Abstract
We report that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Delta spike mutation P681R plays a key role in the Alpha-to-Delta variant replacement during the coronavirus disease 2019 (COVID-19) pandemic. Delta SARS-CoV-2 efficiently outcompetes the Alpha variant in human lung epithelial cells and primary human airway tissues. The Delta spike mutation P681R is located at a furin cleavage site that separates the spike 1 (S1) and S2 subunits. Reverting the P681R mutation to wild-type P681 significantly reduces the replication of the Delta variant to a level lower than the Alpha variant. Mechanistically, the Delta P681R mutation enhances the cleavage of the full-length spike to S1 and S2, which could improve cell-surface-mediated virus entry. In contrast, the Alpha spike also has a mutation at the same amino acid (P681H), but the cleavage of the Alpha spike is reduced compared with the Delta spike. Our results suggest P681R as a key mutation in enhancing Delta-variant replication via increased S1/S2 cleavage.
Keywords: SARS-CoV-2, Delta variant, fitness, spike P681R, S1/S2 cleavage, viral entry, human airway culture
Graphical abstract
It is important to identify mutations that account for the emergence of SARS-CoV-2 variants. Liu et al. show that the Delta spike mutation P681R enhances the cleavage of full-length spike to S1 and S2, which improves cell-surface-mediated virus entry and leads to the Alpha-to-Delta variant replacement.
Introduction
The continuous emergence of new variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) poses the greatest threat to pandemic control, vaccine effectiveness, therapeutic efficacy, and surveillance (Liu et al., 2022). Since its emergence in late 2019, mutations have unceasingly emerged in the circulating viruses, leading to variants with enhanced transmissibility, evasion of therapeutic antibodies, and breakthrough infections in vaccinated individuals (Brown et al., 2021; Chen et al., 2021; Hou et al., 2020; Liu et al., 2021b; Plante et al., 2021; Zhou et al., 2021). Since the viral spike glycoprotein is responsible for binding to the human cellular receptor angiotensin-converting enzyme (ACE2), many mutations have accumulated in the spike gene with the potential to alter viral fitness or to escape immunity. The variants have emerged from different geographic regions and, depending on their biological properties, have spread to other regions. The World Health Organization (WHO) has classified variants as “variants of concern” (i.e., Alpha, Beta, Gamma, Delta, and Omicron) and “variants of interest” (i.e., Eta, Iota, Kappa, and Lambda) (WHO, 2021). The Alpha variant was first identified in the United Kingdom in September 2020 and subsequently became dominant in many parts of the world. Afterward, the Delta variant emerged in India in October 2020 and subsequently spread to over 119 countries, displacing the Alpha variant globally (CDC, 2021; WHO, 2021). From May 2 to July 31 of 2021, the prevalence of the Delta variant in the United States increased from 1.3% to 94.4%, whereas the prevalence of the Alpha variant decreased from 70% to 2.4% (https://covid.cdc.gov/covid-data-tracker/#variant-proportions). More seriously, the Delta variant has been associated with increased transmissibility, disease severity, and breakthrough infections in vaccinated individuals (Brown et al., 2021; Chia et al., 2021; Li et al., 2021; Mlcochova et al., 2021). The mutation(s) that have driven the explosive spread of the Delta variant and its displacement of the Alpha variant remain to be defined. In this study, we used a reverse genetic approach to identify the molecular determinant(s) for the enhanced fitness of Delta variant and its dominance over the Alpha variant.
Results
Delta variant replicates more efficiently than Alpha variant on Calu-3 and primary human airway epithelial cultures
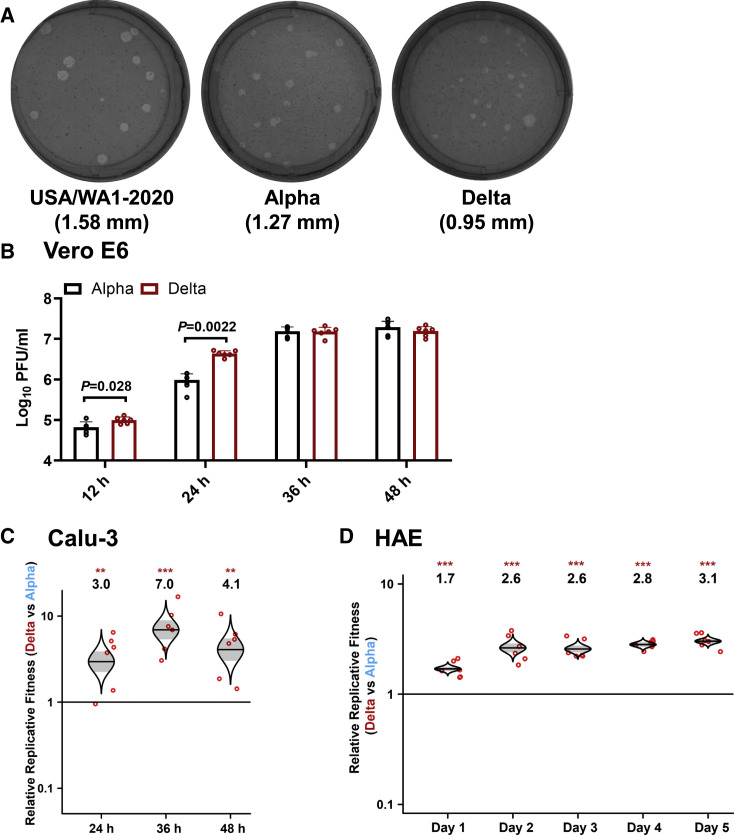
We constructed infectious cDNA clones for the Alpha (GISAIS: EPI_ISL_999340) and Delta (GISAIS: EPI_ISL_2100646) SARS-CoV-2 variants using a previously established protocol (Figure S1) (Xie et al., 2020, 2021). The infectious cDNA clones enabled us to prepare recombinant Alpha and Delta SARS-CoV-2 variants (Figure 1A). Both Alpha and Delta variants rescued from these infectious clones developed smaller plaques on Vero E6 cells than the earlier USA/WA1-2020 (wild-type) strain isolated in January 2020 (Figure 1A). Sequencing analysis showed no undesired mutations in the rescued recombinant virus stocks. On Vero E6 cells, the Delta variant replicated faster than the Alpha variant at 12 and 24 h; both variants produced comparable peak viral titers at 36 and 48 h post infection (Figure 1B). To compare the viral replication fitness more precisely between the Alpha and Delta variants, we performed a competition assay by infecting cells with a mixture of the two viruses at a plaque-forming unit (PFU) ratio of 1:1, followed by quantifying the ratios of the two viral RNA species at different days post infection. Compared with analyzing individual viruses separately, the competition assay has the advantages of (1) a built-in internal control of each viral replication and (2) elimination of host-to-host variation that reduces experimental power. Because of its precision and reproducibility (Liu et al., 2021a), the competition assay has been widely used to study microbial fitness (Bergren et al., 2020; Grubaugh et al., 2016; Wiser and Lenski, 2015), including SARS-CoV-2 (Liu et al., 2021b; Plante et al., 2021; Touret et al., 2021). When infecting human lung adenocarcinoma Calu-3 cells, the RNA ratio of Delta versus Alpha increased to 3.0, 7.0, and 4.1 at 24, 36, and 48 h post infection, respectively (Figure 1C). When infecting primary human airway epithelial (HAE) cultures, the RNA ratio of Delta versus Alpha increased from 1.7 on day 1 to 3.1 on day 5 (Figure 1D). These results indicate that the Delta variant has greater replication fitness compared with the Alpha variant in in vitro respiratory models of SARS-CoV-2 infection.
Figure 1.
Delta variant has an improved fitness over Alpha variant on the Calu-3 and primary human airway epithelial cultures
(A) Plaque morphologies of USA-WA1/2020, Alpha, and Delta viruses. The plaque images were taken on day 2.5 post infection of Vero E6 cells. The average diameters of the plaques are presented in the parentheses.
(B) Viral kinetics on Vero E6 cells. Recombinant Alpha and Delta variants were inoculated onto Vero E6 cells at an MOI of 0.01. After 1 h infection, the cells were washed thrice with DPBS to remove unattached viruses. The viral titers in the culture supernatant were detected at 12, 24, 36, and 48 h post infection. Red dots represent individual cell cultures (n = 6) pooled from 2 independent biological repeats. Data are mean ± SEM. Statistical analysis was performed using nonparametric two-tailed Mann-Whitney test.
(C) Replication competition between Delta and Alpha variants on Calu-3 cells. Culture medium was sampled for Sanger sequencing at 24, 36, and 48 h post infection.
(D) Replication competitions between Alpha and Delta variants on primary human airway epithelial (HAE) cells. Secreted progeny viruses were collected by incubating the apical side of the HAE culture with 300 μL DPBS at 37°C for 30 min from day 1 to 5. For both (C) and (D), equal PFUs of Alpha and Delta SARS-CoV-2s were mixed and inoculated onto Calu-3 and HAE cells at an MOI of 5. At 2 h post infection, the cells were washed thrice with DPBS to remove unattached viruses. Red dots represent individual cell cultures (n = 6) pooled from 2 independent biological repeats. The horizontal lines in each catseye represent the mean. Shaded regions represent standard error of the mean. y axes use a log10 scale. Black numbers above each set of values (catseye) indicate the ratios of two viral RNA species. p values were calculated for group coefficient using linear regression model. ∗∗p < 0.01, ∗∗∗p < 0.001.
Spike gene drives the improved replication of Delta variant over Alpha in HAE cultures
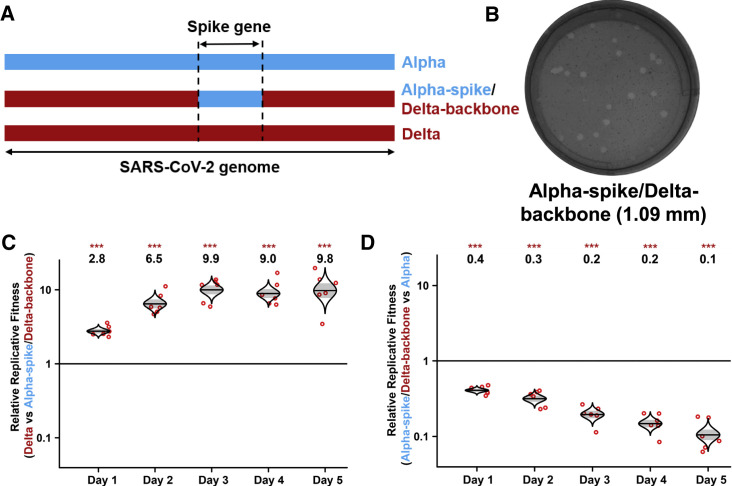
To examine if the spike gene alone determines the improved replication fitness of the Delta variant, we constructed a chimeric Delta SARS-CoV-2 bearing the Alpha spike glycoprotein (i.e., Alpha-spike/Delta-backbone virus; Figure 2A). The plaque size of Alpha-spike/Delta-backbone virus is between those of Alpha and Delta variants (compare Figure 2B with Figure 1A). In a competition assay on HAE culture, the RNA ratio of Delta versus Alpha spike/Delta backbone continuously increased from 2.8 on day 1 to 9.8 on day 5 post infection (Figure 2C), suggesting that (1) the Alpha spike reduces the replication fitness of the Delta variant and (2) the spike gene drives the improved replication of Delta variant. Interestingly, the Alpha-spike/Delta-backbone virus replicated less efficiently than the Alpha variant on HAE culture (Figure 2D), suggesting that, in contrast to Delta spike mutations that enhance replication, mutations outside the spike gene of the Delta variant reduced to some degree of fitness for viral replication. The above HAE competition experiments were quantified by Sanger sequencing (Figures 1D, 2C, and 2D) and subsequently verified by Illumina next-generation sequencing (NGS; Figure S2).
Figure 2.
The spike glycoprotein is responsible for the improved replication fitness of Delta variant in primary human epithelial airway cultures
(A) Schemes of Alpha variant, Delta variant, and Delta variant bearing Alpha spike. The spike gene of Delta variant was swapped with Alpha variant, resulting in a chimeric SARS-CoV-2 of “Alpha-spike/Delta-backbone.”
(B) Plaques of Alpha-spike/Delta-backbone chimeric virus. The plaque images were taken on day 2.5 post infection of Vero E6 cells. The average diameter of the plaques are presented in the parentheses.
(C and D) Replication competitions between Delta and Alpha-spike/Delta-backbone (C) and Alpha-spike/Delta-backbone and Alpha (D) on primary HAE cells. Equal PFU amounts of two viruses were mixed and inoculated onto HAE cells at an MOI of 5. After 2 h incubation, the cells were washed thrice with DPBS and maintained for 5 days. Secreted progeny viruses were collected daily by incubating the apical side of the HAE culture with 300 μL DPBS at 37°C for 30 min. Red dots represent individual cell cultures (n = 6) pooled from 2 independent biological repeats; the horizontal lines in each catseye represent the mean; shaded regions represent standard error of the mean; y axes use a log10 scale. Black numbers above each set of values (catseye) indicate the ratios of two viral RNA species. p values were calculated for group coefficient using linear regression model. ∗∗∗p < 0.001.
Spike mutation P681R enhances Delta viral replication
The Delta spike has accumulated mutations T19R, G142D, E156G, F157-R158 deletion, L452R, T478K, D614G, P681R, and D950N (Figure S1C) (World Health Organization, 2022), among which P681R is located at a furin cleavage site (PRRAR↓S with P681 underlined and “ ↓” indicating furin cleavage) that is absent in other group 2B coronaviruses (Coutard et al., 2020). Since the furin cleavage site was shown to be important for SARS-CoV-2 replication and pathogenesis (Johnson et al., 2021; Peacock et al., 2021), we hypothesized that mutation P681R may improve the furin cleavage efficiency of full-length spike to S1 and S2, leading to a more efficient virus entry into respiratory epithelial cells. To test this hypothesis, we reverted the Delta R681 substitution to wild-type P681 in the Delta SARS-CoV-2 backbone (Figure 3A). The resulting Delta-P681 virus developed plaques slightly bigger than wild-type Delta (compare Figures 3B and 1A). Remarkably, the P681 reversion attenuated Delta variant replication on HAE cultures, as evidenced by the increase in the RNA ratio of wild-type Delta versus Delta-P681 from 2.2 on day 1 to 3.4 on day 5 (Figure 3C). The replication of the Delta-P681 virus was even lower than that of the Alpha variant, as suggested by the decrease in the RNA ratio of Delta-P681 versus Alpha variant from 0.9 on day 1 to 0.6 on day 5 (Figure 3D). These results demonstrate that mutation P681R at the furin cleavage site plays a critical role in enhancing the replication of the Delta variant on primary human airway cultures.
Figure 3.
Delta spike P681 reversion impairs viral fitness
(A) Construction of revertant Delta-P681 SARS-CoV-2. Single nucleotide G-to-C substitution was engineered into the Delta variant to construct Delta-P681 SARS-CoV-2.
(B) Plaques of Delta-P681 virus. The plaque images were taken on day 2.5 post infection of Vero E6 cells. The average diameter of the plaques are presented in the parentheses. For direct comparison of plaque sizes of different viruses, all plaques presented in different figures of this paper were performed from the same set of experiment.
(C and D) Replication competitions between Delta and Delta-P681 (C) and Delta-P681 and Alpha (D) on HAE cultures. Equal PFU inputs of two viruses were mixed and inoculated onto HAE cultures at an MOI of 5. After 2 h incubation, the cells were washed thrice with DPBS and maintained for 5 days. Secreted progeny viruses were collected daily by incubating the apical side of the HAE culture with 300 μL DPBS at 37°C for 30 min. Red dots represent individual cell cultures (n = 6) pooled from 2 independent biological repeats; the horizontal lines in each catseye represent the mean; shaded regions represent standard error of the mean; y axes use a log10 scale. Black numbers above each set of values (catseye) indicate the ratios of two viral RNA species. p values were calculated for group coefficient using linear regression model. ∗∗p < 0.01, ∗∗∗p < 0.001.
Spike mutation P681R improves the cleavage of Delta full-length S to S1 and S2 subunits
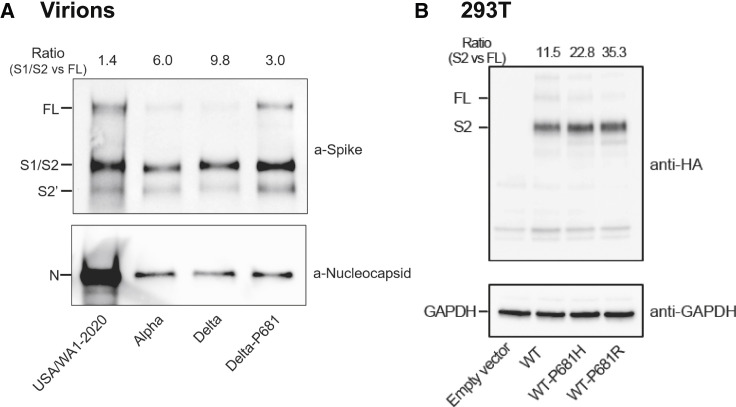
We directly evaluated the spike cleavage of Alpha, Delta, Delta-P681, and wild-type SARS-CoV-2. Virions were prepared from Vero E6 cells expressing TMPRSS2 (Vero-TMPRSS2), a host serine protease that is required for SARS-CoV-2 entry via the ACE2-mediated cell surface mechanism (Murgolo et al., 2021). We chose Vero-TMPRSS2 cells for virion production because the primary HAE cultures did not produce enough virions for sucrose-cushion purification. After virions were purified through the sucrose-cushion ultracentrifugation, pelleted viruses were analyzed for spike cleavage by western blotting. The results showed that spike-processing efficiency of the purified virions occurred in the order of Delta > Alpha > Delta-P681 > wild-type USA/WA1-2020 virions (Figure 4A). On a 4%–20% gradient SDS-PAGE gel, the S1 and S2 bands migrated closely together (S1/S2), and the Delta S2′ band migrated slightly slower than the wild-type USA/WA1-2020 and Alpha S2′ bands (Figure 4A), possibly because of distinct amino-acid substitutions and deletions among the variants. It should be noted that, similar to the Delta P618R substitution, the Alpha variant also has a spike mutation at amino-acid position 681 (P681H), which may contribute to the increase in spike cleavage when compared with the wild-type USA/WA1-2020 virus; however, we previously showed that this mutation alone did not enhance viral fitness or transmission (Liu et al., 2021b). Overall, our results suggest a correlation of improved virion spike cleavage with enhanced viral replication of Delta variant.
Figure 4.
The Delta spike P681R mutation improves spike protein processing
(A) Spike cleavages of purified virions. USA/WA1-2020, Alpha, Delta, and Delta-P681 viruses were purified and analyzed by western blot using polyclonal antibodies against spike and anti-nucleocapsid antibodies. Full-length (FL) spike, cleaved S1/S2, and S2′ proteins were annotated.
(B) Intracellular spike cleavages. C-terminally HA-tagged USA/WA1-2020 wild-type, P681H, and P681R mutant spikes were expressed in 293T cells. Total cell lysates were analyzed by western blot using anti-HA antibody. For both (A) and (B), one representative image of three independent experiments is shown. The densitometry was quantified by ImageLab 6.0.1 (BioRad). The ratios of S1/S2 or S2 subunits over FL spike are indicated at the top of the western blots.
Next, we examined the intracellular spike cleavage by transfecting HEK293T cells with a plasmid expressing the USA/WA1-2020 full-length spike (wild-type) fused with a C-terminal hemagglutinin (HA)-tag. Western blot of cell lysates showed that, compared with the wild-type spike (P681), Alpha H681 and Delta R681 substitution increased the intracellular spike cleavage by approximately 2- and 3-fold, respectively (Figure 4B). The results indicate that Delta P681R substitution and, to a lesser extent, alpha P681H substitution improve the intracellular furin-mediated cleavage of spike into S1 and S2 subunits.
More efficient replication of Delta variant than Alpha is not due to improved spike/hACE2 receptor interaction
To exclude the possibility that enhanced replication of the Delta variant over Alpha was due to an improved spike/ACE2 receptor interaction, we performed a binding assay using recombinant spike receptor-binding domain (RBD) and human ACE2 proteins on a Bio-Layer Interferometry (BLI) system (Figure S3). Within the RBD, the Alpha RBD has a N501Y mutation, whereas Delta RBD has L452R and T478K mutations. The BLI results indicate that the Alpha RBD has a higher binding affinity for ACE2 than Delta RBD, as indicated by >2,000-fold KD improvement (Figure S3). These data argue that the higher replication of Delta variant than Alpha variant is not due to an improved spike/ACE2 receptor binding.
Viral replication and transmission in hamsters
Encouraged by the observation that Delta variant replicated more efficiently than Alpha variant on Calu-3 cells (Figure 1C) and HAE cultures (Figure 1D), we performed competition experiments between the Delta and Alpha variants in a hamster viral transmission model, as we had previously done for D614G and Alpha variants (Liu et al., 2021b; Plante et al., 2021). After being intranasally inoculated with a 1:1 ratio of Alpha- and Delta-variant mixture, the donor hamsters were individually co-housed with naive recipient hamsters for 8 h to allow transmission, after which the animals were separated into individual cages. NGS analysis was performed on nasal washes from both donor and recipient animals to quantify the viral RNA ratio of Alpha versus Delta variants. For donor animals, the Alpha versus Delta RNA ratio increased from 1.9 on day 1 to 3.5 on day 4 post infection (Figure S4A). For recipient animals, the Alpha versus Delta ratio fluctuated between 5.7 and 18.3 from day 1 to 4 post contact (Figure S4B). Thus, the Alpha variant exhibited more efficient replication and transmission than the Delta variant in hamsters. These results suggest that hamster results do not recapitulate the HAE results or the human disease/transmission in this case. Similar discrepant results were recently reported for Omicron variant when analyzed in mouse and hamster models (Halfmann et al., 2022). The better replication of Alpha over Delta variant in hamsters might result from mutation N501Y in the Alpha spike that seems to significantly improve the usage of rodent ACE2 (Niu et al., 2021).
Discussion
Since the emergence of SARS-CoV-2, the virus has accumulated mutations to improve transmission and/or evasion of immunity. A number of mutations have been identified to improve viral fitness and transmission. First, SARS-CoV-2 accumulated a D614G mutation in the spike gene to enhance viral transmission (Hou et al., 2020; Korber et al., 2020; Plante et al., 2021; Zhou et al., 2021). This mutation promotes spike RBD in an “open” conformation to facilitate ACE2 receptor binding (Yurkovetskiy et al., 2020). Second, spike mutation N501Y emerged independently in Alpha, Beta, Gamma, and Omicron variants from different geographic regions. The N501Y mutation further increases the binding affinity between the spike protein and ACE2, leading to additional improvements in viral transmission (Liu et al., 2021b; Mlcochova et al., 2021; Wan et al., 2020). The current study aimed to identify mutation(s) in the Delta variant that are responsible for the Alpha-to-Delta replacement. Our results showed that spike mutation(s) contribute to the enhanced viral fitness of the Delta variant over Alpha. Importantly, the P681R mutation plays a critical role in this fitness advantage and increases the processing of Delta spike to S1 and S2, most likely through an improved furin cleavage when newly assembled virions egress through the trans-Golgi network. Although the original SARS-CoV-2 strain has a functional furin cleavage site with a minimal recognition site of RXXR↓ (Molloy et al., 1992), adjacent residues influence the cleavage efficiency (Krysan et al., 1999). Indeed, the Delta P681R substitution increases the intracellular spike cleavage (Figure 4B). Corroboratively, purified extracellular Delta virions showed an improved spike cleavage than the wild-type USA/WA1-2020 and Alpha virions; reversion of the Delta P681R to wild-type P681 alone reduced the spike cleavage of the purified Delta virion (Figure 4A). These results support the hypothesis that when the Delta variant infects respiratory epithelial cells, it binds to ACE2 receptor via the RBD in S1; already cleaved at the S1/S2 site, the Delta virion facilitates cleavage at S2′ by the cell-surface protease TMPRSS2, leading to an activation of the S2 fusion peptide for viral and plasma membrane fusion (Murgolo et al., 2021). This hypothesis is also supported by other studies reporting that the P681R substitution improves spike cleavage of Delta variant to increase viral replication and cell fusion (Escalera et al., 2022; Saito et al., 2022).
In contrast to the Delta variant, which improves a TMPRSS2-mediated plasma membrane route of virus entry, the recently emerged Omicron variant, which also harbors the same spike P681H substitution as Alpha variant, increases viral replication on human primary nasal epithelial cultures through an efficient endosomal route of virus entry. Consistently, the Omicron spike was less efficiently cleaved than the Delta spike, suggesting an altered TMPRSS2 usage for virus entry. The altered TMPRSS2 usage could impact viral tropism and reduce fusogenicity of infected cells (Meng et al., 2022; Peacock et al., 2022; Yamasoba et al., 2022). Nevertheless, the Omicron variant has a higher transmissibility than the Delta variant in humans (Lyngse et al., 2022). The molecular mechanism of high transmission for Omicron remains to be determined. Besides viral replication, other factors, such as immune evasion of antibody neutralization elicited by vaccination or infection (Xia et al., 2022; Zou et al., 2022), could contribute to Omicron surges around the world.
In summary, using a reverse genetic system and primary human airway cultures, we have identified spike mutation P681R as a determinant for enhanced viral replication fitness of the Delta compared with the Alpha variant. The P681R mutation enhances spike protein processing, most likely through the improved furin cleavage site. As new variants continue to emerge, spike mutations that affect furin cleavage efficiency, as well as other mutations that may increase viral replication, pathogenesis, and/or immune escape, must be closely monitored. In addition, the recent studies on both Delta and Omicron variants underline the importance in functionally analyzing variant mutations to validate sequence-based predictions.
Limitations of the study
One weakness of the current study is the discrepancy between the cell culture Calu-3/HAE results and the hamster transmission results. A similar discrepancy was observed for Omicron variant when analyzed in cell culture and mouse/hamster models (Halfmann et al., 2022). These discrepancies may be caused by the differences in ACE receptors and their distributions in the respiratory tracts of human and animal species. However, it should be noted that the Calu-3 and HAE culture data recapitulated the increased viral replication, transmission, and disease severity of the Delta variant that were observed in humans (Brown et al., 2021; Chia et al., 2021; Li et al., 2021; Mlcochova et al., 2021). More studies are needed to identify the factor(s) responsible for the cell culture and hamster model discrepancy. Another weakness is the lack of data on Delta-spike/Alpha-backbone virus or Alpha-R681 virus (i.e., Alpha virus with a single R681 mutation) to further support our conclusion that spike P681R is a key mutation in enhancing Delta variant replication via increased S1/S2 cleavage.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-GAPDH | Sigma-Aldrich | Cat#G9545; RRID:AB_796208 |
| anti-HA | Cell Signaling Technology | Cat#2367; RRID:AB_10691311 |
| SuperScript IV First-Strand Synthesis System | Thermo Fisher Scientific | Cat#18091300 |
| Platinum SuperFi II DNA Polymerase | Thermo Fisher Scientific | Cat#12361010 |
| E. coli strain Top10 | ThermoFisher Scientific | Cat#C404006 |
| Bacterial and virus strains | ||
| USA-WA1-2020 SARS-CoV-2 | Xie et al., 2020 | N/A |
| Alpha-FL | This paper | N/A |
| Delta-FL | This paper | N/A |
| Alpha-spike/Delta backbone | This paper | N/A |
| Delta-P681 | This paper | N/A |
| WT-P681H | This paper | N/A |
| WT-P681R | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Puromycin | Thermo Fisher Scientific | Cat#A1113802 |
| Polybrene | Sigma-Aldrich | Cat#TR-1003 |
| TRIzol LS Reagent | Thermo Fisher Scientific | Cat#10296028 |
| Human ACE2 protein | Sino Biological | Cat#10108-H08H |
| Human IgG1 Fc-tagged RBD protein | Ku et al., 2021 | N/A |
| Critical commercial assays | ||
| T7 mMessage mMachine kit | Thermo Fisher Scientific | Cat#AM1344 |
| Ingenio Electroporation solution | Mirus Bio LLC | Cat#MIR 50117 |
| Lenti-X Packaging Single Shots (VSV-G) | Takara | Cat#631275 |
| NEBuilder HiFi DNA Assembly kit | New England Biolabs | Cat#E5520S |
| Deposited data | ||
| Raw NGS data for HAE competition | This paper | NCBI Sequence Read Archive database (accession: PRJNA829976) |
| Experimental models: Cell lines | ||
| HEK293T cells | ATCC | Cat#CRL-3216,RRID:CVCL_0063 |
| Vero E6 cells | ATCC | Cat#CRL-1586,RRID: CVCL_0574 |
| Vero-E6 TMPRSS2 | SEKISUI XenoTech, LLC | N/A |
| Calu-3 cells | ATCC | Cat#HTB-55, RRID:CVCL_0609 |
| The EpiAirway system | MatTek Life Science | Cat#EpiAirway |
| Experimental models: Organisms/strains | ||
| Hamster: HsdHan:AURA | Envigo | Cat#8902M |
| Oligonucleotides | ||
| Primers used for SARS-CoV-2 infectious clones’ construction, RT-PCR and sequencing, See Table S1 | This paper (see Table S1) | N/A |
| Software and algorithms | ||
| Prism 9.0 software | GraphPad | N/A |
| QSVanalyser | Insilicase | N/A |
| ImageJ | NIH | N/A |
| Illustrator CC | Adobe | N/A |
| Octet Data Acquisition 9.0 | Sartorius | N/A |
| Octet Data Analysis software V11.1 | Sartorius | N/A |
| R 3.6.1 | The R Project for Statistical Computing | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Lead Contact, Dr. Pei-Yong Shi (peshi@utmb.edu).
Materials availability
Plasmids and virus generated in this study will be made available on request, but we might require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
Experimental model and subject details
Cells
African green monkey kidney epithelial Vero E6 cells and HEK293 cells (ATCC, Manassas, VA, USA) were grown in Dulbecco’s modified Eagle’s medium (DMEM; Gibco/Thermo Fisher, Waltham, MA, USA) with 10% fetal bovine serum (FBS; HyClone Laboratories, South Logan, UT) and 1% penicillin/streptomycin (Gibco). Vero E6 cells expressing human TMPRSS2 (Vero-TMPRSS2) were obtained from SEKISUI XenoTech, LLC (Kansas City, KS) and maintained in 10% fetal bovine serum (FBS; HyClone Laboratories, South Logan, UT) and 1% P/S and 1 mg/ml G418 (Gibco) as previously reported (Matsuyama et al., 2020). Human lung adenocarcinoma epithelial Calu-3 cells (ATCC) were maintained in a high-glucose DMEM containing sodium pyruvate and GlutaMAX (Gibco) with 10% FBS and 1% penicillin/streptomycin at 37°C with 5% CO2. The EpiAirway system is a primary human airway 3D tissue model purchased from MatTek Life Science (Ashland, MA, USA). This EpiAirway system was maintained with the provided culture medium at 37°C with 5% CO2 following manufacturer’s instruction. All other culture medium and supplements were purchased from Thermo Fisher Scientific (Waltham, MA, USA). All cell lines were verified and tested negative for mycoplasma.
Hamsters
Hamster studies were performed in accordance with the guidance for the Care and Use of Laboratory Animals of the University of Texas Medical Branch (UTMB). The protocol (2009087) was approved by the Institutional Animal Care and Use Committee (IACUC) at UTMB. All the hamster operations were performed under anesthesia by isoflurane to minimize animal suffering. Four- to six-week-old male golden Syrian hamsters, strain HsdHan:AURA (Envigo, Indianapolis, IN), were inoculated intranasally with 100 μl SARS-CoV-2. Five donor animals received a mixture containing 5 x 104 PFU of Alpha virus and 5 x 104 PFU of Delta virus. One day later, one infected donor animal was co-housed with one naïve recipient animal for 8 h (5 pairs). Afterwards, the donor and recipient animals were individually housed to separate cages. Nasal washes were collected in 400 μl sterile DPBS at indicated time points. Ratios of Alpha versus Delta RNA were determined via RT-PCR with quantification of NGS.
Method details
Construction of infectious cDNA clones and SARS-CoV-2 mutant viruses
The full-length cDNA clones of Alpha and Delta variants were constructed through mutagenesis of a previously established cDNA clone of USA-WA1-2020 SARS-CoV-2 (Xie et al., 2020, 2021). The previous seven-fragment in vitro ligation method was improved to a three-fragment ligation approach (Figure S1A) to construct the full-length cDNA clones of Alpha and Delta SARS-CoV-2, resulting in Alpha-FL and Delta-FL, respectively. Prior to the three-fragment ligation, mutations from Alpha or Delta variants were engineered into individual fragments of USA-WA1-2020 using a standard mutagenesis method. The sequences for constructing Alpha-FL, Delta-FL and Alpha-spike/Delta-backbone were downloaded from GISAID database, the accession ID for Alpha is EPI_ISL_999340, accession ID for Delta is EPI_ISL_2100646. Individual point mutations for Alpha (NSP3: P153L, T183I, A890D, I1412T; NSP6: SGF106-108del; NSP12: P323L; Spike: HV69-70del, Y145del, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H; ORF8: Q27stop, R52I, Y73C, S84L; N: D3L, R203K, G204R, S235F) and individual point mutations for Delta (NSP2: P129L; NSP3: P822L; H1274Y; NSP4: A446V; NSP6: V149A; NSP12: P323L; V355A; G671S; NSP13: P77L; NSP15: H234Y; Spike: T19R, G142D, E156G, FR157-158del, L452R, T478K, D614G, P681R, D950N; ORF3a: S26L; M: I82T; ORF7a: V82A; L116F; T120I; ORF8: S84L; DF119-120del; N: D63G; R203M; D377Y; R385K) were introduced into subclones of individual fragments by overlapping fusion PCR. For preparing Alpha-spike/Delta-backbone virus, the spike gene of Delta-FL was replaced with the spike gene of the Alpha. For preparing Delta-P681 virus, the P681 reversion was introduced into a subclone containing Delta spike gene by overlapping fusion PCR. All primers used for the construction were listed in Table S1. The full-length infectious clones of SARS-CoV-2 variants were assembled by in vitro ligation of contiguous DNA fragments. In vitro transcription was then performed to synthesize full-length genomic RNA. For recovering recombinant viruses, the RNA transcripts were electroporated into Vero E6 cells. The viruses from electroporated cells were harvested at 40 h post electroporation and served as P0 stocks. All viruses were passaged once on Vero E6 cells to produce P1 stocks for subsequent experiments. All P1 viruses were subjected to next generation sequencing to confirm the introduced mutations without undesired changes. Viral titers were determined by plaque assay on Vero E6 cells. The infectious titers of all recombinant P1 viruses were 1.9 > 107 PFU/ml in this study. All virus preparation and experiments were performed in a biosafety level 3 (BSL-3) facility. Viruses and plasmids are available from the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) at the University of Texas Medical Branch.
RNA extraction, RT-PCR, and sanger sequencing
Cell culture supernatants or nasal wash samples of hamsters were mixed with a five-fold excess of TRIzol™ LS Reagent (Thermo Fisher Scientific, Waltham, MA, USA). Viral RNAs were extracted according to the manufacturer’s instructions. The RNAs were amplified using a SuperScript™ III One-Step RT-PCR kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. The size of desired amplicon was verified with 2 μl of PCR product on an agarose gel. The remaining 18 μl of RT-PCR DNA was purified by a QIAquick PCR Purification kit (Qiagen, Germantown, MD, USA). Sequences of the purified RT-PCR products were generated using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Austin, TX, USA). The sequencing reactions were purified using a 96-well plate format (EdgeBio, San Jose, CA, USA) and analyzed on a 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA). The peak electropherogram height representing each mutation site and the proportion of each competitor was analyzed using the QSVanalyser program.
Plaque assay
Approximately 1.2×106 Vero E6 cells were seeded to each well of 6-well plates and cultured at 37°C, 5% CO2 for 16 h. Virus was serially diluted in DMEM with 2% FBS and 200 μl diluted viruses were transferred onto the cell monolayers. The viruses were incubated with the cells at 37°C with 5% CO2 for 1 h. After the incubation, overlay medium was added to the infected cells per well. The overlay medium contained DMEM with 2% FBS, 1% penicillin/streptomycin, and 1% sea-plaque agarose (Lonza, Walkersville, MD). After 2.5-day incubation, plates were stained with neutral red (Sigma-Aldrich, St. Louis, MO, USA) and plaques were counted on a light box. All plaque morphologies presented in different figures were performed from the same set of experiment.
Next generation sequencing (NGS)
The competition results generated by Sanger sequencing were confirmed using NGS methods. Briefly, viral RNA samples from competition groups of (i) Delta versus Alpha and (ii) Delta versus Alpha-spike/Delta-backbone were used for a specific one-step RT-PCR that containing the A23063T mutation site. Viral RNA samples from competition group of Alpha versus Alpha-spike/Delta-backbone were quantified by the T14444C mutation. The RT-PCR primers were listed in Table S1. The PCR products were purified by a QIAquick PCR Purification kit (Qiagen, Germantown, MD) according to the manufacturer’s protocol. Dual-indexed adapter sequences (New England BioLabs, Ipswich, MA) were added with 5 cycles of PCR. Samples were pooled and sequenced on an Illumina MiniSeq Mid-Output flow cell with the paired-end 150 base protocol. The reads were filtered for Q-scores of 37 at the A23063T and T14444C mutation sites and adjacent bases and counted. The input ratios and output ratios of two viruses were obtained and used for the relative replicative fitness analysis (Table S2). The output virus ratios were normalized by the input virus ratios and presented in Figures 1, 2, 3, and S2.
Viral infection of cell lines
Approximately 3×105 Calu-3 cells were seeded onto each well of 12-well plates and cultured at 37°C, 5% CO2 for 16 h. Equal PFUs of two viruses were inoculated onto Calu-3 cells at a final MOI of 0.1. The mixed viruses were incubated with the cells at 37°C for 2 h. After infection, the cells were washed thrice with DPBS to remove residual viruses. One milliliter of culture medium was added into each well. At each time point, 100 μl of culture supernatants were lysed in TRIzol LS reagent for the detection of competition assay, and 100 μl of fresh medium was added into each well to replenish the culture volume. The cells were infected in triplicate for each group of viruses. All samples were stored at -80°C until analysis.
Primary human airway cultures
The EpiAirway system is a primary human airway 3D mucociliary tissue model consisting of normal, human-derived tracheal/bronchial epithelial cells. Different combinations of mixed viruses for competition assays were inoculated onto the culture at a total MOI of 5 in DMEM with 2% FBS. After 2 h infection at 37°C with 5% CO2, the inoculum was removed, and the culture was washed three times with DPBS. The infected epithelial cells were maintained without any medium in the apical well, and medium was provided to the culture through the basal well. The infected cells were incubated at 37°C, 5% CO2. From day 1 to day 5 post-infection, 300 μl of DPBS were added onto the apical side of the airway culture and incubated at 37°C for 30 min to elute progeny viruses. All virus samples in DPBS were stored at −80°C and quantified by plaque assays on Vero E6 cells.
Virion purification and western blotting
Vero E6 expressing TMPRSS2 were infected with different SARS-CoV-2 at an MOI of 0.01. At 24 h post-infection, the culture medium was collected, purified through a 20% sucrose cushion, and analyzed by Western blot as previously described on a 4–20% gradient SDS-PAGE gel (Johnson et al., 2021). For the intracellular spike cleavage experiments, USA/WA1-2020 spike, P681H mutant, and P681R mutant were inserted to plasmid pVAX1 Vector with a C-terminal HA tag. Two micrograms of each plasmid were transfected and expressed in HEK293T cells for 24 h. Cells were harvested and lysed by RIPA buffer for Western blot analysis (anti-HA, Cell Signaling Technology, 2367; anti-GAPDH, Sigma-Aldrich, G9545). Densitometry was performed to quantify the cleavage efficiency of full-length spike to S1/S2 or S2 subunits using ImageLab 6.0.1 (Bio-Rad #12012931). The average results of three experiments were presented.
Spike RBD and ACE2 binding
The human ACE2 protein was purchased from Sino Biological (Beijing, China; Cat# 10108-H08H) and the human IgG1 Fc-tagged RBD proteins were made in-house using a method as previously described (Ku et al., 2021). The affinity measurement was performed on the ForteBio Octet RED 96 system (Sartorius, Goettingen, Germany). Briefly, the RBD proteins (20 μg/ml) of Alpha or Delta RBDs were captured onto protein A biosensors for 300s. The loaded biosensors were then dipped into the kinetics buffer for 10 s for adjustment of baselines. Subsequently, the biosensors were dipped into serially diluted (from 1.23 to 300 nM) human ACE2 protein for 200 s to record association kinetics and then dipped into kinetics buffer for 400 s to record dissociation kinetics. Kinetic buffer without ACE2 was used to correct the background. The Octet Data Acquisition 9.0 software was used to collect affinity data. For fitting of KD values, Octet Data Analysis software V11.1 was used to fit the curve by a 1:1 binding model using the global fitting method.
Quantification and statistical analysis
Hamsters were randomly allocated into different groups. The investigators were not blinded to allocation during the experiments or to the outcome assessment. No statistical methods were used to predetermine sample size. Descriptive statistics have been provided in the figure legends. For in vitro replication kinetics, Kruskal–Wallis analysis of variance was conducted to detect any significant variation among replicates. If no significant variation was detected, the results were pooled for further comparison. Differences between continuous variables were assessed with a non-parametric Mann–Whitney test. Analyses were performed in Prism version 9.0 (GraphPad, San Diego, CA). For virus competition experiments, relative replicative fitness values for different variants were analyzed according to w=(f0/i0), where i0 is the initial two-virus ratio and f0 is the final two-virus ratio after competition. Sanger sequencing (initial timepoint T0) counts for each virus being compared were based upon average counts over three replicate samples of inocula per experiment, and post-infection (timepoint T1) counts were taken from samples of individual subjects. For cell culture samples, multiple experiments were performed, so that f0/i0 was clustered by experiment. To model f0/i0, the ratio T0/T1 was found separately for each subject in each virus group, log (base-10) transformed to an improved approximation of normality and modeled by analysis of variance with relation to group, adjusting by experiment when appropriate to control for clustering within experiment. Specifically, the model was of the form Log10_CountT1overCountT0 ∼ Experiment + Group. Fitness ratios between the two groups [the model’s estimate of w=(f0/i0)] were assessed per the coefficient of the model’s Group term, which was transformed to the original scale as 10∧coefficient. This modeling approach compensates for any correlation due to clustering within experiment similarly to that of corresponding mixed effect models and is effective since the number of experiments was small. Statistical analyses were performed using R statistical software (R Core Team, 2019, version 3.6.1). In all statistical tests, two-sided alpha = .05. Catseye plots (Cumming, 2014), which illustrate the normal distribution of the model-adjusted means, were produced using the “catseyes” package (Andersen, 2019).
Acknowledgments
P.-Y.S. was supported by National Institutes of Health (NIH) grants HHSN272201600013C, U01AI151801, and UL1TR001439 and awards from the Sealy and Smith Foundation, the Kleberg Foundation, the John S. Dunn Foundation, the Amon G. Carter Foundation, the Gillson Longenbaugh Foundation, and the Summerfield Robert Foundation. Z.A. was supported by Welch Foundation grant AU-0042-20030616 and Cancer Prevention and Research Institute of Texas (CPRIT) grants RP150551 and RP190561. S.C.W. was supported by NIH grant R24 AI120942. V.D.M. was supported by NIH grants AI153602 and 1R21AI145400. J.L. and B.A.J. were supported by the James W. McLaughlin Fellowship Fund.
Author contributions
Conceptualization, Y.L., J.L., S.C.W., V.D.M., X.X., and P.-Y.S.; methodology, Y.L., J.L., B.A.J., H.X., Z.K., C.S., S.G.W., Z.A., and X.X.; investigation, Y.L., J.L., B.A.J., H.X., Z.A., S.C.W., V.D.M., X.X., and P.-Y.S.; resources, H.X., B.A.J., Z.K., Z.A., and X.X.; data curation, Y.L., J.L., B.A.J., H.X., Z.K., C.S., S.C.W., V.D.M., X.X., and P.-Y.S.; writing – original draft, Y.L., J.L, X.X., and P.-Y.S.; writing – review & editing, Y.L., J.L., B.A.J., H.X., Z.K., C.S., S.G.W., Z.A., S.C.W., V.D.M., X.X., and P.-Y.S.; supervision, X.X., S.C.W., V.D.M., and P.-Y.S.; funding acquisition, Z.A., S.C.W., V.D.M., and P.-Y.S.
Declaration of interests
X.X., V.D.M., and P.-Y.S. have filed a patent on the reverse genetic system and reporter SARS-CoV-2.
Published: May 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110829.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. The NGS sequencing data has been deposited to NCBI Sequence Read Archive database (accession: PRJNA829976).
This paper does not report original code.
Any other information required to reanalyze the data reported in this paper are available upon request.
References
- Andersen C. 2019. Catseyes: Create Catseye Plots Illustrating the Normal Distribution of the Means. R package version 0.2.3. [Google Scholar]
- Bergren N.A., Haller S., Rossi S.L., Seymour R.L., Huang J., Miller A.L., Bowen R.A., Hartman D.A., Brault A.C., Weaver S.C. "Submergence" of Western equine encephalitis virus: evidence of positive selection argues against genetic drift and fitness reductions. PLoS Pathog. 2020;16:e1008102. doi: 10.1371/journal.ppat.1008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.M., Vostok J., Johnson H., Burns M., Gharpure R., Sami S., Sabo R.T., Hall N., Foreman A., Schubert P.L., et al. Outbreak of SARS-CoV-2 infections, including COVID-19 vaccine breakthrough infections, associated with large public gatherings — Barnstable county, Massachusetts, July 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:1059–1062. doi: 10.15585/mmwr.mm7031e2. https://www.cdc.gov/mmwr/volumes/70/wr/mm7031e7032.htm?s_cid=mm7031e7032_w&fbclid=IwAR7032WV7036ul_A-l_VN_7015KX7034bedb7038CeLJKRwiDWZ-bIUuWmZMKbs7094xdhTiPLs [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC COVID data tracker. 2021. https://covid.cdc.gov/covid-data-tracker/#variant-proportions
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia P.Y., Ong S.W.X., Chiew C.J., Ang L.W., Chavatte J.-M., Mak T.-M., Cui L., Kalimuddin S., Chia W.N., Tan C.W., et al. Virological and serological kinetics of SARS-CoV-2 Delta variant vaccine2 breakthrough infections: a multi-center cohort study. medRxiv. 2021 doi: 10.1016/j.cmi.2021.11.010. https://www.medrxiv.org/content/10.1101/2021.1107.1128.21261295v21261291 Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming G. The new statistics: why and how. Psychol. Sci. 2014;25:7–29. doi: 10.1177/0956797613504966. [DOI] [PubMed] [Google Scholar]
- Lyngse F.P., Kirkeby C.T., Denwood M., Christiansen L.E., Mølbak K., Møller C.H., Leo Skov R., Krause T.G., Rasmussen M., Sieber R.N., et al. Transmission of SARS-CoV-2 Omicron VOC subvariants BA.1 and BA.2: evidence from Danish households. BioRxiv. 2022 doi: 10.1101/2022.1101.1128.22270044. Preprint at. [DOI] [Google Scholar]
- Escalera A., Gonzalez-Reiche A.S., Aslam S., Mena I., Laporte M., Pearl R.L., Fossati A., Rathnasinghe R., Alshammary H., van de Guchte A., et al. Mutations in SARS-CoV-2 variants of concern link to increased spike cleavage and virus transmission. Cell Host Microbe. 2022;30:373–387.e7. doi: 10.1016/j.chom.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N.D., Weger-Lucarelli J., Murrieta R.A., Fauver J.R., Garcia-Luna S.M., Prasad A.N., Black W.C.T., Ebel G.D. Genetic drift during systemic Arbovirus infection of mosquito vectors leads to decreased relative fitness during host switching. Cell Host Microbe. 2016;19:481–492. doi: 10.1016/j.chom.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann P.J., Iida S., Iwatsuki-Horimoto K., Maemura T., Kiso M., Scheaffer S.M., Darling T.L., Joshi A., Loeber S., Singh G., et al. Consortium Mount Sinai Pathogen Surveillance PSP Study Group SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schafer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.A., Xie X., Bailey A.L., Kalveram B., Lokugamage K.G., Muruato A., Zou J., Zhang X., Juelich T., Smith J.K., et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature. 2021;591:293–299. doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan D.J., Rockwell N.C., Fuller R.S. Quantitative characterization of furin specificity. Energetics of substrate discrimination using an internally consistent set of hexapeptidyl methylcoumarinamides. J. Biol. Chem. 1999;274:23229–23234. doi: 10.1074/jbc.274.33.23229. [DOI] [PubMed] [Google Scholar]
- Ku Z., Xie X., Davidson E., Ye X., Su H., Menachery V.D., Li Y., Yuan Z., Zhang X., Muruato A.E., et al. Author Correction: molecular determinants and mechanism for antibody cocktail preventing SARS-CoV-2 escape. Nat. Commun. 2021;12:4177. doi: 10.1038/s41467-021-24440-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Deng A., Li K., Hu Y., Li Z., Xiong Q., Liu Z., Guo Q., Zou L., Zhang H., et al. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 Delta variant. medRxiv. 2021 doi: 10.1038/s41467-022-28089-y. https://www.medrxiv.org/content/10.1101/2021.1107.1107.21260122v21260122 Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Liu Y., Shan C., Nunes B.T.D., Yun R., Haller S.L., Rafael G.H., Azar S.R., Andersen C.R., Plante K., et al. Role of mutational reversions and fitness restoration in Zika virus spread to the Americas. Nat. Commun. 2021;12:595. doi: 10.1038/s41467-020-20747-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu J., Plante K.S., Plante J.A., Xie X., Zhang X., Ku Z., An Z., Scharton D., Schindewolf C., et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature. 2021;602:294–299. doi: 10.1038/s41586-021-04245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu J., Shi P.Y. SARS-CoV-2 variants and vaccination. Zoonoses. 2022;2:6. doi: 10.15212/zoonoses-2022-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U S A. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng B., Abdullahi A., Ferreira I., Goonawardane N., Saito A., Kimura I., Yamasoba D., Gerber P.P., Fatihi S., Rathore S., et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts tropism and fusogenicity. Nature. 2022;603:706–714. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlcochova P., Kemp S., Dhar M.S., Papa G., Meng B., Mishra S., Whittaker C., Mellan T., Ferreira I., Datir R., et al. SARS-CoV-2 B.1.617.2 Delta variant emergence, replication and sensitivity to neutralising antibodies. bioRxiv. 2021 doi: 10.1101/2021.05.08.443253. Preprint at. [DOI] [Google Scholar]
- Molloy S.S., Bresnahan P.A., Leppla S.H., Klimpel K.R., Thomas G. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J. Biol. Chem. 1992;267:16396–16402. doi: 10.1016/s0021-9258(18)42016-9. [DOI] [PubMed] [Google Scholar]
- Murgolo N., Therien A.G., Howell B., Klein D., Koeplinger K., Lieberman L.A., Adam G.C., Flynn J., McKenna P., Swaminathan G., et al. SARS-CoV-2 tropism, entry, replication, and propagation: considerations for drug discovery and development. PLoS Pathog. 2021;17:e1009225. doi: 10.1371/journal.ppat.1009225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Z., Zhang Z., Gao X., Du P., Lu J., Yan B., Wang C., Zheng Y., Huang H., Sun Q. N501Y mutation imparts cross-species transmission of SARS-CoV-2 to mice by enhancing receptor binding. Signal Transduct. Target. Ther. 2021;6:284. doi: 10.1038/s41392-021-00704-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock T.P., Goldhill D.H., Zhou J., Baillon L., Frise R., Swann O.C., Kugathasan R., Penn R., Brown J.C., Sanchez-David R.Y., et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021;6:899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Irie T., Suzuki R., Maemura T., Nasser H., Uriu K., Kosugi Y., Shirakawa K., Sadamasu K., Kimura I., et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature. 2022;602:300–306. doi: 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock T.P., Brown J.C., Zhou J., Thakur N., Newman J., Kugathasan R., Sukhova K., Kaforou M., Bailey D., Barclay W.S. The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. bioRxiv. 2022 doi: 10.1101/2021.1112.1131.474653. Preprint at. [DOI] [Google Scholar]
- Touret F., Luciani L., Baronti C., Cochin M., Driouich J.S., Gilles M., Thirion L., Nougairede A., de Lamballerie X. Replicative Fitness of a SARS-CoV-2 20I/501Y.V1 Variant from Lineage B.1.1.7 in Human Reconstituted Bronchial Epithelium. mBio. 2021;12:e0085021. doi: 10.1128/mBio.00850-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94:e00127-20. doi: 10.1128/jvi.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Track SARS-CoV-2 variants. 2021. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- Wiser M.J., Lenski R.E. A comparison of methods to measure fitness in Escherichia coli. PLoS One. 2015;10:e0126210. doi: 10.1371/journal.pone.0126210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Coronavirus Disease (COVID-19): Weekly Epidemiological Update. 2022. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---4-may-2022
- Xia H., Zou J., Kurhade C., Cai H., Yang Q., Cutler M., Cooper D., Muik A., Jansen K.U., Xie X., et al. Neutralization and durability of 2 or 3 doses of the BNT162b2 vaccine against Omicron SARS-CoV-2. Cell Host Microbe. 2022;30:485–488.e3. doi: 10.1016/j.chom.2022.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V., et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Lokugamage K.G., Zhang X., Vu M.N., Muruato A.E., Menachery V.D., Shi P.-Y. Engineering SARS-CoV-2 using a reverse genetic system. Nat. Protoc. 2021;16:1761–1784. doi: 10.1038/s41596-021-00491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasoba D., Kimura I., Nasser H., Morioka Y., Nao N., Ito J., Uriu K., Tsuda M., Zahradnik J., Shirakawa K., et al. Virological characteristics of SARS-CoV-2 BA.2 variant. bioRxiv. 2022 doi: 10.1101/2022.02.14.480335. Preprint at. [DOI] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Thao T.T.N., Hoffmann D., Taddeo A., Ebert N., Labroussaa F., Pohlmann A., King J., Steiner S., Kelly J.N., et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature. 2021;592:122–127. doi: 10.1038/s41586-021-03361-1. [DOI] [PubMed] [Google Scholar]
- Zou J., Xia H., Xie X., Kurhade C., Machado R.R.G., Weaver S.C., Ren P., Shi P.Y. Neutralization against Omicron SARS-CoV-2 from previous non-Omicron infection. Nat. Commun. 2022;13:852. doi: 10.1038/s41467-022-28544-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. The NGS sequencing data has been deposited to NCBI Sequence Read Archive database (accession: PRJNA829976).
This paper does not report original code.
Any other information required to reanalyze the data reported in this paper are available upon request.