Abstract
Metabolomics has been used as a promising strategy to evaluate the efficacy of and potential targets for natural products. Alcoholic liver disease (ALD) as a result of chronic ethanol consumption has high morbidity and mortality. Geniposide possesses a hepatoprotective activity against ALD, but its mechanism of action is still not clear. In this study, serum metabolomics based on ultra-performance liquid chromatography-quadrupole time of flight-tandem mass spectrometry (UPLC-Q/TOF-MS) combined with ingenuity pathway analysis was used to explore the therapeutic mechanisms of geniposide. We found that the levels of AST, ALT, MDA, TG, and γ-GT in the geniposide-treated group were significantly decreased, and the level of GSH was significantly increased, compared with the model group. Meanwhile, geniposide effectively inhibits apoptosis and caspase-3 activity in liver tissue. A total of 33 metabolites were identified and related with the model group to illuminate the pathogenesis of ALD, 21 of which are regulated by geniposide, involving the relevant metabolic pathways, such as amino acid metabolism, arachidonic acid metabolism, pyruvate metabolism, TCA cycle, etc. Furthermore, a significant change in amino acid metabolism suggested that it might be a promising mechanism-related target for geniposide against ALD. It also showed that a metabolomic strategy using UPLC-Q/TOF-MS combined with ingenuity pathway analysis is a potentially powerful tool for providing a comprehensive understanding of the therapeutic mechanisms of natural products, but it also offers a theoretical basis for the prevention or treatment of disease.
Metabolomics has been used as a strategy to evaluate the efficacy of and potential targets for natural products.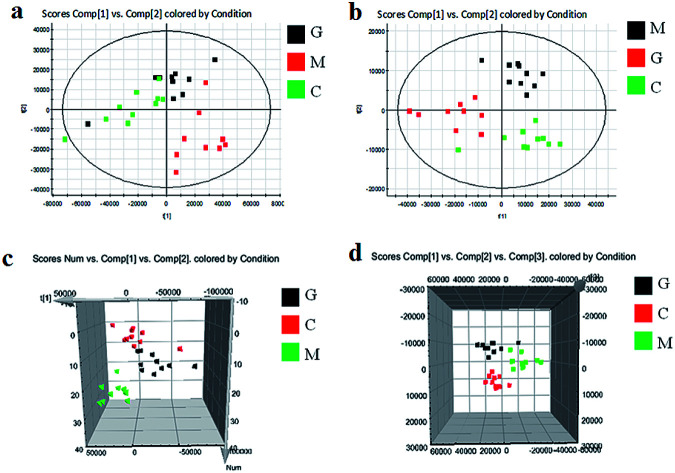
1. Introduction
Alcoholic liver disease (ALD) is one of the most important causes of chronic liver disease worldwide, and has seriously endangered social public health. It begins with a fatty liver, continuing with progressive fibrosis that eventually leads to cirrhosis.1,2 It is a clinical syndrome characterized by rapid hepatic decompensation, such as jaundice, coagulation impairment and encephalopathy.3 The pathogenesis of ALD refers mainly to hepatic steatosis, oxidative stress, acetaldehyde-mediated toxicity, and cytokine or chemokine-induced inflammation.4 Compared with other kinds of liver disease, the distinctive spectrum of alcoholic hepatitis (AH), that is still detected by a liver biopsy test as the gold standard for diagnosis in clinics, can develop any time in the pathological natural history of ALD, but its invasiveness means it has a relatively high cost to sufferers.5,6 The etiology of alcoholic liver disease is clear; however, its specific pathogenesis is complex. If the patients cannot be treated in time, it will cause death in up to 50% of cases.7 The prevention and treatment of ALD and other related diseases lack effective means of intervention and corresponding drugs, and this has become a difficulty and “bottleneck” in the field of public health. Therefore, it is an urgent task to carry out more in-depth research on ALD to find an effective way of discovering new treatment methods.
Traditional Chinese medicine (TCM) has been used in China for a long time, and has been applied to various kinds of disease, such as chronic liver disease.8,9 Yin Chen Hao Decoction originated from “Treatise on Febrile Diseases” is used to control Yang Huang syndrome that is highly correlated with hepatobiliary function and the clinical manifestations of ALD.10,11 The results of previous studies showed that the protective effect of Yin Chen Hao Decoction on ALD, and the chromatographic fingerprint of the samples in rats before and after oral administration were established using the serum pharmacochemistry method of traditional Chinese medicine.12,13 The in vitro and in vivo compounds were respectively detected and, using correlation analysis, the potential pharmacodynamic material basis of Yin Chen Hao Decoction in the treatment of liver injury was explored and it was found that three chemical components including geniposide were closely associated with the metabolism of liver damage.14 As one kind of iridoid glycoside compound, geniposide can reduce the free radical scavenging ability, enhance the ability to scavenge free radicals, inhibit lipid peroxidation, and significantly regulate the metabolic pathways of multiple disorders. In addition to its medicinal use, it can also be applied as a plant growth inhibitor and biological detection agent.15 However, there is insufficient exhaustive information about the medicinal mechanism in the therapeutic process of geniposide. The efficacy and treatment concept of TCM has received more and more attention from the international community. It is puzzling that the methods and theories of Chinese medicine for understanding diseases still lack a suitable modern scientific system of expression.
Metabolomics has come up with a noninvasive means and comprehensive probe for integral metabolic profiling by monitoring small molecules with 1000 Da molecular weight in urine, saliva, blood and other biological fluids and has become a pivotal element in systems biology.16 As the endpoint of the omic cascade, metabolome is the closest point to the phenotype, so that it identifies characteristic changes bound up with any physiological perturbations from in vitro and in vivo interference.17 The connection and dynamical rule between physiological and diseased changes are disclosed using qualitative analysis and quantitative determination of endogenous metabolites coupled with pattern recognition and other chemical detection sciences.18,19 Currently, the application of metabolomics in ALD has been carried out in the research fields of diagnosis, prognosis, pathogenesis and therapeutic effects.20,21 It was suggested that the accumulation of free fatty acids in the liver may lead to the earliest pathological change in ALD. The rise of metabolomics provides a method of integrating and analyzing multi-level and multidimensional data from the system level and expressing the role of TCM in modern scientific language.22 This feature is thoroughly comparable to the probity and systemic traits of TCM, suggesting it has the ability to demonstrate the latent usefulness and therapeutic mechanisms of TCM. This study employed a metabolomics strategy to explore the metabolic profile changes, and then analyze the biomarkers, metabolic pathways and key metabolic enzymes in an alcohol-induced mouse liver injury model. It is expected that it will reveal the pathological mechanism of ALD and seek the key target of geniposide in the treatment of ALD to provide a novel target for drug discovery for an ALD remedy.
2. Experimental method
2.1. Chemicals and reagents
Analytical grade alcohol was bought from Tianjin Chemical Reagent Co. (Shanghai, China). Distilled water was obtained from Watson's Food & Beverage Co. (Guangzhou, China). Leucine enkephalin was obtained from Sigma-Aldrich (USA) with a purity of more than 99%. The reference substance of geniposide was purchased from Shanghai Chemical Reagent Co. (Shanghai, China) of more than 99.7% purity. Saline solution was obtained from Tong Ren Tang Chinese Medicine Co., Ltd. (Beijing, China). The assay kit for alanine amino-transferase (ALT), aspartate aminotransferase (AST), maleic dialdehyde (MDA) and glutathione (GSH) were obtained from the Weifang SanWei Biotechnology Institute (Beijing, China). The assay kits for γ-glutamyl transpeptidase (γ-GT) and triglyceride (TG) were obtained from Cobioer Biosciences Company (Nanjing, China). The assay kits for hepatocyte apoptosis and caspase-3 activity detection were obtained from Roche Applied Science (Roche, Germany).
2.2. Establishment and administration of animal models
Male BALB/c mice (four-weeks-old, weighing 20 ± 2 g) came from the Drug Safety Evaluation Center in Heilongjiang University of Chinese Medicine. All mice were raised under a controllable temperature of 24 ± 2 °C, humidity 50 ± 5%, 12 h light and 12 h dark cycles and free water and food for a one-week acclimation period. Then, thirty mice were split into three groups at random, a control, model and geniposide-treated group, with ten animals in each group. The model of ALD in mice was established for seven consecutive days, during which the model and the geniposide-treated group were intragastrically administered with 60% (v/v) ethanol every day according to the body weight formula of 0.15 mL/10 g, and mice in the control group were administered an equal dose of purified water daily in the same way. The geniposide solution was prepared as follows: 9 mg of geniposide was dispersed in 0.4 mL of water and 0.4 mL of glycerol, followed by 10 min of ultrasonic processing for homogeneous mixing. The geniposide-treated group was given 1 mL/100 g of the geniposide solution daily. The control and the model group were nourished with an equal amount of purified water every day. All groups of mice were fed on a conventional diet and the treatment period was fourteen days. The experimental procedures were approved by the Animal Care and Ethics Committee at Heilongjiang University of Chinese Medicine and all experiments were performed in accordance with the declaration of Helsinki.
2.3. Biochemical index and histopathology detection
After two weeks of treatment, blood from the eyeball in the control, model and geniposide-treated groups was collected on the 22nd day. Subsequently, the liver was immediately removed. Blood samples were centrifuged at 3000 rpm at 4 °C for 10 min, and then the obtained serum was taken and reserved in a refrigerator at −80 °C until use. Serum ALT and AST activities were determined by the continuous monitoring method according to the instructions in the kit. The liver tissue was rinsed with ice-cold saline until there was no blood color and wiped dry with blotting paper. 0.5 g of left liver lobe and 4.5 mL of physiological saline were mixed to prepare a 10% liver tissue homogenate, and then centrifuged at 4500 rpm for 15 minutes at 4 °C. The liquid supernatant was used for a biochemical index assay according to the instructions of the kit, in which the r-GT activity was determined by the continuous monitoring method, the GSH activity was determined by chemical colorimetry, the MDA content was measured by the thiobarbituric acid colorimetric method and the TG content was measured by the glycerol oxidase-end point method. Part of the left lobe of the liver from the mouse was fixed with 10% formalin solution for liver histopathological examination. A cross-section was taken from the middle left lobe of the liver, and samples were taken to carry out HE staining and microscopic examination. Cell pathological changes were recorded, and the main lesion types were observed.
2.4. Hepatocyte apoptosis and caspase-3 activity detection
The liver tissue was firstly fixed with 4% paraformaldehyde, and then the prepared TUNEL reaction solution was added so that the A and B solutions in the kit were mixed in a proportion of 9 : 1 under dark conditions. The cell slice was processed at 37 °C for 1 h followed by a phosphate buffer saline (PBS) solution wash. After staining with Hoechst dye for 5 min, the state of hepatocyte apoptosis was observed under a fluorescence microscope and photographed. Liver tissue sections, after management with ice methanol, 1% Triton and 10% bovine serum albumin (BSA), were added to caspase-3 antibody and goat anti-rabbit immunoglobulin G-fluorescein isothiocyanate (IgG-FIFC) reagent for caspase-3 activity detection in the mouse liver. 1 μg mL−1 4′,6-diamidino-2-phenylindole (DAPI) solution was applied to the stain tissue section for 10 min, according to the instruction of the kits, and E800 immunofluorescence was performed for observation and photographing under a microscope.
2.5. Metabolomics study of serum samples
2.5.1. Handling of serum samples
The serum was thawed at 4 °C and vortexed for 10 s. 200 μL of serum sample was mixed with 800 μL of methanol, then the blend was shaken for 30 s, sonicated for 1 min and centrifuged at 13 000 rpm under 4 °C for 10 min. 800 μL of supernatants were performed in a 40 °C water bath, and then evaporated to dryness under a nitrogen gas stream at 45 °C. The residue was redissolved in 1 mL of 80% methanol and centrifuged at 13 000 rpm at 4 °C for 10 min. The obtained supernatant was delivered into a centrifuge tube and passed through a 0.22 μm membrane for the instrumental analysis. A quality control sample is considered to be an outstanding sample associated with all the constituents of the sample to be resolved and is applied to explore the essential information of the column state, which was prepared in equal volumes of three different groups in a corresponding time.
2.5.2. Chromatography and mass spectrometry analysis
Serum metabolomics analysis was conducted on a Waters ACQUITY UPLC H-Class (Waters Technology Co., Ltd., USA) system furnished with a pump, binary solvent delivery and auto sampler. A 2 μL solution was injected for separation by an UPLC ACQUITY BEH C18 column (1.7 μm, 2.1 × 100 mm) under 0.4 mL min−1 at 45 °C. The optimized elution system consists of solvent A (acetonitrile with 0.1% formic acid) and solvent B (water with 0.1% formic acid). The gradient procedure was optimized as follows: 0–2 min, 1–50% A; 2–4 min, 50–60% A; 4–7 min, 60–80% A; 7–8 min, 80–94% A, 8–9 min, 100% A. To ensure the durability and repeatability of the UPLC-MS system, the QC sample was disclosed every 10 samples from beginning to end. A needle wash cycle was carried out during the metabolomics analysis in order to dislodge the remains and prepare the next injections.
The obtained elution was promptly delivered to a Waters Micromass Q-TOF Micro Synapt High Definition Mass Spectrometer (Manchester, UK) equipped with an electrospray ion source in the positive ion mode and negative ion mode without splitting. The MS condition was set as follows: in positive ion scan mode, the capillary voltage was 3.0 kV; in negative ion mode, the capillary voltage was 2.6 kV; the cone voltage of the samples was set at 20 V; the desolvation gas temperature was set at 350 °C and the desolvation gas flow was set at 800 L h−1. Accurate mass determination and centroided data during the MS analysis was ensured using 0.1 ng mL−1 leucine-enkephalin solution ([M + H]+ = 556.2771 and [M–H]− = 554.2615) as the lock mass solution at 140 μL min−1, during which the parameters settled as follows: fragment voltage of 70 V, collision energy of 30 V, cell accelerator voltage of 20 V. The mass scan range was m/z 50–1200 Da. Nitrogen was used for gas drying and collision.
2.5.3. Data processing and pattern recognition analysis
Preprocessing of the original data includes peak data extraction, peak denoise, peak matching, peak alignment, and data export for non-target metabolomics by Progenesis QI software (Nonlinear Dynamics, 2014, version 1.0). After unit variance scaling and mean-centered method processing, the intensity of each ion was normalized to the total ion count and then a multivariate data matrix consisting of retention time (Rt), m/z value as well as the normalized peak area was produced and exported for further multivariate data analyses using EZinfo software (Waters Corp., Milford, USA). An unsupervised principal component analysis (PCA) was conducted to visualize common clustering and tendency. Orthogonal partial least-squared discriminant analysis (OPLS-DA) was used to ratify the PCA model, in which the parameters containing R2X, R2Y, and Q2 were calculated to evaluate the goodness of fit and predictive ability. Values of R2Y and Q2 close to 1.0 indicate an outstanding model. The S-plots were established following OPLS analysis according to their contribution to the mutation and conjunction rate of the changes in metabolic profiling in three different groups. Differential ions were selected from a VIP-plot arising from the loading plots of the OPLS-DA between the control and model groups. Variables where VIP > 1.0 in the OPLS-DA model, P < 0.05 in ANOVA and t-test and fold change value > 1.5 were selected as potential variables. The MS/MS fragment analysis was executed by the MassFragment™ application manager (Waters MassLynx V 4.1). Subsequently, the latent biomarkers were preliminarily identified by comparing the combination of accurate mass charge ratio and MS/MS fragmentation patterns to an online database such as METLIN or HMDB with a mass error of less than 20 ppm. The metabolic pathway analysis of potential biomarkers of geniposide against ALD was executed with metabolomics pathway analysis (MetPA) in MetaboAnalyst software 3.0 to discover the changed pathways.
2.6. Statistical analysis
The SPSS 20.0 software program (IBM, SPSS, Chicago, IL, USA) was employed for the statistical analysis. Data was presented as mean ± SD and scaled to Pareto variance. The statistical implication between the groups was analyzed by the Student's t-test, which is significant when the p values are less than 0.05, and is extremely significant when the p values are less than 0.01.
3. Results
3.1. Histopathological observations and biochemical analysis
In Fig. S1a,† the hepatic lobule structure of the mice in the control group was integrated, and the hepatocytes were fixed up neatly in the center of the central vein. The HE staining was uniform, and the nucleus that was present was large and the circle was mostly in the center of the cell. The contour of the hepatic sinus was clear. The liver lobule structure in the model group was disordered. Intravenous and hepatic sinus expansion was obvious. Hepatocytes were swollen, crowded and necrotic. In the cells, lipid droplets, fat vacuoles, and a large number of infiltrated inflammatory cells were observed. After treatment with geniposide, the liver lobular structure of the mouse gradually returned to normal and some cells were regularly arranged. The central vein and hepatic sinus were expanded and congested, and the blood vessels in the portal area had proliferated. Liver cells were still swollen and crowded, and the hepatocyte necrosis was reduced.
In Fig. S1b–g,† the liver function indexes, such as ALS and ALT, in mice with alcoholic liver injury were significantly increased, which can better evaluate the degree of alcoholic liver injury. Compared with the control group, the serum levels of AST and ALT in the model group had increased remarkably (P < 0.01). The AST and ALT levels in the geniposide-treated group were significantly lower than those in the model group (P < 0.05). An alcohol-induced liver injury model can easily lead to lipid peroxidation, in which the changed content of GSH and MDA can be detected to reflect the oxidative damage in mice. Compared with the control group, the GSH content in the model group had decreased significantly (P < 0.05), and the MDA content had increased significantly (P < 0.05). Compared with the model group, the GSH content in the geniposide-treated group was significantly up-regulated (P < 0.01) and the MDA content was down-regulated (P < 0.05). The detection of serum TG content can indicate lipid metabolism in the liver. Compared with the control group, the increased TG content in the model group was significant (P < 0.05). Compared with the model group, the TG content in the geniposide-treated group was significantly lower (P < 0.05). When liver injury occurs in mice caused by alcohol, intrahepatic biliary excretion is disordered and γ-GT in serum is significantly elevated, which are good indicators for evaluating alcoholic liver injury. Compared with the control group, the serum γ-GT content in the model group was significantly increased (P < 0.05). Compared with the model group, the γ-GT content in the geniposide-treated group was significantly lower (P < 0.05).
3.2. Hepatocyte apoptosis and caspase-3 activity detection
In Fig. S2a,† a TUNEL in situ terminal apoptosis assay showed that the control group was evenly stained and only a few positive cells were distributed, showing a normal state. The apoptosis cells in the liver tissue of the model group were dense and deeply stained. The geniposide-treated group had a certain degree of reversal effect on hepatocyte apoptosis induced by alcoholic liver injury. In Fig. S2c† the apoplastic cell density had decreased significantly and tended towards the control, which was notably different from the model group (P < 0.01). Caspase-3 as the most vital terminal cleavage enzyme in the process of apoptosis plays an irreplaceable role in metabolism. From the CASPASE-3 activity assay in Fig. S2b,† the model group was heavily stained, indicating the strong activity of CASPASE-3, the wide distribution range, and the high areal density value. After administration of geniposide, the expressive activity of CASSASE was inhibited, which was very different from the model group (P < 0.01) in Fig. S2d.†
3.3. Trajectory and metabolic profiling analysis
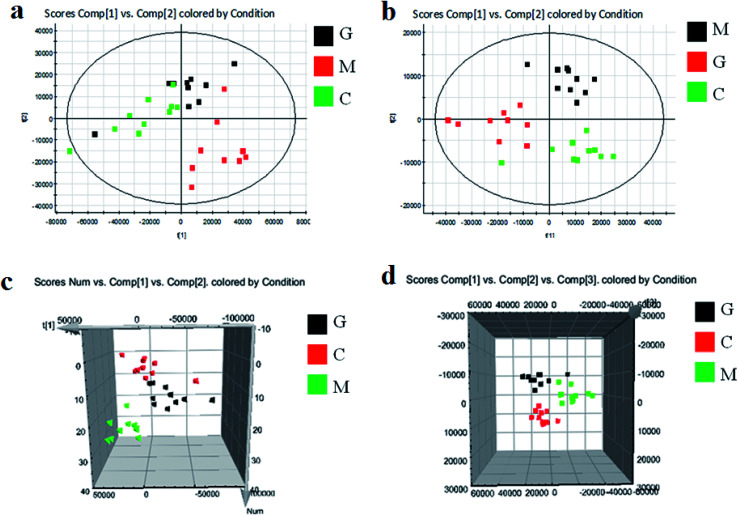
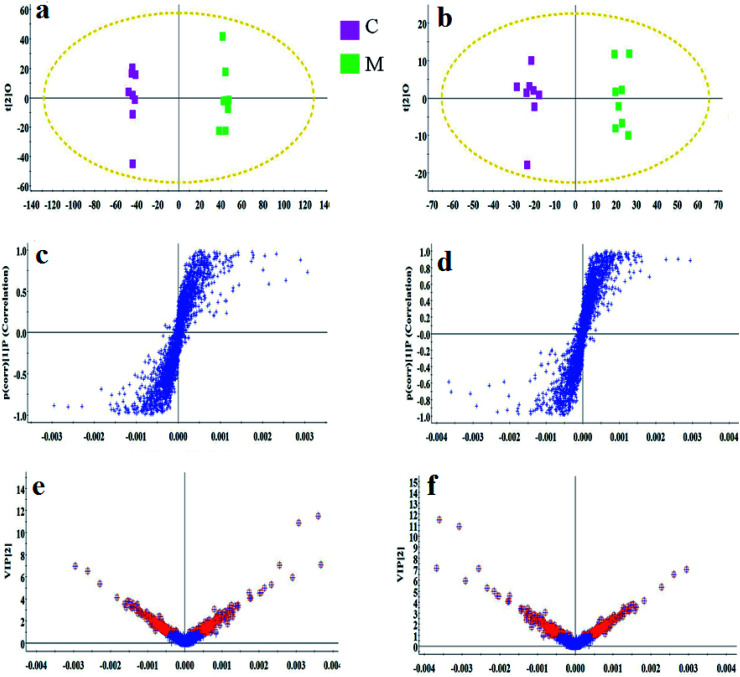
As shown in Fig. S3,† metabolic profiles of the serum samples were obtained using UPLC-Q/TOF-MS in the positive and negative ion modes. The trajectory analysis of the PCA score plots in the control, model and geniposide-treated groups shows clear separations in both positive (Fig. 1a) and negative ion modes (Fig. 1b). 3D PCA score plots are more easily visualized for explaining the liver injury caused by alcohol and the pharmacodynamic activity of geniposide, in which the clustering in the model group showed the maximum separation trend from the control group causing significant metabolic disturbance and pathobiological changes; in addition, the geniposide-treated group was located between the control and the model group, and remained relatively far from the model group in both positive (Fig. 1c) and negative ion modes (Fig. 1d). Significant metabolic differences between the control and the model group indicated that the establishment of the animal model was successful and further multivariate statistical analysis for seeking discriminative metabolites is essential to distinguish the relationships among the three different groups. The OPLS-DA model in both positive (Fig. 2a) and negative ion modes (Fig. 2b) derived from UPLC-Q/TOF-MS data displayed an obvious separation between the control and model groups. The parameters R2Y and Q2 obtained by cross-validation were 0.924 and 0.675 in the positive mode, and 0.908 and 0.707 in the negative mode, which suggested that these models have satisfactory goodness-of-fit and goodness-of-prediction to the data.
Fig. 1. Score plots of PCA analysis in both ion modes of blood samples in the control, model and geniposide-treated groups. (a) and (b) are 2D maps in positive and negative ion modes; (c) and (d) are 3D maps in positive and negative ion modes. C: control group; M: model group; G: geniposide-treated group.
Fig. 2. OPLS-DA score plot, S-plot and VIP-plot of the OPLS-DA model between the control and the model group in the positive and negative ion modes of blood samples. (a) OPLS-DA score plot in the positive ion mode; (c) S-plot in the positive ion mode; (e) VIP-plot in the positive ion mode; (b) OPLS-DA score plot in the negative ion mode; (d) S-plot in the negative ion mode; (f) VIP-plot in the negative ion mode. C: control group; M: model group.
3.4. Metabolite selection and identification
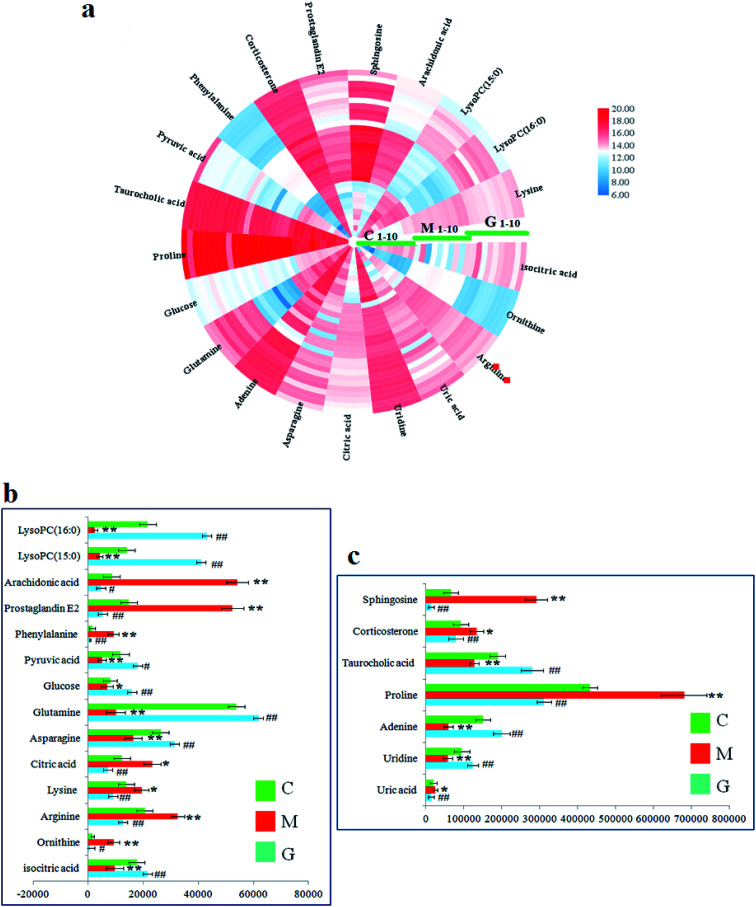
In the S-plot (Fig. 2c and d), the ions further away from the origin make a greater contribution to the ALD disorder in the metabolic trajectory and are likely to be considered as potential metabolites. Identified biomarkers were selected in accordance with their VIP values from the V-plots (Fig. 2e and f) followed by OPLS-DA between the control and the model group. In general, the VIP values reflects the influence of each variable, which the longer the distance, the greater the impact value. Hence, the serum VIP value standard was increased to narrow down the targets. A total of 33 metabolites in the serum from positive and negative ion mode were screened out and identified with VIP values greater than 1.5 and P-values from the t-test of less than 0.05, combined with the accurate mass number, Rt, MS/MS information, standard sample spectra and online database information, which were differentially expressed between the control and ALD model groups (Table S1†). 21 biomarkers are effectively regulated by geniposide, of which 10 metabolites were elevated in content compared with the model group, including isocitric acid, uridine, asparagine, adenine, glutamine, glucose, taurocholic acid, pyruvic acid, LysoPC (15 : 0) and LysoPC (16 : 0). Meanwhile, 11 metabolites were reduced in content compared with the model group, including ornithine, arginine, lysine, uric acid, citric acid, proline, phenylalanine, corticosterone, prostaglandin E2, sphingosine and arachidonic acid. Further, the differentially abundant metabolites are visualized as a heat-map in Fig. 3a, which directly shows the variation in relative concentrations in the identified metabolites of the serum between the control, model and geniposide-treated groups. In Fig. 3b and c, relative signal intensities of identified serum metabolites using UPLC-MS are listed.
Fig. 3. (a) Heatmap visualization of 21 metabolites in serum samples from the control, model and geniposide-treated groups. Different colors show changes in the relative level of metabolites. Red indicates an increase in content, and blue indicates a decrease in content. (b) and (c) Relative signal intensities of the metabolites identified by UPLC-Q/TOF-MS between the control, model and geniposide-treated groups.
3.5. Metabolic pathway analysis
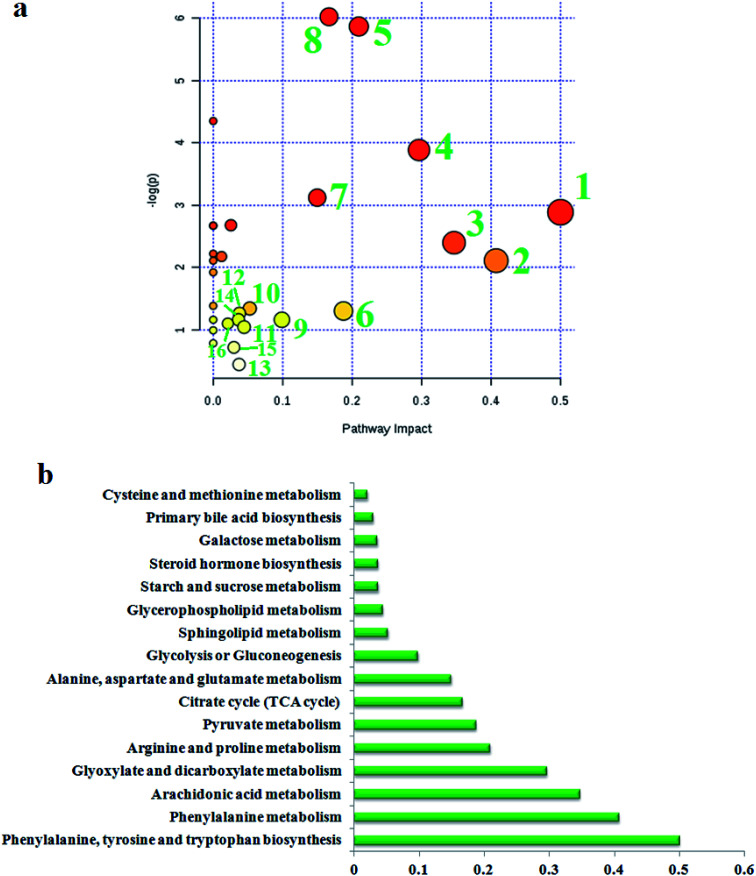
To find the impacts of these selected metabolites and to analyze the possible metabolic pathway of geniposide against ALD, MetPA was built to determine the most pertinent pathways modified by geniposide treatment in Fig. 4a. After using the Pareto method to standardize the data, it was shown that some metabolic pathways in the blood, involved in phenylalanine, tyrosine and tryptophan biosynthesis, phenylalanine metabolism, arachidonic acid metabolism, glyoxylate and dicarboxylate metabolism, arginine and proline metabolism, pyruvate metabolism, citrate cycle (TCA cycle), alanine, aspartate and glutamate metabolism, glycolysis or gluconeogenesis, sphingolipid metabolism, glycerophospholipid metabolism, starch and sucrose metabolism, steroid hormone biosynthesis, galactose metabolism, primary bile acid biosynthesis, cysteine and methionine metabolism, were associated with the process of geniposide acting against ALD. The metabolic pathway influence value is displayed in Fig. 4b, where a metabolic pathway with an influence value greater than or equal to 0.10 can be selected as a potential key metabolic pathway for drugs. Then, integrated network analysis of geniposide in ALD was performed to outline the biochemical relationships. From the KEGG global metabolic network, which can map metabolites in model animals with ALD after geniposide administration and enzymes/KOs (KEGG Orthologs) (Fig. S4a†), it is appropriate to confirm the results from common metabolomics and metagenomics studies, mostly relating to amino acid metabolism, citrate cycle (TCA cycle), aminoacyl-tRNA biosynthesis, glyoxylate and dicarboxylate metabolism, taurine and hypotaurine metabolism, glycolysis or gluconeogenesis, purine metabolism, nitrogen metabolism. A metabolite–metabolite interaction network based on reactions from similar chemical structures and similar molecular activities that highlight potential functional relationships between a wide set of annotated metabolites is shown in Fig. S4b,† where seven metabolites, pyruvic acid, citric acid, l-lysine, l-glutamine, l-arginine, ornithine and adenine, stand out with 230, 131, 114, 103, 102, 93 and 89 degrees. The relationships of genes and the main metabolites of the serum sample under CA exposure are demonstrated in Fig. S4c,† mainly referring to prostaglandin E2, arachidonic acid, l-arginine, citric acid, ornithine, l-lysine, l-glutamine and adenine, respectively.
Fig. 4. (a) Pathway analysis of metabolic changes in serum samples after geniposide treatment. (1) Phenylalanine, tyrosine and tryptophan biosynthesis; (2) phenylalanine metabolism; (3) arachidonic acid metabolism; (4) glyoxylate and dicarboxylate metabolism; (5) arginine and proline metabolism; (6) pyruvate metabolism; (7) citrate cycle (TCA cycle); (8) alanine, aspartate and glutamate metabolism; (9) glycolysis or gluconeogenesis; (10) sphingolipid metabolism; (11) glycerophospholipid metabolism; (12) starch and sucrose metabolism; (13) steroid hormone biosynthesis; (14) galactose metabolism; (15) primary bile acid biosynthesis; (16) cysteine and methionine metabolism. (b) The metabolic pathway influences the value of different pathways.
4. Discussion
ALD is linked to the toxic effects of alcohol and its metabolites on the liver caused by oxidative stress, immune-mediation and cytokines, apoptosis, endotoxin, genetic polymorphism, and the superposition of viruses. Alcohol in the body is oxidized to acetaldehyde and further oxidized to acetic acid by the action of alcohol dehydrogenase.23,24 During the oxidation of ethanol, oxidized coenzyme I is converted to reduced coenzyme I, and the cells are in a redox state. The accumulation of reduced coenzyme I, high concentrations of free radicals and acetaldehyde affect the metabolism of fat, sugar and protein in the liver, resulting in fatty acid metabolism, glycogen synthesis, protein synthesis, secretion, excretion disorders, especially the citrate cycle, amino acid oxidation and oxidative metabolism of fatty acids. The increased synthesis of fatty acids and the level of acceleration of l-glycerol-3-phosphate and fatty acids lead to the increased activity of TG synthesis, which is beyond the secretion capacity of VLDL to synthesize TG, resulting in an accumulation of fat in liver cells and a significantly elevated level of MDA.25,26 The results of biochemical indicators showed that the levels of AST, ALT, TG and γ-GT in the model group were abnormally elevated, indicating that the liver and gallbladder function and lipid metabolism were disordered in animals with alcoholic livers. After treatment with geniposide, the levels of AST, ALT, TG, MDA and γ-GT in the model group decreased, and GSH levels increased.
As the main organ for amino acid metabolism, the liver is rich in enzyme content, leading to an active change in amino acid metabolism. When there is liver injury or pathological change, amino acid metabolism in liver cells decreases on account of the liver function being presented with an obstacle, which brings about a rise in the concentration of amino acid in the blood and a lack of it in the body.27 The liver is the main site of phenylalanine (Phe) metabolism, and Phe as an essential amino acid in the human body must be provided by food. Excessive Phe concentration can cause PKU, nervous system damage, albinism and other diseases, and can lead to mental retardation, stunted growth, coma and even death.28 The study found that Phe levels were abnormally elevated in the model group compared to the control group. Geniposide could improve Phe levels by regulating phenylalanine, tyrosine, tryptophan biosynthesis and phenylalanine metabolism pathways. Damage to the liver by a high content of phenylalanine may be related to lipid peroxidation. The body in a physiological and pathological state can produce oxygen free radicals. When ROS free radicals cannot be cleared in time, they will attack PUFA in the biofilm, giving rise to lipid peroxidation and lipid peroxide formation. A large amount of enzymatic removal agent, such as GSH-PX in the model group, was expended, and then the content of H2O2 and superoxide anion free radicals in the body was increased, eventually causing liver damage.29 After geniposide treatment, the GSH level recovered to normal and was significantly different from that of the model group. Glutamine, the main energy source of intestinal mucosa, lymphocyte and pancreatic acinar cells, provides a substrate for anabolism and promotes cell proliferation. It is abundant in tissues and also involved in the synthesis of the antioxidant glutathione. At present, glutamine is not only considered as a conditional amino acid, but also as a substance with pharmacological effects.30 Arginine is a component of ornithine circulation and has an extremely important physiological function, which is its ability to enhance the enzymatic activity of arginine in the liver, contributing to the conversion of ammonia in the blood to urea and excretion. The body cannot maintain a normal nitrogen balance and physiological functions without arginine. The lack of arginine in patients can lead to high blood ammonia, and even coma. As a precursor of citrulline, proline, and glutamate, ornithine is synthesized in the body from arginine, which promotes the release of growth hormone. It is useful for maintaining a normal immune system and liver function, such as removing ammonia from the liver and repairing internal tissue. In addition, levels of ornithine are high in skin and connective tissue, which helps maintain a healthy skin and restore damaged tissue.31,32 Proline is not only an ideal osmotic regulator, but also can be used for membrane and enzyme protection, and as a free radical scavenger and cytoplasmic osmotic balance regulator.33 The arginine, ornithine and proline levels were down-regulated and the glutamine level was up-regulated in the blood of mice from the model group by regulating arginine and proline metabolism, and alanine, aspartate and glutamate metabolism under geniposide treatment. Pyruvate is one of the intermediate products involved in the basic metabolism of the whole organism. It can realize the mutual conversion of sugars, fats and amino acids in the body through the acetyl CoA and citrate cycle (TCA cycle). Therefore, pyruvate plays an important and pivotal role in the metabolism of the three nutrients.34 The level of pyruvate is obviously reduced in the model group, with reference to the disordered changes of cysteine and methionine metabolism, alanine, aspartate and glutamate metabolism, pyruvate metabolism, glycolysis or gluconeogenesis, and TCA cycle in the model group. Geniposide can relieve the pathological symptoms of ALD patients by regulating the above metabolic pathways.
Arachidonic acid (AA) is a kind of omega-6 polyunsaturated fatty acid, which possesses a series of physiological activities such as cholesterol esterification, increase in vascular elasticity, decrease in blood viscosity and blood cell function regulation. It also plays a vital role in preventing cardiovascular disease, diabetes and cancer. AA is a direct precursor of many bioactive derivatives of cyclic eicosane acids, such as prostaglandin E2 (PGE2), prostaglandin (PGI2), thromboxane A2 (TXA2), and leucocyte triene and C4 (LTC4). These bioactive substances have important regulatory effects on lipid protein metabolism, hemorheology, vascular elasticity, leukocyte function and platelet activation.35 When the liver is injured, the uptake, metabolic inactivation and excretion of AA and its metabolites are bound to be impaired. Compared with the control group, the blood AA and prostaglandin E2 level in the model group are elevated, indicating that the arachidonic acid metabolism is in an abnormal state. Prostaglandin can cause vasodilation, erythema, pain and edema in inflammatory sites, which is closely associated with the main cause of pain in the liver area of clinical ALD patients.36 Bile acids usually retain an equilibrium state of free and bound forms in the peripheral circulation. When liver or intestinal diseases occur, the balance is impaired, especially the synthesis, reabsorption and excretion of bile acids, which may result in abnormal changes in total bile acid levels. Taurocholic acid can reduce the capillary permeability of inflammatory tissues, inhibit inflammatory swelling, and impede the production of NO, PGE2, histamine and other inflammatory mediators.37 In this study, it was found that the content of taurocholic acid in the model group was significantly reduced, which was related to fever and inflammatory reaction in patients with alcoholic livers. Geniposide with its analgesic and anti-inflammatory role can increase the level of taurocholic acid by improving the abnormality of the primary bile acid biosynthesis pathway.
Functional metabolomics as a promising strategy can used for discovering biomarkers,38–49 metabolic mechanisms,50–65 evaluating efficacy,66–78 and revealing the key metabolism pathways as potential targets for natural products.79–86 The metabolic biomarkers and pathways are crucial to revealing the pathological mechanism87–95 and insight into the pharmacological targets of natural products.96–108 The TCA cycle is the ultimate universal oxidation pathway for fats, carbohydrates, and amino acids, and is the key most central pathway linking almost all individual metabolic pathways. In addition, the TCA cycle is closely related to some liver diseases. Some studies reported that lipid accumulation and a loss of insulin action resulted in elevation of the oxidative and anaplerotic pathways of the hepatic TCA cycle in mice, which accelerates insulin to reserve and a fatty liver due to a high-fat diet.109 In our study, citric acid levels in the model group were significantly increased compared with the control group, while isocitric acid and d-glucose levels were significantly decreased, which are involved in galactose metabolism, starch and sucrose metabolism, glyoxylate and dicarboxylate metabolism and the TCA cycle. These changes were associated with hypoglycemia, decreased appetite and other clinical symptoms of ALD illness. After treatment with geniposide, the abnormal condition was improved to close to a normal level. Glycerophospholipids (GPs), as the stores of lipid mediators, act in the membrane protein process, as transporters, receptors, and ion channels. Ethanol-treated mice may go through lipid metabolism disorders that can lead to liver damage. Lysolipids are metabolites of GP metabolism and can be produced briefly during GP reconstruction.110 It has been suggested that LysoPC plays a critical role in the progression of liver injury and its deficiency reflects rapid membrane phosphatidylcholine renewal in the early stages of liver injury. Phosphatidylcholine (PC), as a major supporting nutrient of the liver, has a significant protective activity on liver damage caused by viruses, drugs, alcoholism and other toxic effects.111 In this study, LysoPC (18 : 1), LysoPC (17 : 0), LysoPC (15 : 0), and LysoPC (16 : 0) levels were significantly reduced in the model group, indicating that GP metabolism is down-regulated during ALD processes. By improving the glycerophospholipid metabolism pathway after geniposide treatment, the levels of LysoPC (15 : 0) and LysoPC (16 : 0) tended towards normal. Serum sphingosine levels were significantly elevated in ALD mice in this study, in which the changes in sphingolipid metabolism may be related to inflammatory response and increased ceramide. Ceramide, sphingolipid, free sphingosine and others based on phosphates and complex sphingolipids are considered to be highly diverse lipids. These lipids are involved in complicated signaling processes, including proliferation, differentiation, and apoptosis, and thus may affect the pathogenesis of various diseases, especially ALD.
5. Conclusion
This study aimed to systematically discover the biomarkers of ALD and undertake an investigation into the mechanism of geniposide based on an UPLC-Q/TOF-MS serum metabolomic strategy. PCA analysis of serum metabolic profiles showed that the geniposide-treated group was close to the control group in the three-dimensional space and away from the model group. Meanwhile, a total of 33 biomarkers were associated with ALD, 21 of whose levels were effectively regulated by geniposide. The results of pathway analysis implied that amino acid metabolism is a promising target for geniposide against ALD. In addition, with the help of biochemical analysis and histopathological observation, alcohol-induced ALD was confirmed and geniposide has the ability to ameliorate liver and gallbladder function, antioxidant capacity and lipid metabolism of ALD mice in keeping with the result of metabolomics. The metabolomics technique could offer a thorough opinion of the properties of ALD and also provide better direction for drug discovery and the development of effective herbal medicines in ALD prevention and treatment.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by grants from the Key Program of Natural Science Foundation of State (Grant No. 81830110, 81861168037, 81973745, 81903818, 81903847, 81430093, 81673586, 81703685), National Key Research and Development Program of China (2018YFC1706103), National Key Subject of Drug Innovation (Grant No. 2015ZX09101043-005, 2015ZX09101043-011), TCM State Administration Subject of Public Welfare (Grant No. 2015468004), Major Projects of Application Technology Research and Development Plan in Heilongjiang Province (GA18C004, GX16C003), Natural Science Foundation of Heilongjiang Province (YQ2019H030, LH2019H056, QC2018117, H2016056), University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province (UNPYSCT-2015118, UNPYSCT-2016213, UNPYSCT-2016212), Young Talent Lift Engineering Project of China Association of Traditional Chinese Medicine (QNRC2-B06), Foundation of Heilongjiang University of Chinese Medicine (2018jc01, 2018bs02, 2018bs05, 201809), Nursing Program for Young Scholars with Creative Talents of Heilongjiang University of Chinese Medicine (2018RCQ13, 2018RCD21), Longjiang Scholar Program of Education Department of Heilongjiang Province (Q201916), Heilongjiang Touyan Innovation Team Program.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra09305b
References
- Miller A. M. Horiguchi N. Jeong W. I. et al., Molecular mechanisms of alcoholic liver disease: innate immunity and cytokines. Alcohol.: Clin. Exp. Res. 2011;35(5):787–793. doi: 10.1111/j.1530-0277.2010.01399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrekar P. Epigenetic regulation in alcoholic liver disease. World J. Gastroenterol. 2011;17:2456–2464. doi: 10.3748/wjg.v17.i20.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D. Sgrillo J. Daniels L. H. Managing alcoholic liver disease. Nursing. 2014;44(11):30–40. doi: 10.1097/01.NURSE.0000454950.13699.e0. [DOI] [PubMed] [Google Scholar]
- Seitz H. K. Bataller R. Cortez-Pinto H. et al., Alcoholic liver disease. Nat. Rev. Dis. Primers. 2018;4(1):16. doi: 10.1038/s41572-018-0014-7. [DOI] [PubMed] [Google Scholar]
- Poynard T. Mathurin P. Lai C. L. et al., A comparison of fibrosis progression in chronic liver diseases. J. Hepatol. 2003;38:257–265. doi: 10.1016/S0168-8278(02)00413-0. [DOI] [PubMed] [Google Scholar]
- Chacko K. R. Reinus J. Spectrum of Alcoholic Liver Disease. Clin. Liver Dis. 2016;20(3):419–427. doi: 10.1016/j.cld.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Mellinger J. L. Epidemiology of Alcohol Use and Alcoholic Liver Disease. Clin. Liver Dis. 2019;13(5):136–139. doi: 10.1002/cld.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. Sun H. Wang Z. et al., Metabolomics: towards understanding traditional Chinese medicine. Planta Med. 2010;76(17):2026–2035. doi: 10.1055/s-0030-1250542. [DOI] [PubMed] [Google Scholar]
- Zhang A. Sun H. Wang X. Potentiating therapeutic effects by enhancing synergism based on active constituents from traditional medicine. Phytother. Res. 2014;28(4):526–533. doi: 10.1002/ptr.5032. [DOI] [PubMed] [Google Scholar]
- Zhang T. Zhang A. Qiu S. et al., High-throughput metabolomics approach reveals new mechanistic insights for drug response of phenotypes of geniposide towards alcohol-induced liver injury by using liquid chromatography coupled to high resolution mass spectrometry. Mol. BioSyst. 2017;13(1):73–82. doi: 10.1039/C6MB00742B. [DOI] [PubMed] [Google Scholar]
- Zhang A. Sun H. Qiu S. et al., Advancing drug discovery and development from active constituents of yinchenhao tang, a famous traditional chinese medicine formula. Evid.-Based Complementary Altern. Med. 2013;2013 doi: 10.1155/2013/257909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. Zhang A. H. Yang L. et al., High-throughput chinmedomics strategy for discovering the quality-markers and potential targets for Yinchenhao decoction. Phytomedicine. 2019;54:328–338. doi: 10.1016/j.phymed.2018.04.015. [DOI] [PubMed] [Google Scholar]
- Zhang A., Sun H., Sun W., et al., Metabolomics and Proteomics Annotate Therapeutic Mechanisms of Geniposide, Chinmedomics, Academic Press, 2015, pp. 157–173 [Google Scholar]
- Sun H. Zhang A. Yan G. et al., Proteomics study on the hepatoprotective effects of traditional Chinese medicine formulae Yin-Chen-Hao-Tang by a combination of two-dimensional polyacrylamide gel electrophoresis and matrix-assisted laser desorption/ionization-time of flight mass spectrometry. J. Pharm. Biomed. Anal. 2013;75:173–179. doi: 10.1016/j.jpba.2012.11.025. [DOI] [PubMed] [Google Scholar]
- Wang X. Zhang A. Yan G. et al., Metabolomics and proteomics annotate therapeutic properties of geniposide: targeting and regulating multiple perturbed pathways. PLoS One. 2013;8(8):e71403. doi: 10.1371/journal.pone.0071403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. Sun H. Wang X. Urinary metabolic profiling of rat models revealed protective function of scoparone against alcohol induced hepatotoxicity. Sci. Rep. 2014;4:6768. doi: 10.1038/srep06768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi C. A. Nieman D. C. Signini E. F. et al., Metabolomics-Based Studies Assessing Exercise-Induced Alterations of the Human Metabolome: A Systematic Review. Metabolites. 2019;9(8):E164. doi: 10.3390/metabo9080164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. H. Sun H. Han Y. et al., Ultraperformance liquid chromatography-mass spectrometry based comprehensive metabolomics combined with pattern recognition and network analysis methods for characterization of metabolites and metabolic pathways from biological data sets. Anal. Chem. 2013;85(15):7606–7612. doi: 10.1021/ac401793d. [DOI] [PubMed] [Google Scholar]
- Wu G. Zhang W. Li H. Application of metabolomics for unveiling the therapeutic role of traditional Chinese medicine in metabolic diseases. Ethnopharmacology. 2019;242:112057. doi: 10.1016/j.jep.2019.112057. [DOI] [PubMed] [Google Scholar]
- Zhang A. Sun H. Wang X. Power of metabolomics in biomarker discovery and mining mechanisms of obesity. Obes. Rev. 2013;14(4):344–349. doi: 10.1111/obr.12011. [DOI] [PubMed] [Google Scholar]
- Lelliott C. J. Lopez M. Curtis R. K. et al., Transcript and metabolite analysis of the effects of tamoxifen in rat liver reveals inhibition of fatty acid synthesis in the presence of hepatic steatosis. FASEB J. 2005;19:1108–1119. doi: 10.1096/fj.04-3196com. [DOI] [PubMed] [Google Scholar]
- Zhang A. Liu Q. Zhao H. et al., Phenotypic characterization of nanshi oral liquid alters metabolic signatures during disease prevention. Sci. Rep. 2016;6:19333. doi: 10.1038/srep19333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschke R. Alcoholic Liver Disease: Current Mechanistic Aspects with Focus on Their Clinical Relevance. Biomedicines. 2019;7(3):E68. doi: 10.3390/biomedicines7030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschke R. Alcoholic Liver Disease: Alcohol Metabolism, Cascade of Molecular Mechanisms, Cellular Targets, and Clinical Aspects. Biomedicines. 2018;6(4):E106. doi: 10.3390/biomedicines6040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K. S. Balakrishnan V. Mukherjee S. et al., Evaluation of blood oxidative stress-related parameters in alcoholic liver disease and non-alcoholic fatty liver disease. Scand. J. Clin. Lab. Invest. 2008;68(4):323–334. doi: 10.1080/00365510701673383. [DOI] [PubMed] [Google Scholar]
- Wang M. Zhang X. Ma L. J. et al., Omega-3 polyunsaturated fatty acids ameliorate ethanol-induced adipose hyperlipolysis: A mechanism for hepatoprotective effect against alcoholic liver disease. Biochim. Biophys. Acta, Mol. Basis Dis. 2017;1863(12):3190–3201. doi: 10.1016/j.bbadis.2017.08.026. [DOI] [PubMed] [Google Scholar]
- Tedesco L. Corsetti G. Ruocco C. et al., A specific amino acid formula prevents alcoholic liver disease in rodents. Am. J. Physiol.: Gastrointest. Liver Physiol. 2018;314(5):G566–G582. doi: 10.1152/ajpgi.00231.2017. [DOI] [PubMed] [Google Scholar]
- Swierczynski J. Sledzinski T. Slominska E. et al., Serum phenylalanine concentration as a marker of liver function in obese patients before and after bariatric surgery. Obes. Surg. 2009;19(7):883–889. doi: 10.1007/s11695-008-9521-z. [DOI] [PubMed] [Google Scholar]
- Seitz H. K. Mueller S. The role of Cytochrome P4502E1 in Alcoholic Liver Disease and alcohol mediated carcinogenesis. Z. Gastroenterol. 2019;57(1):37–45. doi: 10.1055/a-0784-8815. [DOI] [PubMed] [Google Scholar]
- Lin Z. Cai F. Lin N. et al., Effects of glutamine on oxidative stress and nuclear factor-κB expression in the livers of rats with nonalcoholic fatty liver disease. Exp. Ther. Med. 2014;7(2):365–370. doi: 10.3892/etm.2013.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuo Y. Imai T. Shibato J. et al., Omic analyses unravels global molecular changes in the brain and liver of a rat model for chronic Sake (Japanese alcoholic beverage) intake. Electrophoresis. 2009;30(8):1259–1275. doi: 10.1002/elps.200900045. [DOI] [PubMed] [Google Scholar]
- Butterworth R. F. L-Ornithine L-Aspartate for the Treatment of Sarcopenia in Chronic Liver Disease: The Taming of a Vicious Cycle. Can. J. Gastroenterol. Hepatol. 2019;2019:8182195. doi: 10.1155/2019/8182195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesgari-Abbasi M. Valizadeh H. Mirzakhani N. et al., Protective effects of di- and tri-peptides containing proline, glycine, and leucine on liver enzymology and histopathology of diabetic mice. Arch. Physiol. Biochem. 2019;9:1–10. doi: 10.1080/13813455.2019.1662453. [DOI] [PubMed] [Google Scholar]
- Ferriero R. Nusco E. De Cegli R. et al., Pyruvate dehydrogenase complex and lactate dehydrogenase are targets for therapy of acute liver failure. J. Hepatol. 2018;69(2):325–335. doi: 10.1016/j.jhep.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T. Lentsch A. B. Changes in arachidonic acid metabolism during liver ischemia triggers induction of inflammatory injury. Hepatology. 2018;68(4):1642–1643. doi: 10.1002/hep.29870. [DOI] [PubMed] [Google Scholar]
- Huang X. P. Wang Y. Chen L. et al., Elevated serum prostaglandin E2 predicts the risk of infection in hepatitis B virus-related acute-on-chronic liver failure patients. Asian Pac. J. Trop. Med. 2017;10(9):916–920. doi: 10.1016/j.apjtm.2017.08.008. [DOI] [PubMed] [Google Scholar]
- Liu Z. Zhang Z. Huang M. et al., Taurocholic acid is an active promoting factor, not just a biomarker of progression of liver cirrhosis: evidence from a human metabolomic study and in vitro experiments. BMC Gastroenterol. 2018;18(1):112. doi: 10.1186/s12876-018-0842-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. Zhang A. H. Song Q. et al., Functional metabolomics discover pentose and glucuronate interconversion pathways as promising targets for Yang Huang syndrome treatment with Yinchenhao Tang. RSC Adv. 2018;8:36831–36839. doi: 10.1039/C8RA06553E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan Y. Zhou X. Liu Q. et al., Serum metabolomics strategy for understanding pharmacological effects of ShenQi pill acting on kidney yang deficiency syndrome. J. Chromatogr. B. 2016;1026:217–226. doi: 10.1016/j.jchromb.2015.12.004. [DOI] [PubMed] [Google Scholar]
- Li X. Zhang A. Sun H. et al., Metabolic characterization and pathway analysis of berberine protects against prostate cancer. Oncotarget. 2017;8:65022–65041. doi: 10.18632/oncotarget.17531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. N. Zhang A. Wang M. et al., Screening the active compounds of Phellodendri Amurensis cortex for treating prostate cancer by high-throughput chinmedomics. Sci. Rep. 2017;7:46234. doi: 10.1038/srep46234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. Sun H. Yan G. et al., Metabolomics in diagnosis and biomarker discovery of colorectal cancer. Cancer Lett. 2014;345:17–20. doi: 10.1016/j.canlet.2013.11.011. [DOI] [PubMed] [Google Scholar]
- Wang X. J. Ren J. L. Zhang A. H. et al., Novel applications of mass spectrometry-based metabolomics in herbal medicines and its active ingredients: Current evidence. Mass Spectrom. Rev. 2019;38(4–5):380–402. doi: 10.1002/mas.21589. [DOI] [PubMed] [Google Scholar]
- Zhang A. Yan G. Han Y. Wang X. Metabolomics approaches and applications in prostate cancer research. Appl. Biochem. Biotechnol. 2014;174:6–12. doi: 10.1007/s12010-014-0955-6. [DOI] [PubMed] [Google Scholar]
- Li Y. Qiu S. Zhang A. H. et al., High-throughput metabolomics to identify metabolites to serve as diagnostic biomarkers of prostate cancer. Anal. Methods. 2016;8:3284–3290. doi: 10.1039/C6AY00127K. [DOI] [Google Scholar]
- Zhang A. Sun H. Wu X. et al., Urine metabolomics. Clin. Chim. Acta. 2012;414:65–69. doi: 10.1016/j.cca.2012.08.016. [DOI] [PubMed] [Google Scholar]
- Xie J. Zhang A. H. Qiu S. et al., Identification of the perturbed metabolic pathways associating with prostate cancer cells and anticancer affects of obacunone. J. Proteomics. 2019;206:103447. doi: 10.1016/j.jprot.2019.103447. [DOI] [PubMed] [Google Scholar]
- Zhang A. Sun H. Yuan Y. et al., An in vivo analysis of the therapeutic and synergistic properties of Chinese medicinal formula Yin-Chen-Hao-Tang based on its active constituents. Fitoterapia. 2011;82:1160–1168. doi: 10.1016/j.fitote.2011.07.014. [DOI] [PubMed] [Google Scholar]
- Zhang A. Yan G. Han Y. et al., Metabolomics approaches and applications in prostate cancer research. Appl. Biochem. Biotechnol. 2014;174:6–12. doi: 10.1007/s12010-014-0955-6. [DOI] [PubMed] [Google Scholar]
- Wang H. Yan G. Zhang A. et al., Rapid discovery and global characterization of chemical constituents and rats metabolites of Phellodendri amurensis cortex by ultra-performance liquid chromatography-electrospray ionization/quadrupole-time-of-flight mass spectrometry coupled with pattern recognition approach. Analyst. 2013;138:3303–3312. doi: 10.1039/C3AN36902A. [DOI] [PubMed] [Google Scholar]
- Zhang A. Sun H. Xu H. et al., Cell metabolomics. OMICS: J. Integr. Biol. 2013;17(10):495–501. doi: 10.1089/omi.2012.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. H. Ma Z. M. Sun H. Zhang Y. Liu J. H. Wu F. F. Wang X. J. High-Throughput Metabolomics Evaluate the Efficacy of Total Lignans From Acanthophanax Senticosus Stem Against Ovariectomized Osteoporosis Rat. Front. Pharmacol. 2019;10:553. doi: 10.3389/fphar.2019.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. Sun H. Han Y. et al., Urinary metabolic biomarker and pathway study of hepatitis B virus infected patients based on UPLC-MS system. PLoS One. 2013;8(5):e64381. doi: 10.1371/journal.pone.0064381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. Sun H. Qiu S. et al., NMR-based metabolomics coupled with pattern recognition methods in biomarker discovery and disease diagnosis. Magn. Reson. Chem. 2013;51(9):549–556. doi: 10.1002/mrc.3985. [DOI] [PubMed] [Google Scholar]
- Wang X. Zhang A. Zhou X. et al., An integrated chinmedomics strategy for discovery of effective constituents from traditional herbal medicine. Sci. Rep. 2016;6:18997. doi: 10.1038/srep18997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Liu P. Li Y. et al., Exploration of metabolite signatures using high-throughput mass spectrometry coupled with multivariate data analysis. RSC Adv. 2017;7:6780–6787. doi: 10.1039/C6RA27461G. [DOI] [Google Scholar]
- Zhang A. Sun H. Wang X. Emerging role and recent applications of metabolomics biomarkers in obesity disease research. RSC Adv. 2017;7(25):14966–14973. doi: 10.1039/C6RA28715H. [DOI] [Google Scholar]
- Wang X. J. Ren J. L. Zhang A. H. Sun H. Yan G. L. Han Y. Liu L. Novel applications of mass spectrometry-based metabolomics in herbal medicines and its active ingredients: Current evidence. Mass Spectrom. Rev. 2019;38(4–5):380–402. doi: 10.1002/mas.21589. [DOI] [PubMed] [Google Scholar]
- Wang X. Zhang A. Sun H. Future perspectives of Chinese medical formulae: chinmedomics as an effector. OMICS: J. Integr. Biol. 2012;16(7–8):414–421. doi: 10.1089/omi.2011.0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H. Zhang A. Zhang H. et al., The application of metabolomics in traditional Chinese medicine opens up a dialogue between Chinese and Western medicine. Phytother. Res. 2015;29(2):159–166. doi: 10.1002/ptr.5240. [DOI] [PubMed] [Google Scholar]
- Liang Q. Liu H. Xing H. et al., Urinary UPLC-MS metabolomics dissecting the underlying mechanisms of huaxian capsule protects against sepsis. RSC Adv. 2016;6:40436–40441. doi: 10.1039/C6RA07987C. [DOI] [Google Scholar]
- Sun H. Zhang A. H. Liu S. B. et al., Cell metabolomics identify regulatory pathways and targets of magnoline against prostate cancer. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2018;1102–1103:143–151. doi: 10.1016/j.jchromb.2018.10.017. [DOI] [PubMed] [Google Scholar]
- Zhang H. Zhang A. Miao J. et al., Targeting regulation of tryptophan metabolism for colorectal cancer therapy: a systematic review. RSC Adv. 2019;9:3072–3080. doi: 10.1039/C8RA08520J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Liu P. Li Y. et al., Exploration of metabolite signatures using high-throughput mass spectrometry coupled with multivariate data analysis. RSC Adv. 2017;7:6780–6787. doi: 10.1039/C6RA27461G. [DOI] [Google Scholar]
- Xiong H. Zhang A. H. Zhao Q. Q. et al., Discovery and screening quality-marker ingredients of Panax quinquefolius using chinmedomics approach. Phytomedicine. 2019;12:152928. doi: 10.1016/j.phymed.2019.152928. [DOI] [PubMed] [Google Scholar]
- Zhang A. Qiu S. Sun H. Zhang T. Guan Y. Han Y. Yan G. Wang X. Scoparone affects lipid metabolism in primary hepatocytes using lipidomics. Sci. Rep. 2016;6:28031. doi: 10.1038/srep28031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H. Zhang A. H. Sun H. et al., High-throughput metabolomics screen coupled with multivariate statistical analysis identifies therapeutic targets in alcoholic liver disease rats using liquid chromatography-mass spectrometry. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2019;1109:112–120. doi: 10.1016/j.jchromb.2019.01.017. [DOI] [PubMed] [Google Scholar]
- Zhang A. H. Sun H. Yan G. L. et al., Chinmedomics: A Powerful Approach Integrating Metabolomics with Serum Pharmacochemistry to Evaluate the Efficacy of Traditional Chinese Medicine. Engineering. 2019;5:60–68. doi: 10.1016/j.eng.2018.11.008. [DOI] [Google Scholar]
- Liu X. Zhang A. Fang H. et al., Serum metabolomics strategy for understanding the therapeutic effects of Yin-Chen-Hao-Tang against Yanghuang syndrome. RSC Adv. 2018;8:7403–7413. doi: 10.1039/C7RA11048K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. Zhang H. L. Zhang A. H. et al., Network pharmacology combined with functional metabolomics discover bile acid metabolism as a promising target for mirabilite against colorectal cancer. RSC Adv. 2018;8:30061–30070. doi: 10.1039/C8RA04886J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. Wang H. Zhang A. et al., Berberine ameliorates nonbacterial prostatitis via multi-target metabolic network regulation. OMICS: J. Integr. Biol. 2015;19(3):186–195. doi: 10.1089/omi.2014.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Li J. Zhang A. H. Urine metabolic phenotypes analysis of extrahepatic cholangiocarcinoma disease using ultra-high performance liquid chromatography-mass spectrometry. RSC Adv. 2016;6(67):63049–63057. doi: 10.1039/C6RA09430A. [DOI] [Google Scholar]
- Zhao Y. Lv H. Qiu S. et al., Plasma metabolic profiling and novel metabolite biomarkers for diagnosing prostate cancer. RSC Adv. 2017;7(48):30060–30069. doi: 10.1039/C7RA04337F. [DOI] [Google Scholar]
- Sun H. Zhang H. L. Zhang A. H. et al., Network pharmacology combined with functional metabolomics discover bile acid metabolism as a promising target for mirabilite against colorectal cancer. RSC Adv. 2018;8:30061–30070. doi: 10.1039/C8RA04886J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. F. Qiu S. Gao L. J. et al., Metabolomic estimation of the diagnosis of hepatocellular carcinoma based on ultrahigh performance liquid chromatography coupled with time-of-flight mass spectrometry. RSC Adv. 2018;8(17):9375–9382. doi: 10.1039/C7RA13616A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T. Zhang A. Qiu S. Yang S. Wang X. Current Trends and Innovations in Bioanalytical Techniques of Metabolomics. Crit. Rev. Anal. Chem. 2016;46(4):342–351. doi: 10.1080/10408347.2015.1079475. [DOI] [PubMed] [Google Scholar]
- Zhang A. Sun H. Qiu S. et al., Metabolomics in noninvasive breast cancer. Clin. Chim. Acta. 2013;424:3–7. doi: 10.1016/j.cca.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Qiu S. Zhang A. Zhang T. Sun H. Guan Y. Yan G. Wang X. Dissect new mechanistic insights for geniposide efficacy on the hepatoprotection using multiomics approach. Oncotarget. 2017;8(65):108760–108770. doi: 10.18632/oncotarget.21897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. H. Yu J. B. Sun H. et al., Identifying quality-markers from Shengmai San protects against transgenic mouse model of Alzheimer's disease using chinmedomics approach. Phytomedicine. 2018;45:84–92. doi: 10.1016/j.phymed.2018.04.004. [DOI] [PubMed] [Google Scholar]
- Zhang A. Sun H. Yan G. et al., Metabolomics study of type 2 diabetes using ultra-performance LC-ESI/quadrupole-TOF high-definition MS coupled with pattern recognition methods. J. Physiol. Biochem. 2014;70(1):117–128. doi: 10.1007/s13105-013-0286-z. [DOI] [PubMed] [Google Scholar]
- Sun H. Zhang A. Yang L. et al., High-throughput chinmedomics strategy for discovering the quality-markers and potential targets for Yinchenhao decoction. Phytomedicine. 2019;54:328–338. doi: 10.1016/j.phymed.2018.04.015. [DOI] [PubMed] [Google Scholar]
- Zhang A. Sun H. Yan G. et al., Serum proteomics in biomedical research: a systematic review. Appl. Biochem. Biotechnol. 2013;170(4):774–786. doi: 10.1007/s12010-013-0238-7. [DOI] [PubMed] [Google Scholar]
- Ren J. L. Zhang A. H. Kong L. et al., Advances in mass spectrometry-based metabolomics for investigation of metabolites. RSC Adv. 2018;8(40):22335–22350. doi: 10.1039/C8RA01574K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. Qiu S. Xu H. et al., Metabolomics in diabetes. Clin. Chim. Acta. 2014;429:106–110. doi: 10.1016/j.cca.2013.11.037. [DOI] [PubMed] [Google Scholar]
- Zhang A. Sun H. Wang X. Recent advances in metabolomics in neurological disease, and future perspectives. Anal. Bioanal. Chem. 2013;405(25):8143–8150. doi: 10.1007/s00216-013-7061-4. [DOI] [PubMed] [Google Scholar]
- Xiao-Hang Z. Zhang A. H. Liang W. et al., Novel chinmedomics strategy for discovering effective constituents from ShenQiWan acting on ShenYangXu syndrome. Chin. J. Nat. Med. 2016;14(8):561–581. doi: 10.1016/S1875-5364(16)30067-X. [DOI] [PubMed] [Google Scholar]
- Zhang A. Sun H. Qiu S. et al., Recent highlights of metabolomics in chinese medicine syndrome research. Evid.-Based Complementary Altern. Med. 2013;2013:402159. doi: 10.1155/2013/402159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. Zhang A. Zou D. et al., Metabolomics coupled with pattern recognition and pathway analysis on potential biomarkers in liver injury and hepatoprotective effects of yinchenhao. Appl. Biochem. Biotechnol. 2014;173(4):857–869. doi: 10.1007/s12010-014-0903-5. [DOI] [PubMed] [Google Scholar]
- Liu J. Sun H. Zhang A. et al., Serum pharmacochemistry combined with multiple data processing approach to screen the bioactive components and their metabolites in Mutan Cortex by ultra-performance liquid chromatography tandem mass spectrometry. Biomed. Chromatogr. 2014;28(4):500–510. doi: 10.1002/bmc.3060. [DOI] [PubMed] [Google Scholar]
- Zou D. Zhang A. Yan G. et al., UPLC-MS coupled with a dynamic multiple data processing method for the comprehensive detection of the chemical constituents of the herbal formula San-Miao-Wan. Anal. Methods. 2014;6(9):2848–2854. doi: 10.1039/C3AY41814F. [DOI] [Google Scholar]
- Song Q. Zhang A. Yan G. et al., Technological advances in current metabolomics and its application in tradition Chinese medicine. RSC Adv. 2017;7(84):53516–53524. doi: 10.1039/C7RA02056B. [DOI] [Google Scholar]
- Sun H. Yang L. Li M. et al., UPLC-G2Si-HDMS untargeted metabolomics for identification of metabolic targets of Yin-Chen-Hao-Tang used as a therapeutic agent of dampness-heat jaundice syndrome. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2018;1081:41–50. doi: 10.1016/j.jchromb.2018.02.035. [DOI] [PubMed] [Google Scholar]
- Wang X. Zhang A. Sun H. et al., Chinmedomics: newer theory and application. Chin. Herb. Med. 2016;8(4):299–307. doi: 10.1016/S1674-6384(16)60055-2. [DOI] [Google Scholar]
- Sun H. Zhang A. Song Q. et al., Functional metabolomics discover pentose and glucuronate interconversion pathways as promising targets for Yang Huang syndrome treatment with Yinchenhao Tang. RSC Adv. 2018;8(64):36831–36839. doi: 10.1039/C8RA06553E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Zhang A. Yan G. et al., UHPLC-MS for the analytical characterization of traditional Chinese medicines. TrAC, Trends Anal. Chem. 2014;63:180–187. doi: 10.1016/j.trac.2014.05.013. [DOI] [Google Scholar]
- Sun H. Li X. Zhang A. et al., Exploring potential biomarkers of coronary heart disease treated by Jing Zhi Guan Xin Pian using high-throughput metabolomics. RSC Adv. 2019;9(20):11420–11432. doi: 10.1039/C8RA10557J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q. Zhang A. Miao J. et al., Metabolomics biotechnology, applications, and future trends: a systematic review. RSC Adv. 2019;9(64):37245–37257. doi: 10.1039/C9RA06697G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. Sun H. Wang P. et al., Modern analytical techniques in metabolomics analysis. Analyst. 2012;137(2):293–300. doi: 10.1039/C1AN15605E. [DOI] [PubMed] [Google Scholar]
- Wang X. Zhang A. Han Y. et al., Urine metabolomics analysis for biomarker discovery and detection of jaundice syndrome in patients with liver disease. Mol. Cell. Proteomics. 2012;11(8):370–380. doi: 10.1074/mcp.M111.016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. Sun H. Han Y. et al., Urinary metabolic biomarker and pathway study of hepatitis B virus infected patients based on UPLC-MS system. PLoS One. 2013;8(5):e64381. doi: 10.1371/journal.pone.0064381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Zhang A. Sun H. et al., Discovery and development of innovative drug from traditional medicine by integrated chinmedomics strategies in the post-genomic era. TrAC, Trends Anal. Chem. 2016;76:86–94. doi: 10.1016/j.trac.2015.11.010. [DOI] [Google Scholar]
- Zhang A. Sun H. Yan G. et al., Recent developments and emerging trends of mass spectrometry for herbal ingredients analysis. TrAC, Trends Anal. Chem. 2017;94:70–76. doi: 10.1016/j.trac.2017.07.007. [DOI] [Google Scholar]
- Wang X. Wang Q. Zhang A. et al., Metabolomics study of intervention effects of Wen-Xin-Formula using ultra high-performance liquid chromatography/mass spectrometry coupled with pattern recognition approach. J. Pharm. Biomed. Anal. 2013;74:22–30. doi: 10.1016/j.jpba.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Zhang A. Sun H. Wang X. Mass spectrometry-driven drug discovery for development of herbal medicine. Mass Spectrom. Rev. 2018;37(3):307–320. doi: 10.1002/mas.21529. [DOI] [PubMed] [Google Scholar]
- Gao H. L. Zhang A. H. Yu J. B. et al., High-throughput lipidomics characterize key lipid molecules as potential therapeutic targets of Kaixinsan protects against Alzheimer's disease in APP/PS1 transgenic mice. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2018;1092:286–295. doi: 10.1016/j.jchromb.2018.06.032. [DOI] [PubMed] [Google Scholar]
- Qiu S. Zhang A. H. Sun H. et al., Overview on metabolomics in traditional Chinese medicine. World J. Pharmacol. 2014;3(3):33–38. doi: 10.5497/wjp.v3.i3.33. [DOI] [Google Scholar]
- Zhang A. Sun H. Wang P. et al., Metabonomics for discovering biomarkers of hepatotoxicity and nephrotoxicity. Pharmazie. 2012;67(2):99–105. [PubMed] [Google Scholar]
- Chu H. Sun H. Yan G. L. et al., Metabolomics analysis of health functions of Physalis pubescens L. using by ultra-performance liquid chromatography/electrospray ionization quadruple time-of-flight mass spectrometry. World J. Tradit. Chin. Med. 2015;1(3):9–20. [Google Scholar]
- Satapati S. Sunny N. E. Kucejova B. et al., Elevated TCA cycle function in the pathology of diet-induced hepatic insulin resistance and fatty liver. J. Lipid Res. 2012;53(6):1080–1092. doi: 10.1194/jlr.M023382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Han X. Gross R. W. Identification of hepatic peroxisomal phospholipase A(2) and characterization of arachidonic acid-containing choline glycerophospholipids in hepatic peroxisomes. FEBS Lett. 2003;546(2–3):247–250. doi: 10.1016/S0014-5793(03)00581-7. [DOI] [PubMed] [Google Scholar]
- Ma A. L. Guo X. Z. Liu X. et al., Efficacy comparison between bicyclol and polyene phosphatidylcholine treatments for alcoholic liver disease. Zhonghua Ganzangbing Zazhi. 2011;19(6):471–472. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.