Abstract
Primary metabolism provides energy for growth and development as well as secondary metabolites for diverse environmental responses. Here we describe an unexpected consequence of disruption of a glycolytic enzyme enolase named LOW EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 2 (LOS2) in causing constitutive defense responses or autoimmunity in Arabidopsis thaliana. The autoimmunity in the los2 mutant is accompanied by a higher expression of about one-quarter of intracellular immune receptor nucleotide-binding leucine-rich repeat (NLR) genes in the genome and is partially dependent on one of these NLR genes. The LOS2 gene was hypothesized to produce an alternatively translated protein c-Myc Binding Protein (MBP-1) that functions as a transcriptional repressor. Complementation tests show that LOS2 executes its function in growth and immunity regulation through the canonical enolase activity but not the production of MBP-1. In addition, the autoimmunity in the los2 mutants leads to a higher accumulation of sugars and organic acids and a depletion of glycolytic metabolites. These findings indicate that LOS2 does not exert its function in immune responses through an alternatively translated protein MBP-1. Rather, they show that a perturbation of glycolysis from the reduction of the enolase activity results in activation of NLR-involved immune responses which further influences primary metabolism and plant growth, highlighting the complex interaction between primary metabolism and plant immunity.
One-sentence summary
Perturbation of glycolysis from reduced enolase activity triggers plant immune responses which in turn influences metabolism and growth.
Introduction
Plant primary metabolism generates energy, reducing equivalents and carbon skeletons which are not only essential for growth and development but also to fuel defense responses against pathogens (Bolton, 2009). Pathogen defenses in plants are carried out by a multi-layered immune system (Chisholm et al., 2006; Jones and Dangl, 2006). Intracellular nucleotide-binding leucine-rich repeat (NLR) proteins directly or indirectly detect effectors secreted from pathogens and mediate one of the critical layers of immune responses (Cui et al., 2015; Lolle et al., 2020). The recognition of pathogens triggers a diverse array of events including Ca2+ spikes, reactive oxygen species (ROS) burst, transcriptional reprograming of defense genes, and generation of defense phytohormones including salicylic acid (SA) (Buscaill and Rivas, 2014; Couto and Zipfel, 2016; Tang et al., 2017; Lolle et al., 2020). The immune signaling triggered by the activation of some NLR proteins requires the ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1)/PHYTOALEXIN-DEFICIENT 4 (PAD4) protein complex (Lapin et al., 2020) which also plays an essential role in boosting SA-related resistance programs (Cui et al., 2017). Primary metabolism has been shown to affect plant immunity via the production of primary metabolites. For instance, higher accumulation of sugars in leaves induces immune responses via interplay with phytohormones including SA (Bolouri Moghaddam and Van den Ende, 2012; Morkunas and Ratajczak, 2014; Trouvelot et al., 2014; Gebauer et al., 2017). Sorbitol in apple (Malus domestica) modulates the resistance to Alternaria alternata by regulating the expression of an NLR gene (Meng et al., 2018). More importantly, primary metabolism generates essential precursors to synthesize defense-related secondary metabolites including SA, pipecolic acid, and camalexin that have critical functions in plant immunity (Delaney et al., 1994; Glawischnig, 2007; Návarová et al., 2012; Shah et al., 2014; Piasecka et al., 2015). Therefore, primary metabolism plays a pivotal and often positive role in modulating immune responses.
Glycolysis is a central primary metabolic pathway conserved in almost all living organisms. The primary function of plant glycolysis is to oxidize hexoses to generate pyruvate, ATP, reductant as well as building blocks for a variety of amino acids, fatty acids, phytohormones, and secondary metabolites (Plaxton, 1996). Glycolysis is amphibolic and is able to utilize various low-molecular weight metabolites to generate hexoses including glucose and fructose via gluconeogenesis. Mutant plants with disrupted glycolysis pathway display drastic defects in growth and development such as stunted vegetative growth, distorted floral morphogenesis, reduced fertility, and dysfunction of guard cells (Rius et al., 2008; Prabhakar et al., 2010; Zhao and Assmann, 2011; Eremina et al., 2015), underscoring the essential function of glycolysis in plant growth and development.
Glycolytic proteins in nonplant organisms were reported to carry out versatile moonlighting functions in addition to their primary enzymatic functions, and their subcellular localizations determine which functions they execute (Pancholi, 2001; Kim and Dang, 2005; Boukouris et al., 2016; Didiasova et al., 2019). For instance, glyceraldehyde-3-phosphate dehydrogenase in human acts as a glycolytic enzyme in cytosol but is a component of transcriptional coactivator complex regulating expression of a histone gene H2B (Zheng et al., 2003). Also, the mammalian α-enolase (ENO1) gene encodes an enolase that catalyzes the conversion of 2-phophoglycerate (2-PG) to phosphoenolpyruvate (PEP) in cytoplasm, while its alternative translation product c-Myc Binding Protein (MBP-1) acts as a transcriptional repressor of the c-myc gene in the nucleus (Ray and Miller, 1991; Feo et al., 2000; Lung et al., 2010). Although moonlighting functions of glycolytic proteins are common in nonplant organisms (Rodríguez et al., 2001; Zheng et al., 2003; Shams et al., 2014; Boukouris et al., 2016), these functions are rarely reported in plants.
One enolase protein in Arabidopsis thaliana has been suggested to have such a moonlighting function. Arabidopsis has three enolase (ENO) genes, two of which (ENO1 and ENO2) encode proteins with demonstrated enolase activities (Andriotis et al., 2010). ENO1 functions in plastids while ENO2 is cytosolic and accounts for the majority of enolase activities throughout plant development (Andriotis et al., 2010). The loss-of-function eno1 mutations or overexpression ENO1 does not confer measurable defects (Andriotis et al., 2010), while knockdown mutants of ENO2 display multiple growth and developmental defects such as reduced shoot and root growth (Eremina et al., 2015). These biochemical and genetic studies indicate that ENO2 is the predominant enolase essential for plant growth and development. The ENO2 gene has also been implicated in plant abiotic stress responses. ENO2 is also known as LOW EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 2 (LOS2) because the los2-1 mutant was isolated based on having a low expression of the abiotic stress response gene RD29A in the Arabidopsis C24 accession (Lee et al., 2002). The los2-1 mutant is hypersensitive to freezing stress (Lee et al., 2002) and salt stress (Barkla et al., 2009) compared to the C24 wild-type (WT) plants. By analogy to its mammalian homolog ENO1, the Arabidopsis LOS2 gene was postulated to encode two proteins, the full-length isoform enolase (named as ENO2 herein) and the N-terminal truncated isoform MBP-1, via distinct translational initiation sites (Kang et al., 2013). The LOS2 function in abiotic stress response was hypothesized to result from the transcriptional repressor activity of MBP-1 or the proposed moonlighting function of enolase as a transcriptional repressor. Recombinant ENO2 protein was shown to bind in vitro to the promoter sequences of ZAT10, which is a negative regulator of the cold-responsive gene RD29A, and the los2-1 mutant has a higher expression of ZAT10 compared to the C24 WT plant (Lee et al., 2002). Additionally, plants that carry the transgene coding for MBP-1 under the strong CaMV 35S promoter exhibit hypersensitivity to ABA (Kang et al., 2013; Liu et al., 2017) and osmotic stress (Kang et al., 2013). However, the MBP-1 protein was not easily detected in plants, and this was attributed to proteasomal protein degradation (Kang et al., 2013). Interestingly, plants overexpressing MBP-1 exhibit stunted growth, reduced fertility, decreased expression of endogenous LOS2 transcripts, and reduced enolase activity similarly to the loss-of-function los2 mutants (Kang et al., 2013; Eremina et al., 2015). It was proposed that LOS2 gene alternatively translates MBP-1 to repress its own expression to maintain enolase homeostasis (Eremina et al., 2015). Despite the study of the effect of MBP-1 overexpression, the existence and function of endogenous MBP-1 have not yet been determined.
In this study, we found an unexpected role of the enolase coding gene LOS2 in repressing plant immunity, and we further investigated the contribution of the canonical function of enolase and the putative endogenous MBP-1 to the role of LOS2 in growth, development, and immunity. We found that the los2 mutants exhibited enhanced disease resistance and autoimmunity, which were largely dependent on NLR and SA signaling. Genetic elimination of the potential production of the MBP-1 protein did not yield measurable effect on plant growth or immunity, while the long-form ENO2 alone accounts for the full LOS2 function observed. In addition, the MBP-1 protein could not be detected from the LOS2 gene under conditions used in this study, and it could be detected at a very low level when sequence encoding only MBP-1 was expressed under the strong 35S promoter. Therefore, MBP-1 is unlikely a bona fide alternative translation product of the LOS2 gene under normal growth conditions. Furthermore, site-directed mutagenesis of a conserved amino acid critical for enolase activity could not recue any of the los2 mutant defects examined, supporting that canonical enolase activity of ENO2 mediates all roles of LOS2 in growth and immunity. Our study strongly indicates the use of enolase function of the LOS2 gene in regulating growth, development, and immunity in Arabidopsis and highlights the large effect that perturbation of glycolysis has on growth and environmental responses, both directly and indirectly.
Results
A smo3/los2 mutation constitutively activates immune responses
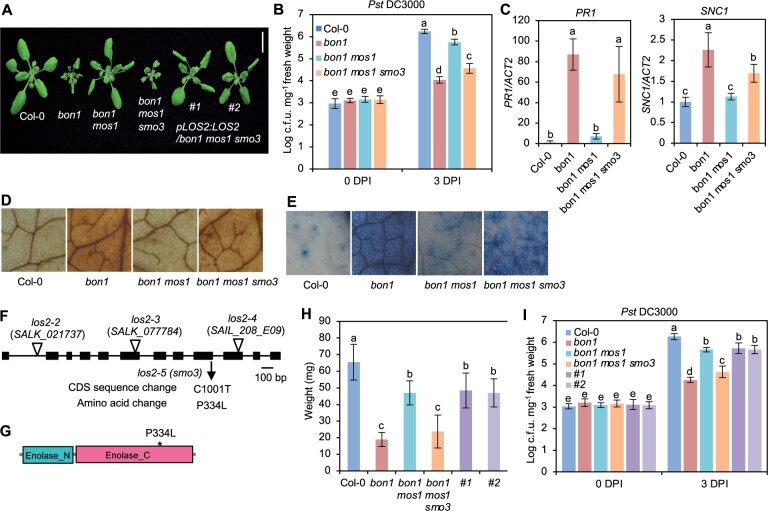
We sought enhancer mutations for immune responses via a sensitized genetic screen using the bon1 mos1 mutant. The loss-of-function mutant bon1 displays autoimmunity phenotypes due to the elevated expression of an NLR gene SUPPRESSOR OF npr1-1, CONSTITUTIVE 1 (SNC1) (Yang and Hua, 2004), and the mos1 mutation restores the SNC1 expression to the WT level and suppresses autoimmunity of bon1 (Bao et al., 2014). An ethylmethylsulfonate mutagenesis screen was performed to identify suppressor mutations that restored autoimmunity to the bon1 mos1 mutant, and those mutated alleles were named as suppressor of mos1 bon1 (smo) (Yang et al., 2020). The smo3 mutation was one of the mutations identified; and the bon1 mos1 smo3 triple mutant was significantly smaller than bon1 mos1 mutant (Figure 1, A and H). In addition, the triple mutant displayed enhanced disease resistance to a virulent bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 compared to bon1 mos1 mutant (Figure 1B). Also, under nonpathogenic conditions, the bon1 mos1 smo3 mutant had increased expression of PR1 (a defense response marker gene) and SNC1, as well as accumulation of H2O2 and cell death as compared to bon1 mos1 mutant (Figure 1, C–E). These results indicate that bon1 mos1 smo3 is an autoimmune mutant, and the smo3 mutation constitutively activates immune responses in the bon1 mos1 mutant.
Figure 1.
A mutation in LOS2 constitutively activates immune responses. A, Morphology of Col-0 WT, bon1, bon1 mos1, bon1 mos1 smo3/los2 and two representative complementation lines of pLOS:LOS2 in bon1 mos1 smo3 (#1 and #2). Scale bar = 1 cm. B–E, Growth of bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 (B), analysis of PR1 and SNC1 gene expression (C), DAB staining (D), and trypan blue staining (E) in Col-0, bon1, bon1 mos1, and bon1 mos1 smo3/los2. F, Position of the point mutation generated from the ethylmethylsulfonate screen and T-DNA insertion mutations in LOS2. The position of T-DNA insertions was adapted from Eremina et al. (2015). G, Predicted functional domains of LOS2 protein by SMART. * indicates smo3/los2-5 mutation. H and I, Quantification of weight (H) and growth of bacterial pathogen Pst DC3000 (I) in Col-0, bon1, bon1 mos1, bon1 mos1 smo3, complementation lines #1 and #2. N ≥ 23 for (H). Six biological replicates were performed for (B) and (I). Three biological replicates were performed for (C). Error bars represent standard deviation (sd). Different letters indicate significant difference tested by one-way ANOVA/Duncan’s multiple range test via R 3.6.3 with “agricolae” package, P < 0.05.
Using Mapping by Sequencing (Zhu et al., 2012; Hua et al., 2017) on the F2 population of a bon1 mos1 smo3 and bon1 mos1 cross, we localized the smo3 mutation to the LOS2 gene (Figure 1F), and the resulting proline-334-to-leucine change was in the C-terminal enolase domain of LOS2 (Figure 1G). To verify that SMO3 is LOS2, a WT genomic fragment of LOS2 (pLOS2:LOS2) was transformed into bon1 mos1 smo3 mutant for complementation test. The majority of the T1 plants showed the bon1 mos1 mutant phenotype, and co-segregation of the bon1 mos1-like plants with the transgene was observed in all six independent T2 lines analyzed (Figure 1, A and H). In addition, these complementation lines were more susceptible to Pst DC3000 compared to bon1 mos1 smo3 mutant, at a similar level to the bon1 mos1 mutant (Figure 1I). These results confirm that LOS2 is SMO3 and that a nonsynonymous mutation in LOS2 activates immune responses in the bon1 mos1 mutant background.
los2 mutants have autoimmune responses
We further characterized the disease-resistance phenotype of the smo3/los2 single mutant, which is hereafter designated as los2-5. At 3 days after inoculation, the growth of the virulent pathogen Pst DC3000 was about 10 times lower in the los2-5 mutant compared to the Col-0 WT, indicating an enhanced disease resistance in the mutant (Supplemental Figure S1A). Even in the absence of pathogen infection, the los2-5 mutant already had elevated expression of PR1, suggesting a defense response induction (Supplemental Figure S1B). These analyses indicate that LOS2 is a negative regulator of plant immunity and that the los2-5 mutant exhibits autoimmunity, or upregulated immune responses in the absence of infection. Because the los2-5 mutant had a weaker growth defect than bon1 mos1 los2 (Figures 1, A and 2, A) and bon1 mos1 has a residual autoimmunity, it is likely that the los2 defect is independent and additive with defects of bon1 mos1. Therefore, the bon1 mos1 mutant provided a sensitized background for isolating weak autoimmune mutants, which has been seen in another SMO gene HOS15 (Yang et al., 2020).
Figure 2.
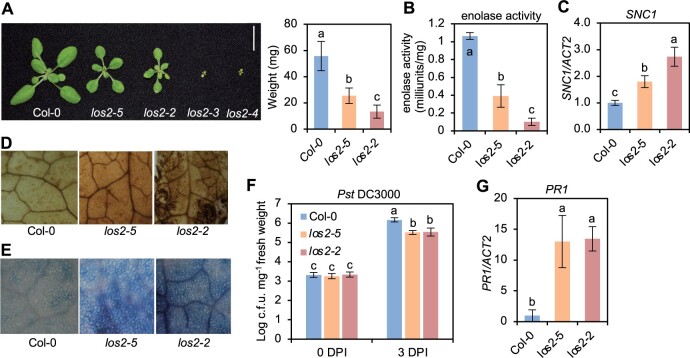
The los2 mutants display constitutively activated immune responses. A, Morphology of Col-0, los2-5, los2-2, los2-3, and los2-4 mutants, and quantification of weight of Col-0, los2-5, and los2-2. Scale bar = 1 cm. N = 20. B–G, Enolase activity (B), analysis of SNC1 gene expression (C), DAB staining (D), trypan blue staining (E), growth of bacterial pathogen Pst DC3000 (F), and analysis of PR1 gene expression (G) in 2-week-old WT, los2-5, and los2-2. Four biological replicates were performed for (B). Three biological replicates were performed for (C) and (G). Six biological replicates were performed for (F). Error bars represent sd. Different letters indicate significant difference tested by one-way ANOVA/Duncan’s multiple range test via R 3.6.3 with “agricolae” package, P < 0.05.
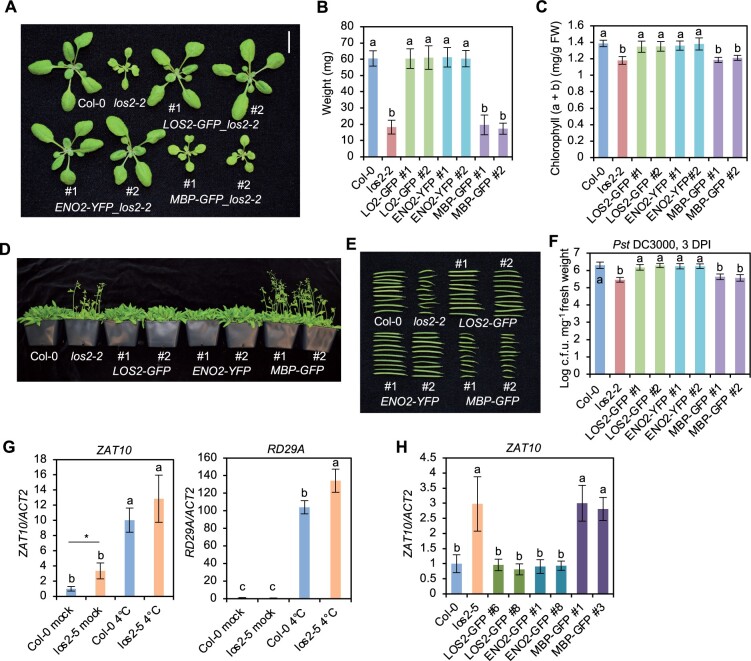
The LOS2 gene has been studied previously for its role in abiotic stress tolerance but has not been characterized in the context of biotic stress responses. We therefore examined three previously reported los2 mutants in the Col-0 background for their immune phenotypes. The los2-2 (SALK_021737) contains a T-DNA insertion in the first intron (Figure 1F) which results in reduced LOS2 transcript accumulation and reduced enolase activity compared to WT (Eremina et al., 2015). Both the los2-3 (SALK_077784) and los2-4 (SAIL_208_E09) alleles, with T-DNAs inserted in exons (Figure 1F), have even lower enolase activities compared to the los2-2 mutant (Eremina et al., 2015). We found that the enolase activity in the los2-5 mutant isolated in this study was reduced to 37% of the WT level, to a less extent as compared to the los2-2 mutant which had 10% of WT enolase activity (Figure 2B; Supplemental Figure S1C). Correlated with the strength of the molecular defects of LOS2 mutations, los2-5, los2-2, and los2-3/los2-4 had increasingly stronger growth defects. Both los-5 and los2-2 had smaller rosette size and exhibited early flowering compared to WT with these defects stronger in los2-2 (Figure 2A; Supplemental Figure S1D). Additionally, the los2-2 mutant had shorter siliques (Supplemental Figure S1E) similarly to previously reported (Eremina et al., 2015), and accumulated less chlorophyll compared to WT (Supplemental Figure S1F). These two defects were not observed in the los2-5 (Supplemental Figure S1, E and F) but were attributed to the defect of the LOS2 gene (see later result), further supporting los2-2 being a stronger allele than los2-5. The other two alleles, los2-3 and los2-4, exhibited more severe defects in growth and development than los2-2 (Eremina et al., 2015; Figure 2A). They were extremely tiny and were not able to set seeds in standard growth conditions. Therefore, we utilized los2-5 and los2-2 for further analyses.
Both the los2-5 and los2-2 mutants exhibited autoimmunity. They had increased expression of SNC1 and accumulated more H2O2 and cell death as compared to WT in the absence of pathogen infection, while the los2-2 mutant had a stronger effect as compared to the los2-5 mutant (Figure 2, C–E). The los2-5 and los2-2 mutants also had elevated resistance to Pst DC3000 as compared to WT; and the level of increase of resistance was similar in the two mutants (Figure 2F). In addition, both mutants showed a similar fold increase in the expression of PR1 compared to the WT (Figure 2G). Taken together, these data indicate that los2-2 mutant is a stronger allele in growth and development than los2-5 mutant while the increase of disease resistance compared to the WT is comparable between these two mutants.
Defense-related genes are upregulated in the los2 mutant
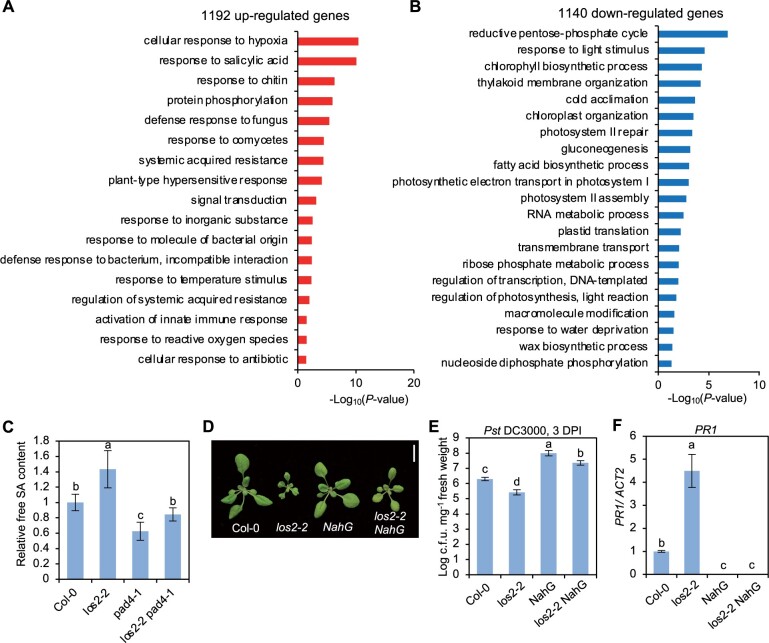
To investigate how LOS2 influences plant immunity, we carried out transcriptomic analyses of the los2-5 mutant and Col-0 WT by mRNA sequencing (RNA-seq). Two-week-old shoots were sampled with three biological repeats for RNA extraction and subsequent sequencing. Differentially expressed genes (DEGs) between los2-5 and the WT were selected with false discovery rate (FDR) set at less than 0.05. The los2-5 mutant had 1192 up-DEGs and 1140 down-DEGs compared to the WT (Supplemental Figure S2A; Supplemental Data Set S1). The up- and down-DEGs were subject to Gene Ontology (GO) enrichment analysis using the GO Term Enrichment tool on TAIR (https://www.arabidopsis.org/tools/go_term_enrichment.jsp). Among up-DEGs for the los2-5 mutant, GO terms related to immune responses such as “response to salicylic acid”, “response to chitin” and “defense response to fungus” were highly enriched (Figure 3A). In particular, genes involved in response to SA were overrepresented (Figure 3A). Genes associated with “plant-type hypersensitive response” and “response to reactive oxygen species” were also enriched (Figure 3A). This was consistent with the H2O2 accumulation and cell death phenotypes observed in the los2 mutants (Figure 2, D and E). In contrast, genes associated with plant growth and development, metabolism and response to abiotic stresses were overrepresented among down-DEGs of los2-5 mutant (Figure 3B). GO terms associated with primary metabolism such as reductive pentose-phosphate cycle, gluconeogenesis, and fatty acid biosynthetic process were enriched in down-DEGs of the mutant versus the WT (Figure 3B). Since LOS2 is a glycolytic enzyme, a defect in glycolysis in the los2-5 mutant is expected to affect plant metabolism. Chlorophyll biosynthetic process was overrepresented in DEGs (Figure 3B), which was consistent with the observation that the los2 mutants had less chlorophyll content compared to WT (Supplemental Figure S1F). Additionally, genes involved in cold acclimation were enriched among downregulated genes in los2-5 mutant (Figure 3B). An altered expression of cold-response genes had also been reported for the los2-1 mutant (Lee et al., 2002). These results support that LOS2 is a negative regulator of plant immunity while being a positive regulator of plant growth and development.
Figure 3.
The activated defense responses in los2 mutants are dependent on salicylic acid pathway. A and B, GO enrichment analysis of 1192 upregulated genes (A) and 1140 downregulated genes (B) in los2-5 mutant as compared to WT. Enriched GO terms were identified by Panther (https://www.arabidopsis.org/tools/go_term_enrichment.jsp). C, Relative amount of free SA in Col-0, los2-2, pad4-1 and los2-2 pad4-1. The amount of salicylic acid in Col-0 was set to “1.” Five biological replicates were performed. D, Morphology of Col-0, los2-2, NahG and los2-2 NahG. Scale bar = 1 cm. E and F, Growth of bacterial pathogen Pst DC3000 (E) and analysis of PR1 gene expression (F) in Col-0, los2-2, NahG and los2-2 NahG. Six and three biological replicates were performed for (E) and (F), respectively. Error bars represent sd. Different letters indicate significant difference tested by one-way ANOVA/Duncan’s multiple range test via R 3.6.3 with “agricolae” package, P < 0.05.
The autoimmunity in los2-2 mutant is dependent on the NLR/SA pathway and reduces plant growth
To assess the involvement of SA in the autoimmunity in the los2 mutant as revealed by the RNA-seq data, we first quantified the SA level in the los2 mutant. The los2-2 mutant accumulated about 40% more free SA than the WT in the absence of pathogen infection (Figure 3C). To assess the role of SA increase in autoimmunity, we introduced into los2-2 mutant the NahG gene which encodes a salicylate hydroxylase to degrade SA. The NahG transgene largely suppressed the growth defect of los2-2, and the los2 nahG plant had significantly larger rosette size compared to the los2-2 mutant (Figure 3D; Supplemental Figure S2B). The NahG transgene also reduced the elevated immune responses in the los2-2 mutant, including enhanced resistance to Pst DC3000 and increased PR1 expression (Figure 3, E and F). This indicates that SA increase is responsible for autoimmunity and partially accounts for growth defects of los2.
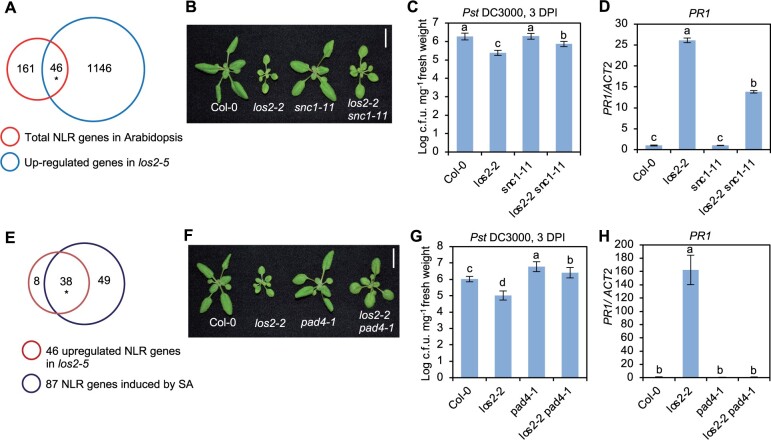
Since los2-5 was identified as a suppressor of bon1 mos1 mutant where upregulation of the NLR gene SNC1 is the key for autoimmunity, we asked whether NLR genes played a role in LOS2-repressed immune responses. In Arabidopsis thaliana Col-0 accession, 207 NLR or NLR-like genes (hereafter, NLR genes) are identified (Meyers et al., 2003). RNA-seq data revealed that 46 NLR genes were upregulated in los2-5 mutant and no NLR genes were downregulated in los2-5 mutant (Figure 4A). The contribution of the SNC1 gene to the elevated immune responses in los2 mutant was examined by introducing the loss of SNC1 mutation snc1-11 into los2-2 (Figure 4B). The los2-2 snc1-11 double mutant had reduced disease resistance and PR1 gene expression compared to the los2-2 mutant (Figure 4, C and D). These results suggest that SNC1, and perhaps other NLR genes, play a critical role in the activated immune responses in the los2 mutants.
Figure 4.
NLR genes and NLR/SA signaling contribute to LOS2-mediated immune responses. A, Venn diagram of 1192 upregulated genes in los2-5 mutant and 207 total NLR genes in Arabidopsis. Asterisk indicates a significant difference tested by Fisher’s exact test, P < 3.158e–14. B, Morphology and quantification of weight of Col-0, los2-2, snc1-11 and los2-2 snc1-11. Scale bar = 1 cm. C and D, Growth of bacterial pathogen Pst DC3000 (C) and analysis of PR1 gene expression (D) in Col-0, los2-2, snc1-11 and los2-2 snc1-11. E, Venn diagram of 46 upregulated NLR genes in los2-5 mutant and 87 SA-induced NLR genes in Arabidopsis. Asterisk indicates a significant difference tested by Fisher’s exact test, P < 6.917e–90. F, Morphology of Col-0, los2-2, pad4-1 and los2-2 pad4-1. Scale bar = 1 cm. G and H, Growth of bacterial pathogen Pst DC3000 (G) and analysis of PR1 gene expression (H) in Col-0, los2-2, pad4-1 and los2-2 pad4-1. Six biological replicates were performed for (C) and (G). Three biological replicates were performed for (D) and (H). Error bars represent sd. Different letters indicate significant difference tested by one-way ANOVA/Duncan’s multiple range test via R 3.6.3 with “agricolae” package, P < 0.05. DPI, days post-inoculation.
SA and NLR signaling are intertwined as NLR activation can induce SA and many NLR genes are induced by SA (Yang et al., 2021). Among the 46 NLR genes upregulated in los2 mutant, 38 NLR genes can be induced by SA (Figure 4E). This suggested an amplification signaling loop from NLR to SA and to NLR in the los2 mutant. To test the involvement of NLR/SA amplification in autoimmunity of los2, we introduced in los2-2 a loss-of-function mutation of PAD4, a critical component in SA- and NLR-mediated immune signaling (Figure 4F). The pad4-1 mutant reduced the accumulation of SA in los2-2 to WT level (Figure 3C), indicating that the over-accumulation of SA in los2-2 mutant was dependent on functional PAD4. Additionally, the enhanced resistance to Pst DC3000 and increased PR1 expression in the los2-2 mutant were largely decreased by the pad4-1 mutation (Figure 4, G and H). This indicates that the los2 autoimmunity is dependent on the function of PAD4.
Because autoimmunity leads to growth defects, we examined its contribution to the defects of growth and development observed in the los2 mutants. We used pad4 and snc1 mutation to block or reduce autoimmunity in los2-2. The double mutant los2-2 pad4-1 displayed similar flowering time and chlorophyll content to the los2-2 mutant (Supplemental Figure 3, A and B). In contrast, both the double mutants los2-2 snc1-11 and los2-2 pad4-1 had a larger rosette size as compared to the los2-2 mutant but were still smaller compared to the WT (Figure 4, B and F; Supplemental Figure S3, C and D). This indicates that the growth defects of the los2 mutants are not entirely due to a deficiency in primary metabolism but rather due to the activated defense responses to a large extent.
The enolase form is important for the LOS2 function in growth and immunity regulation
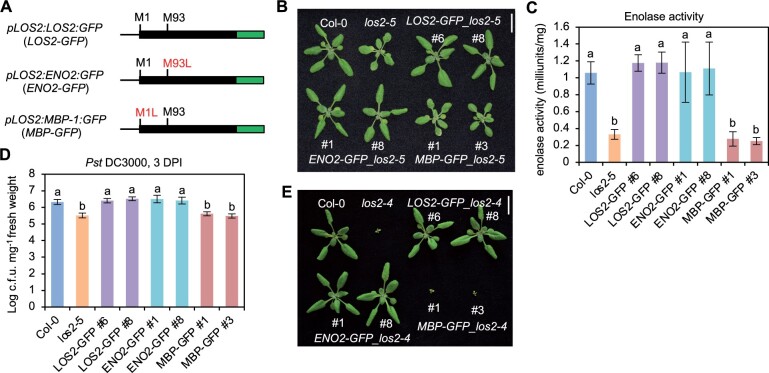
The LOS2 gene was postulated to encode two isoforms, enolase and MBP-1, via different translation initiation sites (Kang et al., 2013), and its function in abiotic stress response was attributed to the transcriptional repressor MBP-1 (Kang et al., 2013; Liu et al., 2017). We tested the contribution of these two alternatively translated proteins to the different roles of the LOS2 gene. For simplicity, we named the long protein form starting from the first methionine as ENO2 (enolase) and the short protein form starting from the second methionine residue +93 as MBP. The LOS2 genomic fragment was in vitro mutagenized so that only one protein form was expected to be produced from the LOS2 gene, and the proteins were tagged by GFP at their C-termini to aid protein detection (Figure 5A). The Met93 mutation to Leu (M93L) was expected to produce only the long form ENO2 (pLOS2:ENO2:GFP or ENO2-GFP), and Met1 mutation to Leu (M1L) was expected to produce only the short form MBP (pLOS2:MBP:GFP or MBP-GFP) (Figure 5A). These constructs were transformed into the los2-5 mutant background, which had no defects in seed setting. A similarly constructed WT LOS2 gene, pLOS2:LOS2:GFP (LOS2-GFP), was also transformed into the los2-5 mutant as a control. Of the 13 independent transgenic lines harboring LOS2-GFP, all had a WT phenotype, indicating that the GFP tag did not compromise the LOS2 activity in rescuing the los2-5 defects. Analysis of two representative transgenic lines (#6 and #8) harboring LOS2-GFP in los2-5 revealed WT enolase activity, biomass and WT level of disease resistance to Pst DC3000 in these lines (Figure 5, B–D; Supplemental Figure S4A). Interestingly, of the 27 transgenic lines containing the ENO2 form, all exhibited a WT growth phenotype, similarly to the LOS2 form. Two representative ENO2-GFP lines in los2-5 (#1 and #8) analyzed also exhibited WT phenotypes in enolase activity, biomass, and disease resistance (Figure 5, B–D; Supplemental Figure S4A). In contrast, MBP-GFP did not rescue growth defects of the los2-5 mutant in any of the 12 transgenic lines analyzed. Two representative MBP-GFP transgenic plants (#1 and #3) analyzed were similar to the los2-5 mutant in enolase activity, biomass, and resistance to Pst DC3000 (Figure 5, B–D; Supplemental Figure S4A). Together with the full rescue of los2-5 by ENO2, this indicates that a lack of MBP production does not compromise the LOS2 function in growth or immunity.
Figure 5.
ENO2, but not MBP-1, contributes to plant growth and immunity regulation. A, Simplified gene structures of fusion proteins LOS2-GFP and two mutated forms ENO2-GFP and MBP-GFP. Genomic DNA was used for fusing with GFP. B–D, Morphology (B), enolase activity (C), and growth of bacterial pathogen Pst DC3000 (D) of Col-0, los2-5 and complementation lines of LOS2-GFP (#6 and #8), ENO2-GFP (#1 and #8) and MBP-GFP (#1 and #3) in los2-5 background. E, Morphology of Col-0, los2-4 and complementation lines of LOS2-GFP (#6 and #8), ENO2-GFP (#1 and #8), and MBP-GFP (#1 and #3) in los2-4 background. Four and six biological replicates were performed for (C) and (D), respectively. Error bars represent sd. Different letters indicate significant difference tested by one-way ANOVA/Duncan’s multiple range test via R 3.6.3 with “agricolae” package, P < 0.05. Scale bar = 1 cm for (B) and (E). ; L, leucine; M, methionine.
Because los2-5 is a weak allele, it remained possible that a minor contribution from MBP was not measurable in this background but could be seen in the stronger allele los2-4 background. As the los2-4 mutant was too weak to be transformed, we introgressed the transgenes of LOS2-GFP, ENO2-GFP, and MBP-GFP from four independent lines in the weak allele los2-5 background to the strong knock-down mutant los2-4. Similar to the observations in los2-5, LOS2-GFP, and ENO2-GFP both completely rescued the los2-4 mutant defects while MBP-GFP did not (Figure 5E; Supplemental Figure S4B). These analyses further indicate that the ENO2 form alone but not MBP is sufficient to account for the function of LOS2 in growth and immunity regulation.
The enzymatic activity but not transcriptional activity is critical for LOS2 function in growth and immunity regulation
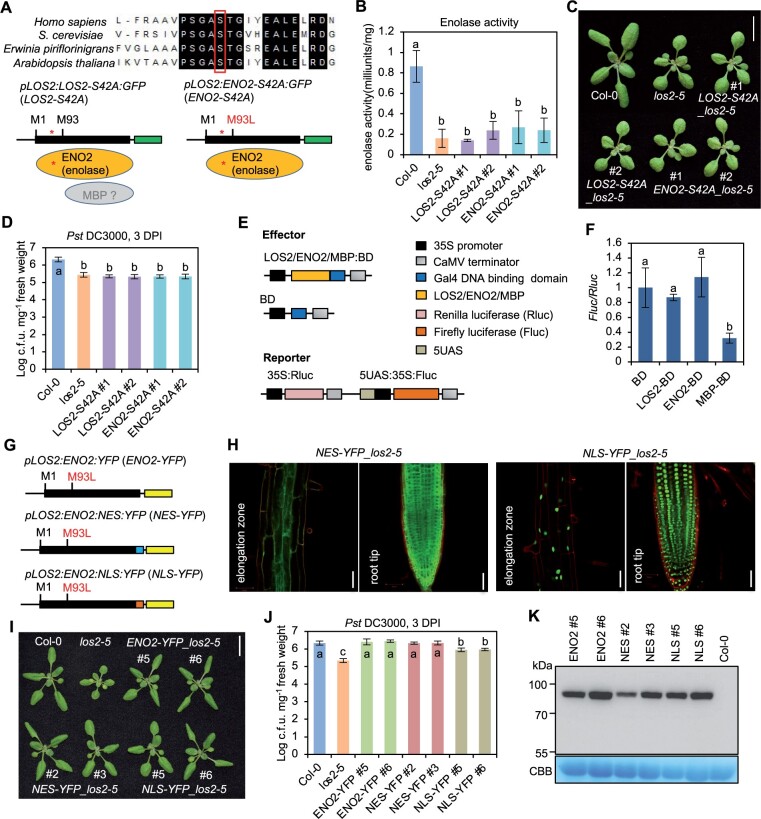
Since the ENO2 form is sufficient for the role of LOS2 in growth and immunity regulation, we tested if the enolase activity of LOS2 gene was critical for the regulation. To this end, we generated mutated forms of LOS2 and ENO2 in which serine at position 42 (S42) was mutated to alanine to abolish enolase activity (Figure 6A). The S42 residue in Arabidopsis is highly conserved among enolases in eukaryotes and prokaryotes (Figure 6A), and its corresponding S40 residue is critical for magnesium binding in human and is essential for enolase activity in yeast (Kang et al., 2008; https://www.uniprot.org/uniprot/P00924). Two constructs pLOS2:LOS2-S42A:GFP (LOS2-S42A) and pLOS2:ENO2-S42A:GFP (ENO2-S42A) containing the S42A mutation in LOS2 and ENO2, respectively, were transformed into the los2-5 background. Two representative transgenic lines for each construct were selected based on the comparable expression levels of fusion proteins (Supplemental Figure S5). The S42A mutation indeed eliminated the enolase activity of LOS2 and ENO2 as neither the LOS2-S42A nor the ENO2-S42A transgene restored the enolase activity in the los2-5 mutant (Figure 6B). Importantly, these catalytic dead transgenes, unlike their WT counterparts, could not complement the growth or immunity defects of the los2-5 mutant (Figure 6, C and D; Supplemental Figure S6A). These results indicate the importance of enolase activity of LOS2 in plant growth and immunity regulation.
Figure 6.
The enzymatic activity of ENO2 contributes to plant growth and immunity regulation. A, Sequence alignment of enolases from human, yeast, bacterium, and Arabidopsis. The partial alignment result is shown. The red box and star indicate the serine at position 42 in Arabidopsis which was mutated into Alanine (S42A). B–D, Enolase activity (B), morphology (C), and growth of bacterial pathogen Pst DC3000 (D) in Col-0, los2-5 and two representative lines of pLOS2:LOS2-S42A:GFP and pLOS2:ENO2-S42A:GFP in los2-5 background. E, Schematics of constructs used for the dual-luciferase reporter assay. F, Firefly luciferase expression level after normalized to renilla luciferase expression. The expression of BD was set to 1. G, Simplified gene structure for fusion proteins ENO2-YFP, NES-GFP, and NLS-GFP. H, Confocal imaging of fusion proteins in Arabidopsis root treated with propidium iodide. Scale bar = 50 μm. I, Morphology of Col-0, los2-5 and complementation lines of ENO2-YFP (#5 and #6), NES-YFP (#2 and #3) and NLS-YFP (#5 and #6) in los2-5 background. J, Growth of bacterial pathogen Pst DC3000 in Col-0, los2-5, ENO2-YFP (#5 and #6), NES-YFP (#2 and #3) and NLS-YFP (#5 and #6) in los2-5 background. K, Immunoblot detecting the fusion proteins by anti-GFP antibody in complementation lines of ENO2-YFP (#5 and #6), NES-YFP (#2 and #3), and NLS-YFP (#5 and #6) in los2-5 background. Col-0 was used as a negative control. Four and three biological replicates were performed for (B) and (F), respectively. Six biological replicates were performed for (D) and (J). Error bars represent sd. Different letters indicate significant difference tested by one-way ANOVA/Duncan’s multiple range test via R 3.6.3 with “agricolae” package, P < 0.05. Scale bar in (C) and (I) is 1 cm. CBB, Coomassie brilliant blue.
Because the C-terminus of ENO2 protein has the same amino acid sequences as the postulated MBP protein, we asked whether the ENO2 protein could execute the transcriptional repressor activity as MBP-1 protein in mammalian cells. We utilized a dual-luciferase reporter assay in Arabidopsis protoplasts to examine the repressor activity (Figure 6E). Effector proteins including LOS2, ENO2, and MBP were fused to Gal4 DNA binding domain (BD), respectively. Each effector construct was co-transformed with a reporter construct into protoplasts, and the expression of firefly luciferase was used as an indicator of the transcription repressor activity of the effector. Protoplasts co-transformed either LOS2-BD or ENO2-BD with the reporter exhibited a similar luciferase activity as compared to that of the empty control BD (Figure 6F). In contrast, protoplasts harboring MBP-BD and reporter showed a significantly decreased expression of luciferase as compared to the control BD (Figure 6F). These data suggest that ENO2 may not have a transcriptional repressor activity.
To further examine the potential function of ENO2 as a transcriptional repressor, we tested the function of a nucleus-excluded ENO2 form in plants. The YFP-tagged ENO2 protein was fused with a nuclear export signal (NES) and transformed into the los2-5 mutant background (Figure 6G). While the ENO2-YFP protein was found in both nucleus and cytosol (Kang et al., 2013), the ENO2-NES-YFP was found solely in cytosol (Figure 6H). As expected, the ENO2-YFP transgene completely rescued the los2-5 growth and immunity defects (Figure 6, I and J; Supplemental Figure S6B). Interestingly, ENO2-NES also fully complement los2 mutant phenotypes in growth and immunity (Figure 6, I and J; Supplemental Figure S6B). This indicates that ENO2 in cytosol is sufficient to carry out all functions of LOS2 in plant growth and immunity regulation, arguing against an involvement of transcriptional repressor activity in this regulation. In addition, we generated a nucleus localized ENO2-YFP by fusing it to a nuclear localization signal (NLS) and transformed it into los2-5. Microscopy on transgenic plants revealed that the florescent signal from the ENO2-NLS-YFP protein was accumulated in the nucleus (Figure 6H). This ENO2-NLS-YFP form, unlike the ENO2-YFP or ENO2-NES-YFP, did not rescue the immunity defects of los2-5, but it did rescue its growth defect (Figure 6, I and J; Supplemental Figure S6B). We excluded the possibility that a lower protein expression of ENO2-NLS-YFP was responsible for the difference because immunoblot analysis showed that ENO2-YFP and ENO2-NLS-YFP had comparable protein levels even with line #2 of ENO2-NES-YFP having the lowest protein expression (Figure 6K). This indicates that the ENO2-NES form has the same activity as ENO2 while ENO2-NLS has a lower activity than ENO2. Therefore, the cytosolic function of ENO2 can account for all activities of LOS2. The partial activity of ENO2-NLS might be due to its transient and small amount of cytosol accumulation.
Autoimmunity in the los2 mutants induces accumulation of sugars and organic acids
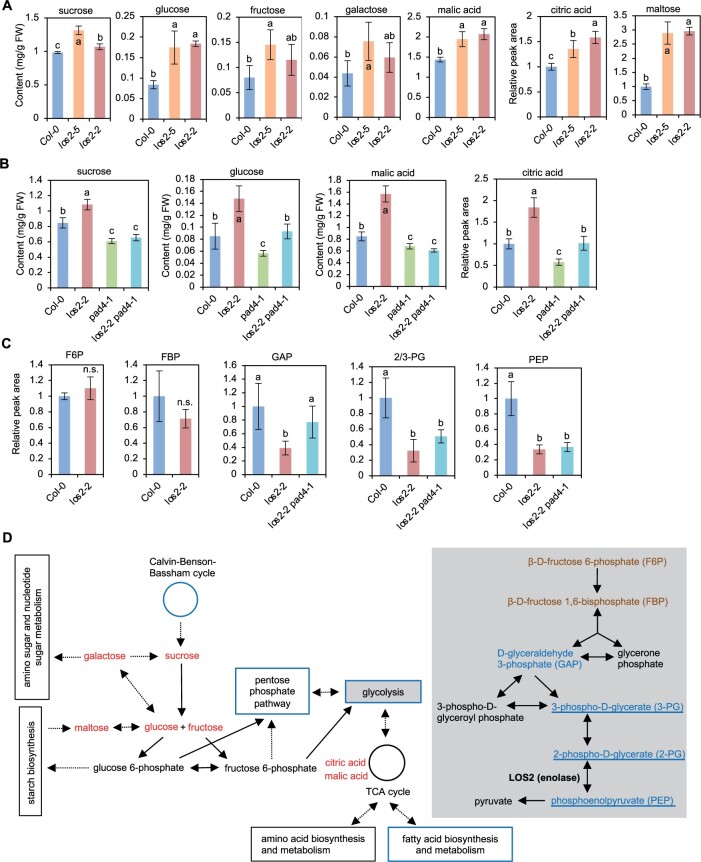
We measured the amount of some core primary metabolites in the los2 mutants to assess how the reduction of glycolysis enzyme affects primary metabolism and whether autoimmunity has an indirect effect on primary metabolism as it does to plant growth. Sucrose, glucose, galactose, fructose, maltose, malic acid, and citric acid were quantified in leaves of Col-0 WT, los2-5, and los2-2 plants by gas chromatography-mass spectrometry (GC-MS) analysis. The los2-5 mutant had a higher accumulation of all these metabolites compared to WT (Figure 7A). Likewise, the los2-2 mutant accumulated more sucrose, glucose, malic acid, citric acid, and maltose (Figure 7A). The difference in fructose and galactose amount between the los2-2 mutant and the WT could not be assessed due to the low amount of these sugars and large measurement variations (Figure 7A).
Figure 7.
The los2 mutants have altered primary metabolite homeostasis. A, Absolute content of sucrose, glucose, fructose, galactose, and malic acid as well as relative peak area of citric acid and maltose in Col-0, los2-5 and los2-2 measured by GC–MS. The relative amount of citric acid and maltose was calculated in respect to the amount of internal control in the samples and values were normalized to Col-0. B, Absolute content of sucrose, glucose, and malic acid as well as the relative peak area of citric acid in Col-0, los2-2, pad4-1 and los2-2 pad4-1 measured by GC–MS. The relative amount of citric acid was calculated in respect to the amount of internal control in the samples and values were normalized to Col-0. C, Relative peak area of glycolytic metabolites in Col-0, los2-2 and los2-2 pad4-1 measured by HPLC. The value of Col-0 WT was set as “1” and the value of others was normalized to the value of Col-0. D, Simplified schematic presentation of the connection between glycolysis and other major primary metabolic pathways. Glycolysis pathway was colored light gray in the background. Solid arrows indicate one single biochemical reaction while dashed arrows represent multiple biochemical reactions. Metabolites increased or decreased in the los2-2 mutant as compared to WT were colored red or blue, respectively; metabolites having comparable amount in the los2-2 mutant and WT were colored mocha. Pathways that were enriched in the GO term analysis of downregulated genes in los2-5 mutant (Figure 3B) were boxed or cycled with blue. Metabolites that were not dependent on functional PAD4 were underlined. Five biological replicates were performed for (A), (B), and (C). Error bars represent sd. Different letters indicate significant difference tested by one-way ANOVA/Duncan’s multiple range test via R 3.6.3 with “agricolae” package, P < 0.05. “n.s.” indicates no significant difference tested by Student t test, P < 0.05. FW, fresh weight. TCA cycle: The tricarboxylic acid cycle.
The amounts of sucrose, glucose, malic acid, and citric acid were measured in the los2-2 pad4-1 mutant where autoimmunity was largely suppressed (Figure 4, F–H). Interestingly, the pad4 mutation completely abolished the higher accumulation of sucrose and malic acid, and partially suppressed the higher accumulation of glucose and citric acid in los2-2 mutant (Figure 7B). Additionally, the pad4-1 single mutant had a reduced accumulation of these metabolites as compared to WT, and the los2-2 pad4-1 mutant had a similar amount of sucrose and malic acid as the pad4 mutant (Figure 7B). The change of primary metabolite amount was not simply a result from the change of plant size, because the los2-2 and los2-5 mutants, with one-fold difference in fresh weight (Figure 2A), had a similar amount change of most of the metabolites measured (Figure 7A). These results suggest that the increased accumulation of primary metabolites in the los2 mutants is largely due to activated immune responses. Interestingly, the pad4 mutant had a slightly lower accumulation of primary metabolites, further suggesting a promotion of some primary metabolites by an increase of defense responses.
Autoimmunity in the los2 mutants perturbs glycolysis
We further quantified the relative amount of six major glycolytic metabolites in Col-0 WT, los2-2, and los2-2 pad4-1 plants by high performance liquid chromatography (HPLC)-mass spectrometry (MS) analysis (Figure 7, C and D). These included β-d-fructose 6-phosphate (F6P), β-d-fructose 1,6-bisphosphate (FBP), d-glyceraldehyde 3-phosphate (GAP), 3-phospho-d-glycerate (3-PG), 2-PG, and PEP. Because 2-PG and 3-PG are structural isomers having the same monoisotopic mass and indistinguishable in the HPLC-MS analysis, the total relative amount of these two metabolites in each genotype was displayed as 2/3-PG. F6P and FBP accumulated to a similar level in WT and los2-2, but GAP, 2/3-PG, and PEP accumulated to a lower level in los2-2 than in the WT (Figure 7C). The reduced GAP in the los2-2 mutant was due to autoimmunity since the pad4-1 mutation elevated the GAP level in los2-2 mutant to the WT level (Figure 7C). By contrast, pad4 mutation did not alter the reduced amount of 2/3-PG and PEP, the substrate and product of enolase, in los2-2 compared to the WT (Figure 7, C and D). These results indicate that the reduction of LOS2 activity perturbs glycolysis and the resulting autoimmunity further affects primary metabolites.
The MBP protein is detected only when overexpressed under a non-native gene context
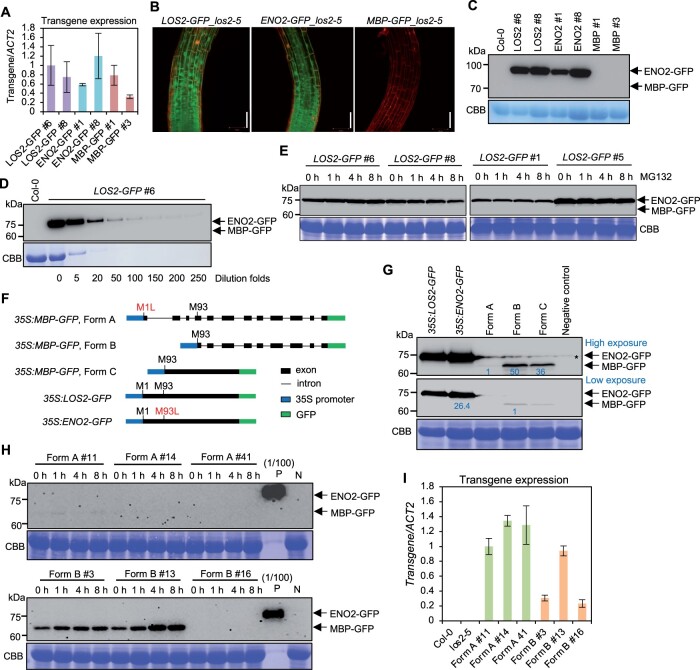
Since the putative MBP form when made from the native content is dispensable for the LOS2 function, we examined the existence of MBP in Arabidopsis using above-mentioned transgenic lines expressing the genes under the native promoter of LOS2. All transgenic plants of LOS2-GFP, ENO2-GFP, and MBP-GFP in los2-5 had comparable LOS2 transcript levels (Figure 8A), indicating low transgene expression was not the reason for phenotypic differences observed in ENO2-GFP and MBP-GFP transgenic plants. Confocal microscopy on roots of transgenic plants revealed that LOS2-GFP was localized to both nucleus and cytoplasm in transgenic plants as previously reported (Lee et al., 2002; Kang et al., 2013; Figure 8B). ENO2-GFP had the same dual localization pattern as LOS2-GFP in transgenic plants (Figure 8B). However, GFP signal was not detectable in the MBP-GFP transgenic plants (Figure 8B). We further examined the accumulation of fusion proteins in two representative transgenic plants for each transgene by immunoblotting. While ENO2-GFP protein was detected at the expected size in the ENO2-GFP plants, no MBP-GFP protein was detected in the MBP-GFP plants by anti-GFP antibody (Figure 8C). Only one protein signal at the ENO2-GFP size could be detected from the LOS2-GFP transgenic plants and no MBP-GFP signal, which is expected to be 10 kDa smaller than the ENO2-GFP, could be detected (Figure 8C). Serial dilution of the protein extracts revealed that the MBP form, if produced, would be at least 250-fold less than the ENO2 form from the LOS2-GFP transgene (Figure 8D). Since MBP protein was reported to be subject to ubiquitin-dependent degradation (Kang et al., 2013), we examined the presence of MBP proteins in LOS2-GFP transgenic lines after MG132 (a ubiquitin-dependent proteasome inhibitor) treatment by immunoblotting. All four independent LOS2-GFP transgenic lines examined produced only the ENO2 form with or without MG132 treatment, and no MBP protein could be detected even with MG132 treatment (Figure 8E). These results indicate the absence or very low abundance of an endogenous MBP protein in plants under the growth condition investigated.
Figure 8.
Endogenous MBP cannot be detected in Arabidopsis under the native LOS2 gene context. A, Analysis of transgene expression in complementation lines of LOS2-GFP (#6 and #8), ENO2-GFP (#1 and #8), and MBP-GFP (#1 and #3). The expression of LOS2-GFP #6 was set to 1. B, Confocal microscopy showing the expression pattern of LOS2-GFP, ENO2-GFP, and MBP-GFP in Arabidopsis root treated with propidium iodide. Scale bar = 50 μm. C, Immunoblot detecting the fusion protein by anti-GFP antibody in complementation lines of LOS2-GFP (#6 and #8), ENO2-GFP (#1 and #8), and MBP-GFP (#1 and #3) in los2-5 background. Col-0 was used as a negative control. D, Immunoblot of LOS2-GFP #6 using anti-GFP antibody. The number under each band indicated the dilution folds of LOS2-GFP #6 total proteins. Col-0 was used as a negative control. E, Immunoblot detecting ENO2-GFP and MBP-GFP proteins by anti-GFP antibody in four independent lines of LOS2-GFP (#6, #8, #1, and #5) after 50-μM MG132 treatment for 1, 4, and 8 h. F, Simplified gene structures of fusion proteins LOS2-GFP, ENO2-GFP, and three forms of MBP-GFP all of which were driven by 35S promoter. CDS of LOS2 and ENO2 were fused with GFP to form LOS2-GFP and ENO2-GFP. LOS2 genomic DNA with the first translation initiation site mutated (M1L) was fused with GFP to generate MBP-GFP (Form A) while LOS2 genomic DNA lacking the N-terminal sequence was fused with GFP for MBP-GFP (Form B). MBP-GFP (Form C) contains MBP CDS fused with GFP. G, Immunoblot detecting the fusion proteins expressed in N. benthamiana using anti-GFP antibody. Negative control was a sample without infiltration. Asterisk indicates a nonspecific band detected in the samples. Protein levels were quantified by ImageJ. For the upper, proteins from Forms B and C were normalized to proteins from Form A; for the lower, ENO2-GFP was normalized to proteins from Form B. Blue letters indicated the fold changes. H, Immunoblot of MBP proteins from transgenic plants of Form A (#11, #14, and #41) and Form B (#3, #13, and #16) treated with 50 μM MG132 for 1, 4, and 8 h. Each form had two representative lines in which MBP could be detected (#11 and #14 from Form A; #3 and #13 from Form B) and one representative line in which MBP could not be detected (#41 from Form A and #16 from Form B) under normal conditions. “(1/100) P,” positive control sample from pLOS2:LOS2:GFP #6 diluted 100 times. The same amounts of protein from positive control and negative control samples were loaded for the upper and lower blots. I, Transgene expression of three independent lines of 35S:MBP (Form A):GFP and 35S:MBP (Form B):GFP. The expression of Form A #11 was set to “1.” Three biological replicates were performed for (A) and (I), and error bars represented sd. For transgene expression analysis, the forward primer targeted LOS2 sequences and the reverse primer targeted GFP sequences. This same primer pair was able to amplify each of the three transgenes. Arrows in (C–E) and (G and H) indicate the expected size of ENO2(enolase)-GFP and MBP-GFP fusion proteins. CBB, Coomassie brilliant blue; L, leucine; M, methionine; N, negative control sample from Col-0.
To increase the chance of detection of MBP, we overexpressed the LOS2 gene transiently in Nicotiana benthamiana. The LOS2 coding sequence (CDS) was fused with GFP at the C-terminus and driven by the 35S promoter to generate the construct 35S:LOS2-GFP (Figure 8F). As a control, ENO2 CDS fused with GFP was similarly expressed under the 35S promoter to generate the construct 35S:ENO2-GFP. Both the 35S:LOS2-GFP and 35S:ENO2-GFP constructs produced ENO2-GFP at the expected size (Figure 8G; Supplemental Figure S7). However, no protein at the MBP-GFP size could be detected from 35S:LOS2-GFP (Figure 8G; Supplemental Figure S7). These results indicate that MBP protein could not be detected even when under the CDS of LOS2 was expressed in N. benthamiana.
A previous study detected MBP protein when sequence encoding only the MBP part of LOS2 gene was expressed under the control of the 35S promoter (Kang et al., 2013; Liu et al., 2017). We therefore examined whether the detection of MBP was due to the deletion of the leader sequences 5′ to the MBP CDS in the earlier overexpression construct. We generated three LOS2 forms varying in the leader sequence but were all expected to express the MBP protein but not ENO2 (Figure 8F). Form A was a LOS2 genomic DNA fragment starting from the first translation initiation site but with this Met codon mutated to Leu. Form B was a LOS2 genomic DNA fragment starting from the putative second translation initiation site. Form C started from the putative second translation site similar to Form B, but having CDS instead of genomic DNA of LOS2. All three variants were fused with GFP at the C-terminus and transiently expressed in N. benthamiana under the control of the 35S promoter (Figure 8, F and G). MBP-GFP could be detected from Form B and Form C, but at a much lower level (<4%) compared to ENO2-GFP that was expressed in N. benthamiana as well (Figure 8G). Intriguingly, MBP-GFP could be detected only under a very long exposure from Form A variant, which best mimicked the endogenous genomic context (Figure 8G). The expression level of MBP protein from Form A was about 1/50 and 1/36 of that from Forms B and C, respectively (Figure 8G). These results indicate that the MBP protein could be produced in small amount when the sequence coding for MBP only was directly overexpressed and that M93 is not an effective alternative translation start site or not a site at all under the native context of the LOS2 gene.
This observation from transient expression in N. benthamiana was also seen in transgenic Arabidopsis plants expressing the Form A and Form B variants respectively. Only 2 out of 24 Form A transgenic lines had detectable MBP expression, while 7 out of 21 Form B transgenic lines had detectable MBP protein. In addition, expressions in Form A lines were much lower than that in Form B lines (Figure 8H), while the transgene transcripts were expressed at a comparable level (Figure 8I). Quantification of the immunoblot signals revealed that the MBP protein from Form B overexpression was at least 100-fold lower in amount than the ENO2 expressed under the control of the LOS2 native promoter (Figure 8H). We further treated transgenic plants with MG132 as it was reported to stabilize MBP (Kang et al., 2013). A moderate increase of MBP protein was observed with MG132 treatment, but MBP remained undetectable in lines where it was not detected without treatment (Figure 8H). Taken together, these results show that the production of MBP protein cannot be observed under the endogenous genomic context in the conditions tested and MBP can be produced at a very low amount when the sequence coding for MBP only (without its 5′-flanking sequences) is expressed under the control of 35S promoter.
MBP protein does not contribute to the LOS2 function in development or expression of cold-response genes
We further asked if the enolase function rather than the putative MBP is responsible for the role of LOS2 in plant development similarly to its role in immunity. LOS2, ENO2, and MBP were each expressed under the control of the LOS2 native promoter, via constructs pLOS2:LOS2:GFP (LOS2-GFP), pLOS2:ENO2:YFP (ENO2-YFP), and pLOS2:MBP:GFP (MBP-GFP), and were introduced in the los2-2 mutant which displayed a more drastic developmental defects compared to los2-5 (Supplemental Figure S1, D–F). Similar to findings with these forms expressed under the LOS2 native promoter (Figure 8, C–E), MBP-GFP could not be detected in the LOS2-GFP or MBP-GFP transgenic plants while ENO2 fusion protein was readily detected in LOS2-GFP and ENO2-YFP transgenic lines (Supplemental Figure S8A), despite a comparable RNA expression of MBP-GFP and LOS2-GFP transgene (Supplemental Figure S8B). Two representative transgenic lines from each transgene were analyzed for their growth, development, and immunity phenotypes. Transgenic lines of LOS2-GFP and ENO2-YFP, but not MBP-GFP, in los2-2 mutant had WT fresh weight, chlorophyll content, flowering time, and silique length (Figure 9, A–E), indicating a full rescue of developmental defects. LOS2-GFP and ENO2-YFP, but not MBP-GFP, also rescued the disease resistance defect in los2-2 (Figure 9F). These results demonstrate that enolase but not MBP contributes to the role of LOS2 in plant growth and development.
Figure 9.
ENO2, but not MBP, rescues developmental defects and ZAT10 gene expression in the los2 mutant. A–F, Morphology (A), weight (B), chlorophyll amount (C), flowering (D), silique (E), and Pst DC3000 growth assay (F) of 2-week-old Col-0, los2-2 and two representative transgenic lines of pLOS2:LOS2:GFP (LOS2-GFP), pLOS2:ENO2:YFP (ENO2-YFP) and pLOS2:MBP:GFP (MBP-GFP) in los2-2 background. Scale bar in (A) is 1 cm and N = 20 for (B). G, Analysis of ZAT10 and RD29A gene expression in Col-0 WT and los2-5 mutant before (mock) and after 4°C treatment for 24 h (4°C). H, Analysis of ZAT10 gene expression in Col-0 WT, los2-5 mutant and complementation lines of LOS2-GFP (#6 and #8), ENO2-GFP (#1 and #8) and MBP-GFP (#1 and #3) in los2-5 background under normal condition. Four and six biological replicates were performed for (C) and (F), respectively. Three biological replicates were performed for (G) and (H). Different letters indicate significant difference tested by one-way ANOVA/Duncan’s multiple range test via R 3.6.3 with “agricolae” package, P < 0.05. Asterisk indicates a significant difference tested by Student’s t test, P < 0.05. FW, fresh weight.
The LOS2 gene was found to contribute to freezing tolerance by modulating the expression of cold-response genes including ZAT10 and RD29A in the Arabidopsis C24 ecotype (Lee et al., 2002). We investigated whether the enolase function of LOS2 is responsible for its role in abiotic stress response. Expression of cold-responsive gene RD29A and its transcriptional repressor ZAT10 was examined in the los2-2 and los2-5 mutants before and after 4°C treatment for 24 h. ZAT10 had a higher expression in both mutants compared to the Col-0 WT before cold treatment (Figure 9G; Supplemental Figure S9A). It was significantly induced by cold treatment in all plants, but ZAT10 had a higher expression in los2-2 but not in los2-5 compared to the WT after cold treatment (Figure 9G; Supplemental Figure S9A). Surprisingly, although RD29A was highly induced by cold as expected, it had a higher expression in both the los2-5 and los2-2 mutants compared to the WT (Figure 9G; Supplemental Figure S9A). This is in contrast to the previous observation that RD29A had a lower expression in los2-1 compared to the C24 WT under cold treatment (Lee et al., 2002). Similar to RD29A, two other cold-response genes COR47 and COR15A did not have a reduced expression in the los2-2 mutant compared to Col-0 WT after cold treatment (Supplemental Figure S9A). Therefore, the los2-2 and los2-5 mutants had higher expression of the repressor gene ZAT10 (similarly to los2-1 reported) but not lower expression of cold-response genes including RD29A after cold treatment (unlike the los2-1 reported) compared to WT. These data do not support the early hypothesis that the increased expression of ZAT10 by the loss of the transcriptional repressor function of LOS2 leads to a reduced expression of cold-response genes. The reason for this expression difference of cold-response genes in los2 mutants between this study and the previous study is not fully understood, and it might be due to the ecotype difference of los2-1 (in C24) versus los2-2 and los2-5 (in Col-0) (Bechtold et al., 2018). Nevertheless, we examined the contribution of MBP to LOS2 function in the expression regulation of cold-response genes in the Col-0 accession. The LOS2 and ENO2 transgene totally reverted the higher than WT expression of ZAT10 and RD29A in the los2 mutants while MBP transgene had no effect on their expression in the los2 mutants (Figure 9H; Supplemental Figure S9B). This result indicates that ENO2 but not MBP contributes to expression regulation of cold-response genes.
Discussion
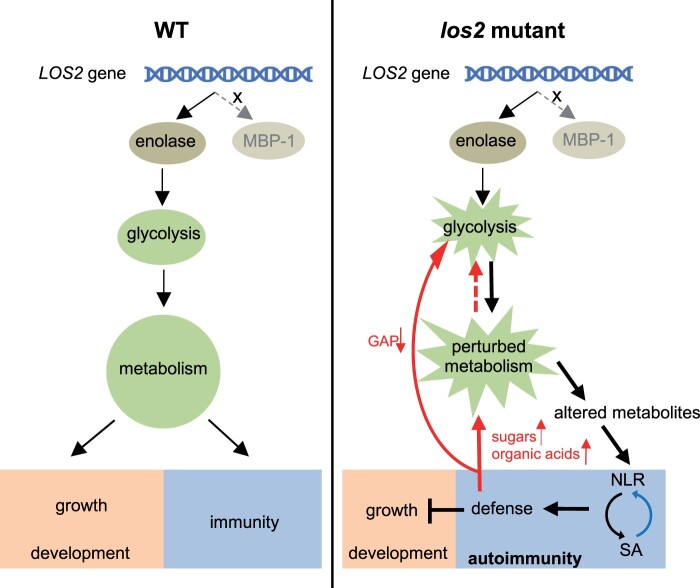
Primary metabolism not only provides essential building blocks and energy for plant growth and development, but also is the source of secondary metabolites for diverse environmental responses including plant immune responses (Figure 10). This study uncovers an unexpected consequence of the perturbation of primary metabolism in activating defense responses rather than reducing immunity (Figure 10). The reduction of the glycolytic enolase function in los2 mutant causes reduced growth, abnormal development, and alteration of cold-response gene expression. Importantly, it triggers an NLR/SA-mediated autoimmunity that is accompanied by high accumulation of SA. Autoimmunity contributes to the repression of plant growth by the los2 mutation, but not the developmental defects in flowering time and silique development. Autoimmunity also influences metabolism by promoting the accumulation of sugars and organic acids and depletion of glycolytic metabolite GAP. Therefore, LOS2 gene is critical for a balanced growth and defense status in plants by maintaining metabolism homeostasis (Figure 10).
Figure 10.
Working model for the role of LOS2 in plant growth and immunity. The Arabidopsis LOS2 gene encodes only a glycolytic enzyme enolase, but not the proposed MBP-1 protein. In WT plants, the homeostasis of glycolysis is critical for unperturbed metabolism and this is required for a balanced status of growth, development, and immunity. In the los2 mutant, enolase activity is reduced, resulting in a perturbation of the glycolysis pathway and consequently of metabolism. The perturbed metabolism causes reduced growth and abnormal development. Importantly, the perturbed metabolism causes altered metabolites which might be sensed by NLR proteins or induce NLR transcription to trigger NLR activation. The activation of NLR proteins initiates downstream immune responses including the accumulation of SA which in turn induces NLR expression (blue arrow), resulting in a positive feedback loop to boost immunity. On one hand, defense responses repress plant growth, but not affect plant development. On the other hand, defense responses further disturb the perturbed metabolism (red arrows) by promoting the accumulation of sugars and organic acids or depleting of glycolytic metabolite GAP, leading to a second positive feedback loop for a robust immune response. Therefore, LOS2 is critical for a balanced growth and defense status in plants by maintaining metabolism homeostasis.
This study reveals that LOS2 affects plant immunity through its canonical function in glycolysis rather than an alternatively translated product MBP, which was postulated to be important for the LOS2 function in abiotic stress tolerance. A mutation at the critical site for the enolase activity abolished the ability of enolase to rescue the los2 mutant defects in growth and immunity (Figure 6, A–D). In addition, a mutated LOS2 gene that cannot produce MBP is fully functional in Arabidopsis (Figure 5). These data indicate that LOS2 functions as the canonical glycolytic enolase and the alternatively translated MBP plays little if any role in Arabidopsis. This study further shows that the LOS2 gene does not produce MBP or produces it at undetectable amounts under the conditions we analyzed in Arabidopsis (Figure 8, B–E). The MBP protein could be detected only at low amount when the sequence coding for MBP only was driven by the 35S promoter (Figure 8, G and H; Supplemental Figure S7). The MBP production was reported in transgenic plants carrying a LOS2-YFP overexpression construct (Kang et al., 2013). As the transgenic line in the previous report was not available, we were unable to re-analyze that line together with transgenic lines generated in this study to determine if the growth conditions could be the reason for the discrepancy. It is possible that MBP is produced under specific conditions, considering that its putative translation start site M93 is conserved among homologs of LOS2 in plants. Further studies using various stress treatment might reveal if and when MBP could be produced from the LOS2 gene in plants. They may also reveal if this production might have some function in certain stress responses although it does not contribute to the role of LOS2 in growth and immunity regulation.
It is important to make a distinction between the possible MBP function from a natively produced protein (from alternative translation from the LOS2 locus) and the consequences of overexpression of a constructed MBP CDS free of its native gene context. Early studies of the role of MBP in abiotic stress responses and the function of MBP as a transcriptional repressor were mostly based on overexpression of the latter (Kang et al., 2013; Liu et al., 2017). It is possible that the MBP produced under the artificial condition could carry out a direct regulation on gene expression in abiotic responses, but it may not be the function of the endogenous LOS2. Intriguingly, overexpression of MBP in early studies induced a similar defect as the loss of LOS2 function (Kang et al., 2013; Eremina et al., 2015). Although this was thought to be a feedback regulation from the repressor activity of MBP, it might result from gene silencing of the endogenous LOS2 by MBP overexpression. Further studies could reveal whether MBP from overexpression has an effect on its own independently of the endogenous ENO2 from LOS2.
Data from this study do not support the hypothesis that the enolase protein has a second function as a transcriptional repressor. In addition, we show that this second function as a transcriptional repressor, if existing, is not critical for the enolase to regulate metabolism or impact growth and immunity. An earlier study found that the recombinant enolase protein can bind to the promoter sequences from the ZAT10 gene in a gel shift assay (Lee et al., 2002). However, no binding of LOS2/ENO2 to DNA in vivo has been attempted, and no transcriptional repressor activity has been examined. We found that the full-length enolase (ENO2), unlike MBP, did not exhibit the transcriptional repressor activity in the reporter assay system (Figure 6F). In addition, the cytosol-localized enolase (ENO2-NES) was fully functional in growth and immunity regulation, supporting the importance of the enolase activity but not the transcriptional repressor activity of LOS2 (Figure 6, I and J). The partial functionality of the ENO2-NLS form might have resulted from small amount of cytoplasm-localized ENO2 protein from cytosol translation before being transported into nucleus. Further supporting the one-activity hypothesis is the full elimination of the LOS2/ENO2 function by the catalytic dead mutation of enolase (Figures 5 and 6, A–D). Therefore, LOS2 functions through its enolase activity in plants and its mutant defects result from perturbation of glycolysis which leads to altered gene expression in immune responses. Nevertheless, the possibility that LOS2/ENO2 might have transcriptional repressor activity under specific conditions or for other biological processes such as cold response is not excluded. However, future studies to explore this repressor possibility should fully consider the potential indirect impact from perturbed metabolism.
This study shows that growth defects of the los2 mutants are not entirely due to the limit in primary metabolism but rather partially due to the activated defense responses. The dwarfism of the los2-2 mutant was largely rescued when the SA-mediated immune signaling was blocked either by a loss of PAD4 function or the NahG transgene (Figures 3, C–F and 4, F–H; Supplemental Figures S2B and S3D) while early flowering and reduced chlorophyll content of the los2-2 mutant were not dependent on PAD4 (Supplemental Figure 3, A and B). This implies that the LOS2 gene has diverse roles, either directly through its glycolysis function or indirectly through its effect on immune activation.
This study raises questions on the function of LOS2 in cold tolerance and perhaps other abiotic stress tolerance. The effect of the loss of LOS2 function on cold response in the Col-0 accession is different from what was reported in the C24 accession. Although the cold-response repressor gene ZAT10 (postulated as a target gene of the LOS2 transcriptional repressor) had higher expression in the los2 mutants compared to WT, the cold-response marker gene RD29A (that is repressed by ZAT10) did not have a lower expression in the Col-0 los2 mutants in contrast to the C24 los2-1 mutant (Figure 9G; Supplemental Figure S9A). This suggests that the Col-0 los2 mutants might not have a similar cold tolerance defect as the C24 los2-1 mutant. It also raises the question of whether lower expression of RD29A, and thus compromised freezing tolerance, in the C24 los2 mutant is through noncanonical transcriptional activity of LOS2 on the expression of genes like ZAT10. As low temperature responses and immune responses are highly connected (Saijo and Loo, 2020), it is important to re-evaluate the LOS2 function in abiotic stress response to differentiate direct effects and indirect effects from metabolism perturbation or immune response activation. It is noted that C24 has unusual tolerance for multiple abiotic and biotic stresses without overt growth deficiency (Bechtold et al., 2018), suggesting a different wiring of growth, immune responses, and abiotic stress responses in C24 compared to the Col-0 accession. Molecular genetic dissection and comparison of the LOS2 function in these accessions will further our understanding of the connection of primary metabolism and environmental responses.
The effect of the reduction of enolase activity on primary metabolism is complex. A decrease of the enolase product PEP was observed regardless of the immunity status (Figure 7C). A decrease was also seen for the total amount of 2-PG (substrate of enolase) and its direct precursor 3-PG in the los2-2 mutant (Figure 7, C and D). This suggests that reducing enolase activity might lead to a decreased flux of glycolysis resulting in decreased substrate and product of enolase in the los2 mutants. Strikingly, autoimmunity, but not reduction of enolase activity itself, resulted in an accumulation of a large number of sugars and organic acids, as well as a depletion of a glycolytic metabolite GAP (Figure 7, B and C). It is not known why and how upregulated defense responses induce such changes in these primary metabolites. It has been reported that sugars can act as ROS scavengers (Keunen et al., 2013). Additionally, glucose, fructose, and sucrose significantly induce the transcription of Pathogenesis-Related (PR) genes in tobacco (Herbers et al., 1996), and pretreatment of sucrose increases resistance to Magnaporthe oryzae infection in rice (Gómez-Ariza et al., 2007). Thus, accumulating more sugars in los2 mutants may potentially reduce ROS stress and/or amplify immune responses. It is unexpected that the los2 has reduced PEP but increased SA levels considering that SA is mainly synthesized through shikimate pathway from chorismate which is converted from PEP (Dempsey et al., 2011). However, it is possible that activated immune responses in the los2 mutant induces a shift of metabolite flux from PEP so that the flux is channeled more to the biosynthesis of SA over some other metabolites despite a reduction of overall PEP. Indeed, it has been shown that enzymes in both the shikimate pathway and the SA biosynthesis pathway are activated at the protein level or induced at gene expression level by pathogen infection or elicitors (Keith et al., 1991; Umeda et al., 1994; Görlach et al., 1995; Ferrari et al., 2007; Yang et al., 2021). Further metabolite analysis will likely reveal the metabolic flux changes in both primary and secondary metabolism with a perturbed glycolysis.
It is not understood how disruption of glycolysis leads to autoimmunity or upregulated defense responses. One hypothesis is that reduced glycolysis reduces energy level and thus reduces pathogen growth. However, similar level of pathogen growth was observed in los2-5 (with 37% of the WT enolase activity) and los2-2 alleles (with 10% of the WT enolase activity) (Figure 2, B and F), suggesting that the energy level is not directly related with defense responses. The other hypothesis is that reduced enolase activity perturbs metabolism homeostasis and certain metabolite changes trigger defense responses. Altered levels of some metabolites might induce expression of some NLR genes and therefore activate plant defense responses, and this has been observed for sorbitol in apples (Meng et al., 2018) and plastidial fatty acid in Arabidopsis (Chandra-Shekara et al., 2007). In mammalian cells, an NLR protein complex TLR4-MD-2 can be directly triggered by metabolites lipopolysaccharide and sulfatides (3-O-sulfogalactosylceramides) (Park et al., 2009; Su et al., 2021). Although it has not been observed in plants, it is not impossible that one or multiple metabolites altered by the los2 mutation might activate an NLR protein or a defense response regulator which in turn triggers SA production and upregulates more NLR genes. The NLR gene SNC1 was shown to be induced by the perturbation of phenylpropanoid metabolism (Huang et al., 2021). It also plays a significant role in conferring autoimmunity in the los2 mutants (Figure 4, B–D; Supplemental Figure S3C). It is yet to be determined whether SNC1 or some other NLR genes could be directly activated by a change of metabolites and therefore as a primary sensor in the los2 mutant or they are induced by SA and therefore as an amplifier of immune response. Future investigation employing metabolomic analysis coupled with molecular genetics will reveal the link between the primary metabolism perturbation and the activation of plant defense responses.
Materials and methods
Plant materials and growth condition
Arabidopsis thaliana accession Col-0 was used as the control for all the experiments performed in this study. Seeds of los2-2 (SALK_021737), los2-3 (SALK_077784), and los2-4 (SAIL_208_E09) were kind gifts from Dr Brigitte Poppenberger (Technical University Munich). For most of the experiments in this study, plants were grown in growth chambers at 22°C under constant light (∼100 μmol m−2s−1 of fluorescent lamp) and 50% humidity conditions, while plants for pathogen growth assay were grown at 22°C under 12-h light/12-h dark cycle and 50% humidity conditions. Soil (LM111 from Lambert)-grown plants were assayed 2 weeks after germination unless otherwise specified.
Genomic DNA of LOS2 gene with about 1.5-kb promoter and 600-bp terminator was used for complementation analysis. This DNA fragment was first cloned into pCR8/GW/TOPO vector (Invitrogen, K250020) and then recombined into binary vector pMDC99 by gateway LR clonase (Invitrogen, 11791-020). For pLOS2:LOS2:GFP, pLOS2:ENO2:GFP, pLOS2:MBP:GFP, and pLOS2:LOS2-S42A:GFP constructs, an LOS2 genomic DNA from 1.5-kb upstream of “ATG” site to “TAA” site (without “TAA”) was first cloned into pDONR222 vector by gateway BP clonase (Invitrogen, 11789020). The resulting pLOS2:LOS2_pDONR222 was used as the template for site mutagenesis by ClonExperess II One Step Cloning kit (Vazyme, C112) or Phusion Site-Directed Mutagenesis Kit (Thermo Scientific, F541) to generate pLOS2:ENO2_pDONR222, pLOS2:MBP_pDONR222, and pLOS2:LOS2-S42A_pDONR222. For constructs pLOS2:ENO2:NES:YFP (ENO2-NES) and pLOS2:ENO2:NLS:YFP (ENO2-NLS), ENO2-NES and ENO2-NLS fusions were made by PCR amplification of the ENO2 gene with chimeric primers containing the 3′-end of the ENO2 sequence (right before the stop codon) fused with the NES coding sequences “cttgctcttaagttggctggacttgatatt” or NLS coding sequences “cctaagaagaagagaaaggtt” (see Supplemental Data Set S2). The fusion genes were cloned into pCR8/GW/TOPO vector and then recombined into gateway binary vector pGWB550 or pGWB540 (Nakagawa et al., 2007). To generate overexpression constructs of LOS2 variants, the LOS2 coding region (LOS2-CDS) was amplified from WT cDNA and cloned into pDONR222 by gateway BP clonase. The resulting LOS2-CDS_pDONR222 was used as a temperate to generate ENO2-CDS_pDONR222 by ClonExperess II One Step Cloning kit. The MBP-CDS was directly amplified from the second transcription site to the last codon before the stop codon “TAA” and cloned into pDONR222 vector by BP clonase to generate MBP-CDS_pDONR222. These entry clones were recombined into gateway binary vector pGWB551 (Nakagawa et al., 2007) or digested by MluI restriction enzyme before recombined into pEarleyGate 103 binary vector by gateway LR clonase. All constructs were sequenced to ensure no PCR errors to cause mutated protein sequences.
Agrobacteria GV3101 (pMP90) strain carrying the desired construct was used for floral dipping to generate transgenic plants. To generate transgenic plants in los2-4 mutant background, selected transgenic lines in los2-5 mutant background were crossed with los2-4 heterozygous plant. Transgenes with los2-4 homozygous mutant background were isolated in F2 populations by genotyping. Transgenic seeds were screened on 1/2 MS plates with respective antibiotics. T1 plants were transferred into soil from selection plates and grown in the growth chamber for getting T2 seeds. Single-copy transgenic lines were selected by screening T2 seeds on 1/2 MS plates with respective antibiotics, and homozygous plants were identified at T3 generation for these single-copy lines. Experiments were done either on homozygous T3 or T4 plants or on T2 plants that were first selected on 1/2 MS plates with respective antibiotics and were then grown on soil. Primers for making constructs and genotyping can be found in Supplemental Data Set S2.
Mapping-by-sequencing
The cloning of SMO3 was from the same sequencing analysis as of SMO1/HOS15 in Yang et al. (2020). Mapping-by-sequencing analysis was performed according to Hua et al. (2017) and Zhu et al. (2012), and further described in Yang et al. (2020).
Pathogen growth assay
Pathogen growth assay in this study was carried out exactly as Yang et al. (2020). Briefly, Pseudomonas syringae pv. tomato (Pst) DC3000 was grown on King’s B medium plates with rifamycin at 28°C for 2 days and re-grown on a new plate for 1 day. Fresh Pst DC3000 was suspended in 10-mM MgCl2 and 0.02% Silwet L-77 to OD600 of 0.05. Two-week-old plants were dipped in the bacterial suspension for 20 s. Pathogen growth was measured at 1-h or 0 day post-inoculation (0 DPI) and 3 days post-inoculation (3 DPI). Six biological replicates were performed for each analysis.
Gene expression analysis
Two-week-old plants were sampled for total RNA extraction by TRIzol Reagent (Invitrogen, 15596026). cDNA synthesis was performed using PrimeScript RT reagent kit with gDNA eraser (Takara, RR047A). The qRT-PCR reactions were run on CFX96 Real-Time System (Bio-Rad) using iQ SYBR Green supermix (Bio-Rad, 1708880). About two to three individual plants from one pot were pooled as one biological replicate. At least three biological replicates from different pots were performed for each gene expression analysis.
RNA-seq analysis
The RNA-seq analysis of los2-5 mutant was performed exactly as described in Yang et al. (2020). Three biological replicates were performed for both Col-0 WT and los2-5 mutants. RNA-seq data of this study can be found at NCBI GEO under the accession number GSE156268. DEGs in los2-5 mutant with FDR < 0.05 can be found in Supplemental Data Set S1.
3,3′-Diaminobenzidine staining
Two-week-old plants were used for 3,3′-diaminobenzidine (DAB) staining according to Daudi and O’Brien (2012). Briefly, approximately six plants were placed into 50 mL tube with staining solution (1-mg/mL DAB with 0.05% Tween-20) and gently vacuumed for 5 min. Tubes were covered with aluminum foil since DAB is light-sensitive. Tubes were then placed on a shaker at 100 rpm shaking speed until a desired staining was obtained. After incubation, staining solution was replaced with bleaching solution (ethanol: acetic acid: glycerol = 3:1:1) and tubes boiled in a water bath at 90°C for 15 min. Bleaching solution was decanted and replaced with fresh bleaching solution. Samples were allowed to stand for 30 min before plants were photographed on a plain white background under uniform lighting.
Trypan blue staining
The staining solution contained one volume of trypan blue-lactophenol solution (trypan blue at a concentration of 2.5 mg/mL in lactophenol which is a mixture of equal volume of lactic acid, glycerol, phenol, and water) and two volumes of ethanol. At least six 2-week-old plants were immersed in staining solution at room temperature for 1–24 h until desired staining was observed. The staining solution was replaced with bleaching solution (lactophenol: ethanol = 1:2) and the samples were incubated overnight on a shaker at 100 rpm shaking speed. The bleaching solution was removed and the tissue was covered in 70% glycerol. Plants were photographed on a plain white background under uniform lighting.
Immunoblotting
Total protein was extracted from 2-week-old plants (extraction buffer: 50 mM Tris–HCl pH 7.5, 1 mM EDTA, 1 mM EGTA, 150 mM NaCl, 10% glycerol, 5 mM DTT, 0.25% Triton-X 100, 2% Polyvinylpolypyrrolidone, 1 mM PMSF and 1× protease inhibitor cocktail). After separating, proteins were transferred into PVDF membrane (Bio-Rad, 1620177). The anti-GFP antibody (Takara, 632381, at a 1:3,000 dilution) and HRP-linked antibody (Cell Signaling, 7076, at a 1:5,000 dilution) were used as the primary and secondary antibody, respectively, for detecting GFP, YFP and mGFP5 fusion proteins using ECL western blotting detection reagents (GE Healthcare, RPN2209). Membranes were photographed with the Bio-Rad ChemiDoc imaging system.
Dual-luciferase reporter assay
pGreenII 0800-LUC (Hellens et al., 2005) was used as the backbone to generate both effector constructs and the reporter construct. For effectors, LOS2 CDS was amplified from cDNA of Col-0 WT and Gal4 BD was subcloned from pDEST-GBKT7. DNA fragments were recombined into pGreenII 0800-LUC backbone without renilla and firefly luciferase sequence to generate LOS2-BD_pGreenII0800-LUC and BD_pGreenII0800-LUC by ClonExperess MultiS One Step Cloning kit (Vazyme, C113) or ClonExperess II One Step Cloning kit (Vazyme, C112), respectively. ENO-BD_ pGreenII 0800-LUC and MBP-BD_ pGreenII 0800-LUC were generated using LOS2-BD_ pGreenII 0800-LUC as the temperate by ClonExperess II One Step Cloning kit (Vazyme, C112). In order to generate 5UAS:35S_ pGreenII 0800-LUC, pGreenII 0800-LUC was first digested with PstI and NcoI, then 5UAS (UAS sequence: cggagtactgtcctccgag, from P2-pACU):35S promoter (from pGreen 0800-LUC) with PstI and NcoI restriction site at each end was ligated into the digested backbone by T4 ligation. Ten micrograms of plasmids of one of the effectors and the reporter were co-transformed into Arabidopsis protoplasts. After 16-h incubation in the dark, total RNA in protoplasts was extracted using TRIzol and extracted RNA was subject to cDNA synthesis and subsequent RT-qPCR analyses. Primers used for cloning were listed in Supplemental Data Set S2.
MG132 treatment
Plants were grown on 1/2 MS plates vertically for 1 week in growth chambers at 22°C under constant light and 50% humidity conditions. Seedlings were transferred to six-well tissue culture plates and immersed in 5 mL 1/2 MS liquid medium with 50-μM MG132 (Sigma, M7449). Then the plates were covered with aluminum foil and sited on a shaker with 50 rpm at room temperature. Samples were collected at 0, 1, 4, and 8 h after MG132 treatment, and then subjected to protein extraction and immunoblotting.
Enolase activity measurement
Enolase activity assay in this study was performed following the instructions of Enolase Activity Assay Kit (Sigma, MAK178) with minor modifications. Enolase activity was determined by a couple enzyme assay in which D-2-phosphoglycerate was converted to PEP, resulting in the formation of an intermediate that reacted with a peroxidase substrate. This generated a colorimetric (570 nm) product proportional to the enolase activity in the samples. One milliunit of enolase was the amount of enzyme that generated 1.0 nmole of H2O2 per minute at pH 7.2 at 25°C. For sample preparation, 10 mg of 2-week-old plants was homogenized in 100-μL ice-cold enolase assay buffer. Centrifuge the samples at 10,000g for 5 min to remove insoluble material and 2 μL of each sample was added into a 96-well plate (Corning Costar, #3696) pre-filled with 23-μL enolase assay buffer in each well. The H2O2 standard for colorimetric detection was made by adding 0, 1, 2, 3, 4, 5 μL of the 1 mM H2O2 solution into water for a total volume of 25 μL. Then 25 μL reaction mix (21-μL enolase assay buffer, 1-μL enolase substrate mix, 1-μL enolase converter, 1-μL enolase developer, and 1-μL peroxidase substrate) was added into each sample and standard in the well. The 96-well plates with reactions were loaded immediately into a Microplate Reader (BioTek Synergy 2). A blank control with 2 μL of samples, 23-μL enolase assay buffer, and 25-μL reaction mix without the enolase substrate was included for each sample to remove potential background reading not from enolase activity. The microplate reader recorded the measurement every 3 min for 2 h with plates under continuous shaking at 25°C.
Measurement of sugars and organic acids by gas chromatography–mass spectrometry (GC–MS) analysis
The measurement of soluble sugars was performed mainly according to Lisec et al. (2006) as modified by Wang et al. (2010). The whole shoots of 2-week-old soil-grown plants in growth chambers at 22°C under constant light and 50% humidity conditions were sampled. Three individual plants were pooled as one biological replicate and five biological replicates were performed for each genotype. Plant tissues were thoroughly ground in liquid nitrogen, and polar metabolites from 100 mg ground tissue were extracted in 1.4 mL 75% ice-cold methanol, with 60 μL 0.2 mg/mL ribitol added as an internal standard. After fractionation of nonpolar metabolites into chloroform, 30 μL of the aqueous phase was dried under vacuum without heat. The dried sample was derivatized with methoxyamine hydrochloride (Santa Cruz, sc-257710A) and N-methyl-N-trimethylsilyl-trifluoroacetamide (Macherey-Nagel, 701270.110) sequentially for analysis on an Agilent 7890A GC/5975C MS (Agilent Technology, Palo Alto, CA, USA).
Targeted metabolic analysis of glycolytic metabolites and salicylic acid by HPLC-MS
The targeted metabolic analysis for the relative quantitation of glycolytic metabolites and SA was performed using Sciex X500B Q-TOF mass spectrometer coupled with the Exion LC system in Proteomics and Metabolomics facility at Cornell University. The extraction method in this study was performed based on t’Kindt et al. (2008) with the best extracting agent MeOH/H2O 80/20 as mentioned. Briefly, 50 mg 2-week-old Arabidopsis plants leaves from each genotype were thoroughly ground in liquid nitrogen, and then 250 μL of 2.5 ppm Pyruvic 13C in 80% methanol as an internal standard were added to the homogenized leaf tissues. Sample solutions were vortexed for 10 s at high speed, sonicated for 10 min and then centrifuged to collect supernatant. The sonication step was repeated for the insoluble part by adding 100 μL of 80% methanol for an additional 10 min, then centrifuging at 16,000g for 10 min at room temperature. The resulting supernatant was collected and pooled together. A total of 200 μL supernatant was transferred into a clean tube for each sample and 100 μL of the supernatant filtered using 0.22-μM spin filter (Costar #8169). The filtrate was transferred into autosampler vial and 30 μL of each sample was injected for separation on an HILIC column (100 × 2 mm i.d., 3 μm from Phenomenex). The X500B Q-TOF was operated in negative ion mode under MRMHR method after optimization of MRM parameters for all glycolytic metabolites and SA with their standards. The raw data were acquired and processed using Sciex OS 2.0 software and the relative quantitation of each glycolytic metabolite and SA was determined by integrating the peak areas of each analyte using MRM transitions after normalization against the internal standard Pyruvic 13C.
Statistical analysis
All Student’s t tests were performed using Excel with P < 0.05. The one-way analysis of variation (ANOVA)/Duncan′s new multiple range tests were performed using R 3.6.3 with “agricolae” package under P < 0.05. Statistical tests and biological replicate numbers were shown in figure legends and methods. Detailed statistical results are provided in Supplemental Data Set S3.
Accession numbers
DNA sequences of all genes in this study were extracted from TAIR with the gene ID listed below: LOS2 (AT2G36530); SNC1 (AT4G16890); PR1 (AT2G14610); BON1 (AT5G61900); MOS1 (AT4G24680); ZAT10 (AT1G27730); RD29A (5G52310); COR15A (AT2G42540); COR47 (AT1G20440); ACTIN2 (AT3G18780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1 . The los2 mutants are autoimmune mutants and have defects in enolase activity and development (Supports Figure 2).
Supplemental Figure S2. Total DEGs in los2-5 mutant and NahG transgene rescues the growth defects of los2-2 mutant (Supports Figure 3).
Supplemental Figure S3. Activated immunity contributes to growth but not developmental defects of los2 mutant (Supports Figure 4).
Supplemental Figure S4. ENO2 but not MBP rescues the growth defects of los2 mutants (Supports Figure 5).
Supplemental Figure S5. Protein expression of pLOS2:LOS2-S42A:GFP and pLOS2:ENO2-S42A:GFP transgenic plants (Supports Figure 6).
Supplemental Figure S6. Enolase activity contributes to the growth regulation of los2 mutants (Supports Figure 6).
Supplemental Figure S7. Protein expression of different LOS2 variants in N. benthamiana (Supports Figure 8).
Supplemental Figure S8. Transgene and protein expression of complementation lines in los2-2 mutant (Supports Figure 9).
Supplemental Figure S9. Enolase but not MBP rescues the cold-response gene expression in los2 mutant. (Supports Figure 9).
Supplemental Data Set S1. Differentially expressed genes (FDR < 0.05) in los2-5 mutant from RNA-seq.
Supplemental Data Set S2. Primers used in this study.
Supplemental Data Set S3. Summary of statistical analyses.
Supplementary Material
Acknowledgments
We thank Dr Brigitte Poppenberger for sharing los2-2, los2-3, and los2-4 mutant seeds, Dr Chun Han for the 5UAS construct (P2-pACU), Drs Adrienne Roeder, Olena Vatamaniuk, Wei Zhang, Melanie Filiatrault, and Shaoqun Zhou for technical advice and assistance. We thank the Genomics Facility, Bioinformatics Facility, and Imaging Facility of the Biotechnology Resource Center of Cornell Institute of Biotechnology for their assistance and service.
Contributor Information
Leiyun Yang, Plant Biology Section, School of Integrative Plant Science, Cornell University, Ithaca, New York 14853, USA.
Zhixue Wang, Plant Biology Section, School of Integrative Plant Science, Cornell University, Ithaca, New York 14853, USA.
Aiqin Zhang, Plant Biology Section, School of Integrative Plant Science, Cornell University, Ithaca, New York 14853, USA.
Ruchika Bhawal, Proteomics and Metabolomics Facility, Cornell University, New York 14853, USA.
Chunlong Li, Horticulture Section, School of Integrative Plant Science, Cornell University, Ithaca, New York 14853, USA.
Sheng Zhang, Proteomics and Metabolomics Facility, Cornell University, New York 14853, USA.
Lailiang Cheng, Horticulture Section, School of Integrative Plant Science, Cornell University, Ithaca, New York 14853, USA.
Jian Hua, Plant Biology Section, School of Integrative Plant Science, Cornell University, Ithaca, New York 14853, USA.
J.H. conceived the study; J.H. and L.Y. designed the experiments; L.Y., Z.W., and A.Z. conducted experiments; R.B. and S.Z. conducted HPLC-MS and its data analyses; C.L. and L.C. assisted the GC-MS analysis; L.Y. and J.H. wrote the manuscript with inputs from all other authors.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is Jian Hua (jh299@cornell.edu).
Funding
This work has been supported by National Science Foundation (NSF) IOS-1353738 and IOS-1946174 to J.H.
Conflict of interest statement. None declared.
References
- Andriotis VM, Kruger NJ, Pike MJ, Smith AM (2010) Plastidial glycolysis in developing Arabidopsis embryos. New Phytol 185: 649–662 [DOI] [PubMed] [Google Scholar]
- Bao Z, Zhang N, Hua J (2014) Endopolyploidization and flowering time are antagonistically regulated by checkpoint component MAD1 and immunity modulator MOS1. Nat Commun 5: 1–10 [DOI] [PubMed] [Google Scholar]
- Barkla BJ, Vera-Estrella R, Hernández-Coronado M, Pantoja O (2009) Quantitative proteomics of the tonoplast reveals a role for glycolytic enzymes in salt tolerance. Plant Cell 21: 4044–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold U, Ferguson JN, Mullineaux PM (2018) To defend or to grow: lessons from Arabidopsis C24. J Exp Bot 69: 2809–2821 [DOI] [PubMed] [Google Scholar]
- Bolouri Moghaddam MR, Van den Ende W (2012) Sugars and plant innate immunity. J Exp Bot 63: 3989–3998 [DOI] [PubMed] [Google Scholar]
- Bolton MD (2009) Primary metabolism and plant defense—fuel for the fire. Mol Plant-Microbe Interact 22: 487–497 [DOI] [PubMed] [Google Scholar]
- Boukouris AE, Zervopoulos SD, Michelakis ED (2016) Metabolic enzymes moonlighting in the nucleus: metabolic regulation of gene transcription. Trends Biochem Sci 41: 712–730 [DOI] [PubMed] [Google Scholar]
- Buscaill P, Rivas S (2014) Transcriptional control of plant defence responses. Curr Opin Plant Biol 20: 35–46 [DOI] [PubMed] [Google Scholar]
- Chandra-Shekara AC, Venugopal SC, Barman SR, Kachroo A, Kachroo P (2007) Plastidial fatty acid levels regulate resistance gene-dependent defense signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 7277–7282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Couto D, Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16: 537. [DOI] [PubMed] [Google Scholar]
- Cui H, Gobbato E, Kracher B, Qiu J, Bautor J, Parker JE (2017) A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol 213: 1802–1817 [DOI] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE (2015) Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 66: 487–511 [DOI] [PubMed] [Google Scholar]
- Daudi A, O’Brien JA (2012) Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio-protocol 2: e263. [PMC free article] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al. (1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Dempsey DMA, Vlot AC, Wildermuth MC, Klessig DF (2011) Salicylic acid biosynthesis and metabolism. The Arabidopsis Book 9: e0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiasova M, Schaefer L, Wygrecka M (2019) When place matters: shuttling of enolase-1 across cellular compartments. Front Cell Dev Biol 7: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eremina M, Rozhon W, Yang S, Poppenberger B (2015) ENO 2 activity is required for the development and reproductive success of plants, and is feedback‐repressed by At MBP‐1. Plant J 81: 895–906 [DOI] [PubMed] [Google Scholar]
- Feo S, Arcuri D, Piddini E, Passantino R, Giallongo A (2000) ENO1 gene product binds to the c‐myc promoter and acts as a transcriptional repressor: relationship with Myc promoter‐binding protein 1 (MBP‐1). FEBS Lett 473: 47–52 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Denoux C, De Lorenzo G, Ausubel FM, Dewdney J (2007) Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol 144: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer P, Korn M, Engelsdorf T, Sonnewald U, Koch C, Voll LM (2017) Sugar accumulation in leaves of Arabidopsis sweet11/sweet12 double mutants enhances priming of the salicylic acid-mediated defense response. Front Plant Sci 8: 1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glawischnig E (2007) Camalexin. Phytochemistry 68: 401–406 [DOI] [PubMed] [Google Scholar]
- Gómez-Ariza J, Campo S, Rufat M, Estopà M, Messeguer J, Segundo BS, Coca M (2007) Sucrose-mediated priming of plant defense responses and broad-spectrum disease resistance by overexpression of the maize pathogenesis-related PRms protein in rice plants. Mol Plant-Microbe Interact 20: 832–842 [DOI] [PubMed] [Google Scholar]
- Görlach J, Raesecke HR, Rentsch D, Regenass M, RoY P, Zala M, Keel C, Boller T, Amrhein N, Schmid J (1995) Temporally distinct accumulation of transcripts encoding enzymes of the prechorismate pathway in elicitor-treated, cultured tomato cells. Proc Natl Acad Sci USA 92: 3166–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbers K, Meuwly P, Métraux JP, Sonnewald U (1996) Salicylic acid-independent induction of pathogenesis-related protein transcripts by sugars is dependent on leaf developmental stage. FEBS Lett 397: 239–244 [DOI] [PubMed] [Google Scholar]
- Hua J, Wang S, Sun Q (2017) Mapping and cloning of chemical induced mutations by whole-genome sequencing of bulked segregants. In Plant Pattern Recognition Receptors. Humana Press, New York, pp 285–289 [DOI] [PubMed] [Google Scholar]
- Huang XX, Wang Y, Lin JS, Chen L, Li YJ, Liu Q, Wang G-F, Xu F, Liu L, Hou BK (2021) The novel pathogen‐responsive glycosyltransferase UGT73C7 mediates the redirection of phenylpropanoid metabolism and promotes SNC1‐dependent Arabidopsis immunity. Plant J 107: 149–165 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kang HJ, Jung SK, Kim SJ, Chung SJ (2008) Structure of human α-enolase (hENO1), a multifunctional glycolytic enzyme. Acta Crystallogr D Biol Crystallogr 64: 651–657 [DOI] [PubMed] [Google Scholar]
- Kang M, Abdelmageed H, Lee S, Reichert A, Mysore KS, Allen RD (2013) At MBP‐1, an alternative translation product of LOS2, affects abscisic acid responses and is modulated by the E3 ubiquitin ligase AtSAP5. Plant J 76: 481–493 [DOI] [PubMed] [Google Scholar]
- Keith B, Dong XN, Ausubel FM, Fink GR (1991) Differential induction of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase genes in Arabidopsis thaliana by wounding and pathogenic attack. Proc Natl Acad Sci USA 88: 8821–8825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keunen ELS, Peshev D, Vangronsveld J, Van Den Ende WIM, Cuypers ANN (2013) Plant sugars are crucial players in the oxidative challenge during abiotic stress: extending the traditional concept. Plant Cell Environ 36: 1242–1255 [DOI] [PubMed] [Google Scholar]
- Kim JW, Dang CV (2005) Multifaceted roles of glycolytic enzymes. Trends Biochem Sci 30: 142–150 [DOI] [PubMed] [Google Scholar]
- Lapin D, Bhandari DD, Parker JE (2020) Origins and immunity networking functions of EDS1 family proteins. Annu Rev Phytopathol 58: 253–276 [DOI] [PubMed] [Google Scholar]
- Lee H, Guo Y, Ohta M, Xiong L, Stevenson B, Zhu JK (2002) LOS2, a genetic locus required for cold‐responsive gene transcription encodes a bi‐functional enolase. EMBO J 21: 2692–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry–based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Liu X, Wei W, Zhu W, Su L, Xiong Z, Zhou M, Zheng Y, Zhou DX (2017) Histone deacetylase AtSRT1 links metabolic flux and stress response in Arabidopsis. Mol Plant 10: 1510–1522 [DOI] [PubMed] [Google Scholar]
- Lolle S, Stevens D, Coaker G (2020) Plant NLR-triggered immunity: from receptor activation to downstream signaling. Curr Opin Immunol 62: 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung J, Liu KJ, Chang JY, Leu SJ, Shih NY (2010) MBP-1 is efficiently encoded by an alternative transcript of the ENO1 gene but post-translationally regulated by proteasome‐dependent protein turnover. FEBS J 277: 4308–4321 [DOI] [PubMed] [Google Scholar]
- Meng D, Li C, Park HJ, González J, Wang J, Dandekar AM, Turgeon G, Cheng L (2018) Sorbitol modulates resistance to Alternaria alternata by regulating the expression of an NLR resistance gene in apple. Plant Cell 30: 1562–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR–encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morkunas I, Ratajczak L (2014) The role of sugar signaling in plant defense responses against fungal pathogens. Acta Physiol Plant 36: 1607–1619 [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Návarová H, Bernsdorff F, Döring AC, Zeier J (2012) Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24: 5123–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancholi V (2001) Multifunctional α-enolase: its role in diseases. Cell Mol Life Sci CMLS, 58: 902–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO (2009) The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature 458: 1191–1195 [DOI] [PubMed] [Google Scholar]
- Piasecka A, Jedrzejczak-Rey N, Bednarek P (2015) Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol 206: 948–964 [DOI] [PubMed] [Google Scholar]
- Plaxton WC (1996) The organization and regulation of plant glycolysis. Annu Rev Plant Biol 47: 185–214 [DOI] [PubMed] [Google Scholar]
- Prabhakar V, Löttgert T, Geimer S, Dörmann P, Krüger S, Vijayakumar V, Schreiber L, Gobel C, Feussner K, Feussner I, et al. (2010) Phosphoenolpyruvate provision to plastids is essential for gametophyte and sporophyte development in Arabidopsis thaliana. Plant Cell 22: 2594–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray R, Miller DM (1991) Cloning and characterization of a human c-myc promoter-binding protein. Mol Cell Biol 11: 2154–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rius SP, Casati P, Iglesias AA, Gomez-Casati DF (2008) Characterization of Arabidopsis lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. Plant Physiol 148: 1655–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez A, Cera TDL, Herrero P, Moreno F (2001) The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae. Biochem J 355: 625–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Loo EPI (2020) Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol 225: 87–104 [DOI] [PubMed] [Google Scholar]
- Shah J, Chaturvedi R, Chowdhury Z, Venables B, Petros RA (2014) Signaling by small metabolites in systemic acquired resistance. Plant J 79: 645–658 [DOI] [PubMed] [Google Scholar]
- Shams F, Oldfield NJ, Wooldridge KG, Turner DP (2014) Fructose-1, 6-bisphosphate aldolase (FBA)–a conserved glycolytic enzyme with virulence functions in bacteria: ‘Ill met by moonlight’. Biochem Soc Trans 42: 1792–1795 [DOI] [PubMed] [Google Scholar]
- Su L, Athamna M, Wang Y, Wang J, Freudenberg M, Yue T, Wang J, Moresco EMY, He H, Zor T, et al. (2021) Sulfatides are endogenous ligands for the TLR4–MD-2 complex. Proc Natl Acad Sci USA 118: e2105316118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang G, Zhou JM (2017) Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29: 618–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- t'Kindt R, Veylder LD, Storme M, Deforce D, Bocxlaer JV (2008) LC-MS metabolic profilin of Arabidopsis thaliana plant leaves and cell cultures: optimization of pre-LC-MS procedure parameters. J Chromatogr B 871: 37–43 [DOI] [PubMed] [Google Scholar]
- Trouvelot S, Héloir MC, Poinssot B, Gauthier A, Paris F, Guillier C, Combier M, Trda L, Daire X, Adrian M (2014) Carbohydrates in plant immunity and plant protection: roles and potential application as foliar sprays. Front Plant Sci 5: 592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda M, Hara C, Matsubayashi Y, Li HH, Liu Q, Tadokoro F, Aotsuka S, Uchimiya H (1994) Expressed sequence tags from cultured cells of rice (Oryza sativa L.) under stressed conditions: analysis of transcripts of genes engaged in ATP-generating pathways. Plant Mol Biol 25: 469–478 [DOI] [PubMed] [Google Scholar]
- Wang H, Ma F, Cheng L (2010) Metabolism of organic acids, nitrogen and amino acids in chlorotic leaves of ‘Honeycrisp’ apple (Malus domestica Borkh) with excessive accumulation of carbohydrates. Planta 232: 511–522 [DOI] [PubMed] [Google Scholar]
- Yang L, Chen X, Wang Z, Sun Q, Hong A, Zhang A, Zhong X, Hua J (2020) HOS15 and HDA9 negatively regulate immunity through histone deacetylation of intracellular immune receptor NLR genes in Arabidopsis. New Phytol 226: 507–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wang Z, Hua J (2021) A meta-analysis reveals opposite effects of biotic and abiotic stresses on transcript levels of Arabidopsis intracellular immune receptor genes. Front Plant Sci 12: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Hua J (2004) A haplotype-specific resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 16: 1060–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Assmann SM (2011) The glycolytic enzyme, phosphoglycerate mutase, has critical roles in stomatal movement, vegetative growth, and pollen production in Arabidopsis thaliana. J Exp Bot 62: 5179–5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Roeder RG, Luo Y (2003) S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114: 255–266 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Mang HG, Sun Q, Qian J, Hipps A, Hua J (2012) Gene discovery using mutagen-induced polymorphisms and deep sequencing: application to plant disease resistance. Genetics 192: 139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.