Abstract
Salmonella is a facultative intracellular pathogen that has co-evolved with its host and has also developed various strategies to evade the host immune responses. Salmonella recruits an array of virulence factors to escape from host defense mechanisms. Previously chitinase A (chiA) was found to be upregulated in intracellular Salmonella. Although studies show that several structurally similar chitinases and chitin-binding proteins (CBP) of many human pathogens have a profound role in various aspects of pathogenesis, like adhesion, virulence, and immune evasion, the role of chitinase in the intravacuolar pathogen Salmonella has not yet been elucidated. Therefore, we made chromosomal deletions of the chitinase encoding gene (chiA) to study the role of chitinase of Salmonella enterica in the pathogenesis of the serovars, Typhimurium, and Typhi using in vitro cell culture model and two different in vivo hosts. Our data indicate that ChiA removes the terminal sialic acid moiety from the host cell surface, and facilitates the invasion of the pathogen into the epithelial cells. Interestingly we found that the mutant bacteria also quit the Salmonella-containing vacuole and hyper-proliferate in the cytoplasm of the epithelial cells. Further, we found that ChiA aids in reactive nitrogen species (RNS) and reactive oxygen species (ROS) production in the phagocytes, leading to MHCII downregulation followed by suppression of antigen presentation and antibacterial responses. Notably, in the murine host, the mutant shows compromised virulence, leading to immune activation and pathogen clearance. In continuation of the study in C. elegans, Salmonella Typhi ChiA was found to facilitate bacterial attachment to the intestinal epithelium, intestinal colonization, and persistence by downregulating antimicrobial peptides. This study provides new insights on chitinase as an important and novel virulence determinant that helps in immune evasion and increased pathogenesis of Salmonella.
Author summary
Chitinases and chitin-binding proteins have been implicated in the pathogenesis of several human pathogens associated with the mucosal barrier. Interestingly, chitinases from the major enteric pathogen, Salmonella enterica, were reported to be upregulated during macrophage and epithelial cell infection. Although Salmonella Chitinase ChiA (encoded by STM14_0022) shares sequence similarity with the pathogenic chitinases, its role as a virulence determinant remained obscured. Here we aim to investigate the role of chitinase in the context of Salmonella pathogenesis using cell culture, mouse, and nematode models. We found that Salmonella requires ChiA to remodel the intestinal epithelium and access the host system. In the phagocytes, chitinase-mediated upregulation of nitric oxide (NO) leads to inhibition of MHC-I bound antigen presentation and CD8+ T cell proliferation. Furthermore, the absence of ChiA impairs bacterial adhesion and colonization in vivo. During the systemic phase in the murine host, Salmonella Typhimurium chitinase prevents immune activation and antimicrobial responses. Additionally, in the Caenorhabditis elegans, Salmonella Typhi chitinase promotes bacterial attachment to the intestinal epithelium and enhances pathogen colonization and persistence in the intestine by downregulating the antimicrobial peptides SPP1 and ABF2. In conclusion, our study provides novel insights into the role of Salmonella chitinase as a novel virulence factor.
Introduction
Salmonella is one of the major foodborne pathogens that cause enteric diseases in humans and other mammals. Although Salmonella-mediated enteric illnesses can be treated, the high occurrences of drug-resistant strains challenge pathogen eradication. The human gastrointestinal tract is covered with two distinct types of glycan layers- mucin and complex oligosaccharides (glycocalyx) that protect the enterocytes from the environment [1]. An enteric pathogen, like Salmonella, should be able to cleave the mucinous layer to gain access to the enterocytes. In various human pathogens, glycoside hydrolases such as sialidases, muraminidases, glucosaminidases, pullulanases, N-acetylgalactosaminidases (GalNAcases), etc. are known to facilitate the bacterial attachment to the host cells [2]. GH18 family protein chitinases and chitin-binding proteins were also found to be involved in the pathogenesis of several human enteric (Vibrio cholerae, Listeria monocytogenes, Serratia marcescens) [3–7] and non-enteric pathogens (Pseudomonas aeruginosa, Legionella pneumophila) [8–10]. During the infections caused by these pathogens, the commonality of a mucin-rich host-pathogen interface hinted towards a potentially significant role of chitinases and chitin-binding proteins in breaching the mucosal barrier. Furthermore STM0018 encoded chitinase from Salmonella Typhimurium strain SL1344 has been implicated in cleaving β1–6 linked LacdiNAc molecules (prevalent on mammalian glycome and invertebrate glycans) along with other chitin-like chains containing β1–4 linkages [11,12]. However, chitinase from Salmonella Typhimurium str. LT2 did not have any effect on pathogenesis [13]. Salmonella causes infection in the gut mucosal region, which also has a protective mucinous layer. A BLAST search revealed that Salmonella Typhimurium exochitinase ChiA (encoded by STM14_0022) showed 20–40% identity with the abovementioned pathogenic proteins. Further, Salmonella Typhi chitinase (ChiA; STY0018) is 98% similar to the S. Typhimurium SL1344 chiA (STM0018) that was reported to be upregulated in the infected macrophages and epithelial cells [14–16].
We infected epithelial cells and phagocytes with the ΔchiA mutant strains, and interestingly, we found that the strains lacking chiA were invasion defective in epithelial cells. Salmonella is known to remodel the host cell surface glycans to facilitate invasion in the epithelial cells [17–19]. We checked the host cell surface glycan modification by lectin-binding assay. Our data suggest that chitinase aids in glycan remodeling by cleaving the terminal sialic acid (Neu5Ac) and Gal-β1,4-GalNAc, thus making the mannose residues accessible to the bacteria for binding. Further, we found that the phagocytes infected with the mutant bacteria produced less antibacterial molecules. Interestingly, the mutants were significantly less virulent, less persistent, and were unable to dampen host antibacterial and immune responses in vivo. Moreover, in this study, we demonstrated a novel role of ChiA in facilitating extra-intestinal colonization of Salmonella Typhi in C. elegans. Together our data indicate that chitinase A plays a multifaceted role in Salmonella pathogenesis ranging from aiding bacterial invasion in the epithelial cells, enhancing antibacterial NO production by the phagocytes ex vivo, to increasing bacterial persistence in the nematodes and regulating cellular and humoral immune responses in vivo.
Results
Chitinase deletion impairs bacterial invasion in human epithelial cells
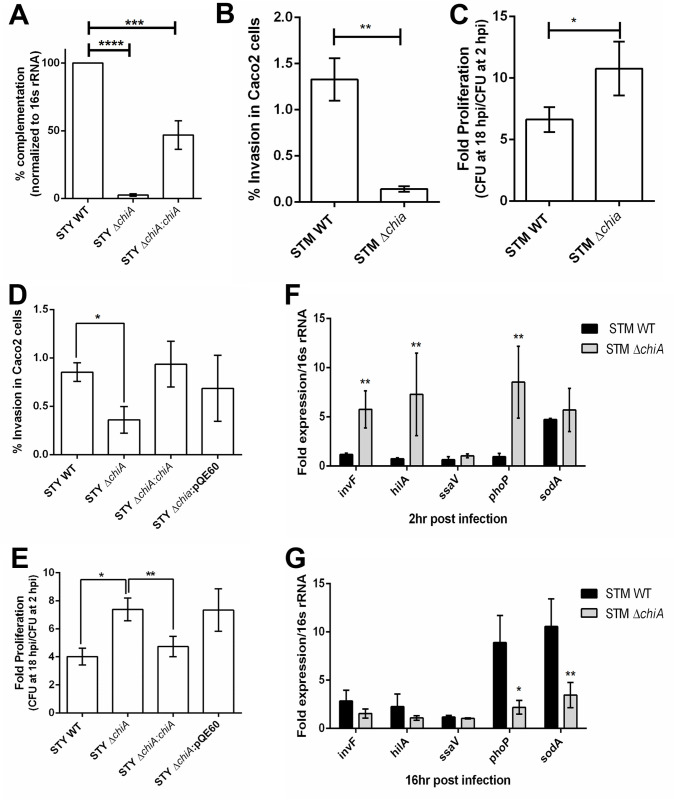
Since STM ChiA and STY ChiA were 19–24% identical with previously reported pathogenic chitinases and chitin-binding proteins (CBPs) [3–10] (S1A Fig), we made isogenic mutants of chiA using the lambda red recombinase method (Tables 1 and 2) [20]. The trans-complemented strain (STY ΔchiA:chiA) showed ~50% complementation of chiA expression (Fig 1A). The mutants and the complemented strain did not show any growth difference in vitro (S1B and S1C Fig), suggesting that Chitinase A is non-essential for the extracellular life of Salmonella sp. We checked bacterial invasion and intracellular proliferation in Caco-2 cells and found that the chiA deletion rendered the bacteria less invasive and hyperproliferative in epithelial cells (Figs 1B–1E and S1D and S1E). Inside the host, Salmonella SPI1-T3SS effectors induce membrane ruffling that facilitates bacterial entry into the epithelial cells, whereas SPI2 effectors are required for intracellular survival and proliferation [21]. SPI1 effectors invF and SPI1 master regulator hilA were significantly upregulated in STM ΔchiA mutant 2 hours post-infection (hpi), whereas SPI2 encoded ssaV expression was similar in both STM WT and STM ΔchiA strains at 16 hpi (Fig 1F and 1G), suggesting that the reduced bacterial invasion in the epithelial cells by ΔchiA mutant is independent of SPI1 gene expression. However, at 16hpi, we also observed a reduction in the expression of sodA that encodes superoxide dismutase. SodA protects from oxygen-dependent microbicidal activity [22]; therefore, reduced SodA expression indicates reduced oxidative burst in the infected cells.
Table 1. List of strains used in this study.
| Strain name | Description | Reference |
|---|---|---|
| S. Typhimurium ATCC 14028S (STM WT) | Wildtype (WT) | Kind gift from Prof. M. Hensel (Division of Microbiology, University of Osnabrück, Germany) |
| STM ΔchiA | Isogenic knockout strain for the gene chiA; Kan | This study |
| STM ΔinvC | Isogenic knockout strain for the gene invC; | Kind gift from Prof. M. Hensel (Division of Microbiology, University of Osnabrück, Germany) |
| S. Typhi CT18 (STY WT) | Wildtype (WT) | PGIMER, Chandigarh |
| STY ΔchiA | Isogenic knockout strain for the gene chiA; Kanr | This study |
| STY ΔchiA:chiA | Isogenic complement strain for ΔchiA expressing chiA under the T5 promoter present in the pQE60 plasmid; Kanr Ampr | This study |
| STY ΔchiA:pQE60 | Isogenic complement strain with empty pQE60 plasmid; Kanr Ampr | This study |
| mCherry tagged strains | Respective strains carrying pFPV-mCherry plasmid, Ampr | This study |
Table 2. List of primers used in this study.
| Gene | Sequence (5’-3’) |
|---|---|
| STM chiA KO FP | TTATGGACCCCGCAGAACGAGCTGCGACAATTTTGAAACGTAAAAGGAAATTTGAAAGTGTAGGCTGGAGCTGCTTC |
| STM chiA KO RP | GGTAAACCAGGGCTTGAATCATGAAGCCCAATACATCGGCTTAATACCGTGTACATATGAATATCCTCCTTAG |
| STM chiA conf FP | GCTGCGACAATTTTGAAAC |
| STM chiA conf RP | GAAGCCCAATACATCGG |
| STY chiA KO FP | GGACCCCGCAGAACGAGCTGCGACAATTTTGAAACGTAAAAGGAAATTTGAAAGTGTAGGCTGGAGCTGCTTC |
| STY chiA KO RP | CCCCGGTAAACCGGGGCTTGAATCATGAAGCCCAATACATCGGCTTAATACCGTGTACATATGAATATCCTCCTTAG |
| STY chiA conf FP | CTGCGACAATTTTGAAACG |
| STY chiA conf RP | CCAATACATCGGCTTAATACC |
| STY chiA:pQE60-chiA FP | TACGCCATGGATGGCTACAAGCAAACTGATTCAAG |
| STY chiA:pQE60-chiA RP | AGTCGGATCCTTAGTAAGCGCCAAGATCGG |
| STM/STY chiA RT FP | CGGAAGAGGAAGAAGAGATT |
| STM/STY chiA RT RP | CATAGACCACCATTTCACCT |
| invF FP | AGATCGTAAACGCTGCGAGT |
| invF RP | CTGCTGCACAAACGACGAAA |
| hilA FP | GCCGGTGACCATTACGAAGA |
| hilA RP | AAGAGAGAAGCGGGTTGGTG |
| ssaV FP | TATTGATAGGCGCGGACGCTA |
| ssaV RP | CGCCTTATGGGCCATGTCTTT |
| phoP FP | GATCTCTCACGCCGGGAATT |
| phoP RP | TGACATCGTGCGGATACTGG |
| sodA FP | CCTGCCGGTTGAAGAACTGA |
| sodA RP | GGTTGCTGCTGCTTTTTCGA |
| STM 16s rRNA FP | GTGAGGTAACGGCTCACCAA |
| STM 16s rRNA RP | TAACCGCAACACCTTCCTCC |
| C. elegans act2 FP | ATCGTCCTCGACTCTGGAGAT |
| C. elegans act2 RP | TCACGTCCAGCCAAGTCAAG |
| C. elegans pmk1 FP | CCAAAAATGACTCGCCGTGA |
| C. elegans pmk1 RP | CTTTTGCAGTTGGACGACGA |
| C. elegans mek1 FP | AGCAGCCAATTCCAGAGAGA |
| C. elegans mek1 RP | CGATCAGTCTGCCAGCAATA |
| C. elegans clec85 FP | CCAATGGGATGACGGAACCA |
| C. elegans clec85 RP | CTTCTGTCCAGCCAACGTCT |
| C. elegans lys7 FP | GTACAGCGGTGGAGTCACTG |
| C. elegans lys7 RP | GCCTTGAGCACATTTCCAGC |
| C. elegans ilys2 FP | TGTTGGATCGCTTTCTTGTG |
| C. elegans ilys2 RP | CATTATGGTTCGGGCCATC |
| C. elegans spp1 FP | TGGACTATGCTGTTGCCGTT |
| C. elegans spp1 RP | ACGCCTTGTCTGGAGAATCC |
| C. elegans abf2 FP | CCGTTCCCTTTTCCTTGCAC |
| C. elegans abf2 RP | GACGACCGCTTCGTTTCTTG |
Fig 1. Chitinase deletion impairs bacterial invasion in human epithelial cells.
(A) chiA expression from STY WT, STY ΔchiA, STY ΔchiA:chiA isolated from infected Caco-2 cells 16 hpi. (B) % invasion, (C) intracellular proliferation of STM WT and STM ΔchiA strains, (D) % invasion and (E) Intracellular proliferation of STY WT, STY ΔchiA, STY ΔchiA:chiA and STY ΔchiA:pQE60 strains in Caco-2 cells by gentamicin protection assay. (N = 3, n = 3). Unpaired Student’s t test was used to analyze the data for B-C and one-way ANOVA was used to analyze the data for D-E. RNA expression level of SPI1 and SPI2 genes in from STM WT and STM ΔchiA during (F) early phase (2 hpi) and (G) late phase (16 hpi) of infection in Caco-2 cells. (N = 3, n = 3); Two-way ANOVA was used to analyze the data.
Chitinase A facilitates bacterial entry into the epithelial cells by cell surface glycan modification
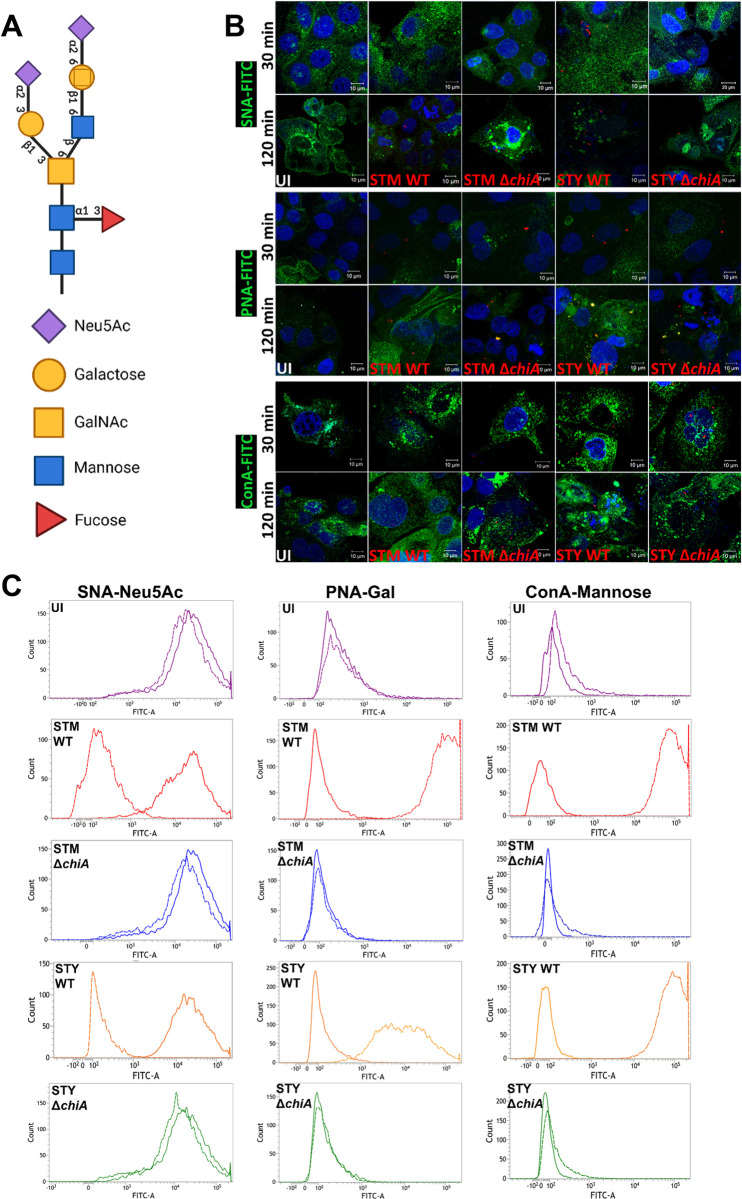
The intestinal epithelial cells are layered with α2–6, α2–3, α1–3, β1–3 or β1–6 linked glycans on the host epithelial cells in a particular array to form the glycocalyx (Fig 2A) [18]. Interestingly, the typhoid toxin from S. Typhi also binds to the terminal Neu5Ac (N-Acetylneuraminic acid) moieties to initiate the bacterial attachment to the intestinal epithelium [23]. Therefore, we quantified various glycosyl molecules present on Caco-2 cells after Salmonella infection by lectin binding assay. Interestingly, 120 min post-infection (mpi), wildtype (WT) bacteria-infected host cell surface showed a significantly decreased sialylation, but not the ΔchiA mutants infected cells, suggesting chitinase is involved in the removal of the terminal sialic acids (Fig 2B; top panel). Subsequently, we observed a significant increase in the detection of Gal-β1,3-GalNAc on the WT infected cells compared to the ΔchiA infected cells (Fig 2B; middle panel). Finally, we observed a substantial increase in the mannose-bound concanavalin A-FITC fluorescence on the surface of WT bacteria-infected cells as compared to the ΔchiA infected cells (Fig 2B; bottom panel). The cell surface glycan-bound lectin fluorescence was further quantified (S1F Fig) and validated by corresponding shift of the glycan-bound FITC lectins using flow cytometry (Fig 2C). Together these data suggested that Salmonella chitinase facilitates host cell surface remodeling.
Fig 2. Chitinase aids in glycan remodeling in host epithelial cells.
(A) Cell surface glycan assembly. (B) Representative confocal images of Caco-2 cells stained with SNA-FITC (top panel), PNA-FITC (middle panel) and ConA-FITC (bottom panel) lectin after indicated time intervals of STM WT, STM ΔchiA, STY WT and STY ΔchiA infection (UI- Uninfected). (C) Representative flow cytometry histogram showing the cell surface Neu5Ac-bound SNA-FITC (first column), Gal bound PNA-FITC (second column) and mannose bound ConA-FITC (third column) lectin (UI- Uninfected). Solid lines represent MFI 30 mpi and dashed lines represent MFI 120 mpi. (N = 2).
Salmonella ChiA is required for stabilization of the SCVs in epithelial cells
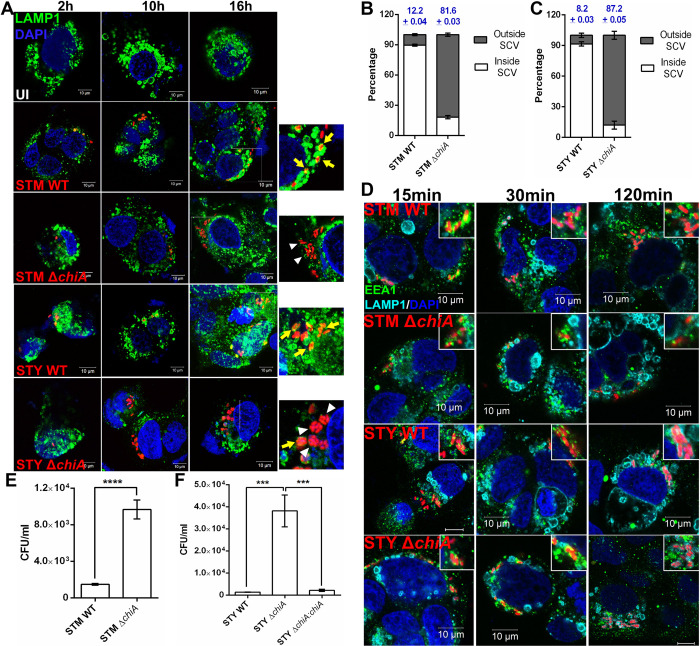
Since several reports suggested that disruption of Salmonella-containing vacuoles (SCVs) leads to bacterial hyperproliferation in the cytoplasm of the epithelial cells [24], we checked the intracellular niche of the bacteria in the infected Caco-2 cells. Early SCVs contain early endosomal markers, such as EEA1, Rab4, Rab5, and transferrin receptors, etc., while the late maturation phase is marked by late endosomal markers LAMP1/2, Rab7, Rab11, and vATPases [25]. Interestingly, ΔchiA mutant bacteria did not colocalize with the late-endosomal marker LAMP1 at 16 hpi (Figs 3A and S1G and S1H), suggesting disruption of SCVs in the ΔchiA mutant bacteria-infected cells. Upon counting the number of SCV-bound and cytoplasmic bacterial population, we found that 81.6±0.03% of STM ΔchiA and 87.2±0.05% of STY ΔchiA quit the vacuolar niche compared to the WT bacteria (STM WT 12.2±0.04%, STY WT 8.2±0.03%; Fig 3B and 3C). We also found that EEA1, an early endosomal marker, remained associated with the SCVs in WT and ΔchiA mutant infected Caco-2 cells at 15–120 mpi, while LAMP1 did not colocalize with the bacteria at early time points (15–30 mpi; Figs 3D and S1I). It is known that the cytoplasmic bacteria that escape xenophagy, can hyper-replicate in the cytosol of the epithelial cells [26]. Therefore, we enumerated the cytosolic population by chloroquine (CHQ) resistance assay. Upon transportation into the SCVs by proton pumps present on the SCV membrane, CHQ increases the vacuolar pH and kills the vacuolar Salmonella, while the cytosolic bacteria remain viable [27]. Notably, we found a significantly higher number of cytosolic mutant bacteria at 16 hpi (Fig 3E and 3F), suggesting that chitinase deletion leads to SCV disruption in epithelial cells and hyper-proliferation in the cytoplasm.
Fig 3. ΔchiA mutants quit SCVs in the epithelial cells and hyper-proliferates in the cytoplasm.
(A) Representative image of Caco-2 cells infected with STM WT, STM ΔchiA, STY WT and STY ΔchiA strains to visualize the intracellular niche of the bacteria. The SCVs were stained for LAMP1 (UI- Uninfected; Yellow arrows- LAMP1+ SCVs, white arrowheads- LAMP1- SCVs). (B) % of STM WT and STM ΔchiA, (C) % of STY WT and STY ΔchiA bacteria inside and outside the LAMP1+ SCVs 16 hpi was calculated. (N = 3). (D) Representative image of Caco-2 cells infected with STM WT, STM ΔchiA, STY WT and STY ΔchiA strains at MOI 50 to visualize EEA1 and LAMP1 recruitment on the SCVs (scale bars- 10μm; insets show SCVs). Absolute CFU/ml values of (E) STM WT and STM ΔchiA, and (F) STY WT, STY ΔchiA and STY ΔchiA:chiA in Caco-2 cells in chloroquine resistance assay 16 hpi. (N = 3, n = 3). One-way ANOVA was used to analyze the data.
Chitinase aids in bacterial survival in phagocytes by suppressing antimicrobial responses
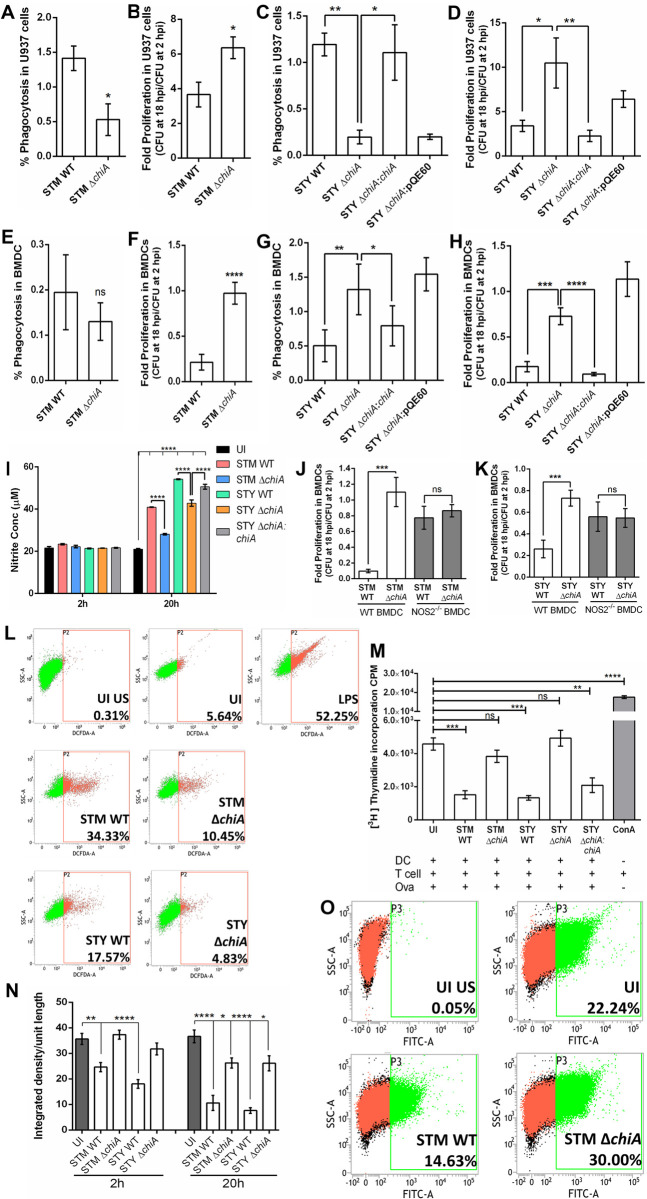
After establishing a successful niche in the epithelial cells, Salmonella transcytoses to the lamina propria (LP) and infects the LP-resident macrophages and dendritic cells [28]. To understand the role of chitinase in phagocytic cell infection, we infected U937 monocytes and bone-marrow derived dendritic cells (BMDCs). Although the ΔchiA mutants were phagocyted less by U937 monocytes, they showed enhanced survival compared to the WT strains (Fig 4A–4D and S2A and S2B). While STM WT and STM ΔchiA were phagocytosed equally, STY ΔchiA showed increased phagocytosis and better survival in BMDCs than STY WT (Figs 4E–4H and S2C and S2D). We detected significantly less nitric oxide (NO) in the spent media from the ΔchiA mutant infected BMDCs (Fig 4I). Interestingly, both WT and ΔchiA mutant bacteria survived equally in NOS2-/- BMDCs (Fig 4J and 4K). Furthermore, ΔchiA infected peritoneal macrophages (PM) showed significantly less ROS level than the WT bacteria-infected cells (Fig 4L), indicating that chitinase might be regulating RNI and ROS levels in the infected cells. To check the effect of NO on antigen presentation and T cell expansion, we quantified CD8+ T cell proliferation using OT1 transgenic mouse (C57BL/6-Tg(TcraTcrb) 1100Mjb/J). The TCR of this transgenic mouse recognizes OVA257-264 when presented by MHC-I molecules. This TCR recognition of MHC-I bound cognate peptide results in CD8+ T cell proliferation that can be measured by incorporation of 3H thymidine in the DNA of the proliferating population. We found that ΔchiA mutant infected BMDCs significantly expanded CD8+ T cells in response to the antigen stimulation (Fig 4M). Since macrophages and DCs possess MHC-I and MHC-II on the cell surface to induce CD8+ and CD4+ T cells population, respectively, we detected the surface MHC-II molecules on activated PMs. We found a significant reduction in the surface MHC-II level with WT infection, but not in ΔchiA infection (Figs 4N and 4O and S2E and S2F), indicating that Salmonella ChiA facilitates pathogen survival by dampening host antimicrobial responses.
Fig 4. Chitinase induces NOS and ROS generation in phagocytic cells and inhibits antigen presentation.
(A) % invasion and (B) fold proliferation of STM WT and STM ΔchiA strains, and (C) % invasion and (D) fold proliferation of STY WT, STY ΔchiA, STY ΔchiA:chiA and STY ΔchiA:pQE60 strains in U937 derived monocytes by gentamicin protection assay. (N = 3, n = 3). (E) % invasion and (F) fold proliferation of STM WT and STM ΔchiA strains, and (G) % invasion and (H) fold proliferation of STY WT, STY ΔchiA, STY ΔchiA:chiA and STY ΔchiA:pQE60 strains in BMDCs by gentamicin protection assay. (N = 3, n = 3). (I) Extracellular NO was estimated by Greiss assay from spent media obtained from STM WT, STM ΔchiA, STY WT, STY ΔchiA and STY ΔchiA:chiA infected BMDCs. (UI- Uninfected). (N = 3, n = 3). Two-way ANOVA was used to analyze the data. Intracellular survival of (J) STM WT and STM ΔchiA and (K) STY WT and STY ΔchiA strains was calculated in WT (NOS2+/+) and NOS2-/- BMDCs by gentamicin protection assay. (N = 3, n = 3). (L) Representative flow cytometry plot for ROS estimation by DCFDA assay from STM WT, STM ΔchiA, STY WT, STY ΔchiA and STY ΔchiA:chiA infected and lipopolysaccharide (LPS; control) treated PMs. (UI US- Unstained and uninfected, UI- Uninfected). (M) 3H thymidine incorporation assay to assess CD8+ T cell proliferation 20 hpi with STM WT, STM ΔchiA, STY WT, STY ΔchiA and STY ΔchiA:chiA (UI- Uninfected, OVA- Ovalbumin, ConA- Concanavalin A). (N = 3, n = 6). (N) Quantification of the MHC-II density per unit length of the cell membrane of STM WT, STM ΔchiA, STY WT, STY ΔchiA and STY ΔchiA:chiA infected PMs after the indicated time (UI- Uninfected). (N = 2). (O) Representative flow cytometry plot showing surface MHC-II level on PMs infected with STM WT and STM ΔchiA for 20 hours (UI US- Unstained and uninfected, UI- Uninfected). Unpaired Student’s t test was used to analyze the data for A, B, E, F, J, K and one-way ANOVA was used to analyze the data for C, D, G, H, M.
Chitinase facilitates in vivo invasion, survival, and pathogenesis of Salmonella Typhimurium
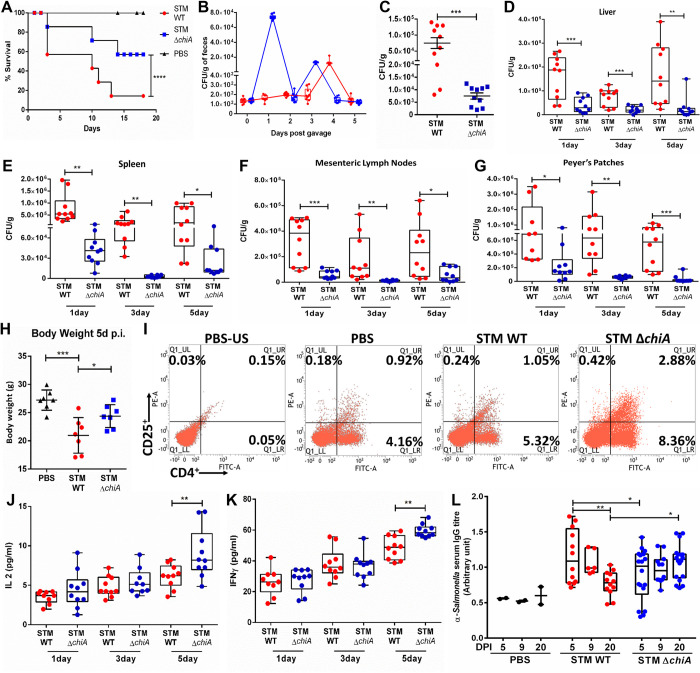
To delineate the role of chitinase in Salmonella infection in vivo, we orally infected C57BL/6J mice with 108 CFU of bacterial strains and monitored animal survival. The STM ΔchiA infected animals showed enhanced survival than the STM WT infected cohort (Fig 5A). We also found that STM ΔchiA bacteria were shed in the feces prior to the STM WT and the ΔchiA mutant was defective in Peyer’s patches (PP) colonization at 2 hpi (Fig 5B and 5C). Further, we orally infected C57BL/6J mice with a sublethal dose of Salmonella strains (107 CFU/animal) and bacterial CFU from the liver, spleen, mesenteric lymph node (MLN), and PP were enumerated. We found that STM ΔchiA mutant infected animals showed less bacterial burden in each organ and bodyweight reduction than the STM WT infected animals (Fig 5D–5H). Additionally, STM ΔchiA infected mice showed a significantly reduced burden till 20 days post-infection (dpi; S3A and S3B Fig). We also found significantly enlarged spleens in STM ΔchiA infected mice 20 dpi (S3C and S3D Fig). We previously showed that STM ΔchiA infected spleens harbored fewer bacteria (Fig 5E); therefore, we hypothesized that STM ΔchiA infection leads to T cell activation and enlargement of the spleens. To validate this hypothesis and our ex vivo data showing the correlation between reduced NO induction and higher T cell activation by ΔchiA mutant, we isolated total splenocytes from the spleens of the STM ΔchiA infected animals and quantified the CD4+CD25+ T cell population by flow cytometry. Interestingly, we found that STM ΔchiA infection leads to a significant increase in the CD4+ and CD25+ T cells, as well as the double-positive CD4+CD25+ T cell population (Fig 5I). Analysis of T cell-mediated cytokine response revealed a significant increase in the pro-inflammatory cytokines IL-2 and IFN-γ in the serum isolated from ΔchiA infected animals (Fig 5J and 5K), while no difference was observed in the anti-inflammatory cytokine levels (S3E and S3F Fig). Previous reports suggested that high IFN-γ can induce B cell proliferation and enhance IgG2a and IgG3 production [29]. Therefore, we quantified the anti-Salmonella IgG titer from infected mice serum. Interestingly, we found a significant increase in the anti-Salmonella IgG titer in the serum obtained from STM ΔchiA infected cohort (Fig 5L). We further used the polyclonal convalescent sera from STM ΔchiA infected mice to probe against STM WT-mCherry whole cell lysate to test the reactivity of the sera. Multiple dense bands against various Salmonella proteins were obtained after incubating the membrane with sera collected from STM ΔchiA mutant infected cohort (S3G Fig). Together these data suggest that Salmonella chitinase A is essential for restricting innate and humoral immune responses in vivo.
Fig 5. Chitinase facilitates in vivo invasion, survival, and pathogenesis of Salmonella Typhimurium.
(A) Survival of the mice infected with a lethal dose of STM WT and STM ΔchiA (PBS = Phosphate Buffered Saline). Data are presented from one independent experiment, representative of 3 independent experiments (N = 3). (B) Bacterial shedding in the feces of STM WT and STM ΔchiA infected animals. Data are presented as mean ± SD of 3 independent experiment; each dot represents an individual animal (N = 3, n = 10). (C) In vivo invasion in PP by STM WT and STM ΔchiA. (N = 3). Bacterial burden in (D) liver, (E) spleen, (F) MLN and (G) PP of the infected mice after the indicated time with a sub-lethal dose of STM WT and STM ΔchiA. (N = 3). Unpaired Student’s t test was used to analyze the data for C-G. (H) Body weight of the infected mice 5 dpi with a sub-lethal dose of STM WT and STM ΔchiA. (N = 3). (PBS- Phosphate Buffered Saline). (I) Flow cytometry analysis of CD4+ and CD25+ T cells from total splenocytes isolated from STM WT and STM ΔchiA infected mice 20 dpi (US-PBS- Unstained splenocytes from PBS treated mouse). Data are presented from one independent experiment, representative of 3 independent experiments (N = 3). Pro-inflammatory cytokines (J) IL-2 and (K) IFN-γ level in serum from STM WT and STM ΔchiA infected mice after the indicated time. (N = 3); One-way ANOVA was used to analyze the data. (L) Serum anti-Salmonella antibody titer was quantified by sandwich ELISA after the indicated time. (N = 3); Two-way ANOVA was used to analyze the data.
Chitinase facilitates Salmonella Typhi colonization in C. elegans
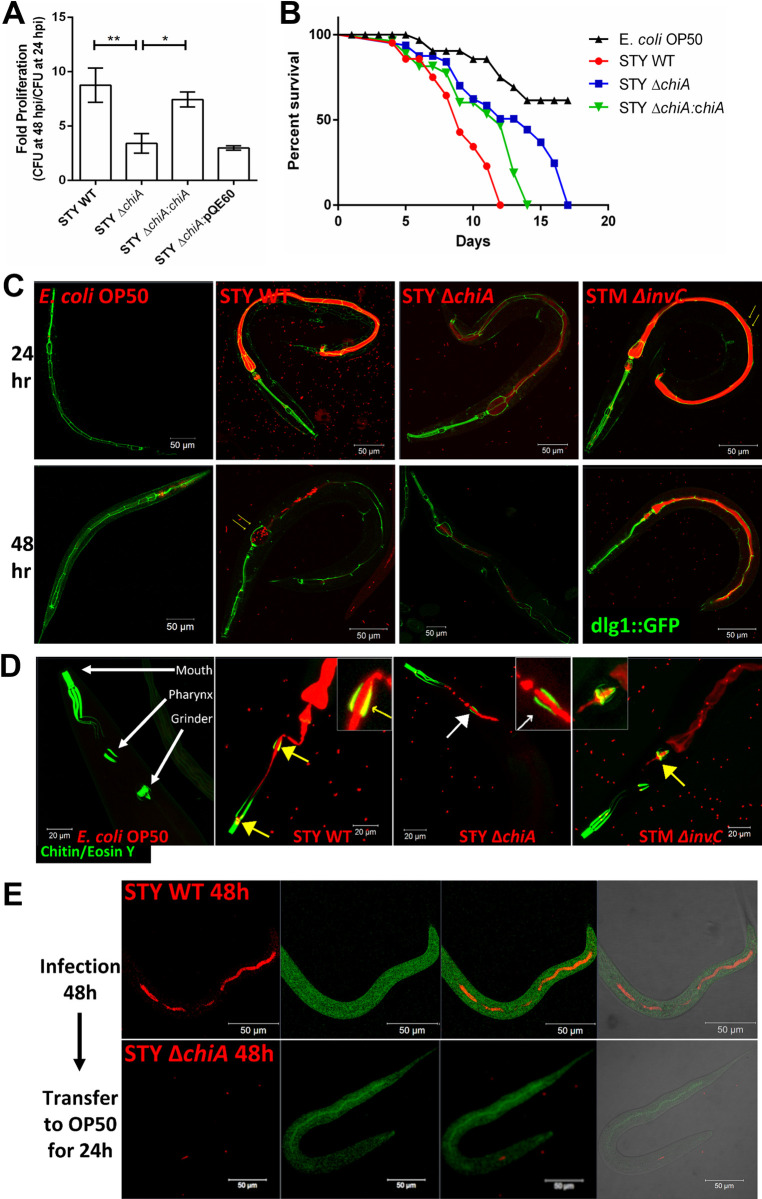
Although Salmonella Typhi is an obligatory human pathogen that does not cause a significant infection in mice because of the presence of TLR11, however use of Tlr11-/- mouse model has been reported to be largely inconsistent [30,31]. Long before the Tlr11-/- mice model came into existence, Labrousse et al. suggested Caenorhabditis elegans could be used as an alternative host to study S. Typhi pathogenesis [32]. Given that C. elegans pharyngeal lumen is rich in chitin and chitinase substrate molecules, it served as a suitable host to study the role of chitinase in bacterial pathogenesis [33]. We checked the bacterial CFU in the infected worms after 24 hours (24hpi) and 48 hours (48hpi) of continuous feeding and found that the STY ΔchiA and STY ΔchiA:pQE60 strains showed a higher bacterial burden at 24hpi (S3H Fig), but the fold proliferation (CFU at 48hpi/CFU at 24hpi) of STY ΔchiA was lesser than that of the STY WT and STY ΔchiA:chiA strains (Fig 6A). Although these Salmonella Typhi strains were lethal to the nematodes, STY ΔchiA infected worms showed slower death (TD50 330±8hrs) as compared to the STY WT (TD50 190±10hrs) and STY ΔchiA:chiA (TD50 270±12hrs) strains (Fig 6B). Together these data suggest that chitinase deletion reduces the virulence of Salmonella Typhi in C. elegans. We further checked bacterial colonization in the worm’s gut using the transgenic worm FT63 strain that expresses GFP in the epithelial cells. S. Typhi ΔchiA showed less colonization than STY WT at 24hpi, while the colonization was significantly reduced at 48hpi (Fig 6C). Several human pathogens such as Salmonella sp. and Pseudomonas aeruginosa have been reported to colonize the nematode gut lumen and cause gut distension [34]. Percent colonization as an indicator of gut distension was measured as the ratio of the diameter of the lumen occupied by the bacteria to the total diameter of the gut (S3I Fig). We next checked if S. Typhi utilizes chitinase to colonize the chitin-rich pharyngeal lumen by infecting N2 wildtype worms with different strains of Salmonella and stained the chitin-rich parts of the worms using eosin Y. Interestingly, after 24 hours of infection, luminal STY WT and STM ΔinvC bacteria, but not STY ΔchiA, colocalized with the chitin-rich regions of the pharyngeal wall and terminal bulb (grinder; Fig 6D), indicating that Salmonella Typhi utilizes chitinase to colonize the chitin-rich pharynx and terminal bulb. Additionally, after 24 hours of feeding on STY ΔchiA followed by 24 hours feeding on E. coli OP50, the STY ΔchiA was unable to persist in the gut, whereas STY WT showed significantly higher colonization in the pharyngeal lumen (S3J Fig). Extended infection for 48 hours, followed by 24 hours of E. coli OP50 feeding revealed that STY WT could profoundly colonize the gut lumen, while STY ΔchiA colonization was diminished (Fig 6E). Interestingly, after 24 hours of continuous feeding, STY WT and STM ΔinvC were found attached to the luminal wall leading to extra-intestinal tissue invasion, but not STY ΔchiA. (Figs 7A and S4A and S4B), suggesting that chitinase might be required to invade extra-intestinal tissues of the worms. To the best of our knowledge, this study is the first report suggesting an extra-intestinal invasion/colonization by Salmonella Typhi in C. elegans.
Fig 6. Chitinase enhances Salmonella Typhi virulence in C. elegans.
(A) Bacterial fold change as the ratio of CFU obtained from the worms after 24 hours and 48 hours continuous feeding on STY WT, STY ΔchiA, STY ΔchiA:chiA and STY ΔchiA:pQE60 strains. (N = 4); One way ANOVA was used to analyze the data. (B) Survival of the worms fed on E. coli OP50, STY WT, STY ΔchiA and STY ΔchiA:chiA. Data are presented from one independent experiment, representative of 4 independent experiments (N = 4). (C) Representative images of bacterial colonization in worms gut as observed by infecting transgenic FT63 worms with mCherry expressing bacteria for the indicated time. Yellow arrows show the presence of intact bacteria in the terminal bulb of the worm. (D) Representative images of bacterial colonization on the chitin-rich organs of the worms as detected by eosin Y staining. Yellow arrows show colocalization of the bacteria (red) and the eosin-stained chitin-containing regions (green). White arrow shows the absence of colocalization of the bacteria and chitin-rich organs. (E) Representative images showing bacterial colonization and persistence in the worms’ gut.
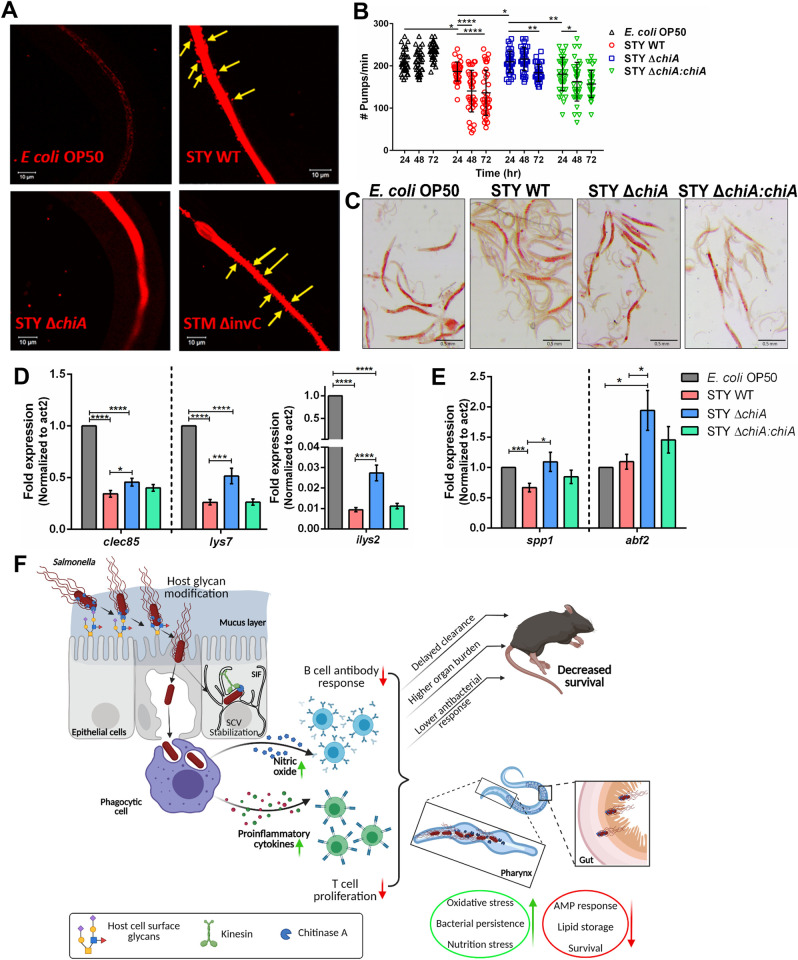
Fig 7. Salmonella chitinase is important for alteration of metabolism and antibacterial defense in C. elegans.
(A) Representative images of bacterial colonization of the worms’ gut at higher magnification. Yellow arrows show the presence of the STY WT and STM ΔinvC bacteria outside the gut lumen. (B) Quantification of no. of pharyngeal pumps/min of the worms at the indicated time. Data are represented as mean ± SD of 3 independent experiments (N = 3); Two-way ANOVA was used to analyze the data. (C) Representative images of Oil Red O (ORO) stained worms fed with different bacterial strains for 48 hours. (D, E) Quantitative RTPCR analysis of the p38 MAPK dependent antimicrobial peptide genes (D) clec85, lys7, ilys2 and (E) spp1 and abf2 in worms fed with different bacterial strains for 48 hours. Fold change was normalized over act2. (N = 4). One-way ANOVA was used to analyze the data. (F) Model depicting the role of Chitinase A in bacterial invasion and regulation of host immune response during Salmonella pathogenesis in mouse and C. elegans host (created with Biorender.com).
Salmonella chitinase is important for alteration of metabolism and antibacterial defense in C. elegans
Since we observed that Salmonella uses chitinase to colonize chitin-rich organs (Fig 6D), such as the terminal bulb, the essential structure that breaks down bacterial cells to provide nutrition to the worms, we next looked into the nutritional state of the worms by counting the number of pharyngeal pumps per min. We found a gradual yet profound reduction in the number of pharyngeal pumps/min after 72 hours of STY WT and STY ΔchiA:chiA infection, beginning as early as 24hpi, while STY ΔchiA infected worms did not show pumping defect until 72hpi (Fig 7B). In vivo oxidative stress due to pathogen infection was quantified using CL2166 worms, that possess oxidative stress-inducible GFP. STY WT and STY ΔchiA:chiA infected worms showed significantly higher oxidative stress and ‘bag of worms’ phenotype (S4C and S4D Fig). Furthermore, we observed significantly less fat storage in worms infected with STY WT and STY ΔchiA:chiA in comparison to STY ΔchiA strain (Figs 7C and S4E). It has been reported that oxidative stress (high ROS), nutritional stress, and pathogen attack can induce the MAPK pathway in the worms, leading to apoptosis [35,36]. Therefore, we checked the level of phosphor-p38 and the RNA expression of several effector genes that are downstream of the MAPK pathway. Although phosphor-p38 MAPK (PMK-1) was upregulated in all three STY strains infected worms, equal transcriptional downregulation of pmk1 and mek1 was observed (S4F and S4G Fig) and PMK-1 regulated antimicrobial peptides were differentially expressed. While the expression of MAPK regulated antimicrobial peptide genes clec85, lys7, ilys2 was downregulated in STY WT, STY ΔchiA and STY ΔchiA:chiA infected worms, their expression was partially rescued in the worms infected with STY ΔchiA bacteria (Fig 7D). Interestingly, STY ΔchiA infection completely rescued spp1 expression and significantly upregulated abf2 expression (Fig 7E), indicating an important function of chitinase in restricting the antimicrobial responses of the host.
Discussion
Salmonella is a facultative intracellular human pathogen that has co-evolved with its host and has also developed various strategies to evade the host’s immune responses. Although Salmonella pathogenesis is governed by classical virulence factors such as adhesins, invasins, and toxins, emerging reports suggest that various unique metabolic proteins are important in various aspects of Salmonella pathogenesis. Several reports suggest that Salmonella can utilize a large pool of chemically diverse host nutrients, such as carbohydrates, lipids, amino acids, etc. [37]. Bacterial chitinases belong to GH18 and GH19, which are getting recognized as bacterial virulence factors along with several other structurally similar glycosidases such as sialidases, muraminidases, N-acetylgalactosaminidases, etc. [2]. Although Salmonella chiA was upregulated during infection, the role of this chitinase in Salmonella pathogenesis remains elusive. To answer this question, we generated isogenic ΔchiA mutant by the one-step gene inactivation method. Interestingly, we found that the mutant was invasion defective in epithelial cells. Salmonella injects several Salmonella pathogenicity island-1 (SPI1) effectors to induce bacterial entry into the epithelial cells [38]. Interestingly, the expression of two major SPI-1 encoded genes invF and hilA was significantly higher in the STM ΔchiA mutant strain, indicating that the bacteria overexpress these effectors to counter the lack of ChiA. Previous reports suggested that Salmonella remodels the host cell surface glycans to facilitate invasion in the epithelial cells [17–19]. Our observations from the lectin-binding assay indicate that chitinase aids in glycan remodeling by cleaving the terminal glycosyl molecules and making the mannose residues accessible to the bacteria for binding. We also found that ΔchiA mutants remain encased in SCVs as seen by EEA1-SCV colocalization at 15 mpi in Caco-2 cells until 10 hpi as seen by LAMP1-SCV colocalization, and after which the mutants escape the SCV and hyper-proliferate in the host cell cytoplasm. One conceivable explanation for this phenomenon could be limited nutrient availability. Salmonella utilizes an extended network of tubular vacuolar structures, known as Salmonella-induced filaments (SIFs), to acquire nutrients from the host cell cytoplasm [39]. SifA, a component of SIFs, interacts with SifA-Kinesin interacting protein (SKIP) to facilitate the recruitment of the motor protein kinesin-1 on the SCVs [40]. Interestingly, kinesin-1 and several other kinesin-like proteins (KIF18A, KIF17b, etc.) are heavily glycosylated, phosphorylated, and sumoylated post-translationally, and often the N-acetylglucosamine (GlcNAc) residues block the phosphorylation sites leading to disruption of the mobility of these motor proteins [41]. It is conceivable that chitinase being a glycoside hydrolase, could remove the GlcNAc residue to facilitate phosphorylation and mobility of the motor proteins. Therefore, in the strain lacking ChiA, this mobility is affected, leading to nutritional stress and quitting of the vacuoles. Additionally, the ΔchiA mutants were protected from phagocytes-mediated bacterial killing since the mutant bacteria-infected phagocytes showed reduced oxidative burst. NO is an important cell signaling molecule, produced against many human pathogens, such as Salmonella, Mycobacterium, Listeria, etc. [42]. Previous studies suggested that a low level of NO enhances T cell survival [43], while very high [NO] inhibit T cell proliferation [44]. Previous literature suggests that mammals also possess several GH18 family enzymes. Among these, chitotriosidase, acidic mammalian chitinase (true chitinases), and BRP-39/YKL-40 (CHI3L1; a chitinase-like protein or CLP) may have chitin-like targets and can modulate host immune responses during infections, allergy, tissue injury, inflammation, and tumor. Furthermore, BRP-39 was found to activate DCs and T cells and induce Th2 inflammatory responses [45]. Interestingly, CHI3L1 neutralization in vivo reduced Salmonella Typhimurium load in the peripheral organs, indicating a definitive role of this CLP in immune modulation during Salmonella pathogenesis [46]. Additionally, Ma et al. showed that CHI3L1 is a potent stimulator of lymphocyte activation gene 3 protein (LAG3), which in turn inhibits T cell activation [47]. Chitinase being a member of the same enzyme class, we can theorize that similar activities could be performed by chitinase as well. We also found that ChiA was important for downregulating the MHC-I molecules on the dendritic cells, leading to the inhibition of CD8+ T cell proliferation and subsequent antigen presentation. In coherence with the available literature [44], the enhanced T cell proliferation could be attributed to the absence of NO induction by the ΔchiA mutant strains. We further showed that the absence of chiA failed to downregulate the surface MHC-II molecules on the activated macrophages, which is a well-known phenomenon during Salmonella infection [48]. Existing literatures suggest that SPI-2 effector SteD stimulates E3 ubiquitin ligase MARCH8 mediated ubiquitination of MHCII, leading to its degradation and suppression of T cell-mediated adaptive immune responses [49,50]. Interestingly, MHCs also have complex glycosylation marks that often end with a terminal sialic acid residue [51]. The glycosylation status of MHC-I can regulate its structure, activity, stability, trafficking and spacing. Glc1Man9GlcNAc2 glycosylation on the MHC-I molecules facilitates its interaction with calnexin and calreticulin and regulates its folding and assembly, whereas improper interactions lead to MHC-I retention in the endoplasmic reticulum [52]. Therefore, further investigation is required to understand whether the glycoside hydrolase activity of chitinase could also contribute to the inhibition of endosomal recycling and MHC replenishment on the cell membrane. In vivo infection in C57BL/6 mice suggested that STM ΔchiA mutant could not invade the PP, leading to an early fecal shedding, a lower bacterial burden in different organs, enhanced pathogen clearance and increased host survival. Additionally, the sustained innate activated IFNγ production could be attributed to iNOS-mediated signaling in ΔchiA mutant infected mice [53]. Bhat et al. suggested that enhanced IFNγ production by cytotoxic CD8+ T cells can facilitate T cell mobility, proliferation, and cytolytic function during viral infection and cancer [54]. Analysis of total splenic lymphocytes by flow cytometry suggested that the ΔchiA mutant infected mice had an increased activated T cell population (CD4+CD25+) in the spleens, suggesting an intensified immune response in these mice. However, this needs to be explored further since Salmonella is known to induce immune tolerance in the chicken intestine by upregulating CD4+CD25+ regulatory T cells [55]. Activation of the adaptive immune responses was corroborated by significant increment in the pro-inflammatory cytokines and anti-STM IgG antibody titer in the STM ΔchiA infected mice sera. Invertebrates also possess a chitinase substrate, LacdiNAc, as cell surface glycans [11,12]. By infecting C. elegans with STY strains, we further showed that chitinase aids in bacterial attachment to the pharyngeal lumen as well as colonization and persistence in the worms. In addition, our data suggest that Salmonella Typhi chitinase might be important for extra-intestinal tissue invasion in the worms. Although the nematodes lack phagocytes-like specialized immune cells, during pathogen infections, C. elegans produce ROS, often localized to the host-pathogen interface [56]. Oxidative stress caused by pathogen infection and nutrient starvation leads to the worm bagging, which is the internal egg hatching [57]. This phenomenon was described by Aballay et al. in the case of Salmonella Typhimurium infection [34]. Our data suggest that Salmonella infection induces oxidative stress, leading to “bag of worms” formation. The host also employs the ROS detox system which is transcriptionally regulated by SKN-1 and DAF-16. SKN-1 induction and its nuclear localization is regulated by the p38 MAPK signaling pathway, which is comprised of NSY-1 (MAPKKK; ASK-1 homolog), SEK-1 (MAPKK) and PMK-1 (MAPK; p38 homolog) [58,59]. Although phospho-p38 (PMK-1) was increased in STY infection, this further validates that ROS induction leads to p38 activation and apoptosis [36]. Furthermore, Salmonella was reported to induce programmed cell death in germline cells by the LPS-Tol1 axis [35]. p38 MAPK pathway and DAF-16 also regulate antimicrobial response in C. elegans as the major defense mechanism. Among these, lysozyme family (LYS), Ascaris suum antibacterial factor family (ABF), saposin-like proteins family (SPP), and C-type lectins family (CLEC) have been shown to play an important role in the induced immune responses to bacterial infection [60]. We found significantly higher expression of fat-responsive antimicrobial peptides genes spp1 and abf2 when the worms were infected with the STY ΔchiA strain. SPP1 and ABF2 are predominantly found in the intestine and on the grinder, respectively [61], both of which are the sites of Salmonella attachment as found in this study. Together these data indicate a potential role of chitinase in modulating the innate immune response in the worms.
In summary, our results reveal that the glycoside hydrolase ChiA plays a wide range of crucial, although not indispensable, functions during Salmonella pathogenesis. Although speculative at this stage, some of our findings can be attributed to the moonlighting activity of chitinase and require further investigation regarding the structural and biochemical properties of this protein. Collectively, we showed that Salmonella Chitinase regulates different aspects of pathogenesis, ranging from aiding in invasion in the epithelial cells, impairing the activity of professional antigen-presenting cells to as diverse as immune response regulation in various hosts (Fig 7F), and emerges as a novel virulence factor.
Materials and methods
Ethics statement
The animal experiments were carried out in accordance with the approved guidelines of the Institutional Animal Ethics Committee at Indian Institute of Science, Bangalore, India (Registration No: 48/1999/CPCSEA). The Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA) is a statutory Committee, which is established under Chapter 4, Section 15(1) of the Prevention of Cruelty to Animals Act 1960. All procedures involving the use of animals were performed according to Institutional Animal Ethics Committee (IAEC) approved protocol by CPCSEA. Ethical clearance approval number for the study is CAF/Ethics/670/2019.
Bacterial strains
All Salmonella Typhimurium strains used in this study are listed below with their genetic description. Salmonella enterica serovar Typhimurium strain 14028S was used as the wildtype strain and the parental background for all the mutant strains used in this study, i.e. ΔchiA and ΔinvC. All strains were grown and maintained in Lennox broth (LB; 0.5% NaCl, 1% casein enzyme hydrolysate and 0.5% yeast extract) at 37°C under shaking conditions (180 rpm). Salmonella enterica serovar Typhi strain CT18 was used as the wildtype strain, and the parental background for the mutant strain used in this study, i.e., ΔchiA. S. Typhi chiA was trans-complemented in pQE60 plasmid in 5’ NcoI-chiA-BamHI 3’ direction. This plasmid was transferred to STY ΔchiA strain to make complement strain. Complemented strain STY ΔchiA:chiA and empty vector strain STY ΔchiA:pQE60 strains were maintained on LBA supplemented with ampicillin (50 μg/ml). The mCherry expressing strains were cultured in Lennox broth with 50μg/ml Ampicillin at 37°C in shaking condition.
Isolation and maintenance of primary cells and cell lines
Human colorectal adenocarcinoma cell line Caco-2 (ATCC HTB-37) was cultured in complete DMEM media (Lonza), whereas human monocyte cell line U937 (ATCC CRL-1593.2) was maintained in complete RPMI 1640 media (Lonza) with 100 μM β-mercaptoethanol and differentiated to macrophages using 20 ng/ml PMA for 24 hours prior to infection. Bone-marrow was isolated from either wildtype (NOS2+/+) C57BL/6J mice or NOS2-/- C57BL/6J mice as described previously [48]. Briefly, the tibia and femur bones were carefully taken out, caps were removed and the marrow was flushed with RPMI 1640 media using a 26G needle. After making single cell suspension, red blood cells (RBCs) were lysed using RBC lysis buffer. Cells were pelleted and grown in complete RPMI 1640 media supplemented with 20 ng/ml mGM-CSF (Peprotech), antibiotics and 100 μM β-mercaptoethanol. After every 2 days, the media was replenished. Once approximately 65–70% of the cells were differentiated to dendritic cells (loosely adherent spheres), the cells were collected and used for further experiments. To obtain peritoneal macrophages (PMs), thioglycolate was injected in the peritoneal cavity of C57BL/6J mice. After 5 days these mice were sacrificed and ice cold PBS was injected in the peritoneum to collect the peritoneal exudate. Any residual erythrocytes were lysed using RBC lysis buffer and the cells were maintained in complete RPMI 1640 media for further experiments.
Generation of deletion mutant
ΔchiA mutant strains were made using one-step deletion strategy as mentioned by Datsenko and Wanner [20]. Briefly, wildtype Salmonella (S. Typhimurium 14028S or S. Typhi CT18) bacteria transformed with a ‘lambda red recombinase’ expressing plasmid under arabinose inducible promoter (pKD46), was grown in LB with 50 μg/ml ampicillin and was induced with 10 mM L-arabinose at 30°C to an OD600 of 0.35–0.4. Electrocompetent cells were prepared by pelleting the bacterial cells and washing the pellet three times with ice cold, sterile MiliQ water and 10% glycerol, followed by resuspension in 50 μl of 10% glycerol. Kanamycin resistance cassette was amplified from pKD4 plasmid using primers containing upstream and downstream sequences of S. Typhimurium chiA gene (STM14_0022) and S. Typhi chiA gene (STY0018) fragment. 500 ng of this PCR product was purified and used for electroporation. Transformants were selected on LB agar containing kanamycin plates and were further confirmed with confirmatory primers, chiA specific RT primers and kanamycin resistance cassette internal primers.
Infection and gentamicin protection assay
Epithelial Caco-2 cell line was infected with mid-log phase culture of bacteria grown in LB (OD600 0.3), whereas phagocytic U937 derived monocytes and BMDCs were infected with overnight culture (OD600 0.3). The multiplicity of infection (MOI) of 10 was used in each case. Bacterial attachment to host cells was enhanced by centrifuging at 600 rpm for 10 min. After 25 min of infection, cells were treated with gentamicin (100 μg/ml in complete media) for 1 hour to remove extracellular bacteria and then maintained with 25 μg/ml gentamicin for the remainder of the experiment. 0.1% Triton-X 100 (v/v in 1x PBS) was used to lyse the cells and the lysate was plated on Salmonella-Shigella (SS) agar for S. Typhimurium strains and Wilson Blair (WB) agar for S. Typhi strains. For invasion assay, cells were lysed after incubation in 100 μg/ml gentamicin treatment (i.e., 1 hour post infection) and percent invasion was calculated with respect to the pre-inoculum used for infection. For intracellular survival assay (ICSA), infected cells were lysed at 2 hours and 18 hours post infection. CFU at 18 hours was divided by CFU at 2 hours to obtain fold replication of the intracellular bacteria. For estimating the cytoplasmic bacterial population, chloroquine resistance assay was performed [62]. Briefly, Caco-2 cells were infected by different bacterial strains as mentioned previously. The infected cells were treated with 800 μM chloroquine 1 hour prior to cell lysis and absolute CFU was calculated by plating the cell lysate on selective media.
Quantitative RT-PCR
Bacterial RNA was isolated from infected cells as described previously by Eriksson et al.[14]. Briefly, Salmonella infected cells were lysed at different time intervals on ice by incubating for 30 minutes with 0.1% SDS, 1% acidic phenol and 19% ethanol in sterile water. Eukaryotic cell debris was removed by centrifuging the cell lysate at 300g for 10 minutes, followed by pelleting bacterial cells at 5000 rpm for 5 minutes. At each timepoint, bacteria were recovered from a 6-well plate of infected Caco-2 and pooled to isolate RNA. In vitro grown bacterial RNA was obtained by growing bacteria at 37°C in DMEM medium, under 5% CO2, without shaking. The bacterial pellet was resuspended in TRIzol reagent (Takara) and stored at -80°C. Young adult hermaphrodites were infected with respective bacterial strains for 48 hr. Infected worms were harvested by washing the plates with M9 buffer and pelleting at 1000g for 1 min. The extracellular bacteria were removed by repeatedly washing the pellet 5–6 times. The worms pellet was resuspended in TRIzol reagent (Takara) and stored at -80°C. RNA was isolated by phase separation method using chloroform. cDNA was synthesized with reverse transcriptase (GCC Biotech). Quantitative PCR was carried out using SYBR Green Q-PCR kit (Takara). Relative expression with respect to control (16s rRNA gene for bacterial genes and act2 for C. elegans genes) was plotted as fold change.
Lectin binding assay for cell surface glycan modification
Human colorectal carcinoma cells Caco-2 were infected with different bacterial strains as mentioned before. For confocal imaging, cells were seeded on coverslips prior to infection. After infection for the specified time, the cells were fixed with 3.5% PFA for 20 min on ice. For flow cytometry, cells were washed with PBS and treated with 1x Trypsin-EDTA (TE) for 15 min, under 5% CO2 at 37°C. After the cells were dislodged from the wells, TE was removed, and the cells were incubated with 1 ml complete media for 20 min under 5% CO2 at 37°C for recovery. To avoid non-specific lectin binding the cells were treated with blocking buffer (PBS+2% FBS) at RT for 15 min. Specific lectins (50μg/ml lectin solution in blocking buffer for every 106 cells) (Vector Laboratories; #FL-1301, #FL-1071, #FL-1001) were added to each samples and incubated for 30 min at RT, followed by washing with blocking buffer. Cells treated with only FITC dye (Merck; #46950) were used as controls.
Flow cytometry, immunofluorescence and immunoblot
Cells were fixed with 3.5% PFA for 20 min on ice. All staining except for the surface markers (MHC-II, CD4 and CD25), were performed in the presence of permeabilizing agent, 0.01% saponin (Sigma) dissolved in 2.5% BSA containing PBS. Flow cytometry analysis was carried out using BD FACSVerse and BD FACSAria and data were analyzed using BD FACSDiva software. Immunofluorescence images were obtained using Zeiss LSM 710 and/or Zeiss LSM 880. The images were analyzed using ZEN Black 2012 platform. For analysis of activated T cell population (CD4+ CD25+) from infected mice spleen, splenocytes were isolated from mice that survived through 20 days of infection. Total splenocytes were fixed using 3.5% PFA on ice for 20 min, followed by incubation for 1 hour at RT with fluorophore conjugated antibody cocktail in dark. The cells were washed with 1x PBS and analyzed by flow cytometry. After 48 hours of infection with indicated strains, C. elegans were harvested and washed 5 times with ice-cold M9 buffer. The worms were resuspended in homogenization buffer (HB: 15 mM HEPES pH 7.6, 10 mM KCl, 1.5 mM MgCl2, 0.1 mM EDTA, 0.5 mM EGTA, 44 mM sucrose and 1:10 PI cocktail) and sonicated with 40% amplitude and 30 sec pulses, 4–5 times, on ice. Equal quantities of proteins were resolved onto a 12% SDS-PAGE gel and transferred to 0.45 μm PVDF membrane using Trans-Blot semi dry transfer cell (Bio-Rad). Immunoblotting was performed to quantify phospho-p38 MAPK using β-actin as loading control. Anti-human LAMP1 (DSHB; #H4A3) and Anti-human EEA1 (CST; #3288) antibody was used for immunofluorescence microscopy. Anti-mouse I-A/I-E (or MHC-II) (clone 2G9) FITC (BD Pharmingen; #553623) antibody was used for immunofluorescence microscopy and flow cytometry. Anti-mouse CD4 FITC (Invitrogen; #11-0041-85), Anti-mouse CD25 PE (Invitrogen; #12-0251-82) antibodies were used for flow cytometry. Anti-human phospho-p38 MAPK (CST; #9211), Anti β-actin HRP (Imgenex; #IMG-5142A) antibodies were used for immunoblotting.
Nitric oxide estimation
Sodium nitrite (Sigma) standards of 100 μM, 50 μM, 25 μM, 12.5 μM, 6.25 μM and 3.13 μM were prepared by diluting 0.1 M stock in deionized distilled water. Conditioned media from infected cells were collected after indicated time intervals for estimation of nitrite by Greiss assay [63]. 1% sulphanilamide solution was made in 5% phosphoric acid. To 50 μl of the standards and the samples (in triplicates), 50 μl acidic sulphanilamide was added and incubated at RT, in dark for 10 min. After incubation, 50 μl of 0.1% NED (N-1-naphthylethylene diamine dihydrochloride) solution was added and incubated for 10 min in dark at RT. OD520 was measured within 30 min of appearance of purple/magenta colored product using TECAN Infinite Pro 200 microplate reader.
ROS measurement
Intracellular ROS was detected by 2’, 7’-dichlorofluorescein diacetate (H2DCFDA; Sigma) staining. Cells were stained with 10μM DCFDA at 37°C in dark. After 30 min, cells were washed with ice cold PBS and harvested followed by flow cytometry analysis at 495/530 nm in BD FACSVerse.
T cell proliferation assay
WT BMDCs were infected by incubating the bacteria with DCs for 90min, followed by removal of the bacteria and incubating the infected cells with 25μg/ml gentamicin. Total splenocytes were isolated from the spleen of C57BL/6-Tg (TcraTcrb) 1100Mjb/J mice by mechanical disruption. Erythrocytes were lysed by RBC lysis buffer (Sigma) and cells were maintained in complete RPMI-1640. Finally, non-adherent cells were collected and were used for mixed lymphocyte proliferation assay. The proliferation of the lymphocytes in response to antigen stimuli, was detected by incorporation of the 3H1 as measured by the scintillation counter.
In vivo experiment
6 weeks old male C57BL/6J mice were used for all the in vivo mice experiments. All animal experiments were approved by the Institutional Animal Ethics Committee (CAF/Ethics/670/2019) and the National Animal Care Guidelines were strictly followed. 108 CFUs of overnight grown STM WT and STM ΔchiA mutant bacteria were used for oral infection for animal survival assay. The control group was orally administered with sterile 1x PBS. Animals were observed for 20 days for survival and body weight was documented. For in vivo invasion, the animals were euthanized after 2 hours of gavage, and the bacterial CFUs in Peyer’s patches (PP) were estimated. To check the bacterial shedding, fecal pellets were collected aseptically from the infected cohorts after the indicated time. Homogenates were plated on SS agar plates and CFUs were counted. For estimating in vivo bacterial burden in different organs, a sublethal dose of 107 CFUs of each bacterial strain was used and bacterial CFUs from liver, spleen. MLN and PP were enumerated after indicated time intervals. Spleens were isolated from the animals after 20 days and the length was measured.
ELISA for serum cytokines and anti-Salmonella IgG
Blood collected from infected animals by cardiac puncture under aseptic conditions, was incubated at RT to facilitate coagulation. Serum was then isolated by centrifugation at 5000 rpm for 10 min at RT and stored at -20°C for further use. Estimation of serum level of different pro-inflammatory cytokines (IL-2 and IFNγ) and anti-inflammatory cytokines (IL10 and IL4) was performed according to the manufacturer’s instructions. Anti-Salmonella IgG titer was measured by sandwich ELISA as mentioned previously [64]. Briefly, wells were coated with Salmonella LPS (200 ng/well; Sigma) at 4°C overnight. Next day, LPS was removed, and the wells were washed with PBST (PBS+0.05% Tween 20), followed by blocking for 1 hour at RT with 5% FBS in PBS to avoid non-specific binding. After blocking, wells were washed with PBST. The serum samples, diluted in blocking buffer, were added to the wells in triplicates and incubated for 2 hours. Subsequently, wells were washed with PBST and anti-mouse IgG (HRP conjugate) was then added to the wells and incubated for 1 hour at RT. Tetramethylbenzidine (TMB; Sigma) was added and the plate was incubated in dark for 20–30 min. The reactions were stopped with 2 N H2SO4 and the absorbance was measured at 450 nm.
In vivo colonization in Caenorhabditis elegans
C. elegans var. Bristol worms wildtype strain N2, FT63 [xnIs17; dlg-1::GFP + rol-6(su1006)], and CL2166 [dvls19 III; dvls (pAF15)gst-4p::GFP::NLS III] strains were maintained on NGM media at 20°C. L4 or Young adult N2 hermaphrodite worms were used for in vivo experiments. 107 CFU of different bacterial strains were seeded on NGM plates and grown for 16 hours. Young adult N2 worms were fed at 20°C with the different bacterial strains for 24 hours or 48 hours to check bacterial colonization in the worms [65]. Bacterial CFU was enumerated by plating worms’ lysate from an equal number of infected worms on WB agar plates. Fold change was calculated as the ratio of CFU after 48 hours to CFU after 24 hours. For confocal analysis of the worm gut colonization, FT63 worms were used. mCherry expressing bacterial strains were used to visualize the gut colonization.
To check worms survivability, 107 CFUs of overnight grown bacterial strains were seeded on 30 mm dishes containing Brain Heart Infusion (BHI) agar media. ~30–40 young adult worms were added at the center of each plate and survival was monitored [66]. Animals were transferred to fresh bacterial plates every day for first 5 days and then after every 5 days. The worms were scored as live or dead at regular intervals throughout the course of the assay. Worms were considered dead when they failed to respond to touch stimulus.
Chitin-rich organs were visualized using Eosin Y stain. After 24 hours of infection, worms were harvested and washed 5 times with M9 buffer, followed by washing the worms pellet with citrate phosphate buffer (0.2 M Na2HPO4, 0.1 M potassium citrate, pH 6.0). The worms were resuspended in 500 μL citrate-phosphate buffer and 15 μL of 5 mg/ml eosin Y (in 70% ethanol) was added. Tubes were incubated at RT, in dark for 10 minutes, followed by centrifugation at 1000g for 1 min for washing. The supernatant was discarded and the pellet was washed with citrate phosphate buffer 5 times to remove excess eosin Y.
The effect of bacterial colonization was determined by infecting CL2166 worms for 48 hours with different strains. CL2166 worms possess oxidative stress inducible GFP. Fluorescence of the infected worms was visualized using Zeiss LSM 880 with Multiphoton mode.
Bacterial persistence assay in C. elegans
Young adult N2 worms were infected as mentioned previously. After 24 hours or 48 hours of infection, the worms were harvested and washed 5 times with M9 buffer. After indicated time, ~30 worms were mounted for confocal imaging. Rest of the worms were transferred to E. coli OP50 plate for further 24 hours. These worms were harvested, washed and imaged as mentioned previously.
Quantification of pharyngeal pumps
The effect of bacterial colonization on the chitin-rich grinder integrity was determined by counting the number of pharyngeal pumps per min. Young adult worms were infected as mentioned previously. After indicated infection time, no. of pharyngeal pumps/min was counted for ~25 worms from each infected plate.
Fat estimation by Oil red O
Neutral lipids present in the worms was estimated by Oil Red O (ORO; Sigma) staining [67]. Briefly, solution of Oil Red O was prepared in isopropanol (5 mg/ml) and diluted to 60% in water before use. Synchronized L4 animals were allowed to feed on E. coli and STY strains for 48 hr. Worms were harvested in M9 buffer, followed by fixing and permeabilizing using MRWB buffer (160 mM KCl, 40 mM NaCl, 14 mM Na2-EGTA, 1 mM spermidine-HCl, 0.4 mM spermine, 30 mM Na-PIPES [Na-piperazine N, N’-bis(2-ethanesulfonic acid); pH 7.4], 0.2% β-mercaptoethanol, 0.2% paraformaldehyde) for 1 hour at RT. The animals were stained with 60% ORO at RT. Excess strain was removed by washing twice with 1x PBST (PBS+0.01% Tween 20). Stained animals were mounted on agar pads.
Statistical analysis
Data were plotted using GraphPad Prism 6 software. Statistical analysis was performed using Student’s t-test or ANOVA as indicated. The results are presented as mean ± SEM, unless mentioned otherwise. p values <0.05 was considered to be significant (p values: ****<0.0001, ***<0.001, **<0.01, *<0.05).
Supporting information
(A) BLAST analysis showing the identity of chitinase A of Salmonella serovars Typhimurium and Typhi with known pathogenic chitinases and chitin-binding proteins. Growth analysis of LB grown cultures of (B) STM WT and STM ΔchiA, (C) STY WT, STY ΔchiA, STY ΔchiA:chiA and STY ΔchiA:pQE60. Absolute CFU/ml values of (D) STM WT and STM ΔchiA, (E) STY WT, STY ΔchiA, STY ΔchiA:chiA and STY ΔchiA:pQE60 in Caco2 cells in gentamicin protection assay after the indicated time. Data are represented as mean ± SEM of 3 independent experiments (N = 3, n = 3). (F) Mean Fluorescence Intensity (MFI) of Neu5Ac-bound SNA-FITC, Gal-bound PNA-FITC and mannose-bound ConA-FITC lectins on Caco2 cells 30 mpi and 120 mpi with STM WT, STM ΔchiA, STY WT and STY ΔchiA (UI- Uninfected). Data are represented as mean ± SEM of 2 independent experiments (N = 2). Two-way ANOVA was used to analyze the data. % Colocalization of mCherry expressing (red) (G) STM WT and STM ΔchiA, (H) STY WT and STY ΔchiA with LAMP1 (green) in Caco-2 cells at 2/10/16 hpi. Data are represented as mean ± SEM of 3 independent experiments (N = 3). Unpaired Student’s t test was used to analyze the data. (I) % Colocalization of mCherry expressing (red) bacteria with EEA1 (green) in Caco-2 cells at 15/30/120 mpi. Data are represented as mean ± SD of 2 independent experiments (N = 2). One-way ANOVA was used to analyze the data.
(TIF)
Absolute CFU/ml values of (A) STM WT and STM ΔchiA, (B) STY WT, STY ΔchiA, STY ΔchiA:chiA and STY ΔchiA:pQE60 in U937 derived monocytes in gentamicin protection assay after the indicated time. Data are represented as mean ± SEM of 3 independent experiments (N = 3, n = 3). Absolute CFU/ml values of (C) STM WT and STM ΔchiA, (D) STY WT, STY ΔchiA, STY ΔchiA:chiA and STY ΔchiA:pQE60 in murine BMDCs in gentamicin protection assay after the indicated time. Data are represented as mean ± SEM of 3 independent experiments (N = 3, n = 3). (E) Representative flow cytometry plot showing surface MHC-II level on PMs infected with STY WT, STY ΔchiA and STY ΔchiA:chiA for 20 hours (UI US- Unstained uninfected, UI- Uninfected, STY Δc:chiA- STY ΔchiA:chiA). (F) Representative images showing surface MHC-II on PMs infected with STM WT, STM ΔchiA, STY WT and STY ΔchiA for the indicated time. PMs were stained for surface MHCII without any permeabilizing agent (UI- Uninfected).
(TIF)
Bacterial CFU in (A) spleen and (B) liver from STM WT and STM ΔchiA infected mice after the indicated time. (C) STM WT and STM ΔchiA infected mice spleen length were measured after 5 days and 20 days of oral infection. Data are presented from 3 independent experiments. One-way ANOVA was used to analyze the data. (D) Representative images of spleens isolated from STM WT and STM ΔchiA infected mice after 20 days. Data are presented from one independent experiment, representative of 3 independent experiments (N = 3). Anti-inflammatory cytokine (E) IL-10 and (F) IL-4 level in serum from STM WT and STM ΔchiA infected mice after the indicated time. Data are presented as mean ± SEM of 3 independent experiments (N = 3). One-way ANOVA was used to analyze the data. (G) Representative immunoblots showing the reactivity of the STM WT and STM ΔchiA infected mice sera against mCherry-tagged Salmonella whole cell lysate. Coomassie Brilliant Blue stained gel shows equal loading in all the lanes. Furthermore, the blot was probed with anti-mCherry antibody. The Data are presented from 2 independent experiments (N = 2). (H) Quantification of absolute bacterial CFU obtained from infected C. elegans after 24 hours and 48 hours continuous feeding on STY WT, STY ΔchiA, STY ΔchiA:chiA and STY ΔchiA:pQE60 strains. Data are represented as mean ± SEM of 4 independent experiments. (I) % colonization of the worms gut after 24 hours of continuous feeding with different bacterial strains. Data are represented as mean ± SEM of 4 independent experiments. One-way ANOVA was used to analyze the data. (J) Representative images showing bacterial colonization and persistence in the worms’ gut after shorter exposure.
(TIF)
Representative images of parts of FT63 worms gut showing extra-intestinal invasion of the STY WT strain after 48 hours of continuous feeding (A) at higher magnification (scale bar- 5 μm) and (B) at lower magnification (scale bar- 10 μm). Yellow arrows show the presence of STY WT and STM ΔinvC bacteria outside the gut lumen (Green). (C) Representative images of CL2166 worms after 48 hours of feeding on STY strains. GFP fluorescence is indicative of oxidative stress. Insets show ‘bag of worms’ resulting from oxidative stress in the STM WT and STM ΔchiA:chiA infected worms. (D) Quantification of GFP MFI in CL2166 worms. Data are represented as mean ± SEM of 3 independent experiments. One-way ANOVA was used to analyze the data. (E) Quantification of the ORO-stained parts of the worms fed with different bacterial strains for 48 hours. Data are represented as mean ± SEM of 3 independent experiments. Two-way ANOVA was used to analyze the data. (F) Immunological detection of phospho-p38 MAPK (PMK-1) from worms fed with E. coli OP50 and STY strains for 48 hours. β-actin was used as loading control. (G) qRTPCR analysis of the p38 MAPK (PMK-1) pathway genes pmk1 and mek1 in worms fed with E. coli OP50 and STY strains for 48 hours. Fold change was normalized over act2. Data represent mean ± SEM of 4 independent experiments. One-way ANOVA was used to analyze the data.
(TIF)
Acknowledgments
We thank the confocal microscopy facility and real-time facility of Dept. of Microbiology and Cell Biology, IISc. We are thankful to SCh lab (MCB, IISc) and VS lab (MRGD, IISc) for kindly gifting the C. elegans strains. We thank RB lab (MRDG, IISc) for their help with the lectin-binding assay. We also thank our laboratory members for their critical input on the manuscript.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the DAE SRC fellowship (DAE00195) and DBT-IISc partnership umbrella program for advanced research in BioSystems Science and Engineering to DC. Infrastructure support from ICMR (Centre for Advanced Study in Molecular Medicine), DST (FIST) to DC, and UGC (special assistance) to KC is acknowledged. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hansson GC (2012) Role of mucus layers in gut infection and inflammation. Curr Opin Microbiol 15: 57–62. doi: 10.1016/j.mib.2011.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frederiksen RF, Paspaliari DK, Larsen T, Storgaard BG, Larsen MH, et al. (2013) Bacterial chitinases and chitin-binding proteins as virulence factors. Microbiology-Sgm 159: 833–847. doi: 10.1099/mic.0.051839-0 [DOI] [PubMed] [Google Scholar]
- 3.Kirn TJ, Jude BA, Taylor RK (2005) A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438: 863–866. doi: 10.1038/nature04249 [DOI] [PubMed] [Google Scholar]
- 4.Leisner JJ, Larsen MH, Jorgensen RL, Brondsted L, Thomsen LE, et al. (2008) Chitin hydrolysis by Listeria spp., including L. monocytogenes. Appl Environ Microbiol 74: 3823–3830. doi: 10.1128/AEM.02701-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, et al. (2006) Intracellular gene expression profile of Listeria monocytogenes. Infect Immun 74: 1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhuri S, Bruno JC, Alonzo F 3rd, Xayarath B, Cianciotto NP, et al. (2010) Contribution of chitinases to Listeria monocytogenes pathogenesis. Appl Environ Microbiol 76: 7302–7305. doi: 10.1128/AEM.01338-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawada M, Chen CC, Arihiro A, Nagatani K, Watanabe T, et al. (2008) Chitinase 3-like-1 enhances bacterial adhesion to colonic epithelial cells through the interaction with bacterial chitin-binding protein. Lab Invest 88: 883–895. doi: 10.1038/labinvest.2008.47 [DOI] [PubMed] [Google Scholar]
- 8.Salunkhe P, Smart CH, Morgan JA, Panagea S, Walshaw MJ, et al. (2005) A cystic fibrosis epidemic strain of Pseudomonas aeruginosa displays enhanced virulence and antimicrobial resistance. J Bacteriol 187: 4908–4920. doi: 10.1128/JB.187.14.4908-4920.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fung C, Naughton S, Turnbull L, Tingpej P, Rose B, et al. (2010) Gene expression of Pseudomonas aeruginosa in a mucin-containing synthetic growth medium mimicking cystic fibrosis lung sputum. J Med Microbiol 59: 1089–1100. doi: 10.1099/jmm.0.019984-0 [DOI] [PubMed] [Google Scholar]
- 10.DebRoy S, Dao J, Soderberg M, Rossier O, Cianciotto NP (2006) Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc Natl Acad Sci U S A 103: 19146–19151. doi: 10.1073/pnas.0608279103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frederiksen RF, Yoshimura Y, Storgaard BG, Paspaliari DK, Petersen BO, et al. (2015) A diverse range of bacterial and eukaryotic chitinases hydrolyzes the LacNAc (Galbeta1-4GlcNAc) and LacdiNAc (GalNAcbeta1-4GlcNAc) motifs found on vertebrate and insect cells. J Biol Chem 290: 5354–5366. doi: 10.1074/jbc.M114.607291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larsen T, Petersen BO, Storgaard BG, Duus JO, Palcic MM, et al. (2011) Characterization of a novel Salmonella Typhimurium chitinase which hydrolyzes chitin, chitooligosaccharides and an N-acetyllactosamine conjugate. Glycobiology 21: 426–436. doi: 10.1093/glycob/cwq174 [DOI] [PubMed] [Google Scholar]
- 13.Frederiksen RF, Leisner JJ (2015) Effects of Listeria monocytogenes EGD-e and Salmonella enterica ser. Typhimurium LT2 chitinases on intracellular survival in Dictyostelium discoideum and mammalian cell lines. FEMS Microbiol Lett 362. doi: 10.1093/femsle/fnv067 [DOI] [PubMed] [Google Scholar]
- 14.Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC (2003) Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol 47: 103–118. doi: 10.1046/j.1365-2958.2003.03313.x [DOI] [PubMed] [Google Scholar]
- 15.Hautefort I, Thompson A, Eriksson-Ygberg S, Parker ML, Lucchini S, et al. (2008) During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell Microbiol 10: 958–984. doi: 10.1111/j.1462-5822.2007.01099.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srikumar S, Kroger C, Hebrard M, Colgan A, Owen SV, et al. (2015) RNA-seq Brings New Insights to the Intra-Macrophage Transcriptome of Salmonella Typhimurium. PLoS Pathog 11: e1005262. doi: 10.1371/journal.ppat.1005262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arabyan N, Park D, Foutouhi S, Weis AM, Huang BC, et al. (2016) Salmonella Degrades the Host Glycocalyx Leading to Altered Infection and Glycan Remodeling. Sci Rep 6: 29525. doi: 10.1038/srep29525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park D, Arabyan N, Williams CC, Song T, Mitra A, et al. (2016) Salmonella Typhimurium Enzymatically Landscapes the Host Intestinal Epithelial Cell (IEC) Surface Glycome to Increase Invasion. Mol Cell Proteomics 15: 3653–3664. doi: 10.1074/mcp.M116.063206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arabyan N, Weis AM, Huang BC, Weimer BC (2017) Implication of Sialidases in Salmonella Infection: Genome Release of Sialidase Knockout Strains from Salmonella enterica Serovar Typhimurium LT2. Genome Announc 5. doi: 10.1128/genomeA.00341-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97: 6640–6645. doi: 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bajaj V, Lucas RL, Hwang C, Lee CA (1996) Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol 22: 703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Yi L, Zhang J, Sun L, Wen W, et al. (2018) Functional analysis of superoxide dismutase of Salmonella typhimurium in serum resistance and biofilm formation. J Appl Microbiol 125: 1526–1533. doi: 10.1111/jam.14044 [DOI] [PubMed] [Google Scholar]
- 23.Galan JE (2016) Typhoid toxin provides a window into typhoid fever and the biology of Salmonella Typhi. Proc Natl Acad Sci U S A 113: 6338–6344. doi: 10.1073/pnas.1606335113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brumell JH, Tang P, Zaharik ML, Finlay BB (2002) Disruption of the Salmonella-containing vacuole leads to increased replication of Salmonella enterica serovar typhimurium in the cytosol of epithelial cells. Infect Immun 70: 3264–3270. doi: 10.1128/IAI.70.6.3264-3270.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorvel JP, Meresse S (2001) Maturation steps of the Salmonella-containing vacuole. Microbes Infect 3: 1299–1303. doi: 10.1016/s1286-4579(01)01490-3 [DOI] [PubMed] [Google Scholar]
- 26.Yu HB, Croxen MA, Marchiando AM, Ferreira RB, Cadwell K, et al. (2014) Autophagy facilitates Salmonella replication in HeLa cells. mBio 5: e00865–00814. doi: 10.1128/mBio.00865-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein JA, Powers TR, Knodler LA (2017) Measurement of Salmonella enterica Internalization and Vacuole Lysis in Epithelial Cells. Methods Mol Biol 1519: 285–296. doi: 10.1007/978-1-4939-6581-6_19 [DOI] [PubMed] [Google Scholar]
- 28.Garai P, Gnanadhas DP, Chakravortty D (2012) Salmonella enterica serovars Typhimurium and Typhi as model organisms: revealing paradigm of host-pathogen interactions. Virulence 3: 377–388. doi: 10.4161/viru.21087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vazquez MI, Catalan-Dibene J, Zlotnik A (2015) B cells responses and cytokine production are regulated by their immune microenvironment. Cytokine 74: 318–326. doi: 10.1016/j.cyto.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathur R, Oh H, Zhang D, Park SG, Seo J, et al. (2012) A mouse model of Salmonella typhi infection. Cell 151: 590–602. doi: 10.1016/j.cell.2012.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song J, Wilhelm CL, Wangdi T, Maira-Litran T, Lee SJ, et al. (2016) Absence of TLR11 in Mice Does Not Confer Susceptibility to Salmonella Typhi. Cell 164: 827–828. doi: 10.1016/j.cell.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labrousse A, Chauvet S, Couillault C, Kurz CL, Ewbank JJ (2000) Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr Biol 10: 1543–1545. doi: 10.1016/s0960-9822(00)00833-2 [DOI] [PubMed] [Google Scholar]
- 33.Heustis RJ, Ng HK, Brand KJ, Rogers MC, Le LT, et al. (2012) Pharyngeal polysaccharide deacetylases affect development in the nematode C. elegans and deacetylate chitin in vitro. PLoS One 7: e40426. doi: 10.1371/journal.pone.0040426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aballay A, Yorgey P, Ausubel FM (2000) Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr Biol 10: 1539–1542. doi: 10.1016/s0960-9822(00)00830-7 [DOI] [PubMed] [Google Scholar]
- 35.Aballay A, Drenkard E, Hilbun LR, Ausubel FM (2003) Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr Biol 13: 47–52. doi: 10.1016/s0960-9822(02)01396-9 [DOI] [PubMed] [Google Scholar]
- 36.Kralova J, Dvorak M, Koc M, Kral V(2008) p38 MAPK plays an essential role in apoptosis induced by photoactivation of a novel ethylene glycol porphyrin derivative. Oncogene 27: 3010–3020. doi: 10.1038/sj.onc.1210960 [DOI] [PubMed] [Google Scholar]
- 37.Steeb B, Claudi B, Burton NA, Tienz P, Schmidt A, et al. (2013) Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLoS Pathog 9: e1003301. doi: 10.1371/journal.ppat.1003301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haraga A, Ohlson MB, Miller SI (2008) Salmonellae interplay with host cells. Nat Rev Microbiol 6: 53–66. doi: 10.1038/nrmicro1788 [DOI] [PubMed] [Google Scholar]
- 39.Knuff K, Finlay BB (2017) What the SIF Is Happening-The Role of Intracellular Salmonella-Induced Filaments. Front Cell Infect Microbiol 7: 335. doi: 10.3389/fcimb.2017.00335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diacovich L, Dumont A, Lafitte D, Soprano E, Guilhon AA, et al. (2009) Interaction between the SifA virulence factor and its host target SKIP is essential for Salmonella pathogenesis. J Biol Chem 284: 33151–33160. doi: 10.1074/jbc.M109.034975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zusev M, Benayahu D (2008) New insights on cellular distribution, microtubule interactions and post-translational modifications of MS-KIF18A. J Cell Physiol 217: 618–625. doi: 10.1002/jcp.21525 [DOI] [PubMed] [Google Scholar]
- 42.Chakravortty D, Hensel M (2003) Inducible nitric oxide synthase and control of intracellular bacterial pathogens. Microbes Infect 5: 621–627. doi: 10.1016/s1286-4579(03)00096-0 [DOI] [PubMed] [Google Scholar]
- 43.Niedbala W, Cai B, Liew FY (2006) Role of nitric oxide in the regulation of T cell functions. Ann Rheum Dis 65 Suppl 3: iii37–40. doi: 10.1136/ard.2006.058446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato K, Ozaki K, Oh I, Meguro A, Hatanaka K, et al. (2007) Nitric oxide plays a critical role in suppression of T-cell proliferation by mesenchymal stem cells. Blood 109: 228–234. doi: 10.1182/blood-2006-02-002246 [DOI] [PubMed] [Google Scholar]
- 45.Lee CG, Da Silva CA, Dela Cruz CS, Ahangari F, Ma B, et al. (2011) Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu Rev Physiol 73: 479–501. doi: 10.1146/annurev-physiol-012110-142250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawada M, Hachiya Y, Arihiro A, Mizoguchi E (2007) Role of mammalian chitinases in inflammatory conditions. Keio J Med 56: 21–27. doi: 10.2302/kjm.56.21 [DOI] [PubMed] [Google Scholar]
- 47.Ma B, Akosman B, Kamle S, Lee CM, He CH, et al. (2021) CHI3L1 regulates PD-L1 and anti-CHI3L1-PD-1 antibody elicits synergistic antitumor responses. J Clin Invest 131. doi: 10.1172/JCI137750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheminay C, Mohlenbrink A, Hensel M (2005) Intracellular Salmonella inhibit antigen presentation by dendritic cells. J Immunol 174: 2892–2899. doi: 10.4049/jimmunol.174.5.2892 [DOI] [PubMed] [Google Scholar]
- 49.Bayer-Santos E, Durkin CH, Rigano LA, Kupz A, Alix E, et al. (2016) The Salmonella Effector SteD Mediates MARCH8-Dependent Ubiquitination of MHC II Molecules and Inhibits T Cell Activation. Cell Host Microbe 20: 584–595. doi: 10.1016/j.chom.2016.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gogoi M, Ravikumar V, Dixit NM, Chakravortty D (2018) Salmonella escapes antigen presentation through K63 ubiquitination mediated endosomal proteolysis of MHC II via modulation of endosomal acidification in dendritic cells. Pathog Dis 76. doi: 10.1093/femspd/ftx125 [DOI] [PubMed] [Google Scholar]
- 51.Ryan SO, Cobb BA (2012) Roles for major histocompatibility complex glycosylation in immune function. Semin Immunopathol 34: 425–441. doi: 10.1007/s00281-012-0309-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ilca FT, Boyle LH (2021) The glycosylation status of MHC class I molecules impacts their interactions with TAPBPR. Mol Immunol 139: 168–176. doi: 10.1016/j.molimm.2021.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yadav J, Dikshit N, Ismaeel S, Qadri A (2020) Innate Activation of IFN-gamma-iNOS Axis During Infection With Salmonella Represses the Ability of T Cells to Produce IL-2. Front Immunol 11: 514. doi: 10.3389/fimmu.2020.00514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhat P, Leggatt G, Waterhouse N, Frazer IH (2017) Interferon-gamma derived from cytotoxic lymphocytes directly enhances their motility and cytotoxicity. Cell Death Dis 8: e2836. doi: 10.1038/cddis.2017.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kogut MH, Arsenault RJ (2017) Immunometabolic Phenotype Alterations Associated with the Induction of Disease Tolerance and Persistent Asymptomatic Infection of Salmonella in the Chicken Intestine. Front Immunol 8: 372. doi: 10.3389/fimmu.2017.00372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chavez V, Mohri-Shiomi A, Maadani A, Vega LA, Garsin DA (2007) Oxidative stress enzymes are required for DAF-16-mediated immunity due to generation of reactive oxygen species by Caenorhabditis elegans. Genetics 176: 1567–1577. doi: 10.1534/genetics.107.072587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mosser T, Matic I, Leroy M (2011) Bacterium-induced internal egg hatching frequency is predictive of life span in Caenorhabditis elegans populations. Appl Environ Microbiol 77: 8189–8192. doi: 10.1128/AEM.06357-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naji A, Houston Iv J, Skalley Rog C, Al Hatem A, Rizvi S, et al. (2018) The activation of the oxidative stress response transcription factor SKN-1 in Caenorhabditis elegans by mitis group streptococci. PLoS One 13: e0202233. doi: 10.1371/journal.pone.0202233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, et al. (2005) The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev 19: 2278–2283. doi: 10.1101/gad.1324805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou M, Liu X, Yu H, Yin X, Nie SP, et al. (2018) Cell Signaling of Caenorhabditis elegans in Response to Enterotoxigenic Escherichia coli Infection and Lactobacillus zeae Protection. Front Immunol 9: 1745. doi: 10.3389/fimmu.2018.01745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bogaerts A, Beets I, Schoofs L, Verleyen P (2010) Antimicrobial peptides in Caenorhabditis elegans. Isj-Invert Surviv 7: 45–52. [Google Scholar]
- 62.Knodler LA, Nair V, Steele-Mortimer O (2014) Quantitative assessment of cytosolic Salmonella in epithelial cells. PLoS One 9: e84681. doi: 10.1371/journal.pone.0084681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Green LC, Wagner DA, Glogowski J, Skipper PL, Wishnok JS, et al. (1982) Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids. Anal Biochem 126: 131–138. doi: 10.1016/0003-2697(82)90118-x [DOI] [PubMed] [Google Scholar]
- 64.Datey A, Shreenivas M, Chandrasekharan G, Joseph J, Sah S, et al. (2018) Rewiring of one carbon metabolism in Salmonella serves as an excellent live vaccine against systemic salmonellosis. Vaccine 36: 7715–7727. doi: 10.1016/j.vaccine.2018.10.079 [DOI] [PubMed] [Google Scholar]
- 65.Tan MW, Mahajan-Miklos S, Ausubel FM (1999) Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A 96: 715–720. doi: 10.1073/pnas.96.2.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Everman JL, Ziaie NR, Bechler J, Bermudez LE (2015) Establishing Caenorhabditis elegans as a model for Mycobacterium avium subspecies hominissuis infection and intestinal colonization. Biol Open 4: 1330–1335. doi: 10.1242/bio.012260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Escorcia W, Ruter DL, Nhan J, Curran SP (2018) Quantification of Lipid Abundance and Evaluation of Lipid Distribution in Caenorhabditis elegans by Nile Red and Oil Red O Staining. J Vis Exp. doi: 10.3791/57352 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) BLAST analysis showing the identity of chitinase A of Salmonella serovars Typhimurium and Typhi with known pathogenic chitinases and chitin-binding proteins. Growth analysis of LB grown cultures of (B) STM WT and STM ΔchiA, (C) STY WT, STY ΔchiA, STY ΔchiA:chiA and STY ΔchiA:pQE60. Absolute CFU/ml values of (D) STM WT and STM ΔchiA, (E) STY WT, STY ΔchiA, STY ΔchiA:chiA and STY ΔchiA:pQE60 in Caco2 cells in gentamicin protection assay after the indicated time. Data are represented as mean ± SEM of 3 independent experiments (N = 3, n = 3). (F) Mean Fluorescence Intensity (MFI) of Neu5Ac-bound SNA-FITC, Gal-bound PNA-FITC and mannose-bound ConA-FITC lectins on Caco2 cells 30 mpi and 120 mpi with STM WT, STM ΔchiA, STY WT and STY ΔchiA (UI- Uninfected). Data are represented as mean ± SEM of 2 independent experiments (N = 2). Two-way ANOVA was used to analyze the data. % Colocalization of mCherry expressing (red) (G) STM WT and STM ΔchiA, (H) STY WT and STY ΔchiA with LAMP1 (green) in Caco-2 cells at 2/10/16 hpi. Data are represented as mean ± SEM of 3 independent experiments (N = 3). Unpaired Student’s t test was used to analyze the data. (I) % Colocalization of mCherry expressing (red) bacteria with EEA1 (green) in Caco-2 cells at 15/30/120 mpi. Data are represented as mean ± SD of 2 independent experiments (N = 2). One-way ANOVA was used to analyze the data.
(TIF)
Absolute CFU/ml values of (A) STM WT and STM ΔchiA, (B) STY WT, STY ΔchiA, STY ΔchiA:chiA and STY ΔchiA:pQE60 in U937 derived monocytes in gentamicin protection assay after the indicated time. Data are represented as mean ± SEM of 3 independent experiments (N = 3, n = 3). Absolute CFU/ml values of (C) STM WT and STM ΔchiA, (D) STY WT, STY ΔchiA, STY ΔchiA:chiA and STY ΔchiA:pQE60 in murine BMDCs in gentamicin protection assay after the indicated time. Data are represented as mean ± SEM of 3 independent experiments (N = 3, n = 3). (E) Representative flow cytometry plot showing surface MHC-II level on PMs infected with STY WT, STY ΔchiA and STY ΔchiA:chiA for 20 hours (UI US- Unstained uninfected, UI- Uninfected, STY Δc:chiA- STY ΔchiA:chiA). (F) Representative images showing surface MHC-II on PMs infected with STM WT, STM ΔchiA, STY WT and STY ΔchiA for the indicated time. PMs were stained for surface MHCII without any permeabilizing agent (UI- Uninfected).
(TIF)
Bacterial CFU in (A) spleen and (B) liver from STM WT and STM ΔchiA infected mice after the indicated time. (C) STM WT and STM ΔchiA infected mice spleen length were measured after 5 days and 20 days of oral infection. Data are presented from 3 independent experiments. One-way ANOVA was used to analyze the data. (D) Representative images of spleens isolated from STM WT and STM ΔchiA infected mice after 20 days. Data are presented from one independent experiment, representative of 3 independent experiments (N = 3). Anti-inflammatory cytokine (E) IL-10 and (F) IL-4 level in serum from STM WT and STM ΔchiA infected mice after the indicated time. Data are presented as mean ± SEM of 3 independent experiments (N = 3). One-way ANOVA was used to analyze the data. (G) Representative immunoblots showing the reactivity of the STM WT and STM ΔchiA infected mice sera against mCherry-tagged Salmonella whole cell lysate. Coomassie Brilliant Blue stained gel shows equal loading in all the lanes. Furthermore, the blot was probed with anti-mCherry antibody. The Data are presented from 2 independent experiments (N = 2). (H) Quantification of absolute bacterial CFU obtained from infected C. elegans after 24 hours and 48 hours continuous feeding on STY WT, STY ΔchiA, STY ΔchiA:chiA and STY ΔchiA:pQE60 strains. Data are represented as mean ± SEM of 4 independent experiments. (I) % colonization of the worms gut after 24 hours of continuous feeding with different bacterial strains. Data are represented as mean ± SEM of 4 independent experiments. One-way ANOVA was used to analyze the data. (J) Representative images showing bacterial colonization and persistence in the worms’ gut after shorter exposure.
(TIF)
Representative images of parts of FT63 worms gut showing extra-intestinal invasion of the STY WT strain after 48 hours of continuous feeding (A) at higher magnification (scale bar- 5 μm) and (B) at lower magnification (scale bar- 10 μm). Yellow arrows show the presence of STY WT and STM ΔinvC bacteria outside the gut lumen (Green). (C) Representative images of CL2166 worms after 48 hours of feeding on STY strains. GFP fluorescence is indicative of oxidative stress. Insets show ‘bag of worms’ resulting from oxidative stress in the STM WT and STM ΔchiA:chiA infected worms. (D) Quantification of GFP MFI in CL2166 worms. Data are represented as mean ± SEM of 3 independent experiments. One-way ANOVA was used to analyze the data. (E) Quantification of the ORO-stained parts of the worms fed with different bacterial strains for 48 hours. Data are represented as mean ± SEM of 3 independent experiments. Two-way ANOVA was used to analyze the data. (F) Immunological detection of phospho-p38 MAPK (PMK-1) from worms fed with E. coli OP50 and STY strains for 48 hours. β-actin was used as loading control. (G) qRTPCR analysis of the p38 MAPK (PMK-1) pathway genes pmk1 and mek1 in worms fed with E. coli OP50 and STY strains for 48 hours. Fold change was normalized over act2. Data represent mean ± SEM of 4 independent experiments. One-way ANOVA was used to analyze the data.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.