Abstract
Background:
Little is known about the association of newborn DNA methylation (DNAm) with asthma acquisition across adolescence and early adult life.
Objective:
We aim to identify epigenetic biomarkers in newborns for asthma acquisition during adolescence or young adulthood.
Methods:
The Isle of Wight Birth Cohort (IOWBC) (n=1456) data at ages 10, 18, and 26 years were assessed. To screen Cytosine-phosphate-Guanine site (CpGs) potentially associated with asthma acquisition, at the genome-scale, we examined differentially methylated regions (DMR) using dmrff R package and individual CpG sites using linear regression on such associations. For CpGs that passed screening, we examined their enrichment in biological pathways using their mapping genes and tested their associations with asthma acquisitions using logistic regressions. Findings in IOWBC were tested in an independent cohort, the Avon Longitudinal Study of Parents and Children (ALSPAC) cohort.
Results:
In total, 2636 unique CpGs passed screening, based on which we identified one biological pathway linked to asthma acquisition during adolescence in females (FDR adjusted p-value=0.003 in IOWBC). Via logistic regressions, for females, 4 CpGs were shown to be associated with asthma acquisition during adolescence, and another 4 CpGs with asthma acquisition in young adulthood (FDR adjusted p-value<0.05 in IOWBC) and these 8 CpGs were replicated in ALSPAC (all p-values<0.05). DNAm at all the identified CpGs was shown to be temporally consistent, and at 6 of the CpGs was associated with expressions of adjacent or mapping genes in females (all p-values<0.05). For males, 622 CpGs were identified in IOWBC (FDR=0.01), but these were not tested in ALSPAC due to small sample sizes.
Conclusion and clinical relevance:
Eight CpGs on LHX5, IL22RA2, SOX11, CBX4, ACPT, CFAP46, MUC4, and ATP1B2 genes have the potential to serve as candidate epigenetic biomarkers in newborns for asthma acquisition in females during adolescence or young adulthood.
Keywords: IOWBC, ALSPAC, DNA methylation, Epigenome-wide, Asthma acquisition
Graphical Abstract
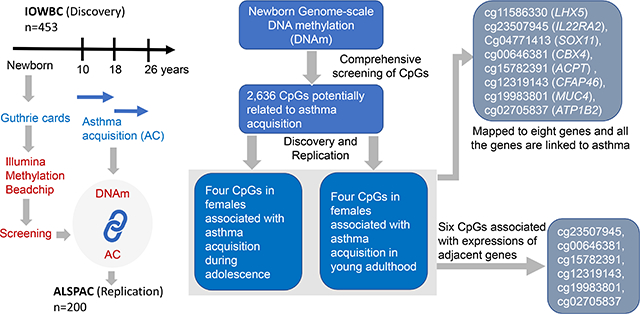
Little is known about the association of newborn DNAm with adolescence and young adulthood asthma acquisitions. In this work, we examined individual CpGs and differentially methylated regions (DMR) at birth at the genome-scale on their association with asthma acquisitions in a discovery and a replication cohort. The eight identified CpGs have a potential to serve as candidate epigenetic biomarker for asthma acquisition in females.
INTRODUCTION
Asthma is the most prevalent non-communicable disease in children and the second most prevalent chronic respiratory disease worldwide, with an estimated 300 million people affected and increasing disease burden in the next few decades1–3. Asthma developmental patterns (or asthma transitions when focusing on status change) involve childhood-onset asthma persistent until adulthood, asthma remission, asthma relapse, and adult-onset asthma4,5. These different developmental patterns are presumably linked to diverse underlying pathophysiological processes6–8. Sex-specific incidence of asthma occurs throughout life. In particular, females have more new-onset asthma during adolescence, while having fewer asthma occurrences during childhood compared to males9. However, to our knowledge, only a few studies focused on asthma patterns from childhood to adulthood with a substantial knowledge gap for asthma status transition from pre- to post-adolescence5,10. Furthermore, adulthood-onset asthma is more likely to have a poorer treatment response and worse prognosis compared to childhood-onset asthma11, emphasizing the importance of studies contributing to new-onset of asthma during adulthood.
Risk factors associated with childhood-onset asthma are generally different from adulthood-onset asthma11. Evidence shows that genetic predisposition plays a stronger role in asthma onset in early childhood with genetic risk factors for adult-onset asthma largely a subset of the genetic risk for childhood-onset asthma but with smaller effect sizes, suggesting a larger role for non-genetic risk factors such as environment in adult-onset asthma12,13. Prospective studies have shown that exposure to perennial allergens or to prenatal smoke are associated with a high risk to childhood-onset asthma14,15. In addition, viral infection and bacterial colonization of the airway also increase the risk of childhood-onset asthma16,17. For onset of asthma in adults, other risk factors such as irritant exposure to environmental pollutants, female sex hormones, upper airway disease, obesity and stress have shown to play a role11. Environmental exposures and developmental changes (e.g., puberty) associated with new onset of asthma in adulthood have been shown to modify the epigenome18,19. DNA methylation (DNAm) is one epigenetic mechanism that is established in utero and modified by environmental exposure and aging, which regulates gene expression and alters pathophysiological process of disease20.
Evidence has shown that DNAm patterns in blood-derived DNA are associated with the status of asthma21. However, the majority of studies have focused on childhood20–24. Some recent studies showed that DNAm in cord blood was associated with asthma status in childhood20–24. Nevertheless, it is unknown whether epigenetic factors at birth were associated with asthma acquisition at different stages of life from childhood to adulthood. Findings from such studies will be beneficial to future studies on predicting asthma acquisition at a much earlier age of life.
This study aimed to identify CpGs in blood at birth where DNAm at those sites was associated with asthma acquisition at different stages of life (from pre-adolescence to young adulthood) using sex-stratified epigenome-wide association studies (EWAS), in two birth cohorts, the Isle of Wight birth cohort (IOWBC; the discovery cohort) and the Avon Longitudinal Study of Parents and Children (ALSPAC; the replication cohort). We hypothesized that newborn DNAm was associated with asthma acquisition patterns from pre- to post-adolescence and to adulthood. For the identified CpGs, we further assessed their biological relevance by examining their association with expression of their mapping genes.
METHODS
Discovery cohort - IOWBC
The Isle of Wight Birth Cohort (IOWBC) is a population-based birth cohort established in 1989–1990 on the Isle of Wight, an island off the south coast of England25. A total of 1456 eligible children with parental consent were included in the cohort (IOWBC-F1 generation). We focused on subjects in the F1 generation with DNAm data available in newborns and who were asthma free at age 10 years (some of them developed asthma at age 18 years or 26 years). Gene expression data in the F2 generation (children of the IOWBC-F1 women and partners of F1-males)25.
Asthma acquisition
Asthma information at ages 10, 18, and 26 years was collected using International Study of Asthma and Allergies in Childhood questionnaires (ISAAC) with a detailed assessment of asthma symptoms and treatment. Asthma was defined by the core questions as: “ever had asthma” and either “wheezing or whistling in the chest during the previous 12 months” or “current treatment for asthma”. Two groups of asthma acquisition were defined based on asthma status at ages 10, 18 and 26 years. Asthma acquisition during adolescence was defined as asthma-free at age 10 and having asthma at ages 18 and 26 years (No-Yes-Yes). Asthma acquisition during young adulthood was defined as asthma-free at ages 10 and 18 years and acquiring asthma by age 26 years (No-No-Yes). A transition pattern involving asthma remission was not considered in the present study, i.e., the pattern of No-Yes-No. Participants without asthma at ages 10, 18, and 26 years (No-No-No) were included as the reference group of no asthma.
DNA methylation (DNAm) and cell type composition
In the F1 generation of IOWBC, a peripheral blood sample from a heel prick was collected on Guthrie cards within the first seven days of age (n=796). DNAm was measured using MethylationEPIC BeadChips. Details of DNAm assessment and preprocessing are in Supplemental Material Methods (S1). Base 2 logit-transformed DNAm β values (M-values) were used in all the analyses.
DNAm measurement is influenced by the heterogeneity of cell-type compositions in cord blood26. To control the influence of cell-type proportions, six cell types, CD4+ T cells, natural killer cells, neutrophil, B cells, monocytes, and eosinophils, were estimated using the R package minfi27,28 and included in the analyses as confounders.
Confounders
Confounding variables potentially associated with DNAm and asthma status included socioeconomic status (SES), secondhand smoke in childhood, breastfeeding duration, and body mass index (BMI) at age 10 years. SES was ascertained as ‘low’, ‘medium’, and’ high’ according to the assessment of parental occupation, level of household income, and the number of rooms in the house29. Secondhand smoke information at childhood was collected from their parents and either of them with smoking was regarded as secondhand smoke to children. Breastfeeding duration data were obtained at the 1- and 2- year follow-ups.
Gene expression in IOW Cohort F2 generation
Gene expression data in the F1 generation at birth was not available. Thus, to assess biological functionality of identified CpGs, DNAm and gene expression in the IOW F2 cohort, both assessed at birth, were utilized. In the F2 generation, RNA was isolated from cord blood samples (n=161) that were collected into PAXgene Bone Marrow RNA kits according to the manufacturer’s instructions (Qiagen, Valencia, CA, USA). RNA quality was determined using the Agilent 2100 BioAnalyzer system. Gene expression levels were measured using SurePrint G3 Human GE v2 8×60K Agilent Microarray (Agilent Technologies). The details on measurement and quality control of gene expression level are described elsewhere30.
Replication cohort – the ALSPAC cohort
The Avon Longitudinal Study of Children and Parents (ALSPAC) is a prospective population-based study established in the Bristol area of the United Kingdom during 1990–9231–33 (details are in the Supplemental Material S2). DNAm in cord blood (n=861) and asthma status at ages 10 (n=7796), 17 (n=5126), and 22 years (n=993), along with related confounders comparable to those in IOWBC, were analyzed34. Procedures for collection of cord blood samples and DNA sample preparation are described elsewhere35,36. DNA methylation in cord blood was measured using the Illumina Infinium HumanMethylation 450K BeadChip assay with over 485,000 CpG sites37. Cell-type proportions for cord blood at birth were estimated using the Houseman method38 with cord blood reference panel39.
Statistical analysis
Statistical analyses in IOWBC
All analyses were separated by sex due to different asthma acquisition patterns from pre-adolescence through young adults in males and females40. Analyses were performed using SAS 9.4 (SAS, Cary, NC, USA) and R packages. To examine whether participants included in the current study reasonably represented the complete study cohort, we evaluated the variables potentially related to asthma acquisition in both sexes. Categorical variables were presented as percentages compared to the complete cohort statistics using a one-sample proportion test. Continuous variables were presented as means and standard deviations and a one-sample t-test was applied to compare the subsamples with the complete cohort. For variables not following normal distribution, median and median absolute deviation (MAD) were present and signed rank tests were applied. A p-value <0.05 was deemed to be statistically significant.
We screened CpGs potentially associated with asthma acquisition from two directions. For both directions, the screening was stratified by sex. In the first direction, a linear regression was applied to each CpG site to assess the association of DNAm (as the dependent variable) with asthma transition status with no asthma at three ages (10, 18, and 26) as the reference group. A CpG with a p-value <0.001 (to be more stringent) was regarded as a candidate CpG. In addition, we applied the dmrff package in R to detect differentially methylated regions (DMRs) with respect to asthma acquisition that has 2 or more CpG sites within 500bp with multiple testing controlled by the Bonferroni approach at experiment-wise p-value <0.0541. CpGs in identified DMRs and CpGs that passed screening via linear regression were included in subsequent analyses for their association with adolescence and young adulthood asthma acquisition using logistic regressions, adjusting for the confounders in both sexes. CpGs in each identified DMR was treated as one CpG unit and the unit’s DNAm was represented by the mean of DNAm at those CpGs. Multiple testing was adjusted by controlling the false discovery rate (FDR) of 0.01, a stringent control of multiple testing in an effort to improve the informativity of identified CpG sites.
Replication analyses in the ALSPAC cohort
CpGs identified in the IOWBC were further tested in the ALSPAC cohort using logistic regression models controlling for comparable covariates. The asthma diagnoses were comparable with those in IOWBC.
Consistency of DNAm over time
The temporal consistency of DNAm would substantiate that asthma acquisition later in life is related to earlier epigenetics at birth. To this end, we applied linear mixed models with repeated measures. Dependent variables were DNAm at age 10 and 18 years, and independent variables were newborn DNAm, age (10 and 18 years), newborn DNAm and age interaction, and sex. In IOWBC, DNAm of newborns (n=796), age 10 (n=330), and age 18 (n=476) was included in the analyses. For ALSPAC, DNAm at birth (n=883), age 7 (n=927), and age 17 (n=688) was analyzed. CpGs with no statistically significant interactions between DNAm and age (p-value >0.05) were regarded as temporally consistent CpGs.
Gene set analysis
To address biological pathways of genes corresponding to the CpGs associated with asthma acquisition in screening via linear regression or DMRs analyses, the gometh function (https://rdrr.io/bioc/missMethyl/man/gometh.html) in the missMethyl R package was applied. The multiple testing correction was conducted at the FDR of 0.05 level.
Biological relevance
For CpGs associated with asthma acquisition in both cohorts, we evaluated the association of newborn DNAm in M values with gene expression (in log-scale) in cord blood on genes within 250kb upstream and downstream of the identified CpGs, following Reese et al.42. Linear regression was applied to assess their relationships. Statistical significance was set at 0.05.
RESULTS
Population characteristics
Cohort members who were asthma free by age 10 years and had newborn DNAm available were included in the study (n=453), of which, 28 developed asthma in adolescence (and persistent in young adulthood; No-Yes-Yes) and 17 participants developed asthma at young adulthood (No-No-Yes) (Table 1). We compared the analytical sample with the cohort members having the defined acquisition patterns (“Whole cohort”; Table 1) on variables potentially related to asthma. No statistically significant difference was found (Table 1). Furthermore, the proportions of acquisition showed no difference between the analytical sample and the whole cohort for asthma acquisition (Table 2). In addition, we assessed maternal status and asthma status at earlier ages (at ages 4,10, 18 years) between those remaining in the cohort and those lost to follow-up. No differences were found (Table S1).
TABLE 1.
Asthma acquisition between the sub-sample and cohort and between the two sexes.
| Asthma acquisition during adolescence | p-value | Asthma acquisition during young adulthood | p-value | |||
|---|---|---|---|---|---|---|
| Males | Whole cohort* | Analytical sample* | 0.65 | Whole cohort* | Analytical sample* | 1.00 |
| Yes | 13 (3.96) | 9 (4.81) | 14 (4.26) | 8 (4.30) | ||
| No | 315 (96.04) | 178 (95.19) | 315 (95.74) | 178 (95.7) | ||
| Females | 1.00 | 1.00 | ||||
| Yes | 30 (7.41) | 19 (7.63) | 16 (4.09) | 9 (3.77) | ||
| No | 375 (92.59) | 230 (92.37) | 375 (95.91) | 230 (96.23) | ||
| p-value | 0.05 | 0.23 | 0.91 | 0.78 | ||
The analytical samples include subjects with newborn DNAm available as well as asthma status data up to young adulthood. The whole cohort samples include subjects with asthma status data up to young adulthood.
TABLE 2.
Characteristics of subjects with available methylation data with their asthma status at ages 10, 18, and 26 years stratified by sex in the IOW cohort.
| Factors | Data for asthma acquisition during adolescence | Data for asthma acquisition during young adulthood | ||||
|---|---|---|---|---|---|---|
| Whole cohort | Analytical sample | p-value | Whole cohort | Analytical sample | p-value | |
| Males | n=328 | n=187 | n=329 | n=186 | ||
| SES# | n (%) | n (%) | 0.48 | n (%) | n (%) | 0.35 |
| High | 33 (10.06) | 16 (8.56) | 32 (9.73) | 15 (8.06) | ||
| Middle | 247 (75.3) | 142 (75.94) | 248 (75.38) | 141 (75.81) | ||
| Low | 39 (11.89) | 27 (14.44) | 39 (11.85) | 28 (15.05) | ||
| Missing | 9 (2.74) | 2 (1.07) | 10 (3.04) | 2 (1.08) | ||
| Secondhand smoke$ | 0.85 | 0.88 | ||||
| Yes | 112 (34.15) | 68 (36.36) | 115 (34.95) | 68 (36.56) | ||
| No | 181 (55.18) | 101 (54.01) | 180 (54.71) | 101 (54.3) | ||
| Missing | 35 (10.67) | 18 (9.63) | 34 (10.33) | 17 (9.14) | ||
| Mean±SD | Mean±SD | Mean±SD | Mean±SD | |||
| BMI at age 10 years | 17.56±2.49 | 17.67±2.59 | 0.59 | 17.53±2.42 | 17.64±2.47 | 0.55 |
| Median (MAD) | Median (MAD) | Median (MAD) | Median (MAD) | |||
| Breastfeeding¥ (weeks) | 12 (12) | 12.5 (12.5) | 0.31 | 12 (11.5) | 12 (12) | 0.35 |
| Females | n=405 | n=249 | n=391 | n=239 | ||
| SES | n (%) | n (%) | 0.63 | n (%) | n (%) | 0.39 |
| High | 39 (9.63) | 25 (10.04) | 38 (9.72) | 25 (10.46) | ||
| Middle | 300 (74.07) | 192 (77.11) | 292 (74.68) | 186 (77.82) | ||
| Low | 59 (14.57) | 30 (12.05) | 54 (13.81) | 27 (11.30) | ||
| Missing | 7 (1.73) | 2 (0.80) | 7 (1.79) | 1 (0.42) | ||
| Secondhand smoke | 0.20 | 0.19 | ||||
| Yes | 144 (35.56) | 74 (29.72) | 135 (34.53) | 68 (28.45) | ||
| No | 212 (52.35) | 136 (54.62) | 208 (53.2) | 133 (55.65) | ||
| Missing | 49 (12.10) | 39 (15.66) | 48 (12.28) | 38 (15.9) | ||
| Mean±SD | Mean±SD | Mean±SD | Mean±SD | |||
| BMI at age 10 years | 18.58±3.08 | 18.49±2.97 | 0.99 | 18.5±3 | 18.41±2.87 | 0.66 |
| Median (MAD) | Median (MAD) | Median (MAD) | Median (MAD) | |||
| Breastfeeding (weeks) | 11.5 (11.5) | 12 (12) | 0.56 | 11.5 (11.5) | 12 (12) | 0.59 |
SES refers to socioeconomic status in childhood.
Participants got secondhand smoke before age 10 years.
The length of time for breastfeeding.
Screening CpGs
The screening of CpGs was conducted for males and females separately as patterns of asthma acquisition in adolescence differ between males and females. We first screened CpGs using linear regression with DNAm at each CpG site as the dependent variable and asthma acquisition status as the independent variable. In total, 900 unique CpGs (463 CpGs for males) for their association with asthma acquisition during adolescence and 1738 unique CpGs (1117 CpGs for males and 624 in females with 3 common CpGs) for asthma acquisition during young adulthood passed screening (Figure 1).
FIGURE 1.
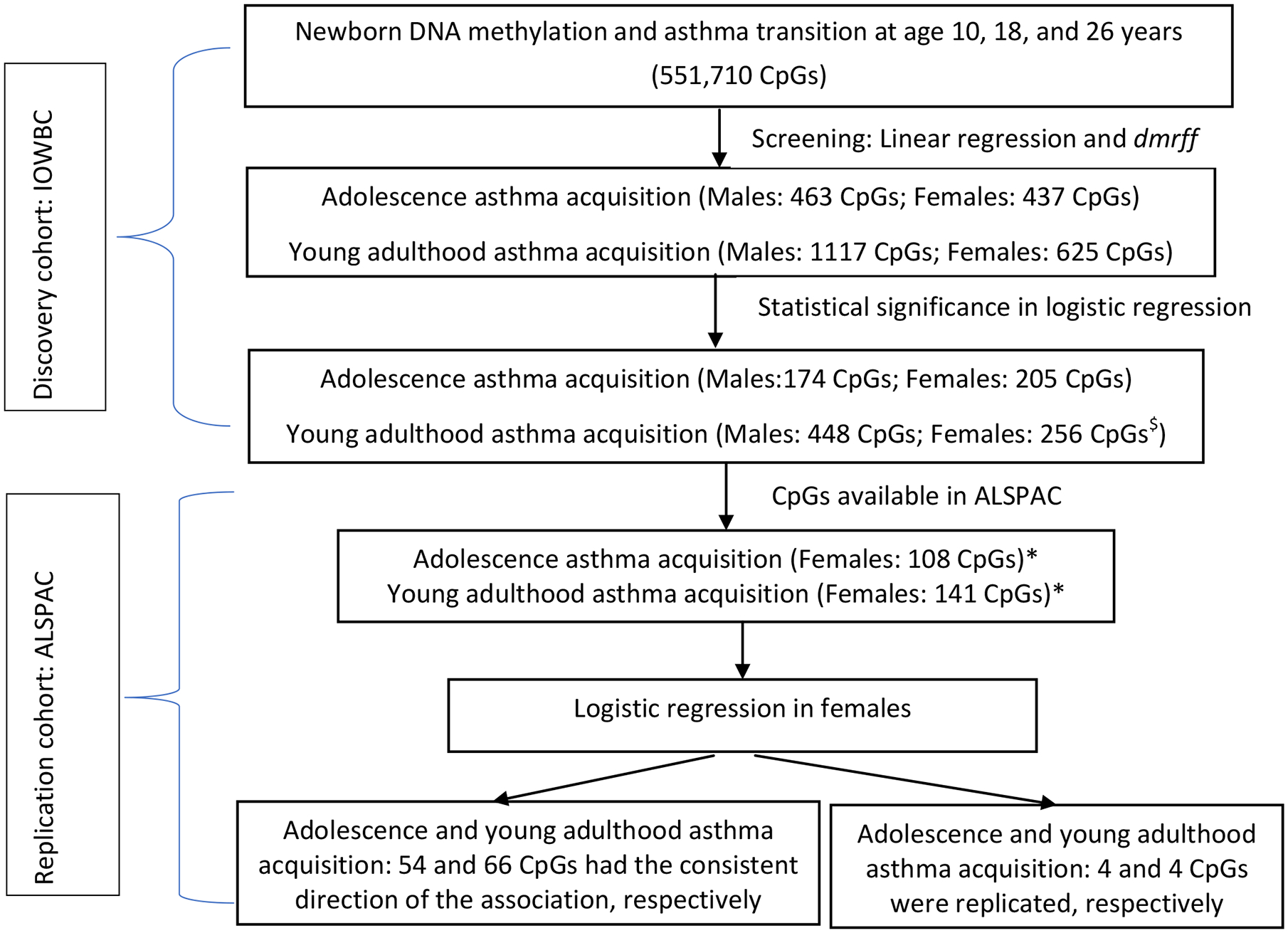
Flow chart of statistical analyses identifying the association of newborn DNAm with asthma acquisition.
* The data in males were not further analyzed in ALSPAC due to small sample sizes. For females, n=196 with 6 having adolescence asthma acquisition and n=194 with 4 having young adulthood asthma acquisition. $ 255 CpG and 1 CpG unit.
Second, we used DMR-based screening. We identified one statistically significant DMR (Bonferroni adjusted p-value=0.042) showing association with young adulthood asthma acquisition in females. This DMR (chromosome 10: 3143354-3143596) spanned two CpG sites (cg09875661, cg16519433) that are annotated to the PFKP gene. These two CpGs were treated as one CpG unit. CpG cg09875661 was among the 1738 candidate CpGs identified through regression. Thus, for young adulthood asthma acquisition, 1738 CpGs/units were included in the full model of logistic regression.
Discovered CpGs in IOWBC
Logistic regression models were applied to evaluate the association of DNAm of the candidate CpGs (including the CpG unit) at birth with each of the two types of asthma acquisition patterns. For males, 174 and 448 CpGs showed statistically significant association with asthma acquisition during adolescence and young adulthood, respectively, after adjusting for multiple testing by controlling the FDR of 0.01 (Figure 1; Table S2–S3). For females, at 205 CpGs and at 255 CpGs plus the CpG unit, DNAm was associated with asthma acquisition during adolescence and young adulthood, respectively, at FDR=0.01. These CpGs identified in IOWBC were further tested in the ALSPAC cohort. No overlap in the identified CpGs was found between males and females.
Replication in ALSPAC
In ALSPAC, for females, 10 participants experienced asthma acquisition during adolescence or during young adulthood, and 190 participants were without asthma at three-time points. For males, the data were not further analyzed due to no participant with asthma acquisition during adolescence and only one young adulthood asthma acquisition participant. For females, of the 205 CpG identified in IOWBC associated with asthma acquisition during adolescence, DNAm at 108 CpGs was available in ALSPAC. Similarly, DNAm at 141 (including the CpG unit) of the 256 discovered CpGs/units for asthma acquisition during young adulthood was available in ALSPAC (Figure 1). For asthma acquisition during adolescence, of the 108 CpGs examined, 54 (50%) CpGs showed consistent directions of association with those in IOWBC (Table S4, Figure 2). Of the 54 CpGs, associations at 4 CpGs were statistically significant (p<0.05) (Table 3, Figure 2); an increase in DNAm at 3 of the 4 CpGs was associated with decreased odds of adolescence asthma acquisition. For young adulthood asthma acquisition, at 66 (47%) of the 141 CpGs/units, consistent directions of association were observed between IOWBC and ALSPAC (Table S5, Figure S1) and at 4 of the 66 CpGs/units statistical significance was observed (p<0.05) (Table 3, Figure S1). At one of these 4 CpGs, increased DNAm was associated with decreased odds of young adulthood asthma acquisition.
FIGURE 2.
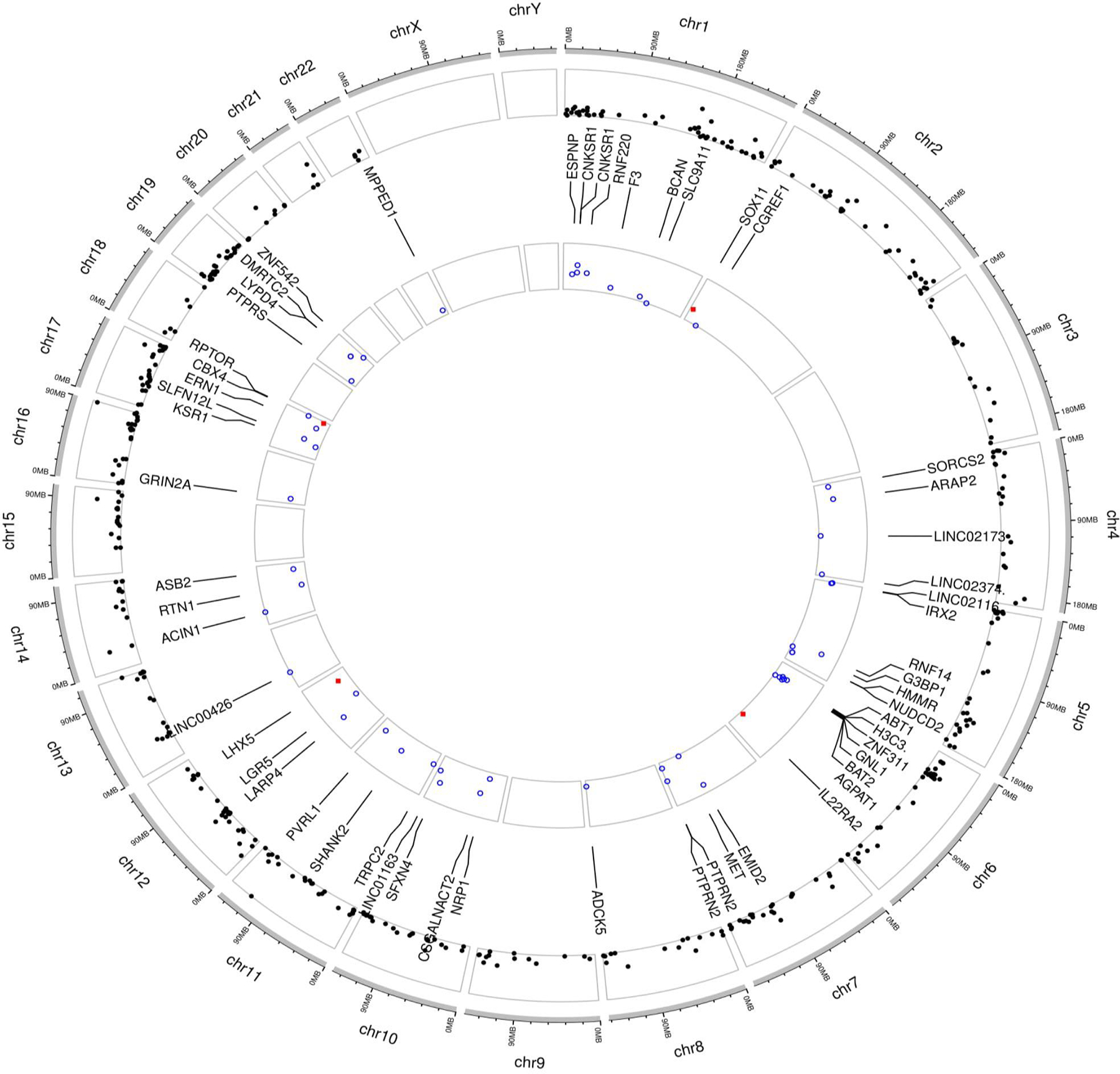
Circos plot of mapping genes of CpGs survived screening via linear regression or identified by DMR and those with consistent directions in two cohorts for adolescence asthma acquisition in females (A circos plot was drawn on the website (https://yimingyu.shinyapps.io/shinycircos/), indicating the location of genes corresponding to the CpGs. The outer track is for the location of mapping genes survived screening or identified by DMR, and the inner track is for the location of mapping genes corresponding to identified CpG with consistent directions between the two cohorts).
On outer track, the black points represent −log10 transformed p-values of CpGs that survived screening via linear regression or identified via DMR. On the inner track, the blue unfilled circles and red filled squares denote −log10 transformed p-values of association with adolescence asthma acquisition for those with consistent direction of associations between the two cohorts (but in ALSPAC the effects were statistically insignificant) and those replicated in ALSPAC (in both cohorts, the directions were consistent and the effects were statistically significant), respectively. Chromosome numbers are shown on outermost circle.
TABLE 3.
CpG sites with DNAm showing associations with adolescence asthma acquisition and young adulthood asthma acquisition in females that are consistent between the discovery cohort, IOWBC and the replication cohort, ALSPAC, with p-value<0.05. Complete results are in Table S4 and S5.
| Asthma acquisition | CpG name | Chr.# | Gene name | Region | IOWBC | ALSPAC | |||
|---|---|---|---|---|---|---|---|---|---|
| Log-odds ratio& | Raw p-value | FDR p-value | Log-odds ratio | p-value | |||||
| In adolescence | cg11586330 | 12 | LHX5 | 3’UTR | −2.06 | 0.0018 | 0.0076 | −1.91 | 0.007 |
| cg23507945 | 6 | IL22RA2 | Body | −0.35 | 0.0036 | 0.0089 | −1.19 | 0.015 | |
| cg04771413 | 2 | SOX11 | Intergenic | −2.44 | 0.0016 | 0.0074 | −1.34 | 0.018 | |
| cg00646381 | 17 | CBX4 | Intergenic | 3.47 | 0.0027 | 0.0079 | 2.22 | 0.018 | |
| In young adulthood | cg15782391 | 19 | ACPT | TSS1500 | 5.32 | 0.0012 | 0.0128 | 2.35 | 0.034 |
| cg12319143 | 10 | CFAP46 | Intergenic | −0.96 | 0.0019 | 0.0128 | −0.59 | 0.036 | |
| cg19983801 | 3 | MUC4 | 5’UTR;1stExon | 3.62 | 0.0006 | 0.0128 | 1.73 | 0.048 | |
| cg02705837 | 17 | ATP1B2 | 1stExon;5’UTR | 1.79 | 0.0025 | 0.0128 | 2.48 | 0.048 | |
Chromosome number.
Log-odds ratios assessing the associations of DNAm level at birth with asthma acquisition, with no asthma at age 10, 18, 26 years as the reference.
Consistency of DNAm over time
Of the 8 CpGs replicated in ALSPAC (p< 0.05), cg23507945 was not available at ages 10 and 18 in IOWBC and was not available at ages 7 and 17 in ALSPAC. Results from the linear mixed models showed that the interaction between time and newborn DNAm was not statistically significant (all p-values>0.05; Table S6; Figure S2–S8), indicating DNAm at these CpG sites was likely to be consistent from birth to age 18 years.
Gene set analysis
Gene set analysis was applied to CpGs that survived screening (2636 unique CpGs) (Table S7–S11). One biological pathway, KEGG pathway Malaria, linked to adolescence asthma acquisition in females was identified (FDR adjusted p-value=0.003) (Table S12), and seven genes, COMP (chromosome [Chr.] 19), IL6 (Chr. 7), LRP1 (Chr. 12), MET (Chr. 7), TGFB2 (Chr. 1), THBS4 (Chr. 5), and TNF (Chr. 6) on our candidate gene lists were involved in this pathway. These seven genes do not overlap with the genes identified (Table 3, Table S13).
DNAm and gene expression
For asthma acquisition during adolescence, DNAm at 2 of the 4 identified CpGs (cg00646381 and cg23507945; all p-values <0.05) was associated with expression of their neighboring genes (Table 4 and Table S14). For asthma acquisition during young adulthood, DNAm at all the 4 identified CpGs (all p-values <0.05) was associated with expression of 8 neighboring genes (Table 4 and Table S15).
TABLE 4.
Association of DNAm with the gene expression on the genes within 250kb upstream and downstream of the CpGs at birth in F2-generation. The complete results on the association of DNAm at the identified CpGs with their neighboring genes are in Tables S6a and S6b.
| Asthma acquisition | CpG name | Gene name$ | Gene probe* | Estimate# | p-value |
|---|---|---|---|---|---|
| Adolescence asthma acquisition | cg23507945 | NHEG1 | NHEG1_A_21_P0000551 | −0.14 | 0.014 |
| cg00646381 | TBC1D16 | TBC1D16_A_33_P3241937 | −0.51 | 0.031 | |
| Young adulthood asthma acquisition | cg15782391 | KLK6 | KLK6_A_24_P236935 | −0.80 | 0.040 |
| cg12319143 | GPR123 | lnc_GPR123_1_A_21_P0006942 | −0.15 | 0.028 | |
| cg19983801 | LINC00969 | LINC00969_A_19_P00316857 | −0.15 | 0.017 | |
| cg02705837 | SHBG | SHBG_A_21_P0000018 | −0.35 | 0.011 | |
| TP53 | lnc_TP53_1_A_21_P0009294 | −0.58 | 0.017 | ||
| POLR2A | POLR2A_A_33_P3218138 | 0.29 | 0.024 | ||
| ATP1B2 & | ATP1B2_A_24_P31275 | 0.36 | 0.025 | ||
| EIF4A1 | EIF4A1_A_23_P137103 | 0.19 | 0.030 | ||
| EIF4A1 | EIF4A1_A_21_P0011135 | 0.15 | 0.039 | ||
| SPEM1 | SPEM1_A_24_P67189 | 0.25 | 0.048 |
Genes within 250kb upstream and downstream of the CpGs, including neighboring and mapped genes.
Gene expression level measured by probes.
Estimate: Regression coefficient estimates. It represents expected log2 transformed gene expression change for one unit increase of M values of DNAm.
Gene ATP1B2 is the gene to which the cg02705837 is mapped.
DISCUSSION
Previous studies have been focusing on the connection of newborn DNAm with childhood asthma20–24. To our knowledge, this is the first epigenome-scale association study on newborn DNAm in whole blood with asthma acquisition at different stages of life. Via single CpG site screening and screening via DMRs, we first identified potentially informative CpGs with respect to asthma acquisition during adolescence or young adulthood in IOWBC, and these CpGS were in-depth tested for such associations using logistic regressions with confounders adjusted. Four CpGs unique to each period discovered in females in IOWBC were replicated in ALSPAC. Our assessment indicated that DNAm at these eight CpG sites was consistent from birth to age 18 years and DNAm at six of the eight CpGs were associated with expressions of their neighboring genes.
Findings in our screening process deserve discussion. One DMR composed of two CpGs related to young adulthood asthma acquisition in females was detected. However, one of the two CpGs, cg16519433, did not show an association with asthma transition by itself. This observation suggests that methylation in multiple adjacent CpGs might have a joint biological association with the diseases43,44, and future studies may focus on investigating CpGs by regions rather than individually. Furthermore, evidence has shown that sex-specific acquisition of asthma occurs during puberty45, and our previous study showed that the association of DNAm changes with asthma acquisition from pre- to post-adolescence was different between males and females46. These findings support our sex-specific screening results and our sex-stratified study design.
In addition, among the genes with DNAm at CpGs that passed screening for asthma acquisition during adolescence in females, seven were enriched in a KEGG pathway, Malaria, and six of the seven genes have been linked to asthma in previous studies47–52. The pathway Malaria is related to immune and inflammatory response to malaria infection53.
For the identified CpGs, our findings on temporal consistency in DNAm strengthen the potential of those CpGs as epigenetic markers in newborns for asthma acquisition later in life. This finding will benefit future effort to detect subjects within days after birth who are at a high risk of asthma acquisition, making primary prevention possible. Temporal consistency in DNAm does not claim biological stability from birth to age 18 years, rather a consistent DNAm changes between birth and age 10 and between birth and age 18. It is worth noting that the CpGs identified in our study do not overlap with those from cross-sectional studies by Xu et al20, although we observed a nice agreement if the same study design (cross-sectional) is applied to our data (Supplemental Material S3). With a longitudinal study design as in the present study, the time-order between DNAm and asthma acquisition is clearer compared to cross-sectional studies, which might explain the unique findings between our study and Xu et al. It is interesting to note that no common CpGs or genes were found between the markers for asthma acquisition during adolescence and those during young adulthood.
Asthma is a complex and heterogeneous disease due to various immune pathways which determine inflammatory pathway54. Five genes (IL22RA2, CBX4, CFAP46, ATP1B2, and SOX11), of the 8 genes to which the 8 identified CpGs were mapped (Table 3), have been shown to be related to asthma or airway diseases. Of these five genes, IL22RA2 was linked to various immune-mediated diseases. This gene is related to two pathways, Innate Immune System and Immune response Antigen presentation by MHC class II55, which play a critical role in asthma pathogenesis56. The other four genes, CBX4, CFAP46, ATP1B2, and SOX11, have been reported to be related to airway and lung diseases via regulating inflammation response57–60. The relation of biological pathways involving the other three genes with asthma is unclear, but SOX11, a member of the SOX C group, regulates interleukin 13 (IL-13) signaling in lung fibroblasts and allergic disease through IL-13/STAT6/SOX11 pathway. Also, among the 8 genes (Table 3), MUC4 encodes a membrane bound mucin that plays an important role in cell proliferation and differentiation of epithelial cells by inducing specific phosphorylation of ERBB261 and whose expression has been shown to be modified by oral corticosteroids62.
Importantly, among all the eight CpGs identified in our study based on two independent cohorts, most of them showed an association with expression of their neighboring genes. In particular, cg02705837 is located in the 1st Exon and 5’ UTR of gene ATP1B2, a Wnt signaling gene shown to be involved in the pathogenesis of impairment in individuals with asthma63. Unlike the other identified CpGs, DNAm at this CpG was associated with the expression of multiple neighboring genes, SHBG, TP53, POLR2A, EIF4A1, and SPEM1, and both SHBG and TP53 have found to be associated with asthma64,65. Of particular interest, in an earlier study by Yuan et al., DNAm of gene TP53, at site chr17:7591672 close to the location of cg02705837 (chr17: 7554666), is suggested as a peripheral blood biomarker to predict late-onset asthma65. In our study, we showed that newborn DNAm of cg02705837 was associated with young adulthood asthma acquisition and with expression of its neighboring gene TP53. The comparison of our findings with that in Yuan et al. suggests that cg02705837, although not located on gene TP53, has a potential to regulate activity of its neighboring gene and, further, cg02705837 is likely to be a stable epigenetic marker for asthma acquisition.
The strengths of our study include the time-ordered study design with newborn DNAm and postnatal asthma acquisition with multi-layered analyses protocols, carefully designed screening process in the detection of candidate CpGs, and the utilization of an independent replication cohort. With this careful design, the findings tend to be conservative and the identified CpGs are likely to be informative with a strong potential of replicability. Some limitations did exist in this study. The sources of DNAm assessment are different between the two cohorts; DNAm was measured in heel prick blood of newborns in IOWBC and in cord blood in ALSPAC. Since our analyses focused on the differences in DNAm between subjects with asthma and no-asthma subjects rather than linear associations between continuous variables, a recent assessment showed that under this context, DNAm between the two sources had a high agreement overall66. In addition, DNAm in both cohorts was based on blood cells and may not reflect DNAm in airway tissues. However, most of the genes corresponding to CpGs identified in our study are expressed in respiratory tissue47–52,57,58,60,62. As a related limitation, our biological relevance assessment of the identified CpGs was carried out in the offspring of IOWBC. Since generation-specific factors might have confounded the underlying associations, interpretation of the CpGs showing potential biological relevance needs to be cautious. It is worth noting that, although they seemed not to be a concern in our study, household smoking during pregnancy, active smoking during pregnancy, season of birth, birth order, maternal asthma history, and maternal BMI, along with other prenatal factors, could potentially confound the findings. Due to rather small sample sizes of asthma acquisition in males in the ALSPAC cohort, we did not further test the IOWBC-discovered CpGs in males in ALSPAC. A larger scale study will be needed to confirm the viability of the discovered CpGs. Nevertheless, the findings from both cohorts indicate a potential of newborn epigenetic markers for asthma acquisition at later ages, and such markers are likely to be different at different stages of life.
CONCLUSION
In female participants, DNAm in newborns or at birth is associated with asthma acquisition after puberty. Our findings suggest that adolescent and young adulthood asthma acquisition possibly do not share common epigenetic markers.
Supplementary Material
Key Messages:
A longitudinal epigenome-scale study was conducted using discovery and replication birth cohorts.
DNA methylation at birth at eight CpGs in females was associated with asthma acquisition later in life.
The identified CpGs have a potential to serve as candidate epigenetic biomarker for asthma acquisition.
Acknowledgements:
We are indebted to all participants and their families who are followed up in the Isle of Wight 1989 birth cohort and the Avon Longitudinal Study of Parents and Children, which include interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, midwives, and nurses. We also appreciate the hard efforts of these two research teams in maintaining the cohorts and collecting data. We also thanks to the High Performance Computing facility at the University of Memphis.
Support statement:
This work was supported by the National Institutes of Health research funds R01AI121226 (MPI: H Zhang and JW Holloway). DNAm analyses of F1 newborns and gene expression analyses of F2-newborns were funded by National Institute of Allergy and Infectious Diseases [R01 AI091905] and National Heart, Lung, and Blood Institute [R01 HL132321] (PI: W Karmaus). The UK Medical Research Council and Wellcome (Grant ref: 217065/Z/19/Z) and the University of Bristol provide core support for ALSPAC. A comprehensive list of grants funding is available on the ALSPAC website (http://www.bristol.ac.uk/alspac/external/documents/grant-acknowledgements.pdf). CR is supported by the Medical Research Council and University of Bristol Integrative Epidemiology Unit (MC_UU_00011/5/MRC). The sponsor had no role in study design, data collection and analysis, or the preparation of the manuscript. This publication is the work of the authors and Hongmei Zhang will serve as guarantor for the contents of this paper.
Footnotes
Conflict of interest: All authors are nothing to disclose.
Ethical approval
For IOWBC, local/national ethics committees approved the recruitment of the birth cohort between January 1989 and February 1990 and subsequent follow-up assessments. For ALSPAC, the ethical approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees.
Data availability:
The data used in the study are available on request from the corresponding author with justification due to privacy and ethical restrictions.
References
- 1.Peters SP, Ferguson G, Deniz Y, Reisner C. Uncontrolled asthma: A review of the prevalence, disease burden and options for treatment. Respiratory Medicine. 2006;100(7):1139–1151. [DOI] [PubMed] [Google Scholar]
- 2.Soriano JB, Kendrick PJ, Paulson KR, et al. Prevalence and attributable health burden of chronic respiratory diseases, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. The Lancet Respiratory Medicine. 2020;8(6):585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zar HJ, Ferkol TW. The global burden of respiratory disease-impact on child health. Pediatric pulmonology. 2014;49(5):430–434. [DOI] [PubMed] [Google Scholar]
- 4.Wu T-J, Wu C-F, Lee YL, Hsiue T-R, Guo YL. Asthma incidence, remission, relapse and persistence: a population-based study in southern Taiwan. Respiratory research. 2014;15(1):135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuchs O, Bahmer T, Rabe KF, von Mutius E. Asthma transition from childhood into adulthood. The Lancet Respiratory Medicine. 2017;5(3):224–234. [DOI] [PubMed] [Google Scholar]
- 6.Arshad SH, Raza A, Lau L, et al. Pathophysiological characterization of asthma transitions across adolescence. Respiratory research. 2014;15(1):153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush A Phenotype specific treatment of asthma in childhood. Paediatric respiratory reviews. 2004;5 Suppl A:S93–101. [DOI] [PubMed] [Google Scholar]
- 8.Martinez FD, Vercelli D. Asthma. Lancet (London, England). 2013;382(9901):1360–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuseini H, Newcomb DC. Mechanisms Driving Gender Differences in Asthma. Curr Allergy Asthma Rep. 2017;17(3):19–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchs O, Bahmer T, Weckmann M, et al. The all age asthma cohort (ALLIANCE) - from early beginnings to chronic disease: a longitudinal cohort study. BMC pulmonary medicine. 2018;18(1):140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Nijs SB, Venekamp LN, Bel EH. Adult-onset asthma: is it really different? European Respiratory Review. 2013;22(127):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouzigon E, Corda E, Aschard H, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. The New England journal of medicine. 2008;359(19):1985–1994. [DOI] [PubMed] [Google Scholar]
- 13.Pividori M, Schoettler N, Nicolae DL, Ober C, Im HK. Shared and distinct genetic risk factors for childhood-onset and adult-onset asthma: genome-wide and transcriptome-wide studies. The Lancet Respiratory medicine. 2019;7(6):509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau S, Illi S, Sommerfeld C, et al. Early exposure to house-dust mite and cat allergens and development of childhood asthma: a cohort study. Multicentre Allergy Study Group. Lancet (London, England). 2000;356(9239):1392–1397. [DOI] [PubMed] [Google Scholar]
- 15.Burke H, Leonardi-Bee J, Hashim A, et al. Prenatal and passive smoke exposure and incidence of asthma and wheeze: systematic review and meta-analysis. Pediatrics. 2012;129(4):735–744. [DOI] [PubMed] [Google Scholar]
- 16.Wu P, Dupont WD, Griffin MR, et al. Evidence of a causal role of winter virus infection during infancy in early childhood asthma. American journal of respiratory and critical care medicine. 2008;178(11):1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bisgaard H, Hermansen MN, Buchvald F, et al. Childhood asthma after bacterial colonization of the airway in neonates. The New England journal of medicine. 2007;357(15):1487–1495. [DOI] [PubMed] [Google Scholar]
- 18.Ho S-M, Johnson A, Tarapore P, Janakiram V, Zhang X, Leung Y-K. Environmental epigenetics and its implication on disease risk and health outcomes. ILAR J. 2012;53(3–4):289–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han L, Zhang H, Kaushal A, et al. Changes in DNA methylation from pre- to post-adolescence are associated with pubertal exposures. Clin Epigenetics. 2019;11(1):176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu CJ, Söderhäll C, Bustamante M, et al. DNA methylation in childhood asthma: an epigenome-wide meta-analysis. The Lancet Respiratory medicine. 2018;6(5):379–388. [DOI] [PubMed] [Google Scholar]
- 21.Qi C, Xu CJ, Koppelman GH. The role of epigenetics in the development of childhood asthma. Expert review of clinical immunology. 2019;15(12):1287–1302. [DOI] [PubMed] [Google Scholar]
- 22.DeVries A, Vercelli D. Early predictors of asthma and allergy in children: the role of epigenetics. Current opinion in allergy and clinical immunology. 2015;15(5):435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bae DJ, Jun JA, Chang HS, Park JS, Park CS. Epigenetic Changes in Asthma: Role of DNA CpG Methylation. Tuberc Respir Dis (Seoul). 2020;83(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeVries A, Wlasiuk G, Miller SJ, et al. Epigenome-wide analysis links SMAD3 methylation at birth to asthma in children of asthmatic mothers. The Journal of allergy and clinical immunology. 2017;140(2):534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arshad SH, Holloway JW, Karmaus W, et al. Cohort Profile: The Isle Of Wight Whole Population Birth Cohort (IOWBC). International Journal of Epidemiology. 2018;47(4):1043–1044i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gervin K, Page CM, Aass HCD, et al. Cell type specific DNA methylation in cord blood: A 450K-reference data set and cell count-based validation of estimated cell type composition. Epigenetics. 2016;11(9):690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics (Oxford, England). 2014;30(10):1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogbuanu IU, Karmaus W, Arshad SH, Kurukulaaratchy RJ, Ewart S. Effect of breastfeeding duration on lung function at age 10 years: a prospective birth cohort study. Thorax. 2009;64(1):62–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ziyab AH, Ewart S, Lockett GA, et al. Expression of the filaggrin gene in umbilical cord blood predicts eczema risk in infancy: A birth cohort study. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2017;47(9):1185–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyd A, Golding J, Macleod J, et al. Cohort Profile: the ‘children of the 90s’--the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2013;42(1):111–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraser A, Macdonald-Wallis C, Tilling K, et al. Cohort Profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int J Epidemiol. 2013;42(1):97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Northstone K, Lewcock M, Groom A, et al. The Avon Longitudinal Study of Parents and Children (ALSPAC): an update on the enrolled sample of index children in 2019. Wellcome Open Res. 2019;4:51–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. Journal of biomedical informatics. 2009;42(2):377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viuff AC, Sharp GC, Rai D, et al. Maternal depression during pregnancy and cord blood DNA methylation: findings from the Avon Longitudinal Study of Parents and Children. Translational Psychiatry. 2018;8(1):244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharp GC, Lawlor DA, Richmond RC, et al. Maternal pre-pregnancy BMI and gestational weight gain, offspring DNA methylation and later offspring adiposity: findings from the Avon Longitudinal Study of Parents and Children. International Journal of Epidemiology. 2015;44(4):1288–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bibikova M, Barnes B, Tsan C, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98(4):288–295. [DOI] [PubMed] [Google Scholar]
- 38.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics. 2012;13(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bakulski KM, Feinberg JI, Andrews SV, et al. DNA methylation of cord blood cell types: Applications for mixed cell birth studies. Epigenetics. 2016;11(5):354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vink NM, Postma DS, Schouten JP, Rosmalen JGM, Boezen HM. Gender differences in asthma development and remission during transition through puberty: The TRacking Adolescents’ Individual Lives Survey (TRAILS) study. Journal of Allergy and Clinical Immunology. 2010;126(3):498–504.e496. [DOI] [PubMed] [Google Scholar]
- 41.Suderman M, Staley JR, French R, Arathimos R, Simpkin A, Tilling K. dmrff: identifying differentially methylated regions efficiently with power and control. bioRxiv. 2018:508556. [Google Scholar]
- 42.Reese SE, Xu CJ, den Dekker HT, et al. Epigenome-wide meta-analysis of DNA methylation and childhood asthma. The Journal of allergy and clinical immunology. 2019;143(6):2062–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eckhardt F, Lewin J, Cortese R, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nature Genetics. 2006;38(12):1378–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hotta K, Kitamoto A, Kitamoto T, et al. Identification of differentially methylated region (DMR) networks associated with progression of nonalcoholic fatty liver disease. Scientific reports. 2018;8(1):13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Withers AL, Green R. Transition for Adolescents and Young Adults With Asthma. Front Pediatr. 2019;7:301–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel R, Solatikia F, Zhang H, et al. Sex-specific associations of asthma acquisition with changes in DNA methylation during adolescence. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rincon M, Irvin CG. Role of IL-6 in asthma and other inflammatory pulmonary diseases. Int J Biol Sci. 2012;8(9):1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mishra A, Yao X, Saxena A, et al. Low-density lipoprotein receptor-related protein 1 attenuates house dust mite-induced eosinophilic airway inflammation by suppressing dendritic cell-mediated adaptive immune responses. The Journal of allergy and clinical immunology. 2018;142(4):1066–1079.e1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuo CH, Guo Y, Pandis I, et al. MMP10 and MET as predictive classifiers of bronchial eosinophilic asthma in UBIOPRED. European Respiratory Journal. 2015;46(suppl 59):PA4890. [Google Scholar]
- 50.Balzar S, Chu HW, Silkoff P, et al. Increased TGF-beta2 in severe asthma with eosinophilia. The Journal of allergy and clinical immunology. 2005;115(1):110–117. [DOI] [PubMed] [Google Scholar]
- 51.Modena BD, Tedrow JR, Milosevic J, et al. Gene expression in relation to exhaled nitric oxide identifies novel asthma phenotypes with unique biomolecular pathways. American journal of respiratory and critical care medicine. 2014;190(12):1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berry M, Brightling C, Pavord I, Wardlaw A. TNF-alpha in asthma. Current opinion in pharmacology. 2007;7(3):279–282. [DOI] [PubMed] [Google Scholar]
- 53.Hisaeda H, Tetsutani K, Imai T, et al. Malaria Parasites Require TLR9 Signaling for Immune Evasion by Activating Regulatory T Cells. The Journal of Immunology. 2008;180(4):2496. [DOI] [PubMed] [Google Scholar]
- 54.Busse WW. Biological treatments for severe asthma: A major advance in asthma care. Allergology International. 2019;68(2):158–166. [DOI] [PubMed] [Google Scholar]
- 55.Muls N, Nasr Z, Dang HA, Sindic C, van Pesch V. IL-22, GM-CSF and IL-17 in peripheral CD4+ T cell subpopulations during multiple sclerosis relapses and remission. Impact of corticosteroid therapy. PloS one. 2017;12(3):e0173780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Finn PW, Bigby TD. Innate immunity and asthma. Proceedings of the American Thoracic Society. 2009;6(3):260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182(8):4994–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Esnault S, Bernau K, Torr EE, Bochkov YA, Jarjour NN, Sandbo N. RNA-sequencing analysis of lung primary fibroblast response to eosinophil-degranulation products predicts downstream effects on inflammation, tissue remodeling and lipid metabolism. Respiratory research. 2017;18(1):188–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu W-P, Zeng Y-Y, Zuo Y-H, Zhang J Identification of novel candidate genes involved in the progression of emphysema by bioinformatic methods. International journal of chronic obstructive pulmonary disease. 2018;13:3733–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mitamura Y, Nunomura S, Nanri Y, et al. Hierarchical control of interleukin 13 (IL-13) signals in lung fibroblasts by STAT6 and SOX11. J Biol Chem. 2018;293(38):14646–14658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bafna S, Kaur S, Batra SK. Membrane-bound mucins: the mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene. 2010;29(20):2893–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Milara J, Morell A, Ballester B, Armengot M, Morcillo E, Cortijo J. MUC4 impairs the anti-inflammatory effects of corticosteroids in patients with chronic rhinosinusitis with nasal polyps. Journal of Allergy and Clinical Immunology. 2017;139(3):855–862.e813. [DOI] [PubMed] [Google Scholar]
- 63.Sharma S, Tantisira K, Carey V, et al. A role for Wnt signaling genes in the pathogenesis of impaired lung function in asthma. Am J Respir Crit Care Med. 2010;181(4):328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arathimos R, Granell R, Haycock P, et al. Genetic and observational evidence supports a causal role of sex hormones on the development of asthma. Thorax. 2019;74(7):633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuan L, Wang L, Du X, et al. The DNA methylation of FOXO3 and TP53 as a blood biomarker of late-onset asthma. Journal of Translational Medicine. 2020;18(1):467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang Y, Wei J, Zhang H, et al. Epigenome wide comparison of DNA methylation profile between paired umbilical cord blood and neonatal blood on Guthrie cards. Epigenetics. 2020;15(5):454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in the study are available on request from the corresponding author with justification due to privacy and ethical restrictions.


