Summary
Valosin-containing protein (VCP, also known as p97/Cdc48) comprises six identical 97 kDa VCP protomers and functions as a master regulator of cellular homeostasis. VCP dodecamer in an apo nucleotide status was recently reported, providing a new framework for studying VCP’s diverse biological functions. Here, we present a detailed protocol for purifying and cryo-EM structurally characterizing VCP dodecamers from both bacterial and mammalian cells. This protocol can also be adapted to yeast Cdc48.
For complete details on the use and execution of this protocol, please refer to Yu et al. (2021).
Subject areas: Molecular Biology, Protein Biochemistry, Protein expression and purification, Structural Biology, Cryo-EM
Graphical abstract
Highlights
-
•
Detailed protocol for purification of VCP dodecamers from H1299 mammalian cells
-
•
Detailed protocol for purification of His-VCP from E.coli and generation of dodecamers
-
•
Step-by-step guide for Cryo-EM data collection of VCP dodecamers
-
•
Use of Coot and Phenix for model building and refinement
Valosin-containing protein (VCP, also known as p97/Cdc48) comprises six identical 97 kDa VCP protomers and functions as a master regulator of cellular homeostasis. VCP dodecamer in an apo nucleotide status was recently reported, providing a new framework for studying VCP’s diverse biological functions. Here, we present a detailed protocol for purifying and cryo-EM structurally characterizing VCP dodecamers from both bacterial and mammalian cells. This protocol can also be adapted to yeast Cdc48.
Before you begin
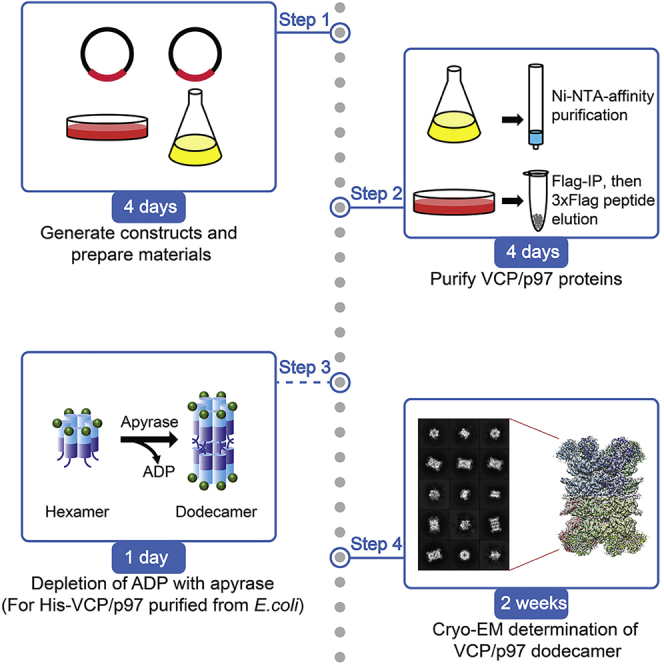
This protocol describes the detailed steps for purifying recombinant VCP dodecamers from bacterial and mammalian cells for high resolution single particle cryo-electron microscopy (cryo-EM) structure determination. Before structure determination, bacterial and mammalian full length VCP-expressing plasmids, transfection reagent polyethylenimine (PEI), and H1299 mammalian cells need to be prepared. For the preparation of VCP protein from mammalian cell, we use FLAG affinity tags for purification, but other tags, such as HA or V5 tag, may also be utilized. We use H1299 lung cancer cells for their fast growth rate and high transfection efficiency, but the current procedure can also be adapted to other mammalian cell types.
VCP-expressing plasmids construction and preparation
Timing: 4 days
-
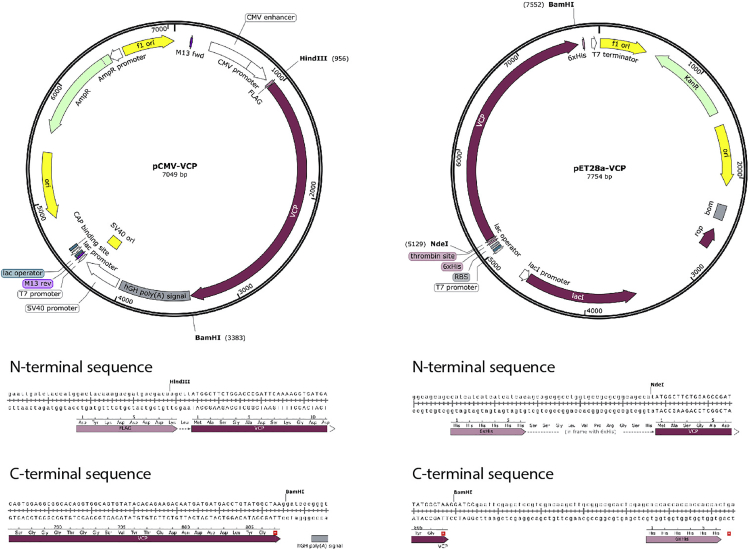
1.Subclone the DNA sequence of VCP (Uniprot: P55072) into the pCMV (Addgene) and pET28a (Novagen) vectors to generate constructs with an N-terminal FLAG-tag for mammalian expression or in-frame with a N-terminal hexa-Histidine tag (His-tag) for expression in E. coli, respectively (Figure 1).
-
a.Amplify the DNA sequence of human VCP (1-806 aa) with VCP primer set1 for pCMV vector and VCP primer set2 for pET28a vector by polymerase chain reaction (PCR) with PfuTurbo DNA Polymerase (Agilent).
-
b.Run a 1% agarose gel to confirm the correct size (∼2.4 kb for VCP) of the PCR product, and Fast DNA Ladder (NEB) was used to assess the size of digested fragments. The amplified VCP DNA fragment was purified using the QIAquick PCR Purification Kit (Qiagen) with at least 80% yield.
-
c.Perform restriction enzyme digestion of purified DNA sequence of VCP and vectors at 37°C for 1 h (HindIII/BamHI double digestion for pCMV-VCP and NdeI/BamHI double digestion for pET28a-VCP). Then, purify the digested products using QIAquick Gel Extraction Kit (Qiagen) with at least 80% yield.
-
d.Ligate the insert (150 ng) and vector (100 ng) using a molar ratio of 4:1 with 1 μL T4 DNA ligase (NEB) and 1 μL 10× T4 DNA ligase buffer in 10 μL reaction volume at 16°C for 12–16 h.
-
e.Transform chemically competent DH5a cells (100 μL) with 6 μL of the above ligation mixture by performing the heat-shock in a water bath at 42°C for 90 s followed by incubation on ice for 3 min, and incubate at 37°C for 1 h after adding 200 μL LB medium.
-
f.Transfer 100 μL of the above cultures to LB agar plates containing 100 μg/mL Ampicillin or 50 μg/mL Kanamycin for pCMV-VCP and pET28a-VCP constructs, respectively, and incubate for 15–18 h at 37°C.
-
g.Pick at least 5 single colonies for each construct and inoculate with 5 mL LB medium containing corresponding antibiotics (100 μg/mL of Ampicillin for pCMV-VCP and 50 μg/mL of Kanamycin for pET28a-VCP), and incubate for 16–24 h at 37°C with shaking at 200 rpm (New Brunswick Innova 44R). Then, extract plasmids DNA using QIAprep Spin Miniprep Kit (Qiagen). The miniprep yield is about 20 μg for pCMV-VCP vector and 5 μg for pET28a-VCP vector.
-
a.
-
2.
Sequence the purified plasmids with T7 promoter primer (TAATACGACTCACTATAGGG) and T7 terminator primer (GCTAGTTATTGCTCAGCGG) for pET28a-VCP, and CMV promoter primer (CGCAAATGGGCGGTAGGCGTG) and pBABE 3′ primer (ACCCTAACTGACACACATTCC) to confirm the subcloning prior to proceeding.
CRITICAL: The full-length sequence of VCP (806 aa) and the reading frame of N-terminal FLAG or His tag should be confirmed before proceeding to the next step.
Pause point: Constructed plasmids DNA could be stored at −20°C until ready to proceed.
-
3.
Inoculate 500 mL LB medium with a pCMV-VCP containing colony with 100 μg/mL of Ampicillin and extract the amplified plasmids using a Plasmid Maxi kit (Qiagen). The Maxi kit yield is about 500 μg for pCMV-VCP vector.
Pause point: Purified pCMV-VCP plasmids (typically around 1,000 ng/μL) of ∼ 500 μL in H2O could be stored at −20°C till use.
Figure 1.
Detailed plasmid map of pCMV-VCP and pET28a-VCP for mammalian and bacterial expression
Restriction enzymes used to ligate VCP into the vectors were shown in the map. The detailed sequence of N-terminal and C-terminal of VCP were shown in bottom. SnapGene Viewer (GSL, Biotech; available at atsnapgene.com) was used to generate the figures.
PEI transfection reagent preparation
Timing: 3 days
-
4.Prepare 1 mg/mL PEI stock solution.
-
a.Dissolve 100 mg of Polyethylenimine, linear (PEI, Polysciences) powder (MW 25,000) in H2O to a final concentration of 1 mg/mL (100 mL total volume).
-
b.Adjust the pH to 7.0 with HCl to completely dissolve PEI at 55°C with stirring, which takes hours (∼4–6 h) to get PEI completely dissolved.
-
a.
-
5.
Filtrate the PEI solution through 0.22 μm filter to sterilize it and to get rid of undissolved PEI particles.
-
6.
Test the transfection efficiency using different Plasmid/PEI ratio to get the best transfection condition for further experiments.
Pause point: Make 1 mL aliquots (100 aliquots in total), and store at −80°C. Avoid repeated frozen-thaw cycles. Thawed aliquot will be kept at 4°C, which will be good for 2 months.
Note: For every batch of new PEI preparation, the transfection efficiency would be tested with different DNA/PEI ratio (w/v, such as 1 μg of DNA with either 1 μL, 2 μL or 3 μL of PEI). We usually use the empty pEGFP vector to test the transfection efficiency for different cell lines such as HEK293, H1299 and HeLa by quantifying the percentage of GFP positive cells.
H1299 cells seeding and passage
Timing: 3 days
-
7.Start a new H1299 cell culture.
-
a.Thaw a vial of cryopreserved H1299 cell rapidly (< 1 min) in a 37°C water bath.
-
b.Transfer thawed H1299 cells into a 10 cm culture dish with DMEM medium supplemented with 10% fetal bovine serum, penicillin (50 units/mL), and streptomycin (50 μg/mL) in a 37°C incubator containing 5% CO2.
-
c.Passage H1299 cells at the log phase to keep cells healthy and > 90% viable.
-
d.Seed H1299 cells at 80% confluence in antibiotic-free DMEM medium supplemented with 10% fetal bovine serum and grow for 16–24 h in 15 cm culture dishes one day before the transfection.
-
a.
CRITICAL: Confirm the confluence before transfection. It is important to avoid overgrowth of the cells.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-FLAG (1:1000) | Sigma-Aldrich | Cat#F1804; RRID: AB_262044 |
| Mouse monoclonal anti-VCP (1:1000) | Thermo Fisher Scientific | Cat#MA3-004; RRID: AB_2214638 |
| Anti-FLAG M2 affinity gel | Sigma-Aldrich | Cat#A2220; RRID: AB_10063035 |
| Bacterial and virus strains | ||
| Escherichia Coli DH5a | Thermo Fisher Scientific | Cat#18265017 |
| Escherichia Coli BL21(DE3) | Thermo Fisher Scientific | Cat#EC0114 |
| Chemicals, peptides, and recombinant proteins | ||
| Tris base | Fisher Scientific | Cat#BP1521; CAS: 77-86-1 |
| Imidazole | Fisher Scientific | Cat#AC30187-2500; CAS: 288-32-4 |
| Tris (2-carboxyethyl) phosphine (TCEP) | Sigma-Aldrich | Cat#C4706-10G; CAS: 51805-45-9 |
| Isopropyl β-D-thiogalactopyranoside (IPTG) | RPI | Cat#I56000-100.0; CAS: 367-93-1 |
| Phenylmethylsulfonyl fluoride (PMSF) | Fisher Scientific | Cat#PI36978; CAS: 329-98-6 |
| 3× FLAG peptides | Sigma-Aldrich | Cat#F4799 |
| Polyethylenimine (PEI) | Polysciences | Cat#23966-2; CAS: 9002-98-6 |
| cOmplete Protease Inhibitor Cocktail | Roche | Cat#05 056 489 001 |
| Ni-NTA agarose | QIAGEN | Cat#30230 |
| PfuTurbo DNA Polymerase | Agilent | Cat#600252 |
| Apyrase | New England Biolabs | Cat#M0398S |
| HindIII | New England Biolabs | Cat#R0104L |
| BamHI | New England Biolabs | Cat#R0136L |
| NdeI | New England Biolabs | Cat#R0111L |
| T4 DNA ligase | New England Biolabs | Cat#M0202L |
| Fast DNA Ladder | New England Biolabs | Cat#N3238S |
| DMEM with glucose | Corning | Cat#10-013-CV |
| Opti-MEM | Thermo Fisher Scientific | Cat#11058021 |
| Penicillin-Streptomycin Solution (100×) | Corning | Cat#MT30002CI |
| Fetal Bovine Serum | Gibco | Cat#26400044 |
| Bovine Calf Serum | Thermo Fisher Scientific | Cat#16777-204 |
| NativePAGE™ 3%–12% | Thermo Fisher Scientific | Cat#BN1003BOX |
| NativePAGE™ Sample Buffer (4×) | Thermo Fisher Scientific | Cat#BN2003 |
| GelCode™ Blue Stain Reagent | Thermo Fisher Scientific | Cat#24590 |
| Phosphatase inhibitor cocktail (100×) | Bimake | Cat#B15002 |
| Graphene oxide | Sigma-Aldrich | Cat#763705-25ML |
| Pyrene | Sigma-Aldrich | Cat#185515; CAS: 129-00-0 |
| Critical commercial assays | ||
| Miniprep plasmid extraction kit | QIAGEN | Cat#27106 |
| Maxiprep plasmid extraction kit | QIAGEN | Cat#12163 |
| QIAquick PCR Purification Kit | QIAGEN | Cat#28104 |
| QIAquick Gel Extraction Kit | QIAGEN | Cat#28106 |
| ADP/ATP Ratio Assay Kit | Sigma-Aldrich | Cat#MAK135-1KT |
| Deposited data | ||
| FLAG-VCP (H1299 cells), dodecamer state, cryo-EM map | (Yu et al., 2021) | EMDB: EMD-22675 |
| FLAG-VCP (H1299 cells), dodecamer state, coordinates | (Yu et al., 2021) | PDB: 7K56 |
| His-VCP (E. coli BL21(DE3)), apyrase treated, dodecamer state, cryo-EM map | (Yu et al., 2021) | EMDB: EMD-22676 |
| His-VCP (E. coli BL21(DE3)), apyrase treated, dodecamer state, coordinates | (Yu et al., 2021) | PDB: 7K57 |
| His-VCP (E. coli BL21(DE3)), apyrase treated, hexamer state, cryo-EM map | (Yu et al., 2021) | EMDB: EMD-22678 |
| His-VCP (E. coli BL21(DE3)), apyrase treated, hexamer state, coordinates | (Yu et al., 2021) | PDB: 7K59 |
| Experimental models: Cell lines | ||
| Human: NCI-H1299 | ATCC | CRL-5803 |
| Oligonucleotides | ||
| VCP primer set1_Forward: CCCAA GCTTATGGCTTCTGGAGCCGATTCA |
(Yu et al., 2021) | N/A |
| VCP primer set1_Reverse: CGCG GATCCTTAGCCATACAGGTCATCATC |
(Yu et al., 2021) | N/A |
| VCP primer set2_Forward: GGAAT TCCATATGGCTTCTGGAGCCGATTCA |
(Yu et al., 2021) | N/A |
| VCP primer set2_Reverse: CGCGG ATCCTTAGCCATACAGGTCATCATC |
(Yu et al., 2021) | N/A |
| Recombinant DNA | ||
| pCMV-VCP | (Yu et al., 2021) | N/A |
| pET28a-VCP | (Yu et al., 2021) | N/A |
| Software and algorithms | ||
| MotionCor2 | (Zheng et al., 2017) | https://emcore.ucsf.edu/ucsf-software |
| Gctf | (Zhang, 2016) | https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/#gctf |
| Gautomatch | Zhang lab | http://www.mrc-lmb.cam.ac.uk/kzhang/ |
| cryoSPARC v2 or v3 | (Punjani et al., 2017) | https://cryosparc.com/ |
| Coot | (Emsley et al., 2010) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Phenix | (Adams et al., 2010; Afonine et al., 2018) | https://phenix-online.org/documentation/reference/real_space_refine.html |
| UCSF Chimera | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera/ |
| Other | ||
| TED PELLA Lacey Carbon, 300 mesh | TED PELLA, INC. | Cat#01895-F |
| PVDF membrane | Bio-Rad | Cat#E2311 |
| Amicon Ultra-0.5 Centrifuge Filter Unit (30K) | Millipore | Cat#UFC503008 |
| Amicon Ultra-15 Centrifuge Filter Unit (30K) | Millipore | Cat#UFC903024 |
| Forma Series II Water-Jacketed CO2 Incubator | Thermo Scientific | Cat#3110 |
| Innova 44R Shaker | New Brunswick | Cat#M1282-0004 |
| ÄKTA pure 25 M | Cytiva | Cat#29018226 |
| Superose® 6 Increase 10/300 GL | MilliporeSigma | Cat#GE29-0915-96 |
| Sorvall RC 6 PLUS Centrifuge | Thermo Scientific | Cat#12121680 |
| Refrigerated Centrifuge | Eppendorf | 5810R |
| Sorvall Legend Micro 21R Microcentrifuge | Thermo Scientific | Cat#75002447 |
| 200 KV Talos F200C transmission electron microscope | Thermo Scientific | N/A |
| 300 KV FEI Titan Krios transmission electron microscope | Thermo Scientific | N/A |
| 64-core AMD Ryzen Threadripper 3,990× Linux computing station equipped with 4 NVIDIA GPU | AMD | N/A |
| Azure imaging system | Azure Biosystems | C500 |
| Labquake rotator | Barnstead|Thermolyne | Cat#3.625.485 |
| Nanodrop | Thermo Scientific | Cat#ND-ONE-W |
Alternatives: In addition to the antibodies presented in the key resources table, VCP and Flag antibodies can also be purchased from Cell Signaling Technology, Santa Cruz Biotechnology, etc. However, their quality has to be verified. Similarly, as an alternative of apyrase from NEB, other companies also have apyrase, such as Sigma (Cat#A2230). Make sure to test enzyme efficiency before use.
Materials and equipment
PCR reaction master mix
| Reagent | Amount |
|---|---|
| DNA template (100 ng/μL) | 1 μL (100 ng) |
| PfuTurbo DNA polymerase (2.5 U/μL) | 1 μL (2.5 U) |
| Primer 1 (100 ng/μL) | 1 μL |
| Primer 2 (100 ng/μL) | 1 μL |
| 10× Pfu reaction buffer | 5 μL |
| dNTPs (10 mM each dNTP) | 1 μL |
| ddH2O | 40 μL |
| Total reaction volume | 50 μL |
PCR cycling conditions
| Steps | Temperature | Time | Cycles |
|---|---|---|---|
| Initial Denaturation | 95°C | 2 min | 1 |
| Denaturation | 95°C | 30 s | 30 cycles |
| Annealing | 55°C | 30 s | |
| Extension | 72°C | 3 min | |
| Final extension | 72°C | 10 min | 1 |
| Hold | 4°C | forever | |
Buffer A: H1299 cell lysis buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 1 M Tris HCl (pH 8.0) | 50 mM | 5 mL |
| 4 M NaCl | 150 mM | 3.75 mL |
| Glycerol | 10% | 10 mL |
| Triton-X-100 | 1% | 1 mL |
| Protease inhibitor cocktail (50×) | 1× | 2 mL |
| Phosphatase inhibitor cocktail (100×) | 1× | 1 mL |
| ddH2O | n/a | 82.25 |
| Total | n/a | 100 mL |
Note: Store at 4°C for up to a year. The protease inhibitor was added prior to use.
Alternatives: To minimize the potential interference of Triton X-100 detergent with BN-PAGE, a reduced detergent concentrations of 0.1%–0.5% can be used.
Buffer B
| Reagent | Final concentration | Amount |
|---|---|---|
| 1 M Tris-HCl pH 7.4 | 50 mM | 25 mL |
| 4 M NaCl | 150 mM | 18.75 mL |
| ddH2O | n/a | 455.25 mL |
| 0.5 M TCEP | 1 mM | 1 mL |
| Total | n/a | 500 mL |
Note: Store at 4°C for up to a year. Add the 1 mM TCEP before use.
Buffer C
| Reagent | Final concentration | Amount |
|---|---|---|
| 0.5 M HEPES pH 7.4 | 20 mM | 20 mL |
| 4 M NaCl | 150 mM | 18.75 mL |
| ddH2O | n/a | 460.25 mL |
| 0.5 M TCEP | 1 mM | 1 mL |
| Total | n/a | 500 mL |
Note: Store at 4°C for up to a year. Add the 1 mM TCEP before use.
FLAG-VCP elution buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 1 M Tris HCl (pH 7.5) | 50 mM | 5 mL |
| 4 M NaCl | 150 mM | 3.75 mL |
| ddH2O | n/a | 91.25 mL |
| 3× FLAG peptide (1 mg/mL) | 0.1 mg/mL | 10 mL |
| Total | n/a | 100 mL |
Note: To minimize the degradation of the 3× FLAG peptide, prepare the 3× FLAG stock solution (1 mg/mL, in buffer B) and store at −20°C for up to a year. Prepare the elution buffer with a final concentration of 100 μg/mL 3× FLAG peptide before use.
E. coli expressed His-VCP lysis buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 1 M Tris HCl (pH 7.6) | 20 mM | 20 mL |
| 4 M NaCl | 150 mM | 37.5 mL |
| 5 M Imidazole | 5 mM | 10 mL |
| ddH2O | n/a | 932.5 mL |
| Total | n/a | 1 L |
Note: Store at 4°C for up to a year.
E. coli expressed His-VCP elution buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 1 M Tris HCL (pH 7.6) | 20 mM | 20 mL |
| 4 M NaCl | 150 mM | 37.5 mL |
| 5 M Imidazole | 500 mM | 100 mL |
| ddH2O | n/a | 842.5 mL |
| Total | n/a | 1 L |
Note: Store at 4°C for up to a year.
Step-by-step method details
Expression and purification of VCP dodecamers from H1299 mammalian cells
Timing: 4 days
The following section describes the detailed steps for purifying recombinant FLAG-VCP dodecamers from mammalian cells. Previously amplified and purified pCMV-VCP plasmids are transfected into H1299 cells using PEI, and allow VCP to express for 48–72 h. Then, cells are collected for anti-FLAG based affinity chromatography purification.
-
1.pCMV-VCP transfection into H1299 cells.
-
a.Add 50 μg pCMV-VCP plasmids into 3.7 mL Opti-MEM and mix gently by vortexing at low speed for 5 s at 25°C.
-
b.Add 50 μL of 1 mg/mL PEI to the plasmids/Opti-MEM and mix gently by vortexing at low speed for 5 s at 25°C.
-
c.Leave the mixture for 5–10 min at 25°C.
-
d.Add the PEI/plasmids/Opti-MEM mixture to H1299 cells in the 15 cm culture dishes and mix gently by moving the dishes from front to back and side to side for at least 5 times to ensure even distribution.
-
a.
-
2.Cell collection and FLAG-VCP purification.
-
a.After 48–72 h post transfection, discard the cell culture medium in the 15 cm culture dishes and wash cells with 10 mL PBS buffer two times by gently swirling the dishes by hand.
-
b.Add 1 mL buffer A into each 15 cm culture dish and incubate on ice for 30 min for cell lysis.
-
c.Collect the suspension and centrifuge at 17,000 g for 15 min at 4°C.
-
d.Collect the supernatant and add 50 μL Anti-FLAG M2 affinity gel per mL.
-
e.Incubate the supernatant-resin mixture at 4°C for 2 h by gently rotating with a Labquake rotator (Barnstead|Thermolyne).
-
f.Pellet at 800 g for 1 min and wash the resin with 1 mL buffer A three times at 4°C.
-
g.Add 500 μL FLAG-VCP elution buffer, incubate at 4°C for 1 h by gently rotating with a Labquake rotator (Barnstead|Thermolyne), and collect the elution supernatant carefully after pelleting the resin.
-
h.Repeat step g one more time.
 Pause point: Add 1 mL FLAG-VCP elution buffer (with protease inhibitor) and elute after incubation with shaking for 16–24 h at 4°C.
Pause point: Add 1 mL FLAG-VCP elution buffer (with protease inhibitor) and elute after incubation with shaking for 16–24 h at 4°C. -
i.Concentrate the 1 mL elution to 50–100 μL with Amicon Ultra-0.5 Centrifuge Filter Unit with 30 KDa cutoff (Millipore, UFC503008) at 17,000 g for 10 min.
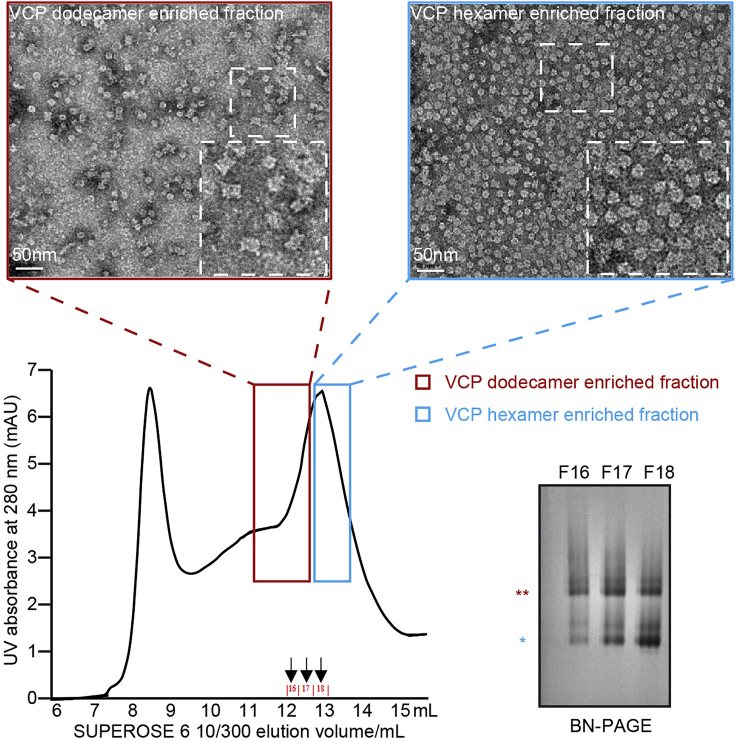
 CRITICAL: Centrifuge filters with correct molecular weight cutoff is critical to concentrate VCP protein and get rid of 3×FLAG peptides.Note: For two 15 cm culture dishes, typical sample concentration is around 1–2 mg/mL. The dodecamer and hexamer ratio could be evaluated by the blue native-PAGE (NativePAGE™ 3%–12%) (Yu et al., 2021) and gel filtration (Superose 6 10/300 GL column) could be performed to improve the population of dodecamers (Figure 2). See below steps 3 and 4 for detailed protocols.
CRITICAL: Centrifuge filters with correct molecular weight cutoff is critical to concentrate VCP protein and get rid of 3×FLAG peptides.Note: For two 15 cm culture dishes, typical sample concentration is around 1–2 mg/mL. The dodecamer and hexamer ratio could be evaluated by the blue native-PAGE (NativePAGE™ 3%–12%) (Yu et al., 2021) and gel filtration (Superose 6 10/300 GL column) could be performed to improve the population of dodecamers (Figure 2). See below steps 3 and 4 for detailed protocols.
-
a.
-
3.Blue native-PAGE to examine dodecamer and hexamer ratio.
-
a.Assemble the blue native-PAGE gel electrophoresis unit according to the manufacturer’s manual (Thermo Fisher).
-
b.7.5 μL VCP sample mixed with 2.5 μL 4× blue native-PAGE sample buffer (Thermo Fisher) was loaded.
-
c.Perform the gel electrophoresis at 150 V for 90 min at 25°C.
-
d.Fix the gel by microwaving in fix solution (40% methanol and 10% acetic acid) on high (950–1,100 watts) for 45 s.
-
e.Shake the gel on an orbital shaker for 15 min at 25°C, and then decant the fix solution.
-
f.Stain the gel with GelCode Blue Stain Reagent (Thermo Fisher) by shaking the gel on an orbital shaker for 1 h.
 Pause point: The gel can be left in the GelCode Blue Stain Reagent for longer time until the desired background is obtained.
Pause point: The gel can be left in the GelCode Blue Stain Reagent for longer time until the desired background is obtained. -
g.Image the gel using Azure C500 imaging system (Azure Biosystems).
-
a.
-
4.Gel filtration to improve the population of dodecamers.
-
a.Equilibrate the Superose 6 10/300 GL column (Millipore Sigma) using buffer C. A flow rate of 0.5 mL/min was used.
-
b.Load 100 μL VCP sample of about 1–2 mg/mL to the column. All the elution fractions starting at 4 mL were collected.
-
c.Examine the elution fractions using SDS-PAGE and blue native-PAGE.
-
a.
Figure 2.
Separation of VCP dodecamers and hexamers using gel filtration
(Bottom left) Gel filtration with Superose 6 10/300 GL column profile. Arrow bars indicate the fractions used for BN-PAGE analysis. (Bottom right) BN-PAGE analysis of elution fractions 16–18. “∗” and “∗∗” mark the positions of hexamers and dodecamers, respectively. (Top) Negative staining TEM examination of elution at 12 mL (VCP dodecamer, left panel) and elution at 13 mL (VCP hexamer, right panel) elution fractions.
Purification of recombinant His-VCP from E.coli and generation of dodecamers
Timing: 4 days
This section describes detailed procedure for bacterially expressed human His-VCP purification and dodecamer generation with apyrase treatment, which removes the prebound nucleotides. Since apyrase eliminates nucleotides in all AAA domains, the resultant VCP dodecamers contains both empty D1 and D2 AAA domains, which controls the N-terminal and C-terminal conformations, respectively (Banerjee et al., 2016; Xia et al., 2016).
-
5.Transformation of pET28a-VCP to BL21(DE3) cells using the 42 °C heat shock method.
-
a.Mix 1 μL of prepared pET28a-VCP plasmid (20 ng/μL) with 50 μL freshly thawed (on ice) chemically competent BL21(DE3) cells and incubate on ice for 30–45 min.Note: Mix by gently tapping the tube instead of pipetting.
-
b.Transfer the tube to a 42°C water or metal bath for 45 s and then quickly cool down by putting on ice for 3 min.
-
c.Add 200 μL LB medium to the tube and shake at 200 rpm (New Brunswick Innova 44R), 37°C for 1 h.
-
d.Spread 50 μL above culture on a LB agar plate containing 50 μg/mL kanamycin.
-
e.Incubate for 16–24 h at 37°C.
-
a.
-
6.Grow the transformed cells and induce His-VCP expression.
-
a.Pick a single colony, inoculate 5 mL LB supplemented with 50 μg/mL kanamycin and culture for 16–24 h at 37°C with shaking at 200 rpm (New Brunswick Innova 44R).
-
b.Transfer the above culture to 400 mL LB (50 μg/mL kanamycin) and culture at 37°C with shaking at 200 rpm (New Brunswick Innova 44R) till OD600 of 0.4–0.6.
-
c.Adjust the shaker incubator temperature to 20°C and add 0.4 mM isopropyl β-D-thiogalactopyranoside (IPTG) to induce protein expression for 18 h.
-
d.Harvest cells by centrifuge (1,000 g, 10 min).
-
a.
-
7.His-VCP purification.
-
a.Resuspend cells (∼2–3 g) in 20 mL His-VCP lysis buffer in the presence of protease inhibitor (1 mM PMSF).
-
b.Lyse cells by sonication (35% power, 1 s/3 s on/off, for a total on time of 3 min) on ice.
-
c.Pellet the cell debris by centrifuge at 4°C, 17,000 g for 30 min.
-
d.Transfer the supernatant to a 50 mL tube and add 1 mL 50% Ni-NTA agarose (prewashed with the His-VCP lysis buffer).
-
e.Incubate and mix gently by rotating with a Labquake rotator (Barnstead|Thermolyne) for 1 h at 4°C.
-
f.Transfer the mixture to a 15 mL chromatography gravity column.
-
g.Wash the resin with 30 mL His-VCP lysis buffer supplemented with 20 mM Imidazole at 4°C.
-
h.Repeat washing with 20 mL His-VCP lysis buffer supplemented with 40 mM Imidazole at 4°C.
-
i.Elute bound His-VCP using 5 mL His-VCP elution buffer (containing 500 mM Imidazole).
-
j.Repeat the elution step one more time.Optional: After a few elution drops, pause, and incubate with the His-VCP elution buffer for 5 min to improve the elution if necessary.
-
k.Concentrate the elution using Amicon Ultra-15 Centrifuge Filter Unit with 30 KDa cutoff (Millipore, UFC903024) to about 1 mL at 1,860 g at 4°C.
 CRITICAL: Centrifuge filters with correct molecular weight cutoff is critical to concentrate VCP protein.Note: Mix the elution by gently pipetting up and down to prevent protein precipitation between centrifuge rounds.
CRITICAL: Centrifuge filters with correct molecular weight cutoff is critical to concentrate VCP protein.Note: Mix the elution by gently pipetting up and down to prevent protein precipitation between centrifuge rounds. -
l.Add 5 mL buffer C and repeat step k.
-
m.Repeat step k and l for 3 more times.
-
n.Concentrate the sample to 500 μL, measure the protein concentration using nanodrop (Thermo Scientific).Note: Typical concentration is about 20–40 mg/mL.
-
o.Prepare 50 μL aliquots and flash frozen in liquid nitrogen. Store at −80°C till use.
-
a.
-
8.Generation of His-VCP dodecamers.
-
a.Dilute His-VCP to 1–2 mg/mL using buffer C.
-
b.Add 0.5 U/mg apyrase (NEB) to treat His-VCP at 25°C for 12–18 h.
 CRITICAL: Removing nucleotides from D2 AAA domain of VCP by apyrase is required to generate His-VCP dodecamers.Note: Usually obvious formation of dodecamers can be observed within 1 h. 12–18 h treatment can be performed to ensure complete elimination of ADP. The treatment time can be shortened by increasing apyrase concentration (e.g., double the concentration to 1 U/mg).Optional: The turnover of ADP to AMP by apyrase can be monitored using an ADP assay kit (Sigma Aldrich) (Yu et al., 2021).
CRITICAL: Removing nucleotides from D2 AAA domain of VCP by apyrase is required to generate His-VCP dodecamers.Note: Usually obvious formation of dodecamers can be observed within 1 h. 12–18 h treatment can be performed to ensure complete elimination of ADP. The treatment time can be shortened by increasing apyrase concentration (e.g., double the concentration to 1 U/mg).Optional: The turnover of ADP to AMP by apyrase can be monitored using an ADP assay kit (Sigma Aldrich) (Yu et al., 2021). -
c.Dodecamer formation can be examined with blue native-PAGE (NativePAGE™ 3%–12%), negative staining TEM and cryo-EM (Yu et al., 2021).
-
d.To remove apyrase post treatment, repurify the sample using the Ni-NTA resin following the steps (7e–7n) described above.
-
a.
TEM grid preparation and sample freezing
Timing: 3–6 h
-
9.
Glow discharge 300-mesh lacey TEM grid (Ted Pella), 15 mA for 1 min.
Alternatives: Grids from other brands could also be used.
-
10.
Dilute graphene oxide (GO, stock solution 2 mg/mL, Sigma) to 0.2 mg/mL with ddH2O. Centrifuge at 300 g for 30 s to remove large precipitates.
-
11.
Dilute pyrene stock solution (∼ 1 mg/mL in methanol) to 1 μg/mL with ddH2O.
-
12.
Apply 5 μL diluted pyrene to the Ted Pella grid, incubate for 5 min at about 25°C and blot dry.
-
13.
Apply 3 μL 0.2 mg/mL GO to the same side, incubate for 1 min, and remove the excess liquid from the side using a filter paper.
-
14.
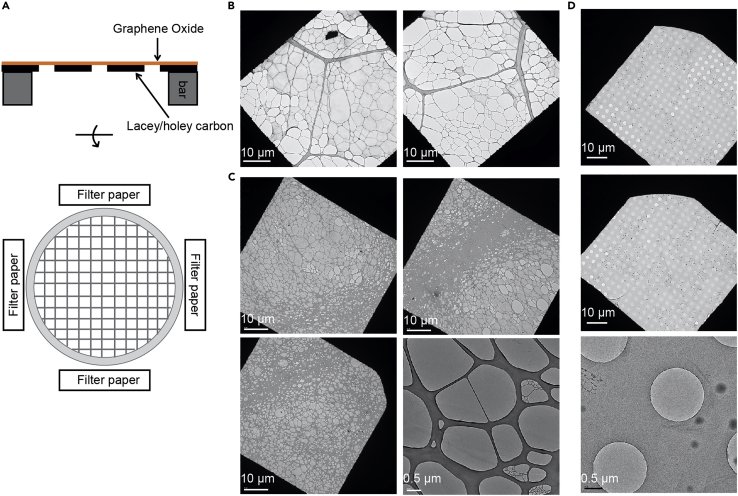
Wash with 5 μL ddH2O for two times and dry the grids on a filter paper in a clean Petri dish for >2 h (Figure 3).
-
15.
3.5 μL purified FLAG-VCP dodecamers from the H1299 cells or apo bacterial recombinant His-VCP at a concentration of about 0.5 mg/mL in buffer C was applied to the GO-coated grid, blotted for 6–9 s at 20°C with a humidity of about 70%–80% and plunge-frozen in liquid ethene using a Cp3 cryo plunger (Gatan).
CRITICAL: When handling the grid, be sure not to damage the grid surface with the pipet tip or bend the grids.
Note: Multiple grids (e.g., 6 grids) with variations in freezing condition (usually the blotting time) are typically prepared for one sample.
Alternatives: Other cryo-EM sample freezing units such as Mark IV vitrobot (Thermo fisher scientific), can also be used, but the freezing condition need to be explored for individual instruments.
Pause point: The frozen grids should be maintained in liquid nitrogen all the time and stored till ready to proceed.
Figure 3.
Coating TEM grids with GO
(A) Overall scheme of GO coating.
(B) Grids coated with GO after one step of GO application and blotting.
(C and D) Lacey and holey grids coated with 2–4 times of GO application and filter paper blotting from different orientations.
Cryo-EM screening, data collection, and image processing
Timing: 1–2 weeks
-
16.
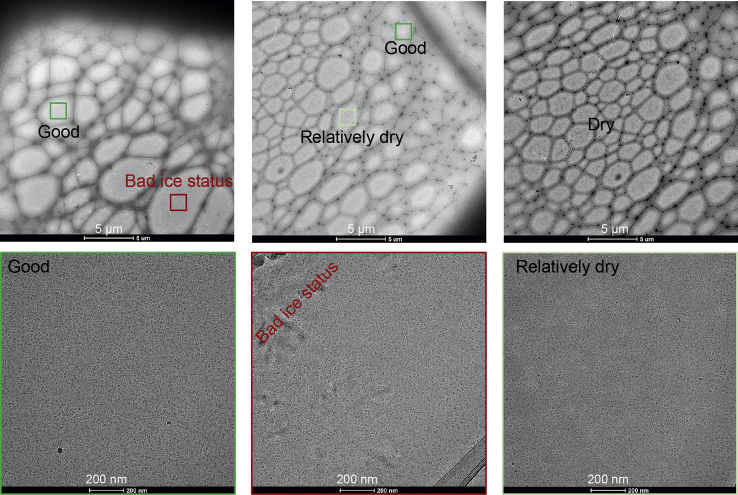
Examine the frozen grids using a lower-level cryo-EM microscope such as 200 KV Talos F200C electron microscope (the low-dose mode was used for the screening). The optimal grid with good ice and uniformly distributed particles is the target (Figure 4).
-
17.
Prepare two more frozen grids using the optimal condition for each sample.
-
18.
Load the grids into a high-end cryo-EM microscope such as a 300 KV FEI Titan Krios TEM for data collection.
-
19.
Record projection images with a direct electron detector (e.g., Gatan K2 Summit). A frame rate of 5 frames per second, a total dose of about 40 e/Å2 and a defocus range of −0.8 to −2 μm are usually used. About 600 and 800 movies were collected for FLAG-VCP and His-VCP, respectively.
Alternatives: Gatan K3 direct electron detector or Falcon 4 (Thermo Fisher Scientific). Other 200 KV or 300 KV microscopes from JEOL could also be used for screening and data collection.
-
20.
Align the frames using MotionCor2 (Dose per frame: 0.96, B factor of -150 and 5 × 5 patches) (Zheng et al., 2017). A 64-core Linux computing station with 4 NVIDIA GPUs was used for all the computation in steps 20–28.
-
21.
Determine the contrast transfer function (CTF) parameters with Gctf (Zhang, 2016) using the summed micrographs without dose-weighting (binning 2). Micrographs with overall resolutions worse than 6 Å were discarded (∼50 discarded).
-
22.
Randomly select about 10% of the micrographs and perform particle picking using Gautomatch (http://www.mrc-lmb.cam.ac.uk/kzhang/) without a template using a particle diameter of 200 Å.
-
23.
Perform 2D class average using RELION or cryoSPARC with about 10,000 particles from step 22 (k=30) and select multiple representative classes (e.g., including both the top/bottom and side views of the particles as shown in Figure 5C) for a second round of template-based particle picking with Gautomatch.
Note: Due to the elongated shape of VCP dodecamers, top and bottom views could be less captured. Include more classes similar to top/bottom views in the template-based particle picking step.
-
24.
Import the picked particles into cryoSPARC v2 (Punjani et al., 2017) for further image processing.
Alternatives: We processed the data using an earlier version of cryoSPARC (v2) but image processing can also be performed using newer cryoSPARC versions such as v3 or other packages such as RELION (Zivanov et al., 2018).
-
25.
For FLAG-VCP, about 40,000 particles (initial particles of about 60,000) remained after cleaning via 2D classification. For the apo His-VCP, about 71,600 particles remained after 2D classification cleaning (initial particles of about 100,000).
-
26.
Build 3 initial models using the ab initio model module in cryoSPARC with default settings.
-
27.
Perform hetero-refinements using the 3 models. An initial low pass filtering of 60 Å was used. For FLAG-VCP, a best class of 27,500 dodecamer particles and two other classes of dodecamer or hexamer particles with reduced quality (8,800 and 3,800 particles, respectively) were revealed. For apyrase-treated His-VCP, a hexamer class of about 38, 300 particles and a dodecamer class of about 33,300 particles were identified (Yu et al., 2021).
Note: This step could be done repeatedly to improve the homogeneity of individual classes when necessary.
-
28.
Perform homo-refinements for the best classes using default settings of cryoSPARC and by imposing D7 symmetry. A dodecamer structure with a resolution of 3.9 Å was obtained for FLAG-VCP (Figure 5). A 4.2 Å apo hexamer structure and a dodecamer structure of 3.7 Å were resolved for His-VCP (Yu et al., 2021).
Figure 4.
Cryo-EM sample freezing conditions screening
The green boxes make areas with ice of optimal thickness and uniformly distributed particles. The light green box indicates an area with too thin ice. The red boxes mark areas with warmed up ice.
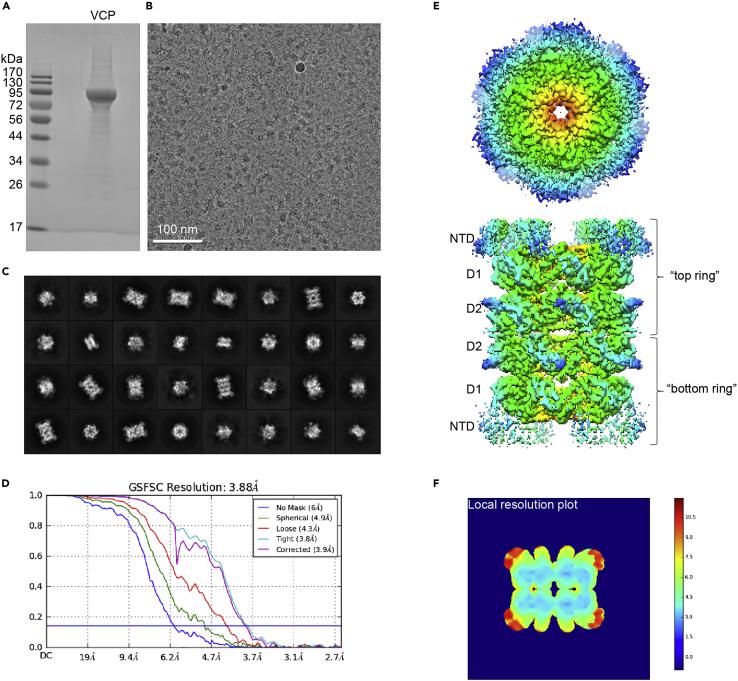
Figure 5.
Cryo-EM characterization of VCP dodecamer
(A) SDS-PAGE of purified FLAG-VCP (∼97 kDa).
(B) A representative cryo-EM micrograph taken at 225,000 magnification.
(C) Selected 2D class averages.
(D) Fourier Shell correlation curves.
(E) Cryo-EM density map. The map was radially colored based on distance from the central axis.
(F) The local resolution analysis of the map.
Model building and refinement
Timing: 1 week
-
29.
Sharpen the maps using phenix.autosharpen (Adams et al., 2010) according to the resolutions of the maps.
-
30.
Fit the ATPγS-bound VCP structure (PDB: 5FTN) into the density map as a rigid body using the ‘fit in map’ function in Chimera, and density for a single subunit was segmented using the “color zone” routine in Chimera (Pettersen et al., 2004).
-
31.
Carry out manual adjustment and real-space refinement of the atomic model against the subunit density in COOT (Emsley et al., 2010).
-
32.
Perform real space refinement in PHENIX using phenix.real_space_refine (Adams et al., 2010; Afonine et al., 2018). The minimization_global, local_grid_search, morphing, simulated_annealing, and adp refinement strategies were used.
Note: Due to the poor density quality of the N-terminal domain, residues 22-206 were refined as a rigid body.
-
33.
Repeat steps 31 and 32 till good fitting of the atomic model with the density, referring to the modeling validation report post phenix refinement (Yu et al., 2021).
-
34.
Generate the oligomer (hexamer or dodecamer) models using the phenix.map_symmetry (input: density map, map resolution) and phenix.apply_ncs using the refined single subunit model in the previous step (Adams et al., 2010).
-
35.
Refinement of the oligomer model against the whole density map using Coot and phenix.real_space_refine with settings used in steps 31–33.
-
36.
Analyze the quality of the final models with MolProbity in Phenix and generate the refinement statistics (Williams et al., 2018). For details on the refinement statistics, please refer to Yu et al. (2021).
Expected outcomes
Typically, 50 μL of FLAG-VCP at around 1–2 mg/mL could be obtained from two 15 cm culture dishes of H1299 cells. After approximately complete removal of ADP, apo hexamers and dodecamers of about equal populations could usually be received. Cryo-EM structures of VCP dodecamers at about 3.7–3.9 Å could be resolved with about 30,000 particles.
Limitations
This protocol has been successfully used for isolation of N-terminal FLAG- and His-tagged VCP from both mammalian and bacterial cells. We envision that other affinity tags may also be utilized for VCP purification. However, tagging of N-terminal domain may affect VCP function and/or protein expression levels. To avoid possible artifacts, only functional TAP-tagged VCP constructs should be used. To rule out the potential effects of N-terminal tagging on dodecamer formation, we have examined the oligomeric states of endogenous VCP and the dodecamer state has also been observed (Yu et al., 2021). Flexibility of the N-terminal domain in the apo VCP structures could limit the quality of the cryo-EM maps.
Troubleshooting
Problem 1
Low yield of FLAG-VCP from mammalian cultures (step 2).
Potential solution
Improve the quality of the plasmids (e.g., higher concentration and purity) and cells (e.g., using cells with healthy morphology), and optimize the transfection protocol to improve efficiency. Owing to the variations in transfection efficiencies, the dodecamer:hexamer ratio of purified FLAG-VCP may vary among batches.
Problem 2
Weak dodecamer band after apyrase treatment (step 8).
Potential solution
Perform a time kinetics experiment to figure out the optimal reaction time. Make sure 5 mM CaCl2 is included in the reaction and use fresh apyrase enzyme.
Problem 3
Low GO coverage of TEM grid (step 14). (Figure 3).
Potential solution
Repeat step 13 and blot with the filter paper from different orientations if the coverage of GO is limited (Figure 3).
Problem 4
Too low or too high particle density on grid (step 16).
Potential solution
Adjust sample concentration (e.g., 0.5–1.5 mg/mL) accordingly.
Problem 5
Preferred view of particles in 3D reconstructions (steps 27–28).
Potential solution
Due to the elongated shape of VCP dodecamers, top and bottom views could be less captured. Include more classes similar to top/bottom views in the template-based particle picking step (step 23). Collect more data to acquire enough particles for these low-frequency views.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Zhong-Yin Zhang (zhang-zy@purdue.edu).
Materials availability
All unique reagents generated in this study will be shared by the lead contact upon request, Dr. Zhong-Yin Zhang (zhang-zy@purdue.edu).
Acknowledgments
This study was supported in part by NIH RO1 CA69202 and the Robert C. and Charlotte Anderson Chair Endowment. We also acknowledge NIH P30CA023168 for supporting Purdue University Center for Cancer Research shared resources. The Cryo-EM and negative staining TEM images were collected using the Purdue Cryo-EM facility and Purdue Electron microscopy facility.
Author contributions
G.Y. and Y.B. performed the experiments. G.Y., Y.B., and Z.Y.Z. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Guimei Yu, Email: guimei.yu@tmu.edu.cn.
Zhong-Yin Zhang, Email: zhang-zy@purdue.edu.
Data and code availability
This paper does not report original code. All the cryo-EM density maps and corresponding atomic models generated have been deposited to the EMDB database (EMDB: EMD-22675, EMD-22676, EMD-22678) and PDB database (PDB: 7K56, 7K57, 7K59) in our previous work (Yu et al., 2021).
References
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P.V., Poon B.K., Read R.J., Sobolev O.V., Terwilliger T.C., Urzhumtsev A., Adams P.D. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Bartesaghi A., Merk A., Rao P., Bulfer S.L., Yan Y., Green N., Mroczkowski B., Neitz R.J., Wipf P., et al. 2.3 A resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016;351:871–875. doi: 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B., et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D., Tang W.K., Ye Y. Structure and function of the AAA+ ATPase p97/Cdc48p. Gene. 2016;583:64–77. doi: 10.1016/j.gene.2016.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Bai Y., Li K., Amarasinghe O., Jiang W., Zhang Z.Y. Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation. iScience. 2021;24:103310. doi: 10.1016/j.isci.2021.103310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This paper does not report original code. All the cryo-EM density maps and corresponding atomic models generated have been deposited to the EMDB database (EMDB: EMD-22675, EMD-22676, EMD-22678) and PDB database (PDB: 7K56, 7K57, 7K59) in our previous work (Yu et al., 2021).