Abstract
Screening functional phenotypes in small animals is important for genetics and drug discovery. Multiphase microfluidics has great potential for enhancing throughput but has been hampered by inefficient animal encapsulation and limited control over the animal’s environment in droplets. Here we demonstrate a highly efficient single-animal encapsulation unit, a liquid exchanger system for controlling the droplet chemical environment dynamically, and an automation scheme for the programming and robust execution of complex protocols. By careful use of interfacial forces, the liquid exchanger unit allows for adding and removing chemicals from a droplet and, therefore, generating chemical gradients inaccessible in previous multiphase systems. Using Caenorhabditis elegans as an example, we demonstrate that these advances can serve to analyze dynamic phenotyping, such as behavior and neuronal activity, perform forward genetic screen, and are scalable to manipulate animals of different sizes. This platform paves the way for large-scale screens of complex dynamic phenotypes in small animals.
Keywords: multiphase microfluidics, droplets, functional phenotyping, high-throughput screen, small animals
Graphical Abstract
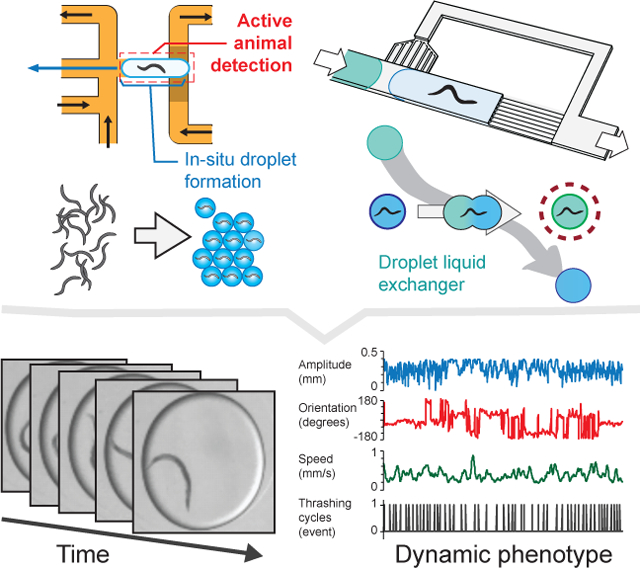
A multiphase microfluidic platform is demonstrated for performing automated functional screens of small animals. The in-situ droplet generator scheme efficiently encapsulates complex objects. The liquid exchanger unit allows for generating chemical temporal profiles previously impossible in droplets. Because the platform operation relies on manipulating droplets rather than directly animals, the platform is scalable and accomodates animals of different sizes.
1. Introduction
Research on small animals is essential for studying functional phenotypes such as behavior and brain activity, inaccessible to single-cell assays. The outcome of these studies is of high interest in the biomedical field for gene [1, 2] and drug [3–5] discovery. Screening these dynamic phenotypes is challenging because of the interplay between genetics and environment that creates a large phenospace to investigate. Moreover, changes in the environment may trigger and alter the animal responses, which renders functional assays sensitive to any perturbations. Therefore, automated screening protocols, extreme care in animal handling, and precise temporal control over the environment are keys to further the development of techniques for screening dynamic phenotypes.
Multi-well plate technology has been the cornerstone of screening techniques for microscopic animals. [6–9] However, the limited ability to change the environment and 3D animal motion make it difficult to assess dynamic phenotypes other than rough motion analysis. In contrast, microfluidics, with relevant length scales down to the animal’s size, offers improved means to handle small animals and control their environment. [10–14] Animals can be confined in 2D or even immobilized, and liquids can be exchanged rapidly. These approaches enable advanced functional assays such as monitoring brain activity [15, 16] or performing high-resolution behavior. [17] However, most of the high-throughput screening techniques are end-point assays based on fluorescence images [18, 19] and do not translate well for phenotyping based on functional imaging. Single-phase microfluidic systems, although powerful, have limitations in the number of animals they can handle [17, 20–23] and the ability to modulate the experimental conditions at will. [16, 23–25]
Multiphase microfluidics has unique attributes that make it well suited to tackle these challenges. These include the ability to partition, create independent containers of various sizes, generate precise chemical environments, perform serial processing, and consume ultra-low amounts of reagents. Multiphase microfluidics has led to advanced screening technologies impacting a broad range of disciplines, from particle synthesis [26, 27] and synthetic biology [28, 29], to drug discovery [30, 31], and research in fundamental biology. [32, 33] It has been applied successfully for numerous single-cell screens [34] and is gaining popularity for multicellular organisms such as spheroids and organoids. [35–38] In contrast, the development of multiphase microfluidics techniques for small animals [39–43] has seen a slow progress because of several technical bottlenecks. For instance, passive encapsulation techniques [44] are not adaptable to motile, multi-cellular organisms, mining the throughput. Second, techniques for controlling the droplet composition post-production rely mainly on merging, [45] which limits the ability to dynamically alter the droplet content. Furthermore, advanced active droplet manipulation tools [46–49] are inadequate as mechanical, thermal, and electrical-induced forces may create undesired stimulation. [50–52] Addressing these limitations is critical to fully exploit the potential of multiphase microfluidics as small-animal screening technology.
Here we describe a multiphase microfluidic platform to screen dynamic phenotypes in small animals. We demonstrate the use of this platform with the nematode C. elegans, a highly-motile and flexible animal, and a prominent model organism for research on chemosensation.[53] We developed a robust method for encapsulating complex organisms and achieving efficient single-animal encapsulation by combining an in situ, on-demand droplet generation scheme and real-time image processing. Furthermore, by carefully designing the channel geometry and balancing Laplacian forces, we developed a liquid exchanger that can swap the liquid content (and therefore the chemical environment) of an animal-containing droplet with that of another droplet. This unit allows for adding and removing chemicals and creating temporal gradients, both previously not possible in droplets. We demonstrate that these advances enable screens based on dynamic phenotypes, such as behavior and neuronal activity, and are scalable to manipulate animals of different sizes. Finally, we show that the unique properties of this multiphase microfluidics screening platform enable unprecedented means to study neurodevelopment in C. elegans larvae, including their behavioral and neuronal activities, which were not technically feasible previously due to the small size and fragility.
2. Results
2.1. System Design
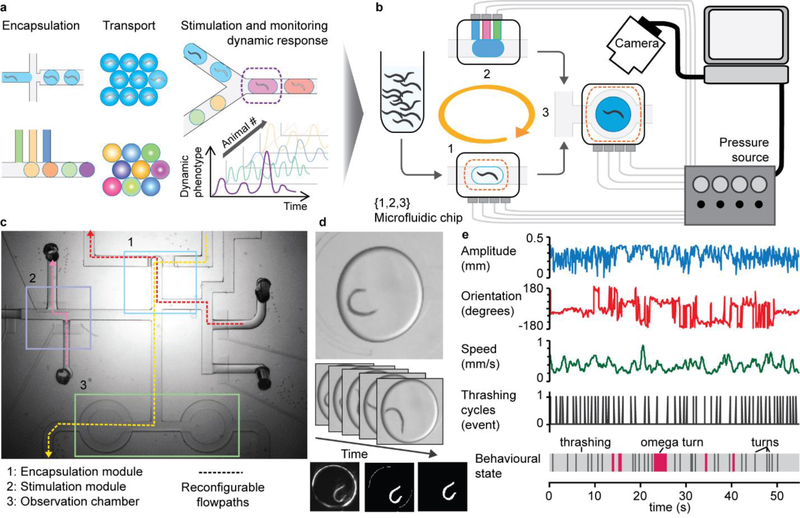
Multiphase microfluidics is ideal for generating independent containers and enabling individualized treatment of animals. However, screening dynamic phenotypes requires continuous monitoring of the animals, and doing so with a traditional droplet strategy based on constant flow would be very difficult (Figure 1a). To circumvent this obstacle, we implemented a droplet-on-demand approach that relies on several key developments: (i) a droplet generator for efficient encapsulation of single animals; (ii) microfluidic units for the dynamic modulation of the chemical environment; (iii) programmable actuation of on-chip valves and real-time image processing for robust execution of complex protocols (Figure 1b,c). This modular system is compatible with several optical modalities – bright-field, darkfield, and fluorescence imaging at low or high resolution – and, coupled with image processing, turns into a versatile technique to monitor and analyze dynamic phenotypes, such as behavior (Figure 1d). When resolution is high enough in the imaging setup, quantitative behavior traits can be extracted (Figure 1e), which is valuable for detecting subtle changes in behavior for screening applications.
Figure 1.
Overall presentation of the droplet-based platform for monitoring dynamic events in small animals. (a) Conceptual view of a droplet flow-cytometry technique applied to small animals. This scheme highlights the intrinsic qualities of droplet-microfluidics screening technology, including single-animal resolution, small reagent consumption, and independent reactors, but its execution faces challenges for characterizing dynamic phenotypes. (b) Implementation of an on-demand droplet system to encapsulate single animals, store droplets, and dynamically control droplet chemical content using computer-assisted automation. (c) Example of a channel network showing several units integrated on the same chip and a sample of the multiple flow paths that are sequentially executed to perform a given task. (d-e) High-content phenotyping of single animals: (d) Time-lapse sequence of bright-field images of an adult C. elegans animal encapsulated in a droplet (top); image processing to identify the animal (bottom). (e) Time course of postural and behavioral parameters for a single C. elegans individual freely swimming in a droplet.
2.2. An Active In-Situ Droplet Production Unit for Efficient Single-Animal Encapsulation
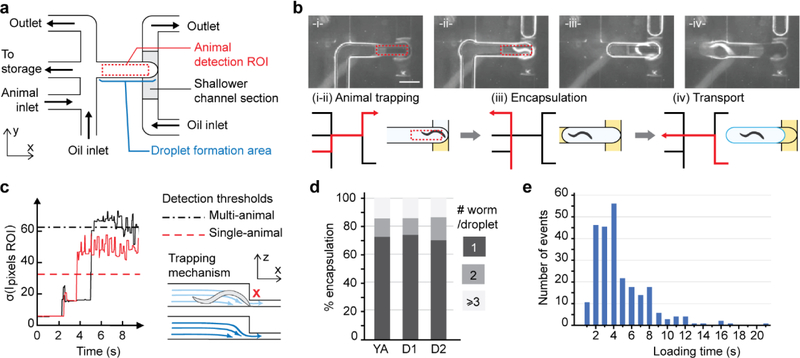
Efficient single-animal encapsulation is a pre-requisite to single-animal behavior assay but is challenging because of the animals’ shape, size, and movement that make them difficult to handle. To achieve this, we designed a method combining passive and active components: a physical constriction for trapping animals and programmed flow segmentation for generating droplets. Figure 2a shows the layout of the channel network. The trapping chamber at the center coincides with the droplet formation area. Multiple flow paths are sequentially activated to isolate and package an animal in a droplet (Figure 2b; Movie S1, Supporting Information). The process starts with the aqueous phase flowing through the chamber, and the shallow restriction acts as a filter for trapping an adult animal (diameter ~80 μm; Figure 2b(i–ii)). Second, a droplet is formed in situ by segmenting the chamber’s front and rear sides with the oil phase (Figure 2b(iii)). Finally, the droplet is pushed backward to the next processing step (Figure 2b(iv)). This encapsulation scheme works robustly as it separates animal isolation and droplet generation and is particularly suited to handle large or complex-shaped objects.
Figure 2.
Active in-situ droplet production unit for efficient single-animal encapsulation. (a) Microfluidic network layout. The sequential flow pattern is controlled using a set of valves upstream of the channels (not shown on the schematics). (b) Top: Representative pictures of the sequential flow routine (scale bar is 800 μm). Bottom: schematic of the encapsulation process. For each step, the active flow path is indicated in red. (i-ii) The aqueous phase flows through the trapping chamber until an animal is detected. (iii)The oil flow segments the aqueous droplet encapsulating the animal. The rear interface is created by actively flowing oil, while, on the front side, the aqueous film in the shallower channel section breaks spontaneously. (iv) The droplet is pushed outside the trapping chamber for further processing. (c) The trapping mechanism is based on the physical constriction of the channel. Real-time image processing of the animal detection ROI allows for the immediate detection of a trapped animal: The signal takes the characteristic shape of a near-zero level, brief peak, plateau, and a sudden prolonged increase corresponding respectively to the oil phase filling the chamber, water-oil interface passing, water-filled chamber state, and finally the presence of a trapped animal. (d) Single animal encapsulation performance; (e) Animal loading time is ~ 4 s long, ensuring a time-efficient encapsulation process.
To achieve efficient single-animal encapsulation requires efficient animal detection and consistent animal suspension. Both requirements are important to avoid empty droplets and multiple-animal encapsulation events. First, active detection is achieved by monitoring a region of interest (ROI) near the restriction through time. We used the standard deviation of the pixel intensity of the ROI as a metric (Figure 2c) and set a threshold to robustly detect an animal. Empty droplets are prevented as the droplet generation is prompted by an animal’s presence in the chamber. In addition, if several animals get trapped simultaneously, another threshold detects these events (see black dotted line in Figure 2c) and triggers a procedure to dispose of the animals. Second, maintaining a homogeneous animal suspension is crucial for achieving continuous performance. To avoid animals settling down in the reservoir, we engineered an off-chip mechanical system that periodically toggles the reservoir containing the worm suspension (Figure S1, Supporting Information). The animal suspension is set at a low density (~200 animals/mL) to favor single-animal trapping and thus single-animal loading. By doing so, we demonstrated over 70 % single-animal encapsulation for adult animals at different ages (Figure 2d), far above previous encapsulation schemes based on continuous flow. [40, 41] Furthermore, the passive trapping mechanism is suitable with high flow rates of the animal suspension, which speeds up animal encapsulation. We demonstrated the loading of several hundred animals with an average trapping time of 4 s (Figure 2e).
2.3. Liquid Exchanger for Generating Dynamic Chemical Gradients in Droplets
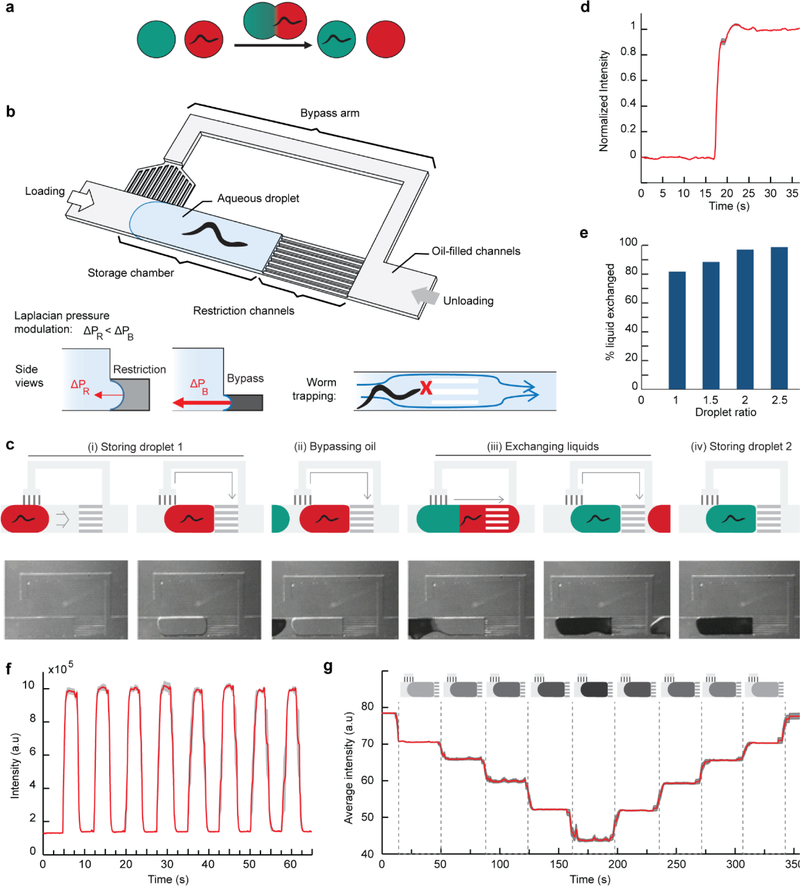
To achieve full dynamic environmental control, one must be able to both add and remove chemicals from the animal’s environment. The main challenges for efficiently exchanging the chemical environment of an encapsulated animal lies in avoiding mixing and preventing the animal’s escape while changing the droplet’s liquid content (Figure 3a). We have developed a Liquid Exchanger unit to exchange the chemical environment of a droplet with another without disturbing the animal. The unit (Figure 3b) is composed of a trapping chamber, restriction channels, and bypass channels, and its operation requires only controlling the flow direction. As a first droplet flows in, the restriction channels trap the droplet in the chamber (Figure 3c(i); Movie S2, Supporting Information). When a second droplet approaches, the bypass drains the oil spacer between the two droplets (Figure 3c(ii)). The two droplets merge, obstructing the bypass. Because the oil flow is not deflected through the bypass anymore, the backpressure increases and pushes the large newly-formed droplet through the restriction channels (Figure 3c(iii)). Importantly, the channel dimensions only allow the liquid to flow through and not the animal (Figure 3b, right insert). When the droplet rear side uncovers the bypass entrance, the backpressure drops. The oil phase flows through the bypass again and applies pressure on the aqueous phase at the exit of the restriction channel, triggering a droplet break-up. The animal finds itself in a new chemical environment (Figure 3c(iv)). This cycle can continue with additional droplets and the same animal or start afresh with a different animal by first emptying the trap using an inverted oil flow.
Figure 3.
Dynamic control of the chemical environment of an animal in a droplet. (a) The liquid exchanger allows swapping the whole chemical environment of an animal encapsulated in a droplet with another droplet. (b) Schematic of the unit. Left insert: The difference in channel cross-sections induces a modulation of the Laplacian forces at the droplet interfaces in the restriction channels and bypass channels. Right insert: The cross-section of the restriction channels is smaller than the animal, preventing its escape. (c) Step-by-step operation of the device for one exchange (i-iv). Oil flow is from left to right. (i) As the red droplet enters the chamber, the oil flow is deflected through the bypass. The pressure on the red droplet decreases and the red droplet remains in the chamber. (ii) The oil spacer flows through the bypass as the green droplet approaches. (iii) Liquid exchange operates as the bypass is blocked and the oil pressure increases to push the merged droplet through the restriction channels. (iv) The green droplet is now trapped in the chamber in a situation similar to the red droplet in step (i). One can perform multiple exchanges by repeating steps (ii-iv) or switch the oil flow from right to left to unload the chamber and start a new assay from step (i). (d) Liquid exchange dynamics. (e) Characterization of liquid exchange efficiency when varying the volume of the second droplet, here reported as the droplet ratio (length of the second droplet over the length of the first droplet, i.e., the chamber length). (d-e) were performed with a single-arm bypass device, Nbypass channels = 12, and Nrestriction channels = 9. (f,g) Generation of chemical profiles previously impossible to achieve in droplets: (f) Square signal achieved using an alternation of droplets with and without dye (n= 5 trials; droplet ratio: 2; grey curves are average ± SEM). (g) Linear up and down gradient obtained via a train of droplets with different dye concentration (n= 10 trials; droplet ratio: 3.2; grey curves are average ± SEM).
Designing a liquid exchanger that operates passively is difficult as the unit must execute multiple steps, some with opposing functions: trapping and releasing, and merging and splitting. To trap droplets (Figure 3c(i)), the Laplacian pressure at the water/oil interface in front of the droplet must compensate for the backpressure pushing the droplet in the storage chamber; this can be achieved using restriction channels with small cross-sections (Figure S2a,b, Supporting Information). However, a system to modulate the backpressure and push the droplet through the restriction channels is required to exchange liquids. Achieving this effect with a single pressure command is the key to obtain a self-regulating system. We addressed this challenge by using a bypass channel and via carefully balancing the Laplacian pressures at the bypass and restriction channels. The bypass channel acts as a pressure toggling system; the pressure at the rear of the droplet (backpressure) varies depending on whether the trapped droplet blocks the oil flow in the bypass. The Laplacian pressures at the bypass and restriction channels determine two threshold pressures above which the droplet will move through the channels (Figure 3b, left insert; Figure S2b, Supporting Information). To achieve the desired function, we designed the bypass channels with smaller cross-sections than for the restriction channels. We also set the command pressure at the oil reservoir so that the backpressure never crosses the threshold pressure for the bypass channels but toggles above and below the threshold pressure of the restriction channels (Figure S2c, Supporting Information). We found that a 2:1 channel cross-section ratio between the bypass and restriction channels allows for a comfortable pressure range to adjust the pressure command meeting these requirements. Finally, because we want the restriction channels to act as a grid preventing the animal from escaping, we used restriction channels with a cross-section slightly smaller than the animal diameter (Figure 3b, left insert).
Optimizing the design allows for switching chemical environments efficiently. We aimed to minimize the hydrodynamic resistances of the restriction channels and bypass channels without disrupting the Laplacian pressure balance. We achieved this by adding restriction channels and/or bypass channels in parallel and used two configurations: one with a single-arm bypass and another with a double arm bypass to test for a large number of bypass channels (Figure S3a, Supporting Information). As expected, increasing the number of restriction channels or bypass channels leads to higher operating speeds (Figure S3b,c, Supporting Information). Using a carrier oil of 10 cSt, the system can operate at 2.5 mm/s. Figure 3d shows the liquid exchange dynamics in the storage chamber. Swapping of the chemical environment happens within 3s, which fulfills our initial goal. If need be, it is possible to go faster, for example, by varying the viscosity of the carrier oil, where we demonstrated operating speeds up to 9 mm/s (Figure S3d, Supporting Information).
Importantly, liquid exchange is performed effectively during the droplet swapping. We characterized the amount of liquid exchange via microscope fluorescence imaging. Figure 3e shows the system’s performance as a function of the size of the second droplet passing through the liquid exchanger and normalized by the size of the chamber. For a droplet matching the chamber size, we observed 80% liquid exchange, and for droplets two times larger than the chamber, we demonstrated 94% liquid exchange. As a comparison, the state-of-the-art microfluidic device with substance exchange in droplets requires eight droplets and more than half a minute to achieve similar performance. [42]
Furthermore, automation and multi-cycle liquid exchange lead to the generation of chemical profiles previously impossible in droplet-based systems. For examples, one can create advanced patterns such as a square signal by alternating droplets of two chemicals or linear step-increase/decrease ramps (Figure 3f,g; Movie S3, Supporting Information). The measured signal is accurate to 5.6 % of the command, which allows for reliably assaying the animals’ response to precise and complex stimuli. Moreover, these patterns are reproducible – we observe a standard deviation of 4.4 % for ten repeats, which is more than necessary for chemotaxis and other chemical assays.
The liquid exchanger constitutes a complementary unit to other droplet-based chemical reactors. Indeed, the most widespread chemical reactor consists of merging two droplets and performs the valuable operation of adding contents. [54] We recently published a version of such a reactor, or Adder, designed for the acute stimulation of small animals. [43] Here, we have simplified its design, further characterized the temporal chemical profile, and demonstrated its operation for the merging of tens of droplets successively (Figure S4, Supporting Information; Movie S4, Supporting Information). Despite these performances, removing chemicals from a droplet is impossible using the Adder. At best, merging a buffer droplet may dilute a chemical concentration but will not remove the chemical from the environment. Therefore, the Liquid Exchanger brings a new function that allows the creation of a temporal chemical gradient out of reach with the Adder. The two systems are complementary and can be combined to create versatile on-chip chemical environments.
2.4. Automation of Platform Operation and Programming
The orchestration of complex protocols in a timely manner faces several challenges. It requires the accurate manipulation of flowing droplets, tracking of their positions on chip, and multiplexing of tasks to increase throughput. To address these requirements, we first automated the actuation of on-chip valves that control the flow. We also positioned all on-chip valves at the periphery to avoid any possible obstruction to animal transport. In addition, we relied on real-time image processing for controlling droplet transportation. In contrast with other recent works with automated droplet-based devices,[55] monitoring the droplet position continuously as it moves through the channel network is not required. Automation can be achieved by simply timing the droplet movement or detecting the droplet’s arrival at a desired location. By strategically positioning regions of interest within the microchannel network, we can detect the water/oil interface of the droplet and the carrier oil and execute advanced droplet manipulation sequences (Figure S5, Supporting Information). For example, for a standard protocol consisting of encapsulation, storing, stimulation, and 60 s long behavior recording, we demonstrate a throughput of 100 worms per hour (Movie S5, Supporting Information). We note that because of the automation, the droplet switching and animal loading take minimal amount of time as compared to the behavior and calcium imaging phenotypes that neurobiologists are interested in. This also demonstrates that by multiplexing, we can achieve behavioral recording of almost two worms simultaneously and continuously. Further multiplexing is possible via the integration of additional chambers; we previously operated manually an array of up to 12 chambers. [43] The bottleneck resides in the compromise between the field of view for imaging all chambers and the resolution necessary to observe a specific animal’s behavior. Finally, to facilitate the user experience, we developed a LabVIEW graphic user interface that takes as an input an Excel file containing the valve on/off state series (Figure S6 and S7, Supporting Information). The use of an Excel file allows for easy encoding of a protocol and re-writing if necessary. Such format suits the platform’s modular design well, where different units can be assembled in different schemes to fill a user’s application needs.
2.5. Versatile Screening of the Behavior and Neuronal Responses to Chemicals
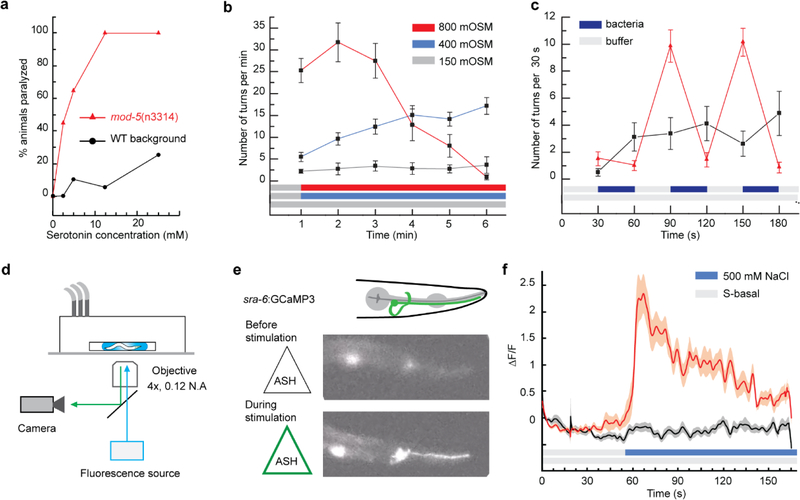
The ability to observe freely swimming animals in droplets opens to an extensive range of behavior-based assays while taking advantage of droplets’ unique properties. To demonstrate the advantage of our automated platform to test a range of conditions with low reagent consumption, we determined the dose-response curve of young adult WT and mutant animals to serotonin 5-HT. Using the Adder chamber, we screened for animal paralysis after 1 min exposure to serotonin in two genotypes, WT background animals and mod-5 mutant animals that are defective in serotonin reuptake. [56] As shown in Figure 4a, the response curves established for five different concentrations (0, 2.5, 5, 12.5, and 25 mM) highlight a higher sensitivity of mod-5 mutants than wild-type animals to 5-HT induced paralysis, which is in good agreement with prior literature. [56] The low reagent consumption (20 animals per condition, 600 nL stimulus doses per animal) demonstrates the unique advantage of using droplets. One limitation of the system lies in the necessary dilution of the stimulus upon merging with the droplet in the Adder unit, but this drawback can be resolved using the Liquid Exchanger module.
Figure 4.
Versatile platform for the screening of behavior and neuronal responses to chemicals in the adult C. elegans. (a-c) Behavior phenotyping. (a) Serotonin 5-HT dose-response assay performed using the Adder and generated with 20 animals of each genotype at each concentration. (b) Aversive response to osmotic upshift carried out with the liquid exchanger (n = 15 for 400 mM and 800 mM conditions, n = 10 for the control, error bars are SEM). (c) Food-exposure response using a train of droplets alternating buffer and bacteria in the liquid exchanger (n = 15 for stimulus, n = 10 for control, error bars are SEM). (d-f) Monitoring neuronal activity. (d) Fluorescence microscopy set-up for calcium transient imaging in freely swimming animals. (e) Fluorescence images of the chemosensory neuron ASH in a rested state (top) and activated state upon stimulation with 500 mM NaCl solution (bottom). (f) Average calcium response of ASH upon stimulation with 500 mM NaCl solution (red curve) and control (black curve). Shaded areas indicate SEM (n = 20 per condition).
To highlight the Liquid Exchanger’s ability to establish accurate environments, we recapitulated an assay testing the animal response to an osmotic upshift for different salt concentrations. [57] We monitored turning events, a typical phenotype for quantifying the animal’s response to an attractive or repulsive cue (Figure 4b). The results closely match the literature. [57] Conditions for the upshifts from 150 to 400 mM, and 150 to 800 mM NaCl solutions, show an increase of turning events in the first few minutes following the stimulation, and the response is more potent with the stimulus strength (Figure 4b). Moreover, the 150 to 400 mM step leads to a continuous increase through the 6 min long assay, while we observe a fall-out with the 800 mM condition on the fourth minute due to decreased mobility of the animals.
Furthermore, the Liquid Exchanger allows for assaying behaviors previously impossible in droplet microfluidics systems. To illustrate this capability, we monitored the response of animals to the removal of biological cues. Figure 4c displays turnings events for WT animals experiencing alternating buffer solution and suspension of OP50 bacteria in a buffer solution. OP50 E. coli is a common food source for C. elegans. As the animals are first exposed to the food, the near absence of reaction is indicative of the animals’ positive response to the change of environment. However, upon removing the bacteria, the animals engage in food search mode, as shown by the increase of turning events (see also Movie S6, Supporting Information). This response is reliably reproduced in each of the cycles alternating between food and buffer. This demonstrates the droplet platform’s ability to exchange the microenvironment dynamically, which allows assaying a single animal repeatedly for robust quantification of the animal’s response. Importantly, the ability to assess response upon the removal of a cue is critical to investigate numerous behaviors, for examples, in chemotaxis when navigating gradients, habituation, and learning.
Besides behavioral studies, our platform can be useful to assess cellular activity. For example, monitoring neuronal activity is essential to understand how the brain functions, how information about the environment flows from sensory neurons to interneurons, and motorneurons to result in specific behaviors. Our droplet platform is compatible with fluorescence imaging in general and calcium transient recording in particular. However, imaging calcium transients in freely swimming animals is challenging as fast thrashing movements and z-motion degrade the signal over noise ratio and create motion artifacts. To circumvent these issues, we used a low-magnification high-numerical-aperture objective (4x, 0.28 NA) and a high-sensitivity camera (Hamamatsu CMOS ORCA Flash 4.0) (Figure 4d). With this configuration, we can monitor the neuronal activity of animals freely swimming in droplets before, during, and after liquid exchange. As a demonstration, we observed the response of the chemosensory neuron ASH upon stimulation with a 500 mM NaCl solution (Figure 4e–f; Movie S7, Supporting Information). As expected, NaCl exposure results in a strong response while the control experiment that uses the same initial buffer solution fails to elicit a response. Because ASH is also a mechanosensory neuron, the control condition also confirms that the fluid exchange process does not mechanically perturb animals. The streamlining of droplet processing and freely moving animals inside the droplets will benefit largely to the study of neuronal circuitry and correlation with behavior.
2.6. Automated Forward Genetic Screening Based on Behavioral Phenotype
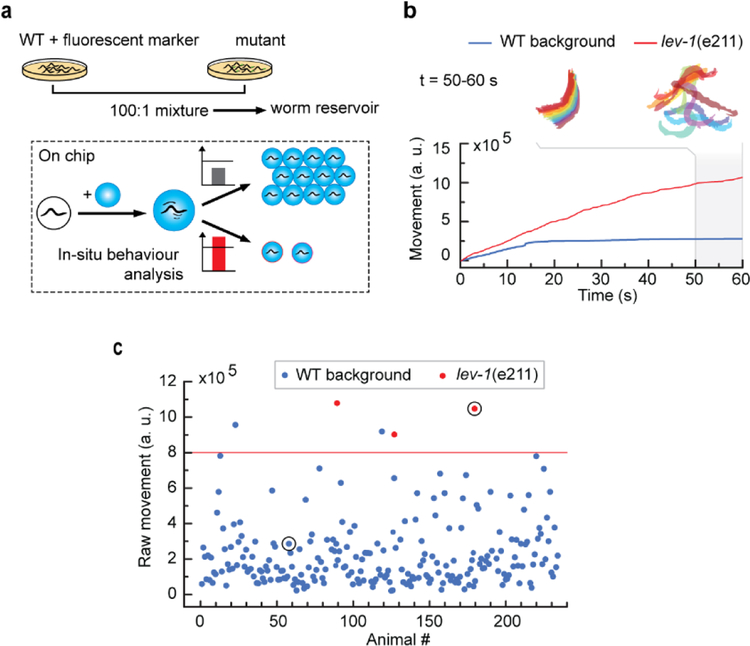
Because we can automate the droplet platform’s operation, it can also enable complex forward genetic screens based on a behavioral phenotype. Forward genetic screens are particularly interesting to find gene functions, such as drug targets, receptors, or effectors of small molecules and, therefore, are essential in deciphering signaling pathways. However, forward genetic screens rely on phenotyping thousands of animals; without automation in operation and also quantification for the phenotyping, this turns into a daunting task. Here we show that the droplet platform can alleviate the pain of performing genetic screens as it enables precise and automated manipulation of single animals, in-situ phenotyping, and selective recovery. As a proof-of-concept, we performed a mock screen for the acute effect of levamisole on C. elegans. We used a fluorescent transgenic strain with a WT N2 background mixed with lev-1 mutant strain in a 1:100 ratio (Figure 5a). Exposure to 10 mM levamisole, causes paralysis in WT animals within a minute, whereas lev-1 mutants can resist to the drug’s effects. [58] We monitored the raw movement of the animal over 60 s following drug exposure (Figure 5b). The WT background animal gets paralyzed within 15 s, as indicated by the blue curve’s plateau. In contrast, the red curve showing the mutant’s behavior displays a steady increase, representative of continuous thrashing, and is clearly distinguishable from the WT response. Based on that criterion, animals displaying a mutant phenotype were sorted in a separate channel. Fluorescence imaging was used to confirm the genotypes (see Figure S8, Supporting Information, for the detailed operation of the whole sequence). Our results demonstrate the successful recovery of all mutant animals and the presence of two false positives (Figure 5c). The presence of false-positive is to be expected as behavioral phenotypes usually exhibit significant heterogeneity. Importantly, no false negative was reported. This method is only bottlenecked by the behavior assessment that may be computationally demanding; however, the success of neuronal network-based programs [59, 60] opens the scope to real-time analysis of complex behaviors. Overall, this proof-of-concept demonstrates the potential of the platform to perform fully automated, reliable screens of behavior and could be extended to other advanced functional phenotypes.
Figure 5.
Automated forward genetic screening based on behavioral phenotype. (a) Screen protocol. Off-chip, a mock mutagenized population is created by mixing a WT background strain and mutant animals. On-chip, an automated routine stimulates every animal, analyzes each animal’s response in situ, takes a picture in fluorescence imaging modality, and selectively recovers the animal if a pre-determined behavioral phenotype is flagged. Fluorescence images are used to establish ground truth. (b) Screening criteria based on the animal’s activity in response to 10 mM levamisole drug treatment: raw behavior integrated over 60 s exposure is chosen to target abnormal responses. Traces correspond to the individuals circled in (c) and are representative of a paralyzed animal and drug-resistant animal. (c) Results of the mock screen highlighting the ability to recover all mutants of interest.
2.7. Scalable Technology – Application to the Manipulation of First Larval Stage C. elegans Animals
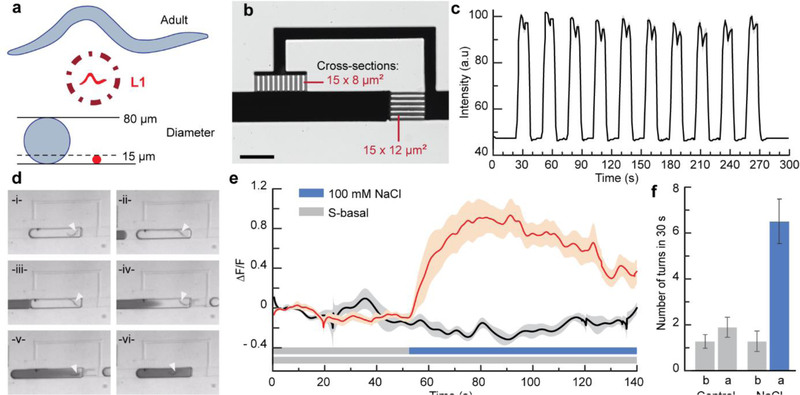
To demonstrate the scalability of the platform, we targeted C. elegans first larval stage. The larvae animals are much smaller than adults (L1 ~ 200 μm long and 15 μm in diameter versus Day 1 adults ~1000 μm long and 80 μm in diameter), very active, flexible, and fragile (Figure 6a). We demonstrate miniaturized units for controlling the chemical environment of L1s (Figure 6b). We set the channel cross-sections to 15 × 8 μm2 and 15 × 12 μm2 for the bypass and restriction channels respectively. The channel dimensions meet the condition on the Laplacian pressures necessarily for proper liquid exchange. Figure 6c shows highly reliable and efficient exchange for a series of 13 s long pulses (Movie S8, Supporting Information). In addition, the restriction channels being slightly smaller than the animals, L1 larvae remain trapped in the droplets during the operation (Figure 6d, white arrows indicate the animal’s position; Movie S9, Supporting Information). Clogging issues are then avoided because the encapsulated animal is transported in channels that are much larger than the animal diameter, while the shallower sections of the network are designed for liquids only. Furthermore, we demonstrate functional neuronal imaging of larvae encapsulated in droplets. We monitored the response of ASH sensory neuron to osmotic shock, a sensory response likely acquired early in life as to avoid hostile environments. Upon presenting a 100 mM NaCl solution, ASH neuronal activity increases rapidly before decaying (Figure 6e; Movie S10, Supporting Information). Looking simultaneously at the behavioral response confirmed the aversive response to the change of environment. Figure 6f shows an increase of turns/reversals of L1 larvae to the osmotic upshift. Altogether these results demonstrate the droplet platform’s utility to research on development and pave the way for future extensive studies on neurodevelopmental plasticity.
Figure 6.
Scalable technology: manipulation and chemical stimulation of first larval stage C. elegans. (a) L1 animals are very challenging to manipulate individually because of their small size, high motility, and flexibility. (b) Scaling-down of the Liquid Exchanger unit to suit the size of L1 animals (scale bar is 200 μm). (c) Square variation of the chemical profile of a droplet trapped in the miniaturized Liquid Exchanger shown in (b) and upon flowing a series of droplets with different dye concentrations. (d) Time-lapse sequence simulating the stimulation of an L1 animal using dye. (e) Functional neuronal imaging of ASH chemosensory neurons in L1s. The red curve is the average response upon stimulation with 100 mM NaCl solution, and the black curve is the average response for the control. Shaded areas indicate SEM (n = 20 for stimulus, n = 10 for control). (f) Behavioral response associated with (e): “b” is for a 30 s period before stimulation, and “a” is for a 30 s period after stimulation.
3. Discussion
We have developed a multiphase-microfluidics platform that offers advanced environmental control on single animals and high-throughput serial processing. To achieve this, we created an efficient strategy to isolate single live animals in droplets and a liquid exchanger system with unprecedented dynamic control over the chemical environment of encapsulated animals. In addition, we developed a general framework for facile programming of multistep protocols and serial processing of up to a hundred animals per hour. The different microfluidic units are designed as independent functional modules, compatible with each other to offer maximal flexibility in creating microfluidic circuits. We demonstrate how our platform uniquely enables experiments with motile animals using C. elegans nematodes at larval and adult stages and performing multiple screens using diverse dynamic phenotypes.
The platform possesses several key unique features that will advance phenotyping capacity in multiple fields and help bridge the gap between genotype and phenotype. The liquid exchanger unit allows for renewing the chemical environment in a single pass, which has not previously been demonstrated and allows for temporal control at the timescale of a few seconds. Coupled with the programming capability of valve-based dispensers,[61–63] virtually any chemical profile is possible. This provides full control over the sensory experience of a freely moving animal, which is invaluable when applied to study neuronal circuits. Combining precise environmental perturbation with optogenetics, calcium imaging, and behavioral read-out will find applications in deciphering circuits, neuronal dynamics, temporal filters using white noise analysis, receptive fields, and correlating whole-brain activity with evoked behaviors. Furthermore, we showed that the platform could recreate different physical environments (Note S1 and Figure S9, Supporting Information). This ability could be further expanded by integrating valves and a Peltier module to perform active mechanical and thermal stimulations for multimodal sensory integration studies.
Another key feature of the platform lies in the efficient encapsulation of single animals and automation that open the possibility to perform traditionally challenging screens for gene and drug discovery. Efficient single animal encapsulation participates in the platform throughput and enables selective recovery. Automation relieves painstaking manual tasks and user bias that may interfere with the animal’s response. We demonstrated continuous operation over several hours, requiring only human intervention for refilling the animal reservoir, and observed consistent performance over several weeks, paving the way for large-scale screens. Importantly, advanced droplet manipulation offers combinatorial capability, of high interest for drug screens. Droplets also provide means to target stochastic phenotypes, such as interneuron activation and most behavioral read-outs, by assessing the same individual multiple times. Finally, compared to screens relying on a batch strategy, single-animal interrogation allows for screening phenotypes that are more demanding in temporal or spatial resolution. Importantly, the nanoliter amount of reagent per animal will benefit screens with rare or expensive reagents, such as pheromones and drugs.[31]
A third quality of the platform consists of effective manipulation of complex, motile, and sometimes fragile animals. The liquid-liquid boundary and fully-closed space of water-in-oil emulsion ensure robust trapping and gentle animal handling. We pushed the platform’s ability using L1s, the smallest of all C. elegans larvae, whose manipulation is challenging even in the hands of trained scientists. We showed precise modulation of the chemical environment of L1s without losing animals or introducing undesired perturbations. We demonstrated for the first time monitoring of L1s’ neuronal activity in response to chemical stimulation. Such functionality was previously impossible to achieve before with any microfluidic or non-microfluidic system. Traditional manual techniques such as gluing animals on a substrate [64] already require extreme care when handling adult animals, and gluing L1s that are a lot more fragile is nearly impossible. Single-phase microfluidic devices are prone to clogging if miniaturized and adapted to L1s and do not provide a robust method for manipulating young larvae. This new functionality is particularly significant as it will enable future investigations of the developing brain. C. elegans nervous system undergoes significant post-embryonic development, including the generation of a third of its neurons, neuronal differentiation, and maturation of neuronal circuits.[65] The platform will allow studying how these events impact neuronal function and computation. Adapting device dimensions and tuning droplet sizes to match the length of the four larval stage and adult animals will enable longitudinal studies through development and aging.
Furthermore, many model organisms used in research, including zebrafish larvae, Drosophila larvae, copepods, daphnia, rotifers, Hydra animals, would benefit from the controlled environment and screening capabilities offered by our multiphase platform. Similar to the experiments we performed, these organisms present a palette of dynamic phenotypes in behavior such as locomotion, feeding, mating, learning, or decision-making, and research into the underlying neuronal processes that drive these activities requires improved tools. The technical bottlenecks addressed in this work – handling motile animals, performing individualized treatments, controlling the environment, achieving selective recovery, and serial processing – are the same that hampers phenotyping of other small animals. The platform principles are not specific to C. elegans animals as the operation focuses on droplet manipulation. We have shown that we were able to manipulate objects from 10 μm to the mm range covering the same order of magnitude as other animals’ sizes. Device dimensions may need to be adapted (SI Note S2, and Figure S10, Supporting Information), but this can be easily achieved through microfabrication techniques such as optical lithography using thicker resists, micro-milling, and 3D printing.[66] Therefore, due to the variety of animals and phenotypes, we envision this platform to have a broad impact in the fields of neuroscience, development, behavior, environmental toxicology, genetics, and drug screens.
Finally, beyond research on small animals, elements of this platform can be used for droplet-based methods for other biological systems and chemical processes. Increasing or downsizing the channel cross-section would allow multicellular organisms, like spheroids or organoids, and large single cells to be processed. For example, studies on brain organoids, such as in a drug screen, would benefit from monitoring and sorting based on neuronal activity while rapidly exchanging the environment. Studies on single cells, such as motile sperm, may benefit from fast environmental changes enabled by the liquid exchanger. In chemical assays, liquid exchange properties and automation can also be of interest in multistep chemical processes for particle synthesis and bead-based chemistry,[67] such as in DNA/RNA extraction.
We demonstrated here a modular platform that uses multiphase microfluidics to screen dynamic phenotypes in small animals. Automation of droplet processing on chip allows for high-precision chemical stimulation and serial processing of single individuals, leading to a powerful technology that will help bridge the gap between environment, genotype, and phenotype.
4. Experimental Section
Microfabrication:
Multi-layer PDMS devices are fabricated using soft lithography protocols. First, a master is made for the valve control channels with 50 μm high structures in SU-8 photoresist. Another master for the droplet channel network is fabricated with a hybrid structure of SU-8 negative photoresist and AZ40XT positive photoresist. Multiple layers of SU-8 resist are applied to modulate channel height (15–200 μm), and a single layer of AX40XT resist (50/100 μm) is used for the valve sections. Second, PDMS blocks containing the droplet channel design are made in 10:1 PDMS:cross-linker mixture, fully cured, with punched access wells to the channel inlets and outlets. A thin membrane (20:1 PDMS:cross-linker) is created over the valve control channel master via spincoating and partially cured at 80 °C for 10 min. Thermal bonding is then achieved by aligning the PDMS block containing the droplet channel over the master with the valve control. After curing at 80 °C overnight, the PDMS blocks are peeled off and cut, and access wells for the valves are punched. The resulting PDMS blocks are thermally bonded again on glass slide spincoated with a 5:1 PDMS:cross-linker mixture. After curing overnight, the devices are primed with 10 cSt silicone oil.
C. elegans Maintenance and Reagent Preparation:
Animals were cultured on nematode growth medium (NGM) plates seeded with OP50 Escherichia coli bacteria using standard methods. We used the following strains: Wild-type N2; PTM336 oxTi721 [eft-3p::tdTomato::H2B::unc- 54 3’UTR]; MT9772 [mod-5(n3314)]; CX10979 kyEx2865 [p-sra-6:GCaMP3; p-ofm-1:GFP]; and CB211 lev-1(e211)IV. Assays using adult animals were performed with age-synchronized population via egg laying. Assays using L1 animals were performed on larvae in L1 arrest by bleaching gravid hermaphrodites and leaving the eggs in suspension in S-basal solution overnight. Prior to starting an experiment, animals were suspended in either M9 or S-basal buffer solution. Ethics approval was not required for the experiments using animals in this study. The Institutional Biosafety Committee’s approved protocols for biosafety were followed.
Stimulus solutions of serotonin 5-HT (Sigmal), NaCl (Sigma), and OP50 bacteria (liquid culture grown overnight) were prepared in the same buffer as the animal suspension. For regular device operation, 10 cSt or 50 cSt silicone oils were used as the continuous phase. The operation speed characterization involved silicone oils over a range of viscosities: 0.65–80 cSt (Clearco, PSF-0.65/10/50cSt Silicone Fluids, and mixtures of PSF-10 and 1,000cSt Silicone Fluids). The viscosities of home-made mixtures were measured using Brookfield DVT3HB-CJ0 rheometer and 2.5 mL sample solution in a circular chamber and spindle CPA-40Z.
Microfluidic Platform Operation:
All liquids were actuated using pressure-driven flow. Liquid reservoirs and on-chip valves were connected to the device via PE tubing and pressurized via a home-made pressure box. Protocols were executed via a home-made LabVIEW interface that allows for manual or automated operation (Figure S6, Supporting Information). Behavior assays were performed on compound scopes using either a Lumenera Infinity 3 camera or a Thorlabs (DC1645C-HQ) camera, and calcium imaging experiments were performed on inverted epifluorescence scope (Olympus IX73) with a 4× 0.28 NA objective, a CMOS camera (Hamamatsu ORCA Flash 4.0 C11440–22CU) and fluorescence light source (Leica EL6000). Toggling of the animal reservoir was performed using a servo motor (LS-0009AF, Metal Gear Digital) controlled with an Arduino system (chipKIT uC32 Arduino – programmable PIC32 Microcontroller board, Digilent) and the LabVIEW GUI (Figure S1&S6, Supporting Information).
Characterization of Chemical Profiles in Droplets:
Fluorescein dye (1 mM) solution was used for the square chemical profile characterization of the Liquid Exchanger unit. Food dye solutions were used to create the other chemical profiles for both Liquid Exchanger and Adder units. Assays were performed on a compound scope with either Infinity 3 camera for fluorescence imaging or Thorlabs camera for bright field imaging. All results were analyzed via Matlab programs.
Behavior Assays and Analysis:
Worm paralysis was scored by comparing the raw movement of an animal to a pre-established threshold. Raw movement is quantified by integrating pixel-to-pixel subtraction of consecutive thresholded images over 60 seconds. Worm turning events are calculated by counting the number of times that the head-tail distance goes under a pre-established threshold.
Calcium Imaging Set-up and Data Analysis:
Once loaded on chip, animals were exposed for 90 s with blue light before recording. Neuronal activity was analyzed using a custom MATLAB program. Fluorescent GCaMP response was calculated using the formula ΔF/F0 where ΔF = FNeuron − FBackground. FNeuron is the average intensity in a region of interest, including the target neuron, and FBackground is the average intensity in a region of interest adjacent to the target neuron (background). F0 is defined as the starting signal baseline (the average ΔF value for the first ten seconds of recording).
Mock Behavioral Screen:
The experimental set-up was slightly modified to perform the mock screen. First, a red LED ring (SuperbrightLEDs, AE80-R48-BK), computer-controlled via a driver board (Ultimarc, Pac-Drive), was used for dark field imaging. Second, a fluorescence source was used with a mechanical shutter actuated with the PAC-drive. A script was integrated in the LabVIEW interface program that runs the microfluidic routine to switch imaging modalities from fluorescence to darkfield, record pictures, and characterize the phenotype of the animal response to a stimulus before sorting.
Data availability:
Software and code for interfacing the platform are publicly available at github at: https://github.com/Guillaume-Gatech/Automated-droplet-platform. The data that support the findings of this study are available from the corresponding author on request.
Supplementary Material
Acknowledgements
The authors acknowledge National Institute of Health (NIH R21NS117066, R01NS096581, and R01AG056436) for funding, and the McGrath lab (Georgia Tech) and Bargmann lab (Rockefeller U.) for strains. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was performed in part at the Georgia Tech Institute for Electronics and Nanotechnology, a member of the National Nanotechnology Coordinated Infrastructure (NNCI), which is supported by the National Science Foundation (Grant ECCS-1542174). The authors also thank Drs. G. Sun, D. Patel, and M. Crane for inputs on the manuscript.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
References
- [1].Patton EE, Zon LI. Nat Rev Genet 2001, 2, 956. [DOI] [PubMed] [Google Scholar]
- [2].Jorgensen EM, Mango SE. Nat Rev Genet 2002, 3, 356. [DOI] [PubMed] [Google Scholar]
- [3].Cully M. Nat Rev Drug Discov 2019, 18, 811. [DOI] [PubMed] [Google Scholar]
- [4].MacRae CA, Peterson RT. Nat Rev Drug Discov 2015, 14, 721. [DOI] [PubMed] [Google Scholar]
- [5].O’Reilly LP, Luke CJ, Perlmutter DH, Silverman GA, Pak SC. Adv Drug Deliver Rev 2014, 69, 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kwok TCY, Ricker N, Fraser R, Chan AW, Burns A, Stanley EF, McCourt P, Cutler SR, Roy PJ. Nature 2006, 441, 91. [DOI] [PubMed] [Google Scholar]
- [7].Lehner B, Tischler J, Fraser AG. Nat Protoc 2006, 1, 1617. [DOI] [PubMed] [Google Scholar]
- [8].Burns CG, Milan DJ, Grande EJ, Rottbauer W, MacRae CA, Fishman MC. Nat Chem Biol 2005, 1, 263. [DOI] [PubMed] [Google Scholar]
- [9].Rihel J, Prober DA, Arvanites A, Lam K, Zimmerman S, Jang S, Haggarty SJ, Kokel D, Rubin LL, Peterson RT, Schier AF. Science 2010, 327, 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kamili F, Lu H. Annual Review of Analytical Chemistry 2018, 11, 245. [DOI] [PubMed] [Google Scholar]
- [11].Yang F, Gao C, Wang P, Zhang GJ, Chen ZG. Lab Chip 2016, 16, 1106. [DOI] [PubMed] [Google Scholar]
- [12].Badhiwala KN, Gonzales DL, Vercosa DG, Avants BW, Robinson JT. Lab Chip 2018, 18 [DOI] [PubMed] [Google Scholar]
- [13].Ramanathan N, Simakov O, Merten CA, Arendt D. Plos One 2015, 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Campana O, Wlodkowic D. Sensor Actuat B-Chem 2018, 257, 692 [Google Scholar]
- [15].Kato S, Kaplan HS, Schrodel T, Skora S, Lindsay TH, Yemini E, Lockery S, Zimmer M. Cell 2015, 163, 656. [DOI] [PubMed] [Google Scholar]
- [16].Chronis N, Zimmer M, Bargmann CI. Nat Methods 2007, 4, 727. [DOI] [PubMed] [Google Scholar]
- [17].Albrecht DR, Bargmann CI. Nat Methods 2011, 8, 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chung KH, Crane MM, Lu H. Nat Methods 2008, 5, 637. [DOI] [PubMed] [Google Scholar]
- [19].Mondal S, Hegarty E, Martin C, Gokce SK, Ghorashian N, Ben-Yakar A. Nat Commun 2016, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kopito RB, Levine E. Lab Chip 2014, 14, 764. [DOI] [PubMed] [Google Scholar]
- [21].Fuad NM, Kaslin J, Wlodkowic D. Sensor Actuat B-Chem 2018, 256, 1131 [Google Scholar]
- [22].Nady A, Peimani AR, Zoidl G, Rezai P. Lab Chip 2017, 17, 4048. [DOI] [PubMed] [Google Scholar]
- [23].Chung K, Zhan M, Srinivasan J, Sternberg PW, Gong E, Schroeder FC, Lu H. Lab Chip 2011, 11, 3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lockery SR, Hulme SE, Roberts WM, Robinson KJ, Laromaine A, Lindsay TH, Whitesides GM, Weeks JC. Lab Chip 2012, 12, 2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gonzales DL, Badhiwala KN, Vercosa DG, Avants BW, Liu Z, Zhong WW, Robinson JT. Nat Nanotechnol 2017, 12, 684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yadavali S, Jeong HH, Lee D, Issadore D. Nat Commun 2018, 9, 1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nightingale AM, Phillips TW, Bannock JH, de Mello JC. Nat Commun 2014, 5, 3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bowman EK, Alper HS. Trends Biotechnol 2020, 38, 701. [DOI] [PubMed] [Google Scholar]
- [29].Trantidou T, Friddin M, Elani Y, Brooks NJ, Law RV, Seddon JM, Ces O. Acs Nano 2017, 11, 6549. [DOI] [PubMed] [Google Scholar]
- [30].Kulesa A, Kehe J, Hurtado JE, Tawde P, Blainey PC. P Natl Acad Sci USA 2018, 115, 6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shembekar N, Chaipan C, Utharala R, Merten CA. Lab Chip 2016, 16, 1314. [DOI] [PubMed] [Google Scholar]
- [32].Han XP, Zhou ZM, Fei LJ, Sun HY, Wang RY, Chen Y, Chen HD, Wang JJ, Tang HN, Ge WH, Zhou YC, Ye F, Jiang MM, Wu JQ, Xiao YY, Jia XN, Zhang TY, Ma XJ, Zhang Q, Bai XL, Lai SJ, Yu CX, Zhu LJ, Lin R, Gao YC, Wang M, Wu YQ, Zhang JM, Zhan RY, Zhu SY, Hu HL, Wang CC, Chen M, Huang H, Liang TB, Chen JH, Wang WL, Zhang D, Guo GJ. Nature 2020, 581, 303. [DOI] [PubMed] [Google Scholar]
- [33].Siebert S, Farrell JA, Cazet JF, Abeykoon Y, Primack AS, Schnitzler CE, Juliano CE. Science 2019, 365, eaav9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Matula K, Rivello F, Huck WTS. Adv Biosyst 2020, 4, 1900188. [DOI] [PubMed] [Google Scholar]
- [35].Frey O, Misun PM, Fluri DA, Hengstler JG, Hierlemann A. Nat Commun 2014, 5, 4250. [DOI] [PubMed] [Google Scholar]
- [36].Sart S, Tomasi RFX, Amselem G, Baroud CN. Nat Commun 2017, 8, 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sart S, Tomasi RFX, Barizien A, Amselem G, Cumano A, Baroud CN. Sci Adv 2020, 6, eaaw7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Liu HT, Wang YQ, Wang H, Zhao MQ, Tao TT, Zhang X, Qin JH. Adv Sci 2020, 7, 1903739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shi WW, Qin JH, Ye NN, Lin BC. Lab Chip 2008, 8, 1432. [DOI] [PubMed] [Google Scholar]
- [40].Shi WW, Wen H, Lu Y, Shi Y, Lin BC, Qin JH. Lab Chip 2010, 10, 2855. [DOI] [PubMed] [Google Scholar]
- [41].Aubry G, Zhan M, Lu H. Lab Chip 2015, 15, 1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wen H, Yu Y, Zhu GL, Jiang L, Qin JH. Lab Chip 2015, 15, 1905. [DOI] [PubMed] [Google Scholar]
- [43].Aubry G, Lu H. Lab Chip 2017, 17, 4303. [DOI] [PubMed] [Google Scholar]
- [44].Collins DJ, Neild A, deMello A, Liu AQ, Ai Y. Lab Chip 2015, 15, 3439. [DOI] [PubMed] [Google Scholar]
- [45].Shang LR, Cheng Y, Zhao YJ. Chem Rev 2017, 117, 7964. [DOI] [PubMed] [Google Scholar]
- [46].Chong ZZ, Tan SH, Ganan-Calvo AM, Tor SB, Loh NH, Nguyen NT. Lab Chip 2016, 16, 35. [DOI] [PubMed] [Google Scholar]
- [47].Jin SH, Jeong HH, Lee B, Lee SS, Lee CS. Lab Chip 2015, 15, 3677. [DOI] [PubMed] [Google Scholar]
- [48].Baroud CN, de Saint Vincent MR, Delville JP. Lab Chip 2007, 7, 1029. [DOI] [PubMed] [Google Scholar]
- [49].Choi K, Ng AHC, Fobel R, Wheeler AR. Annual Review of Analytical Chemistry 2012, 5, 413. [DOI] [PubMed] [Google Scholar]
- [50].Cho YM, Porto DA, Hwang H, Grundy LJ, Schafer WR, Lu H. Lab Chip 2017, 17, 2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rezai P, Siddiqui A, Selvaganapathy PR, Gupta BP. Appl Phys Lett 2010, 96, 153702 [Google Scholar]
- [52].Yoon S, Piao H, Jeon TJ, Kim SM. Anal Sci 2017, 33, 1435. [DOI] [PubMed] [Google Scholar]
- [53].Bargmann CI, Chemosensation in C elegans, in Wormbook, W. The C. elegans Research Community, Editor. 2006. [Google Scholar]
- [54].Kaminski TS, Garstecki P. Chem Soc Rev 2017, 46, 6210. [DOI] [PubMed] [Google Scholar]
- [55].Hebert M, Courtney M, Ren CL. Lab Chip 2019, 19, 1490. [DOI] [PubMed] [Google Scholar]
- [56].Ranganathan R, Sawin ER, Trent C, Horvitz HR. J Neurosci 2001, 21, 5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yu JY, Yang WX, Liu H, Hao YS, Zhang Y. Eneuro 2017, 4, e0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Brenner S. Genetics 1974, 77, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kane GA, Lopes G, Saunders JL, Mathis A, Mathis MW. Elife 2020, 9, e61909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Mathis MW, Mathis A. Curr Opin Neurobiol 2020, 60, 1. [DOI] [PubMed] [Google Scholar]
- [61].Zeng SJ, Li BW, Su XO, Qin JH, Lin BC. Lab Chip 2009, 9, 1340. [DOI] [PubMed] [Google Scholar]
- [62].Zec H, Rane TD, Wang TH. Lab Chip 2012, 12, 3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Woodruff K, Maerkl SJ. Anal Chem 2018, 90, 696. [DOI] [PubMed] [Google Scholar]
- [64].Kerr RA, Imaging the activity of neurons and muscles, in Wormbook, in WormBook. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Altun ZF, Hall DH, Nervous system, general description, in WormAtlas. 2011. [Google Scholar]
- [66].Au AK, Huynh W, Horowitz LF, Folch A. Angew Chem Int Edit 2016, 55, 3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Serra M, Ferraro D, Pereiro I, Viovy JL, Descroix S. Lab Chip 2017, 17, 3979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Software and code for interfacing the platform are publicly available at github at: https://github.com/Guillaume-Gatech/Automated-droplet-platform. The data that support the findings of this study are available from the corresponding author on request.