Abstract
Context.—
Molecular diagnostics play an increasing role in the diagnosis of Ewing sarcoma. The type of molecular testing used in clinical practice has been poorly described.
Objective.—
To describe patterns of translocation testing for newly diagnosed Ewing sarcoma.
Design.—
Children’s Oncology Group (COG) trial AEWS1221 was a phase III randomized trial enrolling patients with newly diagnosed metastatic Ewing sarcoma from 2014 to 2019. Patients were required to have a histologic diagnosis of Ewing sarcoma, but translocation testing was not required. Sites provided types and results of any molecular diagnostics performed.
Results.—
Data from 305 enrolled patients were available. The most common type of molecular testing was fluorescence in situ hybridization (FISH) performed on the primary tumor (236 of 305 patients; 77.4%), with positive testing for an EWSR1 or FUS translocation in 211 (89.4%). Reverse transcription–polymerase chain reaction (RT-PCR) on the primary tumor was performed in 61 of 305 (20%), with positive results in 48 of 61 patients (78.7%). Next-generation sequencing was reported in 7 patients on primary tumor and in 3 patients on metastatic sites. Evaluating all types of testing on either primary or metastatic tumor, 16 of 305 patients (5.2%) had no reported translocation testing. Evaluating all results from all testing, 44 of 305 patients (14.4%) lacked documentation of an abnormality consistent with a molecular diagnosis of Ewing sarcoma.
Conclusions.—
COG sites enrolling in a Ewing sarcoma trial have high rates of testing by FISH or PCR. A small proportion of patients have no translocation testing on either primary or metastatic sites. Next-generation sequencing techniques are not yet commonly used in this context.
Ewing sarcoma is a translocation-associated sarcoma mainly affecting children, adolescents, and young adults. The most common translocation is EWSR1/FLI1, though any translocation of EWSR1 or FUS with a member of the ETS family of transcription factors, in a tumor with characteristic histomorphologic and immunohistochemical features, is considered to be consistent with a molecular diagnosis of Ewing sarcoma.1 In recent years, sarcomas of bone and soft tissue with morphologic and immunohistochemical overlap with Ewing sarcoma but lacking a pathognomonic fusion have been described.2–5 These “Ewing-like sarcomas” are characterized by distinct translocations, potentially driving divergent biology and unique natural histories.
Several approaches are available to determine the translocation status of a suspected Ewing sarcoma. For example, break-apart fluorescence in situ hybridization (FISH) can provide evidence for translocation at the EWSR1 locus, but does not identify the translocation partner. Reverse transcription–polymerase chain reaction (RT-PCR) assays have likewise been established to identify the most common translocations, though less common translocations may not be included.6,7 More recently, DNA- and RNA-based next-generation sequencing (NGS) assays have been developed and these tests usually are able to identify multiple different Ewing-related translocations.8,9
Given these multiple options for translocation testing, current strategies used to apply molecular diagnostics to patients with Ewing sarcoma are not known. Understanding patterns of testing is important, since the diagnosis of Ewing sarcoma is increasingly based on molecular confirmation of a characteristic translocation. Moreover, novel agents in development for Ewing sarcoma may only be relevant for patients with tumors harboring specific translocations.10 Understanding these patterns can also inform strategies to implement routine translocation testing either at the site level or in central laboratories in the context of cooperative group trials. In this context, we analyzed site-reported translocation testing performed in patients with newly diagnosed metastatic Ewing sarcoma enrolling to a recently completed cooperative group randomized phase III clinical trial.
MATERIALS AND METHODS
Study Design and Eligibility
Children’s Oncology Group (COG) trial AEWS1221 (clinical-trials.gov identifier NCT02306161) was a phase III randomized trial for patients with newly diagnosed metastatic Ewing sarcoma. Patients were randomly assigned at study entry to interval compressed chemotherapy analogous to AEWS0031, Regimen B,11 or to that same chemotherapy plus the addition of the anti–insulin-like growth factor-1 receptor (IGF-1R) monoclonal antibody ganitumab at the start of chemotherapy cycles. Enrollment to AEWS1221 has completed, with final outcome results of the primary trial objective focused on IGF-1R inhibition to be reported in a future publication. The objective of the current report is to describe patterns of translocation testing for newly diagnosed Ewing sarcoma reported in the context of AEWS1221.
To be eligible for AEWS1221, patients were required to have a histologic diagnosis of Ewing sarcoma/primitive neuroectodermal tumor as determined by the institutional pathologist. Identification of abnormalities consistent with a molecular diagnosis of Ewing sarcoma was not required. Patients were not eligible if they were diagnosed with round cell sarcomas other than Ewing sarcoma. Given the nature of the current analysis, all patients who enrolled in AEWS1221 and submitted data on translocation testing (see below) were included in the analytic cohort, even if subsequently determined to be ineligible.
AEWS1221 was approved by the National Cancer Institute’s Pediatric Central Institutional Review Board (IRB). Participating sites either acknowledged reliance upon the central IRB or obtained local IRB approval. All patients (or legal guardians for minor subjects) provided informed consent for participation in AEWS1221 at the time of study entry.
Translocation Status
Centralized clinical or research testing for translocation status was not integral to AEWS1221. Instead, participating sites could perform any standard-of-care molecular testing available at their centers or use reference laboratories contracted by their institutions to complete such testing. Sites reported the type of molecular testing and results of testing for enrolled patients on a case report form. These data form the basis for the current analysis.
Statistical Analysis
Descriptive statistics were calculated for proportions of patients with specific types of translocation testing on primary tumor material and on metastatic tumor material. The proportion of patients with identified ETS family translocations and the proportion of patients with no translocation testing performed on any tumor material at diagnosis were calculated.
RESULTS
Patient Characteristics
Three hundred twelve patients enrolled in AEWS1221 from December 2014 to March 2019, including 14 patients who were subsequently determined to be ineligible owing to incorrect diagnosis by site report (n = 8), inadequate correlative sample submission (n = 3), incorrect timing of eligibility procedures (n = 2), or incorrect stage (n = 1). All 298 eligible patients provided data on translocation testing. Of the 14 ineligible patients, 7 provided data on translocation testing as was done for eligible patients. Therefore, the analytic cohort with available translocation data includes 305 patients. Characteristics of these 305 patients are shown in Table 1 and are as expected for a population of patients with newly diagnosed metastatic Ewing sarcoma.
Table 1.
Characteristics of 305 Patients Enrolled in Children’s Oncology Group Trial AEWS1221 With Available Translocation Testing Data
| Characteristic | N (%) |
|---|---|
| Age at enrollment | |
| < 21 y | 259 (84.9) |
| ≥ 21 y | 46 (15.1) |
| Sex | |
| Female | 135 (44.3) |
| Male | 170 (55.7) |
| Race | |
| White | 249 (81.6) |
| Asian | 8 (2.6) |
| Black | 6 (2) |
| Other | 3 (1) |
| Unknown | 39 (12.8) |
| Primary Site | |
| Pelvic bones | 98 (32.1) |
| Extremity bones | 90 (29.5) |
| Other bones | 61 (20) |
| Soft tissue | 56 (18.4) |
| Metastatic pattern | |
| Lung only | 120 (39.3) |
| Not isolated to lung | 185 (60.7) |
Testing Strategy on Primary and Metastatic Tumor
We evaluated the type of testing performed on both primary and metastatic tumor (Table 2). FISH analysis of the primary tumor was the predominant mode of testing, reported in 236 of 305 patients (77.4%). Details of the specific FISH test(s) performed (eg, EWSR1 break-apart) were not available. All other types of testing on the primary tumor were reported in a minority of patients. RT-PCR was performed in 61 of 305 primary tumors (20%) and NGS was rarely used, with only 7 of 305 patients (2.3%) having this testing performed on primary tumor material. Details of the specific RT-PCR test(s) performed (eg, EWSR1//FLI1) were not available.
Table 2.
Frequency of Any Translocation Testing on Primary and Metastatic Tumors in Patients Enrolled in AEWS1221
| Test | Tested on Primary Tumor, N (%) | Tested on Metastasis, N (%) |
|---|---|---|
| FISH | 236/305 (77.4) | 31/305 (10.2) |
| RT-PCR | 61/305 (20.0) | 13/305 (4.3) |
| Cytogenetics/karyotype | 26/305 (8.5) | 8/305 (2.6) |
| Next-generation sequencing | 7/305 (2.3) | 3/305 (1) |
Abbreviations: FISH, fluorescence in situ hybridization; RT-PCR, reverse transcription–polymerase chain reaction.
Translocation testing on metastatic sites was infrequently performed. FISH testing on metastatic sites was performed in 31 of 305 patients (10.2%), RT-PCR in 13 of 305 (4.3%), and NGS in 3 of 305 (1%).
We also investigated the likelihood of using both FISH and RT-PCR on the primary tumor (Table 3). When multiple testing techniques were used in the same patient, neither the order nor the rationale for repeated testing was available. Of the 236 patients who had FISH testing on the primary tumor, only 27 (11.4%) also had RT-PCR performed on the primary tumor. Of the 305 patients in the cohort, only 35 (11.5%) had neither FISH nor RT-PCR performed on the primary tumor.
Table 3.
Number of Patients Whose Primary Tumor Was Tested With FISH or RT-PCR
| Primary Tested by RT-PCR |
||
|---|---|---|
| Primary Tested by FISH | Yes | No |
| Yes | 27 | 209 |
| No | 34 | 35 |
Abbreviations: FISH, fluorescence in situ hybridization; RT-PCR, reverse transcription–polymerase chain reaction.
Results of Testing Performed
We next evaluated the results of the translocation testing that was performed in the context of this trial. Table 4 provides the percentage of patients with an abnormality consistent with a molecular diagnosis of Ewing sarcoma among those reported to have been tested by each approach. FISH testing on the primary tumor resulted in detection of an EWRS1 or FUS rearrangement in 211 of 236 samples tested (89.4%). RT-PCR on primary tumor showed a translocation consistent with Ewing sarcoma in 48 of 61 cases tested (78.7%). For each assay, the percentage of tests revealing a rearrangement was nominally lower when metastatic sites were tested than with testing on primary tumors.
Table 4.
Results of Translocation Testing Performed on Primary and Metastatic Tumors in Patients Enrolled in AEWS1221
| Test | Evidencea of Translocation in Primary Tumor, N/No. Tested (%) | Evidencea of Translocation in Metastasis, N/No. Tested (%) |
|---|---|---|
| FISH | 211/236 (89.4) | 21/31 (67.7) |
| RT-PCR | 48/61 (78.7) | 10/13 (76.9) |
| Cytogenetics/karyotype | 17/26 (65.4) | 4/8 (50.0) |
| Next-generation sequencing | 5/7 (71.4) | 1/3 (33.3) |
Abbreviations: FISH, fluorescence in situ hybridization; RT-PCR, reverse transcription–polymerase chain reaction.
FISH was considered to have evidence of a molecular diagnosis of Ewing sarcoma if it showed EWRS1 or FUS rearrangement. RT-PCR and next-generation sequencing were considered to have evidence of molecular diagnosis of Ewing sarcoma if they showed EWSR1 or FUS translocated with an ETS family member. Cytogenetics/karyotype was considered to have evidence of molecular diagnosis of Ewing sarcoma if a translocation was identified that was consistent with EWSR1 or FUS translocated with an ETS family member.
We also investigated concordance between FISH and RT-PCR among the 27 patients who had both tests performed on the primary tumor (Supplemental Table 1, see supplemental digital content containing 2 tables). In 14 of 27 patients (51.9%), the results were both positive for an abnormality consistent with a molecular diagnosis of Ewing sarcoma. In 5 of 27 patients (18.5%), the results were both negative for an abnormality consistent with a molecular diagnosis of Ewing sarcoma. In 8 of 27 patients (29.6%), the results of FISH and RT-PCR were discordant.
Table 5 provides the spectrum of results reported, including the most specific test result for patients who had multiple tests performed. In 201 of 305 cases (65.9%), the most specific result was detection of an EWSR1 translocation, with fusion partner not identified.
Table 5.
Specific Translocation Results Obtained From 305 Enrolled Patients With Translocation Testing Data Available
| Translocation Test Resulta | Percentage |
|---|---|
| EWSR1 translocation (fusion partner not identified) | 65.9 |
| EWSR1-FLI1 | 16.1 |
| EWSR1-ERG | 2.6 |
| FUS translocation (fusion partner not identified) | 0.7 |
| EWSR1-ETV1 | 0.3 |
| No translocation reportedb | 12.8 |
| Diagnosis other than Ewing sarcoma | 1.6 |
The most specific test result of all available testing is shown.
Either no translocation testing performed or any testing was negative.
Per Patient Analysis
As some patients had multiple tests performed on multiple samples, we next completed a “per patient” analysis that considered each patient once to determine the proportion of patients with no testing and the proportion of patients without documented evidence of an abnormality consistent with a molecular diagnosis of Ewing sarcoma, based upon reported test results (Figure). For both analyses, we began with FISH testing and FISH test results on primary tumor as this was the most common approach. We found that 31 of 305 patients (10.2%) had no reported translocation testing on primary tumor and 16 of 305 patients (5.2%) had no reported translocation testing on either primary or metastatic tumor (Figure, A). Considering all testing types on primary and metastatic tumor material, 44 of 305 patients (14.4%) did not have documented evidence of an abnormality consistent with a molecular diagnosis of Ewing sarcoma (Figure, B).
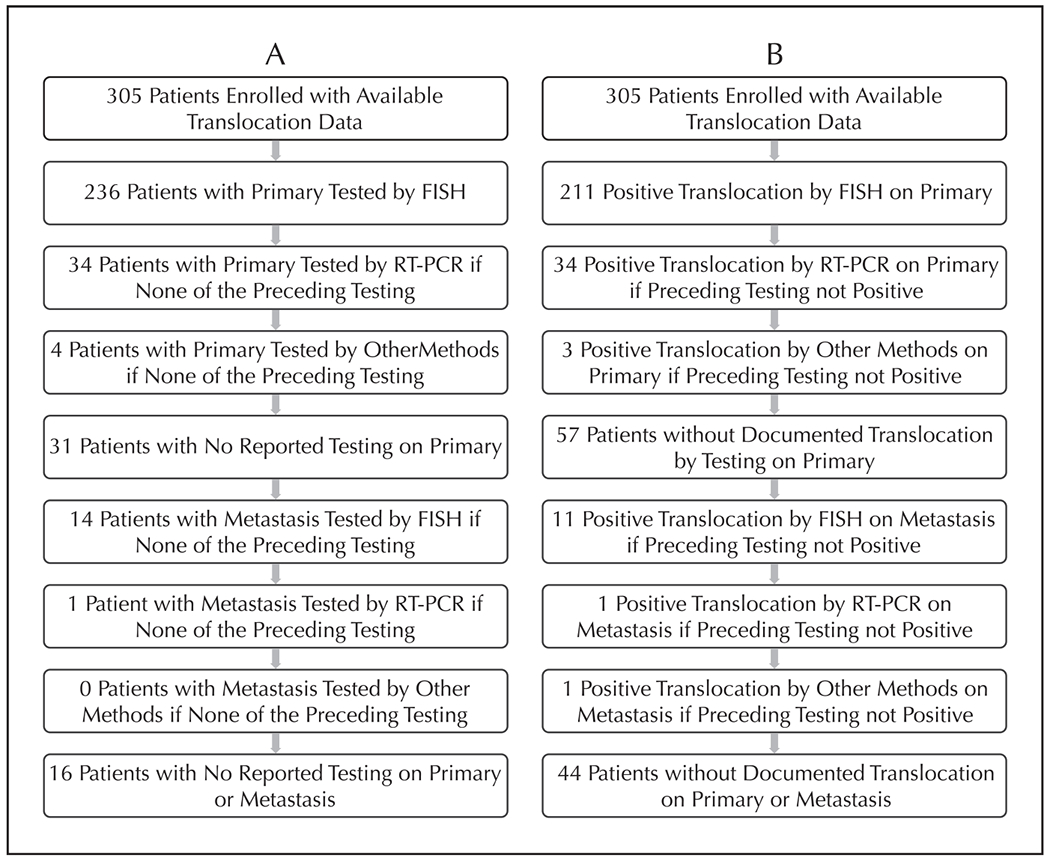
A, Flow diagram of translocation testing performed accounting for all patients enrolled in AEWS1221 with submitted translocation testing data. B, Flow diagram of translocation test results for all patients enrolled in AEWS1221 with submitted translocation testing data. Diagrams consider each patient once, starting with testing by FISH on primary tumor. Only patients who did not have testing (A) or a positive test result (B) in the preceding boxes are considered in subsequent boxes. Abbreviations: FISH, fluorescence in situ hybridization; RT-PCR reverse transcription–polymerase chain reaction.
We also evaluated whether clinical characteristics differed according to translocation status (Supplemental Table 2). Compared to patients with documented evidence of an abnormality consistent with a molecular diagnosis of Ewing sarcoma, patients without such evidence appeared to have lower rates of primary tumors in the bony pelvis and higher rates of soft tissue primary tumors, with other characteristics broadly similar between groups. Likewise, clinical characteristics appeared similar between patients with documented evidence of an abnormality consistent with a molecular diagnosis of Ewing sarcoma who had documentation via FISH alone versus those with documentation via RT-PCR.
DISCUSSION
Our study provides new observational data on practice patterns for translocation testing for patients with newly diagnosed metastatic Ewing sarcoma enrolling to a cooperative group clinical trial. We show that FISH testing on primary tumor is the predominant testing type in this context, which is a cause for concern because EWSR1 translocation by FISH is not specific for the diagnosis of Ewing sarcoma as it can be seen in desmoplastic small round cell tumor and other emerging round cell sarcomas with EWSR1–non ETS fusions.12,13 Other testing strategies were much less commonly used, with NGS of the primary tumor used in only 7 of 305 patients (2.3%). In 27 patients who had both FISH and RT-PCR performed on the primary tumor, results were discordant in 8 patients (29.6%). In 44 of 305 patients (14.4%), there was no documented evidence of an abnormality consistent with a molecular diagnosis of Ewing sarcoma, based on testing performed clinically. These findings highlight the need for reliable testing for translocations specific for Ewing sarcoma in the context of clinical trials for this disease.
To our knowledge, data on translocation testing strategies in Ewing sarcoma are not available from other cooperative group trials. Instead, we place our results in context with single institution studies. In one series of 109 patients with Ewing sarcoma, FISH testing was used universally, with RT-PCR used in 78.9% of cases.7 Despite frequent use of both approaches, 14 of 109 patients (12.8%) with a diagnosis of Ewing sarcoma, based upon morphologic and immunohistochemical features, did not have documented evidence of an abnormality consistent with a molecular diagnosis of Ewing sarcoma. These results are similar to our reported rate of 44 of 305 patients (14.4%). Another single center study reported their experience with cytogenetics, FISH, and RT-PCR in 32 pediatric patients with Ewing sarcoma.14 Molecular evidence of Ewing sarcoma was not found in 6.2% of cases, suggesting that multiple complementary testing approaches may increase the likelihood of detecting a fusion. There are several potential reasons for negative testing, including incorrect diagnosis, technical issues due to sample quantity/quality, or presence of a rare translocation not included as part of the testing strategy (eg, a patient with EWSR1/ETV1-translocated Ewing sarcoma evaluated with RT-PCR probes that only evaluate EWSR1/FLI1, EWSR1/ERG, and FUS/ERG).
Another key finding from our analysis was the paucity of NGS approaches used in the setting of this cooperative group trial. Several groups have reported on the use of these techniques for detection of translocations relevant to Ewing sarcoma. For example, one group reported 96.4% sensitivity and 100% specificity when an RNA-based NGS approach was applied to a cohort of pediatric fusion-positive sarcomas.8 Another group likewise reported 89% sensitivity for detecting an EWSR1 translocation, using a custom RNA-based NGS assay designed to detect fusions, using positive EWSR1 break-apart FISH as the gold standard for their analysis.9
This study provides a large national cohort of patients with available data on translocation testing and results of that testing. We nevertheless acknowledge limitations in our analysis. The data included only patients enrolled in the parent trial (AEWS1221) with an initial institutional diagnosis of Ewing sarcoma and therefore does not capture testing strategy for a broader group of patients with suspected Ewing sarcoma. We did not collect data on subtypes of specific assays (eg, EWSR1 break-apart versus dual-color FISH). Likewise, we did not collect rates of cancelled testing due to insufficient tumor material. The reported data are observational based on site testing strategy and therefore should not be viewed as a head-to-head comparison of diagnostic accuracy of the approaches used by enrolling sites. As above, our reported rates of the use of NGS approaches were low. As these technologies are both rapidly evolving and also becoming more widely available, it is possible that our results may not fully reflect present-day testing strategies. Finally, we captured data at time of enrollment to AEWS1221. We cannot exclude the possibility that additional testing was performed subsequent to enrollment, such as on tumor material obtained at time of postenrollment tumor resection or relapse.
Importantly, this analysis was not intended to identify optimal approach to the molecular diagnosis of Ewing sarcoma. Each approach has potential limitations that may result in diagnostic pitfalls. For example, break-apart FISH for EWSR1 is a commonly used approach but does not provide information about the fusion partner. This introduces the potential for misdiagnosis of other round cell sarcomas as Ewing sarcoma in the setting of a positive EWSR1 translocation when an alternative diagnosis (eg, desmoplastic small round cell tumor) would have been rendered if the fusion partner (eg, WT1) was identified by the testing strategy. In some cases (eg, desmoplastic small round cell tumor), the treatment paradigm would differ by molecular findings,15 while in other cases (eg, an emerging group of round cell sarcoma with EWSR1–non ETS fusions) the optimal therapy may still be unknown.12,13 It will be of interest to determine if patterns of testing evolve as NGS approaches that provide information on both fusion partners become more widely available. Likewise, RT-PCR only evaluates for prespecified translocations, and not all panels will include less common translocations that are nevertheless compatible with a molecular diagnosis of Ewing sarcoma.
In summary, we provide a comprehensive overview of current practice patterns of translocation testing in patients with Ewing sarcoma enrolling to a large cooperative group clinical trial. These results can inform strategies to implement centralized translocation testing in future cooperative group trials relevant to patients with Ewing sarcoma. Molecular confirmation of translocation status in this disease is taking on increasing importance as new therapies are developed that target EWSR1/ETS translocations, or in some cases, specific translocation subtypes such as EWSR1/FLI1.16 As new approaches to translocation ascertainment become increasingly used in the coming years, it will be of interest to reassess these findings.
Supplementary Material
Acknowledgments
AEWS1221 trial conduct was supported by NIH/NCI Grant U10 CA180899 (Children’s Oncology Group Statistics and Data Center), the COG Foundation, NCTN Operations Center Grant U10CA180886, St. Baldrick’s Foundation. Glade-Bender was supported by NIH/NCI CA008748. DuBois has received fees for consulting and advisory board roles from Bayer and Loxo Oncology and has received travel expenses from Loxo Oncology, Roche/Genentech, and Salarius Pharmaceuticals. Lessnick is a Scientific Advisor to Salarius Pharmaceuticals and is listed as an inventor on patents related to NKZ2.2 and GSTM4 in Ewing sarcoma. The other authors have no relevant financial interest in the products or companies described in this article.
Footnotes
A preliminary version of this work was presented at the 2018 Connective Tissue Oncology Society Annual Meeting; November 14–17, 2018; Lisbon, Portugal.
Contributor Information
Steven G. DuBois, Dana-Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston, Massachusetts.
Mark D. Krailo, Children’s Oncology Group Statistics and Data Center, Monrovia, California.
Allen Buxton, Children’s Oncology Group Statistics and Data Center, Monrovia, California.
Stephen L. Lessnick, Center for Childhood Cancer and Blood Diseases, Abigail Wexner Research Institute, Nationwide Children’s Hospital, and The Division of Pediatric Heme/Onc/BMT, The Ohio State University College of Medicine, Columbus.
Lisa A. Teot, Department of Pathology, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts.
Dinesh Rakheja, Department of Pathology, University of Texas Southwestern Medical Center, Dallas.
Brian D. Crompto, Dana-Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston, Massachusetts.
Katherine A. Janeway, Dana-Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston, Massachusetts.
Richard G. Gorlick, Department of Pediatrics, MD Anderson Cancer Center, Houston, Texas.
Julia Glade-Bender, Department of Pediatrics, Memorial Sloan Kettering Cancer Center, New York, New York.
References
- 1.Grunewald TCP, Cidre-Aranaz F, Surdez D, et al. Ewing sarcoma. Nat Rev Dis Primers. 2018;4(1):5. [DOI] [PubMed] [Google Scholar]
- 2.Kawamura-Saito M, Yamazaki Y, Kaneko K, et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35; q13) translocation. Hum Mol Genet. 2006;15(13):2125–2137. [DOI] [PubMed] [Google Scholar]
- 3.Yoshimoto M, Graham C, Chilton-MacNeill S, et al. Detailed cytogenetic and array analysis of pediatric primitive sarcomas reveals a recurrent CIC-DUX4 fusion gene event. Cancer Genet Cytogenet. 2009;195(1):1–11. [DOI] [PubMed] [Google Scholar]
- 4.Cohen-Gogo S, Cellier C, Coindre JM, et al. Ewing-like sarcomas with BCOR-CCNB3 fusion transcript: a clinical, radiological and pathological retrospective study from the Societe Francaise des Cancers de l’Enfant. Pediatr Blood Cancer. 2014;61(12):2191–2198. [DOI] [PubMed] [Google Scholar]
- 5.Pierron G, Tirode F, Lucchesi C, et al. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat Genet. 2012;44(4):461–466. [DOI] [PubMed] [Google Scholar]
- 6.Machado I, Navarro L, Pellin A, et al. Defining Ewing and Ewing-like small round cell tumors (SRCT): the need for molecular techniques in their categorization and differential diagnosis—a study of 200 cases. Ann Diagn Pathol. 2016;22:25–32. [DOI] [PubMed] [Google Scholar]
- 7.Noujaim J, Jones RL, Swansbury J, et al. The spectrum of EWSR1-rearranged neoplasms at a tertiary sarcoma centre: assessing 772 tumour specimens and the value of current ancillary molecular diagnostic modalities. Br J Cancer. 2017; 116(5):669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qadir MA, Zhan SH, Kwok B, et al. ChildSeq-RNA: a next-generation sequencing-based diagnostic assay to identify known fusion transcripts in childhood sarcomas. J Mol Diagn. 2014;16(3):361–370. [DOI] [PubMed] [Google Scholar]
- 9.Krystel-Whittemore M, Taylor MS, Rivera M, et al. Novel and established EWSR1 gene fusions and associations identified by next-generation sequencing and fluorescence in-situ hybridization. Hum Pathol. 2019;93:65–73. [DOI] [PubMed] [Google Scholar]
- 10.Bailey K, Cost C, Davis I, et al. Emerging novel agents for patients with advanced Ewing sarcoma: a report from the Children’s Oncology Group (COG) New Agents for Ewing Sarcoma Task Force. F1000Res. 2019;8:F1000 Faculty Rev-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Womer RB, West DC, Krailo MD, et al. Randomized controlled trial of interval-compressed chemotherapy for the treatment of localized Ewing sarcoma: a report from the Children’s Oncology Group. J Clin Oncol. 2012;30(33):4148–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuda Y, Zhang L, Meyers P, Tap WD, Healey JH, Antonescu CR. The clinical heterogeneity of round cell sarcomas with EWSR1/FUS gene fusions: impact of gene fusion type on clinical features and outcome. Genes Chromosomes Cancer. 2020;59(9):525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salguero-Aranda C, Amaral AT, Olmedo-Pelayo J, Diaz-Martin J, Alava E. Breakthrough technologies reshape the Ewing sarcoma molecular landscape. Cells. 2020;9(4):804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warren M, Weindel M, Ringrose J, et al. Integrated multimodal genetic testing of Ewing sarcoma—a single-institution experience. Hum Pathol. 2013; 44(10):2010–2019. [DOI] [PubMed] [Google Scholar]
- 15.Liu KX, Collins NB, Greenzang KA, et al. The use of interval-compressed chemotherapy with the addition of vincristine, irinotecan, and temozolomide for pediatric patients with newly diagnosed desmoplastic small round cell tumor. Pediatr Blood Cancer. 2020;67(10):e28559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erkizan HV, Kong Y, Merchant M, et al. A small molecule blocking oncogenic protein EWS-FLI1 interaction with RNA helicase A inhibits growth of Ewing’s sarcoma. Nat Med. 2009;15(7):750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


