Abstract
The activity of intestinal stem cells (ISCs) is foremost in maintaining homeostasis and repair of intestines. As a pivotal substrate of RNA and DNA biosynthesis, uridine plays essential roles in nutritional and disease monitoring. Whether uridine influences ISC activity remains undefined. To answer this question, 3-dimensional (3D) mouse intestinal organoids and living mice were used as a model. It was found that uridine causes a significant decrease in the number of crypts per intestinal organoid. Uridine also significantly decreases mRNA expression and protein levels with markers of ISCs in intestinal organoids in a dose-dependent manner, which was instructed via mTOR. In parallel, uridine decreases the expression of marker of ISCs in mouse intestine in vivo. Our findings are the first to demonstrate that uridine is able to govern the functions of ISCs in intestinal organoid and mouse models. Thus, this study may provide a useful reference for developing novel functional food bioactives that maintain intestinal homeostasis.
The activity of intestinal stem cells (ISCs) is foremost in maintaining homeostasis and repair of intestines.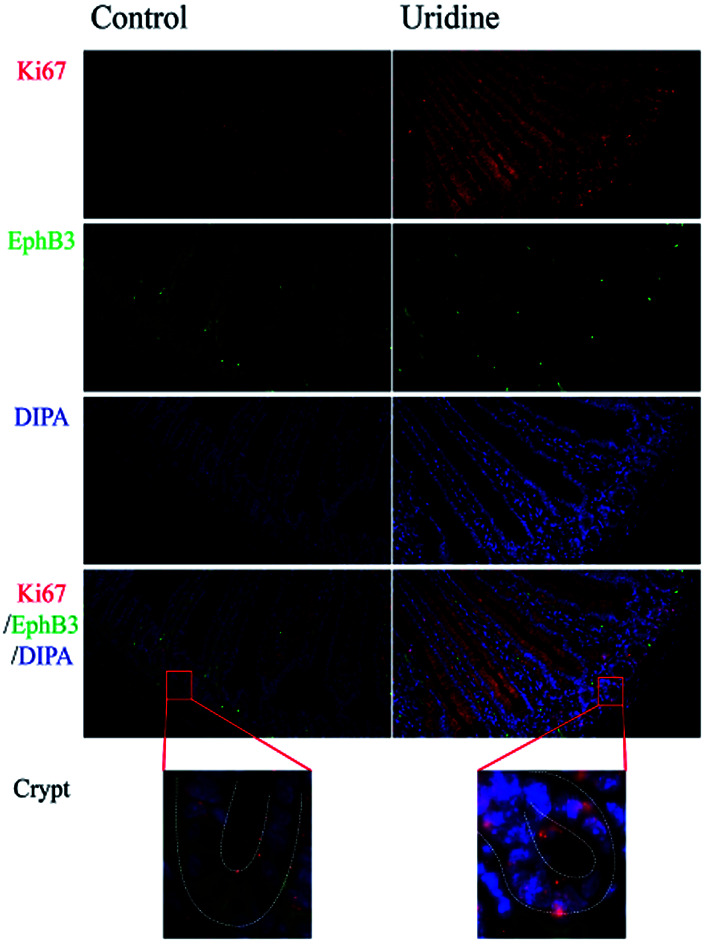
1. Introduction
The epithelial cell layer lining the intestinal surface plays an essential role in maintaining homeostasis by governing the digestion and absorption of nutrients, and functioning as an effective a barrier against pathogenic microorganisms, carcinogens and toxins in the gut lumen.1 As the fastest renewing tissue, the epithelium continuously produces new cells by division of intestinal stem cells (ISCs) at the bottom of the crypts to compensate for the loss of epithelial cells at the tips of the villi.1,2 The ISCs are capable of differentiating into a variety of intestinal cell types including enterocytes, enteroendocrine cells, goblet cells, and Paneth cells.3 Specific ISC markers have been identified comprising a B cell-specific MLV integration site-1 (Bmi1), leucine-rich-repeat-containing G-protein-coupled receptor 5 (Lgr5), achaete scute-like 2 (Ascl2), olfactomedin-4 (Olfm4), musashi-1 (Msi-1), and ephrin type-B receptor 3 (EphB3),4,5 which have been instrumental in our understanding of the ISC function. So far, the lack of effective in vitro models has hampered a proper understanding of the role of uridine in intestinal cell turnover. Two-dimensional (2D) cultures of cell lines fail to precisely mimic the complexity of in vivo tissues. To circumvent the shortcomings of conventional cell models, a 3D intestinal organoid (also called mini-gut or enteroid) model was developed by Sato and Clevers.1 3D intestinal organoids were generated from mouse small intestine comprising villus and crypt domains with multiple intestinal cell types, which were deemed to be superior to cell lines as an in vitro model.1,6 This model is excellently suited for investigations regarding ISCs and intestinal homeostasis.7–9 Thus, the present study was conducted to verify the effects of uridine on ISCs and decipher the underlying molecular mechanism using the intestinal organoid model.
As one of the three pyrimidine nucleosides, uridine is a glycosylated pyrimidine analog consisting of uracil linked to a ribose ring, and plays a crucial role in RNA and DNA biosynthesis.10 The de novo synthesis of uridine is accomplished through several reversible reactions including de-phosphorylation of a uridine monophosphate, deamination of a cytidine, or combination of a uracil and a ribose-1-phosphate, and this process is closely regulated by the mammalian target of rapamycin (mTOR) signaling.11 Uridine is also involved in some nutrition related biochemical processes including protein and lipid glycosylation, and extracellular matrix biosynthesis, etc.10 It has also been shown to tightly regulate liver energy metabolism.12,13 Moreover, uridine has been demonstrated to be pivotal in ameliorating side-effects caused by various anti-cancer and anti-HIV medicines.14 The level of a derivative of urine, uridine monophosphate (UMP), in bovine colostrum is higher than in normal milk, and this derivative is found to enhance innate immunity of newborn calves.15 Emerging evidence indicates that uridine is a key regulator of intestinal homeostasis and physiology.16 As the richest nucleotide in the milk of sows, UMP, as well as uridine, has been demonstrated to be beneficial for the growth performance of piglets and their intestinal morphology development.17–20 An adenine–uridine rich element binding protein, Tis11 closely regulates the activity of intestinal stem cells (ISCs).21 Most recently, it was confirmed that diets containing uridine prevent rotenone-induced gastrointestinal dysfunctions.22 Nevertheless, the precise molecular mechanism underlying the beneficial effect of uridine on intestinal cell turnover and ISC function remained elusive. Here, we present compelling evidence that uridine is capable of inhibiting the stemness of ISCs via mTOR in mouse and 3D intestinal organoids models. The observations in this study may provide a useful reference for developing novel functional food bioactives of maintaining intestinal homeostasis.
2. Materials and methods
2.1. Chemicals
Stocks (0.1 M) of uridine (Meiya Pharm, Hangzhou, China) were dissolved in phosphate-buffered saline (PBS), and rapamycin (2 mg mL−1, Beyotime Biotechnology, Shanghai, China) was dissolved in dimethylsulfoxide (DMSO). All chemicals were stored in 50 μL aliquots and frozen at −80 °C.
2.2. 3D primary mouse intestinal organoid culture
3D culture of mouse intestinal organoids was conducted as previously described.23 Briefly, small intestine was collected from a mouse sacrificed by cervical dislocation, followed by cleaning up the stools with ice-cold PBS. The intestine was longitudinally cut, followed by removal of the villi. To this aim, the intestine was dissected and cleaned with ice-cold PBS. The tissues were exposed to 2.5 mM EDTA for 30 min on ice. Then, the tissue was thoroughly suspended by pipetting it up and down 10 times with a fetal bovine serum (FBS, Gibco) rinsed 10 mL pipette tip to loosen crypts. Suspensions with crypts were filtered through a 70 μL cell strainer (Corning). Crypt suspension was supplemented with 10% FBS (vol/vol) and spun at 300g for 5 min at 4 °C. Next, the supernatant was discarded and crypts were re-suspended in 10 mL of DMEM/F12 media (Gibco™) supplemented with 1% (vol/vol) of GlutaMAX™ supplement (Gibco™, Grand island, USA), 10 mM of HEPES (Gibco™) and 100 U mL−1 of penicillin–streptomycin (P/S, Gibco™), and then the crypts were collected by spinning at 150g for 2 min at 4 °C. Around 500 crypts were suspended in 50 μL of growth factor reduced phenol-red free Matrigel (Corning, Bedford, USA), and seeded in the center of each well of a 24-well plate, and 500 μL of intestinal organoid growth media (the formula is referred to in a previous study24) was added after Matrigel solidification, which was subsequently incubated at 37 °C with 5% CO2. Passaging was performed every 3–4 days with a 1 : 3 split ratio. The organoid in the present study was passage 10 to 20. After passage, the organoid was treated with different concentrations of uridine for 48 h.
2.3. Animals and experiment design
Animal experiments were executed under the guidelines of the Laboratory Animal Ethical Commission of the Chinese Academy of Science and were approved by the Animal Welfare Committee of the Institute of Subtropical Agriculture, Chinese Academy of Sciences (2015-8A). Male C57BL/6J mice were purchased from SLAC Laboratory Animal Central (Changsha, China). All animals had free access to food and drinking water and were housed in a controlled room (temperature, 25 ± 2 °C; relative humidity, 45–60%; lighting cycle, 12 h d−1; 08:00–20:00 for light) during the experimentation. After 1 week of acclimatization, the 20 mice were randomly separated into two groups (10 mice per group): control and uridine groups, respectively. The mice in the control group received a basal diet (D12450B, Research Diets, Inc.), while the mice in the uridine group had access to the same basal diet, but were treated with uridine (Meiya Pharmaceutical co., Hangzhou, China) at a dosage of 1.0 g kg−1 in their drinking water. This treatment lasted for 11 week.
2.4. Quantitative real-time PCR (qRT-PCR)
The total RNA from the intestinal organoid samples was extracted using TRIZOL regent (Invitrogen, USA) supplemented with DNase I (Invitrogen, U.S.A.) according to the manufacturer's instructions, and quantified with a Nanodrop ND-1000 (Wilmington, DE, USA). The cDNA was generated with a reverse transcription system from TAKARA (TAKARA BIO). The qRT-PCR reactions were performed with TB Green™ (TAKARA BIO) according to the manufacturer's instruction. The qRT-PCR was executed using an ABI 7900HT PCR machine (ABI). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an endogenous reference to normalize the quantities of the target mRNA using the formula 2−ΔΔCT (ΔΔCT = ΔCTsample − ΔCTcontrol). Primer sequences are listed in Table 1.
Primers used in the qRT-PR.
| Gene name | Nucleotide sequence | Melting temperatures (°C) | PCR product size (bp) |
|---|---|---|---|
| LGR5-sense | 5′-AGAGCCTGATACCATCTGCAAAC-3′ | 61.9 | 1114 |
| LGR5-antisense | 5′-TGAAGGTCGTCCACACTGTTGC-3′ | 65.8 | |
| ASCL2-sense | 5′-TTTCCTGTGCCGCACCAGAACT-3′ | 68.4 | 108 |
| ASCL2-antisense | 5′-CAGCGACTCCAGACGAGGTGG-3′ | 67.8 | |
| CDKN1-sense | 5′-TCGCTGTCTTGCACTCTGGTGT-3′ | 65.7 | 630 |
| CDKN1-antisense | 5′-CCAATCTGCGCTTGGAGTGATAG-3′ | 65.3 | |
| Bmi1-sense | 5′-ACTACACGCTAATGGACATTGCC-3′ | 62.4 | 222 |
| Bmi1-antisense | 5′-CTCTCCAGCATTCGTCAGTCCA-3′ | 65 | |
| Msi1-sense | 5′-GTTCATCGGAGGACTCAGTTGG-3′ | 63.2 | 852 |
| Msi1-antisense | 5′-CTGGTCCATGAAAGTGACGAAGC-3′ | 65.3 | |
| EphB3-sense | 5′-CCTGTGTCAAGATCGAGGAGGT-3′ | 62.9 | 217 |
| EphB3-antisense | 5′-CTTCAGCGTCTTGATAGCCACG-3′ | 64.4 | |
| Caspase3-sense | 5′-GGAGTCTGACTGGAAAGCCGAA-3′ | 65.1 | 762 |
| Caspase3-antisense | 5′-CTTCTGGCAAGCCATCTCCTCA-3′ | 66.1 | |
| GAPDH-sense | 5′-CATCACTGCCACCCAGAAGACTG-3′ | 66.4 | 153 |
| GAPDH-antisense | 5′-ATGCCAGTGAGCTTCCCGTTCAG-3′ | 68.9 | |
| Rictor-sense | 5′-CAGTGTGAGGTCCTTTCCATCC-3′ | 63.1 | 400 |
| Rictor-antisense | 5′-GCCATAGATGCTTGCGACTGTG-3′ | 65.2 | |
| Rptor-sense | 5′-AGAAGGGTCTCCAAGGACGACT-3′ | 62.7 | 1875 |
| Rptor-antisense | 5′-GCAGGACACAAAGGCAGCATTG-3′ | 67.3 |
2.5. Intestinal organoid lysis, SDS-PAGE and western blotting
Intestinal organoid samples were collected, and the protein levels were determined as described previously.25 Briefly, samples were lysed with RIPA lysis buffer (strong, Beyotime Biotechnology, Shanghai, China), and boiled at 95 °C for 10 min. Then, lysates were subjected to SDS-PAGE, followed by the transferring of proteins to a polyvinylidene difluoride (PVDF) membrane (Immobilon-FL). Then, the membrane was blocked with 5% BAS solution diluted with TBS containing 0.05% Tween 20 (TBST) at room temperature for 1 h, followed by incubation with the corresponding primary antibodies including Lgr5 (Rabbit, Miltenyi Biotec, 1 : 1000), EphB3 (Mouse, SANTA CRUZ, 1 : 1000) and GAPDH (Rabbit, Cell Signaling Technology, 1 : 1000) overnight at 4 °C. Afterwards, the membranes were washed with TBST three times after being incubated with the corresponding secondary antibodies, followed by measurement of the immunoreactive bands using an Odyssey infrared imaging system (Bio-Rad).
2.6. Cytospin preparations and confocal laser scanning microscopy (CLSM) for intestinal organoids
Intestinal organoids were harvested from Matrigel using cold PBS, followed by fixing in 4% paraformaldehyde in PBS at 4 °C for 10 min. Then, the fixed intestinal organoids were attached to slides using a CytoSpin Cytocentrifuge (Yingtai Ltd, Changsha, China), spun at 1500 rpm for 5 min, followed by drying for 15 min. Slides containing intestinal organoids were washed with PBS for 3× 2 min, followed by treatment with 5% (vol/vol) Triton-X100 for 5 min. Then, the slides were washed with PBS for 3× 2 min, followed by incubation with 5% BSA solution for 30 min to block background staining. Then, the slides were incubated in a humidity chamber with anti-EphB3 antibodies (SANTA CRUZ, 1 : 200) diluted in 5% BSA solution at 4 °C overnight. The were washed for 3× 2 min with PBS prior to 1 h of incubation with a 1 : 1000 dilution of anti-rabbit IgG (H + L), F(ab′)2 fragment (Alexa Fluor® 488 Conjugate) secondary antibodies (Abcam). Nuclei were stained with Hoechst (1 : 1000, Beyotime Biotechnology, Shanghai, China). Images were taken using a confocal electroscope (Zeiss, Germany). Fluorescence density was measured using the ImageJ software (https://imagej.nih.gov/ij/).
2.7. Immunofluorescence histochemistry
Mice were sacrificed by cervical dislocation. The abdomen was opened to expose the intestine, and the small intestine was quickly isolated and fixed in 4% paraformaldehyde in PBS (PFA) for 1 h, followed by making paraffin sections. Slides were washed for 3× 5 min with PBS, followed by carrying out antigen retrieval. Slides were then blocked with PBS containing normal donkey serum and 0.3% Triton X-100 for 1 h, followed by incubation with primary antibodies (EphB3, SANTA CRUZ, 1 : 200; Ki67, SANTA CRUZ, 1 : 200) at 4 °C overnight. Tissues were incubated with secondary antibodies for 1 h at room temperature while being shielded from light. The tissues were examined using a fluorescence microscope (Nikon ECLIPSE C1, Tokyo, Japan).
2.8. Statistical analyses
All numerical results are expressed as the mean ± SEM. Statistical comparisons were analyzed using the Mann–Whitney test. P-values of less than 0.05 were considered to be statistically significant. An asterisk (*) is used to indicate P < 0.05 and double asterisks (**) indicate P < 0.01. Analysis was performed using GraphPad Prism Version 5 (GraphPad Software Inc., La Jolla, CA).
3. Results
3.1. Effect of uridine on Ki67 and EphB3 expression on intestinal crypts in vivo
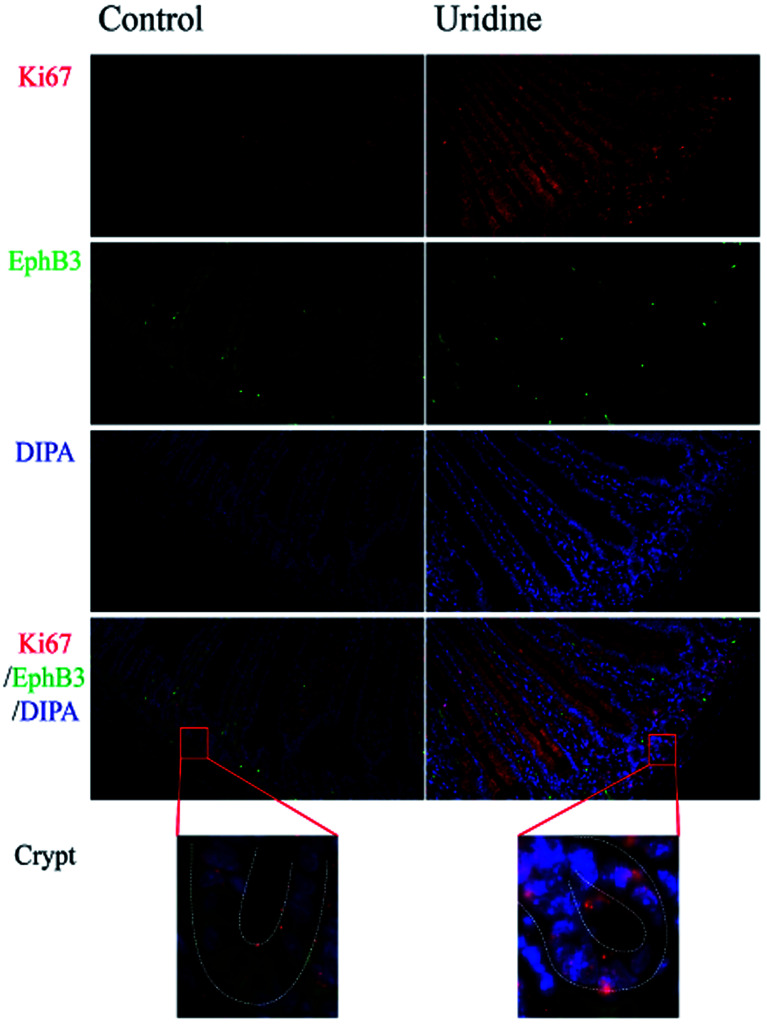
To assess the effect of uridine on the proliferation and stemness of ISCs, an in vivo experiment was performed, in which immunofluorescence staining showed that uridine clearly suppresses the expression of EphB3 in the crypts of mouse jejunum (Fig. 1), which was congruent with the observation in 3D intestinal organoids. At the same time, immunofluorescence staining showed that uridine clearly increased the expression of Ki67 in the crypts of mouse jejunum.
Fig. 1. Uridine treatment increases Ki67 expression and decreases EphB3 expression in the jejunum of mice (n = 4, 40×).
3.2. Effect of uridine on the number of crypts per intestinal organoid
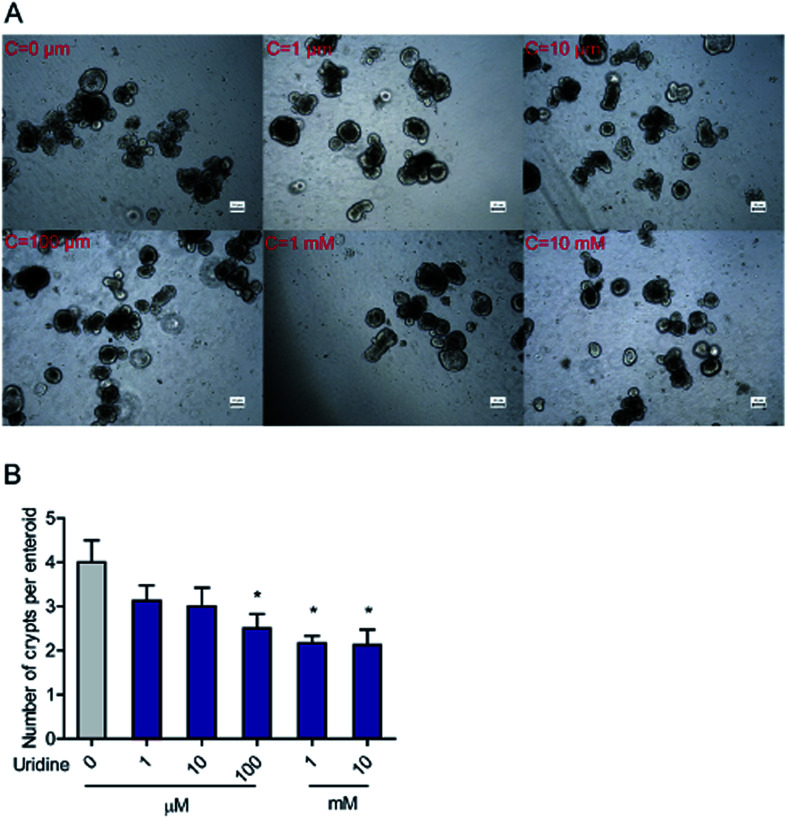
To further evaluate the effect of uridine on activity of ISCs, 3D intestinal organoids were used. Since ISCs are located at the crypts of intestinal organoids, the number of budded crypt structures in intestinal organoids is plausibly a fair indicator of the number of ISCs.26 First, we investigated the effect of exogenous uridine added at different concentrations to a number of crypt structures per intestinal organoid counted under the microscope. The results indicate that uridine significantly decreases the number of crypts per intestinal organoid by 1.50 ± 0.17 (P < 0.05), 1.83 ± 0.33 (P < 0.05), and 1.97 ± 0.15 (P < 0.05) fold at concentrations of 0.1, 1 and 10 mM, respectively (Fig. 2A and B). However, when the incubation time was decreased to 18 h, there was no significant effect on the number of crypts per intestinal organoid (ESI Fig. 1A and B†).
Fig. 2. Uridine decreased the number of crypts per intestinal organoid. (A) Representative images of the morphology of intestinal organoid, (B) quantification of crypts per intestinal organoid (n = 40).
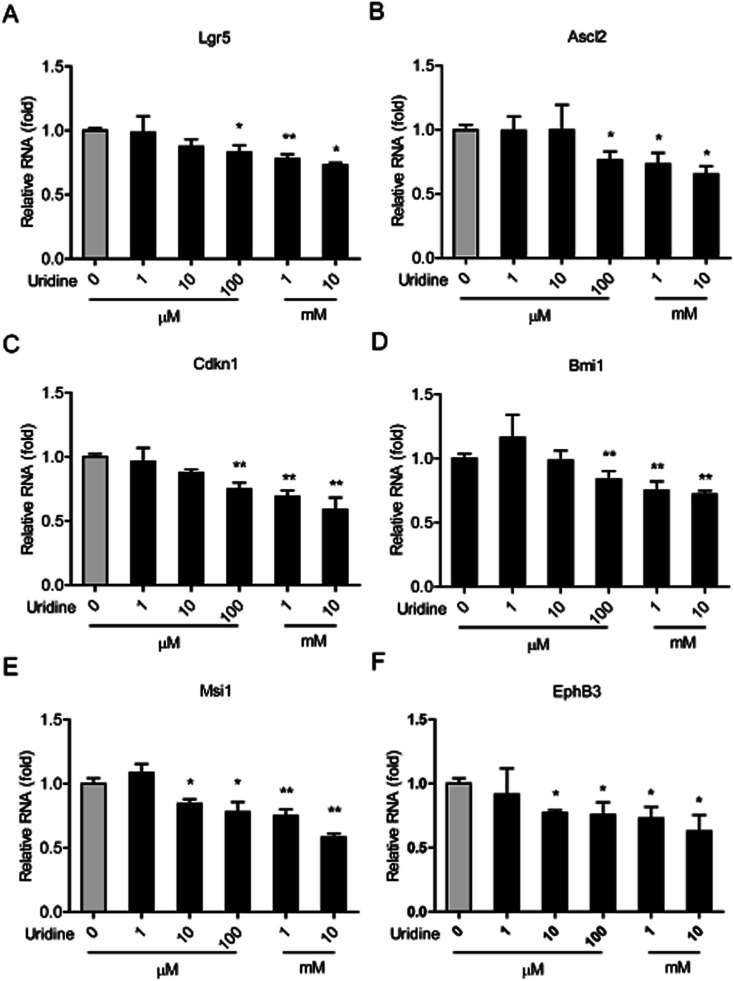
3.3. Effect of uridine on the mRNA expression of marker genes of ISCs
We next investigated the effect of uridine on the expression of marker genes of ISCs in intestinal organoids using qRT-PCR. In line with the results of our morphological analysis, the expression of different ISC marker genes was found to be significantly suppressed by uridine in a dose-dependent manner. Suppression of Lgr5, Ascl2, Cdkn1, and Bmi1 genes became significant at 100 μM uridine, and Msci1 and EphB3 genes became significant at 10 μM uridine. The suppression effect reached a maximum at 10 mM uridine.
3.4. Uridine causes a decrease in the protein levels of the marker genes of ISCs
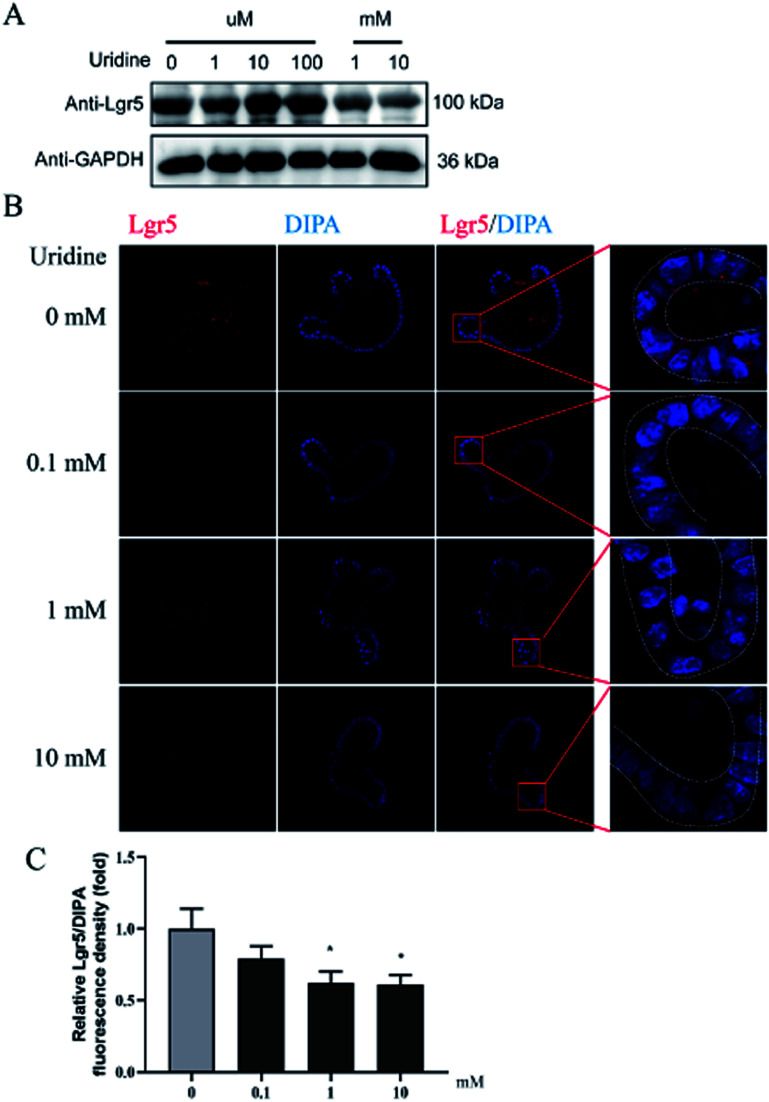
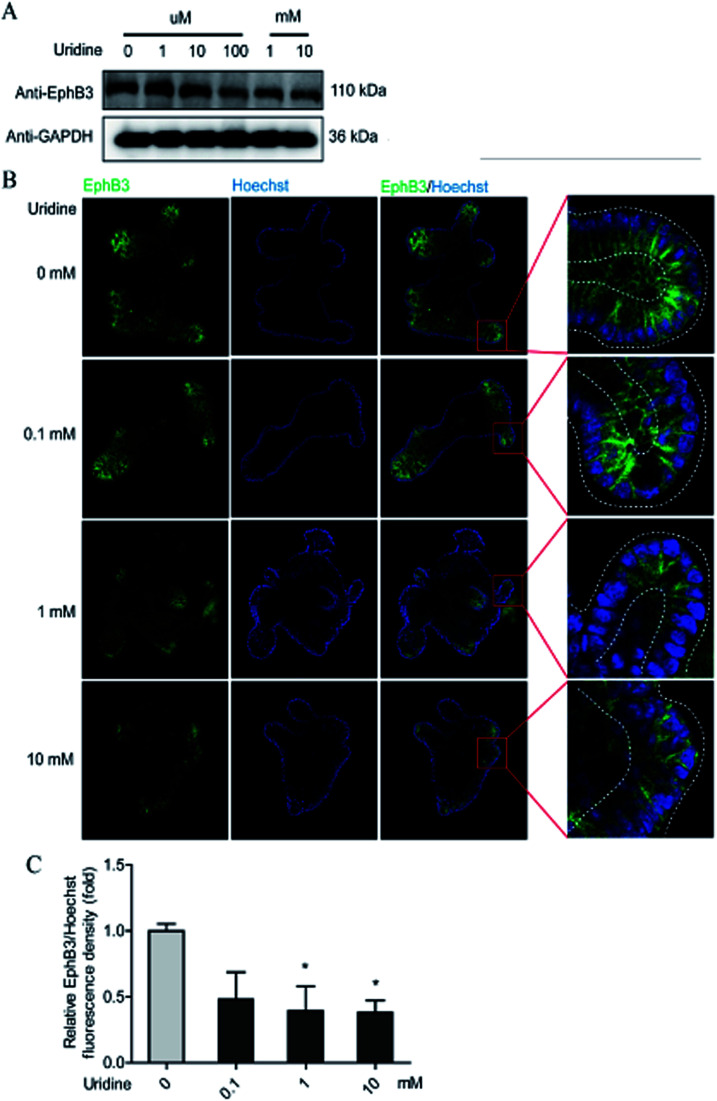
To further probe the effect of uridine on ISCs, western blotting was performed, which showed that uridine reduces the protein levels of EphB3 and Lgr5 in a dose-dependent manner (Fig. 5A and B). Furthermore, an immunofluorescence assay was implemented, which confirmed that EphB3 and LGR5 are specifically expressed at the crypts of intestinal organoids (Fig. 4B and 5B), and showed that 1 and 10 mM UR significantly decrease the protein levels of EphB3 and LGR5 per cell (Fig. 4C and 5C). In addition, when the incubation time was decreased to 18 h, immunofluorescence assay showed that 10 mM UR significantly decreases the protein levels of EphB3 per cell (denoted as EphB3/DIPA), and 0.1, 1 and 10 mM UR significantly decreased the protein levels of LGR5 per cell (denoted as LGR5/DIPA) (ESI Fig. 1C–E†).
Fig. 5. Uridine suppresses the protein level of markers of LGR5 in intestinal organoids (n = 4). (A) Representative gel for LGR5 (upper panel) and GAPDH (lower panel) of intestinal organoid, (B) representative images of LGR5 (red) and DIPA (blue) immunofluorescence of intestinal organoid, (C) quantification of fluorescence intensity of LGR5 in intestinal organoids (n = 4).
Fig. 4. Uridine suppresses the protein level of markers of EphB3 in intestinal organoids. (A) Representative gel for EphB3 (upper panel) and GAPDH (lower panel) of intestinal organoid, (B) representative images of EphB3 (green) and DIPA (blue) immunofluorescence of intestinal organoid, (C) quantification of fluorescence intensity of EphB3 in intestinal organoids (n = 4).
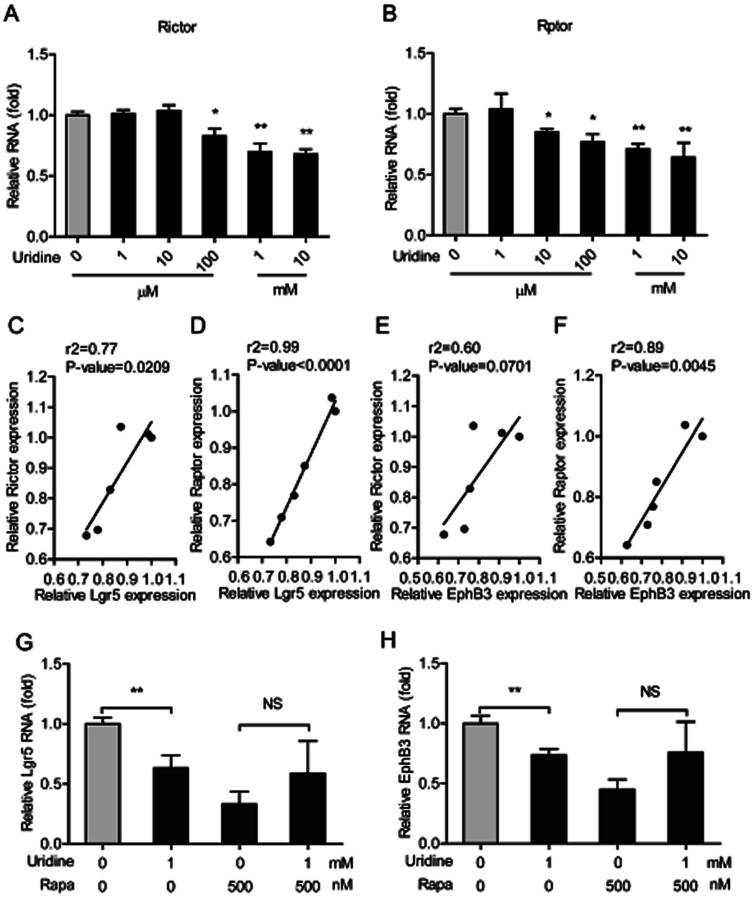
3.5. The inhibitory effect of uridine on the expression of marker genes of ISCs is mediated by mTOR
The mTOR plays a pivotal role in uridine synthesis,11 and it is an important regulator of stem cells.27 Thus, it is plausible that mTOR is involved in the regulatory effect of uridine on the stemness of ISCs. To this end, the effect of uridine on the expression of mTOR including mTOR complex 1 (mTORC1, which is also named as Raptor) and mTOR complex 2 (mTORC2, which is also named as Rictor) were measured by qRT-PCR. The results showed that uridine significantly decreases the mRNA expression levels of Rictor and Raptor (Fig. 6A and B). Subsequently, correlation analysis between the effects of uridine on the expression of Rictor/Raptor and marker genes of ISCs including Lgr5 and EphB3 was performed. Interestingly, the effect of uridine on the expression of Lgr5 showed a significant positive correlation with the expression of Rictor and Raptor, with correlation coefficients (r2) of 0.77 (P = 0.0209) and 0.99 (P < 0.0001), respectively (Fig. 6C and D); likewise, the effect of uridine on the expression of EphB3 was also positively correlated with the expression of Rictor and Raptor with correlation coefficients (r2) of 0.60 (P = 0.0701) and 0.89 (P < 0.0045), respectively (Fig. 6E and F). To further verify the underlying mechanism of action of the inhibitory effect of uridine on ISC marker expression, intestinal organoids were co-treated with uridine and rapamycin (mTOR inhibitor). The outcome showed that the inhibitory effect of uridine on mRNA expression of Lgr5 and EphB3 was completely abolished in the presence of rapamycin (Fig. 6G and H).
Fig. 6. The inhibitory effect of uridine on expression of markers of ISCs involves mTOR down-regulation. The mRNA abundances of (A) Rictor and (B) Raptor were determined by real-time PCR analysis, and relative gene expressions were normalized with GAPDH (n = 5). The correlation of Lgr5 and Rictor (C), Lgr5 and Raptor (D), EphB3 and Rictor (E), and EphB3 and Raptor (F). The mRNA expression of Lgr5 (G) and EphB3 (H) of intestinal organoid by treatment with rapamycin (n = 4).
4. Discussion
We present here a new understanding of how exogenously administered uridine may affect intestinal homoeostasis. The epithelial cell layer lining the intestinal surface plays an essential role in maintaining homeostasis and barrier functions of the intestine, which are thought to be tightly regulated by the stemness and differentiation of ISCs situated at the bottom of intestinal crypts. In most of our experiments we used a 3D primary intestinal organoid model, which contains a variety of cell types and bona fide recapitulates the physiology of in vivo intestinal epithelium, hence offering a new highly suitable platform for intestine-related studies. The outcome showed that uridine governs and suppresses the stemness of ISCs in mouse models and 3D intestinal organoids, and that this effect is mediated by mTOR.
The characteristics of multilineage differentiation of stem cells gives them a central role in maintaining homeostasis and repair of tissues, since the homeostasis requires newly produced cells to be differentiated from stem cells.27 Differentiation of ISCs is influenced by various factors, including immune regulation,28 epigenetic factors,29 transcription factors,30 and mitochondrial pyruvate metabolism.31 Increasing evidence shows that uridine is useful for treating many diseases such as neonatal brain damage,32 cancer,33 and fatty liver.13 Uridine is particularly rich in the plasma of rodents and humans.10 A decrease in plasma uridine induced by feeding has a beneficial effect on glucose metabolism, showing that uridine is closely involved in the regulation of energy homeostasis.10 It was found that suppression of mitochondrial activity in ISCs and suppression of glycolysis in Paneth cells are able to regulate ISC function.34 Moreover, mitochondrial pyruvate metabolism closely controls ISC function and proliferation.31 Thus, it seems that uridine may play a role in regulating ISC functions. In this study, we initially observed that dietary supplementation of uridine in mice in vivo was capable of inhibiting the expression of a marker gene of ISCs and EphB in the intestinal crypts (Fig. 1). Subsequently, we confirmed that uridine decreases the number of crypts per intestinal organoid (Fig. 2) as well as the expression of ISC marker genes in intestinal organoids (Fig. 3 and 4). In line with this finding, Palomo-Guerrero et al. showed that uridine-5′-triphosphate is capable of partially inhibiting differentiation signals in rat Schwann cells.35 However, to the best of our knowledge, the effects of uridine on ISCs or other types of stem cells have not been explored before, and the molecular mechanism underlying such effects is therefore not known.
Fig. 3. Uridine inhibits the mRNA expression levels of markers of ISCs in the intestinal organoids (n = 5).
The mammalian target rapamycin (mTOR) is a serine/threonine kinase that receives inputs from nutrients, growth factors, and environmental cues to regulate several fundamental cellular processes including protein synthesis, growth, metabolism, aging, regeneration, and autophagy. mTOR was also demonstrated to regulate de novo uridine synthesis.11 Moreover, calorie restriction induces mTORC1 to increase the number of ISCs in intestinal organoids and in vivo in the intestine, showing that mTOR also plays a crucial role in the expansion of ISCs.36 In contrast, mTOR disruption causes isolated intestinal crypts to fail to grow into intestinal organoids, which suggests that mTOR is crucial for ISC function.37 Interestingly, a S6K1 (downstream effector of mTOR) inhibitor, PF4708671, and an inhibitor of mTOR itself, rapamycin, abrogates the augmentation of intestinal organoid formation induced by calorie restriction.36 Consistently, rapamycin was found to inhibit the expression of EphB3 in our study (Fig. 6H). Furthermore, we found that uridine down regulated the expression of Rictor (mTOR complex 1) and Raptor (mTOR complex 2). Importantly, the inhibitory effects of uridine on the expression of ISC markers was positively correlated with the down regulation of Rictor and Raptor (Fig. 6C–F). Additionally, we found that rapamycin alone strongly suppresses the expression of the ISC markers EphB3 and Lgr5 and thereby prevents any further inhibitory effect of uridine on the expression of these markers (Fig. 6). Thus, mechanistically, we can show that the inhibitory effect of uridine on the expression of ISC marker genes occurs via inhibition of the mTOR pathway.
In summary, this is the first study demonstrating that uridine has an inhibitory effect on the expression of ISC markers in mouse intestinal organoids and in vivo, mediated by mTOR. Thus, our study provides a useful reference for understanding the effect that uridine has on the stemness of ISCs and may offer a firm basis for the development of functional food bioactives aimed at contributing towards the maintenance of intestinal homeostasis.
Abbreviations
- ISCs
Intestinal stem cells
- Ascl2
Achaete scute-like 2
- Olfm4
Olfactomedin-4
- Msi-1
Musashi-1
- mTOR
Mammalian target of rapamycin
- UMP
Uridine monophosphate
- 2D
Two-dimensional
- DMSO
Dimethylsulfoxide
- FBS
Fetal bovine serum
- qRT-PCR
Quantitative real-time PCR
- PVDF
Polyvinylidene difluoride
- CLSM
Confocal laser scanning microscopy
- GLP-1
Glucagon-like peptide 1
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
Author contributions
The authors' responsibilities were as follows—Xin Wu: conceived and designed the research; Yi-Lin Liu, and Song-Ge Guo: performed experiments and analyzed samples; Yi-Lin Liu: analyzed the data, interpreted the results of the experiments, prepared the figures, and drafted the manuscript; Chun-yan Xie, Kaimin Niu, and Hugo De Jonge: edited and revised the manuscript; and all authors: read and approved the final manuscript, Foreign Expert Project (G20190161004) and CAS President's International Fellowship for Visiting Scientists (2019VBA0015).
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
We thank the National Key Research and Development Program of China (2016YFD0500500), the Science and Technology Projects of Hunan Province (2019RS3020; 2019RS3021), the CSC scholarship, Jiangxi Provincial Innovation and Entrepreneurship projects, and the STS Network Initiative program of the Chinese Academy of Sciences (KFJ-STS-QYZX-031) for providing funding support.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra07742a
References
- Sato T. Vries R. G. Snippert H. J. van de Wetering M. Barker N. Stange D. E. van Es J. H. Abo A. Kujala P. Peters P. J. Clevers H. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Beumer J. Clevers H. Development. 2016;143:3639–3649. doi: 10.1242/dev.133132. [DOI] [PubMed] [Google Scholar]
- Biswas S. Davis H. Irshad S. Sandberg T. Worthley D. Leedham S. J. Pathol. 2015;237:135–145. doi: 10.1002/path.4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T. M. Chia L. A. Kosinski C. M. Kuo C. J. Cell. Mol. Life Sci. 2011;68:2513–2523. doi: 10.1007/s00018-011-0687-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell A. E. Wang Y. Li Y. Poulin E. J. Means A. L. Washington M. K. Higginbotham J. N. Juchheim A. Prasad N. Levy S. E. Guo Y. Shyr Y. Aronow B. J. Haigis K. M. Franklin J. L. Coffey R. J. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y. Zhou D. Front. Cell. Infect. Microbiol. 2018;8:102. doi: 10.3389/fcimb.2018.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y. B. Guo S. G. Wan D. Wu X. Yin Y. L. J. Agric. Food Chem. 2019;67:2421–2428. doi: 10.1021/acs.jafc.8b06908. [DOI] [PubMed] [Google Scholar]
- Yin Y. B. de Jonge H. R. Wu X. Yin Y. L. Drug Discovery Today. 2019;24:1784–1794. doi: 10.1016/j.drudis.2019.06.006. [DOI] [PubMed] [Google Scholar]
- Yin Y. B. de Jonge H. R. Wu X. Yin Y. L. Mol. Nutr. Food Res. 2019:e1801143. doi: 10.1002/mnfr.201801143. doi: 10.1002/mnfr.201801143. [DOI] [PubMed] [Google Scholar]
- Deng Y. Wang Z. V. Gordillo R. An Y. Zhang C. Liang Q. Yoshino J. Cautivo K. M. De Brabander J. Elmquist J. K. Horton J. D. Hill J. A. Klein S. Scherer P. E. Science. 2017;355:eaaf5375. doi: 10.1126/science.aaf5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I. Howell J. J. Asara J. M. Manning B. D. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urasaki Y. Pizzorno G. Le T. T. PLoS One. 2014;9:e99728. doi: 10.1371/journal.pone.0099728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T. T. Urasaki Y. Pizzorno G. PLoS One. 2014;9:e87179. doi: 10.1371/journal.pone.0087179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. E. Go J. Sung J. E. Lee H. A. Yun W. B. Hong J. T. Hwang D. Y. BMC Gastroenterol. 2017;17:21. doi: 10.1186/s12876-017-0576-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiko T. Nagafuchi S. Kanbe M. Obara Y. Hagawa Y. Takahashi T. Katoh K. J. Anim. Sci. 2009;87:1042–1047. doi: 10.2527/jas..2008-1004. [DOI] [PubMed] [Google Scholar]
- Liu Y. Zhang Y. Yin J. Ruan Z. Wu X. Yin Y. Chronobiol. Int. 2019;36:1258–1267. doi: 10.1080/07420528.2019.1637347. [DOI] [PubMed] [Google Scholar]
- Li B. Zhou H. Wu X. Chen Z. Yao J. Yin Y. J. Anim. Sci. 2016;94:82–86. doi: 10.2527/jas.2015-9440. [DOI] [Google Scholar]
- Xie C. Y. Wang Q. Li G. Fan Z. Wang H. Wu X. J. Sci. Food Agric. 2019;99:6108–6113. doi: 10.1002/jsfa.9850. [DOI] [PubMed] [Google Scholar]
- Li G. Xie C. Wang Q. Wan D. Zhang Y. Wu X. Yin Y. Food Funct. 2019;10:4081–4089. doi: 10.1039/C9FO00360F. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Guo S. Xie C. Wang R. Zhang Y. Zhou X. Wu X. Animals. 2019;9:610. doi: 10.3390/ani9090610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland L. Jasper H. Biteau B. Dev. Biol. 2017;426:8–16. doi: 10.1016/j.ydbio.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pardo P. Dodiya H. B. Broersen L. M. Douna H. van Wijk N. Lopes da Silva S. Garssen J. Keshavarzian A. Kraneveld A. D. Nutr. Neurosci. 2018;21:391–402. doi: 10.1080/1028415X.2017.1294555. [DOI] [PubMed] [Google Scholar]
- Yin Y. Dang W. Zhou X. Xu L. Wang W. Cao W. Chen S. Su J. Cai X. Xiao S. Peppelenbosch M. P. Pan Q. Virulence. 2018;9:83–98. doi: 10.1080/21505594.2017.1326443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y. Bijvelds M. Dang W. Xu L. van der Eijk A. A. Knipping K. Tuysuz N. Dekkers J. F. Wang Y. de Jonge J. Sprengers D. van der Laan L. J. Beekman J. M. Ten Berge D. Metselaar H. J. de Jonge H. Koopmans M. P. Peppelenbosch M. P. Pan Q. Antiviral Res. 2015;123:120–131. doi: 10.1016/j.antiviral.2015.09.010. [DOI] [PubMed] [Google Scholar]
- Yin Y. Chen S. Hakim M. S. Wang W. Xu L. Dang W. Qu C. Verhaar A. P. Su J. Fuhler G. M. Peppelenbosch M. P. Pan Q. Antiviral Res. 2018;156:92–101. doi: 10.1016/j.antiviral.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J. M. Calabrese P. P. Miller A. J. Munoz N. M. Grady W. M. Shibata D. Liskay R. M. Proc. Natl. Acad. Sci. U. S. A. 2016;113:12192–12197. doi: 10.1073/pnas.1611980113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. K. Du W. Shelton S. J. Oldham M. C. DiPersio C. M. Klein O. D. Cell Stem Cell. 2017;21:91–106.e6. doi: 10.1016/j.stem.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton M. Haber A. L. Rogel N. Burgin G. Beyaz S. Schnell A. Ashenberg O. Su C. W. Smillie C. Shekhar K. Chen Z. Wu C. Ordovas-Montanes J. Alvarez D. Herbst R. H. Zhang M. Tirosh I. Dionne D. Nguyen L. T. Xifaras M. E. Shalek A. K. von Andrian U. H. Graham D. B. Rozenblatt-Rosen O. Shi H. N. Kuchroo V. Yilmaz O. H. Regev A. Xavier R. J. Cell. 2018;175:1307–1320.e22. doi: 10.1016/j.cell.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaffer K. L. Kim R. Aoki R. Elliott E. N. Schug J. Burger L. Schubeler D. Kaestner K. H. Genes Dev. 2014;28:652–664. doi: 10.1101/gad.230318.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loza-Coll M. A. Southall T. D. Sandall S. L. Brand A. H. Jones D. L. EMBO J. 2014;33:2983–2996. doi: 10.15252/embj.201489050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell J. C. Wisidagama D. R. Bensard C. Zhao H. Wei P. Tanner J. Flores A. Mohlman J. Sorensen L. K. Earl C. S. Olson K. A. Miao R. Waller T. C. Delker D. Kanth P. Jiang L. DeBerardinis R. J. Bronner M. P. Li D. Y. Cox J. E. Christofk H. R. Lowry W. E. Thummel C. S. Rutter J. Nat. Cell Biol. 2017;19:1027–1036. doi: 10.1038/ncb3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren B. Cakir A. Sevinc C. Serter Kocoglu S. Ocalan B. Oy C. Minbay Z. Kahveci N. Alkan T. Cansev M. Brain Res. 2017;1676:57–68. doi: 10.1016/j.brainres.2017.09.010. [DOI] [PubMed] [Google Scholar]
- Field M. S. Lan X. Stover D. M. Stover P. J. Curr. Dev. Nutr. 2018;2:nzy013. doi: 10.1093/cdn/nzy013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Colman M. J. Schewe M. Meerlo M. Stigter E. Gerrits J. Pras-Raves M. Sacchetti A. Hornsveld M. Oost K. C. Snippert H. J. Verhoeven-Duif N. Fodde R. Burgering B. M. Nature. 2017;543:424–427. doi: 10.1038/nature21673. [DOI] [PubMed] [Google Scholar]
- Palomo-Guerrero M. Cosgaya J. M. Gella A. Casals N. Grijota-Martinez C. Neuroscience. 2018;372:255–265. doi: 10.1016/j.neuroscience.2018.01.010. [DOI] [PubMed] [Google Scholar]
- Igarashi M. Guarente L. Cell. 2016;166:436–450. doi: 10.1016/j.cell.2016.05.044. [DOI] [PubMed] [Google Scholar]
- Sampson L. L. Davis A. K. Grogg M. W. Zheng Y. FASEB J. 2016;30:1263–1275. doi: 10.1096/fj.15-278606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.