Maraghy SA, Adel O, Zayed N, et al. (2020) Circulatory miRNA-484, 524, 615 and 628 expression profiling in HCV mediated HCC among Egyptian patients; implications for diagnosis and staging of hepatic cirrhosis and fibrosis. J Adv Res 22: 57–66. DOI: 10.1016/j.jare.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibriel AA, Al-Anany AM, Al-Arab MAE, et al. (2020) Investigating circulatory microRNA expression profiles in Egyptian patients infected with hepatitis C virus mediated hepatic disorders. Meta gene 26: 100792. [Google Scholar]
- 18.Shen K, Cao Z, Zhu R, et al. (2019) The dual functional role of MicroRNA-18a (miR-18a) in cancer development. Clin Transl Med 8: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L, Cai X, Liu E, et al. (2017) MicroRNA-18a promotes proliferation and metastasis in hepatocellular carcinoma via targeting KLF4. Oncotarget 8: 68263–68269. DOI: 10.18632/oncotarget.19293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Motawi TM, Sadik NA, Shaker OG, et al. (2016) Elevated serum microRNA-122/222 levels are potential diagnostic biomarkers in Egyptian patients with chronic hepatitis C but not hepatic cancer. Tumour biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine 37: 9865–9874. DOI: 10.1007/s13277-016-4884-6. [DOI] [PubMed] [Google Scholar]
- 21.Song J, Ouyang Y, Che J, et al. (2017) Potential value of miR-221/222 as Diagnostic, prognostic, and therapeutic biomarkers for diseases. Frontiers in immunology 8: 56. DOI: 10.3389/fimmu.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kutay H, Bai S, Datta J, et al. (2006) Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem 99: 671–678. DOI: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Ali HEA, Abdel Hameed R, Effat H, et al. (2017) Circulating microRNAs panel as a diagnostic tool for discrimination of HCV-associated hepatocellular carcinoma. Clinics and research in hepatology and gastroenterology 41: e51–e62. DOI: 10.1016/j.clinre.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Abo ElAtta AS, Ali YBM, Bassyouni IH, Talaat RM. (2019) Upregulation of miR-221/222 expression in rheumatoid arthritis (RA) patients: correlation with disease activity. Clin Exp Med 19: 47–53. DOI: 10.1007/s10238-018-0524-3. [DOI] [PubMed] [Google Scholar]
- 25.Ahmed SH, Espinoza-Sa´nchez NA, ElDamen A, Fahim SA, Badawy MA, Greve B, et al. (2021) Small extracellular vesicle-encapsulated miR-181b-5p, miR-222-3p and let-7a-5p: Next generation plasma biopsy-based diagnostic biomarkers for inflammatory breast cancer. PLoS ONE 16: e0250642. DOI: 10.1371/journal.pone.0250642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Guendy NM, Helwa R, El-Halawany MS, et al. (2016) The liver MicroRNA expression profiles associated with chronic hepatitis C virus (HCV) genotype-4 infection: A preliminary study. Hepatitis monthly 16: e33881. DOI: 10.5812/hepatmon.33881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atef AA, Abdel-Wahab AA, Ahmed EK, Abdel-Hameed R. (2016) Efficacy of serum miR-106b and miR-222 as non-invasive diagnostic markers for hepatocellular carcinoma in HCV-infected Egyptian patients. Egyptian journal of pure and applied science 54: 09–17. [Google Scholar]
- 28.Ibrahim NH, Abdellateif MS, Kassem SH, Abd El Salam MA, El Gammal MM. (2019) Diagnostic significance of miR-21, miR-141, miR-18a and miR-221 as novel biomarkers in prostate cancer among Egyptian patients. Andrologia 51: e13384. DOI: 10.1111/and.13384. [DOI] [PubMed] [Google Scholar]
- 29.Ibrahim NA, Abdellateif MS, Ismail YM, et al. (2018) The potential prognostic and diagnostic role of MiRNAs as novel biomarkers for prostate cancer. Cancer therapy & oncology international 12(5): 555846. DOI: 10.19080/CTOIJ.2018.12.555846. [DOI] [Google Scholar]
- 30.Zekri A-RN, Youssef ASE-D, Lotfy MM, et al. (2016) Circulating serum miRNAs as diagnostic markers for colorectal cancer. PLoS ONE 11: e0154130. DOI: 10.1371/journal.pone.0154130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khalifa MM, Zaki NE, Nazier AA, et al. (2021) Prognostic significance of microRNA 17–92 cluster expression in Egyptian chronic lymphocytic leukemia patients. Journal of the Egyptian national cancer institute 33: 37. DOI: 10.1186/s43046-021-00097-x. [DOI] [PubMed] [Google Scholar]
- 32.Lee JM, Park JW, Choi BI. (2014) KLCSG-NCC Korea practice guidelines for the management of hepatocellular carcinoma: HCC diagnostic algorithm. Digestive diseases (Basel, Switzerland) 32: 764–777. DOI: 10.1159/000368020. [DOI] [PubMed] [Google Scholar]
- 33.Llovet J, Ducreux M, Lencioni R, et al. (2012) European Association for the Study of the Liver European Organisation for Research and Treatment of Cancer: EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. Journal of hepatology 56: 908–943. [DOI] [PubMed] [Google Scholar]
- 34.Procopet B, Berzigotti A. (2017) Diagnosis of cirrhosis and portal hypertension: imaging, non-invasive markers of fibrosis and liver biopsy. Gastroenterology report 5: 79–89. DOI: 10.1093/gastro/gox012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibriel AA, Tate RJ, Yu Y, et al. (2013) The p.Arg86Gln change in GARP2 (glutamic acid-rich protein-2) is a common West African-related polymorphism. Gene 515: 155–158. [DOI] [PubMed] [Google Scholar]
- 36.Gibriel AA. (2012) Options available for labelling nucleic acid samples in DNA microarray-based detection methods. Briefings in functional genomics 4: 311–318. [DOI] [PubMed] [Google Scholar]
- 37.Bouzid A, Smeti I, Dhouib L, et al. (2018) Down-expression of P2RX2, KCNQ5, ERBB3 and SOCS3 through DNA hypermethylation in elderly women with presbycusis. Biomarkers: Biochemical Indicators of Exposure, Response, and Susceptibility to Chemicals 23: 347–356. DOI: 10.1080/1354750X.2018.1427795. [DOI] [PubMed] [Google Scholar]
- 38.Shelbaya A, Kuznik A, Salem M, et al. (2015) P1265: Estimating the epidemiologic and economic impact of different treatment rates for hepatitis C virus (HCV) in Egypt. Journal of hepatology 62: S832–S833. [Google Scholar]
- 39.Demerdash HM, Hussien HM, Hassouna E, et al. (2017) Detection of microRNA in hepatic cirrhosis and hepatocellular carcinoma in hepatitis C genotype-4 in Egyptian patients. BioMed Research International 2017: 1806069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sohn W, Kim J, Kang SH, et al. (2015) Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Experimental & molecular medicine 47: e184–e184. DOI: 10.1038/emm.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ali NA, Hamdy NM, Gibriel AA, et al. (2021) Investigation of the relationship between CTLA4 and the tumor suppressor RASSF1A and the possible mediating role of STAT4 in a cohort of Egyptian patients infected with hepatitis C virus with and without hepatocellular carcinoma. Archives of virology 166: 1643–1651. DOI: 10.1007/s00705-021-04981-8. [DOI] [PubMed] [Google Scholar]
- 42.Cortez MA, Anfossi S, Ramapriyan R, et al. (2019) Role of miRNAs in immune responses and immunotherapy in cancer. Genes, chromosomes & cancer 58: 244–253. DOI: 10.1002/gcc.22725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye J, Xu M, Tian X, et al. (2019) Research advances in the detection of miRNA. Journal of pharmaceutical analysis 9: 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrés-León E, Rojas AM. (2019) miARma-Seq, a comprehensive pipeline for the simultaneous study and integration of miRNA and mRNA expression data. Methods: 15231–15240. [DOI] [PubMed] [Google Scholar]
- 45.Giannini EG, Testa R, Savarino V. (2005) Liver enzyme alteration: a guide for clinicians. CMAJ 172: 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okonkwo U, Nwosu M, Nnadozie O, et al. (2011) Is liver function test of any diagnostic relevance in patients presenting with hepatocellular carcinoma? Orient Journal of Medicine 23: 17–23. [Google Scholar]
- 47.Bialecki ES, Di Bisceglie AM. (2005) Diagnosis of hepatocellular carcinoma. HPB 7: 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spinella R, Sawhney R, Jalan R. (2016) Albumin in chronic liver disease: structure, functions and therapeutic implications. Hepatology international 10: 124–132. DOI: 10.1007/s12072-015-9665-6. [DOI] [PubMed] [Google Scholar]
- 49.Murakami Y, Tanaka M, Toyoda H, et al. (2010) Hepatic microRNA expression is associated with the response to interferon treatment of chronic hepatitis C. BMC Medical Genomics 3: 48. DOI: 10.1186/1755-8794-3-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morita K, Shirabe K, Taketomi A, et al. (2016) Relevance of microRNA-18a and microRNA-199a-5p to hepatocellular carcinoma recurrence after living donor liver transplantation. Liver Transplantation : Official Publication of the American Association for the Study of Liver Diseases and the International Liver Transplantation Society 22: 665–676. DOI: 10.1002/lt.24400. [DOI] [PubMed] [Google Scholar]
- 51.Mai S, Xiao R, Shi L, et al. (2019) MicroRNA-18a promotes cancer progression through SMG1 suppression and mTOR pathway activation in nasopharyngeal carcinoma. Cell Death & Disease 10: 819. DOI: 10.1038/s41419-019-2060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanchez-Mejias A, Kwon J, Chew XH, et al. (2019) A novel SOCS5/miR-18/miR-25 axis promotes tumorigenesis in liver cancer. International journal of cancer 144: 311–321. DOI: 10.1002/ijc.31857. [DOI] [PubMed] [Google Scholar]
- 53.Liu WH, Yeh SH, Lu CC, et al. (2009) MicroRNA-18a prevents estrogen receptor-α expression, promoting proliferation of hepatocellular carcinoma cells. Gastroenterology 136: 683–693. [DOI] [PubMed] [Google Scholar]
- 54.Li CL, Yeh KH, Liu WH, et al. (2015) Elevated p53 promotes the processing of miR-18a to decrease estrogen receptor-α in female hepatocellular carcinoma. International journal of cancer 136: 761–770. DOI: 10.1002/ijc.29052. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Z, Li JZ, Wei ZW, et al. (2020) Correlation between expression levels of lncRNA UCA1 and miR-18a with prognosis of hepatocellular cancer. European review for medical and pharmacological sciences 24: 3586–3591. DOI: 10.26355/eurrev_202004_20820. [DOI] [PubMed] [Google Scholar]
- 56.Shen J, Wang Q, Gurvich I, et al. (2016) Evaluating normalization approaches for the better identification of aberrant microRNAs associated with hepatocellular carcinoma. Hepatoma research 2: 305–315. DOI: 10.20517/2394-5079.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ogawa T, Enomoto M, Fujii H, et al. (2012) MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut 61: 1600–1609. DOI: 10.1136/gutjnl-2011-300717. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Tian Y. (2016) miRNA for diagnosis and clinical implications of human hepatocellular carcinoma. Hepatology research: The Official Journal of the Japan Society of Hepatology 46: 89–99. DOI: 10.1111/hepr.12571. [DOI] [PubMed] [Google Scholar]
- 59.Wang C, Hu J, Lu M, et al. (2015) A panel of five serum miRNAs as a potential diagnostic tool for early-stage renal cell carcinoma. Scientific Reports 5: 7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garofalo M, Quintavalle C, Romano G, et al. (2012) miR221/222 in cancer: their role in tumor progression and response to therapy. Current molecular medicine 12: 27–33. DOI: 10.2174/156652412798376170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen X, Ba Y, Ma L, et al. (2008) Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Research 18: 997–1006. DOI: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]