Abstract
Reactive oxygen species (ROS) are vital for plant immunity and regulation of their production is crucial for plant health. While the mechanisms that elicit ROS production have been relatively well studied, those that repress ROS generation are less well understood. Here, via screening Brachypodium distachyon RNA interference mutants, we identified BdWRKY19 as a negative regulator of ROS generation whose knockdown confers elevated resistance to the rust fungus Puccinia brachypodii. The three wheat paralogous genes TaWRKY19 are induced during infection by virulent P. striiformis f. sp. tritici (Pst) and have partially redundant roles in resistance. The stable overexpression of TaWRKY19 in wheat increased susceptibility to an avirulent Pst race, while mutations in all three TaWRKY19 copies conferred strong resistance to Pst by enhancing host plant ROS accumulation. We show that TaWRKY19 is a transcriptional repressor that binds to a W-box element in the promoter of TaNOX10, which encodes an NADPH oxidase and is required for ROS generation and host resistance to Pst. Collectively, our findings reveal that TaWRKY19 compromises wheat resistance to the fungal pathogen and suggest TaWRKY19 as a potential target to improve wheat resistance to the commercially important wheat stripe rust fungus.
TaWRKY19 transcriptionally represses the expression of TaNOX10, impairing the host ROS burst and resulting in wheat susceptibility to stripe rust.
Introduction
During their coevolution, plants have evolved an immune system to thwart pathogens. Reactive oxygen species (ROS) are important components of the plant immune system. Many host interactions with incompatible pathogens (avirulent pathogens), including fungi, viruses, and bacteria, involve a burst of ROS production that results in resistance, whereas compatible (virulent) pathogens elicit no such ROS burst (Apel and Hirt, 2004; Camejo et al., 2016). Rapid ROS accumulation in plant tissues in response to avirulent pathogens triggers an early defense response—the hypersensitive response (HR)—which causes localized cell necrosis in the vicinity of the infection site that can inhibit pathogen growth (Shetty et al., 2003; Hakmaoui et al., 2012). The bacterial protein flagellin and fungus-derived polygalacturonase are examples of pathogen-associated molecular patterns (PAMPs), which elicit an instantaneous ROS burst from host plants. Additionally, interaction between a pathogen avirulence (Avr) gene and a host resistance (R) gene can initiate ROS production (Kaku et al., 2006; Torres et al., 2006; Zhang et al., 2007). In addition to inducing the HR, ROS act as signaling molecules that regulate the expression of genes involved in plant immunity, such as those encoding antimicrobial peptides and those that close stomata and other points through which pathogens might invade (Suzuki et al., 2011; Gilroy et al., 2014; Camejo et al., 2016). Stomata, the main channels for water transpiration and gas exchange in plants, are used by some pathogens to invade their hosts (Melotto et al., 2006; Dou and Zhou, 2012; Munemasa et al., 2015). Thus, ROS play an important role in regulating diverse cellular processes involved in plant immunity.
ROS are produced mainly by NADPH oxidases (NOXs) at the plasma membrane and organelle internal membranes (Segal and Wilson, 2017). NOXs are vital for local and systemic immune responses, and they initiate downstream disease resistance mechanisms (Mersmann et al., 2010; Kadota et al., 2014). In plants, NOXs are also known as respiratory burst oxidase homologs (RBOHs); they produce a burst of ROS in a broad range of plant–pathogen interactions. In the model plant Arabidopsis (Arabidopsis thaliana), there are 10 RBOH genes; overexpression or lack of expression of a specific RBOH gene, AtRBOHD, can significantly affect ROS production, thus affecting the immune response of Arabidopsis to pathogens (Torres et al., 2002; Denness et al., 2011). In the tobacco (Nicotiana tabacum) relative Nicotiana benthamiana, NbRBOHD is essential for ROS production in response to oomycete pathogens (Adachi et al., 2015). In agriculturally important crops, RBOH enzymes are also important for ROS production in response to pathogens—for example, HvRRBOHA in barley (Hordeum vulgare) produces ROS in response to powdery mildew; OsRBOHB and OsRBOHH in rice (Oryza sativa) produce ROS in response to rice blast, and StRBOHB in potato (Solanum tuberosum) produces ROS in response to Phytophthora infestans (Trujillo et al., 2006; Kobayashi et al., 2007; Wong et al., 2007; Nagano et al., 2016). Thus, RBOHs are crucial for plant resistance against disease.
Arabidopsis RBOHD is regulated by several molecular mechanisms. RBOHD is activated by a pattern recognition receptor at the plasma membrane upon recognition of a PAMP such as the bacterial flagellin protein or its N-terminal peptide flg22 (Benschop et al., 2007; Kadota et al., 2015). The cytoplasmic kinase PBS1-LIKE 1 (PBL1) contributes to flg22-induced ROS generation by interacting with RBOHD (Kadota et al., 2014). Several additional kinases phosphorylate and activate RBOHD to promote ROS production and enhance immunity (Li et al., 2014; Kadota et al., 2015; Zhang et al., 2018). RBOHD abundance is also negatively regulated via phosphorylation, mediated by PBL13, and by ubiquitination mediated by PIRE (PBL13 interacting RING domain E3 ligase; Lee et al., 2020). It is likely that RBOHs generally establish the signaling specificity of the ROS burst with the help of a collection of diverse upstream regulatory factors.
Little is known about the regulation of RBOH proteins in other species. In potato, elevated intracellular Ca2+ activates the calcium-dependent protein kinase StCDPK, which phosphorylates StRBOHB and leads to ROS generation, contributing to resistance against P. infestans (Kobayashi et al., 2007). In rice, OsRBOHB and OsRBOHH are activated by GTP-bound OsRac1, which promotes ROS generation and contributes to resistance to rice blast (Nagano et al., 2016). These mechanisms promote ROS production by RBOH enzymes; however, as yet little is known regarding mechanisms used to negatively regulate ROS generation during plant–pathogen interactions.
The causal agent for stripe rust, Puccinia striiformis f. sp. tritici (Pst), is a biotrophic fungal pathogen of wheat (Triticum aestivum) that devastates crops and causes substantial economic losses annually (Dean et al., 2012). When wheat is infected by an avirulent Pst strain, it produces ROS for defense against the pathogen (Wang et al., 2007), but the mechanisms regulating ROS generation in this context are largely unknown. There are 15 NOX/RBOH genes in the wheat genome, most of which are expressed specifically during rooting, anthesis, and seed germination, with the exception of TaNOX10, which is expressed throughout plant development (Hu et al., 2018). It is very difficult to study the function and regulation of these genes due to the large and complex wheat genome and low transformation efficiency (Ismagul et al., 2018; Ling et al., 2018). In contrast, purple false brome (Brachypodium distachyon) is closely related to wheat, barley, and other major cereal crops (Barbieri et al., 2011; Karen-Beth et al., 2018) but has a small diploid genome for which high-quality sequence data is available. Brachypodium distachyon also has a short generation time and can easily be transformed and genetically manipulated, making it an ideal model to study gene function in cereal crops (Karen-Beth et al., 2018).
In a preliminary study to identify genes involved in the interaction between cereals and stripe rust pathogens, we identified a candidate gene (BdWRKY67, Bradi1g22680) whose silencing increased resistance to the pathogen Puccinia brachypodii (Pb). Here, we demonstrate that BdWRKY67, which encodes a WRKY transcription factor (TF), plays a negative role in the resistance of B. distachyon to Pb by preventing ROS accumulation during host infection by the fungus. We further identify the homolog of BdWRKY67 in wheat, TaWRKY19. Genetic manipulation of TaWRKY19 levels suggests that TaWRKY19 acts as a repressor of ROS generation during the wheat–Pst interaction. Mechanistically, TaWRKY19 represses transcription of TaNOX10, which is required for ROS production during infection, by binding to the W-box in its promoter region. In agreement, tawrky19 knockout plants are extremely resistant to Pst. These findings reveal that TaWRKY19 is a wheat susceptibility factor whose induction minimizes ROS generation, thus compromising plant immunity against the pathogen.
Results
Silencing of BdWRKY67 confers resistance to Pb
To identify the key genes involved in the interaction between B. distachyon and Pb, we selected six B. distachyon candidate WRKY TF genes for functional analysis by RNA interference (RNAi) (Supplemental Figure S1 and Supplemental Table S1). To obtain WRKY-RNAi plants, we cloned gene-specific fragments into the inducible pOpOff2 vector for dexamethasone (Dex)-inducible expression (pOp6/LhGR) (Figure 1A; Samalova et al., 2005). We transformed the resulting RNAi constructs into B. distachyon accession Bd21-3 to generate WRKY-RNAi plants. We used T1 plantlets for phenotyping. Unlike the other tested WRKY-RNAi plants, BdWRKY67-RNAi plants specifically exhibited strong resistance to Pb (Supplemental Figure S1).
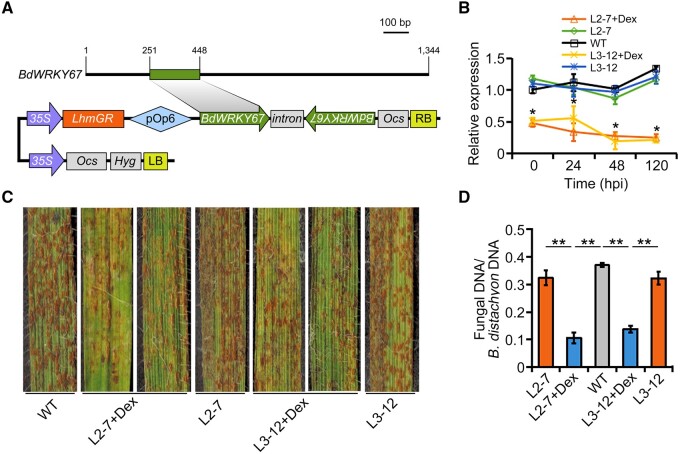
Figure 1.
BdWRKY67-silenced transgenic B. distachyon plants have enhanced resistance to Pb. A, Schematic diagram of the RNAi cassette in the pOpOff2-RNAi construct used to silence BdWRKY67. Analogous cassettes were used to silence the other five WRKY genes. B, Relative BdWRKY67 transcript levels in BdWRKY67-RNAi transgenic plants (L2-7 and L3-12) at various times up to 120 hpi with Pb. BdUBC18 was used as the internal control gene. Dex was used to induce silencing prior to infection. Data are shown as means ± standard deviation (SD) from three biological replicates. Statistical significance was determined by Student’s t test. *P < 0.05. C, WT B. distachyon and two independent lines containing the pOpOff2-RNAi construct (L2-7 and L3-12) were inoculated with Pb and examined 12 dpi. Transgenic lines were either treated with Dex (+ Dex) to induce RNAi expression or water as a control. The rust-colored spots are urediniospore pustules. BdWRKY67-silenced plants (+Dex) had fewer urediniospore pustules than their respective controls. Two independent lines L2-7 and L3-12 were watered with or without 20 mM Dex 3 times per week. D, Fungal biomass, determined as the ratio of fungal/B. distachyon DNA and measured by qPCR at 12 dpi. The Pb RustEF1 gene and B. distachyon BdUBC18 were used as the internal control genes for qPCR. Data are shown as means ± SD from three biological replicates. Statistical significance was determined by Student’s t test. **P < 0.01.
Given this result, we chose to further characterize the phenotypes caused by BdWRKY67 silencing. Accordingly, we identified T2 and T3 transgenic plants by PCR using LhGR-specific primers (LhGS_F and LhGS_R) (Supplemental Data Set S1 and Supplemental Figure S2A). We used two independent lines, L2-7 and L3-12, for further studies. We induced expression of the RNAi construct by root irrigation with 20 mM Dex, as previously described (Samalova et al., 2005). We observed no obvious changes in the morphology or height of BdWRKY67-RNAi plants compared to wild-type (WT) Bd21-3 plants one month after RNAi induction (Supplemental Figure S2, B–D).
To evaluate potential RNAi off-targets in the transgenic lines L2-7 and L3-12, we performed Basic Local Alignment Search Tool for DNA with the RNAi fragment sequence (Supplemental Table S2). We identified no consecutive >21-bp alignments between BdWRKY67 RNAi fragments and other Brachypodium or Pb transcripts. We nevertheless selected 15 putative off-target genes with 1–3 bp mismatches to assess potential off-target effects (Supplemental Data Set S1 and Supplemental Figure S3). Importantly, the expression levels of these genes were comparable in the transgenic lines and WT, as assessed by reverse transcription-quantitative PCR (RT-qPCR; Supplemental Figure S3). We conclude that there are no likely off-target sites for this BdWRKY67-RNAi fragment.
We inoculated the third leaves of L2-7, L3-12, and WT plants with Pb race F-co, which is highly virulent in the WT host. To determine the silencing efficiency of these RNAi lines, we measured BdWRKY67 transcript levels from Dex-induced and noninduced transgenic lines and WT by reverse transcription quantitative PCR (RT-qPCR) at 0, 24, 48, and 120 h post inoculation (hpi) with Pb. Transcript levels of BdWRKY67 decreased by 52%–75% in L2-7 plants and by 44%–81% in L3-12 plants when compared to noninduced lines and WT controls (Figure 1B). Importantly, the Dex-induced BdWRKY67-RNAi lines were significantly resistant to Pb, while the WT and noninduced lines were highly susceptible (P < 0.001; Figure 1C). Together, these data showed that lower BdWRKY67 transcript levels result in enhanced plant resistance against Pb.
To further evaluate the effects of silencing BdWRKY67 on the resistance of B. distachyon to Pb, we examined leaves infected with Pb under the microscope to assess pathogen development. The Dex-induced lines exhibited fewer hyphal branches and shorter hyphae at 24 and 48 hpi (Supplemental Figure S4, A–C), and the infected areas were smaller at 120 hpi compared to WT and noninduced controls (Supplemental Figure S4, D and E). We measured fungal biomass at later stages of infection by evaluating the ratio of fungal genomic DNA to wheat genomic DNA. At 12-day postinoculation (dpi), the ratio of fungal/wheat DNA was lower in BdWRKY67-RNAi lines when compared to uninduced and WT plants (Figure 1D). These data indicate that BdWRKY67 plays a negative role in B. distachyon resistance to Pb.
BdWRKY67 binds to the BdRBOHD promoter to repress its expression
To investigate how BdWRKY67 affects immunity in B. distachyon, we used digital transcriptome deep sequencing (digital RNA-seq) to compare gene expression levels in the BdWRKY67-RNAi line L2-7 and WT plants. First, however, we specifically assessed the expression pattern of BdWRKY67 by RT-qPCR during host–pathogen interaction in the WT. The transcript levels of BdWRKY67 were induced 24 hpi with Pb and peaked at 48 hpi, with about seven-fold higher transcript levels than in uninfected plants. In contrast, BdWRKY67 transcript levels were unchanged when the plants were inoculated with an incompatible pathogen, Pst race Chinese yellow rust race 31 (CYR31) (Supplemental Figure S5). Based on these data, we decided to analyze differentially expressed genes in the BdWRKY67-RNAi line at the 48 hpi time point by RNA-seq.
Kyoto encyclopedia of genes and genomes pathway enrichment analysis showed that the differentially expressed genes in L2-7 after Dex induction are enriched for the plant–pathogen interaction pathway, as illustrated in a targeted heatmap (Figure 2A; Supplemental Figure S6). By applying a false discovery rate cutoff of ≤0.001, we determined that 17 of the 47 genes in the plant–pathogen interaction pathway are significantly upregulated in L2-7 plants relative to the WT (Figure 2A). These genes included five WRKY TF genes, five protein kinase genes, four calcium-binding protein genes, one disease resistance gene (RPM1), one RBOHD, and one predicted gene of unknown function (Supplemental Data Set S2). We were particularly interested in the upregulation of BdRBOHD, which encodes RBHOD (Bradi4g17020) in the BdWRKY67-RNAi lines (Figure 2A), in light of the important role of RBOH genes in pathogen resistance.
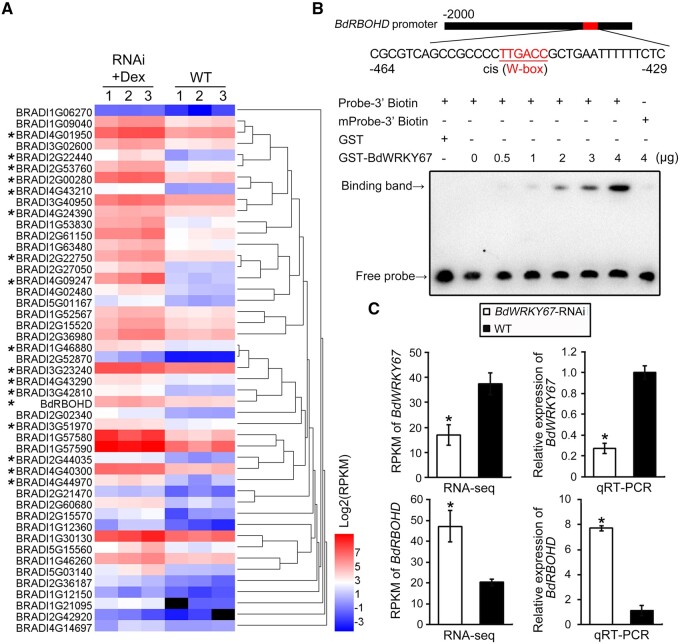
Figure 2.
BdWRKY67 suppresses transcription of BdRBOHD. A, Heatmap of differentially regulated genes of the plant–pathogen interaction pathway in three plants of the L2-7 transgenic line expressing the BdWRKY67-RNAi construct and induced with Dex (RNAi Dex 48) and three WT plants treated with water (WT 48). All plants were inoculated with Pb 48 h after treatment and transcription was measured by digital RNA-seq (with Unique Molecular Identifier per cDNA) with the Illumina HiSeq TM2500 sequencing platform (SeqHealth, Wuhan, China). The reference genome used was Brachypodium_distachyon.v1 (ftp://ftp.ensemblgenomes.org/pub/plants/release39/fasta/brachypodium_distachyon/dna/). The black rectangles stand for clusterings of multiple gene expression levels. Asterisks indicate significantly upregulated genes under FDR cutoff of ≤0.001. B, Binding of recombinant GST-BdWRKY67 protein to the BdRBOHD promoter, analyzed by EMSA with titration. The W-box element is indicated in red. W-box-mutated biotinylated DNA probe (mProbe-3′-biotin) was used as the negative control. Probe-3′-biotin (lanes 1–7) or mProbe-3′-biotin (lane 8) = 0.005 μM of biotinylated DNA probe added to the reaction. The BdWRKY67–GST fusion protein concentrations were 0, 0.5, 1, 2, 3, 4, 4 µg in lanes 2–8. The GST protein concentration was 4 µg in lane 1. C, Reads per kilobase million and relative expression of BdWRKY67 (top) and BdRBOHD (bottom) in WT and BdWRKY67-silenced plants, as determined by digital RNA-seq (left) and RT-qPCR (right) 48 hpi. Data are shown as means ± SD of three biological replicates. Statistical significance was determined by Student’s t test. *P < 0.05.
To investigate how BdWRKY67 might regulate BdRBOHD expression, we analyzed the BdRBOHD promoter region for TF-binding sites using the JASPAR (http://jaspardev.genereg.net/) and PlantCARE databases. The BdRBOHD promoter contained a W-box located 429–464 bp upstream of the start codon (Figure 2B; Supplemental Figure S7A). To determine whether BdWRKY67 binds directly to the W-box, we performed an electrophoretic mobility shift assay (EMSA) in which we incubated a 36-bp biotinylated oligonucleotide probe comprising the BdRBOHD promoter region with a glutathione S transferase (GST)-tagged purified recombinant BdWRKY67 protein or to a control GST protein (Supplemental Figure S7, A and B). As the amount of added GST–BdWRKY67 fusion protein increased, a specific band indicative of binding appeared that is absent in the GST control, while we detected no such binding band when the probe with its W-box element mutated (mProbe) was present (Figure 2B). These data showed that BdWRKY67 can bind directly to the W-box in the BdRBOHD promoter in vitro. Moreover, BdRBOHD expression was inversely correlated with that of BdWRKY67 during Pb infection, consistent with our digital RNA-seq results (Figure 2C). These findings suggested that BdWRKY67 binds to the W-box in the promoter of the respiratory burst oxidase gene BdRBOHD to downregulate its expression.
Silencing the BdWRKY67 homolog TaWRKY19 in wheat enhances ROS accumulation and resistance to Pst
We conducted a BLASTP (https://blast.ncbi.nlm.nih.gov/Blast.cgi) search to identify the BdWRKY67 ortholog in wheat, which revealed that TaWRKY19 shares ∼70% similarity with BdWRKY67 and also contains two WRKY domains (Supplemental Figure S8A). Furthermore, the N-terminal and C-terminal domains of TaWRKY19, BdWRKY67, and other related proteins contained the highly conserved WRKYGQK peptide sequence and a zinc finger motif (C-X4-C-X22-23HXH/C) (Supplemental Figure S8B).
Both TaWRKY19 and BdWRKY67 had a predicted nuclear localization sequence (https://www.predictprotein.org/), as expected for putative TFs. To validate their predicted localization, we generated constructs by cloning the TaWRKY19 and BdWRKY67 coding sequences in-frame and upstream of the green fluorescent protein (GFP) sequence for transient infiltration in N. benthamiana epidermal cells via Agrobacterium (Agrobacterium tumefaciens). Fluorescence microscopy showed a specific nuclear localization for the TaWRKY19–GFP and BdWRKY67–GFP fusion proteins, whereas control-free GFP was distributed throughout the cell (Supplemental Figure S9). We also transiently transfected wheat protoplasts with another set of plasmids expressing similar GFP fusions; we detected green fluorescence specifically in the nucleus, as expected (Supplemental Figure S10). Confirmation of TaWRKY19 and BdWRKY67 nuclear localization was consistent with these proteins functioning as TFs.
The wheat genome encodes three TaWRKY19 paralogs: TaWRKY19-2A, TaWRKY19-2B, and TaWRKY19-2D. We compared the coding sequences of these three TaWRKY19 homeoalleles: they shared 96.9% sequence identity and their encoded proteins were 96.2% identical (Supplemental Figures S11 and S12). RT-qPCR analyses established that TaWRKY19-2A and TaWRKY19-2D are upregulated 5.7- and 3.4-fold, respectively, while TaWRKY19-2B was upregulated 2.7-fold upon infection with virulent Pst CYR31 at 48 hpi (Supplemental Figure S13). This observation suggested that, like their B. distachyon ortholog, TaWRKY19 might influence wheat susceptibility to Pst and that the three TaWRKY19 paralogs may have potential functions during wheat infection.
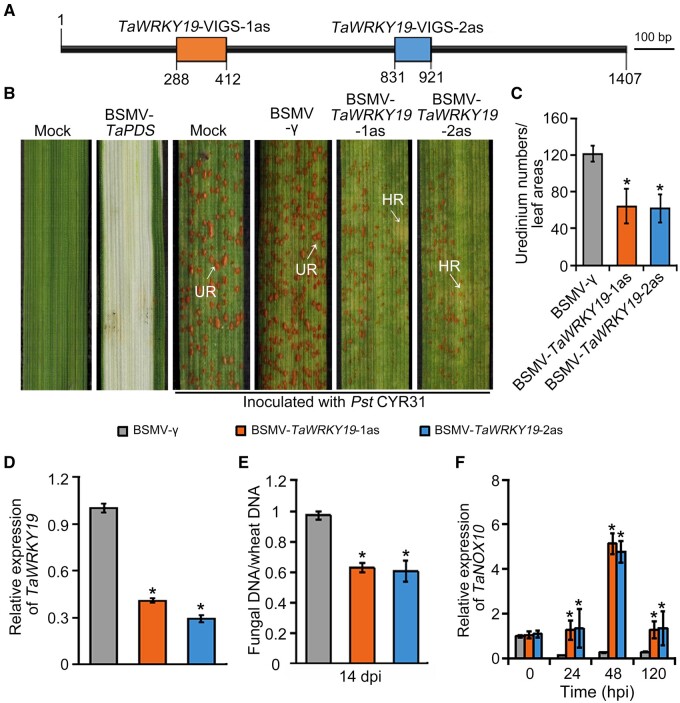
To directly assess the function of TaWRKY19 in wheat susceptibility to Pst, we silenced all three TaWRKY19 copies and examined the effect on wheat–Pst infection. Accordingly, we used barley stripe mosaic virus (BSMV)-induced gene silencing to knockdown TaWRKY19 transcript levels in wheat by expressing two RNAi fragments specific for TaWRKY19 (Figure 3A). The Phytoene desaturase (TaPDS) gene was used as a positive control whose silencing causes a photobleaching phenotype (Figure 3B). Seven days after silencing, we inoculated the fourth leaves of wheat plants with fresh urediniospores of the virulent Pst race CYR31. Fourteen dpi, we scored fewer urediniospore pustules on TaWRKY19-silenced wheat plants than on the BSMV:γ negative controls (Figure 3, B and C). Based on RT-qPCR data, the TaWRKY19-2as fragment reduced TaWRKY19 transcript levels more effectively than the TaWRKY19-1as fragment (70.9% and 59.4% silencing, respectively; Figure 3D). Growth and development of the pathogen were delayed in TaWRKY19-silenced plants, as observed by histological analysis; the number of hyphal branches and haustorial mother cells (HMCs), hyphal length, and colony areas was all smaller in TaWRKY19-silenced plants at 24, 48, and 120 hpi when compared to controls (Supplemental Figure S14, A–E). Moreover, the ratio between fungal genomic DNA and wheat genomic DNA was lower in TaWRKY19-silenced plants at 14 dpi compared to controls (Figure 3E). These results indicated that TaWRKY19 negatively regulates wheat resistance to Pst.
Figure 3.
TaWRKY19 contributes to wheat susceptibility to virulent Pst CYR31. A, Schematic diagram of the specific gene fragments targeted for silencing in the TaWRKY19 gene. B, Wheat leaves were either treated with 1×FES buffer (Mock) or infected with BSMV-γ as negative controls, or they were infected with BSMV-TaPDS (conferring a photobleaching phenotype as positive control for silencing). BSMV-TaWRKY19-1as and BSMV-TaWRKY19-2as were used to silence TaWRKY19. Leaves were inoculated with virulent Pst CYR31 and examined 14 dpi. Urediniospore pustules (UR) and regions of HR are indicated. C, Number of urediniospore pustules per centimeter square on the leaves in (B), counted using ImageJ at 14 dpi. Data are shown as means ± SD from 20 leaves. Statistically significant differences were determined by Tukey’s multiple comparisons test. *P < 0.05. D, Relative TaWRKY19 expression in BMSV-γ, BMSV-TaWRKY19-1as, and BMSV-TaWRKY19-2as infected plants, as assessed by RT-qPCR using TaEF1α as an internal control. Data are shown as means ± sd from three biological replicates. Statistical significance was determined by Student’s t test. *P < 0.05. E, Fungal biomass at 14 dpi with Pst CYR31, as determined by qPCR. PsEF1 and wheat TaEF1α were used as the internal control gene to evaluate Pst and wheat DNA with reference to gene-specific standard curves. Data are shown as means ± SD from three biological replicates. Statistical significance was determined by Student’s t test. *P < 0.05. F, Relative TaNOX10 expression levels in TaWRKY19-silenced plants, as assessed by RT-qPCR at various times postinfection. The TaEF1α gene was used as internal control. Data are shown as means ± SD from three biological replicates. Statistical significance was determined by Student’s t test. *P < 0.05.
To investigate whether TaWRKY19 might downregulate a ROS-generating NOX, as in the case of BdWRKY67 downregulating BdRBOHD, we identified the NOX gene in wheat most closely related to Arabidopsis RBOHD: TaNOX10 (Supplemental Figure S15). Transcript levels were higher in TaWRKY19-silenced plants at 24, 48, and 120 hpi with Pst when compared to controls (Figure 3F). These results suggested a function for wheat TaWRKY19 similar to that of BdWRKY67 in repressing expression of an RBOH-type gene during pathogen infection.
Wheat homeoalleles of TaWRKY19 play redundant roles in the regulation of ROS and Pst resistance
To determine whether all three homeoalleles function during infection of wheat with Pst, we used clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9-mediated gene editing to genetically inactivate TaWRKY19 in the wheat genome. To this end, we cloned single-guide RNAs (sgRNAs) complementary to conserved regions in exon 1 of all three genomic copies of TaWRKY19 (Supplemental Figure S16A) under the control of the promoter of the wheat Ubiquitin 6 (U6) gene. We generated transgenic wheat plants using Agrobacterium-mediated stable transformation of the WT Fielder variety. We then sequenced T1 transgenic wheat lines using six pairs of primers specific for TaWRKY19-2A, TaWRKY19-2B, or TaWRKY19-2D (Supplementary Data Set S1). Two mutant plants, tawrky19-AB and tawrky19-A, contained nucleotide deletions or insertions leading to frameshift mutations in the region targeted by Cas9 in TaWRKY19-2A and TaWRKY19-2B (tawrky19-AB) or TaWRKY19-2A (tawrky19-A) (Supplemental Figure S16B). When inoculated with the virulent Pst race CYR33, both tawrky19-AB and tawrky19-B plants exhibited strong resistance to Pst with few urediniospore pustules, although tawrky19-A plants showed weaker resistance compared to tawrky19-AB (Supplemental Figure 16, C and D). Analysis of fungal biomass by qPCR indicated a reduction of 59.4% and 33.2% in the tawrky19-AB and tawrky19-A knockout plants, respectively, compared to WT at 14 dpi (Supplemental Figure S16E). These results suggested that both the TaWRKY19-2A and TaWRKY19-2B homeoalleles play important roles in the wheat–Pst interaction.
To further uncover the relationship between the homeoalleles in our infection context, we stably expressed each of the three homeoalleles TaWRKY19-2A, TaWRKY19-2B, and TaWRKY19-2D in tawrky19-A to generate tawrky19-A°TaWRKY19-A, tawrky19-A°TaWRKY19-B, and tawrky19-A°TaWRKY19-D complementation plants (Figure 4A). When inoculated with the virulent Pst race CYR33, tawrky19-A plants were resistant and mounted an HR with few urediniospore pustules; in contrast, more urediniospore pustules were produced on tawrky19-A°TaWRKY19-A and tawrky19-A°TaWRKY19-D plants, but only slightly more urediniospore pustules on tawrky19-A°TaWRKY19-B, which did mount an HR (Figure 4A). These data indicated that both the TaWRKY19-2A and TaWRKY19-2D homeoalleles can complement the tawrky19-A Pst resistance phenotype. These data also suggested that the observed resistance of tawrky19-A is due to the specific mutation in the TaWRKY19-2A homeoallele, which can be fully rescued by TaWRKY19-2A or TaWRKY19-2D and only partially by TaWRKY19-2B, indicating more important roles of TaWRKY19-2A and TaWRKY19-2D in this phenotype.
Figure 4.
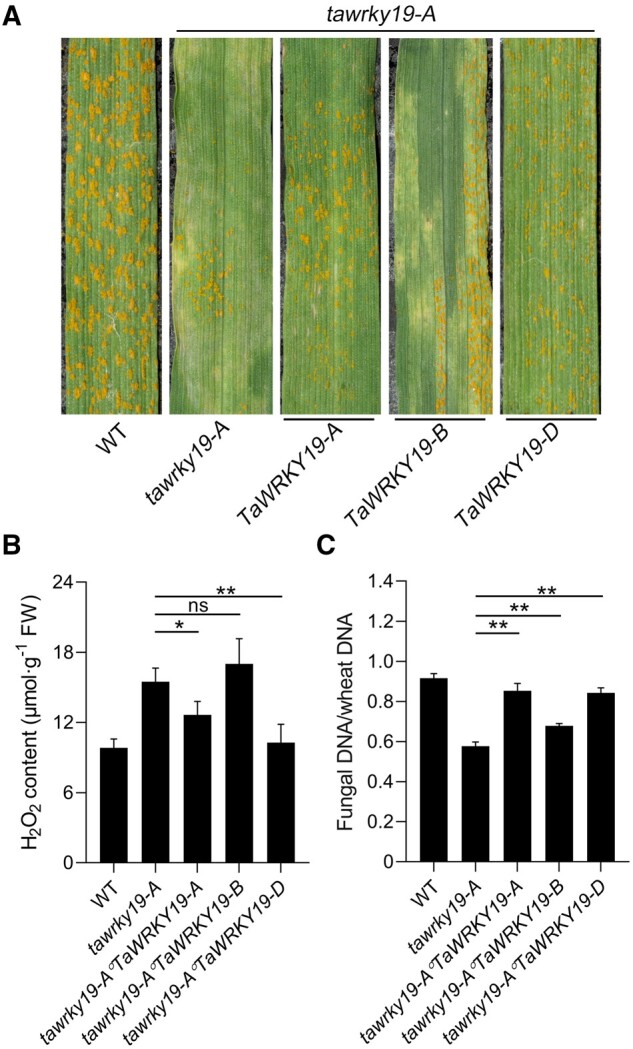
Three homeoalleles of TaWRKY19 in wheat play redundant roles in regulation of ROS production and Pst resistance. A, Transgenic complementation assay using the three wheat homeoalleles of TaWRKY19 (TaWRKY19-2A, TaWRKY19-2B, and TaWRKY19-2D). tawrky19-A transgenic plants were transformed with each homeoallele individually (tawrky19-A°TaWRKY19-A, tawrky19-A°TaWRKY19-B, and tawrky19-A°TaWRKY19-D). The leaves were inoculated with Pst CYR33, which is virulent on the WT Fielder plants and examined at 14 dpi. B, Extracellular H2O2 contents in the plants from (A) at 24 hpi with Pst CYR33. Data are shown as means ± SD from three biological replicates. Statistical significance was determined by Student’s t test. *P < 0.05. **P < 0.01. ns, not significant. C, Fungal biomass at 14 dpi with Pst CYR33, as determined by qPCR. PsEF1 and wheat TaEF1α were used as the internal control gene to evaluate Pst and wheat DNA with reference to gene-specific standard curves. Data are shown as means ± SD from three biological replicates. Statistical significance was determined by Student’s t test. **P < 0.01.
Given the potential role of TaWRKY19 in regulating TaNOX10 and the importance of ROS in the HR, we evaluated ROS production in the knockout and transgenic complementation plants. We assessed production of extracellular ROS in response to Pst infection using an enzyme-linked immunosorbent assay (ELISA), which revealed lower H2O2 contents in the leaves of tawrky19-A°TaWRKY19-A and tawrky19-A°TaWRKY19-D plants as compared to tawrky19-A, while we observed no significant changes in H2O2 contents in tawrky19-A°TaWRKY19-B plants relative to tawrky19-A (Figure 4B).
Quantification of fungal biomass by qPCR confirmed these results, with a 24.7%, 26.9%, and 15.0% increase in Pst growth on tawrky19-A°TaWRKY19-A, tawrky19-A°TaWRKY19-D, and tawrky19-A°TaWRKY19-B plants, respectively, compared to tawrky19-A plants at 14 dpi (Figure 4C). These results confirmed that the TaWRKY19-2A and TaWRKY19-2D homeoalleles play important and redundant roles in the wheat–Pst interaction.
TaWRKY19 represses TaNOX10 expression by binding to its promoter
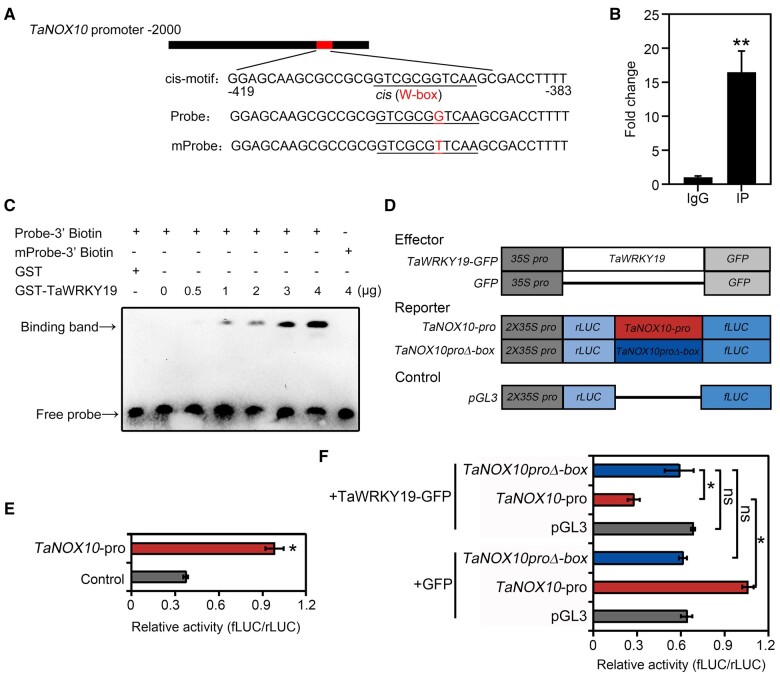
A search for TF-binding sites in the TaNOX10 promoter revealed a W-box element 383- to 419-bp upstream of the ATG start codon in the TaNOX10 promoter, similar to that found in the BdRBOHD promoter (Figure 5A). To determine whether TaWRKY19 binds directly to the TaNOX10 promoter, we performed chromatin immunoprecipitation (ChIP) with a TaWRKY19-specific polyclonal antibody in the context of virulent Pst infection of the WT variety Fielder. We detected a 16.3-fold enrichment for the TaNOX10 promoter region in TaWRKY19 IP DNA when compared to the negative control consisting of a nonspecific IgG antibody (Figure 5B). To further test whether TaWRKY19 regulates expression of TaNOX10 by binding to its promoter, we produced and purified a recombinant GST–TaWRKY19 fusion protein (Supplemental Figure S17), then performed EMSA with a 36-bp biotinylated oligonucleotide probe and a mutated probe (mProbe; Figure 5A). A specific band appeared only when the GST-TaWRKY19 protein was included (Figure 5C); as the GST-TaWRKY19 concentration increased, a band indicative of binding similarly appeared and increased in abundance that is absent in the W-box element-mProbe control (Figure 5C). These data confirmed that TaWRKY19 can bind to the W-box in the TaNOX10 promoter.
Figure 5.
TaWRKY19 directly binds the TaNOX10 promoter. A, Putative TaWRKY19-binding motif GTCGCGGTCAA, located in the TaNOX10 promoter. The biotin-labeled probe and the mProbe contain the binding motif sequence and mutated sequence are shown. B, Binding of TaWRKY19 to the TaNOX10 promoter, as determined by ChIP-qPCR. Data are shown as means ± SD from three biological replicates. Statistical significance was determined by Student’s t test. **P < 0.01. C, EMSA titration, performed as described in Figure 2B, assessing TaWRKY19 binding to the TaNOX10 promoter, which is dependent on the presence of the W box. Probe-3′-biotin (lanes 1 to 7) or mProbe-3′-biotin (lane 8) = 0.0025 µM of biotinylated DNA probe added to the reaction. D, Schematic diagrams of the effector, reporter, and control constructs used in dual-LUC reporter assays. TaNOX10pro consists of 2,000-bp upstream of the start codon of TaNOX10 as in (A); TaNOX10proΔ-box includes a mutation in the W-box element and was used as a negative control. E, TaNOX10 promoter activity. The relative fLUC/rLUC activity was evaluated in wheat protoplasts transformed with TaNOX10pro and pGL3, respectively. TaNOX10pro indicates the intact TaNOX10 promoter. The empty vector pGL3 was used as a negative control. Statistical significance was determined by Student’s t test. Data are shown as means ± SD are from five replicates. *P < 0.05. F, TaWRKY19 compromises TaNOX10 promoter activity. +TaWRKY19-GFP indicates co-expression of TaWRKY19 with TaNOX10pro, TaNOX10proΔ-box, or the pGL3 control. GFP was used as a negative control. In (E) and (F), relative activity (fLUC/rLUC) is the ratio of bioluminescence intensity of fLUC (TaNOX10pro or TaNOX10proΔ-box) to rLUC (35S promoter). LUC activity (fLUC/rLUC) was detected by a Modulus Single Tube Luminometer (Promega, Madison, USA). Statistical significance was determined by Student’s t test. Data are shown as means ± SD are from five replicates. *P < 0.05.
To investigate how TaWRKY19 regulates TaNOX10 expression, we performed dual-luciferase (LUC) reporter assays by transfecting effector, reporter, and control constructs in wheat protoplasts (Figure 5D). We placed the firefly LUC (fLUC) reporter gene under the control of the TaNOX10 promoter (TaNOX10pro). The intact TaNOX10 promoter resulted in higher fLUC/renilla LUC (rLUC) activity compared to that of pGL3 controls (Figure 5E). Notably, relative reporter activity (fLUC/rLUC) decreased when the intact TaNOX10pro:LUC reporter was co-transfected with TaWRKY19-GFP compared to protoplasts expressing the GFP control. In addition, this effect was dependent on the W-box, as we observed no such reduction using the mutated promoter construct (Figure 5F). These data confirm that, like BdWRKY67, TaWRKY19 binding to the TaNOX10 promoter leads to transcriptional repression.
Loss of function of TaNOX10 compromises ROS production and wheat resistance to Pst
To directly determine the function of TaNOX10 in wheat resistance to Pst, we used BSMV-induced gene silencing to knockdown TaNOX10 in wheat We then inoculated plants displaying BSMV-induced mild chlorotic mosaic symptoms with the avirulent Pst race CYR23. The leaves in which TaNOX10 was silenced displayed evident urediniospore pustules, unlike resistant controls (Supplemental Figure S18, A and B). Consistently, fungal biomass increased by over 50% in TaNOX10-silenced wheat plants compared with the controls (Supplemental Figure S18C). These data indicated that TaNOX10 contributes to the natural resistance of wheat to Pst.
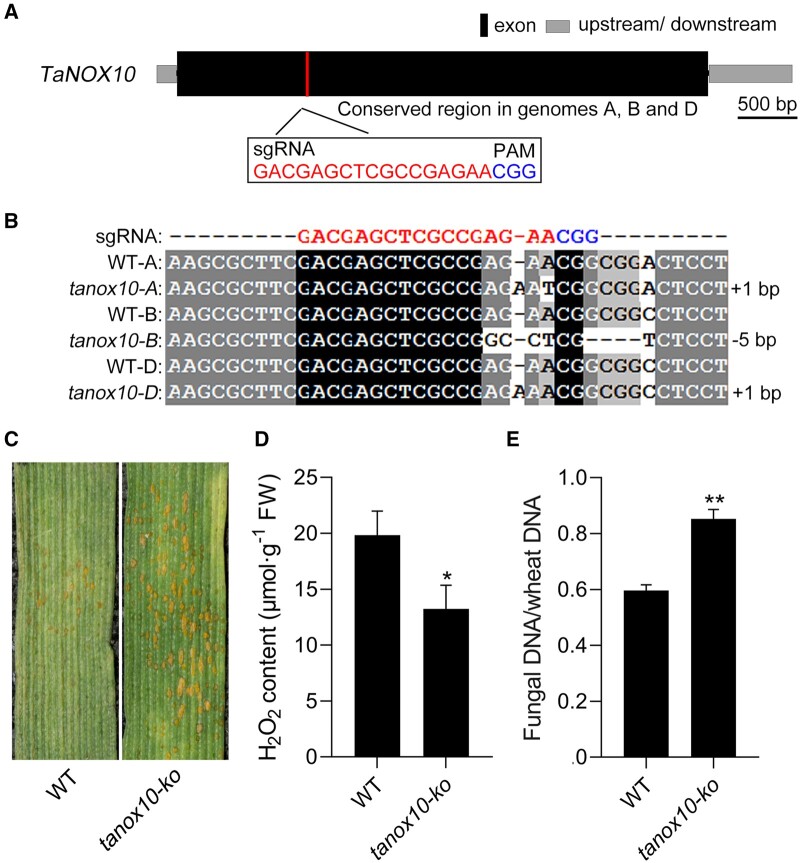
To further validate the function of TaNOX10 in ROS production and wheat resistance to Pst, we used CRISPR–Cas9-mediated gene editing to knockout TaNOX10 in the wheat genome using sgRNAs complementary to conserved regions of all three genomic copies of TaNOX10 (Figure 6A). We generated transgenic wheat lines by particle bombardment-mediated stable transformation of the WT Kenong 199 (KN199) variety, and sequenced the selected transformed wheat lines using six pairs of primers specific for TaNOX10-5A, TaNOX10-5B, or TaNOX10-5D (Supplementary Data Set S1). We obtained one tanox10-ko line in which all three copies of TaNOX10 contained nucleotide deletions or insertions leading to frameshift mutations in the region targeted by Cas9 (Figure 6B). When inoculated with Pst race CYR32, which is avirulent on KN199 (moderate resistance with few to no urediniospore pustules), tanox10-ko plants were instead fully susceptible to the fungus (Figure 6C), confirming that TaNOX10 plays a positive role in wheat resistance to Pst infection and that loss of this protein is sufficient to trigger pathogen susceptibility. We assayed extracellular ROS contents in the tanox10-ko plants, which showed a significant decrease, as expected (Figure 6D). Moreover, fungal biomass increased by 30% compared to that in WT plants (Figure 6E). These data together confirm that TaNOX10 is important for ROS generation and contributes to the resistance of wheat to Pst.
Figure 6.
TaNOX10 plays a positive role in ROS production and wheat resistance to Pst. A, Schematic diagram of TaNOX10 gene structure and the sequences of the two sgRNAs designed to target the three homoeoalleles (A, B, D), of TaNOX10 for editing by CRISPR–Cas9. Black rectangle, exon. Blue letters, PAM. B, Sequences of the WT TaNOX10 and tanox10-knockout (tanox10-ko) plant at the site targeted by sgRNA. The tanox10-ko line contains frameshift mutations in tanox10-A (1-bp insertion), tanox10-B (5-bp deletion), and tanox10-D (1-bp insertion). C, tanox10-ko and the WT KN199 variety were inoculated with Pst race CYR32, and their disease phenotypes were observed at 14 dpi. D, Extracellular H2O2 contents in tanox10-ko and WT plants at 24 hpi with Pst CYR32. Data are shown as means ± SD from three biological replicates. Statistical significance was determined by Student’s t test. *P < 0.05. E, Fungal biomass in tanox10-ko and WT at 14 dpi with Pst CYR32, as estimated by qPCR of PsEF1 and wheat TaEF1α DNAs in infected samples and calculated with reference to gene-specific standard curves. Data are shown as means ± SD from three biological replicates. Statistical significance was determined by Student’s t test. **P < 0.01.
Elevated TaWRKY19 expression represses RBOH-mediated ROS production
To test if the effects of TaWRKY19 on ROS generation are due to its regulation of TaNOX10, we generated transgenic wheat lines overexpressing TaWRKY19 in the wheat cultivar Fielder. The selected TaWRKY19-OE line had 3.2-fold higher transcript levels for TaWRKY19 than the WT (Supplemental Figure S19A). At 14 dpi with the avirulent Pst race CYR23, we observed a strong resistance response with HR on inoculated WT leaves, whereas many urediniospore pustules formed on TaWRKY19-OE leaves (Supplemental Figure S19B). Fungal biomass increased by 33.1% in the TaWRKY19-OE line relative to the WT (Supplemental Figure S19C). These data confirmed that TaWRKY19 is a negative regulator of stripe rust resistance.
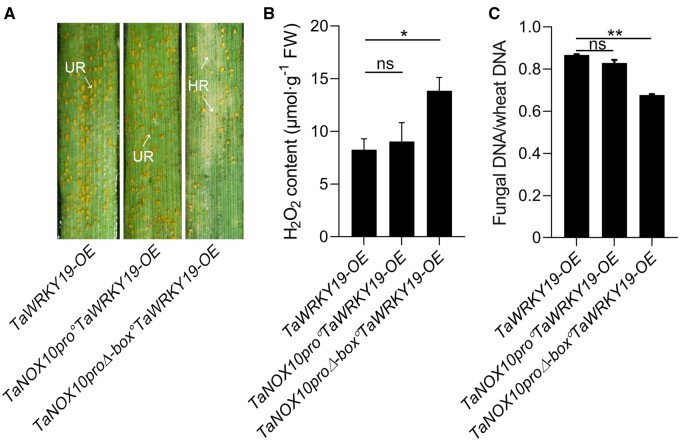
Next, to investigate the functional relevance of TaWRKY19 regulation of TaNOX10 expression, we transiently expressed TaNOX10 under the control of its intact promoter (TaNOX10pro) or a W-box deleted promoter (TaNOX10proΔ-box) in TaWRKY19-OE plants and infected them with Pst race CYR32, which is virulent on TaWRKY19-OE plants. The susceptibility phenotype of TaWRKY19-overexpressing plants was substantially ameliorated by the co-expression of TaNOX10 driven by its promoter without the W-box (Figure 7B), while co-expression of TaNOX10 from its intact promoter did not change susceptibility (Figure 7A). These results suggested that because a TaNOX10 construct lacking the W-box (TaNOX10proΔ-box) in the promoter is no longer subject to repression by TaWRKY19, its introduction into the TaWRKY19-OE plants can rescue their susceptibility to avirulent Pst (Figure 7C). Thus, the transcriptional repression of TaNOX10 by TaWRKY19 is important for the negative role of this TF in plant resistance to Pst. Consistent with the infection data, ROS production was induced specifically by the introduction of the TaNOX10proΔ-box:TaNOX10 transgene in TaWRKY19-OE plants (Figure 7B), suggesting that the effect of TaWRKY19 on H2O2 accumulation depends on regulation of TaNOX10-mediated ROS production.
Figure 7.
Elevated expression of TaWRKY19 represses RBOH-mediated ROS production. A, Transient overexpression assay showing the repression of the TaNOX10 promoter (TaNOX10pro) by TaWRKY19. TaNOX10pro or TaNOX10proΔ-box was transiently expressed in plants overexpressing TaWRKY19 (TaWRKY19-OE) by particle bombardment-mediated transformation, and then inoculated with Pst CYR23. The disease phenotypes were observed at 14 dpi. TaNOX10pro:TaNOX10 includes the full promoter and the coding sequence of TaNOX10. TaNOX10proΔ-box:TaNOX10 includes the promoter with the W-box deleted (see Figure 5A) and the coding sequence of TaNOX10. Urediniospore pustules (UR) and regions of HR are indicated. B, Extracellular H2O2 contents in TaWRKY19-OE, TaNOX10pro°TaWRKY19-OE and TaNOX10proΔ-box°TaWRKY19-OE at 24 hpi with Pst CYR33. Data are shown as means ± SD from three biological replicates. Statistical significance was determined by Student’s t test (compared to the WT). *P < 0.05. C, Fungal biomass in TaWRKY19-OE, TaNOX10pro°TaWRKY19-OE and TaNOX10proΔ-box°TaWRKY19-OE at 14 dpi with Pst CYR33 as estimated by qPCR of PsEF1 and wheat TaEF1α DNAs in infected samples and calculated with reference to gene-specific standard curves. Data are shown as means ± SD from three biological replicates. Statistical significance was determined by Student’s t test (compared to the WT). **P < 0.01.
A knockout in TaWRKY19 confers resistance to Pst by promoting ROS accumulation
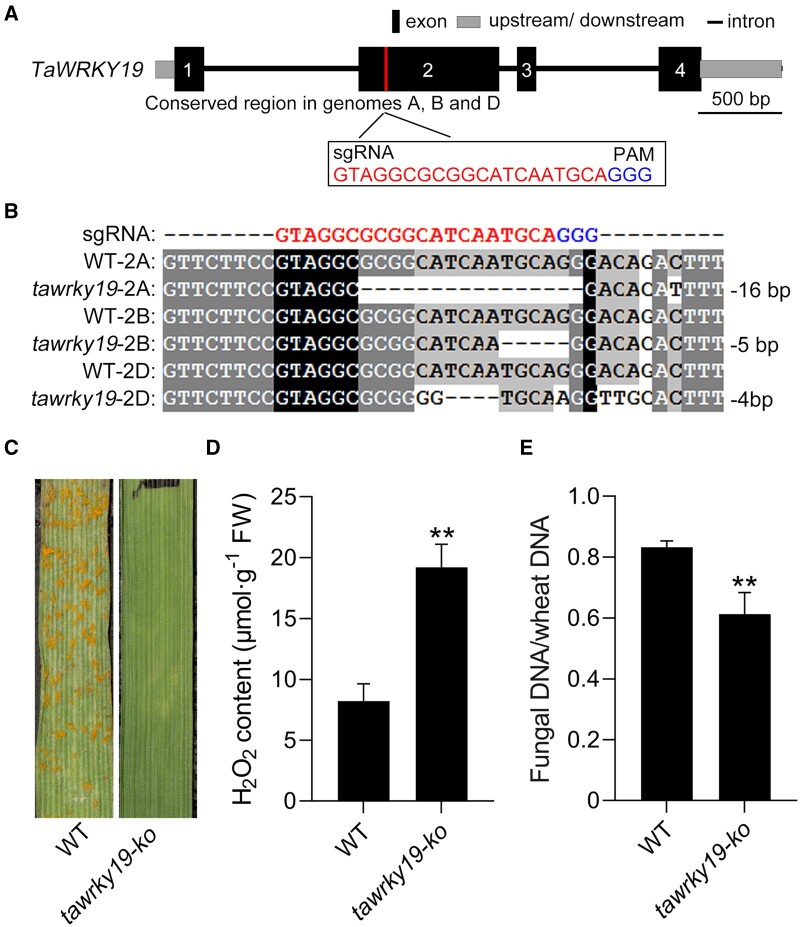
In an effort to develop rust-resistant wheat plants, we used CRISPR–Cas9-mediated gene editing to fully inactivate TaWRKY19 in the wheat genome. To this end, we cloned sgRNAs complementary to conserved regions in exon 2 of all three genomic copies of TaWRKY19 under the control of the promoter of the wheat U6 gene (Figure 8A). We then generated transgenic lines using particle bombardment-mediated stable transformation of the WT KN199 variety. We selected transformed wheat lines and sequenced the sgRNA target sites using six pairs of primers specific for TaWRKY19-2B, TaWRKY19-2D, or TaWRKY19-2A (Supplementary Data Set S1). We obtained a tawrky19-ko mutant line with nucleotide deletions or insertions leading to frameshift mutations in the region targeted by Cas9 in all three genes (Figure 8B). When inoculated with Pst race CYR33, which is virulent on KN199, tawrky19-ko plants exhibited strong resistance with no urediniospore pustules (Figure 8C). These results further confirmed that TaWRKY19 is a negative regulator of wheat resistance to Pst and that inactivation of the three TaWRKY19 genes can render plants extremely resistant to this pathogen.
Figure 8.
Loss of TaWRKY19 confers resistance to Pst. A, Schematic diagram of TaWRKY19 gene structure and the sequences of the two sgRNAs designed to target the three homoeoalleles (A, B, D) of TaWRKY19 for editing by CRISPR–Cas9. Black rectangles, exons. Blue letters, PAM. B, Sequences of the WT TaWRKY19 and TaWRKY19-ko (tawrky19-ko) plant at the sites targeted by the sgRNAs. The tawrky19-ko contains mutations in tawrky19-2A (16-bp deletion), tawrky19-2B (5-bp deletion), and tawrky19-2D (4-bp deletion). C, The tawrky19-ko line and the WT KN199 variety were inoculated with Pst race CYR33, and their disease phenotypes were observed at 14 dpi. D, Extracellular H2O2 contents in tawrky19-ko and WT at 24 hpi with Pst CYR33. Data are shown as means ± SD from three biological replicates. Statistical significance was determined by Student’s t test. **P < 0.01. E, Fungal biomass in tawrky19-ko and WT at 14 dpi with Pst CYR33, as estimated by qPCR of PsEF1 and wheat TaEF1α DNAs in infected samples and calculated with reference to gene-specific standard curves. Data are shown as means ± SD from three biological replicates. Statistical significance was determined by Student’s t test. **P < 0.01.
Finally, to evaluate the production of extracellular ROS in the tawrky19-ko plants, we assessed production of ROS in wheat apoplast fluid in response to Pst infection using ELISA and measured a significant H2O2 increase in infected tawrky19-ko plants compared to WT plants (Figure 8D). To quantify Pst growth in the tawrky19-ko plant, we analyzed fungal biomass by qPCR and observed a 26.3% decrease in the tawrky19-ko line compared to WT plants at 14 dpi (Figure 8E). These data support the notion that the enhanced ROS accumulation in tawrky19-ko plants leads to their resistance to Pst and that TaWRK19 is a promising genetic engineering target to protect wheat from this devastating pathogen.
Discussion
In this study, we set out to identify TF genes that are differentially expressed during infection of wheat and related cereal plants by the commercially important stripe rust fungus, with the goal of identifying genes that confer susceptibility to this pathogen and exploring their functional mechanism to facilitate their use in precise resistance engineering. In this study, we identified a negative immune defense regulator (TaWRKY19) in wheat via rapid screening of WRKY-RNAi lines in B. distachyon as an important positive regulator of wheat susceptibility to Pst by direct repression of RBOHD/NOX-mediated ROS production. Simultaneous modification of the three TaWRKY19 homoeologs by CRISPR–Cas9 conferred enhanced stripe rust resistance to wheat. Our discovery of a negative regulator of ROS production upon infection broadly advances our knowledge of the network of ROS regulators and simultaneously provides germplasm for disease resistance breeding.
ROS are important signaling molecules that can induce programmed cell death and play a vital role in plant defense against phytopathogens (Qi et al., 2017). The apoplast is an important site for PTI, and ROS are generated in the apoplast mainly by plasma membrane-localized RBOHs (Kadota et al., 2015). The mechanism of RBOH-mediated ROS production is a topic of ongoing research, and the regulatory mechanisms of ROS generation are well studied in Arabidopsis. Several lines of evidence indicate that receptor-like kinases and receptor-like cytoplasmic kinases activate Arabidopsis RBOHD-mediated ROS accumulation either by phosphorylating the RBOH enzymes or via direct interaction (Dubiella et al., 2013; Li et al., 2014; Kadota et al., 2015; Zhang et al., 2018). Like AtRBOHD activation in PTI, ROS production is also induced during abscisic acid signaling via OPEN STOMATA 1 phosphorylation and activation of AtRBOHD (Mustilli et al., 2002). Despite the protective role of ROS, their continuous production and constitutive defense responses would negatively affect the growth and development of plants. Thus, precise regulation of the place and time of ROS generation is vital to balance plant growth with stress resistance. Nevertheless, few studies have identified negative regulators that directly affect RBOH-mediated ROS generation.
Wheat employs ROS production as a defense mechanism against attack by avirulent Pst races (Wang et al., 2007). Here, we identified a repressor of ROS generation in wheat plants challenged with virulent Pst. This repressor, the TF TaWRKY19, directly repressed expression of TaNOX10 in wheat protoplasts. Upon infection, tawrky19-ko wheat plants were highly resistant to Pst, exhibiting both reduced fungal growth and massive ROS accumulation. We conclude that the virulent rust pathogen induces expression of a gene (TaWRKY19) whose gene product negatively regulates host ROS accumulation, thus permitting nutrient uptake from host cells, which is necessary to promote infection and fungal growth.
Transcriptional regulation plays a critical role in orchestrating plant immune gene expression, relying on host TFs to activate target genes by binding to specific cis elements in their promoters (Li et al., 2016). WRKY TFs contain a highly conserved amino acid sequence, WRKYGQK, and are classified into three groups (I, II, and III) based on the number of encoded WRKY domains and the features of their zinc-finger like motif (Eulgem et al., 2000). TaWRKY19 contains two WRKY domains and belongs to subgroup I. Transgenic plants overexpressing its closest homolog AtWRKY4 displayed enhanced resistance to the necrotrophic pathogen Botrytis cinerea, but it was also shown to play a negative role in resistance to the biotrophic pathogen Pseudomonas syringae (Lai et al., 2008). Cotton (Gossypium hirsutum) GhWRKY25, another group I WRKY TF, plays a negative role in the response to Botrytis cinerea by reducing the expression of salicylic acid or ethylene signaling-related genes and inducing the expression of genes involved in the jasmonic acid signaling pathway (Liu et al., 2018). Therefore, WRKY proteins, while having diverse roles and directions of effect, commonly orchestrate complex signaling and transcriptional networks of plant defense that require tight regulation and fine-tuning. Here, we revealed that overexpressing TaWRKY19 in wheat enhances susceptibility to Pst, while knocking down or knocking out TaWRKY19 rendered wheat resistant to Pst. Therefore, TaWRKY19 clearly negatively regulates resistance against the biotrophic rust fungus Pst. TaWRKY19 directly bound the W-box cis-element in the TaNOX10 promoter to inhibit ROS production and wheat resistance to Pst. Importantly, when we expressed TaNOX10 with a version of the TaNOX10 promoter lacking the W-box in TaWRKY19-OE plants, the expression of this transgene was sufficient to restore a resistance phenotype, as the lack of the W-box element no longer placed TaNOX10 expression under repression imposed by TaWRKY19. Thus, TaWRKY19-mediated transcriptional repression of TaNOX10 expression is responsible for inhibiting TaNOX10-mediated ROS production and promotes wheat susceptibility to pathogens. These results indicate that group I WRKY TFs can play opposite roles in different pathosystems.
Bread wheat is an allohexaploid plant composed of three subgenomes, meaning that most wheat genes are present in three very similar but not identical copies (Slade et al., 2005). Several studies have uncovered that homoeologous genes from different subgenomes contribute unevenly to particular biological processes, such as plant growth and development and stress responses in polyploid species, indicating that the homoeologous genes have different functional compartmentalization or sub-functionalization (Bekaert et al., 2011). In the case of the mildew resistance locus (MLO), one well-known gene can confer susceptibility to mildew on crops. The three MLO homoeologs in wheat share 98% and 99% identity at the nucleotide and protein levels respectively, and only when all three homoeologs are lost does wheat show strong resistance to powdery mildew (Wang et al., 2014), indicating that all three MLO genes contribute to powdery mildew susceptibility (Elliott et al., 2002). Likewise, the TaWRKY19 gene exists as three homoeoalleles, TaWRKY19-2A, TaWRKY19-2B, and TaWRKY19-2D, in hexaploid wheat, sharing 96% identity at the nucleotide and protein levels. In contrast to mlo, however, we noted differences in the degree of responsiveness to pathogen infection among homoeologs, with the A and D homologs exhibiting stronger expression responses to rust infection than the B homolog. The TaWRKY19-2A and TaWRKY19-2D homoeoalleles appeared to play a more important role than TaWRKY19-2B in resistance to infection, and either TaWRKY19-2A or TaWRKY19-2D can rescue the phenotype of the TaWRKY19-2A mutant, while TaWRKY19-2B can only partially rescue this phenotype. It is important to note that the copy number of TaWRKY19 in these complemented lines might affect their expression levels, with implications for the expression of genes downstream of these TFs. This caveat may contribute to the difference in disease phenotype among the complementation lines; however, it cannot explain the differential induction response of each homolog to pathogen infection. Therefore, our findings remain of interest with regards to broadly understanding the functional redundancy of homeologous genes in polyploid plants.
Cas9-mediated editing of susceptibility genes has been widely used to generate broad-spectrum and durable resistance in plants (Langner et al., 2018; Zaidi et al., 2018; Xu et al., 2019). Thus, in an effort to develop rust-resistant wheat plants, we fully inactivated all three homoeoalleles of TaWRKY19 in the wheat genome. Promisingly, the tawrky19-ko plants were highly resistant to Pst, showing that TaWRK19 is a favorable genetic engineering target to protect wheat from this devastating pathogen.
Another valuable finding from our study is the utility of B. distachyon as a model for the less genetically tractable organism wheat. Model organisms are extensively employed in plant biology (Leonelli and Ankeny, 2013). Arabidopsis has been widely used to investigate the interaction between dicot plants and their pathogens. For monocots, B. distachyon is evolutionarily closely related to wheat, barley, and other major cereal crops, and offers various advantages over rice (Barbieri et al., 2011; Catalán et al., 2012; Karen-Beth et al., 2018). Importantly, most fungal pathogens of wheat also infect B. distachyon (Ayliffe et al., 2013; Figueroa et al., 2013; Fitzgerald et al., 2015), making it an excellent model for studying wheat–pathogen interactions. Our study confirmed that TaWRKY19 and BdWRKY67 have similar functions in plant–rust fungus interactions, which indicates that B. distachyon is a powerful model plant with which to study the molecular interaction of monocots including wheat with rust fungi.
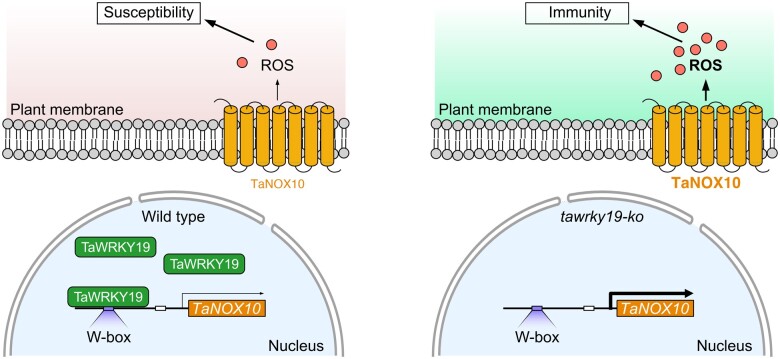
Together, this study provides new insight into the molecular regulation of ROS production during host–rust fungus interactions. In preinfected TaWRKY19-silencing plants, TaNOX10 expression is not altered, but TaNOX10 expression is upregulated in Pst-infected TaWRKY19-silencing plants, suggesting that TaWRKY19 likely does not affect TaNOX10 expression at basal level or prePst infection, but specifically postPst infection. During wheat–Pst interaction, when the expression of TaWRKY19 is induced by virulent Pst, TaWRKY19 directly represses the transcription of TaNOX10, the product of which is required for the host ROS burst, thus compromising host immunity (Figure 9). In contrast, in tawrky19 knockout plants, TaNOX10 expression is no longer repressed by TaWRKY19, thereby resulting in enhanced TaNOX10 transcription and a TaNOX10-mediated ROS burst, which impairs infection and confers host resistance. Artificial elimination of TaWRKY19 via genetic deletion is sufficient to confer resistance to even virulent Pst races.
Figure 9.
Model for the transcriptional regulation of TaNOX10 by TaWRKY19. Infection of wheat virulent Pst (left) induces the expression of TaWRKY19, whose encoded TF binds to the W-box element in the TaNOX10 promoter to repress its expression, resulting in low levels of ROS production that fail to inhibit infection. Whereas, in tawrky19-ko plant (right), no repression of TaNOX10 and thus abundant production of ROS that contributes to host immunity.
Materials and methods
Plant materials and fungal races
The wheat cultivar Suwon 11 is resistant to P. striiformis f. sp. tritici (Pst) CYR23 (inoculation results in ROS accumulation and a typical HR) and susceptible to CYR31 (inoculation fails to trigger ROS accumulation, there is no HR, and urediniospores develop). The wheat cultivar Fielder, which is resistant to Pst race CYR23, was used to generate the TaWRKY19 overexpression lines, tawrky19-A and tawrky19-AB. TaNOX10 and TaWRKY19 gene editing for fully inactivating of all three copies was carried out in the wheat cultivar KN199, which is susceptible to Pst race CYR33 and moderately resistant to Pst race CYR32. Wheat seedlings were grown and inoculated with Pst as previously described (Kang et al., 2002). Briefly, the plants were grown in an illumination incubator set manually at 16°C and 85% relative humidity with 16-h light (500 μmol·m−2·s−1 with incandescent light bulbs)/8-h dark. The inoculated plants were placed at 16°C and 100% relative humidity in the dark for 24 h after infection, and then returned to normal conditions as described above.
The B. distachyon accession Bd21-3 and Pb race F-co were used in the BdWRKY67 functional studies. The WT and RNAi plants of B. distachyon were cultured in an illumination incubator under cycles of 16 h at 25°C in the light and 8 h at 16°C in the dark, with humidity maintained at 80%. Inoculation of B. distachyon seedlings was performed as previously described (Wang et al., 2017). The inoculated seedlings were placed in 100% humidity and cultured as described for wheat.
Generation of BdWRKY67 B. distachyon RNAi lines
A Dex-inducible derivative of the pOpOff2 vector (Dex-inducible pOp6/LhGR promoter system; Wielopolska et al., 2005) was used for Agrobacterium (A. tumefaciens)-mediated transformation, as described previously (Păcurar et al., 2008). The pOpOff2 vector containing a BdWRKY67-specific fragment (251- to 448-bp downstream from the start codon) was transformed into Agrobacterium strain AGL1 (Lazo et al., 1991). Genetic transformation of B. distachyon was performed as previously described (Vogel et al., 2006) with some modifications. Agrobacterium colonies harboring the recombinant plasmid were cultured for 24 h in MG/L liquid medium (containing peptone 5 g·L−1, yeast extract powder 2.5 g·L−1, NaCl 5 g·L−1, mannitol 5 g·L−1, MgSO4 0.1 g·L−1, K2HPO4 0.25 g·L−1, glutamic acid 1.2 g·L−1, pH = 7.2) with 80 mg·L−1 carbenicillin and 60 mg·L−1 spectinomycin. Immature B. distachyon embryos were picked from newly filled seed, and then transfected with an Agrobacterium cell suspension diluted to OD600 = 0.6 with Callus Inducing Medium liquid medium and an Agrobacterium cell suspension (containing Linsmaier and Skoog [LS] basal medium 4.43 g·L−1, sucrose 30 g·L−1 and CuSO4 0.6 mg·L−1, pH = 5.8) supplemented with 200 μM acetosyringone and 0.1% (v/v) synperonic PE/F68. Transgenic plants were then screened on differentiation medium (containing LS 4.43 g·L−1, maltose 30 g·L−1 and phytagel 2 g·L−1 pH = 5.8, supplemented with 0.2 mg·L−1 kinetin) and rooting medium (containing Murashige and Skoog basal medium w/vitamins 4.42 g·L−1, sucrose 30 g·L−1 and phytagel 2 g·L−1 pH = 5.8) containing 40 μg·mL−1 hygromycin.
RT-qPCR and biomass assays
Total RNA was extracted with the MiniBEST Plant RNA Extraction Kit (Takara, Dalian, China) according to the manufacturer’s protocol. The RNase-free DNase Set (Takara, Dalian, China) was used to remove genomic DNA from the total RNA. First-strand cDNA was synthesized with the SMARTScribe Reverse Transcription Kit (Clontech, Palo Alto, CA, USA). Genomic DNA was extracted from 2-week-old B. distachyon and wheat leaves, by the cetyl trimethylammonium bromide method (Rogers and Bendich, 1994) for PCR detection. Genomic DNA was isolated from plant leaves 14 dpi for fungal biomass analysis.
RT-qPCR was used to quantify the expression of selected genes, confirm transgenic plant genotypes, and determine silencing efficiency. TaEF-1α (encoding wheat elongation factor, GenBank No. Q03033) was used as an internal reference gene. RT-qPCR was performed using TB Green Premix Ex Taq II (RR820A) (Takara, Dalian, China). Reactions were carried out as described previously (Wang et al., 2011). A 7500 Real-Time PCR System (Applied Biosystems, Foster City, Foster, USA, USA) was used to quantify gene expression, and the data were analyzed by the comparative 2–ΔΔCt method (Pfaffl, 2001). The quantity of PCR product for rust EF1 and plant (wheat TaEF1α or B. distachyon BdUBC18) internal control in infected samples were calculated using gene-specific standard curves (Supplemental Figure S20) to determine the Pst and wheat gDNA contents, respectively. Each experiment comprised three independent biological replicates.
Subcellular localization of BdWRKY67 and TaWRKY19
The coding sequences of TaWRKY19 and BdWRKY67 were amplified and cloned into the pCAMBIA-1302-GFP and 16318-hGFP vectors, respectively. The resulting plasmids TaWRKY19:1302-GFP, BdWRKY67:1302-GFP and the empty vector control were transformed into Agrobacterium strain GV3101. For infiltration into N. benthamiana leaves, the agrobacteria were suspended in infiltration buffer (containing 150 mM acetosyringone, 10 mM MgCl2 and 10 mM MES, pH 5.6) at a final optical density (OD) at 0.6 and left in the dark for 2 h. Then, the N. benthamiana plants were infiltrated with the agrobacteria suspension using a 1 mL syringe, and the infiltrated plants were maintained in the dark for 24 h. Forty-eight hours after infiltration, an Olympus FV1000 confocal laser microscope (Olympus, Tokyo, Japan) was used to observe GFP fluorescence.
To localize BdWRKY67 and TaWRKY19 in wheat protoplasts, protoplasts were prepared as described (Yoo et al., 2007) and transfected with the plasmids TaWRKY19:16318-hGFP, BdWRKY67:16318-hGFP and the empty vector control by PEG-mediated transformation as in Yoo et al. (2007). The protoplasts were cultured as previously described (Yoo et al., 2007). GFP was detected with an Olympus FV1000 confocal laser microscope using a 488 nm argon laser.
BSMV-mediated gene silencing
To determine the function of TaWRKY19 and TaNOX10 in the wheat–Pst interaction, BSMV-mediated gene silencing was employed to knockdown the expression of TaWRKY19 and TaNOX10. Two fragments were used to silence TaWRKY19 and one to silence TaNOX10. Each BSMV construct (BSMV:TaWRKY19-1as and BSMV:TaWRKY19-2as for silencing TaWRKY19, BSMV:TaNOX10 for silencing TaNOX10, BSMV:γ as empty control, and BSMV:TaPDS as controls) (Supplementary Table S2) was inoculated onto the second leaves as described previously (Holzberg et al., 2002; Hein et al., 2005; Scofield et al., 2005). Mock control (Mock) plants were treated with 1×FES buffer (7.5 g·L−1 glycine, 10.45 g·L−1 K2HPO4 dibasic, 10 g·L−1 sodium pyrophosphate decahydrate, 10 g·L−1 bentonite, and 10 g·L−1 celite). The seedlings were maintained in 100% relative humidity in darkness for 24 h, then placed in an incubator at 25°C for 7 days before phenotypic analysis. When the plants inoculated with BSMV:TaPDS showed the expected photobleaching phenotype, the fourth leaves were sampled to determine the silencing efficiency and were then infected with freshly harvested urediniospores of Pst race CYR31 or CYR23. The disease phenotype of knockdown wheat plants was examined at 14 dpi. The experiments were independently performed 3 times.
Histological observations of host response and fungal growth
Puccinia hyphae were stained with wheat germ agglutinin conjugated to Alexa-488 (Amresco, Solon,OH, USA; Wang et al., 2011). Hyphae length, number of branches, and HMCs were observed with an Olympus BX-53 microscope and quantified with the cellSens Entry software program (Olympus, Tokyo, Japan). Fungal development was observed by epifluorescence microscopy. More than 40 infection sites were observed, and each of 5 randomly selected leaf segments was statistically analyzed.
ChIP and Chip-qPCR
For the ChIP assay, the EpiQuik Plant ChIP Kit was used as per manufacturer’s instructions (Epigentek, Brooklyn, NY, USA). More specifically, 2 g of 3-week-old seedlings of the wheat variety Fielder were used 48 h after inoculation of the leaves with CYR33, as TaWRKY19 is strongly induced at 48 hpi with the virulent race CYR33. First, 1% (w/v) formaldehyde was used for in vitro crosslinking for 40 min, and 2-M glycine solution was added to the sample to quench the reaction. Then, the chromatin was sheared into 200- to 500-bp fragments by ultrasonic disruption (Fisher Scientific, Waltham, MA, USA) with power of 60 W. The pulse length was 5 s on/10 s off, with a total ultrasonic time of 15 min. The anti-TaWRKY19 antibody was produced by Genecreate (Wuhan, China), and the specificity of the antibody was detected by immunoblotting. Anti-TaWRKY19 (2 μg) was added to the fragmented chromatin solution and then incubated at 65°C overnight. Nonspecific DNA was washed from the beads before proceeding to the purification of immunoprecipitated DNA fragments as described in the kit manufacturer’s instructions. Subsequently, immunoprecipitated DNA was quantified by qPCR as described above using locus-specific primers.
Extracellular ROS accumulation detection and quantification
Preparation of the apoplastic fluid from wheat leaves was performed as described (van der Linde et al., 2012) with some modifications. Briefly, 5 g of wheat leaves were collected and cut into about 4-cm segments, and placed in a beaker filled with Tris-buffered EDTA at pH 7.5. Leaves were vacuum-infiltrated using a vacuum pump for 3 × 20 min at 400 mbar. The leaves were then transferred into a 20-mL syringe and the syringe was placed into a 50-mL falcon tube and centrifuged for 15 min at 2,500 g and 4°C to collect the apoplastic fluid. The apoplastic fluid ROS contents were quantified by using the Plant ROS ELISA kit (Trust Specialty Zeal Biological trade, USA) according to the manufacturer’s instructions. In detail, ROS were extracted from plant leaves (>50 mg) in 1 mg·μL−1 PBS (KH2PO4 0.24 g·L−1, Na2HPO4 1.44 g·L−1, NaCl 8 g·L−1, KCl 0.2 g·L−1, pH = 7.4); 50 μL samples were then incubated in the wells of the 96-well plate provided with the kit for 45 min at 37°C. Each well was washed 5 times with 200-µL wash buffer 200, after which 50 μL biotinylated anti-IgG was added to each well and incubated for 30 min at 37°C. After washing as above, 50-μL streptavidin-HRP was added into the plate and incubated at 37°C for 15 min. Finally, chromogen solution was added, followed by the stop solution (provided with the kit) as per manufacturer’s methods. The OD at 450 nm was then measured with a Multiskan Spectrum plate reader (Tecan, Männerdorf, Switzerland). Each experiment comprised three independent biological replicates.
Recombinant protein purification
To obtain BdWRKY67 and TaWRKY19 protein, the full-length cDNAs of BdWRKY67 and TaWRKY19 were amplified by PCR and cloned into the pGEX4T-1 (GST Tag) vector by using the ClonExpress II One-Step Cloning Kit (Vazyme, Nanjing, China) following the manufacturer’s instructions. The resulting plasmids BdWRKY67:pGEX-4T-1 and TaWRKY19:pGEX-4T-1 were transformed into the Escherichia coli strain Rosetta and grown in LB medium containing ampicillin at 37°C. One millimolar isopropyl β-D-1-thiogalactopyranoside was added to the E. coli cultures when an OD600 = 0.5 was reached, followed by an incubation for 16 h at 4°C to induce fusion protein production. Recombinant GST–BdWRKY67 and GST–TaWRKY19 fusion proteins and the control GST protein were purified by using the GST-tag Protein Purification Kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions.
EMSAs
A 1-bp fragment upstream of the start codon (ATG) was selected as the promoter of TaNOX10 and BdRBOHD, respectively. The cis-acting regulatory element W-box in the promoter region was identified by using the PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and JASPAR databases (http://jaspardev.genereg.net/) (Lescot et al., 2002; Vlieghe et al., 2006). For EMSAs, the Light Shift Chemiluminescent EMSA Kit (Thermo, Waltham, MA, USA) was used. In detail, biotinylated (3′-biotin-labeled) DNA probes (Probe and mProbe) (Supplemental Table S1) were synthesized by Sangon Biotech (Shanghai, China). For EMSA titration assays, the GST-BdWRKY67 or GST-TaWRKY19 protein concentrations were 0, 0.5, 1, 2, 3, and 4 µg. Four microgram GST protein was added as the control. The corresponding concentration of purified GST, BdWRKY67-GST or TaWRKY19-GST protein was incubated with each probe in 5×EMSA/Gel-Shift Binding Buffer (provided with the EMSA kit) for 20 min. To demonstrate the specificity of binding, the mutated biotin-labeled probes were added in the reaction as the mProbe control. Binding reaction products were separated by electrophoresis on 6% (w/v) nondenaturing polyacrylamide gels in 0.5×Tris-borate EDTA buffer on ice, transferred to Hybond-N+Nylon Membrane (GE Healthcare, Piscataway, NJ, USA) and crosslinked with ultraviolet light. Washing and visualization were performed according to the manufacturer’s instructions.
Dual LUC reporter assay
The intact promoter of TaNOX10 (TaNOX10pro) and the W-box mutant promoter (TaNOX10proΔ-box) were cloned into the artificially modified pGL3 vector (after removal of the 2 × 35S promoter upstream of fLUC; Figure 5C, Reporter). The basic pGL3 vector contains two expression cassettes, namely rLUC and fLUC (Figure 5C, Control pGL3 vector). The primers used to construct these vectors for dual LUC reporter assays are listed in Supplementary Data Set S1.
For in vivo dual LUC reporter assays, wheat protoplasts were prepared and transformed to determine the relative fluorescence activity (fLUC/rLUC), as previously described (Gu et al., 2013). After 16 h, total proteins were extracted from the transfected protoplasts were extracted using the Dual-LUC reporter kit (Promega, Madison, WI, USA) following the manufacturer’s instructions. A Modulus Single Tube Luminometer (Promega, Madison, USA) was used to detect relative LUC activity (fLUC/rLUC). To detect fLUC activity, 100-μL LUC assay reagent was added to the extract from transfected protoplasts. For detection of rLUC activity, 100 μL rLUC reagent was added to the above reaction. The ratio of fLUC/rLUC was used to determine the activity of the reporter gene (TaNOX10pro and TaNOX10proΔ-box). The experiments were performed 4 times.
Generation and identification of wheat mutants
For construction of the TaWRKY19 overexpression vector, the full-length coding sequence of TaWRKY19 was inserted into the PANIC6E-Ha vector (with the Bar gene conferring Basta resistance) using Gateway technology (Mann et al., 2012). The recombinant constructs were transformed into Agrobacterium strain EHA105 and transgenic wheat lines were generated by Agrobacterium-mediated infiltration of immature wheat embryos as described previously (Ishida et al., 2015). Positive transgenic seedlings were selected with Basta (0.1 mL·L−1).
For CRISPR-Cas9 editing of TaNOX10 and TaWRKY19, sgRNAs were designed to target the three copies of TaNOX10 or TaWRKY19. The fusion-sgRNA–CRISPR–Cas9 vector contains the wheat U6 promoter and the Bar gene conferring Basta resistance. Transgenic wheat lines were generated by particle bombardment of immature wheat embryos as described previously (Wang et al., 2014). Genomic DNA was extracted for PCR analysis from regenerated plants at the three-leaf stage. Positive transgenic seedlings were selected with phosphinothricin (5 mg·L−1), and genomic DNA was extracted for PCR detection and sequencing analysis. All primers for construction and detection are shown in Supplemental Data Set S1.
Complementation assay
For construction of the complementation vector, the full-length coding sequence of each of the three homeoalleles TaWRKY19-2A, TaWRKY19-2B, and TaWRKY19-2D was inserted into the PANIC6E vector using Gateway technology. The recombinant constructs were transformed into Agrobacterium strain EHA105. Transgenic wheat lines were generated through Agrobacterium-mediated infiltration of immature wheat embryos from the tawrky19-A background as described previously (Ishida et al., 2015). Genomic DNA was extracted for PCR detection after the regenerated plants reached the third-leaf stage.
Particle bombardment assay
For particle bombardment assays, the recombinant plasmids containing the TaNOX10 genomic sequence with its intact promoter or promoter lacking the W-box were prepared in large quantities. Leaves from 14-day-old TaWRKY19-OE and Fielder wheat plants (5-cm segments and fixed in an agar medium plate containing 75 μg·mL−1 6-benzylaminopurine) were bombarded using He/1100 particles (Bio-Rad, Hercules, CA, USA) at a bombardment distance of 6 cm as previously described (Wang et al., 2012; Peng et al., 2020). After bombardment, the leaves were cultured at 16°C for 12 h. The Pst-inoculated wheat leave plates were incubated at 16°C with 16 h of light and at 14°C with 8 h of dark as above. Each assay consisted of six shots and was conducted at least twice.
Statistical analysis and quantification
Urediniospore pustule numbers were analyzed with ImageJ software. Statistically significant differences were calculated with SPSS software program (version 19, SPSS Inc., IBM, USA), in which two-group differences were evaluated by Student’s t test, and multiple sample differences were evaluated by Tukey’s multiple comparison test followed by one-way analysis of variance (ANOVA). Data for all statistical analyses are shown in Supplemental Data Set S3.
Accession numbers
The accession number of the sequence data can be found in the GenBank and Ensemble plant database. TaWRKY19 (ACD80362), BdWRKY67 (Bradi1g22680), TaNOX10 (TraesCS5B02G299000.1), and BdRBOHD (Bradi4g17020). The RNA-seq data generated in this study were deposited to the SRA database at the NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number PRJNA788221.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phenotypes of WRKY-RNAi silenced plants.
Supplemental Figure S2. Identification of Agrobacterium-mediated transgenic plants and their growth phenotypes.
Supplemental Figure S3. Expression levels of putative off-target genes are not significantly changed in BdWRKY67-RNAi lines L2-7 and L3-12.
Supplemental Figure S4. Histological observation of Pb growth in BdWRKY67-RNAi lines.
Supplemental Figure S5. Expression of BdWRKY67 in the compatible and incompatible interaction between B. distachyon and rust fungi.
Supplemental Figure S6. Differentially upregulated genes are enriched in plant–pathogen interaction pathway.
Supplemental Figure S7. The BdRBOHD promoter element is predicted to bind BdWRKY67.
Supplemental Figure S8. Sequence alignment of the TaWRKY19 and BdWRKY67 proteins.
Supplemental Figure S9. Subcellular localization of TaWRKY19 and BdWRKY67 in N. benthamiana epidermal cells.
Supplemental Figure S10. Subcellular localization of TaWRKY19 and BdWRKY67 in wheat protoplasts.
Supplemental Figure S11. Alignment of the cDNA sequences of the three TaWRKY19 homoealleles.
Supplemental Figure S12. Alignment of the protein sequences of the three homoealleles of TaWRKY19.
Supplemental Figure S13. All the three homoealleles of TaWRKY19 are induced by a compatible interaction between wheat and Pst.
Supplemental Figure S14. Histological observation of Pst growth and development in TaWRKY19-silencing wheat leaves inoculated with Pst CYR31.
Supplemental Figure S15. Phylogenetic analysis of the RBOH family proteins.
Supplemental Figure S16. Loss of function of TaWRKY19 confers resistance to Pst.
Supplemental Figure S17. SDS-PAGE of purified GST-TaWRKY19 protein.
Supplemental Figure S18. TaNOX10 promotes wheat resistance to Pst.
Supplemental Figure S19. TaWRKY19 negatively regulates wheat resistance to Pst.
Supplemental Figure S20. Standard curves for qPCR expression data.
Supplemental Table S1. Candidate WRKY TF genes highly enriched in B. distachyon and Pb interaction.
Supplemental Table S2. Off-target transcript prediction.
Supplemental Table S3. Accession numbers of the protein sequence included in the phylogenetic tree.
Supplemental Data Set S1. Primers used in this study.
Supplemental Data Set S2. Differentially upregulated genes are enriched in plant–pathogen interaction pathway genes.
Supplemental Data Set S3. Data for all statistical analyses.
Supplemental File S1. Multiple protein alignment used for the phylogenetic tree shown in Supplemental Figure S15.
Supplemental File S2. Newick format of the tree shown in Supplemental Figure S15.
Supplementary Material
Acknowledgments
We thank Prof. Xiaoquan Qi from the Chinese Academy of Sciences for kindly providing the Pb strain F-co, Dr Hua Zhao, Dr Qiong Zhang, and Xiaona Zhou from State Key Laboratory of Crop Stress Biology for Arid Areas, Northwest A&F University (NWAFU) for their assistance of confocal laser microscope and protein purification. We appreciate Prof. Brett M. Tyler from Oregon State University, Prof. Ertao Wang from Chinese Academy of Sciences Center for Excellence in Molecular Plant Sciences, Prof. Qingmei Guan and Prof. Cong Jiang from the NWAFU for helpful suggestion and discussion. We thank Dr Ming Xu from NWAFU for transcriptome data analysis and phylogenetics construction.
Funding
This study was supported by the National Natural Science Foundation of China (31620103913, 31772150, and 31772116) and National Key Research and Development Program of China (2021YFD1401003), Shaanxi Innovation Team Project (2018TD-004), the China Agriculture Research System (CARS-3), International Science and Technology Cooperation Project of Shaanxi Provincial Key R&D Plan-key Project (2020KWZ-009) and 111 Project from the Ministry of Education of China (BP0719026).
Conflict of interest statement. None declared.
Contributor Information
Ning Wang, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, Shaanxi 712100, China; Pioneering Innovation Center for Wheat Stress Tolerance Improvement, State Key Laboratory of Crop Stress Biology for Arid Areas, Northwest A&F University, Yangling, Shaanxi 712100, China; State Key Laboratory of Crop Stress Biology for Arid Areas, College of Life Science, Northwest A&F University, Yangling, Shaanxi 712100, China.
Xin Fan, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, Shaanxi 712100, China; Pioneering Innovation Center for Wheat Stress Tolerance Improvement, State Key Laboratory of Crop Stress Biology for Arid Areas, Northwest A&F University, Yangling, Shaanxi 712100, China.
Mengying He, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, Shaanxi 712100, China; Pioneering Innovation Center for Wheat Stress Tolerance Improvement, State Key Laboratory of Crop Stress Biology for Arid Areas, Northwest A&F University, Yangling, Shaanxi 712100, China.
Zeyu Hu, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, Shaanxi 712100, China; Pioneering Innovation Center for Wheat Stress Tolerance Improvement, State Key Laboratory of Crop Stress Biology for Arid Areas, Northwest A&F University, Yangling, Shaanxi 712100, China.
Chunlei Tang, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, Shaanxi 712100, China; Pioneering Innovation Center for Wheat Stress Tolerance Improvement, State Key Laboratory of Crop Stress Biology for Arid Areas, Northwest A&F University, Yangling, Shaanxi 712100, China.
Shan Zhang, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, Shaanxi 712100, China; Pioneering Innovation Center for Wheat Stress Tolerance Improvement, State Key Laboratory of Crop Stress Biology for Arid Areas, Northwest A&F University, Yangling, Shaanxi 712100, China.
Dexing Lin, State Key Laboratory of Plant Cell and Chromosome Engineering, Center for Genome Editing, Institute of Genetics and Developmental Biology, Innovation Academy for Seed Design, Chinese Academy of Sciences, Beijing, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing, China.
Pengfei Gan, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, Shaanxi 712100, China; Pioneering Innovation Center for Wheat Stress Tolerance Improvement, State Key Laboratory of Crop Stress Biology for Arid Areas, Northwest A&F University, Yangling, Shaanxi 712100, China.
Jianfeng Wang, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, Shaanxi 712100, China; Pioneering Innovation Center for Wheat Stress Tolerance Improvement, State Key Laboratory of Crop Stress Biology for Arid Areas, Northwest A&F University, Yangling, Shaanxi 712100, China.
Xueling Huang, State Key Laboratory of Crop Stress Biology for Arid Areas, Northwest A&F University, Yangling, 712100, China.
Caixia Gao, State Key Laboratory of Plant Cell and Chromosome Engineering, Center for Genome Editing, Institute of Genetics and Developmental Biology, Innovation Academy for Seed Design, Chinese Academy of Sciences, Beijing, China; College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing, China.
Zhensheng Kang, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, Shaanxi 712100, China; Pioneering Innovation Center for Wheat Stress Tolerance Improvement, State Key Laboratory of Crop Stress Biology for Arid Areas, Northwest A&F University, Yangling, Shaanxi 712100, China.
Xiaojie Wang, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, Northwest A&F University, Yangling, Shaanxi 712100, China; Pioneering Innovation Center for Wheat Stress Tolerance Improvement, State Key Laboratory of Crop Stress Biology for Arid Areas, Northwest A&F University, Yangling, Shaanxi 712100, China.
X.W. and N.W. conceived and designed the experiments. N.W., M.H., X.F., Z.H., P.G., and S.Z. performed the experiments. D.L. and C.G. generated tanox10 and tawrky19 knockout plants. N.W. and J.W. developed Brachypodium distachyon materials. X.H. generated the TaWRKY19-overexpression transgenic wheat plants. N.W. and C.T. analyzed the data. N.W., Z.K., and X.W. wrote the paper.
The authors responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is Xiaojie Wang (wangxiaojie@nwsuaf.edu.cn).
References
- Adachi H, Nakano T, Miyagawa N, Ishihama N, Yoshioka M, Katou Y, Yaeno T, Shirasu K, Yoshioka H (2015) WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 27: 2645–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Ayliffe M, Singh D, Park R, Moscou M, Pryor T (2013) Infection of Brachypodium distachyon with selected grass rust pathogens. Mol Plant Microbe Interact 26: 946–957 [DOI] [PubMed] [Google Scholar]
- Barbieri M, Marcel TC, Niks RE (2011) Host status of false brome grass to the leaf rust fungus Puccinia brachypodii and the stripe rust fungus P. striiformis. Plant Dis 95: 1339–1345 [DOI] [PubMed] [Google Scholar]
- Bekaert M, Edger PP, Pires JC, Conant GC (2011) Two-phase resolution of polyploidy in the Arabidopsis metabolic network gives rise to relative and absolute dosage constraints. Plant Cell 23: 1719–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop JJ, Mohammed S, O’Flaherty M, Heck AJ, Slijper M, Menke FL (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics 6: 1198–1214 [DOI] [PubMed] [Google Scholar]
- Camejo D, Guzmán-Cedeno A, Moreno A (2016) Reactive oxygen species, essential molecules, during plant-pathogen interactions. Plant Physiol Biochem 103: 10–23 [DOI] [PubMed] [Google Scholar]
- Catalán P, Müller J, Hasterok R, Jenkins G, Mur LA, Langdon T, Betekhtin A, Siwinska D, Pimentel M, López-Alvarez D (2012) Evolution and taxonomic split of the model grass Brachypodium distachyon. Ann Bot 109: 385–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R, Van Kan JAL, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, et al. (2012) The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13: 414–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denness L, McKenna JF, Segonzac C, Wormit A, Madhou P, Bennett M, Mansfield J, Zipfel C, Hamann T (2011) Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species and jasmonic acid dependent process in Arabidopsis. Plant Physiol 156: 1364–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou DL, Zhou JM (2012) Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12: 484–495 [DOI] [PubMed] [Google Scholar]
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, Romeis T (2013) Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci USA 110: 8744–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott C, Zhou F, Spielmeyer W, Panstruga R, Schulze-Lefert P (2002) Functional conservation of wheat and rice Mlo orthologs in defense modulation to the powdery mildew fungus. Mol Plant Microbe Interact 15: 1069–1077 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Figueroa M, Alderman S, Garvin DF, Pfender WF (2013) Infection of Brachypodium distachyon by formae speciales of Puccinia graminis: early infection events and host-pathogen incompatibility. PLoS One 8: e56857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald TL, Powell JJ, Schneebeli K, Hsia MM, Gardiner DM, Bragg JN, McIntyre CL, Manners JM, Ayliffe M, Watt M, et al. (2015) Brachypodium as an emerging model for cereal-pathogen interactions. Ann Bot 115: 717–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S, Suzuki N, Miller G, Choi WG, Toyota M, Devireddy AR, Mittler R (2014) A tidal wave of signals: calcium and ROS at the forefront of rapid systemic signalling. Trends Plant Sci 19: 623–630 [DOI] [PubMed] [Google Scholar]
- Gu L, Han Z, Zhang L, Downie B, Zhao T (2013) Functional analysis of the 5’ regulatory region of the maize GALACTINOL SYNTHASE2 gene. Plant Sci 213: 38–45 [DOI] [PubMed] [Google Scholar]
- Hakmaoui A, Perez-Bueno ML, García-Fontana B, Camejo D, Jimenez A, Sevilla F, Baron M (2012) Analysis of the antioxidant response of Nicotiana benthamiana to infection with two strains of pepper mild mottle virus. J Exp Bot 63: 5487–5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein I, Barciszewska-Pacak M, Hrubikova K, Williamson S, Dinesen M, Soenderby IE, Sundar S, Jarmolowski A, Shirasu K, Lacomme C (2005) Virus-induced gene silencing-based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol 138: 2155–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzberg S, Brosio P, Gross C, Pogue GP (2002) Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J 30: 315–327 [DOI] [PubMed] [Google Scholar]
- Hu CH, Wei XY, Yuan BO, Yao LB, Ma TT, Zhang PP, Wang X, Wang PQ, Liu WT, Li WQ, et al. (2018) Genome-wide identification and functional analysis of NADPH oxidase family genes in wheat during development and environmental stress responses. Front Plant Sci 9: 906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, Tsunashima M, Hiei Y, Komari T (2015) Wheat (Triticum aestivum L.) transformation using immature embryos. Methods Mol Biol 1223: 189–198 [DOI] [PubMed] [Google Scholar]
- Ismagul A, Yang N, Maltseva E, Iskakova G, Mazonka I, Skiba Y, Bi H, Eliby S, Jataev S, Shavrukov Y, et al. (2018) A biolistic method for high-throughput production of transgenic wheat plants with single gene insertions. BMC Plant Biol 18: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y, Shirasu K, Zipfel C (2015) Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol 56: 1472–1480 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones J, Shirasu K, Menke F, Jones A, et al. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell 54: 43–55 [DOI] [PubMed] [Google Scholar]
- Kaku H, Nishizawa Y, Ishiiminami N, Akimototomiyama C, Dohmae N, Takio K, Minami E, Shibuya N (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci 103: 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang ZS, Huang LL, Buchenauer H (2002) Ultrastructural changes and localization of lignin and callose in compatible and incompatible interactions between wheat and Puccinia striiformis. J Plant Dis Protect 109: 25–37 [Google Scholar]
- Karen-Beth GS, Sonia I, Pilar C, Kranthi KM (2018) Brachypodium: a monocot grass model genus for plant biology. Plant Cell 30: 1673–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Ohura I, Kawakita K, Yokota N, Fujiwara M, Shimamoto K, Doke N, Yoshioka H (2007) Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19: 1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Vinod K, Zheng Z, Fan B, Chen Z (2008) Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol 8: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langner T, Kamoun S, Belhaj K (2018) CRISPR Crops: plant genome editing toward disease resistance. Ann Rev Phytopathol 56: 479–512 [DOI] [PubMed] [Google Scholar]
- Lazo GR, Stein Pascal A, Ludwig RA (1991) A DNA transformation–competent Arabidopsis genomic library in Agrobacterium. Biotechnology 9: 963–967 [DOI] [PubMed] [Google Scholar]
- Lee D, Lal NK, Lin ZD, Ma S, Liu J, Castro B, Toruño T, Dinesh-Kumar SP, Coaker G (2020) Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nat Commun 11:1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonelli S, Ankeny RA (2013) What makes a model organism? Endeavour 37: 209–212 [DOI] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Meng X, Shan L, He P (2016) Transcriptional regulation of pattern-triggered immunity in plants. Cell Host Microbe 19: 641–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li M, Yu LP, Zhou ZY, Liang XX, Liu ZX, Cai ZX, Gao LY, Zhang XJ, Wang YC, et al. (2014) The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RBOHD to control plant immunity. Cell Host Microbe 15: 329–338 [DOI] [PubMed] [Google Scholar]
- Ling HQ, Ma B, Shi X, Liu H, Dong L, Sun H, Gao Q, Zheng S, Li Y, Yu Y, et al. (2018) Genome sequence of the progenitor of wheat a subgenome Triticum urartu. Nature 557: 424–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Ziegler J, Zeier J, Birkenbihl RP, Somssich IE (2018) Botrytis cinerea B05.10 promotes disease development in arabidopsis by suppressing WRKY33-mediated host immunity. Plant Cell Environ 40: 2189–2206 [DOI] [PubMed] [Google Scholar]
- Mann DG, Lafayette PR, Abercrombie LL, King ZR, Mazarei M, Halter M, Stewart CN (2012) Gateway-compatible vectors for high-throughput gene functional analysis in switchgrass (Panicum virgatum L.) and other monocot species. Plant Biotechnol J 10: 226–236 [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Mersmann S, Bourdais G, Rietz S, Robatzek S (2010) Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol 154: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa S, Hauser F, Park J, Waadt R, Brandt B, Schroeder JI (2015) Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr Opin Plant Biol 28: 154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M, Ishikawa T, Fujiwara M, Fukao Y, Kawano Y, Kawai-Yamada M (2016) Plasma membrane microdomains are essential for Rac1-RBOHD/F-mediated immunity in rice. Plant Cell 28: 1966–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Păcurar DI, Thordal-Christensen H, Nielsen KK, Lenk I (2008) A high-throughput agrobacterium-mediated transformation system for the grass model species Brachypodium distachyon. Transgenic Res 17: 965–975 [DOI] [PubMed] [Google Scholar]
- Peng F, Si M, Zizhu Y, Fu Y, Yang Y, Yu Y, Bi C (2020) Rapid quantification of fungicide effectiveness on inhibiting wheat stripe rust pathogen (Puccinia striiformis f. sp. tritici). Plant Dis 104: 2434–2439 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Wang J, Gong Z, Zhou JM (2017) Apoplastic ROS signaling in plant immunity. Curr Opin Plant Biol 38: 92–100 [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ (1994) Extraction of total cellular DNA from plants, algae and fungi. Plant Mol Biol Manual 183–190 [Google Scholar]
- Samalova M, Brzobohaty B, Moore I (2005) pOp6/LhGR: a stringently regulated and highly responsive dexamethasone-inducible gene expression system for tobacco. Plant J 41: 919–935 [DOI] [PubMed] [Google Scholar]
- Segal LM, Wilson RA (2017) Reactive oxygen species metabolism and plant-fungal interactions. Fungal Genet Biol 110: 1–9 [DOI] [PubMed] [Google Scholar]
- Scofield SR, Huang L, Brandt AS, Gill BS (2005) Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol 138: 2165–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty NP, Kristensen BK, Newman MA, Møller K, Gregersen PL, Jørgensen HJL (2003) Association of hydrogen peroxide with restriction of Septoria tritici in resistant wheat. Physiol Mol Plant Pathol 62: 333–346 [Google Scholar]
- Slade AJ, Fuerstenberg SI, Loeffler D, Steine MN, Facciotti D (2005) A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat Biotechnol 23: 75–81 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R (2011) Respiratory burst oxidases: the engines of ROS signalling. Curr Opin Plant Biol 14: 691–699 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JD. (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defence response. Proc Natl Acad Sci USA 99: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JDG, Dangl JL (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo M, Altschmied L, Schweizer P, Kogel KH, Huckelhoven R (2006) Respiratory burst oxidase homologue A of barley contributes to penetration by the powdery mildew fungus Blumeria graminis f. sp. hordei. J Exp Bot 57: 3781–3791 [DOI] [PubMed] [Google Scholar]
- Vlieghe D, Sandelin A, De Bleser PJ, Vleminckx K, Wasserman WW, van Roy F, Lenhard B (2006) A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res 34: 95–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Garvin DF, Leong OM, Hayden DM (2006) Agrobacterium-mediated transformation and inbred line development in the model grass Brachypodium distachyon. Plant Cell Tissue Organ 84: 199–211 [Google Scholar]
- van der Linde K, Hemetsberger C, Kastner C, Kaschani F, van der Hoorn RA, Kumlehn J, Doehlemann G (2012) A maize cystatin suppresses host immunity by inhibiting apoplastic cysteine proteases. Plant Cell 24: 1285–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Wang N, Song N, Wang W, Wang J, Wang XJ, Kang ZS (2017) Overexpression of AtPAD4 in transgenic Brachypodium distachyon enhances resistance to Puccinia brachypodii. Plant Biol 19: 868–874 [DOI] [PubMed] [Google Scholar]
- Wang CF, Huang LL, Buchenauer H, Han QM, Zhang HC, Kang ZS (2007) Histochemical studies on the accumulation of reactive oxygen species (O2− and H2O2) in the incompatible and compatible interaction of wheat-Puccinia striiformis f. sp. tritici. Physiol Mol Plant 71: 230–239 [Google Scholar]
- Wang XD, Feng H, Tang CL, Bai PF, Wei GR, Huang LL, Kang ZS (2012) TaMCA4, a novel wheat metacaspase gene functions in programmed cell death induced by the fungal pathogen Puccinia striiformis f. sp. tritici. Mol Plant Microbe Interact 25: 755–764 [DOI] [PubMed] [Google Scholar]
- Wang XJ, Tang CL, Zhang H, Xu JR, Liu B, LV J, Han DJ, Huang LL, Kang ZS (2011) TaDAD2, a negative regulator of programmed cell death, is important for the interaction between wheat and the stripe rust fungus. Mol Plant Microbe Interact 24: 79–90 [DOI] [PubMed] [Google Scholar]
- Wang YP, Cheng X, Shan QW, Zhang Y, Liu JX, Gao CX, Qiu JL (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32: 947–951 [DOI] [PubMed] [Google Scholar]
- Wielopolska A, Townley H, Moore I, Waterhouse P, Helliwell C (2005) A high-throughput inducible RNAi vector for plants. Plant Biotechnol J 3: 583–590 [DOI] [PubMed] [Google Scholar]
- Wong HL, Pinontoan R, Hayashi K, Tabata R, Yaeno T, Hasegawa K, Kojima CH, Toshioka H, Iba K, Kawasaki T, et al. (2007) Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19: 4022–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Xu X, Gong Q, Li Z, Li Y, Wang S, Yang Y, Ma W, Liu L, Zhu B, et al. (2019) Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol Plant 12: 1434–1446 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zaidi SSEA, Mukhtar MS, Mansoor S (2018) Genome editing: targeting susceptibility genes for plant disease resistance. Trends Biotechnol 36: 898–906 [DOI] [PubMed] [Google Scholar]
- Zhang M, Chiang YH, Toruño TY, Lee D, Ma M, Liang X, Lal NK, Lemos M, Lu YJ, Ma S, et al. (2018) The MAP4 kinase SIK1 ensures robust extracellular ROS burst and antibacterial immunity in plants. Cell Host Microbe 24: 379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Shao F, Li Y, Cui H, Chen L, Li H, Zou Y, Long C, Lan L, Chai J, et al. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 17: 175–185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.