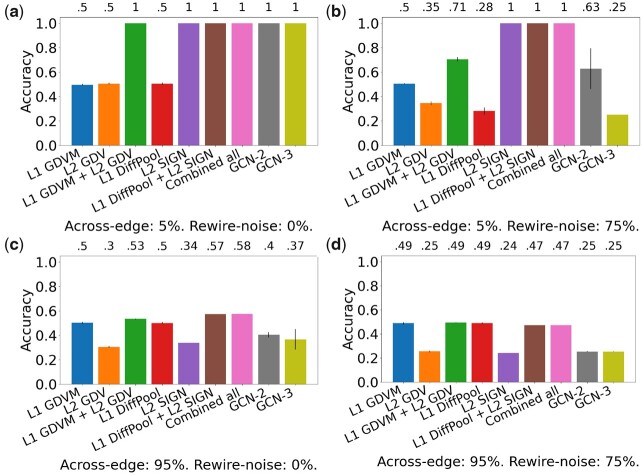
Fig. 3.
Comparison of the nine considered approaches in the task of label prediction for synthetic NoNs with the following parameters: (a) 5% across-edge and 0% rewire-noise amount, (b) 5% across-edge and 75% rewire-noise amount, (c) 95% across-edge and 0% rewire-noise amount and (d) 95% across-edge and 75% rewire-noise amount. ‘Combined all’ refers to L1 GDVM + L2 GDV + L1 DiffPool + L2 SIGN. Accuracy is shown above the bars. SDs are indicated at the top of each bar; some have very small values and are thus not visible. Recall from text that we expect an approach that only uses a single level to have around , or 0.5, accuracy when both across-edge and rewire-noise amount are low. However, L2 SIGN is likely able to achieve an accuracy of 1 in (a) and (b) since it captures clustering structure present in the Level 2 network. Results for other parameter combinations are shown in Supplementary Figures S2–S6