Abstract
Objective
To determine how patients with non-small cell lung cancer (NSCLC) with programmed death-ligand 1 (PD-L1)-negative and/or a low tumor mutation burden status benefit from immune checkpoint inhibitors (ICI).
Methods
We determined the plasma cell-free DNA profiles of 25 patients with PD-L1-negative advanced NSCLC before ICI therapy using low-coverage whole-genome sequencing.
Results
Elevated cell-free copy number variations (CNVs) were associated with progressive disease, with a cutoff CNV score of 0.10 evaluated with an area under the curve of 0.790 in PD-L1-negative NSCLC. CNV changes were also correlated with poor survival. Progression-free survival and overall survival were both significantly shorter in CNVhigh compared with CNVlow patients.
Conclusions
Cell-free CNV may be a useful peripheral blood biomarker for predicting the response to ICIs in patients with NSCLC.
Keywords: Immune checkpoint inhibitor, copy number variation, molecular immune response, cell-free DNA, non-small cell lung cancer, programmed death-ligand 1
Introduction
Immune checkpoint inhibitors (ICIs) targeted to programmed cell death protein 1 (PD-1)/programmed death-ligand 1(PD-L1) have demonstrated clinical activity in many different malignant tumors, including lung cancer and breast cancer. 1 Monoclonal antibodies targeting inhibitory immune checkpoints have been approved as a conventional therapy and have changed the practice of medical oncology. 2 Although immunotherapy has demonstrated evident effects in patients with non-small cell lung cancer (NSCLC), there are obvious differences in responsiveness and efficacy among patients. 3 Establishing predictive biomarkers for immunotherapy is thus key to understanding drug resistance and maximizing the therapeutic effect.
PD-L1 expression has been confirmed as a common biomarker for immunological therapy. 4 However, PD-1/PD-L1 blocking antibodies have been administered based on different PD-L1 statuses and using different immunohistochemical antibodies, and there is currently no gold standard assay for the detection of PD-L1 expression. Pembrolizumab was originally approved for patients with PD-L1 expression ≥50%, and the US Food and Drug Administration subsequently approved its use as second-line therapy in patients with PD-L1 >1%, based on use of the Dako anti-PD-L1 22C3 antibody. 5 Nivolumab was approved for lung cancer, irrespective of PD-L1 expression, with PD-L1 status detected using the Dako 28-8 antibody as a complementary diagnostic method. 6 A relatively high prevalence of PD-L1-negativity of about 30% to 50% was observed in NSCLC patients with PD-L1 expression below 1%.7,8 However, one comprehensive analysis revealed that about 10% of PD-L1-negative patients showed tumor responses to PD-1/PD-L1 monotherapy, 9 indicating that PD-L1 testing alone may mean that some patients lose the opportunity to benefit from immunotherapy.
The tumor mutation burden (TMB) reflects the total number of mutations in tumor cells and may act as a quantitative biomarker for treatment with nivolumab in patients with NSCLC. 10 TMB has been shown to differentiate people likely to benefit from immunotherapy. 11 However, the use of TMB as a biomarker to predict immunotherapy resistance is limited by its cost and the availability of suitable tissues. 12 In addition, dynamic changes over time and tumor heterogeneity also make it an imperfect marker.12,13
Copy number variation (CNV), which is one of the most common and prominent features of solid tumors, was confirmed to be associated with treatment failure and disease recurrence, and accelerated the development of anticancer drug resistance. 14 Previous studies showed that CNV could provide higher levels of genetic diversity in patients, resulting in the emergence of multidrug resistance. 15 Jamal-Hanjani et al. reported that sustained dynamic CNV and genome doubling were associated with tumor heterogeneity and induced parallel evolution of driver somatic copy-number alterations, including BCL11A, CDK4, and FOXA1 in NSCLC. 16 In addition, CNV was more likely to select driving events compared with other mutation processes based on the consistency in variation of mutation levels. CNV therefore enables cells to adapt to the selective pressure generated by therapy and to follow several evolutionary trajectories leading to drug resistance. 17
Chromosome aneuploidy detection of plasma cell-free DNA (cfDNA) has recently been used as a noninvasive tool in prenatal tests, with minimal false positive and false negative results. 18 Similar to fetal tissues, tumors also continue to shed DNA fragments into the bloodstream, referred to as circulating tumor DNA (ctDNA). Although the proportion of ctDNA in plasma is generally low, ctDNA testing technology has been successfully applied in the clinic to detect cancer somatic mutations and other biomarkers, and chromosomal copy number changes were also detected in patients with various cancers including breast, liver, and lung cancer.19–21
In this study, we aimed to detect CNV in the plasma of patients with PD-L1-negative NSCLC treated with second- or third-line therapy, using low-coverage whole-genome sequencing (LC-WGS), to establish a new index for evaluating molecular immune response. We also analyzed the association between ICI survival benefit and CNV score.
Methods
Patients
Patients treated with ICIs in the Department of Oncology at the First Affiliated Hospital of Zhejiang University School of Medicine were enrolled in this study to evaluate the clinical value of CNV in immunotherapy. The eligibility criteria were: (1) age >18 years, (2) diagnosis of NSCLC, (3) presence of either measurable or evaluable disease by imaging before treatment initiation, and (4) negative PD-L1 expression. The exclusion criteria were: (1) loss to follow-up and (2) a history of multiple tumors. Blood samples were collected for cfDNA extraction. Disease progression and therapeutic response were determined by Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1. Radiographic assessments were categorized as progressive disease (PD), partial response (PR), stable disease (SD), and complete response (CR) at the end of the second cycle of immunotherapy. Progression-free survival (PFS) was defined as the time from the start of treatment to the first documentation of PD or death due to any cause, whichever occurred first. Overall survival (OS) was defined as the time from the first dose of immunotherapy to the date of death or the last follow-up. All patients were followed up until 15 March 2020. Blood samples were collected from all patients at baseline (before treatment initiation) and during ICI treatment.
The protocol was approved by the Institutional Review Board of the First Affiliated Hospital of Zhejiang University School of Medicine (No. 2017-873) on 30 December 2017. All recruited subjects signed written informed consent.
Sample preparation
Blood samples were collected before immunotherapy. Ten milliliters of peripheral blood was collected in a cfDNA protection vacuum tube (AmoyDx, Xiamen, Fujian, China) and centrifuged at 2500 ×g for 10 minutes. The supernatant was then transferred to a new tube and the samples were then centrifuged again at 15,800 ×g for 15 minutes. All the collection procedures were performed at 4°C and the plasma supernatant was stored at −80°C. cfDNA was extracted from 4 to 5 mL plasma using a QIAamp Circulating Nucleic Acid kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. For each patient, libraries were prepared with a Kapa Hyper Plus Kit (Roche, Basel, Switzerland), using 20 ng of cfDNA. Amplification was carried out with five to 12 polymerase chain reaction cycles of 98°C for 10 s and 65°C for 75 s, depending on the DNA concentration. Paired-end 150 base-pair (bp) sequencing was carried out according to the manufacturer’s protocol. WGS was performed using an Illumina HiSeq2000 (Illumina) with an average depth of 3×.
PD-L1 expression analysis
Tumor tissues were obtained from biopsy or surgery, fixed, embedded, and then cut into 5-µm sections and stained for PD-L1 (clone 22C3, Dako, Agilent, CA, USA), using the Dako Link 48 platform (Dako). The staining was evaluated by two independent pathologists in a blinded fashion. Positive expression was defined as expression in >1% of the tumor.
Gene-level copy number analyses
Sequencing coverage for each 200 kbp bin was calculated followed by GC normalization. Sequencing coverage was further normalized by a set of control samples. The Z-score for each bin was calculated according to the formula: , where and are the coverages of the bins of testing samples and non-tumor control samples, respectively. The formula calculated the Z value of each genome segment. Samples were excluded if the standard deviation of copy ratios between adjacent bins, genome-wide, was >30, suggesting poor-quality sequence data. The normalized bin values were then sent to the segmentation calls algorithm using the circular segmentation algorithm provided by the R package DNAcopy. 22 CNVs (log ratios and P values) were also reported by DNAcopy. The copy number of a gene was estimated based on the genomic segment where the gene was located. The CNV score was then calculated by summation of all the changed segments according to the following formula: (Formula 1), where is the segment value and is the segment length in units of 200 kbp. This formula was used to calculate the sum of Z-scores of the whole genome.
Statistical analysis
Copy number changes were analyzed using the R package DNAcopy. The absolute segment value was used for further analysis. The predictive ability of the CNV score was estimated by receiver operating characteristic (ROC) curve analysis. Categorical data were analyzed by χ2 test. PFS and OS rates were analyzed according to the Kaplan–Meier method and compared by log-rank tests. The difference in response rates between CNVhigh and CNVlow patients were calculated using the prop trend.test function of R 4.1.2. A P value < 0.05 was considered statistically significant. All statistical analyses were performed using SPSS 22.0 (IBM Corp, Armonk, NY, USA).
Results
Patient characteristics
Twenty-five patients with advanced stage non-small cell lung cancer, including 13 cases of adenocarcinoma and 12 cases of squamous cell carcinoma, were recruited according to the inclusion and exclusion criteria. The median age was 60 years, 28% (7/25) were women, and 72% (18/25) were current or former smokers. About 92% (23/25) of patients received second-line therapy and only 8% (2/25) received third-line therapy. PD-L1 expression was negative in all patients (Supplemental Figure 1). All patients received ICIs, including nivolumab, pembrolizumab, or camrelizumab, with or without chemotherapy. Pretreatments before ICI included chemotherapy, radiochemotherapy, or chemotherapy combined with anti-angiogenesis therapy (Table 1). One patient failed epidermal growth factor-receptor tyrosine kinase inhibitor treatment. Peripheral blood was collected from all patients at baseline before ICI treatment, and post-treatment blood samples during ICI therapy were obtained from two patients.
Table 1.
Patient treatments and responses.
| Patient ID | Age (years) | Sex | Smoking | Histopathologic type | Response | CNV score | Pre-treatment | Treatment | Lines of treatment |
|---|---|---|---|---|---|---|---|---|---|
| P01 | 60 | F | N | A | SD | 0.027 | Chemo + anti-angiogenesis | IO | 2 |
| P02 | 66 | M | Y | S | PR | 0.028 | Chemo + RT | IO | 2 |
| P03 | 59 | M | Y | S | SD | 0.031 | Chemo + RT | IO | 2 |
| P04 | 70 | F | N | A | PD | 0.034 | Chemo | IO | 2 |
| P05 | 65 | M | Y | A | PR | 0.042 | Chemo | IO | 2 |
| P06 | 69 | M | Y | S | PR | 0.043 | Chemo + anti-angiogenesis | IO | 2 |
| P07 | 59 | F | N | A | SD | 0.050 | Chemo | IO | 2 |
| P08 | 74 | M | Y | A | SD | 0.051 | Chemo + anti-angiogenesis | IO | 2 |
| P09 | 61 | F | Y | S | SD | 0.052 | Chemo | IO + chemo | 2 |
| P10 | 60 | M | N | S | SD | 0.059 | Chemo | IO | 3 |
| P11 | 58 | M | Y | S | PR | 0.065 | Chemo | IO + chemo | 2 |
| P12 | 55 | F | N | A | SD | 0.067 | Chemo | IO | 2 |
| P13 | 59 | F | N | A | PR | 0.072 | TKI | IO | 3 |
| P14 | 60 | M | Y | A | SD | 0.078 | Chemo | IO + chemo | 2 |
| P15 | 63 | M | Y | A | PR | 0.084 | Chemo | IO + chemo | 2 |
| P16 | 67 | M | Y | S | PR | 0.089 | Chemo | IO + chemo | 2 |
| P17 | 60 | M | Y | A | PR | 0.090 | Chemo | IO + chemo | 2 |
| P18 | 55 | M | Y | A | PD | 0.101 | Chemo | IO | 2 |
| P19 | 46 | M | Y | S | SD | 0.109 | Chemo + RT | IO | 2 |
| P20 | 74 | M | Y | S | SD | 0.113 | Chemo | IO | 2 |
| P21 | 54 | M | Y | A | PR | 0.122 | Chemo | IO + chemo | 2 |
| P22 | 62 | M | Y | S | SD | 0.142 | Chemo | IO | 2 |
| P23 | 52 | M | Y | S | PD | 0.418 | Chemo | IO | 2 |
| P24 | 60 | F | N | A | PD | 0.538 | Chemo | IO | 2 |
| P25 | 66 | M | Y | S | PD | 30.248 | Chemo + RT | IO | 2 |
M: male; F: female; A: adenocarcinoma; S: Squamous carcinoma; PD: progressive disease; PR: partial response; SD: stable disease; CR: complete response; PFS: progression-free survival; CNV: copy number variation; IO: PD1/PD-L1 immunotherapy; chemo: chemotherapy; RT: radiotherapy.
Whole-genome copy number profiling of cfDNA
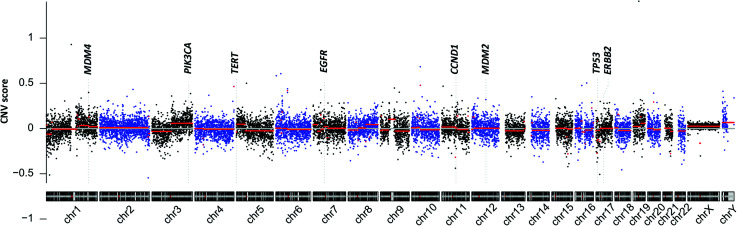
Reads were mapped to the human reference genome hg19. Genomic coverage was counted using samtoolsmpileup, and the average coverage for each 200 kbp bin was then calculated. A circular binary segmentation algorithm was then used to detect significant genomic breakpoints. The overall chromosomal CNVs are shown in Figure 1. Frequent copy number changes were found in chromosome 3q gains, 7p gains, and 17p losses in six (24.0%), 12 (48.0%), and five (20.0%) patients, respectively. The CNV score for each sample was then estimated by the algorithm, as described in the methods.
Figure 1.
Cell-free cancer genome of non-small cell lung cancer.
CNV, copy number variation; chr, chromosome.
Predictive value of CNV in plasma for therapeutic response
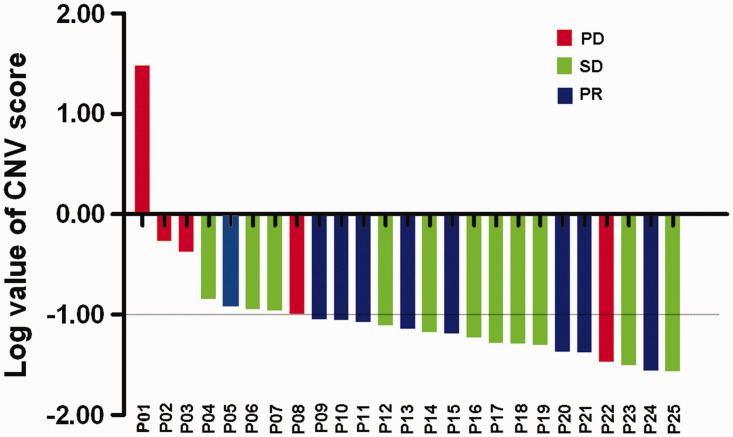
We also investigated the predictive value of CNV score in terms of therapeutic response and disease progression. The patients were divided into two groups: a PD group (n = 5) and a non-PD group (n = 20). The CNV score was able to distinguish the different responses to immunotherapy, with an area under the ROC curve of 0.790 (P = 0.049) and a cut-off value of 0.10. In this case, patients with a high CNV score were defined as a CNVscore ≥0.10 , while a low CNV score was defined as a CNVscore <0.10. Patients were then divided into CNVhigh (n = 8) and CNVlow groups (n = 17) according to the cut-off value. Four patients (50.0%) in the CNVhigh group experienced PD evaluated after second cycle of ICI treatment, while 16 (94.2%) patients in the CNVlow group experienced disease control (SD or PR) (Figure 2).
Figure 2.
Waterfall plots of clinical responses to immunotherapy in patients with non-small cell lung cancer. All patients were ranked on basis of the log values of copy number variation (CNV) score.
PD, progressive disease: SD, stable disease; PR, partial response.
Elevated CNV score in plasma cfDNA predicted worse survival
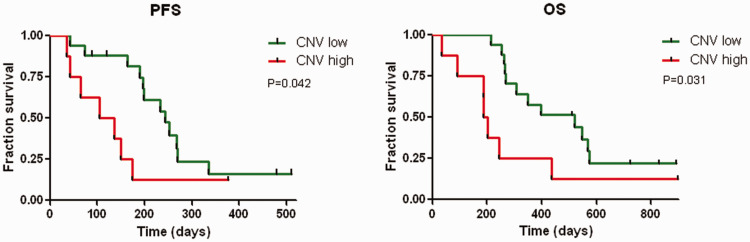
Eight (47.1%) patients with SD and eight (47.1%) patients with PR had low CNV scores before treatment (Table 2), compared with only one (12.5%) patient with PR and three (37.5%) with SD with high baseline CNV scores. These results suggest that plasma cfDNA CNV may be an indicator of the benefits of ICI therapy. We also followed-up the PFS and OS to evaluate the predictive value of CNV assessment. The median follow-up period for all patients was 306 days (range 35–898 days). The median PFS and OS for all patients (n = 25) were 197days (95% confidence interval [CI]: 160–234 days) and 350 days (95% CI: 150–549 days), respectively. Kaplan–Meier analyses revealed that the PFS rate was significantly poorer in the CNVhigh compared with the CNVlow group (P = 0.042; Figure 3a). At the time of data cut-off, 19 patients had died (76.0%, 19/25), including seven in the CNVhigh group (87.5%, 7/8) and 12 in the CNVlow group (70.6%, 12/17). The median OS was significantly poorer in the CNVhigh compared with the CNVlow group (P = 0.031, Figure 3b). Univariate analysis indicated that pretreatment, lines of therapy, histopathology type, and ICI alone or combined with other treatment were not associated with PFS or OS (Table 3).
Table 2.
Clinical response in relation to cell-free tumor DNA concentration.
| Response | High CNV n (%) (n = 8) | Low CNV n (%) (n = 17) |
|---|---|---|
| PD | 4 (50.0) | 1 (5.8) |
| SD | 3 (37.5) | 8 (47.1) |
| PR | 1 (12.5) | 8 (47.1) |
Prop.trend.test, P = 0.049.
CNV: copy number variation; PD: progressive disease; SD: stable disease; PR: partial response.
Figure 3.
Survival curves in the copy number variation (CNV)high and CNVlow groups demonstrated by Kaplan–Meier analyses. Comparisons of (a) progression-free survival (PFS) and (b) overall survival (OS) rates in lung cancer patients with CNVhigh and CNVlow.
Table 3.
Univariate analysis of survival in all patients (n = 25).
| PFS |
OS |
|||
|---|---|---|---|---|
| HR [95%CI] | P value | HR [95%CI] | P value | |
| Age | 0.932 [0.411, 2.113] | 0.867 | 2.089 [0.818, 5.336] | 0.123 |
| >60 years versus ≤60 years | ||||
| Sex | 1.09 [0.448, 2.655] | 0.849 | 0.575 [0.205, 1.61] | 0.292 |
| Female versus male | ||||
| Smoking status | 0.631 [0.255, 1.559] | 0.318 | 2.912 [0.944, 8.978] | 0.063 |
| Ever/current versus never | ||||
| Pre-treatment | 0.192 | 0.598 | ||
| Chemo | ||||
| Chemo+RT | 0.74 [0.23, 2.382] | 0.614 | 0.446 [0.122, 1.629] | 0.222 |
| Chemo+anti-angiogenesis | 0.202 [0.038, 1.069] | 0.060 | 0.549 [0.123, 2.447] | 0.431 |
| Treatment | 0.655 [0.262, 1.633] | 0.364 | 1.984 [0.675, 5.829] | 0.213 |
| IO versus IO plus other treatment | ||||
| Histopathologic type | 1.226 [0.538, 2.794] | 0.628 | 1.412 [0.571, 3.495] | 0.455 |
| Adenocarcinoma versus | ||||
| Squamous carcinoma | ||||
| Lines of chemo | 0.617 [0.136, 2.79] | 0.53 | 0.034 [0, 7.292] | 0.217 |
| 3 versus 2 | ||||
PFS, progression-free survival; OS, overall survival; HR, hazard ratio; CI, confidence interval; Chemo, chemotherapy; RT, radiotherapy; IO, PD1/PD-L1 immunotherapy.
Dynamic changes in plasma CNV score correlated with clinical response to ICIs
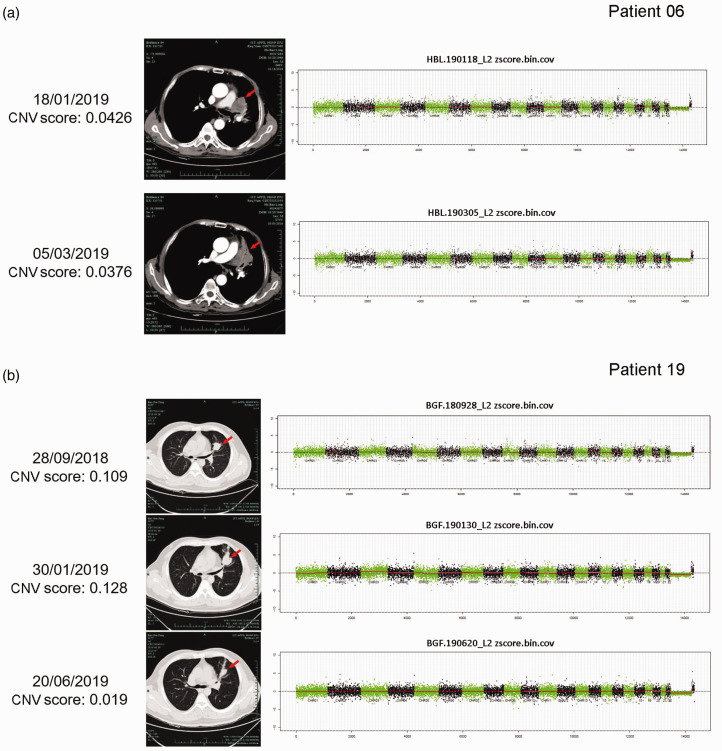
Plasma samples from two patients were analyzed after the first cycle of treatment. The baseline CNV score in Patient 06 was 0.043, which had decreased to 0.038 at the second visit (Figure 4a). Computed tomography (CT) scanning also showed that the tumor lesion had reduced compared with baseline, and was mainly replaced by lung tissue with atelectasis. The baseline CNV score in Patient 19 was 0.109, which had increased slightly to 0.128 at the second visit after ICI treatment and CT showed PD compared with the baseline tumor size. However, the patient’s clinical symptoms were relieved and they continued to receive ICI treatment, and their CNV score had decreased to 0.019 by the eighth month, indicating potential clinical benefits of the ICI treatment. CT scan confirmed that the lesion had shrunk (Figure 4b).
Figure 4.
Dynamic changes in plasma copy number variation (CNV) score correlated with clinical response to immune checkpoint inhibitors. Computed tomography images and CNV scores at different times in (a) Patient 06, (b) Patient 19. Red arrows indicate tumor lesions.
Discussion
ICIs have resulted in unprecedented response rates in various, intractable malignancies. However, some patients fail to respond to ICIs, which may also cause serious side effects. 23 Credible biomarkers for ICI therapy are therefore urgently needed to improve the selection of patients likely to benefit from this therapy. In addition, establishing a predictive index could also help to tailor therapy regimens and clarify drug mechanisms.
Various types of immune biomarkers, including circulating tumor and immune cells, serum proteins, factors associated with the tumor microenvironment, tumor-specific receptor expression patterns, and host genomic factors, have been explored in previous studies. 24 However, credible biomarkers are still lacking because of our poor understanding of how ICIs regulate the immune response to cancer, as well as their effects on the tumor microenvironment, dynamic immune milieu, and the contribution of immune editing. PD-L1 expression in the tumor microenvironment has been shown to have predictive value for the response to immunotherapy in patients with advanced solid tumors. 25 However, many PD-L1-negative patients may also benefit from ICI treatment, and in addition, PD-L1 testing requires the availability of suitable tumor tissue.
In this study, we investigated the whole-genome CNV of ctDNA as a potential biomarker candidate in patients with PD-L1-negative NSCLC. We found that patients with low baseline CNV levels had significantly better survival and better responses than patients with high baseline CNV. In addition, the dynamic changes in CNV levels in two patients showed good concordance with the clinical radiographic assessments. In Patient 19, radiographic evaluation showed PD while the CNV score was only slightly increased, suggesting that the patient may be experiencing pseudoprogression; continued ICI treatment accordingly led to tumor shrinkage, confirmed by CT scan, and an obviously decreased CNV score. CNV score could thus help to differentiate between pseudoprogression and true progression in patients receiving immunotherapy. Several previous studies also evaluated ctDNA at baseline or longitudinal ctDNA dynamics to assess the tumor response to systemic therapy, including chemotherapy and targeted therapy.26–28 Especially in the case of immunotherapy, previous explorative studies have mainly focused on using point mutations to evaluate the blood TMB; however, the suitable panel size and kind of variants that should be included for TMB calling is still controversial.
Several recent studies indicated that chromosomal abnormalities might cause dysregulation of the immune microenvironment, and tumors with low CNV levels displayed more immune infiltration and a better response to ICI treatment.29,30 Detailed analysis of two published clinical trials of melanoma patients treated with immunotherapy showed that the somatic CNV level was a stronger predictive marker of cytotoxic immune cell infiltration than TMB. 29 These findings suggest that plasma CNV score may become an effective and accurate biomarker for predicting resistance to immunotherapeutic agents in patients with lung cancer. Several studies have reported the value of detecting CNV in cfDNA using LC-WGS. Weiss et al. found that CNV scoring using cfDNA could be used as an early predictor of immunotherapy response in patients with advanced solid tumors. 31 Cai and colleagues demonstrated that dynamic plasma CNV levels detected by LC-WGS correlated with tumor burden in patients with hepatocellular carcinoma. 32 We previously performed LC-WGS in 31 patients with advanced stage lung cancer, and showed that cfDNA CNV might be a useful biomarker for predicting intrinsic resistance to epidermal growth factor receptor-targeted therapy. 33 These studies showed that CNV detection by LC-WGS could be developed as a promising noninvasive method for monitoring therapeutic effects, prognosis evaluation, and auxiliary diagnosis. The current results demonstrated that the CNV score detected by LC-WGS could be a good predictive biomarker of ICI response.
LC-WGS is also relatively cost-effective because of the simplified laboratory protocol and low volume of sequencing data, which could increase the feasibility of its clinical application.
This study had several limitations. First, the sample size was limited. Further studies with more patients are therefore needed to validate the utility of ctDNA measurements to assess the therapeutic effect of immunotherapy. Second, TMB testing of patients was not conducted in parallel because of the limited tissue samples, and we were therefore unable to compare the predictive abilities of CNV and TMB. The possible complementarity of these two biomarkers thus remains unclear. Third, in addition to ICI monotherapy, some patients in the study also received a combination of ICIs and chemotherapy, which may have resulted in bias in responding to immunotherapy.
The results of this study thus indicate that CNV score based on ctDNA can be used to predict survival benefit and reflect treatment response to immunotherapy in patients with PD-L1-negative NSCLC. The findings also indicate the potential for using dynamic CNV scores to assess the efficacy of immunotherapy.
Supplemental Material
Supplemental material, sj-jpg-1-imr-10.1177_03000605221093222 for Association between plasma somatic copy number variations and response to immunotherapy in patients with programmed death-ligand 1-negative non-small cell lung cancer by Xiaochen Zhang, Yina Wang, Jingjing Xiang, Pan Zhao, Yanping Xun, Shirong Zhang and Nong Xu in Journal of International Medical Research
Acknowledgements
The authors would like to thank the patients for providing their consent to present the data in this study.
Footnotes
Declaration of conflicting interest: The authors declare that there are no conflicts of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Zhejiang Provincial Natural Science Foundation [grant number LY19H160032] and major project of the Hangzhou Science and Technology Bureau [grant number 20180417A01].
ORCID iD: Shirong Zhang https://orcid.org/0000-0001-9334-1637
Supplemental material: Supplemental material for this article is available online.
References
- 1.Jenkins RW, Barbie DA, Flaherty KT. Mechanisms of resistance to immune checkpoint inhibitors. Br J Cancer 2018; 118: 9–16. DOI: 10.1038/bjc.2017.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma P, Hu-Lieskovan S, Wargo JA, et al. Primary, adaptive, and acquired resistance to Cancer immunotherapy. Cell 2017; 168: 707–723. DOI: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galli G, Proto C, Signorelli D, et al. Characterization of patients with metastatic non-small-cell lung cancer obtaining long-term benefit from immunotherapy. Future Oncol 2019; 15: 2743–2757. DOI: 10.2217/fon-2019-0055. [DOI] [PubMed] [Google Scholar]
- 4.Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012; 366: 2455–2465. DOI: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgensztern D, Herbst RS. Nivolumab and pembrolizumab for non-small cell lung cancer. Clinical Cancer Res 2016; 22: 3713–3717. DOI: 10.1158/1078-0432.CCR-15-2998. [DOI] [PubMed] [Google Scholar]
- 6.Li JX, Huang JM, Jiang ZB, et al. Current clinical progress of PD-1/PD-L1 immunotherapy and potential combination treatment in non-small cell lung cancer. Integr Cancer Ther 2019; 18: 1534735419890020. DOI: 10.1177/1534735419890020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun JM, Zhou W, Choi YL, et al. Prognostic significance of PD-L1 in patients with non-small cell lung cancer: a large cohort study of surgically resected cases. J Thorac Oncol 2016; 11: 1003–1011. DOI: 10.1016/j.jtho.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Zhang M, Li G, Wang Y, et al. PD-L1 expression in lung cancer and its correlation with driver mutations: a meta-analysis. Sci Rep 2017; 7: 10255. DOI: 10.1038/s41598-017-10925-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao B, Zhao H, Zhao J. Efficacy of PD-1/PD-L1 blockade monotherapy in clinical trials. Ther Adv Med Oncol 2020; 12: 1758835920937612. DOI: 10.1177/1758835920937612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015; 348: 124–128. DOI: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019; 51: 202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cyriac G, Gandhi L. Emerging biomarkers for immune checkpoint inhibition in lung cancer. Semin Cancer Biol 2018; 52: 269–277. DOI: 10.1016/j.semcancer.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Ready N, Hellmann MD, Awad MM, et al. First-line nivolumab Plus ipilimumab in advanced non-small-cell lung cancer (CheckMate 568): outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. J Clin Oncol 2019; 37: 992–1000. DOI: 10.1200/JCO.18.01042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuznetsova AY, Seget K, Moeller GK, et al. Chromosomal instability, tolerance of mitotic errors and multidrug resistance are promoted by tetraploidization in human cells. Cell Cycle 2015; 14: 2810–2820. DOI: 10.1080/15384101.2015.1068482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W, Zhang Y, Chen R, et al. Chromosomal instability and acquired drug resistance in multiple myeloma. Oncotarget 2017; 8: 78234–78244. DOI: 10.18632/oncotarget.20829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med 2017; 376: 2109–2121. DOI: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 17.Lee HS, Lee NC, Kouprina N, et al. Effects of anticancer drugs on chromosome instability and new clinical implications for tumor-suppressing therapies. Cancer Res 2016; 76: 902–911. DOI: 10.1158/0008-5472.CAN-15-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith-Bindman R, Miglioretti D. Cell-free DNA analysis for noninvasive examination of trisomy. N Engl J Med 2015; 373: 2581. DOI: 10.1056/NEJMc1509344. [DOI] [PubMed] [Google Scholar]
- 19.Stover DG, Parsons HA, Ha G, et al. Association of cell-free DNA tumor fraction and somatic copy number alterations with survival in metastatic triple-negative breast cancer. J Clin Oncol 2018; 36: 543–553. DOI: 10.1200/jco.2017.76.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang P, Chan CW, Chan KC, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci USA 2015; 112: E1317–E1325. DOI: 10.1073/pnas.1500076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia S, Huang CC, Le M, et al. Genomic variations in plasma cell free DNA differentiate early stage lung cancers from normal controls. Lung Cancer 2015; 90: 78–84. DOI: 10.1016/j.lungcan.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 2007; 23: 657–663. [DOI] [PubMed] [Google Scholar]
- 23.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med 2015; 373: 23–34. DOI: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017; 14: 531–548. DOI: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 25.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012; 366: 2443–2454. DOI: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis AA, Iams WT, Chan D, et al. Early assessment of molecular progression and response by whole-genome circulating tumor DNA in advanced solid tumors. Mol Cancer Ther 2020; 19: 1486–1496. DOI: 10.1158/1535-7163.MCT-19-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang B, Niu X, Zhang Q, et al. Circulating tumor DNA detection is correlated to histologic types in patients with early-stage non-small-cell lung cancer. Lung Cancer 2019; 134: 108–116. DOI: 10.1016/j.lungcan.2019.05.034. [DOI] [PubMed] [Google Scholar]
- 28.Merker JD, Oxnard GR, Compton C, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018; 36: 1631–1641. DOI: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]
- 29.Davoli T, Uno H, Wooten EC, et al. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017; 355: eaaf8399. DOI: 10.1126/science.aaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei Y, Zhang G, Zhang C, et al. The average copy number variation (CNVA) of chromosome fragments is a potential surrogate for tumor mutational burden in predicting responses to immunotherapy in non-small-cell lung cancer. Clin Transl Immunology 2021; 10: e1231. DOI: 10.1002/cti2.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen TJ, Goodman AM, Kato S, et al. Genome-wide sequencing of cell-free DNA identifies copy-number alterations that can be used for monitoring response to immunotherapy in cancer patients. Mol Cancer Ther 2019; 18: 448–458. DOI: 10.1158/1535-7163.mct-18-0535. [DOI] [PubMed] [Google Scholar]
- 32.Zhixiong C, Geng C, Yongyi Z, et al. Comprehensive profiling of circulating tumor DNA and protein biomarkers in long-term follow-up patients with hepatocellular carcinoma. Clin Cancer Res 2019; 25: 5284–5294. DOI: 10.1158/1078-0432.CCR-18-3477. [DOI] [PubMed] [Google Scholar]
- 33.Lucheng Z, Jiafeng L, Bing X, et al. Identification of somatic copy number variations in plasma cell free DNA correlating with intrinsic resistances to EGFR targeted therapy in T790M negative non-small cell lung cancer. J Thorac Dis 2020; 12: 883–892. DOI: 10.21037/jtd.2019.12.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-jpg-1-imr-10.1177_03000605221093222 for Association between plasma somatic copy number variations and response to immunotherapy in patients with programmed death-ligand 1-negative non-small cell lung cancer by Xiaochen Zhang, Yina Wang, Jingjing Xiang, Pan Zhao, Yanping Xun, Shirong Zhang and Nong Xu in Journal of International Medical Research