Abstract
Both plants and animals utilize nucleotide-binding leucine-rich repeat immune receptors (NLRs) to perceive the presence of pathogen-derived molecules and induce immune responses. NLR genes are far more abundant and diverse in vascular plants than in animals. Truncated NLRs, which lack one or more of the canonical domains, are also commonly encoded in plant genomes. However, little is known about their functions, especially the N-terminally truncated ones. Here, we show that the Arabidopsis thaliana N-terminally truncated helper NLR (hNLR) gene N REQUIREMENT GENE1 (NRG1C) is highly induced upon pathogen infection and in autoimmune mutants. The immune response and cell death conferred by some Toll/interleukin-1 receptor-type NLRs (TNLs) were compromised in Arabidopsis NRG1C overexpression lines. Detailed genetic analysis revealed that NRG1C antagonizes the immunity mediated by its full-length neighbors NRG1A and NRG1B. Biochemical tests suggested that NRG1C might interfere with the EDS1–SAG101 complex, which functions in immunity signaling together with NRG1A/1B. Interestingly, Brassicaceae NRG1Cs are functionally exchangeable and that the Nicotiana benthamiana N-terminally truncated hNLR NRG2 also antagonizes NRG1 activity. Together, our study uncovers an unexpected negative role of N-terminally truncated hNLRs in immunity in different plant species.
N-terminally truncated helper nucleotide-binding leucine-rich repeat immune receptors (hNLRs) play unexpected negative roles in plant immunity, antagonizing their full-length counterparts.
Introduction
Plants utilize a sophisticated innate immune system to defend against a plethora of pathogens. They depend on immune receptors to recognize nonself signals and initiate defense responses. Plasma membrane-localized pattern-recognition receptors (PRRs) perceive pathogen-associated molecular patterns (PAMPs), leading to PAMP/pattern-triggered immunity (Couto and Zipfel, 2016). On the other hand, intracellular nucleotide-binding (NB) leucine-rich repeat (LRR) immune receptors (NLRs) are responsible for warding off adapted pathogens by detecting their secreted effectors, thereby activating effector-triggered immunity (Cui et al., 2015).
NLRs are present in both animals and plants. They are members of the signal transduction ATPases with numerous domains family, consisting of a variable N-terminal domain, a central NB motif, and a C-terminal LRR region (Jones and Dangl, 2006). Typical plant sensor NLRs (sNLRs) have either a Toll/interleukin-1 receptor (TIR) or a coiled-coil (CC) domain at their N-termini and are termed TNLs (TIR-NB-LRR) or CNLs (CC-NB-LRR), respectively (Jones et al., 2016). NLRs have evolved to recognize effectors directly or indirectly (Baggs et al., 2017). In contrast, a small subclass of NLRs is genetically required for the immune signaling of diverse sNLRs. These NLRs are termed helper NLRs (hNLRs) (Peart et al., 2005; Bonardi et al., 2011; Collier et al., 2011; Jubic et al., 2019). There are two hNLR gene families in Arabidopsis thaliana, encoding RNLs (Resistance to Powdery Mildew 8 [RPW8] CC-NB-LRR): the ACTIVATED DISEASE RESISTANCE 1 (ADR1) family (Bonardi et al., 2011) and the N REQUIREMENT GENE 1 (NRG1) family (Peart et al., 2005). Besides RNLs, the Solanaceae-specific CNL NRC (NB-LRR protein required for hypersensitive response (HR)-associated cell death) proteins also function as hNLRs, since both cell surface PRRs (Wu et al., 2016; Rathjen and Dodds, 2017) and many sNLRs signal through NRCs (Gabriëls et al., 2006, 2007; Wu et al., 2017a). Compared to the highly divergent sensor TNLs and CNLs, hNLRs are evolutionarily more conserved (Shao et al., 2016; Zhong and Cheng, 2016; Wu et al., 2017a; Adachi et al., 2019; Andolfo et al., 2019).
Plant sNLRs are among the most rapidly evolving genes (Guo et al., 2011; Gao et al., 2018; Van de Weyer et al., 2019). As a result, they are highly polymorphic. NLR allelic diversity mainly depends on the generation of either chimeric LRRs by intra- and intergenic recombination and gene conversion (Serra et al., 2018), or novel LRRs through point mutations (Michelmore and Meyers, 1998; Mago et al., 2015; Lu et al., 2016; Tamborski and Krasileva, 2020). Gene clustering is another feature commonly observed for plant NLRs (Van de Weyer et al., 2019). Two types of cluster patterns exist based on the diversity of NLRs within the clusters (Jacob et al., 2013). Homogenous clusters contain the same type of NLRs derived from tandem duplications. By contrast, heterogeneous clusters contain diverse NLRs generated by ectopic or large-scale segmental duplications (Jacob et al., 2013; Marone et al., 2013; Krasileva, 2019). Pan-genome analysis revealed that NLR cluster patterns vary among A. thaliana accessions. Some clusters have highly conserved copy numbers, while others show huge variations (Lee and Chae, 2020). An increasing number of examples suggest that clustered NLR genes often are co-expressed and may cooperate to function. Clustering sNLRs together has the potential to massively expand pathogen recognition by increasing their oligomerization options (van Wersch and Li, 2019).
Duplication and uneven crossover events may generate NLRs lacking one or more of the canonical domains, yielding truncated NLRs (Wicker et al., 2007; Baggs et al., 2017), which are commonly found in plant genomes (Bai et al., 2002; Meyers et al., 2002; 2003; Yang et al., 2008; Jacob et al., 2013; Van de Weyer et al., 2019; Lee and Chae, 2020). Interesting, most truncated NLRs are expressed (Meyers et al., 2002; Tan et al., 2007; Nandety et al., 2013), suggesting their potential role in plant immunity. Several C-terminally truncated NLRs have been studied. For instance, A. thaliana TIR-only protein RESPONSE TO HopBA1 (RBA1) is specifically required for defense responses mediated by the bacterial effector HopBA1 (Nishimura et al., 2017). Another well-studied example involves the A. thaliana TN (TIR-NB) proteins CHILLING SENSITIVE1 (CHS1) and TN2. CHS1 and TN2 form distinct pairs with full-length TNL SOC3 (SUPPRESSOR of chs1-2, 3) to monitor the homeostasis of the E3 ligase SENESCENCE-ASSOCIATED E3 UBIQUITIN LIGASE 1 (SAUL1) (Tong et al., 2017; Liang et al., 2019). However, little is known about the functions of N-terminally truncated NLRs, including NL (NB-LRR) and LRR proteins.
Although hNLRs may not have effector recognition capabilities, the NRG1-type hNLRs are organized in clusters in various plant species (Andolfo et al., 2019; Jubic et al., 2019; van Wersch and Li, 2019; Wu et al., 2019). In contrast, ADR1s are scattered in the genome (Bonardi et al., 2011; Dong et al., 2016). There are three NRG1 paralogs in the A. thaliana genome: full-length NRG1A and NRG1B, and N-terminally truncated NRG1C (Jubic et al., 2019; Wu et al., 2019). NRG1A and NRG1B are present in the NRG1 cluster across almost all analyzed A. thaliana and A. lyrate accessions (Lee and Chae, 2020). NRC1C encodes a predicted protein missing the RPW8 CC domain and most of the NB domain. Similarly, Nicotiana benthamiana contains a copy of the full-length NRG1 and an N-terminally truncated NRG2. In comparison to the clustered NRG1s in A. thaliana, NRG2 does not cluster with NRG1 and has been assumed to be a pseudogene (Peart et al., 2005). It is unclear whether these N-terminally truncated hNLRs have any biological roles.
Here, we describe the functional analysis of truncated hNLRs. To our surprise, NRG1C negatively regulated autoimmunity mediated by the A. thaliana TNL autoimmune mutant chilling sensitive 3, 2D (chs3-2D), playing an opposite role from its full-length paralogs NRG1A/1B. Plants overexpressing NRG1C phenocopied the nrg1a nrg1b double mutant and the SENESCENCE-ASSOCIATED GENE 101 (sag101) single mutant. Biochemical analysis suggested that NRG1C might function by interfering with the ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1)–SAG101 complex. Interestingly, NRG1Cs from different Brassicaceae species are functionally exchangeable. In addition, the N. benthamiana truncated NRG2 also antagonizes NRG1-mediated cell death and bacterial growth restriction. In summary, our study uncovers an unexpected role of N-terminally truncated hNLRs, which likely evolved to balance the activities of their corresponding full-length hNLRs.
Results
NRG1C is evolutionarily conserved in Brassicaceae and its expression is highly induced in TNL-type autoimmune mutants and upon pathogen infection
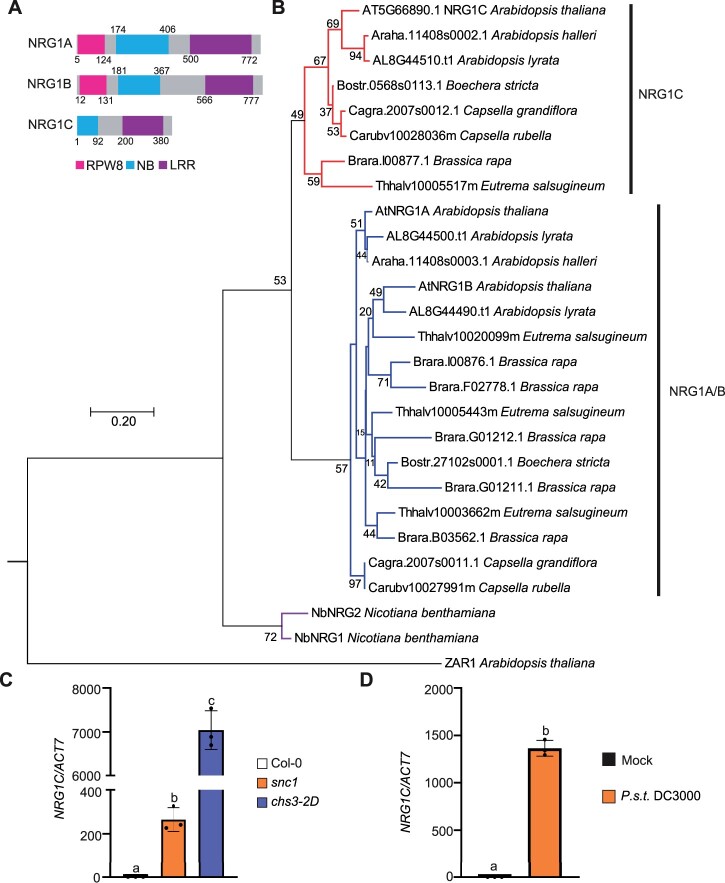
BLASTp analysis revealed that NRG1C-type N-terminally truncated hNLRs are present in all eight sequenced Brassicaceae species and N. benthamiana (Figure 1, A and B, Supplemental Figure S1 and Supplemental Table S1). Additionally, NRG1C can be found in all examined A. thaliana accessions, arguing against it being a pseudogene. However, no NRG1C homolog was found in available protein databases of other land plants. Interestingly, like A. thaliana NRG1C, other Brassicaceae NRG1C homologs tend to cluster with the full-length NRG1 genes in their genomes (Supplemental Table S1). Phylogenetic analysis indicated that all Brassicaceae NRG1Cs came from the same ancestor (Figure 1B). The Brassicaceae NRG1C genes likely originated from either an initial gene duplication of the NRG1A/1B ancestor and a subsequent N-terminal loss, or partial gene duplication. In contrast, NbNRG2 and NbNRG1 belong to a distinct clade from Brassicaceae NRG1, which is suggestive of a different evolutionary pathway between N. benthamiana NRG1s and Brassicaceae NRG1s (Figure 1B; Supplemental Figure S1). In Brassica rapa, there are two close NRG1C homologs found in two gene clusters. Brara.I00877.1 and Brara.G01211.1 share 75.4% and 62.0% amino acid identity, or 86.2% and 75.5% similarity with A. thaliana NRG1C, respectively (Supplemental Figure S1 and Supplemental Table S1). However, phylogenetic analysis indicated that Brara.G01211.1 might not be an NRG1C homolog, as it contains a full NB domain (Supplemental Figure S1) and is distinct from the NRG1C clade (Figure 1B).
Figure 1.
NRG1C is evolutionarily conserved in Brassicaceae, and its transcription is induced in TNL-type autoimmune mutants and upon pathogen infection. A, Protein domain diagrams of NRG1A, NRG1B, and NRG1C. Numbers represent amino acid positions relative to the translation start sites. NRG1C lacks the RPW8 CC and part of the NB domains. B, Phylogenetic tree of NRG1A/1B/1C homologs. Putative NRG1A/1B/1C homologs were obtained from Phytozome (Brassicaceae species) and Sol Genomics Network (N. benthamiana) using A. thaliana NRG1A and NRG1C protein sequences as respective queries. ZAR1 was used as an outgroup to root the tree. MUSCLE was used for sequence alignment, and the Neighbor-joining tree was generated using full-length NRG1C and homologous parts of all other corresponding NRG1 homologs with the JTT model and using 2000 bootstrap replicates in MEGA version 7.0. Numbers at the nodes indicate the bootstrap values as a percentage based on neighbor-joining analysis. C, NRG1C gene expression in 4-week-old soil-grown autoimmune mutants snc1 and chs3-2D, as determined by qRT-PCR (Col-0 served as the WT control, and the transcript level in Col-0 was arbitrarily set at 1.0). Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± standard deviation (sd) (n = 3). Three independent experiments were carried out with similar results. D, qRT-PCR analysis of the expression of NRG1C upon P.s.t. DC3000 infection. Four-week-old soil-grown WT plants were inoculated with 10 mM MgCl2 (Mock) or P.s.t. DC3000 (OD600 = 0.2). Samples were collected at 12 hpi. Values were normalized to the level of ACTIN7, and the transcript level in Mock treatment was set at 1.0. Error bars represent means ± sd (n = 3). Three independent experiments were carried out with similar results.
DA1-RELATED PROTEIN 5 (DAR5, AT5G66630) was proposed to be a chimeric protein resulting from a translocation of NRG1C and a fusion with a member of the nearby cluster carrying DAR homologs, DAR7 (AT5G66610) to DAR3 (AT5G66640) (Meyers et al., 2003). Coincidently, DAR5 carries RPW8 CC, NB, and LIM (Lin11, Isl-1, and Mec-3) domains, but no LRRs. DAR5 shares high similarity with NRG1s in A. thaliana (Andolfo et al., 2019; Castel et al., 2019). However, DAR5 seems to be unique in A. thaliana, as BLAST searches failed to identify putative DAR5 homologs in the genomes of all other sequenced plant species.
To test whether the truncated NRG1C and DAR5 are involved in TNL-mediated immunity, we measured their expression levels in TNL autoimmune mutants. suppressor of npr1-1, constitutive 1 (snc1), and chs3-2D contain gain-of-function mutations in the TNLs SNC1 and CHS3, respectively, resulting in TNL-mediated autoimmunity (Li et al., 2001; Bi et al., 2011). NRG1C transcripts were ∼200- and 7,000-fold upregulated in snc1 and chs3-2D, respectively, compared to Col-0 wild-type (WT) (Figure 1C). However, no significant changes in DAR5 transcript levels were observed (Supplemental Figure S2A). Interestingly, both NRG1C and DAR5 were upregulated in A. thaliana upon infection by the virulent bacterial pathogen Pseudomonas syringae pv. tomato (P.s.t.) DC3000, as revealed in an RNA-sequencing dataset (Howard et al., 2013). However, our qRT-PCR (quantitative Reverse Transcription-PCR) analysis indicated that the transcription of NRG1C, but not DAR5, was highly induced by P.s.t. DC3000 (Figure 1D; Supplemental Figure S2B). These expression analyses suggest that NRG1C in the NRG1 gene cluster may play roles in biotic responses against pathogens.
NRG1C, but not DAR5, negatively regulates chs3-2D- and snc1-mediated autoimmunity
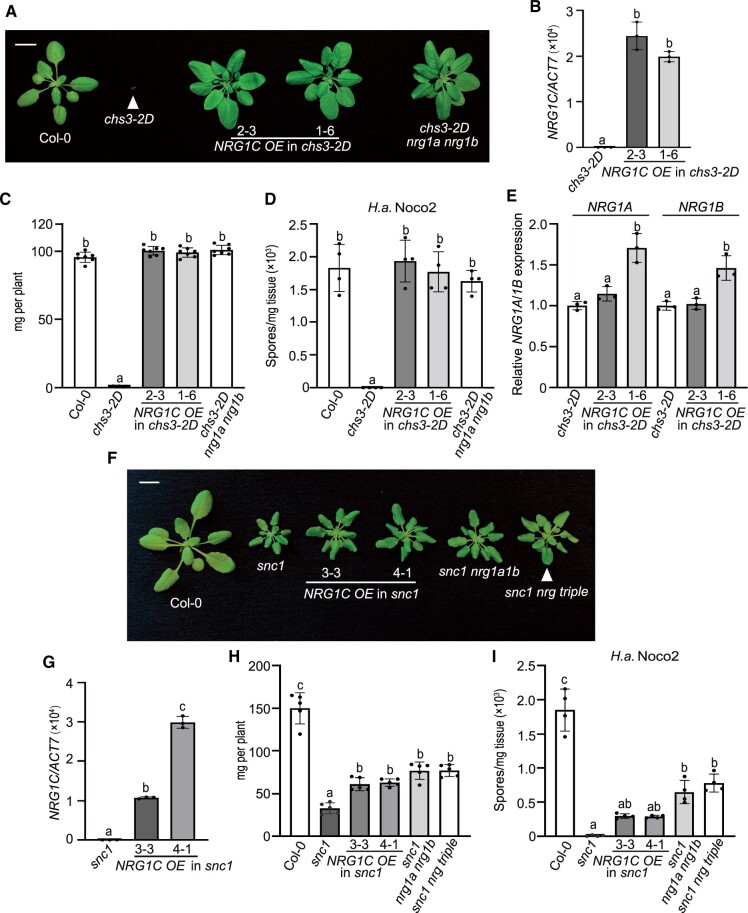
We first used an overexpression approach to examine the role of these truncated NLRs. NRG1C and DAR5 were individually overexpressed in the chs3-2D and snc1 backgrounds. To our surprise, overexpression of NRG1C fully suppressed chs3-2D-mediated dwarfism (Figure 2, A–C) and enhanced disease resistance to the oomycete pathogen Hyaloperonospora arabidopsidis (H.a.) Noco2 (Figure 2D), with no reduction in NRG1A/1B transcript levels (Figure 2E). In contrast, no suppression of chs3-2D-mediated growth defects was observed upon DAR5 overexpression (Supplemental Figure S2, C and D). Furthermore, overexpressing NRG1C partially suppressed snc1-mediated dwarfism to a level similar to that of the nrg1a/1b and nrg1a/1b/1c deletion mutants (Wu et al., 2019) (Figure 2, F–H). Consistently, the enhanced disease resistance to H.a. Noco2 (Figure 2I) in snc1 was partially suppressed upon NRG1C overexpression. However, the snc1 DAR5 OE lines did not show significant differences in growth compared with snc1 (Supplemental Figure S2, E and F). Taken together, these results indicate that NRG1C negatively regulates chs3-2D and snc1-mediated autoimmunity to different degrees. Intriguingly, these NRG1C overexpression phenotypes closely resemble those of the nrg1a nrg1b double mutant and the sag101 single mutant (Xu et al., 2015; Wu et al., 2019).
Figure 2.
NRG1C negatively regulates chs3-2D and snc1-mediated autoimmunity. A, Morphology of 4-week-old soil-grown plants of Col-0, chs3-2D, two independent transgenic lines of NRG1C OE in the chs3-2D background and chs3-2D nrg1a nrg1b. Bar = 1 cm. B, NRG1C gene expression in the indicated 2-week-old plate-grown plants, as determined by qRT-PCR (chs3-2D served as a control whose NRG1C transcript level was set at 1.0). Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 3). Two independent experiments were carried out with similar results. C, Fresh weights of the plants in (A). Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 7). D, Quantification of H.a. Noco2 sporulation in the indicated genotypes 7 days post-inoculation (dpi) with 105 spores per ml water. Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 4). Three independent experiments were carried out with similar results. E, NRG1A/1B gene expression in the indicated 4-week-old soil-grown plants, as determined by qRT-PCR (chs3-2D served as a control whose NRG1A/1B transcript level was set at 1.0). Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 3). Three independent experiments were carried out with similar results. F, Morphology of 4-week-old soil-grown plants of Col-0, snc1, two independent transgenic lines of NRG1C OE in the snc1 background, snc1 nrg1a nrg1b and snc1 nrg triple. Bar = 1 cm. G, NRG1C gene expression in the indicated 2-week-old plate-grown plants, as determined by qRT-PCR (snc1 served as a control whose NRG1C transcript level was set at 1.0). Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 3). Two independent experiments were carried out with similar results. H, Fresh weights of the plants in (F). Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 5). I, Quantification of H.a. Noco2 sporulation in the indicated genotypes at 7 dpi with 105 spores per ml water. Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 4). Three independent experiments were carried out with similar results.
ADR1-L3 is dispensable for snc1-mediated autoimmunity and RPP4-mediated immunity
Similar to NRG1C in the NRG1 family, the A. thaliana ADR1 hNLR family includes a truncated ADR1-L3, missing the N-terminal RPW8 domain (Supplemental Figure S3). In contrast to the broad presence of NRG1C in Brassicaceae, an ADR1-L3 homolog is only found in Drummond’s rockcress (Boechera stricta; Supplemental Figure S3). To test whether ADR1-L3 is involved in immunity conferred by ADR1-dependent sNLRs, we overexpressed ADR1-L3 in the snc1 background. However, snc1-mediated growth defects (Supplemental Figure S4A) and enhanced disease resistance (Supplemental Figure S4C) were not altered upon ADR1-L3 overexpression (Supplemental Figure S4B). Furthermore, ADR1-L3 overexpression lines in the WT background were not susceptible to the oomycete pathogen H.a. Emwa1 (Supplemental Figure S4, D and E), which is recognized by the ADR1-dependent TNL RECOGNITION OF PERONOSPORA PARASITICA 4 (RPP4) (Bonardi et al., 2011; Dong et al., 2016). Taken together, unlike NRG1C, ADR1-L3 does not seem to play any roles in TNL-mediated autoimmunity and defense.
Overexpression of NRG1C has no effect on chs1-2- and chs2-1-mediated autoimmunity, RPS2 and RPS4-mediated immunity, or basal defense
To further test the specificity of NRG1C, we overexpressed NRG1C in additional autoimmune mutants. In chilling sensitive 1, 2 (chs1-2), a missense mutation in a truncated TN protein (Wang et al., 2013) results in constitutive cell death and defense responses at low temperatures (Zbierzak et al., 2013). The chilling sensitive mutant chs2-1 contains a gain-of-function mutation in the TNL RPP4 (Huang et al., 2010). However, neither chs1-2 nor chs2-1-mediated temperature-dependent autoimmunity was suppressed upon NRG1C overexpression (Supplemental Figure S5, A–D). Consistently, the oomycete pathogen H.a. Emwa1 failed to grow in NRG1C OE lines in the WT background (Supplemental Figure S5, E–G).
We also challenged two independent NRG1C OE lines (in the WT Col-0 background) with P.s.t. DC3000 expressing either AvrRps4 or AvrRpt2 effectors, which are recognized by TNL RESISTANT TO P. SYRINGAE 4 (RPS4) or CNL RPS2, respectively. However, no altered disease resistance against these avirulent pathogens was detected (Supplemental Figure S6, A and B). When challenged with P.s.t. DC3000, the NRG1C OE lines exhibited similar bacterial growth as WT, suggesting that overexpression of NRG1C does not compromise the basal defense response either (Supplemental Figure S6C). Therefore, NRG1C seems to function specifically in TNL-mediated immune responses, with different strengths for distinct TNLs. The differential contributions of NRG1C to TNL signaling are reminiscent of the phenotypes of nrg1a nrg1b or sag101 knockout mutants, which show major effects in chs3-2D but relatively mild suppression in snc1 and no effects in chs1-2, chs2-1, RPS4, or RPS2-mediated autoimmunity or bacterial growth restriction (Feys et al., 2005; Xu et al., 2015; Castel et al., 2019; Wu et al., 2019).
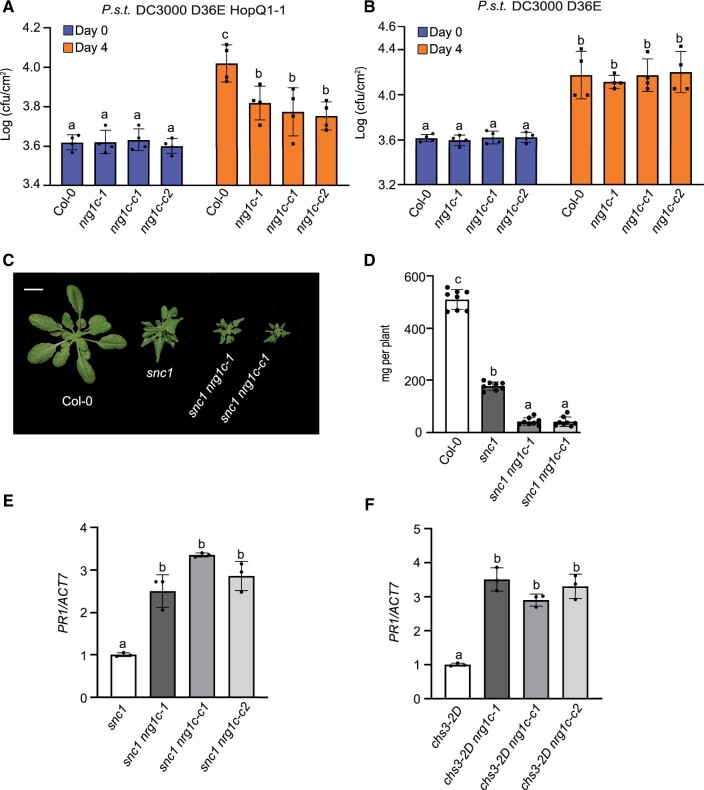
NRG1C is required for HopQ1-1-triggered defense
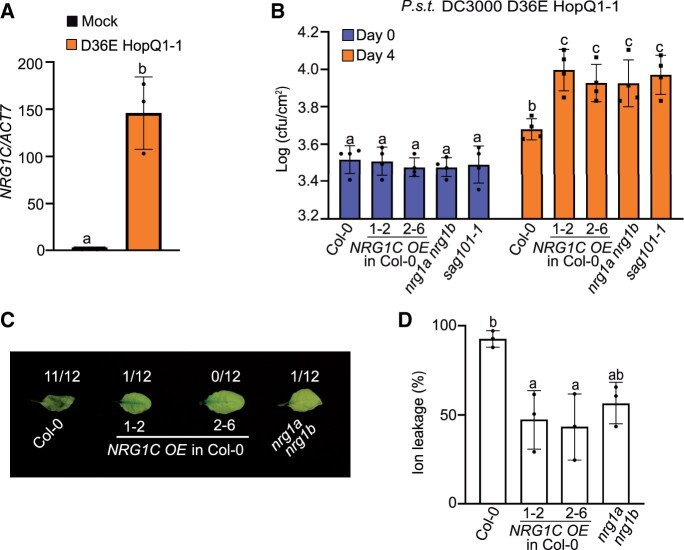
NbNRG1 is essential for immune signaling triggered by the effector HopQ1-1 (Qi et al., 2018). To test whether A. thaliana NRG1s play roles in effector HopQ1-1-triggered defense responses, we performed bacterial infection experiments using the nrg1a nrg1b double mutant and NRG1C OE lines. As P.s.t. DC3000 carries diverse effectors that can mask the effects of HopQ1-1, P.s.t. DC3000 D36E, in which all 36 T3SS (Type III secretion system) effector genes and coronatine biosynthesis genes were deleted, was used as a background to avoid interference from other effectors (Wei et al., 2015). As shown in Figure 3A, WT plants inoculated with P.s.t. DC3000 D36E expressing HopQ1-1 showed induced NRG1C expression, suggesting a potential role for NRG1C in HopQ1-1-triggered defense responses. Additionally, less P.s.t. DC3000 D36E HopQ1-1 bacterial growth was detected in WT plants compared to P.s.t. DC3000 D36E (Supplemental Figure S6D), indicating that there is likely a yet-to-be identified A. thaliana R protein that can respond to the effector HopQ1-1. Indeed, NRG1C OE lines exhibited enhanced disease susceptibility to P.s.t. DC3000 D36E HopQ1-1 to a level similar to that of the nrg1a nrg1b double mutant and sag101 single mutant (Figure 3B). In contrast, no bacterial growth difference was observed among the NRG1C OE lines, nrg1a nrg1b, or sag101-1 compared with WT when challenged with P.s.t. DC3000 D36E (Supplemental Figure S6E). Taken together, these data indicate that NRG1C antagonizes HopQ1-1-triggered NRG1-dependent defense responses in A. thaliana. The phenotypes of the NRG1C overexpression plants seem identical to those of the nrg1a nrg1b double mutant and the sag101 single mutant.
Figure 3.
NRG1C antagonizes HopQ1-1-mediated bacterial growth and AvrRPS4-mediated HR. A, qRT-PCR analysis of the expression of NRG1C upon P.s.t. DC3000 D36E HopQ1-1 infection. 4-week-old soil-grown Col-0 plants were inoculated with 10 mM MgCl2 (Mock) or P.s.t. DC3000 D36E HopQ1-1 (OD600 = 0.2). Samples were collected at 12 hpi. Values were normalized to the level of ACTIN7, and the transcript level in Mock treatment was set at 1.0. Error bars represent means ± sd (n = 3). Three independent experiments were carried out with similar results. B, Growth of P.s.t. DC3000 D36E HopQ1-1 in 4-week-old leaves of the indicated genotypes at 0 and 4 dpi with bacterial inoculum of OD600 = 0.001. Statistical significance is indicated by different letters (P < 0.05). Error bars represent means ± sd (n = 4). Three independent experiments were carried out with similar results. C, HR in leaves infiltrated with Pf0-1 AvrRPS4. 4-week-old A. thaliana leaves of the indicated genotypes were infiltrated with Pf0-1 expressing AvrRPS4 at OD600 = 0.2. Photographs were taken at 36 hpi. The numbers indicate the number of plants displaying HR out of the total number of plants tested. Three independent experiments were carried out with similar results. D, Ion leakage of A. thaliana leaves upon Pf0-1_AvrRPS4 infiltration under the same conditions as for C. Twelve infiltrated leaves were collected as one biological replicate. Individual dots indicate different biological replicates. Error bars represent means ± sd (n = 3). Statistical significance is indicated by different letters (P < 0.01). Three independent experiments were carried out with similar results.
NRG1C antagonizes the AvrRPS4-mediated HR
AvrRPS4 can activate RESISTANT TO RALSTONIA SOLANACEARUM 1 (RRS1)/RPS4 pairs, resulting in HR in the Col-0 background (Saucet et al., 2015). This HR was compromised in the Col-0 nrg1a nrg1b double mutant, although no difference in P.s.t. AvrRPS4 bacterial growth was observed (Supplemental Figure S6A) (Castel et al., 2019; Lapin et al., 2019). NRG1C was significantly upregulated at 6 and 12 h post-inoculation (hpi) with P.s.t DC3000 AvrRPS4, but not at 1 hpi (Howard et al., 2013), suggesting that NRG1C plays roles in AvrRPS4-triggered immunity. We thus tested whether NRG1C regulates the RRS1/RPS4-mediated HR. The effector AvrRPS4 was delivered using the Pseudomonas fluorescens strain Pf0-1 containing the type III secretion system (Pf0-1 EtHAn), but lacking type III effector genes (Thomas et al., 2009). Similar with the phenotype of the nrg1a nrg1b double mutant (Castel et al., 2019; Lapin et al., 2019), RRS1/RPS4-mediated HR was lost in NRG1C OE lines (Figure 3, C and D). The striking overall phenotypic resemblance between NRG1C OE lines and the nrg1a nrg1b double mutant suggests that NRG1C may function antagonistically with NRG1A/1B to regulate defense responses.
NRG1C antagonizes the NRG1–SAG101 module, in parallel with the ADR1-PAD4 module
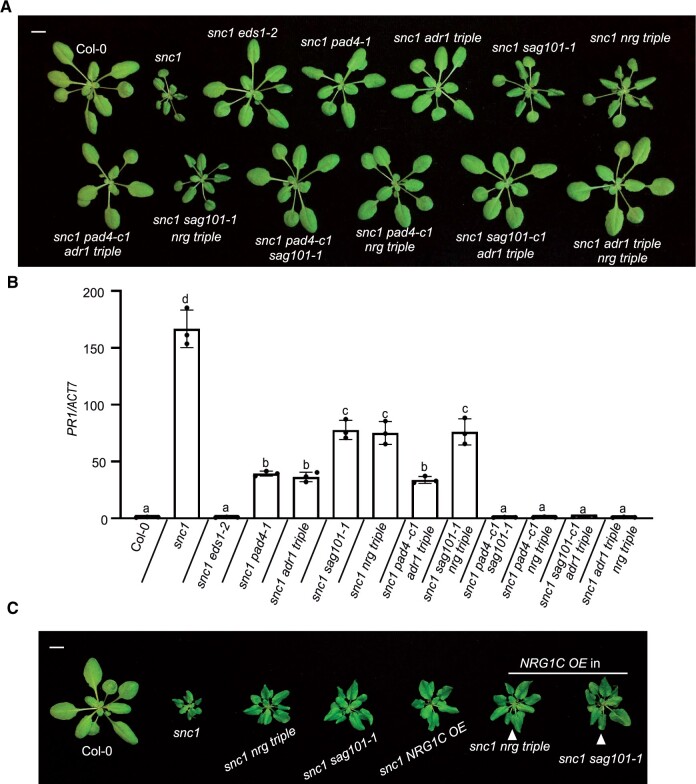
The similar phenotypes of adr1 triple and phytoalexin deficient 4 (pad4) mutants (Dong et al., 2016), and the additive effects between SAG101 and PAD4 (Rietz et al., 2011) or NRG1s and ADR1s (Lapin et al., 2019; Wu et al., 2019; Saile et al., 2020), allude to an EDS1–ADR1–PAD4 signaling module that works in parallel with the EDS1–NRG1–SAG101 module to regulate downstream events in TNL immunity. The existence of the EDS1–PAD4–ADR1 and EDS1–SAG101–NRG1 complexes corroborates the two independent genetic modules (Sun et al., 2021; Wu et al., 2021). To examine the contribution of NRG1C in these modules, we generated higher-order mutants combining loss-of-function mutations from different complexes in the snc1 background. The dwarfism and increased PATHOGENESIS-RELATED 1 (PR1) gene expression of snc1 were partially suppressed by introducing mutations in SAG101 (Feys et al., 2005) or the NRG1 cluster (Wu et al., 2019) (Figure 4, A and B). However, the snc1 sag101-1 nrg triple mutants resembled the snc1 sag101-1 and snc1 nrg triple mutants in terms of morphology (Figure 4A) and PR1 gene expression levels (Figure 4B), confirming the notion that SAG101 functions in the same pathway as NRG1A/1B. By contrast, the increased PR1 gene expression of snc1 was largely, but not fully, suppressed by adr1 triple or pad4-1 (Figure 4B), although the sizes of snc1 adr1 triple and snc1 pad4-1 plants were WT like (Cheng et al., 2011; Dong et al., 2016) (Figure 4A). However, combining adr1 triple and pad4 mutations together did not further reduce PR1 gene expression (Figure 4B; Supplemental Figure S7), confirming the notion that ADR1s and PAD4 work in the same pathway. In contrast, mutants containing mutations in two components from different modules, including snc1 pad4-c1 sag101-1, snc1 pad4-c1 nrg triple, snc1 sag101-c1 adr1 triple, and snc1 adr1 triple nrg triple, exhibited fully restored WT-like levels of PR1 expression (Figure 4, A and B; Supplemental Figure S7). Taken together, these epistasis analyses data further confirm the notion that two distinct modules are present in TNL pathways that act in parallel: one includes PAD4–ADR1s and the other includes SAG101–NRG1A/1B.
Figure 4.
NRG1C antagonizes the EDS1–NRG1–SAG101 module, which functions in parallel with the EDS1–ADR1–PAD4 module. A, Morphology of 4-week-old soil-grown plants of Col-0, snc1, snc1 eds1-2, snc1 pad4-1, snc1 adr1 triple, snc1 sag101-1, snc1 nrg triple, snc1 pad4-c1 adr1 triple, snc1 sag101-1 nrg triple, snc1 pad4-c1 sag101-1, snc1 pad4-c1 nrg triple, snc1 sag101-c1 adr1 triple and snc1 adr1 triple nrg triple. pad4-c1, and sag101-c1 alleles were generated using CRISPR–Cas9-mediated genome editing. Bar = 1 cm. B, PR1 gene expression in the indicated 4-week-old soil-grown plants, as determined by qRT-PCR (Col-0 served as a control whose PR1 transcript level was set at 1.0). Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 3). Three independent experiments were carried out with similar results. C, Morphology of 4-week-old soil-grown plants of Col-0, snc1, snc1 nrg triple, snc1 sag101-1, snc1 NRG1C OE, snc1 nrg triple NRG1C OE, and snc1 sag101-1 NRG1C OE. Bar = 1 cm.
To examine the relationship between NRG1C and the two distinct modules, we overexpressed NRG1C in the snc1 nrg triple, snc1 sag101-1, snc1 pad4-1, or snc1 adr1 triple backgrounds. As shown in Figure 4C, the dwarfism of snc1 nrg triple and snc1 sag101-1 was not further suppressed by NRG1C overexpression. In agreement with this finding, snc1 sag101-1 NRG1C OE and snc1 nrg triple NRG1C OE resembled the snc1 sag101-1 and snc1 nrg triple mutants in terms of PR1 gene expression (Supplemental Figure S8A). Specifically, the increased PR1 gene expression in snc1 was reduced by either nrg triple or sag101, but was not further reduced by NRG1C overexpression (Supplemental Figure S8A). In contrast, the partial suppression of upregulated PR1 gene expression in snc1 by adr1 triple or pad4-1 was further suppressed to WT levels when NRG1C was overexpressed in the snc1 adr1 triple or snc1 pad4-1 background (Supplemental Figure S8B). These comprehensive epistasis analyses support the notion that NRG1C indeed specifically antagonizes the SAG101-NRG1A/1B module.
Loss of NRG1C enhances resistance mediated by some TNLs
As plants overexpressing NRG1C exhibited defects in TNL-mediated defense responses, we further examined the phenotypes of nrg1c knockout mutants. A T-DNA insertion mutant (nrg1c-1, Sail_1265_F08) of NRG1C (Wu et al., 2019) and two independent deletion alleles generated by CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas9 (CRISPR-associated protein 9)-mediated genome editing (Supplemental Figure S7) were used to analyze the knockout phenotypes of nrg1c. Morphologically, these mutant alleles were completely WT-like. When they were challenged with P.s.t. DC3000 D36E HopQ1-1 (Figure 5A), all three nrg1c mutants exhibited enhanced resistance to P.s.t. DC3000 D36E HopQ1-1. However, no altered growth of P.s.t. DC3000 D36E was detected (Figure 5B), which is suggestive of a specific negative role of NRG1C in HopQ1-1-triggered defense responses. Interestingly, under a short-day regime (8-h light/16-h dark), the dwarfism of snc1 was further enhanced when NRG1C was knocked out (Figure 5, C and D). This enhancement was not observed previously when these plants were grown under long-day conditions (16-h light/8-h dark) (Wu et al., 2019). Consistently, increased PR1 gene expression was observed in the snc1 nrg1c double mutants (Figure 5E). In parallel, PR1 gene expression was also upregulated in the chs3-2D nrg1c double mutants compared to their parent chs3-2D (Figure 5F). These data indicate that the loss of NRG1C enhances the defense responses of these two autoimmune mutants. In conclusion, NRG1C plays a negative role in immune regulation and its loss leads to enhanced immunity in autoimmune backgrounds and against P.s.t. DC3000 D36E HopQ1-1.
Figure 5.
Loss of NRG1C enhances TNL-mediated defense responses. A and B, Growth of P.s.t. DC3000 D36E HopQ1-1 (A) or P.s.t. DC3000 D36E (B) in 4-week-old leaves of the indicated genotypes at 0 and 4 dpi with bacterial inoculum of OD600 = 0.001. Statistical significance is indicated by different letters (P < 0.05). Error bars represent means ± sd (n = 4). Three independent experiments were carried out with similar results. C, Morphology of 6-week-old soil-grown plants of Col-0, snc1, snc1 nrg1c-1, and snc1 nrg1c-c1 under short-day conditions (8-h light/16-h dark). Bar = 1 cm. D, Fresh weights of the plants in (C). Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 8). E and F, PR1 gene expression in the indicated 2-week-old plate-grown plants, as determined by qRT-PCR (snc1 (E) and chs3-2D (F) served as controls whose PR1 transcript level was set at 1.0). Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 3). Three independent experiments were carried out with similar results.
NRG1C does not affect NRG1A/1B by transcriptional interference
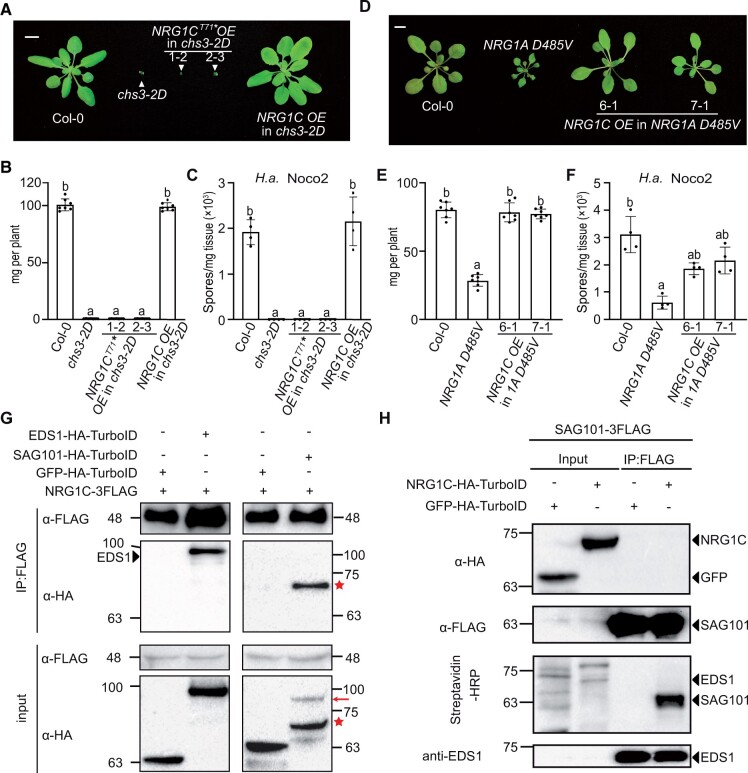
As NRG1C encodes a truncated hNLR that antagonizes the SAG101–NRG1A/1B module, we reasoned that it might interfere with NRG1A/1B transcription via RNA-mediated interference due to their sequence similarity. To test this, we overexpressed NRG1C with a premature stop codon at position 213 of the predicted NRG1C mRNA sequence in the chs3-2D background. Such a mutation would yield a non-functional protein (NRG1CT71*) but a normal transcript. No alterations in chs3-2D-mediated dwarfism (Figure 6, A and B) or enhanced disease resistance to H.a. Noco2 (Figure 6C) were observed upon overexpression of NRG1CT71* (Supplemental Figure S9A). Additionally, no reduction in NRG1A/1B transcript levels was detected upon NRG1C overexpression in either the chs3-2D or WT background (Supplemental Figure S9, B; Figure 2, E). Taken together, these data suggest that NRG1C does not affect mRNA levels of the full-length helper NRG1A/1B. Thus, if NRG1C interferes with NRG1A/1B, the effects are likely at the protein level.
Figure 6.
NRG1C negatively regulates NRG1A/1B-dependent defense responses via its association with the EDS1–SAG101 complex, but not by transcriptional interference of NRG1A/1B. A, Morphology of 4-week-old soil-grown plants of Col-0, chs3-2D, two independent transgenic lines of NRG1CT71* OE in the chs3-2D background and NRG1C OE in the chs3-2D background. NRG1CT71* carries an early stop codon to disrupt protein synthesis, but not transcription. Bar = 1 cm. B and E, Fresh weights of the plants in (A) or (D). Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 7). C and F, Quantification of H.a. Noco2 sporulation in the indicated genotypes at 7 dpi with 105 spores per milliliter water. Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 4). Three independent experiments were carried out with similar results. D, Morphology of 3-week-old soil-grown plants of Col-0, NRG1A D485V, and two independent transgenic lines of NRG1C OE in the NRG1A D485V background. NRG1A D485V was tagged with three FLAG epitopes in frame at its C-terminus. Bar = 1 cm. G, Co-IP of NRG1C-3FLAG with EDS1-HA-TurboID or SAG101-HA-TurboID in N. benthamiana. GFP-HA-TurboID served as a negative control. IP was carried out with anti-FLAG beads. The 3FLAG-tagged proteins were detected using an anti-FLAG antibody. The HA-tagged proteins were detected using an anti-HA antibody. The red arrow denotes SAG101-HA-TurboID, while the stars indicate unknown degradation products from SAG101-HA-TurboID. Three or two independent experiments were carried out with similar results to test the association between NRG1C and EDS1 or SAG101, respectively. H, Co-IP and biotinylation of SAG101-3FLAG with NRG1C-HA-TurboID in WT A. thaliana plants stably transformed with the two transgenes. IP was carried out with anti-FLAG beads. The 3FLAG-tagged proteins were detected using an anti-FLAG antibody. The HA-TurboID-tagged proteins were detected using an anti-HA antibody. EDS1 was detected using an anti-EDS1 antibody (Feys et al., 2001). The biotinylated proteins were detected using Streptavidin-HRP. Molecular mass markers in kiloDaltons are indicated on the left. Three independent experiments were carried out with similar results.
NRG1C antagonizes NRG1A D485V-mediated autoimmunity in A. thaliana and NRG1A D485V-mediated HR in N. benthamiana
To further examine the hypothesis that NRG1C directly interferes with NRG1A/1B, we overexpressed NRG1C in transgenic plants expressing NRG1A D485V, an auto-active version of NRG1A that constitutively activates immunity (Wu et al., 2019). The dwarfism and enhanced resistance of NRG1A D485V were largely suppressed upon NRG1C overexpression (Figure 6, D–F and Supplemental Figure S9, C), while no reduction in NRG1A transcript or protein levels was observed (Supplemental Figure S9, D and E). These results further support the notion that the negative regulation of NRG1A by NRG1C does not function through transcriptional or post-transcriptional repression. In contrast, overexpression of NRG1C did not alter the autoimmunity of ADR1-L2 D484V, an auto-active variant of ADR1-L2 (Roberts et al., 2013) (Supplemental Figure S10, A–D).
Consistent with our observation in A. thaliana, NRG1A D485V-triggered HR in N. benthamiana was alleviated when NRG1C was expressed in these leaves (Supplemental Figure S10, E and F), although the same amount of NRG1A D485V protein accumulated (Supplemental Figure S10G). Taken together, these data indicate that NRG1A-mediated autoimmunity and HR are negatively regulated by NRG1C and that the effects of NRG1C seem to be specific to NRG1-type hNLR-mediated defense responses.
NRG1C does not associate with full-length NRG1A
Since NRG1C does not affect the mRNA or protein levels of full-length NRG1A (Supplemental Figure S9), another hypothesis is that NRG1C interferes with NRG1A/1B through protein-protein interactions. We combined TurboID-based proximity labeling and co-immunoprecipitation (IP) to allow detection of transient protein–protein interactions with low-abundance proteins (Zhang et al., 2019; Wu et al., 2020). As shown in Supplemental Figure S11A, expressing NRG1A-HA-TurboID partially restored the morphological suppression of chs3-2D nrg1a nrg1b, suggesting that the fusion protein is functional. However, when NRG1A-HA-TurboID was co-expressed with NRG1C-3FLAG, NRG1C-3FLAG failed to pull down or be biotinylated by NRG1A-HA-TurboID (Supplemental Figure S11B). This negative interaction result suggests that the NRG1C-mediated negative regulation of NRG1A-mediated defense responses does not function via direct protein interference. However, we cannot exclude the possibility that the in planta interactions between the two proteins could not be detected due to complex conformation constraints.
NRG1C associates with the EDS1–SAG101 complex
As plants overexpressing NRG1C also shared similar phenotypes with the sag101 knockout mutants, and full-length NRG1A can form a complex with EDS1–SAG101 dimers (Sun et al., 2021), it is also possible that NRG1C attenuates NRG1A-mediated defense by interfering with SAG101 or EDS1. In support of this hypothesis, a near 73-fold more enrichment of NRG1C peptides was detected in A. thaliana SAG101-YFP IP-MS (mass spectrometry) analysis compared to full-length NRG1A (Sun et al., 2021). To further verify this observed NRG1C–SAG101 association, we co-infiltrated N. benthamiana leaves with Agrobacterium expressing NRG1C-3FLAG with EDS1-HA-TurboID, SAG101-HA-TurboID, or GFP-HA-TurboID. As shown in Figure 6G, NRG1C-3FLAG was able to pull-down EDS1-HA-TurboID and a degradation product of SAG101-HA-TurboID, but not the negative control GFP-HA-TurboID, although neither EDS1-HA-TurboID nor SAG101-HA-TurboID biotinylated NRG1C-3FLAG (Supplemental Figure S12, A and B). Therefore, NRG1C seems to work together with the EDS1–SAG101 dimers in a complex. The lack of biotinylation observed here is likely due to spatial constraints of the complex.
We further examined such interaction with a reciprocal assay using A. thaliana stable transgenic lines carrying both NRG1C-HA-TurboID and SAG101-3FLAG. Overexpression of NRG1C-HA-TurboID fully suppressed chs3-2D-mediated dwarfism (Supplemental Figure S11C), indicating that the fusion protein was functional. As shown in Figure 6H, although NRG1C-HA-TurboID was not observed in the SAG101-3FLAG pull-down, likely due to its low protein abundance or instability, NRG1C-HA-TurboID successfully biotinylated SAG101-3FLAG, but not EDS1, even though a large amount of EDS1 protein was pulled down. This suggests that SAG101 was in close proximity with the C-terminus of NRG1C, where TurboID was fused. The lack of EDS1 biotinylation by NRG1C could be due to constraints in space in the complex. However, neither EDS1 (Supplemental Figure S13, A and B) nor SAG101 (Supplemental Figure S13, C and D) protein levels were altered by NRG1C overexpression, confirming the notion that NRG1C does not affect the protein abundance of the EDS1–SAG101–NRG1A/1B complex.
Overexpression of the B. rapa NRG1C homolog suppresses chs3-2D-mediated defense
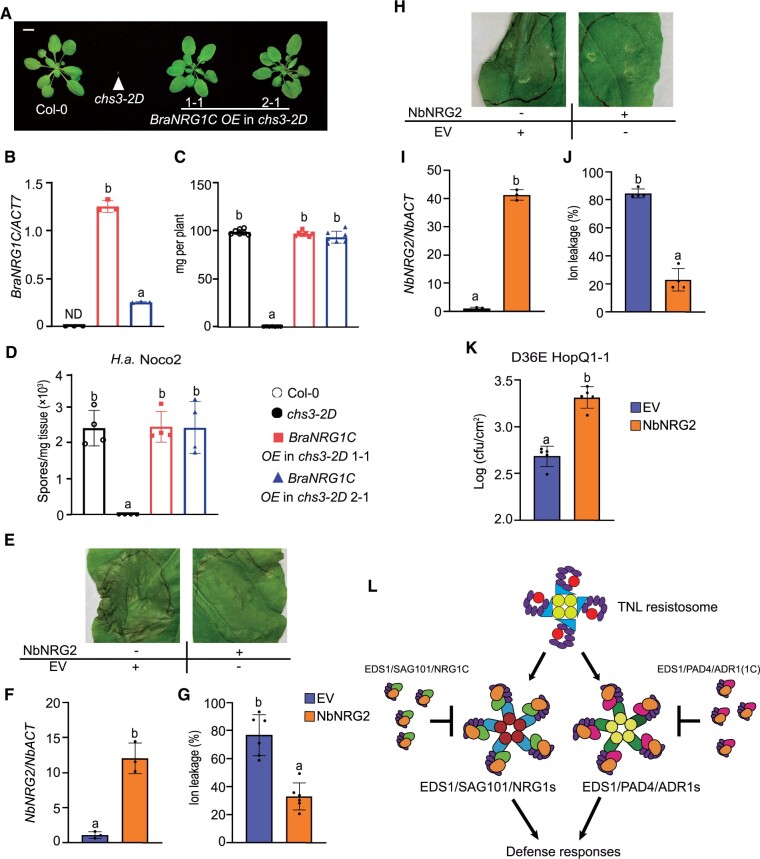
As NRG1C-type N-terminally truncated hNLRs appear to be conserved in Brassicaceae (Figure 1B), we further tested whether they are functionally exchangeable among different Brassicaceae species. To test this, we overexpressed Brara.I00877.1 (BraNRG1C) in the chs3-2D mutant. Consistent with the finding (from phylogenetic analysis) that the Brassicaceae NRG1C family arose from a common ancestor (Figure 1B), overexpression of BraNRG1C fully rescued the chs3-2D-mediated dwarfism (Figure 7, A–C) and enhanced disease resistance to H.a. Noco2 (Figure 7D). Taken together, these results indicate that the NRG1C function is conserved in Brassicaceae species.
Figure 7.
NRG1C-type N-terminally truncated hNLRs from canola (Brassica napus cv. Westar) and N. benthamiana function similarly to A. thaliana NRG1C. A, Morphology of 4-week-old soil-grown plants of Col-0, chs3-2D, and two independent transgenic lines of BraNRG1C OE in the chs3-2D background. Bar = 1 cm. B, BraNRG1C gene expression in 2-week-old plate-grown plants, as determined by qRT-PCR. Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 3). ND, not detectable. Three independent experiments were carried out with similar results. The color legends are included in D. C, Fresh weights of the plants in (A). Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 7). The color legends are included in D. D, Quantification of H.a. Noco2 sporulation in the indicated genotypes at 7 dpi with 105 spores per ml water. Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 4). Three independent experiments were carried out with similar results. E, HR in N. benthamiana leaves expressing the indicated proteins at 48 hpi following the infiltration of P.s.t. DC3000 D36E HopQ1-1 to deliver HopQ1-1 effector at OD600 = 0.2. The photograph was taken at 4 dpi after P.s.t. DC3000 D36E HopQ1-1 infiltration. F, NbNRG2 gene expression in the infiltrated N. benthamiana leaves indicated in E, as determined by qRT-PCR (EV-infiltrated leaves served as a control whose NbNRG2 transcript level was set at 1.0). Samples were collected at 48 hpi with the indicated proteins. Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 3). Three independent experiments were carried out with similar results. The color legends are included in G. G, Ion leakage of N. benthamiana leaves upon P.s.t. DC3000 D36E HopQ1-1 infiltration under the same conditions as in E. Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 6). Three independent experiments were carried out with similar results. H, HR in N. benthamiana leaves expressing the indicated proteins at 48 hpi following the infiltration of Agrobacterium carrying HopQ1-1 effector at OD600 =0.08. The photographs were taken at 2 dpi after Agrobacterium infiltration. Three independent experiments were carried out with similar results. I, NbNRG2 gene expression in the infiltrated N. benthamiana leaves indicated in (H), as determined by qRT-PCR (EV infiltrated leaves served as a control whose NbNRG2 transcript level was set at 1.0). Samples were collected at 48 hpi with the indicated proteins. Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 3). Three independent experiments were carried out with similar results. The color legends are included in K. J, Ion leakage of N. benthamiana leaves upon Agrobacterium infiltration under the same conditions as in H. Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 4). The color legends are included in K. K, Growth of P.s.t. DC3000 D36E HopQ1-1 in N. benthamiana leaves expressing EV-3FLAG or NbNRG2 for 48 h. After 48 h, the leaves were infiltrated with P.s.t. DC3000 D36E HopQ1-1 at OD600 = 0.005 and bacterial growth was counted at 4 dpi. Statistical significance is indicated by different letters (P < 0.01). Error bars represent means ± sd (n = 5). Three independent experiments were carried out with similar results. L, A model depicting the negative regulation of NRG1C downstream of TNL activation in Arabidopsis. An effector-induced TNL resistosome with NADase activity (Horsefield et al., 2019; Wan et al., 2019; Ma et al., 2020; Martin et al., 2020; Tian and Li, 2020) signals through two parallel complexes, EDS1–PAD4–ADR1s and EDS1–SAG101–NRG1A/1B, to activate defense responses (Sun et al., 2021; Wu et al., 2021). To avoid over-activation, N-terminally truncated hNLR NRG1C antagonizes immunity mediated by its full-length neighbors NRG1A and NRG1B, while an engineered truncated ADR1 with a similar length to NRG1C [ADR1(1C)] is able to interfere with EDS1–PAD4 dimer to dampen immunity mediated by ADR1s (Wu et al., 2021). Arrows indicate activation, while the “T’” signs represent suppression.
NbNRG2 negatively regulates HopQ1-1-mediated immunity in N. benthamiana
To further explore the function of truncated NRG1C outside the Brassicaceae, we examined the role of N. benthamiana NRG2 (NbNRG2) in HopQ1-1-triggered NRG1-dependent resistance (Schultink et al., 2017; Qi et al., 2018). When WT N. benthamiana leaves were challenged with P.s.t. DC3000 D36E HopQ1-1, more NbNRG2 transcripts were detected (Supplemental Figure S14A), suggesting that NbNRG2 might play roles in HopQ1-1-trigerred defense responses. In addition, inoculation with P.s.t. DC3000 D36E HopQ1-1, but not P.s.t. DC3000 D36E, resulted in visible cell death in WT N. benthamiana (Supplemental Figure S14, B and C). In contrast, the cell death caused by P.s.t. DC3000 D36E HopQ1-1 was suppressed in the N. benthamiana nrg1-1 mutant (Supplemental Figure S14C), in agreement with the previous finding that HopQ1-1-triggered defense responses are NRG1-dependent (Qi et al., 2018).
To test the function of NbNRG2 in N. benthamiana, we infiltrated leaves with Agrobacteria carrying NbNRG2 or empty vector (EV) for 48 h and delivered the HopQ1-1 effector into the leaves via P.s.t. DC3000 D36E or Agrobacteria (Supplemental Figure S14D). Interestingly, overexpression of NbNRG2 suppressed HopQ1-1-mediated HR, which was introduced by either P.s.t. DC3000 D36E HopQ1-1 (Figure 7, E–G) or Agrobacteria (Figure 7, H–J). Consistently, more P.s.t. DC3000 D36E HopQ1-1 bacterial growth was detected in NbNRG2-expressing leaves than in leaves expressing EV (Figure 7K). To our surprise, significantly more P.s.t. DC3000 D36E grew in NbNRG2-expressing leaves than in leaves expressing EV (Supplemental Figure S14E), suggesting that NbNRG2 might play some roles in basal defense in N. benthamiana. However, the HopQ1-1-mediated HR (Supplemental Figure S14, F–H) and bacterial growth restriction (Supplemental Figure S14, I and J) were not suppressed by AtNRG1C overexpression, indicating that AtNRG1C is not able to inhibit N. benthamiana NRG1-mediated immunity. Additionally, NbNRG2 overexpression failed to suppress chs3-2D-mediated dwarfism (Supplemental Figure S15, A–C) or the enhanced disease resistance to H.a. Noco2 (Supplemental Figure S15D). Since molecular incompatibility among different species or clades was observed in the EDS1–SAG101–NRG1 module (Gantner et al., 2019; Lapin et al., 2019), it is not surprising that NbNRG2 and Brassicaceae NRG1Cs are not functionally exchangeable and that their negative regulation requires molecular compatibility.
Discussion
Truncated NLRs, including TIR only, TN, TIR-unknown domain (TX), CC only, CC-NB (CN), CC-unknown domain (CX), RPW8 only, RPW8-NB (RN), NB-LRR (NL), NB only, and LRR only, are commonly encoded in the genomes of vascular plants (Bai et al., 2002; Meyers et al., 2002, 2003; Yang et al., 2008; Jacob et al., 2013; Nandety et al., 2013; Baggs et al., 2017; Van de Weyer et al., 2019; Lee and Chae, 2020). A genome-wide survey of A. thaliana NLR genes revealed that ∼31% of NLR genes are truncated (Meyers et al., 2003). Among these, ∼90% lack canonical C-terminal domains (Meyers et al., 2002, 2003). Functional characterizations of some C-terminally truncated NLRs have been reported. The TIR-only protein RBA1 can function as an effector sensor and trigger plant immune responses by self-oligomerization (Nishimura et al., 2017). Moreover, the TIR domain in the RBA1 protein possesses NADase activity, which is required to trigger cell death (Wan et al., 2019). Other well-studied TN proteins include TN2 and CHS1. TN2 guards the exocyst complex subunit EXO70B1 (Zhao et al., 2015) and may stabilize the active calcium-dependent protein kinase CPK5 to regulate defense responses (Liu et al., 2017). Interestingly, TNL SOC3 and CHS1 cluster together with TN2 (Zhang et al., 2017; Liang et al., 2019). SOC3 pairs with either CHS1 or TN2 to monitor the absence or overaccumulation of the E3 ligase SAUL1 (Tong et al., 2017; Liang et al., 2019). A recent study found that the LRR-PL (Post-LRR) truncated protein DANGEROUS MIX10 (DM10TueScha-9) shows an incompatible interaction with DANGEROUS MIX11 (DM11Cdm-0), resulting in hybrid necrosis (Barragan et al., 2020). The chili pepper (Capsicum baccatum)-specific truncated CN protein CbCN, missing the LRR domain, positively regulates defense response genes against the fungus Colletotrichum acutatum (Son et al., 2021). Therefore, the C-terminally truncated NLRs can often still play positive roles in NLR activation, as they retain the N-terminal signaling domains such as the TIR and NB regions.
In contrast, ∼10% of truncated NLRs are N-terminally truncated in A. thaliana (Meyers et al., 2003). Although they are widely present in all vascular plant genomes, little is known about their biological significance. In mammalian systems, the Toll-like receptor (TLR) TIR truncations can act as dominant-negative (DN) proteins to dampen immune responses by associating with the same immune activation complexes (Burns et al., 2003; Wells et al., 2006; Padmanabhan et al., 2009). Here, we demonstrated that similar to the truncated TLRs, the N-terminally truncated helper NRG1C lacking the RPW8 CC and part of the NB domain can negatively regulate NRG1A/1B-mediated defense responses. Compared to the DN-TLR examples in the mammalian system, exactly how NRG1C interferes with NRG1A/1B signaling is less clear. As overexpression of a NRG1C variant containing a premature stop codon failed to suppress chs3-2D-mediated autoimmunity (Figure 6, A–C), and the transcript levels of full-length NRG1A/1B were not affected by overexpression of WT NRG1C (Figure 2E; Supplemental Figure S9, B and D), this mechanism unlikely involves transcriptional interference. In addition, NRG1C does not interact with NRG1A/1B (Supplemental Figure S11B), although caution should always be taken with negative protein–protein interaction data.
Sun et al. recently showed that NRG1A associates with the EDS1–SAG101 dimer in an effector-dependent manner (Sun et al., 2021). However, the SAG101–NRG1A/1B signaling branch plays unequal roles in different cases, although EDS1 is required for all tested TNLs (Feys et al., 2005; Xu et al., 2015; Wu et al., 2019, 2021; Sun et al., 2021). For example, SAG101 and NRG1A/1B are fully required for chs3-2D-mediated autoimmunity, but they are not needed for CHS1-SOC3 or RPP4-mediated autoimmunity in saul1-1 or chs2-1 (Wu et al., 2019). Therefore, the EDS1–SAG101–NRG1A/1B complex is specifically required for some, but not all TNL-mediated immunity. Interestingly, NRG1C overexpression phenotypes are identical to those of the nrg1a nrg1b double and the sag101 single mutants. Biochemically, NRG1C can be in complex with EDS1–SAG101 dimers (Figure 6, G and H) (Sun et al., 2021). More NRG1C peptides were detected in the SAG101 IP-MS than NRG1A/1B upon activation of the TNL pair RRS1-RPS4, suggesting that NRG1C might have higher affinity with the EDS1–SAG101 dimer than NRG1A/1B. Taken together, NRG1C may occupy the NRG1A/1B space in the EDS1–SAG101–NRG1A/1B complex, preventing NRG1A/1B interaction with the lipase-like proteins. This would explain the negative interaction data between NRG1A and NRG1C. NRG1C seems to interfere with the EDS1–SAG101 heterodimer via direct association, leading to compromised NRG1-triggered immunity (Figure 7L). Further biochemical and structural analyses of these proteins are needed to clarify the details of these interactions.
Both NRG1C and ADR1-L3 are N-terminally truncated hNLRs. Although NRG1C negatively regulates some TNL-mediated defense responses, no effect on ADR1-dependent TNL-mediated autoimmunity or pathogen growth was observed upon ADR1-L3 overexpression (Supplemental Figure S4). In contrast to NRG1C, ADR1-L3 retains the full NB domain (Supplemental Figures S1 and S3). NRG1C homologs are commonly found in Brassicaceae (Figure 1B), whereas an ADR1-L3 homology is only present in B. stricta (Supplemental Figure S3). Such an evolutionary difference indicates that ADR1-L3 is likely a pseudogene and thus was lost during evolution. Therefore, for N-terminally truncated NLRs in vascular plant genomes, some may be pseudogenes, and some may have negative functions, as with NRG1C. Although an evolutionary examination may provide an initial hint, functional analysis is needed to determine their exact roles.
Besides NRG1C and ADR1-L3, five additional N-terminally truncated NB-LRR genes are present in the A. thaliana Col-0 genome (Supplemental Table S2). AT4G19050, AT5G45510, and AT1G61300 are more closely related to CNLs (Meyers et al., 2003). AT4G19050 is adjacent to the CN gene AT4G19060. AT5G45510 and the head-to-head arranged CNL AT5G45520 form a cluster with two other CN genes, AT5G45490 and AT5G45440. AT1G61310 and the adjacent CNL AT1G61300 are transcribed in the same direction (Supplemental Table S2). Only two NL proteins, AT5G38350 and AT4G09360, share a similar NB motif with TNLs. AT5G38350 clusters together with the TIR-only gene AT5G38344 and the TNL gene AT5G38340, while AT4G09360 resides in the TNL cluster containing TNL AT4G09430 and AT4G09420 (Supplemental Table S2). Therefore, clustering of NL genes with full-length or C-terminally truncated sNLRs can also be detected in A. thaliana. It is possible that the NL genes in these clusters play similar roles as NRG1C in the NRG1 cluster, with negative regulatory functions. Additional investigations into the functions of such NL genes in A. thaliana and other plant species will be of great interest to widen our understanding of NLRs.
Materials and methods
Construction of plasmids
Overexpression constructs of NRG1C and NRG1CT71* were generated in the pCambia1305 vector using the KpnI and SpeI restriction sites. The overexpression constructs of NRG1C in the NRG1A D485V background, BraNRG1C, and NbNRG2 were cloned into the pBASTA vector using the KpnI and SpeI restriction sites. The extracted genomic DNA from Brassica napus cv. Westar and N. benthamiana was used to clone BraNRG1C and NbNRG2 into pBASTA using the DraIII and KpnI/SpeI restriction sites, respectively. DAR5 was cloned into the pHan vector using the SfiI restriction site. For the TurboID-based proximity labeling assay, EDS1, SAG101, NRG1A, and NRG1C were cloned into pBASTA-HA-TurboID using the respective KpnI/SpeI, KpnI/BamHI, KpnI/SpeI, and KpnI/SpeI restriction sites. NRG1C and SAG101 were cloned into pCambia1300-3FLAG using the KpnI/SpeI and KpnI/BamHI restriction sites. All genes were driven by the 35S promoter. pHeeNRG1 (At5g66910-At5g66900) and pHeeNRG1C (At5g66890) were described previously (Wu et al., 2019).
To generate the CRISPR/Cas9 constructs for knocking out PAD4 and SAG101, genomic sequences of PAD4 and SAG101 were subjected to CRISPRscan (http://www.crisprscan.org/?page=sequence) to identify the target sequences. The selected sequences were evaluated with Cas-OFFinder (http://www.rgenome.net/cas-offinder/). The pHEE401E vector was used to generate the CRISPR/Cas9 constructs as previously described (Wang et al., 2015). All primers used are listed in Supplemental Data Set S1.
Plant materials and growth conditions
A. thaliana snc1, chs3-2D, chs1-2, chs2-1, chs3-2D nrg1a nrg1b, sag101-1, snc1 eds1-2, snc1 pad4-1, snc1 adr1 triple, snc1 nrg triple, snc1 adr1 triple nrg triple, chs1-2 nrg triple, chs2-1 nrg triple, ADR1-L2 D484V, and NRG1A D485V were previously described (Li et al., 2001; Huang et al., 2010; Bi et al., 2011; Cheng et al., 2011; Roberts et al., 2013; Zbierzak et al., 2013; Dong et al., 2016; Wu et al., 2019).
The NRG1C transgenic plants in different backgrounds were generated by transforming pCambia1305 NRG1C in the snc1, chs3-2D, Col-0, chs1-2, ADR1-L2 D484V, and chs2-1 backgrounds, pBastaNRG1C in the NRG1A D485V background or pBastaNRG1C-HA-TurboID in chs3-2D. The DAR5 transgenic plants were generated by transforming pHanDAR5 into the snc1 or chs3-2D background. The pBastaADR1-L3, pBastaBraNRG1C, pBastaNRG2 constructs were transformed into either snc1 or chs3-2D to generate the respective transgenic plants. The pBastaNRG1A-HA-TurboID construct was transformed into chs3-2D nrg1a nrg1b to test their functionality.
The snc1 sag101-1 double mutant was generated by crossing snc1 with sag101-1 (Feys et al., 2005). snc1 sag101-1 was crossed with snc1 nrg triple to generate snc1 sag101-1 nrg triple mutant. The CRISPR/Cas9 construct targeting PAD4 was transformed into snc1 adr1 triple (Dong et al., 2016), snc1 sag101-1, or snc1 nrg triple (Wu et al., 2019) to generate snc1 pad4-c1 adr1 triple, snc1 pad4-c1 sag101-1, snc1 pad4-c1 nrg triple mutants, respectively. The snc1 sag101-c1 adr1 triple mutants were generated by transforming the CRISPR/Cas9 construct targeting SAG101 into the snc1 adr1 triple mutant background. Two independent NRG1C deletion alleles were generated by transforming the CRISPR/Cas9 construct (Wu et al., 2019) targeting NRG1C into the Col-0, snc1, and chs3-2D backgrounds. snc1 nrg triple NRG1C OE and snc1 sag101-1 NRG1C OE were generated by crossing snc1 NRG1C OE with snc1 nrg triple and snc1 sag101-1, respectively.
A. thaliana and N. benthamiana plants were grown at 22°C under long day (16-h light/8-h dark) or short day (8-h light/16-h dark) conditions (100 µE; Philips Master TL5 HO 54W/840 bulbs). chs3-2D and its transformants were grown at 28°C to suppress the dwarfism and enable seed production.
Stable transformation in A. thaliana
The binary constructs were introduced into Agrobacterium tumefaciens GV3101 by electroporation and subsequently transformed into A. thaliana by the floral dip method (Clough and Bent, 1998). For most transformations, plants were grown at room temperature. For chs3-2D transformation, plants were grown at 28°C under a 16-h light/8-h dark regime to reduce autoimmunity. Transformants were selected either on soil by spraying with BASTA (Glufosinate ammonium) or on plates with Hygromycin B. At least 10 transformants were selected for each transformation in the T1 generation, and co-segregation analysis in T2 and T3 generations was carried out to make sure the observed phenotypes were due to the transgene expression.
Transient expression in N. benthamiana
Transient N. benthamiana expression assays were performed as previously described (Wu et al., 2017b). In brief, Agrobacteria carrying the respective constructs were co-infiltrated into 4-week-old N. benthamiana leaves. The final OD600 for EDS1-HA-TurboID and NRG1A-HA-TurboID was 0.2. The OD600 value for GFP-HA-TurboID or SAG101-HA-TurboID was 0.6, whereas OD600 = 1.0 was used for strains expressing NRG1C-3FLAG. In Supplemental Figure S14, F–J, NRG1C-3FLAG and EV-3FLAG were infiltrated at OD600 = 1.0. In Figure 7, E–K, NbNRG2 and EV-3FLAG were infiltrated at OD600 = 0.6. In Supplemental Figure S10, E and F, NRG1C-3FLAG and EV-3FLAG were pre-infiltrated in N. benthamiana leaves at OD600 = 1.0. At 36 hpi, NRG1A D485V was infiltrated in the indicated area at OD600 = 0.2. Each infiltration contained Agrobacteria p19 to inhibit gene silencing (Lakatos et al., 2004).
Phylogenetic analysis
Putative NRG1A/1B/1C homologs were obtained from Phytozome (Brassicaceae species) and Sol Genomics Network (N. benthamiana) using A. thaliana NRG1A and NRG1C protein sequences as input, respectively. ADR1 clade hNLRs were excluded from the lists by performing BLAST analysis of ADR1s homologs using AtADR1 protein sequences as input. ZAR1 was used as an outgroup to root the tree. MUSCLE was used for sequence alignment, and a neighbor-joining tree was generated using full-length NRG1C and homologous parts of all other corresponding NRG1 homologs with the JTT model and using 2,000 bootstrap replicates in MEGA version 7.0.
Pathogen infections in A. thaliana
Oomycete and bacterial pathogen infection assays were carried out as described previously (Li et al., 2001). In brief, 2-week-old soil-grown seedlings were sprayed with H.a. Noco2 or Emwa1 conidia spores at a concentration of 105 spores/mL water. After the plants were grown at 18°C for 7 days, sporulation was quantified using a hemocytometer. For bacterial infections, 4-week-old A. thaliana plants were infiltrated with bacterial solution at the designated concentrations (see figure legends). Leaf discs were collected and ground on the day of infection (Day 0) and 3 or 4 days later (Day 3/Day 4) (see figure legends). Colony-forming units were calculated after incubation on LB plates with the appropriate antibiotic selections. For the HR assay in A. thaliana, 4-week-old soil-grown plants were hand infiltrated with Pf0-1 carrying pBS46:AvrRPS4 at OD600 = 0.2. Ion leakage of infected leaves was monitored at 36 hpi.
qRT-PCR analysis
Approximately 0.05 g of tissue from the aboveground parts of 4-week-old soil-grown or 2-week-old plate-grown seedlings was collected. RNA was extracted from the samples using an RNA isolation kit (Bio Basic; Cat#BS82314). ProtoScript II reverse transcriptase (NEB; Cat#B0368) was used to generate cDNA. qRT-PCR was performed using a SYBR premix kit (TaKaRa, Shiga, Japan, Cat#RR82LR) in a Bio-RAD system (Model No. CFX Connect Optics Module) following the manufacturers’ instructions. The primers used are listed in Supplemental Data Set S1.
Agrobacteria infiltration following P.s.t. D36E HopQ1-1 infection or HopQ1-1 Agrobacteria inoculation
Agrobacteria carrying the respective constructs were infiltrated into 4-week-old N. benthamiana leaves expressing EV-FLAG, NRG1C-3FLAG, or NbNRG2. Subsequently, P.s.t. DC3000 D36E or P.s.t. DC3000 D36E HopQ1-1 was infiltrated in the indicated area of each leaf at 48 h after Agrobacteria infiltration at OD600 = 0.2 for HR measurement or OD600 = 0.005 for bacterial growth measurement. Ion leakage and bacterial growth were measured at 4 days after P.s.t. D36E HopQ1-1 infection. Agrobacteria expressing HopQ1-1 were infiltrated into the leaves at 48 h after the initial Agrobacteria infiltration at OD600 = 0.08. Ion leakage was measured at 48 h after HopQ1-1 infiltration.
Ion leakage measurement in N. benthamiana and A. thaliana
After agrobacteria infiltration, P.s.t. DC3000 D36E HopQ1-1 or Pf0-1 AvrRPS4 infection, N. benthamiana leaf discs or whole A. thaliana leaves were collected and washed in 10 mL of MilliQ water overnight at room temperature. Ion leakage was measured with a VWR E C METER Model 2052 conductometer. All samples were then autoclaved to measure the total ion leakage.
TurboID-based proximity labeling and co-IP assay in N. benthamiana
The co-IP assay was performed as described previously (Wu et al., 2020). In brief, N. benthamiana leaves were infiltrated with Agrobacteria containing NRG1C-3FLAG with either EDS1-HA-TurboID, SAG101-HA-TurboID, or NRG1A-HA-TurboID. At 48 hpi, biotin was infiltrated into the leaves. Samples were harvested at 50 hpi and ground into a powder in liquid nitrogen, followed by IP and immunoblot analysis. To test the interaction between NRG1C-3FLAG and SAG101-HA-TurboID, 50 µM of the proteasome inhibitor MG132 was applied with biotin.
TurboID-based proximity labeling and co-IP assay in A. thaliana
Whole 2-week-old plate-grown A. thaliana transgenic seedlings carrying both NRG1C-HA-TurboID and SAG101-3FLAG were soaked in water containing 200 µM biotin. Plants were incubated at room temperature for 2 h for labeling and then harvested, followed by IP and immunoblot analysis.
Protein extraction, IP, and immunoblot analysis
An amount of 100 mg samples of soil-grown A. thaliana plant leaves or N. benthamiana leaves were collected and extracted in extraction buffer (100 mM Tris–HCl pH 8.0, 0.2% SDS, and 2% β-mercaptoethanol). After adding loading buffer to each protein sample, the sample was and boiled for 5 min, followed by immunoblot analysis. Protein abundance was quantified using ImageJ (https://imagej.nih.gov/ij/).
The co-IP assay was performed as previously described (Wu et al., 2017b). In brief, extraction buffer containing 25 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.15% Nonidet P-40, 10% Glycerol, 1 mM PMSF (Phenylmethylsulfonyl fluoride), 1× Protease Inhibitor Cocktail (Roche, Basel, Switzerland; Cat. #11873580001), and 10 mM DTT (Dithiothreitol) was added to each ground tissue sample. To test the interaction between NRG1C-3FLAG and SAG101-HA-TurboID, 50 µM of the proteasome inhibitor MG132 was added to the extraction buffer. The FLAG-tagged NRG1C and SAG101 proteins were immunoprecipitated using 20 µL M2 beads (Sigma, St Louis, Missouri, USA; Cat. #A2220) for co-IP and biotinylation. HA-tagged proteins were detected using anti-HA antibody (Roche Cat. #11867423001). The anti-FLAG antibody was from Sigma (Cat. #F1804). Biotinylated proteins were detected using Streptavidin-HRP (Abcam, Cambridge, UK; Cat. # ab7403). The previously described anti-EDS1 antibody was generously shared by Dr Jane E. Parker at the Max Planck Institute (Feys et al., 2001).
Statistical analysis
Statistical analysis was performed using one-way ANOVA (Analysis of variance) followed by Tukey’s post hoc test. The Scheffé multiple comparison was applied for test correction. Normality tests for all data were performed using SPSS (Statistical Product and Service Solutions). Statistical significance is indicated with different letters. P-values and the number of samples (n) are detailed in the figure legends. ANOVA tables are provided in Supplemental Data Set S2.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: NRG1C (AT5G66890); NRG1A (AT5G66900); NRG1B (AT5G66910); ADR1-L3 (AT5G47280); ADR1-L2 (AT5G04720); ADR1 (AT1G33560); ADR1-L1 (AT4G33300); SNC1 (AT4G16890); CHS3 (AT5G17890); DAR5 (AT5G66630); EDS1 (AT3G48090); SAG101 (AT5G14930); PAD4 (AT3G52430); NbNRG1 (Niben101Scf02118g00018.1), NbNRG2 (Niben101Scf03844g01015.1).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Full-length protein sequence alignment of NRG1C and its homologs in different plant species.
Supplemental Figure S2. DAR5 has no effect on chs3-2D or snc1-mediated autoimmunity.
Supplemental Figure S3. Full-length protein sequence alignment of ADR1-L3 and its homologs in different plant species.
Supplemental Figure S4. No immune-related phenotypes were observed in ADR1-L3 overexpression lines.
Supplemental Figure S5. Overexpression of NRG1C has no effect on chs1-2 or chs2-1-mediated autoimmunity.
Supplemental Figure S6. NRG1C does not affect TNL RPS4, CNL RPS2, P.s.t DC3000, or P.s.t DC3000 D36E-mediated bacterial growth.
Supplemental Figure S7. Schematic description of CRISPR/Cas9 construct design to delete PAD4, SAG101, or NRG1C.
Supplemental Figure S8. NRG1C works in the same pathway with the EDS1–SAG101–NRG1A/1B module.
Supplemental Figure S9. Overexpression of NRG1C does not affect the transcript or protein levels of full-length NRG1A/1B.
Supplemental Figure S10. NRG1C influences the autoimmunity mediated by NRG1A D485V, but not ADR1-L2 D484V.
Supplemental Figure S11. NRG1C does not associate with NRG1A.
Supplemental Figure S12. Biotinylation of NRG1C-3FLAG by EDS1-HA-TurboID or SAG101-HA-TurboID in N. benthamiana leaves.
Supplemental Figure S13. Overexpression of NRG1C does not affect the protein levels of EDS1 or SAG101 in N. benthamiana.
Supplemental Figure S14. AtNRG1C does not influence HopQ1-1-mediated HR in N. benthamiana.
Supplemental Figure S15. NbNRG2 does not influence chs3-2D-mediated autoimmunity.
Supplemental Table S1. The genome arrangements of NRG1C homologs in various plant species.
Supplemental Table S2. The genome arrangements of NL truncated genes in A. thaliana.
Supplemental Data Set S1. List of primers used in this study.
Supplemental Data Set S2. ANOVA tables.
Supplementary Material
Acknowledgments
Drs Jeff Dangl, Jane Parker, Marc Nishimura, Brian Staskawicz, Pingtao Ding, Jonathan Jones, Wei Zhang, and Alan Collmer are cordially thanked for generous sharing seeds of the A. thaliana mutants, the anti-EDS1 antibody, the nrg1-1 N. benthamiana seeds, and Pseudomonas strains. Mr Kevin Ao and Dr Paul Kapos are sincerely thanked for careful reading of the manuscript.
Funding
This work was financially supported by funds from CFI-JELF, the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery and NSERC-CREATE PRoTECT programs to X.Li. and Y.Z. Partial support for the trainees came from scholarships to Z.W. from the Chinese Scholarship Council (CSC) and UBC Dewar Cooper Fund, to L.T. from CSC, to X.Liu. from the MSL Graduate Student Award, and to W.H. from CSC.
Conflict of interest statement. None declared.
Contributor Information
Zhongshou Wu, Michael Smith Laboratories, University of British Columbia, Vancouver, BC V6T 1Z4, Canada; Department of Botany, University of British Columbia, Vancouver, BC V6T 1Z4, Canada.
Lei Tian, Michael Smith Laboratories, University of British Columbia, Vancouver, BC V6T 1Z4, Canada; Department of Botany, University of British Columbia, Vancouver, BC V6T 1Z4, Canada.
Xueru Liu, Michael Smith Laboratories, University of British Columbia, Vancouver, BC V6T 1Z4, Canada; Department of Botany, University of British Columbia, Vancouver, BC V6T 1Z4, Canada.
Weijie Huang, Department of Botany, University of British Columbia, Vancouver, BC V6T 1Z4, Canada.
Yuelin Zhang, Department of Botany, University of British Columbia, Vancouver, BC V6T 1Z4, Canada.
Xin Li, Michael Smith Laboratories, University of British Columbia, Vancouver, BC V6T 1Z4, Canada; Department of Botany, University of British Columbia, Vancouver, BC V6T 1Z4, Canada.
Z.W., Data curation, Validation, Investigation, Methodology, Writing—original draft, Project administration; L.T., Validation, Methodology, Writing; X.Liu, Validation, Methodology; W.H., Validation, Methodology; Y.Z., Formal analysis, Supervision, Funding acquisition; X.L., Conceptualization, Data curation, Formal analysis, Supervision, Funding acquisition, Writing—original draft, Project administration, Writing—revisions. All authors reviewed the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors is (https://academic.oup.com/plcell): Xin Li (xinli@msl.ubc.ca)
References
- Adachi H, Contreras MP, Harant A, Wu CH, Derevnina L, Sakai T, Duggan C, Moratto E, Bozkurt TO, Maqbool A, et al. (2019) An N-terminal motif in NLR immune receptors is functionally conserved across distantly related plant species. Elife 8: e49956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfo G, Villano C, Errico A, Frusciante L, Carputo D, Aversano R, Ercolano MR (2019) Inferring RPW8-NLRs's evolution patterns in seed plants: case study in Vitis vinifera. Planta 251: 32. [DOI] [PubMed] [Google Scholar]
- Baggs E, Dagdas G, Krasileva KV (2017) NLR diversity, helpers and integrated domains: making sense of the NLR IDentity. Curr Opin Plant Biol 38: 59–67 [DOI] [PubMed] [Google Scholar]
- Bai J, Pennill LA, Ning J, Lee SW, Ramalingam J, Webb CA, Zhao B, Sun Q, Nelson JC, Leach JE, et al. (2002) Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res 12: 1871–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barragan AC, Collenberg M, Wang J, Lee RRQ, Cher WY, Rabanal FA, Ashkenazy H, Weigel D, Chae E (2020) A truncated singleton NLR causes hybrid necrosis in Arabidopsis thaliana. Mol Biol Evol 38: 557–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi D, Johnson KC, Zhu Z, Huang Y, Chen F, Zhang Y, Li X (2011) Mutations in an atypical TIR-NB-LRR-LIM resistance protein confer autoimmunity. Front Plant Sci 2: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL (2011) Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci USA 108: 16463–16468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns K, Janssens S, Brissoni B, Olivos N, Beyaert R, Tschopp J (2003) Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J Exp Med 197: 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel B, Ngou PM, Cevik V, Redkar A, Kim DS, Yang Y, Ding P, Jones JDG (2019) Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol 222: 966–980 [DOI] [PubMed] [Google Scholar]
- Cheng YT, Li Y, Huang S, Huang Y, Dong X, Zhang Y, Li X (2011) Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc Natl Acad Sci USA 108: 14694–14699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collier SM, Hamel LP, Moffett P (2011) Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant Microbe Interact 24: 918–931 [DOI] [PubMed] [Google Scholar]
- Couto D, Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16: 537–552 [DOI] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE (2015) Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 66: 487–511 [DOI] [PubMed] [Google Scholar]
- Dong OX, Tong M, Bonardi V, El Kasmi F, Woloshen V, Wünsch LK, Dangl JL, Li X (2016) TNL-mediated immunity in Arabidopsis requires complex regulation of the redundant ADR1 gene family. New Phytol 210: 960–973 [DOI] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J 20: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, Cabral A, Parker JE (2005) Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriëls SH, Takken FL, Vossen JH, de Jong CF, Liu Q, Turk SC, Wachowski LK, Peters J, Witsenboer HM, de Wit PJ, et al. (2006) CDNA-AFLP combined with functional analysis reveals novel genes involved in the hypersensitive response. Mol Plant Microbe Interact 19: 567–576 [DOI] [PubMed] [Google Scholar]
- Gabriëls SH, Vossen JH, Ekengren SK, van Ooijen G, Abd-El-Haliem AM, van den Berg GC, Rainey DY, Martin GB, Takken FL, de Wit PJ, et al. (2007) An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. Plant J 50: 14–28 [DOI] [PubMed] [Google Scholar]
- Gantner J, Ordon J, Kretschmer C, Guerois R, Stuttmann J (2019) An EDS1-SAG101 complex is essential for TNL-mediated immunity in Nicotiana benthamiana Plant Cell 31: 2456–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wang W, Zhang T, Gong Z, Zhao H, Han GZ (2018) Out of water: the origin and early diversification of plant. Plant Physiol 177: 82–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YL, Fitz J, Schneeberger K, Ossowski S, Cao J, Weigel D (2011) Genome-wide comparison of nucleotide-binding site-leucine-rich repeat-encoding genes in Arabidopsis. Plant Physiol 157: 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsefield S, Burdett H, Zhang X, Manik MK, Shi Y, Chen J, Qi T, Gilley J, Lai JS, Rank MX, et al. (2019) NAD + cleavage activity by animal and plant TIR domains in cell death pathways. Science 365: 793–799 [DOI] [PubMed] [Google Scholar]
- Howard BE, Hu Q, Babaoglu AC, Chandra M, Borghi M, Tan X, He L, Winter-Sederoff H, Gassmann W, Veronese P, et al. (2013) High-throughput RNA sequencing of pseudomonas-infected Arabidopsis reveals hidden transcriptome complexity and novel splice variants. PLoS One 8: e74183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Li J, Bao F, Zhang X, Yang S (2010) A gain-of-function mutation in the Arabidopsis disease resistance gene RPP4 confers sensitivity to low temperature. Plant Physiol 154: 796–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Vernaldi S, Maekawa T (2013) Evolution and conservation of plant NLR functions. Front Immunol 4: 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jones JD, Vance RE, Dangl JL (2016) Intracellular innate immune surveillance devices in plants and animals. Science 354: aaf6395. [DOI] [PubMed] [Google Scholar]
- Jubic LM, Saile S, Furzer OJ, El Kasmi F, Dangl JL (2019) Help wanted: helper NLRs and plant immune responses. Curr Opin Plant Biol 50: 82–94 [DOI] [PubMed] [Google Scholar]
- Krasileva KV (2019) The role of transposable elements and DNA damage repair mechanisms in gene duplications and gene fusions in plant genomes. Curr Opin Plant Biol 48: 18–25 [DOI] [PubMed] [Google Scholar]
- Lakatos L, Szittya G, Silhavy D, Burgyán J (2004) Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J 23: 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapin D, Kovacova V, Sun X, Dongus JA, Bhandari D, von Born P, Bautor J, Guarneri N, Rzemieniewski J, Stuttmann J, et al. (2019) A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell 31: 2430–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RRQ, Chae E (2020) Variation patterns of NLR clusters in Arabidopsis thaliana genomes. Plant Commun 1: 100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Clarke JD, Zhang Y, Dong X (2001) Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact 14: 1131–1139 [DOI] [PubMed] [Google Scholar]
- Liang W, van Wersch S, Tong M, Li X (2019) TIR-NB-LRR immune receptor SOC3 pairs with truncated TIR-NB protein CHS1 or TN2 to monitor the homeostasis of E3 ligase SAUL1. New Phytol 221: 2054–2066 [DOI] [PubMed] [Google Scholar]
- Liu N, Hake K, Wang W, Zhao T, Romeis T, Tang D (2017) CALCIUM-DEPENDENT PROTEIN KINASE5 associates with the truncated NLR protein TIR-NBS2 to contribute to exo70B1- mediated immunity. Plant Cell 29: 746–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Kracher B, Saur IM, Bauer S, Ellwood SR, Wise R, Yaeno T, Maekawa T, Schulze-Lefert P (2016) Allelic barley MLA immune receptors recognize sequence-unrelated avirulence effectors of the powdery mildew pathogen. Proc Natl Acad Sci USA 113: E6486–E6495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Lapin D, Liu L, Sun Y, Song W, Zhang X, Logemann E, Yu D, Wang J, Jirschitzka J, et al. (2020) Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 370: eabe3069. [DOI] [PubMed] [Google Scholar]
- Mago R, Zhang P, Vautrin S, Šimková H, Bansal U, Luo MC, Rouse M, Karaoglu H, Periyannan S, Kolmer J, et al. (2015) The wheat Sr50 gene reveals rich diversity at a cereal disease resistance locus. Nat Plants 1: 15186. [DOI] [PubMed] [Google Scholar]
- Marone D, Russo MA, Laidò G, De Leonardis AM, Mastrangelo AM (2013) Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int J Mol Sci 14: 7302–7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R, Qi T, Zhang H, Liu F, King M, Toth C, Nogales E, Staskawicz BJ (2020) Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Science 370: eabd9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Morgante M, Michelmore RW (2002) TIR-X and TIR-NBS proteins: two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J 32: 77–92 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8: 1113–1130 [DOI] [PubMed] [Google Scholar]
- Nandety RS, Caplan JL, Cavanaugh K, Perroud B, Wroblewski T, Michelmore RW, Meyers BC (2013) The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol 162: 1459–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura MT, Anderson RG, Cherkis KA, Law TF, Liu QL, Machius M, Nimchuk ZL, Yang L, Chung EH, El Kasmi F, et al. (2017) TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis. Proc Natl Acad Sci USA 114: E2053–E2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan M, Cournoyer P, Dinesh-Kumar SP (2009) The leucine-rich repeat domain in plant innate immunity: a wealth of possibilities. Cell Microbiol 11: 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart JR, Mestre P, Lu R, Malcuit I, Baulcombe DC (2005) NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr Biol 15: 968–973 [DOI] [PubMed] [Google Scholar]
- Qi T, Seong K, Thomazella DPT, Kim JR, Pham J, Seo E, Cho MJ, Schultink A, Staskawicz BJ (2018) NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc Natl Acad Sci USA 115: E10979–E10987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathjen JP, Dodds PN (2017) Dancing with the stars: an asterid NLR family. Trends Plant Sci 22: 1003–1005 [DOI] [PubMed] [Google Scholar]
- Rietz S, Stamm A, Malonek S, Wagner S, Becker D, Medina-Escobar N, Vlot AC, Feys BJ, Niefind K, Parker JE (2011) Different roles of enhanced disease susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol 191: 107–119 [DOI] [PubMed] [Google Scholar]
- Roberts M, Tang S, Stallmann A, Dangl JL, Bonardi V (2013) Genetic requirements for signaling from an autoactive plant NB-LRR intracellular innate immune receptor. PLoS Genet 9: e1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saile SC, Jacob P, Castel B, Jubic LM, Salas-Gonzáles I, Bäcker M, Jones JDG, Dangl JL, El Kasmi F (2020) Two unequally redundant "helper" immune receptor families mediate Arabidopsis thaliana intracellular "sensor" immune receptor functions. PLoS Biol 18: e3000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucet SB, Ma Y, Sarris PF, Furzer OJ, Sohn KH, Jones JD (2015) Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4. Nat Commun 6: 6338. [DOI] [PubMed] [Google Scholar]
- Schultink A, Qi T, Lee A, Steinbrenner AD, Staskawicz B (2017) Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J 92: 787–795 [DOI] [PubMed] [Google Scholar]
- Serra H, Choi K, Zhao X, Blackwell AR, Kim J, Henderson IR (2018) Interhomolog polymorphism shapes meiotic crossover within the Arabidopsis RAC1 and RPP13 disease resistance genes. PLoS Genet 14: e1007843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao ZQ, Xue JY, Wu P, Zhang YM, Wu Y, Hang YY, Wang B, Chen JQ (2016) Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol 170: 2095–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S, Kim S, Lee KS, Oh J, Choi I, Do JW, Yoon JB, Han J, Park SR (2021) The capsicum baccatum -specific truncated NLR protein CbCN enhances the innate immunity against Colletotrichum acutatum. Int J Mol Sci 22: 7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Lapin D, Feehan JM, Stolze SC, Kramer K, Dongus JA, Rzemieniewski J, Blanvillain-Baufumé S, Harzen A, Bautor J, et al. (2021) Pathogen effector recognition-dependent association of NRG1 with EDS1 and SAG101 in TNL receptor immunity. Nat Commun 12: 3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamborski J, Krasileva KV (2020) Evolution of plant NLRs: from natural history to precise modifications. Annu Rev Plant Biol 71: 355–378 [DOI] [PubMed] [Google Scholar]
- Tan X, Meyers BC, Kozik A, West MA, Morgante M, St Clair DA, Bent AF, Michelmore RW (2007) Global expression analysis of nucleotide binding site-leucine rich repeat-encoding and related genes in Arabidopsis. BMC Plant Biol 7: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas WJ, Thireault CA, Kimbrel JA, Chang JH (2009) Recombineering and stable integration of the Pseudomonas syringae pv. syringae 61 hrp/hrc cluster into the genome of the soil bacterium Pseudomonas fluorescens Pf0-1. Plant J 60: 919–928 [DOI] [PubMed] [Google Scholar]
- Tian L, Li X (2020) Enzyme formation by immune receptors. Science 370: 1163–1164 [DOI] [PubMed] [Google Scholar]
- Tong M, Kotur T, Liang W, Vogelmann K, Kleine T, Leister D, Brieske C, Yang S, Lüdke D, Wiermer M, et al. (2017) E3 ligase SAUL1 serves as a positive regulator of PAMP-triggered immunity and its homeostasis is monitored by immune receptor SOC3. New Phytol 215: 1516–1532 [DOI] [PubMed] [Google Scholar]
- Van de Weyer AL, Monteiro F, Furzer OJ, Nishimura MT, Cevik V, Witek K, Jones JDG, Dangl JL, Weigel D, Bemm F (2019) A species-wide inventory of NLR genes and alleles in Arabidopsis thaliana. Cell 178: 1260–1272.e1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wersch S, Li X (2019) Stronger when together: clustering of plant NLR disease resistance genes. Trends Plant Sci 24: 688–699 [DOI] [PubMed] [Google Scholar]
- Wan L, Essuman K, Anderson RG, Sasaki Y, Monteiro F, Chung EH, Osborne Nishimura E, DiAntonio A, Milbrandt J, et al. (2019) TIR domains of plant immune receptors are NAD + -cleaving enzymes that promote cell death. Science 365: 799–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang Y, Wang Z, Zhang X, Yang S (2013) A missense mutation in CHS1, a TIR-NB protein, induces chilling sensitivity in Arabidopsis. Plant J 75: 553–565 [DOI] [PubMed] [Google Scholar]
- Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ (2015) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei HL, Chakravarthy S, Mathieu J, Helmann TC, Stodghill P, Swingle B, Martin GB, Collmer A (2015) Pseudomonas syringae pv. tomato DC3000 type III secretion effector polymutants reveal an interplay between HopAD1 and AvrPtoB. Cell Host Microbe 17: 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells CA, Chalk AM, Forrest A, Taylor D, Waddell N, Schroder K, Himes SR, Faulkner G, Lo S, Kasukawa T, et al. (2006) Alternate transcription of the Toll-like receptor signaling cascade. Genome Biol 7: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Yahiaoui N, Keller B (2007) Illegitimate recombination is a major evolutionary mechanism for initiating size variation in plant resistance genes. Plant J 51: 631–641 [DOI] [PubMed] [Google Scholar]
- Wu CH, Belhaj K, Bozkurt TO, Birk MS, Kamoun S (2016) Helper NLR proteins NRC2a/b and NRC3 but not NRC1 are required for Pto-mediated cell death and resistance in Nicotiana benthamiana. New Phytol 209: 1344–1352 [DOI] [PubMed] [Google Scholar]
- Wu CH, Abd-El-Haliem A, Bozkurt TO, Belhaj K, Terauchi R, Vossen JH, Kamoun S (2017a) NLR network mediates immunity to diverse plant pathogens. Proc Natl Acad Sci USA 114: 8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Huang S, Zhang X, Wu D, Xia S, Li X (2017b) Regulation of plant immune receptor accumulation through translational repression by a glycine-tyrosine-phenylalanine (GYF) domain protein. Elife 6: e23684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Tong M, Tian L, Zhu C, Liu X, Zhang Y, Li X (2020) Plant E3 ligases SNIPER1 and SNIPER2 broadly regulate the homeostasis of sensor NLR immune receptors. EMBO J 39: e104915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Li M, Dong OX, Xia S, Liang W, Bao Y, Wasteneys G, Li X (2019) Differential regulation of TNL-mediated immune signaling by redundant helper CNLs. New Phytol 222: 938–953 [DOI] [PubMed] [Google Scholar]
- Wu Z, Tian L, Liu X, Zhang Y, Li X (2021) TIR signal promotes interactions between lipase-like proteins and ADR1-L1 receptor and ADR1-L1 oligomerization. Plant Physiol 187: 681–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Zhu C, Cevik V, Johnson K, Liu Y, Sohn K, Jones JD, Holub EB, Li X (2015) Autoimmunity conferred by chs3-2D relies on CSA1, its adjacent TNL-encoding neighbour. Sci Rep 5: 8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Zhang X, Yue JX, Tian D, Chen JQ (2008) Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Genet Genomics 280: 187–198 [DOI] [PubMed] [Google Scholar]
- Zbierzak AM, Porfirova S, Griebel T, Melzer M, Parker JE, Dörmann P (2013) A TIR-NBS protein encoded by Arabidopsis Chilling Sensitive 1 (CHS1) limits chloroplast damage and cell death at low temperature. Plant J 75: 539–552 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang Y, Liu J, Ding Y, Wang S, Zhang X, Liu Y, Yang S (2017) Temperature-dependent autoimmunity mediated by chs1 requires its neighboring TNL gene SOC3. New Phytol 213: 1330–1345 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Song G, Lal NK, Nagalakshmi U, Li Y, Zheng W, Huang PJ, Branon TC, Ting AY, Walley JW, et al. (2019) TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity. Nat Commun 10: 3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Rui L, Li J, Nishimura MT, Vogel JP, Liu N, Liu S, Zhao Y, Dangl JL, Tang D (2015) A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genet 11: e1004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Cheng ZM (2016) A unique RPW8-encoding class of genes that originated in early land plants and evolved through domain fission, fusion, and duplication. Sci Rep 6: 32923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.