Abstract
Neoantigen vaccines aiming to induce tumor-specific T cell responses have achieved promising anti-tumor effects in early clinical trials. However, the underlying mechanism regarding response or resistance to this treatment remains unclear. Here, we observe that neoantigen vaccine-generated T cells can synergize with immune checkpoint blockade (ICB) for effective tumor control. Specifically, we performed single-cell sequencing on over 100,000 T cells and uncovered that combined therapy induces an antigen-specific CD8 T cell population with active chemokine signaling (Cxcr3+/Ccl5+), lower co-inhibitory receptor expression (Lag3−/Havcr2−) and higher cytotoxicity (Fasl+/Gzma+). Furthermore, the generation of neoantigen-specific T cells in the draining lymph node is required for combination treatment. Signature genes of this unique population are associated with T cell clonal frequency and better survival in humans. Our study profiles the dynamics of tumor infiltrated T cells during neoantigen vaccine and ICB treatments, high-dimensionally identifies neoantigen-reactive T cell signatures for future development of therapeutic strategies.
Introduction
T cells play an indispensable role in cancer immunotherapy 1. Therapeutic strategies aiming to restore anti-tumor T cell immunity have seen promising responses in patients, such as immune checkpoint blockade (ICB), neoantigen vaccine, and adoptive T cell transfer 2, 3. However, many patients are still resistant to these treatments clinically. The underlying mechanism of response or resistance remains unclear. In addition, the characteristics of tumor infiltrated T cells in response to treatments have not been well studied.
Among all the tumor-infiltrated lymphocytes (TILs), antigen-specific T cells are the only population that can specifically recognize cancer cells through their unique TCRs 4, initiate cytotoxic pathways to kill the targets and establish T cell memory 5. However, very few tumor antigens bearing immunogenic epitopes have been identified as targets for immunotherapy. Among them, tumor-associated antigens (TAAs) are primarily overexpressed by tumor cells but are also expressed in some normal tissues 6. Cancer vaccines that target TAAs such as NY-ESO-1 and MUC-1 are still challenging with concerns of inducing autoimmunity and the lack of high affinity T cell responses due to thymic negative selection 7. In contrast, neoantigens caused by tumor-specific somatic alterations are more promising therapeutic targets 8. Therapeutic vaccines targeting the highly immunogenic neoantigens have demonstrated promising anti-tumor effects in patients with melanoma in early clinical trials 9–11. However, the neoantigen-specific T cell immune response induced by such vaccines still varies between patients. Furthermore, some evidence suggests that resistance to this treatment can be overcome with adjuvant anti-PD-1 therapy, indicating combination treatment may be required for some patients clinically 10.
Despite these clinical successes, many fundamental immunologic questions regarding neoantigen vaccine remain unclear. First, which T cell subset(s) play an essential role in responders and what are their representative biomarkers; second, what is the mechanism of neoantigen-specific T cell activation or exhaustion in vivo after vaccine treatment; finally, how do neoantigen vaccines synergize with ICB to overcome clinical resistance? Both draining lymph node (dLN) and tumor tissue are involved in the anti-PD-L1 mediated T cell immune response 1, 12. Therefore, reinvigoration of pre-existing T cells or clone replacement from lymphoid tissues may contribute to the therapeutic effect of combination treatment 13.
To address these questions requires a scrutiny of T cell dynamics during neoantigen vaccine therapy, and a thorough investigation on how adjuvant use of ICB treatment specifically remodels the tumor microenvironment (TME). Unfortunately, this task is highly challenging when using human patient samples. It is logistically difficult to obtain biopsies from both lymphoid and tumor tissues simultaneously, especially in early stages of tumorigenesis. Therefore, an investigation that can reveal the phenotype and biomarkers of neoantigen-specific T cells is required to predict responses to immunotherapies and develop therapeutic strategies. With single-cell sequencing technology, it is possible to overcome these limitations by obtaining both transcriptomes and TCR sequences for individual T cell simultaneously 14–16.
In this work, we took advantage of MC38 model that has well-characterized neoantigens 17 to investigate the T cell dynamics during neoantigen vaccination, anti-PD-L1 treatment and the combination. We first reproduced the observation in humans showing that combination treatment can generate better anti-tumor effect than vaccination alone. To investigate the dynamics of neoantigen-specific T cells during these therapies, we implemented single-cell RNA sequencing (scRNA-seq) with targeted V(D)J capture of the T cells in the dLN and tumor tissues. We found that the combination treatment dramatically reshaped the TME by reducing Tregs and dysfunctional CD8 T cells while expanding Th1-like CD4 T cells and CD8 effectors T cells. Lineage tracking through TCRs revealed that combination treatment prevented the terminal fate transition of newly infiltrated T cells. Most importantly, combination treatment generated a Cxcr3+/Lag3− population of CD8 T cells, which was neoantigen-specific and had strong cytotoxicity against tumor cells. Mechanistically, neoantigen vaccine primed T cells in the dLN are required for the generation of this unique subset in the presence of ICB. Finally, our analysis identified discriminative markers of this specific population, which were significantly associated with better clinical outcomes in multiple human cancers.
Results
Neoantigen vaccine synergies with ICB to induce a durable anti-tumor immune response
To investigate neoantigen vaccine induced T cell responses and anti-tumor effects in vivo, we first performed neoantigen vaccine treatment on murine colorectal tumor model MC38, which has well-defined MHC-I restricted immunogenic neoantigens 17. According to the dose and schedule of recent neoantigen vaccine studies11, 17, 18, we choose the 9-mer mutated Adpgk peptide (ASMTNMELM) as the neoantigen and formulated it with two Toll-like receptor (TLR) agonists as adjuvants (Poly-ICLC for TLR-3 and CpG 1826 for TLR-9), both of which can activate dendritic cells and have been used in clinical trials19–21. Two doses of neoantigen Adpgk vaccine displayed moderate anti-tumor effect with delayed tumor growth (Fig. 1a). However, no tumor regression was observed, suggesting a limited anti-tumor effect of neoantigen vaccine alone. We next examined the level of neoantigen-specific T cells in the dLN. As expected, the vaccination efficiently induced tetramer+ cells in the dLN compared with non-dLN at baseline, regardless of whether the mice were tumor bearing (Fig. 1b). However, neoantigen-Adpgk-specific T cells in the tumor tissue were only slightly increased after vaccination and showed an exhaustion phenotype with elevated PD-1 and TIM-3 expression compared to those in the lymph node (Fig. 1c and Extended Data Fig. 1a,b). We also checked the expression of Tox, a critical transcription factor that regulates T exhaustion22, 23. Adpgk-specific T cells in the tumor tissue also expressed higher levels of Tox than those in the dLN (Fig. 1d). Based on these results, we hypothesized that the immunosuppressive TME limits the efficacy of neoantigen vaccination by manipulating T cell functional states, likely through the PD-L1/PD-1 immune checkpoint.
Fig. 1 |. Neoantigen vaccine combined with ICB remodeled TILs to induce a durable immune response.

a, MC38 bearing female C57BL/6J mice were subcutaneously injected with two doses of neoantigen vaccine (10 mcg 9-mer peptide formulated with 50 μg Poly I:C and 50 μg CpG1826) on day 10 and 17 post tumor inoculation. Tumor volume was measured twice per week (n = 4 mice; P < 0.0001). b, WT or MC38 bearing female C57BL/6J mice were treated with neoantigen vaccine subcutaneously (n = 8 mice). The percentage of tetramer+ cells in the draining lymph node and non-draining lymph node was detected by flow cytometry. c,d, MC38 bearing female C57BL/6J mice were treated with neoantigen vaccine subcutaneously. The percentage of tetramer+ cell in the tumor (c) and Tox+ cells in tetramer+ cells (d) were detected by flow cytometry (n = 8 mice). e, Boxplot showing the PD-L1 expression on different myeloid cell populations in lymph node and tumor tissue of MC38 bearing mice. Center line indicates the median value, lower and upper hinges represent the 25th and 75th percentiles, respectively, and whiskers denote 1.5× interquartile range. Each dot corresponds to one cell. f-h, female C57BL/6J mice were subcutaneously inoculated with 1×106 MC38 tumor cells and treated with either neoantigen vaccine, anti-PD-L1 or the combination (n = 5 mice). Tumor volume was measured every three days. Experimental design (f), Tumor growth curve (g), percentage of survival (h) were shown. (i) The experimental flowchart of single-cell RNA sequencing data generation. Data were shown as mean ± s.e.m. from two independent experiments. Statistical analysis was performed by two-way ANOVA with Šídák’s multiple comparisons test (a,b) or Tukey’s multiple comparisons test (g), two-tailed unpaired Student’s t-test (c-d) and Log-rank test (h), ****P ≤ 0.0001.
To test this hypothesis, we first analyzed the scRNA-seq data of TILs from the MC38 model for PD-L1 expression 24. Myeloid cell populations, especially neutrophils and conventional DCs in the tumor showed higher levels of PD-L1 expression than their counterparts in the lymph node (Fig. 1e). In our experiments, treatment of MC38 bearing mice with either anti-PD-L1 or neoantigen vaccine alone showed partial response, while the combination treatment resulted in complete tumor regression and significantly improved survival (Fig. 1f–h), which confirmed the inhibitory role of PD-1/PD-L1 signaling on neoantigen vaccination. In addition, neither adjuvant alone nor its combination with anti-PD-L1 could achieve the same antitumor effect as vaccine plus anti-PD-L1, indicating that the therapeutic effect we observed was due to the neoantigen peptide vaccine (Extended Data Fig. 1c). Recent studies showed that the sequencing of ICB may play a critical role in the phenotype of vaccine-induced T cells25. Hence, we compared and confirmed that anti-PD-L1 treatment given prior to and after vaccination leads to the best antitumor efficacy (Extended Data Fig. 1d), which is consistent with the result of a phase Ib clinical trial11. Therefore, this murine model and our therapeutic regimen is able to reproduce the observations in the human clinical trials and is suitable to investigate the mechanism of the combination therapy. We performed scRNA-seq with targeted TCR capture (scTCR-seq) on T cells collected from tumor, dLN and spleen tissues of untreated mice (at day 10 and day 20) as control, and from mice (day 20) under different therapies to investigate the dynamic changes of T cell populations. In parallel, neoantigen-Adpgk-specific T cells were also enriched by tetramer sorting to perform scTCR-seq, which were used in our downstream analysis to identify the neoantigen reactive T cell clusters (Fig. 1i and Extended Data Fig. 2a ).
High-resolution characterization of T cell populations
In total, we obtained scRNA-seq profiles of 100,899 T cells with 95,138 paired TCR sequences. After quality control, 93,399 T cells were kept for downstream analysis (Supplementary Table 1), of which 70% had at least one productive CDR3 for both TCR α and β chains. We performed unsupervised graph-based clustering and identified 23 clusters (Fig. 2a and Extended Data Fig. 2b). According to the differentially expressed genes (DEG) and tissue distribution pattern, we distinguished these clusters via their representative genes to infer their potential functions (Fig. 2a and Supplementary Table 2). The stability of clusters was also supported by distinct signature genes (Extended Data Fig. 3a–c). The distributions of these clusters were highly heterogeneous between the lymphoid tissues (spleen or lymph node) and tumor tissues in different treatment groups (Extended Data Fig. 3e). In particular, naïve cells (CD8–01-Sell-P.na and CD4–01-Tcf7- P.na), memory cells (CD8–02-Gzmm-P.em and CD8–03-Gpr183-P.cm), Treg cells (CD4–02-Foxp3-P.Treg) and follicular helper cells (CD4–03-Cxcr5-P.Tfh) were enriched in lymphoid tissues. Whereas other CD4 and CD8 clusters were preferentially enriched for tumor infiltrated T cells (Fig. 2b–d). In addition to CD4 and CD8 clusters, there were 3 minor clusters consisting of both CD8+ T cells and CD4+ T cells. Among these clusters, T.Act-Cd5 represents a population of T cells that is being primed due to expressing genes of costimulation like Cd5, Cd6 and Cd82. T.IFNs-Ifit3 represents a population of T cells that is responding to IFN stimulation due to expressing IGS genes like Ifit3. T.Pro-Mki67 is a specific population expressing genes related to cell proliferation like Mki67. In addition, the expression of representative hallmark genes (e.g., Foxp3 for Tregs) has been detected in some unique clusters (Extended Data Fig. 3d). The composition of T cells in the dLN is relatively stable. However, intra-tumoral T clusters displayed much higher heterogeneity across different therapies.
Fig. 2 |. The dynamics of TILs in response to distinct immunotherapies.

a, t-distributed stochastic neighbor embedding (t-SNE) plot showing Seurat-guided unsupervised clustering and distribution of 93,399 T cells from the spleen, lymph node or tumor tissue of MC38 bearing mice (n = 40 mice). Each dot denotes an individual T cell; color denotes cluster origin. There are 23 main clusters, including 9 CD8+ clusters, 7 CD4+ clusters, 4 CD8−CD4− TCRαβ+ double negative (DN) clusters and 3 mixed clusters. We selected one representative signature gene to name each cluster and indicate the potential function (bottom). b, Heat map displaying normalized expression values of discriminative gene sets for CD4 conventional T cells, Tregs and CD8 T cells in the lymphoid (spleen and lymph node) and tumor tissue. c, Heatmap showing the mean expression of representative T cell function-associated genes in each CD4+ and CD8+ cluster. d, Sample preference of each cluster estimated by Ro/e index (Methods). +++ (Ro/e ≥3, P<0.05) represents highly enriched; ++ (2≤ Ro/e <3, P<0.05) represents enriched; + (1.2≤ Ro/e <2, P<0.05) represents slightly enriched; +/− (0.8≤ Ro/e <1.2 or P>0.05) represents non-significant; - (0<Ro/e<0.8, P<0.05) represents deletion. (e-j) MC38 bearing female C57BL/6J mice were treated with either neoantigen vaccine, anti-PD-L1 or the combination. The percentage of TIL subsets were determined by flow cytometry as indicated time points. The percentage of CD62LhiCD44lo CD8 T cells in different treatment groups (e). The percentage of TIM-3+ PD-1+ CD8 T cells in different treatment groups (f). The percentage of TOX+ CD8 T cells in different treatment groups (g). The percentage of CD62LhiCD44lo CD4 T cells in different treatment groups (h). The percentage of Foxp3+ Tregs in different treatment groups (i). The percentage of RANKL+ Th1-like CD4 T cells in different treatment groups (j). Data were shown as mean ± s.e.m. (n=5 (e-f, h-j), n=6 (g) mice) from two independent experiments. Statistical analysis was performed by one-way ANOVA with Tukey’s multiple comparisons test (e-j), ****P ≤ 0.0001.
We next defined the dynamic state changes of CD4 and CD8 T cells in the tumor. CD4 T cells expressed costimulatory factors including Tnfsf11, Tnfsf4, Tnfsf8 and transcription factors like Hopx, Tbx21, Zeb2, Hif1a and Bhlhe40 (Fig. 2b–c and Extended Data Fig. 3a). Cluster CD4–06 was defined as Th-1 like cells according to DEGs like Ifng, Icos and Bhlhe40. Transcription factor Bhlhe40 has been reported to be associated with IFNγ production and favorable responses to immune-checkpoint blockade in patients 15, 26. We have also identified two clusters of Treg cells in the tumor expressing Il2ra, Foxp3 and Ikzf2. Compared to Tregs in the lymphoid tissue (CD4–02), the tumor-infiltrating Tregs (CD4–05, CD4–07) showed higher expression of inhibitory receptors (Ctla4, Havcr2 and Tigit) and transcription factors Hopx and Hif1α (Fig. 2c and Extended Data Fig. 3c). Surprisingly, we found that the expression of Gzmb is much higher in tumor Treg clusters than in any other clusters including peripheral Treg. Studies have shown that Treg derived granzyme B could induce NK and CD8+ T cell death in the TME 27. CD8 T cells in the TME expressed high levels of activation and exhaustion markers, including Ifng, Tnfrsf9, Fasl, Tnf, Pdcd1, Lag3 and Havcr2, while showing low expression of naïve and stem cell markers such as Ccr7, Tcf7 and Sell (Fig. 2c and Extended Data Fig. 3b). T cells in cluster CD8–08 displayed an exhaustion phenotype compared with other TILs, due to elevated expression of multiple inhibitory receptors (Pdcd1, Lag3, Havcr2, Tigit, Ctla4). In addition to typical CD8+ T cell clusters including exhausted T cells (CD8–08), resident memory T cells (CD8–06) and effector T cells (CD8–05), we also identified two unique CD8 clusters that do not share the same gene signature with other well-defined CD8 T cells in the TME. One was cluster CD8–09, which was similar to intraepithelial lymphocyte (IEL). Another one (CD8–07) was only seen in the ICB treated group (Fig. 2d) with a similar exhaustion phenotype to CD8–08 but expressing higher levels of granzyme family genes like Gzmf, Gzmb, Gzmc and lower level of Tigit and Ctla4. According to these gene expression signatures, we defined this cluster as ICB treatment responders (Supplementary Table 2–3). Unexpectedly, we also identified two “naïve-like” clusters in the tumor (CD4–04 and CD8–04). These cells highly express naïve T markers including Ccr7, S1pr1 and Sell and are negative for Cd44. Thus, the combination of ICB and neoantigen vaccine reshaped the tumor microenvironment by inducing distinct T cell clusters.
Combination treatment rejuvenated the TME to favor effector T cell functions
To investigate the dynamics of T cell status during tumor progression, we first compared T cell compartments at early (day 10) and late (day 20) stages. The early-stage TME primarily consisted of T cells with a naïve phenotype, including the CD8–04 and CD4–04 clusters (Fig 2d and Extended Data Fig. 4), which became a minor population in day 20 tumors. In contrast, only a few (4.3%) CD8 TILs at early stage have an exhaustion phenotype (CD8–08), which became the dominant subset in late-stage tumors (48%). Early tumors also contained fewer Tregs (CD4–05) compared to late-stage tumors. Thus, the TME consists of newly infiltrated T cells in the early stage and becomes more immunosuppressive in the late stage.
When looking into the phenotype of TILs in response to different immunotherapies, we first observed that neoantigen vaccine monotherapy reduced the percentage of dysfunctional CD8 T cells while increasing effector CD8 T cells (CD8–05). However, the ratio of Tregs was also increased during treatment, which may explain why neoantigen vaccine treatment only induces limited anti-tumor effect. In contrast to vaccine alone treatment, the combination with ICB dramatically increased effector T cells (70%) and reduced both dysfunctional CD8 T cells and Tregs. At the same time, Th1-like cells (CD4–06) became the dominant CD4 population in tumor tissue (Fig 2d and Extended Data Fig. 4).
To confirm these observations defined by scRNA-seq, we applied flow cytometry to stain the representative markers at the protein level. We first checked the expression of CCR7 on T cells as it is a representative gene of the clusters with a naïve T cell signature. We observed significantly reduced CCR7+ T cells in all late-stage tumor groups (Extended Data Fig. 5). Since CCR7 can also be expressed by central memory T cells, we further checked the expression of CD44 and CD62L to distinguish naïve and memory T cells. Consistently, early-stage tumors contain many T cells with a naïve phenotype (Fig. 2e,h). We also confirmed that tumors treated with combined therapy harbored significantly lower levels of Tregs (Fig. 2i) and CD8 T cells with an exhaustion phenotype as indicated by coinhibitory receptors (PD-1 and TIM-3) (Fig. 2f). Compared to the persistently high expression of multiple inhibitory receptors, the expression of critical transcription factors (e.g., NR4A, TOX, BATF) could better distinguish exhausted T cell from effector T cells. Indeed, the combination treatment also dramatically decreased the percentage of TOX+ CD8 T cells (Fig. 2g). At the same time, CD4-Th1 cells were increased in combination treatment group as indicated by RANKL (a representative marker of CD4–06) expression (Fig. 2j). Based on these results, we concluded that combination treatment generated a TME that is potentially favorable to effector T cell functions.
Lineage tracking of clonal T cell state transitions
Of the 65,058 T cells with full length TCRs, we identified 12,053 clonal T cells (Fig. 3a), which allowed us to track T cell clone expansion, state transition and migration. “Clonal” was defined as more than one T cells carrying identical pairs of TCRs. Most clonal T cells were distributed in the tumor-enriched clusters with dramatic changes in response to different treatments (Fig. 3b). We then used ‘clonal score’ to quantify the degree of clonal expansion (Methods). Of all the CD8+ T cell clusters, CD8–08 is the dominant clonally expanded cluster in all day 20 groups (Fig. 3d). Clonal expansion of CD8–08 was highest in the untreated group, but decreased after immunotherapies, especially combination treatment (Extended Data Fig. 6a). In contrast, combination therapy significantly expanded the effector T cells (CD8–05). Among CD4+ T cells, Tregs (CD4–05) and Th1-like T cells (CD4–06) showed the highest clonal expansion in most tumor samples (Fig. 3e). However, the clonal expansion ability of Tregs was significantly inhibited after combination treatment (Extended Data Fig. 6a). In addition, combination treatment also promoted the clonal expansion of CD4–06. These results suggest that combination treatment can reshape the TME by regulating the expansion ability of T cells.
Fig. 3 |. Lineage tracking of clonal T cell subsets associated with immunotherapies.
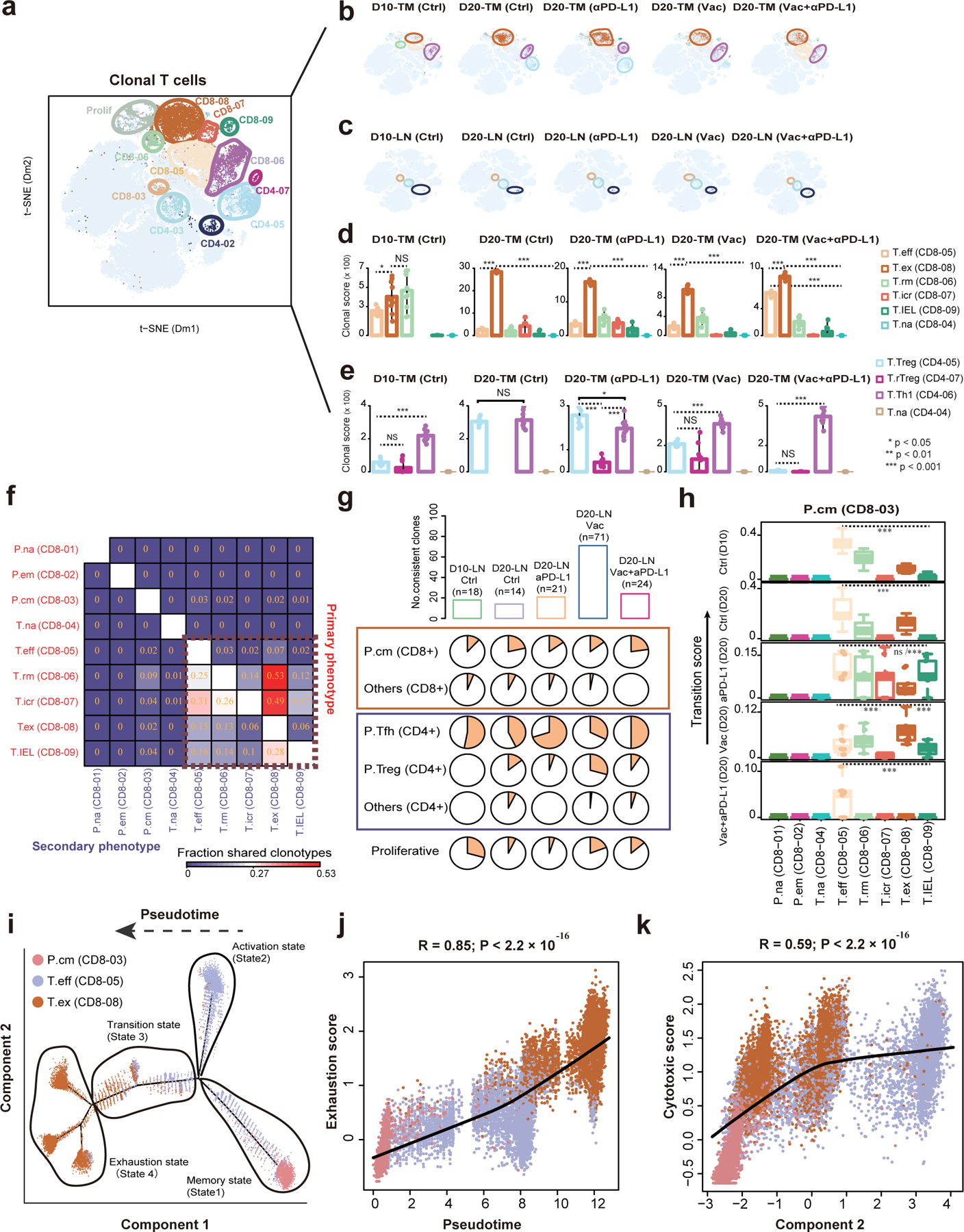
a, The clonal T cells (n=12,053 cells from n=40 mice) were highlighted in the t-SNE plot. Color represents cluster origin. b, Distribution of clonal T cell in the tumor sample of each treatment. The enriched clusters are indicated by colored oval. c, The distribution of clonal T cells in matched lymph nodes of samples in panel (b). d, Clonal expansion levels of 6 tumoral CD8+ T cell clusters quantified by clonal score for each tumor sample. e, Clonal expansion levels of 4 tumoral CD4+ T cell clusters quantified by clonal score for each tumor sample. f, Heat map showing the fraction of T cells with clonotypes belonging to a primary phenotype cluster (rows) that are shared with other secondary phenotype clusters (columns). g, The upper bar plots showing the number of consistent clones in lymph node of each group. The lower pie charts displaying the distribution of these consistent clones in different T cell clusters. h, Boxplots showing the migration potentials of CD8–03-Gpr183-Tcm to other CD8+ T cell clusters. Migration score between two specific clusters is calculated based on the migration level of each member. Center line indicates the median value, lower and upper hinges represent the 25th and 75th percentiles, respectively, and whiskers denote 1.5× interquartile range. Different T cell clusters were randomly downsampled (50%) 10 times for statistical test. Two-sided Wilcoxon rank-sum test were used for multiple groups comparisons (n=10 permutations, ***P ≤ 0.001, exact p values are provided in Source Data Fig. 3 (d-e, h)). i, Monocle-guided cell trajectory of three major CD8+ T cell clusters with high migration relationship. The direction of the inferred pseudotime is indicated by the arrow. The different functional states identified by monocle were ordered along the artificial pseudotime. j, The inferred pseudotime is correlated with the exhaustion feature of CD8+ T cells. The solid line represents the LOESS fitting of the relationship between the pseudotime and exhaustion scores. Dots were colored by their cluster origin. k, The monocle component 2 is correlated with the cytotoxic score of CD8+ T cells. The cytotoxic score was calculated similar to exhaustion score (Methods). P values were calculated by using the two-sided Pearson’s correlation coefficient test (j-k).
We next investigated the clonal sharing of T cell clusters to track their potential migration and state transition. We aggregated all clonotypes in a given cluster (the primary phenotype) and measured the fraction shared with another cluster (the secondary phenotype) 13. In general, clone sharing occurred between different intra-tumoral CD8+ clusters (Fig. 3f), suggesting that CD8+ T cells in the TME usually undergo extensive state transitions. In contrast, the effector T cells had very few clone sharing (2–3%) with other CD8 clusters. A previous study reported clonal replacement of CD8 T cells after ICB treatment 13, and we hypothesized that CD8–05 might primarily come from the lymphoid tissues after combination treatment.
To test this hypothesis, we identified clonal T cells in the lymphoid clusters, including CD8–03, CD4–03 and CD4–02 (Fig. 3c). Substantially, the amount of clonal T cells in the lymphoid tissues is lower than that of TILs. Despite this, we still observed that most CD8 T cell clusters in the tumor share clones with CD8–03 in the lymphoid organs, indicating that cross-tissue migration occurred. We further defined ‘consistent clones’, which were T cells with identical TCRs that can be detected in both tumor and matched lymph node under the same treatment. This definition can rule out cross-animal clonal sharing due to “public” T cells 28. We observed the highest consistent clones in the neoantigen vaccine group (Fig. 3g), supporting our observation that vaccine therapy may promote T cell recruitment from the lymphoid tissues. We also detected a significant enrichment of consistent clones in CD4–03 and CD4–02.
We next defined ‘migration score’, which is weighted entropy of shared clones between two clusters, to characterize the migration between lymphoid and intra-tumoral clusters (Methods). Of all intra-tumoral CD8+ clusters, CD8–05 have the highest migration score to CD8–03 in most treatment groups (Fig. 3h), thus confirming our hypothesis that CD8–05 may come from the dLN. Notably, migration score is also called transition score, as it can also be used to track state transitions within the tumor. We observed the highest transition score between CD8–05 and CD8–08 (Extended Data Fig. 6b), suggesting subsequent phenotypic transition of effector CD8 T cells in the TME.
Following these results, we studied the potential evolutionary trajectory of effector CD8 T cells with unsupervised pseudotime analysis 29 using three CD8+ clusters (CD8–03, CD8–05 and CD8–08). The trajectory originated from state 1, with a peripheral central memory phenotype, then split into two branches (Fig. 3i and Extended Data Fig. 6c–d). The upper branch consisted of cells mainly from the effector T cell population (state 2). The lower one harbored a transitional phase (state 3) with a mixture of effector and dysfunctional T cells, before ending up in a completely exhausted phenotype (state 4). Consistently, T cells along the trajectory demonstrated increased exhaustion and cytotoxicity scores, with decreased level of stemness score (Methods, Fig. 3j,k and Extended Data Fig. 6e–f). These results suggest that combination treatment could prevent the transition of effector T cells to terminal exhausted T cells.
Similarly, for conventional CD4+ T cells, we found significant clonal sharing between two clusters, CD4–03 and CD4–06 (Extended Data Fig. 7g). Pseudotime trajectories revealed differential distribution of functional states for the CD4 T cells. The initial state of the trajectories was peripheral Tfh cells, which transited into Th1-like cells in the tumor. The Th1-like cells were predominantly distributed in the middle state (state 2) in combination treatment but enriched in the terminal state (state 3) in all other treatment groups. The terminal fate (state 3) also showed an exhaustion phenotype, with higher level of inhibitory markers like Lag3, Havcr2, Ctla4 and inhibitory factor Tgfb1 (Extended Data Fig. 7a–c).
Similar to CD8 T cells, the trajectory of Tregs also split into two different branches. The tumor-infiltrating Tregs in the lower branch contained fewer clonal cells and expressed newly-infiltrated markers, such as S1pr1 and Klf2 (Extended Data Fig. 7d,e). Tregs in the combination treatment group were enriched in the lower branch (State 2). In contrast, Tregs in other treatment groups were enriched in the upper branch (State 3–4), which displayed a terminal differentiated phenotype with higher expression of Il10 and Glrx (Extended Data Fig. 7e,f) and higher clonal expansion (Extended Data Fig. 7h). Together, analysis of clonal T cells revealed that combination treatment reshaped the TME by not only promoting the infiltration but also preventing the terminal differentiation of effector T cells.
Neoantigen-specific T cells were enriched in the effector CD8 T cell cluster
The characteristics of tumor-specific T cells are not well defined due to limited identification of immunogenic epitopes. To overcome this hurdle, we first vaccinated naïve mice with neoantigen Adpgk and obtained the TCR sequences of flow-sorted neoantigen-MHC-I tetramer+ T cells (Fig. 1i and Extended Data Fig. 8). Since the process of tetramer staining and flow-sorting can capture a percentage of non-antigen specific T cells 30, we further applied an algorithm iSMART which clusters TCRs according to their similarity to improve the accuracy of neoantigen-specific TCR identification 31. A total of 1,415 TCR groups were identified in the tetramer library after clustering (Supplementary Table 4). We then compared the amount of T cells in each groups to its amount in the lymphoid tissue of untreated mice, and found 20 TCR groups were significantly enriched. After these two steps of quality assurance, we defined these TCRs as high-confidence neoantigen-Adpgk-specific TCRs (Fig. 4a and Supplementary Table 5). We used these TCRs to track neoantigen-specific T cells in the scRNA-seq data and found them enriched in the combination treatment group (Fig. 4b). Specifically, CD8–05 and CD8–04 contained significantly higher percentage of neoantigen-specific T cells than any other intra-tumoral CD8 clusters (Fig. 4c), which was driven by multiple TCR groups (Fig. 4d). Studies have reported that lymph node-like vasculature can mediate the infiltration of naïve T cells in the tumor tissue, which contribute to anti-tumor immunity 32. In contrast, the CD8–08 contained fewer neoantigen-Adpgk-specific T cells. These results indicate that Adpgk-specific T cells infiltrate into the TME at early-stages but are rarely detected in exhausted clusters at later stages. It is also possible that T cells in this cluster mainly recognized other tumor-specific antigens (e.g., Resp1, Dpagt1, etc.) but not Adpgk, since chronic TCR stimulation is generally thought to be a major driver of T cell exhaustion 23, 33. After combination treatment, Adpgk-specific T cells were enriched and had an effector T cell phenotype. Thus, combination treatment may promote the recruitment of Adpgk-specific T cells but not state transition from pre-existing cells.
Fig. 4 |. Identification of neoantigen specific T cell landscape in response to combination treatment.

Tetramer+ T cells were sorted from the spleen and lymph node of neoantigen-Adpgk vaccinated mice, and their CDR3 sequences were clustered using iSMART, revealing 1415 unique iSMART clusters. a, TCR clusters were then compared to clusters from T cells in unvaccinated mouse lymphoid tissues, and the top 20 clusters enriched in neoantigen-Adpgk vaccinated mice are displayed in (a) (n=1802 CDR3 sequences). b, Bar graphics showing the enrichment analysis of tetramer+ neoantigen-specific T cells, which is defined as the T cells with TCRs enriched in any of the iSMART groups in (a). The x-axis represents the ratio of observed cell number over the expected cell number (Ro/e) obtained from two-sided chi-square test, exact p value=3.7×10−30 (b). c, t-SNE plot showing the projection of tetramer+ CD8+ T cells which express the TCRs in the twenty significant iSMART groups (n=461 cells). d, Odds ratio heatmap of twenty tetramer+ TCR groups in six TIL-enriched CD8+ T cell clusters. Statistical significance was evaluated with Chi-square test and FDR correction was performed with Benjamin-Hochberg method on multiple TCR groups. e, Volcano plot showing differentially expressed genes between T cells of CD8–05-T.eff and other CD8+ tumor-infiltrating T cells (n= 6,191 cells in cluster CD8–05). f, Violin plots showing the expression levels of Ccl5, Cxcr3, Fasl, Lag3 and Havcr2 across the tumoral CD8+ T cell clusters. g, Differential pathways enriched for the discriminative markers of CD8–05 T cell subset (n= 6,191 cells) by GSEA.
We further investigated the DEGs of CD8–05 T cells to identify the signature genes of this T cell subset. Cytotoxic effector molecules, such as Gzma, Fasl and Itgb1 were much higher in this cluster. Meanwhile, chemokine and chemokine receptors including Ccl5 and Cxcr3 were also up-regulated, indicating the migration ability of these cells (Fig. 4e). In addition, these cells also expressed the activation maker Cd69 and significantly lower level of co-inhibitory molecules like Lag3 and Havcr2 (Fig. 4f). Notably, two representative genes, Itgb1 (encoding CD29) and Zfp36l2 have been recently reported to participate in the induction of IFNγ 34, 35. These gene expression signatures indicated a strong effector-like phenotype, which was further confirmed by the enrichment of Ifng, Tnfα, chemokines and cytotoxicity (death receptor) signaling pathways (Fig. 4g and Supplementary Table 6).
Combination treatment induced neoantigen-specific effector T cells in vivo
We next conducted experiments to validate the phenotype of effector T cells in vivo. We first performed tetramer staining on TILs after different immunotherapies. As expected, combination treatment dramatically increased the frequency of tetramer+ cells compared to monotherapies (Fig. 5a,b and Extended Data Fig. 9b). Meanwhile, as a functional assessment, the Enzyme-Linked Immunosorbent Spot (Elispot) assay was also performed to confirm that combination treatment could increase the IFNγ-producing tumor-reactive T cells (Extended Data Fig. 9c). This was consistent with scRNA-seq results showing enrichment of the Ifng pathway in the CD8–05 cluster. Therefore, we concluded that ICB treatment coordinates with neoantigen vaccination to promote IFNγ-producing neoantigen-specific T cells in the tumor.
Fig. 5 |. Neoantigen vaccine and ICB coordinately mediated the anti-tumor immune response depending on T cells from draining lymph node.

a-d, female C57BL/6J mice were subcutaneously inoculated with 1×106 MC38 tumor cells and treated with either neoantigen vaccine, anti-PD-L1 or the combination. Experimental design (a), the percentage of tetramer+ CD8 T cells (b), LAG3+ CD8 T cells (c) and CXCR3+ CD8 T cells (d) in the spleen or tumor tissue of different treatments groups was detected by flow cytometry. e-h, female C57BL/6J mice were subcutaneously inoculated with 1×106 MC38 tumor cells and treated with the combination of neoantigen vaccine and anti-PD-L1. 20 μg FTY720 was administrated one day before treatment initiation and then 10 μg every other day for 2 weeks. Experimental design (e), tumor growth curve (f), the percentage of IFNγ producing cell in the tumor tissue was determined by Elispot (g), the percentage of neoantigen-specific CD8+ T cells in the tumor tissue was detected by tetramer staining (h). i-l, female C57BL/6J mice were subcutaneously injected with neoantigen vaccine. Lymphocytes from draining lymph node were harvested at day 6 post vaccination and adoptively transferred to MC38 bearing Rag1−/− mice. Two doses of anti-PD-L1 were given to the recipient mice on day 2 and 5 post adoptive transfer. Schematic of experiment design (i), tumor growth curve (j), the percentage of IFNγ producing cell in the spleen was determined by Elispot (k), the percentage of neoantigen-specific CD8+ T cells in the tumor tissue was detected by tetramer staining (l). m-o, female C57BL/6J mice were subcutaneously inoculated with 1×106 MC38 tumor cells and treated with the combination of neoantigen vaccine and anti-PD-L1. Experimental design (m), representative result of MuLV p15E specific tetramer staining (n), percentage of MuLV p15E specific tetramer+ CD8 T cells (n = 5 mice) (o). Data were shown as mean ± s.e.m. from two independent experiments (n=4 (b, f), n=5 (c-d, h, j, l), n=6 (k) and n=8 (g) mice). Statistical analysis was performed by two-way ANOVA (b, f, j), one-way ANOVA (c-d, g-h, k-l) with Tukey’s multiple comparisons test and two-tailed unpaired Student’s t-test (o), ****P ≤ 0.0001.
To further characterize the neoantigen-specific T cells in vivo, we stained surface markers identified by scRNA-seq analysis. Compared to single treatments, combination therapy dramatically reduced the percentage of LAG3+ CD8 T cell and increased the expression of CXCR3 on TILs (Fig. 5c,d and Extended Data Fig. 9a). This is consistent with our findings in scRNA-seq showing that most T cells within CD8–05 are Lag3 negative and part of them have an increased Cxcr3 expression (Fig. 4f).
Neoantigen specific T cells from the dLN elicit the anti-tumor effect
The lineage tracking of clonal T cells revealed that neoantigen-specific T cells have higher transition scores with central memory T cells (CD8–03) in the dLN and exhausted T cells (CD8–08) in the tumor. We next asked whether the effector T cells induced by combination treatment come from pre-existing T cells in the tumor or newly infiltrated T cells from the lymphoid tissue. To test these hypotheses, we used FTY720, a S1P receptor agonist to block T cell egress from lymph node prior to and during combination therapy (Fig. 5e). The anti-tumor effect was completely abolished by FTY720, indicating that newly infiltrated T cells from dLN are essential (Fig. 5f). Meanwhile, FTY720 treatment dramatically reduced the percentage of IFNγ producing CD8 T cells (Fig. 5g) and tetramer+ T cells (Fig. 5h), which further indicated that lymphoid tissues are the primary source of neoantigen-specific T cells in the tumor. Interestingly, since a higher level of CXCR3 expression on neoantigen-specific T cell was detected by both scRNA-seq and flow cytometry, the CXCL10-CXCR3 pathway may mediate the migration of neoantigen-specific T cells.
To investigate whether T cells from neoantigen vaccine primed dLN were sufficient to mediate a durable anti-tumor immune response in the presence of anti-PD-L1, we vaccinated naïve mice with neoantigen vaccine, and harvested lymphocytes in the dLN after 6 days. The cells were then adoptively transferred to MC38 bearing Rag1−/− mice followed by anti-PD-L1 treatment (Fig. 5i and Extended Data Fig. 9d). Neoantigen vaccine primed lymphocytes but not naïve lymphocytes in the dLN could synergize with anti-PD-L1 to control tumors (Fig. 5j). Furthermore, anti-PD-L1 treatment dramatically increased the percentage of antigen-specific T cells in the spleen and tumor after adoptive transfer of vaccine-primed lymphocytes (Fig. 5k,l and Extended Data Fig. 9e). Finally, we observed that the percentage of T cells specific to another MC38 epitope (MuLV p15E) was also significantly increased after the combination treatment. This result is aligned with a recent work showing that T cell response may undergo epitope-spreading after initial priming by the original tumor antigens 36 (Fig. 5m–o). Based on these results, we concluded that anti-PD-L1 expanded the therapeutic effect of neoantigen vaccine by rejuvenation of neoantigen-specific T cells derived from the dLN.
Neoantigen-specific T cell gene signatures are associated with better clinical outcomes in human cancers
To explore if the neoantigen-specific T cell gene signature of MC38 is reproducible in human cancers, we first defined a gene signature for the neoantigen-specific T cells. Specifically, we selected DEGs for CD8–05 that were also found in the human genome and performed supervised feature selection by using each DEG as a predictor of CD8–05 against all the other CD8 T cells. The average expression of the top 5 most discriminative makers reached a high AUC of 0.92. Therefore, we defined the mean expression of these five markers as Cancer Antigen Specific T cell (CAST) score, which could be a surrogate for the neoantigen-specific T cell fraction (Fig. 6a). We estimated the CAST score for human cancers in the TCGA database and selected 23 cancer types with over 100 patients to investigate its role in prognosis. As CD8 T cell infiltration is a predictor for patient outcome in multiple TCGA cancers 37, we performed Cox proportional hazard regression stratified by CD8 T cell levels, and estimated hazard ratio of CAST score in the univariate Cox model (Fig. 6b). CAST score significantly predicted better overall survival in melanoma, head and neck, cervical and pancreatic cancer patients with high but not low CD8 T cell infiltration (Fig. 6c–j). Therefore, the discriminative markers of mouse neoantigen-specific T cells are predictive of patient survival with high infiltration of CD8 T cells.
Fig. 6 |. The discriminative marker of antigen specific T cells are associated with better survival in human tumors.

a, Receiver operating characteristic (ROC) plots showing the performance in prediction of CD8–05 effector T cells. b, Influence of the CAST score on the overall survival of major human cancers. HR below 1 (horizonal solid line) indicates that patients with higher CAST score exhibit improved survival. The error bar represents the calculated 95% confidence interval (95%CI). For each cancer type, the patients were divided into High CD8 TIL and Low CD8 TIL subgroups. Brain lower grade glioma (LGG, n = 457 patients), bladder urothelial carcinoma (BLCA, n=337 patients), breast invasive carcinoma (BRCA, n=1065 patients), colon adenocarcinoma (COAD, n=450 patients), esophageal carcinoma (ESCA, n=172 patients), glioblastoma multiforme (GBM, n=159 patients), kidney renal clear cell carcinoma (KIRC, n=525 patients), kidney renal papillary cell carcinoma (KIRP, n=254 patients), liver hepatocellular carcinoma (LIHC, n=354 patients), lung adenocarcinoma (LUAD, n=478 patients), lung squamous cell carcinoma (LUSC, n=478 patients), ovarian serous cystadenocarcinoma (OV, n=260 patients), pheochromocytoma and paraganglioma (PCPG, n=178 patients), prostate adenocarcinoma (PRAD, n=421 patients), sarcoma (SARC, n=244 patients), stomach adenocarcinoma (STAD, n=388 patients), thyroid carcinoma (THCA, n=499 patients), uterine corpus endometrial carcinoma (UCEC, n=369 patients), testicular germ cell tumors (TGCT, n=119 patients). c-f, Kaplan-Meier survival curves of patients with high CD8 TIL from skin cutaneous melanoma (SKCM, n = 428 patients) (c), head and neck squamous cell carcinoma (HNSC, n = 497 patients) (d), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC, n = 282 patients) (e) and pancreatic adenocarcinoma (PAAD, n = 163 patients) (f) with respect to high or low CAST score within tumor specimens. g-j, Kaplan-Meier survival curves of patients with low CD8 TIL from SKCM (g), HNSC (h), CESC (i) and PAAD (j). k, The violin plots showing the CAST scores across CD8+ T cell groups with different clone frequency in advanced basal cell carcinoma (BCC) samples. l, t-SNE plot showing CD8+ cells that are color-coded according to whether they belong to large or small clones. m, Three of four BCC patients showed higher CAST scores of CD8+ T cells in post-αPD-1 samples when comparing to pre-treatment counterparts (one-sided Wilcoxon rank-sum test, su005 (p=8.4×10−16), su008 (p=0.00019), su009 (p=5.4×10−11)). Survival distributions were compared using the log-rank test (c-j). Two-sided Spearman’s correlation coefficient test was used with the exact p value=5.0×10−231 (k).
As most TCGA samples were not treated with immunotherapies, we next investigated the performance of the CAST score using human samples under ICB treatment. Clonal expansion is an evidence of antigen-specific T cell response 38, 39. A recent study reported that more “large” CD8+ clones (frequency > 0.5%) was predictive of response to ICB treatment and correlated with effector memory T cell percentage in metastatic melanoma patients 40. Effector T cells (CD8–05) upregulated several cytotoxic markers (Fig. 4e), which were also signature genes of those large clones. To test whether CAST score is predictive of large CD8+ clones in human tumors, we analyzed a public dataset of patients with advanced basal cell carcinoma (BCC) before and after anti-PD-1 therapy 13. Indeed, CAST scores was significantly correlated with CD8+ T cell clonal frequency (Fig. 6k and Extended Data Fig. 10a). Furthermore, large clones with a high CAST score were phenotypically similar (Fig. 6l). Consistently, these human T cell clones upregulated several markers that were also highly expressed in the CD8–05 cluster (Extended Data Fig. 10b). Finally, anti-PD-1 treatment increased the CAST scores in 3 out of 4 patients (Fig. 6m), thus confirming the role of ICB in expanding antigen-specific T cells in the TME.
Discussion
We conducted a mechanistic investigation of ICB combined with neoantigen vaccine treatment. By scRNA-seq of approximately 100,000 T cells, we studied the landscape of TILs at different time points and under combined therapy. The efficacy of personalized vaccines combined with ICB has been demonstrated in patients with melanoma 10, 11. However, the underlying mechanism of how this combination treatment facilitates T cell reactivation has not been defined. Our work complemented the current human clinical studies of neoantigen vaccine and addressed some of the fundamental questions regarding its anti-tumor immune responses.
First, we demonstrated that the anti-tumor effect of neoantigen vaccine could be dramatically improved when combined with ICB. Mechanistically, newly infiltrated T cells from vaccine-primed dLN are essential to promote a durable immune response. Second, a CD8 T cell population emerged after the combination treatment. This T cell subset expressed high levels of cytotoxic genes Gzma and Fasl, and was negative for co-inhibitory receptors Havcr2 and Lag3. The function of this T cell population was further substantiated by elevated IFNγ secretion. This cytotoxic CD8 T cell phenotype is distinct from several reported phenotypes of CD8+ TILs associated with response to immunotherapies, including CD39/TCF1 in responder and non-responder of anti-PD-L1 treatment and CD103+ resident memory T cells 41–43. Third, using an MHC-I tetramer of a well-defined MC38 neoantigen, we were able to confirm that this T cell subset was neoantigen-specific and induced by combination therapy.
A previous study has described the phenomenon of clonal replacement after ICB treatment in patients with BCC 13, but the evidence was observational without functional validations. In this study, we found chemokine receptor Cxcr3 is upregulated in neoantigen-specific T cells, and blocking T cell egress from lymph node by FTY720 dramatically restrained the therapeutic effect of combination treatment. Thus, combination treatment may promote neoantigen-specific T cell infiltration through the CXCL9-CXCR3 pathway. Recent studies have shown that CXCR3 is expressed in memory T cells 44, and the CXCL9-CXCR3 axis in the tumor tissue is required for anti-PD-1 treatment 45. It is possible that CD103+ dendritic cell expressed CXCL9 promoted the interaction with CXCR3+ neoantigen-specific T cells upon anti-PD-L1 blockade to maintain their cytotoxic function 46. Even though ICB therapies including anti-PD-L1 and anti-CTLA4 have received promising anti-tumor effect, the overall response rate is still below 25% for most types of cancers and many responding-patients relapse in a few years. Here, we have demonstrated that increased antigen-specific T cell infiltration by neoantigen vaccination overcomes ICB resistance, which is consistent with our previous study 47.
Although a subset of melanoma patients underwent complete remission after neoantigen vaccine single treatment in clinical trials, some patients were resistant to this therapy. In such scenarios, ICB may be required to overcome the resistance. Indeed, we observed that most antigen-specific T cells expressed PD-1 after neoantigen vaccine treatment, and it is reported that PD-L1 is highly expressed on myeloid cells in the lymphoid and tumor tissues 48, 49. Adoptive transfer of lymphocytes from vaccine-primed dLN still requires anti-PD-L1 to achieve anti-tumor effect, which further demonstrates that ICB preserved the effector function of the neoantigen-specific T cells in the tumor. In addition, combined therapy was able to extensively remodel the TME to promote the survival of effector T cells 50. Hence, we speculate that both rejuvenation of the TME and amelioration of the lymphoid tissues are required to break immune tolerance in advanced tumors.
The sequencing of anti-PD-(L)1 given has been suggested to play a critical role in the phenotype of vaccine-induced T cells. A recent study using murine tumor models has demonstrated that administration of anti-PD-1 prior to vaccination abolished beneficial therapeutic outcomes through the introduction of PD-1+CD38hiCD8+ T cells 25. This observation indicates that T cell priming by vaccine is important for subsequent anti-PD-1 treatment in “cold” tumors which resistant to anti-PD-1 monotherapy. In our study, MC38 is considered a “hot” tumor and responds to anti-PD-(L)1 treatment well. Thus, anti-PD-L1 may improve T cell function in both dLN and tumor tissue 12, 51. In a phase Ib clinical trial, anti-PD-1 given pre and post neoantigen vaccination induced effective immune responses in patients with advanced solid tumors 11, which is consistent with our observations.
One of the biggest challenges of clinical neoantigen vaccine is to acquire personalized neoantigens, which can be expensive and logistically laborious. Our single-cell analysis revealed a discriminative gene signature of neoantigen-specific T cells, including three genes in the IFNγ pathway: Ifngr1, Zfp36l2 and Gimap4 34, 35, 52, and two chemokine related genes: Ccl5 and Cxcr3. This signature can identify clonally expanded T cells in human samples and is predictive of patient outcome. A recent study observed an effector-like CD8 T cell cluster enriched after anti-CD40 treatment on MC38 model. The DEGs and CDR3 sequences of those T cells are very similar to our CD8–05 24, confirming that these signature genes are associated with antigen-specific T cells.
Despite these conclusions, we acknowledge the limitations of our current study. First, additional scRNA-seq data from other qualifying murine models might be required to confirm and improve the definition of signature genes for antigen-specific T cells. Second, even though the use of neoantigen Adpgk was sufficient to map tumor-specific CD8 T cell clusters, T cells against other tumor-specific epitopes remain undefined. Third, our study focused on profiling of the T cell populations but excluded other immune cell types. Nonetheless, our findings suggest certain myeloid cell populations might also play important roles in combination treatment. Finally, additional scRNA-seq data using human cancer biopsies will be needed to verify the conclusions.
We anticipate the cell subsets and biomarkers identified in this work to inform the future development of effective neoantigen vaccine therapies and provide clinical benefits in prognosis of cancer immunotherapies.
Methods
Mice.
6–8 weeks old female C57BL/6J and Rag-1−/− mice were purchased from The Jackson Laboratory and maintained under specific pathogen-free conditions. Animals were housed at 21 ± 2 °C, and humidity was 50–60%. The light cycle consisted of light for 12 h and dark for 12 h. Animal care and experiments were carried out under institutional and National Institutes of Health protocol and guidelines. This study complies with all relevant ethical regulations and has been approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center. The maximal tumor size permitted by the ethics committee was 2 cm. The maximal tumor size in this study was not exceeded.
Cell lines and cell culture.
MC38 cell lines were purchased from Kerafast, Inc (ENH204). All cell lines were routinely tested using mycoplasma contamination kit (R&D) and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, and 100 U/ml streptomycin under 5% CO2 at 37 °C.
Flow Cytometry Analysis.
Single cell suspensions from either spleen or tumor were incubated with anti-FcγIII/II receptor (clone 2.4G2) for 15 minutes to block non-specific binding before staining with the conjugated antibodies or tetramer. 7-AAD Viability Staining Solution or Fixable Viability Dye eFluorTM 506 was used to exclude dead cells. Foxp3 and Tox were stained intracellularly by using True-Nuclear transcription factor buffer set (BioLegend) following the manufacturer’s instructions. All staining steps were conducted at 4 °C in the dark. Data were collected on CytoFLEX flow cytometer (Beckman Coulter, Inc) and analyzed by using CytExpert (Beckman Coulter, Inc) or FlowJo (Tree Star Inc., Ashland, OR) software. Antibodies used were listed in Supplementary Table 7.
IFN-γ Enzyme-Linked Immunosorbent Spot Assay (ELISPOT).
MC38 tumor bearing mice were treated with different immunotherapies on day 10 post tumor inoculation. 10 days after treatment, tumor samples were collected for single-cell suspension preparation. CD45+ TILs were enriched by EasySep™ Biotin Positive Selection Kit II and seeded in each well with either irradiated MC38 tumor cells (3×104) or 5 μg/mL neoantigen peptide (Adpgk) to stimulate the tumor-specific T cells. After 48 hrs culture, the ELISPOT assay was performed using the IFN-γ ELISPOT kit (BD Bioscience) according to the manufacturer’s instructions. IFN-γ spots were enumerated with the CTL-ImmunoSpot® S6 Analyzer (Cellular Technology Limited).
Tumor growth and treatment.
A total of 1×106 MC38 were inoculated subcutaneously into right dorsal flanks of the mice in 100 μl phosphate buffered saline (PBS). Tumor-bearing mice were randomly grouped into treatment groups when tumors grew to around 80–100mm3. For neoantigen vaccine treatment, 10 μg of 9-mer mutated Adpgk peptide (ASMTNMELM) and 50 μg of TLR antagonist (CpG-1826 and Poly I:C) was mixed in 50 μl of PBS, subcutaneously injected on either day 12 (one dose, in combination with anti-PD-L1) or day 10 and 17 (two doses, vaccine alone) post tumor inoculation. For anti-PD-L1 treatment, the antibody was diluted in 200 μl of PBS and intraperitoneally injected on day 10 and 15 post tumor inoculation. For FTY720 treatment, 20 μg FTY720 was intraperitoneally administrated one day before treatment initiation or tumor inoculation and then 10 μg every other day for 2 weeks. Tumor volumes were measured by the length (a), width (b) and height (h) and calculated as tumor volume = abh/2.
Tumor digestion.
Tumor tissues were excised and digested with 2.5 mg/ml Collagenase I (Sigma) and 0.5 mg/ml DNase I (Roche) in the 37°C for 30mins, tumor was then passed through a 70 μm cell strainer to remove large pieces of undigested tumor. Tumor infiltrating cells were washed twice with PBS containing 2 mM EDTA and 2% FBS.
Adoptive immune cell transfer.
Draining lymph node from vaccinated or naïve mice were gently mashed and washed through a 70 μm cell strainer with culture medium. After counting and washing once with PBS, lymphocytes were re-suspended in 100 μl PBS and transferred through tail vein.
Statistics & Reproducibility.
All the experimental data analyses were performed with GraphPad Prism statistical software (v9.0) and shown as mean ± s.e.m.. P value was determined by two-way ANOVA for tumor growth, Log-rank test for survival, t-test and one-way ANOVA for other analysis. Normality and equal variances were tested for t-test and data distribution was found to be normal. A value of p < 0.05 was considered statistically significant. No statistical methods were used to predetermine sample size, but our sample sizes are similar to those reported in previous publications24, 53. No data were excluded from the analyses. For the animal study, mice were randomly assigned to the groups before treatments. Data collection and analysis were not performed blind to the conditions of the experiments.
Single cell suspension preparation and sorting.
MC38 tumors, spleen and tumor draining lymph nodes were harvested at day 10 or day 20 post inoculation. Tumor tissues were cut into approximately 1 mm3 pieces in the RPMI-1640 medium (Invitrogen) with 2% fetal bovine serum (FBS), and enzymatically digested with MACS tumor Dissociation Kit (Miltenyi Biotec) for 40 min on gentleMACS™ Dissociator according to manufacturer’s instruction. The dissociated cells were subsequently passed through a 70 μm cell strainer and centrifuged at 500 g for 10 min. After the supernatant was removed, cells were re-suspended in MACS buffer (1x PBS with 2mM EDTA and 0.5% BSA) and counted. CD45+ cells were isolated with CD45 (TIL) MicroBeads (Miltenyi Biotec). Enriched CD45+ cells were washed and re-suspended in FACS buffer (1x PBS with 2% FBS) and stained for CD3e. CD3+ live cells were sorted by BD FACSMelody™. Spleen and draining lymph node were gently mashed and washed through a 70 μm cell strainer with culture medium. After centrifugation at 500g for 5 minutes, red blood cells in the spleen were removed by ACK lysing buffer. Cell pellets were finally re-suspended in FACS buffer and stained for CD3e. CD3+ live cells were sorted by BD FACSMelody™
Preparation of scRNA-seq libraries and sequencing.
Single-cell RNA-seq were performed by the 10x Genomic single cell 5’ VDJ library platform. Based on FACS analysis, single cells were sorted into 1.5 ml tubes (Eppendorf) and counted manually under the microscope. The concentration of single cell suspensions was adjusted to 900–1100 cells/μl. Cells were loaded between 10,000 and 17,000 cells/chip position using the Chromium Single cell 5’ VDJ Library, Gel Bead & Multiplex Kit and Chip Kit (10x Genomics, V1 barcoding chemistry). Single-cell gene expression and TCR libraries were generated according to the manufacturer’s instructions. All the subsequent steps were performed following the standard manufacturer’s protocols. Purified libraries were analyzed by Nextseq sequencer with 150-bp paired-end reads at a targeted median read depth of 50,000 reads per cell from total gene expression libraries and 5,000 reads per cell for TCR libraries (cycle specifications150:8:0:150 [R1:i7:i5:R2]).
Data processing of single cell RNA-seq and scTCR-seq libraries.
The Cell Ranger (v2.1.1) Software Suite was used to process the raw sequence data into gene expression profiles. The fastq files for each sample were processed with Cell Ranger count, which was used to align to mm10 reference genome, filter and quantify. The recovered-cells parameter was set as 10,000, which is the expected number of cells for each library. The output files of Cell Ranger for each sample included feature barcode and gene expression (in UMI count). Gene expression data were obtained using the cellranger aggr, which performed an inter-sample normalization and merged the results of all samples into one file, to remove the potential technique variability of samples.
For scTCR-seq data, TCR reads were aligned to the mm10 reference genome and TCR annotation was performed using the 10x cellranger vdj pipeline. Overall, 94% of T cells in scRNA-seq data were assigned a TCR and more than 70% had at least one full-length productive CDR3 for both TRA and TRB. The clonotype of each T cell was represented by the paired alpha and beta CDR3 sequences, and clone sizes ranged from 1 cell to 580 cells.
Pre-processing and quality control.
Low quality T cells were filtered if no matched TCR (either α or β chain) was detected or the number of expressed genes (UMI count >0) was smaller than a predefined cutoff value. The cutoff was defined as the median of all cells minus 2 × MAD (Median Absolute Deviation). The Cd3 expression was defined as the average UMI counts of Cd3d, Cd3e, Cd3g, and those cells with Cd3 expression > 0 were kept for subsequent analysis. We identified the CD4 and CD8 status for each cell based on the gene expression data. A small proportion of CD4+ CD8+ (double positive) T cells were filtered out because they are high likely to be doublets, a kind of technical artifact produced during library construction 54. We discarded those genes expressed less than 1% of total cells. The remaining count table of cells was log-normalized implemented in the R package Cell Ranger™ R Kit. A total of 9,881 protein-coding gens and 93,805 cells were retained in the final expression table.
Unsupervised clustering analysis of scRNA-seq dataset.
The Seurat pipeline 55 was applied to the combined expression data. Principle component analysis (PCA) was performed on the scaled data of all 9,881 genes. Then a t-SNE dimensional reduction was performed based on the first 50 PCA components to obtain a two-dimensional projection of the cell states. Clusters were identified using shared nearest neighbor (SNN)-based clustering based on the same principal components. We used the function FindClusters for clustering with resolution from 0.1 to 1.5, leading to 7~35 clusters. For each resolution, the silhouette values were calculated. Such value was used to determine the optional cluster number k. We selected the resolution of 1 by maximizing the average silhouette coefficient, which lead to a total of 25 clusters. Among these clusters, we first excluded one cluster which contains less than 1% of total cells. We also removed the cells in another cluster because they showed unexpected high UMI counts and macrophage-signature gene expression, indicating they may be a subset of macrophage-T doublet cells. After a series of filtering described above, 23 clusters corresponding to 93,399 T cells were retained for downstream analyses.
Identification of Cluster-Specific Genes and Cell cluster annotation.
We used the FindAllmakers and FindMarkers functions with Wilcox ran-sum test to identify the marker genes. For each cluster, only the genes that were expressed by more than 20% of total cells with adjusted p value < 0.05 (Benjamin–Hochberg method) were considered (Supplementary Table 3). The function of each cluster was annotated and manually confirmed based on the expression of well-known cell-type-specific marker genes.
The definition of clonal T cells and consistent clones.
If there are at least two cells sharing an identical productive α-β pair, such α-β pairs are defined as clonal TCRs. Clonal T cells are defined as T cells with clonal TCR. Consistent clones represent the T cells harboring identical TCRs detected in both lymph node and matched tumor sample.
Characterization of tissue or sample enrichment, clonal expansion and state transition of T cell clusters.
We utilized the ratio of observed to expected cell number for tissues/samples in each T cell cluster to measure the enrichment across different tissues/samples. Here, we only described the calculation process of sample preference as an example, which has also been applied for tissue preference. Chi-squared test was used to test whether the distribution of T cell clusters across different samples significantly deviates from random expectations. In addition, the extent of deviation for each combination of T cell cluster and sample is quantified by the Ro/e value:
Where No is the observed number cell for a given sample and cluster combination, while the Ne is the expected number obtained from Chi-square test. Generally, if Ro/e > 1, it suggests that cells classified in the given cluster are more frequent than random expectations for the analyzed sample, and vice versa.
We defined the clonal score to quantify the degree of clonal expansion for T ell clusters. Clonal score (CS) is calculated on the well-described TCR clonality measurement, which uses the normalized Shannon entropy 15, 56:
Where the N is the total number of clonotypes found in the given cluster, pi is the cell frequency of clonotype i in this cluster. Here, a clonotype is determined by identical paired α and β TCR chains. A higher CS (ranging from 0 to 1) indicates higher clonal expansion level.
For T cells with identical TCR clonotypes, they often present in different clusters. Thus, represent a potential transition from one phenotypic state to anther through clonal expansion. In this work, we defined transition score (TS) to characterize the transition potential between two specific clusters. The TS was calculated based on the transition level of each member clonotype. Given two clusters (ex. cluster 1 and cluster 2) in the total T cell clonotypes, the TS of cluster 1 to cluster 2 is calculated by the following formulas:
Where is the ratio of cells with TCR clonotype t in cluster i to the total cell number with clonotype t in clusters 1 and 2, and is the ratio of cells with clonotype t in cluster 1. Similar to clonal score, high values indicate high potential of state transition. When comparing the clonal scores or transition scores of different T cell groups, we randomly downsampled 50% of total T cells for 10 times and re-calculated the clonal/transition score for each group. Wilcox rank-sum test was used to test the statistical significance.
Pseudotime trajectory inference.
To determine the potential lineage differentiation between those T cell populations with high TCR sharing. Monocle (version 2.0) 29 was used to investigate transcriptional and developmental trajectories concerning different CD8+ or CD4+ T cell clusters. The data of raw counts together with cluster annotations were taken as input to monocle, and the default parameters were set to run data normalization and dimension reduction. Next, Monocle leaned the kinetics of gene expression by using the reversed graph embedding approach and placed each cell along an inferred pseudotime trajectory. According to the assumption that the trajectory has a tree structure, functional “State” was identified based on the segment of the tree-like structure.
Stemness, cytotoxic and exhaustion signature scoring.
The stemness, cytotoxic and exhaustion signatures score were derived from variable genes across all CD8+ T cells. The variable genes were identified by the FindVariableFeatures function of Seurat package. The Pearson correlation between reference genes Tcf7 (stemness signature), Fasl (cytotoxic signature) and Lag3 (exhaustion signature) and all CD8+ T enriched genes was computed using the scaled expression values. We selected the top 10 genes with the highest correlation with each reference genes and used them as the signature genes of stemness, cytotoxicity and exhaustion. Individual cell was then scored as the average expression of these signatures.
iSMART groups analysis.
iSMART was implemented to identify TCR specificity 31. We pooled all the productive beta CDR3 sequences derived from scTCR-seq data of pMHC-I tetramer-sorting libraries into a merged matrix with 15,520 CDR3 sequences as the input of iSMART algorithm. Our iSMART performs pairwise comparisons among CDR3s with similar sequence length and filters out the low scoring comparisons and outputs all the CDR3s with empirical group IDs. We performed iSMART with the following conditions: 1) For a given group with high similarity, CDR3 fragments within this group must have no more than one amino acid difference from each other; 2) For CDR3s with different length, iSMART allows no more than one insertion in the comparison. The default values were set for other parameters. We finally detected 1,415 iSMART TCR groups, corresponding to 7,137 different tetramer sorted T cells (Supplementary Table 4). The T cells, which have TCRs in a same group, are expected to recognize a same antigen. We only retained the TCR groups that correspond to more than 20 T cells for further analysis. In order to identify the neoantigen Adpgk specific TCR groups, we used TCRs from untreated mouse spleen and lymph node samples as negative controls. Using Chi-square test followed by Benjamin–Hochberg method, we finally selected 20 TCR groups which showed specifically enrichment in the neoantigen tetramer positive library (FDR<0.05 and OR>1, Fig. 4a, Supplementary Table 5).
Gene set enrichment analysis.
The R packages fgesa and msigdbr were used to perform the gene set enrichment analysis. The pathway gene sets we used were extracted from online databases KEGG, BIOCARTA, REACTOME and curated Hallmark gene sets 57. For a given cluster, GSEA was performed using each pathway gene set against ranked gene lists from comparison with other clusters. Pathways were considered as enriched if the BH adjusted fdr < 0.1, and the normalized enrichment scores (NES) were calculated to represent the enrich level (Supplementary Table 6).
Other statistical analysis.
To uncover the discriminative signatures that can accurately distinguish the CD8 effector T cells (CD8–05) from other CD8+ T cells, we performed a receiver operating characteristics (ROC) analysis on the top 100 differentially expressed genes of CD8–05. We selected five most discriminative markers (AUC > 0.7) and defined the averaged expression of them as Cancer Antigen Specific T cell (CAST) score, which showed a very good prediction power with AUC=0.92 (Fig. 6a). The CAST score was calculated for > 9,000 human tumors from 23 major cancer types with RNA-seq and clinical data > 100 samples in TCGA database. The CD8+ T cell infiltration of each cancer sample in TCGA was estimated in our previous work 37, 58. For each cancer type, the patients were divided into high CD8+ TIL and low CD8+ TIL subgroups based on the median level of CD8+ T cell fraction. Survival analyses were conducted with the use of the Kaplan–Meier and Cox proportional hazards regression methods. Survival distributions were compared across groups with the use of the log-rank test. Other statistical analysis in this work, including Wilcoxon ran-sum tests, chi-square test, odd-ratio test, correlation test and down-sampling analysis, was applied using R programming language (version 3.5.1). All p values from multiple tests were corrected by Benjamini–Hochberg method using the p.adjust function.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The scRNA-seq and scTCR-seq data that support the findings of this study can be accessed through Gene Expression Omnibus under accession code GSE178881. Source data for Figs. 1–6 and Extended data Figs. 1–10 have been provided in Supplementary Tables. TCGA database is available at https://portal.gdc.cancer.gov/. scRNA-seq dataset for Fig. 1e can be accessed at EMBL Nucleotide Sequence Database (ENA) under accession code PRJEB34105 (ref. 24). All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Code availability
The single-cell RNA data was processed using cellranger v.2.1.1 (https://www.10xgenomics.com/) and analyzed with the R package Seurat v.3.1.2 (https://satijalab.org/seurat/). Monocle v2.0 was used to investigate transcriptional and developmental trajectories concerning different CD8+ or CD4+ T cell clusters. iSMART was implemented to identify TCR specificity (https://github.com/s175573/iSMART). The R packages fgesa v1.16.0 (http://bioconductor.org/packages/release/bioc/html/fgsea.html) and msigdbr v7.2.1 (https://cran.r-project.org/web/packages/msigdbr/index.html) were used to perform the gene set enrichment analysis.
Extended Data
Extended Data Fig. 1. Neoantigen vaccine combined with ICB to induce a durable antitumor immune response.

a, Gating strategy for accessing neoantigen specific T cell phenotype. b, MC38 bearing female C57BL/6J mice were treated with two doses of neoantigen vaccine on day 10 and 17 post tumor inoculation. The percentage of PD1+TIM3+tetramer+ CD8 T cells in the draining lymph node and tumor was detected by flow cytometry. c, MC38 bearing female C57BL/6J mice were treated with either anti-PD-L1, adjuvant alone (Adj), the combination of adjuvant and anti-PD-L1 or neoantigen vaccine plus anti-PD-L1. Tumor volume was monitored every 3 days, P=0.0194 (Adj+αPD-L1 vs Vaccine+αPD-L1). d, MC38 bearing female C57BL/6J mice were treated with neoantigen vaccine on day 12 post tumor inoculation. One dose of anti-PD-L1 (200 μg) was given before (day 10) or after (day 15) vaccination. The combination with two doses of anti-PD-L1 (200 μg, day 10 and 15) were used for comparison. Data were shown as mean ± s.e.m. (n=8 (b), n=5 (c) and n=6 (d) mice) from two independent experiments. Statistical analysis was performed by two-way ANOVA with Tukey’s multiple comparisons test (c,d), two-tailed unpaired Student’s t-test (b), *P ≤ 0.05, ****P ≤ 0.0001.
Extended Data Fig. 2. Study design and the distribution of T cell clusters.

a, Gating strategy of single T cell sorting for single-cell sequencing. b, t-SNE plots showing the distribution of CD8+ and CD4+ T cells for each scRNA-seq library.
Extended Data Fig. 3. Expression levels of signature genes in each T cell cluster.

a, Heatmap showing the mean expression of discriminative genes for each cluster of conventional CD4+ T cells (n=29,305 cells). b, Heatmap showing the mean expression of discriminative genes for each cluster of CD8+ T cells (n= 43,453 cells). c, Heatmap showing the mean expression of discriminative genes for each cluster of regulatory T cells (n= 8,058 cells). d, t-SNE plot of expression levels of selected genes in different clusters indicated by the colored oval corresponding to Fig. 2a. e, Bar plots showing the distribution of T cell clusters for each sample (n=93,399 cells).
Extended Data Fig. 4. Single-cell analyses of the dynamic changes of TILs in response to distinct immunotherapies.

Bar plots displaying the dynamics of several major CD8+ T cell clusters (upper panel) and CD4+ T cell clusters (lower panel) in response to different immunotherapies (T.na (n=2,238 cells); T.eff (n=6,191 cells); T.ex (n=7,050 cells); T.na (n=2,046 cells); T.Th1 (n=4,798 cells); T.Treg (n=4,122 cells) ). P values were determined by a chi-square test on counts of T cells, exact p values were provided in Source Data Extended Data Fig. 4.
Extended Data Fig. 5. The dynamic change of major tumor-infiltrated T cell populations in response to distinct immunotherapies.

a, Gating strategies for accessing the major tumor-infiltrated T cell populations by flow cytometry. b, MC38 bearing female C57BL/6J mice were treated with either neoantigen vaccine, anti-PD-L1 or the combination. The percentage of CCR7+ cells in tumor infiltrated CD4 and CD8 T cells were detected by flow cytometry as indicated time points. Data were shown as mean ± s.e.m. (n=5 mice) from two independent experiments. Statistical analysis was performed by two-way ANOVA with Šídák's multiple comparisons test (b), ****P ≤ 0.0001.
Extended Data Fig. 6. Lineage tracking of clonal CD8 T cell subsets associated with immunotherapies.

a, Boxplots showing the clonal score of exhausted T cells (CD8–08), effector T cells (CD8–05), Tregs (CD4–04) and Th1-like T cells (CD4–06) in different treatment groups. b, Boxplot showing the pairwise transition score between CD8–05 and other intra-tumor CD8+ clusters across all tumor samples. Different T cell clusters were randomly downsampled (50%) 10 times for statistical test (n=10 permutations (a,b)). Center line indicates the median value, lower and upper hinges represent the 25th and 75th percentiles, respectively, and whiskers denote 1.5× interquartile range. c, The trajectory of three CD8+ T cell clusters showing the inferred pseudotime along the tree-like structure. d, The trajectory of three CD8+ T cell clusters showing by consistent clones. e, The monocle component 1 correlates with the stemness score of CD8+ T cells. f, The distribution of CD8+ T cells in different transcriptional states identified by monocle across all groups. Two sided Wilcoxon rank-sum test were used for multiple groups comparisons, exact p values were provided in Source Data Extended data Fig. 6 (a,b). Two-sided Pearson’s correlation coefficient test was used to determine the p value, P < 2.2 × 10−16 (e).
Extended Data Fig. 7. Lineage tracking of clonal CD4 T cell subsets associated with immunotherapies.

a, Monocle-guided cell trajectory showing the state transition between two major conventional CD4+ T cell clusters (CD4–03, CD4–06). b, Violin plots showing the expression level of Lag3, Havcr2, Ctla4 and Tgfb1 on the CD4+ T cells in transcriptional state 2–3. c, The distribution of CD4 T cells in the monocle-identified transcriptional states among different groups. d, Monocle-guided cell trajectory of three regulatory T cell (Treg) clusters (CD4–02, CD4–05 and CD4–07). 4 transcriptional states were identified along the inferred trajectory. e, Violin plots showing the expression level of S1pr1, Klf2, Il10 and Glrx in the two terminal transcriptional states (2 and 4). f, The distribution of Treg cells in the monocle-identified transcriptional states among different groups. g, Heat map showing the fraction of clonotypes belonging to a primary phenotype cluster (rows) that are shared with other secondary phenotype clusters (columns). h, The fraction of clonal cells in each functional state of Treg trajectory. The two sided Wilcoxon rank-sum test were used to calculate the p value following the adjustment of the Benjamini-Hochberg method to get the fdr q value, n=6,126 cells (a-c) and n=4,415 cells (d-f). ***represents fdr q value < 0.001 (b,e).
Extended Data Fig. 8. The landscape of neoantigen Adpgk-specific CD8+ T cells.

Female C57BL/6J mice were subcutaneously injected with neoantigen vaccine, the percentage of Adpgk-specific T cell (tetramer+) in the draining lymph node was detected by flow cytometry. Representative data of 6 independent mice was shown.
Extended Data Fig. 9. Neoantigen vaccine and ICB coordinately mediated the anti-tumor immune response depending on T cells from draining lymph node.

a-c, female C57BL/6J mice were subcutaneously inoculated with 1×106 MC38 tumor cells and treated with either neoantigen vaccine, anti-PD-L1 or the combination. Gating strategies for accessing the phenotype of tumor infiltrated CD8 T cells by flow cytometry (a). The percentage of neoantigen-specific T cell in the tumor tissue was detected by flow cytometry. The representative result for Fig. 5b (n=4 independent mice) was shown in (b). The percentage of IFNγ-producing CD8 T cells were detected by Elispot assay (n=5 mice) (c). d-e, C57BL/6J mice were subcutaneously injected with neoantigen vaccine. Lymphocytes from draining lymph node were harvested at day 6 post vaccination and adoptively transferred to MC38 bearing Rag1−/− mice. Two doses of anti-PD-L1 were given to the recipient mice on day 2 and 5 post adoptive transfer. The percentage of tetramer+ cells in the donor draining lymph node was detected by flow cytometry (d). The representative IFNγ+ spots for Fig. 5k were shown (e). Data were shown as mean ± s.e.m. from two independent experiments. Statistical analysis was performed by one-way ANOVA (c), ****P ≤ 0.0001.
Extended Data Fig. 10. The discriminative markers of neoantigen specific T cells are associated with better clinical outcome in human tumors.
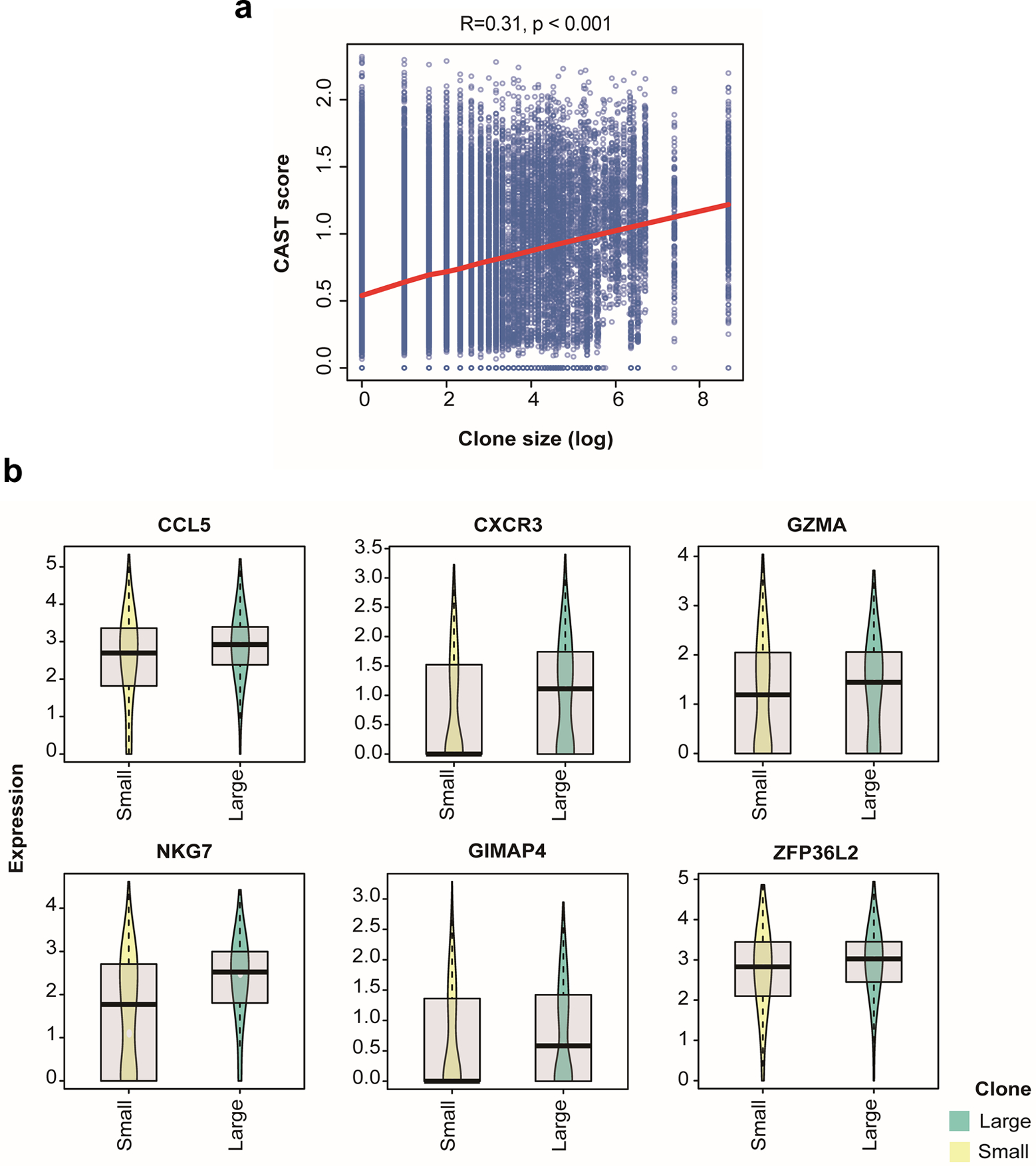
a, The correlation between CAST score and clone size of CD8+ T cells in BCC patients. The solid red line represents LOESS fitting result (n=26,846 cells). b, Boxplots comparing the expression of discriminative marker for CD8–05 in BCC patients’ CD8 T cells with large (n=1,271) or small (n=1,446) clone. Center line indicates the median value, lower and upper hinges represent the 25th and 75th percentiles, respectively, and whiskers denote 1.5× interquartile range. Two-sided Spearman’s correlation coefficient test was used to determine the p value, P < 2.2 × 10−16 (a).
Supplementary Material
Acknowledgments
We thank the UT Southwestern Institutional Animal Care & Use Committee (IACUC) and Animal Resources Center (ARC). Y.-X.F. holds the Mary Nell and Ralph B. Rogers Professorship in Immunology. This work was supported by Cancer Prevention and Research Institute of Texas (CPRIT) grant RR150072 given to Y.-X.F. B.L. is supported by CPRIT RR170079 and NCI 1R01CA245318. We also thank Zhida Liu, Chuanhui Han, Yong Liang, Zhenhua Ren and Anli Zhang for providing experiment materials and helpful discussions.
Footnotes
Competing interests
The authors declare no competing interests.
Reference
- 1.Chen DS & Mellman I Oncology meets immunology: the cancer-immunity cycle. Immunity 39, 1–10 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Sharma P & Allison JP The future of immune checkpoint therapy. Science 348, 56–61 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Waldman AD, Fritz JM & Lenardo MJ A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reinherz EL alphabeta TCR-mediated recognition: relevance to tumor-antigen discovery and cancer immunotherapy. Cancer Immunol Res 3, 305–312 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zamora AE, Crawford JC & Thomas PG Hitting the Target: How T Cells Detect and Eliminate Tumors. J Immunol 200, 392–399 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haen SP, Loffler MW, Rammensee HG & Brossart P Towards new horizons: characterization, classification and implications of the tumour antigenic repertoire. Nat Rev Clin Oncol 17, 595–610 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z, Ott PA & Wu CJ Towards personalized, tumour-specific, therapeutic vaccines for cancer. Nat Rev Immunol 18, 168–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schumacher TN & Schreiber RD Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Sahin U et al. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature 585, 107–112 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Ott PA et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ott PA et al. A Phase Ib Trial of Personalized Neoantigen Therapy Plus Anti-PD-1 in Patients with Advanced Melanoma, Non-small Cell Lung Cancer, or Bladder Cancer. Cell 183, 347–362 e324 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Dammeijer F et al. The PD-1/PD-L1-Checkpoint Restrains T cell Immunity in Tumor-Draining Lymph Nodes. Cancer Cell 38, 685–700 e688 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Yost KE et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med 25, 1251–1259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng GX et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564, 268–272 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Ren X et al. Insights Gained from Single-Cell Analysis of Immune Cells in the Tumor Microenvironment. Annu Rev Immunol 39, 583–609 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Yadav M et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515, 572–576 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Carretero-Iglesia L et al. High Peptide Dose Vaccination Promotes the Early Selection of Tumor Antigen-Specific CD8 T-Cells of Enhanced Functional Competence. Front Immunol 10, 3016 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bode C, Zhao G, Steinhagen F, Kinjo T & Klinman DM CpG DNA as a vaccine adjuvant. Expert Rev Vaccines 10, 499–511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longhi MP et al. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J Exp Med 206, 1589–1602 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheiermann J & Klinman DM Clinical evaluation of CpG oligonucleotides as adjuvants for vaccines targeting infectious diseases and cancer. Vaccine 32, 6377–6389 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott AC et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blank CU et al. Defining ‘T cell exhaustion’. Nat Rev Immunol 19, 665–674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L et al. Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer. Cell 181, 442–459 e429 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Verma V et al. PD-1 blockade in subprimed CD8 cells induces dysfunctional PD-1(+)CD38(hi) cells and anti-PD-1 resistance. Nat Immunol 20, 1231–1243 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu F et al. The transcription factor Bhlhe40 is a switch of inflammatory versus antiinflammatory Th1 cell fate determination. J Exp Med 215, 1813–1821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao X et al. Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity 27, 635–646 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Madi A et al. T-cell receptor repertoires share a restricted set of public and abundant CDR3 sequences that are associated with self-related immunity. Genome Res 24, 1603–1612 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trapnell C et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holman PO, Walsh ER & Jameson SC Characterizing the impact of CD8 antibodies on class I MHC multimer binding. J Immunol 174, 3986–3991 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Zhang H et al. Investigation of Antigen-Specific T-Cell Receptor Clusters in Human Cancers. Clin Cancer Res 26, 1359–1371 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Peske JD et al. Effector lymphocyte-induced lymph node-like vasculature enables naive T-cell entry into tumours and enhanced anti-tumour immunity. Nat Commun 6, 7114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wherry EJ & Kurachi M Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol 15, 486–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salerno F et al. Translational repression of pre-formed cytokine-encoding mRNA prevents chronic activation of memory T cells. Nat Immunol 19, 828–837 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicolet BP et al. CD29 identifies IFN-gamma-producing human CD8(+) T cells with an increased cytotoxic potential. Proc Natl Acad Sci U S A 117, 6686–6696 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lo JA et al. Epitope spreading toward wild-type melanocyte-lineage antigens rescues suboptimal immune checkpoint blockade responses. Sci Transl Med 13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li T et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 77, e108–e110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGranahan N et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Curtsinger JM, Johnson CM & Mescher MF CD8 T cell clonal expansion and development of effector function require prolonged exposure to antigen, costimulation, and signal 3 cytokine. J Immunol 171, 5165–5171 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Fairfax BP et al. Peripheral CD8(+) T cell characteristics associated with durable responses to immune checkpoint blockade in patients with metastatic melanoma. Nat Med 26, 193–199 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 176, 775–789 e718 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Azizi E et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 174, 1293–1308 e1236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Savas P et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med 24, 986–993 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Duckworth BC et al. Effector and stem-like memory cell fates are imprinted in distinct lymph node niches directed by CXCR3 ligands. Nat Immunol 22, 434–448 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Chow MT et al. Intratumoral Activity of the CXCR3 Chemokine System Is Required for the Efficacy of Anti-PD-1 Therapy. Immunity 50, 1498–1512 e1495 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohtani H, Jin Z, Takegawa S, Nakayama T & Yoshie O Abundant expression of CXCL9 (MIG) by stromal cells that include dendritic cells and accumulation of CXCR3+ T cells in lymphocyte-rich gastric carcinoma. J Pathol 217, 21–31 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Tang H et al. Facilitating T Cell Infiltration in Tumor Microenvironment Overcomes Resistance to PD-L1 Blockade. Cancer Cell 29, 285–296 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang H et al. PD-L1 on host cells is essential for PD-L1 blockade-mediated tumor regression. J Clin Invest 128, 580–588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin H et al. Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression. J Clin Invest 128, 805–815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park SL & Mackay LK Bhlhe40 Keeps Resident T Cells Too Fit to Quit. Immunity 51, 418–420 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Peng Q et al. PD-L1 on dendritic cells attenuates T cell activation and regulates response to immune checkpoint blockade. Nat Commun 11, 4835 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heinonen MT, Kanduri K, Lahdesmaki HJ, Lahesmaa R & Henttinen TA Tubulin- and actin-associating GIMAP4 is required for IFN-gamma secretion during Th cell differentiation. Immunol Cell Biol 93, 158–166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu L et al. Rejuvenation of tumour-specific T cells through bispecific antibodies targeting PD-L1 on dendritic cells. Nat Biomed Eng 5, 1261–1273 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolock SL, Lopez R & Klein AM Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst 8, 281–291 e289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kirsch I, Vignali M & Robins H T-cell receptor profiling in cancer. Mol Oncol 9, 2063–2070 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liberzon A et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li B et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 17, 174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq and scTCR-seq data that support the findings of this study can be accessed through Gene Expression Omnibus under accession code GSE178881. Source data for Figs. 1–6 and Extended data Figs. 1–10 have been provided in Supplementary Tables. TCGA database is available at https://portal.gdc.cancer.gov/. scRNA-seq dataset for Fig. 1e can be accessed at EMBL Nucleotide Sequence Database (ENA) under accession code PRJEB34105 (ref. 24). All other data supporting the findings of this study are available from the corresponding author upon reasonable request.


