Letter to the editor
We read with interest the letter by Dimeglio et al. reporting the impact of vaccination and pre-immunity on the proliferation of Omicron BA.1 and BA.2 sublineages in France1. The new emerging Omicron strain of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is currently spreading worldwide. The Omicron strain has multiple spike protein mutations compared with other variants of concern, such as the Alpha and Delta strains2. Consequently, there is concern that serum antibody activity against the Omicron strain in vaccinated or convalescent persons will be weaker than that against previous SARS-CoV-2 strains3 , 4. Because the characteristics of infectivity and treatment response differ among Omicron sublineages5 , 6, it is important to understand the evolutionary process in real time.
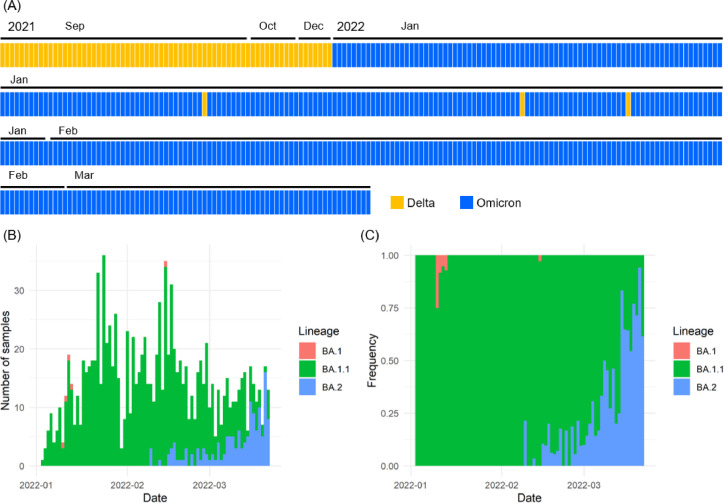
To determine the viral lineage of SARS-CoV-2, we performed whole genome sequencing analyses or TaqMan assays using SARS-CoV-2-positive samples (n = 1298) collected consecutively in Yamanashi, Japan from September 2021 to March 2022 (Supplemental materials)7 , 8 , 9. During this period, we identified Delta strain (n = 159) and Omicron strain (n = 1139). After the first case of Omicron was identified in January 2022, Omicron rapidly replaced Delta as the prevalent strain of SARS-CoV-2 (Fig. 1 A).
Fig. 1.
Changes in Omicron strain prevalence
SARS-CoV-2 strains identified from September 2021 to March 2022. Orange boxes indicate Delta strains, and blue boxes indicate Omicron strains. (B, C) Sublineage of Omicron strains detected from January 2022 to March 2022, indicated by BA.1 (pink), BA.1.1 (green), and BA.2 (blue). The number of samples detected per day (B) and the frequency of detection (C) are shown.
The whole genome sequencing data were analyzed using PANGOLIN (version 3.1.20), and BA.1 (n = 5), BA.1.1 (n = 992), and BA.2 (n = 142) were identified as sublineages of Omicron (Fig. 1B). Sublineage BA.1.1 was the dominant sublineage of Omicron from January to mid-February 2022; however, the incidence of sublineage BA.2 increased from mid-February 2022 onward, with this sublineage becoming dominant by the end of March (Fig. 1B and 1C). The average frequency for the seven-day period from March 8 to March 14 was 62.2% (51/82) for sublineage BA.1.1 and 37.8% (31/82) for sublineage BA.2, whereas from March 15 to March 21 it was 29.3% (27/92) for sublineage BA.1.1 and 70.7% (65/92) for sublineage BA.2. These results indicate an extremely rapid replacement of sublineage BA.1.1 by sublineage BA.2 and a higher transmissibility of sublineage BA.2 compared with sublineage BA.1.1.
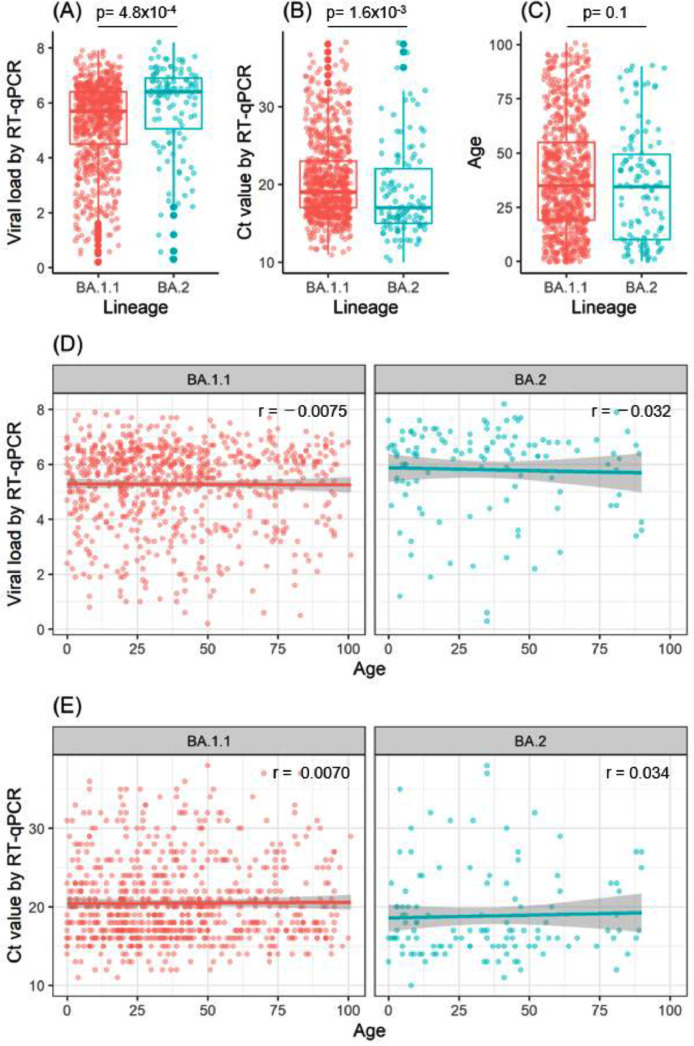
To investigate the underlying factors for the high transmissibility of Omicron sublineage BA.2, we performed an RT-qPCR analysis of the viral load in the nasopharyngeal swabs collected from patients infected with sublineage BA.1.1 (n = 748) or sublineage BA.2 (n = 118). The median viral load (log10 copies/mL) was 5.7 (range: 0.2–7.9) for sublineage BA.1.1 versus 6.4 (range: 0.3–8.2) for sublineage BA.2 (Fig. 2 A). The median Ct value for sublineage BA.1.1 was 19 (range: 11–38) versus 17 (range: 10–38) for sublineage BA.2 (Fig. 2B). There are significant differences in the viral load between cases of sublineage BA.1.1 and sublineage BA.2 (Fig. 2A, p = 4.8 × 10−4, Student's t-test) and Ct value (Fig. 2B, p = 1.6 × 10−3, Student's t-test). However, the median age of infected patients was not significantly different between these sublineages (35 years [range: 0–101 years] for BA.1.1 vs. 34.5 years [range: 0–90 years] for BA.2; p = 0.1, Student's t-test) (Fig. 2C). These results indicate that the viral load in nasopharyngeal swabs is higher for sublineage BA.2 than for sublineage BA.1.1 and that sublineage BA.2 is more contagious.
Fig. 2.
Viral load and age of infected patients for sublineages BA.1. and BA.2.
(A, B) The viral load and Ct values in Omicron sublineages BA.1.1 (n = 748) and BA.2 (n = 118) were analyzed by RT-qPCR. Box plots show the viral load (A) and Ct values (B) in BA.1.1 and BA.2. (C) Box plot shows the age of patients infected with sublineage BA.1.1 or BA.2. (D, E) Relationship between patient age and viral load (D) or Ct value (E). Pearson's correlation coefficient (r) is noted in the figures. The gray background of the regression line indicates the 95% confidence interval.
We next examined whether the viral load varied with patient age. There was no apparent correlation between patient age and viral load or Ct value for either sublineage BA.1.1 or BA.2 (Figure 3D and 3E). The Pearson's correlation coefficients for sublineage BA.1.1 were r = −0.0075 (p = 0.84) for patient age and viral load and r = 0.0070 (p = 0.85) for patient age and Ct value, and those for sublineage BA.2 were r = −0.032 (p = 0.73) for patient age and viral load and r = 0.034 (p = 0.71) for patient age and Ct value (Figs. 2D and 2E). These results indicate that the viral load remained fairly high in Omicron-infected patients regardless of their age.
In summary, this study indicates that after the expansion of the SARS-CoV-2 Delta strain, a rapid spread of the Omicron strain occurred. Sublineage BA.1 was very minor in Japan when Omicron was first discovered. First, sublineage BA.1.1 expanded dominantly and was then gradually replaced by sublineage BA.2. A transition from sublineage BA.1.1 to sublineage BA.2 was clearly observed over approximately one month. The results of the present study show that the amount of viral load in the nasopharyngeal swab was higher for sublineage BA.2 than for sublineage BA.1.1. These epidemiological and viral characteristic results indicate that Omicron sublineage BA.2 is more transmissible than sublineage BA.1.1. Although a high incidence of household COVID-19 infections stemming from young children has been reported10, our results indicate that the Omicron strain retains a fairly high viral load across age groups, which may contribute to the high infectivity of the Omicron strain and its accelerated spread. These data provide insights for determining appropriate COVID-19 prevention and control measures for homes, schools, workplaces, and facilities for the elderly during the spread of Omicron strain viruses.
Declaration of Competing Interest
None.
Acknowledgments
Acknowledgments
We thank all the medical and ancillary hospital staff for their support. We also thank Katie Oakley, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Funding
This work was supported by a Grant-in-Aid for the Genome Research Project from Yamanashi Prefecture (to M.O. and Y.H.), the Japan Society for the Promotion of Science (JSPS) KAKENHI Early-Career Scientists JP18K16292 (to Y.H.), a Grant-in-Aid for Scientific Research (B) 20H03668 (to Y.H.), a Research Grant for Young Scholars (to Y.H.), the YASUDA Medical Foundation (to Y.H.), the Uehara Memorial Foundation (to Y.H.), and Medical Research Grants from the Takeda Science Foundation (to Y.H.).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2022.04.040.
Appendix. Supplementary materials
References
- 1.Dimeglio C., Loubes J.-.M., Migueres M., Sauné K., Trémeaux P., Lhomme S., et al. Influence of vaccination and prior immunity on the dynamics of Omicron BA.1 and BA.2 sub-variants. J Infect. 2022 doi: 10.1016/j.jinf.2022.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Julia L.M., Ginger T., Alaa A.L., Manar A., Marco C., Emily H., et al. outbreak.info.
- 3.Iketani S., Liu L., Guo Y., Liu L., Chan J.F.W., Huang Y., et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022 doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu J., Collier A-rY, Rowe M., Mardas F., Ventura J.D., Wan H., et al. Neutralization of the SARS-CoV-2 Omicron BA.1 and BA.2 Variants. N Engl J Med. 2022 doi: 10.1056/NEJMc2201849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruel T., Hadjadj J., Maes P., Planas D., Seve A., Staropoli I., et al. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat Med. 2022 doi: 10.1038/s41591-022-01792-5. [DOI] [PubMed] [Google Scholar]
- 6.Takashita E., Kinoshita N., Yamayoshi S., Sakai-Tagawa Y., Fujisaki S., Ito M., et al. Efficacy of Antiviral Agents against the SARS-CoV-2 Omicron Subvariant BA.2. N Engl J Med. 2022 doi: 10.1056/NEJMc2201933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirotsu Y., Omata M. Detection of R.1 lineage severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with spike protein W152L/E484K/G769V mutations in Japan. PLoS Pathog. 2021;17(6) doi: 10.1371/journal.ppat.1009619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirotsu Y., Omata M. SARS-CoV-2 B1.1.7 lineage rapidly spreads and replaces R.1 lineage in Japan: serial and stationary observation in a community. Infect, Genet Evol. 2021;95 doi: 10.1016/j.meegid.2021.105088. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirotsu Y., Maejima M., Shibusawa M., Natori Y., Nagakubo Y., Hosaka K., et al. Classification of Omicron BA.1, BA.1.1 and BA.2 sublineages by TaqMan assay consistent with whole genome analysis data. medRxiv. 2022 doi: 10.1101/2022.04.03.22273268. 2022.04.03.22273268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paul L.A., Daneman N., Schwartz K.L., Science M., Brown K.A., Whelan M., et al. Association of age and pediatric household transmission of SARS-CoV-2 infection. JAMA Pediatr. 2021;175(11):1151–1158. doi: 10.1001/jamapediatrics.2021.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.