Abstract
Plant viruses with densely packed genomes employ noncanonical translational strategies to increase the coding capacity for viral function. However, the diverse translational strategies used make it challenging to define the full set of viral genes. Here, using tomato yellow leaf curl Thailand virus (TYLCTHV, genus Begomovirus) as a model system, we identified genes beyond the annotated gene sets by experimentally profiling in vivo translation initiation sites (TISs). We found that unanticipated AUG TISs were prevalent and determined that their usage involves alternative transcriptional and/or translational start sites and is associated with flanking mRNA sequences. Specifically, two downstream in-frame TISs were identified in the viral gene AV2. These TISs were conserved in the begomovirus lineage and led to the translation of different protein isoforms localized to cytoplasmic puncta and at the cell periphery, respectively. In addition, we found translational evidence of an unexplored gene, BV2. BV2 is conserved among TYLCTHV isolates and localizes to the endoplasmic reticulum and plasmodesmata. Mutations of AV2 isoforms and BV2 significantly attenuated disease symptoms in tomato (Solanum lycopersicum). In conclusion, our study pinpointing in vivo TISs untangles the coding complexity of a plant viral genome and, more importantly, illustrates the biological significance of the hidden open-reading frames encoding viral factors for pathogenicity.
Profiling translation initiation sites in plant begomoviruses reveals the complexity and diversity of plant virus coding capacity and uncovers the hidden open-reading frames required for pathogenesis.
IN A NUTSHELL.
Background: When viruses such as begomoviruses infect plants, they hijack the host translation machinery to drive the expression of their proteins. This process causes viral disease, leading to great yield losses in crops such as tomato. To develop effective antiviral strategies, pathogenesis studies are needed to investigate how virus-encoded proteins contribute to the viral lifecycle and disease symptom development.
Question: Plant viruses with densely packed genomes employ noncanonical translational strategies to increase their coding capacity. However, the diverse translational strategies used make it challenging to define the full set of viral genes.
Findings: To obtain a global view of viral genes expressed during the lifecycle of tomato yellow leaf curl Thailand virus, we investigated the sites on viral mRNAs that can initiate protein translation using a technique for globally mapping in vivo translating ribosomes. We found multiple initiation sites that encode viral factors important for disease symptom development in tomato. Our findings suggest that the complexity and diversity of plant virus coding capacity are underappreciated.
Next steps: Our findings raise the next grand questions: How are the newly identified open reading frames induced during viral infection, and how do they contribute to viral pathogenesis? Thus, the next important steps are to elucidate the mechanistic basis of the cis- and trans-regulatory factors involved in viral gene expression and how these new viral factors coordinate the viral lifecycle and function during pathogenesis in plants.
Introduction
Diseases caused by viruses greatly reduce crop yield and quality and are a major issue in crop production. To develop effective antiviral strategies, pathogenesis studies investigating how viruses encode proteins that contribute to the viral life cycle and disease symptom development are critical. Notably, due to the small size of viral genomes and the need to express multiple structural/enzymatic/regulatory viral proteins, plant viruses use diverse noncanonical translational strategies to maximize the coding capacity of their genomes. These strategies include dividing their genomes into smaller segments, producing monocistronic subgenomic RNA, and encoding a polyprotein that is subsequently proteolytically cleaved into individual proteins (Firth and Brierley, 2012; Hull, 2014; Miras et al., 2017; Jaafar and Kieft, 2019). Another key strategy is to initiate translation from noncanonical initiation sites, which can be achieved through internal ribosome entry, leaky scanning, and re-initiation mechanisms and results in AUG- and non-AUG-initiated open-reading frames (ORFs) that are separate from, overlap with, or are nested within annotated coding regions and are densely packed in the viral genome (Firth and Brierley, 2012; Hull, 2014; Stern-Ginossar, 2015; Miras et al., 2017). However, in silico prediction of noncanonical initiation sites, which often lack well-defined sequence signatures, is quite challenging, making it difficult to elucidate the full set of functional genes in viruses.
By integrating sequence conservation and statistical analyses, several plant virus studies have successfully uncovered hidden AUG- and non-AUG-initiated ORFs that overlap with or are located within known genes (Chung et al., 2008; Ling et al., 2013; Smirnova et al., 2015). For example, a small non-AUG-initiated ORF (termed ORF3a) was identified in poleroviruses and luteoviruses through comparative genomics approaches and was found to encode a 45–48 amino acid protein required for systemic movement (Smirnova et al., 2015). Nevertheless, these sequence- and bioinformatics-based strategies depend on the availability of a collection of related virus genomes and/or prior information about viral genes, thus limiting the identification of noncanonical ORFs.
Ribosome profiling, combined with translation initiation inhibitor treatment, is an emerging methodology that facilitates the global mapping of in vivo translation initiation sites (TISs) at single-codon resolution and is used to identify hidden ORFs expressed in vivo (Ingolia et al., 2011; Lee et al., 2012). Studies employing in vivo translation initiation sequencing have uncovered a number of novel AUG/non-AUG-initiated ORFs in mammalian viruses (Ingolia et al., 2011; Fritsch et al., 2012; Lee et al., 2012; Stern-Ginossar et al., 2012; Irigoyen et al., 2016; Machkovech et al., 2019; Finkel et al., 2020). For example, human cytomegalovirus was sequenced 20 years ago and was estimated to encode 165–252 ORFs (Davison et al., 2003); however, 604 translated AUG/non-AUG ORFs were later identified and experimentally validated through ribosome profiling analyses (Stern-Ginossar et al., 2012). Previous ribosome profiling studies in Arabidopsis thaliana and tomato (Solanum lycopersicum) have revealed a great number of unannotated TISs at AUG and non-AUG codons in vivo, which encode different protein isoforms or novel peptides/proteins (Hsu et al., 2016; Bazin et al., 2017; Willems et al., 2017; Wu et al., 2019a; Li and Liu, 2020). These findings indicate that host plants employ noncanonical initiation mechanisms for protein synthesis. As plant viruses hijack host translational machineries for translation of their genes (Walsh et al., 2013; Hoang et al., 2021), they may also potentially use noncanonical initiation sites to translate proteins (Stern-Ginossar and Ingolia, 2015; Finkel et al., 2018). Nevertheless, compared with the fruitful discoveries of unannotated and noncanonical translational events in host plants, the extent to which noncanonical initiation occurs in plant virus genomes remains largely unclear, and studies aimed at obtaining a global and in-depth view of in vivo translated ORFs in plant viruses are limited.
Plant begomoviruses are single-stranded, positive-sense DNA viruses. They primarily infect dicotyledonous crops and cause symptoms including leaf curling/chlorosis and stunting (Scholthof et al., 2011; Leke et al., 2015; Basak, 2016; Prasad et al., 2020). Begomoviruses are either monopartite (with a ∼2.6-kb DNA-A component) or bipartite (with both DNA-A and DNA-B components, each ∼2.6 kb in size). So far, six viral genes in the DNA-A component and two in the DNA-B component have been characterized and shown to play a pathogenic role during viral infection (Fondong, 2013; Hanley-Bowdoin et al., 2013; Prasad et al., 2020). Due to the substantial agricultural losses caused by begomoviruses, systemic identification of the in vivo translated viral elements and exploration of the hidden ORFs are critical for obtaining a mechanistic understanding of how plant begomoviruses infect crops.
Here, employing tomato yellow leaf curl Thailand virus (TYLCTHV; a bipartite begomovirus) as a model system (Basak, 2016; Prasad et al., 2020), we set out to profile the in vivo TISs in a plant viral genome by mapping initiating ribosome positions and to search for hidden ORFs translated during virus infection of tomato. Our results revealed 17 unanticipated AUG and non-AUG start sites, including downstream TISs of ORFs encoding protein isoforms of known genes and novel TISs that direct the translation of distinct viral proteins. We experimentally investigated three unannotated TISs, determining their translation initiation activities, the subcellular localization of the corresponding proteins in plant cells, the translation initiation context of their flanking sequences, their conservation across different begomovirus, and their biological effects on pathogenesis to delineate the molecular basis of their involvement in virus pathogenic mechanisms. Our study, integrating bioinformatics and experimental approaches, provides important insights into the identities of hidden and translated viral elements. In addition, our findings extend our understanding of viral gene expression and pathogenicity in tomato-infecting begomoviruses.
Results
Global profiling of the coding potential of TYLCTHV at single-ORF resolution
To finely resolve the viral genes expressed during TYLCTHV infection, we conducted transcriptomic and translatomic profiling. We first carried out total RNA sequencing to identify the transcribed regions and ribosome profiling combined with treatment with cycloheximide (CHX), a translation inhibitor that prevents translocation of deacylated tRNA from the P (peptidyl) site into the E (exit) site and stabilizes translating ribosomes on mRNAs, to map the translated regions along the genome using total RNAs and ribosome-protected RNAs extracted from TYLCTHV-infected tomato leaves (Ingolia et al., 2009; Stern-Ginossar and Ingolia, 2015; Li and Liu, 2020). The data sets generated from total RNA sequencing and ribosome profiling with CHX treatment are hereafter referred to as the RNA and CHX data sets, respectively. To evaluate the quality of the ribosome profiling data sets, we mapped reads to both host tomato and TYLCTHV genes. In contrast to the RNA reads, the CHX reads were located around the translation start and stop sites and were also distributed in coding regions of tomato genes (Supplemental Figure S1A). The reads mapping to either tomato or TYLCTHV genes were ∼27–30 nts long and displayed significant phasing patterns (i.e. the 3-nt periodicity) (Supplemental Figure S1, B and C), which are consistent with the characteristics of ribosome-protected fragments (RPFs; Ingolia et al., 2009).
In TYLCTHV, the RNA signals were primarily located in annotated genic regions of the viral plus and minus strands of the TYLCTHV DNA-A and -B molecules (Figure 1, A and D); thus, the transcribed regions are in line with previous gene annotations in the TYLCTHV genome and their expression during virus infection (Hanley-Bowdoin et al., 2013; Basak, 2016). At the translational level, the CHX read coverages clearly reflect the translated regions of most genes (Figure 1, B and E). Nevertheless, due to the presence of polycistronic and overlapping viral genes (top panels, Figure 1, A and D) and CHX signals being affected by both translation initiation and elongation rates, it was difficult to finely pinpoint in vivo TISs and reveal the corresponding coding regions at a single-gene resolution.
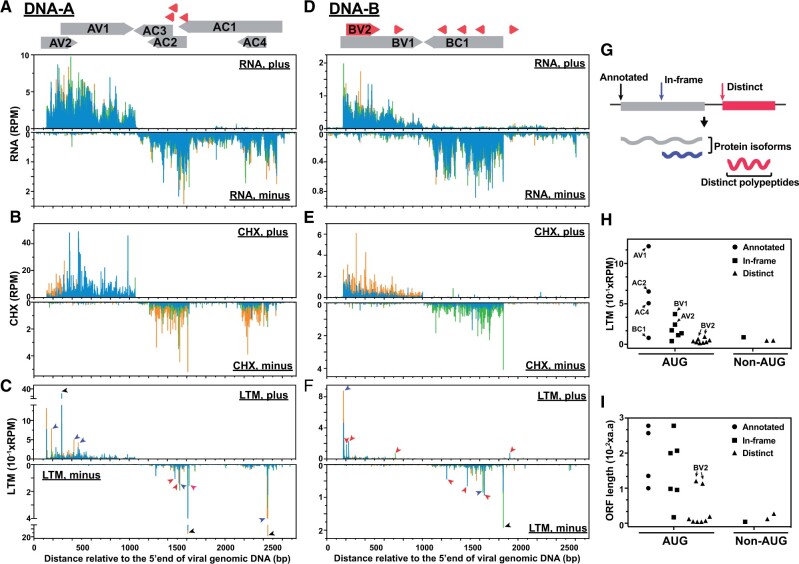
Figure 1.
Transcriptional and translational profiles of TYLCTHV. A–C, Read densities along the viral plus and minus strands (top and bottom panels, respectively) of the DNA-A molecule of TYLCTHV for the RNA (A), CHX (B), and LTM (C) samples. Annotated viral genes (gray boxes with direction of expression indicated) and novel ORFs (red boxes) are shown on the top. Black, purple, and red arrows represent the experimentally derived TISs that were annotated previously (black) or unanticipated, that is either in-frame with annotated TISs (purple) or distinct from annotated TISs (red) as defined in (G). Blue, green and orange bars indicate the reads whose assigned P-sites map to the codon positions 0, 1, and 2 (i.e. phases 1–3, respectively) relative to the 5′ end of the plus strand. D–F, As indicated in (A–C), but for the read densities along the DNA-B molecule. G, Illustration of the types of TISs and the corresponding hypothetical protein products. In-frame (purple): TISs in-frame with and downstream of an annotated TIS that lead to the translation of protein isoforms. Distinct (red): TISs out of frame with the annotated ones that lead to translation of distinct polypeptides. H and I, LTM read intensity (H) and the lengths of TIS-encoded ORFs (I) for the experimentally derived TISs (n=21) grouped based on the TIS codons and TIS types as defined in (G). RPM: reads per million mapped reads.
To address this limitation, we further performed ribosome profiling combined with treatment with lactimidomycin (LTM), a translocation inhibitor that blocks the very first round of elongation, to allow us to pinpoint the in vivo TISs (Schneider-Poetsch et al., 2010; Lee et al., 2012; Gao et al., 2015; Stern-Ginossar and Ingolia, 2015; Li and Liu, 2020); this data set is hereafter referred to as the LTM data set. As expected, the LTM reads displayed the characteristics of RPFs including read lengths of 27–30 nts and 3-nt periodicity (Supplemental Figure S1, B and C). In contrast to the observation that the CHX read coverage was mainly in coding regions, the LTM reads predominantly showed sharp peaks at the annotated TISs of tomato genes (Supplemental Figure S1A) and those of the viral genes, including AV1, AC2, AC4, and BC1 (black arrows, Figure 1, C and F). These results suggest that LTM treatment successfully marks in vivo TISs and allows translated viral elements to be identified at single-gene resolution.
To globally reveal TYLCTHV ORFs expressed during infection, we systematically computed the signal differences between LTM and CHX samples to identify the TIS peaks with higher LTM signals (Lee et al., 2012; Li and Liu, 2020). In total, we identified 21 TIS peaks present in two biological replicates (separate experiments; see “Materials and methods”) and considered these sites to be in vivo TISs used during viral infection (Figure 1; Supplemental Figure S2A and Supplemental Table S1). Among the identified TISs, ∼86% were AUG codons (Figure 1H) and only four of them overlapped with annotated TISs (black arrows in Figure 1, C and F; circles in Figure 1H). These in vivo TISs were classified into three categories: previously annotated TISs (black arrows, Figure 1, C and F; circles, Figure 1, H and I), TISs that are in-frame with annotated TISs that initiate translation of a protein isoform (purple arrows, Figure 1, C and F; squares in Figure 1, H and I), and TISs that are distinct from annotated TISs and lead to a new ORF (red arrows, Figure 1, C and F; triangles, Figure 1, H and I).
These results indicate that, in addition to the eight known viral genes, there was an abundance of uncharacterized AUG start sites, likely encoding previously unidentified ORFs.
AUG87 is not the TIS for AV2 protein synthesis
To further reveal the biological significance of these hidden in vivo TISs, we focused on the downstream TISs in AV2 and BV1, which had predominantly higher LTM signals compared with other TISs (squares indicated by arrows, Figure 1H), and two distinct TISs, which were in-frame with each other and initiate translation of an uncharacterized ORF (referred to as the BV2 gene hereafter; triangles indicated by arrows, Figure 1, H and I). The TISs for the BV1 and BV2 genes will be discussed in detail in subsequent sections.
AV2, which is required for virus pathogenicity, is a movement protein that facilitates cell-to-cell viral movement; it also functions as a suppressor of gene silencing (Fondong, 2013; Hanley-Bowdoin et al., 2013; Basak, 2016). While the annotated start site of AV2 at nucleotide 87 did not show any initiation signals, an in-frame AUG start site at nucleotide 189 was found to have a TIS peak in both replicates (Figure 2A; Supplemental Figure S3A and Supplemental Table S1). In addition, we noticed that an in-frame AUG at nucleotide 135 of AV2 also showed LTM signals (Figure 2A). Also, there were CHX signals (representing the translation status) in the region between AUG135 and AUG189 (gray, Supplemental Figure S3A). We thus hypothesized that both the AUG135 and AUG189 sites may serve as actual in-frame TISs for AV2 expression, with no initiation at AUG87.
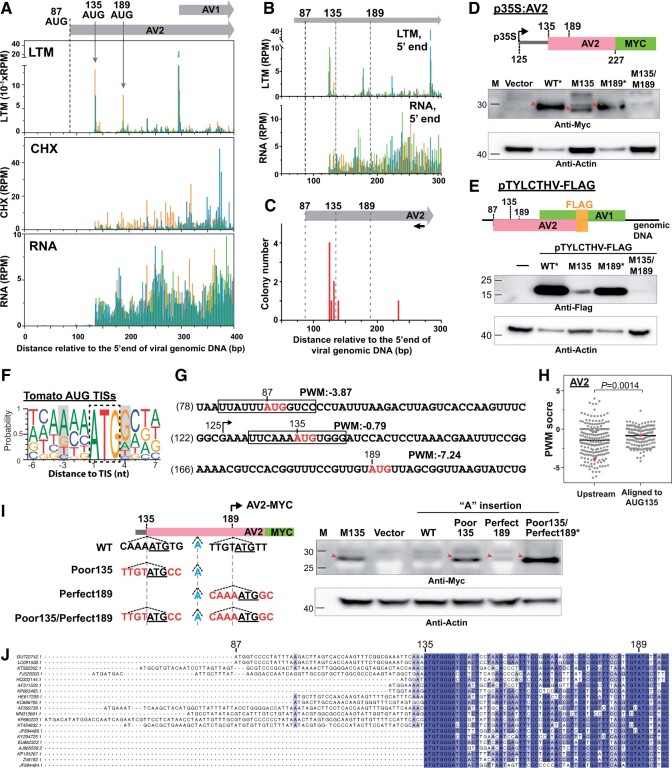
Figure 2.
Characteristics of the in vivo downstream TISs in AV2. A, As described in Figure 1, A–C, but for the LTM, CHX, and RNA read densities along the plus strand of the AV2 genic region. The positions and the corresponding codons of the annotated TIS (AUG87; dashed line) and two downstream in vivo TISs (AUG135 and AUG189; gray arrows) supported by LTM signals are indicated. B, As indicated in (A), but for the positional distributions of the 5′ ends of LTM and RNA reads in order to obtain better resolution of the 5′ transcript ends. C, The positional distributions of the 5′ ends of the AV2 transcripts revealed via 5′ RACE analyses. The numbers of cloned RNA products are shown. The primer position for AV2 cDNA synthesis is marked with a black arrow. D, Immunoblotting analysis of AV2 proteins (red arrows) with translation driven by the WT sequence from the region ranging from 125 to 227 nts or sequences with single or double mutations at nucleotides 135 and/or 189 (M135AUG→GCG and M189AUG→GCG). Proteins were expressed in N. benthamiana leaves using the Agrobacterium-mediated transient expression system. Vector: MYC-containing plasmid without the AV2 fragment. *: to even the protein abundance for presentation clarity, the WT and M189 samples were diluted 20-fold. See Supplemental Figure S3 for the undiluted samples. E, As described in (D), but for FLAG-tagged AV2 proteins expressed during virus infection by co-inoculating N. benthamiana leaves with both the TYLCTHV infectious clone and the TYLCTHV infectious clone with a FLAG protein insertion at the C-terminus of AV2 (pTYLCTHV-FLAG). Results are shown for pTYLCTHV-FLAG with WT sequences or the indicated mutations at nucleotide 135 (M135AUG→GCG) and/or 189 (M189AUG→GCG). −: N. benthamiana leaves only inoculated with the TYLCTHV infectious clone. *: samples were diluted 20-fold as described in (D). F, The probability of occurrence of ATCG nucleotides in sequence regions around the tomato annotated TISs used in vivo, as retrieved from a previous study (Li and Liu, 2020). Gray boxes highlight the −3 and +4 positions of the Kozak sequence. G, PWM scores of TIS sites (red) that represent the sequence similarity between the region surrounding a given TIS (boxed) and those surrounding tomato annotated TISs shown in (F) (see “Materials and methods”). H, PWM scores of AUG sites in begomovirses that were aligned to AUG135 of the TYLCTHV DNA-A molecule and the associated nearest upstream AUG sites. Red: the AUG135 site of TYLCTHV DNA-A and its associated nearest upstream AUG sites. I, As described in (D), but for detecting the AUG189-inititaed AV2-MYC protein. The nucleotide insertions and substitutions in WT sequences of p35S:AV2 described in (D) are highlighted in blue and red. M135: the construct of p35S:M135AUG→GCG described in (D). *: samples were diluted five-fold. J, Plots of nucleotide sequence alignments of the N-terminal regions of annotated AV2 genes among 20 begomoviruses. For the full-length AV2 genes in all 242 begomoviruses, see Supplemental File S1, B and C. The GenBank accession numbers and the corresponding species names are in Supplemental Data Set S1. GU723742.1 (the TYLCTHV used in this study) and the associated annotated TIS at position 87 and two identified TISs at positions 135 and 189 are indicated at the top.
The usage of these downstream TISs could be due to the production of alternative transcripts with different 5′ transcript ends (i.e. the transcript with the 5′ end downstream of the annotated TIS) or the selection of different start sites along the mRNA during translation (Mejia-Guerra et al., 2015; Kurihara et al., 2018). To explore these possibilities, we performed 5′ rapid amplification of cDNA ends (RACE) to identify the 5′ ends of the AV2 transcripts. Its transcript start site was primarily located at nucleotide 125, a position downstream of the annotated TIS (at nucleotide 87) and upstream of the identified TISs at nucleotides 135 and 189 (Figure 2C), consistent with the read coverages observed in the total RNA sample (bottom panel, Figure 2B). These findings suggest that, due to the 5′ transcript ends that were downstream of the annotated TIS, the annotated AUG87 is unlikely to serve as the TIS for AV2 translation.
AUG135 and AUG189 serve as TISs for AV2 protein synthesis
To examine the usage of in-frame AUG135 and AUG189 start sites along the AV2 mRNA for protein translation, we cloned the partial AV2 gene fragment ranging from the 125th to 227th nucleotides under the control of the 35S promoter and fused with a MYC tag (Figure 2D, p35S:AV2). We introduced single mutations at AUG135 (M135AUG→GCG) and AUG189 (M189AUG→GCG) and double mutations at both AUG135 and AUG189 into p35S:AV2 to generate the p35S:M135, p35S:M189, and p35S:M135/M189 constructs and used these constructs to reveal AUG135- and AUG189-initiated AV2-MYC proteins signals in Nicotiana benthamiana leaves transiently expressing these constructs (see “Materials and methods”). We then performed immunoblot analysis with anti-MYC antibodies. Similar protein bands were detected from leaves expressing p35S:AV2 (wild-type [WT]), p35S:M135, and p35S:M189, but not from leaves expressing p35S:M135/M189 (red arrows, Figure 2D). These results suggest that both AUG135 and AUG189 contribute to the initiation of AV2 protein expression.
To further explore whether the distinct AV2 protein isoforms are expressed during viral infection, we fused a FLAG epitope fragment in-frame with the C-terminus of the AV2 gene in a TYLCTHV infectious clone (pTYLCTHV-FLAG, Figure 2E), which allowed us to detect AUG135- and AUG189-initiated AV2-FLAG proteins. Proteins extracted from N. benthamiana leaves co-inoculated with pTYLCTHV-FLAG and the TYLCTHV infectious clone were used to detect endogenous AV2 proteins. Plants expressing the WT AV2 and AV2 with single mutations at AUG135 (M135) and AUG189 (M189) all showed AV2-FLAG protein signals, while those expressing the double mutant (M135/M189) AV2 showed much weaker signals (Figure 2E). Note that the mutation at AUG87 (M87AUG→GCG) did not affect AV2-FLAG protein signals (Supplemental Figure S3D). In addition, compared with the WT, the M189 mutant caused a slight reduction in signals, whereas the mutation at AUG135 substantially reduced AV2-FLAG expression (Figure 2E and Supplemental Figure S3C). These results, together with the finding that the primary 5′ end of the AV2 transcript is located downstream of the annotated AUG87 TIS (Figure 2C), indicate that AV2 gene expression is under the control of alternative transcriptional and translational start sites. Note that we assume that there would be transcripts with 5′ ends upstream of the annotated TIS of AV2 (i.e. AUG87) and thus the transcripts with the detected 5′ ends at nucleotides 125 (Figure 2C) are considered to be transcribed from an alternative transcription start site. Consequently, instead of the annotated TIS, the two downstream in-frame TISs, especially AUG135, initiate AV2 protein synthesis.
To assess whether the preferential usage of AUG135 is influenced by local mRNA sequence elements (Kozak, 1986, 1987; Merchante et al., 2017), we examined the flanking sequence contexts around the regions of AV2 TISs. Compared to AUG87 and AUG189, AUG135 had stronger Kozak sequence contexts, with an A at −3 position and a C and A at the −4 and −2 positions (Figure 2G), which are known to be associated with TIS activity in mammalian and tomato genes (Noderer et al., 2014; Li and Liu, 2020). Supporting this observation, analyses of the flanking sequence similarity between the viral TIS sites and the annotated AUG sites of tomato genes (Figure 2F) (shown as position weight matrix [PWM] scores, see “Materials and methods”) showed that AUG135 had higher PWM scores than AUG189 (Figure 2G). In addition, the AUG135 sites shared among begomoviruses showed higher PWM scores than the nearest upstream AUG sites (Figure 2H, see “Materials and methods”).
To validate the relationship between the flanking sequence contexts and the differential usage of AUG135 and AUG189 sites (Figure 2, D, E, and G), we inserted an additional “A” nucleotide into the region between these two AUG sites in the p35S:AV2 construct so that only the AUG189-initiated AV2 ORF was fused in-frame with MYC tag and could be detected via immunoblot analysis (left panel, Figure 2I). By further introducing mutations in the flanking sequences of AUG135 and AUG189, we found that, compared to the WT sequences, the poor initiation context (including the poor Kozak sequence with the T at −3 and C at +4 positions) of AUG135 (Poor135) led to higher AUG189-initiated AV2-MYC signals (red arrows, right panel in Figure 2I). The most substantial increase in AV2-MYC expression occurred in the double mutant of Poor135 and Perfect189 (i.e. the AUG189 site with perfect initiation contexts including the Kozak sequence with A at position −3 and G at position +4) (Figure 2I). These observations indicate that the usage of AUG189 was influenced by the initiation contexts of both AUG135 and AUG189 and suggest that AUG189 translation initiation can be activated by ribosomes bypassing AUG135 sites via a leaky scanning mechanism. Together, these findings show that the local mRNA sequence was associated with the differential usage of AV2 AUG135 and AUG189 TISs.
To assess the biological significance of alternative TISs in AV2 gene expression in begomoviruses, we examined the evolutionary conservation of these two in-frame TISs among different TYLCTHV isolates and in all begomoviruses (n = 242) listed in the International Committee on Taxonomy of Viruses (ICTV; https://talk.ictvonline.org). The in-frame AUG135 and AUG189 sites were observed across different TYLCTHV isolates found in different countries (i.e. Thailand and Taiwan in Supplemental File S1A). We further found that the AUG135 and AUG189 TISs, but not the annotated AUG87 TIS, were shared in >89% (216 and 213 out of 242) of begomoviruses (Supplemental File S1, B and C and Supplemental Data Set S1), including those that can infect different crop species such as sweet potato, cassava, and spinach (Figure 2J and Supplemental Data Set S1). Together, these results reveal the evolutionary conservation of these two in-frame AUG135 and AUG189 TISs, at least in the genus Begomovirus.
AUG135- and AUG189-encoded AV2 isoforms contribute to virus pathogenicity
Our data suggested that the AV2 gene can encode different protein isoforms via two in-frame TISs, AUG135 and AUG189 (Figure 2). To assess the biological roles of these protein isoforms in virus pathogenicity, we examined the effect of mutations in these in-frame AV2 TISs in TYLCTHV-infected tomato. Compared with the WT TYLCTHV, which induces significant yellow leaf curling symptoms in systemic leaves of tomato plants, mutations in AUG135 (M135AUG→GCG) and AUG189 (M189AUG→GCG) each led to attenuated disease symptoms (Figure 3, A and B and Supplemental Figure S4) and lower accumulation of viral DNA in infected leaves (Figure 3C). On the other hand, TYLCTHV with a mutation in the annotated TIS (M87AUG→GCG) had levels of disease symptoms and viral DNA accumulation similar to those observed for the WT virus (Figure 3, A and C), reflecting the lack of in vivo translation initiation activity at the annotated TIS of AV2 (Figure 2A). The AUG189 mutation (M189AUG→GCG; methionine to alanine) was designed to block the expression of the AUG189-initiated AV2 isoform, but it can also lead to a nonsynonymous mutation that changes the AUG135-initiated AV2 protein sequence. Thus, to minimize the off-target effects of the nonsynonymous AUG189 mutation in AUG135-initiated protein sequence/function, we also mutated the AUG189 site to GUA and UUA, which encode valine and leucine, respectively, with physico-chemical properties similar to those of methionine (Creixell et al., 2012; Machkovech et al., 2019). The different mutants all showed similar phenotypes: attenuated disease symptoms in the host plant (Figure 3, D–F).
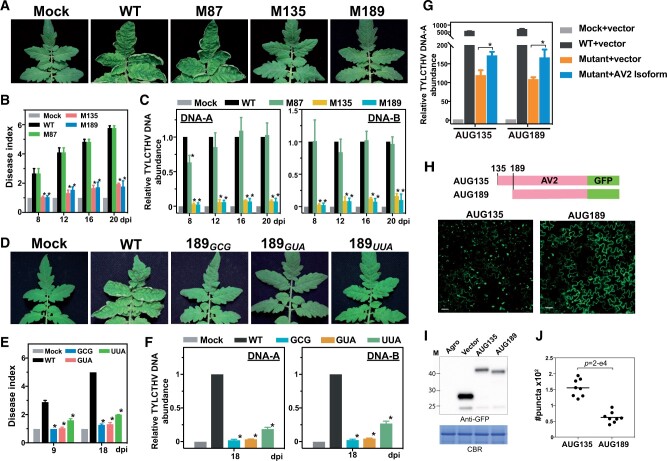
Figure 3.
Mutations of the downstream TISs in AV2 impair virus pathogenicity. A, Tomato plants infected with infectious clones with WT sequences, infectious clones with a mutation in the annotated AV2 TIS (M87AUG→GCG) or downstream TISs (M135AUG→GCG and M189AUG→GCG), and no viral DNAs (Mock). Photographs were taken at 20 dpi. B, Disease indexes representing the quantitative disease severities of the infected plants (indicated in (A)) at the indicated dpi. Data are shown as mean ± se from three biological repeats. In each repeat, three to four plants were included. The disease severity scores are defined in Supplemental Figure S4. *Statistically significant difference (P < 0.05) between plants expressing the WT infectious clone and those expressing infectious clones with mutations, as determined by Student’s t test. C, The relative abundance of DNA-A and DNA-B molecules in infected plants (indicated in (A)) determined by quantitative PCR analyses. D–F, As described in (A–C), but with the AUG189 (WT) site mutated to GCG, GUA, and UUA. G, The relative abundance of the DNA-A molecule as described in (C), but for N. benthamiana leaves inoculated with TYLCTHV infectious clones with AUG135 and AUG189 mutations (left and right groups, respectively), and with overexpression plasmids encoding AV2-MYC isoforms initiated at AUG135 and AUG189 (left and right groups, respectively; see “Materials and methods”). Mock and vector: N. benthamiana leaves infiltrated with agrobacteria without virus clones and with agrobacteria carrying the MYC-containing plasmid without the AV2 fragment (i.e. the plasmid without target gene sequences). *A statistically significant difference (P < 0.05) between mutants with/without AV2 isoform expression, as determined by Student’s t test. H and I, The subcellular localization (H) and expression (I) of the GFP-tagged AV2 proteins encoded by AUG135 and AUG189, revealed through confocal and immunoblot analyses, respectively. Scale bar = 10 µm. Agro and Vector: N. benthamiana leaves infiltrated with agrobacteria without an expression plasmid and with agrobacteria containing the empty expression vector (i.e. the plasmid without target gene sequences). J, Quantification of the AV2-GFP punctate structures in a given confocal image by ImageJ. The values from two leaves, with four different areas chosen for each leaf, are shown. P-values: test of whether the numbers of punctate structures calculated differ between AUG135- and AUG189-initiated protein isoforms (Mann–Whitney U test).
In addition, overexpression of the AUG189-initiated AV2 isoform in N. benthamiana leaves infected by the TYLCTHV M189 mutant (i.e. TYLCTHV infectious clone with the M189AUG→GCG mutation) had significantly higher viral DNA abundance than leaves infiltrated with the control vector and TYLCTHV M189 mutant alone (blue versus orange, Figure 3G). A similar pattern was also observed in leaves transformed with the TYLCTHV M135 mutant overexpressing the AUG135-initiated AV2 isoform (Figure 3G), showing that the expression of either isoform can partly recover the viral pathogenicity of the M135 and M189 virus mutants, respectively. Together, our results support the notion that the distinct AUG135- and AUG189-initiated protein isoforms function in pathogenicity.
To characterize the molecular features of distinct AV2 protein isoforms, we transiently expressed the GFP-tagged AV2 proteins in N. benthamiana leaves to reveal their subcellular locations (Figure 3I). In line with previous studies of AV2 protein localization (Rojas et al., 2001; Moshe et al., 2015), both AUG135- and AUG189-initiatied protein isoforms were localized to the cytosol and nucleus (Supplemental Figure S3E). Intriguingly, while the AUG135-initiated isoform formed significantly more cytoplasmic punctate dots, as revealed in previous studies (Moshe et al., 2015; Zhao et al., 2018), more of the AUG189-initiated isoform was localized at the cell periphery (Figure 3, H and J), showing the differential localizations of these two AV2 isoforms. In addition, the reciprocal complementation tests (i.e. overexpressing the AUG189-initiatied protein isoform in the TYLCTHV M135 mutant and vice versa) showed that the overexpression of a given isoform was unable to recover the viral DNA contents of the viral mutant in question (Supplemental Figure S3F). Together, these findings suggest that AUG135- and AUG189-initiatied protein isoforms may play distinct roles in pathogenicity.
AUG174 but not AUG 147 is required for BV1 translation and viral pathogenicity
BV1 is a nuclear shuttle protein that transports viral single-stranded DNAs between the nucleus and cytoplasm (Fondong, 2013; Hanley-Bowdoin et al., 2013; Basak, 2016). We found that the annotated AUG start site (at position 147) of BV1 did not show initiation activity. On the contrary, a downstream in-frame AUG site at position 174 overlapped with a TIS peak (Figures 1F and 4A; Supplemental Table S1) and had the highest LTM signal among alternative TISs (square indicated by arrow, Figure 1H). These observations suggest that the AUG174 site is likely the in vivo TIS of the BV1 gene. 5′ RACE further showed that the 5′ transcript end of BV1 was mainly located at nucleotide 165, a position between the annotated AUG147 and the downstream in-frame AUG174 TISs (blue, Figure 4C), which is consistent with the read coverages revealed in the total RNA sample (bottom panel, Figure 4B). These results indicate that the usage of the AUG174 site as a TIS was due to the production of a transcript with a 5′ end that differs from that previously annotated.
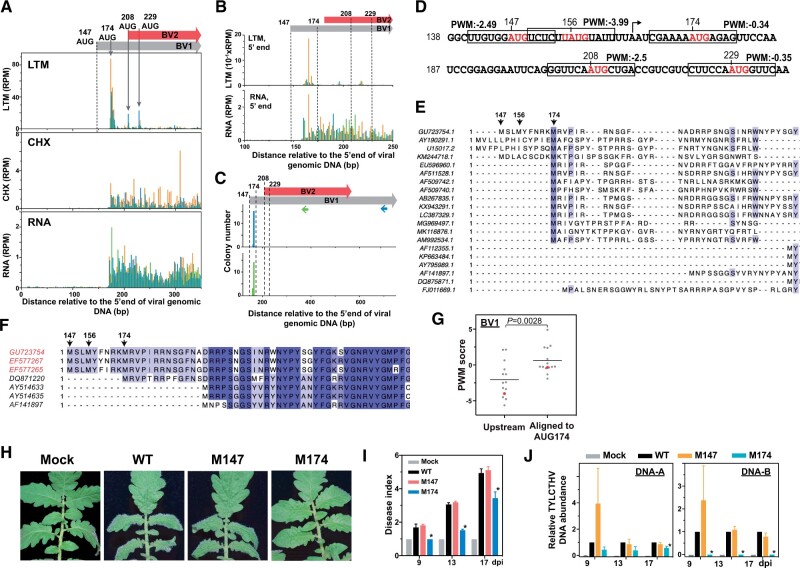
Figure 4.
A downstream TIS is used for BV1 expression and is required for virus infection. A, As described in Figure 1, A–C, but for the LTM, CHX and RNA read densities along the plus strand of the BV1 and BV2 genic regions. The annotated TIS (AUG147; dashed line) and one downstream TIS (AUG174) of BV1 and two TISs (AUG208 and AUG229) of BV2 supported by LTM signals (gray arrows) are indicated. B, As indicated in (A), but for the positional distributions of the 5′ ends of LTM and RNA reads. C, The positional distribution of the 5′ ends of the BV1 and BV2 transcripts (gray and red boxes; top panel) as described in Figure 2A. Two sets of results (middle and bottom panels) generated from two distinct primers targeting BV1 (blue arrow in top panel) and both BV1 and BV2 (green arrow in top panel) are shown. D, As described in Figure 2G, but for PWM scores of AUG sites from the region ranging from 138 to 237 nt. E and F, As described in Figure 2J, but for the amino acid sequence alignments of annotated BV1 genes among 20 begomoviruses (E) and TYLCTHV isolates found in Taiwan (red) and in Thailand (black) (F). For all begomoviruses, see Supplemental File S2. G, As described in Figure 2H, but for the PWM scores of AUG sites of begomovirses that were aligned to AUG174 of TYLCTHV DNA-B molecule and the associated nearest upstream AUG sites. Red: the AUG174 site of TYLCTHV DNA-B and its associated nearest upstream AUG sites. H–J, As described in Figure 3, A–C, but for the phenotypes (H), disease indexes (I), and viral DNA abundances (J) of tomato plants infected with infectious clones with WT sequences, with mutations at the annotated TIS (M147ATG→GCG) or downstream TIS (M174ATG→GCG), and without viral DNAs (Mock). Photographs were taken at 17 dpi. The annotated TIS at position 147, the in vivo TISs at positions 174, 208, and 229 and the AUG site at position 156 that is identified based on sequences are indicated.
The analysis of local mRNA sequences around BV1 TISs showed that compared with the AUG147 (the annotated TIS) and AUG156 sites (the downstream in-frame AUG site) in BV1, AUG174 (the downstream in-frame AUG site with LTM signals) had higher PWM scores (Figure 4D). In addition, sequence alignment revealed that AUG174 start sites were more commonly found than AUG147 in some begomoviruses and among TYLCTHV isolates (Figure 4, E and F; Supplemental File S2). The AUG174 sites shared among begomoviruses showed higher PWM scores than the associated upstream AUG sites (Figure 4G). Collectively, these observations suggest that the annotated AUG147 site is unlikely to serve as a TIS and that the AUG174 site is the in vivo TIS of the BV1 gene.
We then generated mutations in the BV1 AUG147 (M147AUG→GCG) and AUG174 (M174AUG→GCG) TISs to assess their biological roles during TYLCTHV infection. Compared with the WT infectious clone, the clone with a mutation at location 174 (M174AUG→GCG), but not the clone with a mutation at location 147 (M147AUG→GCG), displayed significantly delayed leaf-curling symptom development (Figure 4, H and I) and also led to lower accumulation of viral DNAs, especially the DNA-B molecule (Figure 4J). These data support the hypothesis that the downstream in-frame AUG174 start site is required for BV1 expression and function in virus pathogenicity.
Identification of an unexplored ORF, BV2, during virus infection
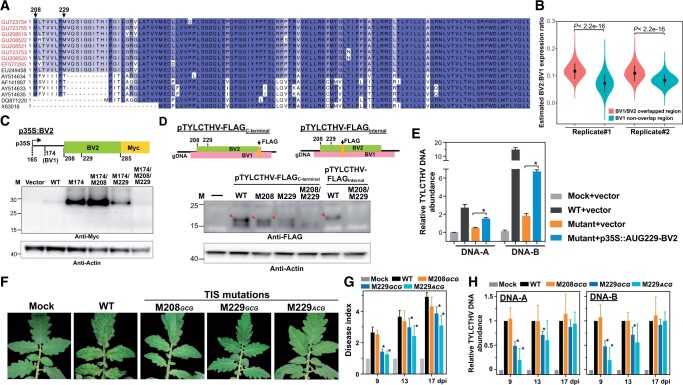
Among the 10 experimentally derived TISs that encode distinct ORFs in TYLCTHV (red arrows, Figure 1, C and F; triangles, Figure 1, H and I), two in-frame TIS peaks were located at a single unannotated ORF and can initiate translation of 120 and 113 amino-acid protein isoforms with alternative N-termini (triangles indicated by arrows, Figure 1, H and I and Supplemental Table S1). The ORF identified here is not a commonly reported ORF among begomoviruses, but has been predicated as the BV2 gene in the TYLCTHV isolates found in Thailand (AY514633, AY514635, and AF141897) (Figure 5A). Nevertheless, the biological significance of BV2 remains unclear.
Figure 5.
Characterization of an unexplored BV2 gene that affects virus pathogenicity. A, As described in Figure 2J, but for the amino acid sequence alignments of the BV2 gene from different TYLCTHV isolates found in Taiwan (red) and in Thailand (black). The identified TISs (208 and 229; black arrows) in TYLCTHV are indicated. B, The estimated BV2/BV1 translation expression ratio for the overlapping regions between BV2 and BV1 (pink) and nonoverlapping region of BV1 (cyan) in two biological replicates shown for the ratios generated by 100,000 bootstrap resampling. P-values are derived from two-tailed Kolmogorov–Smirnov test. C, As described in Figure 2D, but for the protein expression of the BV2 gene with translation driven by the WT sequence from the region ranging from 165 to 285 nt or sequences with single or double mutations at positions 174, 208, and 229 (M174AUG→GCG, M208AUG→GCG, and M229AUG→GCG, respectively). D, As described in Figure 2E, but for FLAG-tagged BV2 proteins (red arrows) expressed during virus infection. The FLAG DNA fragment was inserted into the infectious clone at an internal site or the C-terminal end of the BV2 gene. Results are shown for infectious clones with the WT sequence or the indicated mutations at positions 208 and/or 229. E, As described in Figure 3G, but for the viral DNA-A and DNA-B abundances in N. benthamiana leaves inoculated with TYLCTHV M229AUG→ACG mutant, and with the overexpression plasmid encoding AUG229-initiated BV2-MYC (p35:AUG229-BV2). Mock and Vector: N. benthamiana leaves infiltrated with agrobacteria without virus clones and with agrobacteria carrying the MYC-containing plasmid without the BV2 fragment. F–H, As described in Figure 3, A–C, but for the phenotypes (F), disease indexes (G), and viral DNA abundances (H) of tomato plants infected with infectious clones with WT sequences, with a mutation at the 208 (M208AUG→GCG) or 229 (M229AUG→GCG and M229AUG→ACG) TIS site of BV2, and without viral DNAs (MOCK). Photographs were taken at 13 dpi.
By analyzing the sequence identity among begomoviruses and TYLCTHV isolates, we further found that BV2 was present in TYLCTHY isolates found in Taiwan and Thailand, with amino acid identities ≥ 78% (Figure 5A). To examine whether BV2 exists in other begomoviruses, we first identified the putative AUG-initiated ORFs in the DNA-B molecules of all begomoviruses via sequence prediction (see “Materials and methods”; Supplemental Figure S5A). The pairwise comparisons of amino acid sequences between the predicted AUG-ORFs (n = 48) and BV2 showed sequence identities ranging from 10.5% to 25.8%, with the median being 14.3% (Supplemental Figure S5A), indicating that BV2 was not conserved in begomoviruses.
A coding region generally has characteristic 3-nt periodicity (i.e. phasing patterns), which can be revealed via CHX data sets, while a dual-coding region has mixed phasing patterns affected by the translatability of the overlapping genes (Lulla and Firth, 2020). Thus, by quantifying the differences of phasing in overlapping and nonoverlapping regions (see “Materials and methods”), we found that the relative expression levels of BV1 and BV2 were ∼0.89 and ∼0.11, respectively. This observation indicates that BV2 was translated at ∼11% the level of BV1 in the overlapping regions, which is significantly higher than that estimated from the nonoverlapping regions (Figure 5B). It should be noted that the protein expression assay is expected to provide more accurate quantification than the CHX phasing analysis performed here (Lulla and Firth, 2020).
Together, these observations provide translational evidence for an unexplored BV2 gene, which is evolutionarily conserved among TYLCTHY isolates.
Molecular characterization of BV2 protein synthesis
Since BV2 is a novel and unexplored viral gene, we next aimed to characterize the molecular basis of its gene expression. 5′ RACE analysis showed that the 5′ end of the BV2 transcript was primarily located at nucleotide 165, the same as that of BV1 (green, Figure 4C), suggesting that the BV1 and BV2 proteins can be translated from a single mRNA molecule via alternative translation initiation. Note that BV2 is nested within the BV1 gene, but they are in different reading frames. Thus, the 5′ RACE primer used for BV2 targets both the BV1 and BV2 coding regions (green arrow, Figure 4C), while the 5′ RACE primer used for the BV1 gene specifically targets the BV1 coding region (blue arrow, Figure 4C). In addition, LTM profiling revealed two in-frame AUG TISs at positions 208 and 229 of the genomic DNA-B (Figure 4A and Supplemental Table S1). Collectively, these results suggest that the BV2 protein can be translated from a bicistronic mRNA with two in-frame TISs initiating protein expression.
To test this hypothesis, we cloned the BV1 and BV2 TIS-containing regions (ranging from nucleotides 165 to 285) under the control of the 35S promoter and fused them with MYC (p35S:BV2, Figure 5C). In p35S:BV2, the BV2, but not BV1, TIS sites were in-frame with MYC, thus allowing us to detect AUG208- and AUG229-initiated BV2-MYC proteins. In addition, since the AUG174 TIS site of BV1 showed much stronger LTM signals than the AUG208 and AUG229 TIS sites of BV2 (Figure 4A), we also mutated the AUG174 site to block its TIS activity, thus facilitating the detection of AUG208 and AUG229 TIS activities, if any. Mutations in AUG174 (M174AUG→GCG; the BV1 TIS site), AUG208 (M208AUG→GCG; the BV2 TIS site), and AUG229 (M229AUG→GCG; the BV2 TIS site) were generated in p35S:BV2 to produce mutants with different combinations of single/double/triple mutations, which were then used to reveal BV2-MYC protein signals in N. benthamiana leaves transiently expressing these constructs. Immunoblot analysis using anti-MYC antibodies revealed BV2 signals in leaves expressing the WT, M174, M174/M208, and M174/M229 constructs (Figure 5C); however, no signals were observed in the triple M174/M208/M229 mutant (Figure 5C). These results indicate that both AUG208 and AUG229 can initiate BV2 protein expression from a single mRNA transcript containing both BV1 and BV2. The observation of this bicistronic transcript is similar to the previous findings that AC2 and AC3 are translated from a single transcript in Mungbean yellow mosaic virus-Vigna, a bipartite begomovirus (Shivaprasad et al., 2005; Fondong, 2013).
To further explore whether the distinct BV2 protein isoforms are expressed during viral infection, we inserted a FLAG epitope in-frame with the C-terminus of the BV2 gene in the TYLCTHV infectious clone (pTYLCTHV-FLAGC-terminal, Figure 5D) to detect AUG208- and AUG229-initiated BV2-FLAG proteins. Note that the AUG174 site for BV1 protein synthesis remained intact and was not mutated here. Proteins extracted from N. benthamiana leaves co-inoculated with pTYLCTHV-FLAG and the TYLCTHV infectious clone were used to detect endogenous BV2 proteins during infection. Leaves expressing the WT pTYLCTHV-FLAGC-terminal or pTYLCTHV-FLAGC-terminal with single M208 and M229 mutations had detectable BV2 signals (red arrows, Figure 5D), while those expressing the double M208/M229 mutant form did not. In addition, when the FLAG tag was inserted in-frame into the region downstream of the BV2 TISs (pTYLCTHV-FLAGinternal, Figure 5D), the BV2 signal was clearly not detected in the M208/M229 double mutant (Figure 5D). Lastly, we noticed that the M229 mutation led to much weaker BV2 signals than the M208 mutation, a pattern observed in both the p35S-BV2 and pTYLCTHV-FLAGC-terminal systems (Figure 5, C and D). Supporting this observation, the analysis of flanking sequence contexts showed that AUG229 had stronger Kozak motifs with G at +4 positions and higher PWM scores (Figure 4D).
In summary, our findings demonstrate the presence of an unexplored BV2 gene that is nested within the BV1 gene and expressed mainly from the AUG229 site, which was associated with the surrounding mRNA sequences, during viral infection.
BV2 facilitates virus pathogenicity
To this point, we focused on the molecular characteristics of BV2 gene expression during virus infection (Figure 5). To assess the biological significance of BV2 in virus pathogenicity, we mutated the AUG208 and AUG229 sites to GCG in the TYLCTHV infectious clone to inhibit BV2 expression during virus infection. The M229AUG->GCG TIS mutation, but not the M208AUG->GCG, showed significantly attenuated leaf curling symptoms and decreased the viral DNA abundance compared with WT (Figure 5, F–H). The AUG229 site was also mutated to ACG, which abolished the AUG TIS site of BV2 while leaving the BV1-encoded amino acid sequences in the TYLCTHV infectious clone intact. The M229AUG->ACG mutation showed significantly delayed disease symptoms (Figure 5, F–H). In addition, the overexpression of AUG229-initiated BV2 in N. benthamiana leaves infected by the TYLCTHV M229AUG->ACG mutant led to significantly higher viral DNA abundances compared with leaves infected by the TYLCTHV M229 mutant alone (blue versus orange, Figure 5E). These results, together with the finding that AUG229 plays a major role in BV2 expression (Figure 5, C and D), support the hypothesis that BV2 facilitates TYLCTHV infection in plants.
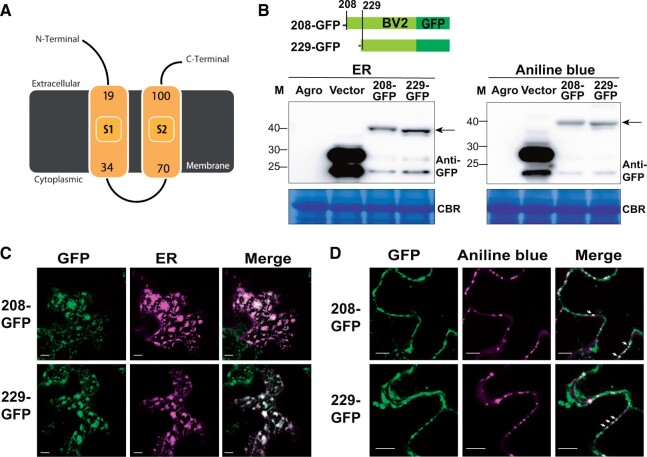
To explore the mechanistic role by which BV2 promotes viral infection/pathogenesis, we analyzed the functional domains of the BV2 protein. Transmembrane domains were predicted in BV2 using Phyre2 and InterPro (Figure 6A; Supplemental Figure S5B; Kelley et al., 2015; Blum et al., 2021). To validate the prediction, we further assessed the subcellular localization of BV2 by fusing the BV2 gene with GFP and transiently expressing the fusion protein in N. benthamiana leaves (Figure 6B). The GFP signals of AUG229-initiated BV2 colocalized with an ER marker (Figure 6C) and formed aggregated structures overlapping with aniline blue signals; aniline blue is a dye that stains callose in plasmodesmata (PD) (arrows, Figure 6D). Similar patterns were also observed for the other BV2 protein isoform initiating from the AUG208 TIS (Figure 6, B–D). Note that the distribution of the two BV2 protein isoforms at the cell periphery (Figure 6D) was likely due to their localization in the ER and sites of ER-PD convergence (Staehelin, 1997; Wang et al., 2017), since the GFP signals at the cell periphery colocalized with an ER marker (Supplemental Figure S5C). These results indicate that BV2 preferentially localizes to the ER and PD, which are parts of the membrane systems necessary for viral replication and cell-to-cell trafficking (Hanley-Bowdoin et al., 2013; Heinlein, 2015; Griffing et al., 2017).
Figure 6.
Subcellular localization of BV2 in the ER and PD. A, Plot of BV2 protein structure showing the predicted transmembrane domains (yellow) analyzed and generated via Phypy2 (http://www.sbg.bio.ic.ac.uk/phyre2). B–D, As described in Figure 3, H and I, but for the protein expression (B) and subcellular localization (C, D) of the GFP-tagged BV2 protein initiated from AUG208 or AUG229. ER marker: CD3-959 (Nelson et al., 2007). Aniline blue: a PD dye. Scale bar = 10 µm.
Discussion
By systematically mapping the in vivo TISs during virus infection, we revealed the TYLCTHV translational profiles at single-gene resolution and uncovered the global coding potential of this viral genome. Specifically, we found that a number of unanticipated AUG TISs are used in vivo and lead to different protein isoforms or encode new ORFs (Figure 1). The use of these identified TISs likely occurs via both alternative transcriptional and translational initiation start sites and was associated with the local mRNA sequences (Figures 2 and 4). We further experimentally confirmed that two protein isoforms are initiated from two downstream TISs of a single AV2 gene and that both are involved in symptom development (Figures 2 and 3). Lastly, we demonstrated that BV2, an annotated but not functionally characterized ORF, is nested within a known genic region and plays a pathogenic role (Figures 5 and 6). These results indicate that the translational start sites used for virus gene expression are more diverse than previously anticipated and are required for initiating the translation of viral factors that function during virus infection of the host.
Our technique employing translation initiation ribosome profiling provides an in-depth experimental investigation of viral TISs used in vivo that is not limited by previous knowledge of annotated genes and complements the previous comparative genomics-based approaches/studies to identify viral ORFs (Chung et al., 2008; Ling et al., 2013; Smirnova et al., 2015). The findings of prevalent hidden ORFs (Figure 1H) indicate that the coding potential of TYLCTHV is higher and more complicated than predicted by sequence-based in silico analyses (Figure 1). In addition, our findings highlight the importance of applying a high-throughput in vivo approach such as initiation ribosome profiling to provide a comprehensive catalog of viral ORFs in genomes and facilitate the identification of viral factors for plant virus studies.
Our findings showing prevalent novel ORFs (Figure 1H) further raise the next grand question: to what extent and how do the newly identified ORFs coordinate TYLCTHV viral processes and function in pathogenesis? Viral short ORFs have been suggested to function as cis-regulatory elements to influence the translation of neighboring genes and/or as trans-acting factors encoding small peptides that regulate different processes during the viral lifecycle (Andrews and Rothnagel, 2014; Hellens et al., 2016; Finkel et al., 2018). For example, in the herpesvirus, the translation of ORF35.1 and ORF35.2 has a cis-regulatory function in the translation of the downstream ORF35 and ORF36 (Arias et al., 2014). A small non-AUG-initiated ORF (ORF3a) in poleroviruses and luteoviruses functions as both a cis-element and trans-factor to alter the protein expression of downstream viral genes along a transcript and also encodes a small protein required for long-distance movement (Smirnova et al., 2015). Our studies suggest that the AV2 protein isoforms and novel BV2 proteins encoded during infection are involved in viral pathogenicity in trans (Figures 2, 3, and 5); however, whether they could also coordinate the expression of the surrounding genes in cis remains to be explored. Further computational/statistical analyses of the results of conservation and experimental investigations will shed light on the functions and mechanisms of the newly identified ORFs, including the AV2 and BV2 genes (Hellens et al., 2016; Finkel et al., 2018).
The preferential usage of TISs in a single viral mRNA can be influenced by noncanonical translational strategies including leaky scanning or cap-independent initiations and are dependent on sequence contexts such as short 5′ UTR length, Kozak motifs, internal ribosome entry site or tRNA-like/mRNA secondary structures (Sedman et al., 1990; Kozak, 1991; Hull, 2014; Miras et al., 2017). The correlation between TIS activity and the flanking mRNA sequence contexts in TYLCTHV AV2/BV1/BV2 genes (Figures 2 and 4) suggests that the local mRNA sequence and the leaky scanning mechanism likely play a role in TIS utilization and further highlights the impact of translational initiation mechanisms on viral gene expression in begomoviruses. Intriguingly, while the AUG135/AUG189 in AV2 mainly initiate protein synthesis in TYLCTHV and are shared in >89% of begomoviruses (Figure 2), we noticed that 9% (22 out of 242) of begomoviruses had an AUG site upstream in-frame to the shared AUG135/AUG189 sites, an observation similar to that for the annotated AUG87 in TYLCTHV (Figure 2J and Supplemental File S1, B and C). A similar observation was also found for the annotated AUG147 sites and the shared AUG174 sites in BV1 in some begomoviruses (Figure 4E); whether some of these AUG sites preferentially serve as TISs and by which mechanisms remain to be determined. Further work on the in vivo TIS activities of these annotated and downstream in-frame AUG sites in various begomoviruses will provide a more comprehensive view of viral gene expression strategies and address the translational regulation mechanisms of TIS usage in plant begomoviruses.
In addition, we identified the in vivo TISs using samples of the systemic leaves with observed disease symptoms and focused on the TISs present in replicates, which could identify the robust in vivo TIS sites, but at the cost of possibility missing the stage-specific TISs. For example, there were few TIS signals in the AC1 and AC3 genes (Figure 1C and Supplemental Figure S2B), which encode replication initiation and enhancer proteins required for viral DNA replication during early stages of infection (Fondong, 2013; Prasad et al., 2020). Since viral factors play distinct roles at different stages of viral lifecycles (Fondong, 2013; Prasad et al., 2020), further work on temporal-specific profiling will provide a more comprehensive and dynamic view of viral gene expression and address the temporal-specific translational regulation mechanisms of TIS usages during viral infection processes.
The AV2 gene acts as a pathogenic determinant by interacting/co-localizing with multiple viral factors including AV2 itself and AV1, which might facilitate virus particle trafficking (Moshe et al., 2015; Zhao et al., 2018). AV2 also interacts with host proteins, including the suppressor of gene silencing 3, CYP1, Argonaute 4, and histone deacetylase 6 to influence host RNA silencing, the hypersensitive response, and DNA methylation to alleviate the host defense response (Glick et al., 2008; Bar-Ziv et al., 2015; Wang et al., 2018; Wang et al., 2019). Our results demonstrate that two distinct protein isoforms are encoded from the AV2 gene, which both function in viral pathogenicity and are located in different subcellular compartments (Figures 2 and 3). Since these two protein isoforms and the N-terminal protein sequences that differ between isoforms did not show any predicted functional domains, it would be worthwhile to investigate whether the AV2 isoforms have similar/distinct biological functions such as silencing suppressor activities. The mechanisms by which the AV2 isoforms are involved in the viral lifecycle and virus–host interactions are also worthy of investigation.
We found that BV2, a previously annotated but not functionally characterized viral ORF, encodes an ER- and PD-localized viral factor that functions in pathogenicity (Figures 4–6). The finding that BV2 is conserved among TYLCTHY isolates (Figure 5), but not in begomoviruses (Supplemental Figure S5A), was not surprising since the BV1 sequence identities between TYLCTHV and other begomoviruses were also low (∼22.2%–68.9% identities, with a median of 26.6%) (Supplemental File S2C) and the DNA-B molecules, in which BV1 and BV2 are located, show greater genetic variations than DNA-A components (Briddon et al., 2010). In addition, the ER and PD are key checkpoints along the membrane pathway that serve as virus replication sites and trafficking routes both intracellularly and intercellularly (Hanley-Bowdoin et al., 2013; Heinlein, 2015; Griffing et al., 2017). Several viral membrane proteins are involved in the virus replication and trafficking process. For example, the p30 movement protein of tobacco mosaic virus is a PD-localized membrane protein that reshapes PD pores and then enables viral spreading in plants (Beachy and Heinlein, 2000). The 6K2 protein of potyvirus and the triple gene block 2 and 3 (TGB2/TGB3) proteins of potato virus X are small integral membrane proteins located in the ER/PD that facilitate replicative vesicle formation and directional trafficking (Grangeon et al., 2013; Tilsner et al., 2013; Wu et al., 2019b). The observation of BV2 localization in the ER and PD (Figure 6) supports its role in facilitating TYLCTHV infection in tomato and suggests the possibility of BV2 functioning in virus spreading and/or the formation of replicative vesicles in hosts. Thus, the stages of the viral lifecycle in which ER- and PD-localized BV2 function and by which mechanisms BV2 facilitates virus infection are worthy of investigation in the future.
In summary, our studies provided much-needed insight into the extent that plant DNA viruses use canonical and noncanonical initiation sites to encode diverse viral factors in small genomes and, more importantly, to enable infection of hosts. Viruses also manipulate elongation and termination processes to trigger frameshift or stop codon read-through in order to expand their protein repertoire (Lewandowski and Dawson, 2000; Hull, 2014; Jaafar and Kieft, 2019). Thus, revealing in vivo TISs represents a key first step (but not the only step) in uncovering the coding potentials of viral genomes. Future studies on ribosomal frameshifting and read-through using ribosome profiling and proteogenomic approaches will further facilitate the identification of the elusive coding features of viral genomes.
Materials and methods
Plant and virus materials
The tomato (S. lycopersicum cv. ANT22) seeds and the TYLCTHV DNA-A/-B infectious clones (GenBank accession: GU723742/GU723754) were obtained from the World Vegetable Center. Approximately 21-day-old tomato plants grown in a growth chamber under a 12-h light (8:00 to 20:00, 150 µmol m−2 s−1 provided by light-emitting diodes [LED] SUN LIGHT Z RGB bulbs [HIPOINT, Taiwan])/12-h dark cycle were inoculated with Agrobacterium tumefaciens LBA4404 containing the infectious TYLCTHV DNA-A and -B clones as described previously (Tsai et al., 2011). To confirm that the viral DNAs that replicated in plants did not contain any unexpected mutations, the genomic DNAs from systemic leaves of infected plants were extracted as described previously (Tsai et al., 2011) and used as templates in polymerase chain reaction (PCR) analysis to amplify the whole virus genome sequence with the primers listed in Supplemental Table S2. Amplified sequences were then analyzed by Sanger sequencing.
Chemical treatments and isolation of RPFs and total RNAs
The systemic leaves of TYLCTHV-infected plants with yellow leaf curl phenotypes were sampled for total RNA and RPF RNA preparation. All tomato leaf samples except for LTM-treated samples were immediately frozen in liquid nitrogen. For LTM treatment, the excised leaves were treated with 30-µM LTM (dissolved in DMSO, Merck#506291) for 30 min before harvesting as described previously (Li and Liu, 2020). Two biological replicates (i.e. two separate sets of plants sampled on different days, each containing pooled leaf tissues from individual plants) were harvested.
RNA samples were prepared for the CHX, total RNA, and LTM datasets as described previously (Li and Liu, 2020). In brief, to purify the RPF samples for the CHX datasets, the polysome complexes were isolated from the plant tissue, which was ground to a powder, using polysome extraction buffer (20-mM HEPES, 100-mM KCl, 5-mM MgCl2, recipe from Li and Liu [2020]) containing 100-µg·mL-1 CHX, centrifuged at 13,000 g at 4°C for 5 min, and digested with RNase I. The purified RPFs were further resolved in a 15% TBE-UREA polyacrylamide gel (Invitrogen) and the gel slices corresponding to the 26–32 nt region were excised for library construction. Total RNAs were purified using a PureLink Plant RNA Reagent (Invitrogen; #12322012) and an Ribo-Zero rRNA depletion kit (MRZ11124C, Illumina) was then used for rRNA removal. To purify the RNA samples for the LTM datasets, the polysome complexes were isolated from LTM-treated plants, which were ground to a powder, using polysome extraction buffer, centrifuged at 13,000g at 4°C for 5 min, subjected to puromycin treatment, and processed for RPF purification as described above for RPF purification from CHX samples. The Illumina Hiseq-2500 (single 75-nt end reads) platform was used for the generation of sequences.
Sequencing data processing
Read mapping and the determination of the P-site of a read for the CHX, LTM, and total RNA samples were performed as described previously (Li and Liu, 2020). The reads with P-site assignment were applied in all figures, except that the positions of LTM and total RNA reads were assigned to the 5′ ends of reads in order to obtain better resolution of the 5′ transcript ends in Figures 2B and 4B (Stern-Ginossar et al., 2012).
The TYLCTHV DNA-A and -B genome sequences and gene models (GU723742/GU723754) were retrieved from GenBank (https://www.ncbi.nlm.nih.gov/genbank/; Supplemental Data Set S1). The S. lycopersicum genome sequences and gene models were based on the genome versions SL3.0 and ITAG3.2 (https://solgenomics.net). The number of mapped reads in each biological replicate is shown in Supplemental Table S3. The data reproducibility was high among replicates of the LTM, CHX, and total RNA samples (Spearman’s rank correlation coefficient > 0.98, P < 2.2 × 10−16) (Supplemental Figure S1D); thus, the reads from replicates were pooled and used to calculate the read coverage in all figures except Supplemental Figures S1, S2A, and S3A, which show signals of reads separately for each replicate.
Identification of in vivo TISs
To identify an LTM peak, which represents an in vivo TIS in a viral genome, a given peak was required to meet the following criteria (pipeline modified from Lee et al. [2012]; Gao et al. [2015]; and Li and Liu [2020]): (1) the position in question has ≥ 10 LTM reads and shows a local maximum of LTM read counts in a 7-nt window (−3, +3) flanking the position in question; (2) the difference between the normalized read intensities (R) of LTM and CHX data is ≥ 0.05. R was calculated as follows: , where X is the number of reads mapping to the position in question and N is the total number of reads mapping to that transcript. When AUGs or near-cognate codons were within 2 nts preceding or succeeding the codon corresponding to the identified TIS peak, the position of the AUG or near-cognate codons was designated as an identified TIS peak. Only the TIS peaks present in both biological repeats were included in downstream analyses (Supplemental Table S1).
5′ RACE
Total RNAs (2–3 μg) were extracted from virus-infected plant leaves using PureLink Plant RNA Reagent (Invitrogen, #12322012) and used for 5′ RACE assays performed according to the manual of the SMARTer RACE 5′ Kit (TakaRa). The PCR products were cloned into the pJET1.2 vector (provided in the kit) and sequenced with gene-specific nested primers using Sanger sequencing to reveal the transcript 5′ ends. Note that the cap structure at the 5′ transcript end can be recognized as guanine (G) during reverse transcription, which leads to additional Gs in the resulting reads (Cumbie et al., 2015; Schon et al., 2018); thus, only the clones with ≥1 additional Gs in the 5′ ends of the resulting reads were included in 5′ RACE data sets to ensure the identification of 5′ transcript ends. The gene-specific primers for cDNA synthesis and gene-specific nested primers for PCR product amplification are listed in Supplemental Table S2.
Generation of protein expression constructs and infectious viral clones with mutations and expression tags
To detect the protein isoforms of AV2 initiated from AUG135 and AUG189 (Figure 2), the genomic region of the AV2 gene ranging from nucleotide 125 (i.e. the 5′ transcript end, Figure 2C) to nucleotide 227, which is located in the N-terminus of the AV2 gene, was chosen. For BV2 protein expression initiated from AUG208 and AUG229 (Figure 5), the genomic region of BV2 ranging from nucleotide 165 (i.e. the 5′ transcript end, Figure 4C) to nucleotide 285 was chosen. The aforementioned DNA fragments were cloned into pCR8 and mutated at the indicated sites, if any, and then fused with 10xMYC in the Gateway pGWB520 plasmid. To reveal expression of the endogenous AV2 and BV2 proteins during virus infection, 3xFLAG was inserted at the end of AV2 and BV2 and in the middle of BV2 at the 264 position via restriction enzyme digestion. The primers used are listed in Supplemental Table S2.
To generate the infectious clones of TYLCTHV DNA-A and -B, a head-to-tail partial dimer of viral genomes was cloned into the pCambia0380 binary vector via restriction enzyme digestion as reported previously (Tsai et al., 2011). To introduce mutations in the infectious clones, two viral DNA fragments were released by enzyme digestion, cloned into the pGEMT/pUC19 vector, mutated via site-direct PCR-mutagenesis, and then cloned back into the pCambia0380 binary vector as described previously (Tsai et al., 2011).
To reveal the subcellular localization of proteins, the viral DNA fragment encompassing the indicated region was amplified by PCR using infectious clones as a template and with primers listed in Supplemental Table S2, and then cloned into the pk7FWG2-eGFP Gateway destination vector.
All site-direct mutagenesis of the tested sites of genes was performed according the manufacturer’s instructions (Q5 Site-Directed Mutagenesis Kit, E0552S, NEB) using the primers listed in Supplemental Table S2.
Quantitative PCR analysis
Genomic DNA was purified from the indicated virus-infected plants as described previously (Tsai et al., 2011) and used for quantification of viral DNAs with qPCRBIO SyGreen Mix (PCR Biosystems Ltd.). The products were analyzed on a BioRad Real-Time PCR System. The relative abundances of viral DNAs were calculated using the ΔCt (threshold cycle) method. The ACTIN gene was used as an internal control. Primers were designed to target regions of viral genomes and are listed in Supplemental Table S2.
Assays for expression analysis, subcellular localization, and functional complementation of viral proteins
To reveal protein expression initiated from a given TIS in a transient expression system (Figures 2, D and I and 5C) and the expression of endogenous proteins encoded by viral genes during virus infection (Figures 2E and 5D), leaves of 3- to 4-week-old N. benthamiana plants grown at 25°C with a 12-h light/12-h dark period were infiltrated with A. tumefaciens strain LBA4404 carrying protein expression constructs and collected after 2–3 days for protein expression assay via immunoblotting. Total protein extraction and detection were performed as described (Liu et al., 2013). The primary MYC-specific (A00173-40, GenScript), GFP-specific (11814460001, Roche), FLAG-specific (F1804, Sigma), and Actin-specific (A0480, Sigma) antibodies were used at concentrations of 1:1,000–1,500, 1:5,000, 1:1,500–3,000, and 10,000, respectively. The anti-mouse (W4021, Promega) and anti-rabbit (W4011, Promega) HRP-coupled secondary antibodies were used at a concentration of 1:10,000–100,000 for detecting ACTIN and 1:5,000–20,000 for detecting MYC/GFP/FLAG fusion proteins via chemiluminescent detection (WBKLS0500, Millipore; 34095, Thermo).
To reveal protein localization, leaves of 3- to 4-week-old N. benthamiana plants grown at 25°C with a 12-h light/12-h dark photoperiod were transformed with GFP-tagged protein expression constructs via Agrobacterium inoculation. For the ER localization assay, GFP-tagged protein expression constructs were co-inoculated with the CD3-959 ER-mCherry marker (Nelson et al., 2007). For aniline blue staining, leaves of N. benthamiana at 38 h after inoculation were infiltrated with 25 µg·mL-1 aniline blue (Biosupplies, 100-1) for 30 min and analyzed under a Zeiss LSM 710 confocal laser microscope. To quantify the punctate-like structures shown in Figure 3J, light signals from images acquired from the N. benthamiana leaves at 2–3 days post inoculation (dpi) with a Zeiss LSM 710 confocal microscope were quantified using ImageJ software with default settings except that (1) the size setting was 50 to infinity; (2) the circularity setting was 0.25 to 1.0, and (3) edges were excluded.
To examine the functional complementation of a given protein isoform, leaves of 3- to 4-week-old N. benthamiana plants grown at 25°C under a 12-h light (8:00 to 20:00, 150 µmol m−2 s−1 provided by LED SUN LIGHT Z RGB bulbs [HIPOINT, Taiwan])/12-h dark period were infiltrated with A. tumefaciens strain LBA4404 carrying TYLCTHV infectious clones with WT sequences or sequences with the indicated mutation sites. At 7 dpi (i.e. after the first inoculation), the systemic leaves were infiltrated with the construct overexpressing the tested protein isoform and collected at 1.5–5 dpi (i.e. after the second inoculation) for subsequent analysis of viral DNA abundance using qPCR (as described above).
At least two independent transient expression assays (i.e. two separate sets of plants on different days) were performed with consistent results.
Phylogenetic analysis
Nucleotide and amino acid sequences of begomoviruses were retrieved from the ICTV (https://talk.ictvonline.org) (Supplemental Data Set S1), and TYLCTHV isolates were retrieved from GenBank. For member species in the genus Begomovirus, the exemplar viruses for each species indicated in ICTV were used as representatives. The putative AUG-initiated ORFs in DNA-B molecules of begomoviruses (Supplemental Figure S5A) were characterized using customized scripts with the criteria: TIS at AUG codon and ORF length > 50 amino acids. When multiple AUG sites were in-frame and could initiate ORFs that overlapped, only the AUG site that encodes the longest ORF was included.
The amino acid and nucleotide sequence alignments of viral genes among different viruses were performed using SnapGene with the Clustal Omega algorithm and plotted via JalView (Waterhouse et al., 2009); sequence identity was calculated using MatGat with BLOSUM50 scoring matrix (Campanella et al., 2003). For the begomoviruses with the annotated AUG TIS site of AV2 and BV1 genes that were aligned with the AUG135 of AV2 and AUG174 of BV1 in TYLCTHV (Figures 2H and 4G), these aligned AUG sites and the AUG site that was nearest upstream to the aligned AUG site in question in the same begomovirus were included for PWM score analyses as described below.
Calculation of PWM scores and the estimated BV2/BV1 translation expression
The degree of sequence similarity between the flanking regions of a given TIS site and the annotated TISs of tomato genes was summarized as PWM scores and calculated as described previously (Reuter et al., 2016; Li and Liu, 2020). Briefly, a PWM matrix generated based on a 13-nt window flanking the annotated TISs of tomato genes with in vivo translation initiation activity was retrieved from a previous study (Li and Liu, 2020). A PWM score for a viral TIS in question was then calculated by inputting the 13-nt sequences flanking the TIS to the PWM matrix to obtain a PWM score (Reuter et al., 2016). A higher PWM score indicates a higher similarity of flanking sequences contexts between a viral TIS and the tomato TISs used in vivo.
The estimated expression ratios of BV1 (in the phase 3) and BV2 (in the phase 1) were determined by quantifying the difference in phasing patterns of CHX signals in the overlapping and nonoverlapping regions as described previously (Lulla and Firth, 2020). To assess the statistical significance, 100,000 bootstrap resamplings of codon positions in the overlapping and nonoverlapping regions, respectively, were performed to calculate the estimated expression ratios of BV1 and BV2.
Statistical analysis
The statistical analysis was performed in either GraphPad Prism version 7 (GraphPad Software; http://www.graphpad.com/) or R version 4.1.1 (https://www.R-project.org/). The statistical tests used and the corresponding results are provided in Supplemental Data Set S2.
Accession numbers
The analytic pipelines for processing the LTM, CHX, and RNA sequencing data sets were retrieved from a previous study (Li and Liu, 2020). The calculation of estimated BV2:BV1 expression ratios was downloaded from a previous study (Lulla and Firth, 2020). The ribosome profiling and total RNA sequencing datasets generated from this study have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE160907.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Characteristics of LTM, CHX, and total RNA-associated reads mapped to tomato and viral genes between biological replicates.
Supplemental Figure S2 . Read densities along the TYLCTHV genome in two biological replicates of LTM, CHX, and total RNA samples.
Supplemental Figure S3. The translation initiation of AV2 at AUG135 and AUG189 and the subcellular localization of the encoded AV2 proteins.
Supplemental Figure S4 . The scale used to score the disease symptoms of virus-infected tomato plants.
Supplemental Figure S5. Characterization of BV2 sequence similarity in begomoviruses and subcellular localization/protein domains.
Supplemental Table S1. The in vivo TISs identified in this study.
Supplemental Table S2. List of primers used in the study.
Supplemental Table S3. Summary of read numbers for LTM, CHX, and mRNA sequencing data.
Supplemental Data Set S1. List of begomoviruses analyzed in this study.
Supplemental Data Set S2. Statistical analysis results.
Supplemental File S1 . Sequence alignments of the AV2 gene in begomoviruses.
Supplemental File S2. Sequence alignments of the BV1 gene in begomoviruses.
Supplementary Material
Acknowledgments
We thank Mr Hong-Jou Tsai for technical support with genomic DNA extraction and Sanger sequencing of viral DNAs; Dr Fu-Chen Hsu for technical support in construct design; Dr Peter Hanson for providing tomato seeds; Dr Lawrence Kenyon for sharing TYLCTHV infectious clones; Dr Ming-Tsair Chan for sharing the LBA4404 strains and pCambia0380 plasmids; Dr Tzyy-Jen Chiou for sharing the pGWB520-Myc and pk7FWG2-eGFP plasmids; the AS-BCST Confocal Microscopy Core Facilities for core services; and Miss Ai-Ping Chen and Miss Shu-Jen Chou in the Genomic Technology Core facility (Institute of Plant and Microbial Biology, Academia Sinica, Taipei, Taiwan) for technical assistance with Ribosome Footprinting sequencing library preparation. We also thank Dr Na-Sheng Lin, Dr Erh-Min Lai, Dr. Yi-Ju Lu and Dr Kuan-Ju Lu for helpful comments on the manuscript; and Dr Melissa Lehti-Shiu for English editing of this article.
Funding
This research was financially supported by a grant from Academia Sinica to M.-J.L.
Conflict of interest statement. None declared.
Contributor Information
Ching-Wen Chiu, Biotechnology Center in Southern Taiwan, Academia Sinica, Tainan 711, Taiwan.
Ya-Ru Li, Biotechnology Center in Southern Taiwan, Academia Sinica, Tainan 711, Taiwan.
Cheng-Yuan Lin, Biotechnology Center in Southern Taiwan, Academia Sinica, Tainan 711, Taiwan.
Hsin-Hung Yeh, Agricultural Biotechnology Research Center, Academia Sinica, Taipei 115, Taiwan.
Ming-Jung Liu, Biotechnology Center in Southern Taiwan, Academia Sinica, Tainan 711, Taiwan; Agricultural Biotechnology Research Center, Academia Sinica, Taipei 115, Taiwan; Graduate Program in Translational Agricultural Sciences, National Cheng Kung University and Academia Sinica, Taiwan.
C.-W.C. performed the construct design, 5′ RACE, virus infections and disease symptom, TIS-driven protein expression assay, and phylogenetic analyses. Y.-R.L. prepared the RNA samples for total RNA and ribosome profiling and performed the protein localization assay. C.-Y.L. performed the TIS-driven protein expression assay. H.-H.Y. contributed to the conception and design of the research and revised the article. M.-J.L. conceived/designed the research, analyzed the sequencing data and mRNA sequence features, and wrote the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Ming-Jung Liu (mjliu@gate.sinica.edu.tw).
References
- Andrews SJ, Rothnagel JA (2014) Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet 15: 193–204 [DOI] [PubMed] [Google Scholar]
- Arias C, Weisburd B, Stern-Ginossar N, Mercier A, Madrid AS, Bellare P, Holdorf M, Weissman JS, Ganem D (2014) KSHV 2.0: a comprehensive annotation of the Kaposi's sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog 10: e1003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Ziv A, Levy Y, Citovsky V, Gafni Y (2015) The tomato yellow leaf curl virus (TYLCV) V2 protein inhibits enzymatic activity of the host papain-like cysteine protease CYP1. Biochem Biophys Res Commun 460: 525–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak J (2016) Tomato yellow leaf curl virus: a serious threat to tomato plants world wide. J Plant Pathol Microbiol 7: 346 [Google Scholar]
- Bazin J, Baerenfaller K, Gosai SJ, Gregory BD, Crespi M, Bailey-Serres J (2017) Global analysis of ribosome-associated noncoding RNAs unveils new modes of translational regulation. Proc Natl Acad Sci USA 114: E10018–E10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachy RN, Heinlein M (2000) Role of P30 in replication and spread of TMV. Traffic 1: 540–544 [DOI] [PubMed] [Google Scholar]
- Blum M, Chang HY, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, et al. (2021) The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 49: D344–D354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briddon RW, Patil BL, Bagewadi B, Nawaz-ul-Rehman MS, Fauquet CM (2010) Distinct evolutionary histories of the DNA-A and DNA-B components of bipartite begomoviruses. BMC Evol Biol 10: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanella JJ, Bitincka L, Smalley J (2003) MatGAT: An application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BY, Miller WA, Atkins JF, Firth AE (2008) An overlapping essential gene in the Potyviridae. Proc Natl Acad Sci USA 105: 5897–5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creixell P, Schoof EM, Tan CS, Linding R (2012) Mutational properties of amino acid residues: Implications for evolvability of phosphorylatable residues. Philos Trans R Soc Lond B Biol Sci 367: 2584–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumbie JS, Ivanchenko MG, Megraw M (2015) NanoCAGE-XL and CapFilter: An approach to genome wide identification of high confidence transcription start sites. BMC Genomics 16: 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AJ, Dolan A, Akter P, Addison C, Dargan DJ, Alcendor DJ, McGeoch DJ, Hayward GS (2003) The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J Gen Virol 84: 17–28 [DOI] [PubMed] [Google Scholar]
- Finkel Y, Stern-Ginossar N, Schwartz M (2018) Viral short ORFs and their possible functions. Proteomics 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel Y, Schmiedel D, Tai-Schmiedel J, Nachshon A, Winkler R, Dobesova M, Schwartz M, Mandelboim O, Stern-Ginossar N (2020) Comprehensive annotations of human herpesvirus 6A and 6B genomes reveal novel and conserved genomic features. eLife 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth AE, Brierley I (2012) Non-canonical translation in RNA viruses. J Gen Virol 93: 1385–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondong VN (2013) Geminivirus protein structure and function. Mol Plant Pathol 14: 635–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch C, Herrmann A, Nothnagel M, Szafranski K, Huse K, Schumann F, Schreiber S, Platzer M, Krawczak M, Hampe J, et al. (2012) Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res 22: 2208–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wan J, Liu B, Ma M, Shen B, Qian SB (2015) Quantitative profiling of initiating ribosomes in vivo. Nat Methods 12: 147–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick E, Zrachya A, Levy Y, Mett A, Gidoni D, Belausov E, Citovsky V, Gafni Y (2008) Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc Natl Acad Sci USA 105: 157–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeon R, Jiang J, Wan J, Agbeci M, Zheng HQ, Laliberte JF (2013) 6K(2)-induced vesicles can move cell to cell during turnip mosaic virus infection. Front Microbiol 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing LR, Lin CP, Perico C, White RR, Sparkes I (2017) Plant ER geometry and dynamics: biophysical and cytoskeletal control during growth and biotic response. Protoplasma 254: 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S (2013) Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat Rev Microbiol 11: 777–788 [DOI] [PubMed] [Google Scholar]
- Heinlein M (2015) Plant virus replication and movement. Virology 479–480: 657–671 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Brown CM, Chisnall MAW, Waterhouse PM, Macknight RC (2016) The emerging world of small ORFs. Trends Plant Sci 21: 317–328 [DOI] [PubMed] [Google Scholar]
- Hoang HD, Neault S, Pelin A, Alain T (2021) Emerging translation strategies during virus-host interaction. Wiley Interdiscip Rev RNA 12: e1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Calviello L, Wu HL, Li FW, Rothfels CJ, Ohler U, Benfey PN (2016) Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proc Natl Acad Sci USA 113: E7126–E7135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull R (2014) Plant Virology. Elsevier/AP, Amsterdam; Boston. [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irigoyen N, Firth AE, Jones JD, Chung BY, Siddell SG, Brierley I (2016) High-resolution analysis of coronavirus gene expression by RNA sequencing and ribosome profiling. PLoS Pathog 12: e1005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaafar ZA, Kieft JS (2019) Viral RNA structure-based strategies to manipulate translation. Nat Rev Microbiol 17: 110–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1986) Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci USA 83: 2850–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1987) At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol 196: 947–950 [DOI] [PubMed] [Google Scholar]
- Kozak M (1991) A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr 1: 111–115 [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Makita Y, Kawashima M, Fujita T, Iwasaki S, Matsui M (2018) Transcripts from downstream alternative transcription start sites evade uORF-mediated inhibition of gene expression in Arabidopsis. Proc Natl Acad Sci USA 115: 7831–7836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB (2012) Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci USA 109: E2424–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leke WN, Mignouna DB, Brown JK, Kvarnheden A (2015) Begomovirus disease complex: emerging threat to vegetable production systems of West and Central Africa. Agric Food Security 4 [Google Scholar]
- Lewandowski DJ, Dawson WO (2000) Functions of the 126- and 183-kDa proteins of tobacco mosaic virus. Virology 271: 90–98 [DOI] [PubMed] [Google Scholar]
- Li YR, Liu MJ (2020) Prevalence of alternative AUG and non-AUG translation initiators and their regulatory effects across plants. Genome Res 30: 1418–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling R, Pate AE, Carr JP, Firth AE (2013) An essential fifth coding ORF in the sobemoviruses. Virology 446: 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MJ, Wu SH, Wu JF, Lin WD, Wu YC, Tsai TY, Tsai HL, Wu SH (2013) Translational landscape of photomorphogenic Arabidopsis. Plant Cell 25: 3699–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulla V, Firth AE (2020) A hidden gene in astroviruses encodes a viroporin. Nat Commun 11: 4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machkovech HM, Bloom JD, Subramaniam AR (2019) Comprehensive profiling of translation initiation in influenza virus infected cells. PloS Pathog 15: e1007518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia-Guerra MK, Li W, Galeano NF, Vidal M, Gray J, Doseff AI, Grotewold E (2015) Core promoter plasticity between maize tissues and genotypes contrasts with predominance of sharp transcription initiation sites. Plant Cell 27: 3309–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C, Stepanova AN, Alonso JM (2017) Translation regulation in plants: An interesting past, an exciting present and a promising future. Plant J 90: 628–653 [DOI] [PubMed] [Google Scholar]
- Miras M, Miller WA, Truniger V, Aranda MA (2017) Non-canonical translation in plant RNA viruses. Front Plant Sci 8: 494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshe A, Belausov E, Niehl A, Heinlein M, Czosnek H, Gorovits R (2015) The tomato yellow leaf curl virus V2 protein forms aggregates depending on the cytoskeleton integrity and binds viral genomic DNA. Sci Rep 5: 9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Noderer WL, Flockhart RJ, Bhaduri A, Diaz de Arce AJ, Zhang J, Khavari PA, Wang CL (2014) Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol Syst Biol 10: 748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A, Sharma N, Hari-Gowthem G, Muthamilarasan M, Prasad M (2020) Tomato yellow leaf curl virus: impact, challenges, and management. Trends Plant Sci 25: 897–911 [DOI] [PubMed] [Google Scholar]
- Reuter K, Biehl A, Koch L, Helms V (2016) PreTIS: A tool to predict non-canonical 5' UTR translational initiation sites in human and mouse. PLoS Comput Biol 12: e1005170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas MR, Jiang H, Salati R, Xoconostle-Cazares B, Sudarshana MR, Lucas WJ, Gilbertson RL (2001) Functional analysis of proteins involved in movement of the monopartite begomovirus, tomato yellow leaf curl virus. Virology 291: 110–125 [DOI] [PubMed] [Google Scholar]
- Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO (2010) Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol 6: 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof KB, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, Hohn B, Saunders K, Candresse T, Ahlquist P, et al. (2011) Top 10 plant viruses in molecular plant pathology. Mol Plant Pathol 12: 938–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon MA, Kellner MJ, Plotnikova A, Hofmann F, Nodine MD (2018) NanoPARE: Parallel analysis of RNA 5' ends from low-input RNA. Genome Res 28: 1931–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedman SA, Gelembiuk GW, Mertz JE (1990) Translation initiation at a downstream AUG occurs with increased efficiency when the upstream AUG is located very close to the 5' cap. J Virol 64: 453–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Akbergenov R, Trinks D, Rajeswaran R, Veluthambi K, Hohn T, Pooggin MM (2005) Promoters, transcripts, and regulatory proteins of Mungbean yellow mosaic geminivirus. J Virol 79: 8149–8163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Firth AE, Miller WA, Scheidecker D, Brault V, Reinbold C, Rakotondrafara AM, Chung BYW, Ziegler-Graff V (2015) Discovery of a small non-AUG-initiated ORF in poleroviruses and luteoviruses that is required for long-distance movement. Plos Pathog 11: e1004868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin LA (1997) The plant ER: A dynamic organelle composed of a large number of discrete functional domains. Plant J 11: 1151–1165 [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar N. (2015) Decoding viral infection by ribosome profiling. J Virol 89: 6164–6166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Ingolia NT (2015) Ribosome profiling as a tool to decipher viral complexity. Annu Rev Virol 2: 335–349 [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, et al. (2012). Decoding human cytomegalovirus. Science 338: 1088–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J, Linnik O, Louveaux M, Roberts IM, Chapman SN, Oparka KJ (2013) Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J Cell Biol 201: 981–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WS, Shih SL, Kenyon L, Green SK, Jan FJ (2011) Temporal distribution and pathogenicity of the predominant tomato-infecting begomoviruses in Taiwan. Plant Pathol 60: 787–799 [Google Scholar]
- Walsh D, Mathews MB, Mohr I (2013) Tinkering with translation: Protein synthesis in virus-infected cells. Cold Spring Harb Perspect Biol 5: a012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yang X, Wang Y, Xie Y, Zhou X (2018) Tomato yellow leaf curl virus V2 interacts with host histone deacetylase 6 to suppress methylation-mediated transcriptional gene silencing in plants. J Virol 92: e00036–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Hawes C, Hussey PJ (2017) Plant endoplasmic reticulum-plasma membrane contact sites. Trends Plant Sci 22: 289–297 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wu Y, Gong Q, Ismayil A, Yuan Y, Lian B, Jia Q, Han M, Deng H, Hong Y, et al. (2019) Geminiviral V2 protein suppresses transcriptional gene silencing through interaction with AGO4. J Virol 93: e01675–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems P, Ndah E, Jonckheere V, Stael S, Sticker A, Martens L, Van Breusegem F, Gevaert K, Van Damme P (2017) N-terminal proteomics assisted profiling of the unexplored translation initiation landscape in Arabidopsis thaliana. Mol Cell Proteomics 16: 1064–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HL, Song G, Walley JW, Hsu PY (2019a) The tomato translational landscape revealed by transcriptome assembly and ribosome profiling. Plant Physiol 181: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XY, Liu JH, Chai MZ, Wang JH, Li DL, Wang AM, Cheng XF (2019b) The potato virus X TGBp2 protein plays dual functional roles in viral replication and movement. J Virol 93: e01635–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Ji Y, Wu S, Ma X, Li S, Sun F, Cheng Z, Zhou Y, Fan Y (2018) Single amino acid in V2 encoded by TYLCV is responsible for its self-interaction, aggregates and pathogenicity. Sci Rep 8: 3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.