Abstract
We report a structural and transcriptional analysis of the pbp5 region of Enterococcus faecium C68. pbp5 exists within a larger operon that includes upstream open reading frames (ORFs) corresponding to previously reported psr (penicillin-binding protein synthesis repressor) and ftsW (whose product is a transmembrane protein that interacts with PBP3 in Escherichia coli septum formation) genes. Hybridization of mRNA from C68, CV133, and four ampicillin-resistant CV133 mutants revealed four distinct transcripts from this region, consisting of (i) E. faecium ftsW (ftsWEfm) alone; (ii) psr and pbp5; (iii) pbp5 alone; and (iv) ftsWEfm, psr, and pbp5. Quantities of the different transcripts varied between strains and did not always correlate with quantities of PBP5 or levels of ampicillin resistance. Since the psr of C68 is presumably nonfunctional due to an insertion of an extra nucleotide in the codon for the 44th amino acid, the region extending from the ftsWEfm promoter through the pbp5 gene of C68 was cloned in E. coli to facilitate mutagenesis. The psr ORF was regenerated using site-directed mutagenesis and introduced into E. faecium D344-SRF on conjugative shuttle vector pTCV-lac (pCWR558 [psr ORF interrupted]; pCWR583 [psr ORF intact]). Ampicillin MICs for both D344-SRF(pCWR558) and D344-SRF(pCWR583) were 64 μg/ml. Quantities of pbp5 transcript and protein were similar in strains containing either construct regardless of whether they were grown in the presence or absence of ampicillin, arguing against a role for PSR as a repressor of pbp5 transcription. However, quantities of psr transcript were increased in D344-SRF(pCWR583) compared to D344-SRF(pCWR558), especially after growth in ampicillin; suggesting that PSR acts in some manner to activate its own transcription.
Penicillin resistance in Enterococcus faecium is associated with production of low-affinity penicillin-binding protein PBP5. The presence of this penicillin-binding protein (PBP) in virtually all clinical E. faecium strains that have been investigated (including those susceptible to clinically achievable levels of penicillin [L. B. Rice, unpublished data]) suggests that it is intrinsic to this species, rather than an acquired gene. Supportive evidence for the role of PBP5 in penicillin resistance is derived from experiments indicating that PBP5-expressing cells replicate when incubated with penicillin at concentrations sufficient to saturate all of the other PBPs, as well as from studies demonstrating that E. faecium strains lacking PBP5 are highly susceptible to penicillin (10–12, 25, 26).
Early studies on Enterococcus hirae 9790 (which until 1985 was considered to be a type strain for Enterococus faecalis [8, 16]) reported that elevated levels of penicillin resistance (to ca. 64 μg/ml) were associated with increased quantities of detectable PBP5. Increased PBP5 production in one resistant mutant (R40) was associated with deletion of the N-terminal portion and some upstream DNA of an open reading frame (ORF) located ca. 1 kb upstream of the pbp5 start codon. Because of its presumed negative impact on PBP5 expression, this upstream ORF was designated psr (penicillin-binding protein synthesis repressor) (16). The presence of intact psr upstream of pbp5 was associated with decreased quantities of PBP5 in E. hirae and when these genes were cloned into E. coli. Importantly, transcriptional studies supporting a repressor role for psr have not been published. More recent publications have described penicillin-resistant mutants of E. hirae that lack detectable deletions in psr (19). Moreover, changes in cell wall carbohydrate concentrations and lytic response in psr-deficient E. hirae suggest that PSR may be a global regulator of cell wall synthesis genes (19). It has been noted previously that psr resembles lytR from Bacillus subtilis, hypothesized to be an attenuator of the N-acetylmuramoyl-l-alanine amidase structural genes (19).
High-level penicillin and ampicillin resistance (128 to 512 μg/ml) in E. faecium has become a grave clinical problem over the past decade, especially since many ampicillin-resistant strains are resistant to vancomycin, which until recently was the only other reliable therapeutic alternative for the treatment of E. faecium infections (24). Studies of clinical isolates expressing high levels of ampicillin resistance suggest that increased production of PBP5 is not the most common mechanism for increased resistance but rather have noted that highly resistant strains encode PBP5 with alterations that result in a still lower affinity for penicillin (17, 23, 28). Since techniques for genetic manipulation are often not readily applicable to E. faecium, virtually all of the information available on ampicillin resistance in this species is based on the analysis of clinical isolates, raising questions about whether unknown differences between strains are also impacting the expression of resistance.
We recently reported the conjugal transfer of pbp5 (along with the vancomycin resistance transposon Tn5382) from clinical isolate E. faecium C68 to ampicillin-susceptible, pbp5-lacking E. faecium recipient strain GE-1 (4). In the present paper, we present the structure of the pbp5 region within C68, a description of gene transcription within that region, and an analysis of the impact of PSR expression on ampicillin resistance.
MATERIALS AND METHODS
Strains and plasmids.
Relevant bacterial strains, cloning vectors, and plasmids are listed in Table 1. E. faecium GE-1 (4, 7) and D344-S (18) are devoid of pbp5, and the MICs of ampicillin for these strains are <0.5 μg/ml. GE-1 is resistant to rifampin and fusidic acid. D344-S was selected for resistance rifampin and fusidic acid for these experiments by plating large inocula serially on fusidic acid (25 μg/ml) and then rifampin (100 μg/ml). C68 is an E. faecium strain isolated from the feces of a patient hospitalized in northeast Ohio (4). It is resistant to high levels of both ampicillin and vancomycin and represents the predominant (more than 50% of area isolates) vancomycin-resistant enterococcal clone in the region (5). pTCV-lac is a gram-negative–gram-positive shuttle vector that can be mobilized from E. coli into gram-positive species by conjugal plasmid pRK24 (21). pCWR558 was constructed by PCR amplification of the region extending from upstream of E. faecium ftsW(ftsWEfm) (containing both identified promoters) to the stop codon of pbp5 (excluding the transcription terminator downstream of pbp5). The amplification product was originally cloned into the multiple-cloning site of pCR-XL-TOPO (Invitrogen) and then subsequently directionally cloned (using SmaI and BamHI) into the multiple-cloning site of pTCV-lac. pCWR583 resulted from performing site-directed mutagenesis on the insert of pCWR558 to regenerate the psr ORF. pCWR561 contains a PCR-generated insert containing only pbp5 with its promoter cloned into pTCV-lac.
TABLE 1.
Bacterial strains and plasmids used in these studies
| Strain or plasmid designation | Resistance trait(s) | Purpose (reference[s]) |
|---|---|---|
| Strains | ||
| E. faecium C68 | Apr Emr Gmr Smr Tcr Vmr | Clinical isolate (3) |
| E. faecium GE-1 | Fusr Rifr Tcr | E. faecium recipient; lacks pbp5 due to a deletion of the entire region (4, 7) |
| E. faecium D344-SRF | Fusr Rifr Emr | E. faecium recipient strain; lacks pbp5 due to a deletion of the entire region (18; this study) |
| E. faecium CV133 | Fusr Rifr Tcr Apr Vmr | Transconjugant from C68 × GE-1 mating (3) |
| E. faecium A1 | Fusr Rifr Tcr Apr Vmr | CV133 mutant with increased AMP MIC |
| E. faecium A2 | Fusr Rifr Tcr Apr Vmr | CV133 mutant with increased AMP MIC |
| E. faecium A3 | Fusr Rifr Tcr Apr Vmr | CV133 mutant with increased AMP MIC |
| E. faecium A4 | Fusr Rifr Tcr Apr Vmr | CV133 mutant with increased AMP MIC |
| Plasmids | ||
| pUC18 | Apr | E. coli cloning vector (BRL) |
| pTCV-lac | Emr Kmr | Conjugative E. coli-E. faecium shuttle plasmid with β-galactosidase reporter construct (21) |
| pRK24 | Apr | Mobilizing plasmid for transfer of pTCV-lac constructs into E. faecium D344-S (29) |
| pCWR558 | Emr Kmr Apr | ftsWEfm (two-promoter)-psr-pbp5 region from C68 cloned into pTCV-lac (this study) |
| pCWR561 | Emr Kmr Apr | pbp5 from C68 with its promoter cloned into pTCV-lac (this study) |
| pCWR583 | Emr Kmr Apr | ftsWEfm (two-promoter)-psr-pbp5 region from C68 cloned into pTCV-lac. In this construct, the intact psr ORF has been regenerated by site-directed mutagenesis (this study). |
| pCWR564 | Kmr Apr | ftsWEfm (two-promoter)-psr-pbp5 region from C68 cloned into pCR-XL-TOPO (this study) |
Antimicrobial susceptibility testing.
MICs were determined by a broth macrodilution technique in brain heart infusion broth (20).
Hybridization experiments.
Genomic DNA was extracted as described previously (4) and digested with restriction enzymes for 1 to 2 h at 37°C according to the specifications of the manufacturer (Promega, Madison, Wis.). Digested DNA was separated on 0.7 to 1% agarose gels overnight. Separated DNA was denatured, neutralized, and transferred to nylon filters using a negative-pressure transfer apparatus (Pharmacia, LKB, Uppsala, Sweden) and baked at 80°C for 1 to 2 h to fix the DNA to the filter. Filters were prehybridized, and this was followed by hybridization with digoxigenin-labeled probes overnight at 68°C and washing under conditions of high stringency according to the specifications of the manufacturer (Boehringer Mannheim, Indianapolis, Ind.).
Cloning of genomic DNA fragments.
Once fragments of interest were identified by hybridization, they were removed from agarose gels and purified using the Qiaquick gel extraction kit (Qiagen, Valencia, Calif.). These fragments were then ligated to like-digested pACYC184 or pUC18 and transformed into E. coli DH5α (13) or E. coli DH10B (Bethesda Research Laboratories, Gaithersburg, Md.) by electroporation (Bio-Rad, Hercules, Calif.). Transformed preparations were inoculated onto plates containing antimicrobial agents selective for the cloning vectors, and colonies with the appropriate inserts were identified by colony hybridization techniques as previously described (4). These colonies were purified and subcloned as necessary for further sequencing.
PCR amplification.
Amplification of genomic DNA was performed on a Perkin-Elmer Cetus 9600 thermal cycler using Taq DNA polymerase according to standard protocols as recommended by the manufacturer (Perkin-Elmer Cetus, Roche Molecular Systems, Branchburg, N.J.). Variations were introduced into each individual protocol depending on the expected size of the amplification product and the specifics of the primers used. A 10-μl aliquot of a total 50-μl PCR mixture was loaded on a 0.7 to 1.2% agarose gel for analysis. PCR products to be cloned were ligated to pCR-XL-TOPO (Invitrogen) and introduced into E. coli DH10B by electroporation. Inserts were removed from pCR-XL-TOPO by restriction digestion and ligated to pTCV-lac using T4 DNA ligase. pCWR558, pCWR561, and pCWR583 were transformed into E. coli HB101 containing pRK24 (mobilizing plasmid) (21) with selection on Luria-Bertani agar plates containing kanamycin (20 μg/ml). Recombinant plasmids were then introduced into E. faecium D344-SRF by conjugation, with selection on kanamycin (1,000 to 1,500 μg/ml) and rifampin (100 μg/ml).
DNA sequence analysis.
DNA sequencing was performed from cloned DNA on double stranded templates using the A.L.F. Automated sequencing kit and fluorescein- or Cy5 indodicarbocyanine dye-labeled forward and reverse primers, or with similarly labeled primers derived from previously determined sequences. DNA was purified for sequencing using the Wizard plus minipreps DNA purification system (Qiagen). Sequence was determined using the ALFExpress automated sequencer (Pharmacia LKB). Sequences were compared for homology with those entered into the GenBank, EMBL, DDBJ, and PDB databases using Blastn and Blastx local alignment search tools (1) and further analyzed using the DNAsis (version 2.0; Hitachi, Ltd.) sequence analysis program.
RNA extraction.
Cells were grown overnight, either with or without antibiotic at various concentrations. In the morning, overnight cultures were freshly inoculated into the same medium at a 1:10 dilution and grown without agitation to an OD600 of 0.7 (approximately 4 to 8 h). At this point cells were centrifuged, the supernatant was discarded, and the pellets were frozen at −80°C until RNA isolation was begun. Total cellular RNA extraction was accomplished using the Rneasy kit (Qiagen). The only deviation from the manufacturer's protocol was that a lysozyme concentration of 10 mg/ml in Tris-EDTA buffer was used. RNA concentration was measured by spectrophotometer prior to precipitation with 100% ethanol and sodium acetate (pH 4.0) and frozen at −80°C until use.
Northern hybridization.
A volume calculated to contain 5 μg of RNA was centrifuged and resuspended in distilled water, and this was followed by dilution with sample buffer (per 300 μl: 180 μl of deionized formamide, 75 μl of deionized formaldehyde, 45 μl of 10× MOPS [morpholinepropanesulfonic acid] buffer). Samples were heated at 65°C for 10 min, chilled on ice and loaded onto 1.2% agarose gels with formaldehyde after addition of ethidium bromide and tracker dye to the sample. RNA was separated overnight at 25 V at 4°C. Gels were washed with sterile water for 15 min four times, which was followed by equilibration (four times) with 20× SSC (1X SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 15 min each. RNA was transferred to nylon membranes by passive capillary action for 48 h. Nylon membranes were then baked for 2 h at 80°C to fix the RNA. Membranes were prehybridized and hybridized at 50°C as detailed by the manufacturer (Boehringer Mannheim). Probes were labeled with digoxigenin either by a random primer method or by PCR as recommended by the manufacturer (Boehringer Mannheim). Hybridized RNA was detected using an antidigoxigenin antibody and a chemiluminescence detection system (Boehringer Mannheim). Densitometry values (in optical density units) were determined using Scion Image, a Windows-compatible version of NIH Image.
Primer extension analysis.
Primer extension analysis used Cy5-labeled primers designed to direct synthesis of the complementary strand upstream of the start of the ftsWEfm, psr, and pbp5 ORFs (Table 1). The primers used were as follows: for pbp5, 5′-TTCTTGATAGTGCTGGTAG-3′; for psr, 5′-TCCATGGATAAACTCAA-3′; and for ftsWEfm, 5′-TAAGAACTGGCACTGTAT-3′. The technique employed high temperatures to minimize complications resulting from secondary structure formation, essentially as described by Yamada et al. (27). In brief, total RNA from 0.1 to 10 μg and 1 pmol of Cy5-labeled primer was added to 10 mM Tris-HCl [pH 8.3], 50 mM KCl, 5 mM MgCl2, 1 mM concentrations of the deoxynucleoside triphosphates, and 20 U of RNase inhibitor in a 0.2 ml microcentrifuge tube, resulting in a final volume of 20 μl. The primer-extension reaction was started with the addition of 5 U of avian myeloblastosis virus reverse transcriptase. The reaction was carried out at 50°C, and the solution was subjected to phenol extraction and ethanol precipitation. The solution was resuspended and the fragment was detected by running the sample on an ALFExpress automated sequencer (Pharmacia) in parallel with a standard sequencing reaction using a plasmid containing the ORF and upstream sequence as a template.
PBP analysis.
PBPs were isolated as described by Williamson et al. (26). In brief, strains were grown in Todd Hewitt broth until they reached an optical density at 600 nm of 0.5. After centrifugation and resuspension in phosphate buffer, cells were exposed to saturating concentrations of [3H]penicillin. After lysis with lysozyme and muramidase 1 samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Relative amounts of PBP5, which binds penicillin with low affinity, were confirmed from samples of the same cultures after separation of PBPs under the same conditions by performing Western blot analysis using specific polyclonal antibodies raised to PBP5 (23). Relative quantities of PBPs were estimated after densitometry scanning of the gels or Western blots using the GEL ANALYST perfect image master 4.01 (Clara vision, France) and recorded to a Kaiser camera (HP Marketing Corporation, N.J.).
Site-directed mutagenesis.
Site-directed mutagenesis to regenerate the psr ORF was performed using the plasmid pCWR564 as a template. Two commercially manufactured oligonucleotides were used (psr1: 5′-CGAAAAGTGTCCCTACCACACTAAATG-3′; psr2: 5′-CATTTAGTGTGGTAGGGACACTTTTCG-3′). Mutagenesis was performed using the Quikchange site-directed mutagenesis kit (Stratagene). Reaction mixtures consisted of 6 ng of pCWR564, 125 ng of primer psr1, 125 ng of primer psr2, 5 μl of 10X reaction buffer, 1 μl of deoxynucleoside triphosphate mix, and water to 50 μl. Pfu Turbo polymerase (2.5 U) was added, and the mixture was incubated for one cycle of 30 s at 95°C and 14 cycles of 95°C (30 s), 55°C (1 min), and 68°C (15 min). After amplification, the preparations were cooled on ice. One microliter of restriction enzyme DpnI was then added, and the mixture was incubated at 37°C for 1 h. The mixture was then electroporated into competent E. coli DH10B with selection for transformants on Luria-Bertani agar plates containing kanamycin (50 μg/ml). Regeneration of the psr ORF was confirmed by dideoxy sequencing reactions (see above) before the insert was removed by digestion with BamHI and SmaI and ligated to pTCV-lac (forming pCWR583).
GenBank accession number.
The sequence of the 5,148-bp region extending from upstream of the ftsWEfm ORF to the left terminus of Tn5382 is entered in the GenBank database under accession number AF11760.
RESULTS
MICs for donors and transconjugants.
Several ampicillin- and vancomycin-resistant transconjugants were obtained from matings between C68 and GE-1 (4). The ampicillin MICs for these transconjugants varied from 8 to 128 μg/ml but never were equivalent to MICs observed in C68 (256 μg/ml). We chose one transconjugant (CV133) (MIC = 16 to 32 μg/ml) for further study because of its low ampicillin MIC relative to that of C68.
Mutants of CV133 expressing greater resistance to ampicillin were readily obtained at a rate of ca. 10−6/CFU after plating high inocula of this transconjugant onto brain heart infusion (BHI) agar plates containing ampicillin (200 μg/ml). Mutants appeared as pinpoint colonies approximately 48 to 72 h after inoculation of the plates. This rate of mutation to resistance is higher than would be anticipated to result from point mutations within pbp5 (attempts to derive point mutations in a susceptible E. faecium pbp5 in a single step were never successful, suggesting a rate of less than 10−10/CFU [Laurent Gutmann, unpublished data]). Four mutants (A1 to A4) were chosen for further study. Broth dilution ampicillin MICs for the resistant mutants were 128 μg/ml (for A1, A3, and A4) or 256 μg/ml (for A2). Ampicillin MICs for GE-1 and D344-S are ≤0.5 μg/ml.
PBP studies.
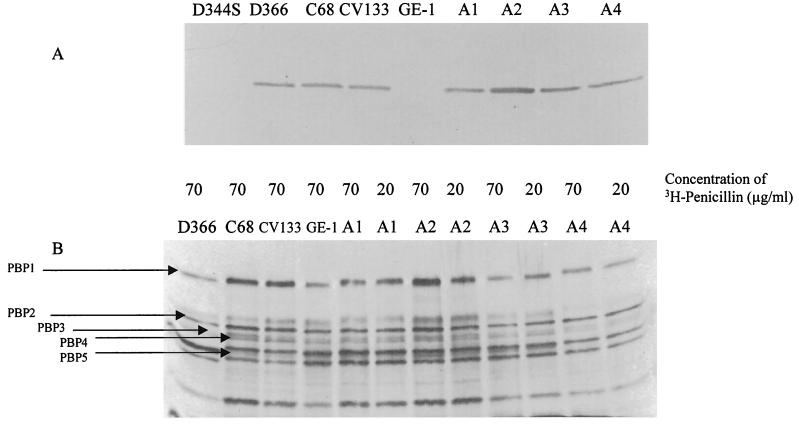
PBP studies using radiolabeled penicillin showed, using an internal standard, a 1.8-fold increase in the amounts of PBP5 in CV133 mutant A2, but not in any other strains (Fig. 1B). These data were confirmed using an anti-PBP5 antibody in a Western blot (Fig. 1A). With the exception of A2, there was no correlation between the ampicillin MIC and the quantities of detectable PBP5 in any of the strains. The Western blot also confirmed the absence of PBP5 from recipient strains GE-1 and D344-S (Fig. 1A), which correlated with the absence of the gene visible on Southern analysis and with the absence of transcript visible on Northern hybridizations (data not shown).
FIG. 1.
PBP studies of the strains described in this paper. (A) Western blot of PBP5 using a polyclonal antibody to PBP5. Strains are indicated above the diagram and described in Table 1. D366 is a low-level-penicillin-resistant E. faecium strain used as a positive control. (B) PBP analysis using [3H]penicillin. The concentration of [3H]penicillin used for each experiment is listed above the figure.
Structure of the region upstream of pbp5.
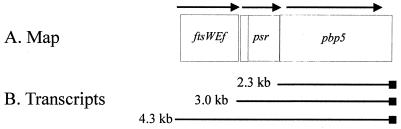
The nucleotide sequence of the 5,148-bp region extending from ca. 3 kb upstream of the pbp5 gene into Tn5382 downstream of the gene was determined. The structure of the region is depicted in Fig. 2. The region contained a pbp5 gene predicting 1-amino-acid difference (F→G at amino acid 497) in comparison to the previously described pbp5 sequence from high-level-ampicillin-resistant E. faecium H80721 (28). This F497G change has not been described in previous pbp5 genes and is of unclear significance. An ORF with homologies to previously described psr genes was observed upstream of the pbp5 ORF (Fig. 2). An added nucleotide in the C68 psr ORF resulted in a frame shift at amino acid 43 and a stop codon at amino acid 103, changes that would be expected to render PSR nonfunctional.
FIG. 2.
Map of genomic area of E. faecium C68. (A) Physical structure. Each boxed area represents an ORF. The psr ORF is split into two parts because it is interrupted in C68 by the nonsense mutation (see text). (B) Composition of transcripts observed in Northern hybridization experiments. The 3.0-kb transcript was seen only in CV133 mutant A2.
An ORF terminating 91 bp upstream of the psr translation initiation codon had significant identity (30 to 70%) with previously described ftsW genes from E. hirae, E. coli, and B. subtilis, among others (3, 6, 14, 15). We have designated this ORF ftsWEfm.
Northern hybridization studies of donor, recipient, and resistant mutants.
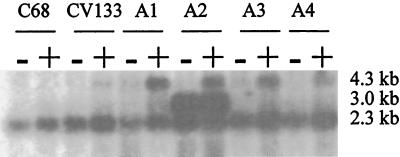
mRNA derived from C68, GE-1, CV133, and mutants A1 to A4 was hybridized with a probe spanning the downstream end of psr into pbp5 (Fig. 3). mRNA was extracted in late log phase after growth in non-selective BHI broth or in BHI broth supplemented with ampicillin (10 μg/ml). Hybridization to a 2.3-kb transcript was observed in all strains (GE-1 had previously been shown to be devoid of sequences homologous to pbp5 [4]). An additional 4.3-kb band detectable only in late log phase was observed in all ampicillin-resistant mutants of CV133 only after incubation with ampicillin. A smaller amount of this extra transcript was also observed in CV133, and even less was observed in C68, after exposure to ampicillin. An additional, very intense 3-kb band was observed in A2 (the mutant that expressed the highest levels of ampicillin resistance and the largest quantity of PBP5 protein) with or without exposure to ampicillin. A very slight increase in the pbp5 2.3-kb transcript was visible to the eye in all strains after exposure to ampicillin, especially for strains for which there was a marked increase in detectable 4.3-kb transcript.
FIG. 3.
Northern hybridization of mRNA derived from strains described in this study. See Table 1 for description of strains. mRNA was extracted from strains grown in the presence (+) or absence (−) of ampicillin (10 μg/ml). The sizes of the different transcripts (see Fig. 2 for description of their contents) are listed to the right of the figure.
Using internal probes for the region upstream of ftsWEfm, the ftsWEfm ORF, the psr ORF, the pbp5 ORF, and the terminus of Tn5382, we were able to determine the genetic regions represented by the transcripts. Results are summarized in Fig. 2. The 2.3-kb transcript contains pbp5 alone. The 3.0-kb transcript visible only in A2 hybridized strongly only with internal psr and pbp5 probes. These data suggest that this transcript originates in the region between the ftsWEfm and psr ORFs. Its absence in the other strains suggested that a mutation had occurred in A2 that either creates or uncovers a powerful promoter of psr transcription, and that no effective transcriptional terminator was present after the psr ORF. The region upstream of psr was cloned, and its sequence was compared to the corresponding region of CV133. The regions were found to be identical (data not shown), excluding the presence of a point mutation or an IS element as the cause of the appearance of the 3-kb transcript in A2.
The ftsWEfm probe hybridized to a 1.4-kb transcript that did not hybridize to any of the other probes. This 1.4-kb transcript is present most abundantly in early log phase, trailing off to nearly undetectable levels as the organisms approach stationary phase (data not shown). The large, 4.3-kb transcript that appears most abundantly in the mutants expressing higher levels of resistance to ampicillin hybridized to ftsWEfm, psr, and pbp5 probes. These data suggest that the 4.3-kb transcript results from transcription originating immediately upstream of ftsWEfm and continuing through both psr and pbp5, terminating at the transcriptional terminator downstream of the pbp5 gene.
Transcription initiation sites.
We used primer extension analysis to identify precisely the transcriptional start sites for the three ORFs in this operon. Three potential transcription initiation sites were identified upstream of ftsWEfm (shown in boldface type below). The furthest upstream was located 351 bp 5′ of the ftsWEfm start codon (GAACTCGTAAA). This was preceded by credible −10 (TATCAT) and −35 (TTGCCT) regions spaced 17 bp apart. The second was located 277 bp upstream of the start codon (CGTCGAAAGTC) and was not preceded by sequences resembling the sigma 70 consensus promoters. The third potential start was located 132 bp upstream of the start codon (GACCTTTTCTTT). It was not preceded by consensus promoter sequences and in fact fell at the downstream end of an inverted repeat. This site may represent not a transcriptional start but rather a fall-off of the avian myeloblastosis virus reverse transcriptase due to secondary structure created by interaction between the two arms of the inverted repeat. The transcriptional start for the 3.0-kb psr-pbp5 message seen only in strain A2 was located 88 bp upstream of the psr start codon (TAAGAGAAACTG). This start site was preceded by credible-10 (AACAAT) and −35 (TTGCCG) regions that were spaced 16 bp apart. The transcription initiation site for the 2.3-kb pbp5 message was located 39 bp upstream of the pbp5 start codon (TAAACAGGTATAAA) and was preceded by possible-10 (TAGAAT) and -35 (TGGATT) sequences spaced 17 bp apart.
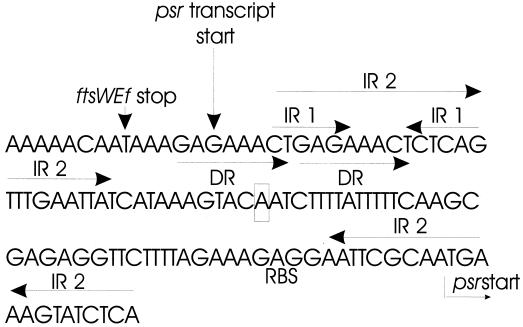
Nucleotide sequence of the region between ftsWEfm and psr.
Examination of the sequence between ftsWEfm and psr in both CV133 and A2 reveals several interesting features (Fig. 4). The distance between the stop codon of ftsWEfm and the start codon of psr is 91 nucleotides. Seven base pairs downstream of the ftsWEfm stop codon lies a perfect 6-bp inverted repeat (inverted repeat 1 [IR1]) followed by three T's. If the last three T's are included within the inverted repeat, it is then a perfect repeat of 9 bp. This sequence could serve as a rho-independent terminator of ftsWEfm transcription. Beginning 2 bp downstream of the ftsWEfm stop codon and terminating at the first base pair of the downstream portion of IR1 lies a perfect 8-bp direct repeat (DR). Similar DRs have been shown in other instances to be sites for binding by proteins (22). There is also a larger imperfect (17 of 22) inverted repeat sequence (IR2) that involves most of IR1 for the upstream arm, and the ribosome binding site and psr start codon for the downstream arm. Interaction between the two arms of IR2 and/or IR1 could obscure any intervening promoter sequences. It is noteworthy that the 3.0-kb psr-pbp5 transcript originates within the upstream arm of the DR and immediately upstream of the two inverted repeats. In summary, this relatively short DNA segment is rich in sequences that could be important in the regulation of transcription initiation or termination.
FIG. 4.
Sequence extending from the ftsWEfm stop codon through the psr start codon. The ftsWEfm stop (translation) and psr starts (transcription and translation) are indicated, as in the psr ribosome binding site (RBS). The two inverted repeats and the DRs are also indicated. The nucleotide corresponding to the 5′ end of the deletion previously reported PBP5 hyper-producer E. hirae R40 is boxed. See text for details.
Impact of PSR on pbp5 and psr transcription and quantities of PBP5.
In order to examine the effect of an intact psr ORF on the expression of ampicillin resistance and transcription of the psr and pbp5 genes, we used site-directed mutagenesis to remove the additional nucleotide in the pCWR558 psr (Table 1), thereby regenerating the psr ORF. The regions extending from ftsWEfm through pbp5 with (pCWR583) or without (pCWR558) the intact psr ORF were conjugated into E. faecium D344-SRF since these constructs were less stable in GE-1. As an mRNA quantity control, we measured the amount of aph-A3 transcript (from the gene conferring kanamycin resistance in pTCV-lac) as well. The ampicillin MIC was the same for both constructs (64 μg/ml). Consistent with this observation, the amount of pbp5 transcript was similar in the two constructs relative to the amount of aph-A3 transcript present (Table 2). These data suggest that PSR does not serve as a repressor of pbp5 transcription. The presence of a psr ORF also did not seem to have an impact on the quantity of the major 2.3-kb pbp5 transcript present after growth in ampicillin (Table 2). In contrast, the psr-pbp5 message (corresponding to the 3.0-kb message seen in A2) was detectable in both strains only after prolonged exposure of the X-ray film (overnight). There was an increase in the quantity of psr transcript in D344-SRF(pCWR583) after exposure to ampicillin in comparison to D344-SRF(pCWR558), suggesting that PSR may serve to amplify its own transcription (and therefore that of pbp5) after exposure to ampicillin.
TABLE 2.
Densitometry measurements of specific mRNA in strains containing plasmid constructs
| Plasmida | Densitometry measurement− (density units) for indicated message
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
pbp5
|
psr
|
aph-A3
|
|||||||
| No AMP | AMPc | AMPc/no AMP | No AMP | AMP | AMP/no AMP | No AMP | AMP | AMP/no AMP | |
| pCWR558 | 11,722 | 11,520 | 0.98 | 1,846 | 2,106 | 1.14 | 30,330 | 29,162 | 0.96 |
| pCWR583 | 14,930 | 16,321 | 1.09 | 1,660 | 5,902 | 3.55 | 27,648 | 32,312 | 1.17 |
Plasmids present in E. faecium D344-SRF.
Absolute number cannot be used to compare the quantities of transcript between blots, since the strengths of the probes and the development time may have differed. Therefore, relative amounts of only those messages included on the same blot and hybridized with the same probe can be compared.
Ampicillin was used at a concentration of 10 μg/ml.
The same strains underwent quantitation of PBP5 by Western blotting. Quantities of PBP5 were equivalent in strains D344-SRF(pCWR583) and D344-SRF(pCWR558) after growth in the presence or absence of ampicillin, consistent with the results of Northern hybridization and MICs and suggesting that the quantities of psr-pbp5 message generated by the two constructs were insufficient to impact overall PBP5 quantities. Quantities of PBP5 detectable from D344-SRF(pCWR561), which lacks the ftsWEfm and psr upstream of pbp5, were less than those observed with the other two constructs (data not shown), consistent with the lower MICs of ampicillin (8 μg/ml) observed for this construct.
DISCUSSION
The data presented in this paper underscore the importance of PBP5 in the expression of ampicillin resistance in E. faecium. Lacking this PBP, E. faecium strains GE-1 and D344-S are highly susceptible to ampicillin. Reintroduction of the pbp5 gene, however, either on a large transposon (as with CV133) or on a transferable plasmid, is associated with expression of increased levels of ampicillin resistance.
Work performed to date suggests that the level of ampicillin resistance expressed by E. faecium strains will be impacted by the quantity and the structure of the PBP5 itself (12, 16, 23). The data presented in this paper are supportive of a relationship between PBP5 quantity and resistance in that mutant strain A2 produces roughly twice the amount of PBP5 as other mutant strains and consistently demonstrated MICs that were approximately twice those for the other mutants. Published data supporting a correlation between the structure of PBP5 and the level of ampicillin resistance are based entirely on analyses of clinical strains and correlation with penicillin binding affinities. At the present time there are no data substantiating the importance of the structural alterations in the C68 PBP5, although several of the mutations present in this pbp5 gene have been observed in other low-affinity PBP5s (28).
The transcriptional studies presented in this paper suggest that the 2.3-kb transcript that includes pbp5 alone is the primary determinant of the quantity of PBP5 produced by most E. faecium strains, as evidenced by the close correlation between transcript and protein quantities among all of the strains except A2. The increased quantities of PBP5 seen in A2 are explainable by the presence of large amounts of the 3-kb psr-pbp5 transcript. The discordance between the presence of large quantities of the ftsWEfm-psr-pbp5 transcript in the CV133 mutants A1, A3, and A4 is more difficult to reconcile but may be explainable by the fact that these transcripts appeared primarily in late log to stationary phase, whereas the PBP5 studies were performed at mid-log phase. Alternatively, secondary structure within the 4.3-kb transcript could impair translation of pbp5 when it is present in this large message.
To state that the quantity and structure of PBP5 are important for the expression of ampicillin resistance in E. faecium is not, however, to suggest that these are the only factors at play in determining the level of resistance expressed by clinical E. faecium isolates. We were unable to document any substantial difference in PBP5 quantities between C68 (ampicillin MIC, 256 to 512 μg/ml); CV133 (ampicillin MIC, 16 to 32 μg/ml), and A1, A3, or A4 (ampicillin MIC, 128 μg/ml). Recent reports of studies performed with Staphylococcus aureus and Streptococcus pneumoniae (2, 9) suggest that levels of methicillin or ampicillin resistance associated with the expression of low-affinity PBPs are significantly impacted by the nature of the peptidoglycan precursors. An important future goal will be to examine peptidoglycan precursors in isogenic E. faecium strains expressing different levels of ampicillin resistance.
The hypothesis that PSR serves as a repressor of PBP5 production originated from experiments in which increased quantities of PBP5 were associated with a deletion of the upstream portion of this ORF in E. hirae. Our data suggest that PSR is neither a transcriptional repressor nor an activator of pbp5 transcription but that it may serve to amplify its own transcription when cells are grown in the presence of ampicillin. psr transcription is virtually undetectable after growth in BHI broth unsupplemented by antibiotics. Even after growth in ampicillin, quantities of psr transcript are very small (data not shown) compared to quantities of pbp5 transcript, which may explain why ampicillin-induced increases in psr-pbp5 transcription in psr-intact strains are not associated with increases in ampicillin resistance. The observation in the prior study that increases in PBP5 quantities were associated with a deletion of the upstream portion of psr (16) may have been due to the elimination of secondary structure that serves to obscure the psr promoter. Loss of this secondary structure could “open up” the psr promoter, resulting in increased pbp5 transcription. In fact, the prior publication had presumed, based on sequence analysis, that part of the promoter for the psr gene had been deleted. Comparing the sequence deleted from that strain with the sequence upstream of the C68 psr, it is clear that the promoter was not deleted in that case, since the transcriptional start site identified in these studies lies upstream of the deleted segment. We observed a variant of this scenario in A2, where dramatically increased psr-pbp5 transcript quantities were associated with increased PBP5 and increased ampicillin MICs. Since no structural changes were observed in the region upstream of psr in A2, we currently hypothesize that increased psr-pbp5 transcription in this mutant is due to the presence of an activator that interacts with the psr promoter region in a way that makes the promoter available to the sigma factor responsible for initiating psr transcription.
The overall relevance of these findings for the mechanisms of ampicillin resistance in clinical E. faecium isolates is not clear. While our experiments describe mechanisms by which transcription of pbp5 could be increased, the majority of these transcriptional changes do not appear to impact the final quantities of translated product or degrees of antibiotic resistance. Despite these limitations, the present study provides important insight into processes that impact the production of PBP5, which is required for the expression of ampicillin resistance. This information will be important for the subsequent interpretation of experiments designed to examine the additional factors that contribute to levels of ampicillin resistance in E. faecium.
ACKNOWLEDGMENT
This work was supported by the Medical Research Service of the Department of Veterans Affairs.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Berger-Bachi B, Strassle A, Gustafson J E, Kayser F H. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1992;36:1367–1373. doi: 10.1128/aac.36.7.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle D S, Khattar M M, Addinall S G, Lutkenhaus J, Donachie W D. ftsW is an essential cell-division gene in Escherichia coli. Mol Microbiol. 1997;24:1262–1273. doi: 10.1046/j.1365-2958.1997.4091773.x. [DOI] [PubMed] [Google Scholar]
- 4.Carias L L, Rudin S D, Donskey C J, Rice L B. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J Bacteriol. 1998;180:4426–4434. doi: 10.1128/jb.180.17.4426-4434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donskey C J, Schreiber J R, Jacobs M R, Shekar R, Smith F, Gordon S, Salata R A, Whalen C, Rice L B. A polyclonal outbreak of predominantly VanB vancomycin-resistant enterococci in Northeast Ohio. Clin Infect Dis. 1999;29:573–579. doi: 10.1086/598636. [DOI] [PubMed] [Google Scholar]
- 6.Duez C, Thamm I, Sapunaric F, Coyette J, Ghuysen J M. The division and cell wall gene cluster of Enterococcus hirae S185. DNA Seq. 1998;9:149–161. doi: 10.3109/10425179809072190. [DOI] [PubMed] [Google Scholar]
- 7.Eliopoulos G M, Wennersten C, Moellering R C., Jr Resistance to β-lactam antibiotics in Streptococcus faecium. Antimicrob Agents Chemother. 1982;22:295–301. doi: 10.1128/aac.22.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farrow J A, Collins M D. Enterococcus hirae, a new species that includes amino acid assay strain NCDO 1258 and strains causing growth depression in young chickens. Int J Syst Bacteriol. 1985;35:73–75. [Google Scholar]
- 9.Filipe S R, Tomasz A. Inhibition of the expression of penicillin resistance in Streptococcus pneumoniae by inactivation of cell wall muropeptide branching genes. Proc Natl Acad Sci USA. 2000;97:4891–4896. doi: 10.1073/pnas.080067697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fontana R, Aldegheri M, Ligozzi M, Lopez H, Sucari A, Satta G. Overproduction of a low-affinity penicillin-binding protein and high-level ampicillin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1994;38:1980–1983. doi: 10.1128/aac.38.9.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fontana R, Cerini R, Longoni P, Grossato A, Canepari P. Identification of a streptococcal penicillin-binding protein that reacts very slowly with penicillin. J Bacteriol. 1983;155:1343–1350. doi: 10.1128/jb.155.3.1343-1350.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fontana R, Grossato A, Rossi L, Cheng Y R, Satta G. Transition from resistance to hypersusceptibility to β-lactam antibiotics associated with loss of a low-affinity penicillin-binding protein in a Streptococcus faecium mutant highly resistant to penicillin. Antimicrob Agents Chemother. 1985;28:678–683. doi: 10.1128/aac.28.5.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:577–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda M, Sato T, Wachi M, Jung H K, Ishino F, Kobayashi Y, Matsuhashi M. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J Bacteriol. 1989;171:6375–6378. doi: 10.1128/jb.171.11.6375-6378.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishino F, Jung H K, Ikeda M, Doi M, Wachi M, Matsuhashi M. New mutations fts-36, lts-33, and ftsW clustered in the mra region of the Escherichia coli chromosome induce thermosensitive cell growth and division. J Bacteriol. 1989;171:5523–5530. doi: 10.1128/jb.171.10.5523-5530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ligozzi M, Pittaluga F, Fontana R. Identification of a genetic element (psr) which negatively controls expression of Enterococcus hirae expression. J Bacteriol. 1993;175:2046–2051. doi: 10.1128/jb.175.7.2046-2051.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ligozzi M, Pittaluga F, Fontana R. Modification of penicillin-binding protein 5 associated with high-level ampicillin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1996;40:354–357. doi: 10.1128/aac.40.2.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mainardi J-L, Legrand R, Arthur M, Schoot B, van Heijenoort J, Gutmann L. Novel mechanism of β-lactam resistance due to bypass of DD-transpeptidation in Enterococcus faecium. J Biol Chem. 2000;275:16490–16496. doi: 10.1074/jbc.M909877199. [DOI] [PubMed] [Google Scholar]
- 19.Massidda O, Kariyama R, Daneo-Moore L, Shockman G D. Evidence that the PBP 5 synthesis repressor (psr) of Enterococcus hirae is also involved in the regulation of cell wall composition and other cell wall-related properties. J Bacteriol. 1996;178:5272–5278. doi: 10.1128/jb.178.17.5272-5278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 3rd ed. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1993. pp. M7–A3. [Google Scholar]
- 21.Poyart C, Trieu-Cuot P. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galatosidase in gram-positive bacteria. FEMS Microbiol Lett. 1997;156:193–198. doi: 10.1111/j.1574-6968.1997.tb12726.x. [DOI] [PubMed] [Google Scholar]
- 22.Rudy C K, Scott J R, Churchward G. DNA binding by the Xis protein of the conjugative transposon Tn916. J Bacteriol. 1997;179:2567–2572. doi: 10.1128/jb.179.8.2567-2572.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rybkine T, Mainardi J-L, Sougakoff W, Collatz E, Gutmann L. Penicillin-binding protein 5 sequence alterations in clinical isolates of Enterococcus faecium with different levels of β-lactam resistance. J Infect Dis. 1998;178:159–163. doi: 10.1086/515605. [DOI] [PubMed] [Google Scholar]
- 24.Sahm D F, Marsilio M K, Piazza G. Antimicrobial resistance in key bloodstream bacterial isolates: electronic surveillance with The Surveillance Network Database-USA. Clin Infect Dis. 1999;29:259–263. doi: 10.1086/520195. [DOI] [PubMed] [Google Scholar]
- 25.Williamson R, Calderwood S B, Moellering R C, Jr, Tomasz A. Studies on the mechanism of intrinsic resistance to β-lactam antibiotic in group D streptococci. J Gen Microbiol. 1983;129:813–822. doi: 10.1099/00221287-129-3-813. [DOI] [PubMed] [Google Scholar]
- 26.Williamson R, LaBouguenec C, Gutmann L, Horaud T. One or two low affinity penicillin-binding proteins may be responsible for the range of susceptibility of Enterococcus faecium to penicillin. J Gen Microbiol. 1985;131:1933–1940. doi: 10.1099/00221287-131-8-1933. [DOI] [PubMed] [Google Scholar]
- 27.Yamada M, Izu H, Nitta T, Kurihara K, Sakurai T. High-temperature, nonradioactive primer extension assay for determination of a transcription initiation site. BioTechniques. 1998;25:72–75. doi: 10.2144/98251st02. [DOI] [PubMed] [Google Scholar]
- 28.Zorzi W, Zhou X Y, Dardenne O, Lamotte J, Raze D, Pierre J, Gutmann L, Coyette J. Structure of the low-affinity penicillin-binding protein 5 PBP5fm in wild-type and highly penicillin-resistant strains of Enterococcus faecium. J Bacteriol. 1996;178:4948–4957. doi: 10.1128/jb.178.16.4948-4957.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]