Summary
Evolutionary history and early association with anthropogenic environments have made Saccharomyces cerevisiae the quintessential wine yeast. This species typically dominates any spontaneous wine fermentation and, until recently, virtually all commercially available wine starters belonged to this species. The Crabtree effect, and the ability to grow under fully anaerobic conditions, contribute decisively to their dominance in this environment. But not all strains of Saccharomyces cerevisiae are equally suitable as starter cultures. In this article, we review the physiological and genetic characteristics of S. cerevisiae wine strains, as well as the biotic and abiotic factors that have shaped them through evolution. Limited genetic diversity of this group of yeasts could be a constraint to solving the new challenges of oenology. However, research in this field has for many years been providing tools to increase this diversity, from genetic engineering and classical genetic tools to the inclusion of other yeast species in the catalogues of wine yeasts. On occasion, these less conventional species may contribute to the generation of interspecific hybrids with S. cerevisiae. Thus, our knowledge about wine strains of S. cerevisiae and other wine yeasts is constantly expanding. Over the last decades, wine yeast research has been a pillar for the modernisation of oenology, and we can be confident that yeast biotechnology will keep contributing to solving any challenges, such as climate change, that we may face in the future.
Saccharomyces cerevisiae typically dominates any spontaneous wine fermentation and. In this article, we review the physiological and genetic characteristics of S. cerevisiae wine strains, as well as the biotic and abiotic factors that have shaped them through evolution. Over the last decades, wine yeast research has been a pillar for the modernisation of oenology, and we can be confident that yeast biotechnology will keep contributing to solving any challengesthat we may face in the future.

Introduction
Alcoholic fermentation is probably the most ancient biotechnological transformation in human history. Chemical analyses of archaeological specimens in China, Iran, Egypt, or Georgia trace the production of wine and other fermented beverages to the origins of agriculture (McGovern et al., 1996, 2004, 2017). However, while we might assume that the domestication of vines and other crops was intentional, the domestication of the agents responsible for wine fermentation has been largely unconscious. The involvement of yeasts in alcoholic fermentation was not generally accepted until the XIX century (Barnett, 2003). Mastering fermentation processes using selected starter cultures is an even younger innovation in the history of winemaking (Kraus et al., 1983); and it did not become a widespread practice until the 1970s. Up to the present, almost all the industrial starters used in winemaking belong to the species Saccharomyces cerevisiae.
This species was adopted early on as a model organism by various branches of biology; microbiology, biochemistry, physiology, genetics or genomics, among others (Barnett, 1998). From the first morphological descriptions or its contribution to the germ theory, S. cerevisiae was synergistically developed both as a biotechnological workhorse and a model organism. It was also the first eukaryotic organism to be completely sequenced (Goffeau et al., 1996) and, with the popularization of NGS and whole‐genome sequencing, it is now also a model for microbial ecology, population genetics and synthetic biology (Goddard and Greig, 2015; Peter et al., 2018; Pretorius and Boeke, 2018).
Nevertheless, there are notable differences between the characteristics of the strains used in the laboratory and the industry, as well as in the growth conditions commonly found in both settings. Therefore, although wine yeast biotechnology can benefit from most of the advanced tools developed in other disciplines, knowledge transfer is not always straightforward.
In addition, it is currently recognized that other yeast species (known as non‐Saccharomyces in the field) might be very relevant for the output of wine alcoholic fermentation (Ciani et al., 2010). Only recently has this finding begun to be industrially exploited. The purpose of this article is to bring together, in a single document, the most relevant information available on the biology of wine strains of S. cerevisiae, including the most up‐to‐date information on genetic and genomic features.
The physico‐chemical environment during wine fermentation
The many styles of wine that exist in the world today differ from each other in the Vitis vinifera cultivars employed, agricultural practices, fermentation conditions, and many different pre‐ and post‐fermentation practices. From the point of view of microbial biotechnology, we must also consider secondary fermentation processes, involving yeasts or lactic acid bacteria. Anyway, most wines are produced following variants of two main workflows, one for red wines and a second one for white wines (Ribéreau‐Gayon et al., 2006). The main difference between red and white fermentation is the presence of grape seeds and skins during most of the fermentation time, for red wines. This is accompanied by physical procedures aimed at improving the extraction of compounds from the solid parts; but also providing extra oxygen for some of the yeast's metabolic pathways. In contrast, white wine is usually fermented with pressed grape must, after the first racking, and in almost completely anaerobic conditions.
The sudden availability of nutrients caused by the crushing or pressing of the grape tends to be rapidly exploited by the epiphytic microbiota in the grape berries, together with the microorganisms already present in the equipment and atmosphere of the winery. Environmental conditions quickly become anaerobic due to the microbial metabolic activity and the use of relatively large containers (low surface‐to‐volume ratios). Nutrient availability is high for sugars, but often limiting for nitrogen sources and some vitamins, like thiamine (Labuschagne and Divol, 2021); a problem that winemakers often tackle by using specific yeast nutrients. The widespread use of sulphiting agents has a great impact on yeast physiology and ecology during wine fermentation. Other factors contributing to selective growth by some yeast species at the expense of others are osmotic pressure (ever‐increasing due to global climate warming), a relatively high total acidity and low pH, or the ethanol released by fermentation. Specific fermentation styles have their own hallmark of factors potentially limiting microbial development (Novo et al., 2012), including suboptimal (low or high) temperatures, high polyphenol content, strong anaerobiosis, extremely high osmotic pressure (noble rot and ice wines), carbon dioxide overpressure (traditional sparkling wines), or aerobic growth on ethanol (biological ageing).
S. cerevisiae is well equipped to overcome many of these hurdles. For example, there are specific response mechanisms to osmotic stress. It involves the overproduction of glycerol as a compatible osmolyte and a dedicated MAP kinase cascade, the HOG pathway (Hohmann, 2015). However, tolerance to osmotic stress depends on many factors and the proper functioning of many cellular components such as mitochondria or the Golgi‐endosome system (Gonzalez et al., 2016). Similarly, tolerance to ethanol or extreme temperatures depends on several cellular systems, including lipid composition and structure of the plasma membrane, antioxidant compounds and enzymes, chaperones, trehalose biosynthesis, plasma membrane proton ATPase, or mitochondrial functions among others (Swan and Watson, 1997; Piper, 1995; Torija et al, 2003; García‐Rios et al., 2017). In addition, signalling pathways involved in the stress response show numerous examples of crosstalk and coordinated regulation (Gustin et al., 1998; Estruch, 2000). Indeed, S. cerevisiae displays an environmental stress response (ESR), which primes stressed cells to better respond to subsequent challenges through the action of the transcriptional regulators Msn2 and Msn4 (Berry and Gasch, 2008). The results of some early transcriptomic studies led several authors to define a coordinated response to fermentation stress (Rossignol et al., 2003). Marks et al. (2008), defined a fermentation stress response (FSR), involving more than two hundred genes showing a minimum of twofold positive change at the final time point. They concern many metabolic pathways. Around 20% of FSR genes are also part of the ESR, and others are common with other laboratory defined stress conditions, like nitrogen starvation, ethanol, osmotic, or oxidative stress.
Yeast ecology of spontaneous wine fermentation
Wine fermentation is a very competitive scenario for microorganisms. Despite the predominant status of S. cerevisiae, many other microorganisms are present and potentially contribute to the development of the sensory characteristics of the wines. Often neglected as spoilage microorganisms or, at most, irrelevant species, the growth of non‐Saccharomyces species during the early stages of fermentation has been known for more than a hundred years. Our current view of the succession of yeast species from vine to wine is supported by decades of observation and yeast isolation efforts by many microbiologists, recently enriched with the help of metataxonomic studies (Setati et al., 2012; Bokulich et al., 2013; Bokulich et al., 2014; Pinto et al., 2015). The dominant yeast microbiota in healthy grapes at harvest is constituted by species that will not survive the first hours of fermentation, including basidiomycetous genera like Cryptococcus or Rhodotorula, and the ascomycetous species Aureobasidium pullulans. Other ascomycetous genera, such as Hanseniaspora/Kloeckera, Candida, Pichia, Torulaspora, Kluyveromyces, Metschnikowia, or Starmerella among others, can survive longer and jointly dominate the process for several hours until S. cerevisiae takes over the alcoholic fermentation. Indeed, these species largely outnumber S. cerevisiae cells in the surface of grape berries. After the initial contact with the grape sugars, they trigger the alcoholic fermentation. However, S. cerevisiae (initially in low numbers) takes advantage of its specific adaptive traits to quickly grow and become the main yeast species from the middle, tumultuous phase of fermentation. Most often, only S. cerevisiae can be readily isolated in the final stages of fermentation. Some physiological adaptations that allow S. cerevisiae strains to thrive during wine fermentation are their preference for fermentative metabolism, their ability to grow strictly anaerobically, and their high tolerance to sulphur dioxide. Moreover, S. cerevisiae is more tolerant to heat stress than other wine yeast species. For some authors, this could also contribute to the dominance of S. cerevisiae in this environment (Goddard, 2008).
Saccharomyces wine yeast starters
For almost the entire history of oenology, spontaneous fermentation has been the only way to transform the must into wine, modulated only by the eventual preparation of a "pied‐de‐cuve". This practice consists of adding a proportionally small volume of fermenting juice to a tank of fresh grape must. Spontaneous fermentation is a technically and conceptually very simple process, which often results in good quality wines. The microbiological control of the process used to be entrusted to sulphur dioxide (equally acting as an antioxidant), in concentrations that inhibit the development of most bacteria and non‐Saccharomyces yeasts. However, this is not enough to prevent fermentation from sometimes going bad, leading to slow or stuck fermentation, excess acidity, or other sensory defects. On the other hand, the variability of the microbiota in the vineyard and cellar introduces a factor of unpredictability into the sensory characteristics of the wines produced each season. The possibilities opened by the discovery of yeasts as transformation agents, the development of pure cultures and, later on, the development of industrial yeast production techniques (especially for bakery and brewery use), did not begin to be exploited by the wine industry until many years later (Gonzalez et al., 2011). Nevertheless, by the end of the 20th‐century inoculation of S. cerevisiae starters was a widespread practice, with dozens of different yeast strains on the market. While in spontaneous fermentation a succession of different yeasts takes place, the use of S. cerevisiae starter cultures usually results in a process dominated almost from the onset by the inoculated strain (Querol et al., 1992). Each strain of S. cerevisiae can contribute differently to the sensory characteristics of the wine, and not all of them are well adapted to all fermentation styles. Accordingly, yeast selection becomes an additional tool in the hands of winemakers to develop and differentiate their products. But the widespread assumption that the best starter cultures for a production region are those isolated from such region is lacking solid scientific support.
A minimal selection criterion for wine yeast starters is good fermentation kinetics, understood as fast fermentation onset and quick sugar consumption kinetics. These traits are directly related to tolerance to osmotic and ethanol stress, and efficiency in the use of available nitrogen sources. Indeed, a large proportion of sugar consumption and alcohol production during wine fermentation takes place in the absence of cell growth. The biomass produced during the initial growth phase must be sufficient to ensure correct fermentation kinetics, and this is highly dependent on nitrogen utilisation (Varela et al., 2004). Indeed, efficient nitrogen utilization is a selection criterion for wine yeast starters, despite the widespread use of nutrient supplementation. S. cerevisiae has developed an exquisite nitrogen source selection system (NCR, for nitrogen catabolite repression), involving nutrient sensing and different transcriptional and post‐transcriptional control mechanisms (Zhang et al., 2018). Nitrogen metabolism by yeast during wine fermentation has a relevant impact on the production of several sensory active molecules, including acetic acid, higher alcohols, and esters (Martínez‐Moreno et al., 2012; Styger et al., 2013; Rollero et al., 2017). It is also worth noting that NCR functioning under laboratory conditions could be very different from oenological fermentation conditions (Vallejo et al., 2020a; Vallejo et al., 2020b).
The killer phenotype is often also considered for starter selection (see below). Similarly, although most strains of wine origin show specific adaptations, it is advised to check the tolerance of candidate strains to sulphur dioxide. Furthermore, winemakers look for yeast starters that show tolerance to high or low temperatures (depending on the specific application); low production of volatile acidity (acetic acid), SO2, or SH2; and, in general, starters that do not contribute any off‐flavours or negatively impact the perception of the wine. Indeed, tasting panels are unavoidable before new wine yeast starters can make their way to the market.
The selection criteria shall not forget that wine yeast starters must be produced and distributed as active dry yeast (ADY). There are other alternatives, primarily for local markets (Fracassetti et al., 2020), but ADY can be produced throughout the year and stored stably until harvest time (typically distributed with a two‐year self‐life), which is a great advantage for a product whose consumption is inherently seasonal. To produce ADY, yeast is usually grown in a fed‐batch process, using diluted molasses as a carbon source, under conditions that favour respiratory metabolism (to maximize biomass yield). The resulting biomass is then dehydrated by a combination of mechanical and thermal processes (Gonzalez et al., 2011). In the cellar, the ADY must be rehydrated to recover viable and metabolically active cells, in a process that is again stressful and deadly for them. Accordingly, the suitability of the selected yeast strains as industrial wine starters depends not only on their behaviour during wine fermentation, but also on their performance during the ADY production process (biomass yield on a substrate, tolerance to thermal and hydric stress), and rehydration (Matallana and Aranda, 2017; Rodriguez‐Porrata et al., 2008).
Non‐Saccharomyces starters
The notable predominance of S. cerevisiae in the inoculated fermentations, and the extensive use of some commercial strains, prompted oenologists and researchers to try to avoid a perceived tendency towards uniformity in the wines. Despite various precedents during the 20th century, it was not until the 21st century that this began to take hold. Currently, commercial non‐Saccharomyces starter cultures are gradually gaining market share (Roudil et al., 2020). The use of non‐Saccharomyces yeasts allows winemakers to recover some of the characteristics associated with spontaneous fermentation while maintaining microbiological control of it. The main recognised impact of non‐Saccharomyces yeasts on wine quality is related to the aroma (Padilla et al., 2016). These yeasts can contribute to the enhancement of the primary aroma of wines, allowing the release of active molecules from precursors present in the must, especially through the production of enzymes with glycosidase activity. Many contributions to the primary aroma, including terpenes and aromatic alcohols, come from molecules mostly present in grapes as odour‐inactive glycosidically‐bound volatile precursors (Hjelmeland and Ebeler, 2015). An important impact on wine aroma is the release of varietal thiols (polyfunctional mercaptans), contributing to the characteristic fruity aroma of wines from Sauvignon Blanc and other aromatic white grape varieties. This depends on a β‐lyase activity (Roncoroni et al., 2011). They also contribute to the complexity of secondary aromas, mainly through the production of aromatic alcohols and esters which add fruity notes (Rojas et al., 2001). Beyond the aroma, the genetic and metabolic diversity provided by this portfolio of yeast species has enabled other oenological applications to be considered, such as acidification or deacidification, higher glycerol or mannoprotein content, better colour stability, lower alcohol content (Charoenchai et al., 1997; Strauss et al., 2001; Rojas et al., 2003; Romano et al., 2003; Bely et al., 2008; Moreira et al., 2008; Viana et al 2008; Ciani et al., 2010; Manzanares et al., 2011; Viana et al., 2011; Gonzalez et al., 2013) or the biological control of spoilage microorganisms (Oro et al., 2014). As with S. cerevisiae, it should be noted that the actual metabolic and technological profile depends not only on the yeast species but also on the strain used.
Saccharomyces spp. are almost the only species in wine able of consuming all the sugars with suitable fermentation kinetics. This prevents stuck fermentation or the predominance of potential spoilage microorganisms. For this reason, alternative yeasts are most often used in combination with S. cerevisiae starter cultures, either sequentially or by simultaneous inoculation. From the point of view of process control, each of these alternatives presents its own challenges. Indeed, the interactions between different starter cultures might be critical for a successful fermentation and represent an interesting field of study (Ciani et al., 2010).
The first commercial wine starter cultures of alternative species belonged to Torulaspora delbrueckii, and this is still the non‐Saccharomyces yeast species with the largest number of wine starters on the market. Schizosaccharomyces pombe is nowadays marketed for deacidifying must by means of malo‐alcoholic fermentation. Other popular non‐Saccharomyces species in wine yeast catalogues are Pichia kluyveri, when looking for improved secondary aroma; Lachancea thermotolerans (formerly Kluyveromyces thermotolerans), attending to lactic acid production; or Metschnikowia pulcherrima, for aroma and biocontrol (Roudil et al., 2020). Although Hanseniaspora isolates have been often related to excessive volatile acidity, Hanseniaspora vineae strains have been extensively studied as a potential wine starter for improved secondary aroma (Martin et al., 2018).
The physiology of S. cerevisiae wine yeast strains
Channelling most of the carbon flux towards fermentative ethanol production, independently of oxygen availability, is a metabolic signature of some yeast species. This is known as the Crabtree effect, and S. cerevisiae is the most prominent representative of the Crabtree‐positive group. Several mechanisms contribute to the Crabtree effect in this species (Barnett and Entian, 2005), including carbon catabolite transcriptional repression of genes required for aerobic respiration. But the rate of sugar consumption plays a critical role since slowing down sugar uptake or a reduced capacity of glycolytic enzymes alleviate the Crabtree effect (Otterstedt et al., 2004; Jansen et al., 2005). High sugar uptake rates result in overflow metabolism at the pyruvate level (Holzer, 1961; Pronk, et al., 1996). The abundance and kinetic properties of mitochondrial pyruvate dehydrogenase complex are not enough to process all the pyruvate produced by glycolysis. Under these circumstances most of the pyruvate is metabolized to acetaldehyde by the activity of pyruvate decarboxylase, and then to ethanol by acetaldehyde dehydrogenase (Pronk, et al., 1996). The Crabtree effect is part of a combination of evolutionary adaptations that constitute a “make‐accumulate‐consume” strategy followed by several yeast lineages (Piskur et al., 2006). Some of the genetic traits that contribute to the Crabtree‐positive character include duplication and specialization of alcohol dehydrogenase coding genes, multiple hexose transporters, or transcriptional control of respiratory and mitochondrial functions (Hagman et al., 2013). Although many yeast species are facultative anaerobes, only a few of them can support a sustained growth under anaerobic conditions. The ability for anaerobic pyrimidine biosynthesis is key to support anaerobic growth. Most eukaryotic dihydroorotate dehydrogenases, catalysing the fourth step of de novo pyrimidine biosynthesis (oxidation of dihydroorotate to orotate), are dependent on enzymes from the respiratory chain for their activity. However, some Saccharomycetaceae acquired a bacterial dihydroorotate dehydrogenase by horizontal gene transfer (HGT) (Hall et al., 2005), using fumarate (which is independent of a functional respiratory chain) as electron acceptor (Gojković et al., 2004). Biosynthesis of unsaturated fatty acids and sterols required by yeast cells is also oxygen dependent. However, the oxygen requirement, in this case, is lower (there is no need for an active electron transport chain); and wine yeasts can incorporate lipids from grape must or winemaking nutrients (Luparia et al., 2004). While the Crabtree effect seems to clearly confer a selective advantage to S. cerevisiae over most other microorganisms during alcoholic fermentation, it poses a problem when producing biomass for ADY preparation. In yeast production, the aim is to maximise the biomass yield. In a batch process, because of the Crabtree effect, yeasts would produce large amounts of ethanol at the expense of biomass. For this reason, yeast production is mostly carried out in fed‐batch cultures, adjusting the substrate supply to yeast uptake, thus ensuring low sugar levels throughout the growth phase to minimise the Crabtree effect (Gonzalez et al., 2011).
The above‐described features of S. cerevisiae seem to be common to most strains of the species, but sulphur dioxide tolerance is restricted to some strains, including most wine isolates. Chromosome rearrangements involving the promoter of SSU1 (coding for a sulphite efflux pump), and resulting in higher levels of the Ssu1 permease, are usually responsible for the enhanced sulphite resistance shown by wine yeast strains (Goto‐Yamamoto et al., 1998; Pérez‐Ortín et al., 2002; Zimmer et al., 2014; García‐Ríos et al., 2019).
Fermentation does more to grape must than just replacing sugars with alcohol, although ethanol does contribute to what is known as the wine's aromatic buffer (Escudero et al., 2004). The microorganisms involved in fermentation, and especially S. cerevisiae catalyse the transformation of some precursors of the primary aroma of grapes such as glycosylated precursors and polyfunctional mercaptans (Swiegers and Pretorius, 2007; Concejero et al., 2016); and above all, they release various volatile organic compounds that constitute the secondary aroma. These include fusel alcohols, which are related to amino acid metabolism via the Ehrlich pathway (Styger et al., 2013), as well as acetate and ethyl esters (Sumby et al., 2010). Glycerol, organic acids (including acetic acid), acetaldehyde, mannoproteins, SH2 and SO2 are further contributions of yeast metabolism to the plethora of compounds involved in the sensory complexity of wine (Styger et al., 2011). There is sufficient metabolic diversity among wine strains of S. cerevisiae for their differential contribution to wine quality to be appreciated by professionals and consumers, regardless of their relative genetic proximity. This justifies the high number of S. cerevisiae strains on the winemaking market, despite some redundancy in trade names (Fernández‐Espinar et al., 2001; Borneman et al., 2016).
S.cerevisiae life cycle
S. cerevisiae is often referred to as "the budding yeast", even though this is the mode of vegetative growth for many other yeast species. Reproduction by budding makes it possible to distinguish mother cells from daughter cells. Mother cells have a finite replicative lifespan which depends on the genotype and the environment, but which is typically only a few tens of divisions (Austriaco, 1996). S. cerevisiae cells can proliferate vegetatively both in haploid and diploid form. However, the dominant form in nature is diploid, and no commercial haploid oenological strains are available. Budding is genetically regulated, and the selection of the budding site depends on the history and mating genotype of the cell (Madden and Snyder, 1998). In addition to the well‐known unicellular ellipsoid form, S. cerevisiae can develop other morphologies (Voordeckers et al., 2012). Adhesins, or flocculins, encoded by the FLO family of genes play determinant roles in growth patterns of S. cerevisiae, as well as on technologically relevant aggregation phenotypes like vellum formation during sherry wine ageing (Fidalgo et al., 2006), or flocculation (Govender et al., 2010).
Under suitable conditions, typically involving starvation, diploid cells might experience meiosis, giving rise to an ascus with a tetrad of ascospores, two of them with an a, and two with an alpha mating‐type. In homothallic strains, which are the most common in nature, a haploid mother cell undergoes a mating‐type switch (starting from the second cell division) after each mitotic division (Haber, 2012). Thus, almost immediately after germination, a new homozygous diploid strain is generated by the fusion of two genetically identical cells. This process is known as haplo‐selfing. In addition, cells can revert to the diploid state by automixis, or hybridisation between spores within the ascus; and amphimixis, or hybridisation between unrelated haploid cells (Knop, 2006). Mating‐type switching is promoted by a DNA double‐strand break catalysed by the HO endonuclease on the active mating‐type locus (MAT), located on chromosome III. After each mitotic division, a different silent mating‐type cassette (HMR or HML), located near one of the two edges of the same chromosome, is used as a template to generate the alternative mating‐type (Haber, 2012).
Many laboratory strains are heterothallic, i.e. they can multiply indefinitely as haploids, due to loss‐of‐function mutations in the ho gene. Some wine strains are heterozygous for the HO/ho locus, and this has provided tools for generating haploid wine yeast derivatives as research tools (Mangado et al., 2018). Conservation of mating‐type switching is in agreement with the “genome renewal” hypothesis for wine yeasts, which will favour homozygote diploids (Mortimer et al., 1994). Experimental evolution results also suggest diploidy as the most stable nuclear content for S. cerevisiae (Gerstein et al., 2006; Mangado et al., 2018). However, the current picture of S. cerevisiae wine yeast strains in nature is more complex than previously anticipated (Fischer et al., 2021). Indeed, one of the most popular industrial wine yeast strains, EC1118, is clearly heterozygous (Muñoz et al., 2009; Novo et al., 2009). There is evidence of mitotic recombination in wine yeasts, but sporulation events seem to be rare under winemaking conditions (Puig et al., 2000). In this context, it is interesting to note the recent discovery that the social wasp intestine is an environment that allows winter survival and hybridisation between yeast strains (Stefanini et al., 2016). Furthermore, S. cerevisiae can form interspecific hybrids with other Saccharomyces species. Hybrids are common for brewer’s yeasts (Libkind et al., 2011) but were discovered more recently for wine yeasts (see below).
Genetic and genomic features of S. cerevisiae wine strains
The haploid genome of S. cerevisiae contains about 12 Mbp and is distributed over 16 chromosomes. The number of protein‐coding genes is about 6000 (Goffeau et al., 1996). The current genome of S. cerevisiae (and a small set of yeast species known as post‐WGD) is the result of a genome duplication event (WGD for whole‐genome duplication) in an ancestral species, which resulted in a tetraploid cell (Wolfe and Shields, 1997). Most of the gene redundancy was subsequently lost, but still about 13% of the proteins of this species constitute pairs derived from this ancient duplication (Wolfe and Shields, 1997). In many cases, this event seems to have allowed the specialisation of at least one of the copies (Kellis et al., 2004). This duplication also seems to underlie an increase in glycolytic flux, which would eventually lead to the Crabtree effect (Conant and Wolfe, 2007; Hagman et al., 2013).
Recent findings suggest the all the Saccharomyces species originated in Asia, with a single out‐of‐China event as the origin of all non‐Chinese S. cerevisiae strains (Peter et al., 2018). There is a strong genetic relationship among wine isolates, which fall in a Wine/European clade on one side of the S. cerevisiae phylogenetic tree. Comparative genomics also indicates that current wine isolates are monophyletic because of a population bottleneck during the domestication process (Borneman et al., 2016; Peter et al., 2018). Although the species can show variation in ploidy as well as aneuploidies, most wine yeast isolates are pure diploids. There are also a few haploid strains, with very few examples of higher ploidy. All those haploid strains are deleted for the HO locus. The authors also found several aneuploid strains (around 15% of wine isolates), most of them carrying an extra copy of a single chromosome (Peter et al., 2018). A summary of the main genomic characteristics of wine yeasts, according to data from Peter et al. (2018) is shown in Table 1.
Table 1.
Main genomic features of wine strains of S. cerevisiae. Information summarised from the supplementary materials of Peter et al. (2018).
| Genomic feature | Number of strains | |
|---|---|---|
| HO deletion | 27 | |
| Plasmid | A | 204 |
| B | 1 | |
| Ploidy | 1 (euploid/homozygous) | 28 (20/28) |
| 2 (euploid/homozygous) | 199 (175/115) | |
| 3 (euploid/homozygous) | 2 (1/0) | |
| 4 (euploid/homozygous) | 1 (0/1) | |
| Copy number | ||
| Plasmid | A (if present) | 1 to 210 (median 25) |
| Extra ORFs* | HGT regions (A, B or C) | Up to 30 (app.) |
| S. paradoxus introgression | 25–50 | |
| Ty copies* | Ty1 | Up to 15 (app.) |
| Ty2 | Up to 30 (app.) | |
| Ty3–5 | <10 each | |
Whenever possible, it refers to 229 strains labelled as “wine” for the “ecological origins” feature in that work. Otherwise, it refers to the 362 Wine/European clade strains used (*).
Pan‐genome analysis revealed more than 900 ORFs introgressed from S. paradoxus, which is the closest relative of S. cerevisiae. Most wine strains carry between 25 and 50 ORFs from this origin (Table 1). In addition, several contributions to the S. cerevisiae pan‐genome from evolutionarily more distant species have been identified. The earliest examples were found in the wine strain EC1118, with three regions named A, B, and C (Novo et al., 2009). The source species of these fragments are, as far as known, Zygosaccharomyces bailii, Torulaspora microellipsoides, and Torulaspora delbrueckii (Novo et al., 2009; Marsit et al., 2015). In some cases, a correlation between HGT‐acquired genes and specific adaptations to fermentation conditions, such as the limitation of nitrogen sources, has been established (Devia et al., 2020). FSY1, from region C, encodes a high‐affinity active fructose transporter, whose expression is induced by ethanol, believed to confer a selective advantage in the final stages of wine fermentation (Galeote et al., 2010).
Many S. cerevisiae strains carry an extrachromosomal element, the 2‐micron plasmid. This is a 6.3 kbp circular DNA with only four coding genes, involved in replication, plasmid maintenance, and copy number control (Chan et al., 2013). Three classes of this plasmid have been identified, A, C, and B, which is the result of recombination between the other two (Strope et al., 2015). Almost all wine strains harbour a variable number of copies of the type A 2‐micron plasmid (Table 1).
In S. cerevisiae spontaneous petit (respiration‐deficient), or rho0 (having lost the entire mitochondrial genome) mutants are found with some ease. However, mitochondria are involved in many essential functions of the cell (McBride et al., 2006), and although the genes encoding most of the mitochondrial proteins have been transferred to the nucleus of S. cerevisiae during evolution (Karlberg et al., 2000), the long‐term survival of these mutants is compromised. They are also sporulation deficient since respiration is required for meiotic entry (Jambhekar and Amon, 2008). The mitochondrial genome of S. cerevisiae is about 86 kbp, is globally low in %GC, and has a large proportion of intergenic regions and group I and II introns (Foury et al., 1998). Most of the genes it encodes are related to oxidative phosphorylation (Freel et al., 2015). The mitochondrial genome has been related in wine yeasts to several characteristics of technological interest, such as tolerance to ethanol and high temperature (Jimenez and Benitez, 1988), cold temperature (Li et al., 2019), or industrial drying to produce ADY (Picazo et al., 2014). Interestingly, the difference in composition of the mitochondrial and nuclear genomes of S. cerevisiae has allowed the development of quick molecular fingerprinting systems for wine strains of this species (Querol et al., 1992).
The killer phenotype in S. cerevisiae is due to the production of toxins against neighbouring microorganisms, mainly yeasts, with different mechanisms (Schmitt and Breinig, 2006). Although they were discovered in S. cerevisiae, it seems that killer toxins are common among different yeast species (Liu et al., 2015). From the four types of killer toxins known for S. cerevisiae, K1, K2, K28 and Klus, (Schmitt and Breinig, 2006; Rodríguez‐Cousiño et al., 2011), mostly K2 and Klus have been found in wine isolates (Maqueda et al., 2012). Indeed, the pH interval of K1 toxin activity falls outside the pH range (around 3.5) of grape juices and wines (van Vuuren and Jacobs, 1992). By the end in industrial wine fermentations prevalence of killer activity ranges from 0% to 100% (van Vuuren and Jacobs, 1992). Toxin production is due to infection by dsRNA viruses of the Totiviridae family, present in the cytoplasm as virus‐like particles. Each killer strain carries a variant of the helper virus ScV‐L‐A, and one of the satellite viruses, specific for each of the known toxins: ScV‐M1, ScV‐M2, ScV‐M28 and ScV‐Mlus. The former encodes the capsid protein and the polymerase, while the latter encodes just for the toxin. Transmission of these viruses is vertical (mother to daughter cell) and through cell fusion (e.g. in mating). There is no known extracellular pathway of killer virus transmission. Apart from the competitive advantage over killer sensitive strains, the presence of these viruses is considered to be asymptomatic. However, recent studies indicate that a specific adaptation to the production of the toxin itself is required (Gier et al., 2020). The exact mechanisms of immunity remain to be elucidated.
Among the non‐Mendelian heritable elements in S. cerevisiae, we also find prion‐like elements (Wickner et al., 2015) and the [GAR+ ] element, one of the most recently discovered, raised expectations for its possible involvement in the adaptation of S. cerevisiae to alcoholic fermentation and interactions with bacteria present during winemaking (Brown and Lindquist, 2009; Jarosz et al., 2014a). This element results in an alleviation of catabolite repression, which according to some authors could allow other carbon sources to be used during fermentation (apart from glucose and fructose) and may even lower alcohol production (Jarosz et al., 2014b). But other authors concluded that the technological relevance of these yeast prions is rather residual and found no evidence of an appreciable decrease in alcoholic strength associated with the use of [GAR+ ] strains (Gonzalez et al., 2019).
The S. cerevisiae genome also harbours long terminal repeat‐retrotransposons from five different families, Ty1 to Ty5 (Carr et al., 2012), with known active elements for at least the first three types. The direct terminal repeats of Ty1 and Ty2 are known as delta elements, and can also be found in isolation, probably because of homologous recombination during ancient transposition events (Curcio et al., 2015). Delta elements have been used to develop molecular typing tools for S. cerevisiae wine strains (Legras and Karst, 2003).
With very few exceptions, such as translocations associated with the SSU1 promoter and their impact on sulphur dioxide tolerance, many traits of technological interest in wine yeasts are quantitative and therefore difficult to deal with from a genetic point of view. In these cases, quantitative trait loci (QTL) mapping is a powerful but traditionally cumbersome tool. The possibility of bulk segregant analysis thanks to the popularisation, initially of genomic microarrays (Marullo et al., 2007), and subsequently of whole‐genome sequencing and mass sequencing experiments, has changed the picture in recent years. Some authors have identified QTLs in crosses between two oenological yeasts, while in other cases the crosses also involve yeasts from other origins, to increase genetic diversity and to be able to identify QTLs that show little variability between wine yeasts, in part because of their relevance to thrive in the wine environment. Some authors have gone up to F13, to increase the resolution of the QTL mapping (García‐Ríos et al., 2017). Examples of QTLs identified in recent years include traits of oenological importance such as acetic acid production (Marullo et al., 2007), several fermentation performance parameters (Ambroset et al., 2011; Marullo et al., 2019), production of various aroma components (Steyer et al., 2012; Eder et al., 2018), stress tolerance (Brion et al., 2013), nitrogen requirement (Brice et al., 2014; Cubillos et al., 2017), dehydration (Lopez‐Martinez et al., 2015), or low‐temperature fermentation (García‐Ríos et al., 2017).
Other Saccharomyces species and natural hybrids in winemaking
Although most wine starters belong to the species S. cerevisiae, S. uvarum, previously known as S. bayanus var. uvarum (Naumov, 2000) has traditionally attracted much interest due to its ability to ferment at low temperatures (Masneuf‐Pomarède et al., 2010). Among the cryophilic Saccharomyces species, we also find the more recently described Saccharomyces kudriavzevii (Naumov et al., 2000). Unlike S. uvarum, this species has not been found spontaneously in winemaking processes. Higher glycerol production and lower alcohol yields have been reported for strains of both species, as well as a differential contribution to the aromatic profile of wines (Pérez‐Torrado et al., 2018). Both form natural hybrids with S. cerevisiae, which have been found in wine, especially in low‐temperature fermentations (González et al., 2006; Boynton and Greig, 2014; Peris et al., 2018). These hybrids often combine the ethanol tolerance of S. cerevisiae with the low‐temperature adaptation of S. uvarum (Querol et al., 2018). In addition to the metabolic particularities of the cryophilic strains, fermentation at low temperatures, especially white wines, allows better retention of volatile compounds and thus more aromatic wines. The combination of characteristics of interspecific hybrids within the genus Saccharomyces has increased the interest in using some of them as starter cultures (Pérez‐Torrado et al., 2017). This has also prompted the development of breeding programmes relying on interspecific hybridisation (see below).
Genetic improvement of wine yeasts
Since the use of starter cultures became established as a common practice, the major source of starter improvement in oenology has been the natural diversity of the S. cerevisiae species. Despite the relative genetic homogeneity of wine strains, selection processes have allowed the development of starter cultures particularly suitable for different sensory profiles (neutral, terpenic, thiolic, and so on), and winemaking styles (white, red, young, aged, sparkling, and so on). However, genomic analysis of the strains currently on the market indicates that a saturation point has been virtually attained (Borneman et al., 2016).
Interest in the genetic improvement of previously selected wine strains began around the 1990s, driven by the rise of genetic engineering (Cebollero et al., 2007). However, the use of genetically modified organisms in wine production is unlikely to be commercially successful in the short term. The two recombinant strains of S. cerevisiae for oenological use that have come onto the market (mainly in the USA and Canada, due to regulatory issues in other countries) did not seem commercially successful, although reliable information on this issue is difficult to obtain. The first of these commercial strains catalyses the biotransformation of malic acid into lactic acid (Husnik et al., 2006). This frees winemakers from the uncertainty of malolactic fermentation, a process traditionally more difficult to control than alcoholic fermentation. The second one helps reduce the amount of urea released by yeasts during arginine metabolism (Coulon et al., 2006). Over time, excess urea reacts with ethanol leading to the formation of ethyl carbamate (Monteiro et al., 1989), considered a potential carcinogen. However, the control of malolactic fermentation, using bacterial starters, has steadily improved for the last decades; while the ethyl carbamate problem appears to be only occasional. The relatively low relevance of the problems they were meant to solve, together with marketing considerations and the limited genetic diversity available (one strain in each case), would explain the low impact of recombinant strains in real oenology.
Reducing alcohol yield is an increasingly common goal in oenological yeast biotechnology (Ciani et al., 2016; Dequin et al., 2017) and was first addressed in the 1990s by genetic engineering approaches, trying to divert carbon flux towards glycerol production or other metabolic endpoints (Dequin and Barre, 1994; Michnick et al., 1997; Cambon et al., 2006). Other authors addressed the expression of extracellular hydrolytic enzymes from different origins, mostly to help the release of primary aroma compounds from odourless precursors and improve mechanical properties of the juice and mass to ease extraction processing (Pérez‐González et al., 1993; Ganga et al., 1999). Another common type of genetic modification was changing the expression levels of S. cerevisiae genes to achieve improvements in secondary aroma, autolysis, or mannoprotein release (Lilly et al., 2000; Lilly et al., 2006; Tabera et al., 2006; Cebollero et al., 2009; González‐Ramos et al., 2009). But none of these efforts directly led to new commercial starter cultures. However, it is worth recognising that the generation of recombinant strains has allowed important advances in the knowledge of wine yeast physiology and the genetic determinants of some industrially relevant traits. Moreover, the information provided by these projects served as a guide to address genetic improvement processes using more conventional methods, some of which have resulted in the development of new commercial strains.
The quest for genetically improved wine strains free of the commercial and regulatory constraints of GMOs led to a burst of breeding efforts using conventional methods. They had been barely explored in this field before the 2000s. These include random mutagenesis, intra‐ and interspecific hybridisation, or adaptive laboratory evolution (ALE), also known as experimental evolution. Besides technical ease, the choice between these technologies must take into account the degree of control over the modification (at the genome sequence), the genetic determination of the trait, and the genetic variability that can be explored in each case (Fig. 1).
Fig. 1.
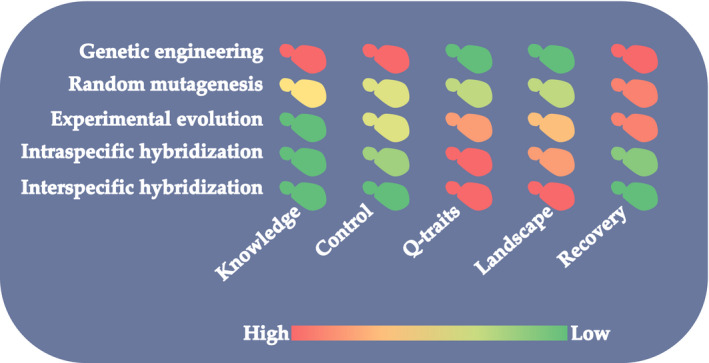
Main characteristics of different technologies available for the genetic improvement of wine yeasts. Knowledge: required level of knowledge about the genetic determination of the trait of interest. Control: degree of control of the technology at the DNA sequence level. Q‐traits: utility for quantitative trait improvement. Landscape: the range of genotypes and phenotypes that could be reached. Recovery: ease of recovering the traits of industrial interest from the original strain.
Induction of random mutagenesis, by physical or chemical agents, and phenotypic selection of mutants is one of the classical breeding techniques for industrial microorganisms. In S. cerevisiae, the most common mutagens are ethyl methane sulfonate, nitrosoguanidine, and UV radiation. Despite their apparent simplicity, the traits to be improved are not always easy to select phenotypically. There are some classic examples collected in a review by Snow (1983), and more recent examples of improvement, by indirect selection, of nitrogen source utilisation (Salmon and Barre, 1998; Long et al., 2018), mannoprotein release (Quirós et al., 2010), reduction of SH2 and SO2 production (Cordente et al., 2009; Walker et al., 2021), or reduction of volatile acidity (Cordente et al., 2013). Direct mutant selection has been used to improve autolysis (Gonzalez et al., 2003; Giovani and Rosi, 2007), or the release of mannoproteins (Gonzalez‐Ramos et al., 2010). Diploidy of wine strains does not favour this approach, which in principle would allow the recovery of dominant or semi‐dominant mutations, but hardly recessive mutations. However, Hashimoto et al. (2005) were able to obtain auxotroph mutants by UV mutagenesis of diploid sake strains; and other authors have been able to obtain spontaneous auxotrophic mutants, using positive selection strategies, from S. cerevisiae wine yeast strains (Pérez‐Través et al., 2012). Auxotrophic strains are very useful for hybrid strain construction, as described below. Furthermore, the connections between amino acid metabolism and the biosynthesis of several secondary aroma compounds (Rollero et al., 2017), opens possibilities for genetic improvement by the selection of auxotrophic mutants. Random mutagenesis can be useful when the trait to be improved depends on one or two genes but may be limited for improving quantitative traits that show a continuous variation within the species.
Sexual hybridisation is an alternative to generate diversity on a genomic scale and is better suited for quantitative traits. Moreover, since there is no prezygotic isolation within the genus, it allows combining genomes of different species of Saccharomyces closely related genera (Santos et al., 2008; Su et al., 2019). Improvement of wine yeasts has sometimes been achieved by obtaining homozygous derivatives from homothallic strains (Ramírez et al., 1999), but the opposite output is also possible (Gimeno‐Alcañiz and Matallana, 2001).
Genetic improvement by hybridisation can be approached by means of spore‐spore crosses, rare mating and mass mating. In the first case, microdissection is used to position together pairs of haploid spores from the original strains. In this way, it is possible to obtain hybrids without selection markers such as auxotrophies or other directly selectable phenotypes, although molecular markers are still required to verify the hybrid nature of the primary zygotes. This technique has been used, for example, to obtain interspecific hybrids suitable for restarting stuck fermentations (Santos et al., 2008), or better adapted to low temperatures and secondary aroma (Kishimoto, 1994); as well as intraspecific hybrids with better fermentative characteristics and mannoprotein production (Pérez‐Través et al., 2015), or improved thermotolerance (Marullo et al., 2009). For rare and mass mating, a selective growth medium is required, in which only the true hybrid strains will grow (Ramirez et al., 1998). Therefore, a first step in the breeding programme is usually to obtain spontaneous auxotrophic derivatives, by some positive selection method such as tolerance to alpha‐aminoadipic acid or fluorotic acid, to obtain respectively auxotrophic strains for lysine or uridine (Pérez‐Través et al., 2012). Spontaneous tolerance to antibiotics or killer factors (Ramirez et al., 1998), ability to use certain carbon sources, or to grow under particular conditions (e.g. thermotolerance) can also be used as hybridisation markers. For mass mating, spores from the two original strains are combined in large numbers on the same medium and allowed to mate. This approach has been used, for example, to obtain interspecific (Bizaj et al., 2012) or intraspecific (Agarbati et al., 2020) hybrids with better aromatic characteristics, especially in relation to hydrogen sulphide production. Rare mating uses vegetative cells and is not dependent on sporulation. With a very low frequency, diploid cells can eventually undergo mating‐type switching and conjugate with other cells. A powerful selection method is required to recover those hybrids. Bellon et al. (2011, 2013) were able to generate wine yeast strains with improved aroma profile by interspecific rare mating. In turn, Pérez‐Través et al. (2015) used intraspecific rare mating to improve mannoprotein release. Another way of combining genomes is the artificial fusion of protoplasts, but in this case, improved strains would fall under GMO regulations according to European legislation. Primary zygotes from spore‐spore and mass‐mating crosses can be relatively stable, depending on the genomic compatibility of the original strains. But allotetraploid strains that are often generated by protoplast fusion or rare mating must become stable through recombination and chromosome loss (Sipiczki, 2008; Pérez‐Través et al., 2012). The advantages of sexual hybridisation are that strains with traits derived from both parental strains can be obtained. In addition, transgressive phenotypes can be obtained for quantitative traits (Marullo et al., 2006). However, it is more difficult to ensure that all the original traits of the parental strain are retained, unless engaging in multiple backcrossing cycles. Availability of genetic markers, like microsatellites or those derived from QTL mapping of the target traits, would be invaluable in those cases (Marullo et al., 2007, 2009).
Adaptive laboratory evolution (ALE) involves maintaining an industrial strain over many generations under a carefully designed selective pressure. The principle is the same as for natural evolution: among the variants that arise spontaneously in each generation, those that leave more offspring will increase their relative frequency in the population, while the negative variants will become replaced after a few generations. To avoid unwanted genetic drift, the evolutionary conditions should mimic those under which the strains are meant to be used, but this is not always feasible. Design is relatively straightforward to improve some traits, such as ethanol tolerance, as done by Novo et al. (2014). But in other cases, indirect selection strategies are required. For example, Tilloy et al. (2014) used osmotic stress as a proxy to select strains with higher glycerol production. ALE shares advantages and limitations with hybridisation and mutagenesis. Like hybridisation, it can be used to improve quantitative traits, as it potentially affects the whole genome, although it does not generate all the variability of a genetic cross, nor does it allow overcoming the species barrier. On the other hand, it shares with mutagenesis a greater potential to preserve the original traits of the strain. However, good design of experimental conditions and final verification of the improved strains are necessary to ensure the recovery of the crucial traits of the original strain. A summary of the merits and drawbacks of the main tools available for the genetic improvement of wine yeasts is shown in Fig. 1.
This revival of traditional methods is not a simple step backwards, as the progress of knowledge and analytical tools for yeast biotechnology, including high‐throughput phenotyping and genotyping tools, renders techniques that were very laborious a few decades ago much easier nowadays. Despite the loss in precision, as compared to genetic engineering, these techniques may be superior when it comes to improving quantitative traits or those for which the genetic basis are not fully elucidated (Fig. 1). For example, in the case of ALE, improved strains may have accumulated mutations in genes not previously linked to that phenotype, due to metabolic trade‐offs or genetic drift.
Finally, some authors see considerable potential in new genome edition techniques (CRISPR/Cas‐based) and synthetic biology as tools for the genetic improvement of wine yeasts in the future (Pretorius, 2017; Vigentini et al., 2017). Tools based on CRISPR/Cas will help solve some of the bottlenecks of conventional genetic engineering, like stacking multiple genetic modifications, or targeting simultaneously all homologous alleles in diploid or aneuploid strains. However, for the time being, it seems that these techniques will continue to face the same challenges that conventional genetic engineering has encountered in most countries so far.
Future perspectives
Our knowledge of S. cerevisiae wine yeast strains keeps increasing in quantity and quality. However, it seems we have reached near saturation regarding the exploitation of natural diversity. Therefore, the tailoring of wine yeast starters to the demands of an ever‐changing market requires the generation of new genotypes. Commercial and regulatory constraints for genetic engineering have pushed wine biotechnology towards alternative techniques, including random mutagenesis, sexual hybridization, or experimental evolution. The power of these technologies has been boosted by recent advances in genomics, NGS based technologies, and systems biology approaches to yeast metabolism. These can help guide backcrossing, for introgression into target starter yeasts of traits from mutant or unrelated strains, as well as the rational design of selection strategies for experimental evolution or random mutagenesis. Strains improved in this way will take advantage of our extensive scientific knowledge on wine yeasts, while benefiting from a non‐GMO status. They are likely to be a major source of innovation in winemaking over the next few years.
Genome edition is, somehow, halfway between these techniques and traditional genetic engineering. It can be considered “cleaner” than any of them, as the genetic modification can be fully targeted while avoiding any unwanted DNA sequences in the final strains. Anyway, its potential cannot be fully exploited without a thorough knowledge of the genetic determination of wine yeast technological traits. There are high expectations among wine biotechnologists that new regulations in Europe, and other wine producing countries, will be less restrictive for CRISPR/Cas‐derived strains. But the multiple regulatory layers affecting the global production and marketing of wine do not warrant easy market entry for genome‐edited strains.
Finally, despite most of our knowledge on wine yeasts comes from studies under axenic conditions, yeasts have been evolutionarily shaped within microbial communities. Moreover, non‐Saccharomyces starters are increasingly used in combination with conventional ones. These considerations have led to a growing interest in the social life of wine yeasts, with the aim of better understanding the different wine yeast species and their ecological context (Jouhten et al., 2016; Conacher et al., 2021). In the short term, the development of multi‐species starter cultures and the study of interactions between wine microorganisms are also likely to become key drivers of oenological innovation.
Funding Information
The author’s work on wine biotechnology is funded by the Spanish Government through grants PID2019‐105159RB‐I00 and PCI2018‐092949 (co‐funded by ERA‐CoBioTech).
Conflict of interest
The authors have no conflict of interest to disclose.
Acknowledgements
The author’s work on wine biotechnology is funded by the Spanish Government through grants PID2019‐105159RB‐I00 and PCI2018‐092949 (co‐funded by ERA‐CoBioTech).
Microbial Biotechnology (2022) 15(5), 1339–1356
Contributor Information
Ramon Gonzalez, Email: rgonzalez@icvv.es.
Pilar Morales, Email: pilar.morales@icvv.es.
References
- Agarbati, A. , Canonico, L. , Comitini, F. , and Ciani, M. (2020) Reduction of sulfur compounds through genetic improvement of native Saccharomyces cerevisiae useful for organic and sulfite‐free wine. Foods 9: 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambroset, C. , Petit, M. , Brion, C. , Sanchez, I. , Delobel, P. , Guérin, C. , et al. (2011) Deciphering the molecular basis of wine yeast fermentation traits using a combined genetic and genomic approach. G3‐Genes Genomes Genet 1: 263–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austriaco, N.R. Jr (1996) To bud until death: the genetics of ageing in the yeast, Saccharomyces . Yeast 12: 623–630. [DOI] [PubMed] [Google Scholar]
- Barnett, J.A. (1998) A history of research on yeasts. 1. Work by chemists and biologists 1789–1850. Yeast 14: 1439–1451. [DOI] [PubMed] [Google Scholar]
- Barnett, J.A. (2003) Beginnings of microbiology and biochemistry: the contribution of yeast research. Microbiology 149: 557–567. [DOI] [PubMed] [Google Scholar]
- Barnett, J.A. , and Entian, K.D. (2005) A history of research on yeasts 9: regulation of sugar metabolism. Yeast 22: 835–894. [DOI] [PubMed] [Google Scholar]
- Bellon, J.R. , Eglinton, J.M. , Siebert, T.E. , Pollnitz, A.P. , Rose, L. , de Barros Lopes, M. , and Chambers, P.J. (2011) Newly generated interspecific wine yeast hybrids introduce flavour and aroma diversity to wines. Appl Microbiol Biotechnol 91: 603–612. [DOI] [PubMed] [Google Scholar]
- Bellon, J.R. , Schmid, F. , Capone, D.L. , Dunn, B.L. , and Chambers, P.J. (2013) Introducing a new breed of wine yeast: interspecific hybridisation between a commercial Saccharomyces cerevisiae wine yeast and Saccharomyces mikatae . PLoS One 8: e62053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bely, M. , Stoeckle, P. , Masneuf‐Pomarede, I. , and Dubourdieu, D. (2008) Impact of mixed Torulaspora delbrueckii‐Saccharomyces cerevisiae culture on high‐sugar fermentation. Int J Food Microbiol 122: 312–320. [DOI] [PubMed] [Google Scholar]
- Berry, D.B. , and Gasch, A.P. (2008) Stress‐activated genomic expression changes serve a preparative role for impending stress in yeast. Mol Biol Cell 19: 4580–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizaj, E. , Cordente, A.G. , Bellon, J.R. , Raspor, P. , Curtin, C.D. , and Pretorius, I.S. (2012) A breeding strategy to harness flavor diversity of Saccharomyces interspecific hybrids and minimize hydrogen sulfide production. FEMS Yeast Res 12: 456–465. [DOI] [PubMed] [Google Scholar]
- Bokulich, N.A. , Ohta, M. , Richardson, P.M. , and Mills, D.A. (2013) Monitoring seasonal changes in winery‐resident microbiota. PLoS One 8: e66437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich, N.A. , Thorngate, J.H. , Richardson, P.M. , and Mills, D.A. (2014) Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci USA 111: E139–E148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman, A. , Forgan, A.H. , Kolouchova, R. , Fraser, J.A. , and Schmidt, S.A. (2016) Whole genome comparison reveals high levels of inbreeding and strain redundancy across the spectrum of commercial wine strains of Saccharomyces cerevisiae . G3‐Genes Genomes Genet 6: 957–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton, P.J. , and Greig, D. (2014) The ecology and evolution of non‐domesticated Saccharomyces species. Yeast 31: 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brice, C. , Sanchez, I. , Bigey, F. , Legras, J.L. , and Blondin, B. (2014) A genetic approach of wine yeast fermentation capacity in nitrogen‐starvation reveals the key role of nitrogen signaling. BMC Genom 15: 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion, C. , Ambroset, C. , Sanchez, I. , Legras, J.L. , and Blondin, B. (2013) Differential adaptation to multi‐stressed conditions of wine fermentation revealed by variations in yeast regulatory networks. BMC Genom 14: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.C.S. , and Lindquist, S. (2009) A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Develop 23: 2320–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambon, B. , Monteil, V. , Remize, F. , Camarasa, C. , and Dequin, S. (2006) Effects of GPD1 overexpression in Saccharomyces cerevisiae commercial wine yeast strains lacking ALD6 genes. Appl Environ Microbiol 72: 4688–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr, M. , Bensansson, D. , and Bergman, C.M. (2012) Evolutionary genomics of transposable elements in Saccharomyces cerevisiae . PLoS One 7: e50978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebollero, E. , Gonzalez‐Ramos, D. , Tabera, L. , and Gonzalez, R. (2007) Transgenic wine yeast technology comes of age: is it time for transgenic wine? Biotechnol Lett 29: 191–200. [DOI] [PubMed] [Google Scholar]
- Cebollero, E. , Gonzalez‐Ramos, D. , and Gonzalez, R. (2009) Construction of a recombinant autolytic wine yeast strain overexpressing the csc1‐1 allele. Biotechnol Prog 25: 1598–1604. [DOI] [PubMed] [Google Scholar]
- Chan, K.M. , Liu, Y.L. , Ma, C.H. , Jayaram, M. , and Sau, S. (2013) The 2 micron plasmid of Saccharomyces cerevisiae: a miniaturized selfish genome with optimized functional competence. Plasmid 70: 2–17. [DOI] [PubMed] [Google Scholar]
- Charoenchai, C. , Fleet, G.H. , Henschke, P.A. , and Tood, B.E.N. (1997) Screening of non‐Saccharomyces wine yeasts for the presence of extra cellular hydrolytic enzymes. Aust J Grape Wine Res 3: 2–8. [Google Scholar]
- Ciani, M. , Comitini, F. , Mannazzu, I. , and Domizio, P. (2010) Controlled mixed culture fermentation: a new perspective on the use of non‐Saccharomyces yeasts in winemaking. FEMS Yeast Res 10: 123–133. [DOI] [PubMed] [Google Scholar]
- Ciani, M. , Morales, P. , Comitini, F. , Tronchoni, J. , Canonico, L. , Curiel, J.A. , et al. (2016) Non‐conventional yeast species for lowering ethanol content of wines. Front Microbiol 7: 642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conacher, C.G. , Luyt, N.A. , Naidoo‐Blassoples, R.K. , Rossouw, D. , Setati, M.E. , and Bauer, F.F. (2021) The ecology of wine fermentation: a model for the study of complex microbial ecosystems. Applied Microbiol Biotechnol 105: 3027–3043. [DOI] [PubMed] [Google Scholar]
- Conant, G.C. , and Wolfe, K.H. (2007) Increased glycolytic flux as an outcome of whole‐genome duplication in yeast. Mol Syst Biol 3: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concejero, B. , Hernandez‐Orte, P. , Astrain, J. , Lacau, B. , Baron, C. , and Ferreira, V. (2016) Evolution of polyfunctional mercaptans and their precursors during Merlot alcoholic fermentation. LWT Food Sci Technol 65: 770–776. [Google Scholar]
- Cordente, A.G. , Cordero‐Bueso, G. , Pretorius, I.S. , and Curtin, C.D. (2013) Novel wine yeast with mutations in YAP1 that produce less acetic acid during fermentation. FEMS Yeast Res 13: 62–73. [DOI] [PubMed] [Google Scholar]
- Cordente, A.G. , Heinrich, A. , Pretorius, I.S. , and Swiegers, J.H. (2009) Isolation of sulfite reductase variants of a commercial wine yeast with significantly reduced hydrogen sulfide production. FEMS Yeast Res 9: 446–459. [DOI] [PubMed] [Google Scholar]
- Coulon, J. , Husnik, J.I. , Inglis, D.L. , van der Merwe, G.K. , Lonvaud, A. , Erasmus, D.J. , and van Vuuren, H.J.J. (2006) Metabolic engineering of Saccharomyces cerevisiae to minimize the production of ethyl carbamate in wine. Am J Enol Vitic 57: 113–124. [Google Scholar]
- Cubillos, F.A. , Brice, C. , Molinet, J. , Tisné, S. , Abarca, V. , Tapia, S.M. , et al. (2017) Identification of nitrogen consumption genetic variants in yeast through QTL mapping and bulk segregant RNA‐seq analyses. G3‐Genes Genomes Genet 7: 1693–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio, M.J. , Lutz, S. , and Lesage, P. (2015) The Ty1 LTR‐retrotransposon of budding yeast, Saccharomyces cerevisiae . In Mobile DNA III. Craig, N. , Chandler, M. , Gellert, M. , Lambowitz, A. , Rice, P. , and Sandmeyer, S. (eds). Washington, DC: ASM Press. [Google Scholar]
- Dequin, S. , and Barre, P. (1994) Mixed lactic acid‐alcoholic fermentation by Saccharomyces cerevisiae expressing the Lactobacillus casei L(+)‐LDH. Nat Biotechnol 12: 173–177. [DOI] [PubMed] [Google Scholar]
- Dequin, S. , Escudier, J.‐L. , Bely, M. , Noble, J. , Albertin, W. , Masneuf‐Pomarède, I. , et al. (2017) How to adapt winemaking practices to modified grape composition under climate change conditions. Oeno One 51: 205–214. [Google Scholar]
- Devia, J. , Bastías, C. , Kessi‐Pérez, E.I. , Villarroel, C.A. , De Chiara, M. , Cubillos, F.A. , et al. (2020) Transcriptional activity and protein levels of horizontally acquired genes in yeast reveal hallmarks of adaptation to fermentative environments. Front Genet 11: art 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder, M. , Sanchez, I. , Brice, C. , Camarasa, C. , Legras, J.L. , and Dequin, S. (2018) QTL mapping of volatile compound production in Saccharomyces cerevisiae during alcoholic fermentation. BMC Genom 19: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero, A. , Gogorza, B. , Melus, M.A. , Ortin, N. , Cacho, J. , and Ferreira, V. (2004) Characterization of the aroma of a wine from Maccabeo. Key role played by compounds with low odor activity values. J Agric Food Chem 52: 3516–3524. [DOI] [PubMed] [Google Scholar]
- Estruch, F. (2000) Stress‐controlled transcription factors, stress‐induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev 24: 469–486. [DOI] [PubMed] [Google Scholar]
- Fernández‐Espinar, M.T. , López, V. , Ramón, D. , Bartra, E. , and Querol, A. (2001) Study of the authenticity of commercial wine yeast strains by molecular techniques. Int J Food Microbiol 70: 1–10. [DOI] [PubMed] [Google Scholar]
- Fidalgo, M. , Barrales, R.R. , Ibeas, J.I. , and Jimenez, J. (2006) Adaptive evolution by mutations in the FLO11 gene. Proc Natl Acad Sci USA 103: 11228–11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, G. , Liti, G. , and Llorente, B. (2021) The budding yeast life cycle: More complex than anticipated? Yeast 38: 5–11. [DOI] [PubMed] [Google Scholar]
- Foury, F. , Roganti, T. , Lecrenier, N. , and Purnelle, B. (1998) The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae . FEBS Lett 440: 325–331. [DOI] [PubMed] [Google Scholar]
- Fracassetti, D. , Massaglia, S. , Viberti, A. , Motta, G. , Foschino, R. , and Vigentini, I. (2020) Wine industry’s attitude towards oenological yeasts: Italy as a case study. Beverages 6: 33. [Google Scholar]
- Freel, K.C. , Friedrich, A. , and Schacherer, J. (2015) Mitochondrial genome evolution in yeasts: an all‐encompassing view. FEMS Yeast Res 15: fov023. [DOI] [PubMed] [Google Scholar]
- Galeote, V. , Novo, M. , Salema‐Oom, M. , Brion, C. , Valerio, E. , Gonçalves, P. , and Dequin, S. (2010) FSY1, a horizontally transferred gene in the Saccharomyces cerevisiae EC1118 wine yeast strain, encodes a high‐affinity fructose/H+ symporter. Microbiology 156: 3754–3761. [DOI] [PubMed] [Google Scholar]
- Ganga, M.A. , Pinaga, F. , Vallés, S. , Ramón, D. , and Querol, A. (1999) Aroma improving in microvinification processes by the use of a recombinant wine yeast strain expressing the Aspergillus nidulans xlnA gene. Int J Food Microbiol 47: 171–178. [DOI] [PubMed] [Google Scholar]
- García‐Ríos, E. , Morard, M. , Parts, L. , Liti, G. , and Guillamón, J.M. (2017) The genetic architecture of low‐temperature adaptation in the wine yeast Saccharomyces cerevisiae . BMC Genom 18: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Ríos, E. , Nuévalos, M. , Barrio, E. , Puig, S. , and Guillamón, J.M. (2019) A new chromosomal rearrangement improves the adaptation of wine yeasts to sulfite. Environ Microbiol 21: 1771–1781. [DOI] [PubMed] [Google Scholar]
- Gerstein, A.C. , Chun, H.J.E. , Grant, A. , and Otto, S.P. (2006) Genomic convergence toward diploidy in Saccharomyces cerevisiae . PLoS Genet 2: e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gier, S. , Simon, M. , Gasparoni, G. , Khalifa, S. , Schulz, M.H. , Schmitt, M.J. , and Breinig, F. (2020) Yeast viral killer toxin K1 induces specific host cell adaptions via intrinsic selection pressure. Appl Environ Microbiol 86: e02446–e2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno‐Alcañiz, J.V. , and Matallana, E. (2001) Performance of industrial strains of Saccharomyces cerevisiae during wine fermentation is affected by manipulation strategies based on sporulation. Syst Appl Microbiol 24: 639–644. [DOI] [PubMed] [Google Scholar]
- Giovani, G. , and Rosi, I. (2007) Release of cell wall polysaccharides from Saccharomyces cerevisiae thermosensitive autolytic mutants during alcoholic fermentation. Int J Food Microbiol 116: 19–24. [DOI] [PubMed] [Google Scholar]
- Goddard, M.R. (2008) Quantifying the complexities of Saccharomyces cerevisiae’s ecosystem engineering via fermentation. Ecology 89: 2077–2082. [DOI] [PubMed] [Google Scholar]
- Goddard, M.R. , and Greig, D. (2015) Saccharomyces cerevisiae: a nomadic yeast with no niche? FEMS Yeast Res 15: fov009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau, A. , Barrell, B.g. , Bussey, H. , Davis, R.w. , Dujon, B. , Feldmann, H. , et al. (1996) Life with 6000 genes. Science 274: 546–567. [DOI] [PubMed] [Google Scholar]
- Gojković, Z. , Knecht, W. , Zameitat, E. , Warneboldt, J. , Coutelis, J.‐B. , Pynyaha, Y. , et al. (2004) Horizontal gene transfer promoted evolution of the ability to propagate under anaerobic conditions in yeasts. Mol Genet Genom 271: 387–393. [DOI] [PubMed] [Google Scholar]
- Gonzalez, R. , Martinez‐Rodriguez, A.J. , and Carrascosa, A.V. (2003) Yeast autolytic mutants potentially useful for sparkling wine production. Int J Food Microbiol 84: 21–26. [DOI] [PubMed] [Google Scholar]
- Gonzalez, R. , Morales, P. , Tronchoni, J. , Cordero‐Bueso, G. , Vaudano, E. , Quirós, M. , et al. (2016) New genes involved in osmotic stress tolerance in Saccharomyces cerevisiae . Front Microbiol 7: 1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, R. , Muñoz, R. , and Carrascosa, A.V. (2011) Production of wine starter cultures. In Molecular Wine Microbiology. Carrascosa, A.V. , Muñoz, R. , and Gonzalez, R. (eds). London, UK: Academic Press, pp. 279–302. ISBN: 978‐0‐12‐375021‐1 [Google Scholar]
- Gonzalez, R. , Quirós, M. , and Morales, P. (2013) Yeast respiration of sugars by non‐Saccharomyces yeast species: a promising and barely explored approach to lowering alcohol content of wines. Trends Food Sci Technol 29: 55–61. [Google Scholar]
- Gonzalez, R. , Tronchoni, J. , Mencher, A. , Curiel, J.A. , Rodrigues, A.J. , López‐Berges, L. , et al. (2019) Low phenotypic penetrance and technological impact of yeast [GAR+] prion‐like elements on winemaking. Front Microbiol 9: 3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González, S.S. , Barrio, E. , Gafner, J. , and Querol, A. (2006) Natural hybrids from Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces kudriavzevii in wine fermentations. FEMS Yeast Res 6: 1221–1234. [DOI] [PubMed] [Google Scholar]
- González‐Ramos, D. , Muñoz, A. , Ortiz‐Julien, A. , Palacios, A.T. , Heras, J.M. , and Gonzalez, R. (2010) A Saccharomyces cerevisiae wine yeast strain overproducing mannoproteins selected through classical genetic methods. Oeno One 44: 243–249. [Google Scholar]
- González‐Ramos, D. , Quirós, M. , and Gonzalez, R. (2009) Three different targets for the genetic modification of wine yeast strains resulting in improved effectiveness of bentonite fining. J Agric Food Chem 57: 8373–8378. [DOI] [PubMed] [Google Scholar]
- Goto‐Yamamoto, N. , Kitano, K. , Shiki, K. , Yoshida, Y. , Suzuki, T. , Iwata, T. , et al. (1998) SSU1‐R, a sulfite resistance gene of wine yeast, is an allele of SSU1 with a different upstream sequence. J Ferment Bioeng 86: 427–433. [Google Scholar]
- Govender, P. , Bester, M. , and Bauer, F.F. (2010) FLO gene‐dependent phenotypes in industrial wine yeast strains. Appl Microbiol Biotechnol 86: 931–945. [DOI] [PubMed] [Google Scholar]
- Gustin, M.C. , Albertyn, J. , Alexander, M. , and Davenport, K. (1998) MAP kinase pathways in the yeast Saccharomyces cerevisiae . Microbiol Mol Biol Rev 62: 1264–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber, J.E. (2012) Mating‐type genes and MAT switching in Saccharomyces cerevisiae . Genetics 191: 33–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman, A. , Sall, T. , Compagno, C. , and Piskur, J. (2013) Yeast “make‐accumulate‐consume” life strategy evolved as a multi‐step process that predates the whole genome duplication. PLoS One 8: e68734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, C. , Brachat, S. , and Dietrich, F.S. (2005) Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae . Eukaryot Cell 4: 1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, S. , Ogura, M. , Aritomi, K. , Hoshida, H. , Nishizawa, Y. , and Akada, R. (2005) Isolation of auxotrophic mutants of diploid industrial yeast strains after UV mutagenesis. Appl Environ Microbiol 71: 312–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmeland, A.K. , and Ebeler, S.E. (2015) Glycosidically bound volatile aroma compounds in grapes and wine: a review. Am J Enol Vitic 66: 1–11. [Google Scholar]
- Hohmann, S. (2015) An integrated view on a eukaryotic osmoregulation system. Current Genet 61: 373–382. [DOI] [PubMed] [Google Scholar]
- Holzer, H. (1961) Regulation of carbohydrate metabolism by enzyme competition. Cold Spring Harbor Symp Quant Biol 26: 277–288. [DOI] [PubMed] [Google Scholar]
- Husnik, J.I. , Volschenk, H. , Bauer, J. , Colavizza, D. , Luo, Z. , and Van Vuuren, H.J.J. (2006) Metabolic engineering of malolactic wine yeast. Metab Eng 8: 315–323. [DOI] [PubMed] [Google Scholar]
- Jambhekar, A. , and Amon, A. (2008) Control of meiosis by respiration. Curr Biol 18: 969–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, M.L.A. , Diderich, J.A. , Mashego, M. , Hassane, A. , de Winde, J.H. , Daran‐Lapujade, P. , and Pronk, J. (2005) Prolonged selection in aerobic, glucose‐limited chemostat cultures of Saccharomyces cerevisiae causes a partial loss of glycolytic capacity. Microbiology 151: 1657–1669. [DOI] [PubMed] [Google Scholar]
- Jarosz, D.F. , Brown, J.C.S. , Walker, G.A. , Datta, M.S. , Ung, W.L. , Lancaster, A.K. , et al. (2014b) Cross‐kingdom chemical communication drives a heritable, mutually beneficial prion‐based transformation of metabolism. Cell 158: 1083–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz, D.F. , Lancaster, A.K. , Brown, J.C. , and Lindquist, S. (2014a) An evolutionarily conserved prion‐like element converts wild fungi from metabolic specialists to generalists. Cell 158: 1072–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez, J. , and Benitez, T. (1988) Yeast cell viability under conditions of high temperature and ethanol concentrations depends on the mitochondrial genome. Curr Genet 13: 461–469. [DOI] [PubMed] [Google Scholar]
- Jouhten, P. , Ponomarova, O. , Gonzalez, R. , and Patil, K.R. (2016) Saccharomyces cerevisiae metabolism in ecological context. FEMS Yeast Res 16: fow080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlberg, O. , Canbäck, B. , Kurland, C.G. , and Andersson, S.G. (2000) The dual origin of the yeast mitochondrial proteome. Yeast 17: 170–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis, M. , Birren, B.W. , and Lander, E.S. (2004) Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae . Nature 428: 617–624. [DOI] [PubMed] [Google Scholar]
- Kishimoto, M. (1994) Fermentation characteristics of hybrids between the cryophilic wine yeast Saccharomyces bayanus and the mesophilic wine yeast Saccharomyces cerevisiae . J Ferment Bioeng 77: 432–435. [Google Scholar]
- Knop, M. (2006) Evolution of the hemiascomycete yeasts: on life styles and the importance of inbreeding. BioEssays 28: 696–708. [DOI] [PubMed] [Google Scholar]
- Kraus, J.K. , Reed, G. , and Villettaz, J.C. (1983) Levures sèches actives de vinification 1re partie: fabrication et charactéristiques. Connaiss Vigne Vin 17: 93–103. [Google Scholar]
- Labuschagne, P.W.J. , and Divol, B. (2021) Thiamine: a key nutrient for yeasts during wine alcoholic fermentation. Appl Microbiol Biotechnol 105: 953–973. [DOI] [PubMed] [Google Scholar]
- Legras, J.L. , and Karst, F. (2003) Optimisation of interdelta analysis for Saccharomyces cerevisiae . FEMS Microbiol Lett 221: 249–255. [DOI] [PubMed] [Google Scholar]
- Li, X.C. , Peris, D. , Hittinger, C.T. , Sia, E.A. , and Fay, J.C. (2019) Mitochondria‐encoded genes contribute to evolution of heat and cold tolerance in yeast. Sci Adv 5: eaav1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libkind, D. , Hittinger, C.t. , Valerio, E. , Goncalves, C. , Dover, J. , Johnston, M. , et al. (2011) Microbe domestication and the identification of the wild genetic stock of lager‐brewing yeast. Proc Natl Acad Sci USA 108: 14539–14544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly, M. , Bauer, F.F. , Lambrechts, M.G. , Swiegers, J.H. , Cozzolino, D. , and Pretorius, I.S. (2006) The effect of increased yeast alcohol acetyltransferase and esterase activity on the flavour profiles of wine and distillates. Yeast 23: 641–659. [DOI] [PubMed] [Google Scholar]
- Lilly, M. , Lambrechts, M.G. , and Pretorius, I.S. (2000) Effect of increased yeast alcohol acetyltransferase activity on flavour profiles of wine and distillates. Appl Environ Microbiol 66: 744–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G.L. , Chi, Z. , Wang, G.Y. , Wang, Z.P. , Li, Y. , and Chi, Z.M. (2015) Yeast killer toxins, molecular mechanisms of their action and their applications. Crit Rev Biotechnol 35: 222–234. [DOI] [PubMed] [Google Scholar]
- Long, D. , Wilkinson, K.L. , Taylor, D.K. , and Jiranek, V. (2018) Novel wine yeast for improved utilisation of proline during fermentation. Fermentation 4: 10. [Google Scholar]
- Lopez‐Martinez, G. , Margalef‐Català, M. , Salinas, F. , Liti, G. , and Cordero‐Otero, R. (2015) ATG18 and FAB1 are involved in dehydration stress tolerance in Saccharomyces cerevisiae . PLoS One 10: e0119606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luparia, V. , Soubeyrand, V. , Berges, T. , Julien, A. , and Salmon, J.M. (2004) Assimilation of grape phytosterols by Saccharomyces cerevisiae and their impact on enological fermentations. Appl Microbiol Biotechnol 65: 25–32. [DOI] [PubMed] [Google Scholar]
- Madden, K. , and Snyder, M. (1998) Cell polarity and morphogenesis in budding yeasts. Annu Rev Microbiol 52: 687–744. [DOI] [PubMed] [Google Scholar]
- Mangado, A. , Morales, P. , Gonzalez, R. , and Tronchoni, J. (2018) Evolution of a yeast with industrial background under winemaking conditions leads to diploidization and chromosomal copy number variation. Front Microbiol 9: 1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanares, P. , Vallés, S. , and Viana, F. (2011) The role of non‐Saccharomyces yeasts in vinification. In Molecular Wine Microbiology. Carrascosa, A.V. , Muñoz, R. , and Gonzalez, R. (eds). London, UK: Academic Press, pp. 85–110. ISBN: 978‐0‐12‐375021‐1 [Google Scholar]
- Maqueda, M. , Zamora, E. , Álvarez, M.L. , and Ramírez, M. (2012) Characterization, ecological distribution, and population dynamics of Saccharomyces sensu stricto killer yeasts in the spontaneous grape must fermentations of southwestern Spain. Appl Environ Microbiol 78: 735–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks, V.D. , Ho Sui, S.J. , Erasmus, D. , van der Merwe, G.K. , Brumm, J. , Wasserman, W.W. , et al. (2008) Dynamics of the yeast transcriptome during wine fermentation reveals a novel fermentation stress response. FEMS Yeast Res 8: 35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit, S. , Mena, A. , Bigey, F. , Sauvage, F.‐X. , Couloux, A. , Guy, J. , et al. (2015) Evolutionary advantage conferred by an eukaryote‐to‐eukaryote gene transfer event in wine yeasts. Mol Biol Evol 32: 1695–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, V. , Valera, M.J. , Medina, K. , Boido, E. , and Carrau, F. (2018) Oenological impact of the Hanseniaspora/Kloeckera yeast genus on wines‐ a review. Fermentation 4: 76. [Google Scholar]
- Martínez‐Moreno, R. , Morales, P. , Gonzalez, R. , Mas, A. , and Beltran, G. (2012) Biomass production and alcoholic fermentation performance of Saccharomyces cerevisiae as a function of nitrogen source. FEMS Yeast Res 12: 477–485. [DOI] [PubMed] [Google Scholar]
- Marullo, P. , Aigle, M. , Bely, M. , Masneuf‐Pomarede, I. , Durrens, P. , Dubourdieu, D. , and Yvert, G. (2007) Single QTL mapping and nucleotide‐level resolution of a physiologic trait in wine Saccharomyces cerevisiae strains. FEMS Yeast Res 7: 941–952. [DOI] [PubMed] [Google Scholar]
- Marullo, P. , Bely, M. , Masneuf‐Pomarède, I. , Pons, M. , Aigle, M. , and Dubourdieu, D. (2006) Breeding strategies for combining fermentative qualities and reducing off‐flavor production in a wine yeast model. FEMS Yeast Res 6: 268–279. [DOI] [PubMed] [Google Scholar]
- Marullo, P. , Durrens, P. , Peltier, E. , Bernard, M. , Mansour, C. , and Dubourdieu, D. (2019) Natural allelic variations of Saccharomyces cerevisiae impact stuck fermentation due to the combined effect of ethanol and temperature; a QTL‐mapping study. BMC Genom 20: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marullo, P. , Mansour, C. , Dufour, M. , Albertin, W. , Sicard, D. , Bely, M. , and Dubourdieu, D. (2009) Genetic improvement of thermo‐tolerance in wine Saccharomyces cerevisiae strains by a backcross approach. FEMS Yeast Res 9: 1148–1160. [DOI] [PubMed] [Google Scholar]
- Masneuf‐Pomarède, I. , Bely, M. , Marullo, P. , Lonvaud‐Funel, A. , and Dubourdieu, D. (2010) Reassessment of phenotypic traits for Saccharomyces bayanus var. uvarum wine yeast strains. Int J Food Microbiol 139: 79–86. [DOI] [PubMed] [Google Scholar]
- Matallana, E. , and Aranda, A. (2017) Biotechnological impact of stress response on wine yeast. Lett Appl Microbiol 64: 103–110. [DOI] [PubMed] [Google Scholar]
- McBride, H.M. , Neuspiel, M. , and Wasiak, S. (2006) Mitochondria: more than just a powerhouse. Curr Biol 16: R551–R560. [DOI] [PubMed] [Google Scholar]
- McGovern, P.E. , Glusker, D.L. , Exner, L.J. , and Voigt, M.M. (1996) Neolithic resinated wine. Nature 381: 480–481. [Google Scholar]
- McGovern, P. , Jalabadze, M. , Batiuk, S. , Callahan, M.P. , Smith, K.E. , Hall, G.R. , et al. (2017) Early neolithic wine of georgia in the south caucasus. Proc Natl Acad Sci USA 114: E10309–E10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern, P.e. , Zhang, J. , Tang, J. , Zhang, Z. , Hall, G.r. , Moreau, R.a. , et al. (2004) Fermented beverages of pre‐and proto‐historic China. Proc Natl Acad Sci USA 101: 17593–17598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michnick, S. , Roustan, J.L. , Remize, F. , Barre, P. , and Dequin, S. (1997) Modulation of glycerol and ethanol yields during alcoholic fermentation in Saccharomyces cerevisiae strains overexpressed or disrupted for GPD1 encoding glycerol 3‐phosphate dehydrogenase. Yeast 13: 783–793. [DOI] [PubMed] [Google Scholar]
- Monteiro, F.F. , Trousdale, E.K. , and Bisson, L.F. (1989) Ethyl carbamate formation in wine: Use of radioactively labeled precursors to demonstrate the involvement of urea. Am J Enol Vitic 40: 1–8. [Google Scholar]
- Moreira, N. , Mendes, F. , Guedes de Pinho, P. , Hogg, T. , and Vasconcelos, I. (2008) Heavy sulphur compounds, higher alcohols and esters production profile of Hanseniaspora uvarum and Hanseniaspora guilliermondii grown as a pure and mixed cultures in grape must. Int J Food Microbiol 124: 231–238. [DOI] [PubMed] [Google Scholar]
- Mortimer, R.K. , Romano, P. , Suzzi, G. , and Polsinelli, M. (1994) Genome renewal: a new phenomenon revealed from a genetic study of 43 strains of Saccharomyces cerevisiae derived from natural fermentation of grape musts. Yeast 10: 1543–1552. [DOI] [PubMed] [Google Scholar]
- Muñoz, R. , Gómez, A. , Robles, V. , Rodríguez, P. , Cebollero, E. , Tabera, L. , et al. (2009) Multilocus sequence typing of oenological Saccharomyces cerevisiae strains. Food Microbiol 26: 841–846. [DOI] [PubMed] [Google Scholar]
- Naumov, G.I. (2000) Saccharomyces bayanus var. uvarum comb. nov., a new variety established by genetic analysis. Microbiology 69: 338–342. [PubMed] [Google Scholar]
- Naumov, G.I. , James, S.A. , Naumova, E.S. , Louis, E. , and Roberts, I.N. (2000) Three new species in the Saccharomyces sensu stricto complex: Saccharomyces cariocanus, Saccharomyces kudriavzevii and Saccharomyces mikatae . Int J Syst Evol Microbiol 50: 1931–1942. [DOI] [PubMed] [Google Scholar]
- Novo, M. , Bigey, F. , Beyne, E. , Galeote, V. , Gavory, F. , Mallet, S. , et al. (2009) Eukaryote‐to‐eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc Natl Acad Sci USA 106: 16333–16338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo, M. , Gonzalez, R. , Bertran, E. , Martínez, M. , Yuste, M. , and Morales, P. (2014) Improved fermentation kinetics by wine yeast strains evolved under ethanol stress. LWT Food Sci Technol 58: 166–172. [Google Scholar]
- Novo, M. , Quirós, M. , Morales, P. , and Gonzalez, R. (2012) Wine technology. In Handbook of Fruits and Fruit Processing. Sinha, N. K. , Sidhu, J. S. , Barta, J. , Wu, J. S. B. , and Cano, M. P. (eds). Oxford, UK: John Wiley and Sons LTD, chapter 28, pp 806‐862. ISBN: 9781118352632 [Google Scholar]
- Oro, L. , Ciani, M. , and Comitini, F. (2014) Antimicrobial activity of Metschnikowia pulcherrima on wine yeasts. J Appl Microbiol 116: 1209–1217. [DOI] [PubMed] [Google Scholar]
- Otterstedt, K. , Larsson, C. , Bill, R.M. , Stahlberg, A. , Boles, E. , Hohmann, S. , and Gustafsson, L. (2004) Switching the mode of metabolism in the yeast Saccharomyces cerevisiae . EMBO Rep 5: 532–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla, B. , Gil, J.V. , and Manzanares, P. (2016) Past and future of Non‐Saccharomyces yeasts: from spoilage microorganisms to biotechnological tools for improving wine aroma complexity. Front Microbiol 7: art 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐González, J.A. , Gonzalez, R. , Querol, A. , Sendra, J. , and Ramón, D. (1993) Construction of a recombinant wine yeast strain expressing beta‐(1, 4)‐endoglucanase and its use in microvinification processes. Appl Environ Microbiol 59: 2801–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Ortín, J.E. , Querol, A. , Puig, S. , and Barrio, E. (2002) Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res 12: 1533–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Torrado, R. , Barrio, E. , and Querol, A. (2018) Alternative yeasts for winemaking: Saccharomyces non‐cerevisiae and its hybrids. Crit Rev Food Sci Nutr 58: 1780–1790. [DOI] [PubMed] [Google Scholar]
- Pérez‐Torrado, R. , Rantsiou, K. , Perrone, B. , Navarro‐Tapia, E. , Querol, A. , and Cocolin, L. (2017) Ecological interactions among Saccharomyces cerevisiae strains: insight into the dominance phenomenon. Sci Rep 7: 43603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Través, L. , Lopes, C.A. , Barrio, E. , and Querol, A. (2012) Evaluation of different genetic procedures for the generation of artificial hybrids in Saccharomyces genus for winemaking. Int J Food Microbiol 156: 102–111. [DOI] [PubMed] [Google Scholar]
- Pérez‐Través, L. , Lopes, C.A. , Gonzalez, R. , Barrio, E. , and Querol, A. (2015) Physiological and genomic characterisation of Saccharomyces cerevisiae hybrids with improved fermentation performance and mannoprotein release capacity. Int J Food Microbiol 205: 30–40. [DOI] [PubMed] [Google Scholar]
- Peris, D. , Pérez‐Torrado, R. , Hittinger, C.T. , Barrio, E. , and Querol, A. (2018) On the origins and industrial applications of Saccharomyces cerevisiae x Saccharomyces kudriavzevii hybrids. Yeast 35: 51–69. [DOI] [PubMed] [Google Scholar]
- Peter, J. , De Chiara, M. , Friedrich, A. , Yue, J.‐X. , Pflieger, D. , Bergström, A. , et al. (2018) Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556: 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picazo, C. , Gamero‐Sandemetrio, E. , Orozco, H. , Albertin, W. , Marullo, P. , Matallana, E. , and Aranda, A. (2014) Mitochondria inheritance is a key factor for tolerance to dehydration in wine yeast production. Lett Appl Microbiol 60: 217–222. [DOI] [PubMed] [Google Scholar]
- Pinto, C. , Pinho, D. , Cardoso, R. , Custódio, V. , Fernandes, J. , Sousa, S. , et al. (2015) Wine fermentation microbiome: a landscape from different Portuguese wine appellations. Front Microbiol 6: 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper, P.W. (1995) The heat shock and ethanol stress responses of yeast exhibit extensive similarity and functional overlap. FEMS Microbiol Lett 134: 121–127. [DOI] [PubMed] [Google Scholar]
- Piskur, J. , Rozpedowska, E. , Polakova, S. , Merico, A. , and Compagno, C. (2006) How did Saccharomyces evolve to become a good brewer? Trends Genet 22: 183–186. [DOI] [PubMed] [Google Scholar]
- Pretorius, I.S. (2017) Solving yeast jigsaw puzzles over a glass of wine: synthetic genome engineering pioneers new possibilities for wine yeast research. EMBO Rep 18: 1875–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretorius, I.S. , and Boeke, J.D. (2018) Yeast 2.0‐connecting the dots in the construction of the world's first functional synthetic eukaryotic genome. FEMS Yeast Res 18: foy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk, J.T. , Steensma, H.Y. , and van Dijken, J.P. (1996) Pyruvate metabolism in Saccharomyces cerevisiae . Yeast 12: 1607–1633. [DOI] [PubMed] [Google Scholar]
- Puig, S. , Querol, A. , Barrio, E. , and Pérez‐Ortín, J.E. (2000) Mitotic recombination and genetic changes in Saccharomyces cerevisiae during wine fermentation. Appl Environ Microbiol 66: 2057–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querol, A. , Barrio, E. , and Ramón, D. (1992) A comparative study of different methods of yeast strain characterization. System Appl Microbiol 15: 439–446. [Google Scholar]
- Querol, A. , Pérez‐Torrado, R. , Alonso‐del‐Real, J. , Minebois, R. , Stribny, J. , Oliveira, B.M. , and Barrio, E. (2018) New trends in the uses of yeasts in oenology. Adv Food Nutr Res 85: 177–210. [DOI] [PubMed] [Google Scholar]
- Quirós, M. , Gonzalez‐Ramos, D. , Tabera, L. , and Gonzalez, R. (2010) A new methodology to obtain wine yeast strains overproducing mannoproteins. Int J Food Microbiol 139: 9–14. [DOI] [PubMed] [Google Scholar]
- Ramírez, M. , Pérez, F. , and Regodón, J.A. (1998) A simple and reliable method for hybridization of homothallic wine strains of Saccharomyces cerevisiae . Appl Environ Microbiol 64: 5039–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez, M. , Regodón, J.A. , Pérez, F. , and Rebollo, J.E. (1999) Wine yeast fermentation vigor may be improved by elimination of recessive growth‐retarding alleles. Biotechnol Bioeng 65: 212–218. [DOI] [PubMed] [Google Scholar]
- Ribéreau‐Gayon, P. , Dubourdieu, D. , Donèche, B. , and Lonvaud, A. (2006) Handbook of Enology. Volume 1. The Microbiology of Wine and Vinifications, 2nd edn. Chichester, UK: John Wiley and Sons Ltd. ISBN: 0‐470‐01034‐7 [Google Scholar]
- Rodríguez‐Cousiño, N. , Maqueda, M. , Ambrona, J. , Zamora, E. , Esteban, R. , and Ramírez, M. (2011) A new wine Saccharomyces cerevisiae killer toxin (Klus), encoded by a double‐stranded RNA virus, with broad antifungal activity is evolutionarily related to a chromosomal host gene. Appl Environ Microbiol 77: 1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Porrata, B. , Novo, M. , Guillamón, J. , Rozes, N. , Mas, A. , and Otero, R.C. (2008) Vitality enhancement of the rehydrated active dry wine yeast. Int J Food Microbiol 126: 116–122. [DOI] [PubMed] [Google Scholar]
- Rojas, V. , Gil, J.V. , Piñaga, F. , and Manzanares, P. (2001) Studies on acetate ester production by non‐Saccharomyces wine yeasts. Int J Food Microbiol 70: 283–289. [DOI] [PubMed] [Google Scholar]
- Rojas, V. , Gil, J.V. , Piñaga, F. , and Manzanares, P. (2003) Acetate ester formation in wine by mixed cultures in laboratory fermentations. Int J Food Microbiol 86: 181–188. [DOI] [PubMed] [Google Scholar]
- Rollero, S. , Mouret, J.R. , Bloem, A. , Sanchez, I. , Ortiz‐Julien, A. , Sablayrolles, J.M. , et al. (2017) Quantitative 13C‐isotope labelling‐based analysis to elucidate the influence of environmental parameters on the production of fermentative aromas during wine fermentation. Microb Biotechnol 10: 1649–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano, P. , Fiore, C. , Paraggio, M. , Caruso, M. , and Capece, A. (2003) Function of yeast species and strains in wine flavour. Int J Food Microbiol 86: 169–180. [DOI] [PubMed] [Google Scholar]
- Roncoroni, M. , Santiago, M. , Hooks, D.O. , Moroney, S. , Harsch, M.J. , Lee, S.A. , et al. (2011) The yeast IRC7 gene encodes a β‐lyase responsible for production of the varietal thiol 4‐mercapto‐4‐methylpentan‐2‐one in wine. Food Microbiol 28: 926–935. [DOI] [PubMed] [Google Scholar]
- Rossignol, T. , Dulau, L. , Julien, A. , and Blondin, B. (2003) Genome‐wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast 20: 1369–1385. [DOI] [PubMed] [Google Scholar]
- Roudil, L. , Russo, P. , Berbegal, C. , Albertin, W. , Spano, G. , and Capozzi, V. (2020) Non‐Saccharomyces commercial starter cultures: scientific trends, recent patents and innovation in the wine sector. Recent Pat Food Nutr Agric 11: 27–39. [DOI] [PubMed] [Google Scholar]
- Salmon, J.M. , and Barre, P. (1998) Improvement of nitrogen assimilation and fermentation kinetics under enological conditions by derepression of alternative nitrogen‐assimilatory pathways in an industrial Saccharomyces cerevisiae strain. Appl Environ Microbiol 64: 3831–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, J. , Sousa, M.J. , Cardoso, H. , Inacio, J. , Silva, S. , Spencer‐Martins, I. , and Leão, C. (2008) Ethanol tolerance of sugar transport, and the rectification of stuck wine fermentations. Microbiology 154: 422–430. [DOI] [PubMed] [Google Scholar]
- Schmitt, M.J. , and Breinig, F. (2006) Yeast viral killer toxins: lethality and self‐protection. Nat Rev Microbiol 4: 212–221. [DOI] [PubMed] [Google Scholar]
- Setati, M.E. , Jacobson, D. , Andong, U.C. , and Bauer, F. (2012) The vineyard yeast microbiome, a mixed model microbial map. PLoS One 7: e52609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki, M. (2008) Interspecies hybridization and recombination in Saccharomyces wine yeasts. FEMS Yeast Res 8: 996–1007. [DOI] [PubMed] [Google Scholar]
- Snow, R. (1983) Genetic improvement of wine yeast. In Yeast Genetics. Spencer, J. F. , Spencer, D. M. , and Smith, A. R. W. (eds). New York, NY: Springer, pp. 439–459. [Google Scholar]
- Stefanini, I. , Dapporto, L. , Berná, L. , Polsinelli, M. , Turillazzi, S. , and Cavalieri, D. (2016) Social wasps are a Saccharomyces mating nest. Proc Natl Acad Sci USA 113: 2247–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyer, D. , Ambroset, C. , Brion, C. , Claudel, P. , Delobel, P. , Sanchez, I. , et al. (2012) QTL mapping of the production of wine aroma compounds by yeast. BMC Genom 13: 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss, M.L.A. , Jolly, N.P. , Lambrechts, M.G. , and van Rensburg, P. (2001) Screening for the production of extracellular hydrolytic enzymes by non‐Saccharomyces wine yeasts. J Appl Microbiol 91: 182–190. [DOI] [PubMed] [Google Scholar]
- Strope, P.K. , Kozmin, S.G. , Skelly, D.A. , Magwene, P.M. , Dietrich, F.S. , and McCusker, J.H. (2015) 2μ plasmid in Saccharomyces species and in Saccharomyces cerevisiae . FEMS Yeast Res 15: fov090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styger, G. , Jacobson, D. , Prior, B.A. , and Bauer, F.F. (2013) Genetic analysis of the metabolic pathways responsible for aroma metabolite production by Saccharomyces cerevisiae . Appl Microbiol Biotechnol 97: 4429–4442. [DOI] [PubMed] [Google Scholar]
- Styger, G. , Prior, B.A. , and Bauer, F.F. (2011) Wine flavor and aroma. J Ind Microbiol Biotechnol 38: 1145–1159. [DOI] [PubMed] [Google Scholar]
- Su, Y. , Gamero, A. , Rodríguez, M.E. , Lopes, C.A. , Querol, A. , and Guillamón, J.M. (2019) Interspecific hybridisation among diverse Saccharomyces species: a combined biotechnological solution for low‐temperature and nitrogen‐limited wine fermentations. Int J Food Microbiol 310: 108331. [DOI] [PubMed] [Google Scholar]
- Sumby, K.M. , Grbin, P.R. , and Jiranek, V. (2010) Microbial modulation of aromatic esters in wine: current knowledge and future prospects. Food Chem 121: 1–16. [Google Scholar]
- Swan, T.M. , and Watson, K. (1997) Membrane fatty acid composition and membrane fluidity as parameters of stress tolerance in yeast. Can J Microbiol 43: 70–77. [DOI] [PubMed] [Google Scholar]
- Swiegers, J.H. , and Pretorius, I.S. (2007) Modulation of volatile sulfur compounds by wine yeast. Appl Microbiol Biotechnol 74: 954–960. [DOI] [PubMed] [Google Scholar]
- Tabera, L. , Muñoz, R. , and Gonzalez, R. (2006) Deletion of BCY1 from the Saccharomyces cerevisiae genome is semidominant and induces autolytic phenotypes suitable for improvement of sparkling wines. Appl Environ Microbiol 72: 2351–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilloy, V. , Ortiz‐Julien, A. , and Dequin, S. (2014) Reduction of ethanol yield and improvement of glycerol formation by adaptive evolution of the wine yeast Saccharomyces cerevisiae under hyperosmotic conditions. Appl Environ Microbiol 80: 2623–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torija, M.J. , Beltran, G. , Novo, M. , Poblet, M. , Guillamón, J.M. , Mas, A. , and Rozes, N. (2003) Effects of fermentation temperature and Saccharomyces species on the cell fatty acid composition and presence of volatile compounds in wine. Int J Food Microbiol 85: 127–136. [DOI] [PubMed] [Google Scholar]
- Vallejo, B. , Matallana, E. , and Aranda, A. (2020a) Saccharomyces cerevisiae nutrient signaling pathways show an unexpected early activation pattern during winemaking. Microb Cell Fact 19: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo, B. , Peltier, E. , Garrigós, V. , Matallana, E. , Marullo, P. , and Aranda, A. (2020b) Role of Saccharomyces cerevisiae nutrient signaling pathways during winemaking: a phenomics approach. Front Bioeng Biotechnol 8: 853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela, C. , Pizarro, F. , and Agosin, E. (2004) Biomass content governs fermentation rate in nitrogen‐deficient wine musts. Appl Environ Microbiol 70: 3392–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana, F. , Belloch, C. , Vallés, S. , and Manzanares, P. (2011) Monitoring a mixed starter of Hanseniaspora vineae/Saccharomyces cerevisiae in natural must: impact on 2‐phenylethyl acetate production. Int J Food Microbiol 151: 235–240. [DOI] [PubMed] [Google Scholar]
- Viana, F. , Gil, J.V. , Genovés, S. , Vallés, S. , and Manzanares, P. (2008) Rational selection of non‐Saccharomyces wine yeasts for mixed starters based on ester formation and enological traits. Food Microbiol 25: 778–785. [DOI] [PubMed] [Google Scholar]
- Vigentini, I. , Gebbia, M. , Belotti, A. , Foschino, R. , and Roth, F.P. (2017) CRISPR/Cas9 system as a valuable genome editing tool for wine yeasts with application to decrease urea production. Front Microbiol 8: 2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordeckers, K. , De Maeyer, D. , van der Zande, E. , Vinces, M.D. , Meert, W. , Cloots, L. , et al. (2012) Identification of a complex genetic network underlying Saccharomyces cerevisiae colony morphology. Mol Microbiol 86: 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vuuren, H.J.J. , and Jacobs, C.J. (1992) Killer yeasts in the wine industry: a review. Am J Enol Vitic 43: 119–128. [Google Scholar]
- Walker, M.E. , Zhang, J. , Sumby, K.M. , Lee, A. , Houlès, A. , Li, S. , and Jiranek, V. (2021) Sulfate transport mutants affect hydrogen sulfide and sulfite production during alcoholic fermentation. Yeast. 10.1002/yea.3553 [DOI] [PubMed] [Google Scholar]
- Wickner, R.B. , Shewmaker, F.P. , Bateman, D.A. , Edskes, H.K. , Gorkovskiy, A. , Dayani, Y. , and Bezsonov, E.E. (2015) Yeast prions: structure, biology, and prion‐handling systems. Microbiol Mol Biol Rev 79: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K.H. , and Shields, D.C. (1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713. [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Du, G. , Zhou, J. , and Chen, J. (2018) Regulation of sensing, transportation, and catabolism of nitrogen sources in Saccharomyces cerevisiae . Microbiol Mol Biol Rev 82: e00040–e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer, A. , Durand, C. , Loira, N. , Durrens, P. , Sherman, D.J. , and Marullo, P. (2014) QTL dissection of lag phase in wine fermentation reveals a new translocation responsible for Saccharomyces cerevisiae adaptation to sulfite. PLoS One 9: e86298. [DOI] [PMC free article] [PubMed] [Google Scholar]


