Summary
Bacterial outer membrane (OM) is a self‐protective and permeable barrier, while having many non‐negligible negative effects in industrial biotechnology. Our previous studies revealed enhanced properties of Halomonas bluephagenesis based on positive cellular properties by OM defects. This study further expands the OM defect on membrane compactness by completely deleting two secondary acyltransferases for lipid A modification in H. bluephagenesis, LpxL and LpxM, and found more significant advantages than that of the previous lpxL mutant. Deletions on LpxL and LpxM accelerated poly(3‐hydroxybutyrate) (PHB) production by H. bluephagenesis WZY229, leading to a 37% increase in PHB accumulation and 84‐folds reduced endotoxin production. Enhanced membrane permeability accelerates the diffusion of γ‐butyrolactone, allowing H. bluephagenesis WZY254 derived from H. bluephagenesis WZY229 to produce 82wt% poly(3‐hydroxybutyrate‐co‐23mol%4‐hydroxybutyrate) (P(3HB‐co‐23mol%4HB)) in shake flasks, showing increases of 102% and 307% in P(3HB‐co‐4HB) production and 4HB accumulation, respectively. The 4HB molar fraction in copolymer can be elevated to 32 mol% in the presence of more γ‐butyrolactone. In a 7‐l bioreactor fed‐batch fermentation, H. bluephagenesis WZY254 supported a 84 g l−1 dry cell mass with 81wt% P(3HB‐co‐26mol%4HB), increasing 136% in 4HB molar fraction. This study further demonstrated that OM defects generate a hyperproduction strain for high 4HB containing copolymers.
In a 7‐l bioreactor fed‐batch fermentation, H. bluephagenesis WZY254 supported a 84 g l‐1 dry cell mass (DCM) with 81wt% P(3HB‐co‐26mol%4HB), increasing 136% in 4HB molar fraction. This study further demonstrated that OM defects generates a hyperproduction strain for high 4HB containing copolymers.
Introduction
Industrial biotechnology based on microbes has been rapidly developed for producing chemicals, materials, biofuels and medicines, showing competitiveness due to its safety, green processes and sustainability compared to chemical industries (Tang and Zhao, 2009; Chen and Kazlauskas, 2011; Chen, 2012). Outer membrane (OM), a characteristic structure of Gram‐negative bacterial envelope, acts as a major component of protective and permeable barrier against bactericidal compounds, controlling the substance exchange strictly (Clifton et al., 2015; Konovalova et al., 2017). As the major molecules on the outer leaflet of the OM, lipopolysaccharides (LPSs) consisting of lipid A, core polysaccharides and O‐antigen (Fig. 1A) (Raetz and Whitfield, 2002; Whitfield and Trent, 2014) have been well studied. However, LPS has severe and non‐negligible negative effects in industrial biotechnology: preventing the substrates and metabolite transportation (Brandt et al., 2012; Wang et al., 2014a), competing for carbon source (Goff et al., 2009) and generating an inflammation‐trigging endotoxin (Wang et al., 2014b). Therefore, the effect of OM defect on bioproduction has been of great interest. In recent years, several studies have claimed that OM deficiency may have a positive impact on cell growth and bioproduction in Cupriavidus necator (Brandt et al., 2012), Escherichia coli (Wang et al., 2020) (Wang et al., 2014a) and Pseudomonas putida EM371 (Dvorak et al., 2020; Martinez‐Garcia et al., 2020).
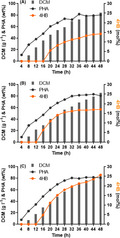
Fig. 1.
Growth and PHB accumulation by Halomonas bluephagenesis deleted with LpxL and LpxM.
A. Major OM component LPS catalyzed by products of genes lpxL and lpxM indicated in blue and red, respectively. Hep, L‐glycero‐D‐manno‐heptose; Glc, glucose; Gal, galactose; Kdo, 3‐deoxy‐D‐manno‐2‐octulosonic acid; GlcN, N‐acetylglucosamine; P, phosphate groups.
B. Growth profiles on 96 plate with MM50 medium.
C. Growth and PHB synthesis by wild type and OM‐defected H. bluephagenesis, respectively. DCM, dry cell mass.
D. PHB (g l−1) and glucose to PHB conversion rate (%) of (C). Data are expressed as the mean ± SD from biologically independent triplicate experiments.
The next generation industrial biotechnology (NGIB), based on a native poly(3‐hydroxybutyrate) (PHB) (Sagong et al., 2018; Pernicova et al., 2019; Wongsirichot et al., 2020) producer, the halophile Halomonas spp., is a rising star because of convenient processes and cost‐effective fermentation in recent years (Chen and Liu, 2021). A well‐studied H. bluephagenesis (Chen and Jiang, 2018; Mitra et al., 2020) has been successfully established as a chassis for NGIB for various productions, including PHB (Weiss et al., 2017; Mitra et al., 2020), biofuels (Amer et al., 2020a, 2020b; Trisrivirat et al., 2020), poly(3‐hydroxybutyrate‐co‐3‐hydroxyvalerate) (Heinrich et al., 2015; de Paula et al., 2017) and poly(3‐hydroxybutyrate‐co‐4‐hydroxybutyrate) (Ye et al., 2018). In our previous studies, OM defect brought positive effects, which remarkably increased the positive properties of H. bluephagenesis on growth and bioproduction (Wang et al., 2021). More importantly, it is revealed that the membrane compactness determines the above‐mentioned advantages rather than membrane thickness (Wang et al., 2021).
LPSs are produced through a multistep and conserved biosynthetic pathway in most Gram‐negative bacteria (Ruiz et al., 2006; Raetz et al., 2009). In the final structural modification (Raetz et al., 2007; Wang and Quinn, 2010), members of a superfamily of lysophospholipid acyltransferases (Takeuchi and Reue, 2009), such as LpxL (Six et al., 2008), LpxM (Dovala et al., 2016), LpxJ (Guillotte et al., 2018) and PagP (Chandler et al., 2020) catalyse the acylation of lipid A precursors to generate mature LPS with 4–7 fatty acids varied among species. Interestingly, the number of the fatty acid chains anchored to lipid A varied with growth temperature, affecting the molecular toxic properties and most importantly, determining the membrane compactness (Anisimov et al., 2007). Therefore, this study aimed on further investigation on how OM defect of H. bluephagenesis affects the related behaviours and productivity. More OM‐defected H. bluephagenesis mutants were constructed and examined by deleting other acyltransferases related to final structures of the lipid A.
Results
Construction of OM deficiency on H. bluephagenesis via CRISPR/Cas9
The well‐studied biosynthetic pathways of LPS in recent decades have revealed that diverse late acyltransferases, e.g. LpxL (Six et al., 2008), LpxM (Dovala et al., 2016), LpxJ (Guillotte et al., 2018) and PagP (Chandler et al., 2020), participate in the last modification of lipid A to generate maturely hexaacylated lipid A with acyl‐carrier protein‐activated fatty acids as co‐substrates (Gorzelak et al., 2021). Besides previously studied LpxL, another lipid A late acyltransferase encoded by lpxM was found by H. bluephagenesis, showing 24% similarity to the LpxM of Escherichia. coli (Wang et al., 2021).
A protein sequence alignment between LpxL and LpxM of E. coli and H. bluephagenesis, respectively, revealed 25 totally conserved residues (Fig. S1). Notably, a conserved catalytic acidic residue (HX4D/E), which is a conserved motif among acyltransferase proteins typically in their active sites (Six et al., 2008), was found in the aforementioned four acyltransferases (Fig. S1). Therefore, lpxM was deleted via CRISPR/Cas9 as described (Fig. 1A, Table S2, see Methods section) for further investigation, resulting in H. bluephagenesis strains WZY19. Meanwhile, lpxM was also deleted in H. bluephagenesis WZY09, resulting in a completely secondary acyltransferase‐defected H. bluephagenesis WZY229 (Fig. 1A and Table S1). Genotyping and sequencing were both utilized to confirm successful gene deletion (data not shown).
Growth and PHB production by OM‐defected H. bluephagenesis strains
Firstly, growth studies of H. bluephagenesis WZY09 and WZY229 were performed in a 96‐well plate containing MM50 medium using H. bluephagenesis TD1.0 as a control. The shake flask studies showed that H. bluephagenesis strains WZY19 and its wild‐type TD1.0 had comparable growth (Fig. 1B). However, H. bluephagenesis WZY229 showed a tardiness on growth but reached to the same level at its stationary phase (Fig. 1B).
Subsequently, growth and polyhydroxyalkanoate (PHA) accumulation were characterized in shake flasks containing MM50 medium supplemented by OM‐defected H. bluephagenesis WZY19 and WZY229, respectively. Surprisingly, both OM‐defected H. bluephagenesis WZY19 and WZY229 showed improved production behaviour. The MM50 medium containing 40 g l−1 glucose supported a growth to 12.17 g l−1 dry cell mass (DCM) containing 82.77wt% PHB by H. bluephagenesis WZY19, and a 13.15 g l−1 DCM with 86.96wt% PHB by H. bluephagenesis WZY229 (Fig. 1C). What is more, it revealed increases of 21% and 37% on PHB production with over 30% glucose to PHB conversion rates by H. bluephagenesis WZY19 and WZY229, compared to that of the wild‐type H. bluephagenesis TD1.0, respectively (Fig. 1D).
Production of P(3HB‐co‐4HB) by OM‐defected H. bluephagenesis grown with γ‐butyrolactone
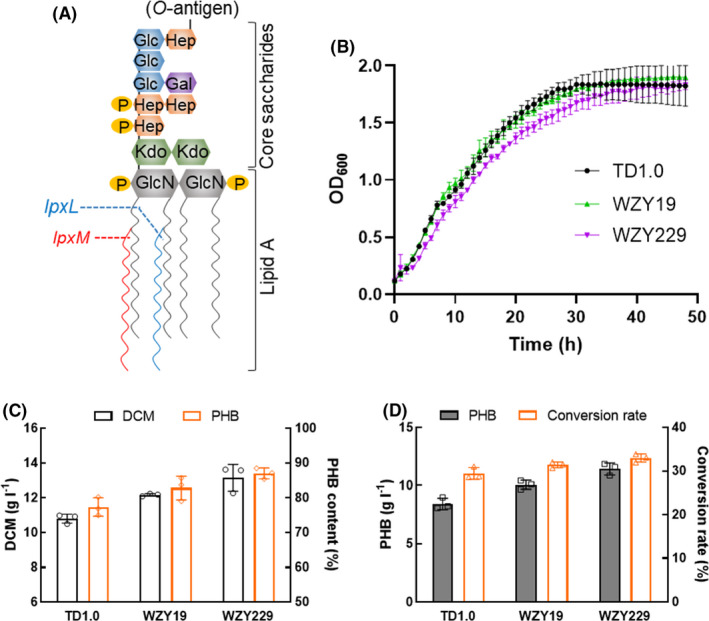
In previous study, a copolyester P(3HB‐co‐19 mol%4HB) was produced by OM‐defected H. bluephagenesis WZY159, generated by deleting lpxL in H. bluephagenesis TDH4ΔphaP1, which could produce large‐granular P(3HB‐co‐4HB), when added with 10 mg l−1 IPTG and 5 g l−1 γ‐butyrolactone in shake flasks containing MM50 medium (Wang et al., 2021). Thus, lpxM was deleted in H. bluephagenesis TDH4ΔphaP1 and WZY159, generating two new OM‐defected H. bluephagenesis strains, namely WZY252 and WZY253, respectively (Table S1).
After 48‐h fermentation in shake flasks supplemented with 10 mg l−1 IPTG and 5 g l−1 γ‐butyrolactone (Fig. 2A and B), two H. bluephagenesis strains WZY252 and WZY253 showed outstanding performance on P(3HB‐co‐4HB) accumulation, especially the latter. Besides the increases of 15% and 32% on DCM, H. bluephagenesis strains WZY252 and WZY253 produced P(3HB‐co‐19.31 mol%4HB) and P(3HB‐co‐21.40 mol%4HB), respectively (Fig. 2C). Furthermore, improvements of 47% and 105% on P(3HB‐co‐4HB) and 4HB accumulation were achieved by H. bluephagenesis strains WZY253 (Fig. 2D).
Fig. 2.
Production of P(3HB‐co‐4HB) by OM‐defected H. bluephagenesis strains cultivated in 500‐ml shake flasks with IPTG induction.
A. H. bluephagenesis TDH4ΔphaP1 has an IPTG‐inducible P Mmp1 promotor for phaCAB expression.
B. Shake flask production of P(3HB‐co‐4HB): 10 mg l−1 IPTG and 5 g l−1 γ‐butyrolactone were added at OD600 = 2 (≈ 4 h) and OD600 = 6 (≈ 8 h), respectively.
C–D. Growth and P(3HB‐co‐4HB) production in MM50 containing IPTG and γ‐butyrolactone. Data are expressed as the mean ± SD from biologically independent triplicate experiments.
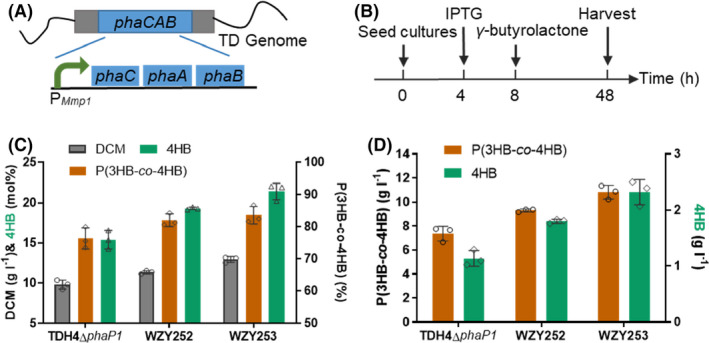
Subsequently, the IPTG‐inducible P Mmp1 of genome‐inserted phaCAB (Zhao et al., 2017) was replaced with a constitutive promoter P porin in H. bluephagenesis WZY252 and WZY253 via CRISPR‐Cas9 (Fig. 3A, Table S2) for stable phaCAB expression and convenience in large scale fermentation, thus generating H. bluephagenesis WZY255 and WZY254, respectively. Similarly, OM‐defected H. bluephagenesis WZY255 and WZY254 showed expectedly excellent performance on both growth and PHA accumulation compared with those of the OM intact H. bluephagenesis TDH4ΔphaP1. In the presence of 5 g l−1 γ‐butyrolactone (Fig. 3B), H. bluephagenesis WZY255 produced a 11.43 g l−1 DCM containing 77.08wt% P(3HB‐co‐21.71 mol%4HB) in shake flasks containing MM50 medium, while WZY254 generated a 13.07 g l−1 DCM containing 81.48wt% P(3HB‐co‐23.28 mol%4HB) (Fig. 3C). Being the best, H. bluephagenesis WZY254 revealed significant improvements on growth and PHA production, showing increases of 74% on DCM (Fig. 3C), as well as 102% and 307% more P(3HB‐co‐4HB) production and 4HB absorption, respectively (Fig. 3D). It was revealed that further increased permeability via OM deficiency allowed easier diffusion of γ‐butyrolactone in the cells for enhanced P(3HB‐co‐4HB).
Fig. 3.
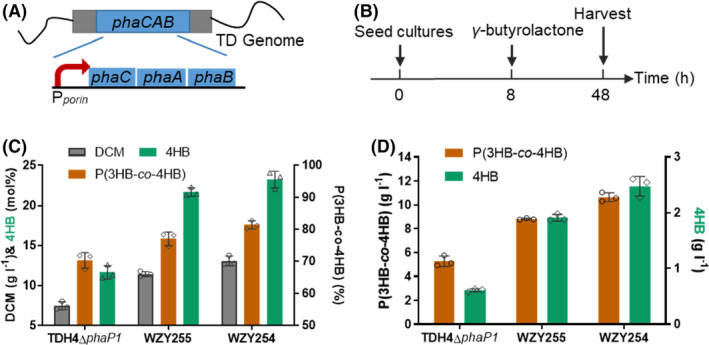
Production of P(3HB‐co‐4HB) by OM‐defected H. bluephagenesis strains cultivated in 500‐ml shake flasks without IPTG induction.
A. H. bluephagenesis TDH4ΔphaP1 has a constitutive Pporin promotor for phaCAB expression.
B. Shake flask procedure of P(3HB‐co‐4HB) production: 5 g l−1 of γ‐butyrolactone was added at OD600 = 6 (≈ 8 h).
C–D. Growth and P(3HB‐co‐4HB) production in MM50 containing IPTG and γ‐butyrolactone. Data are expressed as the mean ± SD from biologically independent triplicate experiments.
Characterizations of OM deficiency in H. bluephagenesis
The cell cultures from the end of shake flasks fermentation by H. bluephagenesis TDH4ΔphaP1, WZY255 and WZY254 were sampled for transmission electron microscopy (TEM) studies to visualize morphology and OM deficiency via ruthenium red staining (see Methods section). Due to phaP1 deletion, images of all the strains showed obviously large and round PHA granules, and the PHB granules accumulated in H. bluephagenesis WZY255 and WZY254 were found larger over those in the H. bluephagenesis TDH4ΔphaP1 under the TEM (Fig. 4A upper panel). When measuring the lengths on their long axis by Image J, H. bluephagenesis WZY255 and WZY254 showed increase of 19% and 22% on cell lengths, respectively (Fig. 4B). In addition, H. bluephagenesis WZY255 and WZY254 were observed thinner and weaker OM compared to their intact and obvious control (Fig. 4A nether panel).
Fig. 4.
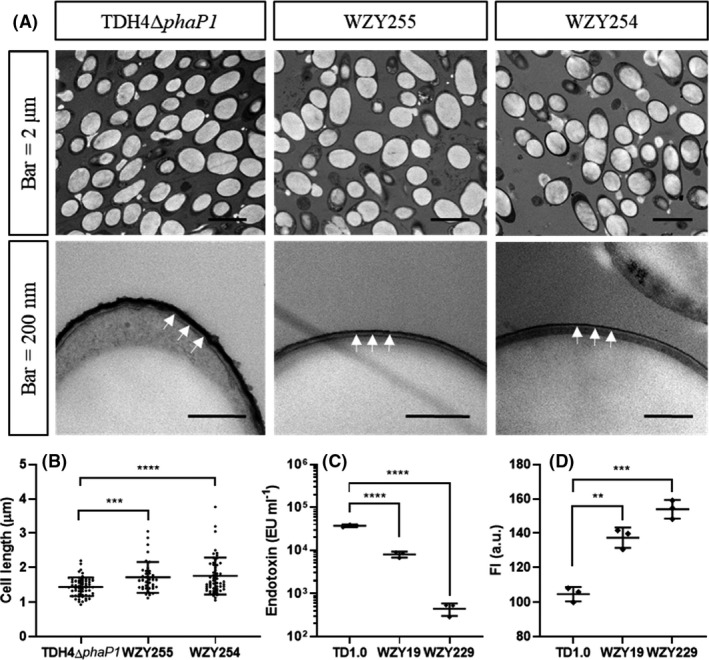
Characterization of outer membrane (OM) defect on H. bluephagenesis.
A. TEM studies of H. bluephagenesis TDH4ΔphaP1, WZY255 and WZY254 with scale bars of 2 μm and 200 nm, respectively. The white arrows indicate stained OM with ruthenium red. AFM studies on membrane morphology (right panels, scale bar 50 nm).
B. Cell length measured from (A) (upper panels) (n TDH0Δ pha P1 = 61, n WZY255 = 39, n WZY254 = 56, two‐tailed Student’s t‐test, P = 0.0002, P < 0.0001).
C. Endotoxin content assayed via LAL (n = 3, two‐tailed Student’s t‐test, P < 0.0001).
D. Membrane permeability tested by NPN (n = 3, two‐tailed Student’s t‐test, P = 0.0015, 0.0002). All the data are expressed as the mean ± SD from biologically independent studies. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Subsequently, the content of endotoxin was assayed via limulus amebocyte lysate (LAL) assay (see Methods section). The results showed that endotoxin of H. bluephagenesis WZY19 and WZY229 decreased 5‐folds and 84‐folds compared with that of the wild‐type TD1.0, respectively (Fig. 4C). Meanwhile, N‐Phenyl‐1‐naphthylamine (NPN) assays were utilized to detect the membrane permeability (see Methods section). It was found that H. bluephagenesis WZY19 and WZY229 were more permeable than H. bluephagenesis TD1.0 (Fig. 4D).
Growth and production of PHB by OM‐defected H. bluephagenesis WZY254 under various salinity
Our previous study showed that H. bluephagenesis WZY09 had a better adaptation to a lower salinity osmotic pressures (Wang et al., 2021). Thus, H. bluephagenesis WZY254 was cultivated in shake flasks with mineral medium (MM) media supplemented 10, 30 and 50 g l−1 NaCl, respectively, to optimize growth and PHB accumulation. H. bluephagenesis WZY254 grown in the MM30 reached the highest 14.12 g l−1 DCM consisting of 84.83wt% PHB, revealing a 36% increase of PHB production, while H. bluephagenesis TDH4ΔphaP1 producing the best 11.44 g l−1 DCM when incubated in the MM50 (Fig. 5A,B). It was obvious that H. bluephagenesis WZY254 preferred low salinity under 30 g l−1 NaCl, compared to the downtrend of H. bluephagenesis TDH4ΔphaP1 from the optimum under 50 g l−1 NaCl (Fig. 5A). When focusing on H. bluephagenesis WZY254, its growth, PHB accumulation and glucose to PHB conversion when cultured in MM30 had slight advantages over those when grown in MM50 (Fig. 5A,B).
Fig. 5.
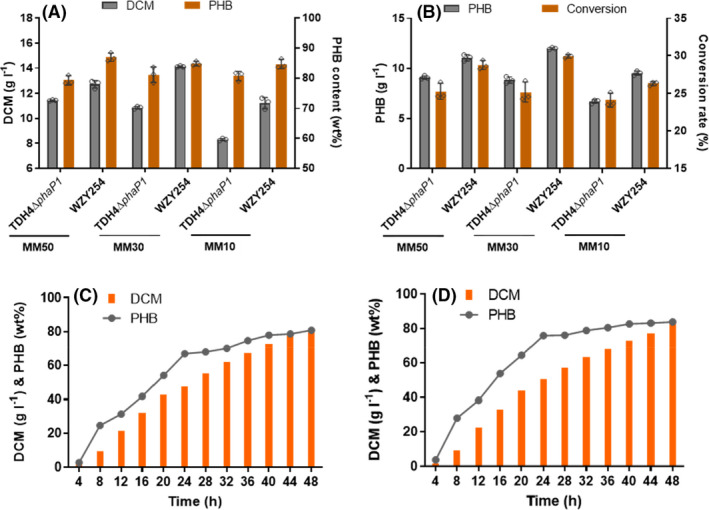
Growth and accumulation of PHB by OM‐defected H. bluephagenesis WZY254 cultured in 500‐ml shake flasks and 7‐l bioreactor, respectively.
A. Growth and PHB accumulation by H. bluephagenesis TDH4ΔphaP1 and WZY254 in shake flasks containing MM50, MM30 and MM10 medium, respectively.
B. PHB production and glucose to PHB conversion analysis from (A).
C–D. Fed‐batch growth and PHB production in a 7‐l bioreactor containing 50 g l−1 (C) and 30 g l−1 (D) NaCl under open unsterile conditions. Final PHB production analysis is presented in Table S4. Data in (A) and (B) are expressed as the mean ± SD from biologically independent triplicate experiments.
Subsequently, fed‐batch fermentations in MM media containing 50 g l−1 and 30 g l−1 NaCl in a 7‐l bioreactor were performed using H. bluephagenesis WZY254 under the same open and unsterile conditions to evaluate their performances, respectively. As expected, H. bluephagenesis WZY254 maintained its superiorities on growth and PHB accumulation in large‐scale fermentations. Both strains cultivated in MM50 and MM30 produced over 80 g l−1 DCM containing over 80wt% PHB after 48 h, still maintaining accelerations of nearly 5 g l−1 DCM in every four hours (Fig. 5C and D). Detailed final PHB production analysis could be found in Table S3. Overall, MM30 did have an advantage over MM50 for growth and PHA production by H. bluephagenesis WZY254, reflecting on a 13% increase of PHB production (Table S2). Collectively, H. bluephagenesis WZY254 had a better adaptation to a lower salinity osmotic pressures due to OM defect and showed great performence in MM30 medium during the fed‐batch growth in the 7‐l bioreactor under open and unsterile conditions. Accordingly, MM30 medium was taken for further P(3HB‐co‐4HB) investigation in shake flasks inoculated with the OM‐defected H. bluephagenesis WZY254.
Hyperproduction of copolyester P(3HB‐co‐4HB) by OM‐defected H. bluephagenesis WZY254 grown with γ‐butyrolactone in 500‐ml shake flask
Previously, a P(3HB‐co‐23.28 mol%4HB) was successfully produced by H. bluephagenesis WZY254 in shake flask supplemented with 5 g l−1 γ‐butyrolactone (Fig. 3C). Taking further increased membrane permeability (Fig. 4D) into consideration, it was assumed that more γ‐butyrolactone could help to further elevate 4HB molar fractions in P(3HB‐co‐4HB). Accordingly, shake flasks studies containing MM30 media were performed for P(3HB‐co‐4HB) production by H. bluephagenesis WZY254 supplemented with 5, 10, 15 and 20 g l−1 γ‐butyrolactone added at OD600 = 6 (≈ 8 h), respectively.
Interestingly, OM‐defected H. bluephagenesis WZY254 produced a 4.06 g l−1 DCM containing 56.07wt% P(3HB‐co‐31.85 mol%4HB), when supplemented with 20 g l−1 γ‐butyrolactone (Fig. 6B). Furthermore, distinct differences were observed between H. bluephagenesis WZY254 and TDH4ΔphaP1, the former showing absolute predominance on PHA production, especially 4HB molar fractions (Fig. 6A and B). It revealed that high concentration of γ‐butyrolactone indeed had serious inhibition on growth, even towards permeability‐increased H. bluephagenesis WZY254.
Fig. 6.
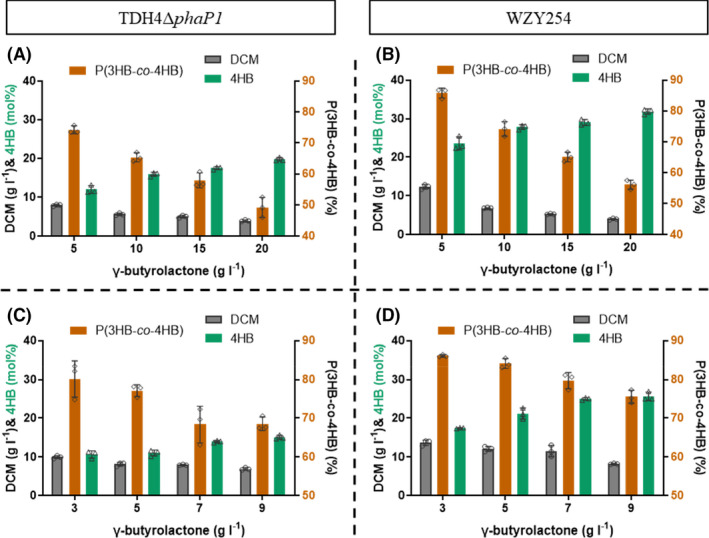
Production of P(3HB‐co‐4HB) by OM‐defected H. bluephagenesis WZY254 cultured in 500‐ml shake flasks.
A–B. Growth and P(3HB‐co‐4HB) production by H. bluephagenesis TDH4ΔphaP1 (A) and WZY254 (B) in shake flasks containing MM30 with γ‐butyrolactone in high concentrations, respectively.
C–D. Tolerance of γ‐butyrolactone studies by H. bluephagenesis TDH4ΔphaP1 (C) and WZY254 (D) in shake flask growths containing MM30.
Subsequently, shake flask cultures containing H. bluephagenesis WZY254 were supplemented with low concentrations of γ‐butyrolactone, ranging from 3 to 9 g l−1, to investigate its tolerance for P(3HB‐co‐4HB) production. Interestingly, the concentration γ‐butyrolactone was positively correlated with 4HB molar fractions but negatively correlated with DCM and P(3HB‐co‐4HB) content (Fig. 6C and D). These results indicated that there should be given and taken on both sides. P(3HB‐co‐4HB) with precise 4HB molar fractions could be produced by controlling the concentration of γ‐butyrolactone.
Fed‐batch studies of hyperproduction of P(3HB‐co‐4HB) by H. bluephagenesis WZY254
H. bluephagenesis TD1.0 has been explored as a platform for NGIB in open and unsterile fermentation (Mitra et al., 2020). Based on the great performance in 4HB‐rich P(3HB‐co‐4HB) production in 500‐ml shake flasks, fed‐batch studies using H. bluephagenesis WZY254 were performed in 7‐l bioreactor for growth and P(3HB‐co‐4HB) production study under the same open and unsterile conditions.
Firstly, H. bluephagenesis WZY254 in fed‐batch fermentation with MM30 medium produced a 84.15 g l−1 DCM containing 80.49wt% P(3HB‐co‐14.24 mol%4HB) when supplemented with γ‐butyrolactone as described (Shen et al., 2018), and the conversion yield of γ‐butyrolactone reached to ~ 80% (Fig. 7A, Table S3). Significantly, γ‐butyrolactone was supplemented in fed batch at OD600 = 120 (≈ 16 h), and nearly 50wt% PHB has been accumulated inside bacteria at this moment (Fig. 7A).
Fig. 7.
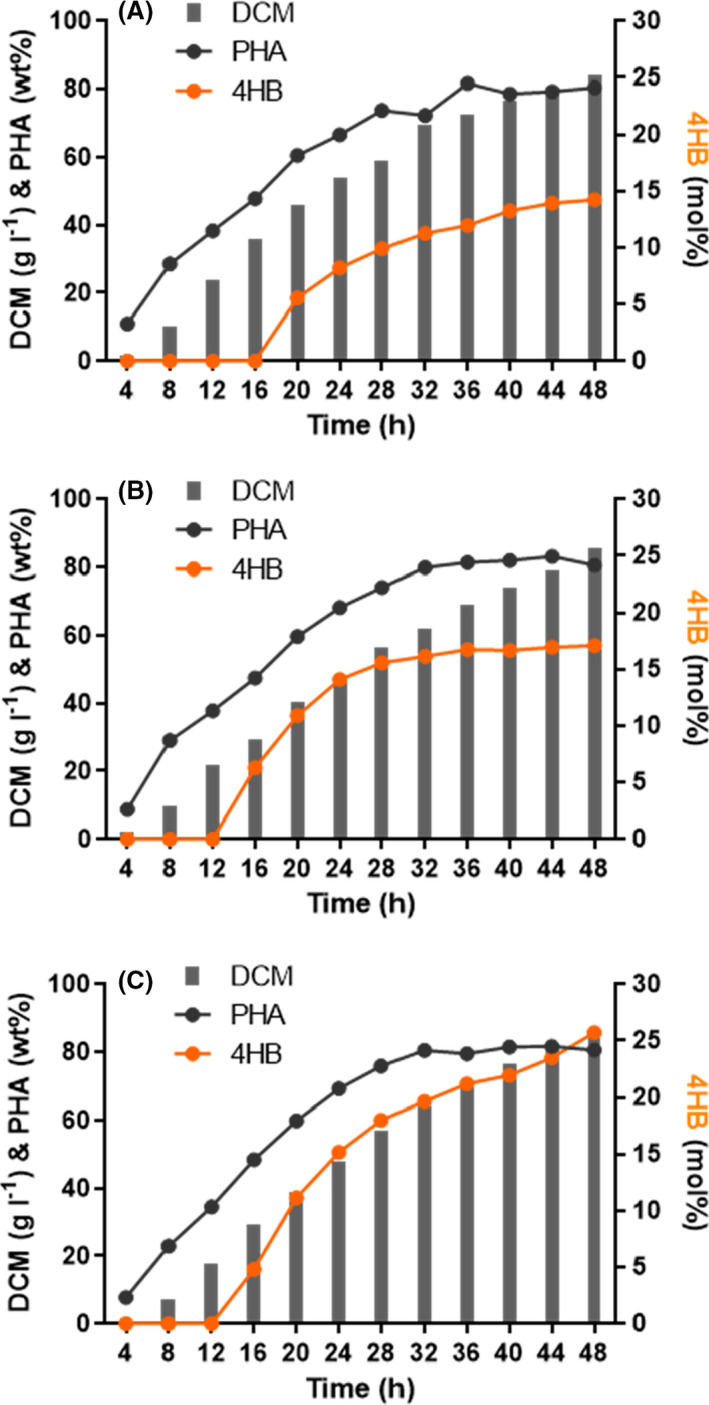
Fed‐batch growth and P(3HB‐co‐4HB) production by OM‐defected H. bluephagenesis WZY254 cultured in a 7‐l bioreactor under open unsterile conditions. In fed‐batch studies of hyperproduction of P(3HB‐co‐4HB) by H. bluephagenesis WZY254 in 7‐l bioreactors with MM30 media, γ‐butyrolactone in various concentrations was supplemented at 16 h (A), 12 h (B and C). Compared to (B), more γ‐butyrolactone was supplemented in (C) to accumulate more 4HB fraction in P(3HB‐co‐4HB). Final P(3HB‐co‐4HB) production and the conversion yield of γ‐butyrolactone were analysed and presented in Table S4.
In order to increase 4HB fraction in P(3HB‐co‐4HB), γ‐butyrolactone was supplemented at the early stage of fermentation. However, significant inhibitions on growth and PHA accumulation were observed when γ‐butyrolactone was added beginning from 6 to 8 h, the moment cells containing 20–30% wt% PHB (Fig. S2). When the supplementation of γ‐butyrolactone was adjusted to start at OD600 = 80 (≈ 12 h), the moment cells having a ~ 25 g l−1 DCM with ~ 40% wt% PHB, H. bluephagenesis WZY254 produced 85.65 g l−1 DCM containing 80.66wt% P(3HB‐co‐17.11 mol%4HB) (Fig. 7B), showing a 20% increase in 4HB fraction. Subsequently, more γ‐butyrolactone supplementation supported a 84.41 g l−1 DCM containing 80.60wt% P(3HB‐co‐25.73 mol%4HB) (Fig. 7C), an increase of 136% on 4HB molar fraction compared to the previous P(3HB‐co‐11 mol%4HB) produced by H. bluephagenesis TDH4 with intact OM in bioreactors (Shen et al., 2018). Taken together, H. bluephagenesis WZY254 showed better absorption and conversion of γ‐butyrolactone based on increased membrane permeability and maintained its superiorities on growth and P(3HB‐co‐4HB) accumulation in large‐scale fermentations.
Discussion
The complicated OM, mostly consisting of LPSs on the external leaflet on Gram‐negative bacterial cell envelops, has always been considered as a strictly permeable barrier protecting cells from bactericidal environments (Konovalova et al., 2017; Guest et al., 2020). Although the disadvantages of OM (or LPS) are obvious, the necessity of OM to bacteria is non‐negligible, and the completely deletion of OM has never been achieved. However, partial deletion of OM or defect on various degrees of LPS was investigated in recent decades and indicated considerable research prospect on future bioproduction. Based on previous results of some positive effects on growth and bioproduction attributing to OM defect (Wang et al., 2021), our further exploration on OM defect in this study using H. bluephagenesis, a chassis for NGIB (Chen and Jiang, 2018; Mitra et al., 2020), revealed more satisfactory findings. Completely loss of the two secondary acyltransferases for lipid A modification, LpxL and LpxM, enabled H. bluephagenesis to further improve cell growth and PHA synthesis (Fig. 1), as well as more advantages for industrial bioproduction, including more enlarged cell size (Fig. 4B), less endotoxin (Fig. 4C) and flexible tolerance to reduced salinity (Fig. 5). Surprisingly, further enhanced membrane permeability accelerates the diffusion of 4HB precursor γ‐butyrolactone, capacitating H. bluephagenesis WZY254 to produce high‐quality and 4HB‐rich P(3HB‐co‐4HB).
The disadvantages of OM (or LPS) allow to investigate the impact of its defect on bioproduction. Being different from the stronger E. coli with truncated structure of LPS (Wang et al., 2020), our previous studies revealed that the membrane compactness benefited H. bluephagenesis rather than membrane thickness (Wang et al., 2021). Besides lpxL, encoding a lauroyltransferase to transfer the secondary C12:0 acyl group to (Kdo)2‐lipid IVA, another secondary acyltransferase gene lpxM, encoding a myristoyltransferase to transfer the secondary C14:0 acyl group (Fig. 1A) was further found and deleted in H. bluephagenesis in this study. Similar to the benefits caused by lpxL deletion on H. bluephagenesis WZY09, lpxM mutant H. bluephagenesis WZY19 also induced improved growth and PHB accumulation similar to the previous strain, while H. bluephagenesis WZY229 with double knock‐out of lpxL and lpxM showed the highest 37% increase on PHB production compared to that of the wild‐type H. bluephagenesis TD1.0. Importantly, lpxM deletion further decreases endotoxin content (Fig. 4C) and increased membrane permeability (Fig. 4D).
As a halophile, the well‐studied Halomonas ssp. has optimized growth in medium containing 60 g l−1 NaCl, but adapting an extensive salinity ranging from 10 to 200 g l−1 NaCl (Sedlacek et al., 2019). In our previous study, OM‐defected H. bluephagenesis WZY09 showed better adaption to lower salinity, performing the best in MM30 medium (Wang et al., 2021). With further defect on membrane compactness, H. bluephagenesis WZY254 also showed better adaption to lower NaCl concentrations than its wild type in shake flasks studies and maintained its superiorities on growth and PHB accumulation in large‐scale fermentations. It has been reported that OM played important roles in maintaining membrane stiffness, turgor pressure and cell morphology in Gram‐negative bacteria (Deng et al., 2011; Rojas et al., 2018). The OM deficiencies caused by lpxL and lpxM deletions in H. bluephagenesis change the membrane stiffness and turgor pressure, leading to enlarged cell sizes (Fig. 4B) and better adaptation to lower salinity osmotic pressures (Fig. 5).
In this study, a P(3HB‐co‐23.28 mol%4HB) was successfully produced by H. bluephagenesis WZY254 in 500 ml−1 shake flask supplemented with 5 g l−1 γ‐butyrolactone (Fig. 3C). It was assumed that further increased membrane permeability (Fig. 4D) promoted γ‐butyrolactone infusion and elevated 4HB molar fraction in P(3HB‐co‐4HB). Indeed, OM‐defected H. bluephagenesis WZY254 produced the highest P(3HB‐co‐31.85 mol%4HB) in shake flasks supplemented with 20 g l−1 γ‐butyrolactone, and the 4HB molar fractions were elevated alone with increased γ‐butyrolactone concentration (Fig. 6B). However, the deletion of lpxL and lpxM in H. bluephagenesis TD68‐194, a P(3HB‐co‐4HB) production strain from glucose directly, revealed no significance on 4HB fractions in P(3HB‐co‐4HB) (data not shown). Taken together, further increased membrane permeability of OM‐defected H. bluephagenesis WZY254 promoted γ‐butyrolactone infusion and elevated 4HB mole fraction in P(3HB‐co‐4HB).
The results of 4HB‐rich P(3HB‐co‐4HB) production in shake flasks by H. bluephagenesis WZY254 revealed that a high concentration of γ‐butyrolactone indeed had serious inhibition on growth (Fig. 6), similar to H. bluephagenesis TDH4 (Shen et al., 2018). In the investigation of tolerance of γ‐butyrolactone on P(3HB‐co‐4HB) production in shake flasks, the concentration of γ‐butyrolactone showed positive correlation with 4HB molar fractions but negatively correlated with DCM and P(3HB‐co‐4HB) content (Figs 5C and 6D), indicating a give and take on both sides. Subsequently, H. bluephagenesis WZY254 produced a P(3HB‐co‐25.73 mol%4HB) (Fig. 7C) by adjusting the amount and initial time of γ‐butyrolactone in supplementation of fed‐batch fermentations in 7‐L bioreactor under open and unsterile conditions. Taken together, H. bluephagenesis WZY254 showed better absorption and conversion of γ‐butyrolactone based on increased membrane permeability and kept its superiorities on growth and P(3HB‐co‐4HB) accumulation in large‐scale fermentations.
In conclusion, further exploration of the OM defect on membrane compactness towards cell growth and bioproduction was studied here using OM defect H. bluephagenesis TD1.0. Completely losing of the two secondary acyltransferases for lipid A modification, LpxL and LpxM, enabled OM‐defected H. bluephagenesis WZY229 to further improve cell growth and PHA production, combined with more advantages for industrial bioproduction, including enlarged cell sizes, reduced endotoxin and flexible tolerances to lower salinity. More importantly, further enhanced membrane permeability accelerates the diffusion of 4HB precursor γ‐butyrolactone, capacitating H. bluephagenesis WZY254 derived from WZY229 to produce the highest P(3HB‐co‐31.85 mol%4HB) in shake flasks. During the fed‐batch fermentation in the 7‐l bioreactor under open and unsterile conditions, H. bluephagenesis WZY254 maintained its superiorities on growth and P(3HB‐co‐4HB) accumulation. This study further explores the impacts and applications of defects on OM compactness and constructs a hyperproduction strain for production of high 4HB containing copolymers in bioreactors.
Experimental procedures
Strains, plasmids and media
All bacterial strains and plasmids used in this study are listed in Table S1. E. coli strain s17‐1 was used for both plasmid construction and conjugation. CRISPR‐Cas9 was used for gene deletion and promotor replacement in H. bluephagenesis, and the gRNA plasmid construction was performed as described (Qin et al., 2018).
For cell growth, LB medium (10 g l−1 tryptone, 5 g l−1 yeast extract and 10 g l−1 NaCl) was used for E. coli s17‐1, while LB60 medium (LB medium supplied with 60 g l−1 NaCl) for H. bluephagenesis strains. For PHA production in 500 ml−1 shake flasks, a MM (Tan et al., 2014) medium supplied with 30 g l−1 glucose, 50 g l−1 NaCl and 0.5 g l−1 yeast extract was used. Especially, 10 mg l−1 IPTG and 5 g l−1 γ‐butyrolactone were added for cultures to produce P(3HB‐co‐4HB). The pH of all the culture media was adjusted to around 8.5–9.0 via 5 M NaOH. Antibiotics for marker selection were added as follows: 50 μg ml−1 kanamycin, 100 μg ml−1 spectinomycin and 25 μg ml−1 chloramphenicol.
Conjugation
The conjugation for plasmid transfer from E. coli to H. bluephagenesis was performed as described (Wang et al., 2021). In brief, overnight E. coli s17‐1 harbouring target plasmids and recipient H. bluephagenesis were grown for 8 h in LB and LB60 media, respectively, followed by centrifuging at 4°C, washing once with fresh medium and resuspending. Subsequently, cells were mixed (1:1 volume) and spread on an antibiotic‐free LB20 agar plate 37°C for 8–10 h incubation. Then, conjugated bacterial lawns suspended with LB60 were spread on LB60 agar plates with relevant antibiotics and incubated at 37°C to obtain single colonies.
Growth analysis
Single colonies of H. bluephagenesis strains were cultured in LB60 medium for 12 h under 600 rpm in a 96‐deep‐well plate (Corning, USA) on a rotary shaker (ThermoFisher Scientific, MB100‐4A, USA), followed by diluting 100‐folds using a fresh medium in a new plate (flat bottom, 200 µl per well). For growth studies, MM media containing 50 g l−1 NaCl were used for online recording of optical density at 600 nm (OD600). The growth monitoring was repeated for three times with three parallel samples.
Shake flask studies
The H. bluephagenesis strains were pre‐cultured overnight in LB60 to obtain seed cultures (OD600 = 3–5) from a fresh single colony. Subsequently, 2.5 ml seed culture was inoculated into 50 ml MM50 medium in 500‐ml shake flasks. The initial pH value of culture media was adjusted to around 8.5–9.0 via 5 M NaOH. After cultivation at 37°C for 48 h at 200 rpm, cell cultures were harvested for DCM and PHA content assays. For P(3HB‐co‐4HB) production, 10 mg l−1 IPTG and 5 g l−1 γ‐butyrolactone were added at OD600 = 2 (≈ 4 h) and OD600 = 6 (≈ 8 h), respectively.
Fermentation studies in 7‐l bioreactor
Studies of fermentation in a 7‐l bioreactor were performed using a fed‐batch process: 300 ml fresh seed cultures in LB60 medium were obtained in 500‐ml shake flasks (OD600 reached 3–5) to be inoculated to a 7‐l bioreactor (NBS Bioflo3000, New Brunswick, NJ, USA). A 3‐l MM50 medium supplemented with 10 g l−1 yeast extract and 20 g l−1 glucose was used as an initial medium. The pH of culture media during fermentations was adjusted to around 8.5–9.0 via 5 M NaOH. During the fermentation, dissolved oxygen (DO%) was maintained at ~ 30% of air saturation by injecting air with a maximum flow rate of 1 VVM (air volume/culture volume/min) via coupling the agitation of less than 800 rpm. Different feeding solutions were prepared for the three‐stage cultivation: 800 g l−1 glucose and 40 g l−1 urea in Feed I, 800 g l−1 glucose and 4 g l−1 urea in Feed II, and 800 g l−1 glucose in Feed III. Glucose concentration was assayed periodically and manually maintained around 6–8 g l−1 throughout the fermentation process. The fed‐batch fermentation was conducted under open unsterile conditions at 37°C. Cultures were sampled for DCM and PHA content assays during the process. In the production of P(3HB‐co‐4HB) by H. bluephagenesis WZY254 in 7‐l bioreactors, γ‐butyrolactone was supplemented in fed batch as described (Shen et al., 2018).
Assays of DCM and PHA content
30 ml cell cultures from shake flasks or bioreactor broth were harvested by centrifuging at 13 400 g (CR 21GIII, HITACHI, Japan) for 5 min, subsequently washed once with distilled water. DCM was calculated by measuring the mass of harvested cells after lyophilization. For PHA content assays, 25–35 mg powdered lyophilized cells were methanolysed under 100°C for 4 h in 2 ml chloroform and 2 ml methanolysis solution (97 wt% methanol, 3 wt% H2SO4, and 1 g l−1 benzoic acid), followed by addition of 1 ml water after cooling to room temperature for extraction and phase separation. The heavy phase was sampled for PHB content calculation using gas chromatography (GC‐2014, Shimadzu, Japan). Twenty milligram of highly pure 3HB or γ‐butyrolactone (A standard from Sigma‐Aldrich) was used as standards.
Microscopy observations
Fresh cells from shake flasks were harvested by centrifuging at 1500 g for 3 min and washed with PBS (phosphate buffer, pH 7.2), and then fixed with 2% glutaraldehyde (pH 7.2) at 4°C for 4 h to prepare for TEM sample preparation. For TEM studies, ruthenium red staining was utilized to visualize OM as described (Wang et al., 2021). In brief, the fixed cells were stained with 0.05% ruthenium red in 0.1 M cacodylate buffer at 4°C for 5–7 h. After being washed with same buffer for once and postfixed in 1% osmium tetroxide solution containing 0.05% ruthenium red for 2 h at 4°C, cells were rinsed in cacodylate buffer and dehydrated using graded ethanol series. After embedding in resin, sections of about 80 nm thickness were cut using an ultra‐microtome (Lecia, Germany) and examined under TEM (H‐7650BA, Japan) at 80 kV.
LAL assays
One millilitre of fresh cells was harvested by centrifugation at 13 400 g for 1 min, washed twice with PBS (phosphate buffer, pH 7.2) and resuspended in de‐ionized water at OD600 ≈ 0.5. This suspension was lysed under 100°C for 30 min, followed by standing at room temperature for 24 h for LPS release. The Genscript ToxinSensorTM Chromogenic LAL Endotoxin Assay Kit (Genscript, Nanjing, China) was utilized to quantify the endotoxin content. The endotoxin contents (EU ml−1) were normalized by OD600 value.
NPN assays
Cell membrane permeability was examined via 1‐N‐Phenyl‐Naphtylamine (NPN) Uptake Assay Kit. One millilitre of fresh bacterial suspension (OD600 ≈ 0.5) was harvested by centrifugation at 13 400 g for 1 min, washed twice with PBS (phosphate buffer, pH 7.2) and resuspended with Reaction Buffer using the same volume. One hundred and ninety‐five microlitre of cell suspension was transferred into a 96‐well plate, added and mixed with 5 μl probe. After incubation at room temperature for 5 min, the plate was detected by a microplate reader for OD600 and fluorescence (excitation at 355 nm, emission at 460 nm). The fluorescence values were normalized by OD600 values.
Conflict of interest
The authors declare no financial or commercial conflict of interest.
Supporting information
Fig. S1. Sequences alignment of LpxL and LpxM of E. coli and H. bluephagenesis, respectively. The consensus residues are emphasized with black asterisks.
Fig. S2. Fed‐batch growth and P(3HB‐co‐4HB) production by H. bluephagenesis WZY254 in 7‐l bioreactor when γ‐butyrolactone was supplemented at early stage of fermentation. The γ‐butyrolactone was supplemented in the fed batch process started at 6 h (A) and 8 h (B), respectively.
Table S1. Strains, plasmids, and genes.
Table S2. sgRNA sequences used in this study.
Table S3. Fed‐batch growth and PHB production by H. bluephagenesis WZY254 cultured in a 7‐l bioreactor for 48 h.
Table S4. Fed‐batch growth and P(3HB‐co‐4HB) production by H. bluephagenesis WZY254 cultured in a 7‐l bioreactor for 48 h.
Acknowledgements
We are grateful to Prof Victor de Lorenzo for kindly donating the SEVA series plasmids. This research was financially supported by grants from the Ministry of Science and Technology of China (Grant No. 2021YFC2101700), National Natural Science Foundation of China (Grant No. 31961133019; No. 21761132013; No. 31870859) and Center of Life Sciences of Tsinghua‐Peking University. This project is also funded by the National Natural Science Foundation of China (grant numbers 31961133017, 31961133018). These grants are part of MIX‐UP, a joint NSFC and EU H2020 collaboration. In Europe, MIX‐UP has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 870294.
Microbial Biotechnology (2022) 15(5), 1586–1597
Funding information
This research was financially supported by grants from the Ministry of Science and Technology of China (Grant No. 2021YFC2101700), National Natural Science Foundation of China (Grant No. 31961133019; No. 21761132013; No. 31870859) and Center of Life Sciences of Tsinghua‐Peking University. This project is also funded by the National Natural Science Foundation of China (grant numbers 31961133017, 31961133018). These grants are part of MIX‐UP, a joint NSFC and EU H2020 collaboration. In Europe, MIX‐UP has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 870294.
Data availability statement
The authors declare that source data processed for figure generation in this study are available within the paper and its Supporting information. Correspondence and requests for raw data should be addressed to G.Q. C. or Z.Y. W.
References
- Amer, M. , Hoeven, R. , Kelly, P. , Faulkner, M. , Smith, M.H. , Toogood, H.S. , and Scrutton, N.S. (2020a) Renewable and tuneable bio‐LPG blends derived from amino acids. Biotechnol Biofuels 13: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amer, M. , Wojcik, E.Z. , Sun, C. , Hoeven, R. , Hughes, J.M.X. , Faulkner, M. , et al. (2020b) Low carbon strategies for sustainable bio‐alkane gas production and renewable energy. Energ Environ Sci 13: 1818–1831. [Google Scholar]
- Anisimov, A.P. , Shaikhutdinova, R.Z. , Pan'kina, L.N. , Feodorova, V.A. , Savostina, E.P. , Bystrova, O.V. , et al. (2007) Effect of deletion of the lpxM gene on virulence and vaccine potential of Yersinia pestis in mice. J Med Microbiol 56: 443–453. [DOI] [PubMed] [Google Scholar]
- Brandt, U. , Raberg, M. , Voigt, B. , Hecker, M. , and Steinbuchel, A. (2012) Elevated poly(3‐hydroxybutyrate) synthesis in mutants of Ralstonia eutropha H16 defective in lipopolysaccharide biosynthesis. Appl Microbiol Biot 95: 471–483. [DOI] [PubMed] [Google Scholar]
- Chandler, C.E. , Harberts, E.M. , Pelletier, M.R. , Thaipisuttikul, I. , Jones, J.W. , Hajjar, A.M. , et al. (2020) Early evolutionary loss of the lipid A modifying enzyme PagP resulting in innate immune evasion in Yersinia pestis . Proc Natl Acad Sci 37: 22984–22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G. , and Liu, X. (2021) On the future fermentation. Microb Biotechnol 14. 10.1111/1751-7915.13674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G.Q. (2012) New challenges and opportunities for industrial biotechnology. Microb Cell Fact 11: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G.Q. , and Jiang, X.R. (2018) Next generation industrial biotechnology based on extremophilic bacteria. Curr Opin Biotechnol 50: 94–100. [DOI] [PubMed] [Google Scholar]
- Chen, G.Q. , and Kazlauskas, R. (2011) Chemical biotechnology in progress. Curr Opin Biotechnol 22: 747–748. [DOI] [PubMed] [Google Scholar]
- Clifton, L.A. , Holt, S.A. , Hughes, A.V. , Daulton, E.L. , Arunmanee, W. , Heinrich, F. , et al. (2015) An accurate in vitro model of the E. coli envelope. Angew Chem Int Ed Engl 54: 11952–11955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y. , Sun, M. , and Shaevitz, J.W. (2011) Direct measurement of cell wall stress stiffening and turgor pressure in live bacterial cells. Phys Rev Lett 107: 158101. [DOI] [PubMed] [Google Scholar]
- Dovala, D. , Rath, C.M. , Hu, Q. , Sawyer, W.S. , Shia, S. , Elling, R.A. , et al. (2016) Structure‐guided enzymology of the lipid A acyltransferase LpxM reveals a dual activity mechanism. Proc Natl Acad Sci 113: E6064–E6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak, P. , Bayer, E.A. , and de Lorenzo, V. (2020) Surface display of designer protein scaffolds on genome‐reduced strains of Pseudomonas putida . ACS Synth Biol 9: 2749–2764. [DOI] [PubMed] [Google Scholar]
- Goff, M. , Nikodinovic‐Runic, J. , and O'Connor, K.E. (2009) Characterization of temperature‐sensitive and lipopolysaccharide overproducing transposon mutants of Pseudomonas putida CA‐3 affected in PHA accumulation. Fems Microbiol Lett 292: 297–305. [DOI] [PubMed] [Google Scholar]
- Gorzelak, P. , Klein, G. , and Raina, S. (2021) Molecular basis of essentiality of early critical steps in the lipopolysaccharide biogenesis in Escherichia coli K‐12: requirement of MsbA, Cardiolipin, LpxL, LpxM and GcvB. Int J Mol Sci 22: 5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest, R.L. , Rutherford, S.T. , and Silhavy, T.J. (2020) Border control: regulating LPS biogenesis. Trends Microbiol 4: 334–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillotte, M.L. , Gillespie, J.J. , Chandler, C.E. , Rahman, M.S. , Ernst, R.K. , and Azad, A.F. (2018) Rickettsia lipid A biosynthesis utilizes the late acyltransferase LpxJ for secondary fatty acid addition. J Bacteriol 200: e00334–e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich, D. , Raberg, M. , and Steinbuchel, A. (2015) Synthesis of poly(3‐hydroxybutyrate‐co‐3‐hydroxyvalerate) from unrelated carbon sources in engineered Rhodospirillum rubrum . Fems Microbiol Lett 362: fnv038. [DOI] [PubMed] [Google Scholar]
- Konovalova, A. , Kahne, D.E. , and Silhavy, T.J. (2017) Outer membrane biogenesis. Annu Rev Microbiol 71: 539–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Garcia, E. , Fraile, S. , Rodriguez Espeso, D. , Vecchietti, D. , Bertoni, G. , and de Lorenzo, V. (2020) Naked bacterium: emerging properties of a surfome‐streamlined Pseudomonas putida strain. ACS Synth Biol 9: 2477–2492. [DOI] [PubMed] [Google Scholar]
- Mitra, R. , Xu, T. , Xiang, H. , and Han, J. (2020) Current developments on polyhydroxyalkanoates synthesis by using halophiles as a promising cell factory. Microb Cell Fact 19: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paula, F.C. , de Paula, C.B.C. , Gomez, J.G.C. , Steinbuchel, A. , and Contiero, J. (2017) Poly(3‐hydroxybutyrate‐co‐3‐hydroxyvalerate) production from biodiesel by‐product and propionic acid by mutant strains of Pandoraea sp. Biotechnol Prog 33: 1077–1084. [DOI] [PubMed] [Google Scholar]
- Pernicova, I. , Kucera, D. , Nebesarova, J. , Kalina, M. , Novackova, I. , Koller, M. , and Obruca, S. (2019) Production of polyhydroxyalkanoates on waste frying oil employing selected Halomonas strains. Bioresource Technol 292: 122028. [DOI] [PubMed] [Google Scholar]
- Qin, Q. , Ling, C. , Zhao, Y. , Yang, T. , Yin, J. , Guo, Y. , and Chen, G.Q. (2018) CRISPR/Cas9 editing genome of extremophile Halomonas spp . Metab Eng 47: 219–229. [DOI] [PubMed] [Google Scholar]
- Raetz, C.R. , Guan, Z. , Ingram, B.O. , Six, D.A. , Song, F. , Wang, X. , and Zhao, J. (2009) Discovery of new biosynthetic pathways: the lipid A story. J Lipid Res 50: S103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz, C.R. , Reynolds, C.M. , Trent, M.S. , and Bishop, R.E. (2007) Lipid A modification systems in gram‐negative bacteria. Annu Rev Biochem 76: 295–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz, C.R. , and Whitfield, C. (2002) Lipopolysaccharide endotoxins. Annu Rev Biochem 71: 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas, E.R. , Billings, G. , Odermatt, P.D. , Auer, G.K. , Zhu, L. , Miguel, A. , et al. (2018) The outer membrane is an essential load‐bearing element in Gram‐negative bacteria. Nature 559: 617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz, N. , Kahne, D. , and Silhavy, T.J. (2006) Advances in understanding bacterial outer‐membrane biogenesis. Nat Rev Microbiol 4: 57–66. [DOI] [PubMed] [Google Scholar]
- Sagong, H.Y. , Son, H.F. , Choi, S.Y. , Lee, S.Y. , and Kim, K.J. (2018) Structural insights into polyhydroxyalkanoates biosynthesis. Trends Biochem Sci 43: 790–805. [DOI] [PubMed] [Google Scholar]
- Sedlacek, P. , Slaninova, E. , Koller, M. , Nebesarova, J. , Marova, I. , Krzyzanek, V. , and Obruca, S. (2019) PHA granules help bacterial cells to preserve cell integrity when exposed to sudden osmotic imbalances. New Biotechnol 49: 129–136. [DOI] [PubMed] [Google Scholar]
- Shen, R. , Yin, J. , Ye, J.W. , Xiang, R.J. , Ning, Z.Y. , Huang, W.Z. , and Chen, G.Q. (2018) Promoter engineering for enhanced P(3HB‐co‐4HB) production by Halomonas bluephagenesis . ACS Synth Biol 7: 1897–1906. [DOI] [PubMed] [Google Scholar]
- Six, D.A. , Carty, S.M. , Guan, Z. , and Raetz, C.R. (2008) Purification and mutagenesis of LpxL, the lauroyltransferase of Escherichia coli lipid A biosynthesis. Biochemistry 47: 8623–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, K. , and Reue, K. (2009) Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am J Physiol Endocrinol Metab 296: E1195–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, D. , Wu, Q. , Chen, J.C. , and Chen, G.Q. (2014) Engineering Halomonas TD01 for the low‐cost production of polyhydroxyalkanoates. Metab Eng 26: 34–47. [DOI] [PubMed] [Google Scholar]
- Tang, W.L. , and Zhao, H. (2009) Industrial biotechnology: tools and applications. Biotechnol J 4: 1725–1739. [DOI] [PubMed] [Google Scholar]
- Trisrivirat, D. , Hughes, J.M.X. , Hoeven, R. , Faulkner, M. , Toogood, H. , Chaiyen, P. , and Scrutton, N.S. (2020) Promoter engineering for microbial bio‐alkane gas production. Synth Biol (Oxf) 5: ysaa022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.L. , Ma, W.J. , Wang, Z. , Li, Y. , and Wang, X.Y. (2014a) Construction and characterization of an Escherichia coli mutant producing Kdo(2)‐lipid A. Marine Drugs 12: 1495–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Ma, W. , Fang, Y. , Zhang, H. , Liang, H. , Li, Y. , and Wang, X. (2020) Truncating the structure of lipopolysaccharide in Escherichia coli can effectively improve poly‐3‐hydroxybutyrate production. ACS Synth Biol 9: 1201–1215. [DOI] [PubMed] [Google Scholar]
- Wang, X. , and Quinn, P.J. (2010) Lipopolysaccharide: biosynthetic pathway and structure modification. Prog Lipid Res 49: 97–107. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Quinn, P.J. , and Yan, A. (2014b) Kdo2‐lipid A: structural diversity and impact on immunopharmacology. Biol Rev 90: 408–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Qin, Q. , Zheng, Y. , Li, F. , Zhao, Y. , and Chen, G.Q. (2021) Engineering the permeability of Halomonas bluephagenesis enhanced its chassis properties. Metab Eng 67: 53–66. [DOI] [PubMed] [Google Scholar]
- Weiss, T.L. , Young, E.J. , and Ducat, D.C. (2017) A synthetic, light‐driven consortium of cyanobacteria and heterotrophic bacteria enables stable polyhydroxybutyrate production. Metab Eng 44: 236–245. [DOI] [PubMed] [Google Scholar]
- Whitfield, C. , and Trent, M.S. (2014) Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem 83: 99–128. [DOI] [PubMed] [Google Scholar]
- Wongsirichot, P. , Gonzalez‐Miquel, M. , and Winterburn, J. (2020) Integrated biorefining approach for the production of polyhydroxyalkanoates from enzymatically hydrolyzed rapeseed meal under nitrogen‐limited conditions. ACS Sustain Chem Eng 8: 8362–8372. [Google Scholar]
- Ye, J. , Hu, D. , Che, X. , Jiang, X. , Li, T. , Chen, J. , et al. (2018) Engineering of Halomonas bluephagenesis for low cost production of poly(3‐hydroxybutyrate‐co‐4‐hydroxybutyrate) from glucose. Metab Eng 47: 143–152. [DOI] [PubMed] [Google Scholar]
- Zhao, H. , Zhang, H.M. , Chen, X. , Li, T. , Wu, Q. , Ouyang, Q. , and Chen, G.Q. (2017) Novel T7‐like expression systems used for Halomonas . Metab Eng 39: 128–140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Sequences alignment of LpxL and LpxM of E. coli and H. bluephagenesis, respectively. The consensus residues are emphasized with black asterisks.
Fig. S2. Fed‐batch growth and P(3HB‐co‐4HB) production by H. bluephagenesis WZY254 in 7‐l bioreactor when γ‐butyrolactone was supplemented at early stage of fermentation. The γ‐butyrolactone was supplemented in the fed batch process started at 6 h (A) and 8 h (B), respectively.
Table S1. Strains, plasmids, and genes.
Table S2. sgRNA sequences used in this study.
Table S3. Fed‐batch growth and PHB production by H. bluephagenesis WZY254 cultured in a 7‐l bioreactor for 48 h.
Table S4. Fed‐batch growth and P(3HB‐co‐4HB) production by H. bluephagenesis WZY254 cultured in a 7‐l bioreactor for 48 h.
Data Availability Statement
The authors declare that source data processed for figure generation in this study are available within the paper and its Supporting information. Correspondence and requests for raw data should be addressed to G.Q. C. or Z.Y. W.


