Abstract
Hemipterans (such as aphids, whiteflies, and leafhoppers) are some of the most devastating insect pests due to the numerous plant pathogens they transmit as vectors, which are primarily viral. Over the past decade, tremendous progress has been made in broadening our understanding of plant–virus–vector interactions, yet on the molecular level, viruses and vectors have typically been studied in isolation of each other until recently. From that work, it is clear that both hemipteran vectors and viruses use effectors to manipulate host physiology and successfully colonize a plant and that co-evolutionary dynamics have resulted in effective host immune responses, as well as diverse mechanisms of counterattack by both challengers. In this review, we focus on advances in effector-mediated plant–virus–vector interactions and the underlying mechanisms. We propose that molecular synergisms in vector–virus interactions occur in cases where both the virus and vector benefit from the interaction (mutualism). To support this view, we show that mutualisms are common in virus–vector interactions and that virus and vector effectors target conserved mechanisms of plant immunity, including plant transcription factors, and plant protein degradation pathways. Finally, we outline ways to identify true effector synergisms in the future and propose future research directions concerning the roles effectors play in plant–virus–vector interactions.
A review of advances in effector-mediated plant–virus–vector interactions suggesting that molecular synergisms sometimes occur that benefit both the virus and insect vector.
Introduction
Plants harvest energy from the sun through photosynthesis in order to produce organic compounds, making them the primary producers at the bottom of the food web in most ecosystems. Occupying the bottom of the food web means that plants face multiple threats, including herbivory and pathogen attack. Plants defend themselves from these threats using preformed defenses, by recognizing elicitors associated with the attacker, and by inducing the appropriate defense responses (Jones and Dangl, 2006; Zhou and Zhang, 2020). Plants recognize many different elicitors associated with attack, including pathogen-associated molecular patterns (PAMPs), damage-associated molecular patterns (DAMPs), and herbivore-associated molecular patterns (HAMPs). Elicitor recognition happens at the cell-surface using receptor-like proteins and receptor kinases and results in pattern-triggered immunity (PTI; Boller and Felix, 2009; Kawai and Akira, 2010; Acevedo et al., 2015; Tang et al., 2017; Tanaka and Heil, 2021). Adapted herbivores and microbes have evolved the ability to secrete effectors, which assist in infestation and infection through defense suppression, nutrient acquisition, and detection avoidance. In response to this, some plants have evolved intracellular receptors with nucleotide-binding and leucine-rich repeat domains (NLRs) that recognize effectors or effector activity, and leads to the induction of defenses, a process termed effector-triggered immunity (ETI; Cui et al., 2015). For more information on the details of plant defense responses to insects and pathogens, elicitors and effectors, we direct readers to these reviews (Kaloshian and Walling, 2005; Jones and Dangl, 2006; Boller and Felix, 2009; Hogenhout and Bos, 2011; Acevedo et al., 2015; Cui et al., 2015; Schmelz, 2015; Couto and Zipfel, 2016).
Another major immune mechanism used by plants is RNA silencing. Plants recognize double-stranded RNA derived from virus genomes and produce small interfering RNAs (siRNAs). siRNAs are used by plants to silence viral RNAs and DNAs via posttranscriptional gene silencing and transcriptional gene silencing (Teresa Ruiz et al., 1998; Li and Ding, 2006). Most viruses have evolved RNA-silencing suppressor effectors as a counter defense to prevent the recognition of siRNA elicitors and the induction of the RNA-silencing pathway (Zhao et al., 2016). Some siRNAs generated from viral genomes are homologous to plant targets, resulting in silencing of the host’s immunity responses (Ramesh et al., 2021). Recent research has shown that plants also use small RNAs (sRNAs) to silence genes from nonviral pathogens and some fungal and oomycete pathogens generate sRNAs that result in the silencing of host immunity genes, but the mechanism of sRNA transport between species is still poorly understood (Qiao et al., 2021). For more details on sRNAs and silencing, we direct readers to several excellent reviews (Brodersen and Voinnet, 2006; Baulcombe, 2015; Sattar and Thompson, 2016; Ye and Ma, 2016).
While our understanding of effectors, elicitors, and plant defense responses has expanded considerably in the past few decades, much less is known about the roles of effectors and elicitors in multi-partite and multi-trophic interactions. Plant microbes and insects often co-exist on plants in seemingly commensal interactions, both introducing elicitors and effectors at the same time as changes in plant immunity are induced. Recent research has demonstrated that these changes in plant immunity can have ecological consequences. For example, rhizosphere-associated microbiomes can increase plant resistance to insect pests via changes in phytohormone biosynthesis and signaling pathways (Murrell et al., 2019; Blundell et al., 2020; French et al., 2021). Whitefly (Bemisia tabaci) feeding on tomato (Solanum lycopersicum) reduces powdery mildew (Erysiphe cichoracearum) abundance (Mayer et al., 2002), and white-backed planthopper (Sogatella furcifera) infestation on rice (Oryza sativa) induces resistance to the rice blast fungus (Magnaporthe oryzae; Kanno et al., 2005). In some cases, the specific insect effectors have been identified that induce systemic acquired resistance and reduce bacterial numbers in the phyllosphere and rhizosphere (Lee et al., 2018). Therefore, even nonpathogenic or nonmutualistic interactions that occur occasionally in nature between insects and microbes can indirectly harm or help the other plant challenger through changes in plant chemistry and immunity. However, for these interactions to be cooperative, there must be a benefit to the other individual and it must have evolved because of this, at least partially (West et al., 2007).
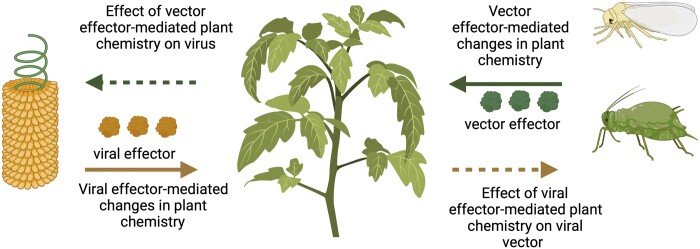
In other cases, there is evidence that plant pathogens and insects have co-evolved together through close associations, where interactions are unavoidable. For example, many bacterial pathogens and most plant viruses depend on insect vectors for transmission among hosts (Whitfield et al., 2015; Casteel and Falk, 2016; Perilla-Henao and Casteel, 2016; Shi et al., 2021), and thus plants must respond to elicitors and effectors from both organisms simultaneously during transmission and acquisition (Figure 1). Numerous studies have demonstrated that virus infection of host plants results in changes in vector behavior and performance in ways that are expected to benefit the virus through increased transmission (Blanc and Michalakis, 2016; Mauck et al., 2018). When the benefit is under the genetic control of the virus (Dawkins, 2016), and subject to natural selection (Poulin and Maure, 2015; Heil, 2016), it is often considered “parasite manipulation.” Detailed studies on the viral effectors that mediate parasite manipulation in plant–virus–vector interactions have recently received significant attention (for several recent reviews see Blanc and Michalakis, 2016; Nalam et al., 2019; Ziegler-Graff, 2020; Naalden et al., 2021; Shi et al., 2021), and in a few systems, the molecular mechanisms and specific plant targets that mediate these changes are known (Ziegler-Graff, 2020; Pan et al., 2021).
Figure 1.
Roles of virus and vector effectors in altering plant chemistry, and the potential effects of plant chemistry on virus and vector performance. Viral effectors are shown as yellow round structures, and vector effectors are shown as green round structures.
In some cases, virus-induced changes in host plants also benefit the insect vector, in addition to the virus, and thus these virus–vector interactions can be considered mutualisms (Casteel and Jander, 2013). These findings have resulted in a more nuanced “vector manipulation hypothesis,” which suggests that vector preferences for infected plants have evolved over time to facilitate the spread of the virus to new hosts (Ingwell et al., 2012). Indeed, aphid vectors often have higher reproduction on virus-infected plants compared to the controls, and aphids that transmit viruses to a new host plant benefit from virus-mediated suppression of plant defenses (Westwood et al., 2013; Casteel et al., 2014; Mauck et al., 2015; Bak et al., 2017; Patton et al., 2020). Insect vectors may also benefit from transmitting plant viruses through host range expansions. For example, whitefly performance is greatly enhanced on begomovirus-infected tobacco (Nicotiana tabacum), which is typically a poor host for some whitefly biotypes (Zhang et al. 2012). These findings suggest that the vector would also benefit by feeding from virus-infected tissue in a targeted manner, such as by releasing effectors to encourage the acquisition or release of viruses, or by specifically selecting virus-infected cells as they feed. Cauliflower mosaic virus rearranges itself within the cell when aphids puncture infected cells during feeding, which promotes attachment to the aphid stylets (Martinière et al., 2013). This may be due to viral recognition of aphid effectors or aphid-induced plant responses. It is also possible that the aphid is actively manipulating the plant to alter plant–virus interactions. While insect vectors can benefit and play an active role in transmission, the underlying genetic factors that mediate this, such as insect effectors, have been largely ignored in plant–virus–vector interactions.
Understanding the functions of effectors and elicitors in complex interactions is critical for deciphering how plant pathogens and insects colonize host organisms and how plant immunity is orchestrated. Here, we propose that molecular synergisms in vector–virus interactions occur in cases where both the virus and vector benefit, as viruses and vectors both use effector proteins to target host pathways and successfully colonize a host. In molecular synergisms, viral or vector effectors might have evolved to benefit both players in the interaction, to only benefit the other player and not the producer, or to have new functions only when effectors from both players are present. We make this case by reviewing the literature and showing that mutualisms in virus–vector interactions are common (Table 1), demonstrating that viral and vector effectors have conserved host targets (Table 2), including transcription, protein turnover, and cellular localization, and proposing how to identify cases where effector synergisms have evolved in plant–virus–vector interactions. Finally, we discuss future research directions regarding the roles of effectors and molecular mechanisms of multi-partite interactions with plants.
Table 1.
Differences in the number of studies reporting positive, neutral, or negative impacts of plant virus infection on vector performancea
| Family of Virus | Virus Vector | Percent of studies reporting |
Positive: Neutral:Negative |
Positive: Neutral |
Positive: Negative |
Number of studies (n) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive impacts |
Neutral impacts |
Negative impacts |
||||||||||
| χ2 value | p-value | χ2 value | p-value | χ2 value | p-value | |||||||
| Geminiviridae | Whitefly, Leafhopper | 52.11 | 28.17 | 19.72 | 16.94 | <0.0001 | 7.14 | 0.0074 | 14.605 | <0.0001 | 71 | |
| Luteoviridae | Aphids | 69.81 | 18.87 | 11.32 | 60.73 | <0.0001 | 29.26 | <0.0001 | 42.168 | <0.0001 | 53 | |
| Potyviridae | Whitefly, Aphids, Mites | 40.48 | 28.57 | 30.95 | 2.38 | 0.30 | 2.17 | 0.10 | 1.2715 | 0.2595 | 42 | |
| Bromoviridae | Aphids | 13.04 | 21.74 | 65.22 | 46.89 | <0.0001 | 1.98 | 0.1595 | 34.16 | <0.0001 | 23 | |
| Closteroviridae | Whitefly, Aphids, Mealybugs | 60.00 | 30.00 | 10.00 | 38.00 | <0.0001 | 10.0 | 0.0016 | 35.71 | <0.0001 | 10 | |
| Reoviridae | Aphids, Leafhopper | 42.86 | 28.57 | 28.57 | 4.08 | 0.13 | 5.05 | 0.0246 | 5.05 | 0.0246 | 7 | |
| Secoviridae | Whitefly, Aphids, Leafhopper, Beetles | 33.33 | 33.33 | 33.33 | 0.00 | 1.00 | 0.00 | 1.0000 | 0.00 | 1.0000 | 3 | |
| Caulimoviridae | Aphids | 0.00 | 0.00 | 100.00 | – | – | – | – | 100.00 | <0.0001 | 2 | |
| Nanoviridae | Aphids | 100.0 | 0.0 | 0.0 | – | – | – | – | – | – | 1b | |
Based on dataset as reviewed by Mauck et al. 2018, Shaded boxes indicate trends for significant differences.
Number of studies too low for a chi-square test.
Table 2.
Summary of putative effectors in virus-vector interactions, cellular targets,and ecological consequences
| Effector | Virus or Vector Origin | Plant Targets and Mechanism | Plant Species | Subcellular Location | Impact on Virus/Vector | References |
|---|---|---|---|---|---|---|
| Target 1: Transcription factors | ||||||
|
| ||||||
| Viral effectors | ||||||
|
| ||||||
| βC1 | Betasatellite of TYLCCNV | Disrupts MYC2 dimerization and glucosinolate defenses downstream of JA | N. tabacum | Nucleus | Increases B. tabaci performance | Li et al. (2014a) |
| βC1 | Betasatellite of TYLCCNV | Interacts with and enhances repressive activity of AS1 attenuating PDF1.2 and PR4 expression | A. thaliana | Nucleus | Increases B. tabaci performance | Yang et al. (2008) |
| βC1 | Betasatellite of TYLCCNV | Disrupts PIF and MYC2 dimerization and reduces terpene synthase and volatile production | A. thaliana | Nucleus | Increases B. tabaci attraction | (Zhao et al., 2021) |
|
βC1 |
Betasatellite of CLCuMuV and betasatellite of TYLCCNV |
Disrupts homeo-dimerization of WRKY20 and WRKY20-ORA59 dimerization to alter glucosinolate profiles in vascular tissue and leaves |
N. benthamiana, A. thaliana, Gossypium barbadense |
Nucleus |
Increases B. tabaci performance, decreases performance of M. persicae and H. armigera |
Zhao et al. (2019) |
|
Vector effectors |
|
|
|
|
|
|
| Bsp9 | B. tabaci | Suppresses DAMP-triggered immunity induced by Pep1; Interacts with WRKY33, and MPK6 | A. thaliana, N. benthamiana, S. lycopersicum | Cytoplasm | Increased B. tabaci and TYLCV performance | Wang et.al. (2019) |
| Bt56 | B. tabaci | Interacts with KNOX transcription factor and increases SA and SA-related transcripts | N. tabacum, N. benthamiana | Nucleus | Increased B. tabaci performance | Li et al. (2014a) |
|
| ||||||
| Target 2: Protein degradation pathways | ||||||
|
| ||||||
| Virus effectors | ||||||
|
| ||||||
| C2 | TYLCV | Interacts with the ubiquitin precursor, RPS27A, to prevent JAZ1 degradation and MYC2 and terpene synthase induction | N. tabacum | Nucleus | Increases B. tabaci performance | Li et al. (2019) |
| 2b, 2a, 1a | CMV | 2b interacts with JAZ proteins to prevent degradation and induction of downstream signaling and volatiles, 2b also suppresses AGO1, which is stabilized by 1a. 2a increases CYP81F2 expression and the production of glucosinolates | A. thaliana | Nucleus, cytoplasm, processing bodies | Increases M. persicae attraction before contact and increases dispersal after contact | Westwood et al. (2013) ; Wu et al. (2017) |
|
| ||||||
| Vector effectors | ||||||
|
| ||||||
| Mp1 | M. persicae | Interacts with and reduces protein levels of the plant trafficking pathway protein VPS52 | N. benthamiana, A. thaliana | Prevacuolar compartments | Increases M. persicae performance | Pitino and Hogenhout (2012); Rodriguez et al. (2017) |
|
| ||||||
| Target 3: Re-localization of proteins | ||||||
|
| ||||||
| Virus effectors | ||||||
|
| ||||||
| NIa-Pro | TuMV, PVY | Localizes outside of nucleus to inhibit plant defenses, increases ethylene production and inhibts callose accumulation |
A. thaliana, N. benthamiana |
Nucleus, vacuole | Increases fecundity of M. persicae | Casteel et al. (2014); Bak et al. (2017) |
| Target 4: Signal transduction | ||||||
|
| ||||||
| Vector effectors | ||||||
|
| ||||||
| Me10 |
M. eurphorbiae, A. gossypii |
Interacts withTFT7 protein, mechanisms unknown |
S. lycopersicum, N. benthamiana |
Cytoplasm, nucleus | Increased M. eurphorbiae fecundity | Atamian et al. (2013); Chaudhary et al. (2014) |
| ApHRCs | A. pisum | Serratia symbiotica induction of ApHRC possibly suppresses Ca2+, ROS, and JA/SA- related transcript induction | M. truncatula | Unknown | Increased A. pisum feeding duration | Wang et al. (2020) |
| BtFer1 | B. tabaci | BtFer1 exhibits Fe2+ binding ability and ferroxidase activity, suppresses H2O2 and, callose production, proteinase inhibitor activation, and JA signaling | S. lycopersicum | Phloem | Increased performance of B. tabaci | Su et al. (2019) |
|
| ||||||
| Target 5: Detoxification of secondary metabolites | ||||||
|
| ||||||
| Vector effectors | ||||||
|
| ||||||
| Me47 | M. eurphorbiae | Me47 encodes a glutathione S-transferase (GST), that was shown based on enzymatic activity to detoxify isothiocyanates | N. benthamiana, S. lycopersicum | Unknown | Increases M. euphorbiae fecundity on tomato, inhibits M. persicae performance on N. benthamiana | Atamian et al. (2013); Chaudhary et al. (2014) |
| AcDXR | A. craccivora | AcDXR is a diacetyl/L-xylulose reductase that detoxifies the plant secondary metabolite methylglyoxal |
V. radiata, P. sativum |
Phloem | Increases A. craccivora fecundity | MacWilliams et al. (2020) |
| Laccase1 | B. tabaci | Laccase 1 is a polyphenol oxidase that might help whiteflies overcome chemical defenses | S. lycopersicum | Unknown | Increased performance of B. tabaci | Yang et al. (2017) |
|
| ||||||
| Target 6: The unknowns | ||||||
|
| ||||||
| Virus effectors | ||||||
|
| ||||||
| P0, P1, P7 | PLRV | Unknown | S. tuberosum | Cytoplasm (P1), nucleus (P0) | Increases performance and preference of M. persicae | Prüfer et al. (1999); Patton et al. (2020) |
| HC-Pro | TuMV | Unknown | N. benthamiana | Cytoplasm | Decreases M. persicae performance | Maia et al. (1996); Casteel et al. (2014) |
| NIa-Pro | TuMV | Increases free amino acid levels in plants |
A. thaliana, N. benthamiana |
Nucleus, vacuole | Unknown | Casteel et al. (2014) |
| 6K1 | TuMV | Unknown | N. benthamiana | Chloroplast | Decreases M. persicae performance | Casteel et al. (2014); Hongguang et al. (2021) |
| VPg | TuMV | Unknown | N. benthamiana | Cytoplasm and nucleus | Decreases M. persicae performance | Schaad et al. (1996); Casteel et al. (2014) |
|
| ||||||
| Vector effectors | ||||||
|
| ||||||
| Mp10 | M. persicae | Induces the hypersensitive response in an SGT1-dependent manner and suppresses flg2- induced PTI | N. benthamiana | Mesophyll cells next to feeding tracks | Over-expression in plants reduces M. persicae fecundity | Bos et al. (2010) |
| Mp56, Mp57, Mp58 | M. persicae | Unknown |
A.thaliana, N. benthamiana |
Unknown | Over-expression in plants reduces M. persicae fecundity | Elzinga et al. (2014) |
| Mp2 | M. persicae | Unknown |
A. thaliana, N. benthamiana |
Unknown | Over-expression in plants reduces M. persicae fecundity | Pitino and Hogenhout (2012) |
| Mp42 | M. persicae | Unknown | N. benthamiana | Unknown | Over-expression in plants reduces M. persicae fecundity | Bos et al. (2010) |
| MpC002, ApC002 |
M. persicae, A. pisum |
Unknown |
Vicia faba, A. thaliana, N. benthamiana |
Sieve elements | Over-expression in plants increases aphid performance/fecundity | Mutti et al. (2008); Bos et al. (2010) |
| Armet | A. pisum | Transient expression induces SA accumulation in plants | N. benthamiana | Probably in sieve elements | Knockdown in A. pisum shortens their lifespan | Wang et al. (2015a) |
| MIF1 | A. pisum | Suppresses callose formation, the hypersensitive response, and defense-related transcript induction | N. benthamiana | Unknown | Over-expression in plants increased A. pisum fecundity | Naessens et al. (2015) |
| Ap25 | A. pisum | Unknown | P. sativum | Unknown | Over-expression increases A. pisum fecundity | Guy et al. (2016) |
| S2G4, 6A10, 2G5 | B. tabaci | The three effectors induced SA-responsive genes PR1a, PR2 | N. benthamiana | Unknown | Unknown on aphids, but suppresses pathogens X. axonopodis pv. vesicatoria and R. solanacearum | Lee et al. (2018) |
| ACEs | A. pisum | Unknown | V. faba | Unknown | Knockdown of ACE1 and ACE2 decreases A. pisum fecundity | Wang et al. (2015b) |
| Me23 | M. eurphorbiae | Unknown | N. benthamiana | Unknown | Over-expression in plants increases M. eurphorbiae fecundity | Atamian et al. (2013) |
Mutualisms are common in plant–virus–vector interactions
Of the plant viruses described to date, over 70% rely on insect vectors for transmission among hosts (Fereres and Raccah, 2015). While beetles (order Coleoptera) and thrips (order Thysanoptera) are important vectors for some viral species, most plant viruses are transmitted by insects in the order Hemiptera, such as aphids, leafhoppers, whiteflies, and mealybugs (Hogenhout et al., 2008). One reason that Hemipterans are such efficient vectors is because they feed with needle-like mouth parts known as stylets, which allow them to feed from individual plant cells and many cell types (Nalam et al., 2019). During the initial host contact, feeding, and virus transmission, hemipterans secrete saliva into the plant. This saliva contains effectors and elicitors. Effectors can inhibit plant defenses, mask detection of the insect, and help insects access the chemical composition and nutrient profile of the plants (Kaloshian and Walling, 2005; Hogenhout and Bos, 2011; Nalam et al., 2019; Naalden et al., 2021). Some hemipterans also deposit gelling saliva during feeding, which forms a sheath around the stylet, protecting it from the apoplast and providing it with a track for movement.
By far the most important hemipteran vectors are whiteflies and aphids, because they transmit over 500 virus species together (Fereres and Raccah, 2015). Whiteflies primarily transmit begomoviruses (Geminiviridae), criniviruses (Closteroviridae), torradoviruses (Secoviridae), and ipomoviruses (Potyviridae; Table 1), although a few rare cases of whiteflies transmitting virus from other families have been documented (Table 1; Zanardo and Carvalho, 2017; Saptarshi et al., 2022). Aphids have been shown to transmit viruses in the family Potyviridae, Bromoviridae, Secoviridae, Caulimoviridae, Closteroviridae, Nanoviridae, and Reovirodae (Table 1; Ng and Perry, 2004; Quito-Avila et al., 2012; Gaafar and Ziebell, 2020).
Although both aphids and whiteflies feed with stylets and secrete effectors and elicitors, their feeding biology is very different. Whiteflies use their stylet to navigate plant cells intracellularly, rarely piercing a cell before they establish phloem feeding sites (Kaloshian and Walling, 2005; Naalden et al., 2021). Aphids, on the other hand, use their stylets to pierce and taste nearly every cell on the pathway to the phloem (Kaloshian and Walling, 2005; Nalam et al., 2019). Both winged whitefly adults and the crawlers that emerge from the eggs are mobile, although after the whitefly crawlers molt into the nymphal stages, they are immobile. This means that whitefly nymphs often feed from a single feeding site for prolonged periods of time (up to 21–30 days; Li et al., 2019). In contrast, all aphid life stages are mobile, and they utilize numerous feeding sites during their lifecycle. For more details on the mechanism of plant–hemipteran interactions, please consult these excellent reviews: (Kaloshian and Walling, 2005; Nalam et al., 2019; Naalden et al., 2021).
Mutualisms, or cooperative interactions between species that benefit both partners, are ubiquitous in nature (Bronstein et al., 2006). For example, plants provide resources to attract pollinators, and mycorrhizal fungi benefit plants by providing various resources (Kiers and Van Der Heijden, 2006; Carol et al., 2014). Mutualisms often involve modifications of morphology, physiology, or behavior of one of the players to provide services for the other, such as the service of transmission among hosts for viruses. In return, the partner provides habitat or food resources, such as increased nutrient content and reduced defenses in virus-infected plants. Although many well-known examples of mutualism involve obligate partners that require each other for survival, other mutualist interactions are not so tightly coupled (Roossinck, 2011; Henry et al., 2015).
While not all virus–vector interactions will be mutualistic, a recent literature review demonstrated that in many cases, the insect vector benefits from interactions with viruses (Mauck et al., 2018), suggesting that these interactions may be adaptive relationships and not just by products of parasite manipulation. In this keystone publication, the findings from 122 journal articles that all examine the impacts of plant virus infection on vector behavior or performance were synthesized. The authors provide evidence for parasite manipulation for particular virus transmission types that span genetically diverse viruses. We re-evaluated this data set, focusing only on aphid- and whitefly-transmitted virus families and only on studies that examined the impacts of these viruses on vector performance (performance = fecundity or population growth; Table 1). We found examples of positive impacts on vectors for all virus families except Caulimoviridae (Table 1). For some virus families, such as Geminiviridae, Closteroviridae, and Luteoviridae, a significantly greater number of studies reported positive impacts on vectors compared to negative impacts (Table 1), while for viruses from the family Bromoviridae, significantly more studies reported negative impacts on vectors compared to positive impacts (Table 1). This suggests that some virus families are more promising systems for the development of viral–vector mutualisms (Table 1). Within viral families where more variation exists, such as Potyviridae (Table 1), cooperation may still occur, but may be dependent on the vector, vector biotype, or host species. This work also highlights bias in research on plant–vector interactions and insect performance to a few families, such as Geminiviridae, Luteoviridae, and Potyviridae, leaving other virus families open as rich resources for discovery by future researchers. Furthermore, some families had fewer than five representative experiments, and thus conclusions on these groups should be taken with caution; more attention to these families in future research will broadly benefit the field.
Effector targets in plant–virus–vector interactions
Effectors secreted from plant pathogens assist in establishing infection by facilitating host entry, overcoming perception, suppressing defenses, and/or modifying the host environment to increase nutrient access or suitability. To accomplish this, many pathogen effectors target conserved cellular processes (Toruño et al., 2016). Here, we demonstrate that this is also true for plant–vector interactions, and we review viral and vector effectors based on the conserved cellular processes they target (Table 2). We focus only on effectors of aphids and whiteflies and the viruses they transmit, since these two Hemipterans transmit nearly 75% of insect-vectored plant viruses (Fereres and Moreno, 2009). We elaborate on mechanisms used by each effector and in cases where the mechanisms are unknown, we synthesize what is known.
Target 1: Transcription factors
Virus effectors that target plant transcription factors
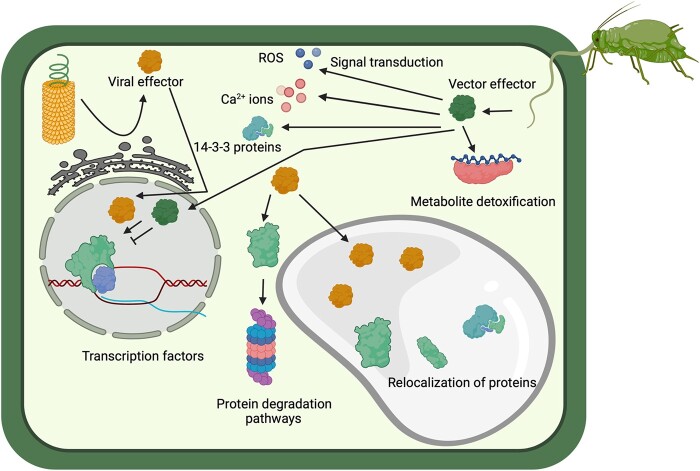
Viral effectors can target plant defense signaling pathways at the transcriptional level by interacting with transcription factors that regulate plant defense responses (Figure 2). For example, some whitefly-transmitted Begomoviruses (Family Geminiviridae) associate with betasatellites; these unrelated ssDNA molecules encode a single protein and depend on the begomovirus for replication, encapsidation, movement, and insect transmission (Gnanasekaran et al., 2019). The betasatellite βC1 determines pathogenicity for Tomato yellow leaf curl China virus (TYLCCNV) and plays a role in enhancing whitefly (B. tabaci) feeding and reproduction by binding to several transcription factors that regulate plant defense responses (Zhang et al., 2012; Li et al., 2014b). βC1 binds to the basic helix–loop–helix transcription factor MYC2, a master regulator of jasmonic acid (JA)-dependent defense responses in Arabidopsis thaliana (Table 2). By binding to MYC2, βC1 prevents MYC2 dimerization and the transcription of genes mediating the production of glucosinolates, which are important anti-insect defense compounds (Li et al., 2014b). βC1 also binds to and enhances the activity of ASYMMETRIC LEAVES 1 (AS1), a negative regulator of plant immunity in A. thaliana (Table 2). AS1 suppresses the expression of the JA- and ethylene (ET)-dependent defense-related gene PLANT DEFENSIN 1.2 (PDF 1.2); Nurmberg et al., 2007; Yang et al., 2008), which was implicated as a component of plant defense against whiteflies (Zarate et al., 2007). Thus, βC1 enhances whitefly performance on infected host plants by inhibiting and enhancing the activities of different plant transcription factors.
Figure 2.
Summary of plant targets of virus and vector effectors and their downstream effects on plant signaling pathways. Viral effectors are shown as yellow round structures, and vector effectors are shown as green round structures.
Whitefly attraction is also increased and terpene synthesis reduced in plants infected with TYLCCNV and βC1 compared to infected plants without the betasatellites (Li et al., 2014b). βC1 mediates increased whitefly attraction and changes in volatile production through interactions with MYC2, as well as the transcription factors PHYTOCHROME-INTERACTING FACTORS (PIFs; Table 2). PIFs positively control light-regulated plant defenses through interactions with MYC2. They also directly bind to the promoters of terpene synthase genes, promoting their transcription. The begomovirus-encoded βC1 inhibits the transcriptional activity of PIFs and MYC2 by disrupting their dimerization via direct interactions (Zhao et al., 2021). Taken together, βC1 binds to multiple transcription factors that are critical for increases in whitefly performance and attraction.
βC1, the betasatellite that associates with TYCCNV, also associates with Cotton leaf curl Multan virus (CLCuMuV) and binds to the vascular-specific transcription factor WRKY20 and prevents its homo-dimerization and WRKY20-ORA59 dimerization (Table 2; Zhao et al., 2019). WRKY20 positively regulates indole-glucosinolate production and negatively regulates aliphatic glucosinolate production, both of which affect generalist caterpillars (Kim et al., 2008). By disrupting WRKY20 binding, indole-glucosinolate production was inhibited in the vascular tissue where whiteflies fed, while aliphatic glucosinolate levels increased in the rest of the leaf. As a result, the nonvector herbivore cotton bollworm caterpillar (Helicoverpa armigera) had reduced performance, while the performance of the vector whiteflies was enhanced on infected plants with the betasatellite compared to those without it (Zhao et al., 2019).
Vector effectors that target plant transcription factors
The whitefly is a prolific vector and pest, transmitting over 300 plant viruses and feeding on numerous plant species (Gilbertson et al., 2015). The whitefly salivary protein Bt56 enhances whitefly performance and feeding by interacting with the KNOX transcription factor NTH202 in host plants (Xu et al., 2019; Table 2). KNOX transcription factors regulate various phytohormone responses, including salicylic acid (SA)- and JA-dependent transcriptional responses (Hake et al., 2004; Bolduc et al., 2012; Tsuda et al., 2014). Xu et al. suggested that Bt56 might facilitate NTH202 turnover, resulting in changes in SA levels. Silencing NTH202 had no impact on constitutive levels of SA; however, whitefly-induced SA production and whitefly performance were enhanced on silenced plants compared to the controls (Xu et al., 2019). As SA regulates plant defense responses against viruses (Carr et al., 2019; Murphy et al., 2020), examining the impact of silencing NTH202 on whitefly-transmitted viral accumulation would be of interest in the future.
One study where the impact of insect effectors on virus accumulation was investigated, was with the whitefly effector Bsp9. Preinfestation of tomato plants with whiteflies increased the accumulation of Tomato yellow leaf curl virus (TYLCV, Family Geminividae) compared to the controls (Wang et al., 2019). Bsp9 is induced at the transcriptional level in viruliferous whiteflies, and Bsp9 expression in host plants increased TYLCV titer threefold compared to the controls, which strongly suggests that whitefly-induced susceptibility to TYLCV is at least partially mediated by the secretion of Bsp9 into host plants (Wang et al., 2019). Bsp9 interacts with the transcription factor WRKY33, which is critical for the induction of various terpene synthase genes (Wang et al., 2019). Although the exact impacts of Bsp9 on WRKY33 activity were not demonstrated, TYLCV titer was higher in wrky33 Arabidopsis mutants compared to the controls. Bsp9 also interacted with MPK6, a MAP kinase that activates downstream defense responses against pathogens (Meng and Zhang, 2013). MAP kinases are induced by TYLCV (Jun-Bo et al., 2011) and therefore, perhaps the interaction of Bsp9 with MPK6 affects virus infection in plants.
Target 2: Protein degradation pathways
Virus effectors that target protein degradation pathways
Accumulating evidence indicates that plant viruses target proteolytic degradation pathways to enhance their own performance, as well as the attraction and performance of their insect vectors (Figure 2; Jia et al., 2016). As mentioned above, TYLCV benefits from whitefly feeding (Wang et al., 2019), and whiteflies also benefit on TYLCV-infected plants due to changes in plant protein turnover (Moriones and Navas-Castillo, 2000; Li et al., 2019). Upon JA perception, the E3 ubiquitin ligase SCFCOi1 recruits JAZs for ubiquitination and degradation by the 26S proteasome pathway, resulting in the upregulation of JA signaling and defense responses (Katsir et al., 2008; Nelson and Millar, 2015). The TYLCV protein C2 interacts with and inhibits the ubiquitin-precursor protein RPS27A, preventing the ubiquitination and degradation of JAZ1 (Table 2). This results in the suppression of MYC2 and JA-responsive terpene synthase genes in plants expressing C2 protein from TYLCV, leading to increased whitefly performance (Li et al., 2019).
Cucumber mosaic virus (CMV, Family Bromoviridae) is transmitted by aphids in a nonpersistent manner. Squash plants (Cucurbita pepo) infected with the Fny strain of CMV emit odors that are attractive to the aphid vector Myzus persicae (Mauck et al., 2010). Increased attraction of M. persicae to CMV-infected plants was attributed to the CMV protein 2b, which interacts directly with several JAZ proteins, including JAZ1. When 2b binds to JAZ1, degradation by the E3 ubiquitin ligase SCFCOi1 is repressed, preventing the induction of JA-dependent defense responses (Table 2; Wu et al., 2017). Earlier, 2b was shown to also interact with and stabilize two other viral proteins, 1a and 2a, to promote vector dispersal from infected host plants (Table 2; Westwood et al., 2013). In CMV-infected A. thaliana plants, 2b represses the ARGONAUTE1(AGO1) protein to suppress silencing, but also positively regulate the expression of the CYP81F2 gene, which is required for the biosynthesis of the aphid deterrent glucosinolate 4-methoxy-indol-3-yl-methylglusionolate (4MI3M; Zhang et al., 2006; Kim et al., 2008; Westwood et al., 2013). The CMV 1a protein stabilizes the repression of AGO1 mediated by 2b, and 2a stimulates CYP81F2 expression, leading to further induction of CYP81F2 and the production of 4MI3M in plants (Westwood et al., 2013). Thus, the viral protein 2b lures vectors to the plant by inducing the emission of plant volatiles and then makes the host unpalatable to the vector to trigger dispersal.
Vector effectors that target protein degradation pathways
No aphid or whitefly effectors have been identified that interact with specific components of the plant protein degradation pathways. However, the salivary protein Mp1 from M. persicae was shown to associate with the plant vesicle transport protein Vacuolar Protein Sorting Protein 52 (VPS52) from Arabidopsis in prevacuolar compartments (Table 2). Myzus persicae feeding and Mp1 expression reduced AtVSP52 protein levels in host plants and increased aphid fecundity (Rodriguez et al., 2017). Overexpression of VPS52 suppressed aphid performance in Arabidopsis, suggesting that VPS52 plays a defensive role in plant–aphid interactions. It is possible that Mp1 targets VPS52 for degradation in order to increase aphid performance. Another significant finding from this study is that VPS52 is preferentially expressed in the inflorescence in Arabidopsis where aphids prefer to feed (Rodriguez et al., 2017).
Target 3: Re-localization of proteins
Virus effectors that target the re-localization of proteins
Intracellular translocation of proteins between organelles is a key regulator of plant defense toward pathogens (dit Frey and Robatzek, 2009). A single viral effector, NIa-Pro, mediates increased performance of M. persicae on Turnip mosaic virus (TuMV)-infected and Potato virus Y (PVY, Family Potyviridae)-infected host plants through changes in plant defense (Casteel et al., 2014, 2015; Bak et al., 2017). While the plant interactants of NIa-Pro are not known, the authors determined that NIa-Pro relocalizes from the nucleus and cytoplasm to the vacuole of the cell when aphids are present and that localization outside of the nucleus is required for NIa-Pro’s ability to inhibit plant defenses and increase aphid fecundity (Figure 2 and Table 2; Bak et al., 2017). It is tempting to speculate that by localizing to the vacuole, NIa-Pro may be targeting plant proteins for degradation. NIa-Pro is the main protease for potyviruses, which cleaves itself from the polyprotein translated from the RNA genomes. NIa-Pro also cleaves the polyprotein at several other places containing conserved amino acid motifs to produce other functional potyvirus proteins (Adams et al., 2005). It is not known whether NIa-Pro also cleaves plant proteins with the same amino acid motifs to regulate localization in the cell or to inhibit plant defenses against aphids.
Vector effectors that target the re-localization of proteins
Insects induce systemic defense responses in plants (Ryan and Pearce, 2003), and since insect vectors of viruses feed on phloem tissue in plants (Kaloshian and Walling, 2005; Nalam et al., 2019), it is possible that vector effectors are mobile and are involved in the re-localization of plant proteins. However, there is no known evidence of such re-localization of plant proteins by vector effectors.
Target 4: Signal transduction
Virus effectors that target plant signal transduction
Although signal transduction is a major artery of downstream defense responses in plants that is targeted by several pathogens and insects (McDowell and Dangl, 2000; Kachroo et al., 2006; Zebelo and Maffei, 2012), a direct link that implicates virus effectors in targeting upstream plant proteins in signal transduction in order to alter plant–vector interactions specifically has not been demonstrated.
Vector effectors that target plant signal transduction
The salivary effector Me10 from potato aphid (Macrosiphum euphorbiae) was identified using RNA-seq analysis of aphid salivary glands and was shown to promote aphid fecundity (Chaudhary et al., 2019). Me10 and its homolog Ag10k from cotton aphid (Aphis gossypii) are secreted into the plant where they interact with the plant 14-3-3 protein TFT7 (Figure 2; Table 2; Chaudhary et al., 2019). The 14-3-3 proteins play important regulatory roles in signal transduction by binding to phosphorylated proteins to modulate their function (Denison et al., 2011). TFT7 associates with plant mitogen activated protein kinase kinase kinase (MAPKKK) protein kinase and its downstream kinase MAPKK, possibly stabilizing the proteins and activating programmed cell death and plant immunity (Oh et al., 2010). While silencing TFT7 in tomato did not affect M. euphoribae interactions, the longevity and fecundity of A. gossypii were enhanced. As A. gossypii is a nonadapted pest of tomato, this finding suggests that M. euphorbiae might produce effectors that compromise defenses downstream of TFT7, though additional studies are still needed. The 14-3-3 protein GF6 increases protein turnover in Arabidopsis; in gf6 mutants, the Plum pox virus titer was significantly reduced compared to wild-type Col-0 plants (Carrasco et al., 2014). Therefore, Me10–TFT7 interactions may also have implications in the performance of viruses transmitted by aphids. It would therefore be interesting in future studies to examine the impact of silencing TFT7 on aphid-transmitted viruses.
Hemipterans carry obligate and facultative symbionts, which convey advantageous traits for aphids in exchange for nutrients. The facultative symbiont Serratia symbiotica increases the heat tolerance of aphids and induces the pea aphids (Acyrthosiphon pisum) to secrete the histidine-rich calcium (Ca2+)-binding protein ApHRC in saliva during feeding (Figure 2 and Table 2). Ca2+ ions are important secondary messengers in signal transduction pathways in plants, which also play roles in plant defense responses in both plant–insect and plant–microbe interactions (Blumwald et al., 1998; Zebelo and Maffei, 2015). Aphids carrying S. symbiotica suppressed plant Ca2+ levels, as well as the levels of JA- and SA-related transcripts and reactive oxygen species (ROS), which are also important signaling molecule, compared to aphids without the symbiont (Wang et al., 2020). When ApHRC was overexpressed in Medicago truncatula, transcription of the JA biosynthesis gene LOX2 (LIPOXYGENASE2) was suppressed, ROS production was attenuated, and there was sustained phloem feeding of the aphids compared to the controls (Wang et al., 2020). Calmodulin is another Ca2+-binding protein that suppresses the production of siRNAs in plants by RNA-DEPENDENT RNA POLYMERASE, leading to reduced host RNA silencing and improved viral performance (Li et al., 2014a). It is not known whether Ca2+ binding by ApHRC also influences calmodulin’s function in RNA silencing or other viral defense responses related to Ca2+ signaling.
Hydrogen peroxide is a ROS and signaling molecule that mediates a wide variety of processes in plants, including early defense responses against insects and pathogens (Marcec et al., 2019). The whitefly effector BtFer1 is a ferretin protein that was shown to have Fe2+ binding ability and ferroxidase activity and to suppress hydrogen peroxide formation in tomato plants during whitefly feeding (Su et al., 2019). BtFer1 overexpression in plants suppressed JA-related transcript production and defense responses and increased the performance of whiteflies on plants compared to the controls (Table 2). The authors silenced BtFer1 in whiteflies using dsRNA feeding assays to demonstrate that BtFer is critical for whitely-induced inhibition of ROS production and defense responses during feeding (Su et al., 2019). The importance of ROS in plant defense responses against viral infection is already known and is two-fold. First, ROS can elicit localized cell death in virus-infected plants; second, they can act as diffusible mobile signals that may confer systemic resistance to the virus (Hernández et al., 2016). Although the work by Su et al. did not examine how BtFer1 affects viral performance (viral population growth or titer), suppression of ROS production in host plants by whitefly salivary effectors may play a significant role in the performance of the viruses transmitted by this insect.
Target 5: Detoxification of secondary metabolites
Virus effectors that target the detoxification of plant molecules
Plant proteins and metabolites that directly affect insects have been studied extensively. However, it is not known if virus effectors are directly involved in detoxifying plant molecules to increase vector performance. From the perspective of molecular synergisms, vector effectors that target plant detoxification of secondary metabolites to increase their own performance may indirectly also benefit the transmission of the virus that they carry. Therefore, in the following section, we review the vector effectors that detoxify plant metabolites and make the case for their significance in the molecular synergisms that occur among plant–virus–vector interactions.
Vector effectors that target the detoxification of plant molecules
Proteomic screening of cowpea aphid (Aphis craccivora) salivary proteins led to the identification of the aphid effector AcDCXR, which is a diacetyl/L-xylulose reductase (MacWilliams et al., 2020). Diacetyl/L-xyluose reductases are multifunctional enzymes that reversibly oxidize xylitol to xylulose and detoxify carbonyls. The levels of methylglyoxal, a toxic carbonyl, are elevated in pea (Pisum sativum) and cowpea (Vigna unguiculata) plants during cowpea aphid feeding (MacWilliams et al., 2020). AcDCXR is able to break down methylglyoxal in vitro, suggesting that this aphid effector plays a role in detoxification for the aphid. Consistent with this hypothesis, transiently overexpressing AcDCXR in pea plants increased aphid fecundity relative to aphids feeding on control plants (Figure 2 and Table 2; MacWilliams et al., 2020). However, the increase in aphid fecundity could also be due to enhanced nutrient quality, as in vitro assays demonstrated that AcDCXR also oxidizes xylitol to xylulose. Methylglyoxal production is also induced in mungbean plants (Vigna radiata) by Mungbean yellow mosaic virus (Melvin et al., 2017). Detoxification of this secondary metabolite may have more far-reaching implications for the performance of the viruses that cowpea aphids transmit; however, additional studies are needed to confirm this notion.
Another example of a vector effector potentially involved in detoxifying plant chemical defenses is the whitefly salivary protein laccase (LAC1; Table 2). LAC1 is secreted by whiteflies during feeding and is a polyphenol oxidase. Polyphenols are thought to be important for digestion and detoxification of plant secondary metabolites for insects. LAC1 expression was higher in whiteflies fed on host plants compared to an artificial diet. Suppression of LAC1 transcripts in the saliva of whiteflies by RNAi resulted in reduced whitefly performance on plants, but not when feeding on an artificial diet (Yang et al., 2017). Although the plant proteins that are targeted by LAC1 are not known, these findings suggest that LAC1 helps whiteflies overcome chemical defenses in plants.
The potato aphid (M. euphorbiae) effector Me47 increases aphid performance, potentially by detoxifying plant defenses, but the mechanisms are largely unclear (Figure 2 and Table 2). Me47 was characterized as a glutathione-S-transferase (GST), an enzyme group associated with detoxification of various defense compounds in insect guts. In vitro experiments demonstrated that Me47 can function as a GST to detoxify isothiocyanates, a type of defensive compound that is present in Arabidopsis but not tomato (Kettles and Kaloshian, 2016). However, overexpression of Me47 in Nicotiana benthamiana and tomato increased the fecundity of M. euphorbiae compared to the controls, whereas in Arabidopsis, Me47 expression suppressed M. persicae performance. Virus infection induces oxidative stress and ROS production during the early stages of plant defense responses, and plant GSTs also accumulate during this time as antioxidants, preventing ROS accumulation and cell damage (Gullner et al., 2018). Attenuation of oxidative stress by plant GSTs increases plant susceptibility to viruses. Therefore, Me47 secretion into the host by aphids may benefit plant viruses transmitted by aphids if the vector effector has antioxidant capacities like other GSTs.
Virus target 6: The unknowns
Virus effectors with unknown plant targets
While the plant targets of the viral effector proteins described above are known, there are still many questions regarding the mechanisms that surround several viral effectors in plant–vector interactions. Several studies have shown that plants infected by Potato leafroll virus (PLRV, family Potyviridae) are preferred by aphids over healthy plants for settling, that insect vectors have higher fecundity on infected plants, and that PLRV inhibits aphid-induced ET, JA, and SA production compared to the controls (Castle and Berger, 1993; Eigenbrode et al., 2002; Srinivasan and Alvarez, 2007; Wu et al., 2014; Patton et al., 2020). Three PLRV proteins, P0, P1, and P7, were shown to all increase aphid performance in N. benthamiana, while aphid preference was increased for plants expressing P0 and decreased for plants expressing P7 and P1, as compared to controls (Table 2; Patton et al., 2020). Although the exact cellular targets of these viral effectors remain unknown, aphid-induced JA and SA production was inhibited in plants expressing P0, P1, or P7 compared to plants expressing the empty vector controls. This could explain the improved performance of aphids, as they are susceptible to SA- and JA-dependent defense responses (Moran and Thompson, 2001; Ellis et al., 2002). P0 is also a known silencing suppressor of other poleroviruses (Baumberger et al., 2007; Cascardo et al., 2015), and this activity might also contribute to changes in plant–aphid interactions. P1 is a self-cleaving protease containing the VPg sequence (similar to NIa-Pro). The other viral effector, P7, reduced ET production in plants and made the plants less preferred as a host by aphids (Patton et al., 2020). These findings, along with a study identifying three viral proteins of CMV (2b, 2a, and 1a; Westwood et al., 2013), highlight the notion that multiple effectors are required to mediate plant–vector interactions.
Aphids feed on the phloem sap of plants, which is rich in sugar but contains low concentrations of amino acids. To augment this lack of amino acids, aphids have endosymbionts to supplement their nitrogen needs (Akman Gündüz and Douglas, 2012). The TuMV effector NIa-Pro mentioned above also increases the pool of free amino acids in N. benthamiana and A. thaliana (Casteel et al., 2014). While the exact mechanism by which NIa-Pro increases the free amino acid levels in host cells is not known, the increased amino acid pool may be beneficial for the performance of aphid vectors. There is also evidence that viral proteins may suppress aphid performance in plants, possibly to increase the movement of vectors off of plants. Three viral proteins from TuMV (HC-Pro, 6K1, and Vpg) all reduced the performance of green peach aphids (M. persicae) when transiently expressed in N. benthamiana (Table 2; Casteel et al., 2014). HC-Pro is a viral protease and silencing suppressor with multiple functions in infected host cells (Maia et al., 1996; Peng et al., 1998; Kasschau and Carrington, 2001; Kasschau et al., 2003). VPg proteins mediate the translation of viral genomes and are transported to the nucleus with NIa-Pro, where they may interfere with host defense responses (Schaad et al., 1996; Beauchemin et al., 2007; Rajamäki and Valkonen, 2009). However, VPg’s individual role in suppressing aphid fecundity in plants is still unknown. Little is known about the function of the viral protein 6K1 except that it plays a role in viral replication (Hongguang and Aiming, 2021). The relatively small size of 6K1 and its instability in virus-infected tissue have posed some challenges to understanding its role in virus–vector–host interactions. More work on the role of 6K1 in altering plant chemistry needs to be done in order to understand how it may suppress aphid performance on plants.
Vector effectors with unknown plant targets
The salivary effector Armet from pea aphid (A. pisum) was shown to be required for sustained feeding on host plants (Table 2; Cui et al., 2019). Expression of Armet had no impact on aphid performance in host plants, although pathogen defenses downstream of the SA pathway were activated in plants expressing Armet or infiltrated with the protein, and resistance to the bacterial pathogen Pseudomonas syringae was enhanced in Armet-infiltrated plants (Wang et al., 2015a; Cui et al., 2019). While it is not known how Armet alters pathogen defense in plants, the transcript abundance of SAMT, a methyltransferase that converts SA to methyl salicylate, was reduced, while that of SABP2, a methyl esterase that converts methyl salicylate to SA, was increased in these plants (Corina Vlot et al., 2009; Cui et al., 2019). Whiteflies have also been shown to induce plant resistance to the leaf pathogen Xanthomonas axonopodis pv. vesicatoria, as well as the soil-borne pathogens Agrobacterium tumefaciens and Ralstonia solanacearum, in pepper (Capsicum annuum) plants (Yang et al., 2011). SA- and JA-related genes were induced in aboveground and belowground tissue by whitefly feeding, indicating that systemic signals traveled from the whitefly feeding site to the roots (Lee et al., 2018). Subsequently, it was determined that three whitefly effectors, 2G4, 2G5, and 6A10, reduced pathogen symptoms in N. benthamiana leaves, while S2G4 and 6A10 also suppressed R. solanacearum accumulation in the roots compared to the controls (Table 2).
As mentioned in the “Introduction”, NLR proteins sense pathogen effectors or effector activity in the host cell and trigger ETI, including one of the most extreme plant defense responses, the hypersensitive response, a form of programmed cell death (Cui et al., 2015). When expressed in host plants, the salivary effector Mp10 from M. persicae inhibited performance and induced chlorosis and local cell death, suggesting that it is recognized by an NLR (Table 2). The activation of NLRs and induction of downstream defenses require molecular chaperones such as HSP90 and co-chaperones such as SGT1 (suppressor of the G2 allele of skp1; Azevedo et al., 2006; Kadota et al., 2010). Mp10-induced chlorosis in N. benthamiana plants requires SGT1, further supporting the notion that Mp10 is recognized by NLRs (Bos et al., 2010). In this study, the authors also found that Mp10 expression suppressed flg22-induced ROS production. Although the exact mechanisms of the induction of ETI and the suppression of PTI by Mp10 are unknown, the authors speculated that additional aphid salivary proteins might mask Mp10 recognition, preventing the induction of cell death.
Vector effectors with unknown plant targets have also been identified that increase vector performance. The salivary proteomes of A. pisum and M. persicae were shown to contain several macrophage migration inhibition (MIF) proteins, including MIF1, which is secreted during feeding (Naessens et al., 2015). When MIF1 was expressed in N. benthamiana, aphid fecundity increased, while the induction of programmed cell death, callose accumulation, and pathogenesis-related transcript accumulation were suppressed. Another M. persicae salivary effector, Mp55, suppressed callose accumulation, as well as the accumulation of hydrogen peroxide and the glucosinolate 4MI3M in host plants compared to the controls (Elzinga et al., 2014). Similar to MIF1, aphid fecundity increased when Mp55 was expressed in host plants compared to the controls. However, the plant targets of Mp55 that mediate changes in plant defense and aphid fecundity are unknown. The zinc metalloproteases ACEs are angiotensin-converting enzymes that regulate blood pressure and electrolyte homeostasis in mammals (Corvol et al., 1995). ACE proteins were shown to be present in the saliva of A. pisum (Macours and Hens, 2004), and two ACE genes are highly expressed in A. pisum salivary glands (Wang et al., 2015b). Knocking down both aphid ACE genes with RNAi resulted in lower aphid fecundity on plants, suggesting they play a role in host plant colonization. Since ACE enzymes remove dipeptides from short oligopeptides (Macours and Hens, 2004), they may be involved in cleaving plant proteins that trigger plant defenses against both aphids and viruses. While some information is known about the above effectors, several salivary effectors have been identified with no similarities to other protiens that increase (Me23, MpC002, ApC002, Mp2, and Ap25) and decrease (Mp42, Mp56, Mp57, and Mp58) aphid performance when expressed in host plants (Mutti et al., 2008; Bos et al., 2010; Atamian et al., 2013; Elzinga et al., 2014; Guy et al., 2016; Boulain et al., 2018). It is not known if any of these aphid effectors have a role in directly altering resistance to plant viruses.
How can we identify effector synergisms in plant–vector–virus interactions?
Due to their long co-evolutionary histories, insect vectors, and vector-borne viruses may produce synergistic effectors that (1) benefit both players through shared targets, (2) benefit only the other player and not the producer, or (3) have new functions that are only active when both effector players are present. It would be difficult to separate synergistic effectors that benefit both players from convergent evolution; however, this could be done using vector populations that are isolated geographically and when a virus is only native to one of the population’s geographic regions. In this case, the salivary proteomes of both vector populations could be examined. Protein effectors that differ between the two populations and benefit both players through shared host targets would be evidence of effector synergism. Some aphid species have developed numerous biotypes that specialize on different host plants, but are considered the same species (Yates and Michel, 2018). For example, there are at least 15 different pea aphid biotypes that specialize on specific hosts (Peccoud et al., 2009, 2010). Recently, it was shown that pea aphid salivary effector expression differs between biotypes and is associated with variation in copy number in the genome (Boulain et al., 2019). The impact of virus–vector interactions in aphid biotype formation would be an interesting area to investigate.
Synergistic effectors that benefit only the other player and not the producer, or effectors that have new functions that are only active when both players are present, could be identified by re-screening vector or virus effector-omes for ecological roles, such as increasing virus titer or vector performance, as was done in Casteel et al. (2014) for all the major TuMV effectors. Evidence for synergistic vector effectors has not been obtained; however, there is evidence that plants have evolved forms of resistance that are only activated by the other player. The plant NLR Vat1 does not convey virus resistance in host plants unless specific aphid vectors are actively feeding on the plant. This suggests that by studying effector synergisms, we may also identify new forms of resistance, further warranting this work. Based on the literature, we already know that certain virus families increase vector performance (Table 1), in addition to altering vector behavior in ways that should increase transmission, and thus these interactions should be considered a form of mutualism. In cases where there is more variation in the impact of viruses on vectors, such as with Potyviridae (Table 1), the relationship between the insect vector and virus and their effectors may be dependent on insect or pathogen host range. For example, viruses with a limited host range or specialist insects that feed only on certain families of plants may result in selection for vector or viral effectors that suppress plant defenses that are intrinsic to that specific plant family.
Future directions and concluding remarks
Over the past decade, our understanding of how viruses modulate plant–vector interactions has significantly progressed, as the molecular mechanisms that underlie how viral effectors influence insect biology, insect behavior, and plant physiology are beginning to be revealed. However, several areas of research need more attention in the future. First, despite recent advances, functional analysis is still required for many of the identified viral effectors that mediate plant–vector interactions, and additional viral–vector–host systems need to be investigated using tools in genetics, molecular biology, and chemical ecology. Second, although functions in virus–plant interactions have been attributed to most plant virus proteins, such proteins are often multi-functional. Their functionality in more ecologically relevant contexts, such as plant–vector interactions, should therefore be re-examined. Third, while research focused on identifying vector effectors and their host targets has also expanded in recent years, additional research on mechanisms used by vector effectors is still needed. Finally, although mutualisms with their insect vectors may be common for some viral families (Table 1), to our knowledge, only one vector effector has been identified that alters plant interactions with the virus it transmits (Wang et al., 2019), underlining the need to re-examine the roles of vector effectors in plant–virus interactions. Below we expand on a few ecological contexts we think are particularly important to explore with the points above in mind.
Developmental changes in vector–virus–plant interactions
Just as plants and insects develop throughout their lifecycles via different stages, virus infection develops over time. Thus, the needs and challenges faced by each organism are likely to change throughout development. The utilization of effector-mediated responses and functionality may also change over time, yet we know little about how this is regulated. Early in the infection process, it would be more beneficial to the virus to attract insect vectors to infected plants and to increase their reproduction, while later in the infection process, repelling insects with increased defenses would be beneficial for viral spread. Consistent with this notion, aphids have increased performance on CMV-infected or Zucchini yellow mosaic virus-infected plants during the early stages of the infection process, whereas both of these unrelated viruses have a negative impact on aphid performance in the late stages of infection (Blua and Perring, 1992). Virus-induced volatile profiles also change during the course of infection and in plants of different ages (Werner et al., 2009). It is reasonable to predict that different viral effectors mediate different plant responses over time and that plant targets change over time, but little research has addressed this. Furthermore, the expression of whitefly effectors changes over the course of insect development (Yang et al., 2017; Su et al., 2019). It is not known whether temporally regulated vector effectors affect whitefly nymphs and adults differently, or if the regulation of vector effectors is related to the ecology of vector–virus interactions. Functional analyses of effectors should involve experiments with plants of different ages, vectors of different ages, and different stages of viral infection.
RNA effectors in vector–virus–plant interactions
There are abundant examples of the secretion of protein effectors into host plants via aphid and whitefly saliva. Aphids and whiteflies also release sRNAs and long noncoding (lnc)RNAs during feeding, which modulate plant suitability for the insect (van Kleeff et al., 2016; Chen et al., 2020). For example, Ya transcripts are introduced into the host plant while M. persicae feeds, and these lncRNAs migrate to other areas of the plant. Ya1 RNA is predicted to alter plant defenses or nutrients, as aphids have higher fecundity on plants stably producing the RNA compared to the controls (Chen et al., 2020). sRNAs from whitefly were also found to be transferred from the insect to tomato plants while feeding, but the impacts of these sRNAs on whitefly biology and changes in the host plant were not examined (van Kleeff et al., 2016). Viruses encode RNA-binding proteins in their genomes to facilitate genome movement from cell to cell (Kasschau and Carrington, 2001) and sRNA-binding proteins to compromise RNA silencing (Kasschau et al., 2003). It is possible that vector-secreted RNAs interact with these viral proteins, facilitating their movement in the plant or masking their perception by the plant. As discussed earlier, siRNA-mediated RNA-induced silencing complexes are important mechanisms that plant viruses use to silence host anti-viral RNA. From the standpoint of a virus–vector mutualism, it would be useful to unravel whether any of these vector noncoding RNAs are directly involved in suppressing plant antiviral RNA.
Vector and nonvector secretions in vector–virus–plant interactions
Potato aphid saliva contains proteins produced by its primary endosymbiont Buchnera aphidicola, such as GroEL, which induces plant defenses against aphids feeding on A. thaliana and tomato plants (Chaudhary et al., 2014). Whiteflies and aphids also deposit honeydew on the plant surface while feeding. Honeydew contains insect and endosymbiont proteins (Sabri et al., 2013) that may serve as effectors or elicitors. In addition to proteins, honeydew from pea aphids also contain SA, which can suppress plant defenses toward aphids and increase insect performance (Schwartzberg and Tumlinson, 2014). Rice planthopper’s honeydew is enriched in microbes, which may induce direct and indirect plant defenses (Wari et al., 2019). Furthermore, oviposition cues from insect egg deposition induce plant responses (Hilker and Fatouros, 2015). It is not known if elicitors or effectors from insect honeydew, eggs, or endosymbionts are altered by plant viruses and their effectors, or if they alter plant–virus interactions. Nonvector insects and nonpathogenic microbes often occur together with viral infections and vectors, and can influence plant–virus–vector interactions (Chisholm et al., 2018; Basu et al., 2021; Lee et al., 2021). The impact of nonvector elicitors and effectors may influence the outcome of plant–vector–virus interactions. For example, nonvector caterpillar herbivores deposit plant proteins such as Endochitinase A and PR4 in their feces, which induces the SA pathway and increases caterpillar performance on the host plant (Ray et al., 2016). While not every interaction can be examined or is biologically relevant, long-term associations that occur in nature and in nonmodel systems deserve additional attention.
Concluding remarks
While significant work has been done on understanding plant responses to virus and vector effectors, we have highlighted the importance of studying these interactions in tandem and in broader ecological contexts, as this will lead to a better understanding of the mechanisms that mediate plant interactions more broadly. From the work reviewed above, we show that mutualisms in virus–vector interactions are common (Table 1), some targets for plant virus and vectors are conserved, and many mechanisms are unknown (Table 2), and we define how to determine in future research which of these effector-mediated interactions are cooperative and have co-evolved together (Table 1).
Acknowledgments
This work is supported by a National Science Foundation Plant Genome Early Career Award to C.L.C (PGRP #1723926). Figures were prepared using Biorender.com. We thank Dr. Chad Nirhanz for valuable comments on earlier versions of this article.
Conflict of interest statement. The authors declare no conflict of interest.
Contributor Information
Swayamjit Ray, School of Integrative Plant Science, Plant Pathology and Plant-Microbe Biology Section, Cornell University, Ithaca, New York 14850, USA.
Clare L Casteel, School of Integrative Plant Science, Plant Pathology and Plant-Microbe Biology Section, Cornell University, Ithaca, New York 14850, USA.
C.L.C. and S.R. conceived the project and wrote the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Clare L. Casteel (ccasteel@cornell.edu).
References
- Acevedo FE, Rivera-Vega LJ, Chung SH, Ray S, Felton GW (2015) Cues from chewing insects - the intersection of DAMPs, HAMPs, MAMPs and effectors. Curr Opin Plant Biol 26: 80–86 [DOI] [PubMed] [Google Scholar]
- Adams MJ, Antoniw JF, Beaudoin F (2005) Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol Plant Pathol 6: 471–487. [DOI] [PubMed] [Google Scholar]
- Akman Gündüz E, Douglas AE (2012) Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc R Soc B Biol Sci 276: 987–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamian HS, Chaudhary R, Cin VD, Bao E, Girke T, Kaloshian I (2013) In planta expression or delivery of potato aphid Macrosiphum euphorbiae effectors Me10 and Me23 enhances aphid fecundity. Mol Plant-Microbe Interact 26: 67–74 [DOI] [PubMed] [Google Scholar]
- Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noël L, Sadanandom A, Casais C, Parker J, Shirasu K (2006) Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J 25: 2007–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak A, Cheung AL, Yang C, Whitham SA, Casteel CL (2017) A viral protease relocalizes in the presence of the vector to promote vector performance. Nat Commun 8: 14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Clark RE, Bera S, Casteel CL, Crowder DW (2021) Responses of pea plants to multiple antagonists are mediated by order of attack and phytohormone crosstalk. Mol Ecol 30: 4939–4948 [DOI] [PubMed] [Google Scholar]
- Baulcombe DC (2015) VIGS, HIGS and FIGS: small RNA silencing in the interactions of viruses or filamentous organisms with their plant hosts. Curr Opin Plant Biol 26: 141–146 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Tsai CH, Lie M, Havecker E, Baulcombe DC (2007) The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol 17: 1609–1614 [DOI] [PubMed] [Google Scholar]
- Beauchemin C, Boutet N, Laliberté JF (2007) Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of Turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E In Planta. J Virol 81: 775–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc S, Michalakis Y (2016) Manipulation of hosts and vectors by plant viruses and impact of the environment. Curr Opin Insect Sci 16: 36–43 [DOI] [PubMed] [Google Scholar]
- Blua MJ, Perring TM (1992) Effects of Zucchini yellow mosaic virus on colonization and feeding behavior of Aphis gossypii (Homoptera: Aphididae) alatae. Environ Entomol 21: 578–585 [Google Scholar]
- Blumwald E, Aharon GS, CH Lam B (1998) Early signal transduction pathways in plant-pathogen interactions. Trends Plant Sci 3: 342–346 [Google Scholar]
- Blundell R, Schmidt JE, Igwe A, Cheung AL, Vannette RL, Gaudin ACM, Casteel CL (2020) Organic management promotes natural pest control through altered plant resistance to insects. Nat Plants 6: 483–491 [DOI] [PubMed] [Google Scholar]
- Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O’Connor D, Grotewold E, Hake S (2012) Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev 26: 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bos JIB, Prince D, Pitino M, Maffei ME, Win J, Hogenhout SA (2010) A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid) PLoS Genet 6: e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulain H, Legeai F, Guy E, Morlière S, Douglas NE, Oh J, Murugan M, Smith M, Jaquiéry J, Peccoud J, et al. (2018) Fast evolution and lineage-specific gene family expansions of aphid salivary effectors driven by interactions with host-plants. Genome Biol Evol 10: 1554–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulain H, Legeai F, Jaquiéry J, Guy E, Morlière S, Simon JC, Sugio A (2019) Differential expression of candidate salivary effector genes in pea aphid biotypes with distinct host plant specificity. Front Plant Sci 10: 1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O (2006) The diversity of RNA silencing pathways in plants. Trends Genet 22: 268–280 [DOI] [PubMed] [Google Scholar]
- Bronstein JL, Alarcón R, Geber M (2006) The evolution of plant-insect mutualisms. New Phytol 172: 412–428 [DOI] [PubMed] [Google Scholar]
- Carol B, Biology O, State TO (2014) Mighty mutualisms: the nature of plant-pollinator interactions. Nat Educ Knowl 3: 37 [Google Scholar]
- Carr JP, Murphy AM, Tungadi T, Yoon JY (2019) Plant defense signals: players and pawns in plant-virus-vector interactions. Plant Sci 279: 87–95 [DOI] [PubMed] [Google Scholar]
- Carrasco JL, Castelló MJ, Naumann K, Lassowskat I, Navarrete-Gómez M, Scheel D, Vera P (2014) Arabidopsis protein phosphatase DBP1 nucleates a protein network with a role in regulating plant defense. PLoS One 9: e90734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascardo RS, Arantes ILG, Silva TF, Sachetto-Martins G, Vaslin MFS, Corrêa RL (2015) Function and diversity of P0 proteins among Cotton leafroll dwarf virus isolates. Virol J 12: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casteel CL, Falk BW (2016) Plant virus-vector interactions: more than just for virus transmission. In Wang A and Zhou X, eds, Current Research Topics in Plant Virology, Springer International Publishing, Cham, Switzerland, pp 217–240 [Google Scholar]
- Casteel CL, Jander G (2013) New synthesis: Investigating mutualisms in virus-vector interactions. J Chem Ecol 39: 809. [DOI] [PubMed] [Google Scholar]
- Casteel CL, De Alwis M, Bak A, Dong H, Whitham SA, Jander G (2015) Disruption of ethylene responses by Turnip mosaic virus mediates suppression of plant defense against the green peach aphid vector. Plant Physiol 169: 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casteel CL, Yang C, Nanduri AC, De Jong HN, Whitham SA, Jander G (2014) The NIa-Pro protein of Turnip mosaic virus improves growth and reproduction of the aphid vector, Myzus persicae (green peach aphid) Plant J 77: 653–663 [DOI] [PubMed] [Google Scholar]
- Castle SJ, Berger PH (1993) Rates of growth and increase of Myzus persicae on virus‐infected potatoes according to type of virus‐vector relationship. Entomol Exp Appl 69: 51–60 [Google Scholar]
- Chaudhary R, Atamian HS, Shen Z, Briggs SP, Kaloshian I (2014) GroEL from the endosymbiont Buchnera aphidicola betrays the aphid by triggering plant defense. Proc Natl Acad Sci USA 111: 8919–8924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary R, Peng HC, He J, MacWilliams J, Teixeira M, Tsuchiya T, Chesnais Q, Mudgett MB, Kaloshian I (2019) Aphid effector Me10 interacts with tomato TFT7, a 14-3-3 isoform involved in aphid resistance. New Phytol 221: 1518–1528 [DOI] [PubMed] [Google Scholar]
- Chen Y, Singh A, Kaithakottil GG, Mathers TC, Gravino M, Mugford ST, van Oosterhout C, Swarbreck D, Hogenhout SA (2020) An aphid RNA transcript migrates systemically within plants and is a virulence factor. Proc Natl Acad Sci USA 117: 12763–12771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm PJ, Sertsuvalkul N, Casteel CL, Crowder DW (2018) Reciprocal plant-mediated interactions between a virus and a non-vector herbivore. Ecology 99: 2139–2144 [DOI] [PubMed] [Google Scholar]
- Corina Vlot A, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Corvol P, Michaud A, Soubrier F, Williams TA (1995) Recent advances in knowledge of the structure and function of the angiotensin I converting enzyme. J Hypertens 13: 3–10 [DOI] [PubMed] [Google Scholar]
- Couto D, Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16: 537–552 [DOI] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE (2015) Effector-triggered immunity: From pathogen perception to robust defense. Annu Rev Plant Biol 66: 487–511 [DOI] [PubMed] [Google Scholar]
- Cui N, Lu H, Wang T, Zhang W, Kang L, Cui F (2019) Armet, an aphid effector protein, induces pathogen resistance in plants by promoting the accumulation of salicylic acid. Philos Trans R Soc B Biol Sci 374: 20180314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins R (2016) The Extended Phenotype: The Long Reach of the Gene, Oxford University Press, Oxford
- Denison FC, Paul AL, Zupanska AK, Ferl RJ (2011) 14-3-3 proteins in plant physiology. Semin Cell Dev Biol 22: 720–727 [DOI] [PubMed] [Google Scholar]
- dit Frey NF, Robatzek S (2009) Trafficking vesicles: pro or contra pathogens? Curr Opin Plant Biol 12: 437–443 [DOI] [PubMed] [Google Scholar]
- Eigenbrode SD, Ding H, Shiel P, Berger PH (2002) Volatiles from potato plants infected with Potato leafroll virus attract and arrest the virus vector, Myzus persicae (Homoptera: Aphididae). Proc R Soc B Biol Sci 269: 455–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Turner JG (2002) Constitutive activation of jasmonate signaling in an Arabidopsis mutant correlates with enhanced resistance to Erysiphe cichoracearum, Pseudomonas syringae, and Myzus persicae. Mol Plant-Microbe Interact 15: 1025–1030 [DOI] [PubMed] [Google Scholar]
- Elzinga DA, De Vos M, Jander G (2014) Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Mol Plant-Microbe Interact 27: 747–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fereres A, Moreno A (2009) Behavioural aspects influencing plant virus transmission by homopteran insects. Virus Res 141: 158–168 [DOI] [PubMed] [Google Scholar]
- Fereres A, Raccah B (2015) Plant virus transmission by insects. In eLS, John Wiley & Sons, Ltd, Chichester [Google Scholar]
- French E, Kaplan I, Iyer-Pascuzzi A, Nakatsu CH, Enders L (2021) Emerging strategies for precision microbiome management in diverse agroecosystems. Nat Plants 7: 256–267 [DOI] [PubMed] [Google Scholar]
- Gaafar YZA, Ziebell H (2020) Aphid transmission of nanoviruses. Arch Insect Biochem Physiol 104: e21668. [DOI] [PubMed] [Google Scholar]
- Gilbertson RL, Batuman O, Webster CG, Adkins S (2015) Role of the insect supervectors Bemisia tabaci and Frankliniella occidentalis in the emergence and global spread of plant viruses. Annu Rev Virol 2: 67–93 [DOI] [PubMed] [Google Scholar]
- Gnanasekaran P, KishoreKumar R, Bhattacharyya D, Vinoth Kumar R, Chakraborty S (2019) Multifaceted role of geminivirus associated betasatellite in pathogenesis. Mol Plant Pathol 20: 1019–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullner G, Komives T, Király L, Schröder P (2018) Glutathione S-transferase enzymes in plant-pathogen interactions. Front Plant Sci 9: 1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy E, Boulain H, Aigu Y, Le Pennec C, Chawki K, Morlière S, Schädel K, Kunert G, Simon JC, Sugio A (2016) Optimization of agroinfiltration in Pisum sativum provides a new tool for studying the salivary protein functions in the pea aphid complex. Front Plant Sci 7: 1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S, Smith HMS, Holtan H, Magnani E, Mele G, Ramirez J (2004) The role of KNOX genes in plant development. Annu Rev Cell Dev Biol 20: 125–151 [DOI] [PubMed] [Google Scholar]
- Heil M (2016) Host manipulation by parasites: cases, patterns, and remaining doubts. Front Ecol Evol 4: 80 [Google Scholar]
- Henry LM, Maiden MCJ, Ferrari J, Godfray HCJ (2015) Insect life history and the evolution of bacterial mutualism. Ecol Lett 18: 516–525 [DOI] [PubMed] [Google Scholar]
- Hernández JA, Gullner G, Clemente-Moreno MJ, Künstler A, Juhász C, Díaz-Vivancos P, Király L (2016) Oxidative stress and antioxidative responses in plant–virus interactions. Physiol Mol Plant Pathol 94: 134–148 [Google Scholar]
- Hilker M, Fatouros NE (2015) Plant responses to insect egg deposition. Annu Rev Entomol 60: 493–515 [DOI] [PubMed] [Google Scholar]
- Hogenhout SA, Bos JIB (2011) Effector proteins that modulate plant–insect interactions. Curr Opin. Plant Biol 14: 422–428 [DOI] [PubMed] [Google Scholar]
- Hogenhout SA, Ammar ED, Whitfield AE, Redinbaugh MG (2008) Insect vector interactions with persistently transmitted viruses. Annu Rev Phytopathol 46: 327–359 [DOI] [PubMed] [Google Scholar]
- Hongguang C, Aiming WAS (2021) Plum pox virus 6K1 protein Is required for viral replication and targets the viral replication complex at the early stage of infection. J Virol 90: 5119–5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingwell LL, Eigenbrode SD, Bosque-Pérez NA (2012) Plant viruses alter insect behavior to enhance their spread. Sci Rep 2: 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Q, Liu N, Xie K, Dai Y, Han S, Zhao X, Qian L, Wang Y, Zhao J, Gorovits R, et al. (2016) CLCuMuB βC1 subverts ubiquitination by interacting with NbSKP1s to enhance geminivirus infection in Nicotiana benthamiana. PLoS Pathog 12: e1005668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jun-Bo L, Jun-Min L, Nélia V, Yong-Liang W, Fang-Fang L, Yan-Yuan B, Chuan-Xi Z, Shu-Sheng L, Xiao-Wei W (2011) Global analysis of the transcriptional response of whitefly to Tomato yellow leaf curl china virus reveals the relationship of coevolved adaptations. J Virol 85: 3330–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo P, Chandra‐Shekara AC, Klessig DF (2006) Plant signal transduction and defense against viral pathogens. Adv Virus Res 66: 161–191 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Shirasu K, Guerois R (2010) NLR sensors meet at the SGT1-HSP90 crossroad. Trends Biochem Sci 35: 199–207 [DOI] [PubMed] [Google Scholar]
- Kaloshian I, Walling LL (2005) Hemipterans as plant pathogens. Annu Rev Phytopathol 43: 491–521 [DOI] [PubMed] [Google Scholar]
- Kanno H, Satoh M, Kimura T, Fujita Y (2005) Some aspects of induced resistance to rice blast fungus, Magnaporthe grisea, in rice plant infested by white-backed planthopper, Sogatella furcifera. Appl Entomol Zool 40: 91–97 [Google Scholar]
- Kasschau KD, Carrington JC (2001) Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 285: 71–81 [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC (2003) P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev Cell 4: 205–217 [DOI] [PubMed] [Google Scholar]
- Katsir L, Chung HS, Koo AJ, Howe GA (2008) Jasmonate signaling: a conserved mechanism of hormone sensing. Curr Opin Plant Biol 11: 428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S (2010) The role of pattern-recognition receptors in innate immunity: Update on toll-like receptors. Nat Immunol 11: 373–384 [DOI] [PubMed] [Google Scholar]
- Kettles GJ, Kaloshian I (2016) The potato aphid salivary effector Me47 is a glutathione-S-transferase involved in modifying plant responses to aphid infestation. Front Plant Sci 7: 1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiers ET, Van Der Heijden MGA (2006) Mutualistic stability in the arbuscular mycorrhizal symbiosis: Exploring hypotheses of evolutionary cooperation. Ecology 87: 1627–1636 [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee BW, Schroeder FC, Jander G (2008) Identification of indole glucosinolate breakdown products with antifeedant effects on Myzus persicae (green peach aphid) Plant J 54: 1015–1026 [DOI] [PubMed] [Google Scholar]
- Lee BW, Basu S, Bera S, Casteel CL, Crowder DW (2021) Responses to predation risk cues and alarm pheromones affect plant virus transmission by an aphid vector. Oecologia 196: 1005–1015 [DOI] [PubMed] [Google Scholar]
- Lee HR, Lee S, Park S, van Kleeff PJM, Schuurink RC, Ryu CM (2018) Transient expression of whitefly effectors in Nicotiana benthamiana leaves activates systemic immunity against the leaf pathogen Pseudomonas syringae and soil-borne pathogen Ralstonia solanacearum. Front Ecol Evol 6: 90 [Google Scholar]
- Li F, Ding SW (2006) Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol 28: 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Huang C, Li Z, Zhou X (2014a) Suppression of RNA silencing by a plant DNA virus satellite requires a host calmodulin-like protein to repress RDR6 expression. PLoS Pathog. 10: e1003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Liu C, Deng WH, Yao DM, Pan LL, Li YQ, Liu YQ, Liang Y, Zhou XP, Wang XW (2019) Plant begomoviruses subvert ubiquitination to suppress plant defenses against insect vectors. PLoS Pathog 15: e1007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Weldegergis BT, Li J, Jung C, Qu J, Sun Y, Qian H, Tee C, van Loon JJA, Dicke M, et al. (2014b) Virulence factors of geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell 26: 4991–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macours N, Hens K (2004) Zinc-metalloproteases in insects: ACE and ECE. Insect Biochem Mol Biol 34: 501–510 [DOI] [PubMed] [Google Scholar]
- MacWilliams JR, Dingwall S, Chesnais Q, Sugio A, Kaloshian I (2020) AcDCXR Is a cowpea aphid effector with putative roles in altering host immunity and physiology. Front Plant Sci 11: 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia IG, Haenni AL, Bernardi F (1996) Potyviral HC-Pro: a multifunctional protein. J Gen Virol 77: 1335–1341 [DOI] [PubMed] [Google Scholar]
- Marcec MJ, Gilroy S, Poovaiah BW, Tanaka K (2019) Mutual interplay of Ca 2+ and ROS signaling in plant immune response. Plant Sci 283: 343–354 [DOI] [PubMed] [Google Scholar]
- Martinière A, Bak A, Macia JL, Lautredou N, Gargani D, Doumayrou J, Garzo E, Moreno A, Fereres A, Blanc S, et al. (2013) A virus responds instantly to the presence of the vector on the host and forms transmission morphs. Elife 2: e00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauck KE, Chesnais Q, Shapiro LR (2018) Evolutionary determinants of host and vector manipulation by plant viruses. In Malmstrom C, ed, Environmental Virology and Virus Ecology, Academic Press, Cambridge, MA, pp 189–250 [DOI] [PubMed] [Google Scholar]
- Mauck KE, De Moraes CM, Mescher MC (2010) Deceptive chemical signals induced by a plant virus attract insect vectors to inferior hosts. Proc Natl Acad Sci USA 107: 3600–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauck KE, De Moraes CM, Mescher MC (2015) Infection of host plants by Cucumber mosaic virus increases the susceptibility of Myzus persicae aphids to the parasitoid Aphidius colemani. Sci Rep 5: 10963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer RT, Inbar M, McKenzie CL, Shatters R, Borowicz V, Albrecht U, Powell CA, Doostdar H (2002) Multitrophic interactions of the silverleaf whitefly, host plants, competing herbivores, and phytopathogens. Arch Insect Biochem Physiol 51: 151–169 [DOI] [PubMed] [Google Scholar]
- McDowell JM, Dangl JL (2000) Signal transduction in the plant immune response. Trends Biochem Sci 25: 79–82 [DOI] [PubMed] [Google Scholar]
- Melvin P, Bankapalli K, D’Silva P, Shivaprasad PV (2017) Methylglyoxal detoxification by a DJ-1 family protein provides dual abiotic and biotic stress tolerance in transgenic plants. Plant Mol Biol 94: 381–397 [DOI] [PubMed] [Google Scholar]
- Meng X, Zhang S (2013) MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 51: 245–266 [DOI] [PubMed] [Google Scholar]
- Moran PJ, Thompson GA (2001) Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol 125: 1074–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriones E, Navas-Castillo J (2000) Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res 71: 123–134 [DOI] [PubMed] [Google Scholar]
- Murphy AM, Zhou T, Carr JP (2020) An update on salicylic acid biosynthesis, its induction and potential exploitation by plant viruses. Curr Opin Virol 42: 8–17 [DOI] [PubMed] [Google Scholar]
- Murrell EG, Ray S, Lemmon ME, Luthe DS, Kaye JP (2019) Cover crop species affect mycorrhizae-mediated nutrient uptake and pest resistance in maize. Renew Agric Food Syst 35: 467–474 [Google Scholar]
- Mutti NS, Louis J, Pappan LK, Pappan K, Begum K, Chen MS, Park Y, Dittmer N, Marshall J, Reese JC, et al. (2008) A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc Natl Acad Sci USA 105: 9965–9969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naalden D, van Kleeff PJM, Dangol S, Mastop M, Corkill R, Hogenhout SA, Kant MR, Schuurink RC (2021) Spotlight on the roles of whitefly effectors in insect–plant interactions. Front Plant Sci 12: 1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naessens E, Dubreuil G, Giordanengo P, Baron OL, Minet-Kebdani N, Keller H, Coustau C (2015) A secreted MIF cytokine enables aphid feeding and represses plant immune responses. Curr Biol 25: 1898–1903 [DOI] [PubMed] [Google Scholar]
- Nalam V, Louis J, Shah J (2019) Plant defense against aphids, the pest extraordinaire. Plant Sci 279: 96–107 [DOI] [PubMed] [Google Scholar]
- Nelson CJ, Millar AH (2015) Protein turnover in plant biology. Nat Plants 1: 15017. [DOI] [PubMed] [Google Scholar]
- Ng JCK, Perry KL (2004) Transmission of plant viruses by aphid vectors. Mol Plant Pathol 5: 505–511 [DOI] [PubMed] [Google Scholar]
- Nurmberg PL, Knox KA, Yun BW, Morris PC, Shafiei R, Hudson A, Loake GJ (2007) The developmental selector AS1 is an evolutionarily conserved regulator of the plant immune response. Proc Natl Acad Sci USA 104: 18795–18800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Pedley KF, Martin GB (2010) Tomato 14-3-3 protein 7 positively regulates immunity-associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKK α. Plant Cell 22: 260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan LL, Miao H, Wang Q, Walling LL, Liu SS (2021) Virus-induced phytohormone dynamics and their effects on plant–insect interactions. New Phytol 230: 1305–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton MF, Bak A, Sayre JM, Heck ML, Casteel CL (2020) A polerovirus, Potato leafroll virus, alters plant–vector interactions using three viral proteins. Plant Cell Environ 43: 387–399 [DOI] [PubMed] [Google Scholar]
- Peccoud J, Ollivier A, Plantegenest M, Simon JC (2009) A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc Natl Acad Sci 106: 7495–7500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccoud J, Simon JC, von Dohlen C, Coeur d’acier A, Plantegenest M, Vanlerberghe-Masutti F, Jousselin E (2010) Evolutionary history of aphid-plant associations and their role in aphid diversification. C R Biol 333: 474–487 [DOI] [PubMed] [Google Scholar]
- Peng YH, Kadoury D, Gal-On A, Huet H, Wang Y, Raccah B (1998) Mutations in the HC-Pro gene of Zucchini yellow mosaic potyvirus: effects on aphid transmission and binding to purified virions. J Gen Virol 79: 897–904 [DOI] [PubMed] [Google Scholar]
- Perilla-Henao LM, Casteel CL (2016) Vector-borne bacterial plant pathogens: interactions with hemipteran insects and plants. Front Plant Sci 7: 1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitino M, Hogenhout SA (2012) Aphid protein effectors promote aphid colonization in a plant species-specific manner. Mol Plant-Microbe Interact 26: 130–139 [DOI] [PubMed] [Google Scholar]
- Poulin R, Maure F (2015) Host manipulation by parasites: a look back before moving forward. Trends Parasitol 31: 563–570 [DOI] [PubMed] [Google Scholar]
- Prüfer D, Kawchuk L, Monecke M, Nowok S, Fischer R, Rohde W (1999) Immunological analysis of Potato leafroll luteovirus (PLRV) P1 expression identifies a 25 kDa RNA-binding protein derived via P1 processing. Nucleic Acids Res 27: 421–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Xia R, Zhai J, Hou Y, Feng L, Zhai Y, Ma W (2021) Small RNAs in plant immunity and virulence of filamentous pathogens. Annu Rev Phytopathol 59: 265–288 [DOI] [PubMed] [Google Scholar]
- Quito-Avila DF, Lightle D, Lee J, Martin RR (2012) Transmission biology of Raspberry latent virus, the first aphid-borne Reovirus. Phytopathology 102: 547–553 [DOI] [PubMed] [Google Scholar]
- Rajamäki ML, Valkonen JPT (2009) Control of nuclear and nucleolar localization of nuclear inclusion protein a of picorna-like potato virus a in nicotiana species. Plant Cell 21: 2485–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh SV, Yogindran S, Gnanasekaran P, Chakraborty S, Winter S, Pappu HR (2021) Virus and viroid-derived small RNAs as modulators of host gene expression: molecular insights into pathogenesis. Front Microbiol 11: 614231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Alves PCMS, Ahmad I, Gaffoor I, Acevedo FE, Peiffer M, Jin S, Han Y, Shakeel S, Felton GW, et al. (2016) Turnabout is fair play: herbivory-induced plant chitinases excreted in fall armyworm frass suppress herbivore defenses in maize. Plant Physiol 171: 694–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez PA, Escudero-Martinez C, Bos JIB (2017) An aphid effector targets trafficking protein VPS52 in a host-specific manner to promote virulence. Plant Physiol 173: 1892–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roossinck MJ (2011) The good viruses: viral mutualistic symbioses. Nat Rev Microbiol 9: 99–108 [DOI] [PubMed] [Google Scholar]
- Ryan CA, Pearce G (2003) Systemins: a functionally defined family of peptide signals that regulate defensive genes in Solanaceae species. Proc Natl Acad Sci USA 100: 14577–14580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabri A, Vandermoten S, Leroy PD, Haubruge E, Hance T, Thonart P, De Pauw E, Francis F (2013) Proteomic investigation of aphid honeydew reveals an unexpected diversity of proteins. PLoS One 8: e74656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saptarshi G, Kanakala S, Lebedev G, Kontsedalov S, Silverman D, Alon T, Mor N, Sela N, Luria N, Dombrovsky A, et al. (2022) Transmission of a new polerovirus infecting pepper by the whitefly Bemisia tabaci. J Virol 93: e00488-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattar S, Thompson GA (2016) Small RNA regulators of plant-hemipteran interactions: Micromanagers with versatile roles. Front Plant Sci 7: 1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad MC, Haldeman-Cahill R, Cronin S, Carrington JC (1996) Analysis of the VPg-proteinase (NIa) encoded by Tobacco etch potyvirus: effects of mutations on subcellular transport, proteolytic processing, and genome amplification. J Virol 70: 7039–7048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA (2015) Impacts of insect oral secretions on defoliation-induced plant defense. Curr Opin Insect Sci 9: 7–15 [DOI] [PubMed] [Google Scholar]
- Schwartzberg EG, Tumlinson JH (2014) Aphid honeydew alters plant defence responses. Funct Ecol 28: 386–394 [Google Scholar]
- Shi X, Zhang Z, Zhang C, Zhou X, Zhang D, Liu Y (2021) The molecular mechanism of efficient transmission of plant viruses in variable virus–vector–plant interactions. Hortic Plant J 7: 501–508 [Google Scholar]
- Srinivasan R, Alvarez JM (2007) Effect of mixed viral infections (Potato virus Y-potato leafroll virus) on biology and preference of vectors Myzus persicae and Macrosiphum euphorbiae (hemiptera: Aphididae). J Econ Entomol 100: 646–655 [DOI] [PubMed] [Google Scholar]
- Su Q, Peng Z, Tong H, Xie W, Wang S, Wu Q, Zhang J, Li C, Zhang Y (2019) A salivary ferritin in the whitefly suppresses plant defenses and facilitates host exploitation. J Exp Bot 70: 3343–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Heil M (2021) Damage-Associated Molecular Patterns (DAMPs) in plant innate immunity: applying the danger model and evolutionary perspectives. Annu Rev Phytopathol 59: 53–75 [DOI] [PubMed] [Google Scholar]
- Tang D, Wang G, Zhou JM (2017) Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29: 618–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teresa Ruiz M, Voinnet O, Baulcombe DC (1998) Initiation and maintenance of virus-induced gene silencing. Plant Cell 10: 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toruño TY, Stergiopoulos I, Coaker G (2016) Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu Rev Phytopathol 54: 419–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Kurata N, Ohyanagi H, Hake S (2014) Genome-wide study of KNOX regulatory network reveals brassinosteroid catabolic genes important for shoot meristem function in rice. Plant Cell 26: 3488–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kleeff PJM, Galland M, Schuurink RC, Bleeker PM (2016) Small RNAs from Bemisia tabaci are transferred to Solanum lycopersicum phloem during feeding. Front Plant Sci 7: 1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Zhao P, Ma Y, Yao X, Sun Y, Huang X, Jin J, Zhang Y, Zhu C, Fang R, et al. (2019) A whitefly effector Bsp9 targets host immunity regulator WRKY33 to promote performance. Philos Trans R Soc B Biol Sci 374: 20180313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Yuan E, Ling X, Zhu-Salzman K, Guo H, Ge F, Sun Y (2020) An aphid facultative symbiont suppresses plant defence by manipulating aphid gene expression in salivary glands. Plant Cell Environ 43: 2311–2322 [DOI] [PubMed] [Google Scholar]
- Wang W, Dai H, Zhang Y, Chandrasekar R, Luo L, Hiromasa Y, Sheng C, Peng G, Chen S, Tomich JM, et al. (2015a) Armet is an effector protein mediating aphid-plant interactions. FASEB J 29: 2032–2045 [DOI] [PubMed] [Google Scholar]
- Wang W, Luo L, Lu H, Chen S, Kang L, Cui F (2015b) Angiotensin-converting enzymes modulate aphid–plant interactions. Sci Rep 5: 8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wari D, Kabir MA, Mujiono K, Hojo Y, Shinya T, Tani A, Nakatani H, Galis I (2019) Honeydew-associated microbes elicit defense responses against brown planthopper in rice. J Exp Bot 70: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner BJ, Mowry TM, Bosque-Pérez NA, Ding H, Eigenbrode SD (2009) Changes in green peach aphid responses to potato leafroll virus-induced volatiles emitted during disease progression. Environ Entomol 38: 1429–1438 [DOI] [PubMed] [Google Scholar]
- West SA, Griffin AS, Gardner A (2007) Social semantics: altruism, cooperation, mutualism, strong reciprocity and group selection. J Evol Biol 20: 415–432 [DOI] [PubMed] [Google Scholar]
- Westwood JH, Groen SC, Du Z, Murphy AM, Anggoro DT, Tungadi T, Luang-In V, Lewsey MG, Rossiter JT, Powell G, et al. (2013) A trio of viral proteins tunes aphid-plant interactions in Arabidopsis thaliana. PLoS One 8: e83066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield AE, Falk BW, Rotenberg D (2015) Insect vector-mediated transmission of plant viruses. Virology 479–480: 278–289 [DOI] [PubMed] [Google Scholar]
- Wu D, Qi T, Li W, Tian H, Gao H, Wang J, Ge J, Yao R, Ren C, Wang X, et al. (2017) Viral effector protein manipulates host hormone signaling to attract insect vectors. Cell Res 27: 402–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Davis TS, Eigenbrode SD (2014) Aphid behavioral responses to virus-infected plants are similar despite divergent fitness effects. Entomol Exp Appl 153: 246–255 [Google Scholar]
- Xu HX, Qian LX, Wang XW, Shao RX, Hong Y, Liu SS, Wang XW (2019) A salivary effector enables whitefly to feed on host plants by eliciting salicylic acid-signaling pathway. Proc Natl Acad Sci USA 116: 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Guo JY, Chu D, Ding TB, Wei KK, Cheng DF, Wan FH (2017) Secretory laccase 1 in Bemisia tabaci MED is involved in whitefly-plant interaction. Sci Rep 7: 3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JW, Yi HS, Kim H, Lee B, Lee S, Ghim SY, Ryu CM (2011) Whitefly infestation of pepper plants elicits defence responses against bacterial pathogens in leaves and roots and changes the below-ground microflora. J Ecol 99: 46–56 [Google Scholar]
- Yang JY, Iwasaki M, Machida C, Machida Y, Zhou X, Chua NH (2008) βCl, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev 22: 2564–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates AD, Michel A (2018) Mechanisms of aphid adaptation to host plant resistance. Curr Opin Insect Sci 26: 41–49 [DOI] [PubMed] [Google Scholar]
- Ye W, Ma W (2016) Filamentous pathogen effectors interfering with small RNA silencing in plant hosts. Curr Opin Microbiol 32: 1–6 [DOI] [PubMed] [Google Scholar]
- Zanardo LG, Carvalho CM (2017) Cowpea mild mottle virus (Carlavirus, Betaflexiviridae): a review. Trop Plant Pathol 42: 417–430 [Google Scholar]
- Zarate SI, Kempema LA, Walling LL (2007) Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol 143: 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebelo SA, Maffei ME (2012) Signal transduction in plant–insect interactions: From membrane potential variations to metabolomics. In Volkov AG, ed, Plant Electrophysiology: Signaling and Responses, Springer Berlin Heidelberg, Berlin, Heidelberg, Germany, pp 143–172 [Google Scholar]
- Zebelo SA, Maffei ME (2015) Role of early signalling events in plant-insect interactions. J Exp Bot 66: 435–448 [DOI] [PubMed] [Google Scholar]
- Zhang T, Luan JB, Qi JF, Huang CJ, Li M, Zhou XP, Liu SS (2012) Begomovirus-whitefly mutualism is achieved through repression of plant defences by a virus pathogenicity factor. Mol Ecol 21: 1294–1304 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH (2006) Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev 20: 3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Yao X, Li R, Cai C, Sun Y, Wang M, Du J, Zou Z, Kliebenstein D, Wang QM, et al. (2019) Viruses mobilize plant immunity to deter nonvector insect herbivores. Sci Adv 5: eaav9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JH, Hua CL, Fang YY, Guo HS (2016) The dual edge of RNA silencing suppressors in the virus-host interactions. Curr Opin Virol 17: 39–44 [DOI] [PubMed] [Google Scholar]
- Zhao P, Zhang X, Gong Y, Wang D, Xu D, Wang N, Sun Y, Gao L, Liu SS, Deng XW, et al. (2021) Red-light is an environmental effector for mutualism between begomovirus and its vector whitefly. PLoS Pathog 17: e1008770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Zhang Y (2020) Plant immunity: danger perception and signaling. Cell 181: 978–989 [DOI] [PubMed] [Google Scholar]
- Ziegler-Graff V (2020) Molecular insights into host and vector manipulation by plant viruses. Viruses 12: 263. [DOI] [PMC free article] [PubMed] [Google Scholar]