Viruses are too small to be seen in a light microscope, yet they cause havoc in plants as well as animal communities. In the midst of a viral pandemic, who would know better than us how much damage this tiny invisible foe can cause. Viruses need to produce numerous proteins for replication, movement, and pathogenesis. Having a tiny genome, viruses use non-conventional strategies like initiating protein synthesis from un-annotated sites to increase their coding capacity (Ho et al., 2021). Accurate annotation of viral genes is crucial to developing antiviral strategies. However, deciphering a full set of viral genes is a great challenge as viruses use hidden open-reading frames (ORFs) with unanticipated translation initiation sites (TISs).
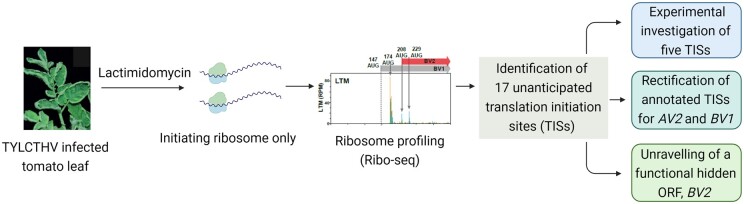
In new work, Ching-Wen Chiu and colleagues (Chiu et al., 2022) experimentally mapped in vivo TISs and identified hidden functional ORFs in tomato yellow leaf curl Thailand virus (TYLCTHV) by employing ribosome profiling (see Figure). Ribosome profiling, a method of deep sequencing of ribosome-protected mRNA fragments, allows us to map and monitor translated regions of a genome (Ingolia et al., 2011). Ribosome profiling combined with lactimidomycin (LTM), which immobilizes ribosomes immediately after translation initiation and thus enriches footprints at initiation codons, can reveal in vivo TISs (Lee et al., 2012). Using LTM-ribosome profiling, Chiu et al. (2022) systematically scanned the genome of TYLCTHV and revealed 21 TISs, of which only four were previously annotated. TYLCTHV is a begomovirus with a bipartite genome containing DNA-A and DNA-B components. AV2, coded in DNA-A, is a movement protein necessary for pathogenicity. Chiu et al. refuted the annotated translation start site AUG87 of AV2 and experimentally proved that, instead, two downstream TISs, AUG135 and AUG189, are responsible for AV2 protein synthesis. These downstream TISs generate two AV2 isoforms that were found to play an essential role in viral pathogenicity. Interestingly, AUG135 and AUG189 TISs, but not the annotated AUG87 TIS, were found to be conserved across begomoviruses.
Figure.
Schematic of the approach employed by Chiu et al. (2022) to identify and characterize un-annotated TISs of TYLCTHV using ribosome profiling combined with lactimidomycin treatment. Figure created with Biorender.com.
Another important gene, BV1, residing in DNA-B, codes for a shuttle protein that transports viral ssDNA between nucleus and cytoplasm. Chiu et al. (2022) further revised the functional TIS of BV1 by identifying and characterizing a novel TIS, AUG174. They found no evidence of translation initiation from the earlier annotated TIS, AUG147. A strain with a mutation in BV1 AUG147 displayed similar disease-causing ability as wild-type infectious TYLCTHV in tomato. In contrast, the strain harboring a mutation at AUG174 exhibited delayed leaf curling symptom development and resulted in lower viral DNA accumulation than wild-type infectious TYLCTHV, indicating that AUG174 is the functional TIS for BV1 expression and required for pathogenicity.
While investigating other identified TISs that are distinct from annotated TISs and encode new ORFs, the authors focused on two closely located sites that can initiate translation of 120 and 113 amino acid protein isoforms from a single unannotated ORF. They obtained translational evidence of this novel ORF and named it BV2. BV2 is nested within the BV1 ORF and produces viral protein via alternative translation initiation. Knock-out mutants of BV2 displayed attenuated disease symptoms in tomato. Additionally, overexpression of BV2 in Nicotiana benthamiana infected with a TYLCTHV BV2 knockout mutant showed higher viral DNA accumulation than in the BV2 mutant-infected leaves without BV2 overexpression. These results indicate that BV2 expedites virus infection. Finally, the authors predicted BV2 as a transmembrane domain protein and showed BV2 is preferentially located in the endoplasmic reticulum and plasmodesmata. The above data suggest that BV2 may play a role in replication and cell-to-cell movement of TYLCTHV.
In summary, this ribosome profiling study by Chiu et al. (2022) provides invaluable insights to revise and rectify previous annotations of a plant DNA virus genome. It also reveals tactics taken by TYLCTHV to expand the coding potential of its small genome and infect its host. This work will facilitate investigations of other plant viruses and aid in devising novel antiviral strategies.
References
- Chiu CW, Lia YR, Lin CY, Yeh HH, Liu MJ (2022) Translation initiation landscape profiling reveals hidden open reading frames required for tomato yellow leaf curl Thailand virus pathogenesis. Plant Cell 34: 1804--1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JSY, Zhu Z, Marazzi I (2021) Unconventional viral gene expression mechanisms as therapeutic targets. Nature 593: 362–371 [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB (2012) Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci USA 109: 14728–14729 [DOI] [PMC free article] [PubMed] [Google Scholar]