Abstract
Opaque2 (O2) functions as a central regulator of the synthesis of starch and storage proteins and the O2 gene is transcriptionally regulated by a hub coordinator of seed development and grain filling, ABSCISIC ACID INSENSITIVE 19 (ZmABI19), in maize (Zea mays). Here, we identified a second hub coordinator, basic Leucine Zipper 29 (ZmbZIP29) that interacts with ZmABI19 to regulate O2 expression. Like zmabi19, zmbzip29 mutations resulted in a dramatic decrease of transcript and protein levels of O2 and thus a significant reduction of starch and storage proteins. zmbzip29 seeds developed slower and had a smaller size at maturity than those of the wild type. The zmbzip29;zmabi19 double mutant displayed more severe seed phenotypes and a greater reduction of storage reserves compared to the single mutants, whereas overexpression of the two transcription factors enhanced O2 expression, storage-reserve accumulation, and kernel weight. ZmbZIP29, ZmABI19, and O2 expression was induced by abscisic acid (ABA). With ABA treatment, ZmbZIP29 and ZmABI19 synergistically transactivated the O2 promoter. Through liquid chromatography tandem-mass spectrometry analysis, we established that the residues threonine(T) 57 in ZmABI19, T75 in ZmbZIP29, and T387 in O2 were phosphorylated, and that SnRK2.2 was responsible for the phosphorylation. The ABA-induced phosphorylation at these sites was essential for maximum transactivation of downstream target genes for endosperm filling in maize.
Basic Leucine Zipper 29 interacts with ABSCISIC ACID INSENSITIVE 19 to regulate Opaque2 expression, and their transactivation activities are enhanced by abscisic acid-induced phosphorylation.
IN A NUTSHELL.
Background: Early seed development and grain filling in maize are two tightly linked developmental stages. The spatial and temporal regulation of the two stages requires the concerted action of several signaling pathways that integrate information from genetic programs, and both hormonal and metabolic signals. Our pioneering work identified a hub coordinator of maize seed development and grain filling, ZmABI19, that directly recognizes and transactivates the central regulator O2 and other critical TFs in the endosperm and Vp1 in the embryo. However, the overall picture of the regulatory networks involved in these processes is far from complete.
Question: Besides ZmABI19, are there more TFs required to coordinate early seed development and grain filling? What are the mechanisms to initiate grain filling by these transcription factors (TFs)?
Findings: We report here that a basic leucine zipper TF, ZmbZIP29, with abundant expression during early endosperm development, interacts with ZmABI19 to regulate the central endosperm-filling regulator O2 gene, and their transactivation capacities, are enhanced by ABA-induced phosphorylation. This work established a mechanistic framework in which ABA-mediated phosphorylation of two hub coordinators ZmABI19 and ZmbZIP29 by SnRK2.2 enhances gene transactivation for endosperm filling.
Next steps: First, besides ABA, what are other signals required to initiate grain filling? Soluble sugars that suddenly increase at the onset of endosperm filling probably function as a signal; second, the transition of early endosperm development to endosperm filling is a critical phase of endosperm development. ZmABI19 and ZmbZIP29 are abundantly expressed during early endosperm development and play crucial roles at this stage. When early endosperm development ends, how do the encoded proteins shift their binding from early targets to genes required for grain filling? We will address these questions in the following years.
Introduction
Abscisic acid (ABA), derived from epoxycarotenoid cleavage, is one of the best-characterized phytohormones in terms of its biosynthesis and catabolism, transport, and signal transduction (Yoshida et al., 2014). ABA regulates many agronomically important aspects of plant growth, including seed protein and lipid storage, desiccation tolerance, and dormancy and regulation of the phase transitions from embryonic to germinative growth and vegetative to reproductive growth (Chen et al., 2020). The ABA receptors PYRABACTIN RESISTANCE1(PYR1)-LIKE (PYL)/PYR1/REGULATORY COMPONENTS OF ABA RECEPTOR (RCAR) have high ABA-binding affinity and interact with the clade A protein phosphatases of type 2Cs (PP2Cs) (Park et al., 2009). ABA stabilizes these ABA receptors and then the sucrose non-fermenting-1 (SNF1)-related protein kinase 2s (SnRK2s) are released from PP2Cs inhibition. The activated SnRK2s phosphorylate downstream transcription factors (TFs) to modulate ABA responses (Cutler et al., 2010). Four main groups of cis-regulatory sequences are required for ABA inducibility, including ABA response elements ABREs (belong to the G-box elements), CE3 (coupling elements)-like sequences, RY elements, and the recognition sequences for MYB DOMAIN PROTEIN (MYB) TFs (Busk and Pagès, 1998; Finkelstein et al., 2002). The identified B3 domain-containing TF ABSCISIC ACID INSENSITIVE 3 (ABI3) and basic leucine zipper (bZIP) TF ABI5 in Arabidopsis recognize the RY element and ABREs, respectively (Parcy et al., 1994; Finkelstein and Lynch, 2000). Mutations of ABI3 and ABI5 result in ABA insensitivity during seed germination and seedling development (Giraudat et al., 1992; Finkelstein and Lynch, 2000).
ABA has been reported to regulate the sink strength and then influence yield and quality through mediating phloem unloading (Jones and Brenner, 1987; Frey et al., 2004). ABA transporters, such as defective grain-filling 1 (DG1) in rice (Oryza sativa), mediate the long-distance transport of ABA from leaves to caryopses (Qin et al., 2021). Reduction of the ABA contents in dg1 results in defective grain filling with a floury endosperm, aberrantly loose starch granules, and decreased grain weight. ABA directly activates the expression of grain filling-related genes to promote starch synthesis, suggesting that ABA contents in grains positively correlate with grain-filling rate and promote reserve accumulation (Yang et al., 2001). Generally, the ABA content of large grains is higher than that of small grains at the grain-filling stage (Kato et al., 1993). However, the mechanism by which ABA positively regulates grain filling in the cereal endosperm remains largely unknown.
The maize endosperm contributes up to 90% of the dry seed weight and its storage compounds mainly consist of starch and storage proteins that account for 70% and 10% of the endosperm dry mass, respectively. Starch consisting of amylopectin and amylose is synthesized by a suite of enzymes that are organized in interacting protein complexes in the starchy endosperm cells. The main storage proteins representing more than 60% of all endosperm proteins are prolamins, called zeins in maize. According to their structure and sequence similarity, zein proteins are divided into four classes, α- (19-kD z1A, z1B and z1D; 22-kD z1C), β- (15-kD), γ- (50-, 27-, and 16-kD), and δ-zeins (18-kD and 10-kD) (Esen, 1987; Coleman and Larkins, 1999). The starch biosynthetic genes and storage-protein zein genes are strictly regulated with a precise temporal and spatial pattern. Opaque2 (O2) is considered as a central regulator of starch and protein synthesis. O2 is an endosperm-specific bZIP TF. Spatially and developmentally controlled expression of O2 is precisely timed, leading to specific expression in the endosperm from 8–10 DAP (days after pollination). The ChIP-seq data revealed that except for 16-kD γ-zein, O2 regulates the expression of all other zein genes (Li et al., 2015; Zhan et al., 2018). Pyruvate orthophosphate dikinase 1 and 2 (PPDKs) and starch synthase III (SSIII) are critical components of the starch biosynthetic enzyme complex. Their genes are transcriptionally regulated by O2 (Zhang et al., 2016). In addition, O2 directly transactivates Shrunken1, Sus1, and Sus2, the three genes encoding sucrose synthase, which converts sucrose plus uridine diphosphate (UDP) to fructose and UDP-glucose for downstream starch synthesis (Deng et al., 2020). O2 also directly regulates the expression of an endosperm-specific GRAS gene (ZmGRAS11) to promote cell expansion during endosperm filling (Ji et al., 2021).
Great complexity of posttranslational modification of O2 was observed during endosperm development in maize, such as ubiquitination and phosphorylation (Ciceri et al., 1997; Li et al., 2020). O2 is ubiquitinated by the E3 ubiquitin ligase ZmRFWD3 to enhance its nuclear localization (Li et al., 2020). An isoelectric focusing assay showed that O2 is phosphorylated in vivo and in vitro, and this phosphorylation pattern is constant throughout endosperm development (Ciceri et al., 1997). As a classic bZIP-type TF, O2 recognizes the G-box or O2 box in the promoters of its own and other downstream target genes (Li et al., 2015; Zhan et al., 2018). ABRE, belonging to the G-box family, is recognized by the ABA-dependent bZIP TFs. However, it is unknown whether O2 expression or O2-mediated transactivation is associated with ABA signaling.
Previous work revealed that a B3 domain-containing TF, ZmABI19, directly binds two RY motifs in the O2 promoter to transactivate O2 (Yang et al., 2021). The zmabi19 mutants displayed developmental defects in both the embryo and endosperm. Moreover, ZmABI19 regulates a series of important grain-filling TF genes, including prolamine-box binding factor 1 (Pbf1), NAC130, ZmbZIP22, and Opaque11 (O11) in the endosperm and Viviparous1 (Vp1) in the embryo. Considering that ZmABI19 expression is high during early seed development and low at the grain-filling stage, ZmABI19 is thought to function as a grain-filling initiation regulator that coordinates endosperm and embryo development. Still, it is unknown whether ZmABI19 expression or ZmABI19-mediated transactivation is associated with ABA signaling.
In this study, we found that O2 and ZmABI19 were both ABA-inducible genes and that transactivation of the O2 promoter by ZmABI19 was markedly enhanced by the addition of ABA. This suggested that the ABA-induced O2 expression depended on the transactivation function of ZmABI19. However, a lessened enhancement of O2 expression by ABA could still be observed in zmabi19, implying the existence of another ABA-inducible TF involved in O2 upstream regulation. Indeed, this observation led to identification of ZmbZIP29, which was found to recognize the ABRE in the O2 promoter for transactivation. ZmbZIP29 and ZmABI19 had protein–protein interaction and synergistically transactivated O2 expression in the presence of ABA. Mutations of ZmbZIP29 resulted in reduced O2 expression and thus decreased starch and zein protein contents. The zmbzip29 seeds were smaller and lighter than those of the wild type and zmbzip29;zmabi19 mutants exhibited even smaller and more flattened seed phenotypes compared with the single mutants. When the two TFs were overexpressed, the kernel weight and storage reserves were significantly increased. We found that ZmbZIP29, ZmABI19, and O2 were phosphorylated by the ABA-induced action of SnRK2.2. Through liquid chromatography–tandem mass spectrometry (LC–MS)/MS analysis, we determined the precise residues for phosphorylation. The phosphorylated ZmbZIP29, ZmABI19, and O2 TFs had enhanced transactivation capacity for the downstream target genes. These studies provide insight into the ABA-based complex regulatory network of maize grain filling and pave the way for improving maize yield and nutritional quality by engineering the expression or phosphorylation of these TFs.
Results
ABA promotes O2 and ZmABI19 expression
To determine the ABA contents of developing endosperms, endosperm samples of the B104 inbred line were collected at different time points and subjected to LC–MS/MS. The measurement revealed that the ABA content before endosperm filling was low, only 5.62 ng/100 mg at 6 DAP and 5.51 ng/100 mg at 8 DAP (Supplemental Figure S1A). When entering the endosperm-filling stage (after 8 DAP), the ABA content was doubled to 12.90 ng/100 mg at 10 DAP, reached the peak at 12 DAP, and then declined afterward (Supplemental Figure S1A). The rapid increase of ABA contents coincided with the initiation of endosperm filling (Sabelli and Larkins, 2009). Since O2 acts as a central hub node for endosperm filling through directly regulating storage-protein zein and starch synthetic genes (Xiong et al., 2017), we tested whether ABA promoted O2 expression. The 10-DAP B104 endosperms were harvested and incubated in half-strength MS (Murashige & Skoog) medium supplemented with different concentrations of ABA for 24 h. Reverse transcription quantitative PCR (RT-qPCR) analysis showed that enhancement of the O2 transcription by ABA depended on an optimal concentration window (100–200 μM) with maximum enhancement at 150 μM, while addition of a low (50 μM) or high (250 μM) concentration of ABA had no apparent effect on O2 transcription (Supplemental Figure S1B). We also examined the expression of the luciferase (LUC) gene driven by the O2 promoter (abbreviated O2pro-LUC) that was bombarded into the B104 endosperms and cultured in the different concentrations of ABA-containing half-strength MS medium for 24 h. Consistently, LUC activities were enhanced by the addition of ABA, with the 150 μM ABA treatment giving rise to the highest enhancement (Supplemental Figure S1C). These results indicate that suitably increased ABA contents promote O2 expression during endosperm filling.
ZmABI19 functions as a grain-filling initiator that regulates a series of TFs, such as O2, Pbf1, and NAC130 in the endosperm, and Vp1 in the embryo (Yang et al., 2021). ZmABI19 recognizes the G-box element in the Vp1 promoter to exert transactivation and the transactivation is remarkably enhanced when 10 μM ABA is applied (Yang et al., 2021). This encouraged us to investigate whether ABA reinforced the ZmABI19-mediated O2 transactivation. Similarly, the dual-LUC reporter (DLR) assay using Arabidopsis mesophyll protoplasts showed that co-expression of ZmABI19 and O2pro-LUC resulted in significantly increased LUC activities compared to the negative control (Supplemental Figure S2A). The activities were further increased when 10-μM ABA was added into the reaction (Supplemental Figure S2A). We also tested whether ABA indirectly promoted O2 transcription through increasing ZmABI19 expression using 10-DAP B104 endosperms cultured on half-strength MS medium containing different concentrations of ABA. RT-qPCR analysis revealed that ZmABI19 expression was dramatically induced when the medium was supplemented with 100–150 μM ABA. Similar to O2, low or high levels of ABA had no apparent effect on ZmABI19 expression (Supplemental Figure S2B). These results demonstrated that ABA could promote O2 expression through increasing ZmABI19 expression, which in turn enhanced ZmABI19-mediated O2 transactivation.
To genetically confirm that ABA is an essential phytohormone to induce ZmABI19 and O2 expression, the w3 and vp5 mutants that had defects in ABA synthesis were analyzed (Robichaud et al., 1979; Hunter et al., 2018). RT-qPCR revealed that transcript levels of ZmABI19 and O2 were significantly reduced in w3 and vp5 endosperms (Supplemental Figure S3, A and B).
To test whether ABA-promoted O2 transactivation depends on the existence of ZmABI19, the B104 wild-type and zmabi19 endosperms at 10 DAP were incubated with 150-μM ABA. RT-qPCR revealed that the O2 transcript level in ABA-treated zmabi19 endosperms was still significantly enhanced compared with that in untreated zmabi19 endosperms, although the extent of enhancement is less than that between ABA-treated and untreated wild-type endosperms (Supplemental Figure S4, A and B). This suggests that, besides ZmABI19, another unknown ABA-inducible TF exists that can transactivate O2 expressions.
Identification of candidate TFs that bind the ABRE in the O2 promoter
Previous work revealed that the sequence between –400 and –500 bp upstream of the start codon in the O2 promoter contained key cis-elements for transactivation (Yang et al., 2021). This 100-bp DNA fragment was predicted to contain nine motifs, of which two motifs, ABRE and O2 box, overlapped (Supplemental Figure S5). The ABRE could be recognized by bZIP TFs for transactivation (Busk and Pagès, 1998). When O2 transcription is activated, O2 recognizes the O2 box and exerts auto-activation during endosperm filling (Yang et al., 2021). However, the TF that binds the ABRE to initiate O2 expression has not been identified. In the maize genome, the bZIP TF family was annotated to contain 128 members (https://www.grassius.org/family.php?family=bZIP&species=Maize). Based on the high-resolution RNA-seq data during early seed development, 83 bZIP genes were detected for expression in all sequenced samples (Yi et al., 2019). Since the candidate bZIP TF might respond to ABA induction and co-transactivate the O2 promoter with ZmABI19, we hypothesized that this bZIP gene and ZmABI19 should share a similar expression pattern. ZmABI19 expression is high during early endosperm development, especially at 6 DAP, and low at the endosperm-filling stage, and remains at basal levels in vegetative tissues (Chen et al., 2014; Yi et al., 2019; Yang et al., 2021). ZmbZIP29 (Zm00001d034571) appeared to be the only candidate that displayed nearly identical expression pattern with ZmABI19 (Supplemental Figure S6; Yang et al., 2021).
We performed a yeast one-hybrid (Y1H) assay to test whether ZmbZIP29 could recognize the O2 promoter. A number of bZIP TFs that showed different expression patterns with ZmABI19 were used as negative controls. A bait fragment containing the ABRE was cloned into the pAbAi plasmid and then integrated into the yeast genome. As a result, only ZmbZIP29 was found to recognize the bait sequence (Supplemental Figure S7).
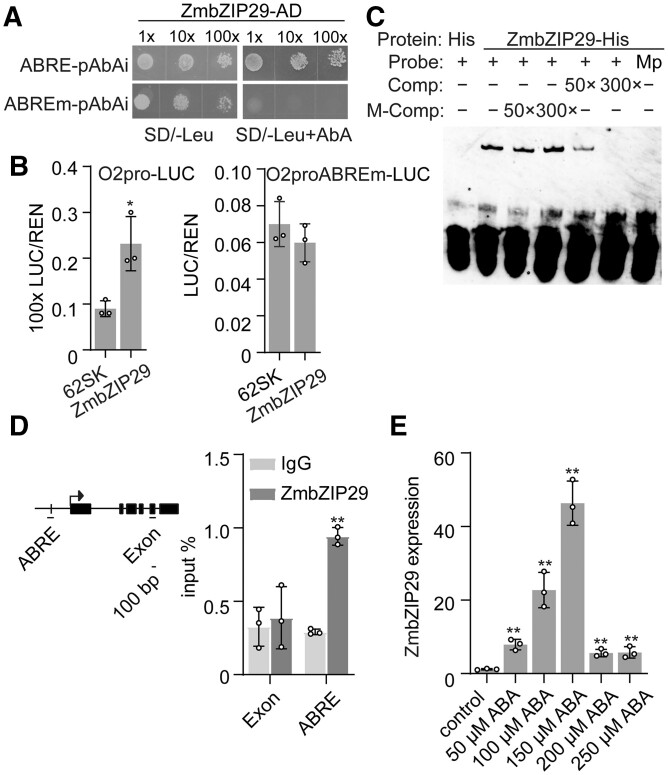
To verify the protein–DNA interaction, fragments with mutations in the ABRE were integrated into the yeast genome and used in Y1H assay. The mutated construct failed to be recognized by ZmbZIP29 (Figure 1A). A DLR assay was performed to test the transactivation activity of the O2 promoter by ZmbZIP29. The full-length CDS of ZmbZIP29 under the control of the CaMV 35S promoter was created as the effector construct. Co-expression of ZmbZIP29 with the reporter O2pro-LUC (created above) resulted in a significantly increased signal compared with the negative control (P = 0.016). In contrast, mutation of the ABRE in the O2 promoter abolished the transactivation ability by ZmbZIP29 (Figure 1B). This result demonstrated that the ABRE was essential for the ZmbZIP29-mediated O2 promoter transactivation.
Figure 1.
ABA-inducible ZmbZIP29 recognizes the ABRE in the O2 promoter for transactivation. A, Y1H assay showing that ZmbZIP29 binds the ABRE in the O2 promoter. The sequences containing the intact and modified ABRE (ABREm) were cloned into the pAbAi plasmid, yielding ABRE-pAbAi and ABREm-pAbAi, respectively. The transformed yeast clones were grown to the cell concentration of OD600 = 1.0. Serial dilutions (1:1, 1:10, 1:100) of the transformed yeast cells were incubated in the plate lacking SD/−Leu with or without AbA. The AbA concentration was 600 ng/mL. B, Transactivation of the O2 promoter by ZmbZIP29. The O2 promoter fragments with intact and modified ABRE (ABREm) were used to drive the firefly LUC gene, creating the reporters O2pro-LUC and O2proABREm-LUC, respectively. The relative ratio of LUC/REN was determined in Arabidopsis mesophyll protoplasts via co-transforming the reporter plasmids with the effector construct. 62SK, empty vector. Error bars represent SD of three biological replicates. Statistical significance was determined with Student’s t test. *P < 0.05. C, EMSA showing the specific binding of ZmbZIP29 to the ABRE in the O2 promoter. The normal and mutant probes (Mp) were labeled with biotin. Unlabeled intact probes were used for competition. Comp, competing probes not labeled with biotin. M-Comp, mutated competing probes not labeled with biotin. D, ChIP-qPCR assay showing ZmbZIP29 binds the ABRE in the O2 promoter. The examined fragments are indicated under the schematic diagrams of the O2 promoter. Arrow indicates the transcription start site regions. Black boxes indicate the exons. Horizontal black line indicates introns. Vertical black line indicates the ABRE. The bar represents 100 bp in length. E, RT-qPCR analysis of ZmbZIP29 expression induced by ABA treatment. All expression levels were normalized to that of ZmActin. Error bars represent SD of three biological replicates. Statistical significance was determined with Student’s t test. **P < 0.01.
To confirm that ZmbZIP29 directly recognizes the ABRE in the O2 promoter to exert transactivation, we performed an electrophoretic mobility shift assay (EMSA). Oligonucleotides containing the ABRE labeled with biotin were used to examine the binding affinity. A retarded band was visualized due to the binding of ZmbZIP29-His fusion protein to the probes in the gel (Figure 1C). When the ABRE in the probes was mutated, the retarded band failed to be visualized. Furthermore, the binding specificity was verified by a competition experiment where the retarded bands were gradually diminished with the increasing unlabeled intact probes added in the reaction. By contrast, the retarded signal was not apparently affected with the increasing amount of unlabeled mutant probes. The chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) assay also revealed the binding of ZmbZIP29 to the ABRE in the O2 promoter (Figure 1D). We then tested whether ZmbZIP29 expression could be induced by ABA. The 10-DAP B104 endosperms were cultured in the half-strength medium containing different ABA concentrations for 24 h. RT-qPCR revealed that ZmbZIP29 expression increased with increased ABA addition and was greatest following addition of 150-μM ABA (Figure 1E). The expression decreased when the ABA concentration was further increased. Collectively, these results demonstrate that ZmbZIP29 is an ABA-inducible TF and directly regulates O2 expression in the endosperm.
Phylogenetic analysis and expression pattern of ZmbZIP29
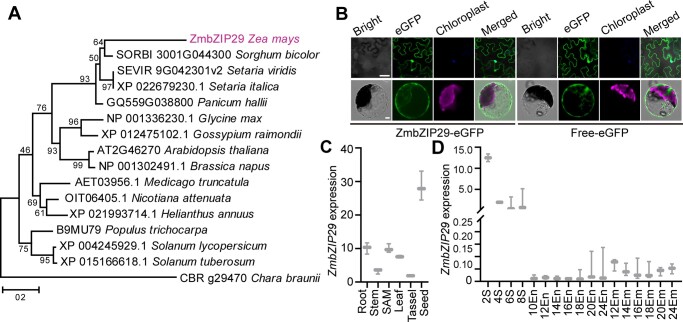
A phylogenetic tree was constructed using the full-length protein sequences of ZmbZIP29 and its orthologues in Panicum hallii, Setaria viridis, Setaria italica, Sorghum bicolor, Medicago truncatula, Glycine max, Gossypium raimondii, Nicotiana attenuate, Solanum lycopersicum, Solanum tuberosum, Helianthus annuus, Arabidopsis thaliana, Brassica napus, Populus trichocarpa, and Chara braunii (as the outgroup, Figure 2A; Supplemental Files S1 and S2). The phylogenetic tree revealed that ZmbZIP29 was conserved in monocots. Sequence alignment analysis showed that the closest ortholog of ZmbZIP29 in Arabidopsis only shared 8.90% sequence identity, indicating a rapid evolutionary divergence since the speciation (Supplemental Figure S8).
Figure 2.
Phylogenetic analysis of ZmbZIP29 and expression pattern of ZmbZIP29. A, Phylogenetic tree of ZmbZIP29 (in purple) and its orthologs. The phylogenetic distance was evaluated using the neighbor-joining algorithm. The numbers at the nodes represent the percentage of 1,000 bootstraps. The C. braunii ortholog protein was used as an outgroup. Scale bar, the average number of amino acid substitutions per site. B, Subcellular localization of ZmbZIP29-eGFP in N. benthamiana leaves (top panel) and Arabidopsis leaf mesophyll protoplasts (bottom panel). Bars = 25 μm. C, RT-qPCR analysis of ZmbZIP29 expression in different tissues from the B104 inbred lines. The 4-DAP seeds were used for mRNA extraction. All expression levels were normalized to that of ZmActin. Error bars represent SD of three biological replicates. D, RT-qPCR analysis of ZmbZIP29 expression during seed development. The numbers under the x-axis indicate the DAP during seed development. S, seed; En, endosperm; Em, embryo. All expression levels were normalized to that of ZmActin. Error bars represent SD of three biological replicates.
To determine the subcellular localization of ZmbZIP29, the full-length CDS of ZmbZIP29 was fused with the sequence encoding the N terminus of the enhanced green fluorescence protein (eGFP) and free eGFP was used as a negative control. Transient expression of the fusion construct in Nicotiana benthamiana leaves and Arabidopsis leaf mesophyll protoplasts showed that the GFP signal was detected both in the cytoplasm and nucleus, similar to the free eGFP control (Figure 2B). The ubiquitous localization of ZmbZIP29 in the cell was similar to that of ZmABI19 (Yang et al., 2021).
To explore the expression pattern of ZmbZIP29, different tissues were analyzed. RT-qPCR analysis showed that ZmbZIP29 was ubiquitously expressed in the root, stem, shoot apical meristem (SAM), leaf, tassel, and seed, of which the seed had the highest expression (Figure 2C). During seed development, ZmbZIP29 was much more highly expressed at the early stage (2–8 DAP) than the grain-filling stage (10–24 DAP) (Figure 2D). The expression of ZmbZIP29 was detected at the highest level at 2 DAP and then decreased, suggesting that ZmbZIP29 plays a critical role during early seed development. After 10 DAP, the endosperm and embryo were separated for mRNA extraction. Transcript levels of ZmbZIP29 were generally lower in the endosperm than in the embryo (Figure 2D), similar to the expression pattern of ZmABI19 (Yang et al., 2021).
When amplifying the full-length cDNA of ZmbZIP29 (423 bp), we characterized a longer transcript isoform of ZmbZIP29 (993 bp, designated ZmbZIP29*) that resulted from the alterative splicing (Supplemental Figure S9, A and B). RT-qPCR analysis was used to quantify the ZmbZIP29* transcript levels in the developing seeds using a primer pair designed within the retained region (Supplemental Figure S9A). This isoform accumulated at low levels (Supplemental Figure S9C), implying that the shorter ZmbZIP29 transcripts are the primary isoforms in the developing seeds. Many studies showed that bZIP TFs often formed homodimers or heterodimers to regulate plant development and growth (Alonso et al., 2009; Zhang et al., 2015; Yang et al., 2016; Feng et al., 2021). Yeast two-hybrid (Y2H) assay revealed that ZmbZIP29 had strong protein–protein interaction with itself and the ZmbZIP29* isoform in vitro (Supplemental Figure S10A). Split LUC (LIC) assay further confirmed that ZmbZIP29 and ZmbZIP29* could interact with themselves and each other (Supplemental Figure S10B). Y1H and EMSA showed that ZmbZIP29* was unable to bind the ABRE in the O2 promoter (Supplemental Figures S7 and S11). These results suggest that the ZmbZIP29* isoform exhibits an altered expression pattern and protein–DNA binding function compared with ZmbZIP29.
Genetic confirmation of ZmbZIP29 function
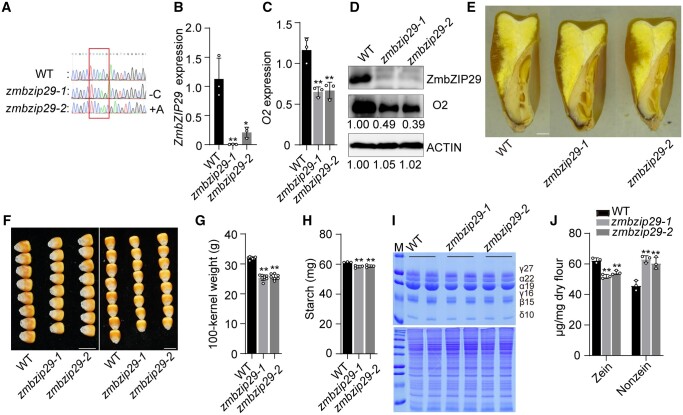
To genetically explore the biological function of ZmbZIP29, null mutants were required to reveal the roles in maize seed development and grain filling. The genome editing approach with clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) was used to edit the first exon of ZmbZIP29 in the B104 inbred line. We identified two independent knockout lines that contained a single-nucleotide “C” deletion and “A” insertion (designated zmbzip29-1 and zmbzip29-2), respectively, both resulting in a frameshift at the target gene (Figure 3A). RT-qPCR analysis revealed that the transcript levels of ZmbZIP29 were significantly reduced in both knockout seeds at 18 DAP (Figure 3B). Immunoblot analysis using anti-ZmbZIP29 antibodies found that ZmbZIP29 was absent in the two events, confirming that zmbzip29-1 and zmbzip29-2 were null alleles (Figure 3D). We examined O2 expression in the zmbzip29 mutants. We found that the O2 transcript levels in zmbzip29-1 and zmbzip29-2 were downregulated to 55.8% and 57.3% of the wild-type level (Figure 3C) and the protein levels were accordingly reduced to 49% and 39% of the wild-type level (Figure 3D), confirming that ZmbZIP29 directly regulates O2 expression genetically.
Figure 3.
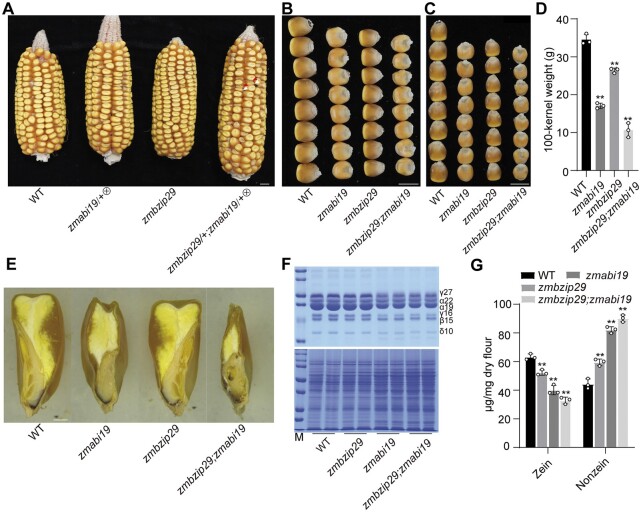
Phenotypes and biochemical analysis of zmbzip29 seeds. A, The CRISPR-Cas9 edited sequence of ZmbZIP29. Two independent knockout lines with a single-nucleotide “C” deletion (zmbzip29-1) and “A” insertion (zmbzip29-2) were recovered in the B104 background, respectively. B and C, RT-qPCR analysis of ZmbZIP29 (B) and O2 (C) expression in 18-DAP wild-type and zmbzip29 seeds. All expression levels were normalized to that of ZmActin. Error bars represent SD of three biological replicates. Statistical significance was determined with Student’s t test. *P < 0.05; **P < 0.01. D, Immunoblot analysis of the protein levels of ZmbZIP29 and O2 in 18-DAP zmbzip29 seeds. Antibodies against ZmbZIP29 and O2 were used at a dilution of 1:1,000 and ACTIN antibodies at a dilution of 1:4,000. The relative protein levels were determined using ImageJ software. E, Longitudinal sections of wild-type and zmbzip29 seeds. Bar = 1 mm. F, Seed phenotypes of the wild type and zmbzip29 mutants. Left panel, kernel width; right panel, kernel length. Bars = 1 cm. G, Measurement of 100-kernel weight. Error bars represent SD of seven biological replicates. Statistical significance was determined with Student’s t test. **P < 0.01. H, Measurement of starch content. Error bars represent SD of three biological replicates. Statistical significance was determined with Student’s t test. **P < 0.01. I, SDS–PAGE analysis of zein (top panel) and nonzein (bottom panel) proteins. The size of each zein protein band is indicated beside it. M, markers; γ27, 27-kD γ-zein; α22, 22-kD α-zein; α19, 19-kD α-zein; γ16, 16-kD γ-zein; β15, 15-kD β-zein; δ10, 10-kD δ-zein. J, Quantification of zein and nonznein proteins. In total, five kernels were ground into fine flour for each replicate and three replicates from three independent ears were performed. Statistical significance was determined with Student’s t test. **P < 0.01.
In contrast to zmabi19 (Yang et al., 2021), the zmbzip29 seeds appeared overall normal but were overtly delayed in endosperm and embryo development (Figure 3E and Supplemental Figure S12, A–C). Paraffin sections of the wild-type and mutant seeds were examined at 6, 8, and 16 DAP, representing the early seed developmental stage, grain-filling initiation, and rapid endosperm-filling stage, respectively. At each time point, zmbzip29 seeds developed overtly slower than the wild type (Supplemental Figure S12, A–C). In the end, the mature zmbzip29 seeds were apparently downsized compared with the wild type, particularly in kernel width and length (Figure 3, E and F). The 100-kernel weight of zmbzip29-1 (25.4 g) and zmbzip29-1 (25.7 g) harvested in Sanya was reduced by 19.6% and 20.5% compared with the wild type (31.9 g, the null segregant was used as the wild-type control) (Figure 3G). The starch content in the mutant seeds was slightly but significantly decreased (P = 0.006–0.007, Figure 3H). O2 is a major TF for regulating storage-protein zein genes. Consistent with the downregulation of O2 expression (Figure 3, C and D), transcript levels of all zein genes except 19-kD z1A were significantly reduced in zmbzip29 endosperms (Supplemental Figure S13). In mature seeds, the zein content was significantly decreased in zmbzip29 mutants compared with the wild type (Figure 3, I and J). Due to proteome rebalancing, the nonzein proteins in zmbzip29 were increased (Figure 3, I and J).
ABA synergistically strengthens transactivation of the O2 promoter by ZmbZIP29 and ZmABI19 interaction
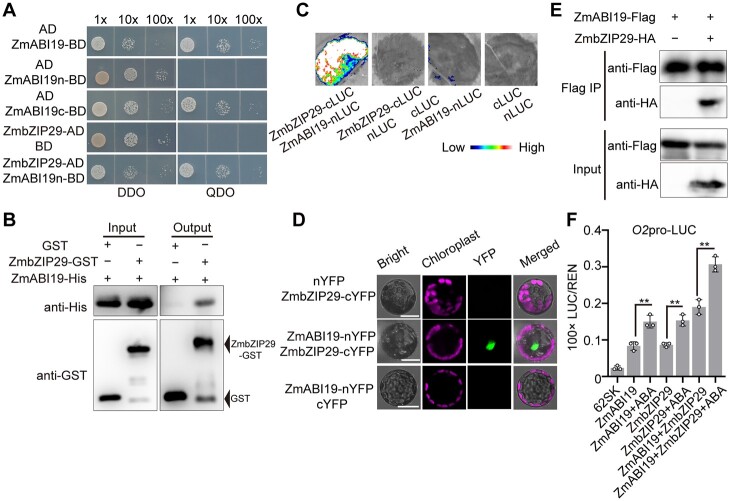
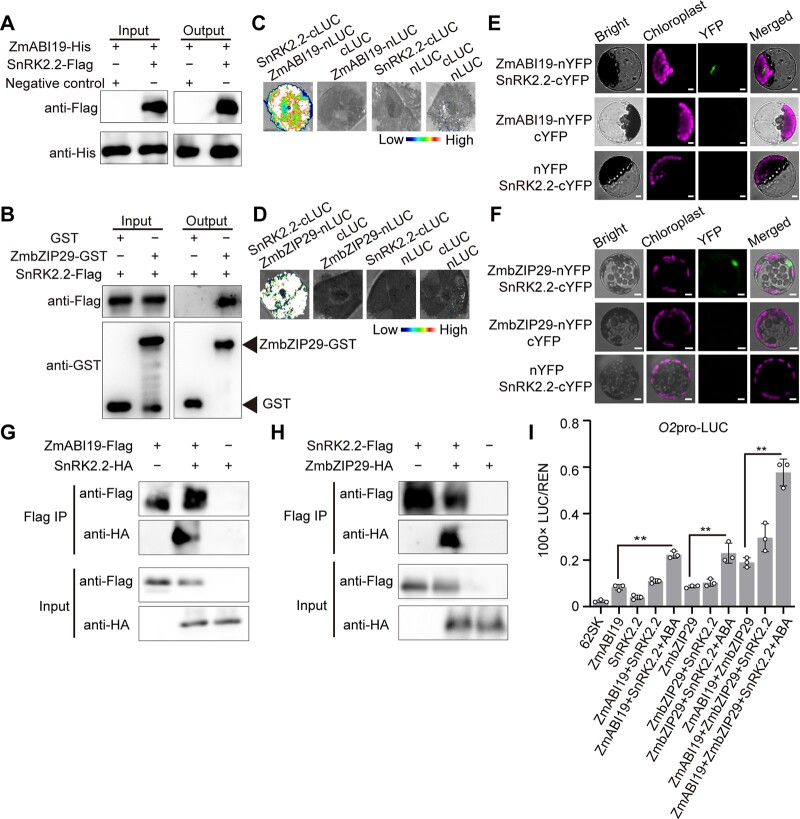
ZmABI19 recognized two RY motifs in the O2 promoter to transactivate O2 expression (Yang et al., 2021). The ABRE is located between the two RY motifs, separated by 25 and 19 bp, respectively, suggesting that ZmbZIP29 and ZmABI19 have protein–protein interaction (Supplemental Figure S5). Considering that the full-length and C-terminal regions (encoding 73–292 amino acids) of ZmABI19 possess transcriptional activity, the N-terminal region (encoding 1–72 amino acids) of ZmABI19 and the full-length CDS of ZmbZIP29 were cloned into a binding domain (BD) vector and an activation domain (AD) vector, respectively (Figure 4A). Y2H assay showed that ZmbZIP29 interacted with the N-terminal region of ZmABI19 (Figure 4A). The full-length CDS of ZmbZIP29 was also fused with the BD vector. Y2H assay showed that the yeast strain with ZmbZIP29-BD failed to grow on the QDO (SD/−Leu/−Trp/−His/−Ade) plate (Supplemental Figure S14A). The full-length, N-terminal, and C-terminal sequences of ZmABI19 were further cloned into the AD vector and transformed into the yeast strain with ZmbZIP29-BD, respectively. The resulting transformation showed that ZmbZIP29 interacted with the full-length and N-terminal regions of ZmABI19 but not the C terminus (Supplemental Figure S14A). A LIC assay was used to confirm the results above (Supplemental Figure S14B). In a pull-down assay, the ZmABI19-His fusion proteins were pulled down by ZmbZIP29-GST (glutathione-S-transferase) but not the GST control, indicating that ZmbZIP29 and ZmABI19 had protein–protein interaction in vitro (Figure 4B). We then used the LIC assay to test whether ZmbZIP29 interacted with ZmABI19 in vivo. We fused ZmbZIP29 to the C terminus of LUC and ZmABI19 to the N terminus of LUC to generate ZmbZIP29-cLUC and ZmABI19-nLUC, respectively. The cLUC and nLUC empty constructs were used as controls. Co-expression of ZmbZIP29-cLUC and ZmABI19-nLUC but not other combinations in N. benthamiana leaves gave rise to strong LUC activities (Figure 4C). We then performed the biomolecular florescence complementation (BiFC) assay to test the interaction, where ZmbZIP29 and ZmABI19 were fused with the C-terminus and N-terminus of split yellow fluorescent protein (YFP), yielding the ZmbZIP29-cYFP and ZmABI19-nYFP constructs, respectively. We co-expressed the two constructs in Arabidopsis leaf mesophyll protoplasts and observed strong fluorescence signals in the nucleus. In control experiments, no visible signals were produced (Figure 4D). At last, we conducted a co-immunoprecipitation (Co-IP) assay to examine the interaction. We created the ZmbZIP29-HA and ZmABI19-Flag fusion proteins and co-injected these proteins into N. benthamiana leaves. The assay showed that an obvious band with the expected mobility of ZmbZIP29-HA was detected in the anti-Flag immunoprecipitates from the leaves co-expressing ZmbZIP29-HA and ZmABI19-Flag fusion proteins (Figure 4E). We also tested the interaction between the ZmbZIP29* isoform and ZmABI19 using Y2H, LIC, and BiFC assays. All three assays demonstrated that ZmbZIP29* could interact with ZmABI19 in vitro and in vivo (Supplemental Figure S15, A–C), suggesting that ZmbZIP29* maintains the ability in protein–protein interaction but has defects in direct binding to the O2 promoter. Taken together, these results confirmed that ZmbZIP29 interacts with ZmABI19 in vitro and in vivo.
Figure 4.
Synergistic transactivation of the O2 promoter by ZmABI19, ZmbZIP29, and ABA. A, Y2H assay showing the interaction between ZmABI19 and ZmbZIP29. The full-length region (1–293 amino acid), N-terminus (1–72 amino acid), and C-terminus (73–293 amino acid) of ZmABI19 were cloned into the pGBKT7 plasmid, yielding ZmABI19-BD, ZmABI19n-BD, and ZmABI19c-BD. The CDS of ZmbZIP29 was cloned into pGADT7, yielding ZmbZIP29-AD. Serial dilutions (1:1, 1:10, 1:100) of transformed yeast cells were grown on the SD/−Leu/−Trp (DDO) and SD/−Leu/−Trp/−His/−Ade (QDO) medium. B, In vitro pull-down assay showing the interaction between ZmABI19 and ZmbZIP29. C, LIC assay showing the interaction between ZmABI19 and ZmbZIP29. The LUC intensity was measured through co-expression of different combinations. Low, weak LUC intensity; High, strong LUC intensity. D, BiFC assay showing the interaction between ZmABI19 and ZmbZIP29. Bars = 50 μm. E, Co-IP showing the interaction between ZmABI19 and ZmbZIP29. Total proteins were extracted from N. benthamiana leaves, where the ZmbZIP29-HA and ZmABI19-Flag fusion proteins were co-expressed. The ZmABI19-Flag proteins were immunoprecipitated with Flag agarose beads and detected with anti-HA and anti-Flag antibodies, respectively. F, Transactivation of the O2 promoter by ZmABI19, ZmbZIP29, and ABA. 10-μM ABA was used in this assay. The relative ratio of LUC/REN was determined in Arabidopsis protoplasts via co-transforming the reporter plasmids with the effector construct. Error bars represent SD of three biological replicates. Statistical significance was determined with Student’s t test. **P < 0.01.
We used the DLR assay to investigate the capacity of ZmbZIP29 and ZmABI19 to activate the transcription of the O2 promoter with or without ABA. The assay showed that ZmbZIP29 and ZmABI19 had a similar effect on transactivation of the O2 promoter (Figure 4F). Co-expression of 35S-ZmbZIP29 and 35S-ZmABI19 with O2pro-LUC resulted in a greater enhancement of the LUC activity compared with single TFs. ZmbZIP29 and ZmABI19 thus exhibit an additive transactivation pattern. When ABA was applied to the reaction, the LUC activity was substantially further elevated, manifesting a synergistic transactivation pattern (Figure 4F).
Previous work demonstrates that in addition to O2, ZmABI19 directly regulates multiple key grain-filling TF genes including Pbf1, ZmbZIP22, and O11 in the endosperm and Vp1 in the embryo (Yang et al., 2021). We tested whether ZmbZIP29 could transactivate their expression. A DLR assay using Arabidopsis leaf mesophyll protoplasts showed enhanced signals when expressing 35S-ZmbZIP29 with Vp1pro-LUC, Pbf1pro-LUC, ZmbZIP22pro-LUC, and O11pro-LUC, respectively (Supplemental Figure S16, A–D). Co-expression of 35S-ZmbZIP29 and 35S-ZmABI19 with these reporters resulted in further enhancement of the LUC activity compared with single TFs, confirming that ZmbZIP29 and ZmABI19 exhibit an additive transactivation pattern (Supplemental Figure S16, A–D).
Phenotypes of the zmbzip29 zmabi19 double mutant
To genetically substantiate the physical interaction between ZmbZIP29 and ZmABI19, the double mutant was created. Since zmabi19 is lethal in seedlings, the double mutant seeds were produced by crossing zmabi19/+ and zmbzip29 plants and then self-pollinating the resulting F1 plants. The ears that segregated zmabi19 seeds were selected to screen for the double mutants. In these ears, we observed that about one sixteenth of seeds (indicated by red arrowheads in Figure 5A) showed more severe phenotypes than the typical zmabi19 seeds (indicated by white arrowheads). We genotyped theses seeds and confirmed the digenic mutations. Although zmbzip29 had milder seed phenotypes compared with zmabi19, the double-mutant seeds were more downsized and flattened than the zmabi19 seeds (Figure 5, A–D). When measuring 100-kernel weight of seeds harvested in Sanya, zmbzip29 (26.4 g), zmabi19 (17.1 g), and zmbzip29;zmabi19 (10.6 g) were 76.4%, 49.4%, and 30.8% of the wild type (34.5 g), respectively (Figure 5E). SDS-PAGE analysis showed that the stacking of zmbzip29 and zmabi19 resulted in a significantly greater reduction in the accumulation of zein proteins compared with the single mutants (Figure 5, F and G). Conversely, nonzein proteins were accordingly increasing in abundance due to proteome rebalancing (Figure 5, F and G).
Figure 5.
Phenotypes and biochemical analysis of the zmbzip29;zmabi19 double mutants. A, Ear phenotypes of the wild type, self-pollinated zmabi19/+, zmbzip29, and self-pollinated zmbzip29/+;zmabi19/+. The white and red arrowheads indicate the monogenic zmabi19 and digenic zmbzip29;zmabi19 mutant seeds, respectively. Bar = 1 cm. B, Kernel width of the wild-type, single and double mutant seeds. Bar = 1 cm. C, Kernel length of the wild-type, single and double mutant seeds. Bar = 1 cm. D, Measurement of 100-kernel weight. Error bars represent SD of three biological replicates. Statistical significance was determined with Student’s t test. **P < 0.01. E, Longitudinal sections of the wild-type, single and double mutant seeds. Bar = 1 mm. F, SDS-PAGE analysis of zein (top panel) and nonzein (bottom panel) proteins in the wild-type, single and double mutant seeds. The size of each zein protein band is indicated beside it. M, markers; γ27, 27-kD γ-zein; α22, 22-kD α-zein; α19, 19-kD α-zein; γ16, 16-kD γ-zein; β15, 15-kD β-zein; δ10, 10-kD δ-zein. G, Quantification of zein and nonzein proteins in the wild-type, single and double mutant seeds. In total, five kernels were ground into fine flour for each replicate. Error bars represent SD of three biological replicates. Statistical significance was determined with Student’s t test. **P < 0.01.
These results demonstrate that zmbzip29 and zmabi19 mutations cumulatively influence seed development and storage protein synthesis.
Overexpression of ZmABI19 and ZmbZIP29 enhances grain filling and yield
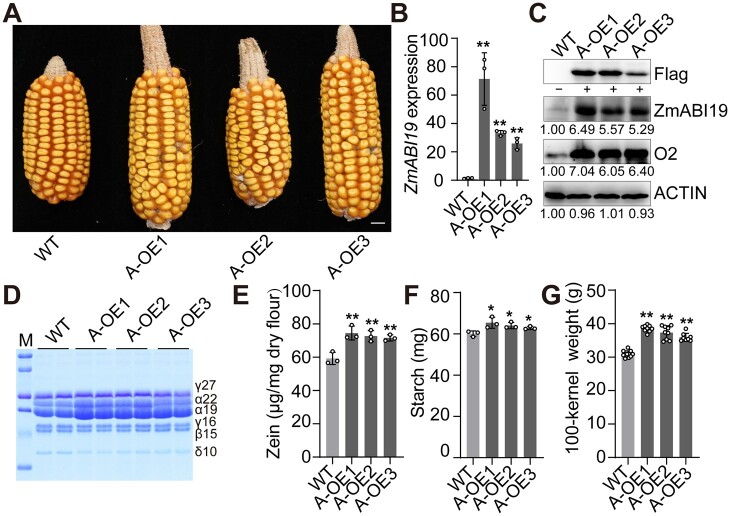
The expression levels of ZmABI19 and ZmbZIP29 were high during early seed development and low at the endosperm-filling stage. We investigated whether enhancing the expression of ZmbZIP29 and ZmABI19 would increase O2 expression and thus promote endosperm filling. To this end, we used a strong and endosperm-filling-specific promoter, the 27-kD γ-zein promoter, to overexpress ZmABI19 and ZmbZIP29 in the endosperm at the filling stage.
Three independent ZmABI19 overexpression (A-OE in abbreviation) lines (the B104 background) were recovered (Figure 6A and Supplemental Figure S17A). RT-qPCR revealed that ZmABI19 expression in 11-DAP A-OE endosperms was increased by 23.76- to 50.98-fold relative to the wild type (Figure 6B). Immunoblot analysis using the anti-Flag antibody showed that a specific band was observed in A-OE endosperms at 11 DAP, but not in the wild type (Figure 6C). Dramatically increased signals for ZmABI19 were also detected using the anti-ZmABI19 antibodies. In the end, the O2 protein in the three A-OE lines was increased to 6.05- to 7.04-fold relative to the wild type (Figure 6C). At 16 DAP, endosperm cells of all A-OE lines appeared to be more densely filled with storage reserves than the wild type (Supplemental Figure S18). At maturity, the starch and zein contents of A-OE seeds were both significantly higher than those of the wild type (Figure 6, D–F). To further examine the kernel phenotypes, we planted the A-OE lines and wild type in Sanya, 2020. Mature A-OE seeds had overtly increased seed size, particularly in seed length, compared with the wild type (Supplemental Figure S19A). Consistent with this, 100-kernel weight of A-OE1, A-OE2, and A-OE3 seeds weighed 38.5, 37.3, and 35.9 g, respectively, all being significantly heavier than the wild-type (null segregant) value (30.9 g) (Figure 6G). We also tested the kernel weight change in Shanghai, 2021, and found that all A-OE lines still had significantly higher 100-kernel weight than the wild type (Supplemental Figure S17B).
Figure 6.
Phenotypes and biochemical analysis of ZmABI19 overexpression seeds. A, Ear phenotypes of ZmABI19 overexpression lines (A-OE1 to A-OE3) harvested in Sanya, 2020. Bar = 1 cm. B, RT-qPCR analysis of ZmABI19 expression in the wild-type and A-OE endosperms at 11 DAP. All expression levels were normalized to that of ZmActin. Error bars represent SD of three biological replicates. Statistical significance was determined with Student’s t test. **P < 0.01. C, Immunoblot analysis of the O2 protein levels in the wild-type and A-OE endosperms at 11 DAP. The total proteins were extracted from 11-DAP endosperms and detected with anti-Flag, anti-ZmABI19, anti-O2, and anti-ACTIN antibodies, respectively. The relative protein levels were determined using ImageJ software. D, SDS-PAGE analysis of zein proteins in the mature wild-type and A-OE seeds. The size of each zein protein band is indicated beside it. M, markers; γ27, 27-kD γ-zein; α22, 22-kD α-zein; α19, 19-kD α-zein; γ16, 16-kD γ-zein; β15, 15-kD β-zein; δ10, 10-kD δ-zein. E, Measurement of zein proteins. Error bars represent SD of three biological replicates. Statistical significance was determined with Student’s t test. **P < 0.01. F, Measurement of starch content. Error bars represent SD of three biological replicates. Statistical significance was determined with Student’s t test. *P < 0.05. G, Measurement of 100-kernel weight. Error bars represent SD of 8–10 biological replicates. Statistical significance was determined with Student’s t test. **P < 0.01.
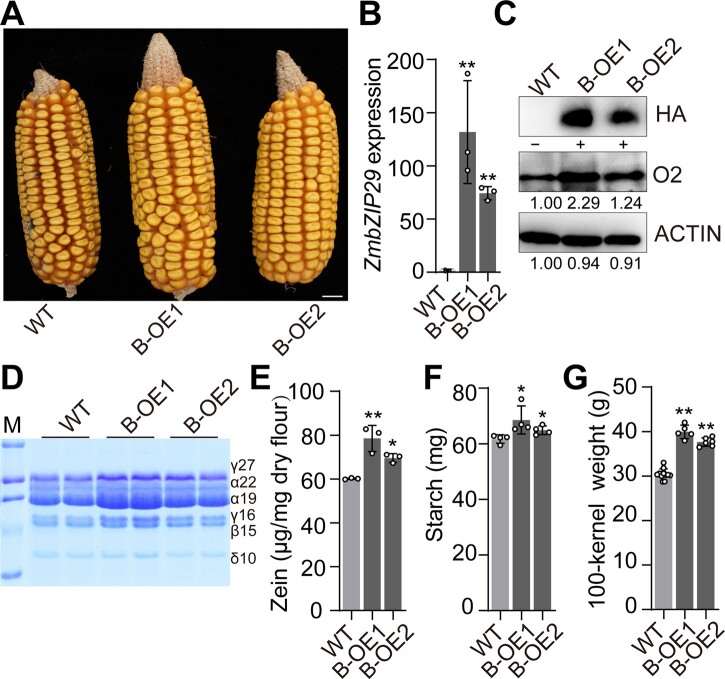
Two independent transgenic events for ZmbZIP29 overexpression (B-OE) were obtained and used for further analysis (Figure 7A and Supplemental Figure 20A). RT-qPCR revealed a dramatic increase of transcript levels of ZmbZIP29 in B-OE endosperms at 11 DAP (Figure 7B). The HA protein indicating the expressed ZmbZIP29-HA recombinant protein was accumulated at a tremendously high level in B-OE endosperms, but was not detected in the wild type (Figure 7C). Similar to A-OE lines, the O2 protein level was remarkably elevated in B-OE endosperms. The developing B-OE endosperms at 16 DAP also appeared to be filled with more storage compounds than the wild type (Supplemental Figure S21). At maturity, B-OE seeds contained significantly more starch and zein proteins than the wild type (Figure 7, D–F). Due to increased seed size in width (Supplemental Figure S19B), 100-kernel weight of B-OE1 and B-OE2 was increased to 37.7 g and 39.9 g, respectively, both being significantly higher than the wild-type (null segregant) value (30.4 g) harvested in Sanya, 2020 (Figure 7G). The batch grown in Shanghai, 2021, also showed a significant increase for the two B-OE lines (Supplemental Figure S20B).
Figure 7.
Phenotypes and biochemical analysis of ZmbZIP29 overexpression seeds. A, Ear phenotypes of ZmbZIP29 overexpression lines (B-OE1 and B-OE2) harvested in Sanya, 2020. Bar = 1 cm. B, RT-qPCR analysis of ZmbZIP29 expression in the wild-type and B-OE endosperms at 11 DAP. All expression levels were normalized to that of ZmActin. Error bars represent SD of three biological replicates. Statistical significance was determined with Student’s t test. **P < 0.01. C, Immunoblotting analysis of the O2 protein levels in the wild-type and B-OE endosperms. The total proteins were extracted from 11-DAP endosperms and detected with anti-HA, anti-O2, and anti-ACTIN antibodies, respectively. The relative protein levels were determined using ImageJ software. D, SDS-PAGE analysis of zein proteins in the mature wild-type and B-OE seeds. The size of each zein protein band is indicated beside it. M, markers; γ27, 27-kD γ-zein; α22, 22-kD α-zein; α19, 19-kD α-zein; γ16, 16-kD γ-zein; β15, 15-kD β-zein; δ10, 10-kD δ-zein. E, Measurement of zein proteins. Error bars represent SD of three biological replicates. Statistical significance was determined with Student’s t test. **P < 0.01, *P < 0.05. F, Measurement of starch content. Error bars represent SD of four biological replicates. Statistical significance was determined with Student’s t test. *P < 0.05. G, Measurement of 100-kernel weight. Error bars represent SD of 5–12 biological replicates. Statistical significance was determined with Student’s t test. **P < 0.01.
These results indicate that overexpression of ZmABI19 and ZmbZIP29 has the potential to improve the yield and nutritional composition of maize.
Identification of candidate ABA-induced kinases that interact with ZmbZIP29 and ZmABI19
Since ABA could enhance the capacity of ZmbZIP29 and ZmABI19 to transactivate the O2 promoter (Figure 4F), we hypothesized that the two TFs might undergo ABA-induced protein modification. In general, SnRK2s are strongly induced in ABA signaling to regulate the activity of ABA-responsive proteins (Raghavendra et al., 2010; Yang et al., 2019). A candidate approach was used to identify possible SnRK2 proteins that interact with ZmbZIP29 and ZmABI19. In maize, SnRK2s contain 14 members (SnRK2.1–SnRK2.14) (Long et al., 2021). We used the Y2H assay to explore which SnRK2s associated with ZmbZIP29 and ZmABI19. To this end, we successfully amplified eight SnRK2 members’ full-length CDSs and fused them to the BD of GAL4. The CDSs of ZmbZIP29 and ZmABI19 were ligated with the sequence encoding the AD of GAL4 in the Y2H system. The assay revealed that ZmbZIP29 interacted with SnRK2.2, SnRK2.4, SnRK2.5, SnRK2.10, and SnRK2.11, and ZmABI19 only interacted with SnRK2.2 (Supplemental Figure S22).
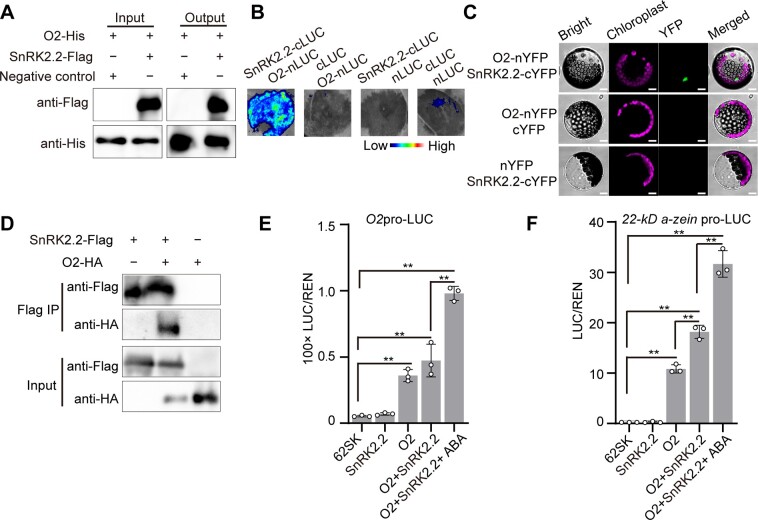
To obtain a snapshot of the complex functions of SnRK2s in ABA-induced activation of ZmbZIP29 and ZmABI19, we only focused on the common candidate SnRK2.2 in the current study (Figure 8, A–H). Consistent with the Y2H assay, a pull-down assay demonstrated that the recombinant SnRK2.2-Flag, but not the negative control, interacted with ZmABI19-His and ZmbZIP29-GST fusion proteins (Figure 8, A and B). Next, we confirmed the interactions by LIC assays in N. benthamiana leaves. The results showed that co-transfection of SnRK2.2-cLUC with either ZmbZIP29-nLUC or ZmABI19-nLUC produced strong complementation signals, while the control experiments, where a testing construct was replaced by an empty plasmid, failed to bring about visible signals (Figure 8, C and D). Likewise, BiFC assays reproduced the interactions in Arabidopsis leaf mesophyll protoplasts (Figure 8, E and F). Co-IP assays further confirmed the interactions between SnRK2.2 and ZmbZIP29 or ZmABI19 in N. benthamiana leaves (Figure 8, G and H). Together, these results confirmed that SnRK2.2 interacted with ZmbZIP29 and ZmABI19 in vitro and in vivo.
Figure 8.
SnRK2.2 interacts with ZmABI19 and ZmbZIP29 to synergistically transactivate O2 expression. A and B, Pull-down assay showing the interaction between SnRK2.2 and ZmABI19 (A) and ZmbZIP29 (B). C and D, LIC assay showing the interaction between SnRK2.2 and ZmABI19 (C) and ZmbZIP29 (D). The LUC intensity was measured through co-expression of different combinations. Low, weak LUC intensity; High, strong LUC intensity. E and F, BiFC assay showing the interaction between SnRK2.2 and ZmABI19 (E) and ZmbZIP29 (F) in Arabidopsis leaf mesophyll protoplasts. Bars = 25 μm. G and H, Co-IP assay showing the interaction between SnRK2.2 and ZmABI19 (G) and ZmbZIP29 (H). Total proteins were extracted from N. benthamiana leaves, where the fusion proteins were co-expressed. The Flag-fused proteins were immunoprecipitated with Flag agarose beads and detected with anti-HA and anti-Flag antibodies, respectively. I, SnRK2.2 plus ABA increases the transactivation capacity of ZmbZIP29 and ZmABI19 on the O2 promoter. 10-μM ABA was used in this assay. The relative ratio of LUC/REN was tested in Arabidopsis mesophyll protoplasts via co-transforming the reporter plasmids with the effector construct. Error bars represent SD of three biological replicates. Statistical significance was determined with Student’s t test. **P < 0.01.
The SnRK2.2 mRNAs were analyzed with 10-DAP B104 endosperms after cultured with different concentrations of ABA. RT-qPCR analysis showed that the expression of SnRK2.2 was induced by ABA, with the highest expression in 100 μM-ABA treated endosperms. With further increasing ABA concentrations, transcript levels of SnRK2.2 rapidly decreased, but remained significantly higher than the control (Supplemental Figure S23).
To determine the effect of SnRK2.2 on the transactivation capacity of ZmbZIP29 and ZmABI19, we performed the DLR assay using the Arabidopsis leaf mesophyll protoplasts. Co-expression of ZmbZIP29 or ZmABI19 with SnRK2.2 showed a limited enhancement in transactivation activity of the O2 promoter compared with expression of each TF alone, whereas co-expression of the two TFs with SnRK2.2 markedly increased the transactivation (Figure 8I). When ABA was added in the presence of SnRK2.2, transactivation activity of the O2 promoter was further elevated, regardless of whether the transactivation was exerted by ZmbZIP29, ZmABI19, or both. In all combinations tested, the reaction containing the two TFs, SnRK2.2 and ABA gave rise to the highest transactivation of the O2 promoter (Figure 8I).
Vp1 is transcriptionally regulated by ZmABI19 and the transcription is enhanced by ABA (Yang et al., 2021). Similarly, transactivation activity of the Vp1 promoter by ZmABI19 was dramatically enhanced by SnRK2.2 and ABA compared with those lacking either or both of them (Supplemental Figure S24).
O2 responds to ABA signaling mediated by SnRK2.2
The ABA-SnRK2.2-ZmABI19/ZmbZIP29 pathway could explain the induction of O2 expression by ABA (Supplemental Figure S1, B and C). Since O2 auto-transactivates its own promoter, we tested whether O2 interacted with SnRK2s as well. Y2H assays showed that, similar to ZmABI19, O2 only interacted with SnRK2.2 in all eight examined SnRK2s (Supplemental Figure S25). Pull-down, LIC, BiFC, and Co-IP assays confirmed the protein–protein interaction between O2 and SnRK2.2 (Figure 9, A–D).
Figure 9.
O2 interacts with SnRK2.2 to promote the expression of the downstream target genes. A, Pull-down assay showing the interaction between SnRK2.2 and O2. B, LIC assay showing the interaction between SnRK2.2 and O2. The LUC intensity was measured through co-expression of different combinations. Low, weak LUC intensity; High, strong LUC intensity. C, BiFC assay showing the interaction between SnRK2.2 and O2 in Arabidopsis leaf mesophyll protoplasts. Bars = 25 μm. D, Co-IP assay showing the interaction between SnRK2.2 and O2. Total proteins were extracted from N. benthamiana leaves, where the SnRK2.2-Flag and O2-HA fusion proteins were co-expressed. The SnRK2.2-Flag proteins were immunoprecipitated with Flag agarose beads and detected with anti-HA and anti-Flag antibodies, respectively. E and F, SnRK2.2 and ABA increase transactivation capacity of O2 on the O2 (E) and 22-kD a-zein (F) promoters. 10-μM ABA was used in this assay. The relative ratio of LUC/REN was determined in Arabidopsis leaf mesophyll protoplasts via co-transforming the reporter plasmids with the effector constructs. Error bars represent SD of three biological replicates. Statistical significance was determined with Student’s t test. **P < 0.01.
We then test whether the transactivation capacity of O2 was enhanced in the presence of SnRK2.2. DLR assays showed that O2 had a substantially higher transactivation effect on the downstream 22-kD α-zein promoter than its own promoter (Figure 9, E and F). Probably because the O2-mediated auto-transactivation of the O2 promoter was relatively low, the increase of the LUC activity by co-transfection of O2 and SnRK2.2 was apparent but not significant compared with that by O2 alone. When the reporter construct was expressed by the 22-kD α-zein promoter, the addition of SnRK2.2 resulted in significantly enhanced LUC activities (Figure 9F). In the presence of ABA and SnRK2.2, LUC activities produced from both O2pro-LUC and 22-kD α-zein pro-LUC constructs were substantially elevated compared with those without them (Figure 9, E and F).
These results demonstrated that O2 associated with SnRK2.2 to increase the expression of its own and downstream target genes, which was induced by ABA signaling.
SnRK2.2 phosphorylates ZmABI19, ZmbZIP29, and O2
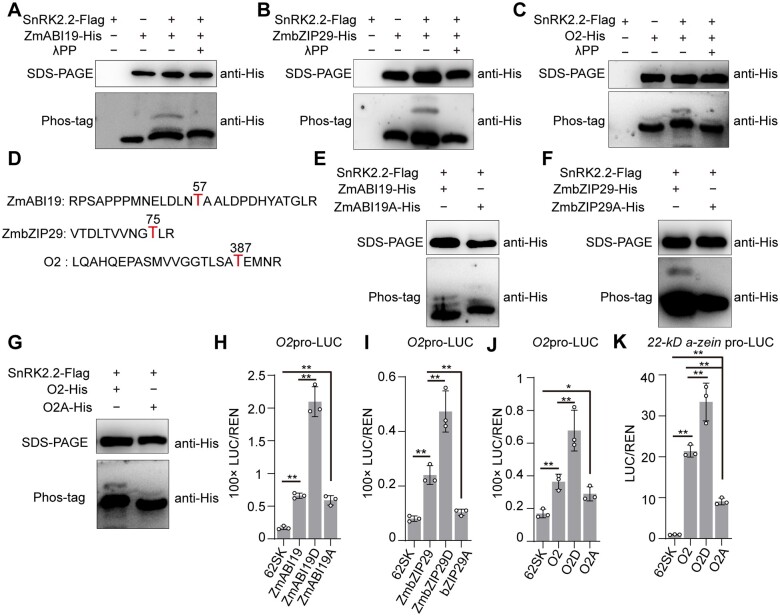
Generally, ABA-induced SnRK2s interact with the downstream ABA-responsive factors for phosphorylation (Finkelstein et al., 2002; Nakashima et al., 2009; Yang et al., 2019). Thus, we hypothesized that SnRK2.2 might target ZmABI19, ZmbZIP29, and O2 for phosphorylation. LC-MS/MS analysis revealed that the purified SnRK2.2-Flag protein isolated from Escherichia coli underwent auto-phosphorylation at the residue serine (S)12 (Supplemental Figure S26). Then, we performed the Phostag mobility shift assay to examine in vitro phosphorylation of purified ZmABI19-His, ZmbZIP29-His, and O2-His fusion proteins by SnRK2.2-Flag. Phostag is able to slow migration of the phosphorylated proteins, thereby resulting in phosphorylated and nonphosphorylated proteins being separated in the gel (Kinoshita et al., 2009). The assays showed that in the absence of SnRK2.2-Flag, the recombinant proteins ZmABI19-His, ZmbZIP29-His, and O2-His migrated as a single band in the gel (Figure 10, A–C). With the addition of SnRK2.2-Flag, a slower band was observed to be migrating behind each of these proteins, indicating the occurrence of protein phosphorylation. When the reactions were treated with lambda protein phosphatase, the retarded band disappeared, confirming that SnRK2.2 phosphorylated O2, ZmABI19, and ZmbZIP29 in vitro.
Figure 10.
Phosphorylation of ZmABI19, ZmbZIP29, and O2 by SnRK2.2 increases transactivation capacity. A, In vitro assay showing phosphorylation of ZmABI19 by SnRK2.2. The phosphorylation of ZmABI19 was assayed in the absence or presence of SnRK2.2-Flag. Lambda protein phosphatase λPP was added to dephosphorylate ZmABI19. B, In vitro assay showing phosphorylation of ZmbZIP29 by SnRK2.2. C, In vitro assay showing phosphorylation of O2 by SnRK2.2. D, The phosphorylated peptides and residues (red) in ZmABI19, ZmbZIP29, and O2. E–G, Substitution of the phosphorylatable threonine with alanine in ZmABI19 (E), ZmbZIP29 (F), and O2 (G) resulted in inhibited phosphorylation. H–K, Altered transactivation capacities of phosphorylation-mimetic (T substituted with D) and dephosphorylated (T substituted with A) forms of ZmABI19 (H), ZmbZIP29 (I), and O2 (J and K). 10-μM ABA was used in these assays. The relative ratio of LUC/REN was tested in Arabidopsis leaf mesophyll protoplasts via co-transforming the reporter plasmids with the effector construct. Error bars represent SD of three biological replicates. Statistical significance was determined with Student’s t test. *P < 0.05; **P < 0.01.
We performed LC-MS/MS to determine the precise phosphorylation residues of ZmABI19, ZmbZIP29, and O2 catalyzed by SnRK2.2. The results revealed that the residues threonine (T)57 in ZmABI19, T75 in ZmbZIP29, and T387 in O2 were phosphorylated by SnRK2.2 in vitro (Figure 10D and Supplemental Figure S27, A–C). To verify this, the three phosphorylation sites were substituted with alanine (A) to mimic a dephosphorylation status. The modified proteins were designated ZmABI19A, ZmbZIP29A, and O2A. For ZmbZIP29A and O2A, in vitro kinase activity assay revealed that the slower migrating bands indicative of protein phosphorylation were blocked by these substitutions (Figure 10, F and G). For ZmABI19A, the phosphorylated bands were reduced but not entirely eliminated compared with those in the intact control, suggesting that ZmABI19 might have other sites phosphorylated by SnRK2.2, which have not been identified by LC-MS/MS in this study (Figure 10E).
These results together confirmed that the identified residues in ZmABI19, ZmbZIP29, and O2 were indeed phosphorylated by SnRK2.2.
Transactivation activity is enhanced by phosphorylation of ZmABI19, ZmbZIP29, and O2
To investigate whether these phosphorylation residues of ZmABI19, ZmbZIP29, and O2 affect the transactivation activity of the downstream target genes, we created phosphorylation-mimetic [T57(aspartic acid)D, T75D, and T387D] and dephosphorylated [T57A, T75A, and T387A] forms of ZmABI19, ZmbZIP29, and O2, respectively.
These modified TF effectors containing the constitutive phosphorylation and dephosphorylation residues were driven by the CaMV 35S promoter. The O2 and 22-kD α-zein promoters were used to create the reporters by fusion with LUC. For the phosphorylation-mimetic substitutions, transactivation activities of the downstream target genes were all significantly elevated compared with the wild-type effectors (Figure 10, H–K). Particularly, co-transfection of ZmABI19D and O2pro-LUC resulted in LUC activity being 2.18-fold higher than those from co-transfection of the wild-type ZmABI19 with the reporter construct (Figure 10H). For the dephosphorylated substitutions, transactivation activity of the O2 promoter was not apparently decreased by ZmABI19A, but those by ZmbZIP29A was significantly reduced compared with their wild-type forms (Figure 10, H and I). Transactivation ability of the O2 promoter was not markedly affected by O2A, but that of the 22-kD α-zein promoter was dramatically reduced compared with the wild-type effector (Figure 10, J and K).
To explore whether the phosphorylated and dephosphorylated residues affect the protein–DNA binding affinity, phosphorylation-mimetic and dephosphorylated proteins of ZmABI19, ZmbZIP29, and O2 were purified. EMSA showed that the phosphorylated proteins had stronger binding affinity for the target promoters, whereas the dephosphorylated proteins lessened the binding affinity compared with the wild-type proteins (Supplemental Figure S28, A–C).
Taken together, these results demonstrated that the phosphorylation of ZmABI19, ZmbZIP29, and O2 by SnRK2.2 was essential for their full-level of transactivation capacity.
Discussion
ABA content increase correlates with endosperm-filling initiation
The initiation of endosperm filling is a critical phase of seed development during which the mitotic division of most endosperm cells ceases, the differentiation of endosperm cells is completed, transportation of photoassimilated products into the grain suddenly increases, and endosperm-filling regulatory TFs are activated, leading to rapid synthesis and accumulation of storage compounds prevailing in the forms of starch and zein proteins in the maize endosperm. Over the last decade, with the development of maize genomics and utilization of different reverse genetics approaches (RNA interference and genome editing), our knowledge of the transcriptional regulation of storage-reserve synthesis has been greatly advanced (Zhang et al., 2015, 2019; Qiao et al., 2016; Yang et al., 2016; Li et al., 2018), but our understanding of endosperm-filling initiation in terms of hormones, metabolic signals, and regulatory factors remains limited (Yang et al., 2021).
ABA mediates the sink strength of kernels and then promotes yield and quality (Frey et al., 2004; Qin et al., 2021). A recent study showed that most of the ABA in rice seeds is transported from leaves and that leaf-to-seed ABA transport is controlled by a multidrug and toxic compound extrusion (MATE) transporter (Qin et al., 2021). Exogenous application of ABA promotes expression of the grain-filling related genes. Therefore, it may be feasible to increase grain yield and quality by precisely manipulating ABA contents during the period of endosperm filling. A hypothetic model by Vreugdenhil suggests that ABA may enhance phloem unloading by increasing passive efflux or reloading restriction (Vreugdenhil, 1983). Another hypothesis is that ABA may stimulate the unloading process through reducing the proton motive force and then enhance the efflux of sucrose into the seed sink (Tanner, 1980). However, the accumulation of ABA in the embryo of viviparous mutants is depressed compared with the wild type in spite of the same supply from the mother plant (Robertson, 1955; McCarty et al., 1991). This observation suggests that the endosperm or embryo may directly control and synthesize ABA at least during the mid and late phases of seed development (Smith and De Koehler, 1980).
The ABA concentration was low during early endosperm development, rapidly increased with the initiation of endosperm filling, and was maintained at relatively high levels during the early period of endosperm filling (Supplemental Figure S1A). Consistent with this, previous studies showed that the ABA contents positively correlate with grain-filling rate at the early grain-filling stage in crops, such as rice and barley (Hordeum vulgare) (Kato et al., 1993; Ji et al., 2011; Seiler et al., 2011). ABA also controls cell-cycle regulation in the barley endosperm and then promotes differentiation and grain filling. A shrunken endosperm-deficient mutant seg8 is caused by the deregulation of ABA levels and thus alteration of grain development (Sreenivasulu et al., 2010). Moreover, ABA deficiency also induces seed abortion, thereby resulting in reduced seed yield and delayed growth of the embryo in dicotyledons, such as N. benthamiana (Frey et al., 2004). RNA-seq analysis revealed that ABA signaling is required for protein reallocation from the endosperm to embryo and this process is mediated by VP1 (Zheng et al., 2019). Globulin1 and Globulin2 are the most abundant storage proteins in the maize embryo. VP1 regulates the transcription of Globulin1 and Globulin2 and their expressions are greatly enhanced in the presence of ABA. At the late grain-filling stage, the highest ABA concentration is detected in the embryo, although the increasing ABA contents are also observed in the seed and endosperm (Jones and Brenner, 1987), indicating that ABA signaling plays an important role in promoting nutrient accumulation in both the endosperm and embryo. However, the detailed molecular mechanism has not been fully understood. We found that the ABA-mediated regulation of endosperm filling through influencing the activities of two hub coordinators of seed development and grain filling, ZmABI19 and ZmbZIP29, and the endosperm-filling central regulator, O2. In this regard, ABA possibly acts as a signal trigger to mediate seed development and grain filling in maize. We used the w3 and vp5 mutants that have defects in ABA synthesis to investigate whether O2 expression was affected by the ABA deficiency. Because w3 and vp5 mutations were lethal in seedlings, the homozygous mutant seeds were generated by self-pollinating heterozygous plants. Due to this, the ABA present in w3 and vp5 seeds at the onset of endosperm filling could be transported from leaves through a MATE-mediated ABA transport mechanism as functioning in rice (Qin et al., 2021). As a result, we observed that the O2 transcription was significantly reduced but not completely repressed in the two mutants (Supplemental Figure S3B).
At 10 DAP, a sharp increase in transcript and protein expression levels of endosperm-filling regulatory TFs and thus starch-synthetic and storage-protein zein genes characterizes the initial period of endosperm filling. Although the ABA content is correlated strongly with endosperm-filling activity, where ABA directly contributes to the induction of O2 gene expression, developmental and metabolic signals, and altered epigenetic modifications must also be involved in controlling endosperm-filling initiation.
ZmbZIP29 and ZmABI19 constitute hub coordinators of seed development and grain filling
Considering that O2 functions as the central regulator for endosperm filling, a better understanding of the O2 upstream regulatory network is critical for yield and quality improvement. Previous work reported that ZmABI19 is highly expressed at early seed development and plays critical roles in initiating grain filling by directly regulating several key factors in the seed including O2, Pbf1, ZmbZIP22, NAC130, and O11 in the endosperm and Vp1 in the embryo (Yang et al., 2021). ZmbZIP29 was found to directly regulate O2 expression and synergistically associate with ZmABI19 to enhance the expression of O2, Pbf1, ZmbZIP22, O11, and Vp1 (Figure 4 and Supplemental Figure S16). Similar to ZmABI19, ZmbZIP29 transcript was more abundant in early developing seeds than vegetative tissue, with the greatest amounts being observed in 2-DAP seeds (Figure 2D), indicating an additional role of ZmbZIP29 in early seed development. RNA-seq data from high temporal resolution of early seed development reveal that ZmbZIP29 occupies the most connections at the group of around double fertilization (Yi et al., 2019). It is predicted that ZmbZIP29 interacts with 13 proteins, including several key factors related to embryogenesis and endosperm development in maize, such as EMP10 and GRAS61 (Cai et al., 2017a, 2017b). Unlike most endosperm-filling regulators that function during a specific stage of endosperm maturation, the upstream regulators ZmABI19 and ZmbZIP29 are required for normal seed development during the early and late phases. In this regard, the two phases are not separated by a precise boundary of gene regulation, but are mechanistically connected by a higher complex regulatory network containing ZmABI19 and ZmbZIP29.
Although ZmABI19 and ZmbZIP29 displayed a similar capacity on transactivation of the O2 promoter for endosperm filling, the zmabi19 and zmbzip29 mutants exhibited specific phenotypes of early seed development, suggesting that the two TFs are not equally essential in the regulation of seed development. This is reminiscent of the observation for abi3 and abi5 in Arabidopsis. Among several ABI TFs and ABRE-BINDING FACTORs tested, ABI3 (B3 TF) and ABI5 (bZIP TF) interact in maturing seeds (Gutierrez et al., 2007). However, mutations of abi5 affect seed maturation, but the effects of such mutations are limited compared with those of abi3 (Lopez-Molina et al., 2002). The ChIP-seq assay reveals that ZmABI19 does not bind the ZmbZIP29 promoter, but ZmbZIP29 transcript is reduced by 2.57-fold in zmabi19 RNA-seq data, suggesting an indirect effect of zmabi19 mutations on ZmbZIP29 expression (Yang et al., 2021). To clarify the genetic relationship of the two TFs in early seed development regulation, cross-complementation experiments are required to test whether mutant phenotypes of one TF gene is rescued by overexpressing the other driven by a maize ubiquitin promoter.
Because ZmABI19 and ZmbZIP29 mRNAs accumulate primarily during early seed development, we overexpressed the two TFs by the endosperm-filling specific 27-kD γ-zein promoter to obtain additional clues about its role in endosperm filling (Figures 6 and 7). The results showed that overexpression of each TF resulted in greatly enhanced O2 expression, and thus increased storage-reserve accumulation and kernel weight, indicating that ZmABI19 and ZmbZIP29 are involved in not only initial transactivation, but also later maintenance of a high level of O2 transcription.
Phosphorylation of O2 by SnRK2.2 promotes endosperm filling
Protein phosphorylation as a posttranslational modification is involved in modulation of protein activity, stability, subcellular localization, and interaction patterns (Umezawa et al., 2013). O2, the central regulator of endosperm filling, is confirmed to be phosphorylated in vivo and in vitro (Ciceri et al., 1997). The phosphorylation pattern of O2 remained constant throughout the endosperm-filling stage, whereas the ratio of hyperphosphorylated to hypophosphorylated O2 polypeptides was dynamic along with diurnal rhythm (Ciceri et al., 1997). Although the phosphorylation of O2 has been known for more than 20 years (Ciceri et al., 1997), the detailed mechanism has not been understood. O2 is a classical bZIP TF that recognizes the G-box or O2 box to regulate the expression of zein and other downstream target genes (Zhang et al., 2016; Yang et al., 2021). An isoelectric focusing assay detects seven to nine isoforms, indicating that O2 has multiple phosphorylation residues involved in regulation of grain filling or response to abiotic stress (Ciceri et al., 1997). In this study, we determined that O2 was phosphorylated at the residue T387 by ABA-induced SnRK2.2 (Figure 10, C, D, and G). The phosphorylated O2 showed enhanced transactivation capacity on the downstream target genes (Figure 10, J and K). An in vitro phosphorylation assay reveals that three putative residues of O2 are phosphorylated by the human CKII in vitro (Ciceri et al., 1997). Thus, more protein kinases and corresponding phosphorylation residues need to be further identified, which may help understand the precise control of O2 activities and then improve yield and quality in maize. Previous work reported that O2 was ubiquitinated by the E3 ubiquitin ligase ZmRFWD3 to positively regulate its transcriptional activity through enhancing its nuclear localization (Li et al., 2020). Various posttranslational modifications in O2, such as ubiquitination and phosphorylation, ensure a precise and efficient regulation of endosperm filling in maize.
Posttranslational modification of ZmABI19 and ZmbZIP29 promotes endosperm filling
Endosperm initiation stage from 8 to 10 DAP is a critical time point to link early endosperm development and grain filling. At this stage, the silencing-related histone marks, such as H3K9me2 and H3K27me2, were substituted with activated H3K4me2 and H3K14ac and then the chromatin was more accessible to DNaseI (Locatelli et al., 2009). O2 is transactivated by ZmABI19 and the newly identified ZmbZIP29 (Yang et al., 2021). ZmABI19 is an ortholog of FUSCA3 (FUS3), the latter acting as a major regulator of seed development in Arabidopsis (Keith et al., 1994). FUS3 is phosphorylated by SnRK1a1, which is involved in energy homeostasis (Chan et al., 2017). The phosphorylated FUS3 is essential for embryogenesis and integration of abiotic stress. In this study, we found that both ZmABI19 and ZmbZIP29 were regulated by ABA signaling and phosphorylated by SnRK2.2 (Figure 10, A, B, and D–F). The phosphorylated ZmABI19 and ZmbZIP29 at T57 and T75, respectively, showed enhanced transactivation capacity on the O2 promoter (Figure 10, H and I). Comparison of the protein–DNA binding affinity for the phosphorylated proteins also showed enhanced effects for ZmABI19, ZmbZIP29, and O2 (Supplemental Figure S28, A–C). However, less-reduced binding affinity and transactivation activity for the dephosphorylated ZmABI19 suggest the existence of other undetected phosphorylated residues in ZmABI19 (Figure 10H and Supplemental Figure S28A).
Overexpression of ZmABI19 and ZmbZIP29 resulted in enhanced O2 expression and increased kernel weight and the contents of starch and zein proteins (Figures 6 and 7). Posttranslational modification of target genes modulates its transcriptional activity. Phosphorylation-null patterns of FUS3 provide weaker rescue of the deficient phenotype than the phosphorylation-mimic lines. Phosphorylation of FUS3 is also essential for plant growth and seed development at high temperatures (Chan et al., 2017). In this study, ABA promoted SnRK2.2-mediated phosphorylation of ZmABI19 and ZmbZIP29 (Figure 10, A and B). Transactivation activities of the downstream target genes by phosphorylated ZmABI19 and ZmbZIP29 were dramatically increased (Figure 10, H and I). Creation of transgenic plants with phosphorylation-mimic ZmABI19 and ZmbZIP29 should have a better potential for yield improvement than the plants overexpressing the wild-type TFs.
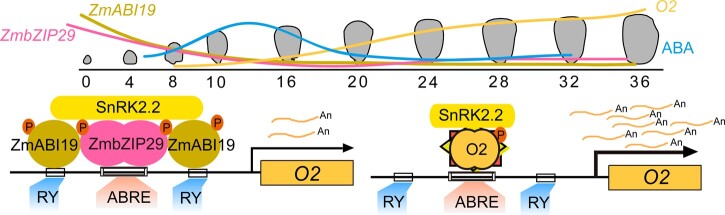
In summary, our findings allow us to formulate a model of transactivation of the central endosperm-filling regulator O2 that is regulated by the interaction between ZmbZIP29 and ZmABI19 (Figure 11). ZmbZIP29 and ZmABI19 function as two hub coordinators connecting early seed development and grain filling. As ABA contents are substantially increasing at 8 DAP, endosperm filling is initiated. ABA promotes ZmABI19 and ZmbZIP29 expression, whose proteins in turn transactivate the expression of O2 and other key grain-filling TF genes. When O2 expression is activated, the O2 protein can auto-transactivate its own promoter. Furthermore, phosphorylation of ZmABI19, ZmbZIP29, and O2 by ABA-induced action of SnRK2.2 confers enhanced transactivation capacity to these TFs. These effects collectively lead to increased O2 expression and thus the expression of downstream genes for endosperm filling in maize.
Figure 11.
A proposed model for O2 transactivation at the initiation of endosperm filling. The expression levels of ZmbZIP29 and ZmABI19 are high at the early seed development and low at the endosperm-filling stage. The encoded proteins act as two hub coordinators connecting early seed development and grain filling. During seed development, ABA contents are substantially increasing at 8 DAP, at which time endosperm filling is initiated. ZmABI19 and ZmbZIP29 co-transactivate O2 expression at the endosperm initiation stage, which is promoted by ABA. When O2 is activated at the endosperm-filling stage, O2 can auto-transactivate its own promoter, which is also mediated by ABA. The ABA-induced action of SnRK2.2 further phosphorylates ZmABI19, ZmbZIP29, and O2, which confers enhanced transactivation capacity to these TFs. Numbers from 0 to 36 indicate serial DAP during seed development. “An” indicates the O2 mRNA. “P” indicates phosphorylation.
Materials and methods
Plant materials and growth conditions
All material plants were grown in the Song Jiang experimental field in Shanghai, China, and the field in Sanya, China. The inbred line B104 was used for particle bombardment assays. Wild-type N. benthamiana and A. thaliana of the Columbia-0 (Col-0) accession were grown in a growth chamber with soil at 22°C and 70% humidity under 16-h light and 8-h dark photoperiod using Philips TLD 36W/865 and 36W/830 bulbs (90 μmol/m2/s).
The maize tissues and seeds were harvested from the B104 inbred line. The collected materials were immediately frozen in liquid nitrogen and stored at –80°C for RNA and protein extraction. Tissue samples were harvested from at least three individual plants for each developmental stage.
The zmbzip29 CRISPR-Cas9 transgenic lines were generated in the B104 background through Agrobacterium tumefaciens-mediated transformation according to previously published protocol (Scott, 2009). The guide RNA editing sequence was chosen to target the first exon of ZmbZIP29 (the target sequence being GTCGCCGCCCGACGTCGTGG). The obtained mutant plants were crossed with B104 and further backcrossed with B104 to generate the BC1 seeds. Then the BC1 seeds were used to screen the heterozygous seedlings without the Cas9 construct by PCR amplification. The resulting plants were self-pollinated and used to screen the wild-type and homozygous plants.
The zmabi19 mutant was created in our previous work and propagated by self-pollinating heterozygous plants (Yang et al., 2021). The double mutant zmbzip29 zmabi19 was generated by crossing zmbzip29 with zmabi19/+ plants, and further self-pollinating the F1 plants. Then, the ears that segregated zmabi19 seeds were selected to screen for the double mutants with a ratio of about 1–16.
For the overexpression transgenic materials, the full-length CDSs of ZmABI19 and ZmbZIP29 were amplified, cloned into pTF102 plasmids, and driven by the 27-kD γ-zein promoter. The ZmABI19 overexpression transgenic lines were generated in the B104 background through A. tumefaciens-mediated transformation. The ZmbZIP29 overexpression transgenic lines were created using F2 immature zygotic embryos of the Hi-II hybrid (A × B) with the same transformation procedures. The recovered positive lines were backcrossed to the B104 inbred line for at least four generations. The null segregants from the self-pollinated overexpression lines were propagated and used as the wild type when comparing with overexpression lines.
Genotyping
Genotyping was performed as described in the literature (Yang et al., 2021). The leaves were collected and genomic DNA was extracted using the cetyl trimethylammonium bromide (CTAB) method. The Cas9-target sites were amplified from the extracted DNA and analyzed by Sanger sequencing. For the overexpression materials, the partial sequences for the target genes and promoters were amplified using the specific primers listed in Supplemental Data Set S1.
ABA treatment assay
For the ABA treatment, the B104 inbred ears at 10 DAP were sterilized with 75% (v/v) ethanol for 10 min. Then, the endosperms were separated using a tweezer and sown on half-strength MS medium containing 50-, 100-, 200-, 250-, and 300-μM ABA. Treatment with ethanol was used as the negative control. After incubation for 24 h in a dark growth chamber at 28°C, the incubated samples were collected, immediately frozen in liquid nitrogen, and stored at –80°C for RNA extraction.
Protein accumulation analysis
The zeins and nonzein proteins were extracted from mature seeds. A total of 50-mg flour for each sample was weighed and incubated with 0.5-mL zein extraction buffer (3.75-mM sodium borate, 2% 2-mercaptoethanol [v/v], 0.3% SDS [w/v], and 70% ethanol [v/v]; pH 10). The nonzein proteins were extracted using nonzein extraction buffer (12.5-mM sodium borate, 2% 2-mercaptoethanol [v/v], and 5% SDS [w/v]; pH 10.0). The proteins were loaded on 15% SDS-PAGE gels and analyzed by Coomassie brilliant blue staining. The protein levels were quantified using the BCA protein assay kit (Thermo Scientific, catalog number: 23227) according to the provided protocols.
Starch measurement
Mature seeds were ground into fine powder by a tissue grinder for evaluating the starch contents. The starch contents were determined by the Megazyme Total Starch Assay Kit according to the manufacturer’s protocols (catalog number: K-TSTA-50A). The starch contents for each sample and the d-glucose control were evaluated at an absorbance of 510 nm. All measurements were performed in at least three biological replicates.
Measurement of ABA content
The immature B104 seeds were harvested at 6, 8, 10, 12, 18, 24, and 30 DAP, respectively, immediately frozen in liquid nitrogen, and stored at –80°C until ABA was extracted. Briefly, 100-mg immature endosperms were ground with 400-μL ethyl acetate with rotation for 30 min. After centrifugation at 15,871 g at 4°C for 10 min, the residual samples were extracted with 400-μL ethyl acetate twice. The extraction was concentrated, eluted in methanol, and analyzed using QTRAP 6500 plus LC-MS/MS system.
Cytological analysis
The wild-type and zmbzip29 seeds at 6, 8, and 16 DAP were collected for paraffin sections. The fresh tissues were fixed in FAA buffer (formaldehyde:acetic acid:ethanol:water = 10:5:50:35, v/v/v/v) and vacuumed twice for 30 min. The samples were embedded in paraffin after graded dehydration using ethanol and vitrification using dimethylbenzene. Sections with 10 μm were dewaxed with dimethylbenzene and rehydrated with graded ethanol. The sections were stained with 0.1% toluidine blue solutions. Then, these sections were observed and photographed using a Leica DM2500 microscope.
For histocytochemical analysis of the ZmbZIP29 and ZmABI19 overexpression endosperms, partial endosperm tissues on the opposite of embryo were fixed in FAA buffer and vacuumed at 4°C for 30 min. After gradient dehydration, the tissues were embedded in epoxide resin for semi-thin sectioning. The sections were stained with 0.1% toluidine blue solution and imaged using an ECLIPSE 80i microscope (Nikon, Tokyo, Japan).
RNA extraction and RT-qPCR
For gene expression analysis, the tissues including root, stem, SAM, leaf, tassel, immature seed, separated endosperms, and embryos from the B104 inbred line were harvested for RNA extraction according to the reported protocol (Yang et al., 2021). Briefly, the harvested samples were suspended with 300-μL RNA extraction buffer (50-mM Tris–HCl, 150-mM LiCl, 5-mM EDTA, and 1% SDS [w/v]). The homogenized samples were further extracted with 1:1 phenol–chloroform and chloroform, 1-mL TRIzol regent (Invitrogen). Purity and concentration of the total RNA were determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific). In total, 1-μg RNA was used to synthesize first-strand cDNA using HiScript II Q RT SuperMix along with DNA elimination (R223, Vazyme). RT-qPCR assay was performed using SYBR Green Premix Pro Taq HS qPCR Kit II (AG, Shanghai Yirui Bio.) on a Bio-Rad CFX-96 PCR thermocycler. The maize ZmActin gene was used as the internal control to normalize the relative quantification of gene expression using the comparative CT method. All primers are listed in Supplemental Data Set S1.
Phylogenetic analysis
The ZmbZIP29 orthologues were searched using the Gramene database (http://www.gramene.org/) and corresponding sequences were downloaded from the National Center for Biotechnology Information nonredundant protein database based on protein–protein alignment tools (https://blast.ncbi.nlm.nih.gov/Blast.cgi) with the ZmbZIP29 protein sequence as a bait. Protein sequences were aligned in the MEGA 6.0 software (https://www.megasoftware.net/) based on the MUSCLE method. Evolutionary distance was calculated using the neighbor-joining algorithm and bootstrap method with 1,000 replicates.
Subcellular localization
To determine the subcellular localization of ZmbZIP29, the CDS of ZmbZIP29 was cloned into the pCAMBIA1300-35S-eGFP plasmid. The constructed plasmid was transferred into Arabidopsis mesophyll protoplasts. After 16-h incubation, the protoplasts were imaged using a Zeiss LSM880 confocal microscope. In addition, the constructs were also transformed into Agrobacterium strain GV3101 and then infiltrated into the N. benthamiana leaves. After incubation for 48 h, the injected leaves were harvested and imaged using a Zeiss LSM880 confocal microscope.
EMSA
The ZmbZIP29-His, ZmABI19-His, and O2-His and modified recombinant proteins were purified using NIA beads (QIAGEN). The oligonucleotide probes were synthesized and labeled with biotin. The purified proteins and probes were mixed in the reaction buffer (1× binding buffer, 5-mM MgCl2, 3% glycerol, 50-ng/mL poly [dI.dC], and 0.05% NP-40). The mixture was incubated at 30°C for 30 min, loaded onto a 6% native PAGE in 0.5× Tris/borate/EDTA buffer. After electrophoresis, the reaction mix was transferred to a nylon membrane and crosslinked to the membrane by UV light. The following procedure was performed using a LightShift Chemiluminescent EMSA kit (Thermo Scientific, catalog number: 20148) and detected on the Tanon 5200 Imaging System (Tanon Science and Technology).
Y1H assay
The partial O2 promoter sequence was synthesized, recombined with pAbAi vector, and then integrated with the genome of the Y1HGold yeast strain. CDSs of the preys were amplified, cloned into the pGADT7-Rec vector, and transferred into the bait yeast strain, respectively. The detailed procedure was according to the Matchmaker Gold Y1H Library Screening System (Clontech). Different concentrations of the aureobasidin A (AbA) antibiotic were used to select the positive bait yeast strain on a medium lacking uracil.
Y2H assay
For the interaction between ZmbZIP29 and ZmABI19, the CDS of ZmbZIP29 was amplified and cloned into the pGADT7 and pGBKT7 plasmids, respectively. The full-length, N-terminal and C-terminal sequences of ZmABI19 were also amplified and cloned into pGADT7 and pGBKT7, respectively. For the interactions between SnRK2s and ZmbZIP29, ZmABI19 or O2, the CDSs of SnRK2s were amplified and inserted into pGBKT7 as the baits, respectively. The coding regions of ZmbZIP29, ZmABI19, and O2 were cloned into pGADT7 as the preys, respectively. The bait and prey constructs were co-transformed into the yeast Y2HGold according to the Yeastmaker Yeast Transformation System 2 (Clontech). The yeast strains were grown on selection medium lacking SD/−Trp/−Leu (DDO) and SD/−Trp/−Leu/−His/−Ade (QDO).
Particle bombardment
The B104 endosperms at 10 DAP were harvested, separated, and then cultured in half-strength MS medium. Generally, 10 μg of plasmids was coated with 60 μg of cleaned gold powder. The gold powder was mixed with the plasmids and then transiently transferred into the cultured endosperm using a Bio-Rad PDS-1000/He biolistic particle delivery system (Bio-Rad, Hercules, CA, USA). After induction for 24 h, total protein was extracted and analyzed on a luminometer (Promega 20/20). The dual-luciferase assay system (Promega) was used to evaluate the ratios between LUC and REN on a luminometer following the manufacturer’s instructions (Promega).
Immunoblotting
The full-length cDNA fragment of ZmbZIP29 was inserted into the prokaryotic expression plasmid pET28a. The anti-ZmbZIP29 antibodies were raised in rabbits by Abclonal, Wuhan, China. The anti-O2 and anti-ZmABI19 antibodies were generated previously (Zhang et al., 2015; Yang et al., 2021). The anti-Flag (Abmart, M20008), anti-HA (Abmart, M20013), anti-His (Abmart, M20001), anti-ACTIN (Abmart, M20009), and anti-GST antibodies (Abmart, M20007) were purchased. The secondary anti-rabbit (Abmart, M21002) and anti-mouse IgG-horseradish peroxidase (HRP) (Abmart, M21001) were used to detect the synthesized antibodies and commercial antibodies, respectively. The total protein for immunoblotting was prepared as described previously (Yang et al., 2021). Briefly, the materials were homogenized in liquid nitrogen and resuspended with lysis buffer in 2-mL tubes (50-mM HEPES pH 7.5, 150-mM NaCl, 1mM EDTA, 1% Triton X-100 [v/v], 1% SDS, and 1× proteinase inhibitor cocktail). After incubation on ice for 30 min, the lysate was centrifuged at 15,871 g at 4°C for 10 min. The supernatants were mixed with 5× SDS loading buffer and boiled at 95°C for 10 min. The proteins were separated on a 12% SDS-PAGE gel and transferred to a polyvinylidene fluoride membrane (BioRad). The primary antibodies were diluted with anti-bZIP29, anti-ZmABI19, and anti-O2 antibodies for 1:1,000, the commercial antibodies for 1:4,000, and the secondary antibodies for 1:10,000. Samples were incubated with primary antibodies for 1 h at room temperature and then the membrane was washed three times with Tris-buffered saline (TBS)-0.1% TWEEN 20 (TBST) buffer. The secondary antibodies were incubated for 1 h and then the membrane was washed four times with the TBST buffer. The signal was visualized using the Tanon-5200 Chemiluminescent Imaging system (Tanon Science and Technology). ImageJ software was used to quantify relative protein levels.
DLR assay
The CDSs of ZmbZIP29, ZmABI19, O2, and SnRK2.2 were inserted into the pGreen II 62SK plasmids driven by the CaMV 35S promoter, respectively. The promoter sequences were amplified and cloned into the pGreen0080 and used as reporters. Mutations of the effector and reporter sequences were created using PCR amplification and validated by Sanger sequencing. The primers used are listed in Supplemental Data Set S1. The Arabidopsis mesophyll protoplasts were isolated and the plasmids were introduced by polyethylene glycol-mediated transformation. After incubation for 16 h, the dual-luciferase assay system (Promega) was used to evaluate the ratios between LUC and REN on the luminometer according to the provided instructions. At least three biological repeats were performed for each experiment.
LIC assay
The full-length CDS, N-terminal, and C-terminal sequences of ZmABI19 were fused with the JW771 (N-terminal half of luciferase, nLUC), respectively. The CDS of ZmbZIP29 was cloned into the JW771 and JW772 (C-terminal half of luciferase, cLUC), respectively. The CDSs of SnRK2.2 and O2 were inserted into the JW772 and JW771, respectively. The constructed plasmids were transferred into Agrobacterium GV3101 cells. Different combinations were transfected into the N. benthamiana leaves. After three days’ accumulation, LUC signals were detected using 0.5-mM luciferin and imaged using the Tanon 5200 chemiluminescent Imaging system.
BiFC assay
The CDSs of ZmbZIP29 and ZmABI19 were cloned into the vectors containing the C-terminus of eYFP and N-terminus of eYFP, respectively. To confirm the interaction between SnRK2.2 and ZmbZIP29, ZmABI19, or O2, the CDS of SnRK2.2 was cloned into the vector containing the C-terminus of eYFP. The CDSs of ZmbZIP29, ZmABI19, and O2 were inserted into the plasmids containing the N-terminus of eYFP, respectively. Equal amounts of the combined plasmids were transformed into the Arabidopsis mesophyll protoplasts. The YFP signals were detected using an excitation wavelength of 488 nm on a Zeiss LSM880 confocal microscope.
Co-IP assay
The CDSs of ZmbZIP29 and ZmABI19 were cloned into pRI101 by the addition of the HA and Flag tags, respectively. To confirm the interactions between SnRK2.2 and ZmABI19, ZmbZIP29, or O2, the CDS of SnRK2.2 was inserted into pRI101 plus Flag or HA tags. The coding regions of ZmbZIP29 and O2 were cloned into pRI101 by addition of the HA tag, respectively. Agrobacterium GV3101 cells harboring different combinations of the Flag- and HA-fusion vectors were co-transformed into the N. benthamiana leaves. Total proteins were extracted from the samples with 0.5-mL lysis extraction buffer (50-mM HEPES, pH 7.5, 150-mM NaCl, 1-mM EDTA, 0.1%SDS, 1%Triton X-100 [v/v], 0.1% deoxycholate [w/v], and EDTA-free protease inhibitor cocktail [#4693132001, Roche]) for 30 min on ice. Then, the extraction suspension was incubated with Flag agarose beads (Abmart, M20018) with rotations for 1 h at 4°C. The beads were washed three times with the lysis buffer for 10 min each time. The 5× SDS loading buffer was added to the reactions and boiled at 98°C for 10 min. The samples were subjected to SDS-PAGE analysis. The anti-Flag (Abmart, M20008, 1:4,000 dilution) and anti-HA antibodies (Abmart, M20013, 1:4,000 dilution) were used to detect the immunoprecipitates, respectively. The signals were detected using the Tanon 5200 chemiluminescent Imaging system.
Pull-down assay
To test the ZmbZIP29 and ZmABI19 interaction, the CDSs of ZmbZIP29 and ZmABI19 were cloned into the pGEX-4T-1 and pET32a plasmids to produce ZmbZIP29-GST and ZmABI19-His, respectively. To test the interaction of SnRK2.2 with ZmbZIP29, ZmABI19, or O2, the CDSs of ZmbZIP29, ZmABI19, and O2 were cloned into pET32a plasmids, and the CDS of SnRK2.2 was cloned into the pET32a, in which the TrxA and His tags were replaced with three repeats of Flag-coding sequences. The recombinant proteins were induced by 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 16 h in E. coli Rosetta cells (DE3, Shanghai Weidi Biotech Co.). Primers used for the vector constructs are listed in Supplemental Data Set S1. The proteins were purified using Ni-NTA agarose beads (Sangon Biotech) or Glutathione Sepharose beads (Sangon Biotech). The recombinant SnRK2.2 protein was purified with Flag agarose beads (Abmart).
In general, 1-μg ZmbZIP29-GST and ZmABI19-His recombinant proteins were incubated in 50-μL GST beads in the GST-binding buffer (20-mM HEPES, 40-Mm KCl, and 1-mM EDTA; pH 7.5). Subsequently, the mixture was washed three times with GST wash buffer (50-mM Tris–HCI, 150-mM NaCl, and 0.1% Triton X-100; pH 8.0) for 10 min. The cleared proteins were eluted by boiling in 1× SDS loading buffer, separated using SDS-PAGE, and detected with anti-GST and anti-His antibodies (1:4,000, Abmart), respectively. Totally, 1-μg SnRK2.2-Flag and 1-μg ZmbZIP29-GST, ZmABI19-His, or O2-His recombinant proteins were incubated in the reaction buffer (25-mM Tris–HCl, 150-mM NaCl, and 1-mM EDTA; pH 7.5) with 50-μL magnetic beads, respectively. The beads-retained proteins were washed six times using washing buffer (25-mM Tris–HCl, 150-mM NaCl, and 0.1% Triton X-100; pH 8.0) and boiled for 10 min in 1× SDS loading buffer. The eluted proteins were analyzed in 12% SDS-PAGE, and then visualized by immunoblot assays with anti-Flag (Abmart, M20008, 1:4,000 dilution), anti-His (Abmart, M20001, 1:4,000 dilution), and anti-GST antibodies (Abmart, M20007, 1:4,000 dilution), respectively.
ChIP-qPCR assay
ChIP assays with ZmbZIP29 antibodies were performed as previously described (Yang et al., 2021). Antibodies against ZmbZIP29 and IgG negative control (#2729, CST) were used for immunoprecipitation. The precipitated DNA samples were recovered using the QIAquick PCR purification kit (Qiagen) and quality was tested using Qubit 3.0. The recovered DNA was diluted by 10 fold and analyzed by real-time qPCR using TB Green Premix Ex TaqTM II (Takara) on the Bio-Rad CFX-96 PCR thermocycler. The values were normalized to the respective input values and the fold changes were calculated based on the relative enrichment in anti-ZmbZIP29 compared with anti-IgG immunoprecipitates. Three replicates were performed using the B104 seeds at 10 DAP.
In vitro phosphorylation assays
The CDS of SnRK2.2 was cloned into the pET32a plasmid by deleting the TrxA and His tags and adding the Flag tag. The CDSs of ZmABI19, ZmbZIP29, O2, and the relative dephosphorylated sequences were cloned into the pET32a plasmid. The fusion proteins were purified from E. coli (DE3) Rosetta cells as described above. For the in vitro kinase assay, the protein samples were mixed in the reaction buffer (50-mM Tris–HCl at pH 7.5, 10-mM MgCl2, 1-mM DTT, and 10-μM ATP). The kinase reaction was conducted at 30°C for 2 h with EDTA-free protease inhibitor cocktail (#4693132001, Roche). The reaction mixtures were terminated by adding 5× SDS protein loading buffer and boiling at 98°C for 10 min. The samples were separated on a 10%–12% polyacrylamide gel containing 50-μM Phos-tag acrylamide (Wako Pure Chemicals). Immunoblot analysis was performed according to the instructions (Wako Pure Chemicals) and detected with the anti-His antibody (Abmart, M20001, 1:4,000 dilution).
Identification of phosphorylation sites
To identify the phosphorylation residues of ZmbZIP29, ZmABI19, and O2, the in vitro kinase reactions were performed as described above. After the reaction process in vitro, the phosphorylated proteins were denatured with 8-M urea, dissolved with 10-mM DTT, alkylated with 100-mM iodoacetamide, and digested with Trypsin at 37°C for 4 h. The concentrated peptides were analyzed using an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). Raw LC-MS/MS data were analyzed using Proteome Discoverer 2.5 (Thermo Fisher Scientific) against specific protein sequences.
Statistical analysis
Microsoft Excel (2016) was used to calculate the P-value by paired two-tailed Student’s t test methods. The raw data and detailed statistical analysis are shown in Supplemental Data Set S2. Quantification of immunoblotting signals was conducted by tracing out the individual band using ImageJ (https://imagej.nih.gov/ij/).
Accession numbers
Sequence data from this article can be found in the MaizeGDB database under the following accession number: ZmbZIP29: Zm00001d034571; O2: Zm00001d018971; ZmABI19: Zm00001d011712; Vp1: Zm00001d042396; and SnRK2.2: Zm00001d022179. The accession numbers for ZmbZIP29 (OL546290) and ZmbZIP29* (OL546289) are available in the NCBI database.
Supplemental data
Supplemental Figure S1. ABA promotes O2 expression.
Supplemental Figure S2. ABA enhances the transactivation capacity of ZmABI19 on the O2 promoter and induces ZmABI19 expression in the endosperm.
Supplemental Figure S3. Decreased expression of ZmABI19 and O2 in w3 and vp5 mutants.
Supplemental Figure S4. ABA induces O2 expression in the wild-type and zmabi19 endosperms.
Supplemental Figure S5. The O2 promoter sequence between –400 and –500 bp upstream of the start cdon.
Supplemental Figure S6. Expression patterns of endosperm-expressed bZIP genes based on the public RNA-seq data.
Supplemental Figure S7. Y1H assay of different bZIP TFs recognizing the O2 promoter.
Supplemental Figure S8. Protein sequence alignment of ZmbZIP29 and its arabidopsis ortholog.
Supplemental Figure S9. Two isoforms of ZmbZIP29 and expression pattern of ZmbZIP29* during seed development.
Supplemental Figure S10. ZmbZIP29 and ZmbZIP29* form homodimers and heterodimers.
Supplemental Figure S11. EMSA showing that ZmbZIP29* is unable to bind the ABRE in the O2 promoter.
Supplemental Figure S12. Light microscopy observation of developing wild-type and zmbzip29 seeds.
Supplemental Figure S13. RT-qPCR analysis of zein expression in 18-DAP wild-type and zmbzip29 endosperms.
Supplemental Figure S14. ZmbZIP29 interacts with the full-length and N-terminal regions of ZmABI19.
Supplemental Figure S15. ZmbZIP29* interacts with ZmABI19.
Supplemental Figure S16. ZmbZIP29 and ZmABI19 synergistically enhance the expression of Vp1, Pbf1, ZmbZIP22, and O11.
Supplemental Figure S17. Ear phenotypes and 100-kernel weight of ZmABI19 overexpression lines (A-OE1, A-OE2, and A-OE3) harvested in Shanghai, 2021.
Supplemental Figure S18. Semi-thin sections of the B104 wild-type and A-OE endosperms.
Supplemental Figure S19. Kernel width and length of ZmABI19 and ZmbZIP29 overexpression lines.
Supplemental Figure S20. Ear phenotypes and 100-kernel weight of ZmbZIP29 overexpression lines (B-OE1 and B-OE2) harvested in Shanghai, 2021.
Supplemental Figure S21. Semi-thin sections of the B014 wild-type and B-OE endosperms.
Supplemental Figure S22. Identification of candidate SnRK2 family members that interact with ZmABI19 and ZmbZIP29 by Y2H assay.
Supplemental Figure S23. RT-qPCR analysis of SnRK2.2 expression induced by ABA treatment.
Supplemental Figure S24. SnRK2.2 and ABA enhance transactivation of the Vp1 promoter by ZmABI19.
Supplemental Figure S25. Identification of candidate SnRK2 family members that interact with O2 by Y2H assay.
Supplemental Figure S26. Identification of the phosphorylated residues of SnRK2.2 by LC-MS/MS.
Supplemental Figure S27. Identification of the phosphorylated residues of ZmABI19, ZmbZIP29, and O2 by LC-MS/MS.
Supplemental Figure S28. Altered binding affinity of phosphorylated and dephosphorylated forms of ZmABI19, ZmbZIP29, and O2 proteins.
Supplemental Data Set S1. Primers used in this study.
Supplemental Data Set S2. Detailed statistical analysis in this study.
Supplemental File S1. Alignment files used for the phylogenetic analysis shown in Figure 2A.
Supplemental File S2. Newick format of the phylogenetic tree in Figure 2A.
Supplementary Material
Acknowledgments
We thank Shanshan Wang, Lianyan Jing, and Xiaoyan Xu from CAS Center for Excellence in Molecular Plant Sciences (CEMPS) for the technical support of LC-MS/MS analysis; Jiqin Li, Xiaosu Gao, and Zhiping Gao from CEMPS for the technical support of histochemical analysis; Shuining Yin and Wenjuan Cai from CEMPS for the technical support of confocal microscopy.
Funding
This research was supported by the Chinese Academy of Sciences (XDB27010201 to Y.W.) and the National Natural Science Foundation of China (91935305, 31830063, and 31925030 to Y.W.).
Conflict of interest statement. None declared.
Contributor Information
Tao Yang, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.
Haonan Wang, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China; University of the Chinese Academy of Sciences, Beijing 100049, China.
Liangxing Guo, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China; University of the Chinese Academy of Sciences, Beijing 100049, China.
Xingguo Wu, Shanghai Key Laboratory of Plant Molecular Sciences, College of Life Sciences, Shanghai Normal University, Shanghai 200233, China.
Qiao Xiao, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China; University of the Chinese Academy of Sciences, Beijing 100049, China.
Jiechen Wang, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.
Qiong Wang, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.
Guangjin Ma, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China; University of the Chinese Academy of Sciences, Beijing 100049, China.
Wenqin Wang, Shanghai Key Laboratory of Plant Molecular Sciences, College of Life Sciences, Shanghai Normal University, Shanghai 200233, China.
Yongrui Wu, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.
T.Y. and Y.W. conceived and designed the experiments, and wrote the manuscript. Y.W. supervised the project. T.Y. and H.W. performed the experiments. L.G. and Q.X. assisted with the field work and material sampling. X.W. performed the histochemical analysis. G.M., Q.W., J.W., and W.W. helped analyze the data.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Yongrui Wu (yrwu@cemps.ac.cn).
References
- Alonso R, Oñate-Sánchez L, Weltmeier F, Ehlert A, Diaz I, Dietrich K, Vicente-Carbajosa J, Dröge-Laser W (2009) A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell 21: 1747–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk PK, Pagès M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37: 425–435 [DOI] [PubMed] [Google Scholar]
- Cai H, Chen Y, Zhang M, Cai R, Cheng B, Ma Q, Zhao Y (2017a) A novel GRAS transcription factor, ZmGRAS20, regulates starch biosynthesis in rice endosperm. Physiol Mol Biol Plants 23: 143–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Li S, Sun F, Sun Q, Zhao H, Ren X, Zhao Y, Tan BC, Zhang Z, Qiu F (2017b) Emp10 encodes a mitochondrial PPR protein that affects the cis‐splicing of nad2 intron 1 and seed development in maize. Plant J 91: 132–144 [DOI] [PubMed] [Google Scholar]
- Chan A, Carianopol C, Tsai AY, Varatharajah K, Chiu RS, Gazzarrini S (2017) SnRK1 phosphorylation of FUSCA3 positively regulates embryogenesis, seed yield, and plant growth at high temperature in Arabidopsis. J Exp Bot 68: 4219–4231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zeng B, Zhang M, Xie S, Wang G, Hauck A, Lai J (2014) Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol 166: 252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Li GJ, Bressan RA, Song CP, Zhu JK, Zhao Y (2020) Abscisic acid dynamics, signaling, and functions in plants. J Integr Plant Biol 62: 25–54 [DOI] [PubMed] [Google Scholar]
- Ciceri P, Gianazza E, Lazzari B, Lippoli G, Genga A, Hoscheck G, Schmidt RJ, Viotti A (1997) Phosphorylation of Opaque2 changes diurnally and impacts its DNA binding activity. Plant Cell 9: 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman CE, Larkins BA (1999). The prolamins of maize. In Shewry PR and Casey R, eds, Seed Proteins. Springer Netherlands, Dordrecht, pp 109–139 [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Deng Y, Wang J, Zhang Z, Wu Y (2020) Transactivation of Sus1 and Sus2 by Opaque2 is an essential supplement to sucrose synthase-mediated endosperm filling in maize. Plant Biotechnol J 18: 1897–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esen A (1987) A proposed nomenclature for the alcohol-soluble proteins (zeins) of maize (Zea mays L.). J Cereal Sci 5: 117–128 [Google Scholar]
- Feng Y, Wang Y, Zhang G, Gan Z, Gao M, Lv J, Wu T, Zhang X, Xu X, Yang S (2021) Group‐C/S1 bZIP heterodimers regulate MdIPT5b to negatively modulate drought tolerance in apple species. Plant J 107: 399–417 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey A, Godin B, Bonnet M, Sotta B, Marion-Poll A (2004) Maternal synthesis of abscisic acid controls seed development and yield in Nicotiana plumbaginifolia. Planta 218: 958–964 [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C (2007) Combined networks regulating seed maturation. Trends Plant Sci 12: 294–300 [DOI] [PubMed] [Google Scholar]
- Hunter CT, Saunders JW, Magallanes-Lundback M, Christensen SA, Willett D, Stinard PS, Li QB, Lee K, DellaPenna D, Koch KE (2018) Maize w3 disrupts homogentisate solanesyl transferase (ZmHst) and reveals a plastoquinone‐9 independent path for phytoene desaturation and tocopherol accumulation in kernels. Plant J 93: 799–813 [DOI] [PubMed] [Google Scholar]
- Ji C, Xu L, Li Y, Fu Y, Li S, Wang Q, Zeng X, Zhang Z, Zhang Z, Wang W, et al. (2021) The O2-ZmGRAS11 transcriptional regulatory network orchestrates the coordination of endosperm cell expansion and grain filling in maize. Mol Plant 11: 1674–2052 [DOI] [PubMed] [Google Scholar]
- Ji X, Dong B, Shiran B, Talbot MJ, Edlington JE, Hughes T, White RG, Gubler F, Dolferus R (2011) Control of abscisic acid catabolism and abscisic acid homeostasis is important for reproductive stage stress tolerance in cereals. Plant Physiol 156: 647–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RJ, Brenner ML (1987) Distribution of abscisic acid in maize kernel during grain filling. Plant Physiol 83: 905–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Sakurai N, Kuraishi S (1993) The changes of endogenous abscisic acid in developing grain of two rice cultivars with different grain size. Jpn J Crop Sci 62: 456–461 [Google Scholar]
- Keith K, Kraml M, Dengler NG, McCourt P (1994) fusca3: A heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 6: 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Koike T (2009) Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat Protoc 4: 1513–1521 [DOI] [PubMed] [Google Scholar]
- Li C, Yue Y, Chen H, Qi W, Song R (2018) The ZmbZIP22 transcription factor regulates 27-kD gamma-zein gene transcription during maize endosperm development. Plant Cell 30: 2402–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Qi W, Liang Z, Yang X, Ma Z, Song R (2020) A SnRK1-ZmRFWD3-Opaque2 signaling axis regulates diurnal nitrogen accumulation in maize seeds. Plant Cell 32: 2823–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Qiao Z, Qi W, Wang Q, Yuan Y, Yang X, Tang Y, Mei B, Lv Y, Zhao H, et al. (2015) Genome-wide characterization of cis-acting DNA targets reveals the transcriptional regulatory framework of Opaque2 in maize. Plant Cell 27: 532–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locatelli S, Piatti P, Motto M, Rossi V (2009) Chromatin and DNA modifications in the Opaque2-mediated regulation of gene transcription during maize endosperm development. Plant Cell 21: 1410–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T, Xu B, Hu Y, Wang Y, Mao C, Wang Y, Zhang J, Liu H, Huang H, Liu Y (2021) Genome-wide identification of ZmSnRK2 genes and functional analysis of ZmSnRK2.10 in ABA signaling pathway in maize (Zea mays L). BMC Plant Biol 21: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32: 317–328 [DOI] [PubMed] [Google Scholar]
- McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK (1991) The viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66: 895–905 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50: 1345–1363 [DOI] [PubMed] [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J (1994) Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6: 1567–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-Y, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Tsz-fung FC (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Z, Qi W, Wang Q, Feng Y, Yang Q, Zhang N, Wang S, Tang Y, Song R (2016) ZmMADS47 regulates zein gene transcription through interaction with Opaque2. PLoS Genet 12: e1005991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P, Zhang G, Hu B, Wu J, Chen W, Ren Z, Liu Y, Xie J, Yuan H, Tu B (2021) Leaf-derived ABA regulates rice seed development via a transporter-mediated and temperature-sensitive mechanism. Sci Adv 7: eabc8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA perception and signalling. Trends Plant Sci 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Robertson DS (1955) The genetics of vivipary in maize. Genetics 40: 745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud C, Wong J, Sussex I (1979) Control of in vitro growth of viviparous embryo mutants of maize by abscisic acid. Dev Genet 1: 325–330 [Google Scholar]
- Sabelli PA, Larkins BA (2009) The development of endosperm in grasses. Plant Physiol 149: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MP (2009) Transgenic Maize: Methods and Protocols. Humana Press c/o Springer Science+ Business Media.
- Seiler C, Harshavardhan VT, Rajesh K, Reddy PS, Strickert M, Rolletschek H, Scholz U, Wobus U, Sreenivasulu N (2011) ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J Exp Bot 62: 2615–2632 [DOI] [PubMed] [Google Scholar]
- Smith J, De Koehler FF (1980) Genetic studies utilizing viviparous mutants of Zea mays indicate that abscisic acid may play a critical role in the induction of seed dormancy. Genetics 94: 100 [Google Scholar]
- Sreenivasulu N, Radchuk V, Alawady A, Borisjuk L, Weier D, Staroske N, Fuchs J, Miersch O, Strickert M, Usadel B (2010) De-regulation of abscisic acid contents causes abnormal endosperm development in the barley mutant seg8. Plant J 64: 589–603 [DOI] [PubMed] [Google Scholar]
- Tanner W (1980) On the possible role of ABA on phloem unloading: Discussion remark following the presentation of Dr. H. Düring. Ber Dtsch Bot Ges 93: 349–351 [Google Scholar]
- Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, Shinozaki K (2013) Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signaling 6: rs8–rs8 [DOI] [PubMed] [Google Scholar]
- Vreugdenhil D (1983) Abscisic acid inhibits phloem loading of sucrose. Physiol Plant 57: 463–467 [Google Scholar]
- Xiong W, Wang C, Zhang X, Yang Q, Shao R, Lai J, Du C (2017) Highly interwoven communities of a gene regulatory network unveil topologically important genes for maize seed development. Plant J 92: 1143–1156 [DOI] [PubMed] [Google Scholar]
- Yang G, Yu Z, Gao L, Zheng C (2019) SnRK2s at the crossroads of growth and stress responses. Trends Plant Sci 24: 672–676 [DOI] [PubMed] [Google Scholar]
- Yang J, Ji C, Wu Y (2016) Divergent transactivation of maize storage protein zein genes by the transcription factors Opaque2 and OHPs. Genetics 204: 581–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang J, Wang Z, Zhu Q, Wang W (2001) Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol 127: 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Guo L, Ji C, Wang H, Wang J, Zheng X, Xiao Q, Wu Y (2021) The B3 domain-containing transcription factor ZmABI19 coordinates expression of key factors required for maize seed development and grain filling. Plant Cell 33: 104–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Gu W, Chen J, Song N, Gao X, Zhang X, Zhou Y, Ma X, Song W, Zhao H, et al. (2019) High temporal-resolution transcriptome landscape of early maize seed development. Plant Cell 31: 974–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Mogami J, Yamaguchi-Shinozaki K (2014) ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol 21: 133–139 [DOI] [PubMed] [Google Scholar]
- Zhan J, Li G, Ryu C-H, Ma C, Zhang S, Lloyd A, Hunter BG, Larkins BA, Drews GN, Wang X, et al. (2018) Opaque-2 regulates a complex gene network associated with cell differentiation and storage functions of maize endosperm. Plant Cell 30: 2425–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yang J, Wu Y (2015) Transcriptional regulation of zein gene expression in maize through the additive and synergistic action of opaque2, prolamine-box binding factor, and O2 heterodimerizing proteins. Plant Cell 27: 1162–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zheng X, Yang J, Messing J, Wu Y (2016) Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis. Proc Natl Acad Sci USA 113: 10842–10847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Dong J, Ji C, Wu Y, Messing J (2019) NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc Natl Acad Sci USA 116: 11223–11228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Li Q, Li C, An D, Xiao Q, Wang W, Wu Y (2019) Intra-kernel reallocation of proteins in maize depends on VP1-mediated scutellum development and nutrient. Plant Cell 31: 2613–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.