Abstract
Magnetic nanoparticles (MNPs) chelating with metal ions can specifically interact with poly-histidine peptides and facilitate immobilization and purification of proteins with poly-histidine tags. Fabrication of MNPs is generally complicated and time consuming. In this paper, we report the preparation of Ni(ii) ion chelated MNPs (Ni-MNPs) in two stages for protein immobilization and purification. In the first stage, organic ligands including pentadentate tris (carboxymethyl) ethylenediamine (TED) and tridentate iminodiacetic acid (IDA) and inorganic Fe3O4–SiO2 MNPs were synthesized separately. In the next stage, ligands were grafted to the surface of MNPs and MNPs with a TED or IDA modified surface were acquired, followed by chelating with Ni(ii) ions. The Ni(ii) ion chelated forms of MNPs (Ni-MNPs) were characterized including morphology, surface charge, structure, size distribution and magnetic response. Taking a his-tagged glycoside hydrolase DspB (Dispersin B) as the protein representative, specific interactions were confirmed between DspB and Ni-MNPs. Purification of his-tagged DspB was achieved with Ni-MNPs that exhibited better performance in terms of purity and activity of DspB than commercial Ni-NTA. Ni-MNPs as enzyme carriers for DspB also exhibited good compatibility and reasonable reusability as well as improved performance in various conditions.
This article reports a novel approach for synthesizing magnetic nanoparticles with a modified surface for purification and immobilization of histidine-tagged proteins.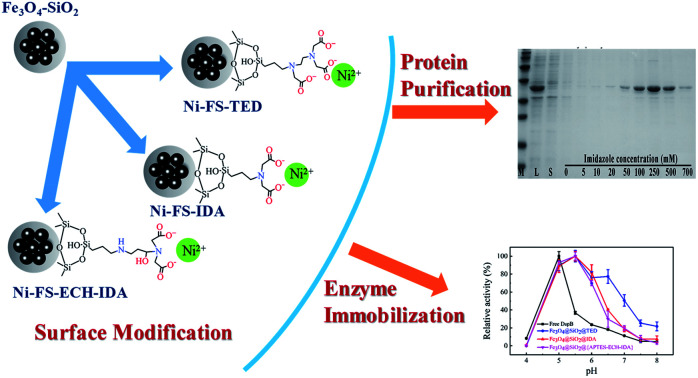
1. Introduction
Protein and enzyme immobilization have extensive applications in the environment and biomedicine.1–4 Current approaches for immobilizing enzymes include embedding, adsorption, crosslinking, covalent bonding.5–8 For example, glucose sensitivity was greatly improved by using glucose oxidase (GOx) and horseradish peroxidase (HRP) co-embedded nanoflowers.5 Cui et al. prepared an electrochemical sensor for the detection of organophosphorus pesticides (OPs) by adsorbing acetylcholinesterase (AChE) to (CSTiO2)–Ti-@-CS/rGO.6 Kim et al. used glutaraldehyde as a cross-linking agent to immobilize Candida rugosa lipase to amino-modified magnetic beads to degrade polycaprolactone (PCL).7 Hosseini et al. covalently immobilized cellulase with epoxy polymer modified Fe3O4, which improved enzyme activity, thermal stability and reusability.8 Reasonable quantity, quality, purity, activity, as well as stability of enzymes are always important issues in immobilizing enzymes.
Immobilized metal ion affinity chromatography (IMAC) is an ideal technique for immobilizing and purifying proteins with poly-histidine tags.9–11 IMAC immobilized transition-metal ions (Cu2+, Ni2+, Co2+, Zn2+) via ligands such as one tridentate iminodiacetic acid (IDA),12–14 two tetradentates nitrilotriacetic acid (NTA),15–17 carboxymethylated aspartic acid (CM-Asp),18,19 pentadentate tris (carboxymethyl) ethylenediamine (TED).20,21 Specific interactions between transition-metal ions and poly-histidine tags (his-tags) on the N- or C-terminals of proteins facilitate protein separation and purification under different ionic strength and pH of solutions. Since only his-tags are involved in protein immobilization, the rest part of his-tagged proteins as well as their functions and characteristics are less influenced during the course of protein immobilization.
Based on IMAC, magnetic nanoparticles (MNPs) with chelated metal ions have been developed for purification of his-tagged proteins.22–26 MNPs are good carriers for proteins owing to their large surface, good dispersion and magnetic response. The use of MNPs in purifying his-tagged proteins is advantageous in improving adsorption capacity, simplifying operating procedures, and reducing time consuming.27,28 MNPs chelating with metal ions enabled one-step immobilization and purification of his-tagged proteins.29 Proteins of enzymes immobilized on MNPs are easily separated from cell lysates or reacting mixtures under the function of external magnetic force.30–32 For example, Zhou and coworkers reported the use of Fe3O4/PMG/IDA-Ni2+ MNPs in on-step purification of his-tagged β-glucosidase (BG).30
For immobilization and purification of proteins, outer layers of MNPs are usually modified and chelated with metal ions. That is, MNPs cores are prepared and followed with chemical modifications that are generally complicated and time consuming. In addition, serious aggregation of MNPs may occur when metal agitators or magnetic instruments are used during preparation procedures. This situation calls for short and simple procedures for MNPs preparation. In the present study, we proposed to synthesize the organic ligand for chelating metal ions and prepare the inorganic magnetic core simultaneously, and then graft the ligand to the surface of the magnetic core. Three types of MNPs were synthesized including Fe3O4–SiO2-TED (FS-TED), Fe3O4–SiO2-IDA (FS-IDA), and Fe3O4–SiO2-{APTES-ECH-IDA} (FS-ECH-IDA) MNPs and then used to generate Ni(ii) ions chelated forms of MNPs (Ni-MNPs). Ni-MNPs based immobilization and purification of his-tagged proteins were investigated by taking advantages of a his-tagged DspB (Dispersin B) as the protein representative. DspB is a biofilm-releasing glycoside hydrolase from the periodontopathogen Actinobacillus actinomycetemcomitans.31 Conditions for immobilization and purification of DspB were examined and optimized including pH, temperature, thermal stability, storage stability, reusability and kinetic characteristics.
2. Materials and methods
2.1. Reagents
Ammonium ferrous sulfate ((NH4)2Fe(SO4)2·6H2O), ferric chloride (FeCl3·6H2O), tetraethyl-orthosilicate (TEOS), ammonium hydroxide (NH3·H2O), nickel sulfate (NiSO4·6H2O), sodium hydroxide (NaOH), epichlorohydrin (ECH) and ethyl alcohol were purchased from Sinopharm Chemical Reagent Co., Ltd. Imidazole and sodium chloroacetate were purchased from Shanghai McLean Biochemical Technology Co., Ltd. Commercial Ni-NTA and isopropyl-β-d-thiogalactopyranoside (IPTG) were purchased from Shenggong Biological Engineering (Shanghai) Co., Ltd. 3-Aminopropyltriethoxysilane (APTES) and N-(2-aminoethyl)-3-aminopropyl-trimethoxysilane (AAPTMS) were purchased from Aladdin Reagent (Shanghai) Co. Ltd. All reagents were used directly without further purification.
2.2. Preparation of Ni-MNPs
Preparation of Fe3O4 MNPs was based on previous reports.33,34 Typically, 1.90 g (NH4)2Fe(SO4)2·6H2O and 2.33 g FeCl3·6H2O were dissolved in 50 mL deionized water at 60 °C, followed by addition of 30 mL concentrated ammonia under nitrogen protection with stirring for 30 minutes. The temperature of the mixture was increased up to 80 °C and kept for 1 hour. As prepared Fe3O4 MNPs were washed several times with ethanol and deionized water.
For preparation of Fe3O4–SiO2 MNPs, 100 mg as prepared Fe3O4 MNPs were dispersed in 100 mL mixed solution of water and ethanol (1 : 4) and then treated with ultrasound for 15 minutes. Then 6 mL ammonia was added and 0.6 mL TEOS was dripped under vigorous mechanical stirring. The reaction was allowed for 12 hours at room temperature. As prepared Fe3O4–SiO2 MNPs were magnetically separated and washed several times with ethanol and deionized water.
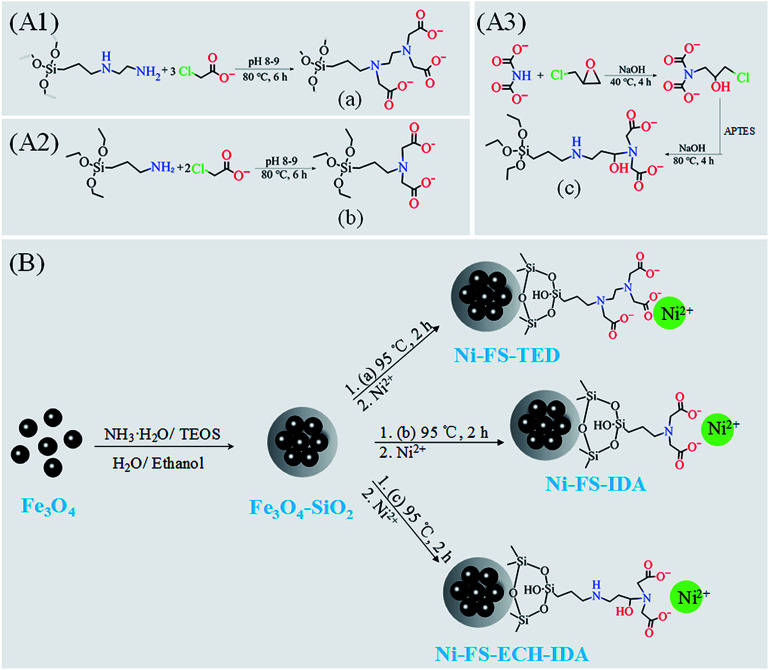
TED-propyl-silane and IDA-propyl-silane were synthesized based on the reaction of amino groups on AAPTMS and APTES with sodium chloroacetate, respectively.35–38 Typically, 0.822 mL AAPTMS or 0.842 mL APTES were dripped into 25 mL deionized water with magnetic stirring and on ice water bath for 1 hour. Then 1.68 g or 1.26 g sodium chloroacetate was added and reacted at 80 °C for 7 hours. The pH of the solution was adjusted between 8.0 and 9.0 by using 5 M NaOH. After the reaction completed, the pH of reacting mixture (transparent light yellow liquid) was adjusted to 3.0 with concentrated hydrochloric acid. TED-propyl-silane or IDA-propyl-silane aqueous solution were obtained (Fig. 1, Panels A1 and A2).
Fig. 1. Preparation of Ni(ii) ions chelated FS-TED, FS-IDA, and FS-ECH-IDA MNPs. Panels (A1)–(A3), synthesis of carboxylated AAPTMS (a) and APTES (b and c). Panel (B), synthesis and functionalization of Fe3O4–SiO2 MNPs to acquire Ni(ii) ions chelated FS-TED, FS-IDA, and FS-ECH-IDA MNPs.
The synthesis of APTES-ECH-IDA was based on previous reports.39–41 Typically, 1.0 g IDA was dissolved in 25 mL deionized water containing 1.2 g NaOH. Then, 1.2 mL epichlorohydrin was added and reacted at 40 °C for 4 hours. After keeping on ice for 5 minutes, 1.76 mL APTES was added and hydrolyzed with magnetic stirring for 1 hour. Then 0.6 g NaOH was added and the reacting mixture was heated up to 80 °C for 4 hours. After the reaction completed, the pH of reacting mixture (transparent light yellow liquid) was adjusted to 3.0 with concentrated hydrochloric acid and APTES-ECH-IDA (Fig. 1, Panel A3) aqueous solution were obtained.
Functionalization of Fe3O4–SiO2 MNPs was started with dispersing 200 mg Fe3O4–SiO2 MNPs into TED-propyl-silane, IDA-propyl-silane, or APTES-ECH-IDA aqueous solution, which was treated with ultrasound for 15 minutes, and then reacted at 95 °C under mechanical agitation for 2 hours. The powder was magnetically separated and washed several times with deionized water to obtain FS-TED, FS-IDA, and FS-ECH-IDA MNPs. MNPs were then added to 0.1 mol L−1 NiSO4 solution, mechanically stirred at room temperature for 2 hours. The obtained Ni(ii) chelated MNPs (Ni-MNPs) were washed several times with deionized water. The whole process was illustrated in Fig. 1, Panel B.
2.3. Characterization of Ni-MNPs
The structure of Ni-MNPs was analyzed by using the FEI Tecnai G2 F20 transmission electron microscope (TEM). Dynamic light scattering (DLS) of sonication dispersed MNPs in deionized water were measured by using the Malvern Instruments Zetasizer Nano ZS90 at 25 °C. Zeta potential of MNPs in 0.01 M KCl was measured by using the Malvern Instruments Zetasizer Nano ZS90 and with pH adjusting by 0.1 M HCl or NaOH.42–44 XRD patterns were recorded by using the rotating target X-ray diffractometer (D/MAX-RB). Fourier transform infrared (FT-IR) spectra were recorded by using a Thermo Fisher Scientific Nicolet iS5. Magnetic measurements were performed on Physical Property Measurement System (PPMS-9T, USA). All data were collected from triplicated measurements.
2.4. Expression, purification and immobilization of his-tagged proteins
Genes encoding his-tagged DspB were cloned into pET-28a vector and expressed in Escherichia coli BL21 (DE3). The transformed single colony was cultured overnight at 37 °C in 5 mL LB medium containing 50 μg mL−1 kanamycin. The culture was inoculated to fresh LB medium at 1 : 100 and cultured at 220 rpm at 37 °C. When the optical density at 600 nm (OD600) reached 0.8, protein expression was induced overnight by IPTG at a final concentration of 0.5 mM. The bacteria were harvested by centrifugation and re-suspended in lysis buffer (50 mM PB, 5 mM imidazole, 500 mM NaCl, pH 8.0) for later use.
For immobilization of DspB, cells were crushed in a high-pressure homogenizer and centrifugated at 8000 rpm, 10 minutes. Typically, supernatant containing DspB from 0.5 mL overnight cell culture was mixed with 5.0 mg Ni-MNPs and incubated at 25 °C for 30 minutes. Magnetic separation of Ni-MNPs was facilitated with a commercial NdFeB magnet (N52, 20 × 10 × 10 mm). Ni-MNPs were washed with washing buffer (50 mM PBS, 0–10 mM imidazole, 500 mM NaCl, pH 8.0). Ni-MNPs with loaded DspB were resuspended in 50 mM PBS, 100 mM NaCl, pH 6.0, and ready for enzymatic assay. Or bound protein DspB were eluded with of elution buffer (50 mM PBS, 250 mM imidazole, 500 mM NaCl, pH 8.0).
The protein concentration was determined by using the Bradford method.45
2.5. SDS-polyacrylamide gel electrophoresis
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) were performed as previously described,46 with 5% stacking gel and 12% running gel for separating proteins and Coomassie Brilliant Blue for staining proteins. All reagents were purchased from Shenggong Biological Engineering (Shanghai) Co., Ltd. After staining and destaining, SDS-PAGE gels were further analyzed by using Gel-Pro Analyzer (Media Cybernetics, USA).
2.6. Activity assay of DspB
For measuring the activity of DspB before and after immobilization, 4-nitrophenyl-N-acetyl β-d-glucosaminide was used as the substrate47 and dissolved in 50 mM PBS, 100 mM NaCl, pH 6.0, to a final concentration of 5 mM. 12 μL DspB was added to 108 μL substrate solution and incubated at 40 °C for 10 minutes. The reaction was terminated by adding with 100 μL 1 M Na2CO3 and followed by measuring light absorption of ρ-nitrophenolate at 405 nm.
2.7. Characterization of interaction between DspB and MNPs
Affinity purified DspB was dialyzed against 50 mM phosphate, pH 7.4, 5 mM imidazole, 500 mM NaCl. Then, 100 μL his-tagged DspB solution (C0) was added with 200 μg Ni-MNPs. After incubation at 25 °C, MNPs with adsorbed DspB were collected by external magnetic force. The adsorption capacity was calculated by eqn (1):
| q = (C0 − C)V/m | 1 |
where, m is the amount of Ni-MNPs (mg). V is the volume of his-tagged DspB solution (mL). C0 (mg mL−1) and C (mg mL−1) are the initial concentration and the concentration in the adsorbed supernatant of his-tagged DspB, respectively.
3. Results and discussion
3.1. Synthesis and characterization of MNPs
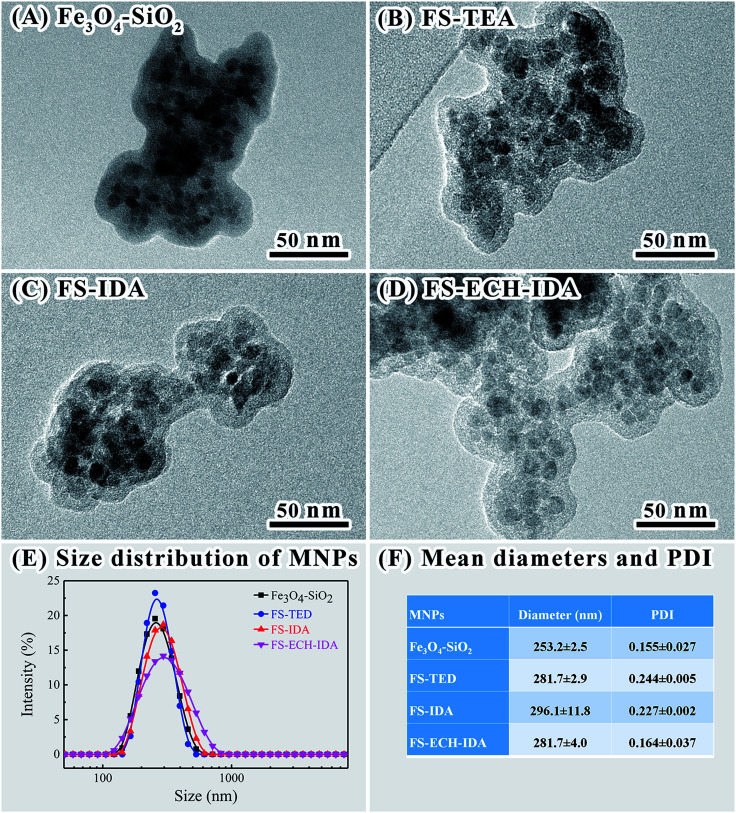
Morphology and structures of MNPs were analyzed by means of TEM (Fig. 2, Panels A–D). It was revealed the formation of silica layer on Fe3O4 MNPs.48,49 Although the size of Fe3O4 MNPs was less than 100 nm, the size distribution of MNPs aggregates measuring by means of dynamic light scattering was between 164.2 and 531.2 nm (Fig. 2, Panels E and F). The mean size of Fe3O4–SiO2 MNPs aggregates was slightly lower than that of derived MNPs including FS-TED, FS-IDA, and FS-ECH-IDA. The size of MNPs aggregates was in the same range of Fe3O4 nanoparticles or derivatives in previous reports.50,51 Reasonable dispersity of MNPs aggregates was proved by PDI (particle dispersion index) values (Fig. 2, Panels F).
Fig. 2. TEM images (A–D) of MNPs and size distributions (E) and mean diameters and PDI (F) of MNPs aggregates revealed by dynamic light scattering.
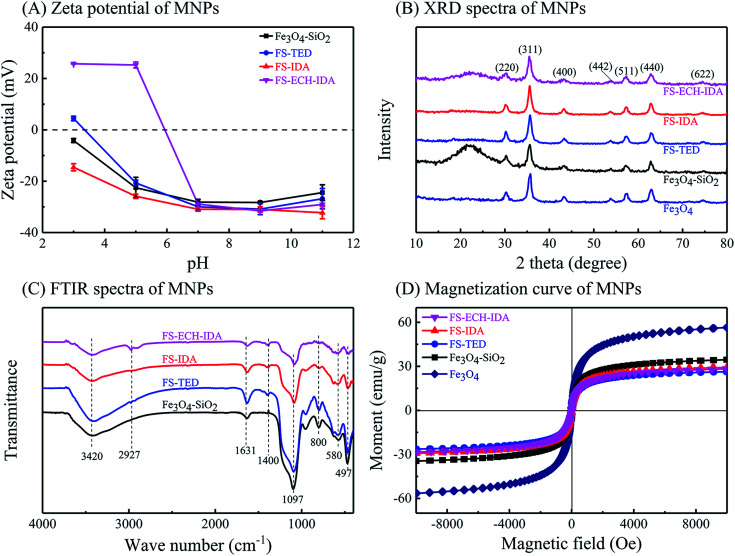
Dispersion stability as well as isoelectric point of MNPs were characterized by zeta potential measurement (Fig. 3, Panel A). Good dispersion stability of the four types of MNPs was proved by zeta potentials between −28.13 and −30.90 at pH 7.0.44 Positive zeta potential values of FS-ECH-IDA MNPs was detected when pH was lower than pH 5.0. This is attributed to the ionization of the imine group (–NH–) on APTES-ECH-IDA. The observed isoelectric point (pI) of Fe3O4–SiO2 MNPs was similar to that of SiO2 (∼2.3).52 The observed pI of FS-TED, FS-IDA, and FS-ECH-IDA MNPs were much lower than that of Fe3O4–SiO2-APTES.53 This was due to that the amino groups were modified and replaced with carboxyl groups on the surface of MNPs.
Fig. 3. Characterization of MNPs. Panels (A), zeta potential; Panel (B), XRD spectra; Panel (C), FTIR spectra; Panel (D), magnetization curves.
Crystallinity of MNPs was examined by XRD measurement. The XRD patterns of all MNPs were consistent with standard Fe3O4 (JCPDS no. 01-076-1849) (Fig. 3, Panel B), which proved cubic spinel structures of these MNPs.54 Good crystallinity was deduced for all prepared MNPs from the XRD patterns which implied that surface modification of TEOS, APTES and AAPTMS groups did not change the crystal structure of Fe3O4. A wide peak around 22° was found for Fe3O4–SiO2 MNPs, which was due to the influence of amorphous SiO2 layer on MNPs surface.55,56
Surface modifications by organic groups on MNPs was examined by FTIR spectrometry (Fig. 3, Panel C). The characteristic peak at 580 cm−1 belongs to the vibration of Fe–O in Fe3O4.57 The wide absorption peak at 1097 cm−1 and the band at 467 cm−1 is due to the vibration of Si–O–Si and the weak band around 800 cm−1 is attributed to Fe–O–Si, suggesting silica modification on Fe3O4 surface.58,59 An absorption peak near 1400 cm−1 was detected for MNPs except Fe3O4–SiO2 MNPs. This peak may be related to the bending vibration of O–H in carboxylic acid.60 Two bands at 3420 cm−1 and 1631 cm−1 can be stretching vibration of –OH groups and related to the water adsorbed on MNPs surface.61,62 A absorption peak near 2927 cm−1 was also observed and ascribed to the stretching vibrations of C–H.62,63 These observations confirmed the introduction of carboxyl groups and silane coupling agents on Fe3O4–SiO2 surface.
Magnetic properties of MNPs before and after surface modification was confirmed and evaluated by analyzing the magnetic hysteresis loops (Fig. 3, Panel D). The saturation magnetization (Ms) of Fe3O4 was observed as 56.51 emu g−1, which was reduced to 26.35–34.53 emu g−1 after surface modification. All MNPs did not exhibit significant remanence, indicating good paramagnetism of these MNPs.
In summary, Fe3O4–SiO2 MNPs and surface modified derivatives were synthesized. The morphology, structure, surface features, dispersity, size, as well as magnetic properties of these MNPs were characterized, which exhibited potentials in magnetic separation and purification of his-tagged proteins.
3.2. Interactions between his-tagged DspB and Ni-MNPs
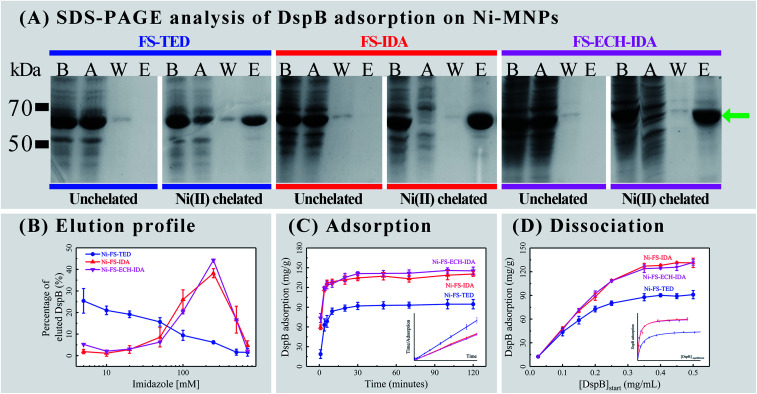
Interactions between DspB and Ni(ii) chelated MNPs (Ni-MNPs) were investigated based on procedures of Ni-NTA affinity chromatography. No his-tagged proteins was found to bind to unchelated MNPs (Fig. 4, Panel A). Purification of his-tagged DspB with high homogeneity was observed by using Ni-MNPs (Fig. 4, Panel A), implying specific interactions between his-tagged proteins and Ni-MNPs. When eluting with stepwise gradient imidazole, eluting profiles of DspB were different when binding to different Ni-MNPs (Fig. S1, see ESI;†Fig. 4, Panel B). Significant amount of DspB binding to Ni-FS-TED MNPs was eluded at 20–50 mM imidazole that was higher than DspB binding to Ni-FS-IDA or Ni-FS-ECH-IDA MNPs, implying week interactions between DspB and Ni-FS-TED MNPs. This might be related to the number of coordination sites of Ni(ii) ions on magnetic nanoparticles. In general, Ni(ii) ions has six coordination sites.64,65 After chelation, the number of remaining sites were reduced to three for IDA and one for TED, which contributed to the difference of binding characteristics between his-tagged proteins and Ni-MNPs.66–68 De Goes et al. stated that “the more polydentate the chelating ligand is, the better the stability of chelate complex, the lower the metal ion leakage, and the higher the selectivity, but on the other hand, the lower the capacity for protein adsorption”.66
Fig. 4. Adsorption of his-tagged DspB on Ni-MNPs. Panel (A), SDS-PAGE analysis, Lanes B and A show cell lysate containing his-tagged DspB before and after adsorption on Ni-MNPs; Lanes W and E show wash and elution fractions. Green arrow points to the position of his-tagged DspB. Panel (B), elution of DspB under different concentrations of imidazole (Fig. S1, see ESI†). Panel (C), adsorption of DspB and pseudo-second-order adsorption kinetic plots (inset). Panel (D), dissociation of DspB and Langmuir isotherm fitting (inset).
The course of interactions between DspB and Ni-MNPs were explored by measuring the adsorption of DspB on Ni-MNPs during two hours (Fig. 4, Panel C; Table S1†). It took about 30 minutes for DspB to complete adsorption on Ni-MNPs and reaching equilibrium, which is consistent with previous reports of adsorption of his-tagged proteins on Ni(ii) ions chelated carriers.69–71 The adsorption kinetics of DspB on Ni-MNPs followed pseudo-second order kinetic model describing by eqn 2.
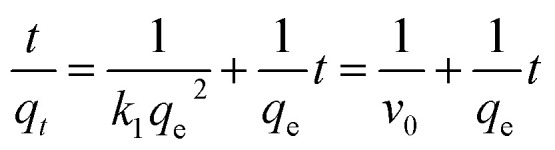 |
2 |
where qt and qe represent the adsorption amount at time t and the adsorption amount at the adsorption equilibrium (mg g−1). k1 is the adsorption rate constant of pseudo-second-order kinetics (g (mg−1 min−1)). v0 represents the initial adsorption rate (mg (g−1 min−1)). qe and k1 can be used to calculate the slope and intercept of t/qtvs. t. The pseudo-second order kinetic model can be used to describe the kinetics of protein adsorption to the surface of carrier.72–74
The k1, qe, and v0, of Ni-FS-IDA or Ni-FS-ECH-IDA MNPs binding to DspB are similar and both are higher than that of Ni-FS-TED MNPs. The v0 of Ni-FS-IDA or Ni-FS-ECH-IDA MNPs are almost 3 times higher than that of Ni-FS-TED MNPs. Since after chelating with Ni(ii) ions, there are three coordination sites remaining on IDA and one on TED, it suggested that the adsorption capacity may be related to number of coordination sites on IDA and TED.
Dissociation of DspB to Ni-MNPs was also measured (Fig. 4, Panel D; Table S2†). Similar curves of Ni-FS-IDA or Ni-FS-ECH-IDA MNPs for adsorbing DspB were observed and exhibited capacity higher than that with Ni-FS-TED MNPs. The dissociation of DspB with Ni-MNPs followed Langmuir isotherm model describing by eqn 3.70,71,75,76
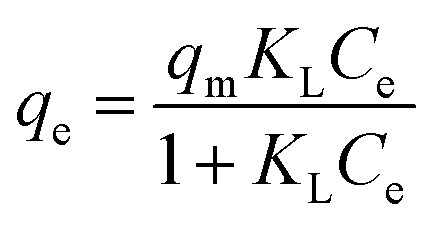 |
3 |
where qe represents the adsorption amount (mg g−1) of DspB at the time of adsorption equilibrium, qm is the maximum adsorption amount (mg g−1) of DspB, and Ce is the equilibrium concentration (mg mL−1) of DspB. KL is the Langmuir constant (mL mg−1) and is related to the adsorption energy.
The calculated adsorption capacities of Ni-MNPs were similar to observed ones, indicating that the adsorption of DspB on Ni-MNPs follows the monolayer adsorption mechanism. It was observed the negative ΔG0 of DspB adsorbing on NI-MNPs, indicating that the adsorption process was spontaneous and exothermic. This is similar to previous reports of Ni or Cu ions chelated matrix in adsorbing his-tagged proteins.77,78
In summary, there were specific interactions between his-tagged proteins and as prepared Ni-MNPs, which may facilitate immobilization and purification of these proteins.
3.3. The use of Ni-MNPs for purification of DspB
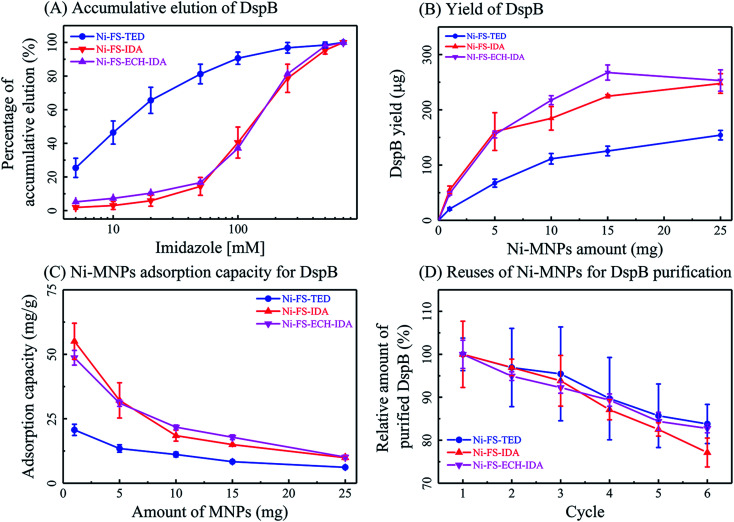
Factors affecting protein purification based on Ni-MNPs were further explored including the eluting agent imidazole, protein purity, binding capacity and reusability of Ni-MNPs. Same amount of non-magnetic commercial Ni-NTA as that of Ni-MNPs was used as a control for evaluating the performance of Ni-MNPs in protein purification. After binding with DspB and washing, Ni-MNPs were subjected to elution with imidazole at stepwise gradient concentrations (Fig. 5, Panel A). Over 60% of DspB bound to Ni-FS-TED MNPs was eluted when increasing imidazole concentration from 0 to 20 mM and over 90% of DspB was eluted when increasing imidazole concentration up to 100 mM. As to DspB bound to Ni-FS-IDA or Ni-FS-ECH-IDA MNPs, the imidazole concentration had to be increased to up to 500 mM to accomplish DspB elution. DspB recovery as well as activity after purification by Ni-MNPs was compared with that purified by commercial Ni-NTA. DspB purified by Ni-MNPs with IDA on surface exhibited similar protein recovery but higher activity than that by commercial Ni-NTA (Table 1). Ni-MNPs with TED on surface exhibited the lowest efficiency in recovering protein as well as activity. In terms of activity, DspB proteins purified by Ni-MNPs were higher than that purified by commercial Ni-NTA, implying better biocompatibility of Ni-MNPs in purifying DspB than commercial Ni-NTA.
Fig. 5. Ni-MNPs based affinity purification of DspB. Accumulative elution of DspB under stepwise gradient elution (A), yield of DspB by different amount of Ni-MNPs (Panel B), Ni-MNPs adsorption capacity for DspB (Panel C), and reusability of Ni-MNPs for DspB purification (Panel D).
Use of different affinity media for purification of DspB.
| Protein source | Protein recovery (mg g−1 Ni-MNPs or Ni-NTA) | DspB activity recovery (U g−1 Ni-MNPs or Ni-NTA) | DspB activitya (U mg−1) | DspB purityb (%) |
|---|---|---|---|---|
| 0.5 mL cell lysatec | 0.71 ± 0.01 (mg) | 0.192 ± 0.004 (U) | 0.27 ± 0.01 | — |
| Ni-FS-TED | 8.37 ± 0.49 | 6.35 ± 0.16 | 0.76 ± 0.04 | 94.3 |
| Ni-FS-IDA | 35.76 ± 1.68 | 28.51 ± 0.88 | 0.80 ± 0.02 | 92.7 |
| Ni-FS-ECH-IDA | 35.61 ± 2.38 | 27.60 ± 0.47 | 0.78 ± 0.05 | 91.2 |
| Commercial Ni-NTA | 36.26 ± 1.68 | 25.25 ± 0.49 | 0.70 ± 0.02 | 85.5 |
DspB activity (U mg−1) = DspB activity recovery (U g−1)/protein recovery (mg g−1).
DspB purity was determined by densitometric scanning of SDS-PAGE gel.
Total amount of proteins (mg) or DspB activity (U) from 0.5 cell lysate was assayed and used for comparison.
The yield and binding capacity of Ni-MNPs was examined. When increasing the amount of Ni-MNPs for protein purification from 1.0 mg to 25.0 mg, the total amount of eluted DspB was increased along with decreases of adsorption capacity of Ni-MNPs (Fig. 5, Panels B and C). This indicates that the adsorption capacity of MNPs relies on its ratio to proteins. Reusability of Ni-MNPs in purifying his-tagged DspB was also examined (Fig. 5, Panel D). After completing each round of protein purification, Ni-MNPs were washed with 50 mM PBS, 500 mM NaCl, and 5 mM imidazole, pH 8.0, and then chelated with Ni(ii) ions. After six cycles, Ni-MNPs remained reasonable activity in purifying DspB with slightly decreases in binding capacity.
In summary, Ni-MNPs acquired in this study exhibited improved biocompatibility in purifying native proteins than that of commercial Ni-NTA. Ni-MNPs with IDA on surface were with higher protein recovery than that with TDA on surface.
3.4. The use of Ni-MNPs as carriers for his-tagged enzymes
Immobilization of enzymes is advantageous in improving stability and activity of enzymes under various conditions, facilitating to recycle and reuse of enzymes, etc. Kinetic characteristics of DspB activity were analyzed before and after immobilization (Table 2). There was decreases of the Michaelis constant (Km) value after immobilization of DspB on Ni-MNPs, indicating improved interactions between substrate and DspB after binding on Ni-MNPs. This may be due to that there was conformational changes of DspB after immobilization to make it more accessible to substrates.79 It is also possible that the negative charged surface of Ni-MNPs makes it more accessible of immobilized enzyme than free enzyme to the positive charged substrate 4-nitrophenyl-N-acetyl β-d-glucosaminide (pKa 12.76) in reaction mixture (pH 6.0). This leads to increased affinity of immobilized DspB to substrate and reduced apparent Michaelis constant of DspB on Ni-MNPs.
Kinetic parameters of free and immobilized DspB.
| DspB samples | K m (mM) | V max (mM min−1) |
|---|---|---|
| Before immobilization | 48.7 | 0.13 |
| Ni-FS-TED | 21.6 | 0.04 |
| Ni-FS-IDA | 32.7 | 0.20 |
| Ni-FS-ECH-IDA | 34.1 | 0.30 |
Decrease of maximum reaction rate (Vmax) was observed when DspB binding on Ni-FS-TED MNPs, indicating TED modified surface might have adverse effect on activity of DspB. Increases of Vmax were detected when DspB binding on Ni-FS-IDA or Ni-FS-ECH-IDA MNPs, indicating a better compatibility of these two types of Ni-MNPs for immobilizing DspB.
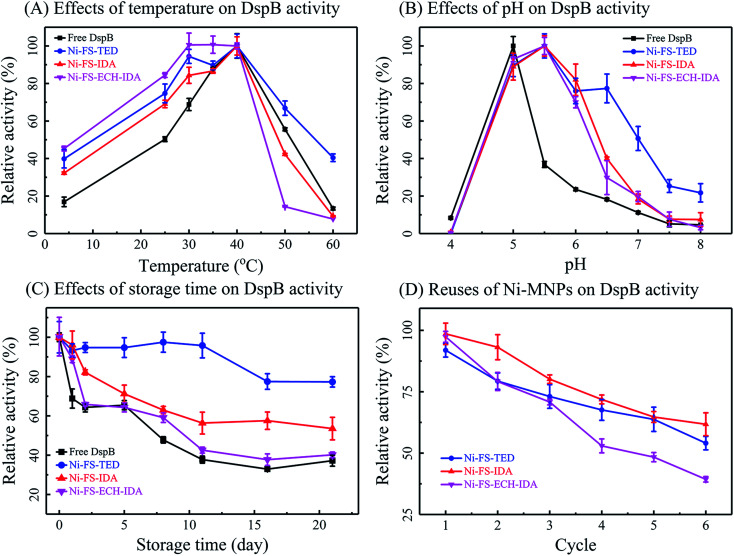
Temperature is critical in practical applications of enzymes. In the present study, effects of temperature on protein immobilization were explored by measuring the activity and thermal stability of immobilized DspB (Fig. 6, Panel A). The optimal temperature was observed as 40 °C for enzymatic activity of DspB before and after immobilization. It was noticed that DspB on Ni-FS-ECH-IDA MNPs had a broad range of optimal temperature from 30 °C to 40 °C and DspB on Ni-FS-TED MNPs retained almost half of its highest activity on 60 °C.
Fig. 6. Enzymatic activity of Ni-MNPs immobilized DspB. Effects of temperature (Panel A), pH (Panel B), storage time (Panel C) and reuses (Panel D) on activities of DspB before and after immobilization on Ni-MNPs.
The activity of immobilized DspB was also tested between pH 4.0 and 8.0 at 40 °C (Fig. 6, Panel B). The optimum pH for DspB with the highest activity shifted from pH 5.0 to pH 5.5 after immobilization. This was due to that negatively charged carboxyl groups on the surface of Ni-MNPs would absorb protons and lead to an acidic microenvironment that facilitates the function of DspB. Therefore, the reaction mixture was not necessary to be that acidic as for un-immobilized DspB. This also contributed to relatively higher activity of immobilized DspB than un-immobilized one as pH was increased.
Storage of immobilized DspB was tested (Fig. 6, Panel C). It was observed that Ni-FS-ECH-IDA MNPs retained relatively high activity in adsorbing DspB for 21 days. While the other two types of Ni-MNPs only exhibited limited effects in retaining activity in adsorbing DspB. Therefore, Ni-FS-ECH-IDA MNPs might be the best in limiting the free moves and conformational changes of DspB, as well as reducing the autolysis of DspB.79,80
One of the most prominent features of immobilized enzymes is the reusability that is of great importance in practical applications. In the present study, the reusability of immobilized DspB was examined by repeating activity assay, magnetic separation, and wash with PBS (Fig. 6, Panel D). The activity of immobilized DspB reduced after washing and reusing. This might be due to denaturation or shedding of the immobilized enzyme during recycling course.78,81 The performance of Ni-FS-IDA and Ni-FS-TED MNPs in DspB reusability is better than that of Ni-FS-ECH-IDA MNPs.
In summary, Ni-MNPs as enzyme carriers for DspB exhibited good compatibility and reasonable reusability as well as improving the performance of DspB in various conditions.
4. Conclusions
In the present study, three types of MNPs with IDA or TED modified surface were synthesized through a two step preparation approach by synthesizing ligands for chelating metal ions and preparing magnetic cores simultaneously, and followed by grafting ligands on the surface of magnetic cores. As prepared MNPs were chelated with Ni(ii) ions and exhibited reasonable size distribution, dispersity, surface charge, and magnetic properties.
Specific interactions were confirmed between his-tagged proteins and Ni-MNPs. The rate of protein adsorption on Ni-MNPs may be related to the number of coordination sites of Ni(ii) ions on the surface.
Ni-MNPs exhibited improved performance in purifying native proteins than commercial Ni-NTA in terms of biocompatibility. Ni-MNPs with IDA modified surface were with higher protein recovery than that with TED modified surface.
Immobilization of DspB on Ni-MNPs facilitated the function of DspB in several ways. Electrostatic attraction between Ni-MNPs and 4-nitrophenyl-N-acetyl β-d-glucosaminide (pKa 12.76) made the substrates were more accessible to immobilized DspB. Immobilization of DspB on Ni-MNPs made DspB more stabile under various conditions including temperature, pH, and storage period. However, the activity of immobilized DspB reduced after several rounds of reuses.
Conflicts of interest
The authors declare there is no conflicts of interest regarding the publication of this paper.
Supplementary Material
Acknowledgments
Supported by National Natural Science Foundation of China (31771032, 5181101987, 51832003).
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra10473a
References
- Sarma R. Islam M. S. Miller A.-F. Bhattacharyya D. ACS Appl. Mater. Interfaces. 2017;9:14858–14867. doi: 10.1021/acsami.7b01999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadam N. H. Salehzadeh S. Rakhtshah J. Moghadam A. H. Tanzadehpanah H. Saidijam M. Int. J. Biol. Macromol. 2019;125:931–940. doi: 10.1016/j.ijbiomac.2018.12.143. [DOI] [PubMed] [Google Scholar]
- Yeroslavsky G. Girshevitz O. Foster-Frey J. Donovan D. M. Rahimipour S. Langmuir. 2015;31:1064–1073. doi: 10.1021/la503911m. [DOI] [PubMed] [Google Scholar]
- Qiao J. Qi L. Yan H. Li Y. Mu X. Electrophoresis. 2013;34:409–416. doi: 10.1002/elps.201200461. [DOI] [PubMed] [Google Scholar]
- Sun J. Ge J. Liu W. Lan M. Zhang H. Wang P. Wang Y. Niu Z. Nanoscale. 2014;6:255–262. doi: 10.1039/C3NR04425D. [DOI] [PubMed] [Google Scholar]
- Cui H.-F. Wu W.-W. Li M.-M. Song X. Lv Y. Zhang T.-T. Biosens. Bioelectron. 2018;99:223–229. doi: 10.1016/j.bios.2017.07.068. [DOI] [PubMed] [Google Scholar]
- Kim M. Park J.-M. Um H.-J. Lee D.-H. Lee K.-H. Kobayashi F. Iwasaka Y. Hong C.-S. Min J. Kim Y.-H. J. Basic Microbiol. 2010;50:218–226. doi: 10.1002/jobm.200900099. [DOI] [PubMed] [Google Scholar]
- Hosseini S. H. Hosseini S. A. Zohreh N. Yaghoubi M. Pourjavadi A. J. Agric. Food Chem. 2018;66:789–798. doi: 10.1021/acs.jafc.7b03922. [DOI] [PubMed] [Google Scholar]
- Zhu Q. Zhuang W. Niu H. Ge L. Hernandez B. V. Wu J. Wang K. Liu D. Chen Y. Zhu C. Ying H. Colloids Surf., B. 2018;164:155–164. doi: 10.1016/j.colsurfb.2018.01.033. [DOI] [PubMed] [Google Scholar]
- Jiang L. D. Ye L. Acta Biomater. 2019;94:447–458. doi: 10.1016/j.actbio.2019.04.056. [DOI] [PubMed] [Google Scholar]
- Takahira I. Fuchida H. Tabata S. Shindo N. Uchinomiya S. Hamachi I. Ojida A. Bioorg. Med. Chem. Lett. 2014;24:2855–2858. doi: 10.1016/j.bmcl.2014.04.096. [DOI] [PubMed] [Google Scholar]
- Li B. Zhang Y. Cao L. Wu Y. Zhao Y. J. Sol–Gel Sci. Technol. 2014;71:380–384. doi: 10.1007/s10971-014-3372-y. [DOI] [Google Scholar]
- Trang H. K. Jiang L. Marcus R. K. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2019;1110:144–154. doi: 10.1016/j.jchromb.2019.02.013. [DOI] [PubMed] [Google Scholar]
- Yin Y. Wei G. Zou X. Zhao Y. Sens. Actuators, B. 2015;209:701–705. doi: 10.1016/j.snb.2014.12.049. [DOI] [Google Scholar]
- Guo H. Wang W. Zhou F. Appl. Phys. A: Mater. Sci. Process. 2019:125. [Google Scholar]
- Uchinomiya S.-H. Nonaka H. Fujishima S.-h. Tsukiji S. Ojida A. Hamachi I. Chem. Commun. 2009;39:5880–5882. doi: 10.1039/B912025D. [DOI] [PubMed] [Google Scholar]
- Auer S. Azizi L. Faschinger F. Blazevic V. Vesikari T. Gruber H. J. Hytonen V. P. Sens. Actuators, B. 2017;243:104–113. doi: 10.1016/j.snb.2016.11.090. [DOI] [Google Scholar]
- Le Devedec F. Bazinet L. Furtos A. Venne K. Brunet S. Mateescu M. A. J. Chromatogr. A. 2008;1194:165–171. doi: 10.1016/j.chroma.2008.03.094. [DOI] [PubMed] [Google Scholar]
- Mourao C. A. Carmignotto G. P. Alves Bueno S. M. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2016;1017:163–173. doi: 10.1016/j.jchromb.2016.01.058. [DOI] [PubMed] [Google Scholar]
- Dabrowski S. Kur J. Protein Expression Purif. 1999;16:96–102. doi: 10.1006/prep.1999.1044. [DOI] [PubMed] [Google Scholar]
- Bresolin I. T. L. Borsoi-Ribeiro M. Tamashiro W. M. S. C. Augusto E. F. P. Vijayalakshmi M. A. Bueno S. M. A. Appl. Biochem. Biotechnol. 2010;160:2148–2165. doi: 10.1007/s12010-009-8734-5. [DOI] [PubMed] [Google Scholar]
- Xie H. Y. Zhen R. Wang B. Feng Y. J. Chen P. Hao J. J. Phys. Chem. C. 2010;114:4825–4830. doi: 10.1021/jp910753f. [DOI] [Google Scholar]
- Zhang Y. Yang Y. Ma W. Guo J. Lin Y. Wang C. ACS Appl. Mater. Interfaces. 2013;5:2626–2633. doi: 10.1021/am4006786. [DOI] [PubMed] [Google Scholar]
- Yang J. Ni K. Wei D. Ren Y. Biotechnol. Bioprocess Eng. 2015;20:901–907. doi: 10.1007/s12257-015-0136-7. [DOI] [Google Scholar]
- Benelmekki M. Xuriguera E. Caparros C. Rodriguez-Carmona E. Mendoza R. Corchero J. L. Lanceros-Mendez S. Martinez L. M. J. Colloid Interface Sci. 2012;365:156–162. doi: 10.1016/j.jcis.2011.09.051. [DOI] [PubMed] [Google Scholar]
- Xu C. J. Xu K. M. Gu H. W. Zhong X. F. Guo Z. H. Zheng R. K. Zhang X. X. Xu B. J. Am. Chem. Soc. 2004;126:3392–3393. doi: 10.1021/ja031776d. [DOI] [PubMed] [Google Scholar]
- Gaedke J. Kleinfeldt L. Schubert C. Rohde M. Biedendieck R. Garnweitner G. Krull R. J. Biotechnol. 2017;242:55–63. doi: 10.1016/j.jbiotec.2016.11.018. [DOI] [PubMed] [Google Scholar]
- Fang W. Chen X. Zheng N. J. Mater. Chem. 2010;20:8624–8630. doi: 10.1039/C0JM02081H. [DOI] [Google Scholar]
- Lo H.-F. Hu H.-Y. Hung C.-P. Chen S.-C. Lin L.-L. Biocatal. Biotransform. 2009;27:318–327. doi: 10.3109/10242420903219167. [DOI] [Google Scholar]
- Zhou Y. Yuan S. Liu Q. Yan D. Wang Y. Gao L. Han J. Shi H. Sci. Rep. 2017;7:41741. doi: 10.1038/srep41741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasubbu N. Thomas L. M. Ragunath C. Kaplan J. B. J. Mol. Biol. 2005;349:475–486. doi: 10.1016/j.jmb.2005.03.082. [DOI] [PubMed] [Google Scholar]
- Zhao X. R. Hong H. F. Cheng X. Z. Liu S. Z. Deng T. Guo Z. W. Wu Z. M. Sci. Rep. 2017;7:6561. doi: 10.1038/s41598-017-06856-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak D. Semenyuk N. Lednicky F. J. Polym. Sci., Polym. Chem. Ed. 2003;41:1848–1863. doi: 10.1002/pola.10728. [DOI] [Google Scholar]
- Zheng J. Dong Y. Wang W. Ma Y. Hu J. Chen X. Chen X. Nanoscale. 2013;5:4894–4901. doi: 10.1039/C3NR01075A. [DOI] [PubMed] [Google Scholar]
- Doustkhah E. Rostamnia S. Hossieni H. G. Luque R. Chemistryselect. 2017;2:329–334. doi: 10.1002/slct.201600624. [DOI] [Google Scholar]
- El Bouchti M. Hannache H. Cherkaoui O. Adv. Polym. Technol. 2013;32:21378. doi: 10.1002/adv.21378. [DOI] [Google Scholar]
- Haggman L. Lindblad C. Oskarsson H. Ullstrom A. S. Persson I. J. Am. Chem. Soc. 2003;125:3631–3641. doi: 10.1021/ja021012i. [DOI] [PubMed] [Google Scholar]
- El-Ashgar N. M. El-Nahhal I. M. Chehimi M. M. Babonneau F. Livage J. Int. J. Environ. Anal. Chem. 2009;89:1057–1069. doi: 10.1080/03067310902717278. [DOI] [Google Scholar]
- Tsirul'nikova N. V. Bolt Y. V. Dernovaya E. S. Fetisova T. S. Russ. J. Gen. Chem. 2016;86:739–741. doi: 10.1134/S1070363216030397. [DOI] [Google Scholar]
- Joseph A. Subramanian S. Ramamurthy P. C. Sampath S. Kumar R. V. Schwandt C. Electrochim. Acta. 2014;137:557–563. doi: 10.1016/j.electacta.2014.06.040. [DOI] [Google Scholar]
- Zhou X. Zhou J. Liu Y. Guo J. Ren J. Zhou F. Fuel. 2018;233:469–479. doi: 10.1016/j.fuel.2018.06.075. [DOI] [Google Scholar]
- Chen J. Feng J. Yan W. J. Colloid Interface Sci. 2016;475:26–35. doi: 10.1016/j.jcis.2016.04.017. [DOI] [PubMed] [Google Scholar]
- Hsu C. Lee D.-J. Hsu J.-P. Wang N. Tseng S. AIChE J. 2014;60:451–458. doi: 10.1002/aic.14276. [DOI] [Google Scholar]
- Ghadimi A. Saidur R. Metselaar H. S. C. Int. J. Heat Mass Transfer. 2011;54:4051–4068. doi: 10.1016/j.ijheatmasstransfer.2011.04.014. [DOI] [Google Scholar]
- Bradford M. M. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Jones G. L., Encyclopedia of Separation Science, 2000, pp. 1309–1315 [Google Scholar]
- Kaplan J. B. Ragunath C. Ramasubbu N. Fine D. H. J. Bacteriol. 2003;185:4693–4698. doi: 10.1128/JB.185.16.4693-4698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M. Ning F. Zhao J. Wei M. Evans D. G. Duan X. J. Am. Chem. Soc. 2012;134:1071–1077. doi: 10.1021/ja2086323. [DOI] [PubMed] [Google Scholar]
- Hou X. Zhao C. Tian Y. Dou S. Zhang X. Zhao J. Chem. Res. Chin. Univ. 2016;32:889–894. doi: 10.1007/s40242-016-6251-y. [DOI] [Google Scholar]
- Yeap S. P. Lim J. Ngang H. P. Ooi B. S. Ahmad A. L. J. Nanosci. Nanotechnol. 2018;18:6957–6964. doi: 10.1166/jnn.2018.15458. [DOI] [PubMed] [Google Scholar]
- Thomas G. Demoisson F. Boudon J. Millot N. Dalton Trans. 2016;45:10821–10829. doi: 10.1039/C6DT01050D. [DOI] [PubMed] [Google Scholar]
- Seo J. Kim J. H. Lee M. Moon J. Yi D. K. Paik U. J. Colloid Interface Sci. 2016;483:177–184. doi: 10.1016/j.jcis.2016.08.041. [DOI] [PubMed] [Google Scholar]
- Liu F. Niu F. Peng N. Su Y. Yang Y. RSC Adv. 2015;5:18128–18136. doi: 10.1039/C4RA15968C. [DOI] [Google Scholar]
- Liu Y. Fu R. Sun Y. Zhou X. Baig S. A. Xu X. Appl. Surf. Sci. 2016;369:267–276. doi: 10.1016/j.apsusc.2016.02.043. [DOI] [Google Scholar]
- Zhao W. Cui B. Peng H. Qiu H. Wang Y. J. Phys. Chem. C. 2015;119:4379–4386. doi: 10.1021/jp512447s. [DOI] [Google Scholar]
- Wang Y. Wang G. Xiao Y. Yang Y. Tang R. ACS Appl. Mater. Interfaces. 2014;6:19092–19099. doi: 10.1021/am505041a. [DOI] [PubMed] [Google Scholar]
- Guo H. Sun H. Su Z. Hu S. Wang X. J. Wuhan Univ. Technol., Mater. Sci. Ed. 2018;33:559–565. doi: 10.1007/s11595-018-1860-6. [DOI] [Google Scholar]
- Fang G. Chen H. Zhang Y. Chen A. Int. J. Biol. Macromol. 2016;88:189–195. doi: 10.1016/j.ijbiomac.2016.03.059. [DOI] [PubMed] [Google Scholar]
- Jiang Y. Guo C. Xia H. Mahmood I. Liu C. Liu H. J. Mol. Catal. B: Enzym. 2009;58:103–109. doi: 10.1016/j.molcatb.2008.12.001. [DOI] [Google Scholar]
- Aygar G. Kaya M. Ozkan N. Kocabiyik S. Volkan M. J. Phys. Chem. Solids. 2015;87:64–71. doi: 10.1016/j.jpcs.2015.08.005. [DOI] [Google Scholar]
- Rashid Z. Ghahremanzadeh R. Nejadmoghaddam M.-R. Nazari M. Shokri M.-R. Naeimi H. Zarnani A.-H. J. Chromatogr. A. 2017;1490:47–53. doi: 10.1016/j.chroma.2017.02.014. [DOI] [PubMed] [Google Scholar]
- Zhang M. Cheng D. He X. Chen L. Zhang Y. Chem.–Asian J. 2010;5:1332–1340. doi: 10.1002/asia.200900463. [DOI] [PubMed] [Google Scholar]
- Lin Z.-A. Zheng J.-N. Lin F. Zhang L. Cai Z. Chen G.-N. J. Mater. Chem. 2011;21:518–524. doi: 10.1039/C0JM02300K. [DOI] [Google Scholar]
- O'Brien J. T. Williams E. R. J. Phys. Chem. A. 2011;115:14612–14619. doi: 10.1021/jp210878s. [DOI] [PubMed] [Google Scholar]
- Alwis K. Ingham B. Mucalo M. Kappen P. Glover C. RSC Adv. 2015;5:15709–15718. doi: 10.1039/C4RA14940H. [DOI] [Google Scholar]
- De Goes L. C. Miranda E. A. Bueno S. M. A. Colloids Surf., A. 2010;369:176–185. doi: 10.1016/j.colsurfa.2010.08.019. [DOI] [Google Scholar]
- Zhang C. M. Reslewic S. A. Glatz C. E. Biotechnol. Bioeng. 2000;68:52–58. doi: 10.1002/(SICI)1097-0290(20000405)68:1<52::AID-BIT6>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Dévédec F. Bazinet L. Furtos A. Venne K. Brunet S. Mateescu M. A. J. Chromatogr. A. 2008;1194:165–171. doi: 10.1016/j.chroma.2008.03.094. [DOI] [PubMed] [Google Scholar]
- Lee J. Lee S. Y. Park S. H. Lee H. S. Lee J. H. Jeong B.-Y. Park S.-E. Chang J. H. J. Mater. Chem. B. 2013;1:610–616. doi: 10.1039/C2TB00232A. [DOI] [PubMed] [Google Scholar]
- Demey H. Tria S. A. Soleri R. Guiseppi-Elie A. Bazin I. Environ. Sci. Pollut. Res. 2017;24:15–24. doi: 10.1007/s11356-015-5758-y. [DOI] [PubMed] [Google Scholar]
- Choe W. S. Clemmitt R. H. Chase H. A. Middelberg A. P. J. J. Chromatogr. A. 2002;953:111–121. doi: 10.1016/S0021-9673(02)00154-1. [DOI] [PubMed] [Google Scholar]
- Li S. Yang K. Zhao B. Li X. Liu L. Chen Y. Zhang L. Zhang Y. J. Mater. Chem. B. 2016;4:1960–1967. doi: 10.1039/C5TB02505B. [DOI] [PubMed] [Google Scholar]
- Zhou H. Yang L. Li W. Shou Q. Xu P. Li W. Wang F. Yu P. Liu H. Ind. Eng. Chem. Res. 2012;51:4582–4590. doi: 10.1021/ie202745c. [DOI] [Google Scholar]
- Ding C. Ma X. Yao X. Jia L. J. Chromatogr. A. 2015;1424:18–26. doi: 10.1016/j.chroma.2015.11.004. [DOI] [PubMed] [Google Scholar]
- Tsai S.-Y. Lin S.-C. Suen S.-Y. Hsu W.-H. Process Biochem. 2006;41:2058–2067. doi: 10.1016/j.procbio.2006.05.013. [DOI] [Google Scholar]
- Liu X. Q. Guan Y. P. Liu H. Z. Ma Z. Y. Yang Y. Wu X. B. J. Magn. Magn. Mater. 2005;293:111–118. doi: 10.1016/j.jmmm.2005.01.051. [DOI] [Google Scholar]
- de Goes L. C. Miranda E. A. Alves Bueno S. M. Colloids Surf., A. 2010;369:176–185. doi: 10.1016/j.colsurfa.2010.08.019. [DOI] [Google Scholar]
- Bayramoglu G. Yilmaz M. Arica M. Y. Bioresour. Technol. 2010;101:6615–6621. doi: 10.1016/j.biortech.2010.03.088. [DOI] [PubMed] [Google Scholar]
- Cao G. Gao J. Zhou L. Huang Z. He Y. Zhu M. Jiang Y. Biochem. Eng. J. 2017;128:116–125. doi: 10.1016/j.bej.2017.09.019. [DOI] [Google Scholar]
- Taheran M. Naghdi M. Brar S. K. Knystautas E. J. Verma M. Surampalli R. Y. Sci. Total Environ. 2017;605:315–321. doi: 10.1016/j.scitotenv.2017.06.185. [DOI] [PubMed] [Google Scholar]
- Wang Q. Cui J. Li G. Zhang J. Huang F. Wei Q. Polymers. 2014;6:2357–2370. doi: 10.3390/polym6092357. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.