Abstract
Replacement of a novel candidate ethanol fermentation-associated regulatory gene, PHO4, from a fast-growing strain MC15, as determined through comparative genomics analysis among three yeast strains with significant differences in ethanol yield, is hypothesised to shorten the fermentation time and enhance ethanol production from sugarcane molasses. This study sought to test this hypothesis through a novel strategy involving the transfer of the PHO4 gene from a low ethanol-producing, yet fast-growing strain MC15 to a high ethanol-producing industrial strain MF01 through homologous recombination. The results indicated that PHO4 in the industrially engineered strain MF01-PHO4 displayed genomic stability with a mean maximum ethanol yield that rose to 114.71 g L−1, accounting for a 5.30% increase in ethanol yield and 12.5% decrease in fermentation time in comparison with that in the original strain MF01, which was the current highest ethanol-producing strain in SCM fermentation in the reported literature. These results serve to advance our current understanding of the association between improving ethanol yield and replacement of PHO4, while providing a feasible strategy for industrially engineered yeast strains to improve ethanol production efficiently.
Replacement of a novel candidate ethanol fermentation-associated regulatory gene, PHO4, from a fast-growing strain through a novel strategy (SHPERM-bCGHR), is hypothesised to shorten fermentation time and enhance ethanol yield from sugarcane molasses.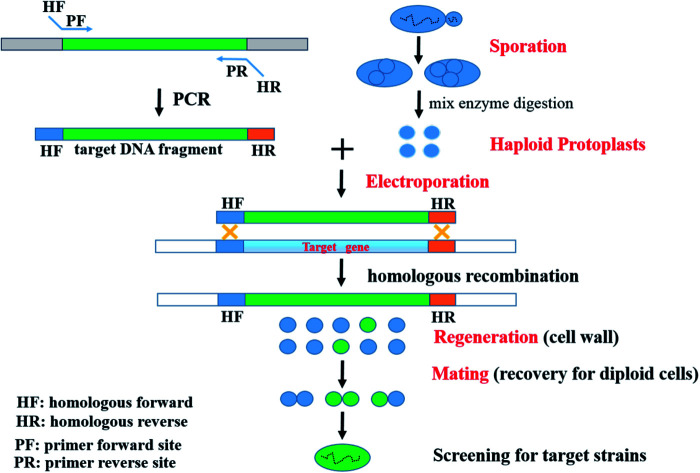
Introduction
The transition from petroleum-derived gasoline to renewable fuels derived from non-food materials is a promising alternative to improve the environment, and the socio-economic status of humans since it potentially reduces the need for fossil fuels and the production of greenhouse gases.1–3 Due to its renewability and large-scale natural production by microbes,4 bioethanol, a largely successful renewable liquid fuel alternative, is increasingly being used to resolve the energy crisis.5,6 Furthermore, the development of non-food bioethanol has emerged as a trend owing to its advantages such as abundant raw materials, low price, and renewability. The non-food bioethanol industry has strategically emerged in China.7
Sugarcane molasses (SCM) is a by-product of sugar production and has a total sugar concentration of approximately 50%.8 Owing to its high sugar content, SCM has become the primary raw material for non-food bioethanol production in China, Brazil, and other nations.9 The annual SCM production in China is approximately 3.8 million tonnes, which accounts for the primary raw material for ethanol fermentation in the major sucrose-producing areas of Guangxi province.10 Using this raw material to produce ethanol has the advantage of a centralised source and low cost. To some extent, it can resolve the issue of direct environmental pollution by the sugar industry, converting waste products into useful resources, thereby potentially improving economic benefits. However, two challenging issues remain in the SCM bioethanol industry, including low ethanol fermentation levels and serious environmental pollution, which are primarily due to the lack of high-performance industrial yeast strains. Saccharomyces cerevisiae is the most commonly employed microbe for industrial production of bioethanol. Various studies have shown that the ethanol content (EC) obtained from SCM fermentation by S. cerevisiae strains is approximately 79.25–96.29 g L−1.11,12 The optimal industrial S. cerevisiae strains in Brazil are CAT1 and PE2, with ECs of 79.25 g L−1 and 77.35 g L−1, respectively.11,13 Further, the EC of M Type wild S. cerevisiae in Scotland is 82.17 g L−1.14
S. cerevisiae is an ideal industrially applicable microbe owing to the ease and precision with which its genome can be manipulated, as its whole genome sequence was reported in 1996. With the advent and development of modern molecular biology methods, numerous approaches have been described to help modify S. cerevisiae strains to improve ethanol production, including the traditional/classical methods and novel modern methods (Table S2†). These methods are categorised into two levels: (1) genetic/pathway engineering and (2) genomic engineering.15 More specifically, these methods utilise the following strategies: (1) genetic engineering (GE), including deletion of gene(s), gene mutation, overexpression of gene(s), artificial zinc finger protein (AZFP), homologous recombination (HR), and yeast surface display system (YSD); (2) metabolic engineering (ME), including overexpression of gene(s) in metabolic pathways and HR; (3) genome modification (GM), including global transcription machinery engineering; (4) genome shuffling (GS); (5) inverse metabolic engineering (IME);16 (6) evolution, including directed evolution (DE), adaptive laboratory evolution (ALE), adaptive evolution (AE); (7) transcriptional engineering (TE); (8) genome replication engineering-assisted continuous evolution (GREACE); (9) gene editing approaches, including RNA interference (RNAi)-assisted genome evolution (RAGE), genome-scale CRISPR interference (CRISPRi), CRISPR/Cas9, and CRISPR-Cas12a (Cpf1)-assisted tag library engineering (CASTLING);17 and (10) the design-test-learn (DTL) cycle of microbial strain engineering, which combines knowledge-based design (X-omics, including comparative genomics, transcriptomics, proteomics, metabolomics, and fluxomics), experimental testing and model-based learning to understand how engineering strategies impact the cell, to promote further cycling to achieve higher product yields.18 Two or more strategies are currently used to modify yeast strains. However, some strategies may introduce antibiotic resistance genes into the wild-type strain, and the potential consumption of certain carbon sources may affect ethanol production. Moreover, genetically modified strains may show genomic instability. This study, a novel strategy (SHPERM-bCGHR) was utilised and it is simple, efficient, and rapid, allowing the construction of genetically modified industrial yeast strains, as it cannot introduce antibiotic resistance genes into the wild-type strain and more importantly, genetically modified strains via this strategy have genomic stability for long-term industrial ethanol production. Indeed, a recombinant industrial yeast strain had been constructed for SCM fermentation via the strategy (SHPERM-bCGHR), and its ethanol-producing capacity was better than that of the wild-type strain (data not shown).
In a previous study, three wild industrial S. cerevisiae strains, MF01, ME13, and MC15, which were screened from old sugar mill waste, are used as typical representatives of high-ethanol-, middle-ethanol-, and low-ethanol-producing strains from SCM fermentation, respectively. The maximum EC of MF01 is 108.61 g L−1, and it has been used for industrial ethanol production from SCM fermentation with an annual output of 50 000 tonnes in Guangxi, China.10 Although MF01 exhibits the highest ethanol yield, it shows a lower growth rate than ME13 and MC15; and although MC15 exhibits the lowest ethanol yield, it shows the highest growth rate compared with that of ME13 and MF01. Indeed, ethanol fermentation in S. cerevisiae proceeds through basic cellular metabolism, which is closely associated with glycolysis and pyruvate metabolism. Furthermore, previous studies have reported that the growth rate and survival time of yeast cells are closely associated with ethanol yield during SCM fermentation. Moreover, we previously determined via comparative genome resequencing analysis among MF01, ME13, and MC15, that MF01 is a heterogeneous diploid yeast species with a significantly higher rate of heterogeneous mutation than MC15 and ME13 (unpublished data). In fact, a common characteristic of industrial yeast is its highly polymorphic chromosomes in comparison with the laboratory strain, S288C.19 Furthermore, several candidate regulatory genes are potentially associated with ethanol fermentation, such as PHO4, as revealed through comparative genomics studies. Moreover, PHO4 gene and protein significantly differ between MF01 and MC15. Hence, MF01 is considered more likely to take up exogenous genes, thus serving as an ideal original modifiable strain to enhance the ethanol fermentation capacity.
Therefore, it has been hypothesised that replacement of the novel candidate ethanol-fermentation-related regulatory gene PHO4 from the fast-growing strain MC15 will shorten the fermentation time and enhance ethanol production from SCM. This study aimed to test this hypothesis using this novel strategy (SHPERM-bCGHR), involving the replacement of PHO4 from MC15 to MF01 via HR, yielding an engineered strain MF01-PHO4. Our results provides insight into the association between the improvement of ethanol production and PHO4 gene and guide the generation of feasible ethanologenic strains with increased ethanol production.
Experimental
Microorganisms
Three wild-type diploid industrial strains of S. cerevisiae, MF01,20 ME13, and MC15, screened from old sugar mill waste, were obtained and cultured on yeast extract peptone dextrose (YPD, containing 10 g L−1 yeast extract, 20 g L−1 peptone and 20 g L−1 glucose). They were then inoculated in YPD broth containing 25% (v/v) glycerol and stored at −80 °C and then sub-cultured on YPD plates. Yeast cells from freshly seeded YPD plates were inoculated in YPD broth and incubated at 30 °C, shaking at 180 rpm overnight. Cells were harvested and used for genomic DNA extraction, haploid protoplast electroporation, or as the inoculum for fermentation experiments.
Genomic DNA extraction
Briefly, total DNA was extracted from 2 mL of MC15 culture. The culture was centrifuged at 4 °C (12 000 rpm) for 2 min and resuspended in 250 μL lysis buffer (1 mM EDTA and 1% (w/v) SDS, pH 8), 100 μL TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8), and 10–20 g quartz sand (60–80 mesh). After thorough mixing at the maximal speed on a high-speed vortex-mixer, the lysates were extracted once with Tris-phenol : chloroform (1 : 1) and centrifuged at 4 °C (12 000 rpm) for 10 min. DNA obtained from the aqueous phase was precipitated with 1 mL ice-cold 95% (v/v) ethanol and 100 μL of 1 M NH4Cl, followed by centrifugation at 12 000 rpm. The white precipitate was then washed with 70% (v/v) ethanol, air-dried, and resuspended in 50 μL ultrapure water, followed by storage at −20 °C.
A novel strategy for the construction of engineered yeast strains
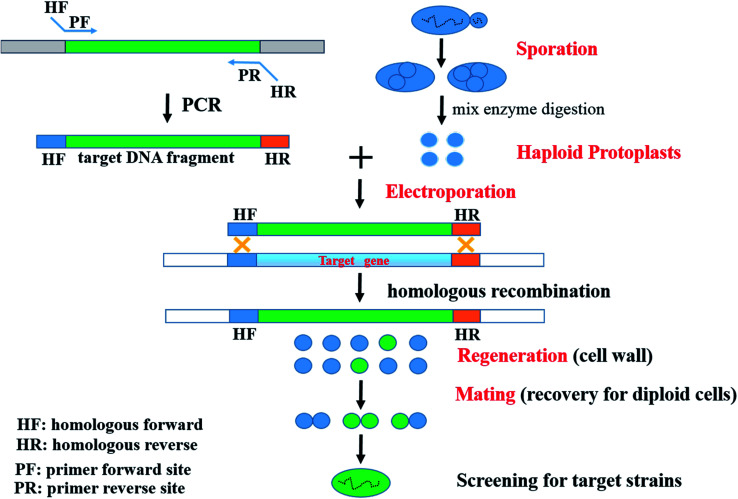
To date, several genome engineering strategies depending on HR in S. cerevisiae have been widely employed and developed. Such sequences used for HR have ranged from 30 bp,21 40 bp,22 and 60 bp (ref. 23) up to 500 bp.22 For instance, the Cre/loxP recombination or Cre/loxP-δ-integration system has been used.24,25 Based on comparative genomics and HR, a novel strategy, which included the processes of Sporulation – Haploid Protoplast formation – Electroporation of haploid protoplasts – Regeneration of cell wall – Mating for recovery of diploid cells (SHPERM-bCGHR), was used to generate engineered strains (Fig. 1).
Fig. 1. A novel strategy for industrially engineered Saccharomyces cerevisiae strains (SHPERM-bCGHR).
PHO4 gene cloning and phylogenetic analysis
PHO4 gene was amplified from the MC15 genome via polymerase chain reaction (PCR) using primers listed in Table S1.† Amplification reactions comprised 50 ng DNA, 5 pmol primers, 10 nmol dNTPs, and 1 U ExTaq DNA polymerase (Takara, Dalian City, China) in 1× PCR buffer (final volume, 25 μL). The cycling conditions were as follows: 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 53.5 °C for 30 s and 72 °C for 1 min, followed by final extension at 72 °C for 10 min using TProfessional (Biometra, Germany). PCR products were separated through electrophoresis on 0.8% (w/v) agarose gels in 0.5× TBE at 7.5 V cm−1 for 30 min (Fig. S1†). Thereafter, a gel imaging system (Bio-Rad, USA) was used for detection, and the amplification products were sent for sequencing (Invitrogen, Guangzhou City, China) for verification. Then, sequences were compared using Align X (Vector NTI, Invitrogen). PCR products of PHO4 gene were purified using the DNA Clean and Concentrator kit (Zymo Research, Epigenetics company, USA), and the purified product was detected via electrophoresis on a 0.8% (w/v) agarose gel and stored at −20 °C. Furthermore, some typical sequences of Pho4 protein were determined and obtained from the NCBI database via BLASTp. Phylogenetic trees based on Pho4 protein were constructed with unambiguously aligned sequences using the Neighbour-Joining (NJ) method in MEGA 5 program, version 5.05. Sequences of Pho4 protein were subjected to multiple alignment using CLUSTALW, and the phylogenetic tree was generated by NJ with a bootstrap set at 1000 replicates.26
Sporulation and haploid protoplast formation
High-yield strain MF01, used as the control strain for gene recombination, was cultivated in YPD overnight at 30 °C, shaking at 200 rpm and centrifuged at 4 °C (12 000 rpm) for 5 min. MF01 cells were washed twice with sterile water and inoculated in 100 mL of McClay medium (g L−1, yeast extract, 2.5; potassium chloride, 1.8; sodium acetate, 8.2 and glucose, 1.0) in a 250 mL Erlenmeyer flask at 28 °C, shaking at 150 rpm for 3–7 d. Vegetative cells were killed in a constant-temperature water bath at 58 °C for 15 min, and thus haploid protoplasts were harvested in a 10 mL tube, centrifuged at 4 °C (12 000 rpm) for 5 min, washed twice with sterile water, digested with the enzyme mix (including 40 g L−1 snails and 30 g L−1 cellulose, 0.22 μm membrane filtration) at 30 °C, shaking at 200 rpm for 6–8 h, and visualised using a microscope every hour for 6–8 h.
Electroporation and regeneration of haploid protoplast, and recovery of diploid cells
Haploid protoplasts of MF01 were harvested as described above via centrifugation at 4 °C (4000 rpm) for 10 min, washed once with 0.7 M KCl solution, washed twice with 1 M sorbitol solution and resuspended in 1 M sorbitol solution to achieve 2 × 109 cells mL−1. Thereafter, 90 μL of cell suspensions were placed in sterile Eppendorf (EP) tubes and treated with 10 μL of purified PHO4 amplified from MC15 (200 ng). The mixed solution (10 μL ddH2O as the control) was transferred into an electroporation cuvette and incubated in an ice bath for 5 min. Electroporation was then conducted using Electro Cell Manipulator (BTX Harvard Apparatus ECM 630, USA) in accordance with the manufacturer's instructions under the following settings: 2 mm cuvette gap, 1.5 kV, 200 Ω, and 25 μF. Thereafter, 900 μL of 1 M sorbitol solution was immediately added into the cuvette. The transformed mixed solution was transferred into a sterile 1.5 mL EP tube and pre-incubated at 30 °C for 2 h and then transferred into 2 mL regeneration medium (52.2 g L−1 potassium chloride, 10 g L−1 yeast extract, 20 g L−1 peptone, and 20 g L−1 glucose) and incubated at 30 °C, shaking at 130 rpm for approximately 24 h until the cell wall was regenerated, as revealed through microscopic observation.
Screening for recombinant strains
After cell wall regeneration, the transformed mixed solution was immediately divided into 20 μL fractions and placed in a 30 mL glass tube containing 3 mL screening medium, YPD4 (10 g L−1 yeast extract, 20 g L−1 peptone, and 40 g L−1 glucose), and incubated at 30 °C, shaking at 200 rpm for approximately 4–8 h. Relatively fast-growing cultures, based on OD600, were spread on predefined screening YPD4 plates. Thereafter, the plates were incubated at 30 °C for 2–3 d. Relatively fast-growing yeast colonies were then selected, sub-cultured on YPD4 plates, and PHO4 in these selected yeast colonies was correspondingly amplified via a rapid preparation protocol for DNA templates (1 mL cell solution was centrifuged at 4 °C and 12 000 rpm for 10 s, washed once with sterile water, suspended in 200 μL sterile water, vortex-mixed with approximately 25 mg quartz sand for 40 s, centrifuged as described above, and the supernatant was placed in a boiling water bath for 40 s with Chelex 100 (Sigma)). PHO4 sequences in these selected yeast colonies were compared using the Align X module of Vector NTI (Invitrogen), and recombinant strains (with PHO4 gene sequence identical to that of MC15) were screened. Furthermore, recombinant strains were cultured up to 30 generations, and PHO4 in generations 10, 20, and 30 of recombinant yeast was correspondingly amplified using the aforementioned method, and the sequencing results were compared to confirm the stability of PHO4 after replacement. Recombinant yeast was screened for diploid cells via PCR.27 Moreover, these recombinant strains were evaluated for relative respiratory intensity, sucrose fermentation, and SCM fermentation for very-high-gravity (VHG) ethanol.
Performance analysis
Accumulation of gas in Durham tubes
The cellular densities of MF01 and MF01-PHO4 were adjusted to prepare an initial inoculum of 1 × 106 cells mL−1 after culturing in YPD at 30 °C, shaking at 200 rpm for 8 h, and then 20 μL of this inoculum was placed in test tubes (180 mm × 18 mm) containing inverted Durham tubes (filled with YPD medium) and 6 mL YPD, and the test tube was placed in the incubator for yeast growth and gas production at 30 °C. Gas accumulation in the Durham tubes was recorded and compared.
Relative respiratory intensity
After MF01, MF01-PHO4, and MC15 cells were cultivated in YPD at 30 °C, shaking at 200 rpm for 8 h, their OD600 was adjusted to 1.5. Cells of these strains were harvested through centrifugation at 4 °C (12 000 rpm) for 2 min, washed twice with sterile water, and inoculated in 5 mL YPD or YPS40 medium (10 g L−1 yeast extract, 20 g L−1 peptone, and 400 g L−1 sucrose) at 30 °C, shaking at 200 rpm for 8 h or 16 h, respectively, and with all media containing 5 mg mL−1 2,3,5-triphenyltetrazolium chloride (TTC, 0.22 μm membrane filtration). Next, 2 mL of each culture medium was harvested through centrifugation at 4 °C (12 000 rpm) for 2 min, washed twice with sterile water, and vibrated thoroughly at maximal speed for 10 min after adding 1 mL 95% (v/v) ethanol for each culture. The supernatant was obtained via centrifugation at 4 °C (12 000 rpm) for 2 min, and its absorbance was measured with a spectrophotometer at 480 nm. The relative respiratory intensity of each strain in their corresponding media was obtained by dividing its OD480 by the OD480 of MF01 and multiplying it by 100%. Thus, the relative respiratory intensity of MF01 for the corresponding medium was defined as 100%.
Sucrose fermentation and SCM fermentation for VHG ethanol
One loopful of S. cerevisiae strains MF01 and MF01-PHO4 was transferred from 1 day YPD plates to a 250 mL Erlenmeyer flask containing 100 mL of YPD broth. Yeast was grown for 24 h at 200 rpm on a rotary shaker at 30 °C. A small volume of each seed culture was inoculated into each 50 mL EP tube (simulating the fermenter used for industrialised ethanol production) containing 20 mL of the fermentation medium (FM), YPS40, to achieve an initial inoculum of 2 × 108 cells mL−1. The EP tubes were shaken at 180 rpm and 30 °C for 24 h, and further incubated at 30 °C to allow for high content sucrose fermentation for VHG ethanol. Samples were withdrawn and assessed periodically to determine the concentration of ethanol and cell biomass.
SCM fermentation with MF01, MF01-PHO4, and MC15 was performed according to the current acidic fermentation process employed by the ethanol industry for SCM fermentation in China. The SCM used for fermentation was the standard molasses, 80–85 degrees Brix (°Bx), from a sugar mill in Guangxi. The SCM was diluted with water, and 20 °Bx SCM and 50 °Bx SCM were used as the initial feedstock for yeast growth and batch feedstock, respectively. To both SCMs, 0.2% (w/w) urea and 0.02% (w/w) phosphoric acid were added, and their pH value was adjusted with sulfuric acid from 3.8 to 4.0. Similarly, for sucrose fermentation, a small volume of seed culture was inoculated into a 50 mL EP tube containing 10 mL of the FM 20 °Bx SCM to create an initial inoculum of 2 × 108 cells mL−1. The EP tubes were shaken at 180 rpm and 30 °C for 24 h, then added to 10 mL of 50 °Bx SCM, shaken for 24 h at the same condition, and transferred to 30 °C condition to induce high content sucrose fermentation of VHG ethanol. Samples were withdrawn periodically to determine the concentration of ethanol every 8 h. Fermentation experiments were conducted in duplicate.
Flow cytometric analysis
The BD Accuri C6 flow cytometer (FCM, BD Biosciences, USA) was used for single-cell light scattering and fluorescence measurements. Scatter characteristics were evaluated as they may serve as indicators of morphological and/or physiological changes. Cells of S. cerevisiae MF01, MF01-PHO4, and MC15 strains were cultivated in YPD or YPS40 medium at 30 °C. These media were diluted 10-fold, and flow cytometric signals were obtained from the FCM performance, with a running time for diluted YPD or YPS40 cultivated medium of 15 s or 10 s, respectively.
Microscopic observation and cell counting
MF01, MF01-PHO4, and MC15 cells were visualised using a Nikon Eclipse 80i microscope (Tokyo, Japan). In addition, cell counts in YPD or fermented liquid were obtained using this microscope and a haemocytometer.
Determination of the content of ethanol and total residual sugars
YPS40 fermented liquid was centrifuged at 12 000 rpm for 2 min, and its supernatant was collected for determination of EC by gas chromatography (Agilent 6890N, USA), using a standard internal method, with 10% acetonitrile (v/v) as the standard. Alternately, the ethanol concentration in SCM fermented liquid was the same as the aforementioned YPS40 method, with an additional process of adding alkaline lead acetate to the liquid before centrifugation. The total residual sugars (TRSs) content in SCM fermented liquid was determined using a 3,5-dinitrosalicylic acid reagent.10,20
Results and discussion
Screening for recombinant engineered MF01-PHO4 strain
The recombinant S. cerevisiae MF01-PHO4 strain was constructed through the novel SHPERM-bCGHR method (Fig. 1). Haploid protoplasts were generated from MF01, and PHO4 was cloned from a low-yield ethanol-producing strain MC15, followed by electroporation of the haploid protoplasts of MF01 with the PHO4 fragment from MC15 (Fig. 2), cell wall regeneration, recovery of diploid cells, and screening for PHO4 gene replaced-strain by a rapid preparation protocol for DNA templates. Accordingly, after mating to recover diploid cells, the fast-growing strains were immediately screened, and recombinant MF01-PHO4 was finally obtained. Moreover, PHO4 was stable after MF01-PHO4 was cultured up to 30 generations (30G).
Fig. 2. Comparison among S. cerevisiae MF01-PHO4, MF01 and S288C strain; (a) PHO4 gene; (b) Pho4 protein.
Five-point mutations were observed in PHO4 gene between MF01-PHO4 and MF01 (or S288C), namely, G476A, A607G, C697T, T912C, and T929G (Fig. 2a); and three-point mutations were observed in Pho4 protein between MF01-PHO4 and MF01 strains (or S288C), namely, R159K, I203V, and V310G (Fig. 2b). On comparing Pho4 protein sequences of MF01-PHO4 and MF01 (S288C), the following differences in the number of amino acid residues were observed (Table S3†): Arg, 17 (5.4%) to 18 (5.8%); Gly, 13 (4.2%) to 12 (3.8%), and Ile, 11 (3.5%) to 12 (3.8%). Comparison among the characteristics of Pho4 protein of MF01-PHO4, MF01, and S288C is shown in Table S4.†
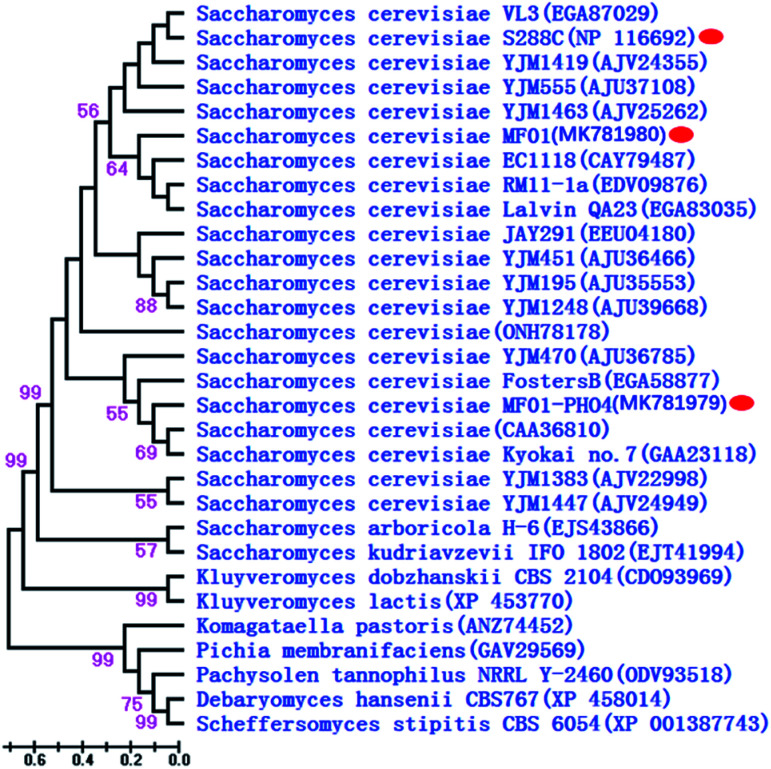
PHO4 sequences of MF01 and recombinant MF01-PHO4 were deposited in GenBank under the accession numbers MK781980 and MK781979, respectively. A phylogenetic tree constructed for 30 representative species on the basis of Pho4 protein is shown in Fig. 3, including 21 species of S. cerevisiae, 2 different Saccharomyces genera and 6 different genera of the phylum “Ascomycota” (Pichia the same as Scheffersomyces). Furthermore, their corresponding accession numbers (provided in parentheses) are shown in Fig. 3. Phylogenetic analysis revealed that Pho4 protein of MF01 was closely associated with that of S288C, rather than that of MF01-PHO4.
Fig. 3. Phylogenetic dendrogram of Pho4 protein based on the NJ method. Branch lengths are in scale to the scale bar shown at lower left corner.
Advantage of engineered MF01-PHO4 in comparison with MF01
Flow cytometric signals (FSC/SSC) of MC15, MF01-PHO4 and MF01 are shown in Fig. S2.† FSC was used to determine the cell size, with larger cells displaying a stronger FSC signal. SSC was used to determine cellular granularity and complexity, with highly granular cells displaying more internal complexity. Moreover, intracellular substances of high granular cells are more complex than that of low granular cells. Flow cytometry analysis revealed that upon culturing in YPD, MF01-PHO4 cells were larger than MF01 cells (Fig. S2(a2) and (a3)†). MF01-PHO4 cells displayed marked changes during YPS40 fermentation for ethanol production (Fig. S2(b2) and (b3)†) with respect to their size, displaying changes in cellular morphology, especially at 48 h, such that cells became more elongated when the EC peaked. Moreover, intracellular substances of MF01-PHO4 cells were more complex than those of MF01 and MC15 cells. Therefore, PHO4 replacement impacted the cellular morphology of the yeast cells.
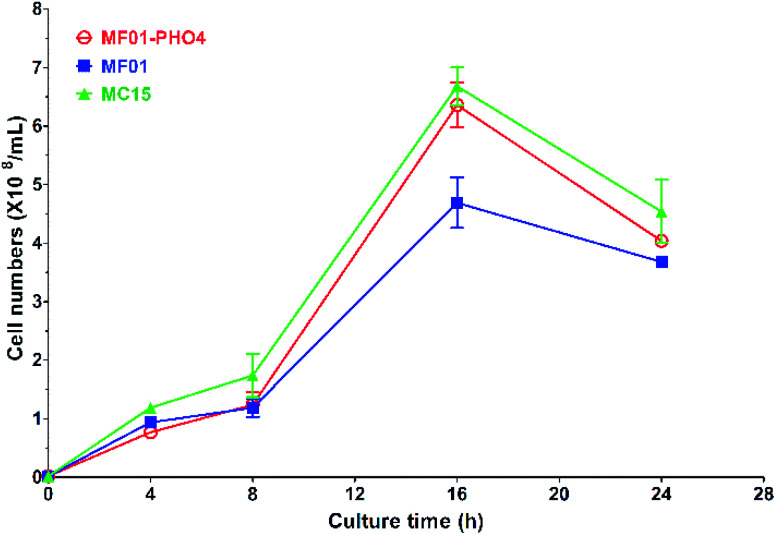
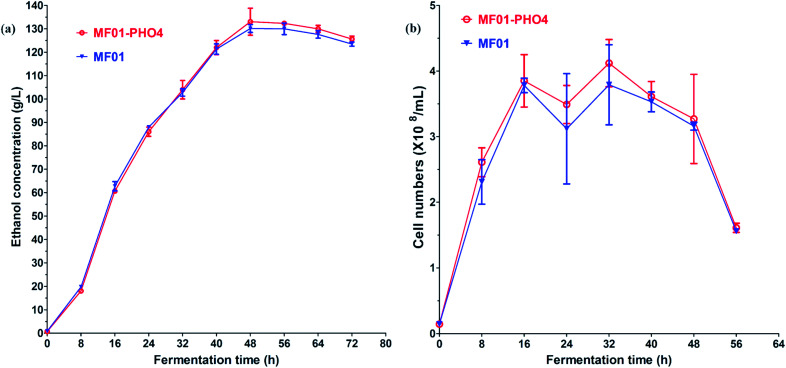
Gas production from MF01 and engineered MF01-PHO4 are shown in Fig. S3.† The rate of gas production was greater with MF01-PHO4 than with MF01. Meanwhile, the growth rate was similar between MF01-PHO4 and MC15 strains, being markedly greater than that of the original strain MF01 (Fig. 4) upon culturing in YPD medium. Upon inoculation in YPS40 medium for ethanol fermentation, cell numbers of MF01-PHO4 were greater than those of MF01 at all fermentation time points (Fig. 5b). Furthermore, the ethanol concentration was slightly greater with the engineered strain MF01-PHO4 than with MF01 (Fig. 5a).
Fig. 4. Growth curve of S. cerevisiae MF01, MF01-PHO4, and MC15 strain in YPD medium.
Fig. 5. YPS40 fermentation of S. cerevisiae MF01 and MF01-PHO4 strain; (a) ethanol production curve; (b) cell number curve.
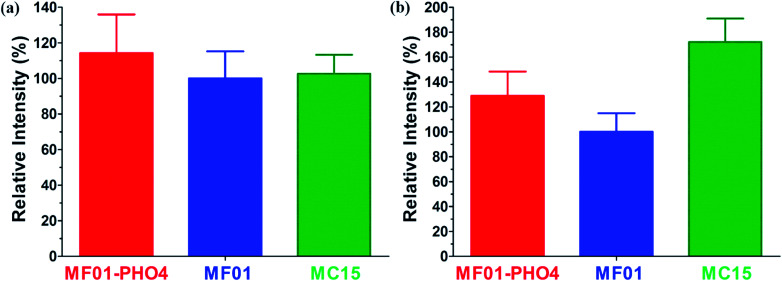
The relative respiratory intensities of MF01, MF01-PHO4, and MC15 are shown in Fig. 6. The relative respiratory intensity was the highest in MF01-PHO4 upon culturing these strains in YPD medium at 30 °C, shaking at 200 rpm for 8 h (Fig. 6a). In other words, the relative respiratory intensity was higher in MF01-PHO4 than in MF01, upon culturing in YPS40 medium at 30 °C, shaking at 200 rpm for 16 h; however, that of MC15 was the highest (Fig. 6b).
Fig. 6. Relative respiratory intensities among S. cerevisiae MF01, MF01-PHO4, and MC15 strain; (a) cultivated in YPD medium at 30 °C, shaking at 200 rpm for 8 h; (b) cultivated in YPS40 medium at 30 °C, shaking at 200 rpm for 16 h.
SCM fermentation by S. cerevisiae for ethanol production
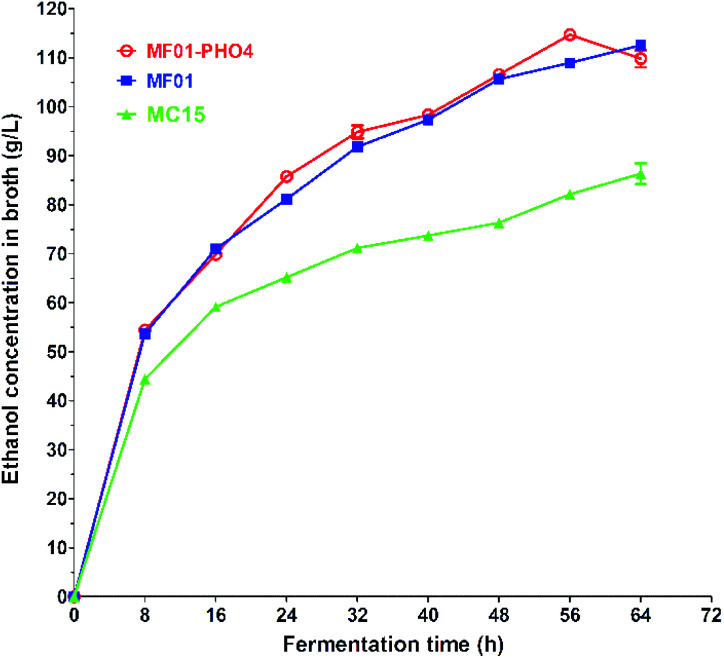
S. cerevisiae is a critical microorganism and the most robust strain used for ethanol production via SCM fermentation. The maximum EC of MF01-PHO4 approached 114.71 ± 0.24 g L−1 at 30 °C for 56 h (Fig. 7), while the maximum EC of MF01 and MC15 was 108.94 ± 0.71 g L−1 and 82.16 ± 0.79 g L−1, respectively, resulting in a 5.30% increase in ethanol yield and 12.5% decrease in the fermentation time compared with those of the original strain MF01.
Fig. 7. Ethanol production curve of S. cerevisiae MF01, MF01-PHO4 and MC15 strain from SCM fermentation.
Numerous studies worldwide, including in Brazil, China, and India,28 have used some optimal yeast for SCM fermentation (Table S5†). The procedure for SCM fermentation is the same as that for industrial ethanol production in China, namely, acidic fermentation, and the pH is adjusted with commercial sulfuric acid; however, sterilisation is not carried out for SCM pre-treatment before fermentation. An industrial mutant strain UAF-1, derived from the native strain S. cerevisiae (SAF-INSTANT) exposed to Co60 gamma radiation, yielded an EC of 87 g L−1 and 96.29 g L−1 upon SCM fermentation, respectively. Furthermore, six industrial strains have proven to be important for ethanol production in Brazil: CAT1, PE2, FT858L, Fermel® (selected by Fermentec), BG1, and SA1; CAT-1 and PE-2 are used in particular, with the ethanol concentration approaching 86.82 g L−1 by the CAT-1 strain at 30 °C. CAT-1 and PE-2 have been utilised by Brazilian distilleries, which account for approximately 60% of the nationwide ethanol production.29 However, the SCM media for these strains ITV-01 (Mexico) and F-514 (Egypt), were steam-sterilised at 121 °C for 15 min before fermentation. Thus, in our previous work, we screened serial industrial S. cerevisiae strains, including high-, middle-, and low-yield ethanol-producing strains for SCM fermentation. MF01, MF02, and MF03 were high-yield ethanol-producing strains, and the overall EC from SCM fermentation with these strains approached more than 106 g L−1. Moreover, the ECs by MF02 and MF03 strains were 114.29 g L−1 and 106.56 g L−1 at 30 °C for 72 h, respectively. Herein, MF01-PHO4 was the current highest ethanol-producing strain in SCM fermentation in the reported literature.
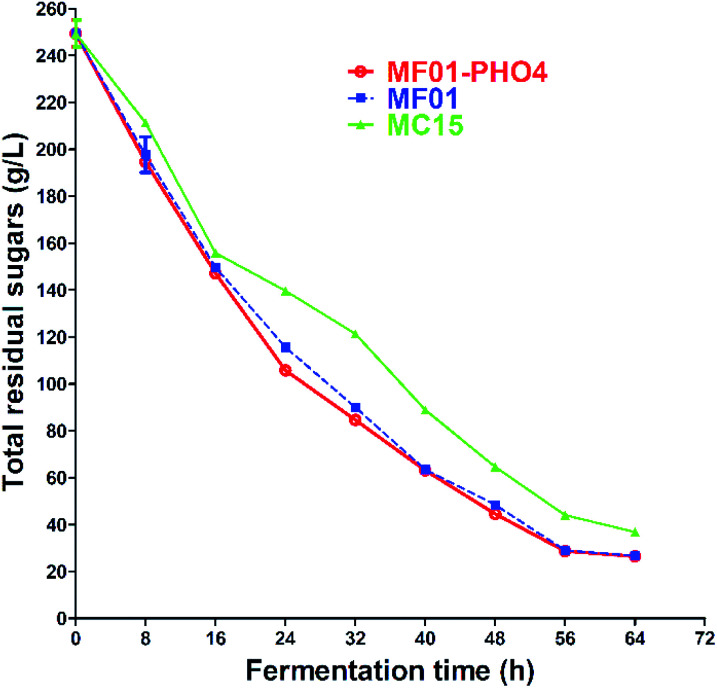
The concentration of TRSs of S. cerevisiae MF01, MF01-PHO4 and MC15 from SCM fermentation were 29.07 ± 0.12 g L−1, 28.68 ± 0.06 g L−1 and 44.05 ± 0.06 g L−1 at 30 °C for 56 h, respectively (Fig. 8). SCM is the primary by-product in the sugar manufacturing process, and its composition is complex, primarily including fermentable sugars, especially sucrose. Fermentable sugars also include approximately 10% (w/w) non-reducing sugars (NRSs) such as raffinose, melibiose, and stachyose. However, wild industrial S. cerevisiae strains cannot directly utilise NRS. As α-galactosidase can hydrolyse NRS and release galactose, we previously constructed a recombinant strain, MF01-3, through ME and HR, which integrated agl3 from T. reesei into the genome of MF01. Compared with the original strain MF01, the highest EC yield of SCM increased to 4.5%, and at the end of SCM fermentation, the concentration of total sugar decreased to 23.6%, while that of reducing sugars increased to 15.05%.30
Fig. 8. Total residual sugars curve of S. cerevisiae MF01, MF01-PHO4 and MC15 strain from SCM fermentation.
Genetically modified S. cerevisiae strains for ethanol production via SCM fermentation
SCM fermentation may be continuous or a batch process. If the fermentation time can be shortened, it would benefit the ethanol industry. Genetically modified S. cerevisiae strains for ethanol production via SCM fermentation are listed in Table 1.
Ethanol production from SCM fermentation by genetically modified Saccharomyces cerevisiae strains.
| Name | Characteristic (approaches) | Result | References |
|---|---|---|---|
| MF01-3b | Integrated agl3 gene from T. reesei (ME) | Improved 4.5% in ethanol yield | 30 |
| Δssk1Δsmp1 a | Double-deletion of ssk1 and smp1 (ME) | 6% increase in ethanol yield, 35% decrease in glycerol yield | 31 |
| ER HAA1-OPb | Overexpression of HAA1 gene (GE) | Showed tolerance to acetate to perform SCM fermentation | 32 |
| PE-2(ΔRIM15ΔMSN2)b | Disruption of RIM15 and MSN2 gene (GE) | Shortened fermentation time (17.1 h) | 13 |
| PE-2(ΔMSN2)b | Disruption of MSN2 gene (GE) | Shortened fermentation time (6.2 h) | 13 |
| PE-2(ΔRIM15)b | Disruption of RIM15 gene (GE) | Shortened fermentation time (13 h) | 13 |
| NF-ybrb | Deletion of YBR019C gene (GE) | Reduced ethanol yield by 9.86% | 33 |
| MF01-PHO4b | Replacement of PHO4 gene (GE: SHPERM-bCGHR) | Enhanced ethanol yield and shortened fermentation time (8 h) | This study |
Laboratory strain (original strain).
Industrial strain (original strain). GE, genetic engineering; ME, metabolic engineering.
Glycerol is a by-product of ethanol production. The ethanol yield can be enhanced by reducing glycerol production, such as deletion of gpd1 or gpd2 gene. Furthermore, genes in the high osmolarity glycerol (HOG) pathway were targeted for increasing ethanol production through SCM, which regulate glycerol synthesis in S. cerevisiae. Strains with smp1 and ssk1 deletions (Δsmp1 and Δssk1) displayed consistent improvements in ethanol yield. Furthermore, strains with double deletions (Δssk1Δsmp1) led to a 35% decrease in glycerol production and a 6% increase in ethanol production.31
During SCM fermentation, acetate is an effective agent in preventing bacterial contamination, which is a major cause for the reduction in ethanol production. However, acetate negatively affects ethanol fermentation with these strains. A HAA1-overexpressing diploid strain (ER HAA1-OP) displayed tolerance not only to acetate but also to lactate, which was derived from an industrial ethanol-producing strain with Ethanol Red (ER), and this tolerance depended on HAA1 upregulation. The ethanol yield of ER HAA1-OP was almost equivalent to that of the ER strain during ethanol production from SCM in the absence of acetate. Acetate supplementation at 0.5% (w/v; pH 4.5) inhibited the fermentation potential of the ER strain but not ER HAA1-OP.32
In S. cerevisiae, Rim15p, a Per-Arnt-Sim kinase, is activated in response to stressors, thus inducing downstream target genes of stress-responsive transcription factors Msn2p and Msn4p (Msn2/4p). Since Msn2/4p targets numerous genes associated with carbohydrate metabolism, regulation of this stress response pathway potentially contributes to metabolic remodelling. PE-2 is one of the most dominant yeast strains in Brazil. Disruption of RIM15 gene in the PE-2 strain accelerated SCM fermentation for ethanol production via enhanced sucrose utilisation. Upon disruption of RIM15 gene, the EC by PE-2 (ΔRIM15) during SCM fermentation approached 77.35 g L−1, increasing to 6.51%. Disruption of RIM15 and MSN2 genes (ΔRIM15ΔMSN2), decreased the fermentation time by 25.1% of that of PE-2.13 Thus, the Rim15p- and Msn2p-mediated stress-response pathways negatively regulate SCM fermentation in S. cerevisiae. Herein, the industrially engineered strain, MF01-PHO4, enhanced the ethanol yield and shortened fermentation time (8 h) from SCM fermentation. Conversely, deletion of YBR019C gene led to reduction in the ethanol yield by 9.86% in comparison with that of the wild strain MF02.33 These results indicate that the YBR019Cp-mediated pathway positively regulates SCM fermentation in S. cerevisiae.
Potential reasons underlying enhanced ethanol production via SCM fermentation through replacement of PHO4
This study reported that the engineered high-yield ethanol-producing yeast MF01-PHO4 had the following advantages in comparison with the original high-yield ethanol-producing yeast MF01: rapid growth (similar to that of the low-yield ethanol-producing strain MC15), rapid gas production, large cells and more abundant cell contents (flow cytometry analysis), high EC from 40% (w/v) sucrose or SCM fermentation, rapid growth and high EC in non-phosphorus conditions, and higher concentration of trehalose (single-cell Raman spectroscopy) synthesised simultaneously with ethanol production at high levels (data not shown).
PHO4 is a positive regulatory gene in the phosphate-responsive signalling (PHO) pathway, which is associated with the cell cycle. Our observations raise the question of how PHO4 replacement from a fast-growing strain MC15 could have such an enhancement effect and promotes high-yield ethanol production from SCM fermentation using industrial S. cerevisiae strains. Based on our findings, three potential reasons were inferred: (1) Pho4 regulates the expression of target genes associated with cell growth and ethanol metabolism by binding to the promoter region at the conserved sequence CACGTG. (2) Transcription factor Pho4 plays a regulatory role through phosphorylation and dephosphorylation processes, and it is speculated that Pho4 might indirectly regulate ATP through these processes, and thereby regulate basic metabolic processes associated with yeast cell growth and ethanol metabolism. (3) With the accumulation of ethanol during SCM fermentation, a high concentration of ethanol can damage or even kill cells. It is speculated that Pho4 regulates the expression of trehalose synthesis pathway genes and indirectly regulates industrial ethanol fermentation from SCM fermentation using yeast. However, these speculations require further verification.
Conclusions
In summary, this study describes the generation of an industrially engineered S. cerevisiae strain via a novel strategy by replacing a novel candidate ethanol-fermentation-associated regulatory gene, PHO4, from a fast-growing strain. The results show that the PHO4-recombinant strain displayed genomic stability, and its maximum yield was 114.71 g L−1, resulting in a 5.30% increase in ethanol yield and 12.5% decrease in the fermentation time compared to those of the original industrial strain, which was the current highest ethanol-producing strain for SCM fermentation in the reported literature. This study serves to advance the current understanding of the association between improving ethanol yield and replacement of PHO4 gene while providing a feasible strategy for industrially engineered yeast strains to efficiently improve ethanol production.
Conflicts of interest
There are no conflicts of interest to declare.
Supplementary Material
Acknowledgments
This work was financially supported by the General Program of the Natural Science Foundation of Guangxi, China (2018GXNSFAA138111, 2018GXNSFAA294118, 2018GXNSFAA294035), Guangxi Science and Technology Program (AB16380024, AB19110041), Guangxi Major Science and Technology Innovation Base Construction Project (2018-15-Z03-1208, 2018-15-Z03-1209), Nanning Science and Technology Program (20133156, 20141001), and the Scientific Research Development Fund Project of Guangxi Academy of Sciences (2018YFJ402).
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra08673k
References
- Lynd L. R. Nat. Biotechnol. 2017;35:912–915. doi: 10.1038/nbt.3976. [DOI] [PubMed] [Google Scholar]
- Libardi N. Soccol C. R. César de Carvalho J. de Souza Vandenberghe L. P. Bioresour. Technol. 2019:42–50. doi: 10.1016/j.biortech.2018.12.088. [DOI] [PubMed] [Google Scholar]
- Serna-Loaiza S. García-Velásquez C. A. Cardona C. A. Biofuels, Bioprod. Biorefin. 2018:1–13. [Google Scholar]
- Peralta-Yahya P. P. Zhang F. del Cardayre S. B. Keasling J. D. Nature. 2012;488:320–328. doi: 10.1038/nature11478. [DOI] [PubMed] [Google Scholar]
- Kalghatgi G. Levinsky H. Colket M. Prog. Energy Combust. Sci. 2018;69:103–105. doi: 10.1016/j.pecs.2018.06.003. [DOI] [Google Scholar]
- Zabed H. Sahu J. N. Boyce A. N. Faruq G. Renewable Sustainable Energy Rev. 2016;66:751–774. doi: 10.1016/j.rser.2016.08.038. [DOI] [Google Scholar]
- Jiao J. L. Li J. J. Bai Y. J. Cleaner Prod. 2018;180:832–845. doi: 10.1016/j.jclepro.2018.01.141. [DOI] [Google Scholar]
- Huang G. Chen X. Wang C. Zheng H. Y. Huang Z. Q. Chen D. Xie H. RSC Adv. 2017;7:47840–47847. doi: 10.1039/C7RA09002A. [DOI] [Google Scholar]
- Lopes M. L. Paulillo S. C. de L. Godoy A. Cherubin R. A. Lorenzi M. S. Giometti F. H. C. Bernardino C. D. de Amorim Neto H. B. de Amorim H. V. Braz. J. Microbiol. 2016;47:64–76. doi: 10.1016/j.bjm.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. Lu Q. Zhang S. S. Chen Y. Huang R. B. Guangxi Sci. 2011;18:385–391. [Google Scholar]
- Neto H. B. de A. Yohannan B. K. Bringhurst T. A. Brosnan J. M. Pearson S. Y. Walker J. W. Walker G. M. J. Inst. Brew. 2009;115:198–207. doi: 10.1002/j.2050-0416.2009.tb00369.x. [DOI] [Google Scholar]
- Arshad M. Hussain T. Iqbal M. Abbas M. Braz. J. Microbiol. 2017;48:403–409. doi: 10.1016/j.bjm.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inai T. Watanabe D. Zhou Y. Fukada R. Akao T. Shima J. Takagi H. Shimoi H. J. Biosci. Bioeng. 2013;116:591–594. doi: 10.1016/j.jbiosc.2013.05.015. [DOI] [PubMed] [Google Scholar]
- de Amorim Neto B. H., Master thesis, University of Abertay Dundee, 2005 [Google Scholar]
- David F. Siewers V. FEMS Yeast Res. 2014;15:1–16. doi: 10.1111/1567-1364.12200. [DOI] [PubMed] [Google Scholar]
- Caspeta L. Coronel J. Montes de Oca A. Abarca E. González L. Martínez A. Biotechnol. Bioeng. 2019;116:2587–2597. doi: 10.1002/bit.27103. [DOI] [PubMed] [Google Scholar]
- Buchmuller B. C. Herbst K. Meurer M. Kirrmaier D. Sass E. Levy E. D. Knop M. Nat. Commun. 2019;10:1–13. doi: 10.1038/s41467-018-07882-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardi M. Gasch A. P. FEMS Yeast Res. 2017;17:1–10. doi: 10.1093/femsyr/fox042. [DOI] [PubMed] [Google Scholar]
- Codón A. C. Benítez T. Korhola M. Appl. Microbiol. Biotechnol. 1998;49:154–163. doi: 10.1007/s002530051152. [DOI] [PubMed] [Google Scholar]
- Lu Q. Zhang S. S. Wu R. Z. Huang R. B. Guangxi Sci. 2010;17:368–372. [Google Scholar]; , 376
- van Leeuwen J. Andrews B. Boone C. Tan G. Cold Spring Harb. Protoc. 2015:853–862. doi: 10.1101/pdb.top084111. [DOI] [PubMed] [Google Scholar]
- Montiel D. Kang H. S. Chang F. Y. Charlop-Powers Z. Brady S. F. Proc. Natl. Acad. Sci. U. S. A. 2015;112:8953–8958. doi: 10.1073/pnas.1507606112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijpers N. G. A. Solis-Escalante D. Bosman L. van den Broek M. Pronk J. T. Daran J. M. Daran-Lapujade P. Microb. Cell Fact. 2013;12:1–13. doi: 10.1186/1475-2859-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener U. Heinisch J. Koehler G. J. Voss D. Hegemann J. H. Nucleic Acids Res. 2002;30:23e–23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H. J. Kim Y. H. J. Microbiol. Biotechnol. 2018;28:826–830. doi: 10.4014/jmb.1802.02004. [DOI] [PubMed] [Google Scholar]
- Lai A. C. K. Wu W. L. Lau S. Y. Guan Y. Chen H. L. FEMS Immunol. Med. Microbiol. 2012;64:205–211. doi: 10.1111/j.1574-695X.2011.00884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E. Genetics. 2012;191:33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soam S. Kumar R. Gupta R. P. Sharma P. K. Tuli D. K. Das B. Energy. 2015;83:307–315. doi: 10.1016/j.energy.2015.02.025. [DOI] [Google Scholar]
- de Souza Paulino J. Dias do Prado C. Eleutherio E. C. A. Bonatto D. Malavazi I. Ferreira da Cunha A. Fungal Biol. 2018;122:583–591. doi: 10.1016/j.funbio.2017.12.006. [DOI] [PubMed] [Google Scholar]
- Chen Y. Chen D. Lu Q. Chen X. L. Lu Z. L. Wu R. Z. Huang R. B. Biotechnology. 2017;27:117–122. [Google Scholar]
- Jagtap R. S. Mahajan D. M. Mistry S. R. Bilaiya M. Singh R. K. Jain R. Appl. Microbiol. Biotechnol. 2019;103:1031–1042. doi: 10.1007/s00253-018-9532-1. [DOI] [PubMed] [Google Scholar]
- Inaba T. Watanabe D. Yoshiyama Y. Tanaka K. Ogawa J. Takagi H. Shimoi H. Shima J. AMB Express. 2013;3:74. doi: 10.1186/2191-0855-3-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. Y. Guo L. Chen D. Long S. Y. Li J. X. Lu Q. Sun L. Huang R. B. Guangxi Sci. 2014;21:108–114. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.