Abstract
Objective
The clinical importance of aberrantly expressed microRNAs (miRNAs) in diagnosing inflammatory bowel disease (IBD) has not been well established, so was investigated in this systematic review and meta-analysis.
Methods
Articles in online databases from inception to March 17, 2021 were retrieved. Random effects meta-analysis was used to obtain sensitivity, specificity, positive (PLRs) and negative likelihood ratios (NLRs), diagnostic odds ratios (DORs), and areas under the curve (AUC) with 95% confidence intervals (CI) for IBD diagnosis.
Results
Of 117 studies reporting altered miRNA expression in IBD included in the systematic review, 15 involving 937 patients with IBD and 707 controls, 22 miRNAs, and two miRNA panels were eligible for meta-analysis. Pooled analyses showed a moderate diagnostic accuracy for miRNAs in the IBD diagnosis, with a sensitivity of 0.80 (95% CI: 0.79–0.82), specificity of 0.84 (95% CI: 0.82–0.86), DOR of 21.19 (95% CI: 13.90–32.31), and AUC of 0.89. Subgroup analyses revealed a better performance in patients with ulcerative colitis (AUC, 0.93) than Crohn’s disease (AUC, 0.84). Consistent upregulation of miR-21, miR-16, and miR-192 in blood with a high-moderate diagnostic accuracy was found in at least two studies.
Conclusions
These findings suggest miRNAs are credible diagnostic biomarkers in IBD.
Keywords: Inflammatory bowel disease, microRNA, meta-analysis, systematic review, diagnosis, area under the curve
Introduction
Inflammatory bowel disease (IBD) involves chronic relapsing inflammation of the gastrointestinal (GI) tract that results from the complex interplay between the immune system, microbes, and the GI tract in genetically susceptible individuals. 1 Ulcerative colitis (UC) and Crohn’s disease (CD) are the two most common entities of IBD. UC is characterized by inflammation of the colonic mucosa, and can affect variable lengths of the colon, 2 while CD features transmural inflammation that may occur in any section of the gut from the mouth to the anus, with a particular predilection for the terminal ileum. 3 Indeterminate colitis or IBD unclassified (IBDU) is an IBD with characteristics overlapping UC and CD. IBD can occur at any age, although incidence peaks in adolescence and early adulthood, and its prevalence has been rising globally. 4 Globally between 1990 and 2017, the total years lived with disability attributed to IBD almost doubled in number, and the disability-adjusted life years decreased significantly. 5 As well as resulting in a poor quality of life, IBD also incurs at least a 3-fold higher direct cost of care. 6
Because the symptoms of IBD are highly variable, a diagnosis must be made from the medical history, together with findings from physical, laboratory, radiologic, endoscopic, and histological examinations. 7 However, these diagnostic techniques are not specific and may produce equivocal results. Therefore, reliable biomarkers for IBD are urgently needed.
Micro (mi)RNAs are single-stranded, noncoding RNAs containing 22 to 24 nucleotides that post-transcriptionally regulate gene expression by blocking mRNA translation or degrading target mRNAs. miRNAs are widely involved in physiological and pathological cellular processes, such as differentiation, proliferation, and apoptosis. Additionally, they are stable, non-invasive, and resistant to degradation by ribonucleases, making them valuable targets in the diagnosis, monitoring, prognosis, and treatment of diseases. 8
Several clinical and preclinical studies have shown that dysregulated miRNAs play a crucial role in IBD development, 9–12 but miRNA expression profiling in patients with IBD has given inconsistent results. Moreover, the diagnostic accuracy of miRNAs for IBD has not been examined by meta-analysis. Therefore, this study systematically reviewed clinical studies investigating miRNAs as diagnostic biomarkers for IBD and assessed their overall diagnostic accuracy by meta-analysis.
Methods
Registration and protocol
This study was retrospectively registered at INPLASY (registration number: INPLASY202220027; DOI number: 10.37766/inplasy2022.2.0027). The review protocol can also be accessed at INPLASY.
Meta-analysis reporting guidelines
This meta-analysis was performed according to the guidelines of the preferred reporting items for systematic reviews and meta-analysis (PRISMA) studies statement. 13
Literature search strategy
We searched articles in online databases including PubMed, EMBASE, Web of Science, the Cochrane Library, and the Cochrane IBD Group Specialized Register from inception to March 17, 2021 using the following medical subject heading terms and keywords: “inflammatory bowel disease*”, “Crohn*”, “ulcerative colitis”, and the combination of “MicroRNA*”, “miRNA*”, and “miR*”. Details of the search terms and combination strategies used in PubMed are reported in Table S1. A manual review of the references from selected articles was also performed to identify relevant studies.
Inclusion criteria
Two reviewers (L.S. and Y.H.) independently screened titles and abstracts and retrieved full-text publications for potentially relevant articles. Disagreements were resolved by a third author (H.Y.) when necessary. Studies were included in this systematic review if they were reports on miRNA expression in patients with IBD, and if they could be used to construct a 2 × 2 contingency table for IBD diagnosis based on miRNA diagnostic accuracy. There were no age restrictions for participants included in the studies.
Exclusion criteria
Studies meeting one or more of the following criteria were excluded: 1) duplicate publications or articles with republished data; 2) reviews, letters, comments, replies, erratum, and conference abstracts; 3) case reports, database, or methodological studies; 4) cell, animal, or microbiological trials; 5) studies focusing on IBD-related diseases; 6) studies of miRNA polymorphisms or methylation; and 7) studies without healthy controls or the use of normal tissues.
Data extraction
Two reviewers (L.S. and H.W.) independently extracted data to a pre-tested Microsoft Excel sheet. Disagreements were resolved by consensus and consultation with a third investigator (Y.H.). Prespecified data parameters included: 1) publication data including the name of the first author and year of publication; 2) demographic data regarding population, number, age, and sex of participants; 3) experimental data including sample source, method of quantifying miRNA expression, control gene for normalization, type of miRNA(s), and cut-off or fold-change value; and 4) statistical data on sensitivity and specificity. If sensitivity or specificity were not reported, we extracted area under the receiver operating characteristic (ROC) curve (AUC) data, and calculated the sensitivity and specificity according to published methods. 14
Quality assessment
The quality of each study was independently evaluated by two reviewers (H.L. and S.L.) according to the Quality Assessment for Studies of Diagnostic Accuracy (QUADAS-2) tool. 15 Disagreements were resolved by a third author (H.Y.) if necessary. QUADAS-2 consisted of four key domains: (1) patient selection; (2) index test; (3) reference standard; and (4) flow and timing. Each domain was assessed in terms of risk of bias, and the first three domains were assessed with respect to applicability. Each item was answered with “yes,” “no,” or “unclear.” A “yes” answer represented a low risk of bias, whereas “no” or “unclear” represented a high risk of bias.
Statistical analysis
Data from individual datasets for meta-analysis were combined using Meta-DiSc v1.4 software (Clinical Biostatistics Unit, Ramóny Cajal Hospital, Madrid, Spain). The number of true positives (TPs), false positives (FPs), false negatives (FNs), and true negatives (TNs) in each study were calculated to obtain a summary (s)ROC; pooled sensitivity, specificity, positive likelihood ratios (PLRs), negative likelihood ratios (NLRs), and diagnostic odds ratios (DORs) with 95% confidence intervals (CIs) were also obtained. Study heterogeneity caused by a threshold effect was assessed by Spearman’s correlation analysis and ROC plane plots. Statistical heterogeneity of non-threshold effects was assessed based on the visual inspection of forest plots, the Higgins’s inconsistency index (I2) statistic, and the P value for the chi-squared test. 16 A random effects model (the DerSimonian–Laird method) was used when heterogeneity presented (I2 > 50% and/or P < 0.05). Otherwise, the fixed effects model (the Mante–Haenszel method) was used. Meta-regression was also conducted to identify potential sources of heterogeneity. The following a priori subgroup analyses were carried out: IBD subtype (UC versus CD), age of participants (pediatric population [<18 years of age] versus adult population [≥18 years of age]), sample source (blood versus others), and method of quantifying miRNA expression (quantitative [q]PCR versus microarray alone). Sensitivity analyses were performed to evaluate the stability of our results. Publication bias was evaluated using STATA v.12 software (Stata Corp, College Station, TX) with Deeks’ funnel plot asymmetry test, in which P < 0.10 indicated statistical significance. 17 All P values were two-sided, with P < 0.05 considered statistically significant. The GRADEpro Guideline Development Tool (https://www.gradepro.org/) was used to assess the certainty of evidence, and shown in GRADE Summary of Findings tables.
Patient and public involvement statement
Neither patients nor the public were involved in the design or conduct of the study.
Results
Literature search and characteristics of included studies
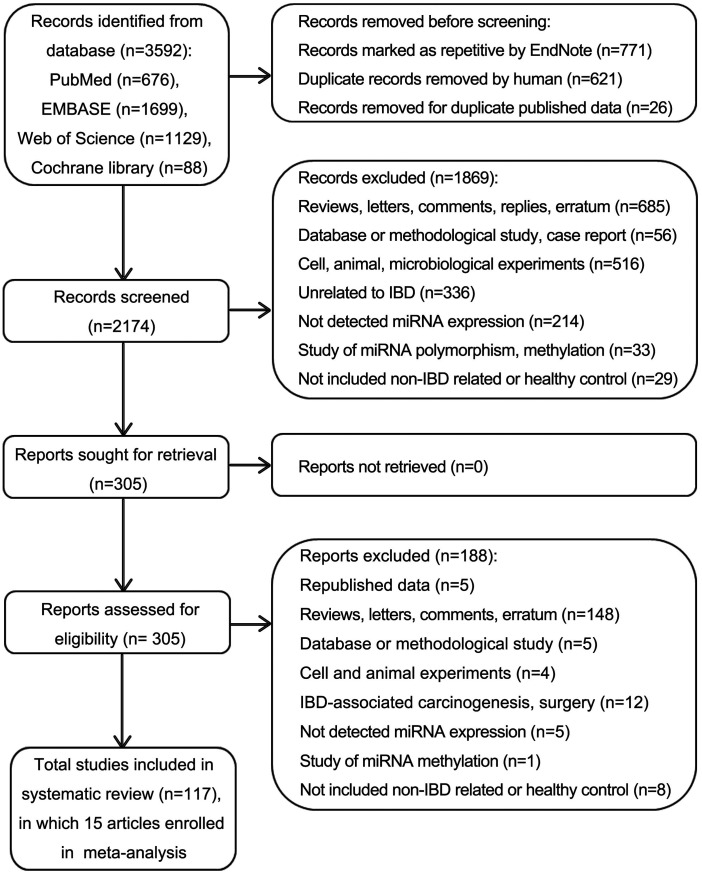
A total of 3592 records were identified in the initial search of PubMed, EMBASE, Web of Science, and Cochrane library databases. After removing duplicates, 2174 records were included (Figure 1). Of these, 1869 were excluded after reviewing the titles and abstracts. The remaining 305 full-text articles were screened and 188 were excluded according to the exclusion criteria. Finally, 117 primary articles of expression profile studies were included in the systematic review, of which 15 studies9,18–31 about diagnostic accuracy were included in the meta-analysis (Table 1). Thirty-one studies only reported altered miRNA expression using various high-throughput miRNA chips (Table S2), and 71 studies detected miRNA expression using qPCR or quantitative in situ hybridization (ISH) (Table S3).
Figure 1.
Flow diagram summarizing the selection of eligible studies.
IBD: inflammatory bowel disease; miRNA: microRNAs; qPCR: quantitative polymerase chain reaction; ISH: in situ hybridization.
Table 1.
Descriptive characteristics of primary studies included in the meta-analysis.
| Study ID | Patients with IBD |
Controls |
Sample type | miRNA assay | Normalization controls | miRNA studied (fold-change, folds) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Disease | Age (years) | Male (%) | No. | Controls | Age (years) | Male (%) | No. | |||||
| Adam M. Zahm 201121 | Active CD | 13.7 | 65.2 | 46 | Healthy controls | 13. | 53.1 | 32 | Serum | qPCR and qPCR LDA platform | cel-miR-54 and cel-miR-238 | miR-16 (8.74), miR-484 (4.5), miR-30e (5.6), miR-106a (4.79), miR-195 (5.67), miR-20a (4.63), miR-21 (2.72), miR-140 (4.51), let-7b (7.49), miR-192 (4.24), miR-93 (4.48) |
| Adam M. Zahm 201427 | Active UC and active CD | CD: 15, UC: 16 | 34.5 | 29 | Non-IBD | 15. | 38.9 | 18 | Serum | qPCR and TaqMan® Array | Exogenous genes | miR-192, miR-142-3p, miR-21 (NA for all) |
| Chenming Sun 201723,† | Active CD | 31 | 41.4 | 29 | Healthy controls | 31 | 51.4 | 37 | Plasma | qPCR | U6 | miR-125a (2.48), miR-125b (1.13, P > 0.05) |
| Christos Polytarchou 201518 | UC | 38 | 38.0 | 46 | Healthy controls | 33 | 61.0 | 21 | Serum | Nanostring | Global levels of miRNA | miR-4454 (NA), miR-223-3p (>2), miR-23a-3p (>2), miR-320e (>7) |
| Katharina Schönauen 201820,† | Active UC and active CD | CD: 34, UC: 43 | 44.7 | CD: 28, UC:10 | Healthy controls | 54 | 33.3 | 15 | Feces | qPCR | cel-miR-39 | miR-223 (17.1) |
| Matthias Hübenthal 201528 | UC and CD | CD: 46, UC: 44 | 54.0 | CD: 40, UC: 36 | Healthy controls | NA | NA | 70 | Whole blood | Geniom miRNA Biochip | NA | miRNA panel: miR-34b-3p, miR-142-5p, miR-205-5p, miR-424-5p, miR-570-3p, miR-885-5p, miR-1301-3p, miR-16-5p, miR-99b-5p (NA) |
| Mengdie Shen 201926,† | Active UC | 46 | 50.0 | 50 | Healthy controls | 43 | 50.0 | 50 | Serum | qPCR | U6 | miR-372 (>30) |
| Michael D. Jensen 201522,† | CD | 32 | 31.4 | 69 | Non-CD | 32 | 31.4 | 33 | Plasma | qPCR and OpenArray system | NA | miR-16 (0.83) |
| Olfat G. Shaker 201919 | UC | 39 | 53.8 | 26 | Healthy controls | 51 | 72.7 | 110 | Serum | qPCR | SNORD68 | miR-106a (0.32) |
| Peng Chen 2019 (training cohort)8 | Active UC and active CD | CD: 28, UC: 42 | 62.0 | 139 | Non-IBD | 32 | 60.9 | 41 | Serum | qPCR | cel-miR-39 | miR-146b-5p (CD: 2.23, UC: 2.27) |
| Peng Chen 2019 (validation cohort)8 | Active CD | 28 | 65.2 | 66 | Non-IBD | 32 | 60.9 | 41 | Serum | qPCR | cel-miR-39 | miR-146b-5p (1.65) |
| Radha Duttagupta 201225 | UC | 50 | 30.0 | 20 | Healthy controls | 43 | 35.0 | 20 | Platelets and microvesicles | qPCR and Affymetrix arrays | RRN18S | miRNA panel: miR-378*, miR-500, miR-378, miR-501-5p, miR-941, miR-140-3p, miR-422a, miR-720, miR-27a*, miR-181b, miR-150*, miR-146b-3p, miR-874, miR-138, miR-92a-1*, miR-1274b, miR-188-5p, miR-769-5p, miR-20b*, miR-769-3p, miR-550*, miR-22, miR-143*, miR-362-5p, miR-532-5p, miR-330-3p, hsa-miR-423-3p, hsa-miR-202 and hsa-miR-1263 (NA) |
| Yulan Ye 201724 | UC, CD and IBDU | CD: 36, UC: 42, IBDU: 48 | 45.8 | 38 | Healthy controls | 49 | 46.4 | 25 | Mucosa | qPCR | U6 | miR-155 (1.79) |
| Elham Ahmed Hassan 202029 | UC | 38 | 40.5 | 37 | Healthy controls and non-UC | 39 | 48.3 | 60 | Plasma | qPCR | The exogenous genes | miR-21 (>1.52) and miR-92a (>1.66) |
| Yuanyuan Tian 202030 | UC | 47 | 41.7 | 187 | Healthy controls | 45 | 45.4 | 152 | Serum | qPCR | U6 | miR-16 (3.78) and miR-195 (4.65) |
| Rui Zhou 202131,† | UC and CD | CD: 34, UC: 36 | 58.5 | 41 | Healthy controls | 30 | 69.6 | 23 | Feces | qPCR | U6 | miR-21 (>7) and miR-16 (>7) |
†Sensitivity and specificity were extracted from AUC.
IBD: Inflammatory bowel disease; UC: Ulcerative colitis; CD: Crohn’s disease; IBDU: IBD unclassified; y: years; SD: Standard deviation; No.: Number; miRNA: microRNAs; qPCR: Quantitative polymerase chain reaction; NA: Not available.
The 15 articles included in the meta-analysis (published from 2009 to 2021) included 937 patients with IBD and 707 controls (healthy controls and those suspected of IBD but subsequently identified as healthy or non-IBD). Two studies focused on the pediatric population (<18 years of age).21,27
Data on altered miRNA expression using miRNA microarray analysis
The 31 studies reporting altered miRNA expression using miRNA microarray chips (Table S2) included 31 cohorts containing 795 patients with IBD and 481 controls. The most common sample resource was tissue, including fresh or frozen or formalin-fixed paraffin-embedded (FFPE) tissues (n = 20, 64.52%). Detailed information on participant characteristics, high-throughput modes of miRNA microarray, number of miRNA probes, expression validation methods, and related clinicopathological factors are shown in Table S2. Several miRNAs were reported to have differential expression between patients with IBD and controls. Cut-off values varied, so miRNAs with a 4-fold change are summarized in Table S2. A total of 135 miRNAs were frequently up-regulated in patients compared with controls, and 53 miRNAs were frequently down-regulated. Among them, 18 miRNAs (13.33%) were up-regulated and three miRNAs (7.55%) were down-regulated in at least two studies. Several studies reported the differential expression of the same miRNAs, which are listed in Table S4.
Data on altered miRNA expression using validated methods
Information about the 71 studies that detected miRNA expression by qPCR or ISH is shown in Table S3. A total of 2908 patients with IBD and 1927 controls were enrolled in 75 cohorts. The samples were mostly collected from the colonic mucosa (n = 50, 66.67%) and blood (n = 13, 17.33%). Most studies (97.18%) detected the expression level of miRNAs by qPCR. U6 (61.33%) was the most common normalization control. Participant characteristics, the miRNA target, and clinicopathological factors are shown in Table S3. Among the 69 miRNAs reported in these studies, 46 (66.67%) were up-regulated, 16 (20.29%) were down-regulated, and the expression of seven (10.14%) was inconsistent. Nineteen miRNAs (27.54%) were up-regulated and four miRNAs (5.80%) were down-regulated in at least two studies (Table S5). The most common significantly up-regulated miRNAs were miR-155 (n = 11 studies), miR-31 (n = 9), miR-21 (n = 9), miR-223 (n = 5), and miR-126 (n = 4) were most frequently reported. The most common significantly down-regulated miRNAs were miR-141 (n = 4), miR-200b (n = 3), miR-200a and miR-200c (both n = 2).
Characteristics of studies included in meta-analysis
Detailed characteristics of the 16 cohorts from 15 studies9,18–31 that fulfilled the criteria for meta-analysis are listed in Table 1. A total of 937 patients with IBD, including 532 (56.78%) with UC, 394 (42.05%) with CD, and 11 (1.17%) with IBDU, were included. A total of 707 controls, including 585 (82.74%) healthy individuals, and 92 (17.26%) suspected of IBD but subsequently identified as healthy or non-IBD, were enrolled. Blood was the most common sample type (n = 12, 75.00%). qPCR was mostly used for miRNA detection (n = 13, 81.25%), but normalization controls varied (Table 1). From the 16 datasets, 22 miRNAs were enrolled in the meta-analysis. Most studies reported up-regulated miRNAs (86.36%). Six miRNAs (27.27%) were reported in at least two studies; miR-223, miR-21, miR-16, miR-195, and miR-192 were consistently up-regulated, while miR-16 and miR-106a showed an inconsistent regulation, which might reflect different IBD types, controls, disease subtypes, or sample resources. Different fold-changes in miRNA expression are shown in Table 1.
Quality assessment of studies included in meta-analysis
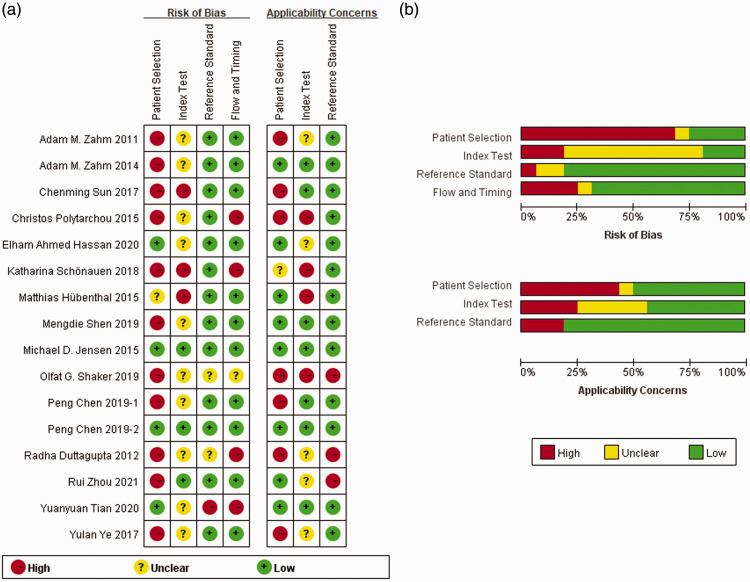
Details of the quality assessment by QUADAS-2 are shown in Figure 2. Only 2 of the 16 cohorts (12.50%) showed no bias in all four parameters assessed. Two studies (12.50%) showed a low risk of bias in three of the four domains, seven (43.75%) showed a low risk of bias in two of the four domains, three (18.75%) showed a low risk of bias in one of the four domains, and two (12.50%) showed an unclear or high risk of bias in all domains. The biases identified in the studies were in “patient selection” and “index test”. In the domain of “patient selection”, most of the included cohorts were case–control designed, without detailed inclusion/exclusion criteria or a description about whether a blind or random method was used, which resulted in a high risk of bias in “patient selection”. Additionally, miRNA results were interpreted after a diagnosis of IBD was made, and the miRNA threshold was set by the ROC curve instead of being pre-specified, leading to a risk of bias in the “index test”.
Figure 2.
Quality assessment of studies selected for analysis. (a) Summary of the risk of bias and applicability concerns: reviewers’ judgments about each domain for each included study. (b) Risk of bias and applicability concerns graph: reviewers’ judgments about each domain presented as percentages across the included studies.
Meta-analysis of diagnostic accuracy
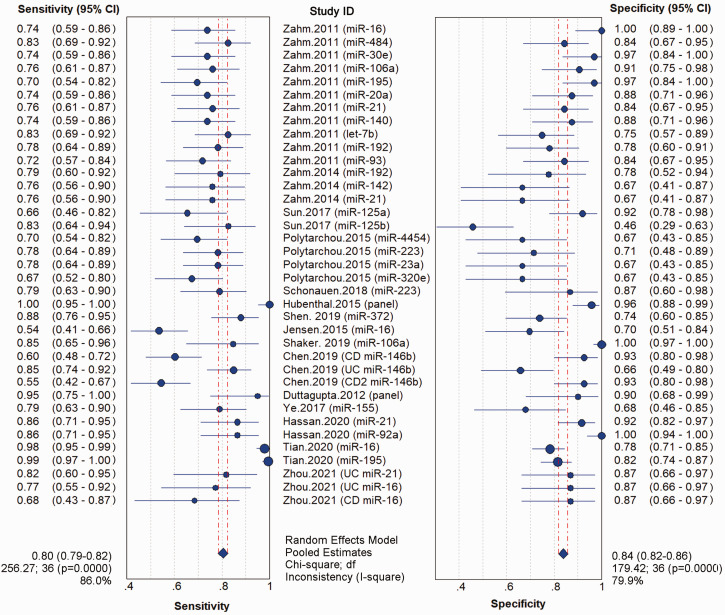
The analysis of heterogeneity showed no “shoulder arm” pattern in the ROC plane (Figure S1A), suggesting the absence of a threshold effect. Spearman’s correlation coefficient was 0.069, also indicating no obvious heterogeneity as a result of the threshold effect. Meta-analysis of the diagnostic odds ratio showed the presence of high heterogeneity with a Q value of 21.19 (P < 0.01) and I2 of 71.7% (Figure S1B). The meta-analysis of overall sensitivity (I2 = 86.0%; P < 0.01), specificity (I2 = 79.9%; P < 0.01), PLR (I2 = 73.3%; P < 0.01), and NLR (I2 = 78.9%; P < 0.01) also showed the presence of high heterogeneity (Figure 3 and Figure S2A). Therefore, a random effects model was applied to each meta-analysis.
Figure 3.
Forest plot for the meta-analysis of aberrant miRNA expression and sensitivity and specificity in IBD. Pooled values and 95% CIs were calculated using a random effects model.
IBD: inflammatory bowel disease; miRNA: microRNAs; CI, confidence interval.
The pooled estimates for sensitivity and specificity of all miRNAs to discriminate IBD from healthy controls or patients without IBD were 0.80 (95% CI: 0.79–0.82) and 0.84 (95% CI: 0.82–0.86), respectively, corresponding to a PLR of 4.43 (95% CI: 3.48–5.63) and an NLR of 0.26 (95% CI: 0.21–0.33), respectively (Figure 3 and Figure S2A). Meta-analysis showed a DOR of 21.19 (95% CI: 13.90–32.31) and an AUC of 0.89 for the diagnosis of IBD versus controls (Figure S1B and Figure S2B). These results indicated the good discriminative ability of miRNAs as diagnostic biomarkers for IBD.
Meta-regression and subgroup analysis
Meta-regression analysis was carried out to identify potential sources that might cause heterogeneity. The covariates considered included IBD subtype, age of participants, sample source, method of quantifying miRNA expression, and controls. Significant heterogeneity was observed in IBD subtype, sample source, method of quantifying miRNA expression, and controls between groups (Table S6). Based on this, subgroup analysis was conducted to improve the precision of the estimated diagnostic value of miRNA in patients with IBD (Table 2). This indicated that miRNAs had a higher diagnostic accuracy in patients with UC (AUC = 0.93) than in those with CD (AUC = 0.84), a higher diagnostic accuracy in distinguishing IBD from healthy controls (AUC = 0.90) than non-IBD controls (AUC = 0.80), and a higher diagnostic accuracy in adults (AUC =0.90) than in children (AUC = 0.85); miRNAs detected by microarray (AUC = 0.67) significantly decreased the diagnostic accuracy.
Table 2.
Subgroup analysis of miRNAs for IBD diagnosis based on different covariates.
| Subgroups | No. of patients | miRNAs | Sensitivity (95% CI) | Specificity (95% CI) | PLR (95% CI) | NLR (95% CI) | AUC | DOR (95% CI) | Heterogeneity (DOR-I2) | Heterogeneity (DOR-P value) |
|---|---|---|---|---|---|---|---|---|---|---|
| IBD subtype | ||||||||||
| UC | 500 | miR-4454, miR-223, miR-23a-3p, miR-320e, miR-372, miR-106a, miR-146b-5p, miR-21, miR-92a, miR-16, miR-195, miRNA panel 1 and 2 | 0.89 (0.87–0.91) | 0.84 (0.81–0.86) | 4.47 (3.25–6.19) | 0.16 (0.09–0.27) | 0.9315 | 40.85 (17.40–95.86) | 81.4% | <0.001 |
| CD | 370 | miR-16, miR-484, miR-30e, miR-106a, miR-195, miR-20a, miR-21, miR-140, let-7b, miR-192, miR-93, miR-125a, miR-125b, miR-223, miR-146b-5p and miRNA panel 1 | 0.72 (0.69–0.75) | 0.87 (0.84–0.89) | 5.67 (3.60–8.94) | 0.32 (0.27–0.39) | 0.8385 | 18.50 (11.38–30.08) | 57.7% | <0.001 |
| IBDU | 67 | miR-192, miR-142-3p, miR-21, miR-155 | 0.78 (0.69–0.85) | 0.70 (0.58–0.79) | 2.51 (1.77–3.54) | 0.32 (0.23–0.46) | 0.8558 | 7.88 (4.15–14.95) | 0.0% | 0.86 |
| Sample source | ||||||||||
| Blood | 820 | miR-16, miR-484, miR-30e, miR-106a, miR-195, miR-20a, miR-21, miR-140, let-7b, miR-192, miR-93, miR-142-3p, miR-125a, miR-125b, miR-4454, miR-223, miR-23a-3p, miR-320e, miR-372, miR-106a, miR-146b-5p, miR-155, miR-92a, miRNA panel 1 and 2 | 0.81 (0.79–0.83) | 0.84 (0.82–0.86) | 4.46 (3.42–5.81) | 0.26 (0.20–0.33) | 0.8952 | 22.37 (13.87–36.09) | 75.2% | <0.001 |
| Others | 117 | miR-223, miR-155, miR-21 and miR-16 | 0.78 (0.70–0.84) | 0.83 (0.74–0.89) | 4.05 (2.59–6.36) | 0.28 (0.20–0.39) | 0.8542 | 15.69 (8.13–30.28) | 0.0% | 0.66 |
| Method of quantifying miRNA | ||||||||||
| qPCR | 815 | miR-16, miR-484, miR-30e, miR-106a, miR-195, miR-20a, miR-21, miR-140, let-7b, miR-192, miR-93, miR-142-3p, miR-125a, miR-125b, miR-223, miR-372, miR-146b-5p, miR-155, miR-92a and miRNA panel 2 | 0.80 (0.78–0.82) | 0.84 (0.82–0.86) | 4.68 (3.62–6.06) | 0.26 (0.21–0.32) | 0.8952 | 23.26 (15.09–35.86) | 68.3% | <0.001 |
| Only microarray | 122 | miR-4454, miR-223, miR-23a-3p, miR-320e and miRNA panel 1 | 0.81 (0.76–0.86) | 0.81 (0.76–0.86) | 3.28 (1.59–6.74) | 0.30 (0.13–0.67) | 0.6677 | 12.09 (3.11–46.97) | 81.4% | <0.001 |
| Controls | ||||||||||
| Healthy controls | 729 | miR-16, miR-484, miR-30e, miR-106a, miR-195, miR-20a, miR-21, miR-140, let-7b, miR-192, miR-93, miR-125a, miR-125b, miR-4454, miR-223, miR-23a-3p, miR-320e, miR-372 and miR-155 | 0.84 (0.82–0.86) | 0.84 (0.81–0.86) | 4.62 (3.51–6.07) | 0.25 (0.19–0.31) | 0.9015 | 24.55 (15.00–40.17) | 71.0% | <0.001 |
| Non-IBD controls† | 171 | miR-192, miR-142-3p, miR-21, miR-16 and miR-146b-5p | 0.66 (0.61–0.71) | 0.78 (0.72–0.83) | 3.05 (2.35–3.97) | 0.42 (0.36–0.50) | 0.8009 | 8.15 (5.46–12.16) | 40.2% | 0.12 |
| Indistinguishable controls† | 37 | miR-21 and miR-16 | 0.86 (0.77–0.93) | 0.96 (0.91–0.99) | 18.97 (8.33–43.18) | 0.15 (0.08–0.26) | NA | 133.11 (43.31–409.18) | 53.9% | 0.14 |
†A fixed effects model was used; miRNA panel 1: miR-34b-3p, miR-142-5p, miR-205-5p, miR-424-5p, miR-570-3p, miR-885-5p, miR-1301-3p, miR-16-5p, hmiR-99b-5p; miRNA panel 2: miR-378*, miR-500, miR-378, miR-501-5p, miR-941, miR-140-3p, miR-422a, miR-720, miR-27a*, miR-181b, miR-150*, miR-146b-3p, miR-874, miR-138, miR-92a-1*, miR-1274b, miR-188-5p, miR-769-5p, miR-20b*, miR-769-3p, miR-550*, miR-22, miR-143*, miR-362-5p, miR-532-5p, miR-330-3p, miR-423-3p, miR-202 and miR-1263.
IBD: Inflammatory bowel disease; UC: Ulcerative colitis; CD: Crohn’s disease; IBDU: IBD unclassified; miRNA: microRNAs; qPCR: Quantitative polymerase chain reaction; No.: Number; 95% CI: 95% confidence interval; PLR: Positive likelihood ratio; NLR: Negative likelihood ratio; AUC: Area under curve; DOR: Diagnostic odds ratio; I2: Higgins’s inconsistency index; NA: Not available.
The potential biomarkers miR-21, miR-16, and miR-192 showed consistent up-regulation in blood samples from patients with IBD compared with controls in at least two studies. The combined effect size and 95% CIs were calculated and used as summary estimates. Heterogeneity was low for miR-192 (I2 = 0.0%) from the meta-analysis of DOR across the two studies, so the fixed effects model was applied. A random effects model was applied for the meta-analysis of miR-21 and miR-16. Heterogeneity test and meta-analysis results are presented in Table 3. miR-21, miR-16, and miR-192 showed a high-moderate diagnostic accuracy with a sensitivity of 0.79 (95% CI: 0.71–0.87), 0.93 (95% CI: 0.89–0.96), and 0.79 (95% CI: 0.68–0.87), respectively, and a specificity of 0.85 (95% CI: 0.77–0.91), 0.82 (95% CI: 0.76–0.87), and 0.78 (95% CI: 0.64–0.88), respectively.
Table 3.
Subgroup analysis of miRNAs for IBD diagnosis.
| Subgroup | miR-21 | miR-16 | miR-192† |
|---|---|---|---|
| No. of studies | 321,27,29 | 221,30 | 221,27 |
| No. of patients | 97 | 233 | 75 |
| No. of controls | 73 | 184 | 50 |
| Sensitivity (95% CI) | 0.79 (0.71–0.87) | 0.93 (0.89–0.96) | 0.79 (0.68–0.87) |
| I2, P value | 0.0%, 0.41 | 96.0%, <0.001 | 0.0%, 0.91 |
| Specificity (95% CI) | 0.85 (0.77–0.91) | 0.82 (0.76–0.87) | 0.78 (0.64–0.88) |
| I2, P value | 67.6%, 0.05 | 92.9%, <0.001 | 0.0%, 0.98 |
| PLR (95% CI) | 4.73 (1.93–11.62) | 5.21 (3.83–7.08) | 3.57 (2.09–6.10), |
| I2, P value | 74.7%, 0.02 | 70.5%, 0.07 | 0.0%, 0.10 |
| NLR (95% CI) | 0.26 (0.16–0.42) | 0.08 (0.05–0.13) | 0.27 (0.17–0.43) |
| I2, P value | 36.8%, 0.21 | 96.5%, <0.001 | 0.0%, 0.93 |
| DOR (95% CI) | 19.55 (5.20–73.49) | 167.39 (61.19–457.87) | 13.06 (5.65–30.17) |
| I2, P value | 69.9%, 0.04 | 0.0%, 0.96 | 0.0%, 0.96 |
†A fixed effects model was used.
No.: Number; 95% CI: 95% confidence interval; I2: Higgins’s inconsistency index; PLR: Positive likelihood ratio; NLR: Negative likelihood ratio; DOR: Diagnostic odds ratio.
Sensitivity analysis and publication bias
Sensitivity analysis showed that the removal of any individual study did not significantly affect the overall outcome (Table S7). The Deeks’ funnel plots revealed a publication bias (P < 0.10) (Figure S2), suggesting the presence of potential publication bias, language bias, inflated estimates by a flawed methodologic design in smaller studies, and/or a lack of publication of small trials with opposite results.
Rating the quality of evidence
The GRADE approach showed the quality of evidence to be low, indicating a limited confidence in the predicted value which might differ from the actual value. The evidence was downgraded in two steps, once for inconsistency and once for publication bias. A summary of these findings is shown in Table 4.
Table 4.
GRADE summary of findings for miRNAs in IBD diagnosis.
| Outcome | No. of studies and patients | Study design | Factors that may decrease certainty of
evidence |
Test accuracy coefficient | ||||
|---|---|---|---|---|---|---|---|---|
| Risk of bias | Indirectness | Inconsistency | Imprecision | Publication bias | ||||
| True positive | 15 studies1644 patients | Cross-sectional (cohort type accuracy study) | Not serious | Not serious | Seriousa | Not serious | Publication bias strongly suspectedb | ⨁⨁◯◯ Low |
| False negative | ||||||||
| True negative | 15 studies1644 patients | Cross-sectional (cohort type accuracy study) | Not serious | Not serious | Seriousa | Not serious | Publication bias strongly suspectedb | ⨁⨁◯◯Low |
| False positive | ||||||||
aMeta-analysis of the overall sensitivity (I2 = 86.0%; P < 0.01) and specificity (I2 = 79.9%; P < 0.01) shows the presence of high heterogeneity.
bThe Deeks’ funnel plots reveal publication bias (P < 0.10) (Figure S2).
Discussion
Our systematic review revealed that several aberrantly expressed miRNAs have been reported in IBD, including 18 up-regulated and three down-regulated miRNAs identified by miRNA microarray analysis, and 19 up-regulated and four down-regulated miRNAs identified by validation analysis. The overall findings of the meta-analysis enrolling 22 miRNAs and two miRNA panels from 15 studies showed that miRNAs could be used in diagnosing IBD, with a moderate level of sensitivity (0.80, 95% CI: 0.79–0.82) and specificity (0.84, 95% CI: 0.82–0.86). Subgroup analysis associated miRNAs in patients with UC (AUC = 0.93) with an increased level of diagnostic accuracy, while miRNAs detected by microarray (AUC = 0.67) were associated with a reduced diagnostic accuracy. Furthermore, miR-21, miR-16 and miR-192 were shown to be consistently up-regulated in the blood of patients with IBD, fulfilling a principal requirement as biomarkers for use in clinical practice. Despite the limited number of included studies, the pooled sensitivity and specificity showed a considerable diagnostic value for miR-21, miR-16, and miR-192 in distinguishing patients with IBD from controls.
Currently, there is no ideal biomarker that can be used to accurately diagnose IBD, although some studies have proposed an “IBD signature” consisting of a panel of biomarkers for IBD assessment. 32 However, the heavy burden on health systems and low diagnostic accuracy makes it challenging for decision makers to recommend it. Therefore, methods for convenient and economical diagnosis and clinical assessment are urgently needed, particularly when considering the reported rising in IBD incidence. 4
Aberrantly expressed miRNAs are involved in various complex diseases such as inflammation, cardiovascular disease, asthma, infections, and cancer. 33 Although systematic reviews and/or meta-analyses have included individual miRNAs showing aberrant expression between patients with IBD and controls, none have reported their diagnostic values.34,35 Thus, this is the first review to systematically report dysregulated miRNAs and to examine their diagnostic accuracy of IBD by meta-analysis.
An enhanced recognition of miRNA expression may aid a diagnosis of IBD and improve overall health outcomes. Detecting miRNAs with altered expression as diagnostic markers for IBD has advantages over other methods. For example, samples from peripheral blood, fresh tissues, or even FFPE tissues can be quickly and conveniently collected, and miRNA expression is consistent in these samples. 12 Additionally, as a mature technique for miRNA testing, qPCR is simple and reliable. An accurate diagnostic tool based on miRNAs will also help divert medical resources toward high-risk populations to improve the cost-effectiveness.
There are several limitations of this study. First, some relevant studies might have been missed because unpublished findings like conference abstracts and sponsor publications were not included, which could have led to publication bias. Additionally, the meta-analysis of miR-21, miR-16, and miR-192 only included a limited number of studies, which could have restricted the power of detection. Second, there was substantial heterogeneity in the data. Although the ROC plane indicated the absence of a threshold effect, meta-regression analysis found significant differences in the effect of IBD subtype, sample source, method of quantifying miRNA expression, and controls between groups. Third, the information obtained from the studies was relatively limited. The studies included in the meta-analysis applied unspecified cut-off values and diverse normalization methods as controls for miRNA quantification. Moreover, some studies failed to provide the most accurate estimate of diagnostic accuracy, so the results might not be sufficiently accurate. These limitations may have impacted our meta-analysis results, but we hope that it will provide the basis for future studies.
In conclusion, our data, representing a quantified synthesis of all published studies, evidenced that the expression of some miRNAs was altered in patients with IBD, and showed moderate diagnostic accuracy in distinguishing patients with IBD from healthy controls or patients with other diseases. Among these miRNAs, miR-21, miR-16, and miR-192 fulfilled a clinical practice requirement through their consistent alteration in the blood of patients with IBD. Additionally, the diagnostic accuracy of miRNAs was more obvious in UC patients and in distinguishing IBD from healthy controls. However, large-scale prospective, multi-center, and blinded studies involving both single- and combined-miRNA assays should be conducted to verify our results and confirm the clinical value of miRNAs in IBD diagnosis.
Supplemental Material
Supplemental material, sj-pdf-1-imr-10.1177_03000605221089503 for MicroRNAs as potential biomarkers for the diagnosis of inflammatory bowel disease: a systematic review and meta-analysis by Lina Sun, Yanan Han, Hua Wang, Huanyu Liu, Shan Liu, Hongbin Yang, Xiaoxia Ren and Ying Fang in Journal of International Medical Research
Author contributions: L.S., Y.H., and Y.F. conceived and designed the study. L.S., Y.H., H.W., H.L., and S.L. collected and analyzed the data. L.S. and H.W. wrote the manuscript and were responsible for language revisions. H.Y. assisted with data analyses. X.R. participated in manuscript writing. Y.F. supervised the project. All authors read and approved the final manuscript.
Declaration of conflicting interest: All authors declare that they have no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and publication of this article: This study was funded by The Basic Research Program of Shaanxi Natural Science Foundation (No. 2020JQ-928), Youth Training Funds by Xi’an Municipal Health Commission (No. 2020qn14), and a research grant from the Xi’an Children’s Hospital Program (Nos. 2019D06, 2020A08). The funding source did not, however, directly participate in the literature search, determination of study eligibility criteria, data analysis; or interpretation, or preparation, review, or approval of the manuscript for publication.
ORCID iD: Lina Sun https://orcid.org/0000-0003-1268-7033
Supplemental material: Supplemental material for this article is available online.
Data availability statement
The data that support the findings of this study are available in the supplementary materials of this article.
The study protocol is available at INPLASY, registration number: INPLASY202220027.
References
- 1.Podolsky D. Inflammatory bowel disease. N Engl J Med 2002; 347: 417–429. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi T, Siegmund B, Le Berre C, et al. Ulcerative colitis. Nat Rev Dis Primers 2020; 6: 74. [DOI] [PubMed] [Google Scholar]
- 3.Roda G, Chien Ng S, Kotze P, et al. Crohn's disease. Nat Rev Dis Primers 2020; 6: 22. [DOI] [PubMed] [Google Scholar]
- 4.Ng S, Shi H, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 2018; 390: 2769–2778. [DOI] [PubMed] [Google Scholar]
- 5.Mehrmal S, Uppal P, Nedley N, et al. The global, regional, and national burden of psoriasis in 195 countries and territories, 1990 to 2017: A systematic analysis from the Global Burden of Disease Study 2017. J Am Acad Dermatol 2021; 84: 46–52. [DOI] [PubMed] [Google Scholar]
- 6.Park K, Ehrlich O, Allen J, et al. The cost of inflammatory bowel disease: an initiative from the Crohn's & colitis foundation. Inflamm Bowel Dis 2020; 26: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandberg K, Yarger E, Saeed S. Updates in diagnosis and management of inflammatory bowel disease. Curr Probl Pediatr Adolesc Health Care 2020; 50: 100785. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura K, Sawada K, Yoshimura A, et al. Clinical relevance of circulating cell-free microRNAs in ovarian cancer. Mol Cancer 2016; 15: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen P, Li Y, Li L, et al. Circulating microRNA146b-5p is superior to C-reactive protein as a novel biomarker for monitoring inflammatory bowel disease. Aliment Pharm Ther 2019; 49: 733–743. [DOI] [PubMed] [Google Scholar]
- 10.Verdier J, Breunig IR, Ohse MC, et al. Faecal micro-RNAs in inflammatory bowel diseases. J Crohns Colitis 2020; 14: 110–117. [DOI] [PubMed] [Google Scholar]
- 11.Felwick RK, Dingley GJR, Martinez-Nunez R, et al. MicroRNA23a overexpression in Crohn's disease targets tumour necrosis factor alpha inhibitor protein 3, increasing sensitivity to TNF and modifying the epithelial barrier. J Crohns Colitis 2019; 14: 381–392. [DOI] [PubMed] [Google Scholar]
- 12.He C, Shi Y, Wu R, et al. MIR-301a promotes intestinal mucosal inflammation through induction of IL-17A and TNF-α in IBD. Gut 2016; 65: 1938–1950. [DOI] [PubMed] [Google Scholar]
- 13.Page MJ, Moher D, Bossuyt PM, et al. PRISMA 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews. BMJ 2021; 372: n160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zamora J, Abraira V, Muriel A, et al. Meta-DiSc: a software for meta-analysis of test accuracy data. Bmc Med Res Methodol 2006; 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whiting P, Rutjes A, Westwood M, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011; 155: 529–536. [DOI] [PubMed] [Google Scholar]
- 16.Higgins J, Thompson S, Deeks J, et al. Measuring inconsistency in meta-analyses. BMJ 2003; 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deeks J, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol 2005; 58: 882–893. [DOI] [PubMed] [Google Scholar]
- 18.Polytarchou C, Oikonomopoulos A, Mahurkar S, et al. Assessment of circulating microRNAs for the diagnosis and disease activity evaluation in patients with ulcerative colitis by using the Nanostring technology. Inflamm Bowel Dis 2015; 21: 2533–2539. [DOI] [PubMed] [Google Scholar]
- 19.Shaker OG, Ali MA, Ahmed TI, et al. Association between LINC00657 and miR-106a serum expression levels and susceptibility to colorectal cancer, adenomatous polyposis, and ulcerative colitis in Egyptian population. IUBMB Life 2019; 71: 1322–1335. [DOI] [PubMed] [Google Scholar]
- 20.Schonauen K, Le N, Von Arnim U, et al. Circulating and fecal microRNAs as biomarkers for inflammatory bowel diseases. Inflamm Bowel Dis 2018; 24: 1547–1557. [DOI] [PubMed] [Google Scholar]
- 21.Zahm AM, Thayu M, Hand NJ, et al. Circulating microRNA is a biomarker of pediatric Crohn disease. J Pediatr Gastr Nutr 2011; 53: 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen MD, Andersen RF, Christensen H, et al. Circulating microRNAs as biomarkers of adult Crohn's disease. Eur J Gastroen Hepat 2015; 27: 1038–1044. [DOI] [PubMed] [Google Scholar]
- 23.Sun CM, Wu J, Zhang H, et al. Circulating miR-125a but not miR-125b is decreased in active disease status and negatively correlates with disease severity as well as inflammatory cytokines in patients with Crohn's disease. World J Gastroentero 2017; 23: 7888–7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye Y, Pang Z, Gu W, et al . Expression of microRNA-155 in inflammatory bowel disease and its clinical significance. Nat Med J China 2017; 97: 3716–3719. [DOI] [PubMed] [Google Scholar]
- 25.Duttagupta R, DiRienzo S, Jiang R, et al. Genome-wide maps of circulating mirna biomarkers for ulcerative colitis. PLoS One 2012; 7: e31241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen M, Meng LN. Peripheral blood miR-372 as a biomarker for ulcerative colitis via direct targeting of NLRP12. Exp Ther Med 2019; 18: 1486–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zahm AM, Hand NJ, Tsoucas DM, et al. Rectal microRNAs are perturbed in pediatric inflammatory bowel disease of the colon. J Crohns Colitis 2014; 8: 1108–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubenthal M, Hemmrich-Stanisak G, Degenhardt F, et al. Sparse modeling reveals mirna signatures for diagnostics of inflammatory bowel disease. PLoS One 2015; 10: e0140155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmed Hassan E, El-Din Abd El-Rehim AS, Mohammed Kholef EF, et al. Potential role of plasma miR-21 and miR-92a in distinguishing between irritable bowel syndrome, ulcerative colitis, and colorectal cancer. Gastroenterol Hepatol Bed Bench 2020; 3: 147–154. [PMC free article] [PubMed] [Google Scholar]
- 30.Tian Y, Cui L, Lin C, et al. LncRNA CDKN2B-AS1 relieved inflammation of ulcerative colitis via sponging miR-16 and miR-195. Int Immunopharmacol 2020; 88: 106970. [DOI] [PubMed] [Google Scholar]
- 31.Zhou R, Qiu P, Wang H, et al. Identification of microRNA-16-5p and microRNA-21-5p in feces as potential noninvasive biomarkers for inflammatory bowel disease. Aging (Albany NY) 2021; 13: 4634–4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pisani L, Moriggi M, Gelfi C, et al. Proteomic insights on the metabolism in inflammatory bowel disease. World J Gastroentero 2020; 26: 696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev 2011; 91: 827–887. [DOI] [PubMed] [Google Scholar]
- 34.Yan H, Zhang X, Xu Y. Aberrant expression of miR-21 in patients with inflammatory bowel disease: A protocol for systematic review and meta analysis. Medicine (Baltimore) 2020; 99: e19693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nijakowski K, Surdacka A. Salivary biomarkers for diagnosis of inflammatory bowel diseases: a systematic review. Int J Mol Sci 2020; 21: 7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-imr-10.1177_03000605221089503 for MicroRNAs as potential biomarkers for the diagnosis of inflammatory bowel disease: a systematic review and meta-analysis by Lina Sun, Yanan Han, Hua Wang, Huanyu Liu, Shan Liu, Hongbin Yang, Xiaoxia Ren and Ying Fang in Journal of International Medical Research
Data Availability Statement
The data that support the findings of this study are available in the supplementary materials of this article.
The study protocol is available at INPLASY, registration number: INPLASY202220027.