Abstract
Background
Automated radiologic analysis using computer-aided detection software (CAD) could facilitate chest X-ray (CXR) use in tuberculosis diagnosis. There is little to no evidence on the accuracy of commercially available deep learning-based CAD in different populations, including patients with smear-negative tuberculosis and people living with human immunodeficiency virus (HIV, PLWH).
Methods
We collected CXRs and individual patient data (IPD) from studies evaluating CAD in patients self-referring for tuberculosis symptoms with culture or nucleic acid amplification testing as the reference. We reanalyzed CXRs with three CAD programs (CAD4TB version (v) 6, Lunit v3.1.0.0, and qXR v2). We estimated sensitivity and specificity within each study and pooled using IPD meta-analysis. We used multivariable meta-regression to identify characteristics modifying accuracy.
Results
We included CXRs and IPD of 3727/3967 participants from 4/7 eligible studies. 17% (621/3727) were PLWH. 17% (645/3727) had microbiologically confirmed tuberculosis. Despite using the same threshold score for classifying CXR in every study, sensitivity and specificity varied from study to study. The software had similar unadjusted accuracy (at 90% pooled sensitivity, pooled specificities were: CAD4TBv6, 56.9% [95% confidence interval {CI}: 51.7–61.9]; Lunit, 54.1% [95% CI: 44.6–63.3]; qXRv2, 60.5% [95% CI: 51.7–68.6]). Adjusted absolute differences in pooled sensitivity between PLWH and HIV-uninfected participants were: CAD4TBv6, −13.4% [−21.1, −6.9]; Lunit, +2.2% [−3.6, +6.3]; qXRv2: −13.4% [−21.5, −6.6]; between smear-negative and smear-positive tuberculosis was: were CAD4TBv6, −12.3% [−19.5, −6.1]; Lunit, −17.2% [−24.6, −10.5]; qXRv2, −16.6% [−24.4, −9.9]. Accuracy was similar to human readers.
Conclusions
For CAD CXR analysis to be implemented as a high-sensitivity tuberculosis rule-out test, users will need threshold scores identified from their own patient populations and stratified by HIV and smear status.
Keywords: tuberculosis, chest X-ray, deep learning, individual patient data meta-analysis, accuracy
An individual patient data (IPD) meta-analysis found the accuracy of commercially available deep learning-based chest X-ray analysis software for detecting tuberculosis varied between studies and by patient characteristics. Diagnostic heterogeneity poses an implementation challenge for this novel technology.
Chest radiography has been used to evaluate individuals for tuberculosis for over a century [1, 2]. As is happening with other radiologic modalities, advances in artificial intelligence are transforming chest X-ray (CXR) interpretation by offering the potential to replace human readers with automated computer analysis, often called computer aided detection (CAD) [3].
Although CAD software that analyze CXR for pulmonary tuberculosis are commercially available, there remains uncertainty surrounding their diagnostic accuracy. A systematic review found the majority of published studies focused on development of CAD algorithms rather than their evaluation in clinical contexts, and that common risks of overestimating diagnostic accuracy included selection bias and use of human CXR-reading rather than microbiologic testing as reference standards [4]. Meta-analysis could not be performed due to methodologic differences [4, 5]. High quality evidence was particularly scarce for deep learning-based CAD—an artificial intelligence method that is highly effective for image recognition [4]. Given the small evidence base, and that most users will have no field experience with this novel technology, it is important to know if accuracy varies between populations and by patient characteristics. There are no published data on whether human immunodeficiency virus (HIV) infection affects accuracy of deep learning-based CXR analysis, and effects of other patient characteristics on sensitivity and specificity were reported in only 1 study [6].
We performed an individual patient data (IPD) meta-analysis to address gaps in the evidence base on the diagnostic accuracy of CXR analysis with CAD for detecting tuberculosis. We focused on the use of CXR to evaluate individuals self-referring for symptoms of tuberculosis. In this context, chest radiography functions as a triage test: when the CXR is abnormal, sputum microbiologic tests are required to diagnose active pulmonary tuberculosis, whereas a normal CXR is sufficient to rule out active disease [7].
METHODS
Our reporting follows PRISMA-IPD recommendations [8].
Objectives
We sought to estimate the diagnostic accuracy of CXR analyzed by deep learning-based, commercially available CAD software for the detection of culture- or nucleic acid amplification test (NAAT)-confirmed pulmonary tuberculosis in symptomatic, self-referred individuals. Our secondary objective was to identify patient characteristics that modify diagnostic accuracy.
Search Strategy, Study Selection, and Quality Assessment
Eligible studies were identified through published systematic reviews [4, 5]. We added 1 study prior to its publication, through referral by its principal investigator (author F. A. K.) [6].
Eligible studies consecutively enrolled individuals self-referring for medical care due to symptoms of pulmonary tuberculosis, estimated the diagnostic accuracy of any commercially available CAD program for the detection of pulmonary tuberculosis, and used either NAAT or mycobacterial culture as the reference test. For eligible studies to be included, investigators had to share de-identified clinical data and digital CXR images. Exclusion criteria are in the Supplementary materials (page 1).
Investigators provided data on age, sex, HIV status, prior tuberculosis history, smear status, and results of culture and/or NAAT, as well as CXR DICOM files. Data management is described in Supplementary materials page 1. We included participants who had available CXRs and conclusive microbiological results. We excluded participants without CXR images; whose CXR could not be analyzed by all 3 CAD programs; and those with growth of nontuberculous mycobacteria in culture. One reviewer (F. A. K.) performed a quality assessment of each study by adapting the QUADAS-2 tool [9].
Ethics
Investigators had local approvals to share data and CXR images. The IPD meta-analysis was approved by the Research Ethics Board of the McGill University Health Centre.
Index Tests
We analyzed each CXR with 3 commercially available deep learning-based CAD programs: CAD4TB version 6 (Delft, Netherlands), Lunit INSIGHT version 3.1.0.0. (Lunit, South Korea), and qXR version 2 (qure.ai, India). Each software was installed and run at the Research Institute of the McGill University Health Centre. After analyzing a CXR image, each software outputs an abnormality score on a 100-point scale (CAD4TB, 0 to 100; Lunit, 0 to 100; qXRv2, 0.00 to 1.00). A threshold score is selected for categorization: if the abnormality score is below the threshold, the CXR is classified as sufficient to rule out pulmonary tuberculosis; otherwise, the CXR is categorized as consistent with pulmonary tuberculosis. Sensitivity and specificity thus depend on the threshold score.
Reference Tests
We classified participants as having pulmonary tuberculosis if at least 1 sputum specimen demonstrated Mycobacterium tuberculosis in culture or NAAT (Xpert MTB/RIF). Among participants not meeting criteria for pulmonary tuberculosis, those with at least 1 sputum specimen negative by culture or NAAT were categorized as not having pulmonary tuberculosis. We classified sputum specimens that grew exclusively nontuberculous mycobacteria as indeterminate.
Data Analysis
We generated within-study and pooled receiver operating characteristic (ROC) curves and estimated area under the ROC curves (AUC). To estimate pooled AUC, we used 1-step parametric linear mixed effects meta-analysis [10, 11], specifying common random intercepts.
We used 3 approaches to select threshold scores for estimating sensitivity and specificity. From an implementation perspective, it would be much easier if software came with a recommended threshold score for universal application. Lunit and qXRv2 come with such developer-recommended threshold scores, whereas CAD4TBv6 does not [12]. To estimate sensitivity and specificity using a universal threshold, we applied: (1) for Lunit and qXRv2, developer-recommended threshold scores; (2) for all 3 software, threshold scores needed to reach a pooled sensitivity of 90%, which we refer to as “meta-analysis-derived threshold scores.” Our third approach to threshold selection was an alternative to using universal threshold scores: (3) the use of threshold scores tailored to each site, which we refer to as “study-specific threshold scores.” We identified study-specific threshold scores by using each study’s ROC curve to find the score with sensitivity of 90% (the minimum recommended by WHO for a tuberculosis triage test) [13].
We first estimated, for each study separately, sensitivity and specificity using developer-recommended, meta-analysis-derived, and study-specific threshold scores. We used forest plots to investigate between-study heterogeneity. Next, using 2-step bivariate random-effects meta-analysis [14, 15], we estimated pooled sensitivity and pooled specificity for developer-recommended and meta-analysis derived threshold scores. We did not estimate pooled sensitivity and specificity using study-specific threshold scores. We estimated pooled negative and positive likelihood ratios, across a range of threshold scores, using a bivariate modelling approach [14].
In addition to estimating unadjusted accuracy, we estimated sensitivity and specificity within predefined subgroups of sex, HIV-status, sputum smear-status, prior tuberculosis history, and age (details in Supplementary materials, page 1). We first identified associations in univariable analyses, within each study, and pooling data across studies. To determine whether associations remained after adjusting for covariates, we performed generalized linear IPD multivariable meta-regression. In these models, parameter estimates are the absolute difference in sensitivity, or specificity, between subgroups adjusted for other variables in the model. We judged absolute differences as statistically significant if 95% confidence intervals (95% CI) excluded 0.
We estimated diagnostic outcomes at varying prevalence of tuberculosis (5%, 17%, 20%) using the meta-analysis-derived threshold scores in hypothetical cohorts of 1000 patients undergoing CXR analysis with these software, stratified by the characteristics that were associated with sensitivity.
In a post-hoc analysis, we sought to compare accuracy of CXR analysis by CAD to interpretation by human readers. For these analyses, we used the categorical CXR categorizations by human readers provided in the original study data to classify images as Abnormal (if any abnormality present) or Normal; we chose this classification as it is known to maximize sensitivity of human-read CXR for tuberculosis (details in Supplementary materials, page 2) [16]. To compare the accuracy of CAD software to human CXR interpretation, we visualized the sensitivity and specificity of human readers on plots of the ROC curves of the 3 software.
In another post hoc analysis, we repeated our search of the literature on 24 April 2020 to identify potentially eligible studies published since our original search.
Statistical analyses were performed with SAS software (version 9.4, SAS Institute, Cary, North Carolina, USA) [17] and R statistical software (RStudio, version 1.2.5033) [18] using packages diagmeta [11] and mada [14].
Role of the Funding Source
The funding source had no input on the design, conduct, analysis, or reporting.
RESULTS
Study Selection and Quality Assessment
The selection of studies for inclusion from the previously published systematic review [4] into the IPD meta-analysis is depicted in Supplementary Figure 1 (Supplementary materials, page 3). Of 54 full-text articles reviewed, 7 met inclusion criteria, of which 3 were excluded because IPD could not be obtained. The IPD meta-analysis includes 4 studies [6, 19–22].
Of 3967 participants for whom IPD were provided, we included 3727/3967 (94%), and we excluded 240 (6%) for the following reasons (Supplementary Table 1, Supplementary materials, page 4): 20 missing CXR; 20 whose CXR could not be analyzed; 140 with nontuberculous mycobacteria in sputum culture; and 60 without reference standard results.
Risks of bias in QUADAS-2 patient selection and flow and timing domains were low for all studies; in the reference standard domain, was low in 3/4 and unclear in 1/4 (Supplementary Figure 2, Supplementary materials, page 5). Applicability concerns were low for 4/4.
Description of Included Participants
The included studies were undertaken in Pakistan [6], South Africa [21, 22], Tanzania [19], and Zambia [20] (Table 1). Age, sex, prior tuberculosis, and smear data were unavailable for 1 study [21, 22]. Women accounted for 47% (1583/3352) of participants, and PLWH for 17% (621/3695). NAAT- or culture-confirmed tuberculosis was diagnosed in 17% (645/3727). Smear-positive disease accounted for 73% (417/573) of confirmed tuberculosis.
Table 1.
Characteristics of 3727 Included Participants
| Study | |||||
|---|---|---|---|---|---|
| Characteristic | Pakistan | South Africa | Tanzania | Zambia | Total |
| N | 2298 | 375 | 712 | 342 | 3727 |
| Age in years | |||||
| Median (IQR) | 33 (23, 49) | Not reported | 38 (30, 50) | 35 (28, 43) | 35 (25, 48) |
| Missing | 0 | 375 | 0 | 5 | 380 |
| Sex | |||||
| Women | 1098 (48%) | Not reported | 353 (50%) | 132 (39%) | 1583 (47%) |
| Men | 1200 (52%) | 359 (50%) | 210 (61%) | 1769 (53%) | |
| Missing | 0 | 375 | 0 | 0 | 375 |
| HIV status | |||||
| Uninfected | 2287 (99%) | 244 (66%) | 400 (56%) | 143 (43%) | 3074 (83%) |
| PLWH | 3 (1%) | 123 (34%) | 308 (44%) | 187 (57%) | 621 (17%) |
| Missing | 8 | 8 | 4 | 12 | 32 |
| Prior TB | |||||
| Yes | 517 (23%) | Not reported | 112 (16%) | 78 (23%) | 707 (21%) |
| No | 1778 (77%) | 600 (84%) | 264 (77%) | 2642 (79%) | |
| Missing | 3 | 375 | 0 | 0 | 378 |
| NAAT or culture positive for MTB | |||||
| Yes | 293 (13%) | 70 (19%) | 188 (26%) | 94 (27%) | 645 (17%) |
| No | 2005 (87%) | 305 (81%) | 524 (74%) | 248 (73%) | 3082 (83%) |
| Smear statusa | |||||
| Negative | 73 (25%) | Not reported | 48 (25.5%) | 35 (38%) | 156 (27%) |
| Positive | 220 (75%) | 140 (74.5%) | 57 (62%) | 417 (73%) | |
| Missing | 0 | 70 | 0 | 2 | 72 |
All numbers are N (%) unless indicated otherwise. Data provided for South Africa study did not include age, sex, history of prior TB, and smear status.
Abbreviations: HIV, human immunodeficiency virus; IQR, interquartile range; MTB, Mycobacterium tuberculosis; NAAT, nucleic acid amplification testing; PLWH, people living with HIV; TB, tuberculosis.
aSmear status in Table 1 is only among individuals who are NAAT- or culture-positive for MTB. Among those without reference-standard confirmed TB, 10/3082 (0.3%) had positive smears.
ROC Analyses
ROC curves are in Supplementary Figures 3 and Supplementary materials, pages 6–7). The software had similar pooled AUC estimates with overlapping confidence intervals: CAD4TBv6, 0.83 (95% CI: .82–.84); Lunit, 0.83 (95% CI: .79–.86); qxRv2, 0.85 (95% CI: .83–.88).
Sensitivity and Specificity Within Each Study
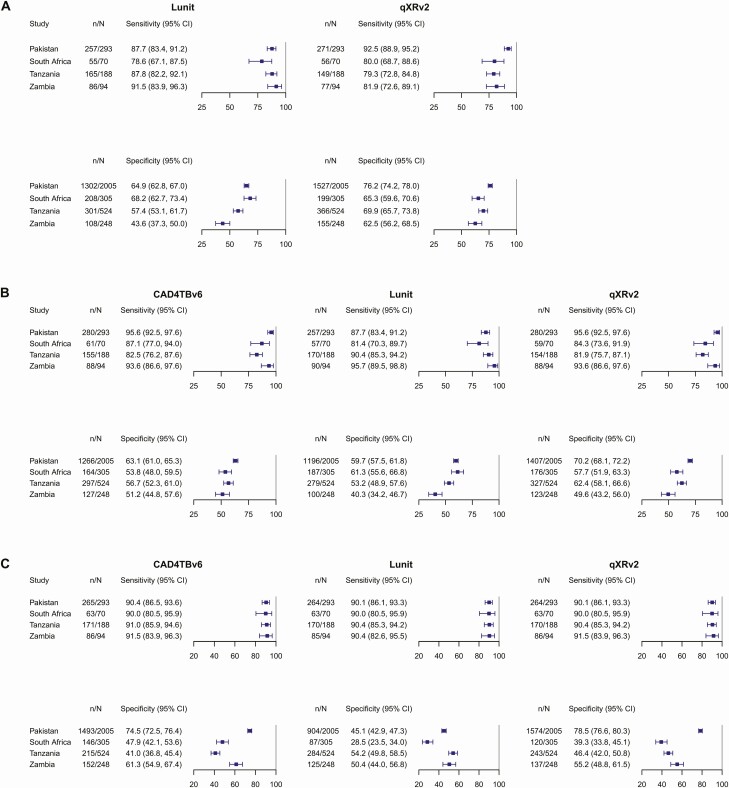
For each software, when the same threshold score was applied in every study, sensitivity and specificity varied from study to study. This between-study variability was observed when using developer-recommended threshold scores (Figure 1A) and also with meta-analysis derived threshold scores (Figure 1B). When study-specific threshold scores were applied, between-study variability in specificity persisted (Figure 1C). The threshold score needed to achieve sensitivity of 90% varied between each study.
Figure 1.
Within-study sensitivity and specificity of chest X-ray analysis with deep learning-based software as a tuberculosis triage test in self-referred, symptomatic individuals. A, Developer-recommended threshold scores. B, Meta-analysis derived threshold scores. C, Study-specific threshold scores. * We used the prespecified developer-recommended threshold score to classify chest X-rays as either consistent with tuberculosis or tuberculosis ruled-out. † For each software, the following threshold scores were applied in all studies: CAD4TBv6, 54; Lunit, 16.68; qXRv2, 0.44. These threshold scores were chosen as each was required to reach a pooled sensitivity of 90%. ‡ For each particular study, we identified the threshold score needed to reach a within-study sensitivity of 90% and estimated its within-study specificity. When no threshold score reached a sensitivity of exactly 90%, we selected the score achieving sensitivity >90%. The following threshold scores were used: CAD4TBv6: Pakistan, 62, South Africa, 50, Tanzania, 48, Zambia, 60; Lunit: Pakistan, 6.52, South Africa, 3.13, Tanzania, 23.11, Zambia, 54.79; qXRv2: Pakistan, 0.62, South Africa, 0.25, Tanzania, 0.27, Zambia, 0.47. Abbreviations: CAD, computer-aided detection software; CI, confidence interval.
Pooled Sensitivity and Specificity Estimated From Individual Patient Data Meta-Analysis
Pooled sensitivity and specificity (Table 2) of developer-recommended threshold scores were: Lunit, 87.7% (95% CI: 82.5–91.5) and 59.2% (95% CI: 48.2–69.3); qXRv2, 84.0% (95% CI: 74.6–90.3) and 69.1% (95% CI: 63.2 to 74.5). At the meta-analysis derived threshold scores that achieved a pooled sensitivity of 90%, pooled specificities were: CAD4TBv6, 56.9% (95% CI: 51.7–61.9); Lunit, 54.1% (95% CI: 44.6–63.3); qXRv2, 60.5% (95% CI: 51.7–68.6). Pooled likelihood ratios across a range of threshold scores are reported in Supplementary Table 2 (Supplementary materials, page 8). At a threshold score close to the maximum abnormality score (95 for CAD4TBv6 and Lunit, and 0.95 for qXRv2), positive likelihood ratios were modest for CAD4TBv6 (5.4, 95% CI: 3.9–7.3) and Lunit (6.3, 95% CI: 3.8–10.2), and high for qXRv2 (20.7, 95% CI: 13.5–30.5).
Table 2.
Pooled Unadjusted Sensitivity and Specificity of Chest X-ray Analysis With Deep Learning-Based Software as a Tuberculosis Triage Test in Self-referred, Symptomatic Individuals, Stratified by Type of Threshold Score and Software
| (a) Developer-Recommended Threshold Scoresa | |||||||
|---|---|---|---|---|---|---|---|
| Software | |||||||
| Lunit | qXRv2 | ||||||
| Measure | Studies | n/N | Pooled Estimate [95% CI] | n/N | Pooled Estimate [95% CI] | ||
| Sensitivity | 4 | 563/645 | 87.7 [82.5–91.5] | 553/645 | 84.0 [74.6–90.3] | ||
| Specificity | 4 | 1919/3082 | 59.2 [48.2–69.3] | 2247/3082 | 69.1 [63.2–74.5] | ||
| (b) Meta-analysis Derived Threshold Scoresb | |||||||
| Software | |||||||
| CAD4TBv6 | Lunit | qXRv2 | |||||
| Measure | Studies | n/N | Pooled Estimate [95% CI] | n/N | Pooled Estimate [95% CI] | n/N | Pooled Estimate [95% CI] |
| Sensitivity | 4 | 584/645 | 90.4 [82.2–95.1] | 574/645 | 90.3 [82.9–94.7] | 581/645 | 90.0 [80.8–95.1] |
| Specificity | 4 | 1854/3082 | 56.9 [51.7–61.9] | 1762/3082 | 54.1 [44.6–63.3] | 2033/3082 | 60.5 [51.7–68.6] |
Pooled sensitivity and specificity estimated using bivariate random effects 2-step individual patient data meta-analysis. Point estimates are not always equivalent to division of numerator by denominator as they were estimated via meta-analysis.
Abbreviation: CI, confidence interval.
aEach software’s threshold score, which we applied in all studies, was prespecified by the software developers.
bEach software’s threshold score, which we applied in all studies, was identified using meta-analysis as the one that reached an unadjusted pooled sensitivity of 90%: CAD4TBv6, 54; Lunit, 16.68; qXRv2, 0.44.
Subgroup Analyses
In within-study univariable analyses (Supplementary Tables 3–5, Figures 3–5, Supplementary materials, pages 9–21), for all 3 approaches to threshold score selection, in at least 1 study, sensitivity was lower among women versus men, lower amongst PLWH versus HIV-uninfected participants, and lower for smear-negative versus smear-positive disease. In at least 1 study, specificity was lower among men versus women, among PLWH versus HIV-uninfected participants, among participants with prior tuberculosis, and in the highest age tertile.
In univariable pooled analyses (Table 3), pooled sensitivity was consistently lower among PLWH versus the HIV-uninfected, for all 3 software; however, differences reached statistical significance only for CAD4TBv6 and qXRv2 (with meta-analysis-derived threshold scores, PLWH: CAD4TBv6, 80.4% [95% CI: 62.4–91.0], qXRv2, 78.9% [95% CI: 61.7–89.7]); versus HIV-uninfected, CAD4TBv6, 94.5% [95% CI: 91.5–96.4], qXRv2: 93.9% [95% CI: 90.4–96.1]. Pooled sensitivity was also consistently lower for smear-negative tuberculosis for all 3 software; however, differences were statistically significant only for Lunit and qXRv2 with the developer-recommended threshold (pooled sensitivity for smear-positive tuberculosis: Lunit, 95.9% [95% CI: 88.4–98.6]; qXRv2, 93.2% [95% CI: 86.4–96.7]; vs smear-negative: Lunit, 74.3% [95% CI: 63.7–82.7], qXRv2, 64.9% [95% CI: 39.8–83.8]). Pooled specificity was significantly lower amongst individuals with prior tuberculosis (with meta-analysis-derived threshold scores, with prior tuberculosis: CAD4TBv6, 26.6% [95% CI: 17.2–38.7], Lunit, 29.7% [95% CI: 22.3–38.4], qXRv2, 33.7% [95% CI: 24.4–44.4]; versus no prior tuberculosis: CAD4TBv6, 66.8% [95% CI: 60.9–72.2], Lunit, 58.0% [95% CI: 45.9–69.2], qXRv2, 69.3% [95% CI: 54.7–80.9]). Pooled specificity was highest in the youngest versus oldest age tertile (with meta-analysis derived threshold scores, in 14–23 year-old group: CAD4TBv6, 73.6% [95% CI: 61.9–82.6], Lunit, 65.9% [95% CI: 53.0–76.9], qXRv2, 74.4% [95% CI: 61.2–84.2]; in 43–90 year-old group: CAD4TBv6, 43.0% [95% CI: 39.4–46.8], Lunit, 45.0% [95% CI: 40.1–50.0], qXRv2, 57.3% [95% CI: 54.2–60.5]).
Table 3.
Subgroup Pooled Sensitivity and Specificity of Chest X-ray Analysis With Deep Learning-Based Software in Self-referred, Symptomatic Individuals, Stratified by Type of Threshold Score and Software
| (a) Developer-Recommended Threshold Scores a | |||||||
|---|---|---|---|---|---|---|---|
| Software | |||||||
| Lunit | qXRv2 | ||||||
| Subgroup | Studies | n/N | Sensitivity [95% CI] | n/N | Sensitivity [95% CI] | ||
| Unadjusted | 4 | 563/645 | 87.7 [82.5–91.5] | 553/645 | 84.0 [74.6–90.3] | ||
| Women | 3 | 201/235 | 82.9 [70.0–91.0] | 192/235 | 78.2 [53.1–91.9] | ||
| Men | 3 | 307/340 | 93.1 [82.3–97.5] | 305/340 | 89.0 [82.8–93.2] | ||
| PLWH | 3 | 144/179 | 80.7 [68.6–88.9] | 123/179 | 69.4 [59.6–77.6] | ||
| HIV uninfected | 4 | 413/460 | 91.2 [83.3–95.6] | 425/460 | 92.1 [89.2–94.3] | ||
| Smear-positive | 3 | 390/417 | 95.9 [88.4–98.6] | 392/417 | 93.2 [86.4–96.7] | ||
| Smear-negative | 3 | 116/156 | 74.3 [63.7–82.7] | 103/156 | 64.9 [39.8–83.8] | ||
| Prior TB | 3 | 71/77 | 90.8 [80.7–95.9] | 67/77 | 86.6 [76.6–92.8] | ||
| No prior TB | 3 | 437/498 | 88.1 [84.6–90.9] | 430/498 | 85.3 [71.4–93.1] | ||
| Age 14–28 years | 3 | 191/211 | 91.3 [85.5–94.9] | 195/211 | 92.6 [88.2–95.5] | ||
| Age 28–43 years | 3 | 193/221 | 86.9 [81.4–91.0] | 178/221 | 81.8 [65.8–91.3] | ||
| Age 43–90 years | 3 | 123/142 | 86.5 [78.8–91.7] | 123/142 | 84.3 [65.8–93.7] | ||
| Subgroup | Studies | n/N | Specificity [95% CI] | n/N | Specificity [95% CI] | ||
| Unadjusted | 4 | 1919/3082 | 59.2 [48.2–69.3] | 2247/3082 | 69.1 [63.2–74.5] | ||
| Women | 3 | 870/1348 | 61.1 [53.4–68.2] | 1033/1348 | 74.3 [68.8–79.1] | ||
| Men | 3 | 841/1429 | 51.5 [36.9–65.8] | 1015/1429 | 66.6 [56.9–75.1] | ||
| PLWH | 3 | 217/439 | 49.8 [34.9–64.7] | 279/439 | 62.9 [55.7–69.6] | ||
| HIV uninfected | 4 | 1681/2614 | 62.4 [56.3–68.1] | 1947/2614 | 71.5 [66.0–76.4] | ||
| Prior TB | 3 | 238/630 | 34.9 [27.7–42.8] | 286/630 | 40.9 [30.9–51.8] | ||
| No prior TB | 3 | 1471/2144 | 62.2 [48.0–74.6] | 1760/2144 | 78.5 [69.9–85.3] | ||
| Age 14–28 years | 3 | 685/914 | 68.7 [52.6–81.3] | 775/914 | 80.8 [70.9–87.9] | ||
| Age 28–43 years | 3 | 529/886 | 52.8 [36.0–69.0] | 637/886 | 67.6 [55.0–78.1] | ||
| Age 43–90 years | 3 | 495/973 | 51.2 [47.5–54.8] | 633/973 | 66.0 [62.3–69.4] | ||
| (b) Meta-analysis-derived Threshold Scoresb | |||||||
| Software | |||||||
| CAD4TBv6 | Lunit | qXRv2 | |||||
| Subgroup | Studies | n/N | Sensitivity [95% CI] | n/N | Sensitivity [95% CI] | n/N | Sensitivity [95% CI] |
| Unadjusted | 4 | 584/645 | 90.4 [82.2–95.1] | 574/645 | 90.3 [82.9–94.7] | 581/645 | 90.0 [80.8–95.1] |
| Women | 3 | 209/235 | 89.2 [74.2–96.0] | 206/235 | 86.2 [78.7–91.4] | 206/235 | 87.9 [63.7–96.8] |
| Men | 3 | 314/340 | 92.8 [80.6–97.6] | 311/340 | 95.3 [81.7–98.9] | 316/340 | 93.1 [87.3–96.4] |
| PLWH | 3 | 140/179 | 80.4 [62.4–91.0] | 152/179 | 86.3 [68.3–94.9] | 137/179 | 78.9 [61.7–89.7] |
| HIV-uninfected | 4 | 439/460 | 94.5 [91.5–96.4] | 416/460 | 92.7 [84.9–96.6] | 438/460 | 93.9 [90.4–96.1] |
| Smear-positive | 3 | 397/417 | 94.8 [86.1–98.2] | 394/417 | 97.4 [87.0–99.5] | 402/417 | 96.3 [89.7–98.7] |
| Smear-negative | 3 | 124/156 | 81.6 [56.2–93.9] | 121/156 | 79.0 [65.4–88.2] | 118/156 | 77.9 [48.8–92.9] |
| Prior TB | 3 | 73/77 | 92.2 [81.8–96.9] | 71/77 | 90.7 [80.8–95.8] | 71/77 | 91.9 [82.8–96.4] |
| No prior TB | 3 | 450/498 | 90.8 [78.9–96.3] | 446/498 | 91.2 [84.8–95.0] | 451/498 | 91.7 [79.1–97.0] |
| Age 14–28 years | 3 | 199/211 | 93.5 [88.2–96.5] | 194/211 | 92.1 [84.2–96.2] | 203/211 | 96.1 [92.5–98.1] |
| Age 28–43 years | 3 | 192/221 | 89.0 [75.7–95.4] | 198/221 | 90.2 [82.6–94.7] | 189/221 | 88.5 [72.9–95.7] |
| Age 43–90 years | 3 | 131/142 | 91.8 [74.9–97.7] | 574/645 | 90.3 [82.9–94.7] | 129/142 | 90.9 [77.4–96.7] |
| Subgroup | Studies | n/N | Specificity [95% CI] | n/N | Specificity [95% CI] | n/N | Specificity [95% CI] |
| Unadjusted | 4 | 1854/3082 | 56.9 [51.7–61.9] | 1762/3082 | 54.1 [44.6–63.3] | 2033/3082 | 60.5 [51.7–68.6] |
| Women | 3 | 879/1348 | 64.6 [60.9–68.2] | 809/1348 | 57.4 [51.0–63.6] | 944/1348 | 65.4 [55.1–74.5] |
| Men | 3 | 811/1429 | 51.9 [42.6–61.1] | 766/1429 | 46.8 [34.1–59.8] | 913/1429 | 58.2 [45.6–69.8] |
| PLWH | 3 | 232/439 | 52.0 [41.0–62.8] | 199/439 | 45.2 [30.6–60.7] | 232/439 | 51.6 [40.1–63.0] |
| HIV uninfected | 4 | 1604/2614 | 58.6 [53.0–64.1] | 1542/2614 | 57.5 [53.8–61.1] | 1782/2614 | 64.4 [58.5–69.9] |
| Prior TB | 3 | 203/630 | 26.6 [17.2–38.7] | 209/630 | 29.7 [22.3–38.4] | 240/630 | 33.7 [24.4–44.4] |
| No prior TB | 3 | 1486/2144 | 66.8 [60.9–72.2] | 1364/2144 | 58.0 [45.9–69.2] | 1615/2144 | 69.3 [54.7–80.9] |
| Age 14–28 years | 3 | 713/914 | 73.6 [61.9–82.6] | 651/914 | 65.9 [53.0–76.9] | 726/914 | 74.4 [61.2–84.2] |
| Age 28–43 years | 3 | 561/886 | 60.3 [51.5–68.5] | 494/886 | 48.9 [32.7–65.3] | 572/886 | 58.6 [42.4–73.2] |
| Age 43–90 years | 3 | 415/973 | 43.0 [39.4–46.8] | 428/973 | 45.0 [40.1–50.0] | 557/973 | 57.3 [54.2–60.5] |
Subgroup estimates with nonoverlapping confidence intervals (CIs) are in bold.
Abbreviations: HIV, human immunodeficiency virus; PLWH, people living with HIV; TB, tuberculosis.
aEach software’s threshold score, which we applied in all subgroups and all studies, was prespecified by the software developers.
bEach software’s threshold score, which we applied in all studies, was identified using meta-analysis as the one that reached an unadjusted pooled sensitivity of 90%: CAD4TBv6, 54; Lunit, 16.68; qXRv2, 0.44.
Adjusted absolute differences in pooled sensitivity and pooled specificity between subgroups estimated using multivariable IPD meta-regression are shown in Table 4. In analyses adjusted for sex, HIV status, and smear status, for all 3 software, there were no significant differences in sensitivity between women and men (absolute difference in sensitivity between women and men: with developer-recommended threshold scores: Lunit, −2.1% [95% CI: −7.3, 2.5]; qXRv2, −2.1% [95% CI: −6.9, 1.5]; with meta-analysis-derived threshold scores: CAD4TBv6, −2.4% [95% CI: −6.4, .7]; Lunit, −1.9% [95% CI: −6.7, 2.4], qXRv2: multivariable model did not converge). HIV status was not associated with sensitivity of Lunit; however, sensitivity was lower among PLWH using CAD4TBv6 and qXRv2 (adjusting for sex and smear status, absolute difference in sensitivity between PLWH and HIV-uninfected: with meta-analysis derived threshold score, CAD4TBv6, −13.4% [95% CI: −21.1, −6.9]; qXRv2 (adjusted for smear-status only), −13.4% [95% CI: −21.5, −6.6]). For all 3 software, sensitivity was lower for smear-negative disease (adjusting for sex and HIV-status, absolute difference in sensitivity between smear-negative and smear-positive participants: with meta-analysis-derived threshold scores: CAD4TBv6, −12.3% [95% CI:−19.5, −6.1]; Lunit, −17.2% [95% CI:−24.6, −10.5], qXRv2 (adjusting for HIV-status): −16.6% [95% CI:−24.4, −9.9]).
Table 4.
Adjusted Absolute Differences in Sensitivity and Specificity Between Subgroups of Sex, HIV Status, and Smear Status, Applying Developer-Recommended Thresholds, or Meta-Analysis-Derived Threshold Scores With Pooled Sensitivity of 90%
| Characteristic | Developer-recommended Threshold Scores | Meta-analysis-derived Threshold Scores | |||
|---|---|---|---|---|---|
| Lunit | qXRv2 | CAD4TBv6 | Lunit | qXRv2 | |
| Difference in Sensitivity [95% CI] | Difference in Sensitivity [95% CI] | Difference in Sensitivity [95% CI] | Difference in Sensitivity [95% CI] | Difference in Sensitivity [95% CI] | |
| Sex | |||||
| Women | −2.1 [−7.3, 2.5] | −2.2 [−6.9, 1.5] | −2.4 [−6.4, .7] | −1.9 [−6.7, 2.4] | Not in modela |
| Men | Ref | Ref | Ref | Ref | Ref |
| HIV status | |||||
| PLWH | −1.0 [−8.1, 4.4] | −17.6 [−26.4, −9.7] | −13.4 [−21.1, −6.9] | 2.2 [−3.6, 6.3] | −13.4 [−21.5, −6.6] |
| Uninfected | Ref | Ref | Ref | Ref | Ref |
| Smear status | |||||
| Negative | −18.6 [−26.4, −11.4] | −23.0 [−31.5, −15.1] | −12.3 [−19.5, −6.1] | −17.2 [−24.6, −10.5] | −16.6 [−24.4, −9.9] |
| Positive | Ref | ref | ref | ref | ref |
| Characteristic | Difference in Specificity [95% CI] | Difference in Specificity [95% CI] | Difference in Specificity [95% CI] | Difference in Specificity [95% CI] | Difference in Specificity [95% CI] |
| Sex | |||||
| Men | −4.8 [−8.2, −1.5] | −3.8 [−6.7, −1.0] | −6.7 [−9.9, −3.6] | −5.7 [−9.1, −2.2] | −5.0 [−8.1, −1.9] |
| Women | Ref | Ref | Ref | Ref | Ref |
| HIV status | |||||
| PLWH | −7.2 [−13.8, −.6] | −7.2 [−14.1, −.9] | −5.8 [−10.5, −1.2] | −6.5 [−12.9, −.1] | −8.5 [−15.1, −2.0] |
| Uninfected | Ref | Ref | Ref | Ref | Ref |
| Prior TB | |||||
| Prior TB | −29.0 [−33.1, −24.8] | −35.3 [−40.0, −31.1] | −34.2 [−38.1, −30.2] | −28.1 [−32.2, −24.0] | −35.7 [−39.9, −31.4] |
| None | Ref | Ref | Ref | Ref | Ref |
| Age tertiles | |||||
| 14–23 years | Ref | Ref | Ref | Ref | Ref |
| 23–43 years | −12.4 [−16.7, −8.2] | −8.8 [−12.3, −5.4] | −12.9 [−16.9, −8.9] | −13.4 [−17.8, −9.0] | −10.8 [−14.8, −6.9] |
| 43–90 years | −21.1 [−25.2, −17.0] | −15.3 [−18.9, −11.8] | −31.7 [−35.6, −27.7] | −24.1 [−28.3, −19.9] | −18.0 [−21.8, −14.1] |
Differences with confidence intervals (CI) that exclude the null value are shown in bold. For sensitivity, N = 567; for specificity N = 2742. For sensitivity we used fixed effects individual patient data multivariable meta-regression, and for specificity we used random effects individual patient data meta-regression.
Abbreviations: HIV, human immunodeficiency virus; PLWH, people living with HIV; TB, tuberculosis.
aModel for differences in sensitivity with qXRv2 did not converge when sex was included. Estimates are the absolute difference in sensitivity (or specificity) comparing subgroups, after adjusting for the other co-variates in the model. For example, the estimate for Lunit with its developer-recommended threshold score for HIV status means that Lunit sensitivity was 1.0% lower among PLWH compared to the HIV-uninfected, after adjusting for sex and smear-status, but the CI and P-value indicate that the difference was not statistically significant.
For all 3 software, in multivariable IPD meta-regression models that included sex, HIV-status, prior TB, and age group, specificity was significantly lower in: men vs women (with meta-analysis-derived threshold scores: CAD4TBv6, −6.7% [95% CI: −9.9, −3.6], Lunit, −5.7% [95% CI: −9.1, −2.2], qXRv2, −5.0% [95% CI: −8.1, −1.9]); PLWH vs HIV-uninfected (CAD4TBv6, −5.8% [95% CI: −10.5, −1.2], Lunit, −6.5% [95% CI:−12.9, −.1], qXRv2, −8.5% [95% CI: −15.1, −2.0]); participants with prior tuberculosis vs with no prior tuberculosis (with meta-analysis-derived threshold scores: CAD4TBv6, −34.2% [95% CI: −38.1, −30.2], Lunit, −28.1% [95% CI: −32.2, −24.0], qXRv2, −35.7% [95% CI: −39.9, −31.4]), and in older age groups vs the youngest (with meta-analysis-derived threshold scores, 23–43 year-old vs 14–23 year-old groups: CAD4TBv6, −12.9% [95% CI: −16.9, −8.9], Lunit, −13.4% [95% CI: −17.8, −9.0], qXRv2, −10.8% [95% CI: −14.8, −6.9]; 43–90 year-old vs 14–23 year-old groups: CAD4TBv6, −31.7% [95% CI: −35.6, −27.7], Lunit, −24.1% [95% CI: −28.3, −19.9], qXRv2, −18.0% [95% CI: −21.8, −14.1]).
Table 5 provides expected outcomes of CXR analysis in hypothetical cohorts at varying tuberculosis prevalence.
Table 5.
Outcomes of Triage Testing Using Chest X-rays Interpreted by Deep Learning-Based Computer-Aided Detection Software in 1000 Individuals Self-Referred for Symptoms of Tuberculosis, Applying Meta-Analysis Derived Threshold Scores With 90% Pooled Sensitivity
| Subgroup | Diagnostic Outcomes per 1000 Patients Tested (95%CI) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 5% Tuberculosis Prevalence | 17% Tuberculosis Prevalence | 20% Tuberculosis Prevalence | |||||||
| CAD4TBv6 | Lunit | qXRv2 | CAD4TBv6 | Lunit | qXRv2 | CAD4TBv6 | Lunit | qXRv2 | |
| HIV-uninfected | |||||||||
| Tuberculosis detected | 47 (46–48) |
46 (42–48) |
47 (45–48) |
161 (156–164) |
158 (144–164) |
160 (154–163) |
189 (183–193) |
185 (170–193) |
188 (181–192) |
| Tuberculosis missed | 3 (2–4) |
4 (2–8) |
3 (2–5) |
9 (6–14) |
12 (6–26) |
10 (7–16) |
11 (7–17) |
15 (7–30) |
12 (8–19) |
| Correctly classified as no tuberculosis | 557 (504–609) |
546 (511–580) |
612 (556–664) |
486 (440–532) |
477 (447–507) |
535 (486–580) |
469 (424–513) |
460 (430–489) |
515 (468–559) |
| Incorrectly classified as tuberculosis | 393 (341–447) |
404 (370–439) |
338 (286–394) |
344 (298–390) |
353 (323–383) |
295 (250–344) |
331 (287–376) |
340 (311–370) |
285 (241–332) |
| PLWH | |||||||||
| Tuberculosis detected | 40 (31–46) |
43 (34–47) |
39 (31–45) |
137 (106–155) |
147 (116–161) |
134 (105–152) |
161 (125–182) |
173 (137–190) |
158 (123–179) |
| Tuberculosis missed | 10 (5–19) |
7 (3–16) |
11 (5–19) |
33 (15–64) |
23 (9–54) |
36 (18–65) |
39 (18–75) |
27 (10–63) |
42 (21–77) |
| Correctly classified as no tuberculosis | 494 (390–597) |
429 (291–577) |
490 (381–599) |
432 (340–521) |
375 (254–504) |
428 (333–523) |
416 (328–502) |
362 (245–486) |
413 (321–504) |
| Incorrectly classified as tuberculosis | 456 (353–561) |
521 (373–659) |
460 (352–569) |
398 (309–490) |
455 (326–576) |
402 (307–497) |
384 (298–472) |
438 (314–555) |
387 (296–479) |
| Smear-positive | |||||||||
| Tuberculosis detected | 47 (43–49) |
49 (44–50) |
48 (45–49) |
161 (146–167) |
166 (148–169) |
164 (152–168) |
190 (172–196) |
195 (174–199) |
193 (179–197) |
| Tuberculosis missed | 3 (1–7) |
1 (0–7) |
2 (1–5) |
9 (3–24) |
4 (1–22) |
6 (2–18) |
10 (4–28) |
5 (1–26) |
7 (3–21) |
| Smear-negative | |||||||||
| Tuberculosis detected | 41 (28–47) |
40 (33–44) |
39 (24–46) |
139 (96–160) |
134 (111–150) |
132 (83–158) |
163 (112–188) |
158 (131–176) |
156 (98–186) |
| Tuberculosis missed | 9 (3–22) |
11 (6–17) |
11 (4–26) |
31 (10–74) |
36 (20–59) |
38 (12–87) |
37 (12–88) |
42 (24–69) |
44 (14–102) |
Prevalence of 17% is shown as this was the prevalence of pulmonary tuberculosis in the present study. Prevalence of 5% and 20% are shown by convention. The same threshold score was applied in each group. Estimates calculated using pooled sensitivity and specificity of the meta-analysis-derived threshold score (CAD4TBv6, 54; Lunit, 16.68; qXRv2, 0.44).
Abbreviations: CAD, computer-aided detection software; HIV, human immunodeficiency virus; PLWH, people living with HIV.
Post hoc Analysis
Results of human CXR readers were available for 2/4 studies, so we did not pool results. In Tanzania, 3 human readers interpreted each CXR (sensitivity range: 83%–97.3%; specificity, 12.0%–58.6%) [19]. In Zambia, a single reader was used (sensitivity, 96.8%; specificity, 48.8%) [20]. In both settings, confidence intervals for each human reader’s sensitivity and specificity intersected with each software’s ROC curves (Supplementary Figure 6, Supplementary materials, page 22), indicating accuracy of CAD and human readers were similar. As was seen for CAD in within-study analysis, sensitivity and specificity of human CXR reading were modified by sex, HIV, sputum smear, prior tuberculosis, and age (Supplementary Table 6, Supplementary materials, page23).
Findings From Updated Literature Search
On 24 April 2020, we repeated our search strategy to identify relevant studies published since our initial search in February 2019. We identified 570 unique records, excluded 557 based on title and abstract screening and 12 after full-text screening, leaving 1 study [23] eligible for inclusion had IPD been available (study selection is summarized in Supplementary Figure 8, Supplementary materials, page 24). The study by Qin et al retrospectively estimated the diagnostic accuracy of CAD4TBv6, Lunit, and qXRv2, against a reference of a single sputum specimen tested by NAAT [23]. Data originated from 2 TB referral centers in Nepal and Cameroon. Among 1196 individuals, 38 (3.2%) were PLWH, and 109 (9.1%) had NAAT-positive sputum of whom 76/109 (69.7%) were sputum smear-positive. AUCs were higher compared to what we reported: CAD4TBv6 (0.92, 95% CI: .90–.95), Lunit (0.94, 95% CI: .93–.96), and qXRv2 (0.94, 95% CI: .92–.97). Sensitivity and specificity stratified by smear and HIV status were not reported. Similar to our study, the authors found that application of the same threshold score resulted in sensitivity and specificity differing between study sites.
DISCUSSION
Through meta-analysis of data from 3727 individuals self-referring for tuberculosis symptoms, we evaluated the diagnostic accuracy of CXR analyzed by commercially available, deep learning-based CAD software, as a triage test for NAAT- or culture-confirmed tuberculosis. For each software, applying the same threshold in all studies resulted in sensitivity and specificity varying from study to study. In adjusted analyses, sensitivity was associated with HIV status for CAD4TBv6 and qXRv2, and with sputum-smear status for all 3 software. For all 3 software, specificity was associated with age, sex, prior tuberculosis, and HIV status. In 2 studies where human interpretation of CXR was reported, accuracies of human readers and CAD software were comparable, and patient characteristics similarly affected human reading.
The high sensitivity and moderate to low specificity of CXR analysis by these software, and our observed associations between certain patient characteristics and software accuracy, are similar to what has been reported for human-read CXR [19, 24–30]. Hence, the evidence suggests that CXR analysis by CAD software would lead to similar diagnostic outcomes for tuberculosis as CXR interpretation by humans. Based in part on our findings, WHO recently issued new guidance, supporting the use of CAD as a replacement to humans for analyzing CXR for tuberculosis. Use of CAD software will improve reliability of CXR analysis by eliminating intra- [31] and inter-reader [25, 31–37] variability that occur with human reading and would also eliminate problems tied to human reader fatigue [38, 39]. However, implementation should take into consideration three important limitations of current software.
First, we found that the accuracy of threshold scores varied between studies, such that users will face uncertainty about the sensitivity and specificity achieved in their particular setting. To reduce uncertainty, users will need estimates of sensitivity and specificity at different threshold scores identified using ROC curve analysis of data from individuals sampled from their own patient population. Developers and other implementation partners will need to provide all de novo users with resources, protocols, and tools to undertake these analyses. Given the importance of minimizing bias when estimating diagnostic accuracy [40], WHO provides a draft protocol for new CAD users to undertake threshold selection for their patient populations [41]. In areas where HIV-associated or smear-negative tuberculosis are epidemiologically important, users should be provided with estimates of accuracy at different thresholds within strata of these variables. We recognize that this represents an important implementation challenge. However, a cautious approach is warranted considering this is a novel technology, and ours is not the only study to report between-setting heterogeneity [23].
A second implementation consideration is that these software do not provide differential diagnoses as would radiologists. Third, none of the software are validated for use in infants and young children. Taken together, these limitations of existing software mean that they cannot yet fully replace human CXR readers, hence their deployment should not preclude efforts to expand access to human readers with expertise in radiologic interpretation.
This study has a number of strengths. First, the quality of the included studies reduces the likelihood of selection or measurement bias. Second, we substantially expanded the evidence base for deep learning-based CAD by reanalyzing CXR images from studies that had initially reported on older, non-deep learning-based programs. Third, through IPD meta-analysis we identified patient characteristics modifying diagnostic accuracy and were able to estimate the associated absolute changes in sensitivity and specificity—which have not previously been reported. Fourth, we conducted our study independently of companies who have a commercial interest in this field. Finally, our evaluation was based on CXR that had not been used for software training, which could have overestimated accuracy [4].
Some limitations should also be considered. First, we did not have data from 3 [25, 27, 42] of 6 eligible studies identified from our initial literature search, nor from the 1 study [23] identified in the updated search. However, we think their inclusion would not have changed our main results of conclusions, for a number of reasons. Two of the studies for which data could not be obtained [25, 42] were conducted by the same investigators and in the same 2 countries as 2 studies that we did include [6, 20]. Another study we could not include [27] evaluated an older CAD4TB version in Bangladesh, against a single NAAT as the reference and without sputum smear data. In a preprint [43], AUCs of CAD4TBv6, Lunit, and qXR on the Bangladesh data set were similar to our estimates from Pakistan (both low HIV prevalence settings). Importantly, our finding of between-site variability in diagnostic accuracy was also reported by Qin et al [23]. A second limitation of our study is that we did not have data on CD4 counts, which could have further explained heterogeneity amongst PLWH. Another limitation is that over 1 year has passed since the updated literature search.
Future research should focus on reducing between-population heterogeneity in accuracy, validation for use in childhood tuberculosis, and re-evaluation in the era of Covid-19, which could reduce CAD specificity for tuberculosis due to shared manifestations.
In summary, among individuals self-referring for pulmonary tuberculosis symptoms, CXR analysis with these deep learning-based CAD software can be a high sensitivity rule-out test. Moreover, tuberculosis diagnostic outcomes when using these software will be similar to those achieved with human CXR readers. However, to reduce uncertainty related to diagnostic heterogeneity, developers should provide de novo users with threshold scores and estimates of accuracy derived from their own patient populations, and stratified by HIV and smear status.
Supplementary Material
Notes
Acknowledgments. The authors thank Delft, Lunit, and qure.ai, for providing technical support with the local installation of the software used in this study. The authors thank Marcel Behr of the McGill International TB Centre for critical feedback on the manuscript.
Financial support. L’Observatoire International Sur Les Impacts Sociétaux de l’Intelligence Artificielle (Fonds de recherche Quebec). The funder had no role in the collection, analysis and interpretation of the data; in the writing of the report; or in the decision to submit the article for publication.
Potential conflicts of interest. M. B. reports grants from European and Development Countries Clinical Trials Partnership, during conduct of the study. A. J. K. has had financial interests in the company Alcela, of which qure.ai is a client, and has been discussions with qure.ai for additional business development since October 2019. A. J. K. had helped conceive and design the included study from Pakistan in 2016 to 2017 but was never directly involved in data collection, analysis, or reporting of that study and his relationship with Alcela arose after the completion of data collection for that study. A. J. K. was not involved in the design, analysis, reporting, writing, editing, or decision to submit the work reported in the present manuscript. C. D. reports working for FIND until April 2019. FIND is a not-for-profit foundation, whose mission is to find diagnostic solutions to overcome diseases of poverty in LMICs. Since leaving FIND, C. D. continues to hold a collaborative agreement with FIND. M. P. reports that he serves on the Scientific Advisory Committee (SAC) of FIND, Geneva. M. P. reports no financial or industry conflicts. F. A. K. reports grants from Fonds de Recherche du Quebec and the Canadian Institutes of Health Research, both are publicly funded government-run research agencies. F. A. K. has no financial or industry conflicts. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Protocol Registration: PROSPERO (CRD42018073016).
References
- 1. Williams FH. The Use of X-Ray examinations in pulmonary tuberculosis. Boston Med Surg J 1907; 157:850–3. [Google Scholar]
- 2. McAdams HP, Samei E, Dobbins J 3rd, Tourassi GD, Ravin CE. Recent advances in chest radiography. Radiology 2006; 241:663–83. [DOI] [PubMed] [Google Scholar]
- 3. Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer 2018; 18:500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harris M, Qi A, Jeagal L, et al. A systematic review of the diagnostic accuracy of artificial intelligence-based computer programs to analyze chest x-rays for pulmonary tuberculosis. PLoS One 2019; 14:e0221339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pande T, Cohen C, Pai M, Ahmad Khan F. Computer-aided detection of pulmonary tuberculosis on digital chest radiographs: a systematic review. Int J Tuberc Lung Dis 2016; 20:1226–30. [DOI] [PubMed] [Google Scholar]
- 6. Ahmad Khan F, Majidulla A, Tavaziva G, et al. Deep learning-based chest X-ray analysis software as triage tests for pulmonary tuberculosis: a prospective study of diagnostic accuracy for culture-confirmed disease. Lancet Digit Health 2020; 2:e573–81. [DOI] [PubMed] [Google Scholar]
- 7. World Health Organization. Chest Radiography in Tuberculosis Detection - Summary of current WHO recommendations and guidance on programmatic approaches, 2016. 2016. [Google Scholar]
- 8. Stewart LA, Clarke M, Rovers M, et al. ; PRISMA-IPD Development Group. Preferred reporting items for systematic review and meta-analyses of individual participant data: the PRISMA-IPD statement. JAMA 2015; 313:1657–65. [DOI] [PubMed] [Google Scholar]
- 9. Whiting PF, Rutjes AW, Westwood ME, et al. ; QUADAS-2 Group. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011; 155:529–36. [DOI] [PubMed] [Google Scholar]
- 10. Steinhauser S, Schumacher M, Rücker G. Modelling multiple thresholds in meta-analysis of diagnostic test accuracy studies. BMC Med Res Methodol 2016; 16:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rücker G, Steinhauser S, Kolampally S, Schwarzer G.. diagmeta: meta-analysis of diagnostic accuracy studies with several R packages. 2019. [Google Scholar]
- 12. CAD4TB 6 White Paper. Available at: https://thirona.eu/wp-content/uploads/2019/05/CAD4TB_6.0.0_WhitePaper.pdf.
- 13. World Health Organization. High-priority target product profiles for new tuberculosis diagnostics: report of a consensus meeting. Geneva, Switzerland: WHO, 2014. Available at: https://apps.who.int/iris/handle/10665/135617. [Google Scholar]
- 14. Doebler P. Meta-analysis of diagnostic accuracy with mada. R Packages. 2015. Accessed 6 June 2020. [Google Scholar]
- 15. Reitsma JB, Glas AS, Rutjes AW, Scholten RJ, Bossuyt PM, Zwinderman AH. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol 2005; 58:982–90. [DOI] [PubMed] [Google Scholar]
- 16. World Health Organization. Systematic screening for active tuberculosis: principles and recommendations. Vol. WHO/HTM/TB/2013.04. Geneva, Switzerland: WHO, 2013. [PubMed] [Google Scholar]
- 17. SAS Institute Inc. Base SAS® 9.4 procedures guide: statistical procedures, 6th ed. Cary: NC SAS Institute Inc, 2016. [Google Scholar]
- 18. R Foundation for Statistical Computing. R: a language and environment for statistical computing. Vienna, Austria, 2013. [Google Scholar]
- 19. Breuninger M, van Ginneken B, Philipsen RH, et al. Diagnostic accuracy of computer-aided detection of pulmonary tuberculosis in chest radiographs: a validation study from sub-Saharan Africa. PLoS One 2014; 9:e106381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muyoyeta M, Maduskar P, Moyo M, et al. The sensitivity and specificity of using a computer aided diagnosis program for automatically scoring chest X-rays of presumptive TB patients compared with Xpert MTB/RIF in Lusaka Zambia. PLoS One 2014; 9:e93757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Melendez J, Sánchez CI, Philipsen RH, et al. An automated tuberculosis screening strategy combining X-ray-based computer-aided detection and clinical information. Sci Rep 2016; 6:25265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Philipsen RHHM, Sánchez CI, Maduskar P, et al. Automated chest-radiography as a triage for Xpert testing in resource-constrained settings: a prospective study of diagnostic accuracy and costs. Sci Rep 2015; 5:12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qin ZZ, Sander MS, Rai B, et al. Using artificial intelligence to read chest radiographs for tuberculosis detection: a multi-site evaluation of the diagnostic accuracy of three deep learning systems. Sci Rep 2019; 9:15000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cohen R, Muzaffar S, Capellan J, Azar H, Chinikamwala M. The validity of classic symptoms and chest radiographic configuration in predicting pulmonary tuberculosis. Chest 1996; 109:420–3. [DOI] [PubMed] [Google Scholar]
- 25. Maduskar P, Muyoyeta M, Ayles H, Hogeweg L, Peters-Bax L, van Ginneken B. Detection of tuberculosis using digital chest radiography: automated reading vs interpretation by clinical officers. Int J Tuberc Lung Dis 2013; 17:1613–20. [DOI] [PubMed] [Google Scholar]
- 26. Pinto LM, Pai M, Dheda K, Schwartzman K, Menzies D, Steingart KR. Scoring systems using chest radiographic features for the diagnosis of pulmonary tuberculosis in adults: a systematic review. Eur Respir J 2013; 42: 480–94. [DOI] [PubMed] [Google Scholar]
- 27. Rahman MT, Codlin AJ, Rahman MM, et al. An evaluation of automated chest radiography reading software for tuberculosis screening among public- and private-sector patients. Eur Respir J 2017; 49(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tamhane A, Chheng P, Dobbs T, Mak S, Sar B, Kimerling ME. Predictors of smear-negative pulmonary tuberculosis in HIV-infected patients, Battambang, Cambodia. Int J Tuberc Lung Dis 2009; 13:347–54. [PubMed] [Google Scholar]
- 29. van Cleeff MR, Kivihya-Ndugga LE, Meme H, Odhiambo JA, Klatser PR. The role and performance of chest X-ray for the diagnosis of tuberculosis: a cost-effectiveness analysis in Nairobi, Kenya. BMC Infect Dis 2005; 5:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilcke JT, Kok-Jensen A. Diagnostic strategy for pulmonary tuberculosis in a low-incidence country: results of chest X-ray and sputum cultured for Mycobacterium tuberculosis. Respir Med 1997; 91:281–5. [DOI] [PubMed] [Google Scholar]
- 31. Graham S, Das GK, Hidvegi RJ, et al. Chest radiograph abnormalities associated with tuberculosis: reproducibility and yield of active cases. Int J Tuberc Lung Dis 2002; 6:137–42. [PubMed] [Google Scholar]
- 32. Balabanova Y, Coker R, Fedorin I, et al. Variability in interpretation of chest radiographs among Russian clinicians and implications for screening programmes: observational study. BMJ 2005; 331:379–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koppaka R, Bock N. How reliable is chest radiography? In: Frieden T. Toman’s tuberculosis case detection, treatment, and monitoring: questions and answers. 2nd ed. Geneva: World Health Organization, 2005:302. [Google Scholar]
- 34. Kosack CS, Spijker S, Halton J, et al. Evaluation of a chest radiograph reading and recording system for tuberculosis in a HIV-positive cohort. Clin Radiol 2017; 72: 519 e1- e9. [DOI] [PubMed] [Google Scholar]
- 35. Pinto LM, Dheda K, Theron G, et al. Development of a simple reliable radiographic scoring system to aid the diagnosis of pulmonary tuberculosis. PLoS One 2013; 8:e54235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ralph AP, Ardian M, Wiguna A, et al. A simple, valid, numerical score for grading chest x-ray severity in adult smear-positive pulmonary tuberculosis. Thorax 2010; 65:863–9. [DOI] [PubMed] [Google Scholar]
- 37. Zellweger JP, Heinzer R, Touray M, Vidondo B, Altpeter E. Intra-observer and overall agreement in the radiological assessment of tuberculosis. Int J Tuberc Lung Dis 2006; 10:1123–6. [PubMed] [Google Scholar]
- 38. Bruno MA, Walker EA, Abujudeh HH. Understanding and confronting our mistakes: the epidemiology of error in radiology and strategies for error reduction. Radiographics 2015; 35:1668–76. [DOI] [PubMed] [Google Scholar]
- 39. Waite S, Kolla S, Jeudy J, et al. Tired in the reading room: the influence of fatigue in radiology. J Am Coll Radiol 2017; 14:191–7. [DOI] [PubMed] [Google Scholar]
- 40. Ahmad Khan F, Pande T, Tessema B, et al. Computer-aided reading of tuberculosis chest radiography: moving the research agenda forward to inform policy. Eur Respir J 2017; 50. [DOI] [PubMed] [Google Scholar]
- 41. World Health Organization. Generic CAD Calibration Study Protocol, 1–20. Available at: https://tdr.who.int/docs/librariesprovider10/cad/cad-calibration-generic-protocol.pdf?sfvrsn=7e62f9da_3. Accessed 18 June.
- 42. Zaidi SMA, Habib SS, Van Ginneken B, et al. Evaluation of the diagnostic accuracy of computer-aided detection of tuberculosis on chest radiography among private sector patients in Pakistan. Sci Rep 2018; 8:12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Qin ZZ, Ahmed S, Sarker MS, et al. Can artificial intelligence (AI) be used to accurately detect tuberculosis (TB) from chest x-ray? a multiplatform evaluation of five AI products used for TB screening in a high TB-burden setting. arXiv preprint arXiv:200605509 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.