Abstract
Plants are continuously exposed to beneficial and pathogenic microbes, but how plants recognize and respond to friends versus foes remains poorly understood. Here, we compared the molecular response of Arabidopsis thaliana independently challenged with a Fusarium oxysporum endophyte Fo47 versus a pathogen Fo5176. These two F. oxysporum strains share a core genome of about 46 Mb, in addition to 1,229 and 5,415 unique accessory genes. Metatranscriptomic data reveal a shared pattern of expression for most plant genes (about 80%) in responding to both fungal inoculums at all timepoints from 12 to 96 h postinoculation (HPI). However, the distinct responding genes depict transcriptional plasticity, as the pathogenic interaction activates plant stress responses and suppresses functions related to plant growth and development, while the endophytic interaction attenuates host immunity but activates plant nitrogen assimilation. The differences in reprogramming of the plant transcriptome are most obvious in 12 HPI, the earliest timepoint sampled, and are linked to accessory genes in both fungal genomes. Collectively, our results indicate that the A. thaliana and F. oxysporum interaction displays both transcriptome conservation and plasticity in the early stages of infection, providing insights into the fine-tuning of gene regulation underlying plant differential responses to fungal endophytes and pathogens.
Keywords: Arabidopsis thaliana, comparative genomics, endophyte, Fusarium oxysporum, host-fungal interactions, pathogen
Over millions of years of coevolution, plants and microbes have established intimate relationships, forming beneficial, neutral, or antagonistic partnerships. Plant pathogens threaten agricultural production and global food security (Dean et al. 2012; Strange and Scott 2005), but beneficial microbes such as rhizobia, mycorrhizae, and endophytes limit plant pests and promote plant growth through nutrient mineralization and availability (Rashid et al. 2016; White et al. 2019). How plants recognize and react differently to friends versus foes is an intriguing topic of research.
Our understanding of plant immunity has been revolutionized by the recent increase in the breadth of genomic data available. The classical, binary view of plant immunity consists of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) (Cui et al. 2015; Jones and Dangl 2006). Plant PTI relies on plasma membrane (PM)-localized pattern recognition receptors (PRRs), which are often receptor-like proteins or kinases (RLPs or RLKs) that sense conserved microbe-associated molecular patterns (MAMPs) or damage-associated molecular patterns and induce downstream defense reactions (Bigeard et al. 2015; Dodds and Rathjen 2010; Jones and Dangl 2006; Zhou et al. 2020). Plant ETI employs an intracellular nucleotide-binding site and leucine-rich repeat domain receptors (NLRs) that recognize specific microbial effectors as a result of ongoing host-pathogen coevolution (Cesari 2018; Cui et al. 2015; Jones and Dangl 2006; Monteiro and Nishimura 2018; W. Wang et al. 2020). The blurry boundary between MAMPs and effectors prompted a new spatial immunity model based on extracellular or intracellular locations of pattern recognition (van der Burgh and Joosten 2019; W. Wang et al. 2020).
This study investigated transcriptome reprogramming of the same plant host and explored how PTI and ETI or extracellular and intracellular immunity are involved in both beneficial and antagonistic interactions. We established the Fusarium oxysporum–Arabidopsis thaliana model system, which includes an endophyte, F. oxysporum Fo47, and a pathogen, F. oxysporum Fo5176. Arabidopsis plants infected by these two F. oxysporum strains display distinctive phenotypes, with Fo5176 causing typical vascular wilt diseases and Fo47 colonizing plants endophytically without any disease symptoms. Their distinct effects on plants, combined with their minimal genetic diversity, should facilitate the identification of meaningful genotype–phenotype correlations.
In addition to being a good model system, F. oxysporum is of great agricultural importance, as it is listed among the top 10 most-researched fungal pathogens for food production (Dean et al. 2012 Collectively, this group of filamentous fungi causes devastating vascular wilt diseases in over 100 crop species, leading to annual yield losses of billions of dollars (Ma et al. 2013). One notorious example is the recent Panama disease outbreak in banana caused by F. oxysporum f. sp. cubense tropical race 4 (Viljoen et al. 2020). Information accumulated over the past 10 years has provided a clear picture of compartmentalization of the F. oxysporum genome: A core genome component that is conserved and vertically transmitted performs essential housekeeping functions, and an accessory genome that is believed to have been initially acquired horizontally mediates unique host-fungal interactions (Armitage et al. 2018; DeIulio et al. 2018; Galazka and Freitag 2014; Hane et al. 2011; Ma et al. 2010, 2013; van Dam et al. 2016; Vlaardingerbroek et al. 2016a and b; Williams et al. 2016; Yang et al. 2020; Zhang et al. 2020).
Using an unbiased approach and taking advantage of two recently released high-quality genome assemblies of Fo47 and Fo5176 (B. Wang et al. 2020; Fokkens et al. 2020), we employed metatranscriptomics to dissect how Arabidopsis plants react to two F. oxysporum isolates with distinct lifestyles during the early course of infection. We demonstrated that endophytic infection suppresses host immunity but activates plant nutrient assimilation. By contrast, pathogenic infection activated defense response but suppressed plant developmental functions. Genome comparison of the two isolates revealed unique accessory chromosomes that harbor genes enriched for fungal virulence and detoxification in Fo5176 and cell signaling and nutrient sensing in Fo47. Our study showed that, while for both plants and F. oxysporum, most genes displayed a similar expression pattern during infections, a small number of genes displayed transcription plasticity between endophytic and pathogenic infections, perhaps leading to the different interaction outcomes.
RESULTS
A pathosystem that reveals both endophytic and pathogenic interactions.
To dissect beneficial versus pathogenic fungal-plant interactions, we inoculated Arabidopsis plants with two F. oxysporum strains, the beneficial (endophytic) strain Fo47 and the pathogenic strain Fo5176. The pathogenic fungus Fo5176, initially isolated in Australia (Chen et al. 2015; Thatcher et al. 2012), causes vascular wilt in several Brassicaceae plants, including A. thaliana (Ma et al. 2010; Thatcher et al. 2009). The endophytic strain Fo47 was originally isolated from disease-suppressing soils (Alabouvette 1999) and has been used as a biocontrol agent to prevent disease from soil-borne pathogens by inducing the production of plant secondary metabolites and priming host resistance (Aimé et al. 2013; Benhamou and Garand 2001; Benhamou et al. 2002; Olivain et al. 2006; Veloso and Díaz 2012).
We adopted a robust and reproducible root-dipping protocol to inoculate 14-day-old Columbia-0 (Col-0) plants with a suspension of fungal spores (Thatcher et al. 2012). Plants inoculated with Fo5176 developed typical yellowing and wilting symptoms, visible at 6 days postinoculation (DPI) (Fig. 1A). Almost all Fo5176-infected plants died within 3 weeks of inoculation (Fig. 1B). By contrast, plants inoculated with Fo47 not only stayed healthy (Fig. 1A and B) but also showed increased aboveground biomass (Wilcoxon rank-sum test, P < 0.001) when compared to mock-inoculated plants (Fig. 1C), suggesting that Fo47 may have a growth-promoting effect.
Fig. 1.
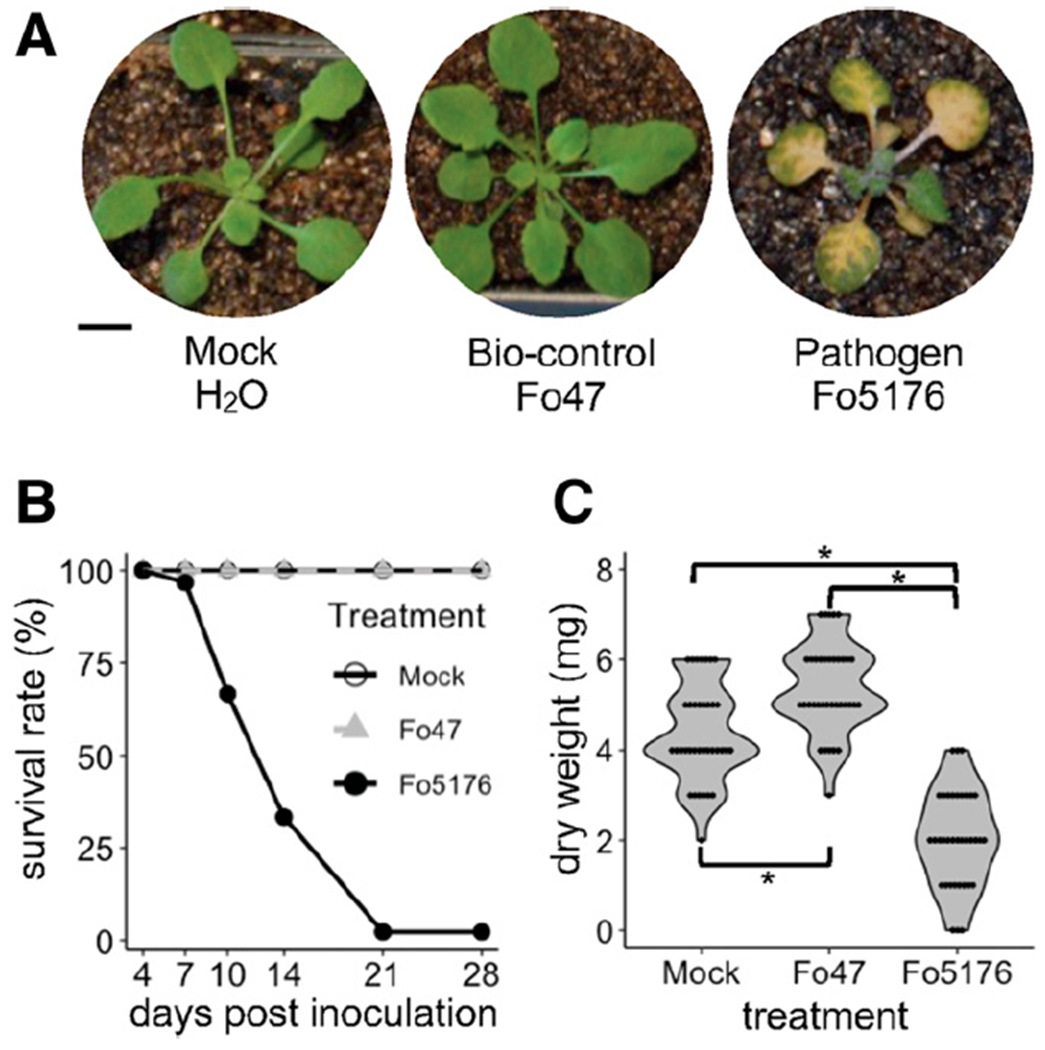
Compatible versus incompatible Arabidopsis interaction with an endophytic (Fo47) versus a pathogenic (Fo5176) Fusarium oxysporum strain. A, F. oxysporum Fo5176 causes chlorosis on Arabidopsis Col-0 plants, while Fo47-infected plants and plants mock-inoculated with water do not exhibit any symptoms. Photographs were taken at 7 days postinoculation (DPI) and representative plants are shown. B, Survival analysis assay illustrating the survival rates of Arabidopsis plants mock-inoculated with water or infected with Fo47 or Fo5176 at six timepoints, from 4 to 28 DPI. Ninety plants were assayed per treatment. C, Summary of shoot dry biomass of Arabidopsis plants mock-inoculated with water or infected with Fo47 or Fo5176 at 6 DPI. Statistical significance was determined by Kruskal–Wallis and Wilcoxon rank-sum tests. Asterisk indicates statistical significance at P < 0.001. Thirty-six plants were assayed per treatment.
By comparison with a sister species, F. verticillioides, accessory chromosomes were identified in both the Fo47 (B. Wang et al. 2020) and Fo5176 (Fokkens et al. 2020) genomes in addition to the 11 core chromosomes (Fig. 2), vertically inherited from the common ancestor shared between these two sister species 10 to 11 million years ago (Ma et al. 2013). The Fo47 genome had one accessory chromosome (chromosome 7, with a length of 4.25 Mb), while the Fo5176 genome had four (chromosomes 2, 14, 16, and 18) (Fig. 2). The combined length of accessory chromosome regions in Fo5176 was 21.63 Mb, including large segments (size >1 Mb) of chromosomes 4, 10, 11, and 13 that shared no syntenic block with the F. verticillioides genome. Fo47 and Fo5176 accessory chromosomes were enriched in repetitive sequences (Supplementary Fig. S1), a common property observed from all accessory chromosomes (Yang et al. 2020). Fo47 accessory genes were significantly enriched for cell signaling and nutrient sensing functions, whereas Fo5176 genes were enriched for functions relating to virulence and detoxification (Supplementary Datasets S1 and S2). As these two genomes share an almost identical core sequence, we hypothesized that distinct accessory chromosomes in each genome may play important roles in the distinct phenotypic outcomes (disease versus growth promotion).
Fig. 2.

Comparative genomics reveals unique sets of accessory chromosomes in Fusarium oxysporum Fo47 and Fo5176. Synteny of genome assemblies between F. verticillioides (Fv) and the two selected F. oxysporum strains (Fo47 and Fo5176). Track a shows distribution of karyotypes of assembled chromosomes; track b, GC density; track c, density of transposable elements (TEs) calculated in 10-kb windows; track d, gene density calculated in 100-kb windows. Track e shows syntenic blocks. Relationships are shown through linking syntenic block genes (gene number >10) in each genome pair. Core chromosomes can be identified through synteny between F. verticillioides and each F. oxysporum strain, whereas accessory chromosomes and regions show no or reduced synteny. Chromosomes 2, 14, and 18, and large segments (size >1 Mb) of chromosomes 4, 10, 11, 13, and 16 in Fo5176, and chromosome 7 in Fo47 show no synteny with the F. verticillioides genome and are thus identified as accessory regions, characterized by their high TE density and low gene density.
Reprogramming of the plant transcriptome in response to a fungal pathogen or endophyte.
To examine the transcriptional regulation underlying the distinct endophytic and pathogenic interactions of the two strains (Fig. 1), we sequenced the fungal and host plant transcriptomes from Arabidopsis plants inoculated with Fo47 or Fo5176. Infected plants were sampled at 12, 24, 48, and 96 h postinoculation (HPI), in parallel with plants mock-inoculated with water at 12 HPI as a control. We harvested root tissues for transcriptome deep sequencing (RNA-seq). Dual RNA-seq data were analyzed using an in-house pipeline to calculate the transcript levels of plant and fungal genes.
About half of all annotated Arabidopsis genes (16,544 of a total of 32,833) were differentially regulated in at least one of 18 comparisons between different timepoints for the same interaction type (12, 24, 48, and 96 HPI; 12 comparisons), between different interaction types at the same timepoint (beneficial versus pathogenic; four comparisons), or between endophytic or pathogenic interactions and the mock control at 12 HPI (two comparisons). These differentially expressed genes (DEGs) revealed several interesting patterns (Fig. 3A).
Fig. 3.

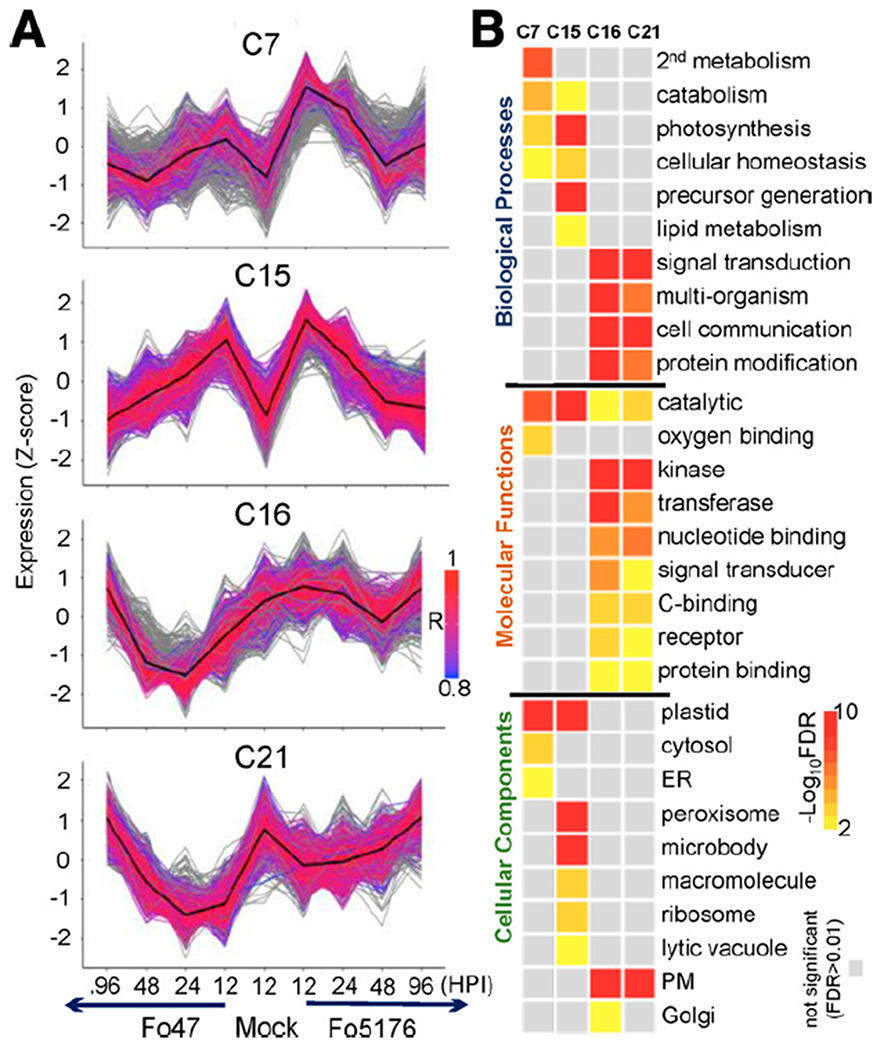
Expression profiling of Arabidopsis roots inoculated with an endophytic versus a pathogenic Fusarium oxysporum strain. A, Extent of correlation between Arabidopsis differentially expressed genes (DEGs) in Fo47- and Fo5176-infected plants across the different timepoints at 12, 24, 48, and 96 h postinoculation (HPI). Correlation coefficients (converted to percentages) are scaled to the sizes and colors of the circles. B, Gene ontology (GO) enrichment of 24 gene clusters from K-means clustering of Arabidopsis DEGs. The color scale of the heatmap represents the significance level of GO enrichment for biological processes related to stimuli response and developmental processes, expressed as –Log10(false discovery rate [FDR]). Four clusters, C7, C15, C16, and C21, highly enriched for stimuli responses and deprived of developmental regulation, are highlighted in red and defined as immunity clusters.
First, we observed a strong correlation between patterns of gene expression for both treatments at the same timepoints, despite clearly distinctive endophytic and pathogenic phenotypes (Fig. 1). The Pearson’s correlation coefficients (PCCs) for the four comparisons between plants infected with either the beneficial or the pathogenic fungal strain at each timepoint were very high, with values of 0.95, 0.94, 0.97, and 0.96 at 12, 24, 48, and 96 HPI, respectively (Fig. 3A, labeled in red), suggesting that a small subset of genes contribute to the observed phenotypic differences. Global clustering analysis using the 16,544 Arabidopsis DEGs yielded 24 co-expression gene clusters (Fig. 3B; Supplementary Fig. S2; Supplementary Dataset S3). A total of 10,014 genes within 12 clusters had similar expression patterns at all timepoints (Supplementary Dataset S3), accounting for 60.5% of all DEGs. Since 16,289 genes were either not expressed or not changed, we concluded that 6,544 (about 20% of all) genes held answers to the transcriptional reprogramming between these two treatments.
We also observed significant transcriptional reprogramming within each interaction over time. For samples inoculated with Fo47, PCC scores decreased from 0.94 (between 12 and 24 HPI) to 0.84 (between 12 and 96 HPI) as infection progressed. Similarly, PCC values dropped from 0.93 (between 12 and 24 HPI) to 0.81 (between 12 and 96 HPI) for Fo5176-inoculated plants. We then compared each fungal interaction pairwise at each timepoint to identify reciprocal DEGs (Supplementary Fig. S3A), yielding 1,009, 642, 59, and 403 genes that were preferentially expressed in Fo47-infected plants and 868, 1,172, 604, and 425 plant genes in Fo5176-infected plants at 12, 24, 48, and 96 HPI, respectively (Supplementary Fig. S3B). Notably, plant genes that were preferentially expressed during the endophytic interaction were enriched in gene ontology (GO) terms such as cell cycle, cell growth, development, response to stimuli, and cellular transport. Moreover, the genes associated with each enriched GO term showed a temporal wave as the infection course progressed, with genes involved in cell cycle highly enriched at the early stages of infection but with a diminishing contribution that was consecutively replaced by genes related to development at around 24 HPI, response to stimuli at 48 HPI, and transport at 96 HPI (Supplementary Fig. S3C). Conversely, genes preferentially induced in response to the pathogenic fungus were consistently enriched in GO terms mainly related to defense responses, with no obvious underlying temporal pattern (Supplementary Fig. S3C).
Second, when compared to the mock-inoculated samples, plants inoculated with Fo47 or Fo5176 both displayed drastic transcriptional reprogramming at the earliest timepoint of this study (12 HPI), as these comparisons had the lowest PCCs of 0.85 for Fo47 and 0.83 for Fo5176. As time from initial inoculation progressed, however, the transcriptomes of all plants became much more similar, with PCCs rising to 0.98 for Fo47 and 0.96 for Fo5176 (Fig. 3A, labeled in green). This observation indicated that the outcome of the plant-host interaction might be decided as early as 12 HPI. To begin to dissect the critical transcriptional reprogramming taking place at 12 HPI in both endophytic and pathogenic interactions, we conducted a careful analysis to identify genes that are not only differentially expressed between the two treatments but also differentially expressed relative to mock-inoculated samples. This analysis resulted in the identification of genes that were specifically upregulated or downregulated in fungus-infected samples. These four plant gene sets consisted of 140 upregulated and 422 downregulated genes specifically in response to Fo47 infection and 286 upregulated and 767 downregulated genes in response to Fo5176 infection.
Functional analysis of these genes using GO enrichment and network analyses (Fig. 4; Supplementary Datasets S4 through S7) confirmed previous observations of fungal-plant interactions but also revealed unexpected findings. As expected, we observed significant suppression of genes related to plant growth by the pathogenic strain Fo5176 (Fig. 4A), including genes associated with the cell cycle, cell-wall organization, plant-type cell-wall biosynthesis, and microtubule-based processes. Genes upregulated early in response to Fo5176 infection were highly enriched in toxin and indole metabolism as well as small molecule biosynthesis (Fig. 4B), possibly reflecting the initial upheaval brought upon by the infection. For the endophytic interaction, we noticed a significant suppression of immunity-related functions, including plant defense and immunity and jasmonic acid response (Fig. 4C). This data therefore also suggested that the endophytic strain Fo47 attenuates plant defenses. Among the genes induced by the endophyte, we were pleased to see that several define a module related to nitrate metabolism and anion transport (Fig. 4D), which would be consistent with the promotion of plant growth by Fo47 (Fig. 1D).
Fig. 4.

A summary of transcriptomic changes occurring at 12 h postinoculation. Gene ontology (GO) enrichment analysis and visualization were performed on four datasets representing up- and downregulation by Fo47 and Fo5176, respectively. Nodes represent the GO categories with enrichment, while edges exist when two GO categories share the same genes. The nodes labeled in the same color represent the GO terms that belong to a master term as labeled. The size of the nodes is scaled to the number of genes within each GO term in each figure section. A, Pathogen suppression: Arabidopsis genes with expression when infected by Fo47 are smaller than when infected by Fo5176 and when infected by water (Fo47 < Fo5176 and Fo47 < mock). B, Pathogen induction: Fo5176 > Fo47 and Fo5176 > mock. C, Endophyte suppression: Fo47 < Fo5176 and Fo47 < mock. D, Endophyte induction: Fo47 > Fo5176 and Fo47 > mock.
A nitrate-CPK (Ca2+-sensor protein kinase)-NLP (Nin-like protein) signaling pathway was previously reported (Liu et al. 2017) that activated the expression of 394 genes and repressed another 79 genes in response to exogenous nitrate treatment. We examined whether our clusters of DEGs (Fig. 3B) showed an overrepresentation of genes differentially regulated by this nitrate signaling pathway. Cluster 21 included the most downregulated genes from this pathway, with eight genes (P value = 2.56e-04, two-sided Fisher’s exact test) that were downregulated in both interactions, with stronger suppression by the endophyte (Supplementary Fig. S2; Supplementary Dataset S8). Of the 394 upregulated genes in the nitrate pathway, 329 were differentially expressed in our dataset, with 251 assigned to clusters. Of those, over half were significantly enriched in five clusters: C5 (20 genes, P value = 2.12e-05), C6 (14 genes, P value = 1.07e-05), C8 (40 genes, P value = 1.46e-14), C16 (16 genes, P value = 7.37e-04), and C23 (38 genes, P value = 6.62e-21). These 251 genes, representing a majority of the genes upregulated in the nitrate signaling pathway, were induced by both the endophyte and the pathogen but exhibited stronger responses in the context of endophytic inoculations (Supplementary Fig. S2).
Notably, cluster C23 included NLP1, encoding a transcription factor involved in the nitrate-CPK-NLP signaling pathway (Liu et al. 2017), as well as NITRATE TRANSPORTER2.1 and 2.2 (NRT2.1, NRT2.2), NITRATE REDUCTASE1 (NIA1), and NITRITE REDUCTASE1 (NIR1), all major components of the pathway that were upregulated when compared to the mock-inoculated sample (Supplementary Fig. S4). Of 24 previously reported transcription factors that control transcriptional regulation of nitrogen-associated metabolism and growth (Gaudinier et al. 2018), 16 were assigned to our clusters (Supplementary Dataset S9), including WUSCHEL RELATED HOMEOBOX14 (WOX14) and LOB DOMAIN-CONTAINING PROTEIN4 (LBD4) in cluster C23. Collectively, this analysis suggests that nitrogen signaling is involved in the F. oxysporum–Arabidopsis interaction and the endophyte may enhance the nitrogen signal and hence change the course of the plant response.
Perturbation of plant immunity.
To better understand how the endophyte and the pathogen perturb plant immunity via shared and distinct responses, we carefully investigated the 24 co-expression clusters, based on their global patterns of expression. Four clusters, C7, C15, C16, and C21, showed enrichment (P < 0.05) for GO terms related to immunity and defense responses; the same clusters also lacked GO terms related to development (Fig. 3C). Compared to plant PTI and ETI networks (consisting of 1,856 PTI-related and 1,843 ETI-related genes) previously constructed using a machine learning algorithm (X. Dong et al. 2015), three clusters (C15, C16, and C21) were enriched for both PTI and ETI genes, whereas cluster C7 was primarily enriched in PTI response genes (two-sided Fisher’s exact test, P < 0.05) (Supplementary Datasets 10 and 11). This suggests a transcriptional plasticity of plant immunity in responding to the endophytic and pathogenic F. oxysporum.
Conserved immune response toward an endophyte and a pathogen.
Cluster C15 comprised 1,290 genes and was the largest immunity-related cluster, with nearly identical plant transcriptome responses following Fo47 and Fo5176 inoculation. Indeed, genes from cluster C15 were initially strongly upregulated at 12 HPI in both interactions and gradually returned to an expression level comparable to that of mock-inoculated plants as infection progressed (Fig. 5A). C15 was most significantly enriched in PTI genes (P value = 2.66e-72), reflecting the general plant perception of fungal signals derived from both pathogenic and symbiotic organisms (e.g., MAMPs). Cluster C15, indeed, included many immunity-related genes involved in fungal perception, signal transduction, and transcriptional regulation, including ERECTA, a RLK that regulates stomatal patterning and immunity (Sopeña-Torres et al. 2018), RECOGNITION OF PERONOSPORA PARASITICA5 (RPP5), which encodes a putative NLR protein that confers resistance to Peronospora parasitica (Noël et al. 1999; Parker et al. 1997), RESPONSIVE TO DEHYDRATION 21A (RD21A), which encodes a cysteine proteinase with peptide ligase and protease activity that is involved in immune responses against the necrotrophic fungal pathogen Botrytis cinerea (Lampl et al. 2013, and NUCLEAR FACTOR Y, SUBUNIT B3 (NF-YB3), which encodes a transcription factor activated by endoplasmic reticulum stress responsible for the regulation of stress responses (Liu and Howell 2010).
Fig. 5.
Expression and gene ontology (GO) enrichment of immunity gene clusters. A, Expression profile of immunity gene clusters. Color scale indicates the correlation of expression between genes and the cluster centroids. Genes that were removed from the clusters before functional analysis, due to the expression correlation with centroid lower than (or equal to) 0.8, are shown in gray. Enrichment of pathogen-associated molecular pattern-triggered immunity and effector-triggered immunity genes (X. Dong et al. 2015) is indicated (in all labeled cases, P < 1E-7, by Fisher’s exact test). Gene number within the four clusters is as follows: C7 = 422, C16 = 615, C21 = 766. B, GO enrichment analysis of immunity gene clusters for biological processes, molecular functions, and cellular components. Color scale of the heatmap represents the false discovery rate. Stimuli responses, developmental processes, and redundant GO terms were removed.
Cluster C15 was also highly enriched in genes with functions related to the chloroplast/plastid (P value = 9.2e-89) (Fig. 5B; Supplementary Dataset S12). An organelle essential for plant photosynthesis, chloroplasts have recently come to the forefront as key players in plant immune responses (Göhre et al. 2012; Serrano et al. 2016), possibly functioning as a signaling hub that links the initial recognition of diverse pathogens at the PM and signal transduction to the nucleus to orchestrate transcriptional reprogramming in response to infection (Chan et al. 2016; de Souza et al. 2017; de Torres Zabala et al. 2015; Liu 2016; Medina-Puche et al. 2020; Wang et al. 2016). It is unlikely that chloroplast-related genes from cluster C15 represent artifacts caused by the manipulation of roots in the light during harvesting, as these genes were expressed at low levels in mock-inoculated plants, although they were subjected to the same inoculation and harvesting procedure. Our observations are also consistent with a previous report in which strain Fo5176 was shown to induce the expression of Arabidopsis genes normally involved in photosynthesis in root tissues at 1 DPI (Lyons et al. 2015). Pathogens may thus interfere with host chloroplast/plastid functions to manipulate host immunity in their favor. We know very little about the possible role played by chloroplasts during endophytic colonization.
Stronger induction of plant PTI responses by the pathogen.
We hypothesized that a subset of plant immune responses against the pathogen and endophyte would differ, given their distinctive phenotypes, even though most clusters showed the same pattern during endophytic and pathogenic responses. Indeed, three immunity-related clusters, C7, C16, and C21, exhibited distinct patterns between the pathogen and the endophyte (Fig. 5A). Cluster C7 (422 genes), which was primarily enriched in genes associated with PTI, exhibited a stronger induction by Fo5176 infection than by Fo47, despite being induced by both strains (Fig. 5A). Several GO terms were shared between clusters C7 and C15, such as chloroplast/plastid-related functions (Fig. 5B), possibly reflecting fine-tuning of the initial recognition of conserved fungal signals.
Genes uniquely induced by the pathogen included PEROXIDASE37 (PRX37), encoding a putative apoplastic peroxidase that generates H2O2 primarily in the vascular bundles for host defense (Pedreira et al. 2011), and PENETRATION2 (PEN2), encoding an atypical tyrosinase required for broad-spectrum resistance to filamentous plant pathogens (Fuchs et al. 2016). Also included in cluster 7 were genes with dual functional roles in immunity against different pathogens. For instance, PATATIN-LIKE PROTEIN2 (PLP2) promotes cell death and facilitates Botrytis cinerea and Pseudomonas syringae infection in Arabidopsis (La Camera et al. 2005), whereas it confers host resistance to cucumber mosaic virus (Camera et al. 2009). KUNITZ TRYPSIN INHIBITOR1 (KTI1), a trypsin inhibitor referenced as an antagonist involved in the negative regulation of programmed cell death that mediates susceptibility in Erwinia carotovora but has an opposite function in Pseudomonas syringae pv tomato DC3000 (Li et al. 2008).
Suppressed plant immunity in the presence of the endophyte.
In contrast to clusters C7 and C15, both clusters C16 (615 genes) and C21 (766 genes) exhibited stronger suppression of expression by the endophyte (Fig. 5A). We also observed a unique and specific suppression of plant immunity by the endophyte from the GO term enrichment and network analyses at 12 HPI described above (Fig. 4C). While clusters C7 and C15 showed minimal overlap of enriched GO terms, clusters C16 and C21 shared many terms, including signal perception and transduction, protein-protein interaction, and PM localization (Fig. 5B). Of 70 genes identified as contributing to danger-sensing and signaling systems (Zhou and Zhang 2020), we detected 12 genes in cluster C16 and another 12 genes in cluster C21 (Table 1; Supplementary Dataset S13). For instance, PRRs and downstream components in cluster C16 include EFR, BAK1, LYK5, and CERK1 and, in cluster C21, PEPR1, PEPR2, FERONIA, and RBOHD. NLRs and downstream signaling components in cluster C16 include ADR1-L1/ADR1-L2 and RPM1, PAD4, and RPS4 in cluster C21. In summary, a strong suppression of diverse immunity-related genes is unique to the endophytic interaction, suggesting that modulation of plant immunity may contribute to the different outcomes.
Table 1.
Genes that play important roles in danger sensing and signaling suppressed by the endophytic interactiona
| PRRs and downstream components | RLCKs | MAP kinase cascades | NLRs and downstream signaling | SA biosynthesis and signaling |
|---|---|---|---|---|
| Cluster C16 | ||||
| EFR | PBL1 | MEKK1 | ADR1-L1 | CBP60g |
| BAK1 | PBL2 | MPK11 | ADR1-L2 | SARD1 |
| LYK5 | LAZ5 | |||
| CERK1 | ZAR1 | |||
| SOBIR1 | ||||
| Cluster C21 | ||||
| PEPR1 | MPK3 | RPM1 | CAMTA3 | |
| PEPR2 | PAD4 | NPR1 | ||
| FER | RPP1 | NPR3 | ||
| RBOHD | RPS4 | |||
| PERK1 | RPS3 | |||
| RFO1 | RPS6 |
PRRs = pattern recognition receptors; RCLKs = receptor-like cytoplasmic kinases; MAP = mitogen-activated protein; SA = salicylic acid.
PTI- and ETI-sensing genes induced by the endophyte and the pathogen.
Overall, we observed strong host immune responses, when challenged with either the endophyte or the pathogen, involving complex signal perception and signal transduction cascades. Distinct responses included the suppression of plant growth and the induction of plant defenses by the pathogenic strain Fo5176 and the attenuation of host immunity with the concomitant induction of nitrogen metabolism by the endophytic strain Fo47 (Fig. 4). To further dissect the plant immunity pathways involved in these two interactions, we conducted a systematic analysis of the expression profiles of genes encoding RLPs or RLKs and NLR proteins (Table 1; Supplementary Datasets S14 and S15).
RLP and RLK genes.
The Arabidopsis genome encodes 533 RLPs and RLKs, as determined by the MAPMAN Mercator annotation (Schwacke et al. 2019). Of those, 311 were assigned to our clusters of DEGs (Supplementary Dataset S14). In addition to the two immunity clusters, C16 (37 genes, P value = 4.76e-11) and C21 (30 genes, P value = 2.11e-05), whose expression is repressed by the endophyte, these RLP and RLK genes were also enriched in clusters C11 (14 genes, P value = 6.31e-04) and C12 (11 genes, P value = 6.31e-04). Their expression appeared to be repressed by both the endophyte and the pathogen to varying degrees. Characterized defense-related RLK genes include SUPPRESSOR OF BIR1 (SOBIR1) and EFR in cluster C16 and RESISTANCE TO FUSARIUM OXYSPORUM1 (RFO1) and PROLINE-RICH EXTENSIN-LIKE RECEPTOR KINASE1 (PERK1) in cluster C21. RFO1 encodes a protein that confers a broad-spectrum resistance to Fusarium spp. (Diener and Ausubel 2005). Cluster C16 also included genes encoding LysM receptor-like kinases CERK1 and LYK5 (Cao et al. 2014), which are essential for the perception and transduction of the chitin oligosaccharide elicitor. Although not significantly enriched, 26 RLP/RLK genes grouped in cluster C15, including FLG22-INDUCED RECEPTOR-LIKE KINASE1 (FRK1) and HERCULES RECEPTOR KINASE1 (HERK1), which are activated in response to both pathogenic and endophytic F. oxysporum strains.
NLR genes.
Many plant NLRs are commonly identified as resistance proteins that act as surveillance molecules recognizing pathogen effectors that target the host machinery. In accordance with the “guard” model, NLRs then trigger an ETI response (Cao et al. 2014). The Arabidopsis genome encodes 160 NLR proteins (Baggs et al. 2020), of which we identified 119 genes among our Dataset of DEGs (84 were assigned to clusters). Among these NLR genes, about 40% clustered across four immunity clusters, cluster C21 (18 genes), C16 (14 genes), C15 (11 genes), and C7 (four genes), and were uniquely enriched in clusters 21 (P value = 2.58e–07) and 16 (P value = 6.82e–06), both of which include genes specifically repressed in the endophyte (Supplementary Dataset S15). The 11 NLR genes in cluster C15 included five known resistance genes (AT1G63880, AT1G61190, RPP4, RPP5, and RPP8) against oomycete and fungal pathogens (Goritschnig et al. 2012; McDowell et al. 2005; Staal et al. 2006; van der Biezen et al. 2002).
Notably, most of the NLR genes that were enriched in endophyte-suppressed clusters C16 and C21 are not functionally characterized. Nevertheless, characterized NLRs represented by genes in cluster C16 included two apoplast/chloroplast-localized ADP-binding immune receptors (ADR1-L1 and ADR1-L2) (Dong et al. 2016). Also belonging to cluster C16 were the two effector-induced resistance genes LAZARUS5 (LAZ5) and HOPZ-ACTIVATED RESISTANCE1 (ZAR1) (Barbacci et al. 2020; Baudin et al. 2017), conferring resistance to a Pseudomonas syringae strain expressing the AvrRPS4 and Hop effectors, respectively. NLR genes in cluster C21 included disease resistance proteins RPS3, RPS4, and RPS6, which provide specific resistance against P. syringae pv. tomato carrying the avirulence genes AvrRPS3, AvrRPS4, and AvrRPS6, respectively (Bisgrove et al. 1994; Kim et al. 2009; Narusaka et al. 2009). Repression of the expression of these NLR genes by the endophyte again supports the idea that the danger-sensing and signaling systems underlying the responses to Fo47 and Fo5176 are distinct.
Accessory chromosomes in two strains harbor genes induced during infection and with distinct biological functions.
The distinct plant responses at both the phenotypic and transcriptome levels, resulting from inoculation with the two F. oxysporum isolates, is no doubt related to genomic differences between the two strains. Even though both strains belong to the same species complex and share an approximately 46-Mb core genome, each strain also carries distinct accessory chromosomes (Fig. 2). The Fo47 accessory chromosome 7 harbored 1,299 predicted genes (7.2% of total predicted genes) (Supplementary Dataset S16), 757 of which were expressed and 160 were strongly induced (false discovery rate [FDR] < 0.05) during one or more timepoints of the infection.
To explore the function of these accessory genes encoded in the endophytic strain Fo47, we next analyzed the functional domains they encoded. After excluding genes that encoded proteins with transposase-like domains or unknown domains, we highlighted five enriched PFAM domains, i.e., the regulator of G-protein signaling domain (PF00615), nitric oxide (NO)-binding membrane sensor involved in signal transduction (PF03707), basic leucine zipper (bZIP) transcription factor (PF00170), chromodomain (PF00385), and bromodomain (PF00439) (Fig. 6A; Supplementary Fig. S5A). The enrichment of functional domains involved in cell signaling and the apparent lack of enrichment for domains related to virulence suggested that the Fo47 accessory chromosome contains genes with functions that are well-suited to a nonpathogenic lifestyle. Transcriptome analysis showed that nine Fo47 genes encoding proteins with the bacterial signaling protein domain (PF03707) are most highly induced at 24 and 48 HPI. Domain PF03707 plays a role in sensing oxygen, carbon monoxide, and NO (Galperin et al. 2001). The Fo47 genome has the highest number of genes encoding a PF03707 domain, with nine genes, compared to other filamentous fungi, such as Aspergillus nidulans (1), Neurospora crassa (1), Pyricularia grisea (1), F. graminearum (2), F. verticillioides (2), and F. solani (3) (Cuomo et al. 2007; Dean et al. 2005; Galagan et al. 2005; Ma et al. 2010), as well as other F. oxysporum species complex members (three to about seven, average of four) (DeIulio et al. 2018). Notably, six of the nine genes reside on accessory chromosome 7, making it a major contributor to the expansion of this gene family within this strain (Supplementary Fig. S6).
Fig. 6.

Distinct biological functions for induced accessory chromosome (AC) genes in the endophyte Fo47 and the pathogen Fo5176. Fold enrichment refers to the ratio of the proportion of genes on the ACs with a specific term over the proportion of genes in the whole genome with a particular term (adjusted P value < 0.05). A, Significantly induced Fo47 accessory genes are represented in five enriched PFAM domains, including regulator of G-protein signaling domain (PF00615), nitric oxide-binding membrane sensor involved in signal transduction (PF03707), bZIP transcription factor (PF00170), chromo domain (PF00385), and bromodomain (PF00439)-containing proteins. B, In Fo5176, six PFAM domains are significantly enriched and induced at different stages of the infection course, i.e., cysteine-rich secretory protein family (PF00188), Calpain family cysteine protease (PF00648), peptidase M16 (PF16187), poly (ADP-ribose) polymerase regulatory domain (PF02877), cyclin C-terminal domain (PF02984), and WGR domain (PF05406).
By contrast, accessory chromosomes and regions from the pathogenic strain Fo5176 contributed 4,136 predicted genes (23% of total predicted genes) (Supplementary Dataset S17), of which 3,502 were expressed and 1,140 were strongly induced during one or more timepoints of the infection. Genes located in accessory regions in Fo5176 encoded proteins that were enriched for 42 PFAM domain terms. We noticed six PFAM domains that were highly enriched at different stages of the infection course and whose encoding genes were highly expressed, namely, cysteine-rich secretory protein family (PF00188), calpain family cysteine protease (PF00648), peptidase M16 (PF16187), poly (ADP-ribose) polymerase regulatory domain (PF02877), cyclin C-terminal domain (PF02984), and WGR domain (PF05406) (Fig. 6B; Supplementary Fig. S5B). Most of these domains are likely associated with microbial pathogenesis or detoxification; their associated genes were induced during infection (Fig. 6B). In particular, members of the cysteine-rich secretory protein (CAP) superfamily (PF00188) have a wide range of biological activities, including fungal virulence, cellular defense, and immune evasion (Schneiter and Di Pietro 2013). For example, the F. oxysporum CAP family protein Fpr1 is a PR-1-like protein that is important for the virulence of strain Fol4287 (Prados-Rosales et al. 2012). The Fo5176 genome encodes 15 CAP family members, significantly more than Fo47 and other comparable fungal species (average five members) (Supplementary Fig. S7). Phylogenetic analysis of CAP proteins showed that four CAP members formed a core group shared by Fo5176 and Fo47. However, a separate clade of six CAP family proteins was expanded in Fo5176 and encoded by Fo5176 accessory chromosomes (Supplementary Fig. S7). These results highlight the distinctive functions of and roles played by accessory chromosomes in the nonpathogenic strain Fo47 and the pathogenic strain Fo5176. These differences might provide the mechanistic basis that allows Fo47 to specialize in host-sensing and benefit its host as an endophyte, while the pathogenic Fo5176 specializes in host invasion and killing.
DISCUSSION
We performed a comparative study of infection by an endophytic (Fo47) and a pathogenic (Fo5176) strain of F. oxysporum in the context of the F. oxysporum–Arabidopsis system that revealed the transcriptional plasticity of plant defense responses. The pathosystem we developed combines the extensive knowledge of plant immunity in Arabidopsis and one of the most damaging fungal pathogens for agriculture, F. oxysporum. Strain-specific interactions with a common host are likely dictated by the accessory chromosomes from each F. oxysporum genome, which allows a comparative study that minimizes genetic differences between strains to address the underlying mechanism that results in distinct phenotypes (growth promotion or disease or even death). Up to 50% of crop losses in the United States can be attributed to soil-borne pathogens (Raaijmakers et al. 2009), and our results provide a foundation for the development of technologies to enhance plant health, sustain a healthy ecosystem, and feed a continuously growing human population.
We employed comparative metatranscriptomics over the course of early infection to systematically capture temporal transcriptional changes in both the host and the interacting microbes. Our results may be summarized along four main axes, as illustrated in Figure 7.
Fig. 7.

Model of transcriptomic plasticity in beneficial and antagonistic plant–fungal interactions. Molecular response of Arabidopsis thaliana plants challenged with an endophyte Fo47 and a pathogen Fo5176, two Fusarium oxysporum strains sharing a core genome of about 46 Mb, in addition to their unique accessory chromosomes. Distinct responding genes depict the transcriptional plasticity, as the pathogenic interaction activates plant stress responses and suppresses plant growth and development-related functions, while the endophyte attenuates host immunity but activates plant nitrogen assimilation. The differences in reprogramming of the plant transcriptome are linked to accessory genes encoded by the two closely related fungal genomes. NO = nitric oxide, TF = transcription factors; MAMPs = microbe-associated molecular patterns, PRRs = pattern recognition receptors, NLR = nucleotide-binding site and leucine-rich repeat domain receptors, and JA = jasmonic acid.
First, host transcriptional responses were strikingly similar at all timepoints, regardless of the obvious phenotypic differences seen after infection of Arabidopsis plants by Fo47 and Fo5176. Of all clusters of DEGs, cluster C15 exhibited a strong and early induction of genes, followed by a return to an expression level comparable to that of control samples. This cluster captured 26 Arabidopsis RLP and RLK genes as well as 269 PTI and 159 ETI response genes, suggesting that both strains initially elicit a similar MAMP response, which would not be surprising, as they belong to the same species.
Second, our data revealed rapid transcriptional reprogramming at the beginning of the interactions. While most plant genes exhibited a common expression pattern during the two infections, a small subset of plant genes displayed divergent gene expression profiles. By far the most striking difference was observed at 12 HPI, when the GO biological processes for genes uniquely induced by Fo47 or Fo5176 reflected almost opposite responses. The endophytic strain Fo47 stimulated nitrogen metabolism and suppressed host immunity, whereas the pathogenic strain Fo5176 stimulated host immune responses and toxin metabolism but repressed functions related to plant growth and development. We propose that this distinct expression profile, reflected in the early divergence of the host transcriptome, is the result of plasticity of the host transcriptome when facing an endophyte or a pathogen. We hypothesize that the perception by the host of distinct fungal signals occurs shortly after inoculation and is followed by the rapid activation of downstream signaling cascades. Our results also stress the importance and necessity of sampling early during the establishment of a fungal-host interaction to better capture the full extent of the underlying temporal dynamics.
Third, Fo47 inoculation resulted in suppression of genes related to plant defense and induced genes related to plant growth, in agreement with the trade-off between growth and defense. It has been reported that plants can channel nitrogen resources towards production of defense-related compounds when confronted with pathogens (Ullmann-Zeunert et al. 2013). For instance, allele polymorphism at the single locus ACCELERATED CELL DEATH6 (ACD6) can dictate a distinct difference between growth and defense among different Arabidopsis ecotypes (Todesco et al. 2010). Further characterizing the F. oxysporum–Arabidopsis pathosystem should illuminate the mechanism or mechanisms by which nutrients are allocated in relation to plant defense.
Finally, we observed an agreement between plant infection phenotypes and distinctive gene functions associated with fungal accessory chromosomes. While upregulated fungal accessory genes were primarily enriched in proteins with roles in cell signaling and nutrient transport in the endophyte Fo47, they were enriched for virulence and detoxification in the pathogen Fo5176, likely contributing to the contrasting phenotypes of plants infected by these two F. oxysporum strains.
In conclusion, time-resolved comparative metatranscriptomics can be used to characterize transcription regulation when the model plant Arabidopsis is challenged with an endophyte and a pathogen of the same fungal species. We showed both the conservation and plasticity of the plant and fungal transcriptomes and how they may relate to the distinctive genomic features associated with each fungal genome. The Arabidopsis and F. oxysporum pathosystem developed here is likely to become an ideal system to characterize plant recognition and response mechanisms against soil-borne root fungi. We believe this system will be pivotal in enriching our understanding of the molecular mechanisms necessary to enhance vascular wilt resistance not only in Arabidopsis but also in crops that are under threat by F. oxysporum pathogens.
MATERIALS AND METHODS
Plant and fungal growth.
F. oxysporum Fo5176 and Fo47 were routinely cultured on potato dextrose agar (BD) at 28°C under a 12-h light AND 12-h dark photoperiod. Fungal spores were collected from 5-day-old cultures in potato dextrose broth (BD) by passing the liquid culture through a double layer of sterile cheesecloth, followed by centrifugation of the flow-through at 3,000 × g for 15 min at room temperature. Fungal spores were mixed with an appropriate volume of sterile deionized water to prepare the spore suspension (concentration 1 × 106 spores/ml) for infection assays.
Plant infection assay.
Seeds of A. thaliana accession Col-0 were obtained from the Arabidopsis Biological Resource Center (Ohio State University) and were surface-sterilized in 1 ml of 70% (vol/vol) ethanol three times, 5 min each, followed by one wash with 50% (vol/vol) bleach for 5 min. After removing the bleach solution, seeds were rinsed with 1 ml of sterile distilled and deionized water and were stratified for 3 to 4 days in darkness at 4°C. Seeds were planted into pots filled with an autoclaved mixture of fine-grain play sand/MetroMix 360/vermiculite in a 1:2:1 ratio, were watered with distilled deionized water, and were covered with a clear plastic lid to retain a high humidity for 3 days in the growth chamber at 24°C, with 14 h of light and 10 h of dark and a light intensity (T8 fluorescent and incandescent bulbs) ranging from 89 to 94 μmol·m−2·s−1. After 3 days, the plastic lid was removed and seedlings were allowed to grow for 11 additional days, prior to inoculation with F. oxysporum microconidia. Plants were 14 days old at the time of inoculation and had at least four fully expanded true leaves. For F. oxysporum infection, the roots of 14-day-old Arabidopsis plants were dipped for 30 s in a 1 × 106 fungal spores/ml suspension of Fo5176 or Fo47 or in sterile distilled H2O for the mock control. Inoculated plants were planted in autoclaved potting mix and moved to a growth chamber set to 28°C with the same photoperiod as above.
RNA preparation, sequencing, and data analysis.
Roots from infected plants at 12, 24, 48, and 96 HPI were harvested from five plants per treatment and timepoint for total RNA isolation. For control samples, roots from the same number of control plants were collected at 12 HPI. Fungal cultures from Fo5176 and Fo47 fungal mycelia were harvested after 5 days from liquid cultures for RNA extraction. Three biological replicates were produced for each treatment. Total RNA was extracted using the ZR Soil/Fecal RNA microprep kit (Zymo Research), following manufacturer protocol, and the RNA quantity and quality were assessed using a NanoDrop 2000 and Agilent 2100 Bioanalyzer. Illumina TruSeq stranded mRNA libraries were prepared and sequenced on an Illumina HiSeq2000 platform at the Broad Institute. One replicate each for infected plant samples inoculated with Fo47 at 12 HPI and Fo5176 at 24 HPI failed, as did one replicate for Fo47 and Fo5176 mycelia samples; these four conditions are therefore only represented by two replicates and were used for downstream processing and analysis.
Paired-end RNA-seq reads were first assessed for quality by FastQC 0.10.1 (Andrews 2010). RNA-seq data were analyzed using the HISAT, StringTie, and DESeq2 pipelines (Love et al. 2014; Pertea et al. 2016). Briefly, reads were mapped to reference genomes of Arabidopsis (annotation version Araport11 [Cheng et al. 2017]), Fo5176 (Fokkens et al. 2020), and Fo47 (B. Wang et al. 2020) using HISAT2 version 2.0.5 (Kim et al. 2015). Mapped reads were used to quantify the transcriptome by string-Tie version 1.3.4 (Pertea et al. 2015). Read count normalization and differential gene expression analysis were conducted using DESeq2 version 1.27.32 with a maximum FDR of 0.05 (Love et al. 2014. Corrplot version 0.84 was used to visualize the correlation in gene expression profiles between different conditions. Read counts of DEGs were first averaged per condition and were then normalized by log transformation as log2(normalized read count +1) and, then, correlations were calculated. Clustering analysis on per-condition averaged, log-transformed, and Z-scaled read counts was performed using the K-means clustering algorithm ‘Lloyd’ (R function K-means) and then visualized in ggplot2 version 3.3.0.
Functional analysis and visualization.
GO enrichment analysis (plant GO slim) of Arabidopsis gene clusters and reciprocal DEG analysis were conducted with the singular enrichment analysis (SEA) tool of agriGO v2 (Du et al. 2010; Tian et al. 2017), using the Arabidopsis TAIR10 annotation. We applied a hypergeometric test, combined with the Hochberg (FDR) multi-test adjustment method to discover enriched GO terms at a significance level of 0.01 with a minimum of three mapping entries. Comparisons of different enrichment results were performed using cross-comparison of SEA (SEACOMPARE). We generated PFAM annotations for F. oxysporum 5176 and 47 by InterproScan, following a standard annotation pipeline (Jones et al. 2014). PFAM enrichment in proteins encoded by fungal genes was performed in TBtools v.1.0692 (Chen et al. 2020) using Fisher’s exact test with FDR < 0.05. The expression heatmaps of PFAM domain were visualized by TBtools. We performed a custom analysis in Metascape (Zhou et al. 2019), with the options minimum overlap of 3, P value cutoff of 0.01, and minimum enrichment of 1.5, for the discovery of GO term enrichment and network visualization of Arabidopsis DEGs at 12 HPI. The top five terms (with the smallest P values) were selected, and the terms that shared the gene entries (forming edges) were visualized. The visualization was further polished in Cytoscape version 3.8.0 (Shannon 2003).
Synteny and phylogenetic tree construction.
Synteny was detected by the Basic Local Alignment Search Tool for nucleotides (BLASTN), with parameters above 50-kb coverage and 98.5% sequence identity, and were visualized as a Circos plot (Krzywinski et al. 2009). MEGA v. 7.0 (Kumar et al. 2016) was used to generate maximum likelihood phylogeny trees for proteins with the PF03707 and PF00188 domain with the JTT (Jones, Taylor, and Thorton) amino acid substitution model. Statistical support for phylogenetic grouping was assessed by 1000 bootstrap re-samplings.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank J. Manners and D. Gardiner of CSIRO for providing the strain Fo5176; C. Joyner, the Superintendent of the College of Natural Sciences Greenhouse at University of Massachusetts, for his helps with all experiments conducted in the greenhouse; and P. Salomé for preparing the summary figure.
Funding:
This project was supported by the Natural Science Foundation (IOS-165241), National Research Initiative Competitive Grants Program (grant no. 2008-35604-18800) and the USDA National Institute of Food and Agriculture grants MASR-2009-04374, MAS00532, and MAS00496. Data were analyzed at the Massachusetts Green High Performance Computing Center (MGHPCC). L. Guo is also supported by a China Postdoctoral Foundation grant (2017 M623188), the National Natural Science Foundation of China (31701739), and the Fundamental Research Fund of Xi’an Jiaotong University (1191329155). L.-J. Ma is also supported by an Investigator Award in Infectious Diseases and Pathogenesis by the Burroughs Wellcome Fund (BWF-1014893) and the National Eye Institute of the National Institutes of Health under award number R01EY030150. H. Yu is also supported by a Lotta M. Crabtree Fellowship. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing of the manuscript.
Footnotes
Accession numbers.
RNA-seq data generated in this study were deposited in the NCBI Short Read Archive (SRA) with accession number GSE87352.
The author(s) declare no conflict of interest.
LITERATURE CITED
- Aimé S, Alabouvette C, Steinberg C, and Olivain C 2013. The endophytic strain Fusarium oxysporum Fo47: A good candidate for priming the defense responses in tomato roots. Mol. Plant-Microbe Interact 26:918–926. [DOI] [PubMed] [Google Scholar]
- Alabouvette C 1999. Fusarium wilt suppressive soils: An example of disease-suppressive soils. Austral. Plant Pathol 28:57. [Google Scholar]
- Andrews S 2010. FastQC: A quality control tool for high throughput sequence data. Babraham Institute, Cambridge, U.K. Published online. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- Armitage AD, Taylor A, Sobczyk MK, Baxter L, Greenfield BPJ, Bates HJ, Wilson F, Jackson AC, Ott S, Harrison RJ, and Clarkson JP 2018. Characterisation of pathogen-specific regions and novel effector candidates in Fusarium oxysporum f. sp. cepae. Sci. Rep 8:13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggs EL, Monroe JG, Thanki AS, O’Grady R, Schudoma C, Haerty W, and Krasileva KV 2020. Convergent loss of an EDS1/PAD4 signaling pathway in several plant lineages reveals coevolved components of plant immunity and drought response. Plant Cell 32:2158–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacci A, Navaud O, Mbengue M, Barascud M, Godiard L, Khafif M, Lacaze A, and Raffaele S 2020. Rapid identification of an Arabidopsis NLR gene as a candidate conferring susceptibility to Sclerotinia sclerotiorum using time-resolved automated phenotyping. Plant J. 103:903–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin M, Hassan JA, Schreiber KJ, and Lewis JD 2017. Analysis of the ZAR1 immune complex reveals determinants for immunity and molecular interactions. Plant Physiol. 174:2038–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamou N, and Garand C 2001. Cytological analysis of defense-related mechanisms induced in pea root tissues in response to colonization by nonpathogenic Fusarium oxysporum Fo47. Phytopathology 91:730–740. [DOI] [PubMed] [Google Scholar]
- Benhamou N, Garand C, and Goulet A 2002. Ability of nonpathogenic Fusarium oxysporum strain Fo47 to induce resistance against Pythium ultimum infection in cucumber. AEM 68:4044–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, and Hirt H 2015. Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8:521–539. [DOI] [PubMed] [Google Scholar]
- Bisgrove SR, Simonich MT, Smith NM, Sattler A, and Innes RW 1994. A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell 6:927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camera SL, Balagué C, Göbel C, Geoffroy P, Legrand M, Feussner I, Roby D, and Heitz T. 2009. The Arabidopsis patatin-like protein 2 (PLP2) plays an essential role in cell death execution and differentially affects biosynthesis of oxylipins and resistance to pathogens. Mol. Plant-Microbe Interact 22:469–481. [DOI] [PubMed] [Google Scholar]
- Cao Y, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A, and Stacey G 2014. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3:e03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S 2018. Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 219:17–24. [DOI] [PubMed] [Google Scholar]
- Chan KX, Mabbitt PD, Phua SY, Mueller JW, Nisar N, Gigolashvili T, Stroeher E, Grassl J, Arlt W, Estavillo GM, Jackson CJ, and Pogson BJ 2016. Sensing and signaling of oxidative stress in chloroplasts by inactivation of the SAL1 phosphoadenosine phosphatase. Plant Biol. 113:E4567–E4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, and Xia R 2020. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13:1194–1202. [DOI] [PubMed] [Google Scholar]
- Chen YC, Wong CL, Muzzi F, Vlaardingerbroek I, Kidd BN, and Schenk PM 2015. Root defense analysis against Fusarium oxysporum reveals new regulators to confer resistance. Sci. Rep 4:5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C-Y, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, and Town CD 2017. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 89:789–804. [DOI] [PubMed] [Google Scholar]
- Cui H, Tsuda K, and Parker JE 2015. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol 66:487–511. [DOI] [PubMed] [Google Scholar]
- Cuomo CA, Guldener U, Xu J-R, Trail F, Turgeon BG, Di Pietro A, Walton JD, Ma L-J, Baker SE, Rep M, Adam G, Antoniw J, Baldwin T, Calvo S, Chang Y-L, DeCaprio D, Gale LR, Gnerre S, Goswami RS, Hammond-Kosack K, Harris LJ, Hilburn K, Kennell JC, Kroken S, Magnuson JK, Mannhaupt G, Mauceli E, Mewes H-W, Mitterbauer R, Muehlbauer G, Munsterkotter M, Nelson D, O’Donnell K, Ouellet T, Qi W, Quesneville H, Roncero MIG, Seong K-Y, Tetko IV, Urban M, Waalwijk C, Ward TJ, Yao J, Birren BW, and Kistler HC 2007. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317:1400–1402. [DOI] [PubMed] [Google Scholar]
- de Souza A, Wang J-Z, and Dehesh K 2017. Retrograde signals: Integrators of interorganellar communication and orchestrators of plant development. Annu. Rev. Plant Biol 68:85–108. [DOI] [PubMed] [Google Scholar]
- de Torres Zabala M, Littlejohn G, Jayaraman S, Studholme D, Bailey T, Lawson T, Tillich M, Licht D, Bölter B, Delfino L, Truman W, Mansfield J, Smirnoff N, and Grant M 2015. Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nat. Plants 1:15074. [DOI] [PubMed] [Google Scholar]
- Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R, Xu J-R, Pan H, Read ND, Lee Y-H, Carbone I, Brown D, Oh YY, Donofrio N, Jeong JS, Soanes DM, Djonovic S, Kolomiets E, Rehmeyer C, Li W, Harding M, Kim S, Lebrun M-H, Bohnert H, Coughlan S, Butler J, Calvo S, Ma L-J, Nicol R, Purcell S, Nusbaum C, Galagan JE, and Birren BW 2005. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434:980–986. [DOI] [PubMed] [Google Scholar]
- Dean R, Van Kan JAL, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, and Foster GD 2012. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol 13:414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeIulio GA, Guo L, Zhang Y, Goldberg JM, Kistler HC, and Ma L-J 2018. Kinome expansion in the Fusarium oxysporum species complex driven by accessory chromosomes. MSphere 3:e00231–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener AC, and Ausubel FM 2005. RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is notrace specific. Genetics 171:305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, and Rathjen JP 2010. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet 11:539–548. [DOI] [PubMed] [Google Scholar]
- Dong OX, Tong M, Bonardi V, El Kasmi F, Woloshen V, Wünsch LK, Dangl JL, and Li X 2016. TNL-mediated immunity in Arabidopsis requires complex regulation of the redundant ADR1 gene family. New Phytol. 210:960–973. [DOI] [PubMed] [Google Scholar]
- Dong S, Raffaele S, and Kamoun S 2015. The two-speed genomes of filamentous pathogens: Waltz with plants. Curr. Opin. Genet. Dev 35: 57–65. [DOI] [PubMed] [Google Scholar]
- Dong X, Jiang Z, Peng Y-L, and Zhang Z 2015. Revealing shared and distinct gene network organization in Arabidopsis immune responses by integrative analysis. Plant Physiol. 167:1186–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, and Su Z 2010. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38:W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokkens L, Guo L, Dora S, Wang B, Ye K, Sánchez-Rodríguez C, and Croll D 2020. A chromosome-scale genome assembly for the Fusarium oxysporum strain Fo5176 to establish a model Arabidopsis-fungal pathosystem. Genes Genom. Genet 10:3549–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs R, Kopischke M, Klapprodt C, Hause G, Meyer AJ, Schwarzländer M, Fricker MD, and Lipka V 2016. Immobilized subpopulations of leaf epidermal mitochondria mediate PENETRATION2-dependent pathogen entry control in Arabidopsis. Plant Cell 28:130–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Cuomo C, Ma L-J, Wortman JR, Batzoglou S, Lee S-I, Baştürkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D’Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Peñalva MÁ, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, and Birren BW 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115. [DOI] [PubMed] [Google Scholar]
- Galazka JM, and Freitag M 2014. Variability of chromosome structure in pathogenic fungi—Of ‘ends and odds.’. Curr. Opin. Microbiol 20:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Gaidenko TA, Mulkidjanian AY, Nakano M, and Price CW 2001. MHYT, a new integral membrane sensor domain. FEMS Microbiol. Lett 205:17–23. [DOI] [PubMed] [Google Scholar]
- Gaudinier A, Rodriguez-Medina J, Zhang L, Olson A, Liseron-Monfils C, Bågman A-M, Foret J, Abbitt S, Tang M, Li B, Runcie DE, Kliebenstein DJ, Shen B, Frank MJ, Ware D, and Brady SM 2018. Transcriptional regulation of nitrogen-associated metabolism and growth. Nature 563:259–264. [DOI] [PubMed] [Google Scholar]
- Göhre V, Jones AME, Sklenář J, Robatzek S, and Weber APM 2012. Molecular crosstalk between PAMP-triggered immunity and photosynthesis. Mol. Plant-Microbe Interact 25:1083–1092. [DOI] [PubMed] [Google Scholar]
- Goritschnig S, Krasileva KV, Dahlbeck D, and Staskawicz BJ 2012. Computational prediction and molecular characterization of an oomycete effector and the cognate Arabidopsis resistance gene. PLoS Genet. 8:e1002502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hane JK, Rouxel T, Howlett BJ, Kema GH, Goodwin SB, and Oliver RP 2011. A novel mode of chromosomal evolution peculiar to filamentous Ascomycete fungi. Genome Biol. 12:R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, and Dangl JL 2006. The plant immune system. Nature 444:323–329. [DOI] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong S-Y, Lopez R, and Hunter S 2014. InterProScan 5: Genome-scale protein function classification. Bioinformatics 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL 2015. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kwon SI, Saha D, Anyanwu NC, and Gassmann W 2009. Resistance to the Pseudomonas syringae effector HopA1 is governed by the TIR-NBS-LRR protein RPS6 and is enhanced by mutations in SRFR1. Plant Physiol. 150:1723–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Gascoyne R, Horsman D, Jones SJ, and Marra MA 2009. Circos: An information aesthetic for comparative genomics. Genome Res. 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, and Tamura K 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Camera S, Geoffroy P, Samaha H, Ndiaye A, Rahim G, Legrand M, and Heitz T 2005. A pathogen-inducible patatin-like lipid acyl hydrolase facilitates fungal and bacterial host colonization in Arabidopsis. Plant J. 44:810–825. [DOI] [PubMed] [Google Scholar]
- Lampl N, Alkan N, Davydov O, and Fluhr R 2013. Set-point control of RD21 protease activity by AtSerpin1 controls cell death in Arabidopsis. Plant J. 74:498–510. [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, and Palva ET 2008. Kunitz trypsin inhibitor: An antagonist of cell death triggered by phytopathogens and fumonisin B1 in Arabidopsis. Mol. Plant 1:482–495. [DOI] [PubMed] [Google Scholar]
- Liu J-X, and Howell SH 2010. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22:782–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Niu Y, Konishi M, Wu Y, Du H, Sun Chung H, Li L, Boudsocq M, McCormack M, Maekawa S, Ishida T, Zhang C, Shokat K, Yanagisawa S, and Sheen J 2017. Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 545: 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y 2016. Chloroplast in Plant-Virus Interaction. Front. Microbiol 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons R, Stiller J, Powell J, Rusu A, Manners JM, and Kazan K 2015. Fusarium oxysporum triggers tissue-specific transcriptional reprogramming in Arabidopsis thaliana. PLoS One 10:e0121902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L-J, van der Does HC, Borkovich KA, Coleman JJ, Daboussi M-J, Di Pietro A, Dufresne M, Freitag M, Grabherr M, Henrissat B, Houterman PM, Kang S, Shim W-B, Woloshuk C, Xie X, Xu J-R, Antoniw J, Baker SE, Bluhm BH, Breakspear A, Brown DW, Butchko RAE, Chapman S, Coulson R, Coutinho PM, Danchin EGJ, Diener A, Gale LR, Gardiner M, Goff S, Hammond-Kosack KE, Hilburn K, Hua-Van A, Jonkers W, Kazan K, Kodira CD, Koehrsen M, Kumar L, Lee Y-H, Li L, Manners JM, Miranda-Saavedra D, Mukherjee M, Park G, Park J, Park S-Y, Proctor RH, Regev A, Ruiz-Roldan MC, Sain D, Sakthikumar S, Sykes S, Schwartz DC, Turgeon BG, Wapinski I, Yoder O, Young S, Zeng Q, Zhou S, Galagan J, Cuomo CA, Kistler HC, and Rep M 2010. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464:367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L-J, Geiser DM, Proctor RH, Rooney AP, O’Donnell K, Trail F, Gardiner DM, Manners JM, and Kazan K 2013. Fusarium pathogenomics. Annu. Rev. Microbiol 67:399–416. [DOI] [PubMed] [Google Scholar]
- McDowell JM, Williams SG, Funderburg NT, Eulgem T, and Dangl JL 2005. Genetic analysis of developmentally regulated resistance to downy mildew (Hyaloperonospora parasitica) in Arabidopsis thaliana. Mol. Plant-Microbe Interact 18:1226–1234. [DOI] [PubMed] [Google Scholar]
- Medina-Puche L, Tan H, Dogra V, Wu M, Rosas-Diaz T, Wang L, Ding X, Zhang D, Fu X, Kim C, and Lozano-Duran R 2020. A defense pathway linking plasma membrane and chloroplasts and co-opted by pathogens. Cell 182:1109–1124. [DOI] [PubMed] [Google Scholar]
- Monteiro F, and Nishimura MT 2018. Structural, functional, and genomic diversity of plant NLR proteins: An evolved resource for rational engineering of plant immunity. Annu. Rev. Phytopathol 56:243–267. [DOI] [PubMed] [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, and Narusaka Y 2009. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 60:218–226. [DOI] [PubMed] [Google Scholar]
- Noël L, Moores TL, Parker JE, and Jones JDG 1999. Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11:2099–2111. [PMC free article] [PubMed] [Google Scholar]
- Olivain C, Humbert C, Nahalkova J, Fatehi J, L’Haridon F, and Alabouvette C 2006. Colonization of tomato root by pathogenic and nonpathogenic Fusarium oxysporum strains inoculated together and separately into the soil. AEM 72:1523–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JE, Coleman MJ, Szabò V, Frost LN, Schmidt R, van der Biezen EA, Moores T, Dean C, Daniels MJ, and Jones JD 1997. The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell 9: 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedreira J, Herrera MT, Zarra I, and Revilla G 2011. The overexpression of AtPrx37, an apoplastic peroxidase, reduces growth in Arabidopsis. Physiol. Plant 141:177–187. [DOI] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, and Salzberg SL 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc 11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, and Salzberg SL 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol 33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prados-Rosales RC, Roldán-Rodríguez R, Serena C, López-Berges MS, Guarro J, Martínez-del-Pozo Á, and Pietro AD 2012. A PR-1-like protein of Fusarium oxysporum functions in virulence on mammalian hosts. J. Biol. Chem 287: 21970–21979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JM, Paulitz TC, Steinberg C, Alabouvette C, and Moënne-Loccoz Y 2009. The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321:341–361. [Google Scholar]
- Rashid MI, Mujawar LH, Shahzad T, Almeelbi T, Ismail IMI, and Oves M 2016. Bacteria and fungi can contribute to nutrients bioavailability and aggregate formation in degraded soils. Microbiol. Res 183:26–41. [DOI] [PubMed] [Google Scholar]
- Schneiter R, and Di Pietro A 2013. The CAP protein superfamily: Function in sterol export and fungal virulence. Biomol. Concepts 4:519–525. [DOI] [PubMed] [Google Scholar]
- Schwacke R, Ponce-Soto GY, Krause K, Bolger AM, Arsova B, Hallab A, Gruden K, Stitt M, Bolger ME, and Usadel B 2019. MapMan4: A refined protein classification and annotation framework applicable to multi-omics data analysis. Mol. Plant 12:879–892. [DOI] [PubMed] [Google Scholar]
- Serrano I, Audran C, and Rivas S 2016. Chloroplasts at work during plant innate immunity. EXBOTJ 67:3845–3854. [DOI] [PubMed] [Google Scholar]
- Shannon P 2003. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopeña-Torres S, Jordá L, Sánchez-Rodríguez C, Miedes E, Escudero V, Swami S, Lóapez G, Piślewska-Bednarek M, Lassowskat I, Lee J, Gu Y, Haigis S, Alexander D, Pattathil S, Muñoz-Barrios A, Bednarek P, Somerville S, Schulze-Lefert P, Hahn MG, Scheel D, and Molina A 2018.YODA MAP3K kinase regulates plant immune responses conferring broad-spectrum disease resistance. New Phytol. 218:661–680. [DOI] [PubMed] [Google Scholar]
- Staal J, Kaliff M, Bohman S, and Dixelius C 2006. Transgressive segregation reveals two Arabidopsis TIR-NB-LRR resistance genes effective against Leptosphaeria maculans, causal agent of blackleg disease. Plant J. 46:218–230. [DOI] [PubMed] [Google Scholar]
- Strange RN, and Scott PR 2005. Plant disease: A threat to global food security. Annu. Rev. Phytopathol 43:83–116. [DOI] [PubMed] [Google Scholar]
- Thatcher LF, Gardiner DM, Kazan K, and Manners JM 2012. A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis. Mol. Plant-Microbe Interact 25:180–190. [DOI] [PubMed] [Google Scholar]
- Thatcher LF, Manners JM, and Kazan K 2009. Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J. 58:927–939. [DOI] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, and Su Z 2017. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45:W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M, Balasubramanian S, Hu TT, Traw MB, Horton M, Epple P, Kuhns C, Sureshkumar S, Schwartz C, Lanz C, Laitinen RAE, Huang Y, Chory J, Lipka V, Borevitz JO, Dangl JL, Bergelson J, Nordborg M, and Weigel D 2010. Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465:632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann-Zeunert L, Stanton MA, Wielsch N, Bartram S, Hummert C, Svatoš A, Baldwin IT, and Groten K 2013. Quantification of growth-defense trade-offs in a common currency: Nitrogen required for phenolamide biosynthesis is not derived from ribulose-1,5-bisphosphate carboxylase/oxygenase turnover. Plant J. 75:417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam P, Fokkens L, Schmidt SM, Linmans JHJ, Kistler HC, Ma L-J, and Rep M 2016. Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ. Microbiol 18:4087–4102. [DOI] [PubMed] [Google Scholar]
- van der Biezen EA, Freddie CT, Kahn K, Parker PJE, and Jones JDG 2002. Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 29:439–451. [DOI] [PubMed] [Google Scholar]
- van der Burgh AM, and Joosten MHAJ 2019. Plant immunity: Thinking outside and inside the box. Trends Plant Sci. 24:587–601. [DOI] [PubMed] [Google Scholar]
- Veloso J, and Díaz J 2012. Fusarium oxysporum Fo47 confers protection to pepper plants against Verticillium dahliae and Phytophthora capsici, and induces the expression of defence genes. Plant Pathol. 61:281–288. [Google Scholar]
- Viljoen A, Ma L-J, and Molina AB 2020. Fusarium wilt (Panama disease) and monoculture banana production: Resurgence of a century-old disease. Pages 159–184 in: Emerging Plant Diseases and Global Food Security. Records A, and Ristaino J, eds. American Phytopathological Society, St Paul, MN, U.S.A. [Google Scholar]
- Vlaardingerbroek I, Beerens B, Rose L, Fokkens L, Cornelissen BJC, and Rep M 2016a. Exchange of core chromosomes and horizontal transfer of lineage-specific chromosomes in Fusarium oxysporum. Environ. Microbiol 18:3702–3713. [DOI] [PubMed] [Google Scholar]
- Vlaardingerbroek I, Beerens B, Schmidt SM, Cornelissen BJC, and Rep M 2016b. Dispensable chromosomes in Fusarium oxysporum f. sp. lycopersici. Mol. Plant Pathol 17:1455–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yu H, Jia Y, Dong Q, Steinberg C, Alabouvette C, Edel-Hermann V, Kistler HC, Ye K, Ma L-J, and Guo L 2020. Chromosome-scale genome assembly of Fusarium oxysporum strain Fo47, a fungal endophyte and biocontrol agent. Mol. Plant-Microbe Interact 33:1108–1111. [DOI] [PubMed] [Google Scholar]
- Wang W, Feng B, Zhou J, and Tang D 2020. Plant immune signaling: Advancing on two frontiers. J. Integr. Plant Biol 62:2–24. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang F, Hong Y, Huang J, Shi H, and Zhu J-K 2016. Two chloroplast proteins suppress drought resistance by affecting ROS production in guard cells. Plant Phy. 172:2491–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JF, Kingsley KL, Zhang Q, Verma R, Obi N, Dvinskikh S, Elmore MT, Verma SK, Gond SK, and Kowalski KP 2019. Review: Endophytic microbes and their potential applications in crop management. Pest Manag. Sci 75:2558–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AH, Sharma M, Thatcher LF, Azam S, Hane JK, Sperschneider J, Kidd BN, Anderson JP, Ghosh R, Garg G, Lichtenzveig J, Kistler HC, Shea T, Young S, Buck S-AG, Kamphuis LG, Saxena R, Pande S, Ma L-J, Varshney RK, and Singh KB 2016. Comparative genomics and prediction of conditionally dispensable sequences in legume-infecting Fusarium oxysporum formae speciales facilitates identification of candidate effectors. BMC Genomics 17:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Yu H, and Ma L-J 2020. Accessory chromosomes in Fusarium oxysporum. Phytopathology 110:1488–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang H, Turra D, Zhou S, Ayhan DH, DeIulio GA, Guo L, Broz K, Wiederhold N, Coleman JJ, O’ Donnell K, Youngster I, McAdam AJ, Savinov S, Shea T, Young S, Zeng Q, Rep M, Pearlman E, Schwartz DC, Di Pietro A, Kistler HC, and Ma L-J 2020. The genome of opportunistic fungal pathogen Fusarium oxysporum carries a unique set of lineage-specific chromosomes. Commun. Biol 3:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Emonet A, Dénervaud Tendon V, Marhavy P, Wu D, Lahaye T, and Geldner N 2020. Co-incidence of damage and microbial patterns controls localized immune responses in roots. Cell 180: 440–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J-M, and Zhang Y 2019. Plant immunity: Danger perception and signaling. Cell 181:978–989. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, and Chanda SK 2019. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun 10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


