Abstract
The concerted effort for bioproduction of higher alcohols and other commodity chemicals has yielded a consortium of metabolic engineering techniques to identify targets to enhance performance of engineered microbial strains. Here, we demonstrate the use of metabolomics as a tool to systematically identify targets for improved production phenotypes in Escherichia coli. Gas chromatography/mass spectrometry (GC/MS) and ion-pair LC-MS/MS were performed to investigate metabolic perturbations in various 1-propanol producing strains. Two initial strains were compared that differ in the expression of the citramalate and threonine pathways, which hold a synergistic relationship to maximize production yields. While this results in increased productivity, no change in titer was observed when the threonine pathway was overexpressed beyond native levels. Metabolomics revealed accumulation of upstream byproducts, norvaline and 2-aminobutyrate, both of which are derived from 2-ketobutyrate (2KB). Eliminating the competing pathway by gene knockouts or improving flux through overexpression of glycolysis gene effectively increased the intracellular 2KB pool. However, the increase in 2KB intracellular concentration yielded decreased production titers, indicating toxicity caused by 2KB and an insufficient turnover rate of 2KB to 1-propanol. Optimization of alcohol dehydrogenase YqhD activity using an ribosome binding site (RBS) library improved 1-propanol titer (g/L) and yield (g/g of glucose) by 38 and 29% in 72 h compared to the base strain, respectively. This study demonstrates the use of metabolomics as a powerful tool to aid systematic strain improvement for metabolically engineered organisms.
Keywords: metabolomics, rate-limiting steps, Escherichia coli, 1-propanol, 2-ketobutyrate, YqhD
Introduction
Engineering microbes to have desired production phenotypes requires optimization of many physiological constraints (Pontrelli et al., 2018). These include the ability to express non-native enzymes, supply of required cofactors, precursor availability, oxygen tolerance, and accumulation of toxic intermediates or products. While rational design strategies prove useful in addressing many of these factors, there often remains a need for further optimization to improve production.
Metabolomics, a technology that involves the global quantitative assessment of metabolites in a biological system, can detect complex biological changes using statistical multivariate pattern recognition methods (chemometrics) (Putri et al., 2013). The system-wide analysis provided by metabolomics can be used to gain a deeper understanding of how various genetic modifications affect production phenotypes. Observations made with metabolomics have led to strain modifications that successfully increased biofuel production titers (Nitta et al., 2017; Ohtake et al., 2017; Fathima et al., 2018). For improvement of 1-butanol production in E. coli, a metabolomics-based approach was used to identify an imbalance of Coenzyme A caused by a Clostridial butanol production pathway. Further genetic modifications based on observations made using metabolomics allowed a production increase from 15 to 18.3 g/L of butanol (Ohtake et al., 2017). Similarly, an orthogonal partial least squares/projections to latent structures (PLS) model-based metabolomics approach was used to identify acetyl-CoA as the rate limiting step of 1-butanol production (Nitta et al., 2017).
In this work, we employed a metabolomics-based approach to identify targets for improving production of 1-propanol in E. coli. 1-Propanol is recognized as an important compound that is industrially valuable for both energy applications and as a C3 chemical feedstock (Liao et al., 2016). Production of 1-propanol has been achieved via the keto acid pathway (Atsumi and Liao, 2008; Shen and Liao, 2013), which offers several advantages owing to its universality and compatibility with the host. 2-Keto acids are intermediates in amino acid biosynthesis pathways that can be found in almost all living organisms. The theoretical yield of 1-propanol was enhanced by combining the native threonine pathway with a heterologous citramalate pathway using glucose as a substrate (Shen and Liao, 2013). Different cofactor requirements are exhibited by each pathway individually. The threonine pathway requires 3 NAD(P)H and 2 ATP as inputs, while the citramalate pathway produces 1 NAD(P)H and 1 ATP as outputs (Figure 1). When the overall net reactions of each pathway are combined, the individual cofactor requirements can complement one another, resulting in a synergistic effect that improves the theoretical maximum yield. The efficacy of this synergistic approach has proven successful in improving the observed 1-propanol yield and productivity (Shen and Liao, 2013).
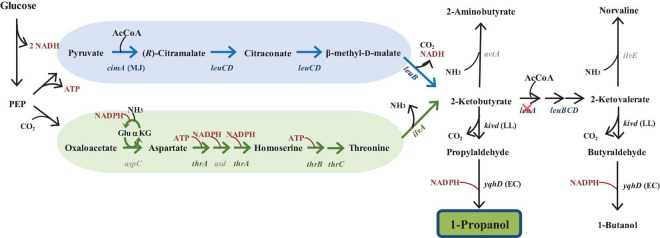
FIGURE 1.
Illustration of the keto-acid pathway for 1-propanol synthesis. Schematic illustration of the threonine pathway (denoted by the green arrow) and the citramalate pathway (denoted by the blue arrow) for the synthesis of 1-propanol in E. coli from glucose as previously described in Shen and Liao (2013). Genes not overexpressed are noted in gray font. Cross mark indicates that the pathway was blocked via gene deletion. The deleted gene encode for 2-isopropylmalate synthase (leuA). PEP, phosphoenolpyruvate; cimA, citramalate synthase from Methanococcus jannaschii (MJ); KivD, alpha-ketoisovalerate decarboxylase from Lactococcus lactis (LL); YqhD, alcohol dehydrogenase from Escherichia coli (EC).
With this production strategy, each pathway converges into a common intermediate, 2-ketobutyrate (2KB), which is then converted into 1-propanol through reactions catalyzed by alpha-ketoisovalerate decarboxylase (KivD) and alcohol dehydrogenase (YqhD) (Figure 1). 2-Ketobutyrate is also a native precursor in the E. coli leucine biosynthesis pathway and subsequently converted into 2-ketovalerate. Because of the promiscuous nature of KivD and YqhD, 1-butanol is also formed as a side product. However, in this work, we focus solely on improving production of 1-propanol.
It has been observed that overexpression of the citramalate pathway in conjunction with the native threonine pathway exhibits an increased productivity (hereafter referred to as strain SYN3). Yet, there is no observed change in titer when the threonine pathway is further overexpressed over basal metabolic levels (hereafter called strain SYN12) (Shen and Liao, 2013). One recent study uses snapshot metabolomic profiling and dynamic metabolic turnover analysis to determine why the presence or overexpression of the citramalate or threonine pathways contributes to this change in productivity (Putri et al., 2018). The strains used in their study were SYN2 (citramalate pathway overexpressed and native threonine pathway deleted), SYN6 (threonine pathway overexpressed), and SYN12 (both pathways overexpressed). Notably, this study identified the importance of the PEP/pyruvate ratio, which affects glucose consumption, and how ammonia assimilation affects the rate of the threonine pathway. Extending these findings, we here build on this study by increasing 1-propanol titer in SYN12. First, derivatives of 1-propanol producing strains were cultivated under the optimal condition for production. Snapshot non-targeted metabolome analysis using gas chromatography/mass spectrometry (GC/MS) and widely targeted metabolome analysis using ion-pair LC/MS/MS were performed. The data sets obtained from GC/MS and ion-pair liquid chromatography (LC)/MS/MS (LC/MS/MS) were subjected to multivariate analyses to extract the most meaningful metabolites corresponding to the phenotype. Following the analysis of important metabolites, the corresponding metabolic pathways were examined, and gene targets were systematically selected to construct strains with improved production phenotypes.
Materials and Methods
Bacterial Strains
Escherichia coli strains, plasmids and primers used in this study are summarized in Table 1 and Supplementary Table 1. XL-1 Blue (Stratagene, La Jolla, CA, United States) was used to propagate all plasmids. Host gene deletion of avtA was achieved with P1 transduction using the Keio collection strains (Baba et al., 2006) as donor and strain CRS59 (Shen and Liao, 2013) as recipient. The kanR inserted into the target gene region was removed with pCP20 (Datsenko and Wanner, 2000) in between each consecutive knock out. Then, removal of the gene segment was verified by colony PCR using the appropriate primers.
TABLE 1.
Strains and plasmids used in this work.
| Name | Relevant characteristics | Source |
| E. coli strain | ||
| BW25113 | rrnBT14 ΔlacZWJ16 ΔhsdR514 ΔaraBADAH33 ΔrhaBADLD78 | Datsenko and Wanner, 2000 |
| JCL16 | BW25113/F’ [ΔtraD36 proAB+ ΔlacIqZΔM15 (Tetr)] | Atsumi et al., 2008 |
| CRS59 | JCL16 ΔilvB ΔilvI ΔleuA | Shen and Liao, 2013 |
| CRS60 | JCL16 ΔilvB ΔilvIΔleuAΔavtA | Shen and Liao, 2013 |
| CRS-SYN3 | CRS59/pSA138+pAFC52 | Shen and Liao, 2013 |
| CRS-SYN12 | CRS59/pAFC66+pCS49+pAFC52 | Shen and Liao, 2013 |
| CRS-SYN14 | CRS60/pAFC66+pCS49+pAFC52 | Shen and Liao, 2013 |
| CRS-SYN17 | CRS59/pAFC66+pCS164+pAFC52 | This study |
| CRS-SYN12-KivD34 | CRS59/pKN2+pCS49+pAFC52 | This study |
| CRS-SYN12-YqhD18 | CRS59/pKN3+pCS49+pAFC52 | This study |
| Plasmid | ||
| pSA138 | PLlacO1::kivd (LL)-yqhD (EC); ColE1 ori; AmpR | Atsumi and Liao, 2008 |
| pAFC52 | PLlacO1::cimA2Δ (MJ)-leuBCD (EC); p15A ori; KanR | Shen and Liao, 2013 |
| pAFC66 | PLlacO1::kivd (LL)-yqhD (EC)-ilvA (CG); ColE1 ori; AmpR | Shen and Liao, 2013 |
| pCS49 | PLlacO1::thrAfbrBC (EC ATCC 21277); pSC101 ori; SpecR | Shen and Liao, 2008 |
| pCS164 | PLlacO1::thrAfbrBC (EC ATCC 21277)-tpiA (EC); pSC101 ori; SpecR | This study |
| pKN2 | PLlacO1::kivd (LL)-yqhD (EC)-ilvA (CG); ColE1 ori; AmpR (Improved kivd RBS strength) | This study |
| pKN3 | PLlacO1::kivd (LL)-yqhD (EC)-ilvA (CG); ColE1 ori; AmpR (Improved yqhD RBS strength) | This study |
| YqhD RBS sequence | ||
| SYN12 | GCATGCAGGAGAAAGGTCAC | This study |
| SYN12-YqhD18 | GCCGCACCTCGCGAGTCAGAAAGGAGGAACCCGC | This study |
Plasmid Construction
Plasmids were constructed by assembling the three PCR fragments (Supplementary Table 1) similar to the previously described protocol (Ohtake et al., 2017).
Ribosome Binding Site (RBS) Library Construction
pAFC66 was used as a template to construct a KivD and YqhD library with total eight variants. They were assembled by amplification of the following primers and assembly of the resulting PCR products (Supplementary Table 1). The KivD library was generated from a degenerate RBS library sequence to create eight variants, while the YqhD library was generated from three degenerate RBS library sequences that were pooled together in an equimolar ratio for eight total variants.
Production Media and Cultivation Condition
Production of 1-propanol was conducted as described previously (Shen and Liao, 2013). For productions which lasted more than 2 days, 18 g/L of glucose was fed into the culture and pH of production culture was re-adjusted back to 7 using 10 M NaOH every day.
Alcohol Dehydrogenase Enzyme Assay
Three independent cultures of SYN12 and SYN12-YqhD18 were grown and induced in production media as described above and harvested at 24 h by centrifugation. The pellets were then lysed by BugBuster (Millipore) and centrifuged at 13,200 rpm for 10 min at 4°C. The supernatant was collected and used as an input for an enzyme assay. The enzyme reaction contained 10 mM propionaldehyde and 1 mM NADPH in 100 mM PBS (pH6.4). The assay was initiated with addition of cell lysate and OD340 was measured over time.
Sample Collection and Sample Preparation for Extracellular and Intracellular Metabolites Analysis
Samples were taken by opening the screw caps of the flasks, and culture broths were centrifuged at 4°C for 5 min, 14,000 rpm to retrieve the supernatant for extracellular metabolite detection. For intracellular metabolome analysis, five OD600 units of cells were collected by fast filtration using a 0.45 μm pore size, 47 mm diameter nylon membrane (Millipore). The cells were immersed in liquid nitrogen immediately for quenching and stored at −80°C until extraction. Extraction procedure for GC/MS and ion-pair LC/MS/MS was done as previously described (Putri et al., 2018).
For GC/MS analysis, extracted metabolites were derivatized by oximation and silylation prior to analysis. The oximation reagent, methoxyamine hydrochloride (Sigma-Aldrich, St. Louis, MO, United States) was first dissolved in pyridine (Wako, Osaka, Japan) to a concentration of 20 mg/mL and 50 μL was added to each sample tube containing the lyophilized extracts. After reaction at 30°C, 1200 rpm for 90 min, 25 μL of N-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA) (GL Sciences, Tokyo, Japan) was added and the silylation reaction was performed at 37°C, 1200 rpm for 30 min. The derivatized samples were transferred to glass vials (Chromacol, Hertfordshire, United Kingdom) and analyzed within 24 h. For ion-pair LC/MS/MS analysis, extracted metabolites were dissolved in 100 μL of ultrapure water (Wako), vortexed, centrifuged at 16,000 × g for 3 min at 4°C and transferred to glass vials.
Quantification of Extracellular Metabolites
Alcohols were quantified as previously described (Ohtake et al., 2017). Briefly, a GC-2010 system (Shimadzu) with a GL Science (Tokyo, Japan) InertCap Pure-WAX capillary column (30 m, 0.25 mm i.d., 0.25 μm film thickness) equipped with a flame ionization detector and an AOC-20i/s auto injector (Shimadzu) was used.
The remaining glucose in the culture was quantified using a F-kit D-glucose (Roche Diagnostics, Manheim, Germany) following the manufacturer’s instructions.
GC/MS Analysis for Snapshot Metabolite Analysis
GC/MS analysis was performed as previous reported (Putri et al., 2018). Briefly, a GCMS-QP2010 Ultra (Shimadzu, Kyoto, Japan) gas chromatograph coupled with quadrupole mass spectrometer equipped with an AOC-20i/s auto injector (Shimadzu) was used with a CP-SIL 8 CB Low Bleed/MS column (30 m × 0.25 mm i.d., film thickness 0.25 mm, Agilent, Santa Clara, CA, United States). The raw data files were converted into netCDF (*.cdf) format according to the ANDI (Analytical Data Interchange Protocol) specification using the proprietary software GCMS solution (Shimadzu) before peak detection, baseline correction and retention time alignment using the freely available data processing tool MetAlign (Lommen, 2009). Data matrices from the alignment were then imported into AIoutput2 ver. 1.29 (Tsugawa et al., 2011) for automated Retention Index-based target compound identification and quantification.
Ion-Pair LC/MS/MS Analysis for Snapshot Metabolite Analysis
Ion-pair LC/MS/MS analysis was performed as previous reported (Ohtake et al., 2017). Briefly, a Shimadzu Nexera UHPLC system coupled with LCMS 8030 Plus (Shimadzu) using a CERI L-column 2 ODS (150 mm × 2.1 mm, particle size 3 μm, Chemicals Evaluation and Research Institute, Tokyo, Japan) was used. The mobile phase (A) was 10 mM tributylamine and 15 mM acetate in water and mobile phase (B) was methanol. The raw data files were converted into *.abf file then imported into MRMPROBS (Tsugawa et al., 2013) to create data matrix.
Multivariate Analysis
Principal component analysis (PCA) was performed using SIMCA-P+ version 14 (Umetrics, Umeå, Sweden). The metabolome data was normalized by an internal standard, mean centered and scaled to unit variance. T-test was performed using MS Excel to determine statistically significant differences.
Results
Systematic Identification of Metabolic Perturbations Caused by Overexpression of the Threonine Pathway
We initiated our study with comprehensive metabolic profiling to understand why overexpression of both the citramalate and threonine pathways did not have any effect on production titers. GC/MS and ion-pair LC/MS/MS analysis for snapshot metabolite analysis were performed on two previously constructed strains (Shen and Liao, 2013): SYN12 has expression of both the threonine and citramalate pathways, while SYN3 has expression of only the citramalate pathway and native expression of the threonine pathway. Each strain also contains an additional leuA knockout to prevent the downstream consumption of 2KB and conversion into 1-butanol or norvaline.
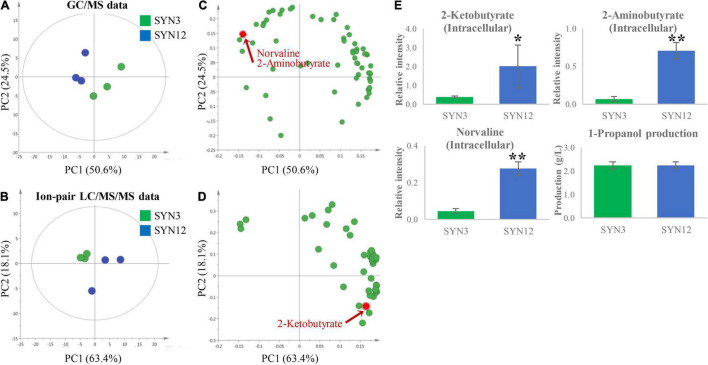
Tentative identification of compounds was performed by comparison with our in-house library. Tentatively identified metabolites from GC/MS consisted of organic acids, sugars, amino acids, and other compounds, whereas the metabolites from ion-pair LC/MS/MS consisted of cofactors and central metabolites belonging to glycolysis, pentose phosphate pathway (PPP) and tricarboxylic acid cycle (TCA cycle) (Supplementary Table 2). The obtained data sets from GC/MS (63 annotated metabolites) and ion-pair LC/MS/MS analyses (41 annotated metabolites) were subsequently subjected to principal component analysis (PCA), an unsupervised multivariate analysis, to extract the most meaningful information from SYN3 and SYN12 strains. The resulting PCA score plot (Figures 2A–D) showed that PC1 separated samples based on different genotypes, while PC2 separated samples based on replicates. PC1 represented 50.6% (GC/MS) and 63.4% (ion-pair LC/MS/MS) of the total variance of the samples, respectively, while PC2 were 24.5% (GC/MS) and 18.1% (ion-pair LC/MS/MS), respectively. Then, compounds contributing to the separation of samples were examined in the loading plot.
FIGURE 2.
PCA analysis of SYN3 and SYN12. The score plot derived from autoscaled (A) GC/MS data. (B) Ion-pair LC/MS/MS data. The corresponding loading plot illustrating metabolites that contributed to the separation on PC1 and PC2 derived from autoscaled. (C) GC/MS data. (D) Ion-pair LC/MS/MS data. All metabolites contribution is shown in Supplementary Figure 1. (E) The result of intracellular key intermediate, by-products and 1-propanol production. Metabolite intensities shown in the y-axis were normalized to an internal standard. Asterisks indicate significant difference from the strain (*p < 0.05, **p < 0.01). The error bars indicate standard deviations obtained from three replicate fermentations.
Based on PCA analysis, several key pathway intermediates with significant perturbations between the two strains were revealed. Norvaline, 2-aminobutyrate, and 2KB were all accumulated at significantly higher concentrations within SYN12 (Figure 2E). Interestingly, the formation of norvaline is not expected, as the gene catalyzing the first step of the reaction from 2KB to 2-ketovalerate (2KV), leuA, has been deleted in both SYN3 and SYN12 strains. This initial finding led to the hypothesis that CimA, a key enzyme within the citramalate pathway and structural homolog to leuA (Atsumi and Liao, 2008), is catalyzing the conversion of 2KB to 2KV, and ultimately to norvaline and 1-butanol. Although the presence of CimA is likely causing this downstream conversion, the buildup of these side products nonetheless suggests that higher titers may be reached by further channeling metabolic flux toward 1-propanol production.
Channeling Pathway Flux Highlights Pathway Bottlenecks
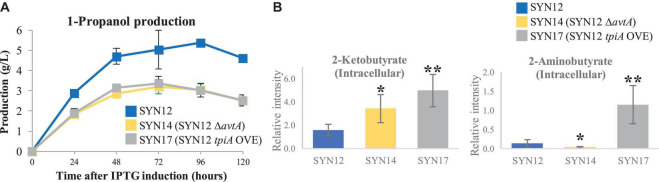
Our initial attempt to improve production titers focused on preventing the formation of the unwanted metabolite 2-aminobutyrate. From SYN12, we knocked out avtA (to yield SYN14) to redirect flux toward 1-propanol production. However, a decrease in 1-propanol production was observed (Figure 3A). As expected, we observed a decrease in 2-aminobutyrate and an increase in 2KB compared to that of SYN12 (Figure 3B). Because 2KB is directly upstream of 1-propanol, the decrease in 1-propanol titer is counterintuitive, and may be a result of 2KB toxicity (Zhang et al., 2016; Fang et al., 2021).
FIGURE 3.
Effect of avtA deletion or tpiA overexpression. (A) 1-Propanol production. (B) Intracellular 2-ketobutyrate and 2-aminobutyrate. Samples were taken after 24 h of fermentation. Metabolite intensities shown in the y-axis were normalized to an internal standard. Asterisks indicate significant difference from the strain (*p < 0.05, **p < 0.01). The error bars indicate standard deviations obtained from three replicate fermentations.
A similar trend was observed in a separate experiment to supply carbon into the 1-propanol pathway. We examined the central metabolism and performed time course sampling in which we took samples every hour from 0 to 7 h after IPTG induction, at which point the cells were already growing in log phase. We performed ion-pair LC/MS/MS-based targeted analysis using 24 annotated metabolites belonging to glycolysis, TCA cycle, PPP, as well as important cofactors as listed in Supplementary Table 3. We found that dihydroxyacetone phosphate (DHAP) levels significantly increased over time (Supplementary Figure 2). To improve flux through glycolysis, we overexpressed tpiA, encoding triose-phosphate isomerase, which catalyzes the reversible reaction to convert DHAP to D-glyceraldehyde 3-phosphate (GAP) (Lee et al., 2013). Overexpression of tpiA in SYN12 (herein referred to as SYN17) resulted in approximately 3-fold increase of 2KB compared to the parental strain, accompanied by over 10-fold increase of downstream by-products derived from 2KB, 2-aminobutyrate (Figure 3B). Similar to SYN14 (SYN12 ΔavtA), a highly accumulated 2KB in the cell was severely detrimental for 1-propanol production (Figure 3A).
Optimization of Alcohol Dehydrogenase Activity for Improved 1-Propanol Titers
Taken together, these results suggest that a bottleneck in 1-propanol biosynthesis may exist in either KivD or YqhD, those that convert 2KB into 1-propanol. We next attempted to improve titers by optimizing the activity of KivD and YqhD. We thus constructed two RBS libraries to vary the expression of KivD or YqhD by altering the RBS translation initiation rate. The Salis RBS Calculator v1.1 (Salis et al., 2009; Espah Borujeni et al., 2014) was used to design 8 variants per library using pAFC66 as a starting template (Table 1). The resulting library was cloned into SYN12 and a minimum of 23 colonies were screened for 1-propanol production after 24 h (Supplementary Figures 3A, 4), which ensured that all variants have been screened at least once with a 95% probability based on the Clarke Carbon formula.
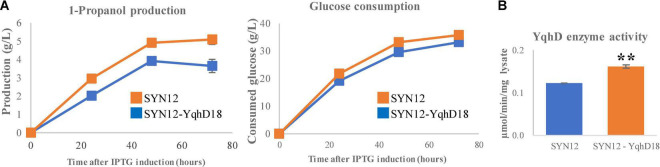
From the screening, one library variant was identified from each that had at least a 30% increased titer compared to SYN12: these were denoted as SYN12-YdhD18 and SYN12-KivD34. We next assessed whether these strains exhibit changes in 1-propanol production. While SYN12-KivD34 did not show increased titers (Supplementary Figure 3B), SYN12-YqhD18 reached titers of 5.1 (g/L) and yield of 0.142 (g/g of glucose), compared to SYN12 that reached titers of 3.7 (g/L) and yield of 0.110 (g/g of glucose) in 72 h (Figure 4A). To verify that the altered RBS conferred an increase in YqhD activity, we measured the specific activity of YqhD in vitro in cell lysate and found a 33% increase in activity (Figure 4B).
FIGURE 4.
Effect of YqhD expression optimization. (A) 1-Propanol production and consumed glucose. (B) The relative specific activity of YqhD within cell lysate with improved RBS sequence. Asterisks indicate significant difference from the strain (**p < 0.01). The error bars indicate standard deviations obtained from three replicate fermentations.
Discussion
The synergistic combination of both the citramalate pathway and threonine pathway to produce 1-propanol has been used previously to demonstrate improved theoretical yields and productivity. However, no change in titer has been observed with the overexpression of the threonine pathway over native levels. Here, we applied metabolomics-based strategies to understand the limitations of the synergistic pathway and to engineer SYN12 for increased 1-propanol titers.
The metabolomics strategies that were applied in this work contributed to a streamlined and logical series of strain improvement steps that ultimately allowed for increased production titers. Initial metabolomics analysis compared SYN3 and SYN12 to reveal an increase in concentrations of 2-aminobutyrate, norvaline, and 2-ketobutyrate in SYN12. SYN12 is additionally modified to overexpress the native threonine pathway. While no increase in titer was observed, the physiological consequences of the pathway overexpression became apparent in the accumulation of these upstream metabolites and byproducts.
We therefore sought to focus efforts on channeling the metabolic flux toward 1-propanol production. However, gene deletions of avtA, aimed at decreasing unwanted byproducts 2-aminobutyrate, had adverse effects on the production titers. Additional experiments aimed at increasing flux through glycolysis by overexpression of tpiA, and subsequently through the 1-propanol pathway, also resulted in a 2KB accumulation with a significant decrease in 1-propanol titers. 2KB is a short-chain alpha keto acid and a structural analog of pyruvate. As an indispensable metabolic intermediate in biological metabolism, 2-KB is found in the biosynthesis of L-isoleucine (Park et al., 2012; Yin et al., 2012), 2-aminobutyrate (Xu et al., 2019), 1-butanol (Atsumi and Liao, 2008), L-homoalanine (Zhang et al., 2010), and so on. However, because of its structural similarity with pyruvate, 2KB competitively inhibits enzymes used in pyruvate metabolism at high concentrations, which causes cellular toxicity (Zhang et al., 2016; Fang et al., 2021). As such, this suggests that enhancing the turnover rate of 2-KB to 1-propanol with KivD and YqhD may relieve toxicity caused by 2KB accumulation.
We then focused on optimizing flux of KivD and YqhD to relinquish this bottleneck. We thus constructed RBS libraries to vary the expression of each enzyme and isolated a variant. While SYN12-KivD34 increased KivD34 specific activity by 69%, it did not show increased titers (Supplementary Figures 3B,C). This might be due to the toxic intermediate propyl aldehyde, which can generate membrane lipid peroxidation (Pérez et al., 2008), thus suggesting that the turnover rate of propyl aldehyde to 1-propanol catalyzed by YqhD was not sufficient. SYN12-YqhD18, with a 33% increase in YqhD specific activity, exhibited a 38% increased titer and a 29% increased yield in 72 h compared to the base strain SYN12, which was the best 1-propanol producing strain constructed in Shen and Liao (2013). These results may imply that there was an insufficient turnover rate of toxic intermediates 2KB and propyl aldehyde into 1-propanol in SYN12. This highlights two potential future directions for further strain improvement. First, since propyl aldehyde is a toxic intermediate, it may be beneficial to simultaneously fine-tune expression of both of these pathway enzymes. However, generating RBS libraries to optimize both enzymes would result in a combinatorial library with a much greater number of variants, and thus, would require screening methods with larger throughput. Second, since turnover and toxicity of 2KB is improved with YqhD18, a future target for further engineering may be to increase flux through the pathway, possibly with overexpression of tpiA as demonstrated in SYN17. These future possibilities further highlight the value of metabolomics in the design-test-build cycle that is endemic to metabolic engineering and strain improvement.
Previous works have aimed at improving 1-propanol production in E. coli with various strategies. In one example, E. coli was modified to contain a keto-acid pathway to convert 2-ketobutyrate to propionyl-phosphate, which is then sequentially converted to propionate, propionyl-CoA, propionyl-aldehyde, and finally to 1-propanol (Choi et al., 2012). A separate study extended succinate dissimilation under anaerobic conditions (Srirangan et al., 2014). The titer reported here is much higher compared to other hosts such as in Corynebacterium glutamicum [12 mM (721 mg/L) from glucose] (Siebert and Wendisch, 2015), Propionibacterium freudenreichii (0.51 g/L from glycerol) (Ammar et al., 2013), and Saccharomyces cerevisiae (180 mg/L from glucose) (Nishimura et al., 2018). The efficient genetic tractability of E. coli allows for more complex genetic modifications than other species, and therefore E. coli has potential to be further engineered for higher 1-propanol production.
Conclusion
The successful strain improvement in this work demonstrates the application of metabolomics-guided metabolic engineering strategies. More importantly, the data generated from metabolomics highlights metabolic perturbations that occur from pathway bottlenecks that may be relevant to the improvement of other production pathways.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
KN, NK, and WL: metabolite measurement and data curation and analysis. TO: data curation and interpretation, and writing the manuscript. SPu: metabolite measurement, data analysis and interpretation, supervision, and writing the manuscript. CS: strain construction, data analysis and interpretation, and revising manuscript. SPo: strain construction, data analysis and interpretation, and writing the manuscript. EF and JL: supervision, project management, and resources. All authors have read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This study will be included in a dissertation submitted by TO to Osaka University in partial fulfillment of the requirement for his doctor degree.
Funding
This research was fully supported by Japan Science and Technology (JST), Strategic International Collaborative Research Program, SICORP for JP-US Metabolomics and National Science Foundation (NSF) MCB-1139318.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.871624/full#supplementary-material
References
- Ammar E. M., Wang Z., Yang S. (2013). Metabolic engineering of Propionibacterium freudenreichii for n-propanol production. Appl. Microbiol. Biotechnol. 97 4677–4690. 10.1007/s00253-013-4861-6 [DOI] [PubMed] [Google Scholar]
- Atsumi S., Cann A. F., Connor M. R., Shen C. R., Smith K. M., Brynildsen M. P., et al. (2008). Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 10 305–311. 10.1016/j.ymben.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Atsumi S., Liao J. C. (2008). Directed Evolution of Methanococcus jannaschii Citramalate Synthase for Biosynthesis of 1-Propanol and 1-Butanol by Escherichia coli. Appl. Environ. Microbiol. 74 7802–7808. 10.1128/AEM.02046-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2 2006.0008. 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. J., Park J. H., Kim T. Y., Lee S. Y. (2012). Metabolic engineering of Escherichia coli for the production of 1-propanol. Metab. Eng. 14 477–486. 10.1016/j.ymben.2012.07.006 [DOI] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espah Borujeni A., Channarasappa A. S., Salis H. M. (2014). Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 42 2646–2659. 10.1093/nar/gkt1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Zhang S., Wang J., Yin L., Zhang H., Wang Z., et al. (2021). Metabolic Detoxification of 2-Oxobutyrate by Remodeling Escherichia coli Acetate Bypass. Metabolites 11:30. 10.3390/metabo11010030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathima A. M., Chuang D., Laviña W. A., Liao J., Putri S. P., Fukusaki E. (2018). Iterative cycle of widely targeted metabolic profiling for the improvement of 1-butanol titer and productivity in Synechococcus elongatus. Biotechnol. Biofuels 11:188. 10.1186/s13068-018-1187-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Kang K. H., Kim E. Y., Chae T. U., Oh Y. H., Hong S. H., et al. (2013). Metabolic engineering of Escherichia coli for enhanced biosynthesis of poly(3-hydroxybutyrate) based on proteome analysis. Biotechnol. Lett. 35 1631–1637. 10.1007/s10529-013-1246-y [DOI] [PubMed] [Google Scholar]
- Liao J. C., Mi L., Pontrelli S., Luo S. (2016). Fuelling the future: microbial engineering for the production of sustainable biofuels. Nat. Rev. Microbiol. 14 288–304. 10.1038/nrmicro.2016.32 [DOI] [PubMed] [Google Scholar]
- Lommen A. (2009). MetAlign: interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal. Chem. 81 3079–3086. 10.1021/ac900036d [DOI] [PubMed] [Google Scholar]
- Nishimura Y., Matsui T., Ishii J., Kondo A. (2018). Metabolic engineering of the 2-ketobutyrate biosynthetic pathway for 1-propanol production in Saccharomyces cerevisiae. Microb. Cell Fact. 17:38. 10.1186/s12934-018-0883-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta K., Laviña W. A., Pontrelli S., Liao J. C., Putri S. P., Fukusaki E. (2017). Orthogonal partial least squares/projections to latent structures regression-based metabolomics approach for identification of gene targets for improvement of 1-butanol production in Escherichia coli. J. Bio. Biosci. Eng. 124 498–505. 10.1016/j.jbiosc.2017.05.015 [DOI] [PubMed] [Google Scholar]
- Ohtake T., Pontrelli S., Laviña W. A., Liao J. C., Putri S. P., Fukusaki E. (2017). Metabolomics-driven approach to solving a CoA imbalance for improved 1-butanol production in Escherichia coli. Metab. Eng. 41 135–143. 10.1016/j.ymben.2017.04.003 [DOI] [PubMed] [Google Scholar]
- Park J. H., Oh J. E., Lee K. H., Kim J. Y., Lee S. Y. (2012). Rational Design of Escherichia coli for L-Isoleucine Production. Synth. Biol. 1 532–540. 10.1021/sb300071a [DOI] [PubMed] [Google Scholar]
- Pérez J. M., Arenas F. A., Pradenas G. A., Sandoval J. M., Vásquez C. C. (2008). Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J. Biol. Chem 283 7346–7353. 10.1074/jbc.M708846200 [DOI] [PubMed] [Google Scholar]
- Pontrelli S., Chiu T. Y., Lan E. I., Chen F. Y. H., Chang P., Liao J. C. (2018). Escherichia coli as a host for metabolic engineering. Metab. Eng. 50 16–46. 10.1016/j.ymben.2018.04.008 [DOI] [PubMed] [Google Scholar]
- Putri S. P., Nakayama Y., Matsuda F., Uchikata T., Kobayashi S., Matsubara A., et al. (2013). Current metabolomics: practical applications. J. Bio. Biosci. Eng. 115 579–589. 10.1016/j.jbiosc.2012.12.007 [DOI] [PubMed] [Google Scholar]
- Putri S. P., Nakayama Y., Shen C., Noguchi S., Nitta K., Bamba T., et al. (2018). Identifying metabolic elements that contribute to productivity of 1-propanol bioproduction using metabolomic analysis. Metabolomics 14:96. 10.1007/s11306-018-1386-0 [DOI] [PubMed] [Google Scholar]
- Salis H. M., Mirsky E. A., Voigt C. A. (2009). Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 27 946–950. 10.1038/nbt.1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C. R., Liao J. C. (2008). Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways. Metab. Eng. 10 312–320. 10.1016/j.ymben.2008.08.001 [DOI] [PubMed] [Google Scholar]
- Shen C. R., Liao J. C. (2013). Synergy as design principle for metabolic engineering of 1-propanol production in Escherichia coli. Metab. Eng. 17 12–22. 10.1016/j.ymben.2013.01.008 [DOI] [PubMed] [Google Scholar]
- Siebert D., Wendisch V. F. (2015). Metabolic pathway engineering for production of 1,2-propanediol and 1-propanol by Corynebacterium glutamicum. Biotechnol Biofuels 8:91. 10.1186/s13068-015-0269-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srirangan K., Liu X., Westbrook A., Akawi L., Pyne M. E., Moo-young M., et al. (2014). Biochemical, genetic, and metabolic engineering strategies to enhance coproduction of 1-propanol and ethanol in engineered Escherichia coli. Appl. Microbiol. Biotechnol. 98 9499–9515. 10.1007/s00253-014-6093-9 [DOI] [PubMed] [Google Scholar]
- Tsugawa H., Arita M., Kanazawa M., Ogiwara A., Bamba T., Fukusaki E. (2013). MRMPROBS: A Data Assessment and Metabolite Identi fi cation Tool for Large-Scale Multiple Reaction Monitoring Based Widely Targeted Metabolomics. Anal. Chem. 85 5191–5199. 10.1021/ac400515s [DOI] [PubMed] [Google Scholar]
- Tsugawa H., Tsujimoto Y., Arita M., Bamba T., Fukusaki E. (2011). GC/MS based metabolomics: development of a data mining system for metabolite identification by using soft independent modeling of class analogy (SIMCA). BMC Bioinform. 12:131. 10.1186/1471-2105-12-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. M., Li J. Q., Zhang B., Liu Z. Q., Zheng Y. G. (2019). Fermentative production of the unnatural amino acid L-2-aminobutyric acid based on metabolic engineering. Microb. Cell Factor. 18:43. 10.1186/s12934-019-1095-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Hu X., Xu D., Ning J., Chen J., Wang X. (2012). Co-expression of feedback-resistant threonine dehydratase and acetohydroxy acid synthase increase L-isoleucine production in Corynebacterium glutamicum. Metab. Eng. 14 542–550. 10.1016/j.ymben.2012.06.002 [DOI] [PubMed] [Google Scholar]
- Zhang C., Qi J., Li Y., Fan X., Xu Q., Chen N., et al. (2016). Production of a-Ketobutyrate Using Engineered Escherichia coli Via Temperature Shift. Biotechnol. Bioeng. 113 2054–2059. 10.1002/bit.25959 [DOI] [PubMed] [Google Scholar]
- Zhang K., Li H., Cho K. M., Liao J. C. (2010). Expanding metabolism for total biosynthesis of the nonnatural amino acid L-homoalanine. Proc. Natl. Acad. Sci. USA 107 6234–6239. 10.1073/pnas.0912903107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.