Abstract
Bacterial wilt caused by Ralstonia solanacearum is a devastating disease of flue-cured tobacco production which poses significant yield losses all around the world. In this study, we evaluated the rhizosphere microbiome of healthy and bacterial wilt-infected (diseased) flue-cured tobacco plants through amplification of V3-V4 and ITS1-5f variable regions of 16S and internal transcribed spacer (ITS) rRNA. The study was based on the location (Qujing, Shilin, and Wenshan), plant components (rhizosphere soil and roots), and sample types (healthy and diseased) to assess the diversity of bacterial and fungal communities. Bacterial and fungal communities present in roots primarily emanated from rhizosphere soil. Healthy flue-cured tobacco plants exhibit high microbial diversity compared to diseased plants. Among three variables, plant components significantly influence the diversity of microbial communities, whereas rhizosphere soil harbors higher microbial diversity than roots. Bacterial phyla Cyanobacteria and Proteobacteria were found in high relative abundance in roots and rhizosphere soil samples, respectively. As far as fungi is concerned, a high relative abundance of Ascomycota and Basidiomycota was found in both rhizosphere soil and root. Bacterial genera such as Bacillus, Bradyrhizobium, Ensifer, Neorhizobium, and Lysobacter related to plant growth promotion and disease suppressing abilities were dominant than fungal genera. Analysis of relative abundance at specie-level revealed that most fungal species are pathogenic to flue-cured tobacco and could provide a conducive environment for wilt infection. In conclusion, R. solanacearum significantly influences the microbial diversity of flue-cured tobacco plants and negatively affects the bacterial community composition. Altogether, our study demonstrates the complexity of bacterial and fungal communities that possibly interact with each other (microbe–microbe) and host (host–microbe). This cross-talk could be helpful for healthy flue-cured tobacco plant growth and to induce resistance against bacterial wilt disease.
Keywords: flue-cured tobacco, disease resistance, locations, microbial diversity, plant components, Ralstonia solanacearum
Introduction
Bacterial wilt disease caused by soilborne pathogenic bacterium Ralstonia solanacearum is a serious threat to flue-cured tobacco (Nicotiana tabacum L.) production worldwide, including China (Cai et al., 2021). Ralstonia solanacearum is widely distributed in tropical and subtropical regions of the world and has a broad host range that infects more than 250 plant species (Paudel et al., 2020). It is a serious threat to important field crops of the Solanaceae family including tomato, potato, tobacco, ginger, eggplant, and pepper, with average yield losses that range from 10 to 55% (Kim et al., 2016).
Ralstonia solanacearum infects all tobacco plant parts (roots, stalk, and leaves) and generally produces symptoms of yellowing and wilting of leaves, discoloration of xylem vessels, and black necrotic spots on the stem. It multiplies systemically in the xylem vessels followed by death of whole plant (Cai et al., 2021). Infected soil acts as a primary source of inoculum, and the pathogen oozes out in the rhizosphere soil of diseased plants from the roots upon completion of life cycle (Wu et al., 2020). The bacterium survives in soil, water, and plant residues for a longer period as saprophytism in the absence of a specific host plant (Li et al., 2014). It spreads from diseased to healthy plants through rain splashes, irrigation water, and mechanical operations (Qi et al., 2020).
As far as losses caused by this disease are concerned, in China, incidence and yield losses vary from region to region, host to host, climatic condition, and pathogenic strain (Jiang et al., 2017). However, incidence and yield losses are recorded between 15 to 35% but can reach up to 75% and 50–60%, respectively, when disease is present with root rot pathogen Phytophtora nicotianae (Jiang et al., 2017). In high humidity and mono-cropping regions, it occurs in epidemic form, and yield losses reach up to 100%. Soil physicochemical properties, climatic conditions, locations, and rhizosphere microbial diversity play an important role on the occurrence of soilborne diseases (Cai et al., 2021).
The plant rhizosphere is considered as one of the most complex ecosystems on earth and hot spot habitat for diverse microbes (Raaijmakers, 2015). Most disease-resistant and developmental mechanisms in plants are directly related to the diversity of rhizosphere microbes (Mendes et al., 2013; Dong et al., 2019). Plant genotype and soil type are the two main factors that are responsible for assembling a healthy rhizosphere microbiome (Dong et al., 2019). Advancements in science and modern sequencing tools made the study of host–microbe interaction easier (Govindasamy et al., 2014; Mhlongo et al., 2018). However, knowledge gaps are still present between host–microbe interactions and their underlying mechanism, which need to be filled (Bulgarelli et al., 2013).
Nowadays, biological control via disease suppressive specific endophytes and rhizobacteria such as Bacillus, Lysobacter, Streptomyces, and Pseudomonas is considered as practical approach to suppress the incidence of many soilborne diseases including bacterial wilt, Fusarium wilt, and clubroot by the mechanism of direct antagonism, reshaping the rhizospheric microbial diversity, and the production of metabolites (Ma et al., 2018; Wu et al., 2020; Zhang et al., 2020a; Wei et al., 2021). The application of bioorganic fertilizer and biochar along with biocontrol agents significantly suppresses the incidence of tobacco bacterial wilt disease (Liu et al., 2013; Zhang et al., 2017; Li et al., 2022). Because of the broad host range, species complexity, wide geographical distribution, and persistent nature, no effective control exists to date against this devastating disease, and it is difficult to completely control the incidence of tobacco bacterial wilt disease (Li et al., 2014; Shen et al., 2018).
Flue-cured tobacco is a major cash crop in Yunnan Province, China. Yunnan is well-known for its unique environment and climatic conditions. It produces high-quality flue-cured tobacco famous for its pure taste, fragrant aroma, and golden color (Li et al., 2019). Yunnan produces about 50% of China’s total tobacco leaf yield, with an annual production of around 750,000 tons and 320,000 ha of agricultural land under tobacco cultivation (Tang et al., 2020). However, R. solanacearum poses significant yield losses every year. Thus, to successfully mitigate the bacterial wilt pathogen, it is necessary to understand the population dynamics and distribution of the microbiome in tobacco plants. Therefore, the present study aims to explore the core microbiota (bacteria and fungi) associated with different locations (Qujing, Shilin, and Wenshan), plant components (rhizosphere soil and roots), and nature of plant (healthy and diseased). We hypothesized that this study helps us to establish a model for studying the naturally occurring flue-cured tobacco microbiome to mitigate the incidence of tobacco bacterial wilt disease.
Materials and Methods
Sample Collection
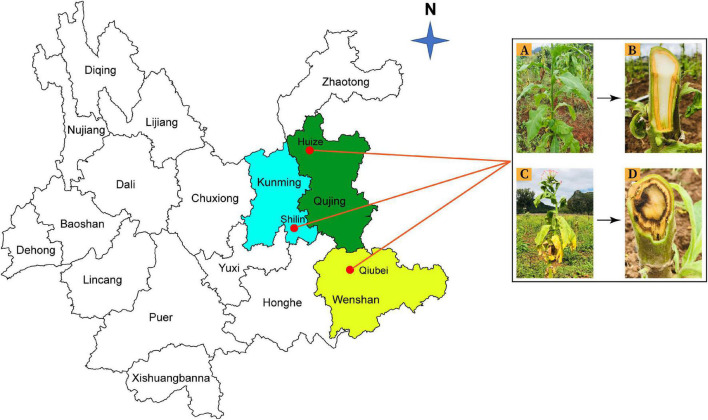
Rhizosphere soil and root samples were collected from healthy and diseased tobacco plants from three locations; Qujing (25.4900°N, 103.7962°E), Shilin (25.0950°N, 121.5246°E), and Wenshan (23.3863°N, 104.2325°E) in Yunnan Province, China, in September 2020 (Figure 1). Tobacco has been continuously grown in those fields for the past 10 years. For sample collection, upper 2–3 cm layer of soil was removed, and tobacco plants were uprooted (three plants per field from three different fields for both healthy and diseased plants). Bulk soil was removed by shaking the roots, and the tiny soil particles attached to roots were collected as rhizosphere soil samples and fibrous roots as root samples. A total of 12 composite samples (three replicates per sample) were collected from three different locations in Yunnan (Supplementary Figure 1). Samples were put in polythene bags and placed in an icebox until delivered to the laboratory and stored at −80°C for further study.
FIGURE 1.
Rhizosphere soil and root samples were collected from three different locations in Yunnan Province: Kunming (Shilin), Qujing (Huize), and Wenshan (Qiubei), from healthy and bacterial wilt infected flue-cured tobacco plants. Healthy flue-cured tobacco plant with green leaves and white-colored xylem vessels (A,B). Disease flue-cured tobacco plant with typical bacterial wilt symptoms (yellowing, wilting of leaves, necrotic lesions on stem, and discoloration of xylem vessels) (C,D).
DNA Extraction and Polymerase Chain Reaction Amplification
Total genomic DNA was extracted from 0.5 g of soil per sample to 1 g of roots per sample using the Soil and Plant DNA Extraction Kit (Zymo Research Corp., Irvine, CA, United States), respectively, following the manufacturer’s instructions. The quality of extracted DNA was quantified at OD260/280 nm 1.7-1.9 using a NanoDrop spectrophotometer (ND2000, Thermo Scientific, Madison, WI, United States), and extracted DNA was stored at -20°C for future use. The V3-V4 and ITS1-5F variable regions of 16S and internal transcribed spacer (ITS) rRNA genes of bacteria and fungi were amplified using two pairs of universal primers 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′), and 1743F (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and 2043R (5′-GCTGCGTTCTTCATCGATGC-3′) for bacterial and fungal diversity analysis, respectively (Zhang et al., 2020a).
Library Preparation and Sequencing
Amplicon library was prepared by the Nextera XT Index Kit (Illumina Inc. Madison, WI, United States) as per 16S and ITS Metagenomic Sequencing Library preparation protocols. Amplicon quality was visualized using gel electrophoresis, and 1X AMPure XP beads were used for amplicons library purification, checked on Agilent DNA1000 chip with Bioanalyzer2100, and quantified by Qubit Fluorometer 2.0 using a Qubit dsDNA assay kit (Life Technologies Cat. No. Q328520) (Gao et al., 2018). The same numbers of purified amplicons were pooled for subsequent sequencing analysis and sequenced on an Illumina MiSeq platform at Novogene Bioinformatics Technology Co. Ltd. (Beijing, China).
Quality Control
Raw data were collected in FASTQ format from DNA sequencing, and Trimmomatic software was used for the cutoff of low-quality reads (score < 20) and preparation of paired-end reads (Bolger et al., 2014). FLASH software was used to assemble paired-end reads with 10 bp/200 bp and 20% minimum/maximum overlapping and maximum mismatch rate, respectively. UCHIME software was used for chimeras removal and production of clean reads (Edgar et al., 2011).
Data Processing
Clean reads were processed with the UPARSE pipeline to generate operational taxonomic units (OTUs) at ≥97% similarity level (Edgar, 2013). For taxonomic information, species annotation was performed for all representative read and OTUs using ribosomal database project (RDP) classifier in SILVA database for bacteria (at 70% confidence threshold) and UNITE database for fungi (Quast et al., 2012; Kõljalg et al., 2013). OTUs were analyzed for relative abundance at genus and phylum levels, and alpha and beta diversity indices were calculated to obtain species richness and uniformity information. A Venn diagram was used for common and unique OTUs among different variables such as sample types, plant components, and locations.
Statistical Analysis
Data were statistically analyzed using a t-test (P < 0.05). All statistical analyses were performed using IBM SPSS Verison 20.0 (SPSS Inc., Chicago, IL, United States). QIIME software (Version 1.9.1) was used to calculate the observed OTUs, Chao1, Shannon, and abundance-based coverage estimator (ACE) indices. The Bray–Curtis dissimilarity was calculated for beta diversity analysis of bacterial and fungal communities and used for principal coordinate analysis (PCoA) with QIIME. The relative abundance bar plots at the phylum level, relative abundance heatmaps at the genera level, and relative abundance bar plots at the species level were generated using R scripts in R software (version 2.15.3) (Dong et al., 2018). Co-occurrence network analysis was conducted using sparcc in R for OTUs at the phylum level (P < 0.05 and correlation coefficient > 0.3). The network properties were calculated and visualized in Gephi 0.9.2. All figures were processed and illustrated using Adobe Illustrator CC 2019 (Adobe Systems Inc., San Francisco, CA, United States).
Results
General Characteristics of Tobacco Microbiome
We explored the bacterial and fungal communities associated with different parts of the tobacco plant (rhizosphere soil and roots), different geographic locations (Qujing, Shilin, and Wenshan), and different sample types (healthy and diseased). Data related to raw reads (#), clean reads (#), and quality control (Q20% and Q30%) through amplification of 16S (V3-V4)/ITS (1-5f) rRNA of bacteria/fungi, respectively, are shown in Supplementary Table 1. After quality control and chimeras filtering, an average of 80,477 bacterial and 92,685 fungal clean reads per sample were obtained with an average length of 412 bps/244 bps per sample by Illumina sequencing (Supplementary Table 1). Rarefaction curves generated from the OTUs demonstrated that high sampling coverage was achieved in all samples for both fungal and bacterial communities (Supplementary Figure 2).
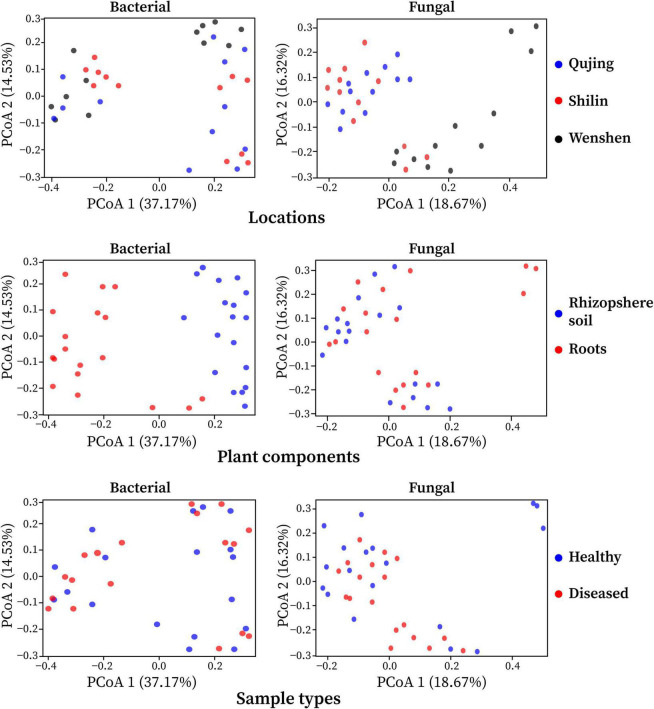
Effects of Different Locations, Plant Components, and Sample Types on Beta Diversity
The effect of variables such as locations (Qujing, Shilin, and Wenshan), plant components (rhizosphere soil and roots), and sample types (healthy and diseased) on bacterial and fungal community composition was analyzed. Bray–Curtis dissimilarity was used to determine the beta diversity (variation in bacterial and fungal communities structure) for all 12 composite samples (Figure 2). Among the three variables, it was observed that plant components (rhizosphere soil and roots) significantly influenced bacterial and fungal community composition. The separation of samples at the one axis for bacterial community composition indicates that the influence of this variable on bacterial community structure was more significant than on fungal community structure. Sample types and locations have little impact on bacterial and fungal communities. A different pattern was observed using PCoA, with a difference of 37.17% and 18.67% in bacterial and fungal community composition, respectively. Distance heatmap graphs based on Weighted UniFrac (based on abundances of taxa) and Unweighted UniFrac (sensitive to rare taxa) of all 12 samples were generated to estimate the beta diversity of bacterial and fungal communities (Supplementary Figure 3).
FIGURE 2.
Principal coordinate analysis (PCoA) was based on the Bray–Curtis dissimilarity metrics showing the beta diversity analysis for all 12 composite samples (three replicates per sample) of flue-cured tobacco plants under three variables.
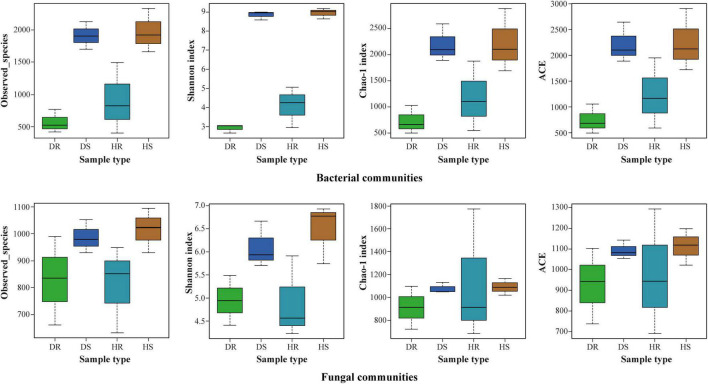
Effects of Different Locations, Plant Components, and Sample Types on Alpha Diversity
The observed species, Shannon, Chao 1, and ACE at cutoff levels of 3% are shown in Figure 3. The locations have little impact on alpha diversity indices of bacterial communities compared with fungal communities (Supplementary Table 2). In plant components (rhizosphere soil and roots), alpha diversity indices of observed species, Shannon, Chao 1, and ACE for rhizosphere soil were found higher than those of roots for both bacterial and fungal communities. It indicates that the number of bacteria and fungi in the rhizosphere was higher than in roots. Among the sample types (healthy and diseased), alpha diversity indices of bacterial and fungal communities were more elevated in healthy samples than in diseased samples.
FIGURE 3.
Boxplot of bacterial (top) and fungal (bottom) showing alpha diversity indexes of flue-cured tobacco plants under three variables. HS, healthy rhizosphere soil; DS, diseased rhizosphere soil; HR, healthy roots; DR, diseased roots.
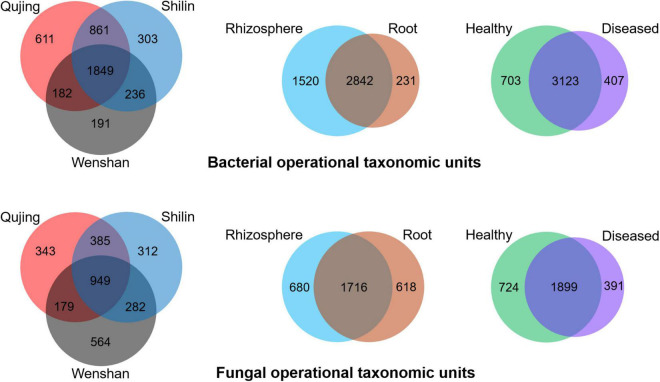
Analysis of Operational Taxonomy Units
Operational taxonomic units count was recorded maximum in rhizosphere soil samples than in root samples. For bacterial and fungal communities, the rhizosphere soil exhibited a high diversity and richness in OTUs compared with roots, and high diversity was found in healthy plant samples than in diseased plant samples (Figure 4). Analysis of OTUs revealed that a total of 4,233 and 3,014 specific OTUs were recovered for both bacterial and fungal communities from different locations (Qujing, Shilin, and Wenshan), respectively, and 1,849 (bacterial) and 949 (fungal) OTUs were found as common OTUs. Plant components (rhizosphere soil and roots) significantly impact the bacterial communities than fungal communities, and fungal OTU count in rhizosphere soil and roots was almost the same. In plant components, the specific bacterial OTUs in rhizosphere soil (1,520) were significantly higher than the specific OTUs (231) in the root tissue, and 2,482 OTUs were common. The specific fungal OTUs in root tissues (680) were slightly higher than specific OTUs (618) in rhizosphere soil, and 1,716 common OTUs were found in rhizosphere soil and roots. Further analysis of OTUs among sample types (healthy and diseased) showed that a total of 703 and 407 unique bacterial OTUs and 3,123 common bacterial OTUs were found among healthy and diseased plants, respectively. The unique fungal OTUs in healthy samples (724) were significantly higher than diseased samples (391), and 1,899 OTUs were found as common. The results indicate that bacterial wilt pathogen had a more significant impact on bacterial communities than fungal communities.
FIGURE 4.
Distribution of bacterial (top) and fungal (bottom) operational taxonomic units in three variables, i.e., locations (Qujing, Shilin, and Wenshan), plant components (rhizosphere soil and roots), and sample types (healthy and diseased).
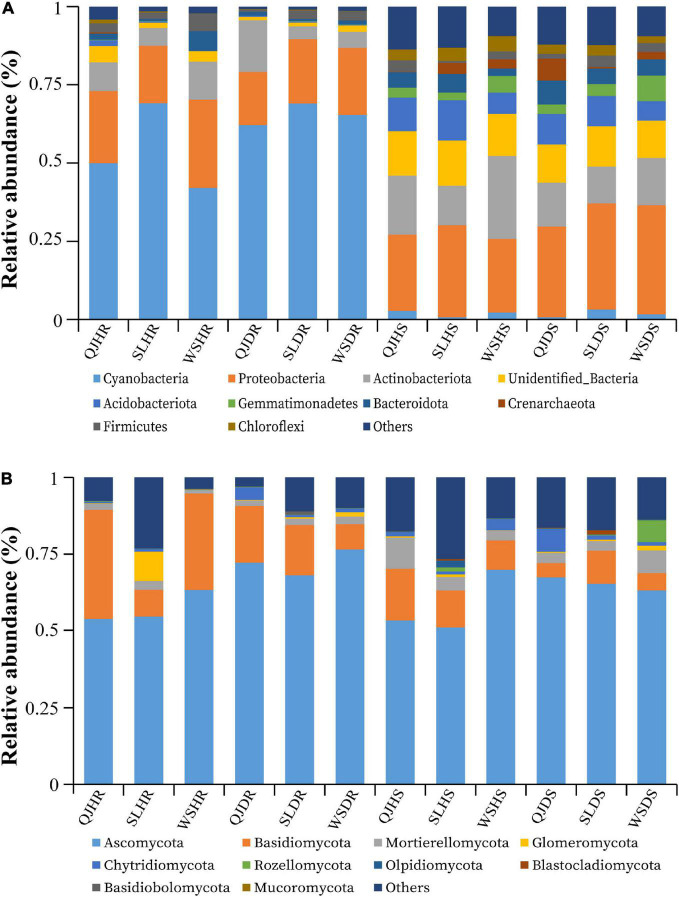
Bacterial and Fungal Community Composition at Phylum Level
The top 10 bacterial and fungal phyla with relative abundance greater than 1% are shown in Figure 5 and Supplementary Table 3. The dominant bacterial phyla in all rhizosphere soil and root samples with a relative abundance greater than 1% are Acidobacteriota, Actinobacteriota, Bacteroidota, Cyanobacteria, Crenarchaeota, Chloroflexi, Gemmatimonadetes, Firmicutes, Proteobacteria, Unidentified_Bacteria, and others (Figure 5A). The dominant fungal phyla in all rhizosphere soil and root samples with a relative abundance greater than 1% are Ascomycota, Basidiomycota, Blastocladiomycota, Basidiobolomycota, Chytridiomycota, Glomeromycota, Mortierellomycota, Mucoromycota, Rozellomycota, Olpidiomycota, and others (Figure 5B). Among the three variables (locations, plant components, and sample types), plant components (rhizosphere soil and roots) significantly influence the bacterial community composition. High relative abundance of phyla Cyanobacteria (average of 59.61%) and Proteobacteria (average of 21.44%) was found in roots; however, phyla Proteobacteria (average of 29.20%) and Actinobacteria (average of 16.46%) were found in high relative abundance in rhizosphere soil (Figure 5A). No significant impact was observed of these three variables on fungal community composition. Fungal phyla such as Ascomycota (average of 65 and 20%) and Basidiomycota (average of 62 and 10%) were present in high relative abundance both in root and rhizosphere soil samples, respectively. However, phylum Basidiomycota was found in low relative abundance in rhizosphere soil (average 10%) than in roots (average 20%) (Figure 5B).
FIGURE 5.
Relative abundance bar plots at phylum level based on the species annotation results in 12 composite samples (average of three replicates per sample) of flue-cured tobacco plants under three variables. (A) Relative abundance at the phylum level in bacterial communities and (B) relative abundance at the phylum level in fungal communities. QJ, Qujing; SL, Shilin; WS, Wenshan; HS, healthy rhizosphere soil; DS, diseased rhizosphere soil; HR, healthy roots; DR, diseased roots.
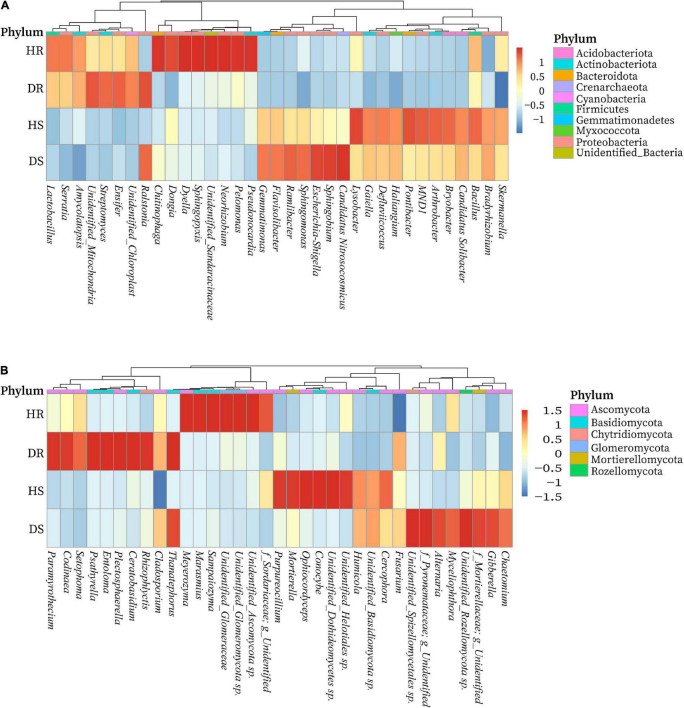
Relative Abundance of Bacterial and Fungal Community Composition at Genera Level
We determined the relative abundance of the top 35 bacterial and fungal genera in the above screened top 10 bacterial and fungal phyla. On the basis of the species relative abundance in all samples, the top 35 bacterial and fungal genera were selected to create a heatmap to determine which genera present in high or low abundance in respective sample type (healthy and diseased). The relative abundance heatmaps of the top 35 bacterial and fungal genera in group-wise comparison under three variables (locations, plant components, and sample types) are shown in Figure 6. Bacterial genera such as Streptomyces, Amycolatopsis, and Ensifer; Chitinophaga, Dyella, Dongia, Neorhizobium, Pelomonas, Pseudonocardia, and Sphingopyxis; Esherichia-Shigella, Sphingomonas, Ramlibacter, Flavisobater, Gemmatimonas, and Sphingobium; and Lysobacter, Arthrobacter, Bryobacter, Bacillus, MND1, Gaiella, and Pontibacter were found in high relative abundance in diseased roots (DR), healthy roots (HR), diseased rhizosphere soil (DS), and healthy rhizosphere soil (HS), respectively. However, a high abundance of genus Ralstonia was present in diseased rhizosphere soil and root samples (Figure 6A). Similarly, fungal genera such as Chaetomium, Gibberella, Myceliophthora, and Alternaria; Ophiocordyceps, Conocybe, Cercophora, Purpureocillium, Humicola, and Mortierella; Codinaea, Rhizophlycits, Ceratobasidium, Entoloma, Paramyrothecium, Setophoma, and Psathyrella; and Marasmius, Meyerozyma, Sampaiozyma, and Unidentified_Ascomycota sp. were found in high relative abundance in DS, HS, DR, and HR, respectively. Whereas genus Thanatephorus was highly abundant with diseased rhizosphere soil and root samples (Figure 6B).
FIGURE 6.
Relative abundance heatmaps at the genus level for top 35 bacterial (A) and fungal (B) genera in group-wise comparison under three variables. HS, healthy rhizosphere soil; DS, diseased rhizosphere soil; HR, healthy roots; DS, diseased roots.
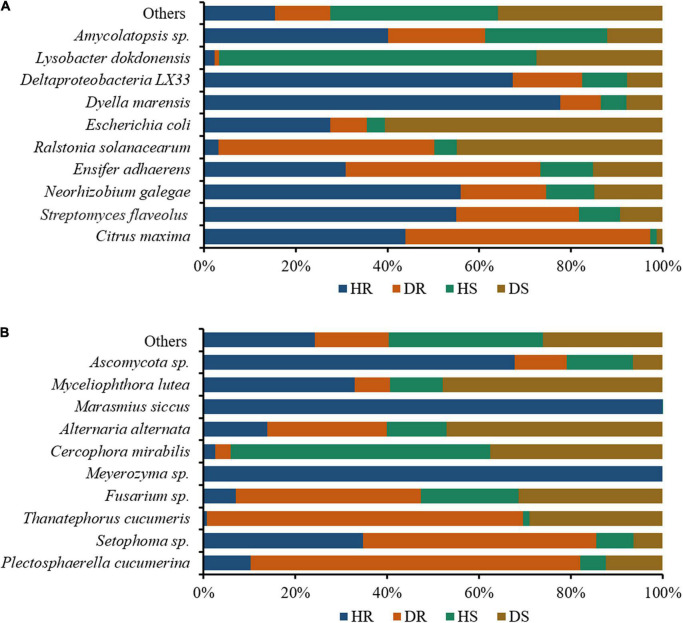
Relative Abundance of Bacterial and Fungal Communities at the Species Level
We assessed the relative abundance of the top 10 bacteria and fungi in the top 35 bacterial and fungal genera. The relative abundance bar plots for the top 10 bacteria and fungi in group-wise comparison under three variables (locations, plant components, and sample types) are shown in Figure 7. A high abundance of bacterial wilt pathogen Ralstonia solanacearum was found in diseased roots and rhizosphere soil. Neorhizobium galegae (well-known nitrogen fixer), Ensifer adhaerens (a bacterial predator of bacteria in the soil), and Lysobacter dokdonensis were found in high abundance in HR, DR, and HS, respectively (Figure 7A). Fungal species such as Alternaria alternata and Thanatephorus cucumeris, disease causing agents of tobacco brown spot and leaf spot disease, respectively, were found in high abundance with diseased rhizosphere soil and root samples. Moreover, higher abundance of Fusarium sp. was found in DR and DS (Figure 7B).
FIGURE 7.
Relative abundance bar plots for top 10 bacterial (A) and fungal (B) species in group-wise comparison under three variables. HS, healthy rhizosphere soil; DS, diseased rhizosphere soil; HR, healthy roots; DS, diseased roots.
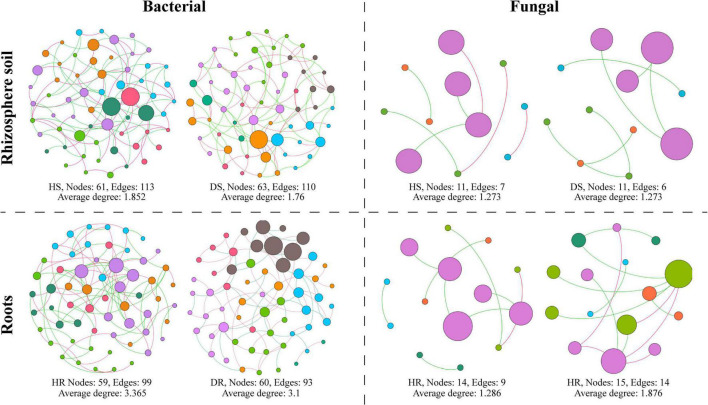
Characteristics of Co-occurrence Network
A microbial co-occurrence network was constructed for bacterial and fungal OTUs at the phylum level for each healthy and diseased sample associated with different plant components (rhizosphere soil and roots) (Figure 8). It was observed that, for bacterial communities, the average degree, number of nodes, and number of edges were found higher in healthy samples compared with diseased samples. In the fungal communities, the average degree, number of nodes, and number of edges were found higher in diseased samples than healthy samples while have no effect on the rhizosphere network. It indicates that the connection degree between members of the bacterial microbial network increased in healthy samples and members interact, whereas the fungal microbial network showed an opposite trend.
FIGURE 8.
Co-occurrence network analysis of bacterial and fungal communities associated with healthy and diseased samples collected from different plant components (rhizosphere soil and roots). HS, healthy soil; DS, diseased soil; HR, healthy roots; DR, diseased roots.
Discussion
Ralstonia solanacearum is a serious threat to flue-cured tobacco production worldwide, including China. It is estimated that approximately 47,133 hm–2 lands are affected by this pathogen, resulting in millions dollars losses every year (Cai et al., 2021). Many integrated disease management strategies are adopted to control the incidence of bacterial wilt pathogen (Jiang et al., 2017). In recent years, biological control via disease suppressive biocontrol agents is considered a promising approach to reduce the incidence of many field crop diseases (Munir et al., 2021; Ahmed et al., 2022). Studies have proven that soil health and rhizosphere microbial diversity are the key factors responsible for soilborne disease occurrence and plant health (Köberl et al., 2013; Dong et al., 2018; Hao and Ashley, 2021).
Soil sickness affects both crop quality and yields when the same crop or its relative species are continuously grown in the same soil under a monocropping system (Zhang et al., 2020b). The rhizosphere microbiome acts as the first line of defense against soilborne pathogen infection and abiotic stress (Bulgarelli et al., 2013; Mendes et al., 2013). In previous studies, flue-cured tobacco bacterial wilt diseases associated with bacterial and fungal communities were studied by direct isolation and high-throughput sequencing technique. However, most studies performed on rhizosphere microbial diversity aimed at bacterial communities with little knowledge about fungal communities of flue-cured tobacco grown on a specific location in pots and field experiments after artificial inoculation of R. solanacearum combined with other biocontrol agents.
In this study, we provided a comprehensive view of the naturally occurring core microbiome of healthy and bacterial wilt–infected (diseased) flue-cured tobacco plants. We presented an in-depth appraisal of bacterial and fungal communities associated with different locations, sample types, and plant components through 16S and ITS1-5f high-throughput sequencing analysis. Our results support the concept that distinct plant components have a significant impact on bacterial and fungal communities, irrespective of sample types and location.
Different plant components have been observed to study the impact of rhizospheric and endophytic bacterial communities on the growth and health of plants with no specific assemblage pattern (Compant et al., 2008; Lundberg et al., 2012; Lebeis et al., 2015). The plant’s rhizosphere acts as the primary host, hot spot habitat, and passage for colonization of microbes into different plant components through roots, especially for bacterial communities (Bulgarelli et al., 2013; Kaushal et al., 2020). In our study, samples were collected from fields where flue-cured tobacco has been continuously grown for the past 10 years as an annual crop for commercial production. Therefore, infected soil and tools used for mechanical operations are primary carriers of bacterial wilt pathogen from one field to another.
Our study demonstrated that rhizosphere soil greatly influences microbial diversity and host more bacterial and fungal communities than roots. Most of the communities present in roots are also present in rhizosphere soil in all locations. The results suggest the microbial communities present in roots were transferred and colonized from the rhizosphere soil. Studies have proven that host plants selectively promote colonization of specific bacterial and fungal communities in roots from rhizosphere soil (Turnbaugh et al., 2007; Lundberg et al., 2012). It was found that rhizosphere soil showed a high microbial diversity than roots. However, healthy tobacco plant rhizosphere soil and root samples host a high diversity of bacterial and fungal communities than bacterial wilt–infected tobacco plant samples. This may be due to disease stress, which is directly linked to the decreased value of available carbon for microbes in the rhizosphere and regulating specific microbe’s growth (Kaushal et al., 2020). Flue-cured tobacco plant resistance and severity of R. solanacearum in different plant components also contribute to the prevalence of microbial communities. Bacterial and fungal OTU count was recorded maximum in all locations and plant components in rhizosphere soil samples than in the root samples.
Host plant environment is known to support or suppress the colonization of certain bacterial and fungal genera with different plant components. Thus, bacterial and fungal communities in these specific plant components are either boosted or exhausted (Kaushal et al., 2020). We observed the core stable bacterial and fungal communities in all sample types that were colonized in different plant components with respect to OTUs distribution. In all rhizosphere soil and root samples for bacterial community composition phylum, Proteobacteria and Cyanobacteria were found in high relative abundance, respectively. Whereas for fungal community composition, phyla Ascomycota and Basidiomycota were found in high relative abundance in both rhizosphere soil and root samples. Results showed that bacterial wilt pathogen has a great impact and significantly influences the bacterial communities compared with fungal communities.
Most of the diverse bacteria and fungi can enhance plant growth, suppress disease incidence, and confer other biological functions that benefit the plants (Santoyo et al., 2016; Gouda et al., 2018). A fraction of core bacterial communities identified in this study are known as plant growth promoters, enhance resistance in plants, and suppress the incidence of disease via production of a specific antibiotic, volatile organic compounds, secondary metabolites, and nitrogen fixation in many fields crops. Bacterial genera such as Amycolatopsis, Bacillus, Streptomyces, Ensifer, Sphingopyxis, Neorhizobium, Pseudonocardia, Sphingomonas, Haliangium, Bradyrhizobium, Arthrobacter, Bryobacter, and Lysobacter were found in high relative abundance, which contribute to plant growth promotion, nutrition acquisition, suppression of soilborne diseases and other plant pathogens. However, a minor fraction of fungal communities such as Gibberella and Purpureocillium was found as plant growth promoters and biocontrol agents, whereas others are saprophytic and pathogenic and cause disease in many important field crops.
Bacterial wilt pathogen R. solanacearum was highly abundant in diseased rhizosphere soil and root. A high abundance of the bacterium was found with nitrogen-fixing ability, producing secondary metabolites, and bacterial predator of bacteria in the soil such as Neorhizobium galegae, Lysobacter dokdonensis, and Ensifer adhaerens. Plants pathogenic fungi such as Plectosphaerella cucumerina, Alternaria alternata, Thanatephorus cucumeris, and Fusarium sp. were found in high relative abundance diseased rhizosphere soil and root. Thus, we conclude that these beneficial bacterial communities are positively related to flue-cured tobacco plant health, whereas the pathogenic fungal communities enhance the population of R. solanacearum and the incidence of bacterial wilt disease. Our results are in accordance with the study of Jiang et al. (2017), who reported that the incidence of tobacco bacterial wilt disease increases when it prevails with other root rot pathogen P. nicotiana. Analysis of co-occurrence networks showed that R. solanacearum has a significant impact on the bacterial community structure compared with fungal communities. Fewer negative correlations were found between bacterial communities in healthy plants compared to diseased plants.
Conclusion
In this study, we conclude that flue-cured tobacco plants host a range of bacterial and fungal communities. Plant components significantly impact bacterial and fungal communities. Rhizosphere soil is enriched with bacterial and fungal communities compared to roots, whereas high diversity was found in healthy plants rather than diseased plants. By comparing the microbial communities with other crops, we suggest that several bacterial communities discovered in the flue-cured tobacco microbiome aid in plant growth promotion and disease suppression. A fraction of fungal communities pathogenic to flue-cured tobacco was found in diseased plants, which correlates with the occurrence of tobacco bacterial wilt disease. However, future work should focus on studying the microbiome of flue-cured tobacco cultivars with resistance to bacterial wilt pathogen both as naturally occurring and artificial inoculation. This will provide experimental and theoretical information, that microbiome of flue-cured tobacco can be engineered to manage the bacterial wilt disease for better yield and quality production.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
ZZ and GJ designed the experiment. WA, QL, SL, and JZ visited the tobacco-growing areas and collected samples. WA, ZD, and JY analyzed data and drew figures. WA, SM, and SK wrote the initial draft of manuscript. WA, SM, GJ, and ZZ wrote, reviewed, and edited final draft of the manuscript. All authors contributed to the final draft of the manuscript and agreed to the published version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Funding
This study was financially supported by the Yunnan Agricultural University Scientific Research Foundation (KX900187), the Science and Technology Platform Plan of Yunnan Province (2019IC005), the National Key R&D Program of China (2019YFD1002000), the Yunnan Ten Thousand Talents Plan Leading Talents of Industrial Technology Project of China (YNWR-CYJS-2019-046), and National Natural Science Foundation of China (No. 32060601).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.845310/full#supplementary-material
References
- Ahmed W., Yang J., Tan Y., Munir S., Liu Q., Zhang J., et al. (2022). Ralstonia solanacearum, a deadly pathogen: revisiting the bacterial wilt biocontrol practices in tobacco and other Solanaceae. Rhizosphere 21:100479. 10.1016/j.rhisph.2022.100479 [DOI] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D., Schlaeppi K., Spaepen S., Van Themaat E. V. L., Schulze-Lefert P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64 807–838. 10.1146/annurev-arplant-050312-120106 [DOI] [PubMed] [Google Scholar]
- Cai Q., Zhou G., Ahmed W., Cao Y., Zhao M., Li Z., et al. (2021). Study on the relationship between bacterial wilt and rhizospheric microbial diversity of flue-cured tobacco cultivars. Eur. J. Plant Pathol. 160 265–276. 10.1007/s10658-021-02237-4 [DOI] [Google Scholar]
- Compant S., Kaplan H., Sessitsch A., Nowak J., Ait Barka E., Clément C. (2008). Endophytic colonization of Vitis vinifera L. by Burkholderia phytofirmans strain PsJN: from the rhizosphere to inflorescence tissues. FEMS Microbiol. Ecol. 63 84–93. 10.1111/j.1574-6941.2007.00410.x [DOI] [PubMed] [Google Scholar]
- Dong C.-J., Wang L.-L., Li Q., Shang Q.-M. (2019). Bacterial communities in the rhizosphere, phyllosphere and endosphere of tomato plants. PLoS One 14:e0223847. 10.1371/journal.pone.0223847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Xu J., Zhang L., Cheng R., Wei G., Su H., et al. (2018). Rhizospheric microbial communities are driven by Panax ginseng at different growth stages and biocontrol bacteria alleviates replanting mortality. Acta Pharmac. Sin. B 8 272–282. 10.1016/j.apsb.2017.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10 996–998. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Hua C., Tong M. (2018). Impact of Dinophysis acuminata feeding Mesodinium rubrum on nutrient dynamics and bacterial composition in a microcosm. Toxins 10:443. 10.3390/toxins10110443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouda S., Kerry R. G., Das G., Paramithiotis S., Shin H.-S., Patra J. K. (2018). Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiol. Res. 206 131–140. 10.1016/j.micres.2017.08.016 [DOI] [PubMed] [Google Scholar]
- Govindasamy V., Franco C. M., Gupta V. V. (2014). “Endophytic actinobacteria: diversity and ecology,” in Advances In Endophytic Research, eds Verma V. C., Gange A. C. (New Delhi: Springer; ), 27–59. [Google Scholar]
- Hao J., Ashley K. (2021). Irreplaceable role of amendment-based strategies to enhance soil health and disease suppression in potato production. Microorganisms 9:1660. 10.3390/microorganisms9081660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G., Wei Z., Xu J., Chen H., Zhang Y., She X., et al. (2017). Bacterial wilt in China: history, current status, and future perspectives. Front. Plant Sci. 8:1549. 10.3389/fpls.2017.01549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal M., Swennen R., Mahuku G. (2020). Unlocking the microbiome communities of banana (Musa spp.) under disease stressed (Fusarium wilt) and non-stressed conditions. Microorganisms 8:443. 10.3390/microorganisms8030443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.-S., French E., Caldwell D., Harrington E. J., Iyer-Pascuzzi A. S. (2016). Bacterial wilt disease: host resistance and pathogen virulence mechanisms. Physiol. Mol. Plant Pathol. 95 37–43. 10.1016/j.pmpp.2016.02.007 [DOI] [Google Scholar]
- Köberl M., Schmidt R., Ramadan E. M., Bauer R., Berg G. (2013). The microbiome of medicinal plants: diversity and importance for plant growth, quality and health. Front. Microbiol. 4:400. 10.3389/fmicb.2013.00400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kõljalg U., Nilsson R. H., Abarenkov K., Tedersoo L., Taylor A. F., Bahram M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22 5271–5277. 10.1111/mec.12481 [DOI] [PubMed] [Google Scholar]
- Lebeis S. L., Paredes S. H., Lundberg D. S., Breakfield N., Gehring J., McDonald M., et al. (2015). Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349 860–864. 10.1126/science.aaa8764 [DOI] [PubMed] [Google Scholar]
- Li C., Ahmed W., Li D., Yu L., Xu L., Xu T., et al. (2022). Biochar suppresses bacterial wilt disease of flue-cured tobacco by improving soil health and functional diversity of rhizosphere microorganisms. Appl. Soil Ecol. 171:104314. 10.1016/j.apsoil.2021.104314 [DOI] [Google Scholar]
- Li L., Feng X., Tang M., Hao W., Han Y., Zhang G., et al. (2014). Antibacterial activity of Lansiumamide B to tobacco bacterial wilt (Ralstonia solanacearum). Microbiol. Res. 169 522–526. 10.1016/j.micres.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Li Y., Ren K., Zou C., Xie J., He X., Chen Y., et al. (2019). Effects of ferrous iron toxicity on agronomic, physiological, and quality indices of flue-cured tobacco. Agron. J. 111 2193–2206. 10.2134/agronj2018.12.0786 [DOI] [Google Scholar]
- Liu Y., Shi J., Feng Y., Yang X., Li X., Shen Q. (2013). Tobacco bacterial wilt can be biologically controlled by the application of antagonistic strains in combination with organic fertilizer. Biol. Fertil. Soils 49 447–464. 10.1007/s00374-012-0740-z [DOI] [Google Scholar]
- Lundberg D. S., Lebeis S. L., Paredes S. H., Yourstone S., Gehring J., Malfatti S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488 86–90. 10.1038/nature11237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Zhang H.-Y., Zhou X.-K., Yang C.-G., Zheng S.-C., Duo J.-L., et al. (2018). Biological control tobacco bacterial wilt and black shank and root colonization by bio-organic fertilizer containing bacterium Pseudomonas aeruginosa NXHG29. Appl. Soil Ecol. 129 136–144. 10.1016/j.apsoil.2018.05.011 [DOI] [Google Scholar]
- Mendes R., Garbeva P., Raaijmakers J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37 634–663. 10.1111/1574-6976.12028 [DOI] [PubMed] [Google Scholar]
- Mhlongo M. I., Piater L. A., Madala N. E., Labuschagne N., Dubery I. A. (2018). The chemistry of plant–microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 9:112. 10.3389/fpls.2018.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir S., Ahmed A., Li Y., He P., Singh B. K., He P., et al. (2021). The hidden treasures of citrus: finding Huanglongbing cure where it was lost. Crit. Rev. Biotechnol. 1–16. 10.1080/07388551.2021.1942780 [DOI] [PubMed] [Google Scholar]
- Paudel S., Dobhal S., Alvarez A. M., Arif M. (2020). Taxonomy and phylogenetic research on ralstonia solanacearum species complex: a complex pathogen with extraordinary economic consequences. Pathogens 9:886. 10.3390/pathogens9110886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi G., Chen S., Ke L., Ma G., Zhao X. (2020). Cover crops restore declining soil properties and suppress bacterial wilt by regulating rhizosphere bacterial communities and improving soil nutrient contents. Microbiol. Res. 238:126505. 10.1016/j.micres.2020.126505 [DOI] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41 D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers J. M. (2015). “The minimal rhizosphere microbiome,” in Principles Of Plant-Microbe Interactions, ed. Lugtenberg B. (Heidelberg: Springer; ), 411–417. [Google Scholar]
- Santoyo G., Moreno-Hagelsieb G., del Carmen Orozco-Mosqueda M., Glick B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183 92–99. 10.1016/j.micres.2015.11.008 [DOI] [PubMed] [Google Scholar]
- Shen G., Zhang S., Liu X., Jiang Q., Ding W. (2018). Soil acidification amendments change the rhizosphere bacterial community of tobacco in a bacterial wilt affected field. Appl. Microbiol. Biotechnol. 102 9781–9791. 10.1007/s00253-018-9347-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Chen L., Chen Z., Fu Y., Sun X., Wang B., et al. (2020). Climatic factors determine the yield and quality of Honghe flue-cured tobacco. Sci. Rep. 10 1–12. 10.1038/s41598-020-76919-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Hamady M., Fraser-Liggett C. M., Knight R., Gordon J. I. (2007). The human microbiome project. Nature 449 804–810. 10.1038/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Yang J., Ahmed W., Xiong X., Liu Q., Huang Q., et al. (2021). Unraveling the association between metabolic changes in inter-genus and intra-genus bacteria to mitigate clubroot disease of chinese cabbage. Agronomy 11:2424. [Google Scholar]
- Wu X., Li H., Wang Y., Zhang X. (2020). Effects of bio-organic fertiliser forti?ed by Bacillus cereus QJ-1 on tobacco bacterial wilt control and soil quality improvement. Biocontrol Sci. Technol. 30 351–369. 10.1080/09583157.2020.1711870 [DOI] [Google Scholar]
- Zhang C., Lin Y., Tian X., Xu Q., Chen Z., Lin W. (2017). Tobacco bacterial wilt suppression with biochar soil addition associates to improved soil physiochemical properties and increased rhizosphere bacteria abundance. Appl. Soil Ecol. 112 90–96. 10.1016/j.apsoil.2016.12.005 [DOI] [Google Scholar]
- Zhang J., Wei L., Yang J., Ahmed W., Wang Y., Fu L., et al. (2020a). Probiotic consortia: Reshaping the rhizospheric microbiome and its role in suppressing root-rot disease of Panax notoginseng. Front. Microbiol. 11:701. 10.3389/fmicb.2020.00701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Fan S., Qin J., Dai J., Zhao F., Gao L., et al. (2020b). Changes in the microbiome in the soil of an American ginseng continuous plantation. Front. Plant Sci. 11:572199. 10.3389/fpls.2020.572199 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.