Abstract
The increase in antibiotic resistance related to microbial biofilms creates an urgent need to search for an alternative and active antimicrobial agent. Recently, nanoparticles have gained considerable attention from scientists due to their potent antimicrobial activity. In the present study, two endosymbiotic actinomycete strains were isolated from marine sponge Crella cyathophora by surface sterilization and incubation of sponge pieces on culture media selective for actinobacteria. The culture filtrate extracts, including the bacterial supernatants (F) and cell filtrate (C) of the two actinomycete strains, were used as the reducing agent for the green biosynthesis of silver nanoparticles. The as-prepared silver nanoparticles were characterized using dynamic light scattering, zeta-potential, UV-Vis spectroscopy, and transmission electron microscopy. The average particle size for synthesized silver nanoparticles was about ∼8.66 ± 2 to 35 ± 2 nm with monodisperse spherical-like shapes and polydispersed shapes, respectively. The synthesized silver nanoparticles exhibited significant antimicrobial activity toward pathogenic microbes, especially with P. aeruginosa and E. cloacae. The effect of silver nanoparticles on the growth curve dynamics of P. aeruginosa and E. cloacae showed that the slope of the bacterial growth curve continuously decreased with increasing nanoparticle concentration. Moreover, the antibiofilm activity of the silver nanoparticles was measured, and the results showed that the silver nanoparticles displayed high biofilm inhibition activity against P. aeruginosa, B. subtilis, and S. aureus. Furthermore, silver nanoparticles exhibited a low to moderate cytotoxic effect against hepatocellular carcinoma cancerous cells, which reflect its possible use in the biomedical field.
Culture filtrate of 2 actinomycetes extracted from marine sponge Crella cyathophora was used for the biosynthesis of AgNPs with a significant anti-microbial and biofilm activity. Also, AgNPs exhibited a low to moderate cytotoxicity against cells.
1. Introduction
With the continuous increase in microbial multidrug resistance and antibiotic therapy limitations, there is an urgent need to develop an efficient antimicrobial agent with new mechanisms of action.1 Nanoparticles have recently gained considerable attention in many fields such as medicine, electronics, food, agriculture, and energy.2 Silver nanoparticles (AgNPs) can be produced either chemically or biologically.3 The chemical synthesis of metal nanoparticles requires the use of a particular set of aggressive and hazardous chemicals to convert metal ions to metal nanoparticles.4
The biological or green synthesis of metal nanoparticles has many more advantages compared to chemical synthesis due to the use of eco-friendly agents as reducing agents instead of hazardous chemicals.5 One of the green nano-factories for synthesis and production of metal nanoparticles are microbes.6 Recently, many strains of bacteria, such as Bacillus sp. and Pseudomonas sp., have been used to convert silver ions to silver nanoparticles through the reduction process.7,8
AgNPs have been proven as efficient metallic nanoparticles due to their promising bioactivities that can include anti-oxidant,9 antifungal,10 antibacterial,11 and anticancer properties.12 The potential antimicrobial property of the silver nanoparticles can be utilized in the medical devices industry to reduce microbial infections and to prevent bacteria colonization on medical equipment such as dental materials,13 catheters,14 vascular grafts,15 stainless steel materials and human skin.16,17 Additionally, silver nanoparticles could be an efficient way to control biofilm formation, thereby reducing the infectious disease transmission, such as hospital nosocomial bacterial infections.18 Recently, bacterial biofilms have been considered one of the public health concerns. Once bacteria form biofilms, it becomes more resistant to antibiotics, which is strong evidence of the critical role of biofilms in the persistence of pathogenic bacteria.19
The present work involves the green biosynthesis of AgNPs as an excellent alternative route and evaluates the antimicrobial, antibiofilm, and anticancer activities of the synthesized silver nanoparticles.
2. Results and discussion
2.1. Collection and isolation of actinomycete from a marine sponge
The marine sponge Crella cyathophora was collected using SCUBA equipment at depth 5 from Makadi Bay South Hurghada at N 26° 59′ 42.87′′, E 33° 54′ 4.02′′. Isolation and purification resulted in a yield of 9 bacterial isolates. The isolates were coded and kept at 4 °C in a culture collection of the Microbial Chemistry Department, National Research Center, Egypt.
2.2. Green biosynthesis of silver nanoparticles
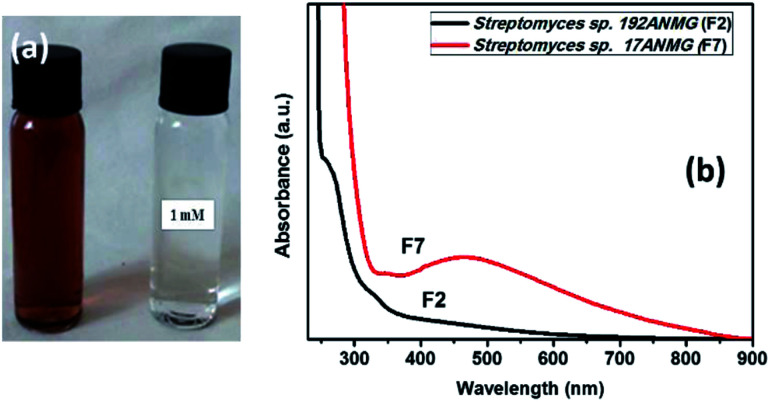
In the present study, silver nanoparticles (AgNPs) were prepared via a green synthesis method using 9 actinomycete isolates including both the bacterial supernatant (F) and their cell filtrate (C) (see Table S1, ESI†). The synthesis was mediated at 32 °C under dark conditions, after the addition of actinomycetes sp. supernatant (i.e., Streptomyces sp. 192ANMG (F2), and Streptomyces sp. 17ANMG (F7)) due to their highest antimicrobial activity as shown in Table S1,† and their ability to form silver nanoparticles compared to others as shown in Fig. 1b and S2.† Typically, a yellowish-brown color was observed in the supernatant after reaction with the Ag+ ions, indicating the metal ion reduction and formation of silver nanoparticles (i.e., Ag0 NPs) (Fig. 1a). AgNPs exhibit a light yellow to brown color due to the excitation of surface plasmon vibrations in the particles, which is confirmed by UV-Vis absorption spectroscopy (Fig. 1b). The as-prepared AgNPs synthesized in the presence of actinomycete bacterial supernatants (i.e., F2 and F7) exhibit a characteristic surface plasmon band (SPR) at 460 nm, and the surface plasmon resonance (SPR) absorption spectral range was from 400 to 550 nm, indicating that AgNPs formed at different time intervals. These results were in agreement with previous studies represented by Składanowski et al.20 and Al-Dhabi et al.21
Fig. 1. (a) Color change due to the reduction of metal ions and the formation of silver nanoparticles. (b) Surface plasmon absorption bands (SPR) of AgNPs formed in the presence of bacterial supernatant of two actinomycete species (192ANMG (F2) and 17ANMG (F7), respectively).
2.3. Characterization of silver nanoparticles
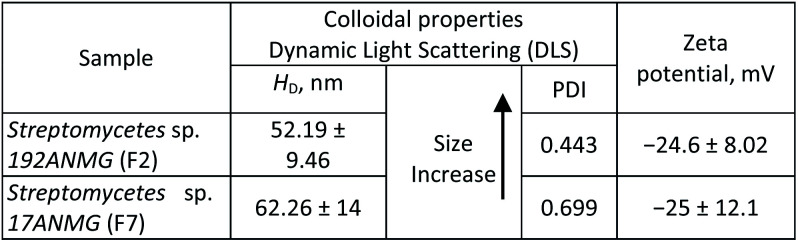
The surface and colloidal stability of as-prepared AgNPs at different intervals were examined via dynamic light scattering (DLS) and zeta potential measurements. It is clear that upon the addition of the actinomycete supernatant and cell filtrate to the aqueous solution of AgNO3, the particle size (i.e., hydrodynamic diameter, HD) increased due to the reduction of Ag+ ions on the Ag nuclei forming big particles (Table 1). Furthermore, the zeta potentials of the two selected AgNP samples, including Streptomyces sp. 192ANMG (F2) and Streptomyces sp. 17ANMG (F7), were −24.6 and −25 mV, respectively. Results indicated that the as-prepared AgNPs tended to have an electrostatic attraction between particles in the solution (Table 1).
Colloidal and surface properties of as-prepared AgNPs.
|
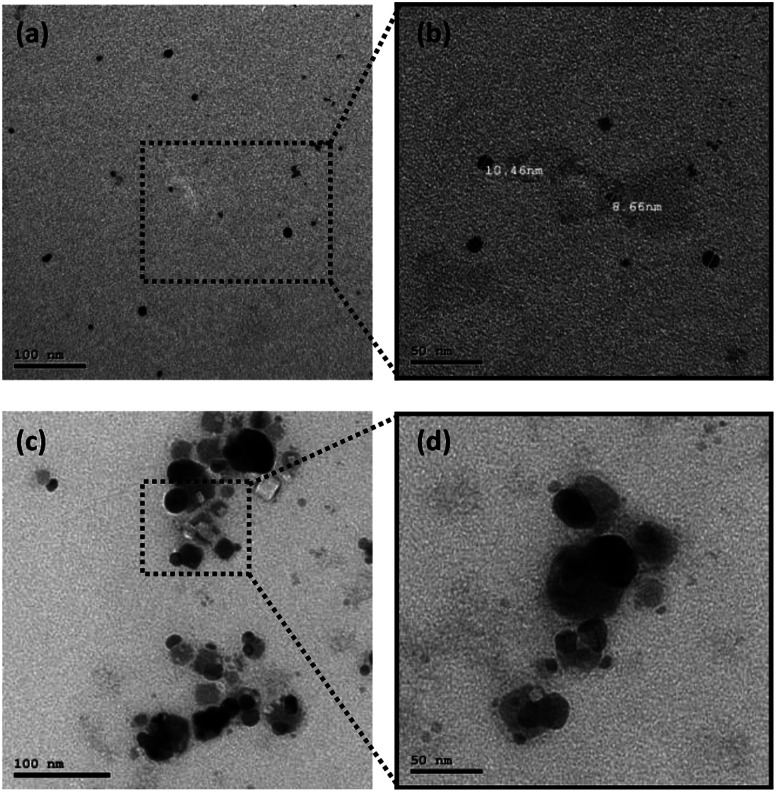
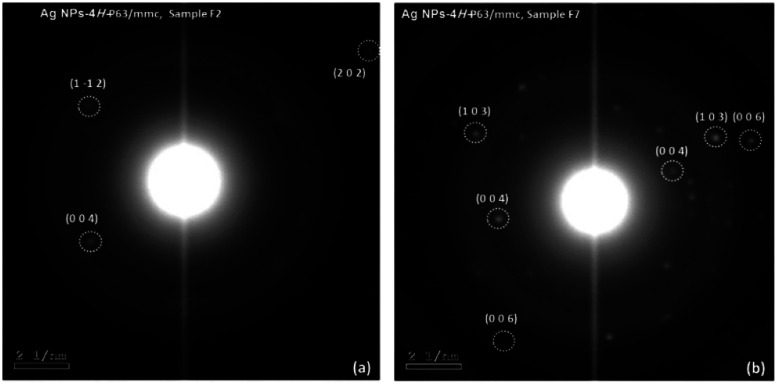
Also, the particle size and shape were examined via transmission electron microscopy (TEM) measurements for two different samples (i.e., Streptomyces sp. 192ANMG, F2 and Streptomyces sp. 17ANMG F7), as shown in Fig. 2. The average particle size for silver nanoparticles (Streptomyces sp. 192ANMG, F2) is about ∼8.66 ± 2 nm with monodisperse spherical-like shapes (Fig. 2a and b). Whereas, the average particle size in the second sample (Streptomyces sp. 17ANMG, F7) is about 35 ± 2 nm with poly-dispersed shapes, including quasi-spherical and cubic-like shapes (Fig. 2c and d), which confirmed the broadening in the UV-Vis absorption spectrum (Fig. 1a, red-line). Also, the as-prepared AgNP samples (i.e., Streptomyces sp. 192ANMG, F2, and Streptomyces sp. 17ANMG, F7) synthesized via two different bacterial extract isolates exhibited a hexagonal crystallographic structure, as shown in Fig. 3a and b.
Fig. 2. TEM micrographs for as-prepared AgNPs with two different bacterial supernatants (i.e., extracts) (a and b) Streptomyces sp. 192ANMG, F2, and (c and d) Streptomyces sp. 17ANMG, F7, respectively, at two different magnifications.
Fig. 3. SAED images for as-prepared AgNPs using two different bacterial isolate extracts (a) Streptomyces sp. 192ANMG (F2), and (b) Streptomyces sp. 17ANMG, (F7).
Based on the chemical compositions of actinomycete isolates, several functional groups such as amino and carboxylic groups (i.e., –NH2 and –COOH) are preset, which facilitate Ag+ ions trapping on the surface of the actinomycetes. In such interactions, Ag+ ions interact with each of the negatively charged carboxylates, and the lone pair of N in NH2 groups exists in enzymes present in the cell wall of mycelia.5 The Ag+ ions are reduced by enzymes present in the cell wall leading to the formation of the silver nuclei due to a decrease in the potential of Ag+/Ag (EAg+/Ag), which promotes the reduction of Ag+, leading to a subsequent growth by further reduction of Ag+ ions and accumulation on these nuclei.22
2.4. Antimicrobial activity of biosynthesized AgNPs
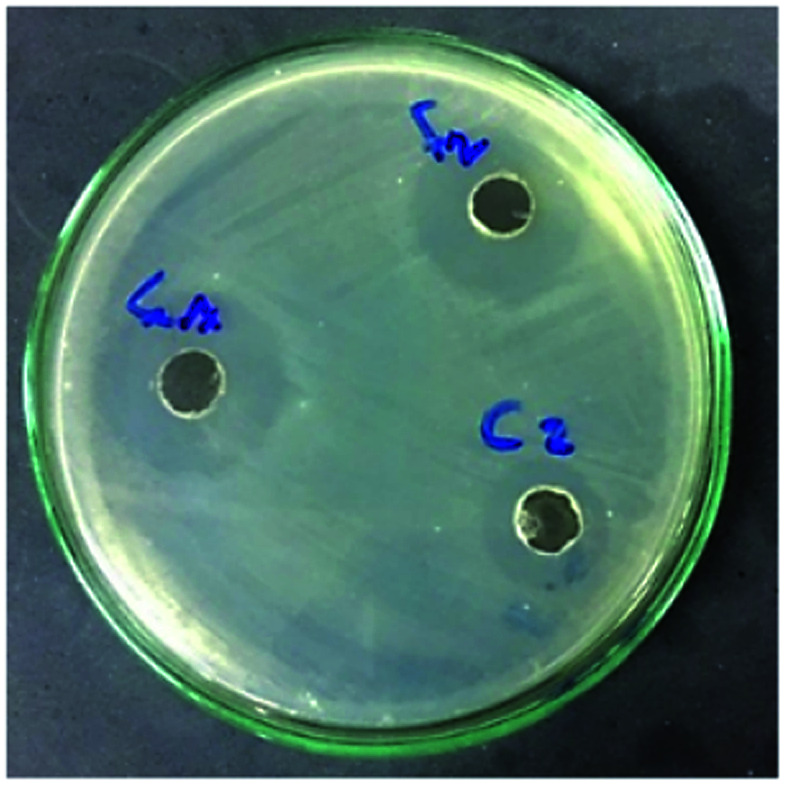
Silver nanoparticles have a detrimental effect on different human pathogens.21 The antimicrobial activity of the produced AgNPs was measured using the agar well diffusion method (Fig. 4). In general, AgNPs biosynthesized by the bacterial supernatant (F) showed greater antibacterial activity compared to those made with the cells filtrate (C). The results, in general, showed that the negative control (the supernatant and cells filtrate of the 9 actinomycetes) did not display any significant activity. Moreover, P. aeruginosa and E. cloacae were the most susceptible bacteria to the antibacterial effect of the AgNP solutions, especially with the two selected Streptomyces isolates (Streptomyces sp. 192ANMG, F2) and (Streptomyces sp. 17ANMG, F7), resulting in an inhibition zone diameter of 1.80 and 1.88 cm, respectively (Table S1†). The rationale for the selection of silver nanoparticles using two Streptomyces isolates (Streptomyces sp. 192ANMG, F2) and (Streptomyces sp. 17ANMG, F7), was attributed to their surface plasmon absorption bands and the antimicrobial activities, as shown in Fig. 1 and Table S1.†A. hydrophila, S. aureus, and Erwinia sp. were the bacterial strains most resistant to the antibacterial effect of the biosynthesized silver nanoparticle solutions.
Fig. 4. Antibacterial effect of as-prepared active silver nanocolloids using bacterial supernatant of Streptomyces sp. 192ANMG (F2), and Streptomyces sp. 17ANMG (F7) and cell filtrate of Streptomyces sp. 192ANMG (C2) on P. aeruginosa nutrient agar (NA) culture.
2.5. Molecular identification of selected actinomycetes
Two selected Streptomyces isolates 17ANMG and 192ANMG were identified by 16S rDNA sequencing. The DNA was extracted, amplified using PCR, sequenced, and aligned with other identified strains in the Gene bank database using an online BLAST tool to determine the similarity score (http://www.blast.ncbi.nlm.nih.gov/Blast). The obtained result confirmed a high similarity of the 16S rDNA gene sequence with more than 99% homology of the two isolates 17ANMG and 192ANMG with Streptomyces sp. The partial 16S rDNA sequences from the Streptomyces isolates identified as Streptomyces sp. 17ANMG and Streptomyces sp. 192ANMG with accession numbers MN339599 and MN365039, respectively. The phylogenetic trees were constructed using the MEGA-X program23 and the neighbor-joining method (Fig. S1†).24
2.6. Monitoring microbial growth curve for P. aeruginosa and E. cloacae in the presence of Streptomyces sp. 192ANMG (F2) and Streptomyces sp. 17ANMG (F7) silver nanoparticles solutions
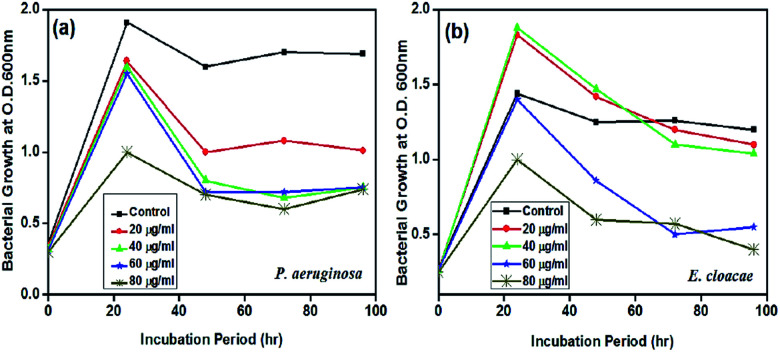
Investigating the effect of F2 and F7 nanoparticle solutions on the growth curve dynamics of P. aeruginosa and E. cloacae individually, the results showed that the slope of the bacterial growth curve continuously decreased with increasing nanoparticle concentration. 1 mL AgNPs (Streptomyces sp. 192ANMG, F2) solution contains 10.3 mg silver nanoparticles; for the P. aeruginosa growth curve, the antibacterial effect of AgNPs increases with increasing AgNPs concentration (Fig. 5a). The maximum inhibitory activity was detected at a 80 μg mL−1 concentration that begun with a dwarfed log period while the most pronounced effect of the AgNPs 80 μg mL−1 concentration is easily visualized at the late logarithmic phase and during the stationary phase of the bacterial growth. 1 mL of AgNPs (Streptomyces sp. 17ANMG, F7) solution contains 13 mg silver nanoparticles, in the case of the E. cloacae growth curve, AgNPs in small concentrations (20 and 40 μg mL−1) showed some growth-promoting activity that changed at the late stationary phase with the rapid decline of the bacterial growth in comparison to the control treatment without AgNPs inoculation. Meanwhile, the concentration of 80 μg mL−1 showed again to be bactericidal for bacterial growth and possessing the highest antibacterial activity during all incubation period intervals (Fig. 5b).
Fig. 5. Growth curve dynamics of P. aeruginosa with Streptomyces sp. 192ANMG (F2) AgNPs (a), and E. cloacae with Streptomyces sp. 17ANMG (F7) AgNPs (b) solutions.
2.7. Antibiofilm activity
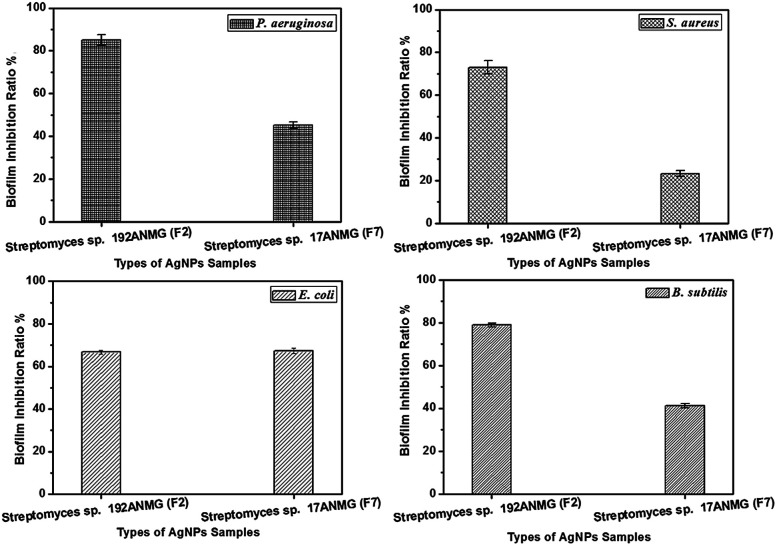
The ability of the Streptomyces sp. 192ANMG (F2) and Streptomyces sp. 17ANMG (F7) AgNPs to eradicate bacterial biofilm formation of four pathogenic bacteria (S. aureus, P. aeruginosa, B. subtilis, and E. coli) was measured using the MTT assay.25 The AgNPs sample F2 displayed potent biofilm inhibition activity against P. aeruginosa, B. subtilis and S. aureus with a biofilm inhibitory ratio of 85.00, 78.81 and 73.04%, respectively, while displaying moderate biofilm inhibition activity against E. coli with a biofilm inhibitory ratio of 66.69%. On the other hand, F7 reduced the biofilm formation of the strain E. coli up to 67.22%. However, P. aeruginosa and B. subtilis biofilm formation was inhibited over 45.20 and 41.19%, respectively. In the case of E. coli, it displayed a very low response to F7 (23.31%) (Fig. 6).
Fig. 6. Biofilm inhibitory ratio% following treatment with 20 μg mL−1 of Streptomyces sp. 192ANMG (F2) and Streptomyces sp. 17ANMG (F7). P. aeruginosa, S. aureus, E. coli, and B. subtilis biofilm was assessed by crystal violet staining. Data presented represent mean ± SD of three independent experiments.
2.8. In-vitro cytotoxicity
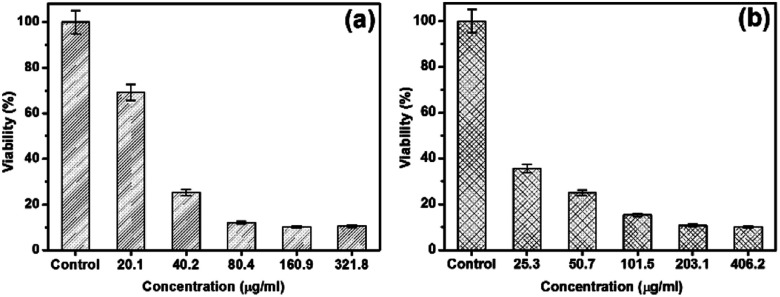
The cytotoxicity of AgNP samples Streptomyces sp. 192ANMG (F2) and Streptomyces sp. 17ANMG (F7) were evaluated by using an MTT assay against hepatocellular carcinoma (HepG-2). Data represented in Fig. 7 showed that silver nanoparticles F2 displayed lower cytotoxicity than F7 while a dosage of 40.2 μg mL−1 of F2 AgNPs was necessary to initiate the toxicity with cell viability of nearly 70% (Fig. 7a). On the other hand, F7 was quite toxic at a low concentration of 25.2 μg mL−1, with cell viability up to 77% (Fig. 7b). The low cytotoxic effect of the green-biosynthesized AgNPs against the human cell line could result from some compounds in the bacterial extracts that have a synergistic effect with the AgNPs.
Fig. 7. Cytotoxicity evaluation at overexposure to different concentrations of silver nanoparticles (a) Streptomyces sp. 192ANMG (F2) and (b) Streptomyces sp. 17ANMG (F7) samples.
3. Experimental section
3.1. Isolation of sponge-associated actinobacteria
Samples were collected from Hurghada, Egypt. The collected marine samples were given codes, photographed, and kept at 4 °C. The sponge tissues were rinsed three times in sterile artificial seawater (SASW) followed by 70% ethanol for 30 seconds, SASW for 1 min, 2% NaOCl for 1 min, and finally washed with SASW three times. After sterilization, the samples were left to dry in the laminar flow. After drying, samples were cut into small pieces and transferred to starch nitrate agar media with the composition (g L−1): starch, 20; K2HPO4, 0.5; KNO3, 1.0; MgSO4·7H2O, 0.5; FeSO4, 0.01; agar, 15. At pH 7, incubation was carried out at 30 °C for seven days at room temperature until growth was observed.26–28 The efficiency of the surface sterilization procedure was evaluated via inoculation of about 4 to 5 drops of the water from the final rinse on starch nitrate agar. Endophytic bacteria should appear only on the organism-inoculated plates and not on the water rinse ones. In such case, water was used as a control to confirm the successful sterilization. Therefore, if the agar plate inoculated with this water showed any colonies, this indicated that the sterilization processes not precise.
3.2. Actinomycete cell-free extract preparation
The isolated actinomycete strains were cultured in 250 mL flasks containing 50 mL of ISP2 media at pH 7.2 with the following composition (g L−1); yeast extract, 4.0; malt extract, 10.0; dextrose, 4.0. The incubation of the inoculated flasks was at 32 °C for six days in a rotary shaker at 150 rpm. After incubation, cultures were centrifuged at 10 000 rpm for 15 min to separate cell pellets of the bacteria from the supernatant. One gram of the collected pellets was re-suspended in sterile double distilled water (25 mL) and incubated under shaking (150 rpm) for 48 h at 28 °C. After incubation, centrifugation at 10 000 rpm for 15 min was carried out to obtain the cell filtrate.
3.3. Green synthesis and characterization of silver nanoparticles
The green biosynthesis of AgNPs was performed by mixing 25 mL of AgNO3 aqueous solution (1.0 mM) with 25 mL of bacterial supernatants (F). The mixture was incubated at 32 °C for five days in a dark rotary shaker (200 rpm). In the same way, the synthetic procedure was repeated under the same conditions but by using the cells filtrate (C). The rationale for using both bacterial supernatant (F), and cell filtrates (C) in the biosynthesis process of Ag NPs was to test the efficiency for each term in the synthesis of Ag NPs. The role of actinomycetes in the biosynthesis of AgNPs was demonstrated as follows; a control experiment was performed by mixing un-inoculated culture media (i.e., ISP-2 broth as control) with the AgNO3 solution. The reduction of silver ions was tested by sampling 2 mL of the incubated solution at different time intervals and monitoring the UV-Vis spectra by using a JASCO V630 spectrophotometer at the range from 200 to 900 nm with an increment of 2 nm with slit width 5 nm. The color of the AgNO3 solution changed from a colorless to yellowish-brown color upon the incubation with actinomycete supernatant. The particle size, shape, and possible crystal structure were characterized via selected area electron diffraction (SAED) using a transmission electron microscope (TEM), a JOEL-JEM 2100, at an operating voltage 200 kV. The size distribution via dynamic light scattering (DLS) technique and the surface charge properties via zeta-potential measurements of as-prepared AgNPs were measured by a Malvern zeta-sizer nano ZS instrument with He/Ne laser (633 nm) at an angle of 173° collecting backscatter optics.
3.4. Determination of the antimicrobial activity of silver nanoparticles
The as-biosynthesized AgNPs from culture supernatant and cell filtrate of 9 actinomycete isolates along with the supernatant and cell filtrate without AgNPs as a negative control were tested individually against different human pathogens including Gram-negative bacteria (Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumonia, Enterobacter cloacae, Aeromonas hydrophila, and Erwinia spp.) and Gram-positive bacteria (Staphylococcus aureus, Bacillus subtilis, Micrococcus luteus, and MRSA) and yeast (i.e., Candida albicans) using the nutrient agar well diffusion method.29,30 A pure microbial colony of each strain was allowed to grow in nutrient broth medium with the following composition (g L−1): peptone, 10.0; beef extract, 10.0; sodium chloride, 5.0, at pH 7.2 ± 0.1 at 37 °C for 24 h in a dark rotary shaker (180 rpm). After incubation, 50 μL of the pathogenic bacterial suspensions were spread on the nutrient agar plates using a sterile cotton swab, and then by using a sterilized cork borer, wells were made with a capacity of 35 μL for each treatment of as-synthesized AgNPs (i.e., 13 μg mL−1) individually against the control samples that were prepared by centrifuging each of the nanoparticles solutions at 10 000 rpm for 5 minutes and separating the supernatant in a sterilizing 1.5 mL Eppendorf. After the loading of the AgNPs and the supernatant treatments in each well separately, the nutrient agar plates were incubated for 24–48 h at 37 °C. By the end of incubation, the diameter of the resulted inhibition zones were measured by zone scale parameters in cm.25 Based on the screening procedures, it was not necessary to determine the exact concentration of AgNPs because of the amount of silver nitrate (i.e., AgNO3) added to each treatment either in the presence of bacterial supernatant (F) or cellular filtrates (C) was the same. In such case, the formation of the AgNPs depends on the presence of a high concentration of reductive proteins and enzymes secreted by the microbe itself either in bacterial supernatant (F) or cellular filtrates (C), so as to total it is only screening for the successful treatments.
Then, detecting the effect of the AgNPs colloidal solution that gave the highest antibacterial activity on the growth curve of the microbe required determining the exact concentrations of AgNPs through serial dilutions needed for the experiment.
3.5. Genomic DNA isolation
According to the antibacterial activity of among all 9 isolates of marine sponge. The potent two Streptomyces sp., Streptomyces sp. 192ANMG (F2) and Streptomyces sp. 17ANMG (F7) were genetically identified by sequencing of the 16S rRNA gene of the selected strains. The DNA extraction was performed using DNeasy Blood & Tissue Kit according to manufacturer instructions.
3.6. PCR amplification
PCR amplification reactions were performed using two universal primers (27F 5′-AGAGTTTGATCCTGGCTCAG-3'; 1492R 5′-GGTTACCTTGTTACGACTT-3′). The final volume of the PCR amplification reaction was as follows: 50 μL (5 μL of 10 × Dream Taq Green PCR buffer, 2 μL of each 10 μmol dm−3 primers, 5 μL of 2 mmol dm−3 dNTP, 0.3 μL Taq DNA polymerase and 0.5 μL of template DNA). The PCR reaction ran under the following conditions: 94 °C for 45 s, 55 °C for 60 s, and 72 °C for 60 s.
3.7. Sequencing
The purified PCR products were sequenced in Macrogen Company, South Korea. The homology and similarity of the 16S rRNA sequences were studied by comparing the resulting sequences with the similar known sequences available in the NCBI database using online BLAST alignment search tools (http://www.ncbi.nlm.nih.gov/BLAST). The phylogenetic trees were constructed using MEGA-X software.23,24
3.8. Effect of different concentrations of silver nanoparticles on P. aeruginosa and E. cloacae growth dynamics
The successful doses/treatment that showed the highest antibacterial activity were subjected to an additional experiment for the determination of the effect of different AgNP concentrations on the microbial growth curves. The concentration of silver nanoparticles in each selected culture filtrate was determined as follows; 1 mL of (Streptomyces sp. 192ANMG, F2), and (Streptomyces sp. 17ANMG, F7) solution was dried overnight at 70 °C for 48 h. Moreover, the calculated net weight was determined for each treatment after evaporation in terms of mg mL−1. The antibacterial effectiveness of different concentrations of both F2 and F7 silver nanoparticles solutions (0, 20, 40, 60, and 80 μg mL−1) was tested to determine its activity on the bacterial growth curve for P. aeruginosa and E. cloacae, respectively. Both P. aeruginosa and E. cloacae bacteria colonies were cultured in nutrient broth medium until reaching the log phase. Then, bacterial cultures were diluted using fresh, sterilized nutrient broth medium until reaching optical density (OD 600) 0.25–0.35.31 Silver nanoparticles were added at different concentrations (0, 20, 40, 60, and 80 μg mL−1) into the inoculated culture medium, and the final volume was 50 mL. The inoculated cultures were incubated at 37 °C at 180 rpm. Bacterial growth was measured after 0, 24, 48, 72, and 96 h by measuring the optical density (OD) at 600 nm (0.1 OD 600 corresponds to 108 cells per milliliter).32
3.9. In vitro antibiofilm activity
The biofilm inhibitory activity of the formed AgNPs was measured using the microtitre plate assay (MTP) in 96 well-flat bottom polystyrene titer plates and four clinical microbes (E. coli, P. aeruginosa, S. aureus, and B. subtilis) according to Christensen et al.33 Each well of the 96 well-plate was filled with 180 μL of lysogeny broth (LB), with the following composition (g L−1): tryptone, 10.0; yeast extract, 5.0; NaCl, 10.0. At pH 7.2, 10 μL of overnight growing test bacteria, 10 μL of the as-synthesized AgNPs at a concentration of 20 μg mL−1 (i.e., Streptomyces sp. 192ANMG (F2) and Streptomyces sp. 17ANMG (F7)) sample, along with the negative control (i.e., filtrate without AgNPs) and then the plate were incubated for 24 h at 37 °C. After incubation, the content of the well was removed, and the floating bacteria were removed by washing each well with 200 μL of phosphate buffer saline pH 7.2. For staining, crystal violet (0.1%, w/v) was added, and after 1 h, the wells were washed with 200 μL per well-distilled water, and then the plate was kept for drying in the laminar flow. 95% ethanol was added to the dried plate, and the optical density (OD) was measured at 570 nm by using (SPECTROstar nano absorbance plate reader – BMG LABTECH).
3.10. Cytotoxic activity of AgNPs on human tumor cell lines
The cytotoxic effect of the produced AgNPs was measured using MTT (3,4,5-dimethylthiazol-2-yl)-2-5-diphenyl tetrazolium bromide. The MTT reduction by the mitochondrial dehydrogenase leads to the formation of a purple formazan product.34 Hepatocellular carcinoma cell line (HepG2) was incubated at 37 °C in a 96-well for 24 h before the treatments. After incubation, the cell lines were subjected to different concentrations of AgNPs samples synthesized by two different actinomycetes species (i.e., 20.1 to 643.6 μg mL−1 for Streptomyces sp. 192ANMG, F2, and 25.2 to 812.4 μg mL−1 for Streptomyces sp. 17ANMG, F7). After the treatment, the content was carefully removed by aspiration, followed by the addition of 100 μL of 0.5 mg mL−1 MTT in cell culture medium to each well and incubated for 2 h. To dissolve the formed formazan crystals, 100 μL of 10% (sodium dodecyl sulfate) SDS was added. The amount of formazan was measured at 560 nm using a microplate reader.
4. Conclusions
From the results obtained, it was concluded that the use of actinomycete extracts for the green biosynthesis of silver nanoparticles is an efficient, less expensive, eco-friendly, and safe strategy.
Moreover, the potential antimicrobial and antibiofilm activity of the green-produced silver nanoparticles indicated its possible use in medical and non-medical applications. Furthermore, the low cytotoxic properties of the produced green AgNPs lower the possible risks to human health and the environment. On the other hand, the spread of microbial resistance to antibiotics establishes an urgent need to search for an alternative and efficient drug to overcome the multi-drug resistant microorganisms. Finally, further studies are needed to determine the antimicrobial and cytotoxic mechanisms of the actions of these particles.
Conflicts of interest
The authors have declared no conflict of interest.
Supplementary Material
Acknowledgments
The authors would like to express their deepest gratitude to the National Research Centre (NRC) for the financial and technical support for this study.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra11021f
Notes and references
- Cantas L. Shah S. Q. A. Cavaco L. M. Manaia C. Walsh F. Popowska M. Garelick H. Bürgmann H. Sørum H. Front. Microbiol. 2013;4:96. doi: 10.3389/fmicb.2013.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biao L. Tan S. Wang Y. Guo X. Fu Y. Xu F. Zu Y. Liu Z. Mater. Sci. Eng. C. 2017;76:73–80. doi: 10.1016/j.msec.2017.02.154. [DOI] [PubMed] [Google Scholar]
- Arya G. Kumari R. M. Gupta N. Kumar A. Chandra R. Nimesh S. Artif. Cells, Nanomed., Biotechnol. 2018;46:985–993. doi: 10.1080/21691401.2017.1354302. [DOI] [PubMed] [Google Scholar]
- Roldán A. Salinas-Garcia J. Alguacil M. Caravaca F. Appl. Soil Ecol. 2005;30:11–20. doi: 10.1016/j.apsoil.2005.01.004. [DOI] [Google Scholar]
- Abdeen S. Geo S. Sukanya S. Praseetha P. Dhanya R. Int. J. Nano Dimens. 2014;5:155–162. [Google Scholar]
- Kaur P. Biotechnology Research and Innovation. 2018;2:63–73. doi: 10.1016/j.biori.2018.09.003. [DOI] [Google Scholar]
- Gopinath V. Velusamy P. Spectrochim. Acta, Part A. 2013;106:170–174. doi: 10.1016/j.saa.2012.12.087. [DOI] [PubMed] [Google Scholar]
- Rajora N. Kaushik S. Jyoti A. Kothari S. L. IET Nanobiotechnol. 2016;10:367–373. doi: 10.1049/iet-nbt.2015.0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshari A. K. Srivastava R. Singh P. Yadav V. B. Nath G. J. Ayurveda Integr. Med. 2018:1–8. doi: 10.1016/j.jaim.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallmann E. J. J. Cunha F. A. Castro B. N. Maciel A. M. Menezes E. A. Fechine P. B. A. Rev. Inst. Med. Trop. Sao Paulo. 2015;57:165–167. doi: 10.1590/S0036-46652015000200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Ouay B. Stellacci F. Nano Today. 2015;10:339–354. doi: 10.1016/j.nantod.2015.04.002. [DOI] [Google Scholar]
- Oves M. Aslam M. Rauf M. A. Qayyum S. Qari H. A. Khan M. S. Alam M. Z. Tabrez S. Pugazhendhi A. Ismail I. M. Mater. Sci. Eng. C. 2018;89:429–443. doi: 10.1016/j.msec.2018.03.035. [DOI] [PubMed] [Google Scholar]
- Maillard J. Y. J. Appl. Microbiol. 2002;92:16S–27S. doi: 10.1046/j.1365-2672.92.5s1.3.x. [DOI] [PubMed] [Google Scholar]
- Yin H. Yamamoto T. Wada Y. Yanagida S. Mater. Chem. Phys. 2004;83:66–70. doi: 10.1016/j.matchemphys.2003.09.006. [DOI] [Google Scholar]
- Edelstein A. S. and Cammaratra R., Nanomaterials: synthesis, properties and applications, CRC press, 1998 [Google Scholar]
- Mock J. Barbic M. Smith D. Schultz D. Schultz S. J. Chem. Phys. 2002;116:6755–6759. doi: 10.1063/1.1462610. [DOI] [Google Scholar]
- Durán N. Marcato P. D. Alves O. L. De Souza G. I. Esposito E. J. Nanobiotechnol. 2005;3:8. doi: 10.1186/1477-3155-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E. Webster T. J. Int. J. Nanomed. 2011;6:1463. doi: 10.2217/nnm.11.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin N. Zheng Y. Opoku-Temeng C. Du Y. Bonsu E. Sintim H. O. Future Med. Chem. 2015;7:493–512. doi: 10.4155/fmc.15.6. [DOI] [PubMed] [Google Scholar]
- Składanowski M. Wypij M. Laskowski D. Golińska P. Dahm H. Rai M. J. Cluster Sci. 2017;28:59–79. doi: 10.1007/s10876-016-1043-6. [DOI] [Google Scholar]
- Al-Dhabi N. Mohammed Ghilan A.-K. Arasu M. Nanomaterials. 2018;8:279. doi: 10.3390/nano8050279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frattini A. Pellegri N. Nicastro D. De Sanctis O. Mater. Chem. Phys. 2005;94:148–152. doi: 10.1016/j.matchemphys.2005.04.023. [DOI] [Google Scholar]
- Kumar S. Stecher G. Tamura K. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N. Nei M. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Rafińska K. Pomastowski P. Buszewski B. Sci. Total Environ. 2019;661:120–129. doi: 10.1016/j.scitotenv.2018.12.139. [DOI] [PubMed] [Google Scholar]
- El-Shafei H. MohamedAbdel-Aziz S. Ghaly M. Afr. J. Biotechnol. 2010;6:125–142. [Google Scholar]
- Singh S. P., Thumar J. T., Gohel S. D., Kikani B., Shukla R., Sharma A. and Dangar K., in Marine Enzymes for Biocatalysis, Elsevier, 2013, pp. 191–214 [Google Scholar]
- Mohammed A. Ahmed A. Mohammed F. El-Shafei H. J. Appl. Sci. Res. 2012;8:3707–3716. [Google Scholar]
- Hamed A. A. Abdel-Aziz M. S. Fadel M. Ghali M. F. Bull. Natl. Res. Cent. 2018;42:22. doi: 10.1186/s42269-018-0022-5. [DOI] [Google Scholar]
- Hathout A. El-Nekeety A. Hamed A. Sabry B. Abdel-Aziz M. Ghareeb M. Aly S. J. Appl. Pharm. Sci. 2016;6:001–010. [Google Scholar]
- Parthiban E. Manivannan N. Ramanibai R. Mathivanan N. Biotechnology Reports. 2019;21:e00297. doi: 10.1016/j.btre.2018.e00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti S. Krishnan D. Barman G. Ghosh S. K. Laha J. K. J. Anal. Sci. Technol. 2014;5:40. doi: 10.1186/s40543-014-0040-3. [DOI] [Google Scholar]
- Christensen G. D. Simpson W. A. Younger J. Baddour L. Barrett F. Melton D. Beachey E. J. Clin. Microbiol. 1985;22:996–1006. doi: 10.1128/JCM.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.