Abstract
Somatostatin and its related peptides (SSRPs) form an important family of hormones with diverse physiological roles. The ubiquitous presence of SSRPs in vertebrates and several invertebrate deuterostomes suggests an ancient origin of the SSRP signaling system. However, the existence of SSRP genes outside of deuterostomes has not been established, and the evolutionary history of this signaling system remains poorly understood. Our recent discovery of SSRP-like toxins (consomatins) in venomous marine cone snails (Conus) suggested the presence of a related signaling system in mollusks and potentially other protostomes. Here, we identify the molluscan SSRP-like signaling gene that gave rise to the consomatin family. Following recruitment into venom, consomatin genes experienced strong positive selection and repeated gene duplications resulting in the formation of a hyperdiverse family of venom peptides. Intriguingly, the largest number of consomatins was found in worm-hunting species (>400 sequences), indicating a homologous system in annelids, another large protostome phylum. Consistent with this, comprehensive sequence mining enabled the identification of SSRP-like sequences (and their corresponding orphan receptor) in annelids and several other protostome phyla. These results established the existence of SSRP-like peptides in many major branches of bilaterians and challenge the prevailing hypothesis that deuterostome SSRPs and protostome allatostatin-C are orthologous peptide families. Finally, having a large set of predator–prey SSRP sequences available, we show that although the cone snail’s signaling SSRP-like genes are under purifying selection, the venom consomatin genes experience rapid directional selection to target receptors in a changing mix of prey.
Keywords: somatostatin, neuropeptides, conotoxins, evolution, allatostatin-C, cone snails, hormones, venom
Introduction
Small, secreted peptides form a large and diverse group of signaling molecules that are essential for fundamental biological processes. Many of these signaling peptides originated early in animal evolution, with descendants found in both protostomes and deuterostomes, the two major groups of bilaterians (Jékely 2013; Mirabeau and Joly 2013). Related (homologous) peptides that originated from a common ancestor gene can be further divided into paralogs if the divergence started as a gene duplication event within the same species and orthologs if the split happened as a result of a speciation event. Since orthologous signaling peptides typically retain the same function in the course of evolution, their study can provide fundamental insight into the shared biology of species that span diverse phyla, for example, from protostome model organisms, such as the fruit fly, the nematode Caenorhabditis, and the sea hare Aplysia, to deuterostomes, including zebrafish and humans. Examples of ancient signaling peptides include the insulin family, neuropeptide Y peptides, and oxytocin/vasopressin-like peptides (Elphick et al. 2018).
The evolutionary origin of signaling peptides can be challenging to uncover. Because of their small size, signaling peptides often lack sufficient sequence conservation to establish homology, especially if peptides are compared between distantly related species. Additionally, many signaling peptide precursors do not contain well-defined structural or functional domains that could help determine their relatedness. Repetitive sequence regions of varying frequency and lengths are also common, further hampering efforts to establish relatedness. Thus, many homologous signaling peptides have diversified to a point beyond recognition, even though they share a common ancestry.
There is alternative evidence that can be gathered to establish homology. For example, conserved structures of the peptide-encoding genes (the number, positions, and phases of introns) can, in some cases, be used to determine homology (Mair et al. 2000; Yañez-Guerra et al. 2020; Zhang et al. 2020). Clustering-based approaches have similarly been indispensable in uncovering neuropeptide evolution (Jékely 2013; Thiel et al. 2021). Furthermore, comparative sequence analysis of the target receptors, which are often G protein-coupled receptors (GPCRs), can assist in establishing homology. Because of their larger size and well-defined sequence structures, compared with their peptide ligands, GPCRs are more amendable to phylogenetic analyses. GPCR receptor–ligand pairs are typically stably associated throughout evolution. Thus, homology between divergent signaling peptide ligands may be inferred from the homology of their receptors (Mirabeau and Joly 2013). This approach has been successfully used to find common ancestry of, for example, deuterostome gonadotropin-releasing hormone and protostome adipokinetic hormone, even though the peptide ligands themselves only share a single amino acid (Grimmelikhuijzen and Hauser 2012). Although these methods can be used to provide secondary evidence of homology between signaling peptides, the evolutionary history of many signaling systems remains unknown.
In this paper, we demonstrate the usefulness of venom peptides for tracing the relatedness of signaling systems across diverse phyla. The venom peptides used here are produced by a highly biodiverse marine lineage, the predatory cone snails (Conus). Cone snail venoms are extremely complex, with hundreds of different components per species, which can be grouped into gene families based on their conserved signal sequence (Woodward et al. 1990). Within each gene family, the mature secreted peptide is subject to accelerated evolution, resulting in the venom of each species having its own distinctive complement of peptides (Li et al. 2017). Endogenous signaling peptides that are widely distributed across all mollusks can be recruited for expression in cone snail venom. This phenomenon has previously been demonstrated for oxytocin (Cruz et al. 1987) and insulin (Safavi-Hemami et al. 2016). In a recent paper, we identified another family of signaling peptide-like toxins in Conus venom with significant sequence similarity to somatostatin (SS) (Ramiro et al. 2022). Whether these SS-like cone snail toxins (consomatins) evolved from an endogenous SS-like signaling peptide was not addressed.
SS is a highly conserved chordate hormone that was initially discovered as a mammalian inhibitor of growth hormone release from the hypothalamus (Brazeau et al. 1973). Since then, SS has been found to have several other physiological functions, including the inhibition of insulin and glucagon release from the pancreas, regulation of gut motility, and as an analgesic, anxiolytic, and anticancer agent (Martinez 2013). SS was the first identified member of a multigene family of peptides that also includes cortistatin (CST), urotensin-II (UII), and urotensin-II-related peptide (URP) (fig. 1A). Whereas SS and CST are inhibitors of hormone secretion, inflammation, and pain, UII, and URP are important regulators of cardiovascular activity (Møller et al. 2003; Vaudry et al. 2010). Several studies have also suggested partially overlapping effects of these peptides. For example, UII is primarily known as a potent vasoconstrictor, but it also shares several functions with SS, such as inhibition of insulin secretion and regulation of food intake (Ong et al. 2008). In this article, we collectively refer to these peptides as somatostatin and its related peptides (SSRPs).
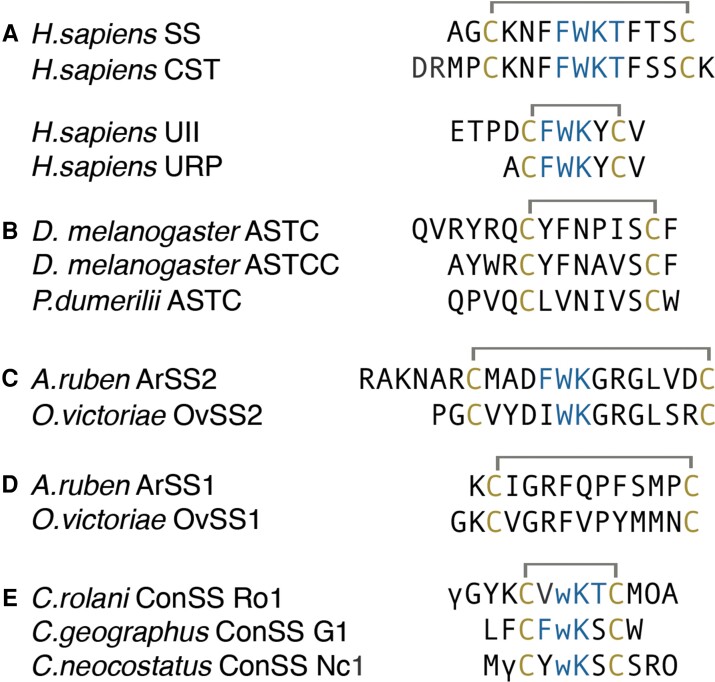
Fig. 1.
(A) Human SSRP, (B) protostome ASTC, (C) echinoderm SS2 peptide, (D) SS1 peptide, and (E) consomatins from fish-hunting cone snails. Cysteines and amino acids of the core SS receptor binding motif are shown in yellow and blue, respectively. Modifications: γ, γ-carboxyglutamate; w, d-tryptophan; O, hydroxyproline.
SSRPs exert their function through activating various members of the GPCR family. In humans, both SS and CST activate the five SS receptors, somatostatin receptors (SSTs)1–5, with comparable potency (Møller et al. 2003). In addition, CST also activates the ghrelin receptor and Mas-Related G Protein-Coupled Receptor-X2 (MRGPRX2) (Deghenghi et al. 2001; Robas et al. 2003). UII and URP both bind to and activate the urotensin receptor (UT) (Vaudry et al. 2010). As with their ligands, the SST1–5 and UT are thought to have originated from a common ancestor during chordate evolution (Tostivint et al. 2014).
In chordates, it is believed that SSRPs evolved from an ancestral gene that underwent a local duplication giving rise to the ancestral SS and UII genes (Tostivint et al. 2006, 2014). These genes further duplicated in multiple rounds, such that there are four paralogous peptides related to SS in mammals (SS, CST, UII, and URP) (fig. 1A) (Tostivint et al. 2014). The duplication event that gave rise to the ancestral SS and UII genes likely took place in Vertebrata, as both SS and UII genes are found in lampreys (Conlon et al. 1995; Waugh et al. 1995), but only a single gene is present in cephalochordates (Mirabeau and Joly 2013).
As for many other vertebrate signaling peptide families, SSRPs have been suggested to have a protostome ortholog, allatostatin-C (ASTC). ASTC was first discovered in the tobacco worm Manduca sexta (phylum Arthropoda) as an inhibitor of juvenile hormone biosynthesis and has now been found in most protostome phyla (Kramer et al. 1991; Thiel et al. 2021; Veenstra 2009). Although the peptide shows limited sequence similarity to SS besides a disulfide loop (fig. 1B) (Kreienkamp et al. 2002; Veenstra 2009), several phylogenetic and clustering analyses have shown close similarities between the receptors of these peptides, the ASTC receptors and the SSTs (Jékely 2013; Mirabeau and Joly 2013; Elphick et al. 2018). This, in addition to the fact that SSRPs were only found in chordates and ASTC only in protostomes, led to the conclusion that SS and ASTC are orthologous peptides.
However, the evolutionary history of these peptides appears to be more complicated than initially thought. Recently, two peptides, ArSS1 and ArSS2, with similarities to ASTC and SS, respectively, were discovered in the starfish Asterias rubens (phylum Echinodermata, group deuterostomes) (fig. 1C and D) (Semmens et al. 2016; Zhang et al. 2020). The coexistence of an SS- and ASTC-like peptide in echinoderms suggested that SS and ASTC might, in fact, represent paralogous rather than orthologous peptide families (Zhang et al. 2022). Furthermore, our recent discovery of consomatins in the venom of fish-hunting cone snails that selectively activate different subtypes of the human SSTs suggested the existence of an SSRP-like signaling system in these protostomes that may have given rise to the consomatin family (fig. 1E) (Ramiro et al. 2022).
Here, by tracing the evolution of consomatins we unravel the existence of an SSRP-like peptide system in diverse protostome phyla. Based on several lines of evidence presented in this study, we propose that these SSRP-like signaling peptides are orthologs of chordate SSRPs, challenging the hypothesis that SSRPs and ASTC are orthologous families. Specifically, we find that consomatins are derived from SSRP-like signaling peptides expressed in neuroendocrine tissue of mollusks, annelids, and several other protostome phyla. The signaling SSRP-like genes experience purifying selection to target a conserved endogenous receptor, but once duplicated and recruited into venom, diversified to target receptors in a shifting mix of prey and, potentially, predators and competitors. Collectively, our findings shed light on the broad existence and early evolution of the SSRP-like and ASTC signaling systems in Bilateria and provide a new paradigm for the use of animal toxins to track the evolution of important signaling systems across diverse prey taxa.
Results
Evolution of Diverse Cone Snail Toxins with Similarity to Chordate SSRPs
Recently, we identified and characterized a novel family of somatostatin-like toxins (consomatins) in fish-hunting cone snails of the Asprella and Gastridium clade that target the SS signaling system in vertebrates (Ramiro et al. 2022). Consomatins share several conserved residues with chordate SSs, selectively activate different subtypes of the human SS receptors, and display a minimized disulfide scaffold and a d-amino acid also seen in synthetic SS drug analogs, such as Octreotide and Lanreotide. In our previous study, we specifically searched for toxins that share at least three of the four residues important for activating the human SSTs or have conservative substitutions in these positions (residues 7–10, Phe-Trp-Lys-Thr (FWKT), numbering based on human SS-14). Here, to elucidate the full diversity of consomatins that not only share sequence similarity with SS but also other members of the SSRP family, we more broadly searched for sequences that display the WK motif (residues 8–9) and disulfide loop characteristic for these peptides.
By searching the genomes and venom gland transcriptomes of a phylogenetically diverse set of 25 cone snail species, we identified 52 consomatin sequences containing these features (fig. 2A and supplementary fig. S1, Supplementary Material online, see supplementary file S1, Supplementary Material online for accession numbers of all datasets interrogated in this study and supplementary file S2, Supplementary Material online for sequences). All sequences belong to the C toxin gene superfamily that is defined by a conserved N-terminal signal sequence involved in translocation to the endoplasmic reticulum and secretory pathway.
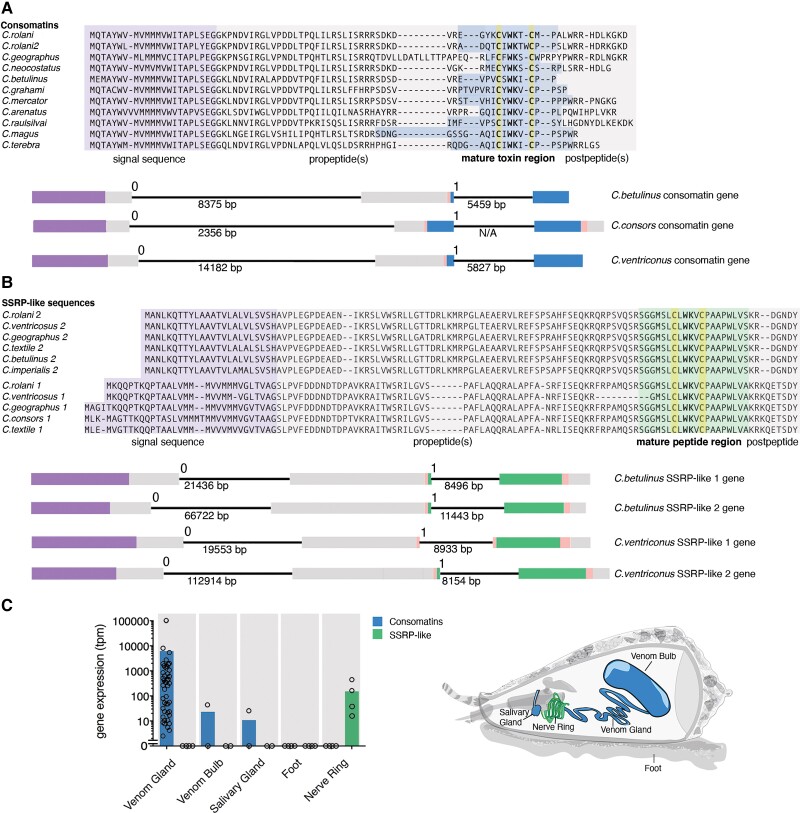
Fig. 2.
Consomatins and signaling SSRP-like peptides are homologs with contrasting expression patterns. Comparative sequence alignments and gene structures of selected (A) consomatins and (B) signaling SSRP-like sequences retrieved from transcriptome and genome data. The position of the signal sequence (purple), propeptides (gray), mature toxin region (blue), mature signaling SSRP-like peptide region (green), and postpeptides (gray) are shown below alignments. Gene structures are shown using the same color codes. The phases of the introns are indicated above the introns, whereas the length is noted below. (C) Mean gene expression levels of consomatins and signaling SSRP-like sequences in the venom gland (n = 46), venom bulb (n = 2), salivary gland (n = 2), foot (n = 4), and circumoesophageal nerve ring (n = 4). Circles represent individual datapoints.
Consomatin genes contain three exons (fig. 2A): the first exon covers the signal sequence and N-terminal part of the proregion, the second exon encodes the C-terminal part of the proregion, the N-terminal processing site, and the first 1–5 amino acids of the mature peptide. The last exon spans the remaining part of the mature peptide and, in most cases, a C-terminal processing site and the remaining postpeptide region. Introns can be positioned in three phases: phase 0 is located between codons, and phases 1 and 2 are located after the first or second nucleotide of a codon, respectively. The two introns of consomatins are of phases 0 and 1, respectively, with the first intron being longer.
Consomatins Evolved from an Ancestral SSRP-like Precursor Gene
The high sequence similarity of consomatins with chordate SSRPs suggested that these toxins may have originated from a molluscan SSRP-like gene involved in neuroendocrine signaling. To investigate this possibility, we sequenced the transcriptomes of the circumoesophageal nerve ring, an arrangement of nerve ganglia known to secrete neuroendocrine signaling peptides (Safavi-Hemami et al. 2016), from four divergent cone snail species, Conus rolani, Conus geographus, Conus textile, and Conus imperialis. Homology searching for an SSRP-like gene led to the identification of two highly expressed sequences encoding prepropeptides that share the WK motif and disulfide loop with consomatins and, therefore, also with vertebrate SSRPs (fig. 2B, supplementary files S1 and S3, Supplementary Material online). The two SSRP-like genes (SSRP-like 1 and SSRP-like 2) have common characteristics of peptide preprohormones (N-terminal signal sequence and canonical proteolytic processing sites) and are expressed specifically in the nerve ring. Consomatins, on the other hand, are expressed primarily in the venom gland and at much lower levels in the venom bulb, a muscular extension of the gland that plays a role in venom delivery, and the salivary gland, an accessory gland located in close proximity to the venom gland (fig. 2C).
To specifically determine whether the consomatin and the SSRP-like signaling genes evolved from a common, ancestral SSRP-like predecessor, we interrogated the available genomes of Conus betulinus, Conus consors, and Conus ventricosus. We found that the organization of the signaling genes mirrors that of the venom genes, including the number, site, and phases of the introns and the location of the mature peptides within the genes (fig. 2A and B). These highly conserved structural features corroborate that consomatins evolved by gene duplication of a signaling SSRP-like gene expressed in the snail’s neuroendocrine system. Whether consomatins emerged from only one or both SSRP-like signaling genes could not be determined. However, as evident by phylogenetic analysis, following the duplication event(s), consomatin genes neofunctionalized and proliferated, leading to a great diversification of this venom peptide family (fig. 3A).
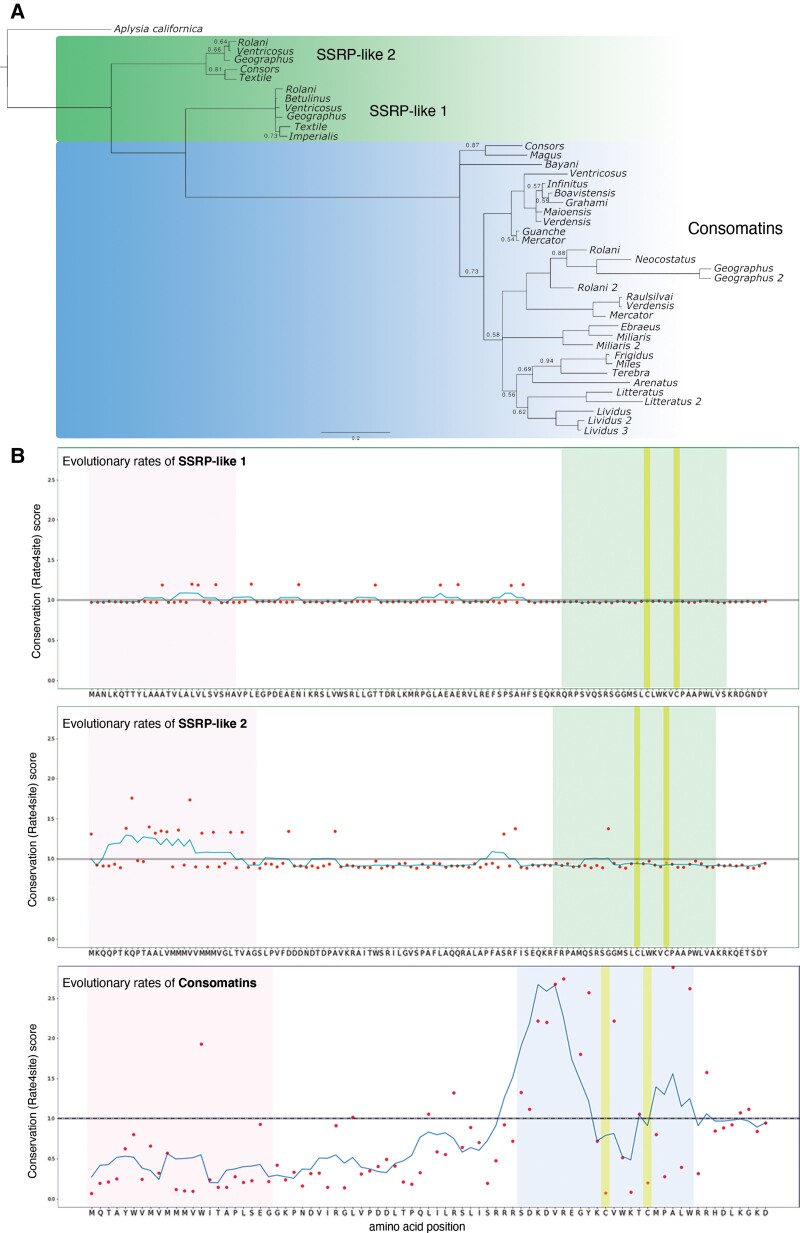
Fig. 3.
Consomatins and signaling SSRP-like sequences show contrasting evolutionary patterns. (A) Bayesian gene tree of consomatin and nerve ring SSRP-like sequences from diverse cone snail species. Branch lengths are proportional to estimated amino acid change, highlighting the speed with which consomatins have diverged, as compared with the nerve ring sequences. Only posterior probabilities values below 0.95 are depicted for visual clarity. (B) Position-specific evolutionary rates of SSRP-like 1 (top), SSRP-like 2 (middle), and consomatin (bottom) sequences as determined using rate4site. Amino acids of the corresponding sequences from Conus rolani are shown for clarity. Color codes are the same as in fig. 2.
Contrasting Evolutionary Patterns between Consomatins and Signaling SSRP-like Genes
In contrast to the hyperdiverse consomatin family, the two signaling SSRP-like genes are almost identical to one another both within and between the four different cone snail species (mean amino acid sequence identity of 50.3% between the genes and 96.6% and 91.5% between species for each of the two genes). This is further highlighted by their close grouping and short branches in phylogenetic analyses (fig. 3A; supplementary file S4, Supplementary Material online). To determine whether the signaling genes have been subject to negative/purifying selection, we estimated the pairwise dN/dS ratios between genes from different cone snails (supplementary file S5, Supplementary Material online). The dN/dS ratio is the rate of nonsynonymous to synonymous mutations, where a ratio above 1 indicates positive selection, whereas a ratio below 1 indicates purifying selection. Based on estimates of pairwise dN/dS ratios, the signaling genes appear to have been subject to negative/purifying selection (SSRP-like gene 1: mean dN/dS = 0.34, range: 0.08–1.20; SSRP-like gene 2: mean dN/dS = 0.57, range: 0.11–1.02). This is also confirmed by comparing M7 and M8 models in the program PAML, which estimates the likelihood that some sites evolved under positive selection. This analysis showed that none of the sites appear to diverge from negative or neutral selection in any of the two genes (supplementary file S6, Supplementary Material online).
To estimate the evolutionary rate of the consomatin genes, we randomly selected 20 transcriptome sequences. Pairwise comparison gave average dN/dS = 3.04 with 88% of comparisons above 1 (supplementary file S5, Supplementary Material online). Tests for positive selection (M7 vs. M8) strongly favor a model where 26% of the 62 aligned codons have evolved with an average dN/dS ratio of 6.94, in contrast to a model constrained to negative or neutral evolution at all sites (model statistics can be seen in supplementary file S6, Supplementary Material online). Positive selection is particularly evident at sites around the mature toxins, except for the cysteine residues that form the disulfide loop and the WK-motif, which are all highly conserved.
Plotting the conservation (Rate4site) scores for consomatins can further be used to visualize how the different regions of the toxins precursor evolve at different rates. Whereas the signal sequence and proregions are all well conserved, amino acids within the mature toxins are highly divergent, apart from the two cysteines and the WK motif. In stark contrast, the signaling SSRP-like sequences are highly conserved all throughout the preprohormone (fig. 3B).
Signaling SSRP-like Genes are Ubiquitously Found in All Classes of Mollusca
We hypothesized that the two SSRP-like genes found in the circumoesophageal nerve ring of cone snails encode secreted peptides involved in neuroendocrine signaling. Since signaling peptides are generally well conserved within phyla, we searched for similar genes in other molluscs. This led to the identification of highly similar genes in all molluscan classes (Gastropoda, Bivalvia, Polyplacophora, Cephalopoda, Scaphopoda, Aplacophora, and Monoplacophora, see supplementary files S1, S3, and S7, Supplementary Material online). In particular, we found that the cone snail SSRP-like genes are orthologous to a previously described gene from the sea hare Aplysia californica that encodes a secreted, bioactive peptide expressed in various ganglia (buccal, cerebral, pleural-pedal, and abdominal) (Romanova et al. 2012). Based on its sequence similarity to vertebrate UII, this peptide was named Aplysia UII (apUII). apUII shares 40% identity with the predicted cone snail signaling SSRP-like peptide. This finding confirms that the SSRP-like gene family, which gave rise to consomatins, encodes a molluscan neuroendocrine peptide that shares sequence similarity with members of the chordate SSRP family. Whereas the A. californica genome assembly was not of sufficient quality to interrogate the structure of the apUII-encoding gene, analysis of four other molluscan genomes, including that of the golden apple snail, Pomacea canaliculata, confirmed the same exon/intron structure as identified for the consomatin and the cone snail signaling SSRP-like genes (supplementary fig. S2, Supplementary Material online).
Consomatins Expressed in Worm-Hunting Species Unravel the Existence of an Orthologous Signaling System in Annelids
Previously, we showed that two consomatins from fish-hunting cone snails of the Asprella and Gastridium clade share several important amino acids with vertebrate SSs and selectively activate the human SST1,4 and SST2 with low micromolar and low nanomolar potency, respectively (Ramiro et al. 2022). We further noticed that consomatins were also expressed in some cone snail species that prey on annelid worms, which strongly suggested the existence of an SSRP-like signaling system in annelids. Here, by searching the genomes and transcriptomes of 19 species of worm-hunting cone snails, we first confirm the wide distribution of consomatins in various clades of worm hunters (48 sequences from 19 species belonging to 10 clades). We then proceeded to use the molluscan signaling SSRP-like sequences to query similar genes in a wide range of annelid genomes and transcriptomes. By doing so, we found 19 SSRP-like sequences from 14 species belonging to both classes of Annelida (Polychaeta and Clitellata), demonstrating the wide distribution and expression of SSRP-like genes in this large protostome phylum (supplementary files S1 and S8, Supplementary Material online). Notably, SSRP-like sequences are not restricted to marine species but are, for example, also present in the earthworm Lumbricus rubella.
Several, but not all, annelid SSRP-like genes have gene structures that are identical to the molluscan counterpart: three exons that are separated by a phase 0 and phase 1 intron with the signal sequence located on exon 1 and with the mature peptide starting at the 3′ end of exon 2 and extending into exon 3, where the conserved WK-motif and disulfide loop is located (supplementary fig. S2, Supplementary Material online). This gene structure and the high similarity to the molluscan genes strongly suggest that these systems are orthologous. However, not all annelids strictly adhere to this canonical SSRP-like gene structure. In Dimorphilus gyrociliatus, an annelid with a miniaturized genome (Martín-Durán et al. 2021), two of the three SSRP-like genes lack the second intron, whereas Streblospio benedicti, an annelid with an unusually large genome (Zakas et al. 2022), has a third intron interrupting the signal sequence (supplementary fig. S2, Supplementary Material online).
Protostome SSRP-like Peptides Activate Receptors That Share Sequence Similarity with Chordate SSRP Receptors
To further investigate the function of protostome SSRP-like sequences, we synthesized the predicted mature peptides of Ct-SSRPL1 from the genome and whole-body transcriptome of the annelid Capitella teleta and Cb-SSRPL1 from the genome of C. betulinus and tested them at their predicted receptor pairs; an annelid candidate receptor from the transcriptome of the annelid C. teleta (Ct-SSTL1), and a predicted molluscan receptor from the genomic DNA assembly of the mollusc C. betulinus (Cb-SSTL1). These orphan receptors clustered with the family of GPCRs that contains human SSRPs in maximum likelihood tree analyses, suggesting that they are members of an invertebrate SSRP-like signaling system (fig. 4A; sequences and alignment can be found in supplementary files S9 and S10, Supplementary Material online). However, we note that the molluscan and annelid receptors with the highest sequence similarity to the chordate SS receptors are ASTC receptors (fig. 4A). This observation is further discussed below.
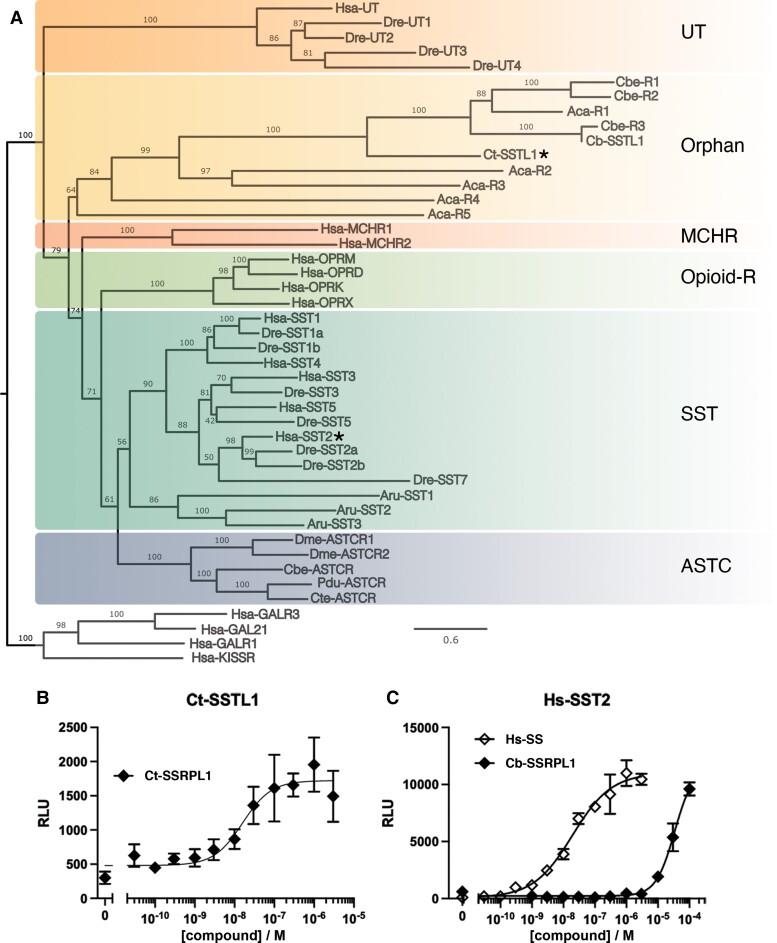
Fig. 4.
Ct-SSTL1 clusters with human SSRP and related receptors and is activated by Ct-SSRPL1. (A) Maximum likelihood tree analysis of human and zebrafish SSRP receptors (SST and UT) and closely related receptors (opioid receptors and melanin-concentrating hormone receptor [MCHR]), ASTC-R, echinoderm SS2 receptor, and orphan receptors from Conus betulinus, Aplysia californica, and Capitella teleta. The tree was rooted with human galanin and kisspeptin receptors. The receptors tested in B and C are denoted with a star. (B) Representative concentration–response curve of Ct-SSRPL1 at the Ct-SSTL1 receptor in the PRESTO-Tango assay (error bars represent SD). The experiment was performed in four independent repeats, and the mean EC50 value was 13.1 nM (pEC50 ± CI95 = 7.88 ± 0.65). (C) Representative concentration–response curve of human SS-14 and the C. betulinus nerve ring SSRP-like peptide (Cb-SSRPL1) at the human SS receptor 2 (SST2) in the PRESTO-Tango assay (error bars represent SD), showing an activation by Cb-SSRPL1 at micromolar concentrations. The experiment was performed in three independent repeats, and the mean EC50 value for Hs-SS was 4.0 nM (pEC50 ± CI95 = 8.40 ± 0.62). Species codes: Hsa: Homo sapiens, Dre: Danio rerio, Cbe: C. betulinus, Aca: A. californica, Cte: C. teleta, Aru: Asterias rubens, Dme: Drosophila melanogaster, Pdu: Platynereis dumerilli.
The C. teleta peptide, Ct-SSRPL1, showed strong activation of its predicted receptor, Ct-SSTL1, with an EC50 of 13.1 nM (pEC50 ± CI95 = 7.88 ± 0.65) (fig. 4B). This finding suggests that the Ct-SSTL1 receptor is indeed the functional target of this peptide. In contrast, the cone snail peptide, Cb-SSRPL1, showed no robust response at its predicted receptor (supplementary fig. S3, Supplementary Material online). To investigate if the lack of response was due to insufficient expression of the receptor on the cell surface, we performed an enzyme-linked immunosorbent assay (ELISA). The surface expression of both invertebrate receptors was very low, with Cb-SSTL1 between that of Ct-SSTL1 and human SST4 (supplementary fig. S3, Supplementary Material online), two receptors for which concentration–response curves can be readily obtained, despite their low expression. This suggests that, albeit being expressed at the cell surface, the cone snail receptor may not be functional in the mammalian expression system used, potentially due to an incorrect prediction of its cDNA sequence from the C. betulinus genome. Alternatively, the predicted receptor does not represent the biological target of Cb-SSRPL1. However, whereas not active at the cone snail receptor, when applied to the C. teleta receptor (Ct-SSTL1), Cb-SSRPL1 showed ∼50% activation at the highest concentration tested (at 30 µM, supplementary fig. S3, Supplementary Material online) and was also able to activate the human SST2 with micromolar potency (fig. 4C). Collectively, these findings indicate that, although we were not able to observe any activity of Cb-SSRPL1 at the predicted cone snail receptor, the native target receptor of this peptide likely shares some functional properties with human SST2 and the deorphanized C. teleta receptor.
SSRP-like Peptides are Present in Many but Not All Protostome Phyla
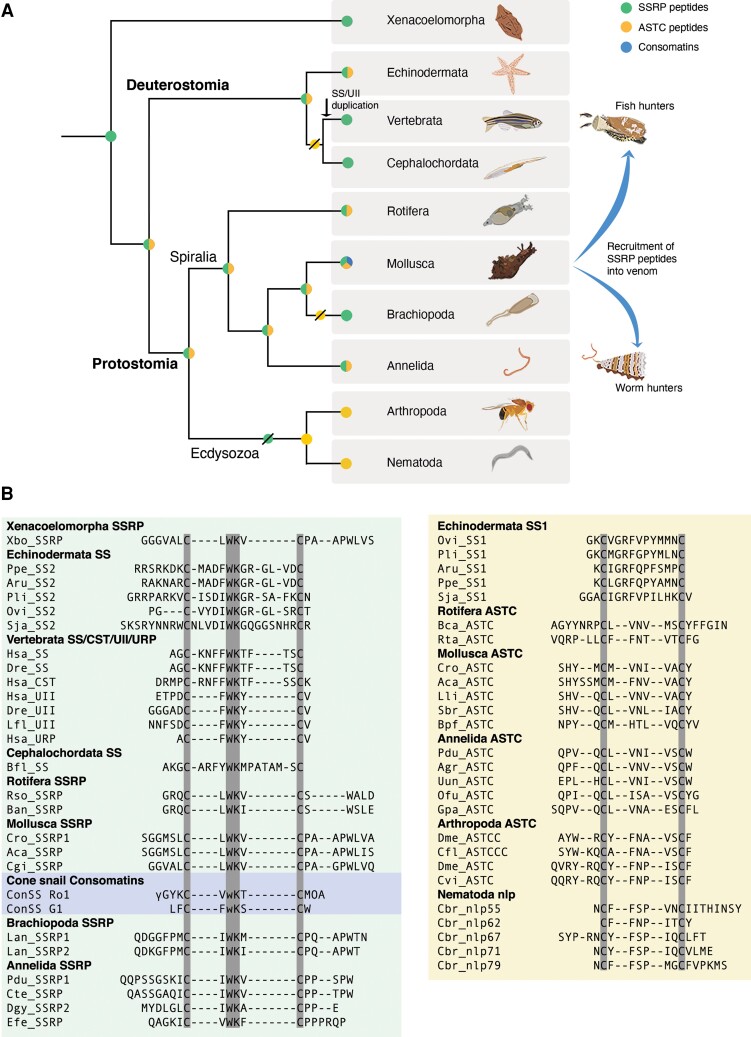
The discovery of SSRP-like sequences in molluscs and annelids prompted us to look for similar sequences in other metazoan phyla. This search led to the identification of SSRP-like sequences in three additional phyla: Brachiopoda, Rotifera, and Xenacoelomorpha (fig. 5 and supplementary files S1 and S11, Supplementary Material online, and where available, their conserved genes, supplementary fig. S2, Supplementary Material online). The phylogenetic position of xenacoelomorphs is still debated (Kapli and Telford 2020), but the current consensus is that xenacoelomorphs are bilaterians that diverged before the major Protostomia–Deuterostomia split (Cannon et al. 2016; Hejnol and Pang 2016; Jondelius et al. 2019).
Fig. 5.
Distribution and alignment of SSRP-like and ASTC genes in diverse bilaterian phyla. (A) Cladogram showing proposed origin, retention, and loss of the SSRP-like (green), ASTC (yellow), and consomatin (blue) gene across diverse animal phyla. SSRP-like genes were lost in ecdysozoa, whereas ASTC was lost in the chordate lineage. SSRP-like peptides were also likely lost in Nemerteans, Brachiopods, and Phoronids, where ASTC was also lost. (B) Alignment of SSRPs (left panel) and ASTC (right panel) from representative species of deuterostomes and protostomes. Residues common in all sequences are highlighted in gray. Peptide key: Somatostatin (SS), Somatostatin and related peptides (SSRP), Cortistatin (CST), Urotensin (UII), Urotensin-related peptide (URP), Allatostatin-C (ASTC). Species key: Echinodermata (Ppe: Patiria pectinifera, Aru: Asterias rubens, Pli: Paracentrotus lividus, Ovi: Ophionotus victoriae, Sja: Stichopus japonicus), Vertebrata (Hsa: Homo sapiens, Dre: Danio rerio, Lfl: Lampetra fluviatilis), Cephalochordata (Bfl: Branchiostoma floridae), Annelida (Pdu: Platynereis dumerelii, Cte: Capitella teleta, Dgy: Dinomorphus gyrociliatus, Agr: Amynthas gracilis, Efe: Eisenia fetida, Uun: Urechis unicinctus, Ofu: Owenia fusiformis, Gpa: Glossoscolex paulistus), Mollusca (Cro: Conus rolani, Aca: Aplysia californica, Cgi: Crassostrea gigas, Lli: Lithophaga lithophaga, Sbr: Scapharca broughtonii, Bpf: Biomphalaria pfeifferi), Brachiopoda (Lan: Lingula anatine), Rotifera (Rso: Rotaria sordida, Ban: Brachionus angularis), Xenacoelomorpha (Xbo: Xenoturbella bocki), Arthropoda (Dme: Drosophila melanogaster, Cfl: Camponotus floridanus, Cvi: Carabus violaceus), and Nematoda (Cbr: Caenorhabditis briggsae).
The group of SSRP-like peptides from other metazoan phyla all share a common WK-motif and cysteine loop (fig. 5B). In SSRPs from chordates and some echinoderms, an aromatic residue is typically found at the position preceding this motif (FWK or YWK), whereas aliphatic amino acids are found in SSRPs of the remaining echinoderm SSRPs and protostome SSRP-like peptides (IWK, VWK, or LWK). The cysteine loop size is conserved with four amino acids in UII, URP, and protostome SSRP-like peptides. Still, it possesses significant flexibility in SS/CST and echinoderm SS2 ranging from 10 to 15 residues. Protostome SSRP-like peptides are N- and C-terminally elongated compared with deuterostome peptides—commonly with multiple proline residues and a single tryptophan in the C-terminus and N-terminal glycine and serine residues.
SSRP-like peptides appear to be missing in ecdysozoans, a group of protostomes that includes nematode worms and arthropods, and are also absent in nemerteans and phoronids. A single nematode peptide, nlp-56, shows some similarities to SSRPs but contains an FK-motif rather than WK (McKay et al. 2022). However, the origin of nlp-56 is uncertain, and this peptide may not represent a member of the SSRP-like peptide family (this is also suggested in the clustering analysis, as shown in figure 6, where nlp-56 peptides form a solitary cluster). In conclusion, SSRP-like sequences are found across many but not all branches of bilaterians.
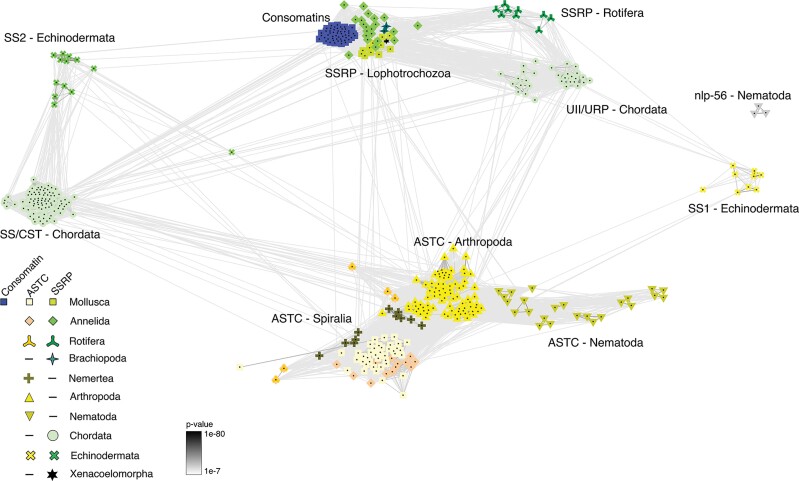
Fig. 6.
BLOSUM62 cluster map of SSRP, SSRP-like, and ASTC peptide precursors. The depicted notes represent individual precursor molecules, edges correspond to BLAST P-values > 1e−7. Nodes are labeled with phylum-specific colors indicated in the legend, but landmarks are also placed within the map for phyla or superphyla (Lopohotrochozoa: Mollusca, Annelida, Brachiopoda. Spiralia: Mollusca, Annelida, Rotifera, Nemertea).
Broad Presence of SSRP-like Peptides in Protostomes Challenges the SS-ASTC Hypothesis
The finding of SSRP-like peptides in several metazoan phyla challenges the hypothesis that ASTC is the protostome ortholog of SS as most of these animals also possess ASTC peptides. In addition to known ASTC sequences from protostomes, we identify an ASTC transcript in the nerve ring of C. rolani (Cr-ASTC) and the ASTC encoding gene from the genome of C. ventricosus (Cv-ASTC) (supplementary files S1 and S12, Supplementary Material online). An alignment of ASTC sequences shows that these peptides display significant variation with no strictly conserved motif (fig. 5B). However, most ASTC peptides have a common asparagine residue within the cysteine loop, and many of these peptides further display an FxP-motif within the cysteine loop. The latter is particularly true for ecdysozoan ASTCs and echinoderm SS1. Aliphatic amino acids are abundant in these peptides, and the first amino acid within the cysteine loop is either aliphatic (molluscan and annelid ASTC and all echinoderm SS1s) or aromatic (arthropod and nematode peptides). The size of the cysteine loop in ASTC is much more conserved compared with SSRP-like peptides and, in protostomes, always consists of six amino acids. The echinoderm SS1 peptide diverges with a length of 10 residues. As in previous analyses, we observe no clear similarities between the SSRP-like peptides and ASTCs besides the presence of a cysteine loop, suggesting that these peptides form two distinct families.
A comparison of the positions and phases of introns of SSRP-like peptides and ASTCs shows that all the genes investigated possess a common phase 0 intron in the proregion of the precursor (supplementary figs. S2 and S4, Supplementary Material online). Apart from this feature, there is a high level of variability in the number, position, and phases of the introns. Chordate SS/CST and echinoderm SS2 only possess this single phase 0 intron; UT/URP have two or three additional phase 1 introns; Echinoderm SS1 has one additional phase 1 intron, as seen in the SSRP-like genes, but at different sites. Spiralian ASTC genes have a phase 0 intron, but ecdysozoan ASTC genes have an additional phase 2 intron in the proregion. Thus, the gene structures show no distinguishing characteristics that can be used as an indicator of the evolutionary relationship between these peptides.
Finally, we performed a cluster-based analysis (CLANS) of the peptide precursors (fig. 6 and supplementary file S13, Supplementary Material online), which is a superior method to identify neuropeptide families across distant phyla compared with phylogenetic methods (Jékely 2013). We recovered three clear clusters with peptides from multiple phyla: (1) the protostome ASTCs; (2) lophotrochozoan (Mollusca, Annelida, and Brachiopoda) SSRP-like precursors, rotifer sequences, and chordate UII/URP peptides; and (3) chordate SS/CST and echinoderm SS2 sequences. Interestingly, a single echinoderm SS2 sequence from Stichopus japonicus, which encodes an unusually long SS2 peptide, is placed in an intermediate position between the other SS2 sequences and the ASTC cluster. Whereas the echinoderm SS1 peptides have recently been suggested to belong to the ASTC-family (Zhang et al. 2022) based, in part, on clustering analysis, we find that these peptides form as many connections to the UII/protostome SSRP-like cluster as to the ASTC cluster.
As observed for sequence alignments, clustering analysis shows a multitude of connections between SSRP-like sequences and chordate UII/URP precursors, clearly suggestive of common ancestry of SSRP-like peptides and members of the chordate SSRP family. The gene models are, however, inconclusive regarding the evolutionary relationships between SSRP, SSRP-like peptides, and ASTC.
Consomatins Evolve to Target the SSRP-like Signaling System in Prey
Collectively, our findings suggest that early in the evolution of cone snails or of an ancestral member of the conoidean superfamily of venomous snails, a duplicated SSRP-like gene that functions in neuroendocrine signaling in many bilaterians was recruited for expression in the venom gland. As cone snails radiated and diversified ecologically, the venom gene greatly proliferated and diverged in sequence.
The prey of ancestral cone snails were polychaete worms (Duda et al. 2001; Puillandre et al. 2014). Some cone snail lineages later shifted to prey on molluscs, and others on fish (Kohn 1956; Puillandre et al. 2014; Olivera et al. 2015). Having established the existence of SSRP-like sequences in these diverse prey taxa allowed us to test if the great diversification of consomatins occurs in response to biotic interactions with different prey. In other words, if consomatins target the SSRP-like receptor of prey species, then they would be expected to resemble the native (signaling) SSRP-like peptides of those prey species. To investigate this hypothesis, we performed principal component analysis (PCA) of vertebrate SSRPs (including fish and human sequences), signaling SSRP-like sequences from molluscs and annelids, as well as a large set of consomatins. PCA transforms a large group of correlated variables into a smaller set of uncorrelated variables (the principal components) that are more easily interpretable. Here, the scored variables included length, aromaticity, molecular weight, isoelectric point, and count of the 150 most common 1–3mers (see Materials and Methods). Since we established that the mature region of consomatins is almost exclusively located on a single exon (exon 3), we were able to utilize recent exon capture data of this exon from 247 distinct species of cone snails representing all major branches of the Conus phylogenetic tree (Phuong et al. 2019) (supplementary file S1, Supplementary Material online).
Using these data sets, we identified 502 consomatin genes of which 71 and 431 were from 30 fish-hunting and 131 were from worm-hunting species, respectively (supplementary file S14, Supplementary Material online). Consistent with transcriptome analysis, consomatin genes greatly proliferated in some lineages (e.g., Africonus, Virroconus, Dauciconus, and Lividoconus) and were seemingly lost in others (e.g., Textilia and Tesseliconus) (supplementary fig. S5, Supplementary Material online). Interestingly, consomatin genes could not be detected in any of the 22 snail-hunting species analyzed. This may point to a loss of this gene during the shift from worm- to snail-hunting behavior, which is believed to have only occurred once (Duda et al. 2001).
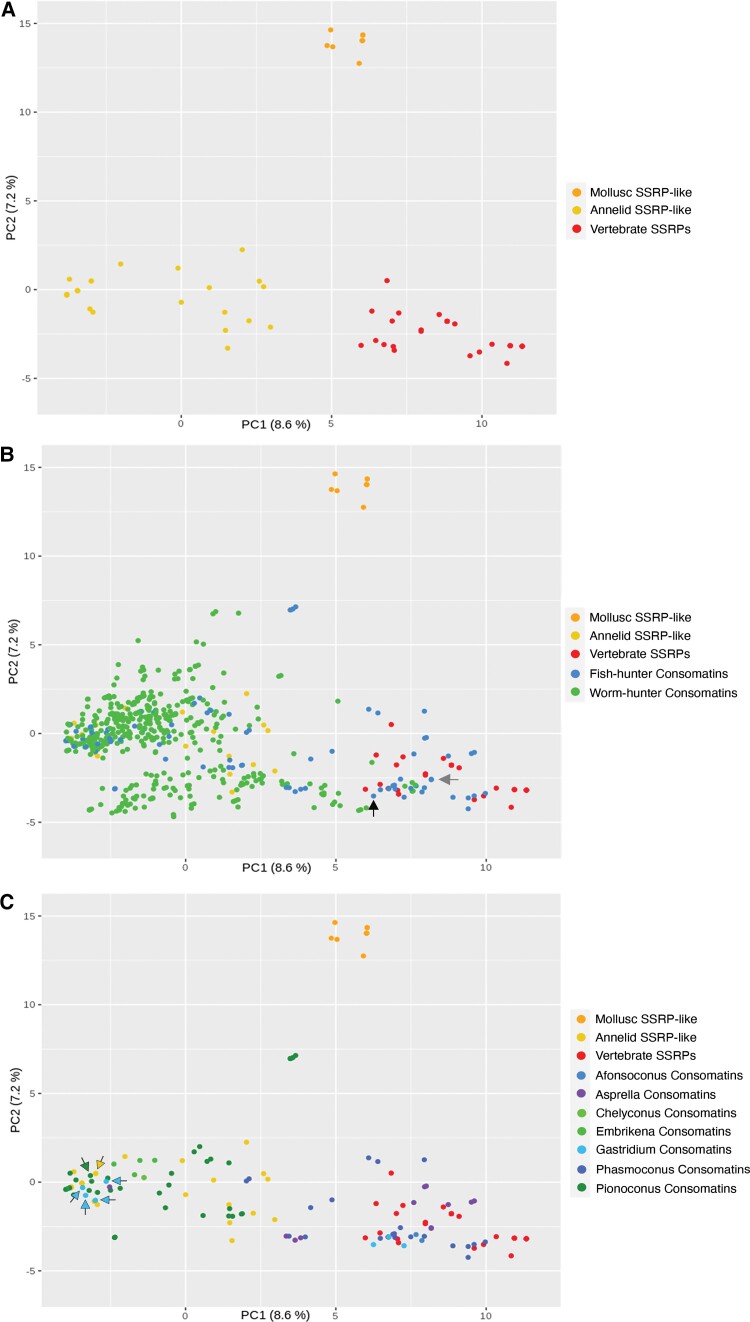
In analyses of the signaling SSRP-like gene alone, the first two principal components separate sequences from molluscs, annelids, and vertebrates into distinct clusters (fig. 7A). In analyses of the signaling SSRP-like peptides together with the consomatin sequences, consomatins noticeably group away from the molluscan signaling genes they derived from and toward the annelid and vertebrate sequences (fig. 7B). This pattern is consistent with the hypothesis that consomatins diverged to target SSRP-like receptors in prey. Indeed, consomatin sequences from fish hunters that were previously shown to selectively target different subtypes of the human SST receptors, closely group with vertebrate SSRPs, including those from fish and humans (see black and gray arrows for consomatin G1 and Ro1, respectively, fig. 7B). In contrast, consomatins identified from worm hunters cluster almost exclusively toward the annelid SSRP-like sequences, again suggesting that consomatins evolve to target the SSRP-like signaling system of specific prey. Notably, a small number of consomatins from the worm hunter, Conus glaucus, and several worm-hunting species of the Africonus clade are placed in the vertebrate region of the plot potentially indicating that these species may also prey on fish (as observed for some other worm hunters, Aman et al. 2015) or may use these toxins for defensive purposes.
Fig. 7.
Diversity of consomatins and their correlation with prey peptides. (A) PCA of characteristics of vertebrate SSRPs, annelid, and molluscan SSRP-like sequences. (B) Same analysis as in (A) including consomatins from fish- and worm-hunting cone snails. (C) PCA showing SSRPs and SSRP-like signaling peptides from vertebrates, annelids, and molluscs as well as consomatins from fish hunters separated into clades.
Except for these sequences, all other consomatins that group with vertebrate SSRPs are from fish-hunting species. This not only includes consomatins from C. rolani (Asprella clade) and C. geographus (Gastridium clade) known to activate the human SS receptors (Ramiro et al. 2022) but also sequences retrieved from other species belonging to these two clades (fig. 7C). Additionally, sequences from two other phylogenetically distinct clades of fish hunters, Phasmoconus and Afonsoconus, also group closely with vertebrate SSRPs. Thus, PCA analysis and comparative sequence analysis highlight how some, but not all fish hunters adapted the duplicated signaling SSRP-like gene to specifically target the receptors of their fish prey.
Notably, a large number of consomatins from fish hunters do not cluster with chordate SSRPs but with those from annelids. Most of these sequences belong to a single clade, Pionoconus, that comprises some of the best-studied cone snail species. Interestingly, this clade includes Conus magus, a species that is known to prey on annelids as a juvenile (Nybakken and Perron 1988), indicating that its consomatin sequence may have adapted to target the SSRP-like receptor in annelid prey, and possibly, that other members of this clade also prey on annelids as juveniles. Grouping of the C. magus peptide with SSRP-like sequences identified in annelids that are closely related to the prey of juvenile C. magus strongly supports this hypothesis (see green and yellow arrows in fig. 7C representing the C. magus and the annelid C. teleta sequence, respectively). Sequence alignment of C. magus and C. consors consomatins with annelid SSRP-like peptides clearly show, that these toxins share a higher similarity with the annelid peptides than the Conus signaling peptides they evolved from (supplementary fig. S6, Supplementary Material online).
Finally, we note that the position of consomatins from fish hunters in the PCA plot also appears to correlate with predation strategy. Species of the Pionoconus clade are all taser-and-tether hunters, a predation strategy characterized by a rapid immobilization of prey facilitated by the action of toxins that rapidly modulate ion channels of the nervous and locomotor systems (Olivera et al. 2015). In contrast, species of the Asprella clade, and potentially also the Phasmoconus clade, are ambush-and-assess hunters, a hunting strategy that is characterized by a very slow onset of action of their toxins (Ramiro et al. 2022). The third predation strategy, net-hunting, in which toxins are released into the water, has only been observed in Gastridium clade and suggested to be used by some Afonsoconus species (Ahorukomeye et al. 2019). The PCA analysis reveals a clear pattern: sequences derived from taser-and-tether hunters group with annelid SSRP-like sequences, whereas toxins from net or ambush-and-assess hunters group with vertebrate SSRPs (fig. 7C). PCA grouping according to the predation strategy, rather than phylogeny alone, is best illustrated by two closely related species of the Gastridium clade: consomatins from Conus obscurus, a taser-and-tether hunter (Olivera et al. 2015), closely groups with the annelid sequences (light blue arrows in fig. 7C), whereas consomatins from the net-hunter C. geographus group with vertebrate SSRPs. Based on this pattern it can be hypothesized that many, if not all, taser-and-tether hunters prey on annelids in their early developmental stages.
Discussion
Our investigations into the evolutionary origin of consomatins, a family of SS-mimicking conotoxins, exposed the existence of neuroendocrine SSRP-like peptides in cone snails and diverse protostome phyla. It is a well-described phenomenon that endogenous proteins can be recruited to the venom system (Fry et al. 2009; Safavi-Hemami et al. 2016; Sachkova et al. 2020). However, here, we show that these toxins can be used as missing links that can uncover a previously undescribed signaling system in diverse phyla.
The evolution of consomatins closely resembles that of cone snail venom insulins, toxins that induce dangerously low blood sugar by activating the insulin receptor of prey (Safavi-Hemami et al. 2016). Once recruited into venom, both cone snail insulins and consomatins experienced strong positive selection resulting in a hyperdiverse family of weaponized hormones. As observed for venom insulins, consomatins of fish hunters tend to resemble the signaling SSRPs of fish. Likewise, consomatins expressed in worm hunters tend to be similar to SSRP-like peptides found in annelids. This pattern is consistent with the hypothesis that many if not all venom insulins and consomatins are used in prey capture. Consequently, they are strongly selected to interact with receptors that regularly appear in a snail’s diet. Furthermore, because of their specialized role in predation, these weaponized hormones display certain properties that their signaling counterparts may not: unlike human insulin, weaponized insulins from Conus tulipa and C. geographus do not self-associate and can therefore act more rapidly than the human hormone (Menting et al. 2016; Ahorukomeye et al. 2019), and, unlike human somatostatin, consomatins from C. rolani and C. geographus are highly stable and selective for certain SS receptor subtypes (Ramiro et al. 2022). Thus, weaponized hormones evolved advantageous properties that can inform on the design of new drug leads for hormone-associated diseases such as neuroendocrine tumors and diabetes (Xiong et al. 2020, 2022; Ramiro et al. 2022).
Whereas investigating the evolutionary origin of consomatins, we discovered that the SSRP-like neuroendocrine peptides that gave rise to the consomatin family are present in several spiralian phyla (Mollusca, Annelida, Rotifera, and Brachiopoda) and Xenacoelomorphs. Several lines of evidence suggest that this previously undetected signaling system shares a common origin with chordate SSRPs.
First, all protostome sequences discovered here share the conserved WK motif known to be of critical importance for SST1–5 and UT activation (fig. 5) (Møller et al. 2003). We note that the residue preceding this motif is almost always aromatic (Phe or Tyr) in deuterostomes but aliphatic (Ile or Leu) in protostomes and many echinoderms. Furthermore, like chordate SSRPs, the predicted mature peptides are all located as a single copy at the C-terminus of the preprohormone. Second, all sequences contain two cysteine residues that can form a disulfide bond. Notably, the size of the cysteine loop is conserved in all protostomes (four residues including the WK motif) but varies in deuterostomes: whereas UII and URP share the loop size with protostome SSRP-like peptides, SS and CST display extended loops ranging from 10 to 15 residues in chordates and echinoderms (fig. 5). If the protostome SSRP-like and deuterostome SSRP systems are indeed related, following multiple rounds of whole-genome duplication in chordates, UII and URP retained the ancestral loop size, whereas SS and CST did not. We suggest that loop size correlates with structural stability but does not directly correlate with biological activity. Shorter loop sizes have been successfully introduced in therapeutic SS analogs to improve the in vivo half-life of these analogs, whereas retaining, and in some cases improving, the biological activity at some SS receptor subtypes (Pless 2005). Third, clustering analysis of precursor molecules showed multiple connections between SSRP-like sequences and chordate UII/URP precursors, supporting the hypothesis that these systems are related (fig. 6). Fourth, an orphan SSRP-like peptide receptor from the model annelid C. teleta, Ct-SSTL1, clusters with human UII, SS, MCH, and opioid receptors, demonstrating sequence similarities between the protostome SSRP-like receptors and the chordate receptor family that includes the SSRP receptors. Ct-SSTL1 can be potently activated by the C. teleta SSRP-like peptide, Ct-SSRPL1, unambiguously deorphanizing this receptor as the molecular target of an SSRPL peptide (fig. 4A and B). Fifth, when screened against the five subtypes of the human SS receptors, a molluscan SSRP-like peptide exhibited micromolar activity at the human SST2, suggesting that the ligands of the chordate SS receptor and the molluscan SSRPL receptors are functionally related (fig. 4C). Finally, the ability of consomatins from some fish-hunting cone snails to activate various subtypes of the human SS receptor (Ramiro et al. 2022) strongly suggests that the molluscan, and thereby protostome SSRPs-like peptides are not only related to chordate SSRPs but can be adapted to potently and selectively activate the chordate SS receptors.
The discovery of SSRP-like peptides in spiralians is striking as we also identified ASTC in most of these phyla. ASTC has been believed to be the ortholog of chordate SSRPs, but the cooccurrence with SSRP-like peptides challenges this hypothesis. The strong sequence similarities of the protostome SSRP-like peptides and chordate SSRPs suggest that this peptide family, not ASTC, is ortholog to chordate SSRPs. Additional evidence supporting orthology was provided by CLANS clustering analysis, which showed that SSRP-like peptides formed a well-connected cluster with the chordate SSRP peptide UII (fig. 6). The recovered receptor phylogeny revealed that the SSRP-like receptors indeed grouped with the SS and related receptors but also showed closer similarities between the chordate SS receptors and protostome ASTC receptors that formed a monophyletic group. We note that, whereas receptor similarity is strong evidence of orthologous signaling systems, it is not a perfect correlation. For example, a prolactin-releasing peptide (PrRP) identified in echinoderms (Yañez-Guerra et al. 2020) was clearly demonstrated to be orthologous to vertebrate PrRP and protostome short neuropeptide F. However, phylogenetic analyses of the respective receptors failed to recover the expected phylogenetic relationship of these receptors. As the authors suggest, this could be due to either fast diverging sequences reflected in long branches (something we also observe for SSRP and SSRP-like peptides as shown in fig. 4) or alternatively to multiple instances of gene duplication and loss (Yañez-Guerra et al. 2020). This might also be the case for the identified SSRP-like peptide receptors. It is also possible that there are additional receptors of these peptides, including both GPCRs and ligand-gated ion channels. Collectively, our findings strongly suggest that SSRP-like peptides are orthologs to chordate SSRPs and that ASTC peptides form a paralogous family.
The coexistence of SSRP-like peptides and ASTC in spiralians mirrors recent findings in the deuterostome phylum Echinodermata, where both an SSRP- and ASTC-like peptides, SS1 and SS2, coexist. This observation also led to the suggestion that SSRPs and ASTC are not orthologs but paralogs (Zhang et al. 2022). Here, we not only corroborate this finding but likely identify the proposed protostome orthologs of SSRPs. Based on these findings, we propose a new model of the origin and evolution of peptides of the SSRP, SSRPL, and ASTC families (fig. 5): at some point in early bilaterian evolution, before the protostome–deuterostome split, gene duplication formed the ancestral SSRP and ASTC genes. The ASTC gene was subsequently lost in the chordate lineage, whereas SSRP-like peptides were lost in ecdysozoans, including nematodes and arthropods. In cone snails or in an ancestral lineage of venomous molluscs, the SSRP-like gene duplicated and was recruited into venom, where it formed the highly diversified family of consomatins. During chordate evolution, an ancestral SSRP-like gene gave rise to the SS/CST and UII/URP families that form the vertebrate SSRP superfamily.
Whereas this is the most parsimonious model of evolution for this peptide superfamily, we acknowledge that there are alternative models that could explain some of these findings. It is, for instance, possible that the ancestral duplication that formed the SS/CST and UII/URP families did not happen in chordates but earlier in bilaterian evolution. In such a scenario, the protostome SSRP-like peptides would only be orthologous to the chordate UII/URP peptides. However, such a model would have to assume many additional cases of gene losses and is, consequently, a less likely model according to parsimony principles.
If, as we propose here, the protostome SSRP-like signaling peptides and consomatins are indeed orthologs of chordate SSRPs, some of these peptides could potentially be new ligands of chordate receptors known to be activated by members of the SSRP family, namely the SST1–5, the UT, the ghrelin receptor and the MRGPRX2. This may particularly be the case for members of the highly diversified consomatin family. Previously, we showed that some consomatins from fish hunters activate different subtypes of the human SS receptor and provide new drug leads for pain and neuroendocrine tumors (Ramiro et al. 2022). Here, we provide >500 additional consomatin sequences from diverse species of cone snails for future receptor target identification and biomedical exploration.
Additionally, we provide a set of new SSRP-like signaling sequences from various bilaterian phyla, including some invertebrate animal models such as Platynereis dumerilli. Based on several observations, we believe that this signaling system not only exists in these animals but also plays important physiological roles. First, SSRP-like sequences were recovered from several transcriptome datasets demonstrating that these genes are actively expressed. Second, all these sequences have an N-terminal signal sequence and are secreted to likely function in neuroendocrine signaling events. Third, the broad presence and targeted use of consomatins by worm-hunting cone snails strongly suggest an important functional role of the SSRP-like signaling system in annelids. Finally, the existence and expression of an annelid SS-like receptor (Ct-SSTL1) that can be potently activated by the C. teleta SSRP-like 1 peptide further support an important physiological role of this signaling system.
One member of the SSRP-like family, namely, apUII from A. californica, has already been shown to be secreted in the nervous system, where it exerts inhibitory effects on motor programs involved in feeding (Romanova et al. 2012). Recently, Zhang et al. (2022) suggested that loss of ASTC in chordates and SSRPs in ecdysozoans might be due to functional redundancy of the peptides as inhibitors of physiological processes. The preliminary finding that SSRP-like peptides exhibit inhibitory functions in A. californica and the well-described inhibitory effect of ASTCs suggest that this is not the case in molluscs. Future investigation of the function and expression patterns of SSRP-like peptides and ASTC in spiralian species are likely to further elucidate the biological role of these signaling peptides. Similarly, elucidating the function of SSRP-like peptides in xenacoelomorphs may provide fundamental insight into the biological role of the ancestral SSRP-like signaling system in animals which may ultimately expand our current understanding of the diverse physiological roles of SSRP in humans.
Finally, we provide a new paradigm for the discovery of novel signaling systems based on peptide toxins that specifically evolved to target such systems and their use in homology detection. With the growing availability of large peptide toxin databases by transcriptome and genome sequencing, we anticipate that this approach will be used to identify other important signaling systems in the future.
Materials and Methods
Transcriptome Sequencing
Specimens of C. textile, C. striatus, and C. imperialis were collected in Oahu, HI, USA. Conus rolani and C. geographus specimens were collected in Cebu, Philippines under the Department of Agriculture—Bureau of Fisheries and Aquatic Resources-issued gratuitous permit no. GP-0084-15. Total RNA was extracted from the circumoesophageal nerve rings, salivary gland, venom bulbs, and foot using the Direct-zol RNA extraction kit (Zymo Research), with on-column DNase treatment, according to the manufacturer’s instructions. Library preparation and sequencing were performed by the University of Utah High Throughput Genomics Core Facility as previously described (Ahorukomeye et al. 2019). Briefly, paired-end sequencing was performed on an Illumina HiSeq2500 or NovaSeq 6000 instrument. Adapter trimming of demultiplexed raw reads was performed using fqtrim (v0.9.4), followed by quality trimming and filtering using prinseq-lite (Schmieder and Edwards 2011). Error correction was performed using the Bbnorm ecc tool, part of the Bbtools package (open-source software, Joint Genome Institute). Trimmed and error-corrected reads were assembled using Trinity (version 2.2.1) (Grabherr et al. 2011; Haas et al. 2013) with a k-mer length of 31 and a minimum k-mer coverage of 10. Expression levels were calculated as transcripts per million (tpm). The tpm values were calculated using the RSEM program (Li and Dewey 2011). Raw reads have been deposited to the NCBI short sequence read archive (SRA) under accession number SRR16493587-99 (supplementary file S1, Supplementary Material online).
Transcriptome Assemblies and Analyses
Raw sequencing data of publicly available cone snail venom gland transcriptomes were retrieved from the NCBI, DDBJ, and CNGB sequence repositories and assembled using the same methods as described for tissues sequenced in this study. The list of all transcriptomes used in this study is provided in supplementary file S1, Supplementary Material online. Consomatin sequences were identified from these data sets based on tblastn with sequences from Ramiro et al. (2022) using an e-value of 0.1. Sequences with the C-terminal C.WK.C-motif were identified as consomatins.
Gene Structure Analysis
The location, size, and phases of introns were identified using the online version of Splign (Kapustin et al. 2008). Signal sequences were calculated using either command-line or online versions of SignalP 5.0 (Almagro Armenteros et al. 2019).
Phylogenetic Analysis
Multiple sequence alignments of protein sequences were performed using MAFFT v 7.310 (Katoh and Standley 2013). The tree in figure 3A was constructed using a Bayesian analysis of phylogeny with Mr. Bayes v.3.2.6 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) and mixed amino acid models. The analysis was performed with two runs of four Markov chains for 1,000,000 generations, at which point the two chains had converged. The first 25% of samples were discarded as burn-in. The tree uses the signaling SSRP-like precursor from A. californica as an outgroup. The alignment used for the analysis can be found in supplementary file S4, Supplementary Material online. The tree in figure 4A was constructed using maximum likelihood with the program IQ-TREE v1.6.1 (Nguyen et al. 2015; Chernomor et al. 2016) run on a single thread on an M1 chip with eight cores @ 3.2 GHz with seed 889704. The program chose the model of evolution according to the Bayesian information criterion, which was LG + F+I + G4. The tree was rooted with human galanin and kisspeptin receptors, which form a known outgroup of the SSRP receptors. The receptor sequences can be found in supplementary file S9, Supplementary Material online and the alignment is given in supplementary file S10, Supplementary Material online.
Selection Analysis
Signaling SSRP-like genes were extracted from the nerve ring transcriptomes of C. rolani, C. geographus, C. textile, and C. imperialis and from the genomes of C. ventricosus and C. betulinus. Twenty random toxin genes were obtained from NCBI GenBank (supplementary file S2, Supplementary Material online). The three groups of the toxin genes and the two signaling genes were all analyzed separately and identically as follows: first, the amino acid sequences were aligned using ClustalO v 1.2.4 (Sievers et al. 2011). The amino acid alignment was then converted to the corresponding nucleotide alignment using program PAL2NAL (Suyama et al. 2006) with the flag “-nogap”. The synonymous and nonsynonymous substitutions were calculated from this codon alignment with program codeml (v4.9) from PAML (Yang 2007) under the F3 × 4 model of evolution. We calculated the likelihood of several different models of evolution, including pairwise comparisons of all sequences, a beta distribution of dN/dS between 0 and 1 (M7), and a beta distribution of dN/dS with positive selection at some sites (M8). Conventional likelihood ratio tests were used by comparing the M7 and M8. The sites with positive selection were estimated with the Bayes empirical Bayes analysis.
The conservation scores for the different groups (signaling SSRP-like and consomatins) were calculated with the program rate4site (Mayrose et al. 2004) using an alignment generated by ClustalO v 1.2.4 and a maximum likelihood tree generated by IQ-TREE using the same alignment. The conservation score for each site is visualized with a sliding window of five amino acids in the figure.
Clustering Analysis
To investigate the relationship between chordate SSRPs, echinoderm SS1 and SS2, and protostome ASTC precursors, we generated a database of these peptides from representative species belonging to Chordata, Echinodermata, Mollusca, Annelida, Brachiopoda, Rotifera, Nemertea, Nematoda, and Arthropoda (the sequences can be seen in supplementary file S13, Supplementary Material online). We performed a clustering-based approach using CLANS (Frickey and Lupas 2004). CLANS randomly initialize the individual sequences as nodes and perform an all-against-all BLASTP. The negative logarithm of the resulting BLAST P-values is used as an attractive force in concert with a uniform repulsive force between the nodes. We used the BLOSUM62 scoring matrix, where we extracted e-values up to 1e−2 using the web tool https://toolkit.tuebingen.mpg.de/tools/clans. The clustering was first done in 3D and subsequently collapsed to 2D for >200,000 rounds, at which point the clustering had converged. The resulting clans file is supplied in supplementary file S13, Supplementary Material online. In the figure, only P-values below 1e−7 were kept.
Principal Component Analysis
The SRA-data sets of cone snails from Bioproject PRNJ526781 were downloaded from NCBI in September 2021 with fastq-dump 2.8.2 (SRA toolkit development team). The individual data set was then preprocessed with fastp v.0.20.1 (Chen et al. 2018) to filter out low-quality reads and were converted to fasta-format with EMBOSS seqret v. 6.6.0.0 (Madeira et al. 2019). To identify reads encoding consomatins, we used the toxin sequences from earlier identified consomatins as queries in a tblastn search against the database using an e-value of 10. The hits were then assembled using Trinity v.2.13.2 (Grabherr et al. 2011; Haas et al. 2013) as single reads. The generated contigs were translated in all reading frames using EMBOSS getorf v.6.6.0.0 (Madeira et al. 2019). The putative toxins were extracted with the regular expression “[^C]{0,2}C[^C]WK[^C]C[^RKC]{0,6}” and only contigs with 5-fold coverage were kept for the subsequent analyses. The pool of extracted toxins from the SRA-data were used as tblastn queries in two additional iterations of toxin identification.
In addition to the identified toxins, we also used signaling SSRP-like peptides from annelids, molluscs, and vertebrates in the analysis. These were modified to only have two amino acid residues preceding the cysteine-ring to match the structure of the toxin sequences in this analysis.
We extracted the length, molecular weight, aromaticity, isoelectric point, and the count for the 150 most common 1–3mers across all the sequences. Principal components were calculated using the sklearn library v 0.24.1 (Pedregosa et al. 2011) in Python 3.6.9 on data scaled to remove the mean and unit variance. The two first principal components were used to visualize the sequences using R 3.6.3 with the library tidyverse 1.3.1 (Wickham et al. 2019). PCA loadings can be seen in supplementary file S15, Supplementary Material online.
Peptide Synthesis
Ct-SSRPL1 predicted from the whole-body transcriptome of the annelid C. teleta was synthesized by solid-phase peptide synthesis in 0.1 mmol scale using preloaded Fmoc-Trp Tentagel R HMPA resin from Rapp Polymere (Tuebingen, Germany). The sequence was H-ZASSGAQICIWKVCPPTPW-OH, where Z represents a pyroglutamic acid and Cys9 and Cys14 are connected by a disulfide bond. Fmoc-protected amino acids, coupling reagents, and solvents used for the synthesis were purchased from Iris Biotech (Marktredwitz, Germany). The synthesis was performed on a Syro I instrument from Biotage (Uppsala, Sweden). Coupling conditions were room temperature (RT) for 2 × 120 min using 5.2 equiv amino acids, 4.7 equiv N-[(1H-benzotriazol-1-yl)(dimethylamino)methylene]-N-methylmethanaminium hexafluorophosphate N-oxide, 5.2 equiv 1-hydroxy-7-azabenzotriazol, and 8 equiv N,N-diisopropylethylamine in dimethylformamide (DMF) relative to resin. Deprotection was performed at RT with 40% piperidine in DMF for 3 min followed by 20% piperidine in DMF for 15 min. Washing steps were with 2 × N-methyl-2-pyrrolidone, 1 × dichloromethane (DCM), and 1 × DMF. After completion of the peptide assembly, the peptidyl resin was washed with 3 × DCM and dried. The peptide was released from ∼0.05 mmol resin using a mixture (3 ml) of 95% trifluoroacetic acid (TFA), 2.5% triethylsilyl, and 2.5% water over 2.5 h. Cold diethylether (15 ml at −20 °C) was added, and the mixture was further cooled to −85 °C for 30 min before the peptide was isolated by centrifugation. The peptide was redissolved in a mixture of acetic acid, water, and acetonitrile (ACN) in volume ratios of 1:10:4 and freeze-dried to remove unwanted carboxylation of Trp. The freeze-dried compound was resuspended in 1% vol/vol TFA and water. Phosphate buffer (0.1 M) and appr. ACN (3 ml) was added to 10 ml to this mixture. The pH was adjusted to 6 using 1 M hydrochloride acid. Dimethyl sulfoxide (1.0 ml) was added (10%, V/V), and the mixture was stirred for 20 h at RT to allow disulfide bond formation. The mixture was filtered and directly purified on a Dionex Ultimate 3000 HPLC system (Thermo Fisher, Waltham, USA) using a Luna C18(2) column from Phenomenex (Torrance, USA, 5 μm, 100 Å, 250 × 10 mm). A gradient of 5–100% ACN in water/0.1% formic acid was applied. The product was isolated and lyophilized by freeze-drying to yield ∼3.5 mg (0.0017 mmol). The final product was analyzed by liquid chromatography (LC)–mass spectrometry (MS) on a Dionex Ultimate 3000 ultrahigh-performance LC system from Thermo Fisher connected to an Impact HD mass spectrometer (Bruker, Bremen, Germany). The calculated monoisotopic MH+1 of the purified product was 2052.97 (detected monoisotopic MH+1: 2053.00) (supplementary fig. S7A, Supplementary Material online).
Cr-SSRPL1 predicted from the nerve ring transcriptome of the cone snail C. rolani was custom synthesized by GenScript (Leiden, Netherlands). The sequence was H-SGGMSLCLWKVCPAAPWLVS-OH, where Cys7 and Cys12 are connected by a disulfide bond. The folded peptide was purified to >95% by HPLC analysis and verified by MS. The calculated monoisotopic MH+1 of the purified product was 2,103.02 (detected monoisotopic MH+1: 2,103.39) (supplementary fig. S7B, Supplementary Material online).
GPCR Assays
Unless otherwise stated, all chemicals used for cell culture were purchased from Sigma–Aldrich (Burlington, MA, USA), and all media and buffers were purchased from ThermoFisher Scientific (Waltham, MA, USA). All predicted receptors were codon-optimized for mammalian expression and synthesized de novo by Twist Biosciences (San Francisco, CA, USA) into the PRESTO-Tango construct backbone at the same position as other GPCRs in this collection (Addgene, Watertown, MA, USA). The PRESTO-Tango receptor assay was performed essentially as previously described (Kroeze et al. 2015).
Briefly, HTLA cells (a kind gift from Prof. Hans Bräuner-Osborne) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Biowest, Nuaillé, France), 100 U/ml penicillin, 100 µg/ml streptomycin, 100 µg/ml hygromycin B, and 2 µg/ml puromycin (growth medium) in a water-jacketed incubator maintaining 5% CO2 and 37 °C. The cells were detached by washing with Ca2+- and Mg2+-free phosphate-buffered saline (PBS) (Substrate Department, University of Copenhagen, DK). One million cells per well were seeded in six-well plates in 2 ml of growth medium on day 1 and transfected with PolyFect (Qiagen, Düsseldorf, DE, Germany) on day 2 using the manufacturer’s protocol (changing the medium to growth medium without hygromycin B and puromycin prior to transfection). On day 3, 15,000 transfected cells per well were seeded in poly-d-lysine-coated white clear-bottomed 384-well plates (Corning New York, NY, USA) in 40 µL of DMEM supplemented with 1% dialyzed FBS (Thermo Fisher) (assay medium) and incubated overnight. On day 4, the medium was changed to 40 μL of fresh assay medium. 10 μL of Hanks’ balanced salt solution supplemented with 1 mM CaCl2, 1 mM MgCl2, 20 mM (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH adjusted to 7.4 with 10 M NaOH (assay buffer), and 0.1% bovine serum albumin (BSA) and sterile-filtered (0.2 μM) was added with indicated compounds at 5 × final concentration, and the cells were incubated overnight. On day 5, the compounds were removed, and the cells were incubated for 20 min at RT in the dark in 20 µL of assay buffer with 0.01% pluronic F-68 (Thermo Fisher) and 5% BrightGlo (Promega, Madison, WI, USA) before reading luminescence on a Molecular Devices SpectraMax iD5 with a 1 s integration time. Data were analyzed using GraphPad Prism 9 (San Diego, CA, USA). Concentration–response curves were fitted to a four-parameter dose–response curve {}, from which EC50 values were extracted. All assays were repeated at least three times independently, and mean EC50 values were calculated from the pEC50 values.
Enzyme-Linked Immunosorbent Assay
Unless otherwise stated, all chemicals used for cell culture were from Sigma–Aldrich. Cells were maintained and transfected as described for “GPCR assays” until day 3, where 50,000 cells per well were seeded in 100 µL of assay medium in black, clear-bottomed 96-well plates (PerkinElmer, Waltham, MA, USA) using only the 60 inner wells. On day 4, the cells were washed with 200 µL per well of PBS, fixated in 125 µL per well of 4% paraformaldehyde in PBS for 20 min on ice, washed twice with 200 µL per well of PBS, and blocked in 0.1% BSA in PBS (blocking buffer) for 30 min at RT. Cells were then incubated with 75 µL per well of 2–5 mg/L M1 mouse antiflag antibody (Sigma, F3040) in blocking buffer for 1 h at RT, washed four times with 200 µL per well of PBS, followed by a 30-min incubation with 75 µL per well of 200 µg/L horseradish peroxidase-conjugated goat antimouse secondary antibody (Sigma AP124P) in blocking buffer, and washed four times with 200 µL per well of PBS. About 60 µL of PBS per well was added, Pierce ELISA Femto Maximum Sensitivity Substrate was mixed 1:1 with enhancer solution (Thermo Fisher Scientific), and 20 µL per well of this mix was added. The plate was shaken softly for 60 s before reading luminescence on a Molecular Devices SpectraMax iD5 with 1 s integration time. Six technical replicates were used, and all experiments were performed in two independent repeats. Data were analyzed using GraphPad Prism 9 and presented as background subtracted (background being cells transfected with an empty pcDNA3.1[+] vector, showing ∼5,000–8,000 counts), and normalized to the highly expressing SST2 (∼40,000 counts).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors thank Kasper Kildegaard Sørensen for help with peptide synthesis, Noel Saguil for assistance with field collections, Maren Watkins for insightful discussions, and the High Throughput Genomics Core Facility at the University of Utah, USA for library preparation and transcriptome sequencing. This work was supported by a Villum Young Investigator Grant (19063 to H.S.-H.), a Starting Grant from the European Commission (ERC-Stg 949830 to H.S.-H.), and a National Institutes of Health Grant (GM048677 to B.M.O.).
Data availability
All accession numbers and sequences used in this study are available in the supplementary information.
References
- Ahorukomeye P, Disotuar MM, Gajewiak G, Karanth S, Watkins M, Robinson SD, Flórez Salcedo P, Smith NA, Smith BJ, Schlegel A, et al. . 2019. Fish-hunting cone snail venoms are a rich source of minimized ligands of the vertebrate insulin receptor. eLife 8(Feb):e41574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman JW, Imperial JS, Ueberheide B, Zhang M-M, Aguilar M, Taylor D, Watkins M, Yoshikami D, Showers-Corneli P, Safavi-Hemami H, et al. . 2015. Insights into the origins of fish hunting in venomous cone snails from studies of Conus tessulatus. Proc Natl Acad Sci U S A 112(16):5087–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armenteros JJA, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. Signalp 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 37(4):420–423. [DOI] [PubMed] [Google Scholar]
- Brazeau P, Vale W, Burgus R, Ling N, Butcher M, Rivier J, Guillemin R. 1973. Hypothalamic polypeptide that inhibits the secretion of immunoreactive pituitary growth hormone. Science 179(4068):77–79. [DOI] [PubMed] [Google Scholar]
- Cannon JT, Vellutini BC, Smith J, Ronquist F, Jondelius U, Hejnol A. 2016. Xenacoelomorpha is the sister group to nephrozoa. Nature 530(7588):89–93. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one fastq preprocessor. Bioinformatics 34(17):i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 65(6):997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon JM, Bondareva V, Rusakov Y, Plisetskaya EM, Mynarcik DC, Whittaker J. 1995. Characterization of insulin, glucagon, and somatostatin from the river lamprey, lampetra fluviatilis. Gen Comp Endocrinol. 100(1):96–105. [DOI] [PubMed] [Google Scholar]
- Cruz LJ, de Santos V, Zafaralla GC, Ramilo CA, Zeikus R, Gray WR, Olivera BM. 1987. Invertebrate vasopressin/oxytocin homologs. Characterization of peptides from Conus geographus and Conus straitus venoms. J Biol Chem. 262(33):15821–15824. [PubMed] [Google Scholar]
- Deghenghi R, Papotti M, Ghigo E, Muccioli G. 2001. Cortistatin, but not somatostatin, binds to growth hormone secretagogue (ghs) receptors of human pituitary gland. J Endocrinol Invest. 24(1):RC1–RC3. [DOI] [PubMed] [Google Scholar]
- Duda TF J, Kohn AJ, Palumbi SR. 2001. Origins of diverse feeding ecologies within conus, a genus of venomous marine gastropods. Biol J Linnean Soc. 73(4):391–409. [Google Scholar]
- Elphick MR, Mirabeau O, Larhammar D. 2018. Evolution of neuropeptide signalling systems. J Exp Biol. 221(Pt 3):jeb151092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickey T, Lupas A. 2004. Clans: a java application for visualizing protein families based on pairwise similarity. Bioinformatics 20(18):3702–3704. [DOI] [PubMed] [Google Scholar]
- Fry BG, Roelants K, Champagne DE, Scheib H, Tyndall JDA, King GF, Nevalainen TJ, Norman JA, Lewis RJ, Norton RS, et al. . 2009. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genom Hum Genet. 10:483–511. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. . 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmelikhuijzen CJP, Hauser F. 2012. Mini-review: the evolution of neuropeptide signaling. Regul Pept. 177(Suppl):S6–S9. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. . 2013. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. 8(8):1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejnol A, Pang K. 2016. Xenacoelomorpha's significance for understanding bilaterian evolution. Curr Opin Genet Dev. 39:48–54. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17(8):754–755. [DOI] [PubMed] [Google Scholar]
- Jékely G. 2013. Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc Natl Acad Sci U S A. 110(21):8702–8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jondelius U, Raikova OI, Martinez P. 2019. Xenacoelomorpha, a key group to understand bilaterian evolution: morphological and molecular perspectives. In: Pontarotti P, editor. Evolution, origin of life, concepts and methods. Cham: Springer International Publishing. p. 287–315. [Google Scholar]
- Kapli P, Telford MJ. 2020. Topology-dependent asymmetry in systematic errors affects phylogenetic placement of ctenophora and xenacoelomorpha. Sci Adv. 6(50):eabc5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapustin Y, Souvorov A, Tatusova T, Lipman D. 2008. Splign: algorithms for computing spliced alignments with identification of paralogs. Biol Direct. 3(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. Mafft multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn AJ. 1956. Piscivorous gastropods of the genus conus. Proc Natl Acad Sci U S A. 42(3):168–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer SJ, Toschi A, Miller CA, Kataoka H, Quistad GB, Li JP, Carney RL, Schooley DA. 1991. Identification of an allatostatin from the tobacco hornworm Manduca sexta. Proc Natl Acad Sci U S A. 88(21):9458–9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreienkamp H-J, Larusson HJ, Witte I, Roeder T, Birgül N, Hönck H-H, Harder S, Ellinghausen G, Buck F, Richter D. 2002. Functional annotation of two orphan g-protein-coupled receptors, drostar1 and -2, from drosophila melanogaster and their ligands by reverse pharmacology*. J Biol Chem. 277(42):39937–39943. [DOI] [PubMed] [Google Scholar]
- Kroeze WK, Sassano MF, Huang X-P, Lansu K, McCorvy JD, Giguère PM, Sciaky N, Roth BL. 2015. Presto-tango as an open-source resource for interrogation of the druggable human gpcrome. Nat Struct Mol Biol. 22(5):362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. Rsem: accurate transcript quantification from rna-seq data with or without a reference genome. BMC Bioinform. 12(1):323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Barghi N, Lu A, Fedosov AE, Bandyopadhyay PK, Lluisma AO, Concepcion GP, Yandell M, Olivera BM, Safavi-Hemami H. 2017. Divergence of the venom exogene repertoire in two sister species of turriconus. Genome Biol Evol. 9(9):2211–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, et al. . 2019. The EMBL-EBI search and sequence analysis tools apis in 2019. Nucleic Acids Res. 47(W1):W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair GR, Halton DW, Shaw C, Maule AG. 2000. The neuropeptide F (NPF) encoding gene from the cestode, Moniezia expansa. Parasitology 120(1):71–77. [DOI] [PubMed] [Google Scholar]
- Martín-Durán JM, Vellutini BC, Marlétaz F, Cetrangolo V, Cvetesic N, Thiel D, Henriet S, Grau-Bové X, Carrillo-Baltodano AM, Gu W, et al. . 2021. Conservative route to genome compaction in a miniature annelid. Nat Ecol Evol. 5(2):231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez V. 2013. Somatostatin. In: Kastin AJ, editor. Handbook of biologically active peptides. Chap. 180. Academic Press. p. 1320–1329. [Google Scholar]
- Mayrose I, Graur D, Ben-Tal N, Pupko T. 2004. Comparison of site-specific rate-inference methods for protein sequences: empirical bayesian methods are superior. Mol Biol Evol. 21(9):1781–1791. [DOI] [PubMed] [Google Scholar]
- McKay FM, McCoy CJ, Crooks B, Marks NJ, Maule AG, Atkinson LE, Mousley A. 2022. In silico analyses of neuropeptide-like protein (NLP) profiles in parasitic nematodes. Int J Parasitol. 52(1):77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menting JG, Gajewiak J, MacRaild CA, Chou DH-C, Disotuar MM, Smith NA, Miller C, Erchegyi J, Rivier JE, Olivera BM, et al. . 2016. A minimized human insulin-receptor-binding motif revealed in a Conus geographus venom insulin. Nat Struct Mol Biol. 23(10):916–920. [DOI] [PubMed] [Google Scholar]
- Mirabeau O, Joly J-S. 2013. Molecular evolution of peptidergic signaling systems in bilaterians. Proc Natl Acad Sci U S A. 110(22):E2028–E2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller LN, Stidsen CE, Hartmann B, Holst JJ. 2003. Somatostatin receptors. Biochim Biophys Acta. 1616(1):1–84. [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. Iq-tree: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken J, Perron FE. 1988. Ontogenetic change in the radula of Conus magus (gastropoda). Mar Biol. 98:239–242. [Google Scholar]
- Olivera BM, Seger J, Horvath MP, Fedosov AE. 2015. Prey-capture strategies of fish-hunting cone snails: behavior, neurobiology and evolution. Brain Behav Evol. 86(1):58–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong KL, Wong LYF, Cheung BMY. 2008. The role of urotensin ii in the metabolic syndrome. Peptides 29(5):859–867. [DOI] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. . 2011. Scikit-learn: machine learning in python. J Mach Learn Res. 12(Oct):2825–2830. [Google Scholar]
- Phuong MA, Alfaro ME, Mahardika GN, Marwoto RM, Prabowo RE, von Rintelen T, Vogt PWH, Hendricks JR, Puillandre N. 2019. Lack of signal for the impact of conotoxin gene diversity on speciation rates in cone snails. Syst Biol. 68(5):781–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless J. 2005. The history of somatostatin analogs. J Endocrinol Invest. 28(11 Suppl International):1–4. [PubMed] [Google Scholar]
- Puillandre N, Bouchet P, Duda TF Jr, Kauferstein S, Kohn AJ, Olivera BM, Watkins M, Meyer C. 2014. Molecular phylogeny and evolution of the cone snails (gastropoda, conoidea). Mol Phylogenet Evol. 78:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro IBL, Bjørn-Yoshimoto WE, Imperial JS, Gajewiak J, Salcedo PF, Watkins M, Taylor D, Resager W, Ueberheide B, Bräuner-Osborne H, et al. . 2022. Somatostatin venom analogs evolved by fish-hunting cone snails: From prey capture behavior to identifying drug leads. Sci Adv. 8(12):eabk1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robas N, Mead E, Fidock M. 2003. Mrgx2 is a high potency cortistatin receptor expressed in dorsal root ganglion. J Biol Chem. 278(45):44400–44404. [DOI] [PubMed] [Google Scholar]
- Romanova EV, Sasaki K, Alexeeva V, Vilim FS, Jing J, Richmond TA, Weiss KR, Sweedler JV. 2012. Urotensin ii in invertebrates: from structure to function in Aplysia californica. PLoS One 7(11):e48764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574. [DOI] [PubMed] [Google Scholar]
- Sachkova MY, Landau M, Surm JM, Macrander J, Singer SA, Reitzel AM, Moran Y. 2020. Toxin-like neuropeptides in the sea anemone Nematostella unravel recruitment from the nervous system to venom. Proc Natl Acad Sci U S A. 117(44):27481–27492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavi-Hemami H, Lu A, Li Q, Fedosov AE, Biggs J, Showers Corneli P, Seger J, Yandell M, Olivera BM. 2016. Venom insulins of cone snails diversify rapidly and track prey taxa. Mol Biol Evol. 33(11):2924–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics (Oxford, England) 27(6):863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semmens DC, Mirabeau O, Moghul I, Pancholi MR, Wurm Y, Elphick MR. 2016. Transcriptomic identification of starfish neuropeptide precursors yields new insights into neuropeptide evolution. Open Biol. 6(2):150224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. . 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol Syst Biol. 7(1):539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. Pal2nal: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server issue):W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel D, Yañez-Guerra LA, Franz-Wachtel M, Hejnol A, Jékely G. 2021. Nemertean, brachiopod, and phoronid neuropeptidomics reveals ancestral spiralian signaling systems. Mol Biol Evol. 38(11):4847–4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tostivint H, Joly L, Lihrmann I, Parmentier C, Lebon A, Morisson M, Calas A, Ekker M, Vaudry H. 2006. Comparative genomics provides evidence for close evolutionary relationships between the urotensin ii and somatostatin gene families. Proc Natl Acad Sci U S A. 103(7):2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tostivint H, Ocampo Daza D, Bergqvist CA, Quan FB, Bougerol M, Lihrmann I, Larhammar D. 2014. Molecular evolution of GPCRS: Somatostatin/urotensin II receptors. J Mol Endocrinol. 52(3):T61–T86. [DOI] [PubMed] [Google Scholar]
- Vaudry H, Do Rego J-C, Le Mevel JC, Chatenet D, Tostivint H, Fournier A, Tonon M-C, Pelletier G, Conlon JM, Leprince J. 2010. Urotensin II, from fish to human. Ann N Y Acad Sci. 1200:53–66. [DOI] [PubMed] [Google Scholar]
- Veenstra JA. 2009. Allatostatin c and its paralog allatostatin double c: the arthropod somatostatins. Insect Biochem Mol Biol. 39(3):161–170. [DOI] [PubMed] [Google Scholar]
- Waugh D, Youson J, Mims SD, Sower S, Conlon JM. 1995. Urotensin II from the river lamprey (Lampetra fluviatilis), the sea lamprey (Petromyzon marinus), and the paddlefish (Polyodon spathula). Gen Comp Endocrinol. 99(3):323–332. [DOI] [PubMed] [Google Scholar]
- Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, Grolemund G, Hayes A, Henry L, Hester J, et al. . 2019. Welcome to the tidyverse. J Open Source Softw. 4:1686. [Google Scholar]
- Woodward SR, Cruz LJ, Olivera BM, Hillyard DR. 1990. Constant and hypervariable regions in conotoxin propeptides. EMBO J. 9(4):1015–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Blakely B, Kim JH, Menting J, Schafer I, Schubert H, Agrawal R, Gutmann T, Delaine C, Zhang Y, et al. . 2022. Symmetric and asymmetric receptor conformation continuum induced by a new insulin. Nat Chem Biol. 1–9. doi: 10.1038/s41589-022-00981-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Menting JG, Disotuar MM, Smith NA, Delaine CA, Ghabash G, Agrawal R, Wang X, He X, Fisher SJ, et al. . 2020. A structurally minimized yet fully active insulin based on cone-snail venom insulin principles. Nat Struct Mol Biol. 27(7):615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yañez-Guerra LA, Zhong X, Moghul I, Butts T, Zampronio CG, Jones AM, Mirabeau O, Elphick MR. 2020. Echinoderms provide missing link in the evolution of PrRP/sNPF-type neuropeptide signalling. eLife 9:e57640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zakas C, Harry ND, Scholl EH, Rockman MV. 2022. The genome of the poecilogonous annelid streblospio benedicti. Genome Biol Evol. 14(2):evac008. doi: 10.1093/gbe/evac008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yañez Guerra LA, Egertová M, Zampronio CG, Jones AM, Elphick MR. 2020. Molecular and functional characterization of somatostatin-type signalling in a deuterostome invertebrate. Open Biol. 10(9):200172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yañez-Guerra LA, Tinoco AB, Escudero Castelán N, Egertová M, Elphick MR. 2022. Somatostatin-type and allatostatin-c–type neuropeptides are paralogous and have opposing myoregulatory roles in an echinoderm. Proc Natl Acad Sci U S A. 119(7):e2113589119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All accession numbers and sequences used in this study are available in the supplementary information.