Abstract
SRY-related high-mobility group box 9 (SOX9) is an indispensable transcription factor that regulates multiple developmental pathways related to stemness, differentiation, and progenitor development. Previous studies have demonstrated that the SOX9 protein directs pathways involved in tumor initiation, proliferation, migration, chemoresistance, and stem cell maintenance, thereby regulating tumorigenesis as an oncogene. SOX9 overexpression is a frequent event in breast cancer (BC) subtypes. Of note, the molecular mechanisms and functional regulation underlying SOX9 upregulation during BC progression are still being uncovered. The focus of this review is to appraise recent advances regarding the involvement of SOX9 in BC pathogenesis. First, we provide a general overview of SOX9 structure and function, as well as its involvement in various kinds of cancer. Next, we discuss pathways of SOX9 regulation, particularly its miRNA-mediated regulation, in BC. Finally, we describe the involvement of SOX9 in BC pathogenesis via its regulation of pathways involved in regulating cancer hallmarks, as well as its clinical and therapeutic importance. In general, this review article aims to serve as an ample source of knowledge on the involvement of SOX9 in BC progression. Targeting SOX9 activity may improve therapeutic strategies to treat BC, but precisely inhibiting SOX9 using drugs and/or small peptides remains a huge challenge for forthcoming cancer research.
Keywords: Breast cancer, SOX9, microRNA, Chemoresistance, Therapy
Graphical Abstract
1. Introduction
Breast cancer (BC) is the leading cause of cancer incidence and the second leading cause of cancer mortality in women [1]. Despite advances in early detection and treatment, about 50% of patients with BC will either fail to respond to initial chemotherapy or will rapidly acquire resistance to chemotherapeutic agents; these data correspond to global or entire world patients. Drug resistance remains a major obstacle to the successful treatment of BC. Chemoresistance and metastasis are responsible for ~90% of deaths in patients with solid tumors, including those originating in the breast [2]. To more effectively tailor treatments to individual patients; it is important to identify potent targets at the molecular level.
In the early 1990s, a novel transcription factor was discovered and found to be involved in testis determination. The gene encoding this transcription factor, termed the sex-determining region Y (SRY) gene due to its location on the Y chromosome, has a distinctive DNA-binding domain [3, 4]. With its conserved high-mobility group (HMG) domain, the SRY protein always binds to specific DNA sequences. Since then, 20 different so-called SOX genes, which contain the HMG domain, have been identified in the mouse and human genomes based on their sequences and functions [5]. These genes have been divided into eight subgroups: SOX A-H, with 1–3 members each [6]. SOX9 belongs to subgroup E, along with SOX8 and SOX10. The subgroup E proteins share high sequence similarity and mediate cell fate by regulating diverse functions that maintain pluripotency, terminal differentiation, cell lineage restriction, and tissue homeostasis, depending on the cell and tissue types in which they are expressed [5]. In recent years, mounting evidence has found that SOX9 can regulate diverse cellular processes, including cell proliferation [7], apoptosis [8], migration [9], invasion [10], chemoresistance [11], stem cell [12], autophagy [13], angiogenesis [14], immune escape [15] and metastasis [16], by regulating the expression of several targeted genes. Extensive studies also demonstrated the involvement of SOX9 in the development of various cancers, such as bladder cancer [17], brain cancer [18], BC [19], cervical cancer [20], colon cancer [21], chondrosarcoma [22], esophageal cancer [23], endometrial cancer [24], gastric cancer [25], head and neck cancer [26], liver cancer [27], lung cancer [28], melanoma [16], ovarian cancer [29], prostate cancer [30], pancreatic cancer [31], renal cell carcinoma [32], and thyroid cancer [33], suggesting that it has a general role in tumor development and progression. SOX9 has also been discovered to play a role in regulating multiple signaling pathways during cancer progression [34–56] (Figs 1 and 2A). In this review, we discuss recent advancements regarding the role of SOX9 in BC pathogenesis through its mediation of important mechanisms, including tumor initiation and proliferation, apoptosis, migration, invasion, angiogenesis, chemoresistance, immune escape, stem cell maintenance, and regulation of the tumor microenvironment (Fig 2B). We also discuss the prognostic value of SOX9 expression and the potential of targeting SOX9 for BC therapy.
Figure 1.
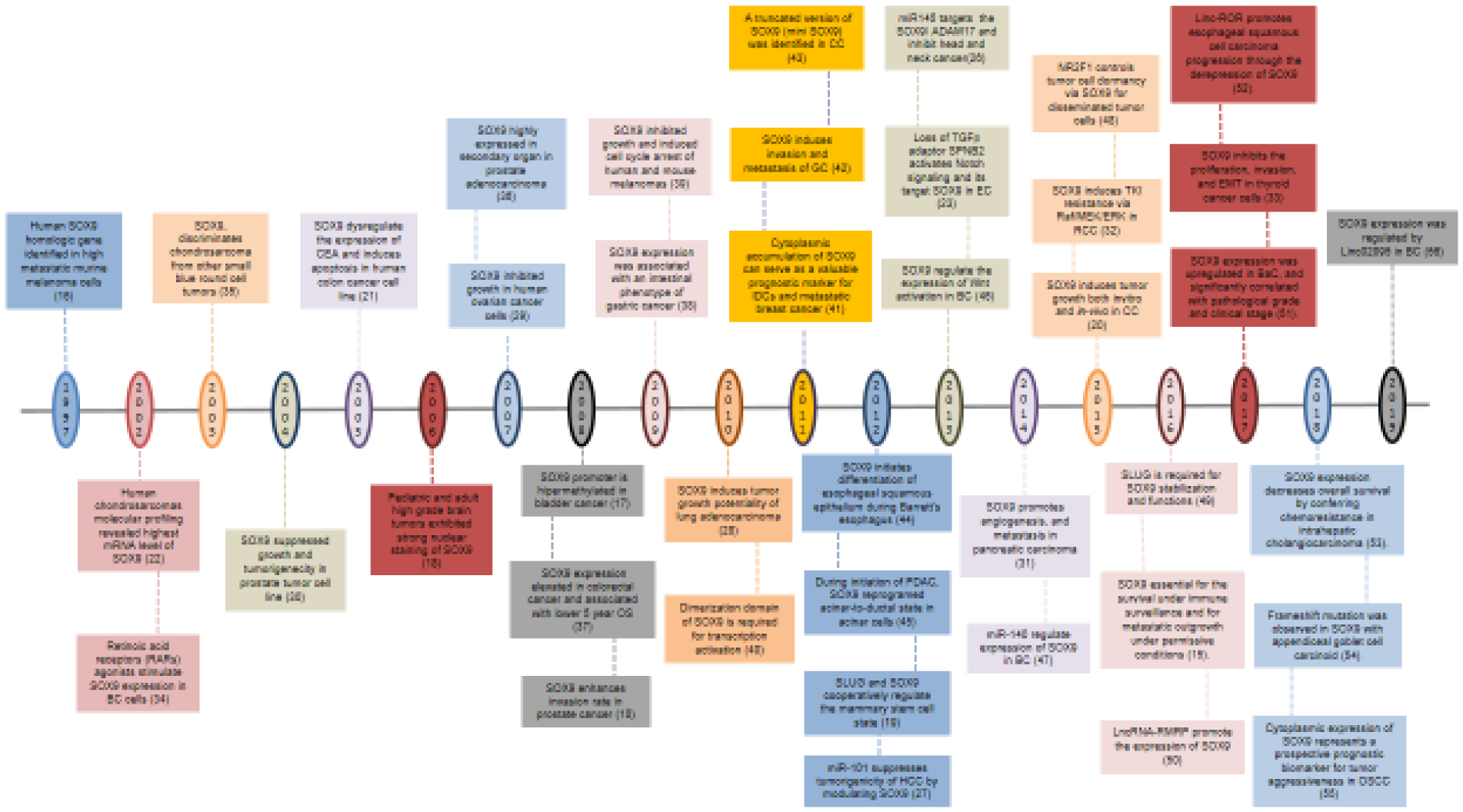
Timeline of the most important findings on the role of SOX9 in tumor progression since its discovery.
Figure 2.
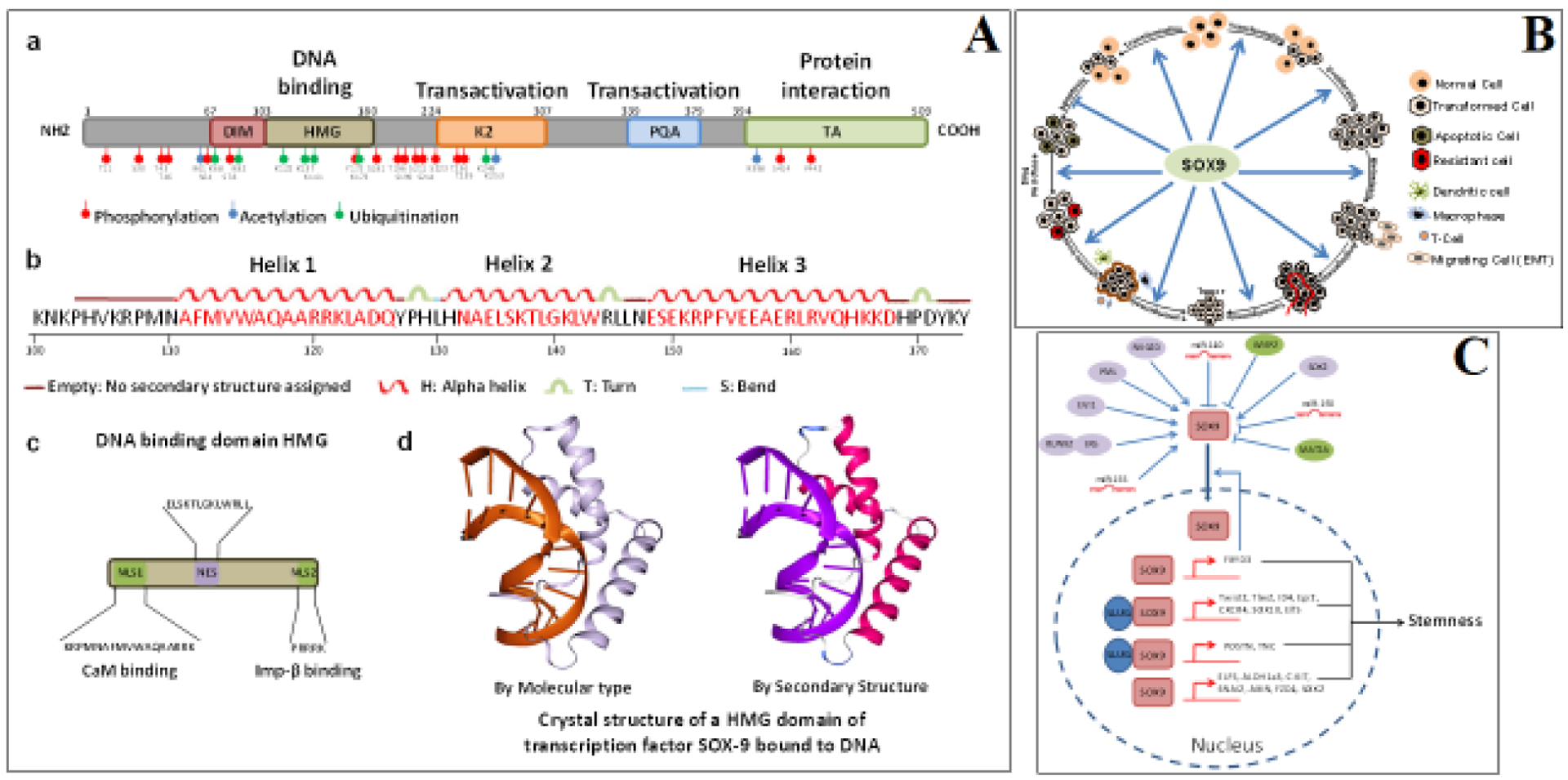
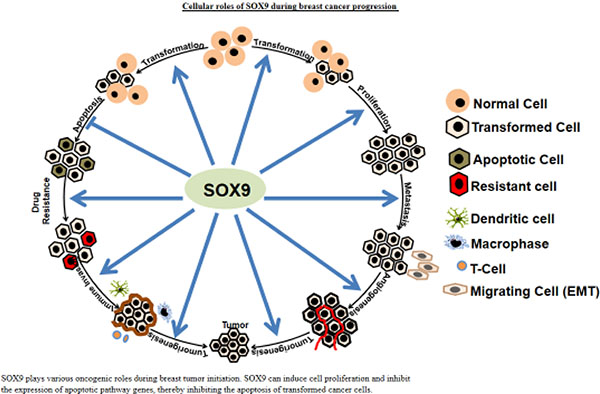
(Panel A) Schematic drawing and crystal structure of SOX9 Schematic structures of SOX9 protein. SOX9 protein has five different domains: the dimerization domain (DIM), followed by the DNA-binding high-mobility group (HMG) domain, two transactivation domains (K2 and PQA) located in a central position, and one at the C-terminal domain (TA). Post-translational modifications identified by phosphorylation sites (red), acetylation sites (blue), and ubiquitination/sumoylation sites (green) are highlighted (a). Schematic diagrams of the SOX9 DNA-binding HMG domain, showing the amino acid sequence involved in the production of its secondary helix structure (b), two independent nuclear localization signal (NLS) sequences that interact with calmodulin (CaM) and importin-β, and nuclear export signal (NES) sequences (c). Crystal structural illustrations of the SOX9 HMG domain (PDB ID: 4EUW) bound to DNA (d). (Panel B) Cellular roles of SOX9 during BC progression SOX9 plays various oncogenic roles during breast tumor initiation. SOX9 can induce cell proliferation and inhibit the expression of apoptotic pathway genes, thereby inhibiting the apoptosis of transformed cancer cells. Moreover, SOX9 induces metastatic signaling involved in progression of tumorigenesis. SOX9 contributes to tumorigenesis (both metastasis and chemoresistance) by regulating BC stem cells. Further, SOX9 contributes to tumorigenesis by promoting angiogenesis and the immune evasion of tumor cells. (Panel C) SOX9 proteins are involved in the induction of stemness of BC cells Upstream regulators and targets of SOX9 in the regulation of the stem cell properties of BC. RUNX2, RUNX family transcription factor 2; ERα, estrogen receptor alpha; EVI1, ecotropic virus integration 1 site protein; PML, promyelocytic leukemia; NKG2D, natural killer group 2D; CCN5, cellular communication network factor 5; SOX2, SRY-box transcription factor 2; MAT2A, methionine adenosyl-transferase 2A; miR-155, miR-140, miR-190. FXYD3, FXYD domain-containing ion transport regulator 3; TWIST2, twist family bHLH transcription factor 2; TBX2, T-box transcription factor 2; ID4, inhibitor of DNA binding 4, HLH protein; EGR2, early growth response 2; CXCR4, C-X-C motif chemokine receptor 4; SOX10, SRY-box transcription factor 10; ELF5, E74-like ETS transcription factor 5; POSTN, periostin; TNC, tenascin-C; ALDH1a3, aldehyde dehydrogenase 1 family member A3; cKit, KIT proto-oncogene, receptor tyrosine kinase; SNAI2 (Slug), snail family transcriptional repressor 2; AXIN, axin 1; FZD4, frizzled class receptor 4.
2. SOX9 structure and functions
To understand the molecular mechanisms underlying the role of SOX9 in development and the progression of various diseases, it is essential to establish a comprehensive picture of the structure and function of the protein. The expression of SOX9, a SRY family gene located a 3-Mb region on chromosome 17, is directed in a complex manner by individual enhancers in different tissues [57]. Generally, SOX9 contains a SRY-related HMG domain with three α helices (Fig 2A) with ~50% amino acid similarity [58]. SOX9 exhibits a high rate of translocation between the nucleus and cytoplasm, as its DNA-binding HMG domain contains two nuclear localization signal (NLS) sequences [59] and one nuclear export signal (NES) sequence enriched with leucines [60] (Fig 2A). During development, this structural arrangement ensures the translocation of SOX [61]. The HMG domain contains a conserved, 79 amino acid-long DNA-binding motif that binds with the minor groove of DNA containing the consensus sequence (A/TA/TCAAA/TG), forming an L-shaped complex [62]. SOX9 is in subgroup E of the SRY family, along with SOX8 and SOX10. These proteins share the highest level of similarity within the HMG domain and contain two other functional domains: 1) a dimerization domain (DIM), which enables homodimerization through dimerization with the HMG box of another SOX [63]; and 2) another C-terminal domain called the transactivation domain (TAC), which interacts with transcription machinery or co-activators associated with transcription. Like other SOX proteins, SOX9 undergoes post-translational modifications like phosphorylation, acetylation, and ubiquitination at different amino acid sequences (Fig 2A).
SOX9 protein, like other proteins in its subgroup, functions by interacting with various partners and shows stimulation and suppression activities in different cell types [58]. SOX9 was first reported in relation to campomelic dysplasia (CMPD), a severe skeleton malformation syndrome in which it is mutated [64]. Differential expression of SOX9 has also been associated with testis development disorder and sex reversal [65], and it is also known to play a critical role in chondrogenesis [66] and Sertoli cell differentiation [67]. SOX9 is also known to control the development of hair follicles [68], the central nervous system [69], the retina [70], the lungs [71], the pancreas [72], the heart [73], and the kidney [74]. In summary, SOX9 is important for cell lineage determination during various developmental processes. Recent studies have also found that SOX9 plays an essential role in the development of several tumor types, including BC. The present review focuses on the role of SOX9 in BC growth and progression.
3. Molecular mechanisms of SOX9 regulation
3.1. Transcriptional regulation
3.1.1. Epigenetic
Anomalous DNA methylation, which may aberrantly regulate transcriptional activity, is a hallmark of cancer [75]. DNA in cancers may be hypermethylated or hypomethylated, dictating the under- and overexpression of genes, respectively, depending on the coverage of the CpG islands in their promoter regions. Previous work has demonstrated that the methylation status of the SOX9 promoter region is dysregulated in several progressing tumor types. The very first study was done in bladder cancer and revealed that SOX9 was hypermethylated in 56.4% of cases and correlated significantly with advanced grade and poor overall survival (OS) [17]. Since then, several similar studies have been conducted in different cancers [76–80]. However, few studies have been performed to assess methylation in BC. One study revealed that several genes associated with BC, but not SOX9 or NFE2L3, were hypermethylated in BC [81]. Another interesting study revealed that the stem cell-associated genes ALDH1A, WNT5A, and SOX9 were significantly hypomethylated after neoadjuvant chemotherapy in all samples tested, including BC cells and tumor tissues [82]. Therefore, methylation may prove to be a valuable regulator that can modulate the expression of SOX9 in BC, but this remains to be evaluated.
3.1.2. Transcription factor
It is well documented that SOX9 expression is elevated in most types of cancer, including BC. Transcription factors that directly activate SOX9 expression have been identified in several cancers, including BC. For example, using a global transcriptomic approach, it was demonstrated that HDAC9 (histone deacetylase 9) regulates the expression of SOX9 [83]. Previous work demonstrated that PML (promyelocytic leukemia) protein was highly expressed in BC-initiating cells, and particularly in highly aggressive BC cells. This work also determined that SOX9 is the downstream modulator of PML via close proximity binding to its promoter region as because it has no canonical DNA-binding domain [84]. In addition, whole-genome EVI1 ChIP data documented the correlation of EVI1 within SOX9 binding sites, and EVI1 exhaustion reduced the expression of SOX9 in BC cell lines [85]. Additionally, Jeselsohn R et al. found that the expression of SOX9 in BC cells is modulated by the RUNX2-ER complex, resulting in induction of the stemness-mediated endocrine resistance of BC cells [86].
3.2. Post-transcriptional regulation
3.2.1. microRNA (miRNA)-mediated
miRNAs are short 22bp noncoding RNAs that regulate the expression of genes by directly binding to their 3’UTR sequences, modulating various cellular pathways, including those involved in proliferation, migration, and development [87]. Emerging studies show that several miRNAs contribute to BC pathogenesis by dysregulating the expression of various genes [88, 89]. Several studies have demonstrated that SOX9 expression is regulated by multiple miRNAs in various cancers, including BC [26, 90]. miR-101 was the first miRNA reported to regulate the expression of SOX9 by directly binding to its 3’UTR sequence in human hepatocellular carcinoma [27]. In BC, miR-140 was the first miRNA found to regulate the expression of SOX9 by direct binding, resulting in reduced self-renewal of cancer stem cells (CSCs) and tumorforming capacity of BC cells [47]. In contrast, another study found that miR-3134, in combination with the RNA binding protein HuR, stabilized the AU-rich element (ARE)-bearing transcript SOX9 in a BC cell line [91]. Furthermore, based on a study by Gernapudi R et al., it was shown that SOX9 is dysregulated by the preadipocyte-derived exosomal miRNA, miR-140 [92]. In addition, an interesting study revealed that miR-206 expression was dysregulated particularly in TNBC (triple negative BC) and regulated the expression of SOX9, along with VEGF and MAPK3. Thus, the downregulation of miR-206 promoted TNBC invasion and angiogenesis [14]. In contrast, CSCs and chemoresistant cells were found to secrete exosomal miR-155, inducing the expression of stem cell factor SOX9, but the molecular mechanisms behind this effect is still unknown [93]. In another study using in vitro and in vivo models, it was revealed that SOX9 expression is regulated by tumor-suppressive miR-133b, which reduces the cell proliferation, migration, and invasion abilities of BC cell. This study also revealed that the simultaneous expression with miR-133b reduced the oncogenic defects of SOX9 [94]. Consistent with these findings, it is known that miR-511 plays an important role in breast tumor growth and metastasis reduction by directly regulating the expression of SOX9 and deactivated the oncogenic PI3K/Akt signaling pathway [95]. Yu Y et al. further explored the expression of SOX9 in BC. They revealed that the expression of miR-190 was downregulated in BC samples, and the enforced expression of miR-190 inhibited the Wnt/β-catenin signaling cascade by directly modulating the expression of SOX9, increasing the sensitivity of BC cells to tamoxifen (TAM) in vitro and in vivo [96]. Additionally, the role of chemotherapy drug-induced extracellular vesicles (EV)-contained miRNA secretion as a positive regulator of SOX9 was revealed by Shen M et al. They observed that drug-treated BC cells secreted EV-contained miR-9-5p, miR-195-5p, and miR-203a-3p, which dysregulated the expression of ONECUT2 by direct binding, resulting in upregulated expression of CSC phenotype and stem cells factors, including NOTCH1, SOX9, NANOG, OCT4, and SOX2 in BC cells [97]. Finally, Gao JB et al. found that enhancing the expression of the tumor suppressor miR-215-5p could decrease the aggressiveness of BC cells by regulating the expression of SOX9 [98]. These findings suggest that miRNAs can be considered potent regulators of SOX9 expression (Table 1), providing a promising strategy to reduce SOX9 expression in BC.
Table 1.
SOX9 Targeting miRNAs in Human Breast Cancer
| miRNA | Patient samples | In-vitro model | In-vivo model | Functions | Refs |
|---|---|---|---|---|---|
| miR-140 | 43 tumor samples and 5 normal samples | MCF10A, MCF10DCIS, SUM225CWN and SUM102PT cells | Immuno-deficient nude female mice | miR-140 is significantly downregulated in cancer stem-like cells and enforced expression reduced CSC self-renewal and tumor formation by directly targeting ALDH1 and SOX9 | [47] |
| miR-3134 | MCF7 cells | miR-3134 (ARE binding miRNA) induced the expression of SOX9 in collaboration with ARE binding protein HuR | [91] | ||
| miR-140 | MCF10DCIS and mouse preadipocytes (3T3L1, MBA1) | Immuno-deficient Nu/Nu female mice | miRNA-140 was upregulated after shikonin treatment and reduced the expression of SOX9 | [92] | |
| miR-206 | 40 tumor samples | MCF7, MDA-MB361, MDA-MB231, MDA-MB435, and HCC1395 | miR-206 expression was downregulated in TNBC. Ectopic expression of miR-206 inhibited cell invasion and angiogenesis of TNBC by downregulating the expression of VEGF, MAPK3, and SOX9 | [14] | |
| miR-155 | MCF7 and MDA-MB231 | miR-155 was induced in exosomes isolated from CSCs and resistant cells and that exosomes induced the expression of SOX9 | [93] | ||
| miR-133b | 38 paired tissues | BT549, MDA-MB231, BT474, SKBR3, HCC1937 | BALB/c female nude mice, SCID/beige mice | miR-133b was downregulated in tumor and associated with advanced grade. Forced expression of miR-133b inhibited cell growth, invasion both in-vitro and in-vivo by directly modulating SOX9 | [94] |
| miR-511 | 51 pair | MDA-MB231, MCF7 | BALB/C nude mice | miR-511 was downregulated in BC tissues and cell lines and associated with lymph node metastasis and tumor stage. Ectopic expression inhibited cell growth and metastasis invite and also attenuated tumor growth in-vivo by directly targeting SOX9 mediated PI3K/Akt pathway | [95] |
| miR-190 | 30 paired tissues | T47D, MCF7, MDA-MB231, MDA-MB435, MDA-MB468 | Female BALB/c nude mice | miR-190 induced TAM sensitivity of BC cells both in-vitro and in-vivo by targeting SOX9 resulted introverted Wnt/β-catenin signaling. The expression of miR-190 inversely correlated with SOX9 in BC samples | [96] |
| miR-9-5p, miR-195-5p, and miR-203a-3p | 12 pre and post NT | MDA-MB231, MCF7, BT474 | NOD/SCID/IL2R γ-null (NSG) mice | Chemotherapy induced EV miRNA which targeted ONECUT2 resulting induction of CSC phenotype through induction of NOTCH1, SOX9, NANOG, OCT4, and SOX2 | [97] |
| miRNA-215-5p | 39 pair of breast carcinoma tissues | MCF10A, MDA-MB468, MDA-MB231 and MCF7 | BALB/c nude mice | miR-215-5p was downregulated in BC tissue samples. Upregulation of miR-215-5p inhibited BC cells growth and metastasis both in-vitro and in-vivo by directly targeting SOX9 | [98] |
BC, breast cancer; TNBC, triple negative BC; NT, neoadjuvant therapy
3.2.2. Long noncoding RNA (lncRNA)-mediated
lncRNAs are noncoding RNAs longer than 200 nucleotides. [99]. Numerous reports have revealed that lncRNAs play crucial roles in all cellular processes, including proliferation, differentiation, apoptosis, migration, invasion, and metabolism, by regulating the expression of various genes and miRNAs [99, 100]. In the very first report of lncRNA-mediated SOX9 regulation, Meng Q et al. showed that an oncogenic lncRNA, LncRNA-RMRP, induced the expression of KRAS, FMNL2, and SOX9 by inhibiting the expression of miR-206 [50]. Since then, many lncRNAs have been found to deregulate the expression of SOX9 in several cancers, including some that induce the expression of SOX9 [52, 56, 101] and others that reduce the expression of SOX9 [102, 103]. However, a recent study by Tariq A et al. revealed the regulatory function of lncRNAs on SOX9 in BC, stated that lncRNA linc02095 and SOX9 are expressed in basal type BC cell lines in a co-regulated manner. They also discovered that linc02095 induced the SOX9 transcription by regulating the tenancy of RNA pol II at the SOX9 gene and the induced linc02095 help in the export process of SOX9 mRNA to cytoplasm [56]. More studies will be required to uncover the mechanisms through which SOX9 is regulated by lncRNAs during BC progression.
4. Role of SOX9 in BC tumorigenesis
Emerging studies have revealed that SOX9 can control several important phenomena in BC during tumorigenesis, including proliferation, metastasis, drug resistance, stem cell maintenance, immune evasion, and modulation of the tumor microenvironment (Figs 2B and C). In this section, we describe each parameter that is regulated by SOX9 during BC progression.
4.1. BC initiation and proliferation
SOX9 is one of the most important genes upregulated during early tumor formation [45, 104]. The contribution of SOX9 to tumor initiation is associated primarily with cell cycle progression and cell proliferation. Contrasting results have been reported regarding the role of SOX9 in cell cycle regulation during BC progression. The very first report on this topic revealed that SOX9 is involved in the G0/G1 arrest of T47D BC cell line [34]. The tumor-suppressive role of SOX9 was further supported by the finding that the SOX9 mediates the HES-1 expression in BC cells during treatment with anti-proliferative retinoic acid (RA) [105]. In contrast, the involvement of SOX9 in BC cell proliferation was first observed by Chakravarty G et al., who demonstrated that cytoplasmic SOX9 expression was highly associated with the expression of the proliferative maker Ki67, specifically in invasive ductal breast tumors [106]. Additionally in basal-like BC cells, the inhibition of SOX9 expression reduced proliferation by controlling the expression of LRP6 and TCF4 [46]. Moreover, SOX9 expression reduced by fucoxanthin and fucoxanthinol in MDA-MB231 cells, but not in MCF7 cells, and may be involved in cell viability [107].
Interestingly, it was also revealed that SOX9 is an HDAC9 target gene, and the expression of both genes was highly correlated in the basal subtype of BC. The study also revealed the significance of SOX9 in maintaining the mitogenic activity of HDAC9, resulting in the dysregulation of cell proliferation [83]. Also, it was claimed that EVI1 and SOX9 cooperatively regulate transcriptional reprogramming, promoting the upregulation of pathways involved in BC tumor initiation [85]. Meanwhile, it was discovered that SOX9 was upregulated and promoted the proliferation of endocrine-resistant BC cells through the RUNX2-ER complex [86]. Interestingly, a recent study determined that the expression of SOX9 was higher in tumor spheres than in adherent BC cells and may be associated with their proliferation status [108]. Notably, one study showed that reducing the expression of SOX9 inhibited the proliferation of BC cells [94]. Furthermore, SOX9 was found to induce the activity of PI3K/Akt signaling pathway thus promoting the proliferation of BC in vitro and in vivo [95]. Recently, Domenici G et al. also found that reducing the expression of SOX9 could decrease the soft agar colony formation of TAM-resistance BC cells in vitro and tumorigenicity of TAM-resistance BC cells in vivo. They also observed that the soft agar colony formation, invasion, and invasion from spheroid were decreased upon lower the expression of SOX9 in TNBC cell lines [109]. Additionally, Yu Y et al. confirmed the involvement of SOX9 in BC cell proliferation; they observed the reversal of the proliferative effects of SOX9 on BC cells by co-treatment with tumor-suppressive miR-190 [96]. Consistent with these findings, SOX9 was shown to be involved in cell proliferation and colony formation in BC cells [98]. In conclusion, SOX9 has a potential role in controlling cell proliferation and may be a crucial molecular target to hamper the proliferative nature of BC cells.
4.2. BC apoptosis
Tumor cell apoptosis is a vital step in cellular development. Apoptosis is stimulated intrinsically and extrinsically through two different pathways that trigger cell death by activating various proteins, resulting in the elimination of damaged, aged, or autoimmune cells [110]. The initiation and progression of tumor growth is supported by the reduction of apoptosis in tumor cells. An increasing number of studies have shown that reduced SOX9 expression is correlated with apoptosis and that SOX9 may function as an essential regulator of cancer cell death. The apoptosis of BC cells was promoted by fucoxanthin and fucoxanthinol, and fucoxanthinolmediated apoptosis was more significant than others in MDA-MB231 cells. In the same report, researchers described the dysregulated expression of nuclear SOX9 after treatment with this compound [111]. Another study by the same group revealed reduced expression of members of the NFκB pathway and SOX9 after treatment with fucoxanthinol, which they associated with the apoptosis-inducing effect of this compound [107]. In addition, SOX9 may inhibit apoptosis by inducing the activity of its target gene HDAC9 (83). SOX9 has been consistently shown to be involved in reducing apoptosis in BC, resulting high rates of growth and metastasis rate [95]. From these studies, it is becoming clear that SOX9 may play an anti-apoptotic role in BC by targeting the apoptotic pathway.
4.3. BC migration and invasion
Metastasis was the most essential sequel in the advancement of BC and represents the main cause of mortality in BC patients, accounting for ~90% of deaths in patients within solid tumors [2]. The phenomenon of metastasis involves a series of sequential and organized events; however, migration and invasion play central roles in metastasis. Important evidence has highlighted the role of SOX9 in these complex processes. Interestingly, SOX9 has been identified as a pro-metastatic gene by Endo Y et al. They demonstrated that SOX9 is involved in the transcriptional regulation of vascular endothelial (VE)-cadherin by directly binding to its promoter region after RA treatment, thereby regulating the endothelial-like differentiation of BC cells and suggesting that SOX9 probably supports the incorporation of cells into a growing organ or tumor [112]. It was found that SOX9 expression was increased in estrogen receptor (ER)-negative and higher-grade human breast tumors, and its cytoplasmic accumulation was associated with the lymph node metastasis of invasive ductal carcinomas [106]. Meanwhile, SOX9 was discovered to induce micro- and macro-metastases within the lungs during BC progression, and abating SOX9 reduced the rate of macro-metastases to the lungs [20]. In addition, it was documented that SOX9 was positively regulated the metastatic outgrowth of latency-competent cancer (LCC) cells from BC cell lines during tolerant conditions [15]. Furthermore, the role of SOX9 as a positive regulator of metastasis was confirmed by Mateo F et al. They found that the dysregulation of SOX9 directly mediated the metastatic signature of BC cells to the lungs [85]. Similarly, the ability of restored miR-133b expression to inhibit BC invasion was halted by the subsequent overexpression of SOX9, suggesting that SOX9 is involved in maintaining micro-metastases and the weight of the lungs during BC progression [94]. Furthermore, the inhibition of SOX9 was shown to suppress the migration and invasion capacity of BC cells [95]. In addition, the induction of invasion caused by SOX9 was elucidated by Gao JB et al., who found that the dysregulation of SOX9 was directly involved in the regulation of BC migration and invasion [98]. Thus, SOX9 dysregulation might be associated with the process of metastasis, though further investigation is needed to understand the mechanisms underlying this association during BC progression.
4.4. BC angiogenesis
Tumor angiogenesis, the formation of new blood vessels, is an important hallmark [113] that is significant for the growth and metastasis of cancers, including BC [114]. The association between angiogenesis and tumor growth has become a major interest in the field of cancer research. Substantial work has demonstrated that SOX9 might support the development and progression of cancers through its role in angiogenesis. VE-cadherin, one of the most important among many endothelial genes, has been associated with angiogenesis [115]. Endo Y et al. found that SKBR3 cells undergo network formation in matrigel, forming mixed structures when co-cultured with human umbilical vein endothelial cells during RA treatment. They also found increased expression of VE-cadherin and SOX9 after the same treatment. In addition, they observed that the induction of VE-cadherin expression by the SOX9-ER81 transcriptional complex resulted in morphological changes that may lead to angiogenesis throughout RA treatment [112]. TNBC has higher rates of primary and secondary metastases compare to other BC subtypes due to greater angiogenic potential [116]. More recently, SOX9 has been proposed to be involved in TNBC invasion and metastasis [14]. Collectively, whether SOX9 enhances angiogenesis and may be appealing as a therapeutic target in BC remains to be explored further.
4.5. BC drug resistance
About 80% of BCs are ER-positive, constituting the predominant subtype of BC. The main treatment strategies to treat ER-positive BC generally involve endocrine therapy. Unfortunately, not all patients with ER-positive BC are responsive to anti-estrogen therapy, and many acquire resistance [117]. Several studies have established the significant role of SOX9 in drug resistance. The very first report of SOX9-mediated drug resistance was on endocrine resistance, demonstrated by Jeselsohn R et al. They observed upregulation of RUNX2 in TAM-resistant BC cells, and the resistance against TAM was mediated by a set of genes transcribed by RUNX2 – ER complex. They also observed the most abundant gene transcribed by this complex was SOX9, which contributed to the development of endocrine resistance [86]. For instance, SOX9 was found to be significantly upregulated in doxorubicin (DOX)- and paclitaxel (PTX)-resistant MCF7 BC cells. The same study also revealed that the expression of SOX9 was upregulated in MCF7 cells that received exosomes from CSCs and DOX- and PTX-resistant MCF7 cells [93]. Moreover, upregulated SOX9 has also been observed in tumor initiating cells (TICs) and ALDH+ cells, and correlated with the expression of FXYD3. It has been also found that SOX9 directly regulating the expression of FXYD3 in a positive manner and that FXYD3 play an important role in chemoresistance of ER+ BC cells [118]. Furthermore, SOX9 was elevated in patient’s samples, and its expression level was significantly high in ER negative tissue samples than ER positive tissue, and its downregulation reduced the growth of TAM-resistant BC cells in vivo [109]. Interestingly, a recent study identified that SOX9 is the primary modulator of the Wnt/β-catenin signaling pathway, increasing resistance of BC cells against anti-estrogen therapy [96]. Consistent with these results, a recent study reported that EVs secreted by BC cells treated with chemotherapeutic agents (docetaxel and DOX) induced the expression of SOX9 in recipient BC cells and led to drug resistance [97]. The studies mentioned above focused primarily on TAM resistance (Table 2a). Nevertheless, increasing evidence indicates that BC patients are distressed from multi-drug resistance, the molecular mechanisms of which remain to be explored.
Table 2a.
Summary of SOX9-Mediated Resistance to Therapy
| Cancer cell subtype | SOX9-mediated resistance to therapy | Major upstream/downstream target involved | Refs |
|---|---|---|---|
| Luminal and basal type | Inhibition of Rapamycin (mTOR) | EVI1, RHEB, mTOR, RAPTOR, FSCN1, and SPARC | [85] |
| Luminal type | Endocrine resistance | RUNX2, ERα, and NCOA3 | [86] |
| Luminal and basal type | Doxorubicin and Paclitaxel | miR-155 | [93] |
| Luminal type | Endocrine resistance (TAM) | FXYD3, pERK, Src, and ERα | [118] |
| Luminal type | Endocrine resistance (TAM) | SOX2, Estrogen (negative regulation) ALDH1A3, and Wnt signalling | [109] |
| Luminal and basal type | Endocrine resistance | ZEB1, ERα, miR-190, β-Catenin, c-Myc, CD44, TCF4 | [96] |
| Luminal and basal type | Doxorubicin and docetaxel | ONECUT2, Notch1, Nanog, Oct4, SOX2 | [97] |
TAM, tamoxifen; EVI1, ecotropic viral integration site-1; mTOR, mammalian target of rapamycin; RAPTOR, regulatory associated protein of mTOR complex 1; FSCN1, fascin actin-bundling protein 1; SPARC, secreted protein acidic and cysteine rich; RUNX2, RUNX family transcription factor 2; ERα/ESR1, estrogen receptor 1; NCOA3. nuclear receptor co-activator 3; FXYD3, FXYD domain containing ion transport regulator 3; pERK, phosphor extracellular-signal-regulated kinase; SRC, proto-oncogene tyrosine-protein kinase Src; SOX2, SRY-box transcription factor 2; ALDH1A3, aldehyde dehydrogenase 1 family member A3; ZEB1, zinc finger E-box binding homeobox 1; cMyc, MYC proto-oncogene; TCF4, transcription factor 4; ONECUT2, one cut homeobox 2; NOTCH1, notch receptor 1; NANOG, Nanog homeobox; OCT4/ POU5F1, POU class 5 homeobox 1
4.6. BC stem cells
CSCs are important players assumed to have significant roles in tumor formation, chemoresistance, metastasis, and disease recurrence [119]. It has been well documented that breast tumors contain a population of self-renewing CSCs [120] that are involved in predetermining BC cells fate [121]. First, Guo W et al. identified that SOX9, collaboratively with Slug, regulates the mammary stem cell (MaSC) state, and inhibition of either protein inhibit MaSC activity. Conversely, the ectopic expression of both proteins induces the conversion of luminal cells into MaSCs by regulating luminal progenitor genes like cKit, CXCR4, ELF5, and LBP [19]. For instance, SOX9 was found to be significantly upregulated in DCIS (ductal carcinoma in situ) cancer stem-like cells and involved in maintaining their mammosphere-forming capacity [47]. Furthermore, the expression of SOX9 was induced in WISP2/CCN5-knockdown BC cells and may be involved in maintaining stem cell properties and tumor growth [122]. In addition, it was acknowledged that SOX9 is positively regulated by NKG2D (natural killer group 2 member D) and stimulates cancer cell plasticity by inducing stem cell phenotypes [123]. Additionally, it was found that SOX9 was increased in lymphangioleiomyomatosis (LAM), a neoplasm characterized by proliferative ERα-negative and PR-positive smooth muscle-like cells with lung-metastatic capabilities. The study revealed that the biomarkers associated with LAM to BC stemness and lung metastasis [124]. As the very first study on this topic determined, Slug and SOX9 are two transcription factors that are important for regulating MaSCs. Consistent with these results, Fazilaty H et al. showed that SOX9 and Slug cooperatively regulate the expression of tenascin-C and periostin, two essential factors associated with tumor initiation and metastatic niche formation [125]. At the same time, Malladi S et al. found that SOX9 and SOX2 are involved in maintaining the stem cell-like characteristics of latency competent cancer (LCC) cells derived from human BC cell lines, resulting in the sustained tumor-instigation properties of LCC cells during the latent metastatic stage [15]. Interestingly, it was also announced that PML protein positively regulates the expression of the stem cell factor SOX9 by directly binding to its promoter region in BC initiating cells, thus regulating cancer initiation and metastatic seeding abilities [84]. Furthermore, the induction of EVI1 was shown to regulate the expression of SOX9 and maintain stem cell-like phenotypes, and cooperatively with EVI1, SOX9 also increased the expression of the mTOR cascade components REHB and RAPTOR, as well as the lung metastasis mediators and FSCN1 and SPARC, in BC cells [85]. Notably, the function of SOX9 in lineage maintenance was observed by Wang C et al., who found that a distinct population of SOX9-positive stem cells always developed into and maintained an ER-negative lineage of cells [126]. Meanwhile, SOX9 was discovered to associate with cadherins, particularly CDH4 and CDH17, and to maintain stem cell phenotypes in TNBC [127]. Meanwhile, it was also discovered that RUNX2-ER complexes regulate the expression of the stem cell factor SOX9 in endocrine-resistant BC cells, and the induction of SOX9 controlled resistance against TAM [86]. Also, it was claimed that human BC cell-derived M13HS hybrid clone cells and their parental cells express SOX9 and Slug and possess CSC-like properties [128]. Moreover, it has been observed that SOX9 is more highly expressed in tumor spheres than in adherent BC cell lines [108]. Interestingly, a recent study determined that FXYD3 (an estrogen-inducible gene) was notably overexpressed in luminal CSCs, particularly the ER-positive subtype. This study also revealed that the stem cell factor SOX9 directly regulates the expression of FXYD3, which regulates the nuclear translocation of SOX9 in BC (118). Additionally, Domenici G et al. confirmed the involvement of SOX9 in human breast CSCs and breast luminal progenitor cell maintenance. They observed that SOX2 is an upstream modulator of SOX9 and that SOX9 was involved in modulating luminal progenitor cell content and expression of ALDH1A3 in breast CSCs. They also observed that CRISPR-mediated knockout of SOX9 diminished the growth of TAM-resistant breast tumors in vivo [109]. Meanwhile, in another study, it was proclaimed that SOX9 rescues TAM resistance and the stemness properties of BC cells treated with tumor-suppressive miR-190. The study also acknowledged that the tumor-suppressive properties of miR-190 were due to its regulation of SOX9 activity by direct targeting [96]. Strekalova E et al. disclosed that the restriction of methionine in TNBC cells repressed mammosphere formation and the CSC population by regulating MAT2A/SOX9 axis [129]. Recently, Shen M et al. found enhanced expression of SOX9 and other stem cell-related transcription factors in BC cells treated with EVs secreted by chemotherapy-treated BC cells. They also found that this effect was due to EV miRNA-mediated regulation of ONECUT2 [97]. Taken together, these results endorsed the functional importance of SOX9 in BC progression and stem cell maintenance (Fig 2C).
4.7. BC immunomodulation
Recent studies have demonstrated that immune cells play an important role in the initiation and progression of cancer by suppressing immune rejection and thus supporting enhanced tumor growth and metastasis [130]. The pivotal role of SOX9 in immune evasion was described for the first time by Malladi et al. in 2016. They observed that SOX2 and SOX9 maintained the long-term survival and tumor-initiating properties of LCC by maintaining stem cell-like characteristics. They also observed that these two transcription factors are essential in LCC cells for maintaining a quiescence state at secondary metastasis sites and evading immune surveillance under immune-tolerant conditions [15].
4.8. BC microenvironment
Recently, it has been shown that the tumor microenvironment plays a crucial role in cancer development/progression and treatment efficacy, which are associated with advanced stages and drug resistance [131]. A previous study implicated preadipocytes in breast tumor initiation and metastasis through their exosomes-mediated communication with BC cells. The study also revealed the role of preadipocyte-secreted exosomal miR-140 in the regulation of stem cell renewal, cell migration, and tumor formation during the early stages of BC. Furthermore, based on the same study, it was proclaimed that the SOX9 is the main regulator of these processes in the tumor microenvironment and thus promoted tumorigenesis in vivo [92].
5. Clinical significance
In the clinic, a simple biomarker to improve early disease detection and prognosis is highly desirable. There have been numerous reports on the prognostic roles of various genes in BC; however, no presently used biomarkers can perfectly predict outcomes in BC. Critical evidence has demonstrated that the aberrant expression of SOX9 in BC, supporting the role of SOX9 in various cancer-associated pathways. BC tumor tissues overexpress SOX9, whereas normal tissues have very low expression of SOX9. This study also revealed that the expression of SOX9 was directly associated with hormone receptor expression and advanced grade, with higher expression in ER-negative BC. The higher expression was also associated with shorter overall survival (OS). In addition, it was observed that the cytoplasmic accumulation of SOX9, which was linked to the elevated proliferation of IDC (invasive ductal carcinoma) and metastatic BC, also contributes to poor clinical outcomes for patients with invasive BC [106]. Guo W et al. confirmed that the overexpression of SOX9 and Slug in BC patient samples was associated with poor OS. Their study also revealed that the co-expression of SOX9 and Slug promotes the tumorigenicity and metastatic seeding abilities of BC cells [19]. In another study using immunohistochemistry, it was revealed that SOX9 is an aggressive basal-like BC signature gene. Its expression was associated with histological grade and may function as an independent factor indicating the poor prognosis of patients with TNBC [46]. Meanwhile, SOX9 was discovered within the stroma of BC patients, and strong expression of SOX9 within the stroma after chemotherapy was associated with shorter OS [132]. Moreover Sox9 expression was closely related with expression of ER, PR, Ki67, p53, and lymph node metastasis. High expression of SOX9 was also associated with BC stem cells and both were correlated with poor overall and disease free survival and related with worst prognosis [133]. Additionally, one group found that SOX9 was highly expressed in patient with TNB, but it was not significantly correlated with disease outcome [134]. Subsequently, Richtig G et al. found that the expression of SOX9 may be a strong indicator of poor 5-year, relapse-free survival [108]. Furthermore, Kündig P et al. reported that the cytoplasmic expression of SOX9 was significantly associated with histological grade and may function as a prognostic marker of BC patients [135]. All of the above studies indicate the potential of SOX9 as a prognostic marker (Table 2b). These studies opened a new way of thinking about SOX9 as a diagnostic marker, and more studies will be needed to confirm this activity.
Table 2b.
Molecular Prognostic Effect Associated with SOX9
| Prognostic role | SOX9 expression | Sample size | Patients sample type | Effect | Refs |
|---|---|---|---|---|---|
| Poor | Up | >200 | Tissue | Associated with higher grade and ER negative tumor. Significantly shorter OS. Cytoplasmic accumulation associated with high proliferation for IDC and metastatic BC | [106] |
| Poor | Up | 306 | Tissue | Co-expression of Slug and Sox9 associated with poor overall survival | [19] |
| Poor | Up | 114 | Tissue | Associated with tumor subtype (high expression in TNBC) and histological grade | [46] |
| Poor | Up | 84 | Tissue (stroma) | Strong stromal expression after chemotherapy associated with shorter overall survival | [132] |
| Poor | Up | 420 | Tissue | High expression with high stem cell population associated with poor OS and DFS | [133] |
| Up in TNBC | 617 | Tissue | No significant association | [134] | |
| Poor | Up | 3951 | Tissue | Strong indicator of poor 5-years relapse free survival | [108] |
OS, overall survival; DFS, disease free survival; BC, breast cancer; IDC, invasive ductal carcinoma; TNBC, triple negative BC
6. Conclusions
SOX9 plays a crucial role in the regulation of several different processes during cancer pathogenesis, such as cell growth, apoptosis, migration, invasion, stemness, drug resistance, and immune escape. The expression of transcriptional factor SOX9 is significantly induced in BC patient samples. However, one investigation using a BC cell line determined that SOX9 was involved in growth retardation. In most cases, the activity of SOX9 during BC progression is maintained by tumor-suppressive miRNA, but a few proteins can also regulate its expression. Various studies have demonstrated that SOX9-related breast carcinomas exhibit treatment failure due to drug resistance and induction of stemness; therefore, SOX9 contributes to a poorer prognosis for patients with BC. Considering the functional activities of SOX9 during BC pathogenesis, it is apparent that SOX9 may represent a critical target for drug development. Additionally, measurable evidence for the expression of SOX9 protein within the exosome might be imperative from a prognostic point of view. However, several vital questions must be addressed to elucidate the influence of SOX9 during BC development and to inform the design of SOX9-targeting drugs, which may open a new era for combinatorial treatment with existing endocrine therapies.
Acknowledgements:
This work was supported in part by grants from the United States Department of Defense (W81XWH-16-1-0641) and the National Cancer Institute of the National Institutes of Health (P30CA33572). Funding from the Beckman Research Institute of City of Hope is also acknowledged.
Abbreviations:
- 3’UTR
3’untranslated region
- ALDH1A
aldehyde dehydrogenase 1 family member A1
- ALDH1A3
aldehyde dehydrogenase 1 family member A3
- ARE
AU-rich element
- BC
breast cancer
- CDH17
cadherin 17
- CDH7
cadherin 7
- cKit
KIT proto-oncogene, receptor tyrosine kinase
- CMPD
campomelic dysplasia
- CSCs
cancer stem cells
- CXCR4
C-X-C motif chemokine receptor 4
- DCIS
ductal carcinoma in situ
- DIM
dimerization domain
- DOX
doxorubicin
- ELF5
E74-like ETS transcription factor 5
- ER
estrogen receptor
- ER81/ETV1
ETS variant transcription factor 1
- ERα/ESR1
estrogen receptor 1
- EV
extracellular vesicle
- EVI1
ecotropic viral integration site-1
- FMNL2
formin-like 2
- FSCN1
fascin actin-bundling protein 1
- FXYD3
FXYD domain-containing ion transport regulator 3
- HDAC9
histone deacetylase 9
- HMG
high-mobility group
- HuR/ELAVL1
ELAV-like RNA binding protein 1
- IDC
invasive ductal carcinoma
- KRAS
KRAS proto-oncogene
- LAM
lymphangioleiomyomatosis
- LBP
lipopolysaccharide binding protein
- LCC
latency-competent cancer
- lncRNAs
long noncoding RNAs
- LRP6
LDL receptor related protein 6
- MAPK3
mitogen-activated protein kinase 3
- MaSC
mammary stem cell
- MAT2A
methionine adenosyl-transferase 2A
- miRNAs
microRNAs
- NANOG
Nanog homeobox
- NES
nuclear export signal
- NFE2L3
nuclear factor, erythroid 2-like 3
- NKG2D
natural killer group 2 member D
- NLS
nuclear localization signal
- NOTCH1
notch receptor 1
- OCT4/ POU5F1
POU class 5 homeobox 1
- ONECUT2
one cut homeobox 2
- OS
overall survival
- PI3K
phosphoinositide 3-kinase
- PML
promyelocytic leukemia
- PR
progesterone receptor
- PTX
paclitaxel
- RA
retinoic acid
- RAPTOR
regulatory associated protein of mTOR complex 1
- RUNX2
RUNX family transcription factor 2
- SNAI2 (Slug)
snail family transcriptional repressor 2
- SOX2
SRY-box transcription factor 2
- SOX9
SRY-related high-mobility group box 9
- SPARC
secreted protein acidic and cysteine rich
- SRY
sex-determining region Y
- TAC
transactivation domain C-terminus
- TAM
tamoxifen
- TCF4
transcription factor 4
- TF
transcription factor
- TNBC
triple-negative breast cancer
- VEGF
vascular endothelial growth factor A
- WISP2/CCN5
cellular communication network factor 5
- WNT5A
Wnt family member 5A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests:The authors declare that they have no competing interests.
Availability of data and materials: Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
References
- 1.Siegel RL, Miller KD, Jemal A (2019). Cancer statistics, 2019. CA Cancer J Clin,69(1), 7–34. [DOI] [PubMed] [Google Scholar]
- 2.Gupta GP, Massagué J (2006). Cancer metastasis: building a framework. Cell, 127(4), 679–95. [DOI] [PubMed] [Google Scholar]
- 3.Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Münsterberg A, et al. (1990). A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature, 346(6281), 245–50. [DOI] [PubMed] [Google Scholar]
- 4.Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, et al. (1990). A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature, 346(6281), 240–4. [DOI] [PubMed] [Google Scholar]
- 5.Wegner M (2010). All purpose Sox: The many roles of Sox proteins in gene expression. Int J Biochem Cell Biol,42(3), 381–90 [DOI] [PubMed] [Google Scholar]
- 6.Weina K, Utikal J (2014). SOX2 and cancer: current research and its implications in the clinic. Clin Transl Med, 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dy P, Wang W, Bhattaram P, Wang Q, Wang L, Ballock RT, et al. (2012). Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev Cell, 22(3), 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yagi R, McBurney D, Horton WE Jr. (2005). Bcl-2 positively regulates Sox9-dependent chondrocyte gene expression by suppressing the MEK-ERK1/2 signaling pathway. J Biol Chem, 280(34), 30517–25. [DOI] [PubMed] [Google Scholar]
- 9.Finzsch M, Stolt CC, Lommes P, Wegner M (2008). Sox9 and Sox10 influence survival and migration of oligodendrocyte precursors in the spinal cord by regulating PDGF receptor alpha expression. Development, 135(4), 637–46. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Leav I, Ibaragi S, Wegner M, Hu GF, Lu ML, et al. (2008). SOX9 is expressed in human fetal prostate epithelium and enhances prostate cancer invasion. Cancer Res, 68(6), 1625–30. [DOI] [PubMed] [Google Scholar]
- 11.Chen H, Garbutt CC, Spentzos D, Choy E, Hornicek FJ, Duan Z (2017). Expression and therapeutic potential of SOX9 in chordoma. Clin Cancer Res, 23(17), 5176–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung M, Briscoe J (2003). Neural crest development is regulated by the transcription factor Sox9. Development, 130(23), 5681–93. [DOI] [PubMed] [Google Scholar]
- 13.Sugiyama M, Yoshizumi T, Yoshida Y, Bekki Y, Matsumoto Y, Yoshiya S, et al. (2017). p62 promotes amino acid sensitivity of mTOR pathway and hepatic differentiation in adult liver stem/progenitor cells. J Cell Physiol, 232(8), 2112–24. [DOI] [PubMed] [Google Scholar]
- 14.Liang Z, Bian X, Shim H (2016). Downregulation of microRNA-206 promotes invasion and angiogenesis of triple negative breast cancer. Biochem Biophys Res Commun, 477(3), 461–6. [DOI] [PubMed] [Google Scholar]
- 15.Malladi S, Macalinao DG, Jin X, He L, Basnet H, Zou Y, et al. (2016). Metastatic latency and immune evasion through autocrine inhibition of WNT. Cell, 165(1), 45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tani M, Shindo-Okada N, Hashimoto Y, Shiroishi T, Takenoshita S, Nagamachi Y, et al. (1997). Isolation of a novel Sry-related gene that is expressed in high-metastatic K-1735 murine melanoma cells. Genomics, 39(1), 30–7. [DOI] [PubMed] [Google Scholar]
- 17.Aleman A, Adrien L, Lopez-Serra L, Cordon-Cardo C, Esteller M, Belbin TJ, et al. (2008). Identification of DNA hypermethylation of SOX9 in association with bladder cancer progression using CpG microarrays. Br J Cancer, 98(2), 466–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kordes U, Hagel C (2006). Expression of SOX9 and SOX10 in central neuro epithelial tumor. J Neurooncol, 80(2), 151–5. [DOI] [PubMed] [Google Scholar]
- 19.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. (2012). Slug and Sox9 cooperatively determine the mammary stem cell state. Cell, 148(5), 1015–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang HY, Lian P, Zheng PS (2015). SOX9, a potential tumor suppressor in cervical cancer, transactivates p21WAF1/CIP1 and suppresses cervical tumor growth. Oncotarget, 6(24), 20711–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jay P, Berta P, Blache P (2005). Expression of the carcinoembryonic antigen gene is inhibited by SOX9 in human colon carcinoma cells. Cancer Res, 65(6), 2193–8. [DOI] [PubMed] [Google Scholar]
- 22.Söderström M, Böhling T, Ekfors T, Nelimarkka L, Aro HT, Vuorio E (2002). Molecular profiling of human chondrosarcomas for matrix production and cancer markers. Int J Cancer, 100(2), 144–51. [DOI] [PubMed] [Google Scholar]
- 23.Song S, Maru DM, Ajani JA, Chan CH, Honjo S, Lin HK, et al. (2013). Loss of TGF-β adaptor β2SP activates notch signaling and SOX9 expression in esophageal adenocarcinoma. Cancer Res, 73(7), 2159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saegusa M, Hashimura M, Suzuki E, Yoshida T, Kuwata T (2012). Transcriptional up-regulation of Sox9 by NF-κB in endometrial carcinoma cells, modulating cell proliferation through alteration in the p14(ARF)/p53/p21(WAF1) pathway. Am J Pathol, 181(2), 684–92. [DOI] [PubMed] [Google Scholar]
- 25.Santos JC, Carrasco-Garcia E, Garcia-Puga M, Aldaz P, Montes M, Fernandez-Reyes M, et al. (2016). SOX9 elevation acts with canonical WNT signaling to drive gastric cancer progression. Cancer Res, 76(22), 6735–46. [DOI] [PubMed] [Google Scholar]
- 26.Yu CC, Tsai LL, Wang ML, Yu CH, Lo WL, Chang YC, et al. (2013). miR145 targets the SOX9/ADAM17 axis to inhibit tumor-initiating cells and IL-6-mediated paracrine effects in head and neck cancer. Cancer Res, 73(11), 3425–40. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Guo X, Xiong L, Kong X, Xu Y, Liu C, et al. (2012). MicroRNA-101 suppresses SOX9-dependent tumorigenicity and promotes favorable prognosis of human hepatocellular carcinoma. FEBS Lett, 586(24), 4362–70. [DOI] [PubMed] [Google Scholar]
- 28.Jiang SS, Fang WT, Hou YH, Huang SF, Yen BL, Chang JL, et al. (2010). Upregulation of SOX9 in lung adenocarcinoma and its involvement in the regulation of cell growth and tumorigenicity. Clin Cancer Res, 16(17), 4363–73. [DOI] [PubMed] [Google Scholar]
- 29.Malki S, Bibeau F, Notarnicola C, Roques S, Berta P, Poulat F, et al. (2007). Expression and biological role of the prostaglandin D synthase/SOX9 pathway in human ovarian cancer cells. Cancer Lett, 255, 182–93. [DOI] [PubMed] [Google Scholar]
- 30.Drivdahl R, Haugk KH, Sprenger CC, Nelson PS, Tennant MK, Plymate SR (2004). Suppression of growth and tumorigenicity in the prostate tumor cell line M12 by overexpression of the transcription factor SOX9. Oncogene, 23(26), 4584–93. [DOI] [PubMed] [Google Scholar]
- 31.Camaj P, Jäckel C, Krebs S, De Toni EN, Blum H, Jauch KW, et al. (2014). Hypoxia-independent gene expression mediated by SOX9 promotes aggressive pancreatic tumor biology. Mol Cancer Res, 12(3), 421–32. [DOI] [PubMed] [Google Scholar]
- 32.Li XL, Chen XQ, Zhang MN, Chen N, Nie L, Xu M, et al. (2015). SOX9 was involved in TKIs resistance in renal cell carcinoma via Raf/MEK/ERK signaling pathway. Int J Clin Exp Pathol, 8(4), 3871–81. [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J, Guo L (2017). Knockdown of SOX9 inhibits the proliferation, invasion, and EMT in thyroid cancer cells. Oncol Res, 25(2), 167–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afonja O, Raaka BM, Huang A, Das S, Zhao X, Helmer E, et al. (2002). RAR agonists stimulate SOX9 gene expression in breast cancer cell lines: evidence for a role in retinoid-mediated growth inhibition. Oncogene, 21(51), 7850–60. [DOI] [PubMed] [Google Scholar]
- 35.Wehrli BM, Huang W, De Crombrugghe B, Ayala AG, Czerniak B (2003). Sox9, a master regulator of chondrogenesis, distinguishes mesenchymal chondrosarcoma from other small blue round cell tumors. Hum Pathol, 34(3), 263–9. [DOI] [PubMed] [Google Scholar]
- 36.Acevedo VD, Gangula RD, Freeman KW, Li R, Zhang Y, Wang F, et al. (2007). Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell, 12(6), 559–71. [DOI] [PubMed] [Google Scholar]
- 37.Lü B, Fang Y, Xu J, Wang L, Xu F, Xu E, et al. (2008). Analysis of SOX9 expression in colorectal cancer. Am J Clin Pathol, 130(6), 897–904. [DOI] [PubMed] [Google Scholar]
- 38.Yasui W, Oue N, Sentani K, Sakamoto N, Motoshita J (2009). Transcriptome dissection of gastric cancer: identification of novel diagnostic and therapeutic targets from pathology specimens. Pathol Int, 59(3), 121–36. [DOI] [PubMed] [Google Scholar]
- 39.Passeron T, Valencia JC, Namiki T, Vieira WD, Passeron H, Miyamura Y, et al. (2009). Upregulation of SOX9 inhibits the growth of human and mouse melanomas and restores their sensitivity to retinoic acid. J Clin Invest, 119(4), 954–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coustry F, Oh CD, Hattori T, Maity SN, de Crombrugghe B, Yasuda H (2010). The dimerization domain of SOX9 is required for transcription activation of a chondrocyte-specific chromatin DNA template. Nucleic Acids Res, 38(18), 6018–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Moroz K, Makridakis NM, Lloyd SA, Galvez SE, Canavello PR, et al. (2011). Prognostic significance of cytoplasmic SOX9 in invasive ductal carcinoma and metastatic breast cancer. Exp Biol Med, 236(2), 145–55. [DOI] [PubMed] [Google Scholar]
- 42.Zhou CJ, Guo JQ, Zhu KX, Zhang QH, Pan CR, Xu WH, et al. (2011). Elevated expression of SOX9 is related with the progression of gastric carcinoma. Diagn Cytopathol, 39(2), 105–9. [DOI] [PubMed] [Google Scholar]
- 43.Abdel-Samad R, Zalzali H, Rammah C, Giraud J, Naudin C, Dupasquier S, et al. (2011). MiniSOX9, a dominant-negative variant in colon cancer cells. Oncogene, 30(22), 2493–503. [DOI] [PubMed] [Google Scholar]
- 44.Clemons NJ, Wang DH, Croagh D, Tikoo A, Fennell CM, Murone C, et al. (2012). Sox9 drives columnar differentiation of esophageal squamous epithelium: a possible role in the pathogenesis of Barrett’s esophagus. Am J Physiol Gastrointest Liver Physiol 303(12), G1335–46. [DOI] [PubMed] [Google Scholar]
- 45.Kopp JL, von Figura G, Mayes E, Liu FF, Dubois CL, Morris JP 4th., et al. (2012). Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 22(6), 737–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H, He L, Ma F, Regan MM, Balk SP, Richardson AL, et al. (2013). SOX9 regulates low density lipoprotein receptor-related protein 6 (LRP6) and T-cell factor 4 (TCF4) expression and Wnt/β-catenin activation in breast cancer. J Biol Chem, 288(9), 6478–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Q, Yao Y, Eades G, Liu Z, Zhang Y, Zhou Q (2014). Downregulation of miR-140 promotes cancer stem cell formation in basal-like early stage breast cancer. Oncogene, 33(20), 2589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sosa MS, Parikh F, Maia AG, Estrada Y, Bosch A, Bragado P, et al. (2015). NR2F1 controls tumour cell dormancy via SOX9-and RARβ-driven quiescence programmes. Nat Commun, 6, 6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luanpitpong S, Li J, Manke A, Brundage K, Ellis E, McLaughlin SL, et al. (2016). SLUG is required for SOX9 stabilization and functions to promote cancer stem cells and metastasis in human lung carcinoma. Oncogene, 35(22), 2824–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meng Q, Ren M, Li Y, Song X (2016). LncRNA-RMRP acts as an oncogene in lung cancer. PLoS One, 11(12), e0164845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wan YP, Xi M, He HC, Wan S, Hua W, Zen ZC, et al. (2017). Expression and clinical significance of SOX9 in renal cell carcinoma, bladder cancer and penile cancer. Oncol Res Treat, 40(1–2), 15–20. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, Yu X, Zhang Z, Pang L, Xu J, Jiang J, et al. (2017). Linc-ROR promotes esophageal squamous cell carcinoma progression through the derepression of SOX9. J Exp Clin Cancer Res, 36(1), 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan X, Li J, Coulouarn C, Lin T, Sulpice L, Bergeat D, et al. (2018). SOX9 expression decreases survival of patients with intrahepatic cholangiocarcinoma by conferring chemoresistance. Br J Cancer, 119(11), 1358–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wen KW, Grenert JP, Joseph NM, Shafizadeh N, Huang A, Hosseini M, et al. (2018). Genomic profile of appendiceal goblet cell carcinoid is distinct compared to appendiceal neuroendocrine tumor and conventional adenocarcinoma. Hum Pathol, 77, 166–74. [DOI] [PubMed] [Google Scholar]
- 55.Sumita Y, Yamazaki M, Maruyama S, Abé T, Cheng J, Takagi R, et al. (2018). Cytoplasmic expression of SOX9 as a poor prognostic factor for oral squamous cell carcinoma. Oncol Rep, 40(5), 2487–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tariq A, Hao Q, Sun Q, Singh DK, Jadaliha M, Zhang Y, et al. (2019). LncRNA-mediated regulation of SOX9 expression in basal sub-type breast cancer cells. RNA. doi: 10.1261/rna.073254.119. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Symon A, Harley V (2017). SOX9: A genomic view of tissue specific expression and action. Int J Biochem Cell Biol, 87, 18–22. [DOI] [PubMed] [Google Scholar]
- 58.Kamachi Y, Kondoh H (2013). Sox proteins: regulators of cell fate specification and differentiation. Development, 140(20), 4129–44. [DOI] [PubMed] [Google Scholar]
- 59.Südbeck P, Scherer G (1997). Two independent nuclear localization signals are present in the DNA-binding high-mobility group domains of SRY and SOX9. J Biol Chem, 272(44), 27848–52. [DOI] [PubMed] [Google Scholar]
- 60.Gasca S, Canizares J, De Santa Barbara P, Mejean C, Poulat F, Berta P, et al. (2002). A nuclear export signal within the high mobility group domain regulates the nucleocytoplasmic translocation of SOX9 during sexual determination. Proc Natl Acad Sci U S A, 99(17), 11199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malki S, Boizet-Bonhoure B, Poulat F (2010). Shuttling of SOX proteins. Int J Biochem Cell Biol, 42(3), 411–6. [DOI] [PubMed] [Google Scholar]
- 62.Jo A, Denduluri S, Zhang B, Wang Z, Yin L, Yan Z, et al. (2014). The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis, 1(2), 149–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang YH, Jankowski A, Cheah KS, Prabhakar S, Jauch R (2015). SOXE transcription factors form selective dimers on non-compact DNA motifs through multifaceted interactions between dimerization and high-mobility group domains. Sci Rep, 5, 10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, et al. (1994). Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell, 79(6), 1111–20. [DOI] [PubMed] [Google Scholar]
- 65.Hiramatsu R, Matoba S, Kanai-Azuma M, Tsunekawa N, Katoh-Fukui Y, Kurohmaru M, et al. (2009). A critical time window of Sry action in gonadal sex determination in mice. Development, 136(1), 129–38. [DOI] [PubMed] [Google Scholar]
- 66.Lefebvre V, de Crombrugghe B (1998). Toward understanding SOX9 function in chondrocyte differentiation. Matrix Biol, 16(9), 529–40. [DOI] [PubMed] [Google Scholar]
- 67.De Santa Barbara P, Bonneaud N, Boizet B, Desclozeaux M, Moniot B, Sudbeck P, et al. (1998). Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Müllerian hormone gene. Mol Cell Biol, 18(11), 6653–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vidal VP, Chaboissier MC, Lützkendorf S, Cotsarelis G, Mill P, Hui CC, et al. (2005). Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr Biol, 15(15), 1340–51. [DOI] [PubMed] [Google Scholar]
- 69.Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M (2003). The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev, 17(13), 1677–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Poché RA, Furuta Y, Chaboissier MC, Schedl A, Behringer RR (2008). Sox9 is expressed in mouse multipotent retinal progenitor cells and functions in Müller glial cell development. J Comp Neurol, 510(3), 237–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perl AK, Kist R, Shan Z, Scherer G, Whitsett JA (2005). Normal lung development and function after Sox9 inactivation in the respiratory epithelium. Genesis. 41(1):23–32. [DOI] [PubMed] [Google Scholar]
- 72.Seymour PA, Freude KK, Tran MN, Mayes EE, Jensen J, Kist R, et al. (2007). SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci USA, 104(6), 1865–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lincoln J, Kist R, Scherer G, Yutzey KE (2007). Sox9 is required for precursor cell expansion and extracellular matrix organization during mouse heart valve development. Dev Biol, 305(1), 120–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reginensi A, Clarkson M, Neirijnck Y, Lu B, Ohyama T, Groves AK, et al. (2011). SOX9 controls epithelial branching by activating RET effector genes during kidney development. Hum Mol Genet, 20(6), 1143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heichman KA, Warren JD (2012). DNA methylation biomarkers and their utility for solid cancer diagnostics. Clin Chem Lab Med, 50(10), 1707–21 [DOI] [PubMed] [Google Scholar]
- 76.Sun M, Uozaki H, Hino R, Kunita A, Shinozaki A, Ushiku T, et al. (2012). SOX9 expression and its methylation status in gastric cancer. Virchows Arch, 460(3), 271–9. [DOI] [PubMed] [Google Scholar]
- 77.Wu JH, Liang XA, Wu YM, Li FS, Dai YM (2013). Identification of DNA methylation of SOX9 in cervical cancer using methylated-CpG island recovery assay. Oncol Rep, 29(1), 125–32. [DOI] [PubMed] [Google Scholar]
- 78.Sun L, Mathews LA, Cabarcas SM, Zhang X, Yang A, Zhang Y, et al. (2013). Epigenetic regulation of SOX9 by the NF-κB signaling pathway in pancreatic cancer stem cells. Stem Cells, 31(8), 1454–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheng PF, Shakhova O, Widmer DS, Eichhoff OM, Zingg D, Frommel SC, et al. (2015). Methylation-dependent SOX9 expression mediates invasion in human melanoma cells and is a negative prognostic factor in advanced melanoma. Genome Biol, 16, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang H, Dong S, Feng J (2019). Epigenetic profiling and mRNA expression reveal candidate genes as biomarkers for colorectal cancer. J Cell Biochem, 120(6), 10767–76. [DOI] [PubMed] [Google Scholar]
- 81.Rauscher GH, Kresovich JK, Poulin M, Yan L, Macias V, Mahmoud AM, et al. (2015). Exploring DNA methylation changes in promoter, intragenic, and intergenic regions as early and late events in breast cancer formation. BMC Cancer, 15, 816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luo Y, Huang J, Tang Y, Luo X, Ge L, Sheng X, et al. (2019). Regional methylome profiling reveals dynamic epigenetic heterogeneity and convergent hypomethylation of stem cell quiescence-associated genes in breast cancer following neoadjuvant chemotherapy. Cell Biosci, 9, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lapierre M, Linares A, Dalvai M, Duraffourd C, Bonnet S, Boulahtouf A, et al. (2016). Histone deacetylase 9 regulates breast cancer cell proliferation and the response to histone deacetylase inhibitors. Oncotarget, 7(15), 19693–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martín-Martín N, Piva M, Urosevic J, Aldaz P, Sutherland JD, Fernández-Ruiz S, et al. (2016). Stratification and therapeutic potential of PML in metastatic breast cancer. Nat Commun, 7, 12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mateo F, Arenas EJ, Aguilar H, Serra-Musach J, de Garibay GR, et al. (2017). Stem cell-like transcriptional reprogramming mediates metastatic resistance to mTOR inhibition. Oncogene, 36(19), 2737–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jeselsohn R, Cornwell M, Pun M, Buchwalter G, Nguyen M, Bango C, et al. (2017). Embryonic transcription factor SOX9 drives breast cancer endocrine resistance. Proc Natl AcadSci U S A, 114(22), E4482–E4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bartel DP (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116(2), 281–97. [DOI] [PubMed] [Google Scholar]
- 88.Jana S, Sengupta S, Biswas S, Chatterjee A, Roy H, Bhattacharyya A (2017). miR-216b suppresses breast cancer growth and metastasis by targeting SDCBP. Biochem Biophys Res Commun, 482(1), 126–133. [DOI] [PubMed] [Google Scholar]
- 89.Chatterjee A, Jana S, Chatterjee S, Wastall LM, Mandal G, Nargis N, et al. (2019). MicroRNA-222 reprogrammed cancer-associated fibroblasts enhance growth and metastasis of breast cancer. Br J Cancer, 121(8), 679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang YJ, Xu F, Zhang YJ, Li HB, Han JC, Li L (2015). miR-206 inhibits non small cell lung cancer cell proliferation and invasion by targeting SOX9. Int J Clin Exp Med, 8(6), 9107–13. [PMC free article] [PubMed] [Google Scholar]
- 91.Sharma S, Verma S, Vasudevan M, Samanta S, Thakur JK, Kulshreshtha R (2013). The interplay of HuR and miR-3134 in regulation of AU rich transcriptome. RNA Biol, 10(8), 1283–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gernapudi R, Yao Y, Zhang Y, Wolfson B, Roy S, Duru N, et al. (2015). Targeting exosomes from preadipocytes inhibits preadipocyte to cancer stem cell signaling in early-stage breast cancer. Breast Cancer Res Treat, 150(3), 685–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Santos JC, Lima NDS, Sarian LO, Matheu A, Ribeiro ML, Derchain SFM (2018). Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci Rep, 8(1), 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang QY, Zhou CX, Zhan MN, Tang J, Wang CL, Ma CN, et al. (2018). MiR-133b targets Sox9 to control pathogenesis and metastasis of breast cancer. Cell Death Dis, 9(7), 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao Y, Pang W, Yang N, Hao L, Wang L (2018). MicroRNA-511 inhibits malignant behaviors of breast cancer by directly targeting SOX9 and regulating the PI3K/Akt pathway. Int J Oncol, 53(6), 2715–26. [DOI] [PubMed] [Google Scholar]
- 96.Yu Y, Yin W, Yu ZH, Zhou YJ, Chi JR, Ge J, et al. (2019). miR-190 enhances endocrine therapy sensitivity by regulating SOX9 expression in breast cancer. J Exp Clin Cancer Res. 38(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shen M, Dong C, Ruan X, Yan W, Cao M, Pizzo D, et al. (2019). Chemotherapyinduced extracellular vesicle miRNAs promote breast cancer stemness by targeting ONECUT2. Cancer Res, 79(14), 3608–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gao JB, Zhu MN, Zhu XL (2019). miRNA-215–5p suppresses the aggressiveness of breast cancer cells by targeting Sox9. FEBS Open Bio, 9(11):1957–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang QY, Liu GF, Qian XL, Tang LB, Huang QY, Xiong LX (2019). Long non-coding RNA: dual effects on breast cancer metastasis and clinical applications. Cancers (Basel), 16, 11(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yao N, Fu Y, Chen L, Liu Z, He J, Zhu Y, Xia T, Wang S (2019). Long non-coding RNA NONHSAT101069 promotes epirubicin resistance, migration, and invasion of breast cancer cells through NONHSAT101069/miR-129–5p/Twist1 axis. Oncogene, 38(47), 7216–33. [DOI] [PubMed] [Google Scholar]
- 101.Cui Y, Zhang F, Zhu C, Geng L, Tian T, Liu H (2017). Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/β-catenin signaling pathway. Oncotarget, 8(11), 17785–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mondal T, Juvvuna PK, Kirkeby A, Mitra S, Kosalai ST, Traxler L, et al. Sense-Antisense lncRNA Pair Encoded by Locus 6p22.3 Determines Neuroblastoma Susceptibility via the USP36-CHD7-SOX9 Regulatory Axis. Cancer Cell, 33(3), 417–434.e7. [DOI] [PubMed] [Google Scholar]
- 103.Yan S, Shan X, Chen K, Liu Y, Yu G, Chen Q, et al. (2018). LINC00052/miR-101-3p axis inhibits cell proliferation and metastasis by targeting SOX9 in hepatocellular carcinoma. Gene, 679, 138–49. [DOI] [PubMed] [Google Scholar]
- 104.Thomsen MK, Ambroisine L, Wynn S, Cheah KS, Foster CS, Fisher G, et al. (2010). Transatlantic Prostate Group. SOX9 elevation in the prostate promotes proliferation and cooperates with PTEN loss to drive tumor formation. Cancer Res, 70(3), 979–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Müller P, Crofts JD, Newman BS, Bridgewater LC, Lin CY, Gustafsson JA, et al. (2010). SOX9 mediates the retinoic acid-induced HES-1 gene expression in human breast cancer cells. Breast Cancer Res Treat, 120(2), 317–26. [DOI] [PubMed] [Google Scholar]
- 106.Chakravarty G, Moroz K, Makridakis NM, Lloyd SA, Galvez SE, Canavello PR, et al. (2011). Prognostic significance of cytoplasmic SOX9 in invasive ductal carcinoma and metastatic breast cancer. ExpBiol Med (Maywood), 236(2), 145–55. [DOI] [PubMed] [Google Scholar]
- 107.Rwigemera A, Mamelona J, Martin LJ (2014). Inhibitory effects of fucoxanthinol on the viability of human breast cancer cell lines MCF7 and MDA-MB231 are correlated with modulation of the NFκB pathway. Cell Biol Toxicol, 30(3), 157–67. [DOI] [PubMed] [Google Scholar]
- 108.Richtig G, Aigelsreiter A, Schwarzenbacher D, Ress AL, Adiprasito JB, Stiegelbauer V, et al. (2017). SOX9 is a proliferation and stem cell factor in hepatocellular carcinoma and possess widespread prognostic significance in different cancer types. PLoS One, 12(11), e0187814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Domenici G, Aurrekoetxea-Rodríguez I, Simões BM, Rábano M, Lee SY, Millán JS, et al. (2019). A Sox2-Sox9 signalling axis maintains human breast luminal progenitor and breast cancer stem cells. Oncogene, 38(17), 3151–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hickman JA (1992). Apoptosis induced by anticancer drugs. Cancer Metastasis Rev, 11(2), 121–39. [DOI] [PubMed] [Google Scholar]
- 111.Rwigemera A, Mamelona J, Martin LJ (2015). Comparative effects between fucoxanthinol and its precursor fucoxanthin on viability and apoptosis of breast cancer cell lines MCF7 and MDA-MB231. Anticancer Res, 35(1), 207–19. [PubMed] [Google Scholar]
- 112.Endo Y, Deonauth K, Prahalad P, Hoxter B, Zhu Y, Byers SW (2008). Role of Sox-9, ER81 and VE-cadherin in retinoic acid-mediated trans-differentiation of breast cancer cells. PLoS One, 3(7), e2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hanahan D, Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell, 144(5), 646–74. [DOI] [PubMed] [Google Scholar]
- 114.Kerbel RS (2008). Tumor angiogenesis. N Engl J Med, 358(19), 2039–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Carmeliet P, Collen D (2000). Molecular basis of angiogenesis. Role of VEGF and VE-cadherin. Ann N Y Acad Sci, 902, 249–62. [DOI] [PubMed] [Google Scholar]
- 116.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. (2007). Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res, 13(15 Pt 1), 4429–34. [DOI] [PubMed] [Google Scholar]
- 117.Acar A, Simões BM, Clarke RB, Brennan K (2016). A role for Notch signalling in breast cancer and endocrine resistance. Stem Cells Int, 2016, 2498764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xue Y, Lai L, Lian W, Tu X, Zhou J, Dong P, et al. (2019). SOX9/FXYD3/Src Axis Is Critical for ER+ Breast Cancer Stem Cell Function. Mol Cancer Res, 17(1), 238–49. [DOI] [PubMed] [Google Scholar]
- 119.Visvader JE, Lindeman GJ (2008). Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer, 8(10), 755–68. [DOI] [PubMed] [Google Scholar]
- 120.O’Brien CS, Farnie G, Howell SJ, Clarke RB (2011). Breast cancer stem cells and their role in resistance to endocrine therapy. Horm Cancer, 2(2), 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.García Bueno JM, Ocaña A, Castro-García P, Gil Gas C, Sánchez-Sánchez F, Poblet E, et al. (2008). An update on the biology of cancer stem cells in breast cancer. Clin Transl Oncol, 10(12), 786–93. [DOI] [PubMed] [Google Scholar]
- 122.Ferrand N, Gnanapragasam A, Dorothee G, Redeuilh G, Larsen AK, Sabbah M (2014). Loss of WISP2/CCN5 in estrogen-dependent MCF7 human breast cancer cells promotes a stem-like cell phenotype. PLoS One, 9(2), e87878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cai X, Dai Z, Reeves RS, Caballero-Benitez A, Duran KL, Delrow JJ, et al. (2014). Autonomous stimulation of cancer cell plasticity by the human NKG2D lymphocyte receptor coexpressed with its ligands on cancer cells. PLoS One, 9(10), e108942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ruiz de Garibay G, Herranz C, Llorente A, Boni J, Serra-Musach J, Mateo F, et al. (2018). Lymphangioleiomyomatosis biomarkers linked to lung metastatic potential and cell stemness. PLoS One, 13(11), e0207586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fazilaty H, Gardaneh M, Akbari P, Zekri A, Behnam B (2016). SLUG and SOX9 cooperatively regulate tumor initiating niche factors in breast cancer. Cancer Microenviron, 9(1), 71–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang C, Christin JR, Oktay MH, Guo W (2017). Lineage-biased stem cells maintain estrogen-receptor-positive and -negative mouse mammary luminal lineages. Cell Rep, 18(12), 2825–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang C, Zhao X, Cui N, Liang Y (2017). Cadherins associate with distinct stem cell-related transcription factors to coordinate the maintenance of stemness in triple-negative breast cancer. Stem Cells Int, 2017, 5091541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gauck D, Keil S, Niggemann B, Zänker KS, Dittmar T (2017). Hybrid clone cells derived from human breast epithelial cells and human breast cancer cells exhibit properties of cancer stem/initiating cells. BMC Cancer, 17(1), 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Strekalova E, Malin D, Weisenhorn EMM, Russell JD, Hoelper D, Jain A, et al. (2019). S-adenosylmethionine biosynthesis is a targetable metabolic vulnerability of cancer stem cells. Breast Cancer Res Treat, 175(1), 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mittal D, Gubin MM, Schreiber RD, Smyth MJ (2014). New insights into cancer immuno-editing and its three component phases--elimination, equilibrium and escape. Curr Opin Immunol, 27, 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mittal S, Brown NJ, Holen I (2018). The breast tumor microenvironment: role in cancer development, progression and response to therapy. Expert Rev Mol Diagn, 18(3), 227–43. [DOI] [PubMed] [Google Scholar]
- 132.Riemenschnitter C, Teleki I, Tischler V, Guo W, Varga Z (2013). Stability and prognostic value of Slug, Sox9 and Sox10 expression in breast cancers treated with neoadjuvant chemotherapy. Springerplus, 2, 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lei B, Zhang Y, Liu T, Li Y, Pang D (2016). Sox9 upregulation in breast cancer is correlated with poor prognosis and the CD44+/CD24−/low phenotype. Int J Clin Exp Pathol, 9(7), 7345–51. [Google Scholar]
- 134.Pomp V, Leo C, Mauracher A, Korol D, Guo W, Varga Z (2015). Differential expression of epithelial–mesenchymal transition and stem cell markers in intrinsic subtypes of breast cancer. Breast Cancer Res Treat, 154(1), 45–55. [DOI] [PubMed] [Google Scholar]
- 135.Kündig P, Giesen C, Jackson H, Bodenmiller B, Papassotirolopus B, Freiberger SN, et al. (2018). Limited utility of tissue micro-arrays in detecting intra-tumoral heterogeneity in stem cell characteristics and tumor progression markers in breast cancer. J Transl Med, 16(1), 180. [DOI] [PMC free article] [PubMed] [Google Scholar]


