Abstract
Coculture of human melanocytes with keratinocytes upregulates CCN3, a matricellular protein critical to maintenance of normal homeostasis of melanocytes in the skin. CCN3 affects two fundamental features of melanocyte physiology: it inhibits melanocyte proliferation and stimulates their adhesion to the basement membrane. Here we report that expression of CCN3 is downregulated in advanced melanomas. Aggressive melanoma cell lines did not respond to treatment with CCN3 inducers, such as interleukin-1β (IL-1β), while less aggressive melanoma cell lines responded similarly to melanocytes. Immunostaining analyses revealed that CCN3 was present in melanoma cells close to the epidermal–dermal interface, but not in melanoma cells that had invaded deep into the dermis or had metastasized to lymph nodes. Contrary to our expectations, overexpression of CCN3 in 1205Lu metastatic melanoma cells did not affect their adhesion to collagen IV. However, CCN3 decreased the transcription and activation of matrix metalloproteinases and suppressed the invasion of 1205Lu melanoma cells. These results suggest that the lack of CCN3 in advanced melanoma cells contributes to their invasive phenotype. Whereas major matricellular proteins, such as osteopontin, tenascin or secreted protein acidic and rich in cysteine (SPARC), are strongly upregulated in melanoma cells; CCN3 is the first member of this family that is downregulated.
Introduction
During development, melanocyte precursors migrate from the neural crest toward the epidermis, where they are arrested when they come into contact with keratinocytes. Differentiated human melanocytes remain tightly localized at the basement membrane and cannot survive within upper epidermal layers unless transformed to form nevi or melanomas. In turn, melanocyte homeostasis is strictly controlled by keratinocytes. Dysregulation of that homeostasis may disturb the balance of the ‘epidermal melanin unit’ and may trigger the continuous proliferation of melanocytes, which may lead to the development of melanoma. It is likely that melanoma cells escape from keratinocyte control through (1) downregulation of receptors important for their communication with and adhesion to keratinocytes (for example, E-cadherin); (2) upregulation of receptors and signaling molecules not found on melanocytes but important for melanoma–melanoma and melanoma–fibroblast interactions (for example, N-cadherin, melanoma cell adhesion molecule, zonula occludens protein-1) and (3) loss of anchorage to the basement membrane because of altered expression of extracellular matrix-binding proteins (Haass et al., 2005).
CCN3 (nephroblastoma overexpressed) is a matricellular protein that shares structural properties of insulin-like growth factor (IGF)-binding protein, von Willebrand factor type C repeats, thrombospondin type 1 repeats and secreted regulatory factors containing cysteine knot motifs with five other family members (Perbal, 2001). CCN3 is aberrantly expressed in several malignancies and is associated with the progression of prostate cancer (Maillard et al., 2001), renal cell carcinoma (Glukhova et al., 2001) and Ewing’s sarcoma (Manara et al., 2002), whereas in rhabdomyosarcoma and cartilage tumors, CCN3 expression correlates with tumor differentiation. Extensive studies have indicated that the biological properties of CCN3 are dependent upon the cellular context (Perbal et al., 2003). Thus, it is not surprising that CCN3 has diverse effects on tumorigenesis in different types of cancer. For example, CCN3 has antiproliferative activities in gliomas (Gupta et al., 2001) and in chronic myeloid leukemia cells (McCallum et al., 2006), whereas it promotes migration and invasion of Ewing’s sarcoma cells.
Recently we reported that CCN3 protein is upregulated in melanocytes after coculture with keratinocytes and that it affects two fundamental features of melanocyte physiology: it inhibits the proliferation of melanocytes and is required for the three-dimensional organization of the melanocyte network on the basement membrane of human skin (Fukunaga-Kalabis et al., 2006). Furthermore, CCN3 stimulates the adhesion of melanocytes to basement membrane collagen type IV but not to dermal collagen type I. Growth inhibition and basement membrane localization conferred by CCN3 are important, if not essential, function for maintaining melanocyte homeostasis in normal skin. Therefore, we hypothesized that dysregulation of CCN3 may be one of the initial events that disrupts the normal balance between melanocytes and keratinocytes and thus confers melanocytes with the capacity to escape that control that is one of the earliest contributors to transformation. In this study, we investigated the involvement of CCN3 in melanomas as well as in melanocytic nevi, which are nonmalignant melanocytic lesions often seen as precursors of malignant transformation. We observed that in vivo, CCN3 expression correlates inversely with melanoma invasion. Furthermore, CCN3 overexpression dramatically decreases the invasion of aggressive melanoma cells by inhibiting matrix metalloproteinase (MMP) expression.
Results
CCN3 expression is lost in invasive melanomas
To investigate the potential involvement of CCN3 in melanoma development and progression in situ, we performed immunohistochemical analyses on 19 melanoma and 21 melanocytic nevus specimens, including 9 superficial spreading melanomas (SSM), 5 nodular melanomas (NM), 5 lymph node metastases, 7 compound nevi, 12 intradermal nevi and 2 atypical nevi. HMB45 staining, which is specific for melanosomes, was used to identify the melanocytic tumors. CCN3 expression was identified in 16/21 nevi, 11/14 primary melanomas and 2/5 metastatic (Met) melanomas, and the percentage of CCN3-positive cells was scored (Supplementary Tables 1 and 2). In most compound nevi and SSM, strong staining was observed at the epidermal–dermal junction (Figure 1) and diffuse, weak staining was seen in dermal areas of the lesions. The majority of intradermal nevi (9/12) and NMs (4/5) presented diffuse faint staining (<10% of cells). None of the lymph node metastases revealed strong staining. Strikingly, CCN3 positivity was inversely correlated with Breslow’s depth (Breslow, 1970), one of the best indicators of melanoma prognosis. A total of eight melanomas less than 1 mm in thickness showed strong CCN3 expression. In contrast, CCN3 was absent in three melanomas more than 5 mm in thickness.
Figure 1.
Distribution of CCN3 expression in benign melanocytic nevi and in malignant melanoma. Immunohistochemistry was performed with antibodies specific for CCN3. Melanocytic cells were identified with HMB45 staining and count CCN3 positive cells in HMB45-positive tumor clusters. CCN3 is expressed at the epidermal–dermal junction in junctional nevi and in superficial spreading melanomas (SSM). Faint CCN3 expression is seen in intradermal nevi, in the dermal invasion portion of SSM, in nodular melanomas (NM) and in melanomas metastatic to lymph nodes (LN metastasis). Control: staining with rabbit sera (original magnification × 400, scale bar 100 μm).
CCN3 expression is downregulated in invasive melanoma cell lines
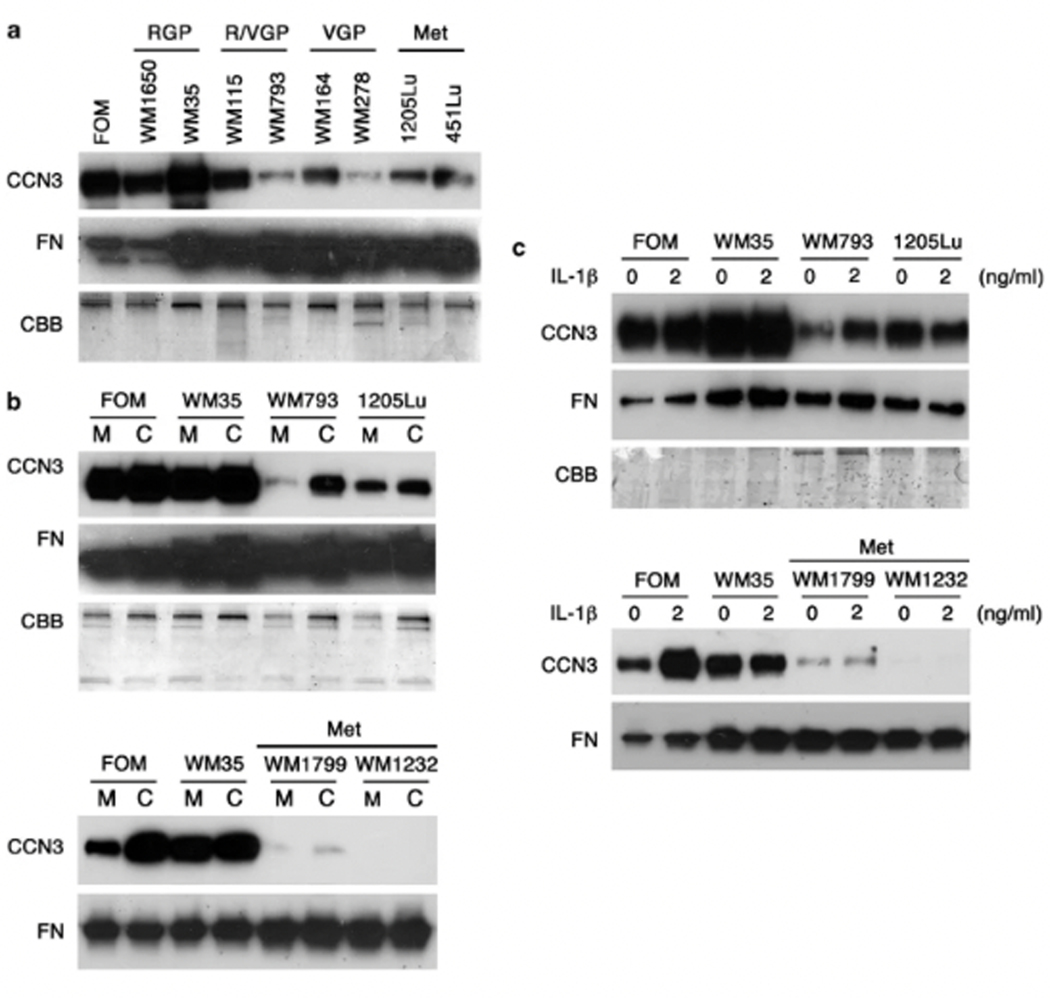
Next we examined whether CCN3 is expressed in melanoma cell lines. Since CCN3 is a secreted protein, conditioned medium from eight melanoma cell lines derived from different stages of progression were tested using western blotting against CCN3 (Figure 2a). Normal primary human melanocytes (foreskin melanocytes (FOM)) were used as a positive control. Compared with the level of CCN3 expression in melanocytes, nontumorigenic radial growth phase (RGP) melanoma cell lines also secreted robust levels of CCN3. As expected, decreased levels of CCN3 were observed in all invasive (vertical growth phase (VGP) and Met) melanoma cell lines, which corresponded with the results of staining in vivo. The proinflammatory cytokine interleukin-1β (IL-1β) produced by keratinocytes upregulates CCN3 in melanocytes (Fukunaga-Kalabis et al., 2006). CCN3 was slightly induced in VGP primary melanoma cell line WM793 cocultured with keratinocytes or treated with IL-1β, while Met melanoma cell lines (1205Lu, WM1799 and WM1232) weakly responded or did not respond at all with increased levels of CCN3 (Figures 2b and c). These data suggest that CCN3 expression is downregulated and can not be recovered to the normal melanocyte level by keratinocyte-derived factors in advanced melanomas.
Figure 2.
CCN3 expression in melanocytes and in melanoma cells. (a) Western blot analysis for levels of CCN3 secreted by human melanocytes or by melanoma cell lines. Decreased levels of CCN3 in melanoma cell lines derived from biologically advanced lesions compared with normal melanocytes. FOM, foreskin melanocytes; RGP, radial growth phase melanoma; R/VGP, radial and vertical growth phase melanoma; VGP, vertical growth phase melanoma; Met, metastatic melanoma. (b) CCN3 protein expression in melanocytes or in melanoma cells in monoculture (M) versus coculture (C) with normal human keratinocytes. Results are from Western blotting of culture medium. (c) Immunoblot of conditioned media from monocultured melanocytes and melanoma cells treated with IL-1β (2 ng ml−1) for 48 h. Fibronectin immunoblot (FN) and Coomassie blue staining (CBB) of Bis-Tris gels were performed as loading controls.
CCN3 overexpression decreases the invasion of melanoma cells
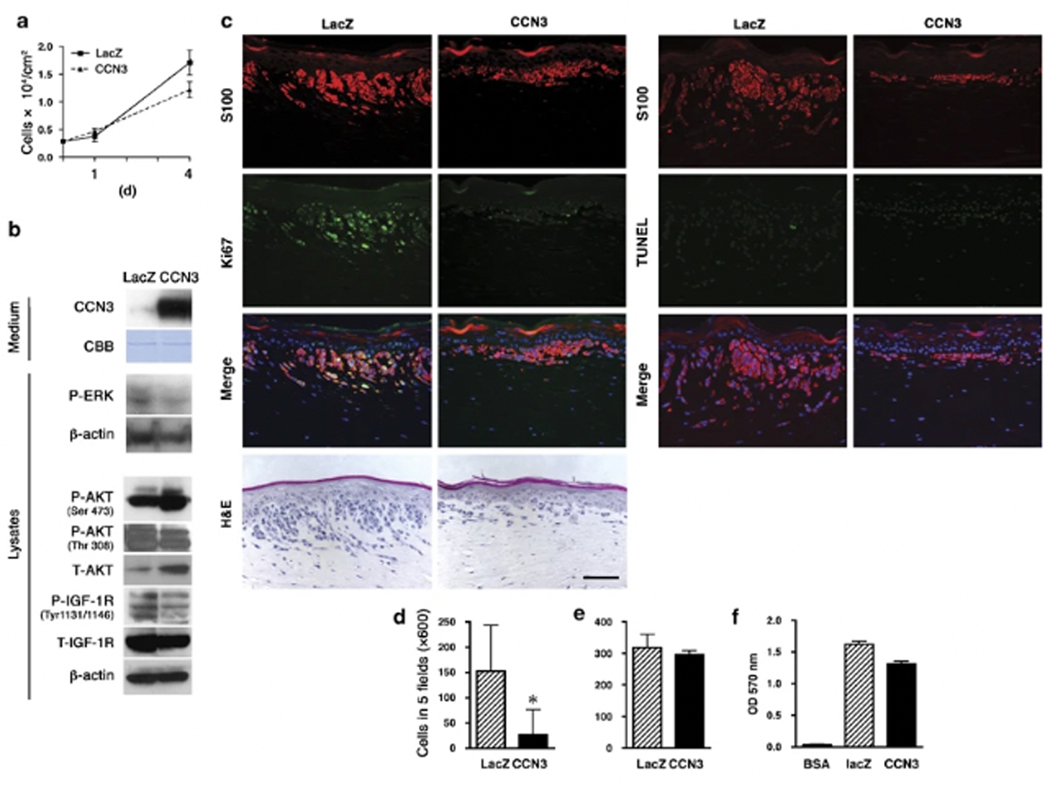
Since CCN3 has an antiproliferative effect on normal melanocytes and on several types of cancer, we hypothesized that CCN3 might also control melanoma cell growth, and we designed a lentiviral vector to overexpress it in melanoma cells. The 1205Lu melanoma cell line was selected because it has been exhaustively characterized in regards to its proliferative, invasive and Met properties, and its endogenous level of CCN3 expression is low. Cells infected with lentiviral vectors for CCN3 (CCN3–1205Lu) or LacZ as a control (LacZ-1205Lu) were selected using blasticidin (10 μg ml−1) and were characterized for CCN3 protein levels by western blotting. The effect of CCN3 overexpression on cellular growth in the presence of serum over a time course of 4 days was assessed by a proliferation assay. CCN3–1205Lu cells showed only a moderate (20–30%) decrease in growth compared to LacZ-1205Lu cells (P=0.042) (Figure 3a), despite the fact that phospho-extracellular-responsive kinase (P-ERK) was downregulated in CCN3–1205Lu cells (Figure 3b). It has been reported that the CCN family member CCN6 inhibited activation of IGF-1 receptor (IGF-1R) and decreased ERK phosphorylation (Kleer et al., 2004). In CCN3-overexpressing cells, IGF-1R phosphorylation was downregulated (Figure 3b). Activation of IGF-1R transduces signals to the mitogen-activated protein kinase pathway as well as to the Akt pathway, therefore it is conceivable that CCN3 modulates ERK signaling through IGF-1R. Surprisingly, in CCN3-overexpressing cells, Akt Ser473 phosphorylation was upregulated, whereas Thr308 phosphorylation was not changed. Total Akt levels were also increased in CCN3-overexpressing cells. These data suggest that upregulation of Akt Ser473 phosphorylation may be due to upregulation at the total expression level of Akt. Previous reports indicate that CCN family proteins regulate migration and invasion in diverse cell types (Lake et al., 2003; Laurent et al., 2003; Benini et al., 2005; Nguyen et al., 2006), and therefore we examined the invasion/migration of CCN3–1205Lu cells under physiological conditions by incorporating the transduced cells into three-dimensional skin reconstructs. Strikingly, control LacZ-1205Lu cells invaded deep into the dermis, whereas CCN3–1205Lu cells spread only horizontally and superficially into the dermis (Figure 3c, S100 staining and hematoxylin and eosin (H&E) staining). In three-dimensional growth conditions, CCN3–1205Lu cells were not only less invasive, but also less proliferative as they showed decreased expression of Ki67 (Figure 3c, left two columns). TdT-mediated dUTP nick end labelling (TUNEL) staining demonstrated that CCN3–1205Lu melanoma cells did not undergo apoptosis in skin reconstructs (Figure 3c, right two columns). Matrigel invasion assays showed a strong inhibition of invasion by CCN3–1205Lu cells (Figure 3d), while no significant difference was revealed in migration assays in the absence of Matrigel (Figure 3e). Since CCN3 influences the localization of melanocytes through collagen IV adhesion, we next hypothesized that CCN3–1205Lu cells might be trapped in Matrigel, which contains collagen type IV as a major component. Thus we tested whether CCN3 expression affected the adhesion of 1205Lu cells to collagen IV. Cells were plated on wells coated with collagen IV, as per the manufacturer’s instructions. After 90 min, the number of adherent cells was determined after incubation with crystal violet. Contrary to our expectations, overexpression of CCN3 in 1205Lu melanoma cells did not affect their adhesion to collagen IV (Figure 3f). These data indicate that CCN3 decreases the invasion of 1205Lu cells via other downstream effectors.
Figure 3.
Overexpression of CCN3 in melanoma cells inhibits their growth and invasion. (a) Growth of 1205Lu melanoma cells transduced with either LacZ or CCN3. Cells were counted on days 1 and 4. Data represent means±s.d. with n=4 (*P<0.05, Student’s t-test). (b) Immunoblot of conditioned medium and cell lysates from 1205Lu melanoma cells transduced with control LacZ or CCN3 using a lentiviral vector. β-actin immunoblot and Coomassie blue staining (CBB) indicate equal loading of medium and lysates. (c) Effect of CCN3 overexpression on melanoma invasion in three-dimensional skin reconstructs. Cells transduced with control LacZ or CCN3 were incorporated into the epidermis of skin reconstructs as described in the ‘Materials and methods’ section. Co-staining of melanoma cells for S100 (red) and Ki67 (green) was performed to identify proliferation in skin reconstructs (left two columns). Co-staining of melanoma cells for S100 (red) and TUNEL (green) was performed to identify apoptosis in skin reconstructs (right two columns). Merged images also show DAPI staining (blue) for the identification of all nuclei (original magnification × 200; scale bar 100 μm). (d and e) Overexpression of CCN3 leads to decreased invasion capacity of 1205Lu cells. Cells transduced with control LacZ or CCN3 lentiviral vectors were seeded onto a Transwell with (d) or without (e) Matrigel. Number of cells migrating through filters from five separate fields were counted; data are means±s.d. with n=3. (f) Adhesion of 1205Lu cells transduced with control LacZ or CCN3 was analysed on collagen type IV as substrate. Data represent means±s.d. of triplicates.
CCN3 overexpression decreases MMP-2/−9 expression
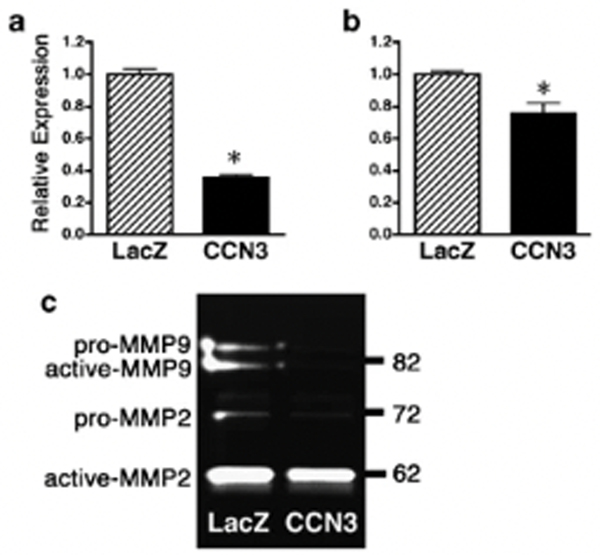
Elevated expression of MMPs has been associated with increased invasiveness of melanoma cells (Hofmann et al., 1999). Especially MMP-2 and −9 play critical roles in invasion because they degrade type IV collagen, which is a major component of the basement membrane. Therefore we first quantified MMP-2 and −9 expression in CCN3–1205Lu cells by real-time quantitative RT-PCR normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Compared with control LacZ transfectants, MMP-2 and −9 expression was significantly lower in CCN3 transfectants (Figures 4a and b) (P=0.0001, P=0.016, respectively). Gelatin zymography assays showed that active-MMP-2 (62 kDa), pro-MMP-2 (72 kDa), active-MMP-9 (82 kDa) and pro-MMP-9 (~90 kDa) exist in supernatants of control LacZ-1205Lu cells (Figure 4c). In agreement with real-time quantitative RT-PCR data, the enzymatic activities of pro-MMP2, pro-MMP-9 and active-MMP-9 were decreased in CCN3–1205Lu cells (Figure 4c).
Figure 4.
Overexpression of CCN3 in melanoma cells decreases MMP-2/−9 activities. (a and b) Downregulated MMP-2 (a) and MMP-9 (b) mRNA expression in 1205Lu cells transduced with CCN3 lentiviral vector as compared to LacZ-transduced cells. Results are presented as relative levels to those of GAPDH by quantitative RT-PCR. The data are expressed as means±s.d. with n=4 (*P<0.05, Student’s t-test). (c) Serum-free conditioned media from 1205Lu cells transduced with control LacZ or CCN3 were analysed by collagen zymography. Shown is a representative zymogram of three independent experiments.
Discussion
Clinical and pathological features of melanoma indicate that the progression of this tumor can be defined in five distinct stages (Clark, 1991). The first stage is characterized as hyperplasia of melanocytes, forming common acquired and congenital nevi. Dysplastic nevi with cytological and architectural atypia define the second stage. The third stage is RGP melanoma, where tumor cells exist within the epidermis or individually invade into the superficial dermis but show little capacity to leave the primary site. In the fourth stage, called VGP, a population of melanoma cells invades deep into the dermis and the subcutaneous tissue as an expanding cluster, increasing the risk for systemic dissemination. Finally, metastasis is the most advanced stage of melanoma. During this process, the development of melanoma can be explained as a disruption of normal homeostatic mechanisms in the skin. Such mechanisms control when and how cells proliferate, differentiate and undergo apoptosis in the epidermal melanin unit (Haass et al., 2004). The disruption of homeostatic controls can lead to the progression of melanoma where cell–cell and cell–matrix cross-talk play key roles.
Our previous results demonstrated that the matricellular protein CCN3 mediates intricate and essential communications between keratinocytes and melanocytes in maintaining normal epidermal homeostasis (Fukunaga-Kalabis et al., 2006). CCN3 interacts with fibulin 1C, Notch1, connexin 43 (Cx43), integrins α5β1, α6β1 and αvβ3. Among those partners, normal melanocytes in situ express integrin α6β1 (Kuphal et al., 2005). In neonatal foreskin, Cx43 was expressed abundantly in suprabasal layers (Matic et al., 2002). These observations are consistent with the fact that CCN3 is expressed primarily at the epidermal–dermal junction where melanocytes and keratinocytes have contact with each other. In this study, we characterized CCN3 as the first member of the matricellular protein family that is downregulated in aggressive melanoma cells. In contrast, other major proteins in the same family, such as osteopontin (Zhou et al., 2005), tenascin C (Herlyn et al., 1991) or SPARC (Massi et al., 1999), are strongly upregulated in melanoma cells compared with normal melanocytes. Our data suggest that qualitative and quantitative shifts of matricellular proteins contribute to melanoma progression similar to the cadherin class switch. The cadherin switch is characterized by the downregulation of E-cadherin in melanoma cells when compared to melanocytes and by the upregulation of N-cadherin, which allows melanoma cells to escape from control by keratinocytes (Hsu et al., 1996).
We found that the lack of CCN3 expression in advanced melanoma cells is well correlated to Breslow’s depth of invasion as one of the most important prognostic markers in melanoma, suggesting that CCN3 expression could be a potential marker for a relatively good prognosis in this most serious type of skin cancer. In this study, we tested whether regaining expression of CCN3 in advanced melanoma cells could change their biological phenotype using lentivirus-mediated gene transduction. CCN3 decreased melanoma invasion through Matrigel. The activation of MMPs is essential in the progression of malignant tumors. MMP-mediated degradation and rearrangement of the extracellular matrix (ECM) is an important early step in tumor invasion, since the ECM functions as a biochemical and mechanical barrier against cell movement in the tumor microenvironment. Among various MMPs, type IV collagenases or gelatinases MMP-2 and −9 are critical for cell migration leading to the invasion and metastasis of melanoma (Seftor et al., 1999).
CCN3 transduction of the aggressive 1205 Lu melanoma cells inhibited the transcription of MMP-2/−9 and resulted in reduced tumor invasion. Recently, abundant evidence has been presented that matricellular proteins are involved in the regulation of MMPs. For example, melanoma-associated matricellular proteins such as osteopontin, tenascin C and SPARC are known to upregulate either MMP-2 or −9 (Gilles et al., 1998; Philip et al., 2001; Kalembeyi et al., 2003; Robert et al., 2006). Here we demonstrated CCN3 as the first member of matricellular proteins that downregulates MMP-2/−9 expression in melanoma cells. This clearly indicates the potential role of CCN3 to oppose melanoma-associated matricellular proteins and to protect against melanoma progression. Previous reports suggested that MMP-2/−9 activity or their expression is regulated via the MAP kinase pathway (Welch et al., 2000; Denkert et al., 2002; Tanimura et al., 2003). Therefore, it is possible that CCN3 downregulates MMP-2/−9 through the inhibition of ERK. Furthermore, it has been suggested that after secretion of the full-length CCN3 and cleavage of the extracellular compartment, the C-terminal portion of the protein could reenter the cell and be routed to the nucleus via a nuclear localization signal-dependent pathway (Planque et al., 2006). Previous reports also strongly indicate that nuclear CCN proteins may regulate transcription, in complex with other regulatory proteins, in a similar manner as general corepressors of transcription (Planque et al., 2006). Therefore it is also conceivable that CCN3 regulates the transcription of MMP-2/−9 as a repressor. Although CCN3 suppressed the phosphorylation of ERK and the proliferation of 1205Lu cells in vitro, a significant reduction of tumor growth was not observed in vivo (data not shown). This finding contrasts with previous studies that identified CCN3 as having an antiproliferative role (Gupta et al., 2001; Benini et al., 2005). The discrepancy might reflect variability in molecular mechanisms underlying CCN3 signaling in different cellular contexts. Indeed, normal melanocyte growth and adhesion is regulated by CCN3 in human skin (Fukunaga-Kalabis et al., 2006). Thus, the biological functions of CCN3 in melanoma cells may depend, in part, on the cellular context at a given stage of tumor progression. Such a mechanism remains to be investigated. The activity of the CCN3 molecule appears to occur in the basement membrane zone, a location with enriched collagen IV, which is a substrate of MMP-2/−9. In order to see if CCN3 blocks cancer cells from escaping the ECM trap and initiating tumor invasion, grafting of skin reconstructs to mice could be considered.
In conclusion, we have shown that CCN3 expression is downregulated during the progression stages of melanoma. Immunohistochemical staining of melanoma lesions revealed that CCN3 expression is inversely correlated with tumor thickness. CCN3 transduction in advanced melanomas suppressed MMP-2 and −9 activities and strongly reduced tumor invasion. These results suggest that the lack of CCN3 expression in advanced melanomas contributes to an invasive phenotype. CCN3 is the first member of the matricellular protein family that is downregulated in melanoma cells, whereas other major proteins in the same family, such as osteopontin, tenascin C or SPARC, are upregulated. Further studies elucidating the mechanism underlying the switch of matricellular proteins will help discover potential therapeutic targets for preventing melanoma progression.
Materials and methods
Cell culture
Human melanoma cell lines (WM1650, WM35 (RGP), WM164, WM793 (RGP/VGP), WM115, WM164, WM278 (VGP), 1205Lu, WM1799, WM1232 and 451Lu (Met)) derived from different stages of melanoma progression were isolated and cultured as previously described (Satyamoorthy et al., 1997). Normal human primary melanocytes, keratinocytes and fibroblasts were isolated from human epidermis of neonatal foreskins and were cultured as described (Hsu et al., 1998; Fukunaga-Kalabis et al., 2006). 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS). For coculture experiments, melanocytes/melanoma cells were cultured with keratinocytes at a 1:3 ratio in EpiLife medium for 2 days. As a control, monocultured samples—melanocytes/melanoma cells and keratinocytes at a 1:3 ratio—were cultured separately for 2 days.
Immunoassays
For western blot analyses to detect secreted CCN3, proteins from conditioned medium corresponding to 5 × 105 melanocytes per melanoma cells were collected with heparin-sepharose beads after overnight incubation at 4 °C, as described previously (Fukunaga-Kalabis et al., 2006). For immunoblotting of whole cell lysates, equal amounts of protein (20 μg per lane) were solubilized in NuPAGE LDS sample buffer (Invitrogen). Samples were separated on 4–12% Bis-Tris gels and were then transferred to polyvinylidene difluoride membranes and probed with K19M: anti-CCN3 rabbit sera, anti-fibronectin (BD Biosciences, San Jose, CA, USA), anti-phospho ERK (Cell Signaling, Danvers, MA, USA), anti-phospho Akt (Ser473) (Cell Signaling), anti-phospho Akt (Thr308) (Cell Signaling), anti-Akt (Cell Signaling), anti-phospho IGF-1 receptor (Tyr1131/Tyr1146) (Cell signaling), anti-IGF-1 receptor (Cell Signaling) or anti β-actin (Sigma-Aldrich, St Louis, MO, USA). For immunohistochemistry, formalin-fixed and paraffin wax-embedded biopsy samples were sliced in 7 μm thick sections and were processed by deparaffinization and rehydration followed by endogenous peroxidase blocking (1% H2O2 in methanol for 20 min) and antigen retrieval (boiled in 10 mM citrate buffer for 10 min). Tissue sections were blocked with 2% goat serum (Vector Laboratories, Burlingame, CA, USA) and were incubated with K19M, anti-CCN3 rabbit sera or control rabbit sera for 1 hour at room temperature, and then with biotinylated secondary antibodies (Vector Laboratories). Immunoreactivity was detected using the ABC Elite kit (Vector Laboratories). We used AEC as the final chromogen and hematoxylin as the nuclear counterstain. After adding the chromogen and counterstaining with Mayer’s hematoxylin (Sigma-Aldrich), samples were evaluated using a Nikon E600 upright microscope. To detect melanocytic cells, immunofluorescent staining was performed on serial sections of biopsy samples with anti pmel 17/HMB-45 (Dako, Carpinteria, CA, USA) as primary antibodies. H&E-stained slides of all biopsy samples were reviewed by a pathologist and were classified according to criteria outlined by the World Health Organization. Staining results were semiquantitatively expressed as follows: (−) no staining in tumor cells, (+) up to 10% of tumor cells were positive, (++) 11–50% of tumor cells were positive and (+++) over 50% of tumor cells were positive.
Cell assays
For cell growth experiments, melanocytes were plated in quadruplicate in 24-well plates at a density of 5.66 × 103 cells per cm2. Cell growth was monitored by counting cells in five randomly selected fields at high power. For cell invasion assays, 5 × 104 cells per chamber were used for each invasion assay. The upper parts of the 24 mm Transwell (Corning, Corning, NY, USA, 8.0-μm pore) were coated with 50 μl of a 1:2 mixture of Matrigel (BD Bioscience): phosphate-buffered saline. Then 100 μL of cell suspension and 500 μl of melanoma culture medium were added to the upper and lower compartments, respectively, and the plate was incubated for 48 h at 37 °C. Cells that had invaded the lower surface of the membranes were fixed with 2% paraformaldehyde and stained with hematoxylin. Random fields were counted by light microscopy at high power ( × 600). For adhesion assays, cells were suspended in serum-free MCDB153 medium (6 × 105 cells ml−1) and transferred in triplicate to CytoMatrix cell adhesion strips coated with human collagen type IV (Chemicon, Temecula, CA, USA) and were incubated for 90 min at 37 °C. After washing to remove unattached cells, the attached cells were stained with 0.2% crystal violet. The cell-bound stain was solubilized and the optical density (570 nm) was determined. All cell assays were performed at least three times.
Skin reconstructs
Skin reconstructs were prepared as described (Hsu et al., 1998) with modifications. A total of 3 μl of fibroblasts (7.5 × 104 cells ml−1) in a 4:1 mixture of bovine type I collagen (Organogenesis, Canton, MA, USA): Matrigel (BD Bioscience) was added to each insert of tissue culture trays (Organogenesis) and were allowed to constrict in DMEM with 10% FBS for 7 days at 37 °C. For epidermal reconstruction, keratinocytes were mixed with melanoma cells at a ratio of 5:1 in epidermal growth medium composed of three parts DMEM and 1 part Ham’s F-12 supplemented with 2.4 M CaCl2, 0.18 mM adenine, 4 mM glutamine, 10 mg ml−1 selenium, 10 μM ethanolamine, 0.1 mM O-phosphoryl ethanolamine, 10 μg ml−1 insulin, 10 μg ml−1 transferrin, 20 pM tri-iodothyronine, 0.5 μg ml−1 hydrocortisone and 4 pM progesterone. A total of 5 × 106 cells were seeded on each contracted collagen gel. Cultures were kept submerged in medium containing 1 ng ml−1 EGF and 0.1% dialysed newborn calf serum for 2 days, then in 0.2 ng ml−1 EGF and 0.1% dialysed newborn calf serum for another 2 days, and then were raised to the air–liquid interface via feeding from below with medium containing 2% dialysed newborn calf serum. After 14 days, skin reconstructs were fixed with 4% paraformaldehyde and were embedded in paraffin. The invasive capacity of melanoma cells was determined by morphological evaluation using H&E staining. Cellular identification of apoptotic cells in skin reconstructs was made by TUNEL staining (Roche Applied Science, Indianapolis, IN, USA) according to the manufacturer’s instructions, and sections were subsequently stained with anti-S100 (Invitrogen) to identify melanoma cells. To analyse proliferation of melanoma cells in skin reconstructs, double-immunofluorescent staining was performed on sections of skin reconstructs with anti-S100 and anti-Ki67 (Invitrogen).
Quantitative reverse transcriptase–polymerase chain reaction
Total RNAs were reverse transcribed into first-strand cDNAs for quantitative reverse transcriptase–polymerase chain reaction. Gene-specific primers were designed as follows: MMP-2, 5′-CCGCAGTGACGGAAAGATGT-3′ and 5′-CACTTGCGGTCGTCATCGTA-3′; MMP-9, 5′-GGACGATGCCTGCAACGT-3′ and 5′-CAAATACAGCTGGTTCCCAATCT-3′; GAPDH, 5′-ATGGAAATCCCATCACCATCTT-3′ and 5′-CGCCCCACTTGATTTTGG-3′. ABsoluteQPCR SYBR Green Mixes (ABgene, Epsom, UK) were used with 1 ng ml−1 cDNA and with 70 nM primers for the evaluation of GAPDH and CCN3 expression. A negative control without the cDNA template was run with each assay. Amplifications were performed in an ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Thermal cycler conditions were 95 °C for 15 min, then 40 cycles of 15 s at 95 °C followed by 1 min at 60 °C. All experiments were performed in triplicate, and mean values were used to determine mRNA levels. At the end of each PCR, baseline and threshold values (CT) for these genes were set using the ABI 7000 Prism Software, and the CT values calculated were exported to Microsoft Excel for analysis. The relative expression of each mRNA was calculated using the comparative CT method. All samples were normalized to relative levels of GAPDH.
Zymography
Culture supernatants obtained from 1205Lu melanoma cells transduced with CCN3 or LacZ expression vectors were resolved (equal amounts of protein per lane) on 10% Novex zymogram Gels (Invitrogen) that contained 0.1% gelatin (Invitrogen). Each gel was soaked in Novex zymogram renaturing buffer for 30 min at room temperature, then incubated in Novex zymogram developing buffer overnight at 37 °C. The gels were subsequently stained with Coomassie blue. Zones of proteolysis appeared as clear bands against a blue background.
Recombinant lentiviruses
For gene transfer, we constructed lentiviral vectors. The PCR-amplified coding sequence for human CCN3 was cloned into pLenti6/V5-D-TOPO (Invitrogen). This vector was co-transfected into 293FT cells with ViraPower packaging mix to generate the lentivirus. Generated plasmids were confirmed by DNA sequencing. The 1205Lu melanoma cells were transduced with the lentivirus and stable cell lines were generated by selecting with blasticidin. Control cells were stably transduced with the lentivirus produced by pLanti6/V5-GW/lacZ control plasmid (Invitrogen).
Statistics
Data reported in cell growth assays are expressed as means±s.d. Data for migration, invasion, quantitative RT-PCR, in vivo tumor growth and colony formation assays are presented as means±s.e. and were analysed by two-tailed Student’s t-test. A P-value of less than 0.05 is considered significant.
Supplementary Material
Acknowledgements
We thank Kate M Belser for technical assistance. This work was supported by grants from the National Institutes of Health (CA76674, CA80999, CA47159, CA76674, CA25874 and CA10815) and was partially supported by funds from the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health.
References
- Benini S, Perbal B, Zambelli D, Colombo MP, Manara MC, Serra M et al. (2005). In Ewing’s sarcoma CCN3(NOV) inhibits proliferation while promoting migration and invasion of the same cell type. Oncogene 24: 4349–4361. [DOI] [PubMed] [Google Scholar]
- Breslow A. (1970). Thickness, cross-sectional areas and depth of invasion in the prognosis of cutaneous melanoma. Ann Surg 172: 902–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark WH . (1991). Tumour progression and the nature of cancer. Br J Cancer 64: 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denkert C, Siegert A, Leclere A, Turzynski A, Hauptmann S. (2002). An inhibitor of stress-activated MAP-kinases reduces invasion and MMP-2 expression of malignant melanoma cells. Clin Exp Metastasis 19: 79–85. [DOI] [PubMed] [Google Scholar]
- Fukunaga-Kalabis M, Martinez G, Liu ZJ, Kalabis J, Mrass P, Weninger W et al. (2006). CCN3 controls 3D spatial localization of melanocytes in the human skin through DDR1. J Cell Biol 175: 563–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles C, Bassuk JA, Pulyaeva H, Sage EH, Foidart JM, Thompson EW . (1998). SPARC/osteonectin induces matrix metalloproteinase 2 activation in human breast cancer cell lines. Cancer Res 58: 5529–5536. [PubMed] [Google Scholar]
- Glukhova L, Angevin E, Lavialle C, Cadot B, Terrier-Lacombe MJ, Perbal B et al. (2001). Patterns of specific genomic alterations associated with poor prognosis in high-grade renal cell carcinomas. Cancer Genet Cytogenet 130: 105–110. [DOI] [PubMed] [Google Scholar]
- Gupta N, Wang H, McLeod TL, Naus CC, Kyurkchiev S, Advani S et al. (2001). Inhibition of glioma cell growth and tumorigenic potential by CCN3 (NOV). Mol Pathol 54: 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass NK, Smalley KS, Herlyn M. (2004). The role of altered cell–cell communication in melanoma progression. J Mol Histol 35: 309–318. [DOI] [PubMed] [Google Scholar]
- Haass NK, Smalley KS, Li L, Herlyn M. (2005). Adhesion, migration and communication in melanocytes and melanoma. Pigment Cell Res 18: 150–159. [DOI] [PubMed] [Google Scholar]
- Herlyn M, Graeven U, Speicher D, Sela BA, Bennicelli JL, Kath R et al. (1991). Characterization of tenascin secreted by human melanoma cells. Cancer Res 51: 4853–4858. [PubMed] [Google Scholar]
- Hofmann UB, Westphal JR, Waas ET, Zendman AJ, Cornelissen IM, Ruiter DJ et al. (1999). Matrix metalloproteinases in human melanoma cell lines and xenografts: increased expression of activated matrix metalloproteinase-2 (MMP-2) correlates with melanoma progression. Br J Cancer 81: 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MY, Shih DT, Meier FE, Van Belle P, Hsu JY, Elder DE et al. (1998). Adenoviral gene transfer of beta3 integrin subunit induces conversion from radial to vertical growth phase in primary human melanoma. Am J Pathol 153: 1435–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MY, Wheelock MJ, Johnson KR, Herlyn M. (1996). Shifts in cadherin profiles between human normal melanocytes and melanomas. J Investig Dermatol Symp Proc 1: 188–194. [PubMed] [Google Scholar]
- Kalembeyi I, Inada H, Nishiura R, Imanaka-Yoshida K, Sakakura T, Yoshida T. (2003). Tenascin-C upregulates matrix metalloproteinase-9 in breast cancer cells: direct and synergistic effects with transforming growth factor beta1. Int J Cancer 105: 53–60. [DOI] [PubMed] [Google Scholar]
- Kleer CG, Zhang Y, Pan Q, Merajver SD . (2004). WISP3 (CCN6) is a secreted tumor-suppressor protein that modulates IGF signaling in inflammatory breast cancer. Neoplasia 6: 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuphal S, Bauer R, Bosserhoff AK . (2005). Integrin signaling in malignant melanoma. Cancer Metastasis Rev 24: 195–222. [DOI] [PubMed] [Google Scholar]
- Lake AC, Bialik A, Walsh K, Castellot JJ Jr . (2003). CCN5 is a growth arrest-specific gene that regulates smooth muscle cell proliferation and motility. Am J Pathol 162: 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent M, Martinerie C, Thibout H, Hoffman MP, Verrecchia F, Le Bouc Y et al. (2003). NOVH increases MMP3 expression and cell migration in glioblastoma cells via a PDGFR-alpha-dependent mechanism. FASEB J 17: 1919–1921. [DOI] [PubMed] [Google Scholar]
- Maillard M, Cadot B, Ball RY, Sethia K, Edwards DR, Perbal B et al. (2001). Differential expression of the ccn3 (nov) proto-oncogene in human prostate cell lines and tissues. Mol Pathol 54: 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manara MC, Perbal B, Benini S, Strammiello R, Cerisano V, Perdichizzi S et al. (2002). The expression of ccn3(nov) gene in musculoskeletal tumors. Am J Pathol 160: 849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massi D, Franchi A, Borgognoni L, Reali UM, Santucci M. (1999). Osteonectin expression correlates with clinical outcome in thin cutaneous malignant melanomas. Hum Pathol 30: 339–344. [DOI] [PubMed] [Google Scholar]
- Matic M, Evans WH, Brink PR, Simon M. (2002). Epidermal stem cells do not communicate through gap junctions. J Invest Dermatol 118: 110–116. [DOI] [PubMed] [Google Scholar]
- McCallum L, Price S, Planque N, Perbal B, Pierce A, Whetton AD et al. (2006). A novel mechanism for BCR-ABL action: stimulated secretion of CCN3 is involved in growth and differentiation regulation. Blood 108: 1716–1723. [DOI] [PubMed] [Google Scholar]
- Nguyen N, Kuliopulos A, Graham RA, Covic L. (2006). Tumor-derived Cyr61(CCN1) promotes stromal matrix metalloproteinase-1 production and protease-activated receptor 1-dependent migration of breast cancer cells. Cancer Res 66: 2658–2665. [DOI] [PubMed] [Google Scholar]
- Perbal B. (2001). NOV (nephroblastoma overexpressed) and the CCN family of genes: structural and functional issues. Mol Pathol 54: 57–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perbal B, Brigstock DR, Lau LF . (2003). Report on the second international workshop on the CCN family of genes. Mol Pathol 56: 80–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip S, Bulbule A, Kundu GC . (2001). Osteopontin stimulates tumor growth and activation of promatrix metalloproteinase-2 through nuclear factor-kappa B-mediated induction of membrane type 1 matrix metalloproteinase in murine melanoma cells. J Biol Chem 276: 44926–44935. [DOI] [PubMed] [Google Scholar]
- Planque N, Long Li C, Saule S, Bleau AM, Perbal B. (2006). Nuclear addressing provides a clue for the transforming activity of amino-truncated CCN3 proteins. J Cell Biochem 99: 105–116. [DOI] [PubMed] [Google Scholar]
- Robert G, Gaggioli C, Bailet O, Chavey C, Abbe P, Aberdam E et al. (2006). SPARC represses E-cadherin and induces mesenchymal transition during melanoma development. Cancer Res 66: 7516–7523. [DOI] [PubMed] [Google Scholar]
- Satyamoorthy K, DeJesus E, Linnenbach AJ, Kraj B, Kornreich DL, Rendle S et al. (1997). Melanoma cell lines from different stages of progression and their biological and molecular analyses. Melanoma Res, (7 Suppl 2): S35–S42. [PubMed] [Google Scholar]
- Seftor RE, Seftor EA, Hendrix MJ . (1999). Molecular role(s) for integrins in human melanoma invasion. Cancer Metastasis Rev 18: 359–375. [DOI] [PubMed] [Google Scholar]
- Tanimura S, Asato K, Fujishiro SH, Kohno M. (2003). Specific blockade of the ERK pathway inhibits the invasiveness of tumor cells: down-regulation of matrix metalloproteinase-3/−9/−14 and CD44. Biochem Biophys Res Commun 304: 801–806. [DOI] [PubMed] [Google Scholar]
- Welch DR, Sakamaki T, Pioquinto R, Leonard TO, Goldberg SF, Hon Q et al. (2000). Transfection of constitutively active mitogen-activated protein/extracellular signal-regulated kinase kinase confers tumorigenic and metastatic potentials to NIH3T3 cells. Cancer Res 60: 1552–1556. [PubMed] [Google Scholar]
- Zhou Y, Dai DL, Martinka M, Su M, Zhang Y, Campos EI et al. (2005). Osteopontin expression correlates with melanoma invasion. J Invest Dermatol 124: 1044–1052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.