Abstract
Grain chalkiness reduces the quality of rice (Oryza sativa) and is a highly undesirable trait for breeding and marketing. However, the underlying molecular cause of chalkiness remains largely unknown. Here, we cloned the F-box gene WHITE-CORE RATE 1 (WCR1), which negatively regulates grain chalkiness and improves grain quality in rice. A functional A/G variation in the promoter region of WCR1 generates the alleles WCR1A and WCR1G, which originated from tropical japonica and wild rice Oryza rufipogon, respectively. OsDOF17 is a transcriptional activator that binds to the AAAAG cis-element in the WCR1A promoter. WCR1 positively affects the transcription of the metallothionein gene MT2b and interacts with MT2b to inhibit its 26S proteasome-mediated degradation, leading to decreased reactive oxygen species production and delayed programmed cell death in rice endosperm. This, in turn, leads to reduced chalkiness. Our findings uncover a molecular mechanism underlying rice chalkiness and identify the promising natural variant WCR1A, with application potential for rice breeding.
The chalkiness gene WHITE-CORE RATE 1 regulates redox homeostasis in rice endosperm by interacting with the metallothionein MT2b, thereby affecting grain quality.
IN A NUTSHELL.
Background: With improved living standards, the demand for better grain quality has become a challenge in rice production areas. Rice chalkiness, that is, endosperm containing opaque regions, seriously reduces the appearance quality of grain, making it unpopular for consumers and marketers. Gene cloning and molecular breeding have become important techniques for improving rice quality, but the improvement of chalkiness still faces many problems, including the scarcity of excellent alleles and unclear molecular mechanisms. Reactive oxygen species (ROS) play important roles in the formation of chalkiness. However, it is unclear which molecules are involved in this process.
Question: We wanted to identify the chalkiness gene by map-based cloning, analyze its natural variation, and find useful alleles. We also wondered what role this gene plays in the formation of chalkiness.
Findings: We cloned the rice chalkiness gene WHITE-CORE RATE 1 (WCR1), encoding an F-box protein that negatively regulates grain chalkiness. A functional A/G variation in the promoter region of WCR1 affects the binding of the transcriptional activator OsDOF17 to its promoter. We propose a model for the role of WCR1 in regulating chalkiness in which WCR1 promotes the elimination of excess ROS, maintains redox homeostasis, and delays programmed cell death in starchy endosperm by positively affecting the transcription of the metallothionein gene MT2b and suppressing the 26S proteasome-mediated degradation of MT2b.
Next steps: We will focus on the precise mechanism underlying the role of WCR1 in regulating MT2b transcription and the protein level of MT2b. We are also interested in using WCR1 to improve rice grain quality.
Introduction
Rice (Oryza sativa), one of three major food crops, supports half the world’s population (He et al., 1999; Wing et al., 2018). Due to improved living standards, the demand for better grain quality has become a challenge in rice production areas (Tian et al., 2009). Rice grain quality is a complex character that includes appearance quality, nutritional quality, milling quality, and eating and cooking quality (ECQ; Wang et al., 2007; Gong et al., 2017). Chalkiness, that is, endosperm containing opaque regions, is an undesirable trait for the appearance quality of rice and hence for consumers and marketing (Lisle et al., 2000; Siebenmorgen et al., 2013). Moreover, chalkiness has adverse impacts on milling quality: the higher the chalkiness, the lower the head rice yield (Fitzgerald et al., 2009). Consequently, the pursuit of reduced chalkiness is extremely important in rice breeding.
Chalkiness is a complex quantitative trait determined by polygenes and environmental factors such as high temperature and nutrition (Siebenmorgen et al., 2013; Jagadish et al., 2015; Kaneko et al., 2016; Tang et al., 2019). This trait is described by various terms, such as white-belly, white-core, white-back, and floury endosperm, depending on the distribution of opaqueness in the grain. In maize (Zea mays), opaque endosperm is commonly associated with altered protein bodies (PBs) and/or starch granules (SGs; Zhang et al., 2018b). Round and loosely packed SGs in chalky rice endosperm are completely different from the polyhedral and densely packed SGs in the preferred transparent endosperm (Guo et al., 2011).
Many quantitative trait loci (QTL) for chalkiness have been mapped on all 12 rice chromosomes (Zhou et al., 2009; Peng et al., 2014b; Gao et al., 2016; Zhu et al., 2018), but only one major white-belly QTL, Chalk5, has been cloned. Chalk5 encodes a vacuolar H+-translocating pyrophosphatase (V-PPase) that might modulate the pH homeostasis of the endomembrane transport system to affect the biogenesis of PBs and other vesicle-like structures (Li et al., 2014). Studies of rice mutants revealed that the cause of floury endosperm is related to the biosynthesis of storage materials and amyloplast development (Kang et al., 2005; Fujita et al., 2007; Ryoo et al., 2007; She et al., 2010; Zhang et al., 2018a; Teng et al., 2019; Wu et al., 2019). Yet, more investigations are still needed to unearth the natural variations and molecular mechanisms underlying chalkiness.
F-box proteins, containing a highly conserved F-box domain with approximately 50 amino acids at the N-terminal region, mediate protein–protein interactions in eukaryotes (Kipreos and Pagano, 2000). These proteins were first found to interact with Cullin and SKP1 to form an SCF (SKP1, Cullin, and F-box) complex that functions as a ubiquitin E3 ligase involved in the ubiquitin–26S proteasome degradation pathway (Schulman et al., 2000). F-box proteins have also been shown to function in non-SCF pathways to mediate protein interactions, transcription elongation, and translation inhibition in yeast, nematode (Caenorhabditis elegans), and humans (Kipreos and Pagano, 2000). The F-box superfamily in rice consists of 687 potential members classified into 10 subfamilies based on their C-terminal domains. These proteins play pivotal roles in vegetative growth and reproductive development (Jain et al., 2007). OsFBK12, which belongs to the FBK subfamily, functions in the 26S proteasome pathway to regulate leaf senescence and seed size as well as grain number (Chen et al., 2013). MEIOTIC F-BOX interacts with rice SKP1-like Protein1 and functions as a component of SCF E3 ligase to regulate meiotic progression (He et al., 2016). Little is known about the roles of F-box proteins in rice endosperm or in non-SCF pathways in plants.
Programmed cell death (PCD) is an indispensable process for the growth and development of multicellular organisms (Schindler et al., 1995; Quirino et al., 2000; Gunawardena et al., 2001; Watanabe and Lam, 2009; Domínguez and Cejudo, 2014; Petrov et al., 2015; Zheng et al., 2019). PCD plays an essential role in starch accumulation and seed maturation in the endosperm of cereal seeds (Young et al., 1997; Young and Gallie, 1999; Chen et al., 2012; Kobayashi et al., 2013). PCD in starchy endosperm might be induced by reactive oxygen species (ROS) including hydrogen peroxide (H2O2), superoxide anion (O2−), and hydroxyl radicals (OH) upon the completion of SG packaging (Xu et al., 2010; Domínguez and Cejudo, 2014). To avoid the over-accumulation of ROS, cells remove these molecules by producing enzymes and antioxidants including catalase, ascorbate peroxidase, and superoxide dismutase (Fath et al., 2001). Metallothioneins (MTs), a family of low molecular weight, cysteine-rich proteins, are involved in ROS scavenging and are widely distributed in animals, plants, and microorganisms (Thornalley and Vašák, 1985; Hussain et al., 1996; Cobbett and Goldsbrough, 2002). MT2b, belonging to the type II MT subfamily, participates in disease responses and delays PCD of root epidermal cells, stem parenchyma cells, and tapetal cells in rice by eliminating ROS (Wong et al., 2004; Steffens et al., 2011, 2012; Yi et al., 2016). However, the roles of PCD and MTs in regulating grain chalkiness in rice are unclear.
In this study, we identified the white-core gene WHITE-CORE RATE 1 (WCR1) in rice through map-based cloning and showed that it functions as a negative regulator of WCR by performing expression analysis and transgenic verification. Natural variation analyses revealed that a functional A/G variation in the promoter region of WCR1 is associated with its expression level and WCR diversity in Asian cultivated rice accessions. OsDOF17 specifically binds to the cis-element in the WCR1A promoter, as determined by yeast one-hybrid (Y1H) assays, electrophoretic mobility shift assay (EMSA), and transient expression assays. We describe a possible mechanism regulating chalkiness by uncovering the relationship between WCR1 and MT2b.
Results
Map-based cloning of WCR1
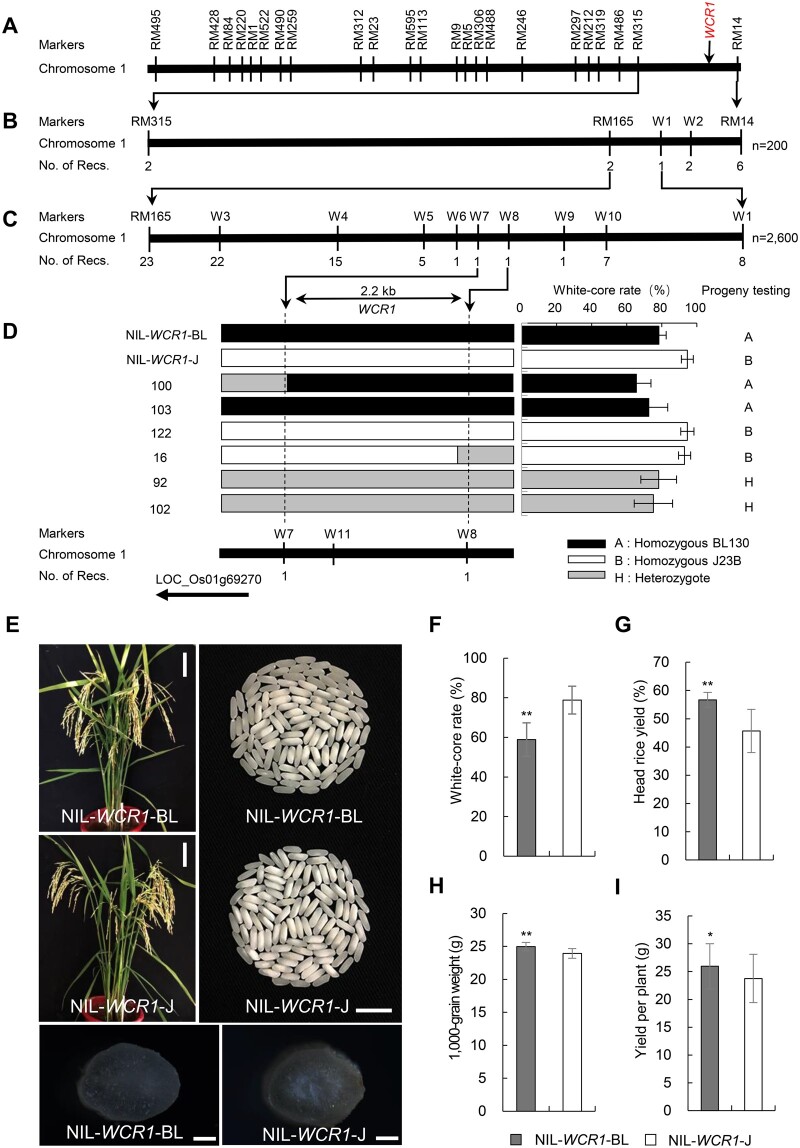
We previously detected a QTL for WCR between markers RM315 and RM14 on the long arm of rice chromosome 1 (Yun et al., 2016) and named it WCR1. This QTL explained 26.1% of the phenotypic variation in this trait in a BC4F2 population derived from a cross between donor parent Beilu130 (BL130, a japonica cultivar) and recurrent parent Jin23B (J23B, an indica cultivar) (Figure 1A; Supplemental Figure S1). Here, to fine-map WCR1, we developed two BC5F2 populations, consisting of 200 (Population 1) and 2,600 (Population 2) individuals. Eight recombinants between RM315 and RM14 identified in Population 1 narrowed WCR1 to a 220 kb segment between markers RM165 and W1 (Figure 1B). Thirty-one recombinants in Population 2 confirmed by progeny testing further delimited WCR1 to a 2.2 kb region between markers W7 and W8, and the locus co-segregated with marker W11 (Figure 1C; Supplemental Table S1). The target region overlapped with the promoter region of LOC_Os01g69270 (Figure 1D; Supplemental Figure S2), which is annotated to encode an F-box protein belonging to the FBO subfamily and containing F-box and MYND-type zinc finger domains (Supplemental Figure S3). Phylogenetic analysis showed that this gene shares homology with the F-box gene AT1g67340 in Arabidopsis thaliana, and the homologs are also found in crop species such as barley, wheat, sorghum, and maize (Supplemental Figure S4). Hence, LOC_Os01g69270 was identified as the candidate gene underlying WCR1.
Figure 1.
Map-based cloning of the QTL WCR1 and agronomic traits of the NILs. A, Location of WCR1 (marked in red) in the genetic map. B and C, Fine mapping of the WCR1 region using Population 1 (B) and Population 2 (C). No. of Recs. indicates the number of recombinants between WCR1 and flanking molecular markers. D, Genotypes and phenotypes of recombinants, each of which was confirmed by progeny test (Supplemental Table S1). E, Plant and seed characteristics of the NILs. Scale bars, 10 cm (plants), 10 mm (grains), and 500 µm (transverse sections of brown rice grains). F–I, Comparisons of WCR, head rice yield, 1,000-grain weight, and yield per plant between NIL-WCR1-BL and NIL-WCR1-J. Data are means (n = 30) with error bars, sem. P-values were determined by two-tailed t tests, *P ≤ 0.05, **P ≤ 0.01.
To evaluate the effects of WCR1 on chalkiness and agronomic traits, we used a pair of near-isogenic line (NILs) derived from a BC7F3 population: NIL-WCR1-BL and NIL-WCR1-J, which carried the WCR1 locus from BL130 and J23B, respectively, in the J23B background (Figure 1E). Cross-sections of brown rice revealed notably opaque endosperm in the center of NIL-WCR1-J (Figure 1E). Compared with NIL-WCR1-J, NIL-WCR1-BL displayed an ∼20% lower WCR value, a higher perfect grain rate (the percentage of grains with no chalkiness), and a similar level of white-belly (Figure 1F; Supplemental Figure S5, A–B). NIL-WCR1-BL had significantly higher values for head rice yield, grain size (grain length, width, and thickness), 1,000-grain weight, and yield per plant than NIL-WCR1-J (Figure 1, G–I; Supplemental Figure S5, C–E). There were no differences in plant height, heading date, panicle number, panicle length, secondary branch number, number of filled grains, or seed setting rate between the NILs (Supplemental Figure S5, F–L). Therefore, the WCR1 locus from BL130 conferred the pleiotropic effects of decreased grain chalkiness and enhanced milling quality and yield.
Confirmation of WCR1
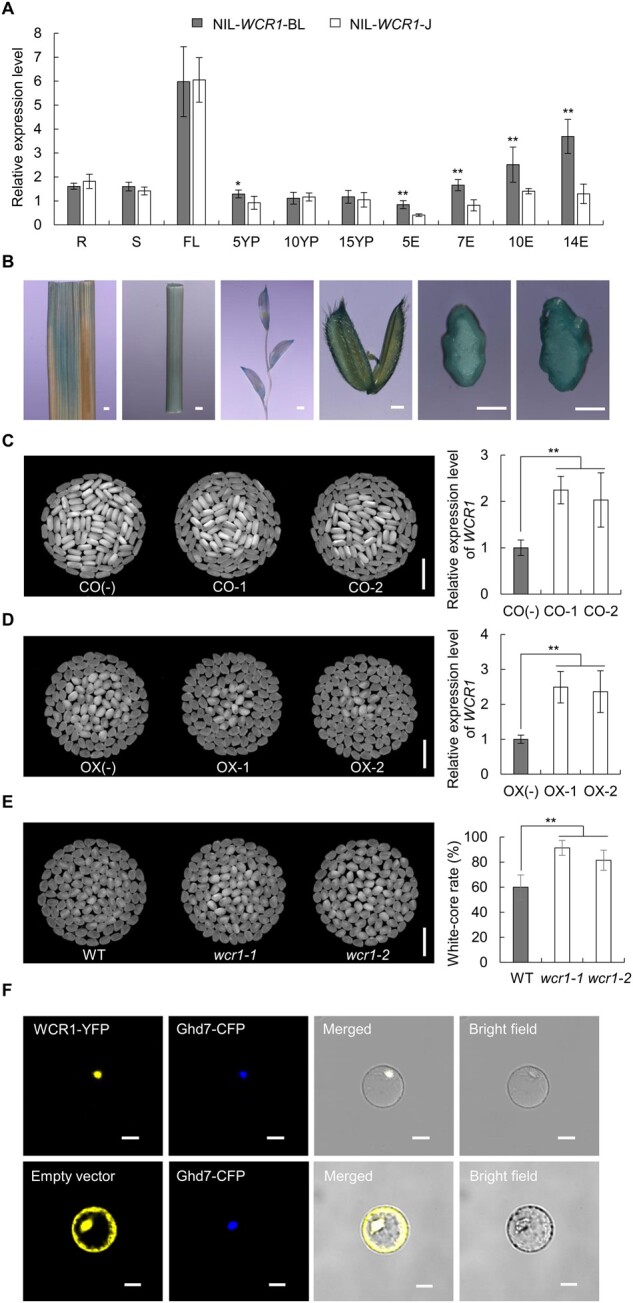
Since the candidate region overlapped with the promoter region of WCR1, we performed quantitative real-time PCR (qRT-PCR) using the NILs to measure WCR1 expression levels. WCR1 was expressed in all tested tissues, and the transcript levels in NIL-WCR1-BL were markedly higher in 5-cm panicles and endosperm at 5–14 days after flowering (DAF) than in NIL-WCR1-J (Figure 2A). GUS staining of plants harboring GUS driven by the WCR1 promoter showed that WCR1 was expressed in flag leaf, stem, young panicle, glume, and developing endosperm tissue (Figure 2B). We, therefore, reasoned that different expression levels of WCR1 were responsible for the phenotypic differences between the NILs.
Figure 2.
Expression patterns, transgenic verification, and subcellular localization of WCR1. A, Spatiotemporal expression patterns of WCR1 in the NILs. R, S, and FL: roots, stems and flag leaves at the heading stage; 5YP, 10YP, and 15YP: young panicles 5, 10, and 15 cm long; 5E, 7E, 10E, and 14E: grain endosperm at 5, 7, 10, and 14 DAF. Samples were obtained from at least five different plants. Significant differences were based on two-tailed t tests (n = 5), *P ≤ 0.05, **P ≤ 0.01. Error bars, sem. B, Expression analysis of WCR1 by GUS staining. From left to right, photographs of a flag leaf, stem, young panicle, glume, and transverse sections of caryopsis from plants harboring GUS driven by the WCR1 promoter at 7 DAF and 10 DAF. Scale bars, 1 mm. C and D, Grain chalkiness and expression level of WCR1 in CO and OX lines. CO-1/CO-2 and OX-1/OX-2 refer to positive complementation and OX lines, respectively. CO(−) and OX(−) represent transgene-negative lines. Scale bars, 10 mm. Significant differences were based on two-tailed t tests (n = 4), **P ≤ 0.01. Error bars, sem. E, Grain chalkiness of KO lines. wcr1-1 and wcr1-2 represent homozygous mutants with a 27- and 1-bp deletion in the third exon of WCR1, respectively. Scale bars, 10 mm. Significant differences were based on two-tailed t tests (n = 25), **P ≤ 0.01. Error bars, sem. F, WCR1-YFP and Ghd7-CFP (nucleus marker) co-localize in the nucleus. The empty vector pM999-YFP was used as a negative control. Scale bars, 20 μm.
To confirm that WCR1 is responsible for the white-core phenotype, we transformed J23B plants with the complementation (CO) vector pWCR1BL:cWCR1J (WCR1 coding region from J23B driven by the WCR1 promoter from BL130) (Supplemental Figure S6A) and transformed japonica variety Zhonghua11 (ZH11) with the overexpression (OX) vector pUbi:cWCR1J (WCR1 coding region from J23B driven by the maize ubiquitin promoter; Supplemental Figure S6B). Transgene-positive plants CO-1 and CO-2 displayed significantly lower WCR values and higher WCR1 expression levels than transgene-negative plants CO(−) in both the T1 and T2 generations (P < 0.01) (Figure 2C; Supplemental Table S2). Similar results for WCR values and expression levels were obtained for OX positive plants OX-1, OX-2 and negative plants OX(−) (Figure 2D; Supplemental Table S2). We also generated knockout (KO) plants in the ZH11 background via clustered regularly interspaced short palindromic repeats-associated protein 9 (Cas9) genome editing (Supplemental Figure S6C). The wcr1-1 and wcr1-2 mutants displayed higher WCR values than the wild-type (WT; P < 0.01; Figure 2E). We also evaluated the head rice yield and found that positive lines of CO and OX exhibited significantly increased values relative to CO(−) and OX(−), respectively, whereas the opposite result was obtained for KO lines (Supplemental Figure S7). These results confirm the notion that LOC_Os01g69270 is the causal gene underlying WCR1, which functions as a negative regulator of WCR.
Analysis of the deduced amino acid sequence of WCR1 revealed two nuclear localization signal peptides (KRKR and RRKR) at the N-terminal region (Supplemental Figure S3). Transient expression of WCR1-YFP (WCR1 fused to yellow fluorescent protein) and the nucleus marker Ghd7-CFP in rice protoplasts showed that these fusion proteins co-localized to the nucleus (Figure 2F). We also transformed Nicotiana benthamiana leaf epidermal cells with WCR1-GFP and nucleus/membrane markers and achieved the same result (Supplemental Figure S8).
Effects of WCR1 on grain storage components
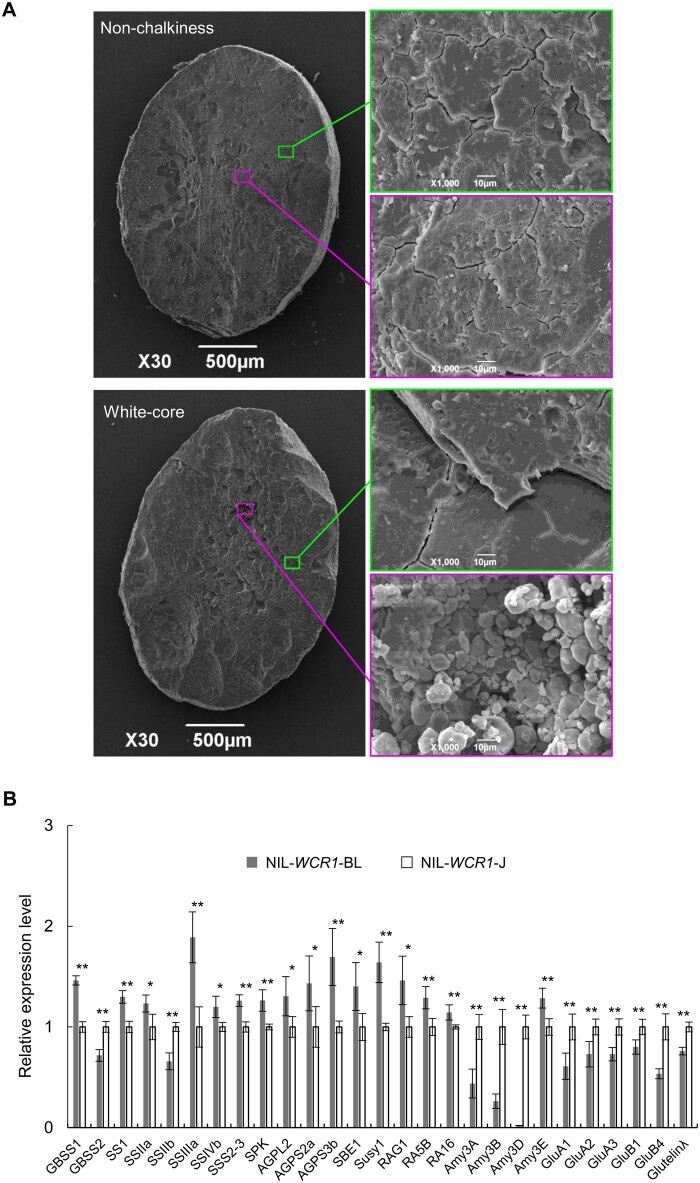
Chalkiness is associated with changes in endosperm structure and the contents of grain storage components (Ryoo et al., 2007; She et al., 2010; Li et al., 2014; Zhang et al., 2018a). Therefore, we observed SGs in the endosperm of the NILs by scanning electron microscopy. SGs in nonchalky grains were arranged very densely and regularly, without spaces, in the center and edge of the endosperm. In contrast, the central endosperm in white-core grains was filled with small, spherical SGs with large air spaces, whereas the SGs in the peripheral endosperm were similar to those in nonchalky grains (Figure 3A). These results indicate that white-core, like other forms of chalkiness, results from the altered morphology and spatial distribution of SGs.
Figure 3.
WCR1 affects SG morphology and the transcript levels of genes involved in endosperm development and metabolism. A, observation of transverse sections of mature seeds from NIL-WCR1-BL and NIL-WCR1-J by scanning electron microscopy. Magenta and green rectangles represent the center and edge positions of endosperm, respectively. Scale bars: 500 µm (left), 10 µm (right). B, Expression analysis of genes related to storage components in 14 DAF endosperm. Endosperm samples were obtained from different plants. Significant differences were based on two-tailed t tests, *P ≤ 0.05, **P ≤ 0.01. Error bars, sem.
We then measured the starch, protein, and lipid contents and taste value of the NILs and transgenic lines. NIL-WCR1-BL, CO(+), and OX(+) had substantially higher total starch, amylose, and total lipid contents coupled with lower glutenin, albumin, and total protein contents than NIL-WCR1-J, CO(−), and OX(−) respectively, whereas the results for wcr1-1 and WT were the opposite in each case (Table 1). Taste value, a comprehensive assessment of ECQ affected by the viscosity, hardness, and appearance of cooked rice, was significantly higher for NIL-WCR1-BL versus NIL-WCR1-J, as well as for OX(+) versus OX(−) (Table 1). Hence, WCR1 has pleiotropic effects on storage components and enhances ECQ.
Table 1.
Grain quality traits of the NILs and the transgenic plants
| Lines | N | Total Starch Content (%) | Amylose Content (%) | Total Protein Content (%) | Albumin Content (mg/g) | Glutelin Content (mg/g) | Globulin Content (mg/g) | Prolamin Content (mg/g) | Total lipid Content (mg/g) | Taste Value |
|---|---|---|---|---|---|---|---|---|---|---|
| NIL-WCR1-BL | 5 | 84.1 ± 1.5 | 26.4 ± 0.8 | 7.1 ± 0.9 | 2.6 ± 0.5 | 61.3 ± 9.1 | 4.5 ± 0.4 | 2.6 ± 0.5 | 34.0 ± 0.3 | 64.0 ± 1.3 |
| NIL-WCR1-J | 5 | 79.8 ± 1.6 | 23.8 ± 1.0 | 9.3 ± 1.0 | 3.5 ± 0.3 | 82.0 ± 9.6 | 4.3 ± 0.3 | 2.8 ± 0.2 | 32.7 ± 0.9 | 60.6 ± 1.9 |
| P-value | **2.4 × 10−3 | **2.1 × 10−3 | **6.0 × 10−3 | **9.0 × 10−3 | **8.1 × 10−3 | 4.1 × 10−1 | 4.3 × 10−1 | *1.8 × 10−2 | *4.6 × 10−2 | |
| CO(+) | 5 | 85.9 ± 3.6 | 25.0 ± 0.9 | 8.2 ± 0.3 | 2.3 ± 0.4 | 72.0 ± 3.4 | 5.0 ± 0.7 | 2.6 ± 0.5 | 38.8 ± 1.7 | – |
| CO(−) | 5 | 80.6 ± 0.8 | 23.5 ± 0.6 | 10.0 ± 1.0 | 3.0 ± 0.1 | 89.5 ± 10.0 | 4.6 ± 0.3 | 2.9 ± 0.3 | 35.9 ± 0.9 | – |
| P-value | *1.3 × 10−2 | *1.2 × 10−2 | **3.9 × 10−3 | **5.1 × 10−3 | **5.8 × 10−3 | 3.1 × 10−1 | 4.0 × 10−1 | *1.1 × 10−2 | – | |
| OX(+) | 5 | 93.0 ± 2.0 | 17.0 ± 1.2 | 4.7 ± 0.6 | 2.5 ± 0.1 | 38.6 ± 6.0 | 3.4 ± 0.5 | 2.6 ± 0.3 | 35.6 ± 2.8 | 79.2 ± 1.4 |
| OX(−) | 5 | 87.5 ± 2.4 | 15.2 ± 0.7 | 6.2 ± 0.8 | 3.0 ± 0.1 | 52.1 ± 7.8 | 3.5 ± 0.2 | 3.0 ± 0.1 | 32.4 ± 0.9 | 74.7 ± 0.8 |
| P-value | **4.0 × 10−3 | *2.0 × 10−2 | **9.0 × 10−3 | **9.0 × 10−4 | *1.5 × 10−2 | 5.9 × 10−1 | 5.1 × 10−2 | *3.6 × 10−2 | *2.9 × 10−2 | |
| wcr1-1 | 5 | 84.4 ± 2.1 | 15.9 ± 0.3 | 8.0 ± 0.5 | 3.1 ± 0.2 | 70.9 ± 5.3 | 3.5 ± 0.3 | 2.6 ± 0.5 | 33.0 ± 0.8 | – |
| WT | 5 | 90.6 ± 2.8 | 18.7 ± 0.7 | 6.1 ± 0.9 | 2.6 ± 0.1 | 52.2 ± 8.6 | 3.2 ± 0.3 | 2.8 ± 0.2 | 34.9 ± 1.3 | – |
| P-value | **4.1 × 10−3 | **2.6 × 10−5 | **3.2 × 10−3 | **2.8 × 10−4 | **3.3 × 10−3 | 2.0 × 10−1 | 2.4 × 10−1 | *3.8 × 10−2 | – |
n, number of plants; (+) and (−), transgene positive and negative plants, respectively; taste value, a comprehensive assessment of the viscosity, hardness, and appearance of cooked rice. Samples consisted of at least 300 grains per genotype. All data are means ± sem. P-values were determined by two-tailed t tests, *P ≤ 0.05, **P ≤ 0.01.
To further analyze the reason for the changes in storage components, we examined the expression levels of 49 genes in 14 DAF endosperm. The expression levels of starch synthesis and α-amylase inhibitor genes such as GBSS1, SS1, SSIIa. SSIIIa, SSIVb, SS2-3, SPK, AGPL2, AGPS3b, SBE1, Susy1, RAG1, RA5B, and RA16 were generally higher in NIL-WCR1-BL than in NIL-WCR1-J, whereas the expression levels of genes for starch decomposition (Amy3A, Amy3B, and Amy3D) and protein synthesis (GluA1, GluA2, GluA3, GluB1, GluB4, and Glutelinλ) were markedly lower in NIL-WCR1-BL (Figure 3B; Supplemental Figure S9). These results suggest that WCR1 promotes starch synthesis, but reduces starch decomposition and storage protein synthesis, by affecting the expression of related genes, ultimately resulting in increased starch accumulation and decreased levels of storage protein components.
Natural variation in WCR1
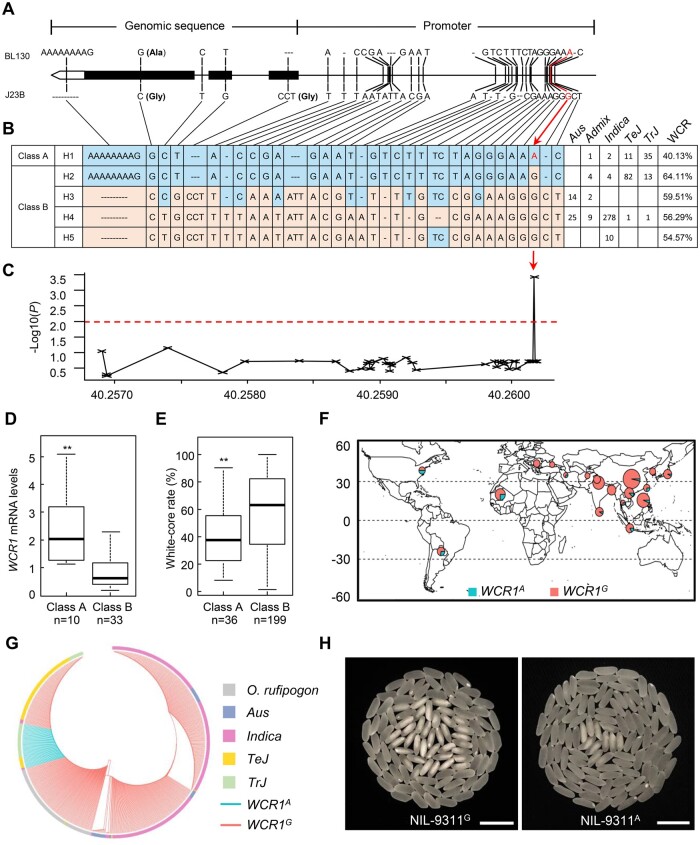
To uncover the functional variation underlying WCR1, we compared the full-length sequences of the two alleles from BL130 and J23B. Twenty-eight polymorphisms were detected in the 2-kb upstream promoter region, including 7 insertion/deletions (InDels) and 21 single-nucleotide polymorphisms (SNPs) leading to changes in transcription factor (TF) binding sites, such as the T/G-box, RY/G-box, ARR1-binding element, and DOFCOREZM (Figure 4A; Supplemental Table S3). We also detected a 9-bp InDel in the 3′-untranslated region and three SNPs and a 3-bp InDel in the open reading frame, causing an amino acid substitution and an amino acid deletion, both of which were located outside of key domains (Figure 4A; Supplemental Figure S3).
Figure 4.
Natural variation in WCR1. A, DNA sequence polymorphisms of WCR1 in BL130 and J23B. B, Natural variation and haplotype analysis of WCR1 in 492 Asian cultivated rice accessions with reference to the sequences of the parents. The five haplotypes were divided into class A and B based on the functional SNP (A/G) at −1,696 bp (red arrow). Frequencies in taxonomic groups are shown on the right. C, Association testing of WCR and 33 variants in the 2-kb promoter and CDS of WCR1. The red dotted line and the red arrow represent the threshold and predicted functional site, respectively. The x-axis represents the position in the genome. D, Relative WCR1 expression levels of the two classes of haplotypes in endosperm at 10 DAF. The data distribution of Class A includes H1 (2.57), and Class B includes H2 (0.63), H3 (0.43), H4 (1.39), and H5 (0.67). E, WCR phenotypes in class A and B. The data distribution of Class A includes H1 (40.13%), and Class B includes H2 (64.12%), H3 (59.51%), H4 (56.29%), and H5 (54.57%). In (D and E), the number below each bar represents the number of analyzed accessions. P-values were determined by two-tailed t tests, **P ≤ 0.01. Error bars, sem. F, Geographic distributions of the 492 cultivated rice accessions. Cyan and red sectors in the circles represent the proportions of rice accessions with WCR1A and WCR1G, respectively. G, Phylogenetic relationship of WCR1 generated from 492 cultivated and 99 wild rice accessions. Cyan and red lines represent the accessions with WCR1A and WCR1G, respectively. Different colored boxes represent rice subspecies. H, Grain chalkiness of NIL-9311G and NIL-9311A. Scale bars,10 mm.
We then analyzed variation in the genomic region of WCR1 in 492 Asian cultivated rice accessions representing a broad range of rice germplasm (Supplemental Data Set 1). Five haplotypes were classified based on the nucleotide variations between the two parents (Figure 4B). We also evaluated WCR in 235 accessions after removing accessions with floury or colored brown rice (Supplemental Data Set 1). Based on WCR phenotype and nucleotide variation, we predicted a functional SNP (A/G) at −1,696 bp by association analysis and divided the five haplotypes into two classes: Haplotype 1 (H1) in Class A with an A base-allele (WCR1A), and H2–5 in Class B with a G base-allele (WCR1G; Figure 4, B and C). Significantly higher WCR1 expression and lower WCR were observed in the endosperm of Class A versus Class B accessions (Figure 4, D and E). Analysis of cis-elements near the A/G site revealed that the binding core site T/AAAAG of the DOF TF changed from Class A: AAAAG to Class B: AGAAG. This finding indicates that the A/G SNP likely alters the binding of DOF TFs to the WCR1 promoter, thereby affecting transcriptional regulation.
An analysis of the geographical distribution of these accessions showed that WCR1G was mainly distributed in accessions from middle latitudes, whereas WCR1A was mainly distributed in accessions from lower latitudes (Figure 4F). Phylogenetic analysis of WCR1 based on 492 cultivated and 99 wild rice accessions suggested that WCR1G originated from Oryza rufipogon and that WCR1A possibly arose in temperate and tropical japonica (TeJ and TrJ) subpopulations (Figure 4G). The proportion of WCR1A was 12% among the 94 TeJ accessions, but 71% among the 49 TrJ accessions (Figure 4B), suggesting that WCR1A first appeared in TrJ accessions and was subsequently introgressed into other subpopulations during domestication.
To further evaluate its breeding value, we introduced WCR1A into the widely planted indica accession 9311. The WCR for NIL-9311A (WCR1A) was 25.3% lower than that for NIL-9311G (WCR1G) (P < 0.01), and grain length, grain thickness, and 1,000-grain weight were significantly higher (Figure 4H; Supplemental Table S4), suggesting that WCR1A represents a promising selection target for rice breeding.
OsDOF17 binds to the WCR1 promoter
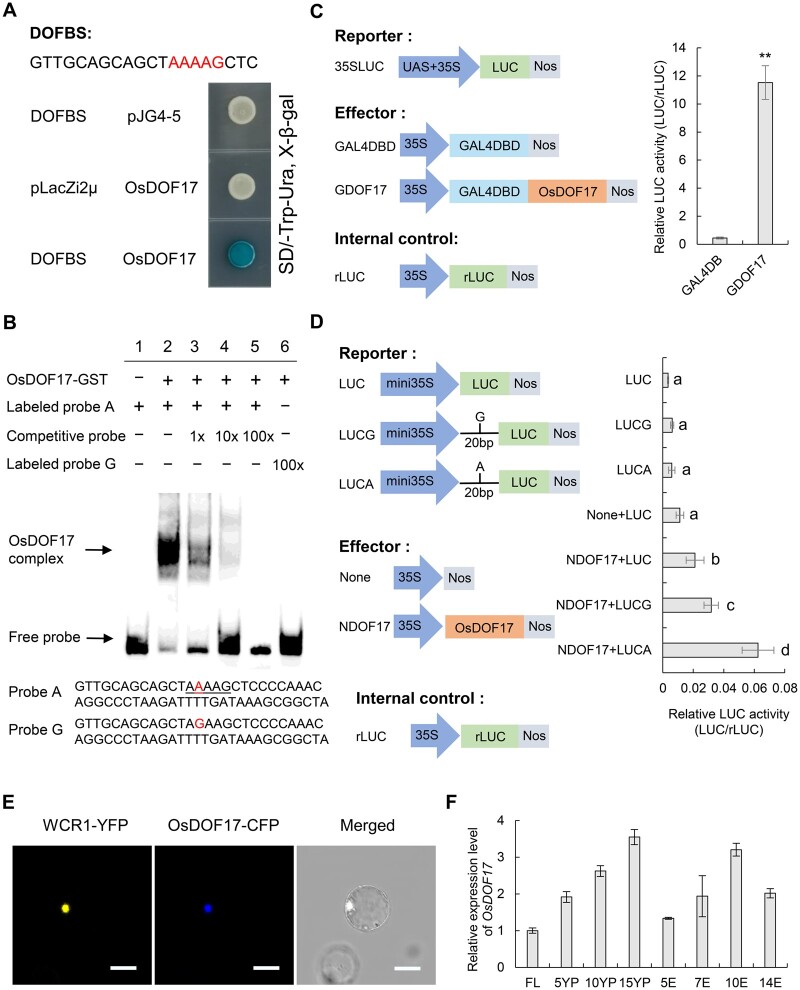
To identify possible DOF TFs that bind to the AAAAG motif of WCR1 and regulate its transcription, we performed Y1H assays using a 20-bp probe with the AAAAG motif from BL130. Among members of the OsDOF family that are predominantly expressed in rice seeds or panicles, only OsDOF17 bound to the probe (Figure 5A; Supplemental Figure S10), which was further verified by an EMSA. A slower migrating band was detected when the glutathione S-transferase (GST)–OsDOF17 fusion protein was incubated with the 55-bp Probe A with the AAAAG motif. In contrast, the band was gradually abolished by the presence of 1-, 10-, and 100-fold molar excess of unlabeled competitive probe. In addition, no migrating band was detected in the presence of 100-fold labeled Probe G and OsDOF17 (Figure 5B). These results suggest that OsDOF17 had differential binding activities to these two probes.
Figure 5.
WCR1 is regulated by OsDOF17. A, Y1H assay. DOFBS represents a 20-bp probe containing the AAAAG motif from BL130. Yeast strains were cultured on SD/-Trp-Ura + X-Gal selection medium. B, EMSA of OsDOF17 protein with the AAAAG motif in the WCR1 promoter. Probe A, a labeled 55-bp sequence containing a AAAAG motif from the WCR1 promoter (−1,737 to −1,683); Probe G, a labeled 55-bp sequence with a single nucleotide mutation (A–G; shown in red). The AAAAG motif is underlined. The OsDOF17-GST fusion protein was used for EMSA with the probes. The unlabeled competitive probe was added in 1-, 10-, and 100-fold molar excess. Lane 1, no protein; lanes 2–6, fusion protein with Probe A, competitive probe, and Probe G, as indicated. C and D, Transient expression assays of the transcriptional activity of OsDOF17 on the A/G site of WCR1 promoter in vivo. In (C), Left, the reporter vector contained the GAL4 binding cis-elements (UAS), the 35S promoter and the firefly LUC gene. The effectors consisted of the negative vector containing a GAL4 DNA-binding domain (GAL4DBD) and the positive vector GDOF17 containing a GAL4DBD and full-length OsDOF17. The internal control was the rLUC gene. Right, relative LUC activity (LUC/rLUC). P-value was determined by a two-tailed t test, **P ≤ 0.01. In (D), Left, the reporter vector containing the mini35S promoter and the LUC gene was inserted in the 20-bp sequence containing A (LUCA) or G (LUCG). The effectors consisted of the negative vector None and positive vector NDOF17 containing the full-length OsDOF17. Right, relative LUC activity. Significant differences were determined by Duncan’s test. All data in (C and D) were based on three replicate experiments. Error bars, sem. E, WCR1-YFP and OsDOF17-CFP co-localize in the nucleus. Scale bar, 20 μm. F, Spatiotemporal expression patterns of OsDOF17. FL: flag leaves at the heading stage; 5YP, 10YP and 15YP: young panicles 5, 10, and 15 cm long; 5E, 7E, 10E, and 14E: grain endosperm at 5, 7, 10, and 14 DAF. Samples were obtained from different plants. Data are means (n = 4) with error bars, sem.
To detect the transcriptional effects of OsDOF17 on the A/G site of the WCR1 promoter, we performed a self-activation experiment in yeast and a transient expression assay in rice protoplasts, finding that OsDOF17 had strong transcriptional activation activity (Supplemental Figure S11; Figure 5C). Moreover, the relative firefly luciferase (LUC) activity of LUCA was markedly higher than that of LUCG after adding the NDOF17 effector in another transient expression assay (Figure 5D), suggesting that OsDOF17 had stronger ability to combine with the A versus G site of the WCR1 promoter. Co-expression of the WCR1-YFP and OsDOF17-CFP fusion proteins in rice protoplasts showed that both proteins were located in the nucleus (Figure 5E). We then examined the mRNA levels of OsDOF17 in different tissues and found that OsDOF17 was mainly expressed in young panicles and endosperm (Figure 5F). These results indicate that OsDOF17 is a transcriptional activator that binds to the WCR1 promoter and that its binding ability is affected by the A/G variation.
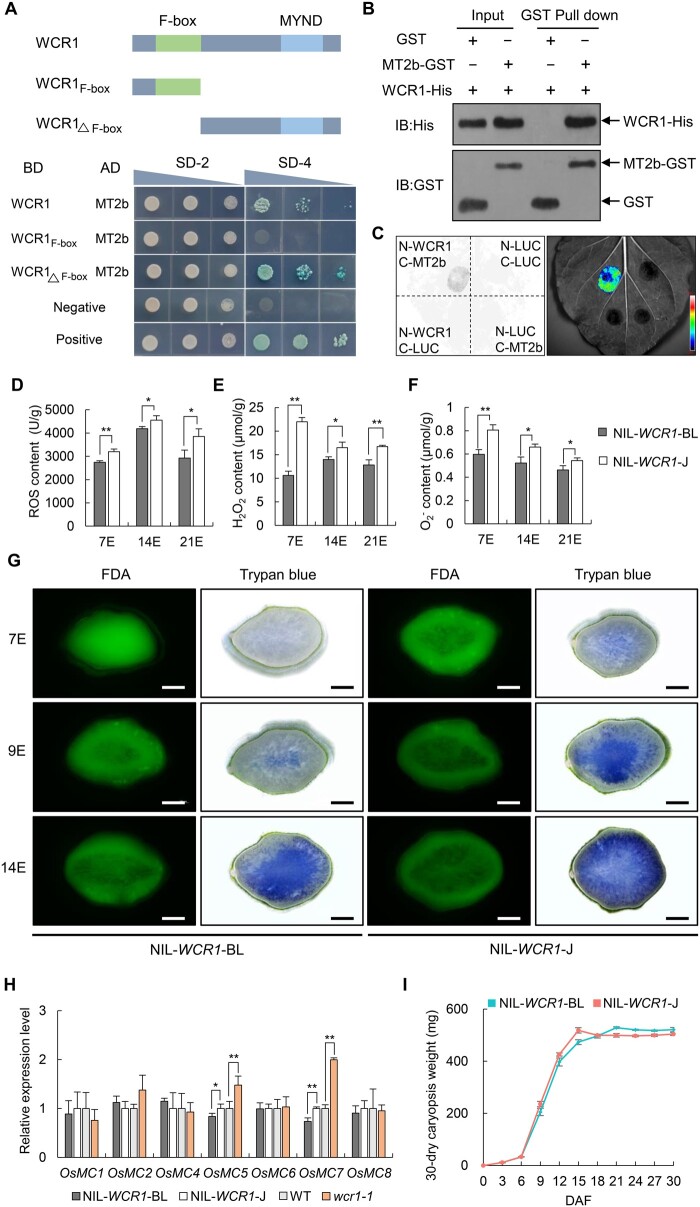
WCR1 interacts with MT2b to delay PCD in developing endosperm
To uncover the mechanism underlying chalkiness, we conducted yeast two-hybrid (Y2H) assays to screen for WCR1-interacting proteins from a cDNA library of rice endosperm and identified MT2b, an MT (Figure 6A). Domain truncation analysis indicated that the C-terminus of WCR1 could bind to MT2b rather than the N-terminus (Figure 6A). The interaction between WCR1 and MT2b was further confirmed in vitro by pull-down assays (Figure 6B) and in vivo by split-LUC assays in N. benthamiana leaves and bimolecular fluorescence complementation (BiFC) assays in rice protoplasts (Figure 6C; Supplemental Figure S12).
Figure 6.
WCR1 interacts with MT2b to scavenge ROS and delay PCD in developing endosperm. A, WCR1 interacts with MT2b in yeast cells. The N terminus of WCR1 including the F-box domain and the C terminus including the MYND domain are shown. SD-2 and SD-4 represent SD/–Trp–Leu and SD/–Trp–Leu–His–Ade + X-Gal selection medium, respectively. Negative, empty vectors pGBKT7 and pGADT7. Positive, the OsMADS1 gene, which formed a homodimer, was linked to pGBKT7 (BD-OsMADS1) and pGADT7 (AD-OsMADS1), respectively, and yeast harboring these two vectors grew on SD-4 medium. B, Pull-down assay. Proteins were pulled down by GST agarose beads and immunoblotted using His and GST antibodies. C, Split-LUC assay. nLUC-tagged WCR1 and cLUC-tagged MT2b were co-transformed into N. benthamiana leaves. Color bar shows the intensity of LUC signals. D–F, Detection of ROS, H2O2, and contents in endosperm at 7, 14, and 21 DAF (7E, 14E, and 21E). Each endosperm sample was obtained from 30 grains with three repeats. G, Trypan blue and FDA staining of transverse sections of grains at different developmental stages for NIL-WCR1-BL and NIL-WCR1-J. Blue areas and green fluorescence represent areas of dead and living cells, respectively. Scale bars, 500 μm. H, Relative expression levels of PCD-related OsMC genes in 9 DAF endosperm. Endosperm samples were taken from at least four plants. In (D–F and H), significant differences were determined by two-tailed t tests, *P ≤ 0.05, **P ≤ 0.01. Error bars, sem. I, Dynamic changes in mean dry weights of caryopses. Data show the total weights of 30 dry dehulled caryopses with three biological repeats from the upper parts of panicles from different plants. Error bars, sem.
As MT2b functions in ROS scavenging and delays PCD in roots, stems, and anthers (Steffens et al., 2011, 2012; Yi et al., 2016), we wondered whether WCR1 plays a similar role in endosperm via MT2b. We measured ROS, H2O2, and O2− levels in developing endosperm of the NILs and found that they were significantly lower in NIL-WCR1-BL than in NIL-WCR1-J at 7, 14, and 21 DAF (Figure 6, D–F). Staining of endosperm with trypan blue, which penetrates the membranes of dead cells, indicated that PCD began at the center of 7 DAF endosperm in NIL-WCR1-J, spread to surrounding areas, and eventually affected the entire section except for the aleurone layer by 14 DAF. In contrast, in NIL-WCR1-BL, PCD began at 9 DAF, and far fewer stained cells were present in this NIL than in NIL-WCR1-J at the same time point (Figure 6G).
To verify the difference in cell death in starchy endosperm of the NILs, we used the fluorescent probe fluorescein diacetate (FDA), which is commonly used to detect active cells. Green fluorescence from FDA was observed in throughout the starchy endosperm at 7 DAF in NIL-WCR1-BL, whereas fluorescence was absent from the central region of endosperm in NIL-WCR1-J, indicating that cells in the central region of endosperm of NIL-WCR1-J died earlier than those of NIL-WCR1-BL. Subsequently, the area of dead starchy endosperm at 9 and 14 DAF spread outwardly more rapidly in NIL-WCR1-J than in NIL-WCR1-BL (Figure 6G). In addition, an examination of the expression of PCD-related genes (OsMC, encoding rice metacaspase) in 9 DAF endosperm showed that OsMC5 and OsMC7 displayed significantly higher transcript levels in NIL-WCR1-J and wcr1-1 than in NIL-WCR1-BL and WT, respectively (Figure 6H). These results suggest that PCD of endosperm occurred earlier in NIL-WCR1-J than in NIL-WCR1-BL.
The degradation of nuclear DNA into multiples of 180–200 bp fragments is a typical feature of PCD in endosperm cells of wheat and maize (Young et al., 1997; Young and Gallie, 1999), whereas it has not been detected in rice (Li et al., 2004; Kobayashi et al., 2013). To examine whether the degradation of nuclear DNA occurs in rice endosperm, we performed DNA fragmentation analysis over the course of endosperm development. A ladder of DNA fragments in multiples of 180–200 bp was not detected in developing starchy endosperm of NIL-WCR1-J or NIL-WCR1-BL (Supplemental Figure S13A). We then observed nuclear DNA from NIL-WCR1-J endosperm by 4′,6-diamidino-2-phenylindole (DAPI) staining. Nuclear DNA was detected throughout developing endosperm, and even in both the peripheral and central regions of mature endosperm (Supplemental Figure S13B). Meanwhile, a terminal deoxynucleotidyl transferase TdT-mediated dUTP nick end labeling (TUNEL) assay failed to detect the degradation of nuclear DNA in 7, 9, or 14 DAF starchy endosperm of both NIL-WCR1-BL and NIL-WCR1-J (Supplemental Figure S13C). Hence, we conclude that PCD of rice endosperm is not accompanied by the severe degradation of nuclear DNA, but is instead associated with the disintegration and decrease in permeability of the cell membrane, which is different from that of wheat and maize.
PCD of endosperm is essential for seed development; the premature induction of PCD limits the deposition of reserves (Young et al., 1997; Young and Gallie, 1999, 2000; Chen et al., 2012; Kobayashi et al., 2013). We, therefore, investigated dynamic changes in seed reserves in the NILs during grain filling. Caryopsis dry weight increased more rapidly in NIL-WCR1-J than in NIL-WCR1-BL from 6 to 15 DAF, the former reaching a peak at 15 DAF compared to 21 DAF for the latter (Figure 6I). Collectively, these results suggest that WCR1 promotes ROS degradation by MT2b and thereby delays PCD during early endosperm development, leading to the increased accumulation of storage components.
WCR1 functions upstream of MT2b in the regulatory pathway
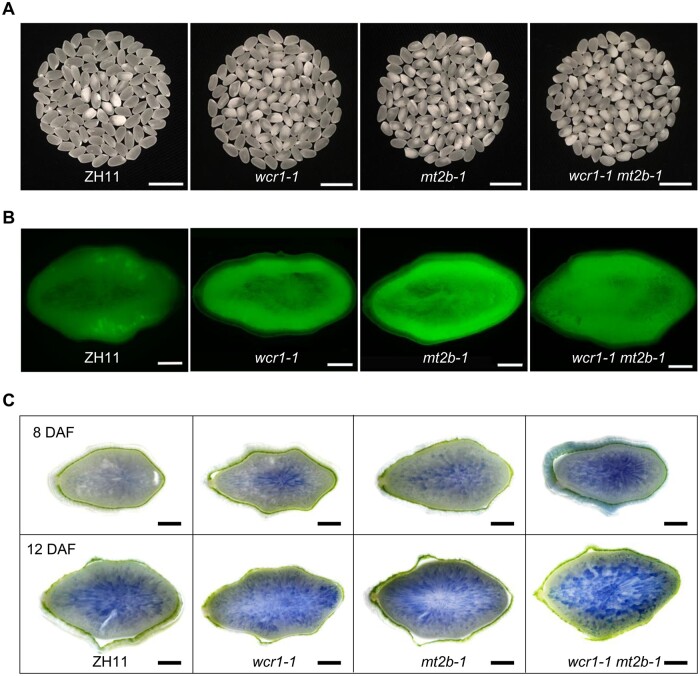
To further clarify the roles of WCR1 and MT2b in regulating chalkiness, we knocked out MT2b in the ZH11 background by CRISPR/Cas9 and obtained the mt2b mutants mt2b-1 and mt2b-2 (Supplemental Figure S14A). T1 generation mutant plants displayed markedly higher WCR than ZH11 (Supplemental Figure S14B). Double mutant (wcr1-1 mt2b-1) plants produced in the ZH11 background displayed a similar level of WCR to mt2b-1 plants, which was significantly higher than the levels in ZH11 and wcr1-1 plants (Figure 7A; Supplemental Figure S15A). Staining with 2,7-Dichlorodi-hydrofluorescein diacetate (DCFH-DA, a fluorescent dye reflecting endogenous ROS level) showed that the ROS contents of 14 DAF endosperm were higher in mt2b-1 and wcr1-1 mt2b-1 than in ZH11 and wcr1-1 (Figure 7B; Supplemental Figure S15B). Trypan blue staining indicated that the number of dead cells was markedly higher in mt2b-1 and wcr1-1 mt2b-1 endosperm than in wcr1-1 and was markedly lower in ZH11 (Figure 7C). Taken together, these results indicate that WCR1 functions upstream of MT2b to regulate ROS, PCD, and ultimately WCR.
Figure 7.
WCR1 functions upstream of MT2b in regulating chalkiness, ROS accumulation, and PCD. A, Grain chalkiness in ZH11, wcr1-1, mt2b-1, and wcr1-1 mt2b-1 lines. wcr1-1 and mt2b-1 indicate grains from homozygous mutants with the 27-bp deletion in WCR1 and the 2-bp deletion in MT2b, respectively. Scale bars, 10 mm. B, DCFH-DA staining of transverse sections of 14 DAF grains in ZH11 and the mutants. DCFH-DA is a fluorescent probe with membrane permeability, reflecting the level of endogenous ROS. Scale bars, 500 μm. C, Trypan blue staining of endosperm in transverse sections of grains from ZH11 and mutant grains at 8 (upper) and 12 (lower) DAF. Scale bars, 500 µm.
WCR1 positively regulates MT2b
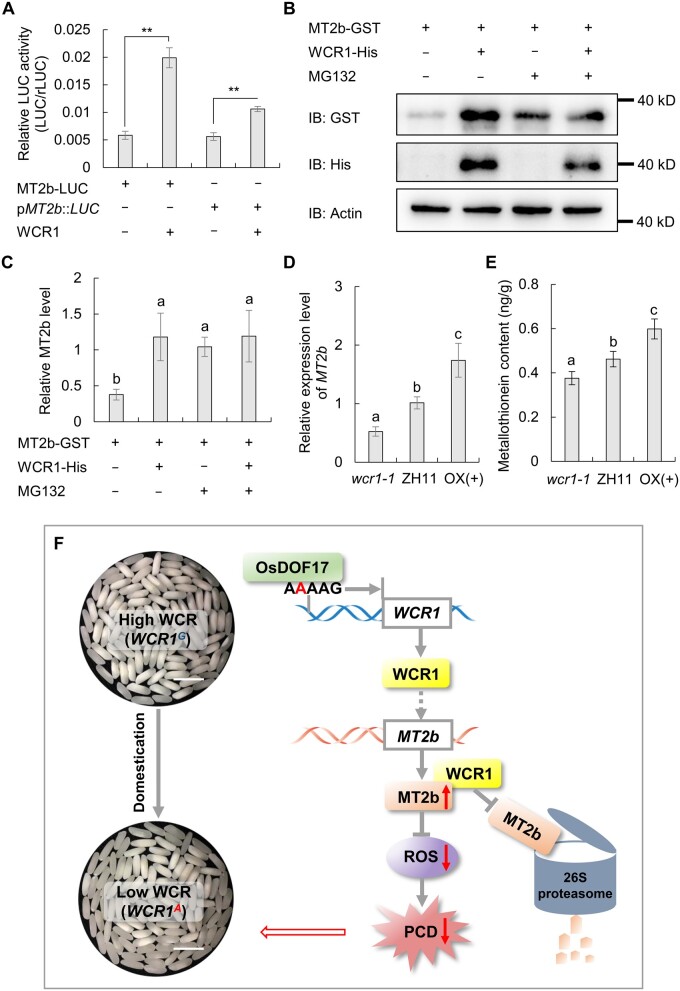
To test the effect of WCR1 on MT2b, we performed a transient expression assay in rice protoplasts using MT2b-LUC fusion protein and pMT2b:LUC as the reporters. Compared to the controls, LUC activities were significantly elevated in the presence of the effector WCR1, suggesting that WCR1 positively affects MT2b protein stability and MT2b transcript levels (Figure 8A). To confirm that WCR1 stabilizes MT2b protein, we performed in vitro degradation assays. When the recombinant fusion proteins MT2b-GST and WCR1-His were incubated with crude extracts from developing seeds of WT plants, the abundance of MT2b-GST markedly increased compared to samples without WCR1-His. Simultaneously, the degradation of MT2b-GST was inhibited by treatment with MG132, a 26S proteasome inhibitor (Figure 8, B and C). Finally, we analyzed the expression level of MT2b and MT contents in 7 DAF endosperm of ZH11 and WCR1 transgenic plants. The mRNA level of MT2b and the MT contents were significantly upregulated in OX(+) and downregulated in wcr1-1 compared to the control (Figure 8, D and E). Together, our findings suggest that WCR1 suppresses the 26S proteasome-mediated degradation of MT2b and improves the transcription of MT2b.
Figure 8.
WCR1 positively regulates MT2b. A, Transient expression assays. MT2b-LUC was constructed as a fusion protein and pMT2b:LUC contained a 1.5-kb promoter region of MT2b to drive the LUC gene. Relative LUC activity is expressed relative to rLUC (n = 4). Significant differences were based on two-tailed t tests, **P ≤ 0.01. B and C, In vitro degradation assay of MT2b. Purified MT2b-GST and/or WCR1-His proteins were added to total protein from WT developing seeds. MG132, proteasome inhibitor. Immunoblot analysis using anti-GST, anti-His, and anti-Actin (1:10,000). In (C), quantitative analysis was based on three replications using ImageJ. D, MT2b transcript levels in 7 DAF endosperm of ZH11 and the WCR1 transgenic lines. Endosperm samples were obtained from different plants (n = 4). E, MT contents in 7 DAF endosperm of ZH11 and the WCR1 transgenic lines. Each endosperm sample was obtained from 30 grains with five repeats. In (C, D, and E), significant differences were obtained by Duncan’s tests (P < 0.05). In all graphs, error bars, sem. F, Proposed model of the role of WCR1 in regulating chalkiness. WCR1A of tropical japonica was derived from WCR1G in wild rice via a mutation that arose during domestication. OsDOF17 binds to the cis-element AAAAG of the WCR1A promoter and increases its transcription level. WCR1 promotes the elimination of excess ROS, maintains redox homeostasis, and delays PCD in starchy endosperm by positively affecting the transcription of MT2b and suppressing the 26S proteasome-mediated degradation of MT2b, which ultimately results in the increased accumulation of storage components and a lower WCR value. Scale bars, 10 mm.
Discussion
Grain chalkiness affects the appearance and end-use quality of rice. However, to date, only a few quantitative trait loci (QTL) affecting chalkiness have been fine mapped or cloned (Zhu et al., 2018). In this study, we cloned the chalkiness gene WCR1, which negatively regulates white-core and is a positive factor in grain quality and yield. A functional A/G SNP variation of WCR1 located in the promoter region was associated with WCR1 expression level and WCR diversity among a panel of Asian cultivated rice accessions (Figure 4C). Notably, functional variations of many genes regulating grain quality and yield in rice are located in gene promoter regions, including Chalk5, as well as OsAAP6 and OsGluA2 for grain protein content and GS5 and GW7/GL7 for grain size (Li et al., 2011, 2014; Peng et al., 2014a; Wang et al., 2015; Yang et al., 2019). This indicates that CRISPR/Cas9 genome editing of the promoters of these genes could be used in breeding to produce predicted effects on quantitative variation in grain quality and size (Rodríguez et al., 2017). Superior quality and high yield are important breeding goals. A negative association between these objectives can be attributed to two causes based on previous studies. One is the tight linkage between genes for grain quality and yield. For example, the Chalk5 allele for high white-belly is closely linked with the GS5 and gw5 alleles, which increase grain width (Li et al. 2014). The other reason is the multiple effects of some single genes on grain quality and yield. The major alleles increasing grain size, such as gw2, GW8/OsSPL16, and GS2, produce larger grains and higher yields but are accompanied by increased chalkiness (Song et al., 2007; Wang et al., 2012; Hu et al., 2015). In this study, we identified the minor allele WCR1A, which positively influences grain quality, including appearance and processing quality as well as taste value (Figure 1, F and G; Table 1). Simultaneously, 1,000-grain weight and yield per plant were slightly enhanced by this allele (Figure 1, H and I; Supplemental Table S4). Perhaps, this is related to the influence of WCR1A on grain filling by prolonging the period of storage component accumulation and the maintenance of redox homeostasis. Therefore, the use of WCR1A likely represents a novel way to overcome the negative association between quality and yield for variety improvement, especially for indica varieties widely planted in Southern China such as J23B and 9311.
In light of global warming, high temperatures will increasingly influence grain chalkiness by affecting redox homeostasis (Liu et al., 2010; Yu et al., 2017; Nevame et al., 2018). H2O2 contents are significantly higher under heat stress (30°C) than under control (25°C) conditions during grain filling, and both ROS enrichment and removal are involved in the formation of grain chalkiness (Suriyasak et al., 2017). We found that under high temperatures (>30°C) in the field, both ROS and WCR were significantly higher in NIL-WCR1-J than in NIL-WCR1-BL, and WCR values were generally over 50% (Figures 1, F and 6, D–F). However, WCR values for both NIL-WCR1-BL and NIL-WCR1-J were ˂10% at field temperatures ˂25°C (Supplemental Figure S16). Therefore, we speculate that ROS accumulation at high temperatures can promote the formation of white-core. Geographic distribution and phylogenetic analyses revealed that the functionally acquired variant A in the promoter region of WCR1 arose in TrJ accessions, which are mainly distributed in tropical regions with higher temperatures (Figure 4, F and G). It is likely that the WCR1A allele improves the anti-oxidative ability of rice, thus enhancing adaptation to higher temperatures during long-term domestication and selection.
F-box proteins are core members of SCF complexes that participate in ubiquitin-mediated proteolysis (Abd-Hamid et al., 2020). F-box proteins are also involved in non-SCF pathways, such as protein interactions and transcription elongation. The F-box protein Elongin A interacts with Elongin B and Elongin C to form an Elongin (SIII) heterotrimer to promote the transcription elongation of RNA polymerase II in mammalian, yeast, and human cells (Aso et al., 1995; Kipreos and Pagano, 2000). However, many functions of F-box proteins in plants are still unknown. In this study, we found that WCR1 interacts with MT2b and promotes its stability (Figures 6, A–C and 8, A–C). Moreover, WCR1 positively affects the transcript level of MT2b (Figure 8, A and D). These results imply that WCR1 participates in non-SCF pathways, but the mechanism is still unknown. Perhaps, WCR1 is involved in the ubiquitin pathway or transcription elongation to affect protein stability and the transcription process. Additionally, phylogenetic analysis suggested that WCR1 shares high homology with AT1g67340 (Supplemental Figure S4), encoding a protein that interacts with ASK1, ASK2, ASK4, ASK11, and ASK13, as revealed by Y2H (Gagne et al., 2002). Therefore, WCR1 might play a similar function in combination with SKP1 proteins. In addition to affecting chalkiness formation and seed development, WCR1 may also be involved in biotic stress responses. MT2b is downregulated by OsRac1, thus potentiating the formation of ROS, which function as signals in the plant response to rice blast (Wong et al., 2004). In this study, WCR1 showed high expression levels in the flag leaves and influenced ROS content by regulating MT2b, suggesting that WCR1 might play a role in disease resistance.
In summary, we propose a model for a mechanism regulating chalkiness. In TrJ accessions, the base variant A in the WCR1 promoter was acquired from WCR1G from wild rice during domestication. OsDOF17 functions as a transcriptional activator that binds to the cis-element AAAAG in the WCR1A promoter and increases its transcription. In turn, WCR1 affects the transcription of MT2b and inhibits the 26S proteasome-mediated degradation of MT2b. These process promote the elimination of excess ROS, maintain redox homeostasis, and delay PCD in starchy endosperm, which results in the increased accumulation of storage components and a lower WCR value (Figure 8F). These finding shed light on the role of WCR1 in chalkiness and point to the application potential of WCR1A for breeding.
Materials and methods
Plant materials and phenotyping
Rice (O. sativa) populations were planted under natural field conditions at the experimental stations of Huazhong Agricultural University at Wuhan (N 30.49°, E 114.36°) and Lingshui (N 18.51°, E 110.04°), China. Twelve 30-day-old seedlings of each line were transplanted with 16.5-cm spacing in single row plots in the field; rows were 26.4-cm apart. Field management followed local practices. Ten plants from the middle of each row were harvested individually for trait measurement.
All grains and effective panicles of each plant were used to measure the number of filled grains, seed setting rate, panicle number, panicle length, and secondary branch number. The statistics of panicle length and secondary branch number were taken from the three longest panicles of each plant. Threshed seeds from each plant were air dried and stored at room temperature for 3 months before further phenotyping. Fully filled grains on each plant were used to measure grain size (length, width, and thickness), 1,000-grain weight, and yield per plant. Head rice yield was estimated as the percentage weight of intact milled grains to total grain weight. White-core phenotype was scored by the visual assessment of the percentage of white-core grains in random samples of more than 100 dehulled grains from each plant. Flour ground from milled grain was used to measure total starch content, amylose content (Bao et al., 2006), and total protein content (Kieldahl Nitrogen Analyzer). Glutelin, prolamin, globulin, and albumin contents in the flour were measured based on previously published methods (Kumamaru, 1988). The brown rice was ground into flour to analyze total lipid content by gas chromatography-mass spectrometry (GC–MS) (Wu et al., 2005). Taste scores for cooked rice were evaluated with a rice taste analyzer kit (SATAKE, STA1B-RHS1A-RFDM1A, Japan) that included taste, appearance, hardness, and viscosity analyzers.
Map-based cloning
BC5F2 populations consisting of 200 (Population 1) and 2,600 (Population 2) individuals were planted in winter 2014 in Hainan and summer 2015 in Wuhan, respectively. To fine map the WCR1 locus, we developed new molecular markers (SSR, InDel, and SNP markers) for genotyping in the region containing WCR1 according to the Nipponbare reference sequence. All recombinant plant genotypes identified by fine-mapping were confirmed by progeny testing.
RNA extraction and qRT-PCR
Total RNA was extracted from plant tissues using an RNA extraction kit (TRIzol, Invitrogen 15596-026, USA). First-strand cDNA was synthesized in 20 µL of reaction medium containing 2 µg of RNA and 200 U of M-MLV reverse transcriptase (Invitrogen C28025-014, USA). qRT-PCR was performed on a QuantStudio6 Flex machine using SYBR Green PCR reagent according to the manufacturer’s instructions. Young panicles and endosperm at different stages were obtained based on flowering time. For each endosperm sample, more than 30 grains were taken from the upper parts of panicles of each plant. The data for each sample were based on 3–5 biological replications from different plants, and three technical replications were performed for each biological replication. The rice ACTIN1 gene served as the internal control to normalize gene expression.
Construct preparation and transformation
To prepare the CO, a 3.5-kb promoter fragment of WCR1 from BL130 was fused with the 1.5-kb coding region from J23B. The WCR1 promoter and coding region were obtained by PCR and confirmed by sequencing. The two fragments were inserted into the plant binary vector pCAMBIA1301 using the one-step connection method. To prepare the OX construct, the WCR1 coding region of J23B was inserted into binary vector PU1301 under the control of the maize ubiquitin promoter. To prepare the GUS construct, a 2.8-kb promoter fragment of WCR1 from BL130 was inserted into vector DX2181 to drive GUS gene. The CRISPR/Cas9 KO construct was prepared by inserting a 20-bp target region from the third exon of WCR1 into the intermediate vector pER8-Cas9-U6 and cloning it into pCXUN-Cas9 as described in (Gao and Zhao, 2014). The constructs were introduced into Agrobacterium tumefaciens strain EHA105 and transferred into the relevant materials by Agrobacterium-mediated transformation (Toki, 1997).
Subcellular localization of WCR1 and OsDOF17 proteins
Subcellular localization in rice protoplasts: cDNAs obtained by reverse transcription were used to amplify the coding sequences of WCR1 and OsDOF17, which were then inserted into pM999-YFP and pM999-CFP to generate the WCR1-YFP and OsDOF17-CFP fusion plasmids, respectively. WCR1-YFP and the nucleus marker Ghd7-CFP (Xue et al., 2008), as well as WCR1-YFP and OsDOF17-CFP, were transiently expressed in rice protoplasts as described below.
Subcellular localization in N. benthamiana: the coding sequence of WCR1 was inserted into the GFP vector pCAMBIA1301S to generate WCR1-GFP (Zhang et al., 2011). WCR1-GFP and nucleus and cell membrane markers were expressed in N. benthamiana leaves (Voinnet et al., 2003).
Imaging of fluorescent proteins was conducted under a confocal laser scanning microscope (Leica TCS SP2). All fluorescence assays were repeated at least 3 times.
Scanning electron microscopy
Milled rice grains were transversely cut at the mid-section and coated with gold under a vacuum condition. SG morphology at the central and peripheral parts of the endosperm was examined under a scanning electron microscope (JSM-6390LV, JEOL) at an accelerating voltage of 10 kV and spot size of 30 nm. The analysis involved three biological replications of mounted specimens from different mature grains. All procedures were carried out according to the manufacturer’s protocol.
Haplotype assays and phylogenetic analysis
The genomic sequences of WCR1 in 492 cultivated and 99 wild rice accessions were acquired from RiceVarMap (http://ricevarmap.ncpgr.cn/) and OryzaGenome (http://viewer.shigen.info/oryzagenome/), respectively. Haplotypes of the cultivated accessions were constructed from SNPs and InDels (frequencies >0.05) in WCR1 from BL130 and J23B. Predicted amino acid sequences were aligned using MUSCLE, and a phylogenetic tree was constructed by the neighbor-joining method with 1,000 bootstrap replicates in MEGA version 7 software (Kumar et al., 2016). The alignment files are available in Supplemental File S1.
Y1H assays
Y1H assays were performed according to the manufacturer’s instructions (Clontech Mountain View, CA, USA). The expression data for OsDOF were obtained from http://ricevarmap.ncpgr.cn/v2/. The coding sequence of OsDOF and the 20-bp probe containing the AAAAG motif were cloned into pJG4-5 and pLacZi2μ (Clontech), respectively, and co-transformed into yeast stain EGY48. Interaction was examined on SD/-Trp/-Ura medium supplemented with 40% galactose, 40% raffinose, BU salts, and X-beta-Gal.
EMSA
The coding region of OsDOF17 was amplified and cloned into pGEX-6P plasmid for fusion with GST tag as OsDOF17-GST fusion protein. The fusion protein was expressed in Escherichia coli (BL21) and purified in vitro using a GST Fusion Protein Purification kit (Sangon Biotech, Shanghai, China). For EMSA, we synthesized two 55-bp DNA probes, Probe A and Probe G (mutant), based on the WCR1 promoter sequence of BL130 and labeled them at the 5′-ends with biotin. We used an unlabeled 55-bp probe A for competition assays. Gel-shift assays were performed according to the protocol supplied by the manufacturer (IGENEBOOK Biotech, Wuhan, China).
Transient expression assays in rice protoplasts
To verify the transcriptional activity of OsDOF17, we used the reporter plasmid 35SLUC containing the firefly LUC gene, driven by a fused promoter with a GAL4 binding element and 35S promoter. Full-length OsDOF17 was fused with GAL4-DNA binding domain (GAL4DBD) as the effector GDOF17.
To verify the effect of the A/G SNP on gene expression without/with OsDOF17, 20-bp sequences containing the SNP were amplified from BL130 (containing AAAAG) and Nipponbare (containing AGAAG), respectively, and the fragments were inserted into the reporter plasmid 190LUC. Full-length OsDOF17 was inserted into the None vector as the effector NDOF17.
To verify the effects of WCR1 on the protein stability and transcription level of MT2b, we introduced the coding sequence region and 1.5-kb promoter of MT2b into 190LUC plasmid to construct the reporters MT2b-LUC and pMT2b:LUC. At the same time, the effector WCR1 was constructed using the pM999 vector.
The 35S-driven Renilla LUC gene (rLUC) was used as an internal control. For each co-transfection assay, 2 μg of reporter plasmid, 2 μg of effector plasmid, and 500 ng of internal control plasmid were used. Different combinations of vectors were co-transfected into rice protoplasts by PEG-mediated transformation (Zhang et al., 2011). After incubation for 16–18 h in the dark at 28°C, relative LUC activity was measured using the Dual-LUC Reporter Assay System (Promega; Hao et al., 2010).
Y2H assays
A cDNA library was constructed from endosperm, and Y2H assays were performed according to the manufacturer’s instructions (Clontech). The full-length and truncated sequences of WCR1 were amplified and cloned into pGBKT7, whereas MT2b was amplified and cloned into pGADT7 (Clontech). The AH109 yeast stain was used in this experiment. Empty pGBKT7 and pGADT7 vectors were co-transformed in parallel as negative control. The OsMADS1 gene, which formed a homodimer, was linked to pGBKT7 (BD-OsMADS1) and pGADT7 (AD-OsMADS1), respectively, and these two vectors were co-transformed in parallel as a positive control. Interactions were examined on SD/–Trp/–Leu/–His/–Ade medium supplemented with X-alpha-Gal (Clontech).
Split-LUC assay
The coding regions of WCR1 and MT2b were amplified and cloned into pCAMBIA 1300-nLuc and pCAMBIA 1300-cLuc, respectively. The recombinant vectors were co-transformed into A. tumefaciens strain EHA105. After centrifugation, the bacteria were collected and resuspended in solution (10-mM MES, 10-mM MgCl2, and 200-μM acetosyringone) for infiltration (at optical density (OD)600 = 0.6). The prepared suspensions were injected into N. benthamiana leaves and grown for 3–4 days. The leaves were then sprayed with 5-mM luciferin and kept in the dark for 10 min to quench the fluorescence. A cooling CCD imaging apparatus (Tanon-5200) was used for image capture.
Pull-down assays
The coding regions of WCR1 and MT2b were amplified and cloned into pET28a and pGEX-6P for separate fusion with 6×His and GST tags as WCR1-His and MT2b-GST fusion proteins. Both fusion proteins were expressed in E. coli BL21 and purified in vitro using His-Tag Protein Purification and GST Fusion Protein Purification kits (Sangon Biotech, Shanghai, China). Pull-down assays were conducted as follows: 50 μL equilibrated Glutathione High Capacity Magnetic Agarose Beads (Sigma, St. Louis, MO, USA) was mixed with 1 mg of recombinant protein in 600 μL of pull-down buffer (20 mM Tris–HCl, pH 8.0, 1 mM EDTA, pH 8.0, 1% Nonidet P-40, 150-mM NaCl, Protease Inhibitor) at 4°C for 6 h. The bound proteins together with the beads were collected, washed 3 times with pull-down buffer, eluted twice with 50-μL GST elution buffer, and detected by anti-GST and anti-His antibodies (Mouse; Fine Biotech, Wuhan, China) (1:5,000 dilution), respectively.
Detection of ROS and cell death
Trypan blue was used to detect dead cells. Transverse sections of fresh caryopses at different times postflowering were treated with 0.1% trypan blue for 2 min, washed 5 times with 0.1 M phosphate buffer, and examined by light microscopy. FDA (Coolaber, Beijing, China) was usually to detect active cells. Transverse sections of fresh caryopses were treated with 2-μg/mL FDA for 15 min, washed 5 times with 0.1-M phosphate buffer, and examined by fluorescence microscopy (emission wavelength 520 nm).
The degradation of nuclear DNA was examined with DAPI and TUNEL. Transverse sections of fresh caryopses were fixed in 4% paraformaldehyde for 30 min and washed twice with PBS. The sections were treated with 20-μg/mL proteinase K for 20 min and washed 3 times with PBS. The positive control was additionally treated with 20 U/mL DNase I for 10 min and washed 3 times with PBS. Degraded nuclear DNA was detected using a TUNEL BrightGreen Apoptosis Detection Kit according to the manufacturer’s instructions (Vazyme Biotech Co., Ltd.). To test for the presence of nuclear DNA in developing starchy endosperm, the caryopsis sections were treated with 2-μg/mL DAPI in the dark for 30 min and washed 3 times with PBS. Degraded nuclear DNA was examined by fluorescence microscopy using emission wavelengths of 520 nm (green) for TUNEL and 460 nm (blue) for DAPI.
DCFH-DA (Sigma) was used to estimate the levels of ROS in cells. The sections were treated with 10-μM DCFH-DA for 30 min at 37°C and washed 5 times with 0.1-M phosphate buffer before microscopic investigation using excitation and emission wavelengths of 450–490 nm and 520 nm (green), respectively. Relative fluorescence intensity (IOD/area) of images at the same parameter settings was estimated by ImageJ. All images were photographed with a Nikon microscope system (NI-E).
DNA extraction and ladder detection
Twenty seed collected from the NILs at various stages throughout development were ground in liquid nitrogen. About 8 mL extraction buffer (100-mM Tris–HC1 pH 9.0, 20-mM EDTA, 200-mM NaC1, 1% [w/v] sarcosyl, and 10-μL/mL β-mercaptoethanol) was added to the samples, followed by 8 mL of phenol:chloroform (1:1, v/v). The samples were centrifuged at 8,000 g for 15 min, and the supernatant was transferred to a new tube. Total nucleic acids were precipitated from the supernatant by adding a 1/10 volume of 3 M sodium acetate, pH 6.0 and an equal volume of isopropanol. The precipitated nucleic acid was dissolved in 1-mL extraction buffer without vortexing, reprecipitated in 2-mL tubes, washed 3 times with 1 mL of 70% (v/v) ethanol, and dissolved in TE (10-mM Tris–HC1, pH 8.0, and 5-mM EDTA). RNase (DNase-free) was added to a final concentration of 100 μg/mL, and the sample was incubated for 1 h at 37°C. The samples were then extracted with an equal volume of phenol: chloroform (1:1, v/v) and centrifuged at 12,000 g, and DNA was re-precipitated from the supernatant and dissolved in TE. DNA concentrations were determined spectrophotometrically. The DNA was then resolved on a 1.8% agarose gel, stained with Goldview, and photographed on a UV light box.
BiFC assay
The coding sequence of WCR1 was amplified into pVYNER, and the coding sequence of MT2b was cloned into pVYCER plasmids. The recombinant vectors and nucleus marker Ghd7-CFP (Xue et al., 2008) were transiently expressed in rice protoplasts (Zhang et al., 2011). Imaging of fluorescent proteins was conducted under a confocal laser scanning microscope (Leica TCS SP2).
In vitro degradation assays
To test the effect of WCR1 on MT2b stability, total protein was extracted from WT seeds with extraction buffer (50-mM Tris–HCl pH 7.5, 100-mM NaCl, 100-mM MgCl2, 5-mM DTT, 5-mM ATP and protease inhibitor) and mixed with purified MT2b-GST protein (0.9 μg) in a total volume of 100 μL before adding recombinant WCR1-His protein and/or MG132 (100 μM). Following incubation for 2 h at 4°C, 30 μL samples were removed, heated at 95°C for 5 min in 4 × sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) sample buffer (10 μL), and subjected to 10% SDS–PAGE. Immunoblotting was performed using horseradish peroxidase-conjugated antibodies: anti-GST (Abbkine, Wuhan, China), anti-His (Abbkine), and anti-Actin (Sangon Biotech), 1:10,000 dilution.
Primers
The primers used in this study are listed in Supplemental Tables S5–S7.
Statistical analysis
Data are presented as the means ± standard deviation. Significant differences between two groups were analyzed by two-tailed t tests. For multiple comparisons, significant differences were obtained by Duncan’s tests (P < 0.05) using SPSS software. Quantitative analysis of gray values and fluorescence intensity in the images was performed using ImageJ software.
Accession numbers
Sequence data from this article can be found at RAPDB under the following accession numbers: WCR1 (LOC_Os01g69270), MT2b (LOC_Os05g02070), OsDOF2 (LOC_Os01g15900), OsDOF4 (LOC_Os01g17000), OsDOF8 (LOC_Os02g49440), OsDOF10 (LOC_Os02g15350), OsDOF11 (LOC_Os03g38870), OsDOF14 (LOC_Os03g16850), OsDOF15 (LOC_Os03g55610), OsDOF17 (LOC_Os04g58190), OsDOF18 (LOC_Os04g47990), OsDOF24 (LOC_Os08g38220), OsDOF25 (LOC_Os09g29960), OsDOF27 (LOC_Os10g35300), Chalk5 (LOC_Os05g06480), OsMC1 (LOC_Os03g27120), OsMC2 (LOC_Os03g27210), OsMC4 (LOC_Os05g41660), OsMC5 (LOC_Os05g41670), OsMC6 (LOC_Os01g58580), OsMC7 (LOC_Os11g04010), and OsMC8 (LOC_Os03g27190).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Plant architecture and grain chalkiness of BL130 and J23B.
Supplemental Figure S2. The candidate region overlapping with the promoter region of LOC_Os01g69270.
Supplemental Figure S3. Deduced amino acid sequences of the candidate WCR1 protein in BL130 and J23B.
Supplemental Figure S4. Phylogenetic tree of LOC_Os01g69270.
Supplemental Figure S5. Chalkiness and agronomic traits of the NILs.
Supplemental Figure S6. Vector construction for transgenic verification.
Supplemental Figure S7. Head rice yield in WCR1 transgenic lines.
Supplemental Figure S8. Subcellular localization of WCR1 in N. benthamiana leaves.
Supplemental Figure S9. Expression analysis of genes related to storage components in 14 DAF endosperm. Endosperm samples were obtained from different plants.
Supplemental Figure S10. Y1H assay. DOF binding site (DOFBS) represents a 20-bp probe containing the AAAAG motif from BL130.
Supplemental Figure S11. Self-activation experiment of OsDOF17 in yeast.
Supplemental Figure S12. BiFC assay of interactions between WCR1 and MT2b in rice protoplasts.
Supplemental Figure S13. Degradation of nuclear DNA does not occur at detectable levels in developing starchy rice endosperm.
Supplemental Figure S14. Construction of the mt2b mutant and WCR detection in the T1 generation.
Supplemental Figure S15. WCR and relative ROS levels in ZH11 and mutant grains from a segregating population.
Supplemental Figure S16. Grain chalkiness in NIL-WCR1-BL and NIL-WCR1-J samples produced at field temperatures ˂25°C.
Supplemental Table S1. Progeny tests of the six recombinants in interval W7–W8.
Supplemental Table S2. Relationship between phenotypes and genotypes in the T1 and T2 generations for the CO and OX lines.
Supplemental Table S3. Putative cis-regulatory elements identified in variant regions of the BL130 and J23B promoters.
Supplemental Table S4. WCR, grain size, and 1,000-grain weight of plants in the CO and 9,311 backgrounds.
Supplemental Table S5. Primers used for qRT-PCR of genes involved in PCD.
Supplemental Table S6. Primers used for qRT-PCR of 49 genes involved in the metabolism of storage components.
Supplemental Table S7. Primers used for map-based cloning and functional analysis of WCR1.
Supplemental Data Set 1. WCR1 genotypes and white-core phenotypes of 492 Asian cultivated rice accessions.
Supplemental File S1. Alignment used for phylogenetic analysis.
Funding
This work was supported by grants from the Ministry of Science and Technology (Grant 2021YFF1000200), Natural Science Foundation of China (U21A20211 and 91935303), the science and technology major program of Hubei Province (2021ABA011 and 2020BBB051), and earmarked fund for the China Agriculture Research System (CARS-01-03).
Conflict of interest statement. The authors declare no conflict of interest.
Supplementary Material
Contributor Information
Bian Wu, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Peng Yun, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Hao Zhou, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Duo Xia, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Yuan Gu, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Pingbo Li, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Jialing Yao, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Zhuqing Zhou, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Jianxian Chen, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Rongjia Liu, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Shiyuan Cheng, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Hao Zhang, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Yuanyuan Zheng, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Guangming Lou, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Pingli Chen, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Shanshan Wan, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Mingsong Zhou, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Yanhua Li, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Guanjun Gao, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Qinglu Zhang, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Xianghua Li, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Xingming Lian, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
Yuqing He, National Key Laboratory of Crop Genetic Improvement and Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei 430070, China.
B.W. conducted most of the experiments, including fine mapping, gene cloning, genetic transformation, expression analysis, electron microscopy, staining assays, protein expression assays, and other functional analyses. P.Y. contributed to fine mapping. H.Z. and D.X. conducted some statistical analyses. Y.G. contributed to protein expression assays. J.Y. and Z.Z provided yeast and staining reagent. P.L., J.C., Y.Z., R.L., S.C., H.Z., G.L., P.C., S.W., M.Z., and Y.L. contributed to phenotypic analyses. G.G., Q.Z., and X.L. participated in field management and logistics. X.L. provided germplasm. Y.H. designed and supervised the study. Y.H. and B.W. analyzed the data and wrote the paper.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Yuqing He (yqhe@mail.hzau.edu.cn).
References
- Abd-Hamid NA, Ahmad-Fauzi MI, Zainal Z, Ismail I (2020) Diverse and dynamic roles of F-box proteins in plant biology. Planta 251: 68. [DOI] [PubMed] [Google Scholar]
- Aso T, Lane WS, Conaway JW, Conaway RC (1995) Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science 269: 1439–1443 [DOI] [PubMed] [Google Scholar]
- Bao J, Shen S, Sun M, Corke H (2006) Analysis of genotypic diversity in the starch physicochemical properties of nonwaxy rice: apparent amylose content, pasting viscosity and gel texture. Starch Stärke 58: 259–267 [Google Scholar]
- Chen Y, Xu Y, Luo W, Li W, Chen N, Zhang D, Chong K (2013) The F-box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice. Plant Physiol 163: 1673–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang J, Xie P, Zhou W, Chen J, Wei C (2012) Programmed cell death in wheat starchy endosperm during kernel development. Afr J Agric Res 7: 6533–6540 [Google Scholar]
- Cobbett C, Goldsbrough P (2002) Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu Rev Plant Biol 53: 159–182 [DOI] [PubMed] [Google Scholar]
- Domínguez F, Cejudo FJ (2014) Programmed cell death (PCD): an essential process of cereal seed development and germination. Front Plant Sci 5: 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath A, Bethke PC, Jones RL (2001) Enzymes that scavenge reactive oxygen species are down-regulated prior to gibberellic acid-induced programmed cell death in barley aleurone. Plant Physiol 126: 156–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald MA, McCouch SR, Hall RD (2009) Not just a grain of rice: the quest for quality. Trends Plant Sci 14: 133–139 [DOI] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Kondo T, Saito K, Utsumi Y, Tokunaga T, Nishi A, Satoh H, Park JH, Jane JL (2007) Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol 144: 2009–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Liu C, Li Y, Zhang A, Dong G, Xie L, Zhang B, Ruan B, Hong K, Xue D (2016) QTL analysis for chalkiness of rice and fine mapping of a candidate gene for qACE9. Rice 9: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhao Y (2014) Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol 56: 343–349 [DOI] [PubMed] [Google Scholar]
- Gong J, Miao J, Zhao Y, Zhao Q, Feng Q, Zhan Q, Cheng B, Xia J, Huang X, Yang S (2017) Dissecting the genetic basis of grain shape and chalkiness traits in hybrid rice using multiple collaborative populations. Mol Plant 10: 1353–1356 [DOI] [PubMed] [Google Scholar]
- Gunawardena AH, Pearce DM, Jackson MB, Hawes CR, Evans DE (2001) Characterisation of programmed cell death during aerenchyma formation induced by ethylene or hypoxia in roots of maize (Zea mays L.). Planta 212: 205–214 [DOI] [PubMed] [Google Scholar]
- Guo T, Liu X, Wan X, Weng J, Liu S, Liu X, Chen M, Li J, Su N, Wu F (2011) Identification of a stable quantitative trait locus for percentage grains with white chalkiness in rice (Oryza sativa). J Integr Plant Biol 53: 598–607 [DOI] [PubMed] [Google Scholar]
- Hao YJ, Song QX, Chen HW, Zou HF, Wei W, Kang XS, Ma B, Zhang WK, Zhang JS, Chen SY (2010) Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 232: 1033–1043 [DOI] [PubMed] [Google Scholar]
- He P, Li S, Qian Q, Ma Y, Li J, Wang W, Chen Y, Zhu L (1999) Genetic analysis of rice grain quality. Theor Appl Genet 98: 502–508 [Google Scholar]
- He Y, Wang C, Higgins JD, Yu J, Zong J, Lu P, Zhang D, Liang W (2016) MEIOTIC F-BOX is essential for male meiotic DNA double-strand break repair in rice. Plant Cell 28: 1879–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Wang Y, Fang Y, Zeng L, Xu J, Yu H, Shi Z, Pan J, Zhang D, Kang S (2015) A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant 8: 1455–1465 [DOI] [PubMed] [Google Scholar]
- Hussain S, Slikker W Jr, Ali SF (1996) Role of metallothionein and other antioxidants in scavenging superoxide radicals and their possible role in neuroprotection. Neurochem Int 29: 145–152 [DOI] [PubMed] [Google Scholar]
- Jagadish S, Murty M, Quick W (2015) Rice responses to rising temperatures–challenges, perspectives and future directions. Plant Cell Environ 38: 1686–1698 [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP (2007) F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143: 1467–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko K, Sasaki M, Kuribayashi N, Suzuki H, Sasuga Y, Shiraya T, Inomata T, Itoh K, Baslam M, Mitsui T (2016) Proteomic and glycomic characterization of rice chalky grains produced under moderate and high-temperature conditions in field system. Rice 9: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HG, Park S, Matsuoka M, An G (2005) White‐core endosperm floury endosperm‐4 in rice is generated by knockout mutations in the C4‐type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J 42: 901–911 [DOI] [PubMed] [Google Scholar]
- Kipreos E, Pagano M (2000) The F-box protein family. Genome Biol 1: REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Ikeda T M, Nagata K (2013) Spatial and temporal progress of programmed cell death in the developing starchy endosperm of rice. Planta 237: 1393–1400 [DOI] [PubMed] [Google Scholar]
- Kumamaru T (1988) Mutants for rice storage proteins 1. Screening of mutants semidwarfism-related proteins and glutelin seed protein in rice (Oryza sativa L.). Theor Appl Genet 83: 153–158 [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Lan S, Xu Z (2004) Studies on the programmed cell death in rice during starchy endosperm development. Agr Sci China 3: 663–670 [Google Scholar]
- Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L, Shao D, Xu C, Li X, Xiao J (2011) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet 43: 1266–1269 [DOI] [PubMed] [Google Scholar]
- Li Y, Fan C, Xing Y, Yun P, Luo L, Yan B, Peng B, Xie W, Wang G, Li X (2014) Chalk5 encodes a vacuolar H+-translocating pyrophosphatase influencing grain chalkiness in rice. Nat Genet 46: 398. [DOI] [PubMed] [Google Scholar]
- Lisle A, Martin M, Fitzgerald M (2000) Chalky and translucent rice grains differ in starch composition and structure and cooking properties. Cereal Chem 77: 627–632 [Google Scholar]
- Liu X, Guo T, Wan X, Wang H, Zhu M, Li A, Su N, Shen Y, Mao B, Zhai H (2010) Transcriptome analysis of grain-filling caryopses reveals involvement of multiple regulatory pathways in chalky grain formation in rice. BMC Genomics 11: 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevame AYM, Emon RM, Malek MA, Hasan MM, Alam MA, Muharam FM, Aslani F, Rafii MY, Ismail MR (2018) Relationship between high temperature and formation of chalkiness and their effects on quality of rice. Biomed Res Int 2018: 1653721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B, Kong H, Li Y, Wang L, Zhong M, Sun L, Gao G, Zhang Q, Luo L, Wang G (2014a) OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice. Nat Commun 5: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B, Wang L, Fan C, Jiang G, Luo L, Li Y, He Y (2014b) Comparative mapping of chalkiness components in rice grain using five populations across two environments. BMC Genet 15: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov V, Hille J, Mueller-Roeber B, Gechev TS (2015) ROS-mediated abiotic stress-induced programmed cell death in plants. Front Plant Sci 6: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino BF, Noh YS, Himelblau E, Amasino RM (2000) Molecular aspects of leaf senescence. Trends Plant Sci 5: 278–282 [DOI] [PubMed] [Google Scholar]
- Rodríguez LD, Lemmon ZH, Man J, Bartlett ME, Lippman ZB (2017) Engineering quantitative trait variation for crop improvement by genome editing. Cell 171: 470–480. e478 [DOI] [PubMed] [Google Scholar]
- Ryoo N, Yu C, Park C, Baik M, Park I M, Cho M, Bhoo SH, An G, Hahn T, Jeon JS (2007) Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa L.). Plant Cell Rep 26: 1083–1095 [DOI] [PubMed] [Google Scholar]
- Schindler T, Bergfeld R, Schopfer P (1995) Arabinogalactan proteins in maize coleoptiles: developmental relationship to cell death during xylem differentiation but not to extension growth. Plant J 7: 25–36 [DOI] [PubMed] [Google Scholar]
- Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP (2000) Insights into SCF ubiquitin ligases from the structure of the Skp1–Skp2 complex. Nature 408: 381–386 [DOI] [PubMed] [Google Scholar]
- She KC, Kusano H, Koizumi K, Yamakawa H, Hakata M, Imamura T, Fukuda M, Naito N, Tsurumaki Y, Yaeshima M (2010) A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell 22: 3280–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenmorgen TJ, Grigg BC, Lanning SB. (2013) Impacts of preharvest factors during kernel development on rice quality and functionality. Annu Rev Food Sci Technol 4: 101–115 [DOI] [PubMed] [Google Scholar]
- Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39: 623–630 [DOI] [PubMed] [Google Scholar]
- Steffens B, Geske T, Sauter M (2011) Aerenchyma formation in the rice stem and its promotion by H2O2. New Phytol 190: 369–378 [DOI] [PubMed] [Google Scholar]
- Steffens B, Kovalev A, Gorb S N, Sauter M (2012) Emerging roots alter epidermal cell fate through mechanical and reactive oxygen species signaling. Plant Cell 24: 3296–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suriyasak C, Harano K, Tanamachi K, Matsuo K, Tamada A, Mari II, Ishibashi Y (2017) Reactive oxygen species induced by heat stress during grain filling of rice (Oryza sativa L.) are involved in occurrence of grain chalkiness. J Plant Physiol 216: 52–57 [DOI] [PubMed] [Google Scholar]
- Tang S, Zhang H, Liu W, Dou Z, Zhou Q, Chen W, Wang S, Ding Y (2019) Nitrogen fertilizer at heading stage effectively compensates for the deterioration of rice quality by affecting the starch-related properties under elevated temperatures. Food Chem 277: 455–462 [DOI] [PubMed] [Google Scholar]
- Teng X, Zhong M, Zhu X, Wang C, Ren Y, Wang Y, Zhang H, Jiang L, Wang D, Hao Y (2019) FLOURY ENDOSPERM16 encoding a NAD-dependent cytosolic malate dehydrogenase plays an important role in starch synthesis and seed development in rice. Plant Biotechnol J 17: 1914–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley PJ, Vašák M (1985) Possible role for metallothionein in protection against radiation-induced oxidative stress. Kinetics and mechanism of its reaction with superoxide and hydroxyl radicals. Bba-protein Struct M 827: 36–44 [DOI] [PubMed] [Google Scholar]
- Tian Z, Qian Q, Liu Q, Yan M, Liu X, Yan C, Liu G, Gao Z, Tang S, Zeng D, et al. (2009) Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc Natl Acad Sci USA 106: 21760–21765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S (1997) Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol Biol Rep 15: 16–21 [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wang J, Wan X, Li H, Pfeiffer W H, Crouch J, Wan J (2007) Application of identified QTL-marker associations in rice quality improvement through a design-breeding approach. Theor Appl Genet 115: 87–100 [DOI] [PubMed] [Google Scholar]
- Wang S, Li S, Liu Q, Wu K, Zhang J, Wang S, Wang Y, Chen X, Zhang Y, Gao C (2015) The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat Genet 47: 949–954 [DOI] [PubMed] [Google Scholar]
- Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44: 950. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Lam E (2009) Bax inhibitor-1, a conserved cell death suppressor, is a key molecular switch downstream from a variety of biotic and abiotic stress signals in plants. Int J Mol Sci 10: 3149–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing R A, Purugganan M D, Zhang Q (2018) The rice genome revolution: from an ancient grain to Green Super Rice. Nat Rev Genet 19: 505–517 [DOI] [PubMed] [Google Scholar]
- Wong H L, Sakamoto T, Kawasaki T, Umemura K, Shimamoto K (2004) Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol 135: 1447–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Truksa M, Datla N, Vrinten P, Bauer J, Zank T, Cirpus P, Heinz E, Qiu X (2005) Stepwise engineering to produce high yields of very long-chain polyunsaturated fatty acids in plants. Nat Biotechnol 23: 1013–1017 [DOI] [PubMed] [Google Scholar]
- Wu M, Ren Y, Cai M, Wang Y, Zhu S, Zhu J, Hao Y, Teng X, Zhu X, Jing R (2019) Rice FLOURY ENDOSPERM 10 encodes a pentatricopeptide repeat protein that is essential for the trans‐splicing of mitochondrial nad1 intron 1 and endosperm development. New Phytol 223: 736–750 [DOI] [PubMed] [Google Scholar]
- Xu S B, Yu H T, Yan L F, Wang T (2010) Integrated proteomic and cytological study of rice endosperms at the storage phase. J Proteome Res 9: 4906–4918 [DOI] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40: 761. [DOI] [PubMed] [Google Scholar]
- Yang Y, Guo M, Sun S, Zou Y, Yin S, Liu Y, Tang S, Gu M, Yang Z, Yan C (2019) Natural variation of OsGluA2 is involved in grain protein content regulation in rice. Nat Commun 10: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, Moon S, Lee YS, Zhu L, Liang W, Zhang D, Jung KH, An G (2016) Defective tapetum cell death 1 (DTC1) regulates ROS levels by binding to metallothionein during tapetum degeneration. Plant Physiol 170: 1611–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TE, Gallie DR (1999) Analysis of programmed cell death in wheat endosperm reveals differences in endosperm development between cereals. Plant Mol Biol 39: 915–926 [DOI] [PubMed] [Google Scholar]
- Young TE, Gallie DR (2000) Programmed cell death during endosperm development. In Fukuda H, Greenberg J, eds, Programmed Cell Death in Higher Plants–Lam E, Kluwer Academic Publishers, Netherlands, pp 283–301 [Google Scholar]
- Young TE, Gallie DR, DeMason DA (1997) Ethylene-mediated programmed cell death during maize endosperm development of wild-type and shrunken2 Genotypes. Plant Physiol 115: 737–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Liu Y, Lu L, Zhang Q, Chen Y, Zhou L, Chen H, Peng C (2017) Ascorbic acid deficiency leads to increased grain chalkiness in transgenic rice for suppressed of L-GalLDH. Plant Physiol 211: 13–26 [DOI] [PubMed] [Google Scholar]
- Yun P, Zhu Y, Wu B, Gao G, Sun P, Zhang Q, He Y (2016) Genetic mapping and confirmation of quantitative trait loci for grain chalkiness in rice. Mol Breeding 36: 162 [Google Scholar]
- Zhang L, Zhao L, Lin L, Zhao L, Liu Q, Wei C (2018a) A novel mutation of OsPPDKB, encoding pyruvate orthophosphate dikinase, affects metabolism and structure of starch in the rice endosperm. Int J Mol Sci 19: 2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhan J, Yadegari R (2018b) Maize opaque mutants are no longer so opaque. Plant Reprod 31: 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Li J, Ma L, Wang H, Zhou H, Ni E, Jiang D, Liu Z, Zhuang C (2019) OsAGO2 controls ROS production and the initiation of tapetal PCD by epigenetically regulating OsHXK1 expression in rice anthers. Proc Natl Acad Sci USA 116: 7549–7558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Chen L, Jiang L, Zhang W, Liu L, Liu X, Zhao Z, Liu S, Zhang L, Wang J (2009) Fine mapping of the grain chalkiness QTL qPGWC-7 in rice (Oryza sativa L.). Theor Appl Genet 118: 581–590 [DOI] [PubMed] [Google Scholar]
- Zhu A, Zhang Y, Zhang Z, Wang B, Xue P, Cao Y, Chen Y, Li Z, Liu Q, Cheng S (2018) Genetic dissection of qPCG1 for a quantitative trait locus for percentage of chalky grain in rice (Oryza sativa L.). Front Plant Sci 9: 1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.