ABSTRACT
Incidences of nosocomial infections mediated by New Delhi metallo-β-lactamase (NDM) enzyme-producing Enterobacterales are increasing globally, resulting in a great burden to public health. The carbapenem-resistant Enterobacterales (CRE) were collected from Henan, China during 2013–2016. The blaNDM-positive strains were characterized using PCR, antimicrobial susceptibility testing, conjugation assay, S1 nuclease pulsed-field gel electrophoresis (S1-PFGE), Southern blot, whole-genome sequencing (WGS), and bioinformatics analysis. Eighty-one NDM-producing strains were identified among 391 nonduplicate CRE strains. Among them, four strains cocarried mcr and blaNDM genes, and two carried blaIMP-4 and blaNDM genes. The coexistence of blaNDM-5 and mcr-9 in Enterobacter hormaechei was found for the first time. In total, four blaNDM subtypes were identified. Among them, blaNDM-1 and blaNDM-5 were predominant. There was an obvious increasing trend in blaNDM-5 from 2013 to 2016. Thirteen different bacterial species were found among the 81 strains, and Escherichia coli was the dominant strain. blaNDM genes were located on nine different Inc-type plasmids, most of them on the IncX3 plasmids, except for the Pr-15-2-50 strain, which was located on the chromosome. We characterized two novel plasmids: the IncHI5-like plasmid carrying blaNDM-9 found in K. pneumonia, and the IncI1 blaNDM-5-positive plasmid. These findings provide the genomic basis for the widespread transmission of blaNDM and pave the way for the formulation of more effective monitoring and control methods.
IMPORTANCE To control the emergence and transmission of CRE, it is important to perform retrospective genomic investigations. It is important to evaluate the plasmid diversity, genetic environment, and evolutionary relationships of the blaNDM-positive clinical strains in the early transmission stages. This study conducted an in-depth analysis of blaNDM-positive pathogens during a 4-year period using different methods for observing the high prevalence and active transmission of blaNDM-positive CRE. Moreover, we also explored the coexistence of the blaNDM and mcr, a clinically important mobile colistin resistance gene. This study shows that the prevalence of blaNDM-positive pathogens in Henan is high and the isolation rates increase each year. Moreover, plasmid-mediated horizontal transfer plays an important role in blaNDM dissemination. The co-occurrence of multiple resistance genes highlighted a long-lasting evolutionary pathway. Therefore, we have suggested the long-term continuous surveillance of clinical pathogens carrying blaNDM to learn the future transmission trend and curb the public health risk caused by CRE.
KEYWORDS: Enterobacterales, plasmid diversity, bla NDM , molecular epidemiology, nanopore sequencing
INTRODUCTION
Carbapenem antibiotics are β-lactam antibiotics with a broad antibacterial spectrum and strong antibacterial activity. They are the most important antibiotics for the treatment of multidrug-resistant (MDR) Gram-negative bacterial infections (1). However, the clinical use of these drugs leads to the emergence of carbapenem-resistant Enterobacterales (CRE) (2) and makes clinical medication selection difficult. In 2013, the Centers for Diseases Control and Prevention in the U.S. reported that more than 9,000 health care-related infections were caused by CRE each year. It ranked CRE in the highest threat level. Moreover, the China CRE Monitoring Network showed that the hospital mortality rate of CRE was 33.5% (222/662) (3). It also showed that the mortality rate increased with the length of hospital stay.
Carbapenem-inactivating carbapenemases are predominantly divided into Classes A, B, and D according to the Ambler classification. Classes A and D belong to serine enzymes, and B belongs to metallo-β-lactamases (MBLs). NDM is a typical member of the B1 class of MBLs. It is capable of hydrolyzing all β-lactams, except monobactams (4). It recruits mobile genetic elements, such as plasmids belonging to different replicon or Inc types (IncFII, IncHI2, IncN, and IncX3), insertion sequences (ISAba125, ISCR1), and transposons (Tn125) (5). blaNDM genes have already spread to various species of bacteria worldwide, including Enterobacterales and nonfermenting Gram-negative bacilli (6). The increasing prevalence of NDM-producing pathogens has seriously compromised the efficacy of carbapenems in clinical settings, and it poses a great threat to public health. According to current reports, 28 NDM variants have been identified in multiple species of Enterobacterales, Acinetobacter, and Pseudomonas. NDM-1 and NDM-5, which were encoded mainly by IncX3 plasmids, were the most frequently detected variants in Enterobacterales. However, NDM-5 was more prevalent compared to NDM-1 in Escherichia coli. Our previous study revealed only NDM-1, and no other variants were detected in NDM-producing Enterobacterales isolated from the Henan province between 2011 and 2012. Moreover, the IncA/C plasmids with broad-host-range were the predominant vehicles for blaNDM compared to the narrow-host range IncX3 plasmids (7). These differences indicate the changes in the prevalence and evolution of blaNDM-bearing plasmids. Therefore, we continuously monitored the NDM-producing CRE strains in a teaching hospital in Zhengzhou University over a 4-year period (2013–2016). We tried to elucidate the molecular mechanisms for the blaNDM gene transfers, and study the evolution of the epidemic blaNDM plasmids and their clones.
RESULTS
Overview of NDMs-producing CRE isolates.
From 2013 to 2016, 391 nonduplicate CRE isolates belonging to 13 different species were collected from a teaching hospital in the Zhengzhou University for screening carbapenemase genes using PCR and Sanger sequencing. The result showed 291 Klebsiella pneumoniae strains (74.42%) carrying the blaKPC-2 gene and another 81 (20.72%) belonging to various species carrying the blaNDM (Table 1). This illustrated that K. pneumoniae and E. coli were the main clinical CREs. KPC and NDM were the primary carbapenem-inactivating enzymes in CRE recovered from the Henan province. It was well recognized that blaNDM genes were mainly carried by Gram-negative Enterobacterales, including E. coli, K. pneumoniae, Citrobacter freundii, and Enterobacter cloacae (8–10). The prevalence of blaNDM in different Enterobacterales was 49.38% (40/81), 14.81% (12/81), 13.58% (11/81), 7.41% (6/81), and 4.94% (4/81) in E. coli, K. pneumoniae, Enterobacter hormaechei, C. freundii, and Citrobacter portucalensis, respectively. There was also 1.23% (1/81) in each Citrobacter braakii, Klebsiella aerogenes, Klebsiella pasteurii, Klebsiella oxytoca, Raoultella ornithinolytica, Serratia marcescens, Proteus mirabilis, and Providencia rettgeri. This indicates that E. coli was the most common host for blaNDM, followed by K. pneumoniae and E. hormaechei. Sanger sequencing of blaNDM genes identified four blaNDM subtypes, including blaNDM-1 (n = 41), blaNDM-5 (n = 38), blaNDM-4 (n = 2), and blaNDM-9 (n = 1) (Table 1). Among them, blaNDM-5 was the most prevalent subtype in E. coli (33/40, 82.5%), and the majority of K. pneumoniae carried blaNDM-1 (9/12, 75%). However, carbapenemase gene blaIMP-4 was only detected in two NDM-producing strains (KA-14-61 and KO-14-71).
TABLE 1.
Basic information of the 81 blaNDM-bearing strains
| Isolate | Species | MLSTa | Collection date | Age/sex | Specimen type | Ward | Prognosis | Conjugation frequency | NDM-type | Plasmid type carrying blaNDM | NDM-positive plasmid size (kb) | Grouping of IncX3 blaNDM-positive plasmids |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KP-13-8 | K. pneumoniae | ST494 | 2013.01.06 | 61yr/female | blood | Gastroenterology dept | discharge | -b | NDM-5 | IncX3 | 46 | B |
| EC-13-1 | E. coli | ST40 | 2013.01.25 | 6days/male | blood | ICU | discharge | - | NDM-1 | IncX3 | 54 | B |
| KP-13-11 | K. pneumoniae | ST35 | 2013.04.25 | 2mo/female | sputum | ICU | death | 3.6 × 10−4 | NDM-1 | IncX3 | 54 | C |
| CR-13-12 | E. hormaechei | ST419 | 2013.05.06 | 89yr/female | sputum | ICU | discharge | 3.3 × 10−4 | NDM-1 | IncFII | 87 | - |
| EC-13-22 | E. coli | ST361 | 2013.08.05 | 41yr/female | drainage liquid | gynecology | discharge | 2.6 × 10−6 | NDM-1 | IncC | 213 | - |
| EC-13-31 | E. coli | ST167 | 2013.09.04 | 68yr/male | blood | gynecology | discharge | - | NDM-5 | IncX3 | 46 | B |
| ECL-13-2 | E. hormaechei | ST177 | 2013.09.04 | 53yr/female | urine | urology | discharge | 1.8 × 10−7 | NDM-1 | IncFII-IncFIB | 138 | - |
| EC-13-30 | E. coli | ST167 | 2013.09.18 | 35yr/male | secreta | endocrinology | discharge | - | NDM-5 | IncX3 | 46 | B |
| KP-13-7 | K. pneumoniae | ST1 | 2013.09.26 | 37yr/male | bile | hepatological surgery | discharge | 1.33 × 10−4 | NDM-1 | IncX3 | 54 | C |
| EC-13-33 | E. coli | ST540 | 2013.10.06 | 68yr/male | blood | gynecology | discharge | 3 × 10−4 | NDM-1 | IncFII-IncN | 78 | - |
| ECL-13-4 | E. hormaechei | ST88 | 2013.10.17 | 48yr/male | blood | ICU | death | 2.8*10−4 | NDM-5 | IncX3 | 46 | B |
| CF-13-34 | C. portucalensis | ST328 | 2013.10.19 | 23yr/male | secreta | hematology | death | 1.1 × 10−4 | NDM-1 | IncX3 | 54 | E |
| EC-13-49 | E. coli | ST167 | 2013.11.07 | 78yr/female | urine | kidney internal | discharge | 3.5 × 10−6 | NDM-1 | IncC | 215 | - |
| ECL-13-37 | E. hormaechei | ST231 | 2013.11.14 | 37yr/male | urine | urology | discharge | - | NDM-5 | IncX3 | 46 | B |
| KP-13-14 | K. pneumoniae | ST782 | 2013.11.23 | 21days/male | wound | pediatric surgery | discharge | - | NDM-9 | IncHI5 | 358 | - |
| CR-13-36 | E. hormaechei | ST419 | 2013.12.05 | 47yr/female | urine | kidney internal | discharge | 1.2 × 10−5 | NDM-1 | IncFII | 87 | - |
| PM58 | P. mirabilis | NA | 2013.12.15 | 3yr/female | urine | rehabilitation medicine | discharge | 9.4 × 10−6 | NDM-1 | - | 85 | - |
| KP-14-2-131 | K. pneumoniae | ST345 | 2014.01.23 | 44yr/male | urine tube tip | neurosurgery | discharge | 1.4 × 10−6 | NDM-1 | IncHI5 | 358 | - |
| KOR-14-72 | R. ornithinolytica | NA | 2014.02.15 | 71yr/female | sputum | ICU | discharge | 3.9 × 10−5 | NDM-1 | IncX3 | 46 | C |
| KO-14-71 | K. pasteurii | NA | 2014.02.20 | 67yr/female | sputum | ICU | discharge | 3.2 × 10−3 | NDM-1 | IncX3 | 54 | C |
| EC-14-2-77 | E. coli | ST410 | 2014.03.30 | 66yr/male | drainage liquid | hepatological surgery | discharge | 2.5 × 10−3 | NDM-4 | IncX3 | 54 | C |
| ECL-14-58 | E. hormaechei | ST177 | 2014.05.12 | 10yr/male | pus | pediatric surgery | discharge | 9.5 × 10−5 | NDM-1 | IncX3 | 54 | C |
| ECL-14-60 | E. hormaechei | ST696 | 2014.06.05 | 62yr/male | blood | ICU | death | - | NDM-1 | IncC, IncX3 | 171-54 | D |
| EC-14-55 | E. coli | ST410 | 2014.06.06 | 14yr/female | blood | Pediatric medicine | death | - | NDM-4 | IncX3 | 46 | C |
| KA-14-61 | K. aerogenes | NA | 2014.08.30 | 33yr/male | secreta | Department of Burn Repair and Reconstruction | discharge | 2.2 × 10−4 | NDM-1 | IncX3 | 46 | B |
| EC-14-54 | E. coli | ST167 | 2014.08.30 | 51yr/male | sanies | intestine surgery | discharge | - | NDM-5 | IncX3 | 46 | B |
| CF-14-50 | C. freundii | ST22 | 2014.09.20 | 44yr/male | urine | urology | discharge | 1.9 × 10−4 | NDM-1 | IncX3 | 54 | D |
| ECL-14-56 | E. hormaechei | ST171 | 2014.11.02 | 45yr/male | blood | ICU | death | - | NDM-1 | IncX3 | 54 | C |
| KP-14-6 | K. pneumoniae | ST76 | 2014.11.13 | 10days/female | blood | Infectious disease | discharge | 2.9 × 10−4 | NDM-1 | IncC | 200 | - |
| EC-14-2-134 | E. coli | ST101 | 2014.11.17 | 31yr/male | swab | Burn Repair and Reconstruction | discharge | 5.9 × 10−5 | NDM-5 | IncX3 | 46 | B |
| EC-14-2-92 | E. coli | ST167 | 2014.11.27 | 50yr/male | blood | oncology | death | 4 × 10−5 | NDM-5 | IncX3 | 46 | B |
| EC-14-2-94 | E. coli | ST167 | 2014.12.10 | 44yr/female | urine | urology | discharge | - | NDM-5 | IncX3 | 46 | B |
| EC-14-2-9 | E. coli | ST167 | 2014.12.19 | 51yr/female | sputum | Rheumatology | discharge | - | NDM-5 | IncX3 | 46 | B |
| EC-15-2-5 | E. coli | ST167 | 2015.01.16 | 26yr/male | sputum | ICU | death | - | NDM-5 | IncX3 | 54 | D |
| EC-15-2-14 | E. coli | ST2083 | 2015.01.25 | 23yr/female | sputum | Rheumatology | discharge | - | NDM-5 | IncX3 | 46 | B |
| CF-15-43 | C. portucalensis | ST17 | 2015.02.21 | 47yr/female | urine | urology | discharge | 3 × 10−5 | NDM-1 | IncX3 | 54 | E |
| CF-15-2-98 | C. portucalensis | ST17 | 2015.02.21 | 47yr/female | urine | urology | discharge | 6.8 × 10−5 | NDM-1 | IncX3 | 54 | E |
| KP-15-2-113 | K. pneumoniae | ST1083 | 2015.03.08 | 2mo/male | sputum | neonatology | discharge | 3.3 × 10−5 | NDM-1 | IncX3 | 46 | C |
| EC-15-10 | E. coli | ST540 | 2015.03.14 | 74yr/male | sputum | ICU | discharge | - | NDM-5 | IncX3 | 54 | D |
| EC-15-3 | E. coli | ST6388 | 2015.03.23 | 53yr/female | urine | urology | discharge | - | NDM-1 | IncFII | 110 | - |
| CF-15-61 | C. freundii | ST22 | 2015.04.08 | 75yr/female | drainage liquid | gastrointestinal surgery | discharge | 1.7 × 10−4 | NDM-1 | IncX3 | 54 | C |
| CF-15-33 | C. freundii | NA | 2015.05.20 | 7yr/male | blood | Pediatric medicine | discharge | 1.7 × 10−5 | NDM-1 | IncX3 | 54 | C |
| EC-15-34 | E. coli | ST746 | 2015.05.22 | 60yr/male | blood | urology | death | - | NDM-5 | IncX3 | 46 | B |
| KP-15-35 | K. pneumoniae | ST17 | 2015.05.22 | 10days/male | blood | neonatology | death | 1.4 × 10−5 | NDM-1 | IncX3 | 54 | C |
| EC-15-2-35 | E. coli | ST540 | 2015.06.27 | 75yr/female | urine | urology | discharge | - | NDM-5 | IncX3 | 54 | D |
| EC-15-2-56 | E. coli | ST167 | 2015.06.27 | 52yr/male | urine | urology | discharge | - | NDM-5 | IncX3 | 46 | A |
| EC-15-2-24 | E. coli | ST540 | 2015.06.29 | 75yr/female | urine | urology | discharge | 2.5 × 10−5 | NDM-5 | IncX3 | 54 | D |
| EC-15-2-47 | E. coli | ST540 | 2015.06.29 | 67yr/male | urine | urology | discharge | - | NDM-5 | IncX3 | 54 | D |
| SM-15-2-16 | S. marcescens | NA | 2015.07.01 | 27yr/female | sputum | respiratory medicine | discharge | 2.5 × 10−5 | NDM-1 | IncX3 | 46 | B |
| EC-15-2-132 | E. coli | ST410 | 2015.07.15 | 38ye/female | blood | Hematology dept | discharge | - | NDM-5 | IncX3 | 46 | D |
| EC-15-2-51 | E. coli | ST617 | 2015.07.21 | 9mo/male | urine | ICU | discharge | - | NDM-5 | IncX3 | 46 | B |
| EC-15-2-65 | E. coli | ST6388 | 2015.07.30 | 65yr/male | urine | ICU | discharge | 6.8 × 10−5 | NDM-5 | IncX3 | 46 | C |
| KP-15-2-62 | K. pneumoniae | ST490 | 2015.07.30 | 2yr/female | blood | ICU | death | - | NDM-5 | IncX3 | 46 | D |
| KP-15-2-52 | K. pneumoniae | ST1440 | 2015.08.03 | 66yr/male | urine | urology | death | 5.5 × 10−6 | NDM-5 | IncX3 | 46 | B |
| CF-15-2-29 | C. freundii | ST22 | 2015.08.04 | 1mo/female | sputum | ICU | discharge | 8.8 × 10−4 | NDM-1 | IncX3 | 54 | C |
| EC-15-2-26 | E. coli | ST167 | 2015.08.08 | 49yr/female | urine | gynecology | discharge | - | NDM-5 | IncX3 | 46 | B |
| Pr-15-2-50 | P. rettgeri | NA | 2015.08.13 | 19yr/female | joint fluid | internal medicine | discharge | - | NDM-1 | - | - | - |
| EC-15-2-1 | E. coli | ST167 | 2015.10.28 | 67yr/female | urine | cardiac surgery | discharge | - | NDM-5 | IncX3 | 54 | B |
| EC-15-2-2 | E. coli | ST617 | 2015.10.28 | 56yr/male | drainage liquid | hepatological surgery | discharge | 3.3 × 10−5 | NDM-5 | IncX3 | 54 | D |
| KP-15-2-6 | K. pneumoniae | ST11 | 2015.11.05 | 78yr/male | sputum | respiratory medicine | discharge | 1.1 × 10−5 | NDM-1 | IncX3 | 54 | C |
| EC-15-2-152 | E. coli | ST405 | 2015.12.02 | 59yr/female | blood | ICU | death | - | NDM-5 | IncX3 | 46 | B |
| EC-15-2-153 | E. coli | ST405 | 2015.12.04 | 61yr/male | drainage liquid | gastrointestinal surgery | discharge | - | NDM-1 | IncX3 | 46 | B |
| EC-15-2-159 | E. coli | ST167 | 2015.12.08 | 23yr/male | urine | urology | discharge | - | NDM-5 | IncX3 | 46 | B |
| CF-15-2-165 | C. portucalensis | NA | 2015.12.11 | 79yr/male | urine | urinary surgery | discharge | 3.3 × 10−4 | NDM-1 | IncX3 | 54 | E |
| EC-16-7 | E. coli | ST167 | 2016.01.08 | 52yr/female | urine | kidney internal | discharge | 4.3 × 10−4 | NDM-1 | IncX3 | 54 | B |
| ECL-16-5 | E. hormaechei | ST51 | 2016.01.08 | 82yr/male | sputum | ICU | death | 3.3 × 10−4 | NDM-1 | IncX3 | 54 | B |
| EC-16-10 | E. coli | ST1193 | 2016.03.03 | 79yr/male | blood | ICU | death | - | NDM-5 | IncI1 | 93 | - |
| CF-16-17 | C. freundii | ST18 | 2016.07.08 | 70yr/male | secreta | endocrinology | discharge | 1.8 × 10−5 | NDM-1 | IncX3 | 54 | E |
| KO-16-21 | K. oxytoca | NA | 2016.07.10 | 83yr/male | sputum | ICU | discharge | 3.8 × 10−5 | NDM-1 | IncHI5 | 370 | - |
| EC-16-20 | E. coli | ST617 | 2016.07.10 | 48yr/male | ascites | Infectious disease | discharge | - | NDM-5 | IncX3 | 46 | B |
| EC-16-35 | E. coli | ST167 | 2016.07.16 | 10yr/female | ascites | pediatric surgery | discharge | 3.2 × 10−5 | NDM-5 | IncX3 | 46 | B |
| EC-16-37 | E. coli | ST46 | 2016.07.18 | 51yr/female | urine | urology | discharge | 1.5 × 10−6 | NDM-5 | IncFII-IncFIA-IncFIB | 159 | - |
| EC-16-52 | E. coli | ST410 | 2016.07.21 | 63yr/female | urine | pediatric surgery | discharge | - | NDM-5 | IncX3 | 46 | B |
| KP-16-57 | K. pneumoniae | ST716 | 2016.07.26 | 10yr/male | sputum | ICU | discharge | 7.3 × 10−4 | NDM-1 | IncC | 180 | - |
| CF-16-58 | C. braakii | NA | 2016.07.27 | 57yr/male | urine | respiratory medicine | discharge | 5.9 × 10−4 | NDM-1 | IncX3 | 54 | C |
| EC-16-59 | E. coli | ST167 | 2016.07.29 | 45yr/male | tissue | kidney internal | discharge | - | NDM-5 | IncX3 | 46 | B |
| EC-16-60 | E. coli | ST167 | 2016.07.29 | 2mo/female | sputum | ICU | discharge | 6.3 × 10−4 | NDM-5 | IncX3 | 54 | C |
| CF-16-61 | C. freundii | ST22 | 2016.07.30 | 50yr/male | blood | ICU | discharge | 8.8 × 10−5 | NDM-1 | IncX3 | 54 | C |
| ECL-16-74 | E. hormaechei | ST93 | 2016.08.03 | 45yr/male | drainage liquid | Liver transplantation | discharge | - | NDM-5 | IncX3 | 46 | C |
| EC-16-76 | E. coli | ST2172 | 2016.08.06 | 58yr/male | urine | emergency internal medicine | discharge | - | NDM-5 | IncX3 | 54 | C |
| ECL-16-79 | E. hormaechei | ST51 | 2016.10.20 | 54yr/female | bile | intervention department | discharge | 8.1 × 10−4 | NDM-5 | IncX3 | 46 | B |
MLST, multilocus sequence typing; NA, not available.
-, not detected.
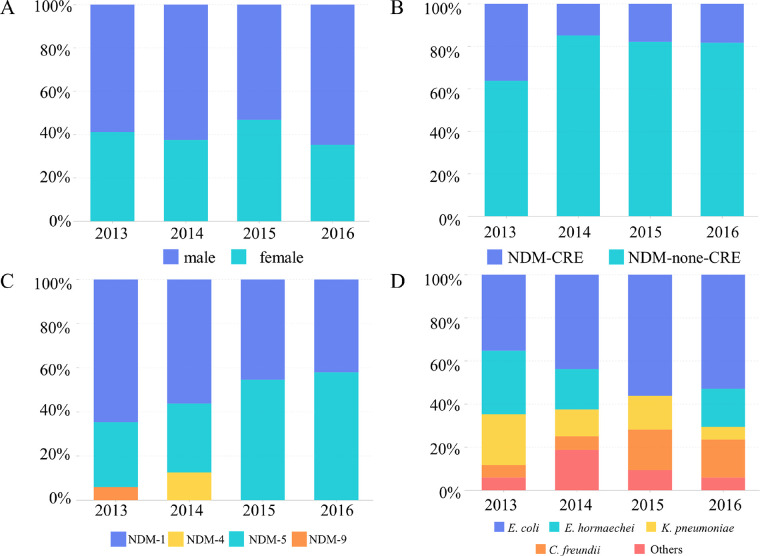
We analyzed the clinical features of these 81 blaNDM carriers (Table 1). We found that most blaNDM-positive strains were isolated from medical Intensive Care Units (ICUs). ICU patients usually have longer hospital stays, which increases the risk of infections and evolution of CRE pathogens. Comparatively, higher NDM-positive rates were also obtained among the Urinary Surgery and Pediatrics wards. The number of male patients was slightly higher compared with female patients (Fig. 1). We observed a wide age gap among these patients, ranging from 6 days to 89 years old; however, maximum cases (49.38%) were concentrated in the 50–79 age group. The mortality among the NDM-positive patients was 18.52%, which was lower compared to our previous report (7).
FIG 1.
Epidemiological description and impact factors of the 81 blaNDM-positive strains used in this study. (A) The proportion carrying the NDM by gender in different years. (B) Isolation rates of NDM among CRE in different years. (C) Proportion of NDM subtypes isolated in different years. (D) Proportion of different species isolated in different years.
Resistance phenotype, determinants, and bacterial genotyping.
Antimicrobial susceptibility testing revealed that all the 81 blaNDM-positive strains were MDR strains, and they were resistant to multiple categories of antimicrobials (n ≥ 3) (Table S2 in the supplemental material). Therefore, each isolate carried at least three categories of resistance genes associated with the resistance phenotype (Fig. 2 and Fig. S1). The MIC values of meropenem or imipenem were distributed between 16 and 64 μg/mL. Given that most NDM-producing isolates (92.59%) were resistant to aztreonam, we detected β-Lactamase encoding genes other than carbapenemase. Therefore, various AmpC (CMY, ACT, DHA) and ESBL (CTX-M, TEM, SHV, VEB, SFO, OXA) genes were identified in different species (Fig. 2 and Fig. S1). Moreover, four strains (EC-15-3, CF-15-2-29, ECL-16-5, and ECL-16-79) also contained plasmid-borne colistin resistance genes (mcr-1 or mcr-9). The abundance of antibiotic resistance genes in strains increases the risk of blaNDM cotransmission. To evaluate the transferability of blaNDM genes, conjugation assays were performed for the 81 blaNDM-positive strains with E. coli (EC600 or J53). The blaNDM genes carried by 46 strains were successfully transferred to the recipient, suggesting that the blaNDM genes carried by these 46 strains were located in conjugative plasmids or other mobilizable genetic elements. The conjugation frequencies ranged from 2.5 × 10−3 to 1.8 × 10−7 (Table 1).
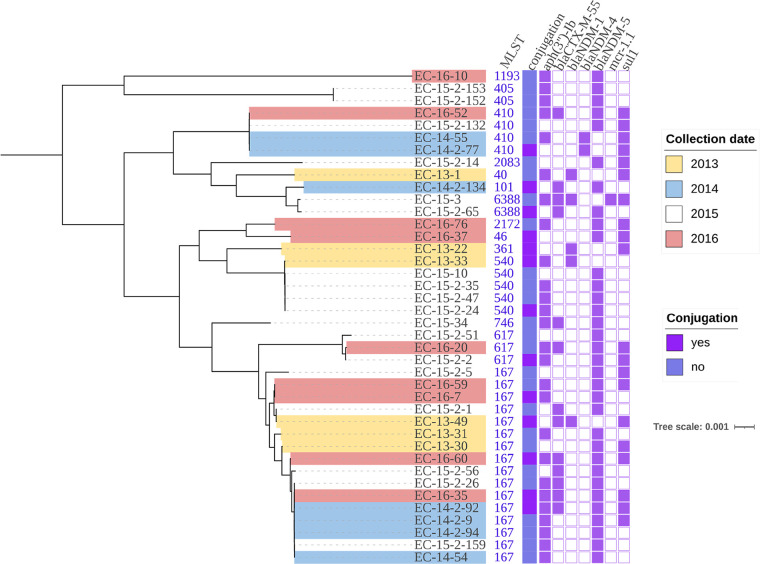
FIG 2.
Phylogenetic tree of all 40 blaNDM-positive E. coli isolates from 2013–2016. Resistance genes are indicated by squares: solid square indicates has; hollow square indicates does not have.
As the most abundant species carrying blaNDM, evolutionary relationships between the 40 E. coli isolates were investigated and a phylogenetic tree based on SNPs of the core genome data was constructed (Fig. 2). These isolates were assigned to 14 distinct sequence types (STs), and ST167 (16/40, 40%) was the most prevalent ST (Table 1). This finding was in agreement with the previous results (11), which suggest that ST167 appears to be the predominant type of blaNDM-positive E. coli in China. To further investigate the evolutionary relationship between these ST167 E. coli and other ST167 E. coli collected from the NCBI database (Table S3), a phylogenetic tree based on SNPs of the core genomes was constructed. ST167 E. coli carrying blaNDM were mainly found in humans. However, they are also found in pets and environmental samples (Fig. S2). blaNDM-5 was dominant in this subtype. Observation of diverse STs in E. coli indicated plasmids or other horizontal mobile elements to be considered as the main vehicles for blaNDM transmission. Similarly, four STs were identified among the eight C. freundii. Moreover, K. pneumoniae (n = 12) and E. hormaechei (n = 11) contained 12 and 8 different STs, respectively. The wide distribution of NDM-producing strains illustrates that in inter- and intraspecies, horizontal gene transfer plays the most important role in the transmission of blaNDM genes.
Systematic analysis of the predominant IncX3 blaNDM-bearing plasmids.
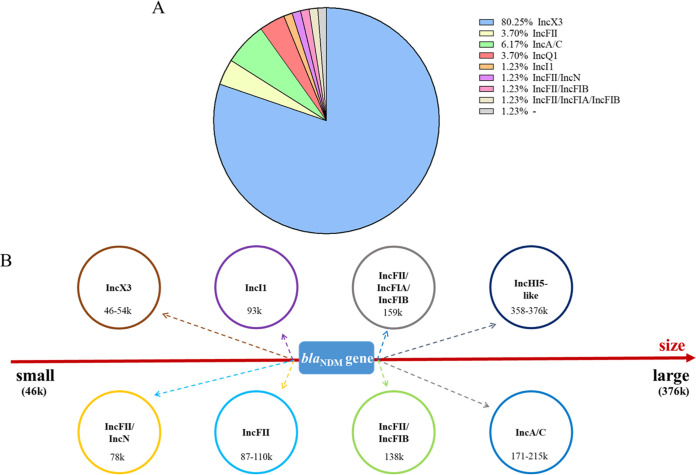
S1-PFGE and Southern blot analysis showed 77 blaNDM-positive strains located on plasmids. The Pr-15-2-50 was an exception, encoding a chromosomal blaNDM gene, and four strains (KA-14-61, EC-14-2-92, EC-15-34, and EC-15-2-153) failed to produce a visible band; however, they were confirmed on plasmids during the transfer experiments and whole-genome sequencing (WGS) analysis. Notably, two different blaNDM-bearing plasmids, pECL-14-60-NDM-1-IncAC (IncC, 171,038 bp) and pECL-14-60-NDM-1 (IncX3, 53,023 bp), were identified in the strain ECL-14-60. These 81 blaNDM-harboring plasmids were categorized into nine different replicon types (Fig. 3A) with sizes ranging from ∼46 to ∼370 kb (Fig. 3B). The isolated Inc types of plasmids carrying blaNDM genes were different each year; however, IncX3 blaNDM-positive plasmids were dominant through the period (Fig. 4 and Table 1). The bacteria carrying blaNDM-positive IncX3-type plasmids were diverse. Sixty-five NDM-producing IncX3 type plasmids with different sizes 54 kb and 46 kb (lacking the blaSHV-12-bearing segment) were found in 10 different bacterial species.
FIG 3.
The distribution of different Inc group plasmids in all blaNDM-positive strains. (A) The percentage of Inc groups found in all blaNDM-positive strains. (B) Diversity of blaNDM-bearing plasmids in terms of replicon types and sizes. Eight different plasmids with various replicon combinations were identified, and each of them was labeled in different circle colors with plasmid types and sizes highlighted.
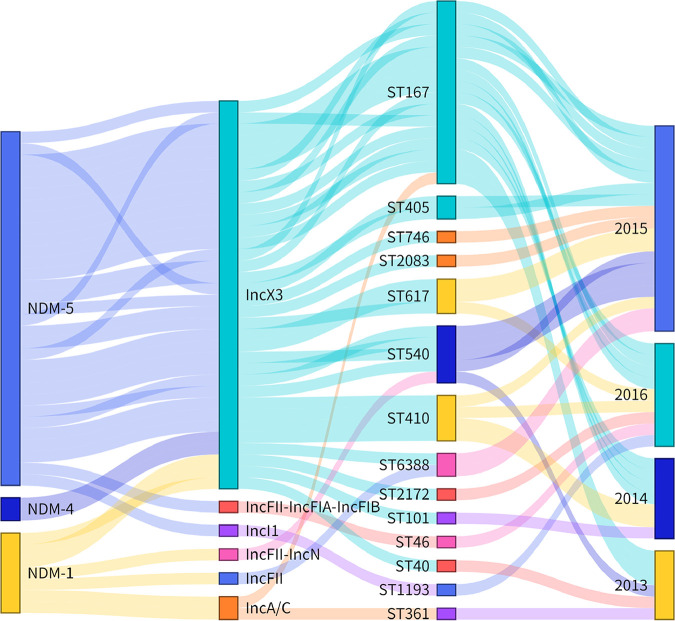
FIG 4.
Sankey diagram combining different NDM subtypes, plasmid Inc types, ST types, and collection date. The diameter of the line is proportional to the number of isolates, which is also labeled at the consolidation points.
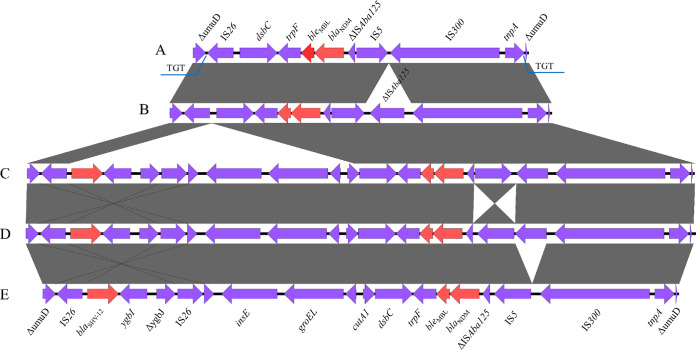
In total, the environment around the blaNDM gene located on the IncX3 plasmid can be classified into five major groups. These regions carrying the blaNDM genes were all inserted into the umuD gene, and a 3-bp (TGT) direct repeat sequence formed at the insertion site. Group A (n = 1) is the simplest among several groups (Fig. 5). Compared with group A, group B (n = 29) had one more ISAba125 insertion downstream from the blaNDM. Group C (n = 20) had more 7,874 bp regions carrying the blaSHV gene downstream from the IS26 compared with group B. Group D had the reverse IS5 arrangement compared with group C. Compared with group D, the region in group E lost the ISAba125 gene downstream from the blaNDM.
FIG 5.
Different blaNDM gene core genetic environments of the IncX3-type plasmids. A total of five (A–E) major types of blaNDM-bearing genetic contexts among the 42 blaNDM-bearing plasmids. Red arrows represent resistance genes.
By connecting the blaNDM subtypes of E. coli to plasmid types, ST types as well as the year of isolation (Fig. 4), we illustrated a complex combination of multiple genetic vehicles and diverse hosts in the spreading of the blaNDM gene. Most of the blaNDM-5 genes were distributed on the IncX3 plasmids. Moreover, blaNDM-1 and blaNDM-4 were also found on the IncX3 plasmids. According to Fig. 4, IncX3 plasmids are the main blaNDM-positive plasmids isolated each year, and these plasmids are distributed in many different STs of E. coli. However, compared with other Inc-type NDM positive plasmids (Fig. S3), IncX3 type plasmids carried only a few antibiotic resistance genes, which may incur a low fitness cost to the host.
Characterization of novel Inc-type and hybrid blaNDM-bearing plasmids.
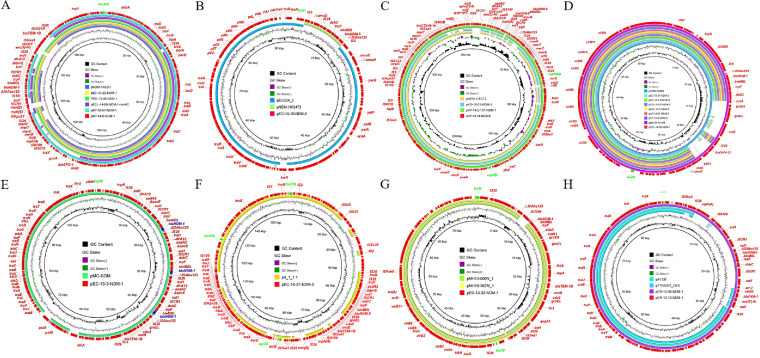
In addition to IncX3 plasmids, other Inc-types of NDM-bearing plasmids were also detected in these strains (Fig. 6). To the best of our knowledge, the IncI1 plasmid pEC-16-10-NDM-5 characterized in this study is a novel blaNDM-bearing plasmid (Fig. S4 and Fig. 6). Plasmid pEC-16-10-NDM-5 was 92,260 bp in size and had an average G+C content of 50.6%. The BLAST comparison against the GenBank database showed that pEC-16-10-NDM-5 exhibited similarities to IncI1 plasmid pEC224_2 (CP018946). The main difference is that plasmid pEC-16-10-NDM-5 has an additional 8,698 bp complex transposon structure composed of two IS5 and a blaNDM-5-bearing region (IS5-hp-hp-ΔumuD-IS26-dsbC-trpF-bleMBL-blaNDM-5-ΔISAba125-IS5). This additional transposon structure is similar to the IncX3 plasmid pNDM-HK3473 (MH234506) carrying the blaNDM-5 gene. It is flanked by 15 bp inverted repeats (TAGGGAAGGTGCGAA) on either side. This phenomenon indicates that the blaNDM-5 could be transferred through this complex transposon and integrated into the IncI1 plasmid (Fig. S5).
FIG 6.
Circular comparison of different blaNDM-bearing plasmids with similar online plasmids. A–H represent different blaNDM-bearing plasmids with various replicons IncC, IncI1, IncHI5-like, IncX3, IncFII, IncFIA-IncFIB-IncFII, IncFII-IncN, and IncFII(p14).
Five IncFII blaNDM-bearing plasmids were also identified among the 22 plasmids with complete and circular sequences using Nanopore sequencing (Table 2). The pMLST of the pEC-15-3-NDM-1 plasmid was F2:A-:B-, and the size of the plasmid is 109,944 bp. BLASTn analysis of the pEC-15-3-NDM-1 plasmid showed that it had 99% nucleotide identity at more than 95% coverage to pMC-NDM (HG003695). The main difference between them was the copy number of blaNDM-1. Three 10,461 bp region repeats were found on the pEC-15-3-NDM-1 plasmid, which carried a variety of resistance genes, including blaNDM-1, dfrA12, aadA2, sul1, and bleMBL. According to the result of BLASTn, two Y2:A-:B- (pCR-13-12-NDM-1 and pCR-13-36-NDM-1) plasmids of IncFII were similar to the pA1137 (NZ_MF190369) and pTTHS031_GES (NZ_LC589514) plasmids in the NCBI database. In contrast to the plasmids in this study, they all lacked regions carrying the blaNDM gene, implying that the regions carrying the blaNDM may insert progenitors before forming these plasmids. Moreover, two-hybrid plasmids pEC-16-37-NDM-5 (IncFII-IncFIA-IncFIB) and pEC-13-33-NDM-1 (IncFII-IncN) were also found. The pEC-16-37-NDM-5 plasmid was similar to the online IncFII-IncFIA-IncFIB plasmid p4_1_1.1 (NZ_CP023845) in E. coli. The plasmid pEC-13-33-NDM-1 was similar to both of the online IncFII-IncN plasmids pMH13-009N_1 (AP018566) and pMH16-367M_1 (AP018565) found in Proteus mirabilis and Morganella morganii, respectively. The core genetic environment of blaNDM-5 in pEC-16-37-NDM-5 is ISCR1-dsbC-trpF-bleMBL-blaNDM-5-ΔISAba125-IS26. Although there are only four IncFII-type plasmids carrying blaNDM-1, the gene environment around blaNDM-1 could be divided into three categories: TnAs3-groEL-cutA-dsbC-trpF-bleMBL-blaNDM-1-ΔISAba125-IS1294 (pEC-13-33-NDM-1), ISCR1-dsbC-trpF-bleMBL-blaNDM-1-ΔISAba125-IS26 (pEC-15-3-NDM-1 and pCR-13-12-NDM-1), and ISCR1-dsbC-trpF-bleMBL-blaNDM-1-ΔISAba125-ISCR1 (pCR-13-36-NDM-1).
TABLE 2.
Basic information of 22 blaNDM-bearing plasmids resolved by Illumina and Nanopore long-read sequencing
| Plasmid | Strain | Status | Size (bp) | Inc-type | Assembly method | Sequencing technology | Accession no. | Resistance genes |
|---|---|---|---|---|---|---|---|---|
| pCR-13-12-NDM-1 | CR-13-12 | complete | 86 619 | IncFII | Unicycler | Oxford Nanopore MinION, Illumina | NZ_MN175388 | aac(6′)-Ib, arr-3, blaNDM-1, blaOXA-1, bleMBL, catB3, mph(A), sul1 |
| pCR-13-36-NDM-1 | CR-13-36 | complete | 86 619 | IncFII | Unicycler | Oxford Nanopore MinION, Illumina | MZ857202 | aac(6′)-Ib, arr-3, blaNDM-1, blaOXA-1, bleMBL, catB3, mph(A), sul1 |
| pEC-13-22-NDM-1 | EC-13-22 | complete | 212 551 | IncC | Unicycler | Oxford Nanopore MinION, Illumina | MZ836796 | aac(3)-IId, aph(3′)-VI, blaNDM-1, blaSFO-1, blaTEM-1, bleMBL, dfrA12, mph(A), mph(E), msr(E), sul1 |
| pEC-13-31-NDM-5 | EC-13-31 | complete | 49 021 | IncX3 | Unicycler | Oxford Nanopore MinION, Illumina | MZ836797 | blaNDM-5, bleMBL |
| pEC-13-33-NDM-1 | EC-13-33 | complete | 74 978 | IncFII-IncN | Unicycler | Oxford Nanopore MinION, Illumina | MZ836798 | blaNDM-1, blaTEM-1, bleMBL, qepA1, rmtB1, tet(A) |
| pEC-13-49-NDM-1 | EC-13-49 | complete | 214 323 | IncC | Unicycler | Oxford Nanopore MinION, Illumina | MZ836799 | aac(3)-IId, aph(3′)-VI, blaNDM-1, blaSFO-1, blaTEM-1, bleMBL, dfrA12, mph(A), mph(E), msr(E), sul1 |
| pEC-14-2-9-NDM-5 | EC-14-2-9 | complete | 46 161 | IncX3 | Unicycler | Oxford Nanopore MinION, Illumina | MZ836800 | blaNDM-5, bleMBL |
| pEC-15-3-NDM-1 | EC-15-3 | complete | 109 944 | IncFII | Unicycler | Oxford Nanopore MinION, Illumina | NZ_MN061455 | aadA2, blaNDM-1, blaTEM-1, bleMBL, dfrA12, rmtB1, sul1 |
| pEC-16-10-NDM-5 | EC-16-10 | complete | 92 260 | IncI1 | Unicycler | Oxford Nanopore MinION, Illumina | MZ836801 | blaNDM-5, bleMBL |
| pEC-16-37-NDM-5 | EC-16-37 | complete | 157 578 | IncFII-IncFIA-IncFIB | Unicycler | Oxford Nanopore MinION, Illumina | MZ836802 | aadA2, blaNDM-5, blaTEM-1, bleMBL, dfrA12, erm(B), mph(A), rmtB1, sul1 |
| pEC55-NDM4 | EC-14-55 | complete | 54 035 | IncX3 | Unicycler | Oxford Nanopore MinION, Illumina | NZ_KX470734 | blaNDM-4, blaSHV-12, bleMBL |
| pECL-13-37-NDM-5 | ECL-13-37 | complete | 46 161 | IncX3 | Unicycler | Oxford Nanopore MinION, Illumina | MZ836804 | blaNDM-5, bleMBL |
| pECL-13-4-NDM-5 | ECL-13-4 | complete | 46 161 | IncX3 | Unicycler | Oxford Nanopore MinION, Illumina | MZ836803 | blaNDM-5, bleMBL |
| pECL-14-60-NDM-1-IncAC | ECL-14-60 | complete | 171 038 | IncC | Unicycler | Oxford Nanopore MinION, Illumina | MZ836805 | aac(6′)-Ib, aph(3′)-Ia, armA, arr-3, blaNDM-1, blaOXA-1, bleMBL, catB3, mph(E), msr(E), qnrA7, sul1, sul2 |
| pECL-14-60-NDM-1 | ECL-14-60 | complete | 53 023 | IncX3 | Unicycler | Oxford Nanopore MinION, Illumina | NZ_MN061454 | blaNDM-1, blaSHV-12, bleMBL |
| pKA-14-61-NDM-5 | KA-14-61 | complete | 46 161 | IncX3 | Unicycler | Oxford Nanopore MinION, Illumina | MZ836806 | blaNDM-5, bleMBL |
| pKO-16-21-NDM-1 | KO-16-21 | complete | 376 570 | IncHI5-like | Unicycler | Oxford Nanopore MinION, Illumina | MZ836807 | aac(3)-IId, aadA16, aph(3′')-Ib, aph(6)-Id, arr-3, blaNDM-1, blaTEM-1, bleMBL, dfrA27, mph(A), qnrB6, sul1, sul2 |
| pKP-13-14-NDM-9 | KP-13-14 | complete | 358 655 | IncHI5-like | Unicycler | Oxford Nanopore MinION, Illumina | NZ_MN175386 | aac(3)-IId, aadA2, aph(3′')-Ib, aph(6)-Id, blaCTX-M-14, blaNDM-9, blaTEM-1, bleMBL, dfrA12, mph(A), tet(D), sul1, sul2 |
| pKP-13-8-NDM-5 | KP-13-8 | complete | 46 161 | IncX3 | Unicycler | Oxford Nanopore MinION, Illumina | NZ_MN175389 | blaNDM-5, bleMBL |
| pKP-14-2-131-NDM-1 | KP-14-2-131 | complete | 358 158 | IncHI5-like | Unicycler | Oxford Nanopore MinION, Illumina | MZ836808 | aac(3)-IId, aadA2, aph(3′')-Ib, aph(6)-Id, blaCTX-M-14, blaNDM-1, blaTEM-235, bleMBL, dfrA12, mph(A), tet(D), sul1, sul2 |
| pKP-14-6-NDM-1 | KP-14-6 | complete | 199 120 | IncC | Unicycler | Oxford Nanopore MinION, Illumina | NZ_MN175387 | aac(3)-IId, blaNDM-1, blaSFO-1, blaTEM-1, bleMBL, dfrA12, mph(A), mph(E), msr(E), sul1 |
| pKP-16-57-NDM-1 | KP-16-57 | complete | 180 309 | IncC | Unicycler | Oxford Nanopore MinION, Illumina | MZ836809 | aac(3)-IId, aadA2, blaNDM-1, blaSFO-1, bleMBL, dfrA12, mph(A), sul1 |
The characteristics of the five NDM-positive plasmids (171 kb–215 kb) of the IncC type were also analyzed (Fig. 6). The blaNDM subtypes carried by these plasmids were all blaNDM-1, and most of them shared similar backbones. Despite their similar backbones, there are three types of genetic environments around blaNDM-1: ISCR1-dsbC-trpF-bleMBL-blaNDM-1-ΔISAba125-IS1R (pECL-14-60-NDM-1 and pKP-14-6-NDM-1), ISCR1-dsbC-trpF-bleMBL-blaNDM-1-ISAba125 (pEC-13-22-NDM-1 and pEC-13-49-NDM-1), and ISCR1-dsbC-trpF-bleMBL-blaNDM-1-ΔISAba125-IS26 (pKP-16-57-NDM-1). Among these five IncC plasmids, pEC-13-22-NDM-1 and pEC-13-49-NDM-1 were isolated from E. coli, pKP-16-57-NDM-1 and pKP-14-6-NDM-1 from K. pneumoniae, and pECL-14-60-NDM-1 from E. hormaechei. BLASTn comparison with the NCBI database showed similarities to pNDM-TAEC1 (NZ_MH001166) found in E. coli.
It is worth noting that three plasmids belonged to the recently discovered IncHI5-like plasmids (Fig. 6). The three blaNDM-harboring IncHI5-like plasmids ranged from 358 to 376 kb and possessed the same plasmid backbone structure. The BLAST comparison against the GenBank database showed that plasmid pKO-16-21-NDM-1 from K. oxytoca exhibited similarities to the same Inc-type plasmids pKP19-3023-374k (CP063748) and pKP19-3088-375k (CP063149), which were collected from K. pneumoniae. The core genetic environment of blaNDM (ISCR1-sul1-ΔqacE-blaNDM-1-bleMBL-trpF-dsbC-ISCR1) carried on the plasmid pKO-16-21-NDM-1 was similar to the pKP19-3023-374k plasmid. This is the first time that a blaNDM-positive IncHI5-like plasmid has been found in K. oxytoca. The pKP-13-14-NDM-9 plasmid that was isolated from K. pneumoniae was 358,655 bp in size. Although IncHI5-like plasmids were reported to carry blaNDM-1 in previous studies (12, 13), pKP-13-14-NDM-9 was the first IncHI5-like plasmid positive for blaNDM-9. The core genetic environment of blaNDM-9 is IS26-ΔISAba125-blaNDM-9-bleMBL-trpF-mocA-cutA-ISCR1, and a similar genetic environment (IS26-ΔISAba125-blaNDM-1-bleMBL-trpF-mocA-cutA-ISCR1) was found in pKP-14-2-131-NDM-1.
Four of the 81 strains were found to carry both blaNDM and mcr genes (mcr-1, n = 1, mcr-9, n = 3). The mcr-1 gene was located on a 60,961 bp plasmid designated as pEC-15-3-mcr-1 in the incompatibility group IncI2 (Fig. S6). The plasmids similar to pEC-15-3-mcr-1 in the public database were the E. coli plasmid pAH62-1 (NZ_CP055260) and Salmonella plasmid pS304_2 (NZ_CP061128), which showed 100% coverage and identity. Moreover, three strains were found (CF-15-2-29, ECL-16-5, and ECL-16-79) carrying the mcr-9. Online BLAST (Fig. S7) showed that mcr-9-positive plasmids all belonged to IncHI2A-IncHI2 and showed similarities to the pBSI034-MCR9 (NZ_MN937241) plasmid. Strains carrying mcr-9 were usually resistant to polymyxin; however, ECL-16-79 was susceptible to polymyxin. It has been reported that the deletion of the two-component system qseCB may silence the mcr-9 gene (14). However, the ECL-16-79 strain contains the two-component system qseCB, and other genes or molecules may regulate the expression of mcr-9. Further investigations are needed to decipher the underlying molecular mechanisms.
Two tandem copies of blaNDM-1 in the chromosome.
In addition to the plasmid-mediated blaNDM genes, we also found the blaNDM-1 on the chromosome of the P. rettgeri strain Pr-15-2-50. The size of the genome was 4,648,900 bp, with 40.3% GC content. Two copies of blaNDM-1 were found on the chromosome of the Pr-15-2-50. On comparing the Pr-15-2-50 chromosome with FZB001 (CP060821) and AR0156 (CP021852), we found a 40,775 bp Tn7-like transposon structure carrying the blaNDM gene, inserted into the chromosomal region (Fig. S8). This Tn7-like transposon had an average GC content of 48.8% and similarly to the p2BJAB07104 (CP003907) plasmid, it was surrounded by 11 bp inverted repeats (ACAAAATAGAT), implying that the transposon could translocate between chromosomes and plasmids. However, this plasmid lacked the blaNDM-bearing region. The 5,250 bp blaNDM-carrying region (ISCR1-dsbC-trpF-bleMBL-blaNDM-1-ΔISAba125-Δsul1) may be incorporated because of the ISCR1-mediated insertion, similar to previous reports(15). Moreover, a 4,390 bp integron (intI2-lnu(F)-dfrA1-aadA1-ΔqacE-sul1) was found downstream to the blaNDM-1 gene. Despite these reports, ISCR1-mediated copies of blaNDM have been found on these chromosomes. However, the ISCR1-mediated transposable units on P. aeruginosa MMA83 (ISCR1-aph(3′)-VIa-ISAba125-blaNDM-1-sul1), E. coli Y5 (ISCR1-traF-bleMBL-blaNDM-1-ΔISAba125-catB3-arr-3-ΔqacE-sul1), and P. mirabilis XH1653 (sul1-arr-3-cat-blaNDM-1-bleo-ISCR1) are different from the Pr-15-2-50 (ISCR1-dsbC-trpF-bleMBL-blaNDM-1-ΔISAba125-Δsul1) strain in this study (15–17). The ble and sul1 genes were also detected in these transposable units. This suggests that blaNDM may cotransfer with other resistance genes.
DISCUSSION
blaNDM-1 was discovered in 2009. Since then, CRE strains carrying blaNDM-1 and its variants have spread in more than 55 countries worldwide. Asian countries such as India, Pakistan, and China are considered major reservoirs of blaNDM (6). The blaNDM-1-positive strains were first isolated in clinical stool samples in China in 2010, followed by an increasing number of blaNDM-positive strains. In 2013, 17 blaNDM-positive strains (38.64%) were obtained from 44 CRE strains isolated from hospitals in Henan, which was an increase compared to 2011–2012 (7). However, the positive rate decreased to 18.89% (17/90) and 17.13% (31/181) in 2014 and 2015, respectively. This may be the result of the effective clinical infection control measures. However, there was an increase in 2016, with the isolation rates reaching up to 21.79% (17/78).
ST11 is the most common type of blaNDM-positive K. pneumoniae that was reported (18, 19). Moreover, ST11 K. pneumoniae often had hypervirulent and/or multidrug resistant phenotypes (20). However, only one strain of ST11 K. pneumoniae was found in this study. The ST types of K. pneumoniae were more diverse, indicating that K. pneumoniae carrying blaNDM in Henan is not clonally transferred. Moreover, we found diverse E. coli STs, and ST167 was predominant among them. This phenomenon is similar to previous domestic reports (21, 22). ST167 NDM-producing E. coli strains are not only widely disseminated in China (11, 23); they also cause infections worldwide (6, 24, 25), which has gained much attention. Consistent with this study, the blaNDM-5 gene is mainly carried by E. coli of ST167 (26, 27), suggesting that ST167 E. coli is an important repository of blaNDM-5. More importantly, ST167 blaNDM-positive E. coli strains have been found in companion animals (28, 29), which suggests that the ST167 E. coli carrying blaNDM-5 gene could be transmitted between animals and humans.
Four NDM subtypes (NDM-1, NDM-4, NDM-5, and NDM-9) were found in 81 NDM-producing strains; however, from 2011 to 2012, all blaNDM-positive strains isolated from Henan were blaNDM-1 (7). Since the isolation of blaNDM-5 in Henan in 2013, the detection rate has gradually increased. It has now become the main subtype of blaNDM. The blaNDM-1 detection rates have been decreasing each year; however, it remains the main epidemic subtype. Previous studies have shown that NDM-5 exhibits higher hydrolytic activity toward carbapenems and cephalosporins compared with NDM-1 (30). This may be caused by the increase in the usage of carbapenems in clinical treatment. It has been shown that IncX3 plasmids could promote the transmission of NDM-5, and the plasmids carrying NDM-5 demonstrated high stability (31, 32). In this study, most of the blaNDM-5 genes were carried by IncX3 plasmids, which led to a higher prevalence. The increasing prevalence of blaNDM-5-positive strains should be of high concern.
Carbapenem and colistin are considered the last line of defense in the treatment of severe infections caused by extensively drug-resistant bacteria. Only a few articles have previously reported the coexistence of blaNDM-1 and mcr-9 genes (33, 34). Four strains with the coexistence of blaNDM and mcr genes were found in this study. This phenomenon greatly increases the risk of treatment failure. The blaIMP-4-producing Enterobacterales have been reported sporadically in China (11, 35). Only two strains harboring the blaIMP-4 gene were found among the 81 blaNDM-positive strains. Moreover, multiple resistance genes are often present on plasmids carrying blaNDM genes, which greatly increases the risk of cotransmission of multiple resistance genes.
Except for the strain Pr-15-2-50, the blaNDM gene was located on the plasmids, which might be the main mode of blaNDM transmission. A variety of blaNDM-positive plasmids with different Inc types and sizes were found in the 80 strains, mainly IncX3 type, which is similar to previous reports. In this study, the ST type of NDM-producing strains carrying the IncX3 plasmid was mainly ST167. This highly prevalent ST and plasmid type promotes the transmission of blaNDM further and seriously threatens public health. We also discovered a novel blaNDM-bearing plasmid pEC-16-10-NDM-5 (IncI1). IncI1 plasmid belongs to the narrow-host range plasmid type (36) and was only found in Enterobacterales. Several articles have pointed out that IncI1 plasmids frequently carry genes encoding antibiotic resistance, especially the extended-spectrum beta-lactamase genes (37–39). These plasmids are widely distributed in animals and patients worldwide (40, 41). The IncHI plasmid has a wide host range and plays an important role in the transmission of resistance genes (42, 43). Previously, it was shown that a variety of carbapenemase genes were found on the IncHI5 plasmids (44), which poses a great threat to clinical treatment. The two IncHI5-like plasmids, carrying both carbapenem and tigecycline resistance genes, were found in our recent study (12), severely restricting the clinical treatment options. In this study, the new blaNDM core genetic environment was found in the IncHI5-like plasmids, suggesting that this plasmid has evolved as a novel MDR plasmid and needs to be continuously monitored.
Conclusion.
To date, NDM is the predominant mechanism for CRE in humans. Carbapenem, polymyxin, and tigecycline are regarded as the last line of defense in the clinical treatment of MDR infections. In recent years, several studies have found that blaNDM coexists with mobile colistin (mcr) and tigecycline resistance genes (tet(X) and tmexCD-toprJ), making clinical treatment extremely difficult. Therefore, continuous long-term surveillance for pathogens that clinically harbor blaNDM is important. This study conducted an in-depth analysis of blaNDM-positive clinical strains and confirmed that the vast majority of blaNDM genes were distributed on plasmids of different Inc types, and are transmitted by horizontal transfer of plasmids. The emergence of Enterobacterales carrying both blaNDM and other resistance genes, such as mcr, is worrying. These isolates can seriously limit clinical treatment options. Therefore, there is an urgent need for large-scale monitoring and the development of effective control measures.
MATERIALS AND METHODS
Bacterial isolates.
The samples in this study were obtained between 2013 and 2016 at an affiliated hospital of Zhengzhou University. This study did not exclude patients based on age, gender, or symptoms. Moreover, the samples collected were nonduplicate isolates from different patients. CRE was defined as Enterobacterales resistant to at least one carbapenem (meropenem or imipenem). A total of 391 CRE strains were collected from blood, urine, sputum, wound, tissue, pus, swab, drainage liquid, secreta, bile, ascites, sanies, joint fluid, and urine tube tips. Clinical data of each patient were collected from the clinical and medical record system. Extracted clinical information included the date of collection, patient age, sex, source of isolate, ward type, and outcome (alive or dead). The blaNDM-positive strains were screened and confirmed using PCR and Sanger sequencing, respectively. All blaNDM-positive isolates were sent to Zhengzhou University for subsequent experiments. This study was approved by the Ethics Committee of Zhengzhou University with a waiver of informed consent because of the retrospective nature of the study.
PCR screening and antimicrobial susceptibility testing.
The presence of carbapenem resistance genes (blaNDM, blaIMP, blaKPC, blaVIM, and blaOXA-48) and other important resistance genes (mcr-1, blaSHV, and blaTEM) was investigated using PCR with the primers (Table S1). The PCR amplified products were confirmed using gel electrophoresis and Sanger sequencing. All CRE species identification was carried out by the automated Vitek 2 system. Antimicrobial susceptibility testing of clinical strains was performed against 17 antimicrobials by determining the MICs using the broth microdilution method, and E. coli ATCC 25922 was used as the quality control. All antibiotic breakpoints were interpreted according to CLSI guidelines (45); however, tigecycline (>2 mg/L) was interpreted according to the EUCAST criteria.
Conjugation, S1-PFGE, and Southern blot.
The conjugation experiment was performed with each of the blaNDM-positive strains using a rifampicin-resistant E. coli EC600 or sodium azide-resistant E. coli J53 recipients. The donor and recipient were mixed in a ratio of 1:1 and incubated statically in an LB broth at 35°C for 24 h. Transconjugants on the LB agar plates containing double antibiotics (meropenem 2 mg/L and rifampicin 100 mg/L, or meropenem 2 mg/L and sodium azide 200 mg/L) were selected and confirmed using PCR and PFGE, respectively. Transfer frequencies were calculated as the number of transconjugants/total number of recipients.
S1-PFGE and Southern blot analyses were performed to determine the plasmid sizes and genomic positions of blaNDM. To elucidate the genetic environments of blaNDM genes, 22 representative blaNDM-carrying plasmids were selected based on the plasmid replicon types and sizes to perform Nanopore sequencing to obtain the complete plasmid sequences.
WGS procedures and analyses.
We characterized the genetic features and resistomes of the blaNDM-positive CRE. The genomes of all blaNDM-positive strains were extracted with the FastPure bacterial DNA isolation minikit (catalog no. DC103; Vazyme) and evaluated using 1% (wt/vol) agarose gel electrophoresis. The concentration and purity were quantified using the Qubit 4 Fluorometer and Nanodrop. The genomic DNA samples were sequenced using the Illumina Hiseq 2500 platform generating 2 × 150 bp paired-end reads. Twenty-two representative strains were sequenced with the Nanopore long-read sequencing platform according to resistant phenotypes and genotypes (46). The Rapid Barcoding Kit RBK004 was used to construct the long-read sequencing libraries, which were subjected to Nanopore sequencing in MinION R9.4.1 flow cells.
The Illumina paired-end reads were de novo assembled using the SPAdes version 3.14.0, and contigs less than 200 bp in length were removed (47). Unicycler v. 0.4.8 was used for hybrid assembly of genomes with the combination of Illumina short reads and Nanopore long reads with default parameters (48). For intricate regions that could not be resolved using the hybrid assembly method, Nanopore sequencing data were assembled using the long-read assembler Flye v. 2.4.2 to acquire accurate structures of complex genomic regions (49). The genomes were annotated using the online tool RAST (http://rast.nmpdr.org/). ResFinder and PlasmidFinder (http://cge.cbs.dtu.dk/services/) were used to identify antimicrobial resistance genes and plasmid replicon types with default parameters. The virulence factors in the assembled genome sequences were identified using the Kleborate software (50) and the virulence factor database (last updated 14th October 2020) in abricate v.1.0.1 (https://github.com/tseemann/abricate) with default parameters. Multilocus sequence typing (MLST) of the 81 blaNDM-positive isolates was conducted using mlst (https://github.com/tseemann/mlst). The plasmid comparison maps were constructed and displayed by using BRIG v. 0.95 and Easyfig v. 2.2.3 (51, 52), respectively. The core genes in the genomes of blaNDM-positive CRE were identified using Roary (53). The phylogenetic trees of blaNDM-positive strains were constructed using FastTree (54) based on the core single-nucleotide polymorphism (SNP) alignments with default parameter settings and visualized using iTOL (https://itol.embl.de).
Data availability.
The sequence data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession numbers PRJNA752009, and individual accession numbers of 22 blaNDM-bearing plasmids are listed in Table 2.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (no. U2004125 and 31872523) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
We have no conflicts of interest or financial conflicts to disclose.
Footnotes
Supplemental material is available online only.
Contributor Information
Ruichao Li, Email: rchl88@yzu.edu.cn.
Shangshang Qin, Email: qinshangshang@126.com.
Rebekah M. Martin, Labcorp
REFERENCES
- 1.van der Bij AK, Pitout JDD. 2012. The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J Antimicrob Chemother 67:2090–2100. doi: 10.1093/jac/dks214. [DOI] [PubMed] [Google Scholar]
- 2.Karaiskos I, Giamarellou H. 2014. Multidrug-resistant and extensively drug-resistant Gram-negative pathogens: current and emerging therapeutic approaches. Expert Opin Pharmacother 15:1351–1370. doi: 10.1517/14656566.2014.914172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y, Wang Q, Yin Y, Chen H, Jin L, Gu B, Xie L, Yang C, Ma X, Li H, Li W, Zhang X, Liao K, Man S, Wang S, Wen H, Li B, Guo Z, Tian J, Pei F, Liu L, Zhang L, Zou C, Hu T, Cai J, Yang H, Huang J, Jia X, Huang W, Cao B, Wang H. 2018. Epidemiology of carbapenem-resistant Enterobacteriaceae infections: report from the China CRE network. Antimicrob Agents Chemother 62:e01882-17. doi: 10.1128/AAC.01882-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol 19:588–595. doi: 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Poirel L, Bonnin RA, Boulanger A, Schrenzel J, Kaase M, Nordmann P. 2012. Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob Agents Chemother 56:1087–1089. doi: 10.1128/AAC.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu W, Feng Y, Tang G, Qiao F, McNally A, Zong Z. 2019. NDM metallo-beta-Lactamases and their bacterial producers in health care settings. Clin Microbiol Rev 32:e00115-18. doi: 10.1128/CMR.00115-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin S, Fu Y, Zhang Q, Qi H, Wen JG, Xu H, Xu L, Zeng L, Tian H, Rong L, Li Y, Shan L, Xu H, Yu Y, Feng X, Liu HM. 2014. High incidence and endemic spread of NDM-1-positive Enterobacteriaceae in Henan Province, China. Antimicrob Agents Chemother 58:4275–4282. doi: 10.1128/AAC.02813-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Zhou Z, Jiang Y, Yu Y. 2011. Emergence of NDM-1-producing Acinetobacter baumannii in China. J Antimicrob Chemother 66:1255–1259. doi: 10.1093/jac/dkr082. [DOI] [PubMed] [Google Scholar]
- 9.Zhou G, Guo S, Luo Y, Ye L, Song Y, Sun G, Guo L, Chen Y, Han L, Yang J. 2014. NDM-1-producing strains, family Enterobacteriaceae, in hospital, Beijing, China. Emerg Infect Dis 20:340–342. doi: 10.3201/eid2002.121263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q, Wang X, Wang J, Ouyang P, Jin C, Wang R, Zhang Y, Jin L, Chen H, Wang Z, Zhang F, Cao B, Xie L, Liao K, Gu B, Yang C, Liu Z, Ma X, Jin L, Zhang X, Man S, Li W, Pei F, Xu X, Jin Y, Ji P, Wang H. 2018. Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016). Clin Infect Dis 67:S196–S205. doi: 10.1093/cid/ciy660. [DOI] [PubMed] [Google Scholar]
- 11.Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, Li Y, Liao K, Chen S. 2017. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine 19:98–106. doi: 10.1016/j.ebiom.2017.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin S, Peng J, Deng R, Peng K, Yan T, Chen F, Li R. 2021. Identification of two plasmids coharboring carbapenemase genes and tmexCD1-toprJ1 in clinical Klebsiella pneumoniae ST2667. Antimicrob Agents Chemother 65:e00115-18. doi: 10.1128/AAC.00625-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Chen R, Xu P, Wang Z, Li R. 2021. Characterization of a blaNDM-1-bearing IncHI5-like plasmid from Klebsiella pneumoniae of infant origin. Front Cell Infect Microbiol 11:738053. doi: 10.3389/fcimb.2021.738053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieffer N, Royer G, Decousser JW, Bourrel AS, Palmieri M, Ortiz De La Rosa JM, Jacquier H, Denamur E, Nordmann P, Poirel L. 2019. mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in Escherichia coli, and its origin. Antimicrob Agents Chemother 63:e00965-19. doi: 10.1128/AAC.00965-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen P, Yi M, Fu Y, Ruan Z, Du X, Yu Y, Xie X. 2017. Detection of an Escherichia coli sequence type 167 strain with two tandem copies of blaNDM-1 in the chromosome. J Clin Microbiol 55:199–205. doi: 10.1128/JCM.01581-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He J, Sun L, Zhang L, Leptihn S, Yu Y, Hua X. 2021. A novel SXT/R391 integrative and conjugative element carries two copies of the blaNDM-1 gene in Proteus mirabilis. mSphere. doi: 10.1128/mSphere.00588-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jovcic B, Lepsanovic Z, Begovic J, Rakonjac B, Perovanovic J, Topisirovic L, Kojic M. 2013. The clinical isolate Pseudomonas aeruginosa MMA83 carries two copies of the blaNDM-1 gene in a novel genetic context. Antimicrob Agents Chemother 57:3405–3407. doi: 10.1128/AAC.02312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain A, Hopkins KL, Turton J, Doumith M, Hill R, Loy R, Meunier D, Pike R, Livermore DM, Woodford N. 2014. NDM carbapenemases in the United Kingdom: an analysis of the first 250 cases. J Antimicrob Chemother 69:1777–1784. doi: 10.1093/jac/dku084. [DOI] [PubMed] [Google Scholar]
- 19.Yoon EJ, Yang JW, Kim JO, Lee H, Lee KJ, Jeong SH. 2018. Carbapenemase-producing Enterobacteriaceae in South Korea: a report from the National Laboratory Surveillance System. Future Microbiol 13:771–783. doi: 10.2217/fmb-2018-0022. [DOI] [PubMed] [Google Scholar]
- 20.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW, Shu L, Yu J, Zhang R, Chen S. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 21.Zong Z, Fenn S, Connor C, Feng Y, McNally A. 2018. Complete genomic characterization of two Escherichia coli lineages responsible for a cluster of carbapenem-resistant infections in a Chinese hospital. J Antimicrob Chemother 73:2340–2346. doi: 10.1093/jac/dky210. [DOI] [PubMed] [Google Scholar]
- 22.Xu L, Wang P, Cheng J, Qin S, Xie W. 2019. Characterization of a novel blaNDM-5-harboring IncFII plasmid and an mcr-1-bearing IncI2 plasmid in a single Escherichia coli ST167 clinical isolate. Infect Drug Resist 12:511–519. doi: 10.2147/IDR.S192998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bi R, Kong Z, Qian H, Jiang F, Kang H, Gu B, Ma P. 2018. High prevalence of blaNDM variants among carbapenem-resistant Escherichia coli in Northern Jiangsu Province, China. Front Microbiol 9:2704. doi: 10.3389/fmicb.2018.02704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakraborty T, Sadek M, Yao Y, Imirzalioglu C, Stephan R, Poirel L, Nordmann P. 2021. Cross-border emergence of Escherichia coli producing the carbapenemase NDM-5 in Switzerland and Germany. J Clin Microbiol 59:e02238-20. doi: 10.1128/JCM.02238-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mari-Almirall M, Cosgaya C, Pitart C, Vines J, Munoz L, Campo I, Cusco A, Rodriguez-Serna L, Santana G, Del Rio A, Francino O, Ciruela P, Pujol I, Ballester F, Marco F, Martinez JA, Soriano A, Vila J, Roca I, Group MES, MERCyCAT Study Group . 2021. Dissemination of NDM-producing Klebsiella pneumoniae and Escherichia coli high-risk clones in Catalan healthcare institutions. J Antimicrob Chemother 76:345–354. doi: 10.1093/jac/dkaa459. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Fernandez A, Villa L, Bibbolino G, Bressan A, Trancassini M, Pietropaolo V, Venditti M, Antonelli G, Carattoli A. 2020. Novel insights and features of the NDM-5-producing Escherichia coli sequence type 167 high-risk clone. mSphere 5:e00269-20. doi: 10.1128/mSphere.00269-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang P, Xie Y, Feng P, Zong Z. 2014. blaNDM-5 carried by an IncX3 plasmid in Escherichia coli sequence type 167. Antimicrob Agents Chemother 58:7548–7552. doi: 10.1128/AAC.03911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cole SD, Peak L, Tyson GH, Reimschuessel R, Ceric O, Rankin SC. 2020. New Delhi metallo-beta-lactamase-5-producing Escherichia coli in companion animals, United States. Emerg Infect Dis 26:381–383. doi: 10.3201/eid2602.191221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong JS, Song W, Park HM, Oh JY, Chae JC, Han JI, Jeong SH. 2019. First detection of New Delhi metallo-beta-lactamase-5-producing Escherichia coli from companion animals in Korea. Microb Drug Resist 25:344–349. doi: 10.1089/mdr.2018.0237. [DOI] [PubMed] [Google Scholar]
- 30.Mei YF, Liu PP, Wan LG, Liu Y, Wang LH, Wei DD, Deng Q, Cao XW. 2017. Virulence and genomic feature of a virulent Klebsiella pneumoniae sequence type 14 strain of serotype K2 harboring blaNDM-5 in China. Front Microbiol 8. doi: 10.3389/fmicb.2017.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu WJ, Wang X, Qin JX, Liang W, Shen Z. 2020. Dissemination and stability of the blaNDM-5-carrying IncX3-type plasmid among multiclonal Klebsiella pneumoniae isolates. Msphere 5:e00917-20. doi: 10.1128/mSphere.00917-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma T, Fu J, Xie N, Ma S, Lei L, Zhai W, Shen Y, Sun C, Wang S, Shen Z, Wang Y, Walsh TR, Shen J. 2020. Fitness cost of blaNDM-5-carrying p3R-IncX3 plasmids in wild-type NDM-free Enterobacteriaceae. Microorganisms 8:377. doi: 10.3390/microorganisms8030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun L, Zhao X, Wang L, Guo X, Shi X, Hu L. 2021. Coexistence of mcr-9 and blaNDM-1 in a multidrug-resistant Enterobacter hormaechei strain recovered from a bloodstream infection in China. J Glob Antimicrob Resist 24:440–442. doi: 10.1016/j.jgar.2021.02.011. [DOI] [PubMed] [Google Scholar]
- 34.Ding M, Shi J, Ud Din A, Liu Y, Zhang F, Yan X, Li Q, Bai J, Chen W, Zhou Y. 2021. Co-infections of two carbapenemase-producing Enterobacter hormaechei clinical strains isolated from the same diabetes individual in China. J Med Microbiol 70. doi: 10.1099/jmm.0.001316. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Chen D, Xu G, Huang W, Wang X. 2018. Molecular epidemiology and drug resistant mechanism in carbapenem-resistant Klebsiella pneumoniae isolated from pediatric patients in Shanghai, China. PLoS One 13:e0194000. doi: 10.1371/journal.pone.0194000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rangnekar VM, Banker DD, Jhala HI. 1983. Antimicrobial resistance and incompatibility groups of R plasmids in Salmonella typhimurium isolated from human sources in Bombay from 1978 to 1980. Antimicrob Agents Chemother 23:54–58. doi: 10.1128/AAC.23.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bortolaia V, Guardabassi L, Trevisani M, Bisgaard M, Venturi L, Bojesen AM. 2010. High diversity of extended-spectrum beta-lactamases in Escherichia coli isolates from Italian broiler flocks. Antimicrob Agents Chemother 54:1623–1626. doi: 10.1128/AAC.01361-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J, Voets GM, van den Munckhof MP, van Essen-Zandbergen A, Platteel T, Fluit AC, van de Sande-Bruinsma N, Scharinga J, Bonten MJ, Mevius DJ, National ESBL surveillance group . 2011. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect 17:873–880. doi: 10.1111/j.1469-0691.2011.03497.x. [DOI] [PubMed] [Google Scholar]
- 39.Diestra K, Juan C, Curiao T, Moya B, Miro E, Oteo J, Coque TM, Perez-Vazquez M, Campos J, Canton R, Oliver A, Navarro F, Red Espanola de Investigacion en Patologia Infecciosa . 2009. Characterization of plasmids encoding blaESBL and surrounding genes in Spanish clinical isolates of Escherichia coli and Klebsiella pneumoniae. J Antimicrob Chemother 63:60–66. doi: 10.1093/jac/dkn453. [DOI] [PubMed] [Google Scholar]
- 40.Carattoli A, Villa L, Fortini D, Garcia-Fernandez A. 2021. Contemporary IncI1 plasmids involved in the transmission and spread of antimicrobial resistance in Enterobacteriaceae. Plasmid 118:102392. doi: 10.1016/j.plasmid.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Chong Y, Shimoda S, Shimono N. 2018. Current epidemiology, genetic evolution and clinical impact of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Genet Evol 61:185–188. doi: 10.1016/j.meegid.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Cain AK, Hall RM. 2012. Evolution of IncHI2 plasmids via acquisition of transposons carrying antibiotic resistance determinants. J Antimicrob Chemother 67:1121–1127. doi: 10.1093/jac/dks004. [DOI] [PubMed] [Google Scholar]
- 43.Rozwandowicz M, Brouwer MSM, Fischer J, Wagenaar JA, Gonzalez-Zorn B, Guerra B, Mevius DJ, Hordijk J. 2018. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J Antimicrob Chemother 73:1121–1137. doi: 10.1093/jac/dkx488. [DOI] [PubMed] [Google Scholar]
- 44.Zhu Y, Liu W, Schwarz S, Wang C, Yang Q, Luan T, Wang L, Liu S, Zhang W. 2020. Characterization of a blaNDM-1-carrying IncHI5 plasmid from Enterobacter cloacae complex of food-producing animal origin. J Antimicrob Chemother 75:1140–1145. doi: 10.1093/jac/dkaa010. [DOI] [PubMed] [Google Scholar]
- 45.Clinical and Laboratory Standards Institute. 2018. M100: Performance standards for antimicrobial susceptibility testing, 27th ed. CLSI, Wayne, PA. [Google Scholar]
- 46.Li R, Lu X, Liu Z, Liu Y, Xiao X, Wang Z. 2020. Rapid detection and characterization of tet(X4)-positive Escherichia coli strains with nanopore sequencing. J Antimicrob Chemother 75:1068–1070. doi: 10.1093/jac/dkz528. [DOI] [PubMed] [Google Scholar]
- 47.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 50.Lam MMC, Wyres KL, Judd LM, Wick RR, Jenney A, Brisse S, Holt KE. 2018. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med 10:77. doi: 10.1186/s13073-018-0587-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM02156-21_Supp_1_seq11.pdf, PDF file, 1.3 MB (1.3MB, pdf)
Data Availability Statement
The sequence data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession numbers PRJNA752009, and individual accession numbers of 22 blaNDM-bearing plasmids are listed in Table 2.