Abstract
Preoperative MRI is one of the most important clinical results for the diagnosis and treatment of glioma patients. The objective of this study was to construct a stable and validatable preoperative T2-weighted MRI-based radiomics model for predicting the survival of gliomas.
A total of 652 glioma patients across three independent cohorts were covered in this study including their preoperative T2-weighted MRI images, RNA-seq and clinical data. Radiomic features (1731) were extracted from preoperative T2-weighted MRI images of 167 gliomas (discovery cohort) collected from Beijing Tiantan Hospital and then used to develop a radiomics prediction model through a machine learning-based method. The performance of the radiomics prediction model was validated in two independent cohorts including 261 gliomas from the The Cancer Genomae Atlas database (external validation cohort) and 224 gliomas collected in the prospective study from Beijing Tiantan Hospital (prospective validation cohort). RNA-seq data of gliomas from discovery and external validation cohorts were applied to establish the relationship between biological function and the key radiomics features, which were further validated by single-cell sequencing and immunohistochemical staining.
The 14 radiomic features-based prediction model was constructed from preoperative T2-weighted MRI images in the discovery cohort, and showed highly robust predictive power for overall survival of gliomas in external and prospective validation cohorts. The radiomic features in the prediction model were associated with immune response, especially tumour macrophage infiltration.
The preoperative T2-weighted MRI radiomics prediction model can stably predict the survival of glioma patients and assist in preoperatively assessing the extent of macrophage infiltration in glioma tumours.
Keywords: prognostic prediction, macrophage, radiomic, machine learning, glioma
Using pre-operative T2-weighted MRI from a large cohort of glioma patients, Li et al. develop a radiomics model that shows robust predictive power for overall survival. The model reveals associations between radiomic and molecular features, particularly tumour macrophage infiltration.
Introduction
Glioma is the most common primary cancer in the CNS and a highly lethal disease.1,2 Despite the same standardized treatment, the prognosis varies in different patients. Therefore, evaluation of the prognosis is of great significance for the guidance of postoperative treatment of glioma. Although some molecular pathological findings, such as isocitrate dehydrogenase 1 (IDH1) mutation and chromosome 1p/19q co-deletion status, are known to be predictors of prognosis, accurate detection of these factors requires enough surgical specimens, professional technical staff, and expensive equipment and materials.3,4 These shortcomings are the main barriers for wide application of prognosis and chemosensitivity prediction by molecular pathological factors.
MRI has the highest degree of confidence in glioma diagnosis and is widely used for identifying the location and size of glioma. Radiomics, quantitative features extracted from radiographic medical images by data-characterization algorithms, is designed to develop prognostic prediction tools and treatment decision support tools in cancers.5-7 In addition, the original state of the tumour and tumour microenvironment are well-reflected by preoperative radiomic features (RFs), especially T2-weighted MRI-derived RFs, which allow evaluation of the tumour’s biological characteristics and microenvironment.8,9 Previous studies have shown that MRI RFs could potentially be used as prognostic or predictive biomarkers in glioma.10,11 Although some prognostic biomarkers or prediction models have yet to be developed in gliomas, a more reliable and easy-to-use predictive model is still needed for clinical practice.
Therefore, the aim of this study was to construct and validate a radiomics prediction model based on preoperative T2-weighted MRI of glioma patients. The stability of this radiomics model was validated in independent and prospective validation cohorts. Subsequently, biological interpretation of the prognostic RFs was performed and validated by single-cell sequencing and immunohistochemical staining from the prospective cohort. In short, a radiomics prediction model that incorporated the clinical prognosis prediction and tumour immune microenvironment assessment was established to change the current clinical management of patients with gliomas.
Materials and methods
Patient enrolment and tumour sequencing
Three independent cohorts of a total of 652 glioma patients with preoperative T2-weighted MRI image data, tumour transcriptome sequencing data, clinicopathological characteristics and follow-up information were included in this study (Supplementary Table 1). The preoperative imaging data of 167 patients in the discovery cohort were collected retrospectively from the imaging system of Beijing Tiantan Hospital and the corresponding transcriptomic data of these patients were obtained from the Chinese Glioma Genome Atlas (CGGA) database (http://www.cgga.org.cn/, accessed 1 December 2021). A total of 261 patients from The Cancer Genome Atlas (TCGA) database with available baseline preoperative imaging data and corresponding transcriptomic data were used as an external validation cohort (https://portal.gdc.cancer.gov/, accessed 1 December 2021). In the prospective validation cohort, 438 glioma patients were consecutively enrolled in this study from November 2016 to August 2019 at Beijing Tiantan Hospital and 214 patients were excluded according to the exclusion criteria. These patients (n = 224) were followed-up trimonthly by telephone or clinic for an average of 709 days (range 254–1232 days). The clinicopathological information of glioma patients in this study is summarized in Supplementary Tables 2–4.
In the discovery cohort, tumour samples obtained during surgery were immediately placed in liquid nitrogen for storage. Transcriptome data of patients were generated by Illumina platform. The pathological diagnosis of tumour samples was completed by two neuropathologists. Molecular pathology was performed at the Molecular Pathology Testing Center of Beijing Neurosurgical Institute.
In the prospective cohort, the acquisition of tumour samples (abnormal hyperintense signals of the T2 image) was carefully designed before surgery and was completed under the guidance of intraoperative neuronavigation. Fresh tumour specimens were collected at the time of resection and the presence of malignant cells was confirmed by fast frozen pathology of nearby tissue during operation. The single-cell RNA-sequencing (scRNA-seq) library was constructed according to the single-cell tagged reverse transcription sequencing (STRT-seq) protocol as previously described.12,13 Single-cell sequencing was performed on an Illumina 4000 platform.
Sample collection and data analyses were approved by Beijing Tiantan Hospital institutional review board and written informed consent was obtained from each participate.
Radiomic features extraction
The tumour region of interest was segmented on T2-weighted MR images, because this sequence is well-accepted in the identification of regions of gliomas. Regions of interest were manually delineated by two neuroradiologists (both with more than 10 years of experience in neuroradiology) using MRIcron software (http://www.mccauslandcenter.sc.edu/mricro). Regions of interest on the T2 image were defined as abnormal hyperintense signals and cerebrospinal fluid signals were avoided. The range of regoin of interest did not refer to signals in other sequences of MRI. For each patient, a total of 1731 RFs were extracted using the ‘PyRadiomics’ package implemented in Python.14 The extracted features were divided into four groups: (i) first-order statistics: n = 18; (ii) shape and size features: n = 13; (iii) textural features derived from texture matrices including grey-level co-occurrence matrix, grey-level run length matrix, grey-level size zone matrix, grey-level dependence matrix: n = 68; and (iv) filter-derived features: filter ‘wavelet’: n = 688; filter ‘LoG’: n = 258; filter ‘LBP’: n = 258; other filter (‘square’, ‘squareroot’, ‘logarithm’, ‘exponential’, ‘gradient’): n = 86 × 5 = 430. The detailed calculation formula for each RF is provided on the official website (https://pyradiomics.readthedocs.io, accessed 1 December 2021).
Machine learning-based radiomics prediction model construction
The risk prediction model was constructed based on RFs. To ensure the stability of the prediction model, the RFs were strictly screened in the discovery cohort with two steps. First, we randomly used 50% of the samples as the training set and the remaining 50% of the samples as the test set. To test the robustness of RFs selection in building the prediction model, we randomly split the samples in the discovery cohort into a training set and a test set at a ratio of 3:7, 4:6, 6:4, and 7:3, respectively. On the training set, we first performed a preselection step to keep the top significant features correlated with overall survival (univariate Cox model, likelihood ratio test, P < 0.05). Second, we applied the risk score formulation (risk score = ∑ feature values × Cox efficient of feature) using the top significant features selected in the first step to calculate an RF score value for each sample in the test set, followed by separating the test set into high and low groups by the median of the RF score. If the overall survivals of these two groups were significantly different (Kaplan–Meier analysis, log rank P < 0.05), the features used in the RF score formulation were chosen. We repeated the above procedure 1000 times and selected the features which were chosen in more than 85% of the total of 1000 procedures.
LASSO-based feature selection
A standard multivariate approach, Cox-LASSO (least absolute shrinkage and selection operator), was also applied for RFs selection in building the prediction models (Supplementary Fig. 1). First, univariate Cox regression analysis was applied to extract the features that were statistically significantly associated with survival (adjusted P-value < 0.01). For the prognostic features, a Cox proportional hazards model (iteration = 1000) with a LASSO penalty was used to find the best RF model utilizing an R package called ‘glmnet’. A total of nine features were obtained.
Random survival forests–variable hunting feature selection
A random forest survival analysis was performed to screen RFs for predictive model building. Specifically, univariate Cox proportional hazards regression analysis was performed to screen out those RFs with a significant relationship with patients’ overall survival in the discovery cohort (adjusted P-value < 0.01). Then, the random survival forests–variable hunting (RSFVH) algorithm was applied to filter prognostic RFs. Finally, we obtained nine features.
Deep learning models
Three widely used deep learning models were built and trained in discovery cohort, followed by independent evaluation in the TCGA cohort and the prospective cohort. Specifically, a stringent criterion was adopted to select the prognostic RFs with the use of univariate Cox proportional hazards regression analysis in the discovery cohort (adjusted P-value < 0.005). A total of 25 prognostic RFs were extracted. The grouping results derived from hierarchical k-means clustering using prognostic RFs were labelled as 0 and 1, respectively. The prognostic RFs in the discovery cohort were used as training data to train the deep learning model. The input data were Z-score-transformed RFs to avoid a gradient disappearance problem. The first deep learning model (deep learning model 1) was built with one hidden layer including eight nodes. The second one (deep learning model 2) was built with two hidden layers with each containing 16 and 8 nodes, respectively. The third one (deep learning model 3), the LSTM (long short-term memory) deep learning model, was built with two hidden layers, including two LSTM layers, each layer containing 16 and 4 nodes, respectively. Sigmoid function was chosen as neuron activation function, mean squared error as the loss function and Adam (adaptive movement estimation algorithm) as the iterative optimizer. The maximum number of iterations was set as 1000. The initial connection weights and biases of each layer were randomly generated and end up reaching stable parameters through training iterations. After determining the framework of the model, cross-validation was a necessary step. The training data were separated into two sections randomly with the proportion of training and testing sets as 6 to 4. The training set was used to train the model to determine the unknown parameters, while the test set was used to validate the effect of the predicted parameters. To obtain the optimal model, the above process was carried out 300 times. Kaplan–Meier survival analysis was operated each time to see if the model can divide the samples into two groups with a statistically significant survival difference. Only groups with a P-value lower than or equal to the threshold of 0.05 were regarded as statistically significant. Among 300 times trials, the more significant stratifications, the more stable our model is. The model with fixed parameters corresponding to the lowest P-value was selected as the optimal model. To test the performance of the optimal model, the TCGA cohort and prospective cohort were used as external test data, respectively. The optimal model divided patients in each cohort into long- and short-term survival clusters. Kaplan–Meier analysis was conducted between the long- and short-term survival clusters in each cohort to test the predictive performance of the optimal model for glioma.
Functional annotation of radiomic features
Functional annotation of RFs was performed by Gene Set Variation Analysis (GSVA) and Pearson correlation analysis. First, the biological process and pathway activation scores of each patient were calculated by GSVA analysis based on tumour transcriptome sequencing data. The gene sets of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were downloaded from Gene Set Enrichment Analysis (GSEA) web portals (http://software.broadinstitute.org/gsea/index.jsp, accessed 1 December 2021). Second, the correlation between biological process and pathway activation scores and RF values was calculated by Pearson correlation analysis. Functions and pathways, significantly correlated with RF values (P < 0.05), were used to annotate RFs. Classification of biological functions was performed according to the classification in the AmiGO 2 portal (http://amigo.geneontology.org/amigo, accessed 1 December 2021).
Single-cell RNA sequencing data analysis
Raw read counts were obtained from scRNA-seq data. Cells with an abundance of reads count > 1000 were kept for further analysis. The imputation of single cells was applied to scRNA-seq data by Markov affinity-based graph imputation of cells.15 Then, ComBat16 was performed to remove the batch effect of single-cell data. Seurat was used to analyse the single-cell sequencing data with default options (normalized with LogNormalize, variable features found with vst, and 5000 high variable features kept). Canonical cluster analysis as implemented in the Seurat package was then performed on the 5000 common genes identified in this manner. Non-linear dimensional reduction t-distributed stochastic neighbour embedding (tSNE) was applied on the scale data to visualize and explore these datasets. Lastly, we used the cell markers previously reported to determine the identification of the cell types. Specifically, CD45+ was used to separate immune cells from non-immune cells. Immune cells with CD11b+, CD14+, CD16+, CD68+, CD86+ and CD163+ were macrophages. Immune cells with CD14+, CD16+ and CD163– were monocytes. Immune cells with CD8+, CD3+ and GZMA+ were CD8+ T cells. Immune cells with CD3+ were CD3 T cells.
Normalized enrichment score of immune cell signatures
We curated a total of 295 different gene signatures for immune cells and CNS from literature.17–21 To evaluate the enrichment of each immune cell type in each sample, we used the normalized enrichment score of the Mann-Whitney Gene Set test.22,23 The normalized enrichment score (NES) was determined as follows:
| (1) |
| (2) |
where m is the number of genes in a gene set, n is the number of genes outside the gene set, and T is the sum of the ranks of the genes in the gene set. Given a gene signature, the gene expression data of a sample were separated into two sections comprising genes expressed in the gene signature and the rest of the genes, respectively. The Wilcoxon rank-sum test was then applied to calculate the normalized enrichment score.
Immunohistochemical staining for macrophage markers
Tumour samples for immunohistochemical staining were obtained from patients in the discovery cohort (n = 62). The surgically removed tumour tissues were stored in formalin immediately and embedded in paraffin within 3 days. The immunohistochemical staining and image capture were performed as previously described.24 The primary antibody for the detection of macrophage markers were as follows: MS4A4A (Sigma-Aldrich, HPA029323), STAB1 (Abcam, ab101035) and COLEC12 (Invitrogen, PA5-30835). Immunohistochemical staining was performed as per the manufacturer’s protocol with recommended concentration. The proportion of positive cells was counted using ImageJ (v1.52) software.
Statistical analyses
Statistical analyses and drawings were performed by software environment R (v3.5.0), SPSS software (v25.0, IBM) and Office 2016 (Microsoft). The Mann-Whitney U-test was used to validate differences between two variables. The chi-square test was used to assess the composition ratio differences between two groups. The log-rank test was used to assess the statistical significance between survival groups in Kaplan–Meier survival analysis. P-values less than 0.05 were considered statistically significant.
Data availability
All datasets used and/or analysed in this study have been uploaded. The sequencing data, clinical and follow-up information of glioma patients were uploaded to the CGGA portal (http://cgga.org.cn/, accessed 1 December 2021). The method has been uploaded to GitHub (https://github.com/zhangjbig/RadioML, accessed 1 December 2021).
Results
Clinical characteristics
Patient clinical characteristics in the discovery (Tiantan), external validation (TCGA) and prospective validation (Beijing Tiantan Hospital) cohorts are shown in Supplementary Table 1. The composition of patients (especially age, IDH1 and 1p/19q status) was significantly distinct among different cohorts and the main reason for this inconsistency was the difference in tumour grade. The distribution of tumour grade was similar between the discovery cohort and the prospective validation cohort, while the majority of tumours in the external validation cohort were grade IV gliomas. As the radiomics prediction model was constructed from preoperative T2-weighted MRI images independent of tumour grade and molecular features, the prediction efficiency might not be significantly affected by the difference between the discovery and validation cohorts.
MRI radiomic features extraction and prediction model construction
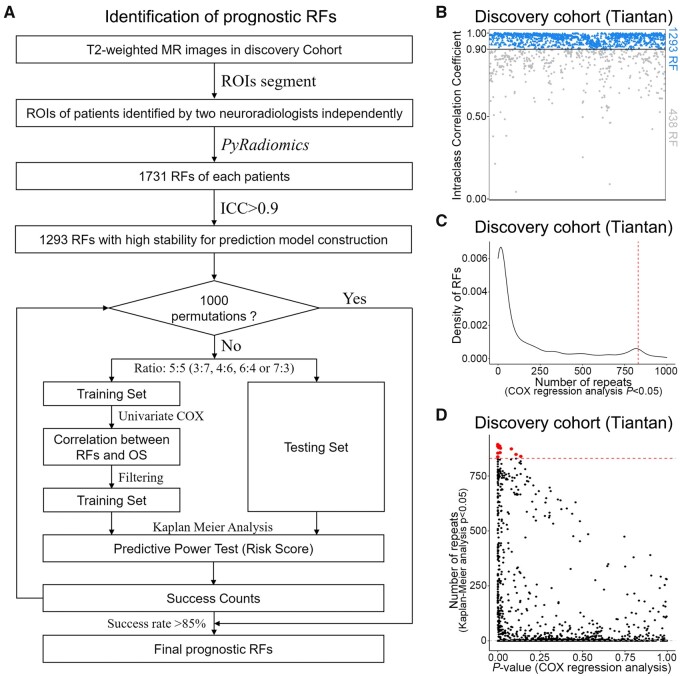
Two neuroradiologists independently reviewed the T2-weighted MRI images and then delineated the tumour contour with mutual concordance. Then, 1731 RFs were retrieved for the tumour area from the T2-weighted MRI images, among which 1293 RFs with high intraclass correlation coefficient (>0.9) were retained in the downstream analysis. A permutation-based machine learning method was applied to screen RFs associated with overall survival of gliomas from 167 glioma patients in the discovery cohort (Tiantan; Fig. 1A and B). Fourteen RFs were identified as significantly associated with overall survival of gliomas (Fig. 1C and D). To test the robustness of RFs selection in building the prediction model, we randomly split the samples in the discovery cohort into a training set and a test set at different ratios of 4:6, 3:7, 7:3, and 6:4, respectively. We discovered that the features selected at different scenarios were highly consistent (Supplementary Fig. 1A–C). A risk prediction model was constructed based on the 14 prognostic RFs. The predictive power of the risk prediction model was validated in an external data cohort consisting of 261 glioma patients from the TCGA database. Two hundred and twenty-four glioma patients were recruited in the prospective cohort from Beijing Tiantan Hospital for further validating the performance of the RF-based risk prediction model. The association between biological functions and the RF-based model was established through GSEA, followed by experimental validation through single-cell sequencing of 1733 cells from four gliomas and immunohistochemical staining of 62 samples in the discovery set.
Figure 1.
Work flow and radiomic feature screening. (A) Work flow of the machine learning method. (B) A total of 1293 RFs showed high intraclass correlation coefficient (ICC) between tumours sketched by radiologist or neurosurgeon. (C) The frequency of RFs chosen (univariate Cox model, likelihood ratio test, P < 0.05, both in the feature value and the median of RF score) in the prediction models. (The peaks are in the 850 times.) (D) The RFs of Kaplan–Meier P-values in the test groups and frequency. OS = overall survival; ROIs = regions of interest.
Performance of the radiomics prediction model in prognosis prediction in the retrospective analysis
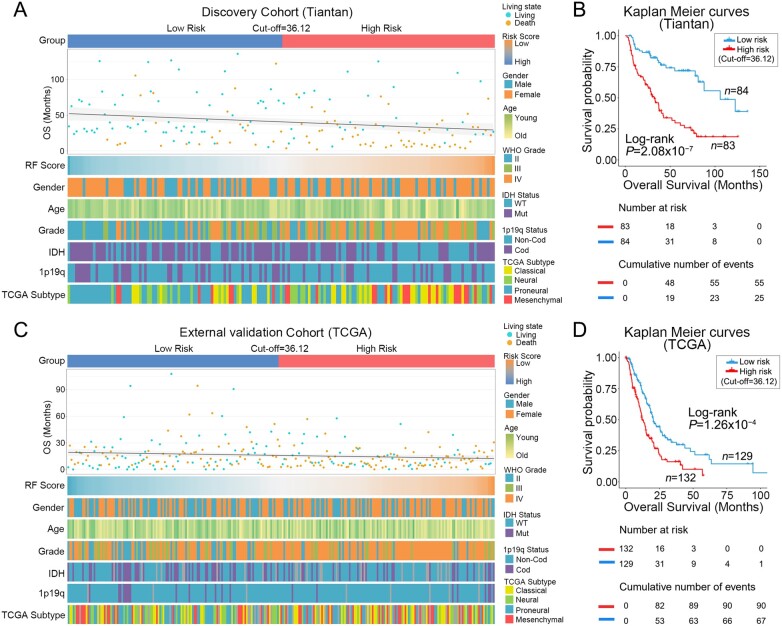
The relationship between the RF scores of radiomics prediction model and the clinicopathological features of patients is shown in the heat maps (Fig. 2A and C). Patients were ranked in ascending order of RF scores in the discovery and external validation cohorts. The median RF score (36.12) in the discovery cohort was used as the cut-off value of risk subgroups in this study. Patients with an RF score greater than 36.12 were classified into the high-risk subgroup and those with lower than 36.12 were classified into the low-risk subgroup. WHO tumour grade and IDH1 mutation status in the discovery cohort and external validation cohort showed asymmetry distribution in different risk subgroups. However, age, gender and 1p/19q status did not differ significantly between the two subgroups (Supplementary Tables 2 and 3). Subsequently, Kaplan–Meier survival analysis in the discovery and external validation cohorts showed that patients in the high-risk subgroup had shorter overall survival than those in the low-risk subgroup (Fig. 2B and D). Univariate and multivariate Cox regression analysis demonstrated that RF score was an independent prognostic factor after adjusting for other prognostic factors in patients of both the Tiantan discovery cohort and the TCGA validation cohort (Table 1).
Figure 2.
Clinicopathological and survival differences in different risk subgroups. (A and C) The differences of survival and clinical molecular pathology in patients in different risk groups are shown in heat maps. Patients in the discovery and external validation cohorts are arranged in ascending order of RF scores of RFs. (B and D) Kaplan–Meier curves show the overall survival of patients in low-risk and high-risk groups. Overall survival of patients in the high-risk group is significantly shorter in both discovery and external validation cohorts.
Table 1.
Cox regression analysis of prognostic factors in discovery cohort and validation cohort
| Variable | Discovery cohort (Tiantan) | External validation cohort (TCGA) | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||
| HR (95% CL) | P-value | HR (95% CL) | P-value | HR (95% CL) | P-value | HR (95% CL) | P-value | |
| RF scorea | 1.092 (1.054–1.132) | 1.07 × 10−6 | 1.049 (1.002–1.098) | 0.0421 | 1.046 (1.017–1.076) | 1.84 × 10−3 | 1.047 (1.009–1.085) | 0.0136 |
| Agea | 1.058 (1.039–1.077) | 2.14 × 10−9 | 1.032 (1.014–1.050) | 4.59 × 10−4 | 1.056 (1.042–1.071) | 1.11 × 10−15 | 1.035 (1.019–1.052) | 1.51 × 10−5 |
| WHO gradeb | 6.66 × 10−14 | 0.0030 | 5.80 × 10−8 | 0.1765 | ||||
| III versus II | 2.387 (1.203–4.734) | 0.0128 | 1.372 (0.653–2.883) | 0.4033 | 24.425 (3.182–187.509) | 0.0021 | 1.00 × 105 (1.63 × 10−35−6.17 × 1044) | 0.8054 |
| IV versus II | 9.714 (5.232–18.035) | 5.94 × 10−13 | 3.195 (1.487–6.863) | 0.0029 | 77.776 (10.535–574.192) | 1.97 × 10−5 | 5.02 × 104 (8.16 × 10−36–3.09 × 1044) | 0.8169 |
| IDH1 statusb Mutant versus wild-type |
0.163 (0.102–0.262) | 6.74 × 10−14 | 0.567 (0.266–1.208) | 0.1418 | 0.050 (0.020–0.124) | 7.87 × 10−11 | 0.171 (0.045–0.651) | 0.0096 |
| 1p/19q statusb Cod versus Non-Cod |
0.390 (0.219–0.696) | 1.43 × 10−3 | 0.650 (0.353–1.199) | 0.1680 | 0.098 (0.024–0.396) | 1.13 × 10−3 | 0.162 (0.017–1.574) | 0.1166 |
| TCGA subtypeb | 4.26 × 10−13 | 0.4225 | 8.93 × 10−7 | 0.5494 | ||||
| Mes versus Cla | 1.344 (0.757–2.388) | 0.3128 | 1.132 (0.604–2.120) | 0.6989 | 1.031 (0.689–1.542) | 0.8817 | 1.175 (0.723–1.909) | 0.5156 |
| Neu versus Cla | 0.214 (0.108–0.425) | 1.01 × 10−5 | 0.605 (0.282–1.297) | 0.1964 | 0.386 (0.228–0.654) | 3.95 × 10−4 | 0.850 (0.457–1.581) | 0.6084 |
| PN versus Cla | 0.173 (0.096–0.314) | 7.80 × 10−9 | 0.634 (0.279–1.440) | 0.2764 | 0.344 (0.210–0.564) | 2.32 × 10−5 | 1.374 (0.768–2.456) | 0.2845 |
Cla = classical; Cod = co-deletion;: Mes = mesenchymal; Neu = neural; PN = proneural.
Numerical variables.
Categorical variables.
Performance of the radiomics prediction model in prognosis prediction in the prospective analysis
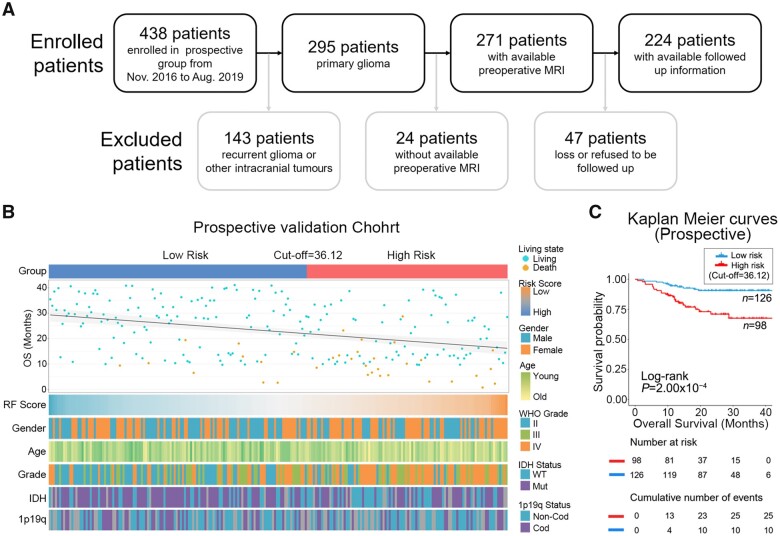
To further validate the concordance and reproducibility of the radiomics prediction model, a single-institutional prospective analysis was performed at Beijing Tiantan Hospital. Two hundred and twenty-four of 438 glioma patients from November 2016 to August 2019 were enrolled in the prospective cohort (Fig. 3A). Based on the RF score of the radiomics prediction model, patients were also divided into high- and low-risk subgroups by the same method and cut-off value from the discovery cohort. The clinicopathological features of patients in different risk subgroups are presented in Fig. 3B. The difference analysis of clinicopathological factors found that there are significant differences between age, WHO tumour grade and IDH1 mutation status in the high- and low-risk groups (Supplementary Table 4). Survival analysis indicated that patients in the high-risk subgroup showed significantly shorter overall survival than those in the low-risk subgroup (Fig. 3C). In addition, the radiomics prediction model particularly proved to be an independent prognostic risk factor in patients from the prospective cohort by multivariate Cox regression analysis (Table 2).
Figure 3.
The stability of the radiomics prediction models was validated in the prospective validation cohort. (A) Flow diagram of glioma patients in the prospective group. A total of 224 glioma patients eligible for the study were screened from the sample of 438 glioma patients from November 2016 to August 2019. (B) The heat map shows clinicopathological information of patients in different risk groups in the prospective validation cohort. (C) Kaplan–Meier curves show the overall survival of patients in the high-risk group is significantly shorter than those in low-risk group in the prospective validation cohort.
Table 2.
Cox regression analysis of prognostic factors in prospective cohort
| Variable | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CL) | P-value | HR (95% CL) | P-value | ||
| RF scorea | 1.110 (1.046–1.178) | 5.93 × 10−4 | 1.088 (1.010–1.173) | 0.0271 | |
| Agea | 1.039 (1.011–1.069) | 7.18 × 10−3 | 1.020 (0.989–1.053) | 0.2122 | |
| WHO gradeb | 4.52 × 10−5 | 0.2390 | |||
| III versus II | 1.751 (0.503–6.096) | 0.3788 | 0.900 (0.215–3.759) | 0.8847 | |
| IV versus II | 6.949 (2.624–18.397) | 9.53 × 10−5 | 2.158 (0.600–7.766) | 0.2390 | |
| IDH1 statusb Mutant versus wild-type |
0.115 (0.050–0.269) | 5.71 × 10−7 | 0.209 (0.079–0.553) | 0.0016 | |
| 1p/19q statusb Cod versus Non-Cod |
0.020 (3.69 × 10−4–1.038) | 0.0522 | |||
Cod = co-deletion
Numerical variables.
Categorical variables.
The prognostic radiomic features were in close correlation with tumour-infiltrating macrophages
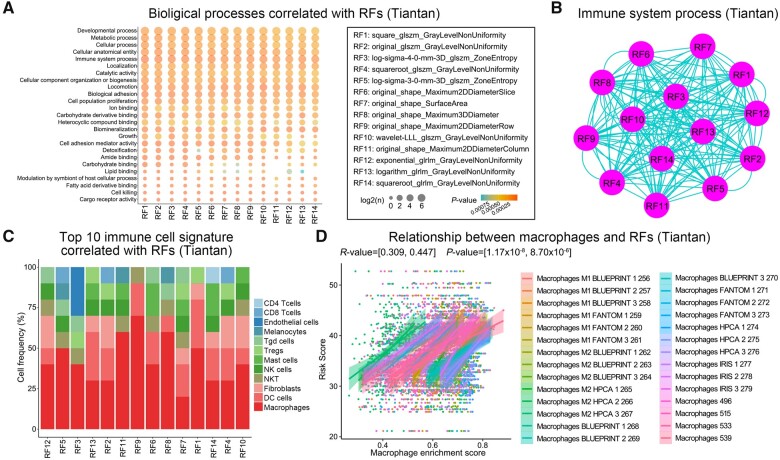
To understand the relationship between the 14 prognostic RFs and the biological functions, we calculated the enrichment score of each biological function for each patient in the discovery cohort. Pearson correlation analysis demonstrated that the immune system process was significantly related to the 14 RFs (Fig. 4A). The GO terms of the immune system process were also highly shared among the 14 RFs (Fig. 4B). To identify which immune cells may be associated with the RFs, we curated the gene signatures for 295 immune cells from the literature. We then performed normalized enrichment score analysis on tumour transcriptome data to predict the enrichment of immune cells in each patient, and found that macrophages showed a distinct and strong correlation with the prognostic RFs (Fig. 4C and D). Even if there were disparities in demographic and tumour-grade distribution between Tiantan and TCGA databases, we still observed a weak but non-negligible association between RFs and macrophages in the TCGA validation cohort (Supplementary Fig. 2). In conclusion, the radiomics prediction model from preoperative T2-weighted MRI images could help assess the tumour-infiltrating macrophages in glioma patients.
Figure 4.
The relationship between prognostic RFs and tumour cell functions in the discovery cohort. (A) The Pearson correlation between RFs and tumour biological processes. (B) The networks among the features on the immune system process: the nodes are features and the edges are the counts of immune system process overlap between the features. (C) The top 10 significant correlation between RFs and cell fractions. (D) The Pearson correlation between RFs and macrophage cell signatures.
Verification of the relationship between radiomic features and tumour macrophage infiltration
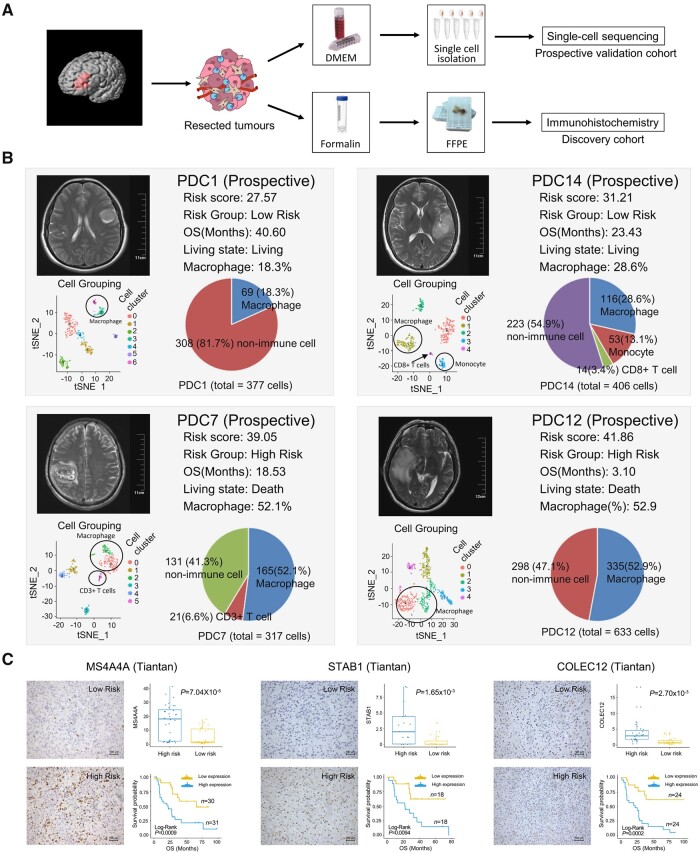
To further validate the biological annotations of the prognostic RFs, scRNA-seq and immunohistochemical staining were performed in representative patients of the prospective and discovery cohort, respectively (Fig. 5A). Specifically, we performed scRNA-seq on isolated cells of surgical specimens from four glioma patients (named as PDC1, PDC7, PDC12, and PDC14) and a total of 1733 cell gene expression profiles were included in the analysis (Fig. 5B).
Figure 5.
Experimental validation of RF-related tumour macrophage infiltration. (A) Scheme of the experimental workflow. (B) t-Distributed stochastic neighbour embedding (tSNE) plot shows clustering of each patient’s cells based on gene expression. Point coordinates are based on tSNE dimensionality reduction of the top principal components calculated from the 5000 most informative genes. Cell colour specifies assignment of cells to these clusters inferred using shared nearest neighbour clustering. Pie charts demonstrate the distribution of the identified cell types across samples in each patient and histograms show the macrophage cell abundance between high-risk and low-risk patients. (C) Immunohistochemical staining displays the RF-related macrophage markers MS4A4A, STAB1 and COLEC12. The scatter diagram shows the expression level of these markers in high-risk and low-risk samples. Kaplan–Meier survival analysis was performed between the samples with high and low expression of macrophage markers.
To characterize the cell identity of the obtained clusters, we applied the immune cell marker curated from the literature. More concretely, CD45+ cells were immune cells and CD45– cells were non-immune cells. Immune cells include macrophages (CD11b+, CD14+, CD16+, CD68+, CD86+, CD163+), CD8 T cells (CD8+, CD3+, GZMA+), CD3+ T cells and monocytes (CD14+, CD16+, CD68–, CD163–). We confirmed that there were more tumour-infiltrating macrophages in patients with higher RF scores through imputation for scRNA-seq data (Supplementary Figs 3–6). It was further indicated that tumour samples harbouring a high macrophage cell gene signature conferred a poorer survival than those with low macrophage signature (Supplementary Fig. 7A and B). The high-risk patients, PDC7 and PDC12, have more abundance of macrophage cell fractions compared with low-risk patients, PDC1 and PDC14 (Fig. 5B). In addition, the selected markers of MS4A4A, STAB1 and COLEC12 reflecting tumour-infiltrating macrophages were detected by immunohistochemical staining in the discovery cohort. The results reconfirmed that tumour-infiltrating macrophages were highly enriched in patients with higher RF scores and the increased expression of these macrophage-specific markers were also indicators of poor prognosis in patients with gliomas (Fig. 5C and Supplementary Fig. 7C–E). Furthermore, the patients were divided into lower grade glioma (WHO II and III) and glioblastoma (WHO IV) groups according to the tumour grade. The results showed that tumour-infiltrating macrophages were enriched in the high-risk group in both lower grade glioma and glioblastoma patients (Supplementary Figs 8 and 9).
Discussion
MRI is one of the most important clinical data for patients with gliomas. Preoperative MRI plays a central role in glioma diagnosis and intraoperative neuronavigation-guided tumour resection. With the development of radiomics analysis, some studies have found that MRI can be used to predict the biological and genomic features of tumours, such as therapeutic response,25 tumour recurrence,26 p53 mutation27 and other molecular markers.28–30 However, a radiomics prediction model for the tumour microenvironment of gliomas is still in development at present. Among the clinical routinely used MRI sequences, the T2-weighted sequence is superior in identifying the tumour boundary and detecting tumours and the surrounding tumour microenvironment.10,27,30,31 In our previous studies and other imaging studies, the identification of tumour regions of interest was mostly based on T2-weighted sequences, even in multimodal and glioblastoma imaging studies.32–34 In this study, we constructed and prospectively validated a prognostic radiomics model based on preoperative T2-weighted MRI images of glioma patients for potential clinical application. Importantly, this prediction model was in close relationship with tumour-infiltrating macrophages, providing an explanation for patient survival benefit from the current clinical management.
Radiomics analysis underwent remarkable progress along with advances in radiological imaging, most notably in CNS tumours, which would be a promising direction to advance personalized medicine. In previous studies by us and others, some radiomic-based glioma prediction models have been established.10,35,36 Our analysis builds on these studies in that we performed a rigorous screening of RFs and a comprehensive validation of the prediction model. A valuable prediction model was usually based on crucial predictors, and variable factors were excluded by two steps in our study. The first step excluded the influence of variations between neuroradiologists on the extraction of RFs. The second step ensured the reproductivity of the prediction model in different populations and different image resources (Tiantan and TCGA databases). In the verification of our prediction model, we not only set up an independent external validation cohort but also designed a prospective validation cohort. All above results demonstrated that the radiomics prediction model is a prognostic factor independent of traditional prognostic factors (patient age, WHO tumour grade, IDH1 and 1p/19q status), and can be used together with these factors to predict the prognosis of glioma patients. Furthermore, the radiomics prediction model has the advantages of non-invasive, economical and can guide the clinical treatment of glioma before surgery. With the wide clinical application, this easy and feasible model is a supplement to the existing classic markers.
Many methods have been tried for the construction of the radiomics prediction model. We applied a standard multivariate approach, Cox-LASSO, to select RFS to construct a prediction model. We also evaluated RSFVH to select RFs for model building. We found the RFs selected by Cox-LASSO and RSFVH were quite different from those selected by our prediction model. We barely found an association between macrophage enrichment and RFs derived from Cox-LASSO or RSFVH (Supplementary Fig. 1A–C). In addition to traditional algorithms, deep learning algorithms have also been used to build predictive models. Based on 25 RFs with prognostic value, three predictive models have been built by deep learning methods in the discovery cohort. The prediction results of the deep learning prediction models consistently show that risk group is an independent prognostic factor in discovery and TCGA cohorts, but not in the prospective cohorts (Supplementary Figs 10–12), suggesting deep learning models fit data with a larger sample size.
At present, functional annotation of RFs is still a scientific challenge in the current radiomics research. Sun et al.6 have constructed a prediction model to assess tumour-infiltrating CD8 cells and immunotherapy response in cancers by conjoint analysis of CT images and RNA-seq genomic data. Grossmann et al.37 have examined the correlation between RFs and pathway scores, obtained from GSEA, to define radiomic–pathway–clinical relationships. Combining the advantages of reported algorithms, functional annotation of the 14 predictive RFs was performed based on the enrichment scores of 5917 biological processes and pathways obtained from RNA-seq data. Consistent with previous reports in other tumours,6,37,38 the predictive RFs were closely related to immune response, especially tumour macrophage infiltration in gliomas. There were significant differences in patient composition between Tiantan and TCGA databases and a weak but non-negligible association between RF model and macrophage was observed in the TCGA database. In addition, tumour samples from patients of the prospective and retrospective cohort were, respectively, collected for single-cell sequencing and immunohistochemical staining, and the RFs-related macrophage infiltration could be reconfirmed in patient-derived cells and surgical specimens. Most interestingly, the correlation between predictive RFs and tumour macrophage infiltration was identified for the first time in glioma patients. This result indicated that the distinct survival benefits in glioma patients with the same diagnosis and treatment may be due to the various tumour microenvironments. The close relationship between tumour microenvironments and RFs was most likely to be an intrinsic mechanism by which the radiomics prediction model could accurately predict prognosis in glioma patients. T2-weighted MRI of gliomas usually reflects the features of the tumours and surrounding areas, which is the most commonly used non-invasive tool to exhibit the diversity of tumour microenvironment, while the underlying mechanisms need to be further elucidated. Most importantly, our finding suggested that the radiomics prediction model might also provide potential clinical guidance for future immunotherapy of gliomas.
In conclusion, we constructed an MRI radiomics model by machine learning to predict clinical outcomes in glioma patients. This prediction model has great potential to guide clinical prognosis prediction and decision-making for immunotherapy in the future.
Supplementary Material
Acknowledgements
We thank Ms Shuqing Sun and Hua Huang for tissue sample collection and clinical data retrieval.
Funding
This work was supported by grants from National Natural Science Foundation of China (No. 82072768; W.Z.), The Medical and Health Technology Innovation Project of the Chinese Academy of Medical Sciences (2020-I2M-C&T-A-024; W.Z.), Sino-German Center Cooperation and Exchanges Program (M-0020; W.Z.), The Pilot Project for the Development and Reform of Medical Research Institutes of the Beijing Municipal Health Commission (PXM2019_026280_000002; W.Z.), the Youth Thousand Scholar Program of China (J.Z.), Program for High-Level Overseas Talents, Beihang University (J.Z.), National Natural Science Foundation of China (NSFC)/Research Grants Council (RGC) Joint Research Scheme (81761168038; T.J.) and Beijing Municipal Administration of Hospitals’ Mission Plan (SML20180501; T.J.).
Competing interests
The authors declare no potential conflicts of interest.
Supplementary material
Supplementary material is available at Brain online.
Abbreviations
- RFs
radiomic features
- scRNA-seq
single-cell RNA-sequencing
- TCGA
The Cancer Genome Atlas
References
- 1. Jiang T, Nam DH, Ram Z, et al. ; Asian Glioma Genome Atlas (AGGA) network . Clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2021;499:60–72. [DOI] [PubMed] [Google Scholar]
- 2. Liang S, Fan X, Zhao M, et al. . Clinical practice guidelines for the diagnosis and treatment of adult diffuse glioma-related epilepsy. Cancer Med. 2019;8(10):4527–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beiko J, Suki D, Hess KR, et al. . IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro Oncol. 2014;16(1):81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hegi ME, Diserens AC, Gorlia T, et al. . MGMT gene silencing and benefit from temozolomide in glioblastoma. New Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 5. Lu H, Arshad M, Thornton A, et al. . A mathematical-descriptor of tumor-mesoscopic-structure from computed-tomography images annotates prognostic- and molecular-phenotypes of epithelial ovarian cancer. Nat Commun. 2019;10(1):764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sun R, Limkin EJ, Vakalopoulou M, et al. . A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: An imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19(9):1180–1191. [DOI] [PubMed] [Google Scholar]
- 7. Dohan A, Gallix B, Guiu B, et al. . Early evaluation using a radiomic signature of unresectable hepatic metastases to predict outcome in patients with colorectal cancer treated with FOLFIRI and bevacizumab. Gut. 2019;69(3):531–539. [DOI] [PubMed] [Google Scholar]
- 8. Huang YQ, Liang CH, He L, et al. . Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34(18):2157–2164. [DOI] [PubMed] [Google Scholar]
- 9. Xu X, Zhang HL, Liu QP, et al. . Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol. 2019;70(6):1133–1144. [DOI] [PubMed] [Google Scholar]
- 10. Liu X, Li Y, Qian Z, et al. . A radiomic signature as a non-invasive predictor of progression-free survival in patients with lower-grade gliomas. Neuroimage Clin. 2018;20:1070–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beig N, Patel J, Prasanna P, et al. . Radiogenomic analysis of hypoxia pathway is predictive of overall survival in Glioblastoma. Sci Rep. 2018;8(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Islam S, Kjallquist U, Moliner A, et al. . Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21(7):1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10(11):1096–1098. [DOI] [PubMed] [Google Scholar]
- 14. van Griethuysen JJM, Fedorov A, Parmar C, et al. . Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Dijk D, Sharma R, Nainys J, et al. . Recovering gene interactions from single-cell data using data diffusion. Cell. 2018;174(3):716–729.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haghverdi L, Lun ATL, Morgan MD, Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol. 2018;36(5):421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aran D, Hu Z, Butte AJ. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bindea G, Mlecnik B, Tosolini M, et al. . Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. [DOI] [PubMed] [Google Scholar]
- 19. Charoentong P, Finotello F, Angelova M, et al. . Pan-cancer immunogenomic analyses reveal genotype–immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. [DOI] [PubMed] [Google Scholar]
- 20. Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tirosh I, Izar B, Prakadan SM, et al. . Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Frattini V, Pagnotta SM, Tala, et al. . A metabolic function of FGFR3-TACC3 gene fusions in cancer. Nature. 2018;553(7687):222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang J, Caruso FP, Sa JK, et al. . The combination of neoantigen quality and T lymphocyte infiltrates identifies glioblastomas with the longest survival. Commun Biol. 2019;2:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu H, Mu Q, Bao Z, et al. . Mutational landscape of secondary glioblastoma guides MET-targeted trial in brain tumor. Cell. 2018;175(6):1665–1678.e18. [DOI] [PubMed] [Google Scholar]
- 25. Richard R, Thomassin I, Chapellier M, et al. . Diffusion-weighted MRI in pretreatment prediction of response to neoadjuvant chemotherapy in patients with breast cancer. Eur Radiol. 2013;23(9):2420–2431. [DOI] [PubMed] [Google Scholar]
- 26. Zhang LL, Huang MY, Li Y, et al. . Pretreatment MRI radiomics analysis allows for reliable prediction of local recurrence in non-metastatic T4 nasopharyngeal carcinoma. EBioMedicine. 2019;42:270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Y, Qian Z, Xu K, et al. . MRI features predict p53 status in lower-grade gliomas via a machine-learning approach. Neuroimage Clin. 2018;17:306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gupta A, Young RJ, Shah AD, et al. . Pretreatment dynamic susceptibility contrast MRI perfusion in glioblastoma: Prediction of EGFR gene amplification. Clin Neuroradiol. 2015;25(2):143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qian Z, Li Y, Fan X, et al. . Molecular and clinical characterization of IDH associated immune signature in lower-grade gliomas. Oncoimmunology. 2018;7(6):e1434466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li Y, Liu X, Qian Z, et al. . Genotype prediction of ATRX mutation in lower-grade gliomas using an MRI radiomics signature. Eur Radiol. 2018;28(7):2960–2968. [DOI] [PubMed] [Google Scholar]
- 31. Kinoshita M, Sakai M, Arita H, et al. . Introduction of high throughput magnetic resonance T2-weighted image texture analysis for WHO Grade 2 and 3 gliomas. PLoS One. 2016;11(10):e0164268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ricard D, Kaloshi G, Amiel-Benouaich A, et al. . Dynamic history of low-grade gliomas before and after temozolomide treatment. Ann Neurol. 2007;61(5):484–490. [DOI] [PubMed] [Google Scholar]
- 33. Li Y, Liu X, Xu K, et al. . MRI features can predict EGFR expression in lower grade gliomas: A voxel-based radiomic analysis. Eur Radiol. 2018;28(1):356–362. [DOI] [PubMed] [Google Scholar]
- 34. Sun Z, Li Y, Wang Y, et al. . Radiogenomic analysis of vascular endothelial growth factor in patients with diffuse gliomas. Cancer Imaging 2019;19(1):68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu X, Li Y, Li S, et al. . IDH mutation-specific radiomic signature in lower-grade gliomas. Aging (Albany NY). 2019;11(2):673–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou H, Vallieres M, Bai HX, et al. . MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol. 2017;19(6):862–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grossmann P, Stringfield O, El-Hachem N, et al. . Defining the biological basis of radiomic phenotypes in lung cancer. Elife. 2017;6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. De Feyter HM, Behar KL, Corbin ZA, et al. . Deuterium metabolic imaging (DMI) for MRI-based 3D mapping of metabolism in vivo. Sci Adv. 2018;4(8):eaat7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets used and/or analysed in this study have been uploaded. The sequencing data, clinical and follow-up information of glioma patients were uploaded to the CGGA portal (http://cgga.org.cn/, accessed 1 December 2021). The method has been uploaded to GitHub (https://github.com/zhangjbig/RadioML, accessed 1 December 2021).