Abstract
The solute carrier (SLC) superfamily encompasses >400 transmembrane transporters involved in the exchange of amino acids, nutrients, ions, metals, neurotransmitters and metabolites across biological membranes. SLCs are highly expressed in the mammalian brain; defects in nearly 100 unique SLC-encoding genes (OMIM: https://www.omim.org) are associated with rare Mendelian disorders including developmental and epileptic encephalopathy and severe neurodevelopmental disorders.
Exome sequencing and family-based rare variant analyses on a cohort with neurodevelopmental disorders identified two siblings with developmental and epileptic encephalopathy and a shared deleterious homozygous splicing variant in SLC38A3. The gene encodes SNAT3, a sodium-coupled neutral amino acid transporter and a principal transporter of the amino acids asparagine, histidine, and glutamine, the latter being the precursor for the neurotransmitters GABA and glutamate. Additional subjects with a similar developmental and epileptic encephalopathy phenotype and biallelic predicted-damaging SLC38A3 variants were ascertained through GeneMatcher and collaborations with research and clinical molecular diagnostic laboratories. Untargeted metabolomic analysis was performed to identify novel metabolic biomarkers.
Ten individuals from seven unrelated families from six different countries with deleterious biallelic variants in SLC38A3 were identified. Global developmental delay, intellectual disability, hypotonia, and absent speech were common features while microcephaly, epilepsy, and visual impairment were present in the majority. Epilepsy was drug-resistant in half. Metabolomic analysis revealed perturbations of glutamate, histidine, and nitrogen metabolism in plasma, urine, and CSF of selected subjects, potentially representing biomarkers of disease.
Our data support the contention that SLC38A3 is a novel disease gene for developmental and epileptic encephalopathy and illuminate the likely pathophysiology of the disease as perturbations in glutamine homeostasis.
Keywords: biallelic, SLC38A3, glutamate transporter, glutamate/GABA-glutamine cycle
Marafi et al. identify SLC38A3, encoding SNAT3—a transmembrane transporter in the glutamate/GABA-glutamine cycle—as a novel developmental and epileptic encephalopathy gene in seven unrelated families, and show the likely pathophysiological mechanism to be perturbation of glutamine homeostasis.
Introduction
Mutations in genes encoding proteins involved in ion channels and synaptic transmission have frequently been identified as underlying aetiologies for developmental and epileptic encephalopathies (DEEs).1,2 Advances in molecular technology and genetic and genomic approaches to disease research have subsequently implicated causative genes for DEE that encode proteins from diverse regulatory and developmental pathways.2
The solute carrier (SLC) family of proteins is a superfamily of transmembrane transporters that includes more than 400 members involved in the exchange of amino acids, nutrients, ions, metals, neurotransmitters and metabolites across various biological membranes.3,4 More than 280 SLC genes are expressed in the brain, playing an important role in energy metabolism in neurons and astrocytes, homeostasis (e.g. glutamate/GABA-glutamine cycle) and synaptic vesicle and neurotransmitter release.3 Defects in several SLC-encoding genes have been shown to cause DEE, including SLC1A2 (DEE41; MIM# 617105), SLC12A5 (DEE34; MIM# 616645), SLC13A5 (DEE25; MIM# 615905), SLC25A12 (DEE39; MIM# 612949), SLC25A22 (DEE3; MIM# 609304), SLC35A2 (DEE22; MIM# 300896).5,6 Defects in SLC-encoding genes are also implicated in more than a dozen severe neurodevelopmental disorders with or without epilepsy, e.g. SLC2A1 (GLUT1 deficiency syndromes 1 and 2; MIM# 606777 and MIM #612126), SLC6A1 (myoclonic-atonic epilepsy; MIM# 616421), SLC6A8 (cerebral creatine deficiency syndrome 1; MIM# 300352), SLC18A2 (infantile parkinsonism-dystonia syndrome 2; MIM# 618049), and SLC16A1 (monocarboxylate transporter 1 deficiency; MIM# 616095).3,5 Yet, despite the growing number of SLC genes implicated in neurodevelopmental disorders and DEE, 45–70% of the DEE cases do not receive a specific aetiological molecular diagnosis due to the variable availability of gene panels, exomes, and genomes.7,8 The molecular yield in DEE also varies significantly by age of onset, and the use of unbiased non-panel approach can aid in gene discovery.7,9,10 Yet, even for those patients for whom a specific SLC gene is identified, the resultant perturbations from biological homeostasis, that potentially underlie the disease, remain unclear.11
Here, we describe 10 individuals with DEE from seven unrelated families from six different countries (Azerbaijan, Egypt, Kuwait, Saudi Arabia, USA and Yemen) that by genomic studies were found to have biallelic predicted-damaging variants in SLC38A3. SLC38A3 encodes a sodium-coupled neutral amino acid transporter (SNAT3), a transporter of asparagine, histidine and glutamine and a major homeostatic regulator of the glutamate/GABA-glutamine cycle in the brain.12 Untargeted metabolomic analyses of patients’ biofluids showed evidence for abnormalities in glutamine/glutamate, histidine, asparagine, and nitrogen metabolism, tricarboxylic (TCA) cycle, and glucose homeostasis, further supporting a significant role for SNAT3 in amino acid transport and utilization in the human brain and possibly revealing metabolic markers of the disease.
Material and methods
Participants and ethical approval
Subjects were enrolled under Institutional Review Board (IRB) approved protocols that include Baylor Hopkins Center for Mendelian Genomics (BHCMG, BCM IRB H-29697; Families 1, 2 and 7), King Faisal Specialist Hospital & Research Centre (KFSRHC RAC#2121053; Family 3), the Institute of Neurology, University College London (IoN UCL 07/Q0512/26; Families 4 and 5), and University of California, San Diego (UCSD IRB; Family 6).
Exome sequencing, variant interpretation, and phenotyping methods
Through family-based genomic studies, exome sequencing, and rare variant analysis on a cohort of more than 300 consanguineous families with neurodevelopmental disorders from the Middle East and Turkey, and by applying our previously described variant parsing and prioritization workflow,13 we identified two siblings from one family (Family 1 in Fig. 1) with a rare homozygous splice site variant (NM_006841.6:c.855+1G>T) in SLC38A3. Briefly, the variant needed to be shared homozygous between the two affected siblings, rare (minor allele frequency ≤ 0.001), absent in homozygous state from Genome Aggregation Database (gnomAD v2.1.1)14 and internal control database, predicted deleterious by three or more prediction models, well-conserved across species and has a CADD score ≥ 15. Only three variants survived our stringent analysis criteria: SLC38A3: NM_006841.6:c.855+1G>T, SCN9A: NM_002977:c.G1336A p.(Glu446Lys), and PAX2: NM_000278:c.G478A p.(Ala160Thr). The SLC38A3 variant was prioritized based on high expression in the brain and biological evidence supporting its role in replenishing the neurotransmitter pool. Homozygous variants in SCN9A are associated with autosomal recessive congenital insensitivity to pain and hereditary sensory and autonomic neuropathy type IID (MIM# 243000) while heterozygous variants in PAX2 are associated with autosomal dominant papillorenal syndrome (MIM# 120330) and focal segmental glumerulosclerosis 7 (MIM# 616002), none of which could explain the phenotype observed in the siblings.
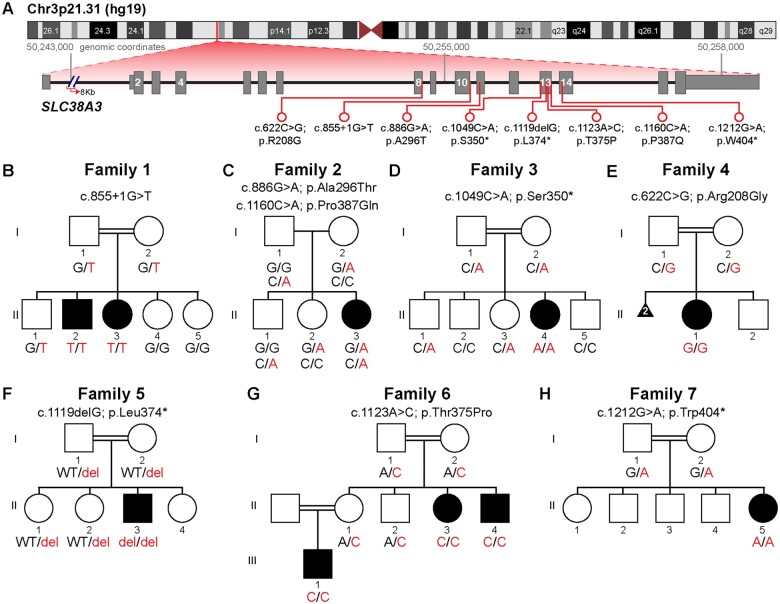
Figure 1.
Chromosome 3 ideogram, pedigrees and genotypes in Families 1–7 with biallelic SLC38A3 variants. (A) Chromosome 3 ideogram showing the position of the SLC38A3 gene on Chr3p21.31. SLC38A3 (NM_006841.6) has 15 coding exons (16 exons, in total). The location of each identified variant is indicated by a red line. (B–H) Pedigrees of Families 1–7 showing segregation of the variants in SLC38A3 in these families. The corresponding genotype is displayed under each individual. Affected individuals are shaded in black on each pedigree.
Four families were identified through collaborations with clinical diagnostic laboratories (Family 2 from Baylor Genetics, Family 5 from Centogene, and Family 7 from GeneDx) and with other research laboratories (Family 6), while two additional subjects from two unrelated families (Families 3 and 4) were identified through GeneMatcher.15 One of the families (Family 3) was previously published with SLC38A3 as a novel candidate recessive gene locus with limited phenotypic data in the context of a large gene discovery cohort.16
Bioinformatic analyses (SIFT, PolyPhen-2, CADD, and PhyloP) were utilized to predict the potential likely damaging deleterious effect of the variants on protein function and evolutionary conservation. BafCalculator (https://github.com/BCM-Lupskilab/BafCalculator), an in-house developed bioinformatic tool that extracts the calculated B-allele frequency from unphased exome data, was used to calculate the absence of heterozygosity (AOH), a surrogate measure for runs of homozygosity (ROH) and genomic intervals identical-by-descent, in the two siblings from Family 1.17-19 The ROH/AOH in Family 7, II-5 was determined by clinical SNP array with a cut-off of 5 Mb to call an AOH/ROH interval. Sanger sequencing was performed on identified variants in all families for variant validation and segregation studies. The referring physicians provided the relevant clinical information on all affected individuals for deep phenotyping. All available brain MRI and head CT images were uniformly reviewed by the same board-certified paediatric neuroradiologist (J.V.H).
Protein modelling
Models of SNAT3 (encoded by SLC38A3) were generated from Phyre2 and I-Tasser.20,21 Based upon unanimous poor prediction scores, three extracellular regions were removed (residues 1–68, 245–282, 494–504). These were energy minimized with Pyrosetta, and due to good score and similarity to the main template, PDB: 6C08 (SLC38A9), the Phyre model was brought forward.22–24 The model was modified to contain glutamate, based upon the bound substrate arginine from PDB:6C08, sodium, based upon the observations from PDB:6C08, and three docked phosphates, based upon the observation that there are strong positively charged pockets at the cytosolic interface, a potential sign of organophosphate binding (phospholipids or PIP3).24–26 Inorganic phosphate was chosen because it is not known which organophosphate may bind. The membrane orientation of the protein was taken from the OPM entry for PDB:6C08.27 The ligand-bound protein was further energy minimized using the RosettaMP framework and the franklin2019 score function,28 mutated, and the 10 Å neighbourhood around the mutation energy minimized to calculate the difference in Gibbs free energy (ΔΔG). Interactive protein view was made in Michelanglo.29 Data available at https://doi.org/10.5281/zenodo.5012607.
Untargeted metabolomic analyses
Clinical untargeted metabolomics was performed under IRB-approved research protocol (H-35388) by Baylor Genetics and Metabolon, Inc, as described.30–34 Briefly, 1 ml of residual CSF (from Family 1, Individual II-3), previously collected and stored at −20°C at a clinical laboratory, was transferred to Baylor Genetics overnight in frozen condition (−80°C). Residual plasma (500 μl) that was extracted from peripheral blood collected in EDTA-coated tubes from the same individual and her affected sibling (Family 1, Individuals II-2 and II-3) was also studied. Plasma was retrieved from a research laboratory where it had been stored at −20°C for a year. A fresh plasma sample (500 μl) and two independent urine (5 ml) samples from two different time intervals were also collected from Family 2, Individual II-3 and shipped on dry ice to Baylor Genetics overnight in frozen condition (−80°C). Small molecules (<1000 Da) were extracted from the patient samples (100 µl) in an 80% methanol solution containing recovery standards. The purified supernatant was divided into five aliquots, one for each of four individual LC/MS analyses and a backup sample. The samples were then briefly evaporated to remove organic solvents and stored under liquid nitrogen overnight before analysis. CSF, urine, and plasma were experimentally analysed as independent sample sets, as previously described.30,31,34,35
Metabolomics network and enrichment analysis
Metabolomics datasets for each biofluid were filtered to identify metabolites that were altered in patient biofluids with z-score >+1.5 or <−1.5, typically representing molecules that fall into the <5% or >95%, respectively, of the control reference population. For biofluids with multiple untargeted datasets, mean values for all significantly altered metabolites were used for downstream analyses. Metabolite enrichment and network analyses were performed by using MetaboAnalyst 5.0 (http://www.metaboanalyst.ca/faces/home.xhtml). Network analysis was performed by MetaboAnalyst 5.0 in three different modes: metabolite-biofluid interaction network, metabolite–KEGG interaction network, and metabolite–metabolic pathway network.
MetaboAnalyst 5.0 performs metabolite set enrichment analysis (MSEA) that contains human and mammalian metabolite sets, as well as chemical class metabolite sets. The analysis is based on several libraries containing ∼9000 biologically meaningful metabolite sets collected primarily from human studies including >1500 chemical classes. All analyses require a Human Metabolome Databases (HMDB) identifier. Not all molecules have this identifier, and not all molecules with an HMDB identifier are mapped within known metabolic pathways.
Data availability statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Results
Molecular characterization
Molecular findings included seven variants: six homozygous variants [NM_006841.6: c.855+1G>T, c.1049C>A; p.(Ser350*), c.622C>G; p.(Arg208Gly), c.1119delG; p.(Leu374*), c.1123A>C; p.(Thr375Pro), c.1212G>A; p.(Trp404*)] identified in six families reporting parental consanguinity and compound heterozygous variants [NM_006841.6: c.886G>A; p.(Ala296Thr) and c.1160C>A; p.(Pro387Gln)] in one family with no reported consanguinity (Fig. 1 and Table 1). The families were from diverse ethnic backgrounds and countries of origin including Kuwait, USA, Saudi Arabia, Azerbaijan, Egypt and Yemen. All eight SLC38A3 rare variants were absent in the homozygous state from the Genome Aggregation Database (gnomAD v2.1.1),14 as well as from our in-house control databases of BHCMG, KFSRHC and Queen Square, UCL. Bioinformatic analyses supported likely pathogenicity for each variant allele (Table 1). The pedigrees and genotypes from segregation studies of the variants of the affected individuals are shown in Fig 1. Two siblings (Family 1, Individuals II-2 and II-3) had a total AOH/ROH of 350–530 Mb. Their SLC38A3 homozygous splicing variant was located within an AOH/ROH block ranging from 4.9 to 5.5 Mb. The total AOH/ROH in Family 7, Individual II-5 was 270 Mb.
Table 1.
Variant information of subjects with biallelic variants in SLC38A3
| Family 1 | Family 2 | Family 3 | Family 4 | Family 5 | Family 6 | Family 7 | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| II-2 | II-3 | II-3 | II-4a | II-1 | II-3 | II-3 | II-4 | III-1 | II-5 | |
| Country of origin | Kuwait | USA | Saudi Arabia | Azerbaijan | Saudi Arabia | Egypt | Yemen | |||
| Consanguinity | + (1st cousins) | – | + (1st cousins) | + (1st cousins) | + (1st cousins) | + (1st cousins) | + (2nd cousins) | + (1st cousins) | ||
| Position (GRCh37\hg19) | Chr3:50255273 | Chr3:50255379 | Chr3:50256037 | Chr3:50254743 | Chr3:50256106 | Chr3:50256111 | Chr3:50256286 | |||
| Chr3:50256148 | ||||||||||
| Nucleotide (NM_006841.6); protein | c.855+1G>T |
|
|
|
|
|
|
|||
| ||||||||||
| Type of variant | Splicing | Missense | Nonsense | Missense | Stop-gain | Missense | Nonsense | |||
| Zygosity | Hmz | Comp Het | Hmz | Hmz | Hmz | Hmz | Hmz | |||
| Allele count/Zygosity (gnomAD) | 0 Het-0 Hmz | 2 Het-0 Hmz | 0 Het-0 Hmz | 0 Het-0 Hmz | 0 Het-0 Hmz | 0 Het-0 Hmz | 0 Het-0 Hmz | |||
| 1 Het-0 Hmz | ||||||||||
| CADD Score GRCh37 v.1.6 (PHRED) | 32 | 19.4 | 20 | 22.1 | 16.39 | 18.7 | 18.8 | |||
| 17 | ||||||||||
| PolyPhen-2 | – | Probably damaging/probably damaging | – | Possibly damaging | – | Possibly damaging | – | |||
| SIFT | – | Damaging/damaging | – | Damaging | – | Damaging | – | |||
| Conservation (mamPhyloP) | 2.36 | 2.3 | 2.37 | 2.49 | – | 1.97 | ||||
| 2.43 | ||||||||||
| AOH block around gene (Mb) | 4.9 | 5.1 | N/A | N/A | 5.5 | N/A | N/A | N/A | ||
| Total AOH (Mb) | 530 | 350 | N/A | N/A | N/A | N/A | N/A | 270 | ||
CADD = Combined Annotation Dependent Depletion; comp het = compound heterozygous; Het = heterozygous; Hmz = homozygous; N/A = not available; SIFT = Sorting Intolerant from Tolerant prediction score.
Previously published with limited clinical data.
Protein modelling
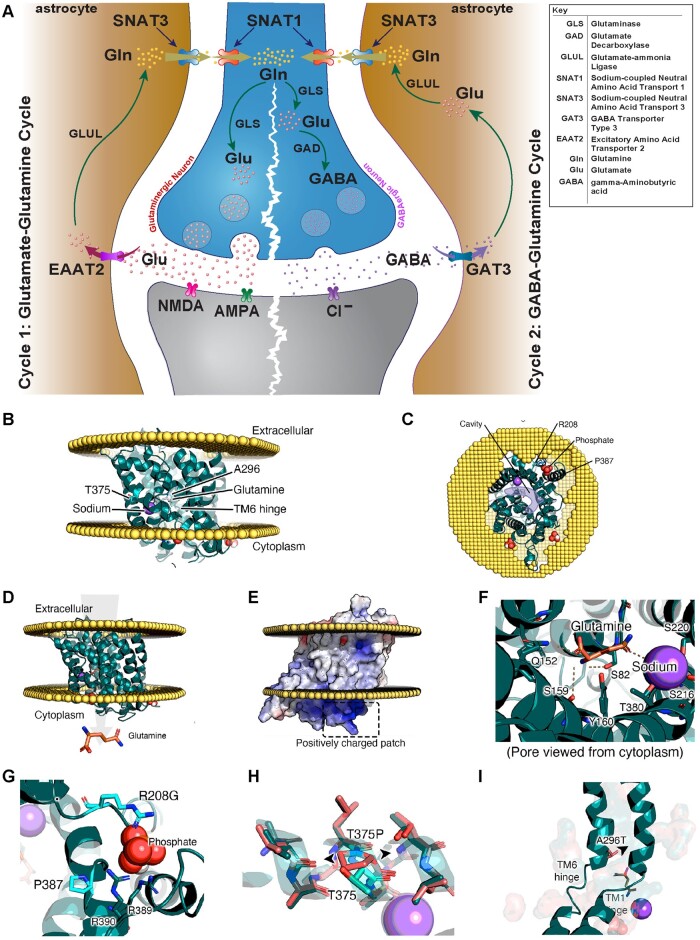
A predicted model of SNAT3 based upon the structure of zebrafish Snat9 (encoded by slc38a9) shows the deleterious variants interfere with the activity [p.(Arg208Gly), p.(Ala296Thr), p.(Pro387Gln)] or stability [p.(Ser350*), p.(Leu374*), p.(Thr375Pro), splicing variant c.855+1G>T] of the SNAT3 protein (Fig. 2B–I; interactive page: https://michelanglo.sgc.ox.ac.uk/r/SLC38A3). For example, the p.(Ala296Thr) is on the hinge region likely preventing the conformational change required for transport, while p.(Thr375Pro) destabilizes the protein (ΔΔG: +20 kcal/mol) more than any of the variants found in the human population (gnomAD). Additionally, the protein may possess cytosolic phospholipid-binding pockets, one of which is predicted to be broken by p.(Arg208Gly). Such pockets are found in some membrane proteins endowing them with a heterogeneous distribution across the plasma membrane by binding particular lipid rafts, which could be speculated to also apply to SNAT3. The splicing variant (c.855+1G>T) and nonsense variants [p.(Ser350*), p.(Leu374*), and p.(Trp404*)] are truncating and predicted to undergo nonsense-mediated mRNA decay (NMD) or result in a non-functional unfolded protein.
Figure 2.
An illustration of the glutamate/GABA-glutamine cycle, SNAT3 protein modelling and effect of amino acid change of identified SLC38A3 variants. (A) An illustration of the glutamate/GABA-glutamine cycle which takes place between neurons and astrocytes to replenish the glutamate [Glu] in the glutaminergic neurons (cycle 1) and GABA pool in the GABAergic neurons (cycle 2). In both cycles, the cycle starts with the synthesis of glutamate from glutamine in the neurons via glutaminase (GLS). In cycle 1 (glutamate-glutamine cycle), glutamate is packed into synaptic vesicles and is transported and released into the synaptic cleft upon neuronal activation. Glutamate then acts on the postsynaptic glutamate receptors (NMDA, AMPA or KAR) before it is taken up to the astrocytes (via excitatory amino acid transporter 2; EAAT2) to be recycled back into glutamine via glutamate-ammonia ligase (GLUL). In cycle 2, glutamate is a converted into GABA via glutamate decarboxylase (GAD) in an additional step. Similarly, GABA is then packed into synaptic vesicles and is released into synaptic cleft upon activating signal. GABA is then taken up by neighbouring astrocytes through GABA transporter 3 (GAT3) to be recycled back into glutamine (Gln). In both cycles, sodium-coupled neural (system N) amino-acid transporter 3 (SNAT3) is then responsible for the transport of glutamine into the extracellular fluid for uptake into neurons via system A transporter 1 (SNAT1) to replenish the glutamate or GABA pools. Loss-of-function of SNAT3 is thus expected to result in depletion of glutamate and GABA neurotransmitter pools in neurons. Note that defects in GAD1, EAAT2, GLS, and GLUL as part of this cycle are known to cause developmental and epileptic encephalopathy. (B–I) show protein modelling SNAT3 and effect of implicated SLC38A3 variants. (B and C) Model of SNAT3 with direction of glutamine transport displayed in arrow. (D) Model of SNAT3 and its embedding in a membrane. (E) Example of one of the positively charged regions at the membrane interface with the cytosol, which may be an organophosphate pocket (see G). (F) Glutamine and sodium added to the model and their interacting residues. (G) Residues P387Q and R208G in the suspected organophosphate pocket. (H) Distortion created in the transmembrane helix caused by T375P (wild-type in teal, mutant in coral). (I) A296T is near the TM1 and TM6 hinges, which shift to allow the channel to open on the other side, even the slightest reduction in this pocket will likely affect the dynamic behaviour.
Interestingly, in gnomAD v3.1, there is a single missense variant, p.(Ala327Thr), detected in homozygous states identified in a single control subject (non-neuro). This variant is predicted to be benign and tolerated by PolyPhen-2 and SIFT, respectively, and is neutral in terms of protein stability.
Clinical findings
The clinical phenotypes of ten affected individuals from seven unrelated families with deleterious biallelic SLC38A3 variants are provided in Table 2. The morphological features, neuroimaging data, EEG and head circumference charts are displayed in Fig. 3 and summarized in Table 3. Additional data and course of illness can be found in the Supplementary material.
Table 2.
Basic perinatal and clinical neurological data of subjects with biallelic variants in SLC38A3
| Family 1 | Family 2 | Family 3 | Family 4 | Family 5 | Family 6 | Family 7 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| II-2 | II-3 | II-3 | II-4a | II-1 | II-3 | II-3 | II-4 | III-1 | II-5 | ||
| Basic clinical information and perinatal history | |||||||||||
| Gender | Male | Female | Female | Female | Female | Male | Female | Male | Male | Female | |
| Current age | 6 y | 5 y | 5 y | 6 y | 4 y | 3 y | 8 y | 3 y 2 mo | 1 y 3 mo | 13 mo | |
| Delivery | SVD, FT | SVD, PT (35) | SVD, FT | SVD, FT (38) | CS, FT | CS, FT | CS, FT (40) | CS, FT (39) | CS, FT | CS, FT (39) | |
| Perinatal complications | None | IUGR, HMD, jaundice | Low foetal movement | None | None | None | None | None | None | None | |
| OFC at birth | N/A | 30 cm | 32 cm | N/A | N/A | N/A | 31.5 cm | 32.5 cm | 33 cm | N/A | |
| (z-score) | (−3.26 SD) | (−1.9 SD) | (−2.2 SD) | (−1.5 SD) | (−1.3 SD) | ||||||
| Weight-birth | N/A | 3 kg | 2.8 kg | 2 kg | 4.3 kg | 3.5 kg | 3.2 kg | 3 kg | 3.2 kg | N/A | |
| (z-score) | (−0.8 SD) | (−1.2 SD) | (−2.7 SD) | (+1.8 SD) | (−0.1 SD) | (−0.4 SD) | (−0.9 SD) | (−0.6SD) | |||
| Neurological, developmental, and other clinical features | |||||||||||
| Age at last exam | 3 y | 5 y | 5 y | 5 y | 3 y 4 mo | 2 y | 7 y | 2 y 2 mo | 1 y 3 mo | 13 mo | |
| OFC at last exam | 46.5 cm | 44 cm | 47 cm | 42 cm | 51 cm | 43 cm | 46 cm | 43.8 cm | 44 cm | 43 cm | |
| (z-score) | (−2.5 SD) | (−4.5 SD) | (−2.4 SD) | (−6 SD) | (+1.4 SD) | (−4 SD) | (−4.4 SD) | (−3.4 SD) | (−2.4 SD) | (−1.82 SD) | |
| Axial hypotonia | + | + | + | + | + | + | + | + | + | + | 10/10 |
| Absent speech | + | + | + | + | + | + | + | + | + | + | 10/10 |
| GDD/ID | + | + | + | + | + | + | + | + | + | + | 10/10 |
| Visual impairment | + (CRD/CB) | + (CRD/CB) | + | − | + | + (CRD/CB) | + | + | + | + (CB) | 9/10 |
| Microcephaly | + | + (P)b | + (P) | + | − | + | + (P) | + (P) | + (P) | – | 8/10 |
| Seizures (onset) | + (4 mo) | + (10 mo) | + (15 mo) | + (1 w) | − | + (5 mo) | + (6 mo) | + (5 mo) | − | + (4 mo) | 8/10 |
| DRE (current ASMs) | + (PB, LEV, CLZ, VGB, keto) | + (PB, CLB, VGB, keto) | − (OXC, CLZ) | + (TPM, VPA) | − | + (TPM, VPA, CLZ) | − (VPA, LEV) | − (LEV) | − | + (PB, CLB, keto) | 5/10 |
| Seizure type and frequency | Tonic/4–11per day | ‘gelastic’/0–2 per day | Focal status epilepticus/4 life-time | GTC/daily | N/A | GTC | GTC/5 life-time (febrile; last 4 y ago) | Focal with generalization/once | N/A | Generalized myoclonic and tonic/∼10 per day | |
| Epileptic encephalopathy | + | + | + | + | − | − | − | − | − | + | 5/10 |
| Peripheral hypertonia | + | + | + | − | − | + | + | + | + | + | 8/10 |
| Constipation | + | + | + | + | − | − | + | + | + | + | 8/10 |
| Dysphagia | + (GT) | + (GT) | − | + | − | + | + | + | − | + (GT) | 7/10 |
| FTT | − | + | − | + | − | + | + | − | − | − | 4/10 |
| Hyperreflexia | + | + | − | − | − | + | − | − | − | + | 4/10 |
| Movement disorder | + | + | − | − | − | − | − | − | + | − | 3/10 |
| Other features | Behavioural problems | Behavioural problems | GERD; hepatomegaly; situs inversus totalis; ASD | Behavioural problems | GERD; behavioural problems | Behavioural problems | Recurrent stridor; Abnormal supra-glottis; s/p supra-glottoplasty | ||||
ASD = atrial septal defect; ASM = anti-seizure medication; CB = cortical blindness; CLB = clobazam; CLZ = clonazepam; CS = Caesarean section; CRD = cone-rod dystrophy; DRE = drug-resistant epilepsy; FT = full term; FTT = failure to thrive; GDD = global developmental delay; GERD = gastroesophageal reflex disease; GTC = generalized tonic clonic; GT = gastric tube; HMD = hyaline membrane disease; IUGR = intrauterine growth restriction; keto = ketogenic diet; LEV = levetiracetam; N/A = not available; OFC = occipitofrontal circumference; P = postnatal/progressive; PB = phenobarbital; PT = preterm; VGB = vigabatrin; SVD = spontaneous vaginal delivery; TPM = topiramate; VPA = valproic acid; w/mo/y = week/month/year; ZNS = zonisamide.
Previously published with limited clinical data.
Based on an OFC of 30 cm (−3.26 SD) at birth and 43.5 (−3SD) at 2.5 years of age
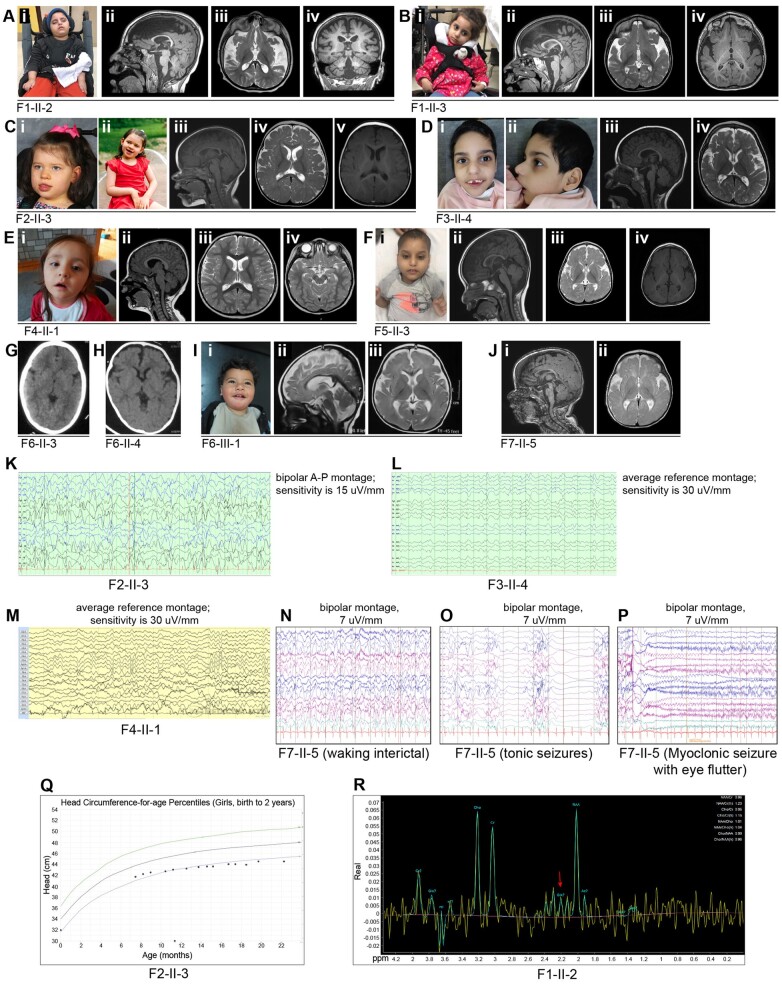
Figure 3.
Facial features, brain images, neurophysiologic studies, and head circumference chart of affected individuals from Families 1–7 with biallelic variants in SLC38A3. [A(i)] Photograph of Individual II-2 (Family 1) at 6 years of age. [A(ii–iv)] MRI brain of Individual II-2 from Family 1 at 6 years of age showing foreshortening of corpus callosum (CC), moderate to severe cerebral volume loss, thin brainstem and cervical spine [A(ii) T1 mid-sagittal sequence], under-opercularization and widening Sylvian fissure, abnormal myelination for age [A(iii) T2 axial sequence], mild (infero-lateral), and cerebellar atrophy [A(iv) T1 coronal sequence]. [B(i)] Photograph of Individual II-3 (Family 1) at 4 years of age. [B(ii–iv)] MRI brain of Individual II-3 from Family 1 at 4.5 years of age showing foreshortening of CC, moderate cerebral and mild cerebellar volume loss [B(ii) T1 mid-sagittal sequence], under-opercularization and widening Sylvian fissure, abnormal myelination for age [B(iii) T2 axial sequence, and B(iv); T1 axial sequence]. [C(i and ii)] Photographs of Individual II-3 (Family 2) at 5 years of age. [C(iii–v)] MRI brain of Individual II-3 from Family 2 at 9 months of age showing hypo-intensity of splenium of CC secondary to recent seizure or a remote infarct [C(iii) T1 mid-sagittal sequence], and normal myelination for age [C(iv) T2 axial sequence, and C(v) T1 axial sequence]. [D(i and ii)] Facial photographs of Individual II-4 (Family 3) at 6 years of age. [D(iii and iv)] Brain MRI of Individual II-4 from Family 3 at 2.5 years of age showing mild cerebral volume loss, slight foreshortening of CC [D(iii) T1 mid-sagittal sequence], under-opercularization and widening Sylvian fissure, and delayed myelination for age [D(iv) T2 axial sequence]. [E(i)] Photograph of Individual II-1 (Family 4) at 4 years of age. [E(ii)] Brain MRI (mid-sagittal T1 sequence) of Individual II-1 from Family 4 at 3 years of age showing normal brain structures [E(iii and iv)] axial sequences showing normal myelination for age. [F(i)] Photographs of Individual II-3 (Family 5) at 3 years of age. [F(ii–iv)] MRI brain of Individual II-3 from Family 5 at 21 months of age showing borderline low-normal body of CC, and mild cerebral volume loss [F(ii)] T1 mid-sagittal sequence), under-opercularization and widening Sylvian fissure, and abnormal myelination for age. [F(iii)] T2 axial sequence, and (iv) T1 axial sequence. (G) Head CT of Individual II-3 from Family 6 showing normal brain parenchyma. (H) Head CT of Individual II-4 from Family 6 showing under-opercularization with normal brain parenchyma. [I(i)] Photographs of Individual III-1 (Family 6) at 15 months of age. [I(ii and iii)] MRI brain of Individual III-1 (Family 6) showing mild cerebral atrophy, thin CC, mild superior vermian volume loss and delayed myelination for age [I(ii) T2 mid-sagittal sequence and (iii) T2 axial sequence]. [J(i and ii)] MRI brain of Individual II-5 from Family 7 at 6 months of age showing under-opercularization and widening Sylvian fissure, and abnormal myelination for age [J(i) T1 mid-sagittal sequence, and (ii) T2 axial sequence]. (K) EEG epoch (11 s) of Family 2, Individual II-3 (bipolar montage; sensitivity 15 μV/s) showing background slowing and abundant multifocal epileptiform activity. (L) EEG epoch (17 s) of Family 3, Individual II-4 (average reference montage; sensitivity 30 μV/s) showing background slowing and multifocal epileptiform activity. (M) EEG epoch (10 s) of Family 4, Individual II-1 (average reference montage; sensitivity 30 μV/s) showing a mild background slowing. (N) EEG epoch (10 s) of Family 7, Individual II-5 (bipolar montage; sensitivity 7 μV/s) at 12 months showing diffuse background slowing and near-continuous high amplitude spike and slow-wave epileptiform activity over posterior regions. (O) EEG epoch (11 s) of Family 7, Individual II-5 (bipolar montage; sensitivity 7 μV/s) at 12 months during a tonic seizure. (P) EEG epoch (10 s) of Family 7, Individual II-5 (bipolar montage; sensitivity 7 μV/s) at 12 months during a myoclonic seizure. (Q) CDC head circumference chart from birth to 2 years of age for Family 2, Individual II-3 showing a progressive/postnatal microcephaly pattern. (R) MRS of Family 1, Individual II-2. The red arrow points to the low glutamate-glutamine (Gx) peak. A-P = anterior-posterior.
Table 3.
Clinical investigations (MRI, EEG and metabolic laboratory tests) of subjects with biallelic variants in SLC38A3
| Family 1 | Family 2 | Family 3 | Family 4 | Family 5 | Family 6 | Family 7 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| II-2 | II-3 | II-3 | II-4a | II-1 | II-3 | II-3 | II-4 | III-1 | II-5 | ||
| Neurophysiological studies (EEG) | |||||||||||
| Diffuse background slowing | + | + | + | + | − | + | − | N/A | − | + | 6/10 |
| Epileptiform activity | + | + | + | + | − | − | − | + | − | + | 6/10 |
| LTM-EEG | Frequent tonic seizures with eyelid myoclonia over 48 h | 20 GTC and 4 focal seizures over 24 h | N/A | N/A | N/A | N/A | N/A | N/A | N/A | Frequent generalized myoclonic and tonic seizures over 48 h | |
| Brain imaging | |||||||||||
| Study type | Brain MRI | Brain MRI | Brain MRI | Brain MRI | Brain MRI | Brain MRI | Head CT | Head CT | Brain MRI | Brain MRI | |
| Cerebral atrophy | + | + | − | + | − | + | − | − | + | − | 5/10 |
| Cerebellar atrophy | + | + | − | − | − | − | − | − | − | − | 2/10 |
| CC abnormality | + | + | + | + | − | + | − | − | + | − | 6/10 |
| Under-opercularization | + | + | − | + | − | − | − | + | + | + | 6/10 |
| Abnormal myelination | + | + | − | + | − | + | − | − | + | + | 6/10 |
| Others | Thin brainstem | Generous extra-axial CSF spaces | – | Dolichocephaly, mild posterior plagiocephaly | Generous extra-axial CSF spaces; mild superior vermian hypoplasia | Generous extra-axial CSF spaces | |||||
| Brain MRS findings | Low NAA and Glx peaks | Lactate peak | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| Metabolic laboratory studies | |||||||||||
| Plasma lactate and pyruvate | Normal | Transient elevation of lactate | Transient metabolic acidosis (due to TPM use) | Transient elevation of lactate and pyruvate | Transient elevation of lactate and pyruvate | Normal | Slightly elevated lactate | Transient elevation of lactate; normal pyruvate | Elevated lactate | Transient elevation of lactate | |
| Plasma ammonia | Normal | Elevated (62 µmol/l) | N/A | N/A | Normal (25 µmol/l) | N/A | Elevated (52 µmol/l) | Elevated (56 µmol/l) | Elevated (87 µmol/l) | Normal (44 µmol/l) | |
| PAA | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | Normal | |
| UOA | Normal | Normal | Normal | N/A | Normal | Normal | Normal | Normal | Normal | Normal | |
| CSF amino acids | Normal | Normal | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| CSF neurotransmitters | Normal | Normal | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| CSF lactate | N/A | Normal | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
CC = corpus callosum; c/w = consistent with; Glx = glutamate-glutamine; GTC = generalized tonic-clonic; LTM-EEG = long-term monitoring EEG; MRS = magnetic resonance spectroscopy; NAA = N-acetylaspartate; N/A = not available; PAA = plasma amino acids; TPM = topiramate; UOA = urine organic acids.
Previously published with limited clinical data.
All 10 subjects had global developmental delay (GDD), were non-verbal, and had moderate to severe axial hypotonia rendering them either non-ambulatory or wheelchair dependent (9/10) or ambulatory only for a short distance without assistance and with ataxic gait (1/10) (Family 4, Individual II-1). The GDD was profound in nine subjects who also had early-onset seizures in the first 2 years of life and microcephaly (except for Family 6, Individual III-1 who had microcephaly but had not had seizure onset by 15 months of age). Individual II-1 in Family 4 was the least severely affected in that she had moderate to severe GDD but lacked the early-onset seizures and microcephaly. Occipitofrontal circumference (OFC) at birth, available in only five subjects, was borderline low to low (z-score: −1.3 to −3.3) with subsequent progression to microcephaly (z-score: −2.4 to −4.5) later in life consistent with a postnatal/progressive microcephaly pattern e.g. Individual II-3 in Family 2 (Fig. 3Q). In one subject (Family 1, Individual II-3), the progressive microcephaly pattern was also evident on serial postnatal head measurements in which OFC progressed from a z-score of −3 standard deviations (SD) at 2.5 years of age to −4.5 SD at 4 years of age. Visual impairment was present in all affected individuals except for Family 3, Individual II-4, in the form of cone-rod retinal dystrophy (3/9; Family 1, Individuals II-2 and II-3, and Family 5, Individual II-3) without or with cortical blindness (2/9; Family 1, Individuals II-2 and II-3), isolated cortical blindness in 1/9 (Family 7, Individual II-5), or unclassified visual impairment (5/9). Peripheral hypertonia (8/9), constipation (8/9) and dysphagia (7/9), were also common features. Five of ten subjects (50%) had drug-resistant epilepsy. Three (3/5) of these individuals (Family 1, Individuals II-2 and II-3, and Family 7, Individual II-5) were managed with a ketogenic diet, and two of these were siblings (Family 1, Individuals II-2 and II-3) that were also treated with vigabatrin and benzodiazepines, including clobazam and clonazepam. Vigabatrin was considered most effective in improving seizure control in the siblings, per parental report, followed by benzodiazepines. One of these two siblings (Family 1, Individual II-3) had seizure exacerbation (to six seizures/day) after weaning vigabatrin, with improved seizure control down to one to two seizures/day immediately after reintroducing vigabatrin. A remote long-term EEG in this individual (Family 1, Individual II-3) had documented up to 24 seizures per day in the past. The ketogenic diet was reportedly partially effective in the siblings (Family 1, Individuals II-2 and II-3) but ineffective in the third subject (Family 7, Individual II-5).
Less common features included failure to thrive in four subjects (Family 1, Individual II-3; Family 3, Individual II-4; Family 5, Individual II-3; Family 6, Individual II-3) and movement disorders (chorea and/or oro-motor dyskinesia) that were present in siblings Individuals II-2 and II-3 from Family 1 and in Individual III-1 from Family 6 and was not thought to be related to medication use.
Brain imaging was available for all subjects including eight brain MRIs and two head CTs (Fig. 3 and Table 3). Abnormalities included corpus callosum abnormalities (6/10) [in the form of foreshortening (3/6), low-normal thickness (1/6), thinning (1/6) or hypo-intensities (1/6)], under-opercularization and widening of the Sylvian fissure (6/10), abnormal myelination for age (6/10) and various degrees of cerebral atrophy (5/10). Other less frequent findings included mild cerebellar atrophy (2/10) and a thin brainstem (1/10). Brain magnetic resonance spectroscopy (MRS) performed in two affected siblings (Family 1, Individuals II-2 and II-3) showed an isolated elevated lactate peak in the proband (Family 1, Individual II-3; per report) and low N-acetylaspartate (NAA) and low glutamate-glutamine (Glx) peak in the sibling (Family 1, Individual II-2; Fig. 3R). Additional sequences or images were not obtained at the time to isolate the glutamine from the glutamate peak.
EEG findings in five subjects with epilepsy (Family 1, Individuals II-2 and II-3, Family 2, Individual II-3, Family 3, Individual II-4, and Family 7, Individual II-5) were consistent with epileptic encephalopathy (Fig. 3K, L and N and Table 2). One subject had a burst-suppression during sleep in infancy (Family 1, Individual II-2) while two others had a hypsarrhythmia pattern (Family 2, Individual II-3 and Family 7, Individual II-5). The hypsarrhythmia pattern subsequently evolved in one of the subjects into a slow (<2 Hz) generalized spike and slow wave pattern compatible with Lennox-Gastaut syndrome (Family 2, Individual II-3). Reported seizure types included generalized tonic-clonic seizures (3/8), tonic seizures (2/8), focal seizures (2/8) with secondary generalization or with evolution into focal status epilepticus, myoclonic seizures (1/8) and ‘gelastic’ seizures (1/8). The associated electrographic patterns of the tonic and myoclonic seizures in one subject (Family 7, Individual II-5) are displayed in Fig. 3O and P.
Biochemical and metabolic screening
The biochemical and metabolic findings are summarized in Table 3. Plasma amino acids profile was reportedly normal in all 10 affected individuals. Urine organic acids analysis was also reported normal in nine subjects and unavailable in one subject (Family 3, Individual II-4). CSF amino acids including GABA, glutamine, and a neurotransmitters profile including 5-hydroxyindoleacetic acid, homovanillic acid and 3-O-methyldopa performed in two siblings (Family 1, Individuals II-2 and II-3) were reported normal, while CSF lactate, alpha aminoadipic semialdehyde, succinyladenosine, tetrahydrobiopterin, and neopterin performed only in one child (Family 1, Individual II-3) were also normal. Plasma ammonia level was elevated in four children (Family 1, Individual II-3, and Family 6, Individuals II-3, II-4 and III-1) and normal in three subjects (Family 1, Individual II-2, Family 4, Individual II-1 and Family 7, Individual II-5). Interestingly, eight subjects had intermittent metabolic acidosis with transient elevation of serum lactate with or without elevation of pyruvate (Family 1, Individual II-3; Family 2, Individual II-3; Family 3, Individual II-4; Family 4, Individual II-1; Family 6, Individuals II-3, II-4, and III-1; and Family 7, Individual II-5) that was not associated with seizure activity.
Untargeted metabolomic profiles show altered amino acid and nitrogen metabolism
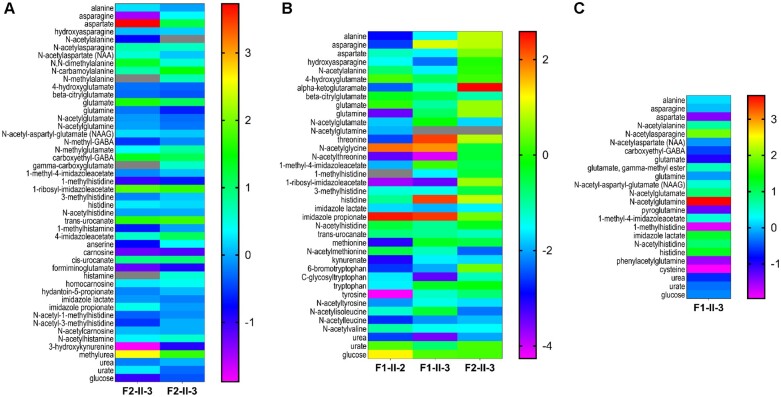
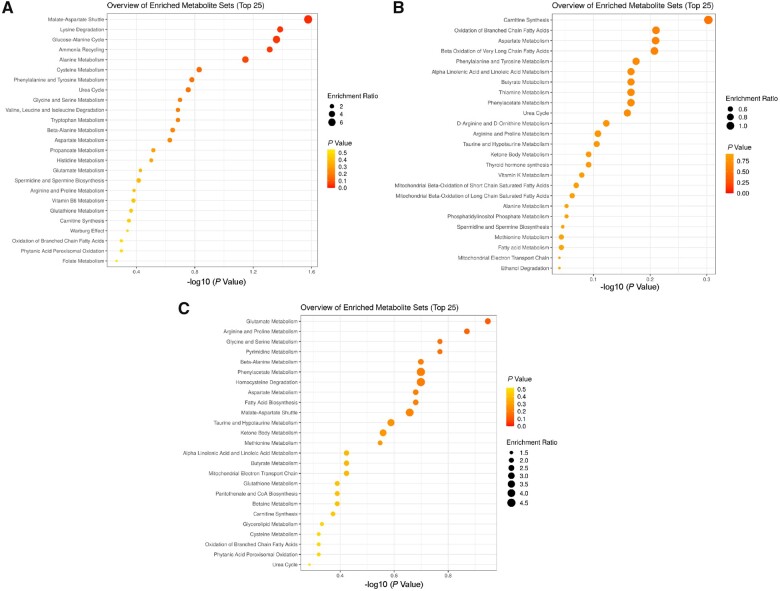
Untargeted metabolomic analyses were performed on biofluids available from a total of three subjects (Family 1, Individuals II-2 and II-3, and Family 2, Individual II-3) (Figs 4, 5, and Supplementary Table 1). Analysis of plasma from the three subjects, including two siblings (Family 1, Individuals II-2 and II-3, and Family 2, Individual II-3), CSF of one of the siblings (Family 1, Individual II-3), and the urine of one subject (Family 2, Individual II-3) to screen for possible underlying associated metabolic perturbations revealed alterations in the levels of amino acids known to be transported by SNAT3 (Figs 4 and 5). Plasma samples yielded an average of 676 z-scored molecules, and yields were 630 molecules in urine and 241 molecules in CSF (Supplementary Table 1). No consistent pattern was observed in all samples from all subjects; however, significantly elevated N-acetylglutamine (z-score: +3.65; top 1% of samples analysed), N-acetylasparagine (z-score: +1.89, top 4% of samples analysed), and 1-palmitoylglycerol (16:0) (z-score: +3.96; top 2.5% of samples analysed) and significantly low levels of aspartate and cysteine were observed in the only available CSF sample (Figs 4, 5 and Supplementary Table 1). Other lysophospholipids, phosphatidylethanolamines, and phosphatidylcholines in the CSF sample were normal, as were CSF glutamine (z-score: −0.16) and glutamate (z-score: −0.99). Additionally, plasma glutamine (z-score: −0.67), glutamate (z-score: −1.0), and N-acetylglutamate (z-score: −0.16) in the same subject (Family 1, Individual II-3) were normal, while plasma N-acetylglutamine was not detected. Plasma glutamine [z-score: −3.47; ranked ninth lowest (bottom 0.33%) among 2707 plasma sample] and N-acetylglutamine (z-score: −1.82) were significantly low in the affected sibling (Family 1, Individual II-2). While glucose was normal in all subjects tested, the plasma and urine of a third unrelated subject (Family 2, Individual II-3) revealed abnormalities in pentose phosphate pathway metabolism (Fig. 5 and Supplementary Table 1). Abnormalities in urea (z-score: −3.5 to −2 in plasma) and nitrogen metabolism were also observed in all patient samples (Figs 4 and 5). The observed abnormalities in glutamate metabolism may represent potential inter-compartmental metabolic signatures but could not be evaluated in other subjects due to lack of accessible biological fluids for analysis. The isolated 1-palmitoylglycerol (16:0) elevation in the CSF of Family 1, Individual II-3 is of unclear significance and is suspected to be secondary to diet.
Figure 4.
Metabolomic analysis reveals altered glutamine, histidine, asparagine, and tryptophan metabolism. Metabolites of amino acids transported by SNAT3 transporter: glutamine, glutamate, aspartate, asparagine, and histidine are depicted. Also shown are altered metabolites of tryptophan, alanine, tyrosine, methionine, proline, threonine, leucine, valine, isoleucine, urea, urate, and glucose. (A) Urine metabolomic assays of Family 2, Individual II-3 (two independent samples collected at different intervals from same subject). (B) Plasma assays of patients Family 1, Individuals II-2 and II-3, Family 2, Individual II-3; and (C) CSF assay of patient Family 1, Individual II-3. The z-scores illustrate whether a given metabolite is significantly increased (positive z-score) relative to the control reference population. In general, a z-score greater than +2 or lower than −2 is considered significantly different from the control. Higher z-scores are yellow to red in colour, lower z-scores are blue to purple in colour, as shown in legend bar. Analytes which were not detected are depicted in grey.
Figure 5.
Deficiency of SNAT3 alters multiple metabolic pathways. Perturbations of biochemical pathways altered by SNAT3 deficiency are illustrated. Enrichment analyses of metabolite alterations in biofluids from patients with SNAT3 deficiency are shown. Glutamine, asparagine, and histidine pathways are significantly affected as illustrated in A. CSF (n = 1), B. Plasma (n = 3), and C. Urine (n = 2). SNAT3 and its role in glutamine, tryptophan, histidine, and aspartate transport are critical, illustrating the systemic need for glutamine homeostasis. Data analysed with MetaboAnalyst 5.0, as described in the ‘Materials and methods’ section.
Discussion
Genetic, genomic, bioinformatic analyses and database mining identified 10 individuals from seven families who each share biallelic predicted-pathogenic variants in SLC38A3. Detailed clinical characterization of these 10 individuals implicates this gene as a cause of DEE. An early potential emerging genotype–phenotype correlation suggests that the five subjects with potential complete loss-of-function (splicing and nonsense variant alleles) have the most severe phenotype as evidenced by the degree of cerebral and cerebellar volume loss (4/5 and 2/5, respectively) and under-opercularization (5/5) on neuroimaging and the exclusive presence of epilepsy refractory to anti-convulsant therapies (5/5). Three out of these five subjects also had cone-rod dystrophy. The three subjects with either the compound heterozygous missense variants c.886G>A; p.(Ala296Thr) and c.1160C>A; p.(Pro387Gln) or the homozygous missense variant c.1123A>C; p.(Thr375Pro) have drug-responsive (controlled) epilepsy (or no epilepsy by 15 months of age in Family 6, Individual III-1) and less volume loss on neuroimaging, while the missense variant c.622C>G; p.(Arg208Gly) results in the least severe clinical phenotype (ambulation with assistance, no epilepsy or microcephaly). This may be consistent with the structure and function perturbations predicted by protein modelling: the missense variant p.(Arg208Gly) is predicted to have a weaker affinity for phosphate and, by extension, possibly a native unknown organophosphate target that may control its localization, while the other missense variants either affect the closing of the cytosolic opening [p.(Ala296Thr), p.(Pro387Gln)] or the stabilization of SNAT3 [p.(Thr375Pro)]. The splicing and nonsense variants are predicted to result in a non-functional truncated protein or an unstable mutant mRNA subject to nonsense-mediated decay.
SLC38A3 encodes SNAT3, a bidirectional neutral (system N) amino acid (Asn, His, Gln) transporter expressed in brain, liver, kidney, retina, and pancreas.12 In the brain, SNAT3 localizes to peri-synaptic astrocytes and plays an important role in replenishing glutamate and the GABA neurotransmitter pool through the glutamate/GABA-glutamine cycle (Fig. 2A).12Snat3-deficient (Slc38a3−/−) mice have reductions in brain glutamate and GABA neurotransmitter pools (∼50% and ∼75% of normal levels detected in homogenized brain tissue, respectively).36 GABA is a major inhibitory neurotransmitter, and depletion/reduction of GABA or abnormal GABA signalling, ‘GABA-pathies’, results in a wide range of DEE.37 Although GABA surrogate analytes in plasma samples and a single CSF sample of select subjects in our study were normal (Supplementary Table 1), we cannot rule out a reduction or dysregulation of GABA in specific neuronal cell types. Glutamate, in contrast, is the main excitatory neurotransmitter, and excessive glutamate is pro-convulsive and neurotoxic, but the effect of glutamate depletion on the neurons is not fully understood.38
Defects in multiple proteins involved in glutamate/GABA-glutamine cycle are known to cause DEE and severe neurodevelopmental disorders. These include glutamate decarboxylase (GAD1 ‘GAD67’, MIM* 605363), excitatory amino acid transporter 2 (SLC1A2/EAAT2, MIM* 600300), glutaminase (GLS) ‘previously known as phosphate-activated glutaminase’ (MIM* 138280) and glutamate-ammonia ligase (GLUL) ‘also known as Glutamine synthetase (GS)’ (MIM* 138290) (Fig. 2A).5 The proposed pathophysiology of these disorders ranges from depletion of GABA (as in GAD1 deficiency) to depletion of glutamate and excess glutamine (as in GLS deficiency) or through glutamate toxicity due to reduced uptake of extracellular glutamate (as in SLC1A2/EAAT2 defects).6,39–41
In addition to the prominent role of SNAT3 in brain function, SNAT3 is also involved in urea formation in the liver, pH regulation in the kidney, and insulin secretion in the pancreas.12 Urea was consistently low in all available biological samples (as low as -3.5 to -2 SD in plasma), and plasma ammonia was elevated in four subjects consistent with a role for SNAT3 in hepatocytes in providing glutamine for ammonia detoxification and urea formation.12 Interestingly, Snat3-deficient mice, in contrast, had “unexpected” elevation of plasma urea with normal ammonia levels.12,36 The absence of reported protein intolerance in our subjects is highly dependent on the type of diet consumed.
The role of SNAT3 in providing glutamine for ammoniagenesis in the renal proximal tubules during metabolic acidosis could potentially explain the transient metabolic acidosis observed in eight subjects with potential loss-of-function of SNAT3.42 Additionally, nine subjects had visual impairment; three had cone-rod retinal dystrophy, which is potentially compatible with retinal expression of SNAT3.43 These eye findings in two subjects (Family 1, Individuals II-2 and II-3) preceded the use of vigabatrin which, in contrast to the central vision loss in cone-rod dystrophy, may result in loss of peripheral vision and increased retinal thickness.44 None of our subjects had reported liver or endocrine problems at the ages studied.
SNAT5 has a similar expression pattern and performs many of the functions provided by SNAT3; yet, the abnormalities observed in our subjects and Snat3-knockout mice suggest that SNAT3 function cannot be fully compensated.12 Snat3-deficient mice also show stunted growth, lethargy and uncoordinated ‘ataxic’ gait, altered plasma amino acids, normal plasma glutamine, abundance of brain glutamine, and early lethality.36 Four of our subjects had stunted growth, but none had any detectable altered plasma amino acid profile per report; yet, it is possible that subtle abnormalities not fitting a specific disorder may have existed but were interpreted as ‘non-significant or normal”’. Alternatively, since available biochemical assays assess non-cellular fluids, it is possible that the astrocytes are retaining glutamine and the redundancy of SNAT5 and the possible upregulation of choroid plexus LAT2 could have moderated the glutamine phenotype.12,45
Additionally, similar to the null mice, severe gait impairment and hypotonia were prominent neurological features we observed in association with human SLC38A3 potential loss-of-funtion alleles. Microcephaly and early-onset seizures were additional features documented only in the human subjects. Early lethality in mice may have possibly masked some of these latter features and the natural course of illness observed in human subjects.
Glutamine is a precursor for α-ketoglutarate, which enters the TCA cycle to generate substrates for gluconeogenesis, the process of producing glucose for cells.12 Additionally, SNAT3 is involved in energy metabolism through providing glutamine for the glutamine-induced insulin secretion in the β-cells in the pancreas. Snat3-deficient mice were reported to have hypoglycaemia; however, while none of our subjects had documented hypoglycaemia, this is highly dependent on the type of diet, feeding intervals, and compensatory mechanisms. Abnormalities of the pentose phosphate pathway (PPP) observed in the plasma of one affected individual (Family 2, Individual II-3) could be reflective of altered glucose metabolism leading to activation of PPP as an alternative pathway to generate energy.46,47
Untargeted metabolomic analysis–a semi-quantitative study of all small metabolites in a given biological specimen–is a promising technique to clinically unravel novel metabolic biomarkers in various neurodevelopmental and neurometabolic disorders.31,48,49 Exploring such biomarkers resulting from perturbations in underlying biochemical pathways are not only important for early screening and diagnostic purposes but can also lay the groundwork for development of precise treatment and monitoring of disease course and treatment response.49,50 Biochemical profiling in SLC-related disorders and epilepsies have aided in identifying novel metabolic signatures.51 CSF metabolomics in one subject yielded a markedly elevated N-acetylglutamine, the acetylated form of glutamine found in all human biofluids including urine, and normal plasma glutamine, while N-acetylglutamine and glutamine were low in the plasma of the affected sibling.52 SNAT3 is expressed in the CSF-facing membrane of the choroid plexus epithelium and is thought to play a role in the influx of glutamine into the brain through the blood–brain barrier (Fig. 2A).45 Yet, the aforementioned observations are consistent with the finding of markedly elevated glutamine in whole brain tissue of Snat3-knockout mice and support the role of SNAT3 in the export rather than the import of glutamine into the brain through the blood–brain barrier.36 Notably, abnormalities in glutamine were also reported in two other conditions resulting from enzyme defects involved in glutamate/GABA-glutamine cycle, including low serum and CSF glutamine in GLUT deficiency and significantly elevated plasma glutamine (up to z-score of 11.7) in GLS deficiency.41
Additionally, one subject interestingly had a low pooled glutamate-glutamine peak on MRS. It is not clear if both or one of the two metabolites (and which one) derived this pooled effect; yet, this observation indicates diminished glutamate-glutamine cycling in the brain.53 Significantly low NAA and Glx peaks have been reported in the brain of subjects with autism and in sclerotic hippocampus of subjects with epilepsy.54,55
The two siblings from Family 1 were started on vigabatrin prior to the molecular finding of SLC38A3 variant, with a favourable response in both children and a three-fold reduction in seizure frequency in one of the siblings. These two siblings were reportedly on a ketogenic diet at time of sample collection, although the result of metabolomics analysis did not show the expected metabolomics signature, which typically shows low glucose and elevated ketones in subjects on ketogenic diet.50 We suspected that this discrepancy is related to poor compliance to the diet regimen. A favourable response to benzodiazepines was also reported in the same siblings. Yet, both siblings continued to have severe neurological impairment and remained at baseline neurological function apart from seizure improvement. These observations are compatible with the mechanism of action of these antiseizure medications and the proposed pathophysiology of the disease. Vigabatrin acts as an irreversible inhibitor of GABA transaminase, the primary GABA-degrading enzyme, which subsequently increases GABA levels at the GABAergic synapses in the brain.56 Benzodiazepines, such as clonazepam and clobazam, are positive allosteric regulators of the GABAA receptor subset and thus exhibit their anti-seizure properties through promoting GABA-induced CNS depression.57 Although we had no direct measure of GABA, the plasma metabolomics of the two siblings showed an increased level (z-scores of 3.4 to 4.7) of the vigabatrin-associated metabolite, 2-pyrrolidinone, as a surrogate measure of GABA-transaminase inhibition (Supplementary Table 1).58 Increased GABA levels or activity may partially restore some of the expected GABA reduction or dysregulation associated with SNAT3 defects. Similar improvement in seizure control with vigabatrin was also observed in biallelic GAD1 variants resulting in GABA depletion through interruption of the GABA-glutamine cycle.39 The retrospective nature of the study, however, limited the ability to assess this response further in additional subjects. Follow-up studies to further explore the response to these medications are warranted.
In summary, we have characterized the clinical features in ten subjects from seven unrelated families with biallelic deleterious variants in SLC38A3 and provide compelling evidence that functional deficiency of SNAT3 can cause DEE. The abnormalities in glutamate metabolism including low plasma glutamine, N-acetylglutamine, and urea and elevated CSF N-acetylglutamine in select subjects may indicate the role for SNAT3 across the blood–brain barrier and potentially represent metabolic markers of the disease. Further studies on the metabolomic profile in CSF and plasma of other individuals with this rare disease are essential to consolidate and further explore these findings. Identifying additional subjects will also unravel the full spectrum of variant alleles and biology of this primarily brain dysfunction and seizure disorder. Finally, further studies on the effect of the variants identified and the mechanistic link between a decreased neurotransmitter pool and epilepsy could improve our understanding of this disorder and other related seizure disorders and DEEs.
Supplementary Material
Acknowledgements
We thank all families for their participation in this study. We also would like to thank Ms Theresa Wilsons, M.S, R.D. for her assistance in the sample collection and transportation for the metabolomics studies.
Funding
This study was supported in part by the U.S. National Human Genome Research Institute (NHGRI) and National Heart Lung and Blood Institute (NHBLI) to the Baylor-Hopkins Center for Mendelian Genomics (BHCMG, UM1 HG006542, J.R.L); NHGRI grant to Baylor College of Medicine Human Genome Sequencing Center (U54HG003273 to R.A.G.), U.S. National Institute of Neurological Disorders and Stroke (NINDS) (R35NS105078 to J.R.L.) and Muscular Dystrophy Association (MDA) (512848 to J.R.L.). Qatar National Research Fund (QNRF) also supported this study (NPRP11S-0110–180250 to K.F.). Family 4 was collected as part of the SYNaPS Study Group collaboration funded by The Wellcome Trust and strategic award (Synaptopathies) funding (WT093205 MA and WT104033AIA). This research was also supported in part by the Queen Square Genomics group at University College London, supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre (MR/S01165X/1, MR/S005021/1, G0601943), as well as Rosetree Trust, Ataxia UK, MSA Trust, Brain Research UK, Sparks GOSH Charity, Muscular Dystrophy UK (MDUK), and Muscular Dystrophy Association (MDA USA). D.M. was supported by a Medical Genetics Research Fellowship Program through the United States National Institute of Health (T32 GM007526-42). M.P.F and J.C.T are supported by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre Programme and a Wellcome Trust Core Award [203141/Z/16/Z]. T.M. is supported by the Uehara Memorial Foundation. D.P. is supported by International Rett Syndrome Foundation (IRSF), Grant/Award Number: #3701–1. J.E.P. was supported by NHGRI K08 HG008986. S.D. is supported by Isabelle Rapin Scholar Recipient for clinical research in autoimmune encephalitis.
Competing interests
J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Genetics Center, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics (BG). C.B. and V.K. are employees of Centogene. E.T. and S.V.M. are employees of GeneDx, Inc. C.G is an employee of BG. All other authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Abbreviations
- AOH
absence of heterozygosity
- DEE
developmental and epileptic encephalopathy
- ROH
runs of homozygosity
- SNAT3
sodium-coupled neutral amino acid transporter 3
- SLC
solute carrier
References
- 1. Klassen T, Davis C, Goldman A, et al. . Exome sequencing of ion channel genes reveals complex profiles confounding personal risk assessment in epilepsy. Cell. 2011;145(7):1036–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McTague A, Howell KB, Cross JH, Kurian MA, Scheffer IE. The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol. 2016;15(3):304–316. [DOI] [PubMed] [Google Scholar]
- 3. Hu C, Tao L, Cao X, Chen L. The solute carrier transporters and the brain: Physiological and pharmacological implications. Asian J Pharm Sci. 2020;15(2):131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang Y, Zhang Y, Sun K, Meng Z, Chen L. The SLC transporter in nutrient and metabolic sensing, regulation, and drug development. J Mol Cell Biol. 2019;11(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43(Database issue):D789–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guella I, McKenzie MB, Evans DM, et al. ; Deciphering Developmental Disorders Study. De Novo mutations in YWHAG cause early-onset epilepsy. Am J Hum Genet. 2017;101(2):300–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jang SS, Kim SY, Kim H, et al. . Diagnostic yield of epilepsy panel testing in patients with seizure onset within the first year of life. Front Neurol. 2019;10:988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Møller RS, Dahl HA, Helbig I. The contribution of next generation sequencing to epilepsy genetics. Expert Rev Mol Diagn. 2015;15(12):1531–1538. [DOI] [PubMed] [Google Scholar]
- 9. Punetha J, Mackay-Loder L, Harel T, et al. . Identification of a pathogenic PMP2 variant in a multi-generational family with CMT type 1: Clinical gene panels versus genome-wide approaches to molecular diagnosis. Mol Genet Metab. 2018;125(3):302–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Palmer EE, Sachdev R, Macintosh R, et al. . Diagnostic yield of whole genome sequencing after nondiagnostic exome sequencing or gene panel in developmental and epileptic encephalopathies. Neurology. 2021;96(13):e1770–e1782. [DOI] [PubMed] [Google Scholar]
- 11. Brnich SE, Rivera-Muñoz EA, Berg JS. Quantifying the potential of functional evidence to reclassify variants of uncertain significance in the categorical and Bayesian interpretation frameworks. Hum Mutat. 2018;39(11):1531–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rubio-Aliaga I, Wagner CA. Regulation and function of the SLC38A3/SNAT3 glutamine transporter. Channels (Austin). 2016;10(6):440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karaca E, Harel T, Pehlivan D, et al. . Genes that affect brain structure and function identified by rare variant analyses of Mendelian neurologic disease. Neuron. 2015;88(3):499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karczewski KJ, Francioli LC, Tiao G, et al. ; Genome Aggregation Database Consortium. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36(10):928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Monies D, Abouelhoda M, Assoum M, et al. . Lessons learned from large-scale, first-tier clinical exome sequencing in a highly consanguineous population. Am J Hum Genet. 2019;104(6):1182–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karaca E, Posey JE, Coban Akdemir Z, et al. . Phenotypic expansion illuminates multilocus pathogenic variation. Genet Med. 2018;20(12):1528–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rehder CW, David KL, Hirsch B, Toriello HV, Wilson CM, Kearney HM. American College of Medical Genetics and Genomics: Standards and guidelines for documenting suspected consanguinity as an incidental finding of genomic testing. Genet Med. 2013;15(2):150–152. [DOI] [PubMed] [Google Scholar]
- 19. Duan R, Saadi NW, Grochowski CM, et al. . A novel homozygous SLC13A5 whole-gene deletion generated by Alu/Alu-mediated rearrangement in an Iraqi family with epileptic encephalopathy. Am J Med Genet A. 2021;185(7):1972–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chaudhury S, Lyskov S, Gray JJ. PyRosetta: A script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics. 2010;26(5):689–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Conway P, Tyka MD, DiMaio F, Konerding DE, Baker D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 2014;23(1):47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lei HT, Ma J, Sanchez Martinez S, Gonen T. Crystal structure of arginine-bound lysosomal transporter SLC38A9 in the cytosol-open state. Nat Struct Mol Biol. 2018;25(6):522–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Falkenburger BH, Jensen JB, Dickson EJ, Suh BC, Hille B. Phosphoinositides: Lipid regulators of membrane proteins. J Physiol. 2010;588(Pt 17):3179–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yen H-Y, Hoi KK, Liko I, et al. . PtdIns(4,5)P2 stabilizes active states of GPCRs and enhances selectivity of G-protein coupling. Nature. 2018;559(7714):423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: Orientations of Proteins in Membranes database. Bioinformatics. 2006;22(5):623–625. [DOI] [PubMed] [Google Scholar]
- 28. Alford RF, Fleming PJ, Fleming KG, Gray JJ. Protein Structure Prediction and Design in a Biologically Realistic Implicit Membrane. Biophys J. 2020;118(8):2042–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ferla MP, Pagnamenta AT, Damerell D, Taylor JC, Marsden BD. MichelaNglo: Sculpting protein views on web pages without coding. Bioinformatics. 2020;36(10):3268–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kennedy AD, Pappan KL, Donti TR, et al. . Elucidation of the complex metabolic profile of cerebrospinal fluid using an untargeted biochemical profiling assay. Mol Genet Metab. 2017;121(2):83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miller MJ, Kennedy AD, Eckhart AD, et al. . Untargeted metabolomic analysis for the clinical screening of inborn errors of metabolism. J Inherit Metab Dis. 2015;38(6):1029–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ford L, Kennedy AD, Goodman KD, et al. . Precision of a Clinical Metabolomics Profiling Platform for Use in the Identification of Inborn Errors of Metabolism. J Appl Lab Med. 2020;5(2):342–356. [DOI] [PubMed] [Google Scholar]
- 33. Alaimo JT, Glinton KE, Liu N, et al. . Integrated analysis of metabolomic profiling and exome data supplements sequence variant interpretation, classification, and diagnosis. Genet Med. 2020;22(9):1560–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Berner J, Elsea S. Cerebrospinal fluid metabolomics uniquely identifies pathways suggesting risk for anesthesia reactions during electroconvulsive therapy for bipolar disorder. Bipolar Disord. 2020;22(8):874–875. [DOI] [PubMed] [Google Scholar]
- 35. Kennedy AD, Miller MJ, Beebe K, et al. . Metabolomic profiling of human urine as a screen for multiple inborn errors of metabolism. Genet Test Mol Biomarkers. 2016;20(9):485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chan K, Busque SM, Sailer M, et al. . Loss of function mutation of the Slc38a3 glutamine transporter reveals its critical role for amino acid metabolism in the liver, brain, and kidney. Pflugers Arch. 2016;468(2):213–227. [DOI] [PubMed] [Google Scholar]
- 37. Gataullina S, Bienvenu T, Nabbout R, Huberfeld G, Dulac O. Gene mutations in paediatric epilepsies cause NMDA-pathy, and phasic and tonic GABA-pathy. Dev Med Child Neurol. 2019;61(8):891–898. [DOI] [PubMed] [Google Scholar]
- 38. Barker-Haliski M, White HS. Glutamatergic mechanisms associated with seizures and epilepsy. Cold Spring Harb Perspect Med. 2015;5(8):a022863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chatron N, Becker F, Morsy H, et al. ; EuroEpinomics-RES consortium AR working group. Bi-allelic GAD1 variants cause a neonatal onset syndromic developmental and epileptic encephalopathy. Brain. 2020;143(5):1447–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Neuray C, Maroofian R, Scala M, et al. . SYNaPS Study Group. Early-infantile onset epilepsy and developmental delay caused by bi-allelic GAD1 variants. Brain. 2020;143(8):2388–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rumping L, Büttner B, Maier O, et al. . Identification of a loss-of-function mutation in the context of glutaminase deficiency and neonatal epileptic encephalopathy. JAMA Neurol. 2019;76(3):342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lister A, Bourgeois S, Imenez Silva PH, et al. . NRF2 regulates the glutamine transporter Slc38a3 (SNAT3) in kidney in response to metabolic acidosis. Sci Rep. 2018;8(1):5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Umapathy NS, Dun Y, Martin PM, et al. . Expression and function of system N glutamine transporters (SN1/SN2 or SNAT3/SNAT5) in retinal ganglion cells. Invest Ophthalmol Vis Sci. 2008;49(11):5151–5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Foroozan R. Vigabatrin: Lessons learned from the United States experience. J Neuroophthalmol. 2018;38(4):442–450. [DOI] [PubMed] [Google Scholar]
- 45. Dolgodilina E, Camargo SM, Roth E, et al. . Choroid plexus LAT2 and SNAT3 as partners in CSF amino acid homeostasis maintenance. Fluids Barriers CNS. 2020;17(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yoo HC, Yu YC, Sung Y, Han JM. Glutamine reliance in cell metabolism. Exp Mol Med. 2020;52(9):1496–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dienel GA. Brain glucose metabolism: Integration of energetics with function. Physiol Rev. 2019;99(1):949–1045. [DOI] [PubMed] [Google Scholar]
- 48. Wangler MF, Hubert L, Donti TR, et al. . A metabolomic map of Zellweger spectrum disorders reveals novel disease biomarkers. Genet Med. 2018;20(10):1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Glinton KE, Elsea SH. Untargeted metabolomics for autism spectrum disorders: Current status and future directions. Front Psychiatry. 2019;10(647):647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cappuccio G, Pinelli M, Alagia M, et al. . Biochemical phenotyping unravels novel metabolic abnormalities and potential biomarkers associated with treatment of GLUT1 deficiency with ketogenic diet. PLoS One. 2017;12(9):e0184022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bainbridge MN, Cooney E, Miller M, et al. . Analyses of SLC13A5-epilepsy patients reveal perturbations of TCA cycle. Mol Genet Metab. 2017;121(4):314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sugahara K, Zhang J, Kodama H. Liquid chromatographic-mass spectrometric analysis of N-acetylamino acids in human urine. J Chromatogr B Biomed Appl. 1994;657(1):15–21. [DOI] [PubMed] [Google Scholar]
- 53. Ramadan S, Lin A, Stanwell P. Glutamate and glutamine: A review of in vivo MRS in the human brain. NMR Biomed. 2013;26(12):1630–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. DeVito TJ, Drost DJ, Neufeld RW, et al. . Evidence for cortical dysfunction in autism: A proton magnetic resonance spectroscopic imaging study. Biol Psychiatry. 2007;61(4):465–473. [DOI] [PubMed] [Google Scholar]
- 55. Pan JW, Williamson A, Cavus I, et al. . Neurometabolism in human epilepsy. Epilepsia. 2008;49 (Suppl 3(0 3)):31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ben-Menachem E. Mechanism of action of vigabatrin: Correcting misperceptions. Acta Neurol Scand Suppl. 2011;124:5–15. [DOI] [PubMed] [Google Scholar]
- 57. Ochoa JG, Kilgo WA. The role of benzodiazepines in the treatment of epilepsy. Curr Treat Options Neurol. 2016;18(4):18. [DOI] [PubMed] [Google Scholar]
- 58. Kennedy AD, Pappan KL, Donti T, et al. . 2-Pyrrolidinone and succinimide as clinical screening biomarkers for GABA-transaminase deficiency: Anti-seizure medications impact accurate diagnosis. Front Neurosci. 2019;13:394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.