Abstract
Of 94 clinical isolates of Staphylococcus aureus (n = 51) and coagulase-negative staphylococci (CNS) (n = 43), mutations in the quinolone resistance-determining region of topoisomerases GrlA, GrlB, GyrA, and GyrB together with MICs of six quinolones were analyzed. Amino acid substitutions at identical residues (GrlA residues 80 and 84; GyrA residues 84 and 88) were found in S. aureus and CNS. Active efflux, as suggested by blocking by reserpine, contributed substantially to the resistance phenotype in some strains. Among ciprofloxacin, clinafloxacin, levofloxacin, nalidixic acid, trovafloxacin, and sparfloxacin, a 0.5-μg/ml concentration of sparfloxacin discriminated best between strains with two or three mutations and those with no mutations.
Considerable information about the mechanisms of quinolone resistance is available for Staphylococcus aureus (2, 5, 7, 12, 16, 17, 20, 23); however, less is known for Staphylococcus epidermidis (9, 19) and other coagulase-negative staphylococci (CNS) (21, 27). In the present study, we analyzed 94 unique clinical isolates with regard to the MICs of various quinolones and to the combinations of ciprofloxacin (CIP)-reserpine (RES) and trovafloxacin (TVA)-RES for these isolates. The MICs were correlated with mutations in the quinolone-resistance determining region (QRDR) of the grlA and gyrA genes (all strains) and grlB and gyrB genes (S. aureus and S. epidermidis).
All strains (except methicillin-resistant [Metr] S. aureus) were consecutive isolates collected from individual patients at the Institute for Medical Microbiology, University of Regensburg, between 1995 and 1998 and included 27 methicillin-susceptible (Mets) S. aureus isolates, 24 (Metr) S. aureus isolates (each with a unique pattern in pulsed-field gel electrophoresis [24]), 12 Mets S. epidermidis isolates, 19 Metr S. epidermidis isolates, 8 Mets CNS (1 Staphylococcus haemolyticus isolate, 5 Staphylococcus hominis isolates, and 2 Staphylococcus capitis isolates), and 4 Metr CNS (3 S. haemolyticus isolates and 1 Staphylococcus simulans isolate). CNS were isolated from normally sterile sites. Isolates were identified by a latex agglutination test (Slidex Staph-Kit; bioMérieux sa, Marcy-l'Étoile, France) and by biochemical reactions (ID 32 STAPH; bioMérieux sa). Antimicrobial agents were provided by the manufacturers: CIP (Bayer AG, Leverkusen, Germany), clinafloxacin (Parke-Davis Pharmaceutical Research, Freiburg, Germany), levofloxacin (Hoechst Marion Roussel, Frankfurt, Germany), sparfloxacin (SPX) (Rhone-Poulenc-Rohrer, Köln, Germany), and TVA (Pfizer, Karlsruhe, Germany). Nalidixic acid was purchased from Sigma (Deisenhofen, Germany) (catalog no. N8878). MICs were determined by the agar dilution method on Mueller-Hinton agar (Oxoid, Wesel, Germany) according to NCCLS guidelines (13). In two determinations the effect of RES (catalog no. R0875; Sigma) on MICs of TVA and CIP was evaluated on Mueller-Hinton agar plates with and without RES (20 μg/ml) and Etest strips (AB BIODISK, Solna, Sweden) and was expressed as the change of dilution steps. If the MIC exceeded the maximum concentration on the strip, double the concentration was arbitrarily used for further calculations. Protocols for the amplification of grlA, grlB, gyrA, and gyrB of S. aureus and grlA of CNS as published previously were used (3, 21, 26). Primers and PCR conditions for amplification of grlB, gyrA, and gyrB genes of CNS are listed in Table 1. Three units of Expand High Fidelity Taq polymerase (precast solution; Roche Molecular Biochemicals, Mannheim, Germany) was used for all amplifications. The PCR products were purified with a PCR purification kit (QIAQuick; Qiagen, Hilden, Germany). Complementary strands were sequenced on a 310 DNA sequencer (Perkin-Elmer, Foster City, Calif.) using PCR primers (6 μmol). Sequences were compared with published wild-type sequences of S. aureus (gyrA and gyrB: GenBank accession number M86227; grlA and grlB: GenBank accession number D67075). SPSS 10.0 for Windows was used for calculation of the chi-square and Mann-Whitney U test results. The partial sequences of the grlA gene of S. haemolyticus, S. hominis, S. capitis, and S. simulans and of the gyrA gene of S. hominis, S. capitis, and S. simulans appear in the GenBank nucleotide sequence database under accession numbers AF159150, AF159151, AF159152, AF159153, AF159154, AF159155, and AF159156, respectively. Partial sequences of the grlB and gyrB genes of S. epidermidis are listed under accession numbers AF314403 and AF314404, respectively.
TABLE 1.
Primers and PCR conditions used in this study
| Target | Oligonucleotide (5′→3′) | Positiona | Tanb (°C) | MgCl2 (μmol) |
|---|---|---|---|---|
| grlA (S. epidermidis, CNS) | GTTTAAAACCAGTACAACG | 2141–2159 | 49 | 2.5 |
| CGCAATGTGACTTCGATTC | 2418–2400 | |||
| gyrB (S. epidermidis) | CAGCGTTAGATGTAGCAAGC | 1531–1550 | 53 | 2 |
| CCGATTCCTGTACCAAATGC | 1780–1761 | |||
| grlB (S. epidermidis) | AAGCACAACAAGCAAGCGAGGCTG | 1526–1549 | 65 | 4 |
| TTAAAGTCAGTACCAACACCAGCACCAA | 1850–1823 |
Numbering according to S. aureus sequence.
Annealing temperature.
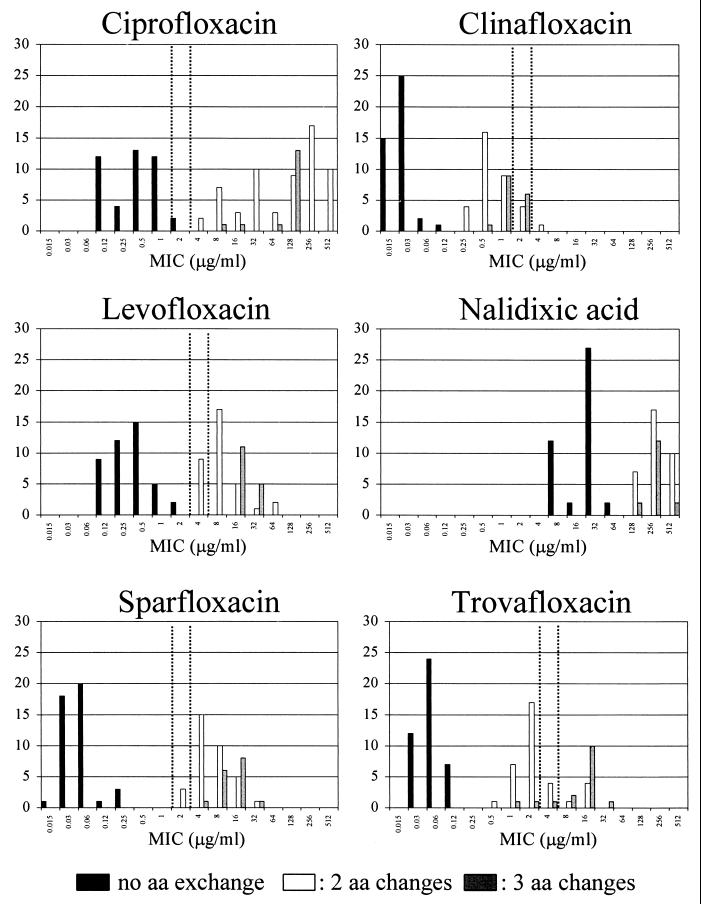
The MICs at which 50 and 90% of S. aureus and CNS strains tested were inhibited were comparable to those found in previous studies (1, 4, 8, 18). The distribution of the MICs for the different groups of staphylococci is shown in Fig. 1. In both S. aureus and CNS, resistance to methicillin and to quinolones was highly correlated (P < 0.0001 [chi-square test]).
FIG. 1.
MICs for 51 S. aureus isolates and 43 CNS with 0, 2, and 3 amino acid (aa) alterations in GrlA (residues 80 and 84) and GyrA (residues 84 and 88). Dotted vertical lines indicate NCCLS breakpoints.
In S. aureus, resistance to quinolones was correlated with the number of point mutations in grlA and gyrA, leading to amino acid changes in residues Ser80 and/or Glu84 of GrlA and Ser84 and/or Glu88 of GyrA. The association of the GrlB432 alteration with resistance is unclear (seen in two strains; MICs of SPX 8 and 16 μg/ml), because the MICs for strains with identical alterations in GyrA and GrlA ranged from 4 to 32 μg/ml (20, 22). Amino acid exchanges at position Ile45 or Pro144 of GrlA did not appear to affect MICs. No mutations in gyrB of S. aureus were observed.
The degree of similarity in nucleotide and amino acid sequences in the QRDRs of GrlA, GrlB, GyrA, and GyrB between quinolone-susceptible S. aureus and CNS is shown in Table 2. In comparison to S. aureus, all quinolone-susceptible CNS had an aspartate residue at position 84 instead of the glutamate present in S. aureus. This has also been reported for S. epidermidis by Li et al. (9). In the QRDR of GyrA, S. epidermidis, S. capitis, and S. simulans had amino acid sequences identical to that of S. aureus. Both S. haemolyticus and S. hominis differ from the other CNS at codon 88 in that they have a conservative change from glutamate to aspartate (21). MICs of SPX for these strains were 0.06 to 0.25 μg/ml, indicating no apparent effect of the Glu88Asp change in GyrA of S. haemolyticus or S. hominis.
TABLE 2.
Degree of similarity of nucleotide and amino acid sequences in the QRDRs of GyrA, GyrB, GrlA, and GrlB of staphylococci.
| Species | % Similarity to S. epidermidis sequence
|
|||||||
|---|---|---|---|---|---|---|---|---|
| GyrA
|
GyrB
|
GrlA
|
GrlB
|
|||||
| nta | aab | nt | aa | nt | aa | nt | aa | |
| S. haemolyticus | 85.8 | 95 | —c | — | 90.8 | 100 | — | |
| S. hominis | 86.7 | 95 | — | — | 87.5 | 100 | — | — |
| S. capitis | 90.5 | 100 | — | — | 83.3 | 100 | — | — |
| S. simulans | 87.5 | 100 | — | — | 89.2 | 100 | — | — |
| S. aureus | 83.3 | 100 | 83 | 95 | 87.5 | 97.5 | 97 | 97 |
nt, nucleotide.
aa, amino acid.
—, not done.
Identical to S. aureus, elevated MICs for CNS were found in strains with amino acid changes in residues Ser80 and Asp84 of GrlA and residues Ser84 and Glu88 of GyrA. In GrlA of S. epidermidis only Ser80Phe or Ser80Tyr changes were found by Li et al. (9) and in the present study, while S. hominis and S. haemolyticus had Ser80Val or Ser80Leu amino acid exchanges (21). No mutations in gyrB or grlB in any strain of S. epidermidis were found. In CNS other than S. epidermidis, only the grlA and gyrA genes were analyzed. Different primers designed for amplification of the grlB and gyrB genes of S. aureus or S. epidermidis, tested under various nonstringent conditions, did not yield any product in S. capitis, S. hominis, S. haemolyticus, and S. simulans.
In strains with identical amino acid changes but different MICs, additional resistance mechanisms may be active. We investigated whether inhibition of efflux pump systems (presumably NorA [11, 14; F. J. Schmitz, B. Hertel, B. Hoffmann, S. Scheuring, J. Verhoef, A. C. Fluit, H. P. Heinz, K. Kohrer, and M. E. Jones, Letter, J. Antimicrob. Chemother., 42:561–563, 1998]) by the alkaloid RES would abolish such differences or whether efflux-mediated resistance was associated with certain species or strains subgrouped according to quinolone resistance. RES decreased MICs of CIP by a median of 1.0 dilution step (range, −4 to 6 [not significant]), and TVA MICs by a median of 1.4 dilution steps (range, −3 to 4.4 [not significant]). This indicates a major contribution of efflux pumps to the resistance phenotype in some strains. Similar findings have been reported for S. aureus and pneumococci (10, 15). In CIP-susceptible (CIP-S) S. aureus the decrease was a median of 3.0 dilution steps (P < 0.001 [Mann-Whitney test] compared to CIP-resistant [CIP-R] S. aureus) when testing CIP plus RES. This indicates a basic efflux activity in wild-type S. aureus. In CIP-R S. aureus the observation may be lost because of the high level of resistance conferred by topoisomerase mutations. Unexpectedly, the combination of RES and CIP produced an increase of MIC in nine strains, both S. aureus (n = 2) and CNS (n = 7), ranging from 1 to 4 dilution steps. We have no explanation for this; however, RES may function as an inducer of efflux pumps in these strains. Also, RES affected the change of MIC differently for CIP and TVA (Table 3), since the changes of MIC induced by CIP-RES and TVA-RES did not correlate, except for CIP-R CNS (chi-square test, r2 = 0.87). No correlation was found between the RES-inhibitable mechanism(s) and MICs for strains with identical amino acid changes in GrlA or GyrA (data not shown). Therefore, other efflux systems or other, as yet unknown mechanisms of resistance are likely to exist (17, 20, 25). Ince and Hooper (6) have recently suggested the extension of the range of the QRDR of grlA of S. aureus.
TABLE 3.
Changes in MICs of CIP and TVA for S. aureus and CNS on Mueller-Hinton agar with RES (20 μg/ml)
| Strains | No. | Change in MIC (dilution steps) of:
|
r2a | |||||
|---|---|---|---|---|---|---|---|---|
| CIP
|
TVA
|
|||||||
| Mean | Median | Range | Mean | Median | Range | |||
| All | 94 | 1.4 | 1 | −4–6 | 1.2 | 1.4 | −3–4.4 | 0.39 |
| S. aureus | 51 | 1.9 | 2 | −1–4.4 | 1.2 | 1.4 | −3–4.4 | 0.06 |
| CIP-S | 23 | 3b | 3b | 2–4.4 | 1.7 | 1.4 | 1–3 | 0.09 |
| CIP-R | 28 | 1.1b | 1b | −1–4.4 | 0.8 | 1 | −3–4.4 | 0.07 |
| CNS | 43 | 0.8 | 1 | −4–6 | 1.2 | 1.4 | −2–4.4 | 0.83 |
| CIP-S | 20 | 0.9 | 1 | 0–3 | 1.4 | 1.5 | 1–2.4 | <0.01 |
| CIP-R | 23 | 0.7 | 1 | −4–6 | 1 | 1.4 | −2–4.4 | 0.87 |
r2, linear regression coefficient (chi-square test).
P < 0.001 (Mann-Whitney test).
From an epidemiological rather than from a clinical point of view, susceptibility testing not only should indicate whether a substance is suitable for treatment but also should identify strains that have the potential to become resistant. Since resistance to quinolones is acquired in a stepwise fashion, detection of the first step might be important. The distribution of MICs for staphylococci with different numbers of amino acid alterations in the QRDRs of GrlA and GyrA is shown graphically in Fig. 1. MICs of CIP of >2 μg/ml (gap of 2 dilution steps), MICs of TVA of >0.5 μg/ml (gap of 1 dilution step), MICs of levofloxacin of >2 μg/ml, and MICs of SPX of >1 μg/ml separated all strains with two or three alterations in GrlA or GyrA from strains without alterations. According to NCCLS criteria TVA classified 26 strains with two or three amino acid changes as susceptible. SPX at 0.5 μg/ml separated all strains with no changes in topoisomerases from those with two or more changes and could therefore act as a predictor of quinolone resistance mechanisms, in the same way that penicillin and methicillin are used in the prediction of susceptibility to other β-lactam antibiotics.
In conclusion, similar resistance mechanisms were found in S. aureus and CNS. Since in staphylococcal infections the use of highly active quinolone agents is likely to increase, prudent use may be guided by the prediction of resistance mechanisms rather than MIC data.
Acknowledgments
The results of typing Metr S. aureus by pulsed-field gel electrophoresis were kindly provided by Michaela Metz. We thank Markus Bollwein and Christine Irtenkauf for excellent technical assistance. Nucleotide sequence determination was performed by Holger Melzl and Josef Köstler.
REFERENCES
- 1.Ednie L M, Jacobs M R, Appelbaum P C. Comparative activities of clinafloxacin against gram-positive and -negative bacteria. Antimicrob Agents Chemother. 1998;42:1269–1273. doi: 10.1128/aac.42.5.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferrero L, Cameron B, Crouzet J. Analysis of gyrA and gylA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:1554–1558. doi: 10.1128/aac.39.7.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fey P D, Climo M W, Archer G L. Determination of the chromosomal relationship between mecA and gyrA in methicillin-resistant coagulase-negative staphylococci. Antimicrob Agents Chemother. 1998;42:306–312. doi: 10.1128/aac.42.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuchs P C, Barry A L, Brown S D. In vitro activities of clinafloxacin against contemporary clinical bacterial isolates from 10 North American centers. Antimicrob Agents Chemother. 1998;42:1274–1277. doi: 10.1128/aac.42.5.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gootz T D, Zaniewski R P, Haskell S L, Kaczmarek F S, Maurice A E. Activities of trovafloxacin compared with those of other fluoroquinolones against purified topoisomerases and gyrA and grlA mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43:1845–1855. doi: 10.1128/aac.43.8.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ince D, Hooper D C. Mechanisms and frequency of resistance to premafloxacin in Staphylococcus aureus: novel mutations suggest novel drug-target interactions. Antimicrob Agents Chemother. 2000;44:3344–3350. doi: 10.1128/aac.44.12.3344-3350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito H, Yoshida H, Bogaki-Shonai M, Niga T, Hattori H, Nakamura S. Quinolone resistance mutations in the DNA gyrase gyrA and gyrB genes of Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2014–2023. doi: 10.1128/aac.38.9.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones M E, Visser M R, Klootwijk M, Heisig P, Verhoef J, Schmitz F J. Comparative activities of clinafloxacin, grepafloxacin, levofloxacin, moxifloxacin, ofloxacin, sparfloxacin, and trovafloxacin and nonquinolones linezolid, quinupristin-dalfopristin, gentamicin, and vancomycin against clinical isolates of ciprofloxacin-resistant and -susceptible Staphylococcus aureus strains. Antimicrob Agents Chemother. 1999;43:421–423. doi: 10.1128/aac.43.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Deguchi T, Yasuda M, Kawamura T, Kanematsu E, Nishino Y, Ishihara S, Kawada Y. Alteration in the gyrA subunit of DNA gyrase and the parC subunit of DNA topoisomerase IV in quinolone-resistant clinical isolates of Staphylococcus epidermidis. Antimicrob Agents Chemother. 1998;42:3293–3295. doi: 10.1128/aac.42.12.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markham P N. Inhibition of the emergence of ciprofloxacin resistance in Streptococcus pneumoniae by the multidrug efflux inhibitor reserpine. Antimicrob Agents Chemother. 1999;43:988–989. doi: 10.1128/aac.43.4.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muñoz-Bellido J L, Manzanares M A, Martínez Andrés J A, Gutiérrez Zufiaurre M N, Ortiz G, Segoria Hernández M, García-Rodríguez J A. Efflux pump-mediated quinolone resistance in Staphylococcus aureus strains wild type for gyrA, gyrB, grlA, and norA. Antimicrob Agents Chemother. 1999;43:354–356. doi: 10.1128/aac.43.2.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muñoz Bellido J L, Alonso Manzanares M A, Yagüe Guirao G, Gutiérrez Zufiaurre M N, Toldos M C, Segovia Hernández M, García-Rodríguez J A. In vitro activities of 13 fluoroquinolones against Staphylococcus aureus isolates with characterized mutations in gyrA, gyrB, grlA, and norA and against wild-type isolates. Antimicrob Agents Chemother. 1999;43:966–968. doi: 10.1128/aac.43.4.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. NCCLS publication no. M7–A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 14.Ng E Y, Trucksis M, Hooper D C. Quinolone resistance mediated by norA: physiologic characterization and relationship to flqB, a quinolone resistance locus on the Staphylococcus aureus chromosome. Antimicrob Agents Chemother. 1994;38:1345–1355. doi: 10.1128/aac.38.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitz F J, Fluit A C, Luckefahr M, Engler B, Hofmann B, Verhoef J, Heinz H P, Hadding U, Jones M E. The effect of reserpine, an inhibitor of multidrug efflux pumps, on the in-vitro activities of ciprofloxacin, sparfloxacin and moxifloxacin against clinical isolates of Staphylococcus aureus. J Antimicrob Chemother. 1998;42:807–810. doi: 10.1093/jac/42.6.807. [DOI] [PubMed] [Google Scholar]
- 16.Schmitz F J, Hofmann B, Hansen B, Scheuring S, Luckefahr M, Klootwijk M, Verhoef J, Fluit A, Heinz H P, Kohrer K, Jones M E. Relationship between ciprofloxacin, ofloxacin, levofloxacin, sparfloxacin and moxifloxacin (BAY 12–8039) MICs and mutations in grlA, grlB, gyrA and gyrB in 116 unrelated clinical isolates of Staphylococcus aureus. J Antimicrob Chemother. 1998;41:481–484. doi: 10.1093/jac/41.4.481. [DOI] [PubMed] [Google Scholar]
- 17.Schmitz F J, Jones M E, Hofmann B, Hansen B, Scheuring S, Luckefahr M, Fluit A, Verhoef J, Hadding U, Heinz H P, Kohrer K. Characterization of grlA, grlB, gyrA, and gyrB mutations in 116 unrelated isolates of Staphylococcus aureus and effects of mutations on ciprofloxacin MIC. Antimicrob Agents Chemother. 1998;42:1249–1252. doi: 10.1128/aac.42.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seifert H. Comparative in-vitro activities of trovafloxacin, ciprofloxacin, ofloxacin, and broad-spectrum beta-lactams against aerobe blood culture isolates. Zentbl Bakteriol. 1998;288:509–518. doi: 10.1016/s0934-8840(98)80070-4. [DOI] [PubMed] [Google Scholar]
- 19.Sreedharan S, Peterson L R, Fisher L M. Ciprofloxacin resistance in coagulase-positive and -negative staphylococci: role of mutations at serine 84 in the DNA gyrase A protein of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 1991;35:2151–2154. doi: 10.1128/aac.35.10.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi H, Kikuchi T, Shoji S, Fujimura S, Lutfor A B, Tokue Y, Nukiwa T, Watanabe A. Characterization of gyrA, gyrB, grlA and grlB mutations in fluoroquinolone-resistant clinical isolates of Staphylococcus aureus. J Antimicrob Chemother. 1998;41:49–57. doi: 10.1093/jac/41.1.49. [DOI] [PubMed] [Google Scholar]
- 21.Takahata M, Yonezawa M, Matsubara N, Watanabe Y, Narita H, Matsunaga T, Igarashi H, Kawahara M, Onodera S, Oishi Y. Antibacterial activity of quinolones against coagulase-negative staphylococci and the quinolone resistance-determining region of the gyrA genes from six species. J Antimicrob Chemother. 1997;40:383–386. doi: 10.1093/jac/40.3.383. . (Erratum, 41:317, 1998.) [DOI] [PubMed] [Google Scholar]
- 22.Tanaka M, Onodera Y, Uchida Y, Sato K. Quinolone resistance mutations in the GrlB protein of Staphylococcus aureus. Antimicrob Agents Chemother. 1998;42:3044–3046. doi: 10.1128/aac.42.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka M, Onodera Y, Uchida Y, Sato K, Hayakawa I. Inhibitory activities of quinolones against DNA gyrase and topoisomerase IV purified from Staphylococcus aureus. Antimicrob Agents Chemother. 1997;41:2362–2366. doi: 10.1128/aac.41.11.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed- field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang T, Tanaka M, Sato K. Detection of grlA and gyrA mutations in 344 Staphylococcus aureus strains. Antimicrob Agents Chemother. 1998;42:236–240. doi: 10.1128/aac.42.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamagishi J, Kojima T, Oyamada Y, Fujimoto K, Hattori H, Nakamura S, Inoue M. Alterations in the DNA topoisomerase IV grlA gene responsible for quinolone resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:1157–1163. doi: 10.1128/aac.40.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamamoto T, Takubo S, Fujita K, Oguri T, Yokota T. Cloning and restriction analysis of DNA conferring new quinolone antimicrobial agent resistance from Staphylococcus aureus and other coagulase-negative Staphylococcus species. FEMS Microbiol Lett. 1990;56:335–339. doi: 10.1111/j.1574-6968.1990.tb13961.x. [DOI] [PubMed] [Google Scholar]