SUMMARY
Pan-neuronally expressed genes, such as genes involved in the synaptic vesicle cycle or in neuropeptide maturation, are critical for proper function of all neurons, but the transcriptional control mechanisms that direct such genes to all neurons of a nervous system remain poorly understood. We show here that six members of the CUT family of homeobox genes control pan-neuronal identity specification in C. elegans. Single CUT mutants show barely any effects on pan-neuronal gene expression or global nervous system function, but such effects become apparent and progressively worsen upon removal of additional CUT family members, indicating a critical role of gene dosage. Overexpression of each individual CUT gene rescued the phenotype of compound mutants, corroborating that gene dosage, rather than the activity of specific members of the gene family, is critical for CUT gene family function. Genome-wide binding profiles as well as mutation of CUT homeodomain binding sites by CRISPR/Cas9 genome engineering show that CUT genes directly control the expression of pan-neuronal features. Moreover, CUT genes act in conjunction with neuron-type specific transcription factors to control pan-neuronal gene expression. Our study, therefore, provides a previously missing key insight into how neuronal gene expression programs are specified and reveals a highly buffered and robust mechanism that controls the most critical functional features of all neuronal cell types.
eTOC blurb:
How do genes expressed by all neurons in a nervous system acquire their pan-neuronal specificity? Leyva-Díaz and Hobert address this long-standing question, demonstrating that pan-neuronal genes which encode, e.g., synaptic vesicle proteins, are controlled via a robust regulatory mechanism that deploys six members of the CUT homeobox gene family.
Graphical Abstract
INTRODUCTION
To understand nervous system development, it is of critical importance to decipher the mechanisms that control the expression of neuronal gene batteries. Apart from ubiquitous housekeeping genes expressed in all tissue types, neuronal gene batteries fall into two categories: (1) Genes selectively expressed in specific neuron classes; these include neurotransmitter synthesis pathway genes, individual neuropeptides genes, ion channels, signaling receptors and many others (Figure 1A)1,2. (2) Pan-neuronally expressed genes that execute functions shared by all neurons, but not necessarily other cell types; these genes encode proteins involved in a number of generic neuronal processes, including synaptic vesicle release (e.g., RAB3, SNAP25, RIM), dense core biogenesis and release (e.g., CAPS), molecular motors (e.g. kinesins) or neuropeptide processing enzymes (e.g., endo- and carboxypeptidases, monooxygenases)2,3. Great strides have been made in understanding the regulation of the first category of genes, neuron type-specific gene batteries, in the nervous system of many species 4–6. However, the regulatory programs that orchestrate pan-neuronal gene expression have remained elusive in any species to date 3. Basic helix-loop-helix transcription factors that act as proneural factors to establish neuronal identity during development are usually only transiently expressed and are therefore not good candidates to initiate and maintain pan-neuronal gene expression throughout the life of a neuron 7.
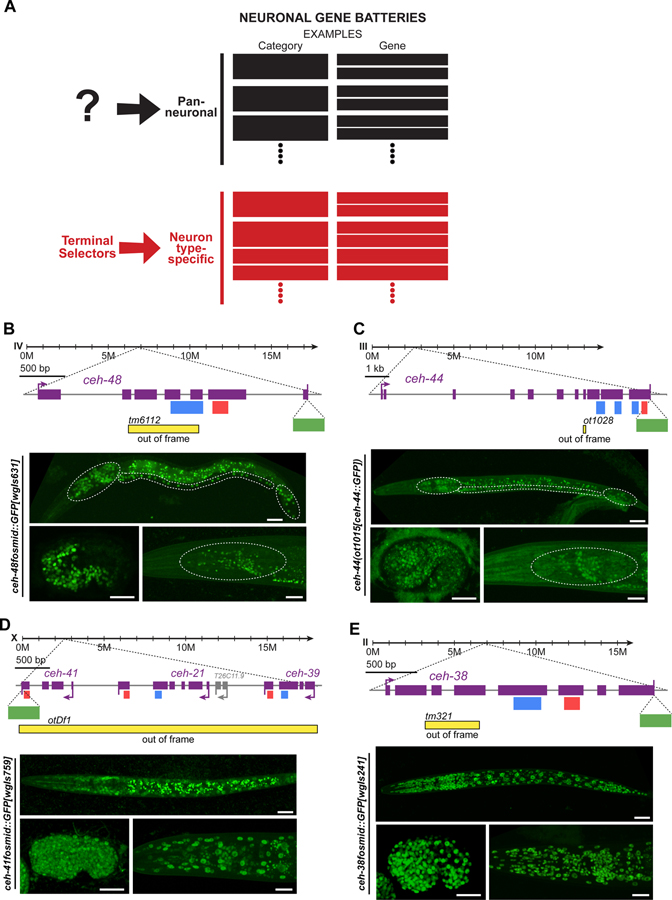
Figure 1. CUT genes are expressed pan-neuronally.
(A) Schematic illustration of two main components of neuronal gene batteries, pan-neuronally expressed genes, for which no current regulator is known, and neuron type-specific gene batteries that are controlled by terminal selector-type transcription factors 1. Examples for genes in each category are provided.
(B-E) Schematic representation of ceh-48 (B), ceh-44 (C), ceh-41, ceh-21 and ceh-39 (D), and ceh-38 (E), gene loci showing mutant alleles, GFP tags, and CUT and Homeodomain motifs location. Reporter expression at the comma embryonic stage (bottom left, lateral view), L1 larval stage (top, full worm lateral views) and young adult stage (bottom right, lateral view of the head) showing ceh-48 (ceh-48fosmid::GFP[wgIs631]). (B) and ceh-44 (ceh-44(ot1015[ceh-44::GFP])) (C) pan-neuronal expression, and ceh-41, ceh-21 and ceh-39 (D), ceh-38 (ceh-38fosmid::GFP[wgIs241]) (E) ubiquitous expression. We use a fosmid reporter for ceh-41 (ceh-41fosmid::GFP[wgIs759]), the last gene in the operon of three ONECUT genes, which provides a read-out for expression of all genes in the operon. The embryonic comma stage is the stage when neurons are born. Head ganglia, ventral nerve cord, and tail ganglia outlined in L1 images, and head ganglia outlined in young adult images for ceh-48 and ceh-44 reporters. Asterisks (*) indicate autofluorescence in L1 (ceh-48 and ceh-44) and Comma (ceh-44) images. See Figure S1 for a comparison between CRISPR reporters expression for the different CUT genes. Note that the ceh-44(ot1028) allele was design to introduce a frameshift in the CUT homeobox isoform of the Y54F10AM.4 locus (isoform a) and does not affect the b isoform of this locus, which generates a different, non-homeodomain containing isoform, homologous to CASP protein 15.
YA, young adult; Scale bars 15 μm.
In the nematode Caenorhabditis elegans, and other organisms as well, the expression of neuron type-specific genes during terminal differentiation is controlled by neuron-type specific combinations of terminal selector transcription factors 1,4,6. However, genetic removal of a terminal selector does not generally affect the expression of pan-neuronal identity features 1,4. For example, loss of the LIM homeobox gene ttx-3 or the EBF-type unc-3 zinc knuckle transcription factor results in the loss of all known neuron type-specific identity features of the cholinergic AIY interneuron or the cholinergic ventral nerve cord motorneurons, respectively, while the expression of pan-neuronal genes remains unaffected 8,9. Similarly, in mice, BRNA3 and ISL1 control neuron type-specific, but not pan-neuronal features of sensory neurons of the trigeminal ganglion and dorsal root ganglia 10. In attempts to decipher the apparent parallel acting gene regulatory programs of pan-neuronal gene expression, we have previously isolated cis-regulatory enhancer elements from pan-neuronally expressed genes 3. However, genetic screens for mutants that affect expression of these cis-regulatory elements have remained unsuccessful 11.
In this paper, we describe the discovery that six members of a specific family of homeobox genes, the CUT homeobox genes, jointly control pan-neuronal gene expression. CUT genes are expressed in all neurons and bind to the regulatory control regions of pan-neuronal genes. Deletion of the CUT homeodomain binding motif from pan-neuronal genes, using CRISPR/Cas9 genome engineering, disrupts expression and function of pan-neuronal genes. Removal of individual CUT genes reveals a dosage-sensitive function of these genes in controlling pan-neuronal gene expression and neuronal function. These phenotypes can be rescued by the expression of individual CUT factors, indicating that these factors act redundantly. A more extensive neuronal transcriptional profiling in neurons lacking all neuronal CUT genes reveals that these factors are required for the expression of large cohorts of neuronal genes. Further genetic loss of function analysis reveals that pan-neuronally expressed CUT genes cooperate with neuron type-specific terminal selectors to control pan-neuronal gene expression. Our studies reveal an exceptionally robust regulatory architecture of pan-neuronal gene expression, which contrasts the regulation of neuron-type specific genes, which depend on fewer regulatory inputs. Our findings may have implications for the evolution of neuronal cell type diversity.
RESULTS
CUT homeobox genes are expressed in all neurons
Our recently reported genome-wide analysis of the expression of all homeobox genes, critical regulators of neuron-type specific identity programs, uncovered a clue for potentially solving the riddle of pan-neuronal gene expression. Using both fosmid-based reporters as well as CRISPR/Cas9-engineered reporter alleles, in which we inserted gfp reporter transgenes in endogenous gene loci, we found that two homeobox genes, ceh-44 and ceh-48, are restricted to all neurons of the adult nervous system (Figure 1B and 1C)12. The only non-neuronal cells that express one of these two genes (ceh-48) are the secretory uv1 uterine cells, whose neuronal characters, including expression of synaptic vesicular machinery and the neurotransmitter tyramine, have been noted before 3,13. Expression of ceh-44 and ceh-48 commences right after the birth of neurons in the embryo, slightly preceding the onset of various other markers of pan-neuronal identity 3, and they are continuously expressed throughout the life of the organism (Figure 1B and 1C).
ceh-44 and ceh-48 are members of the CUT family of homeobox genes, defined by the presence of a homeodomain and one or more CUT domains 14. Based on the presence of multiple CUT domains, ceh-44 is the sole representative of the CUX subclass of the CUT family in C. elegans, while ceh-48 is a member of the ONECUT subclass, characterized by the presence of a single CUT domain 15. The DNA binding sites of CUX and ONECUT homeodomain proteins are very similar 16. In addition to ceh-48, the C. elegans genome encodes five additional ONECUT genes, three of which are located in a single operon (Figure 1D). While ceh-48 is pan-neuronally expressed, four of these additional ONECUT genes are ubiquitously expressed in all tissues at all stages (Figure 1D and 1E), while one ONECUT gene (ceh-49) is only expressed in the early embryo before neurogenesis. ceh-49 was not considered further here. Comparison of the expression level of all CUT gene loci, assessed with CRISPR/Cas9-engineered reporter alleles, shows that ceh-38 is the most highly expressed CUT family member (Figure S1).
Binding sites for CUT homeodomain proteins are required for pan-neuronal gene expression.
The pan-neuronal expression of ceh-44 and ceh-48 made us consider these CUT family genes as potential regulators of pan-neuronal identity. Supporting this notion we find that the many pan-neuronal genes whose cis-regulatory control regions we had previously defined to contribute to pan-neuronal gene expression 3 contain predicted CUT homeodomain binding sites (as mentioned above, the DNA binding sites of CUX and ONECUT proteins appear to be very similar 16 and from hereon, we refer to these sites as “CUT homeodomain binding sites”)(Figures 2A and S2). Moreover, animal-wide chromatin immunoprecipitation (ChIP) of CEH-48 conducted by the modENCODE consortium revealed binding of CEH-48 to these cis-regulatory elements (Data S1A) 17.
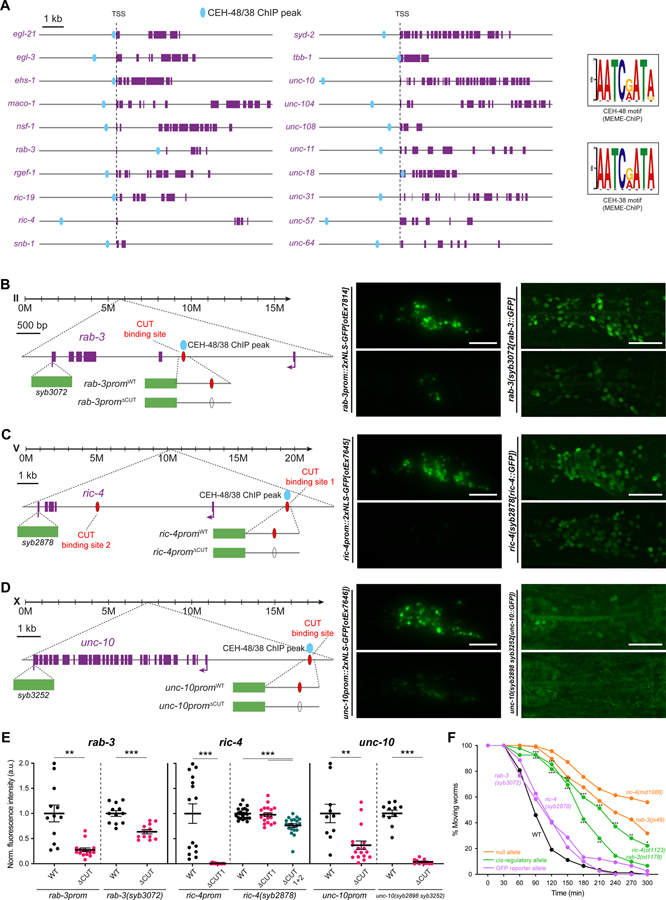
Figure 2. CUT genes binding is required for pan-neuronal gene expression.
(A) Schematic representation of 20 pan-neuronal genes and the location of CEH-48 and CEH-38 peaks found in the ChIP-seq datasets. CEH-48 and CEH-38 peaks overlap for all genes except in maco-1 and tbb-1, which only contain CEH-38 peaks, and ric-19, which only contains a CEH-48 peak. Scale represents 2kb for unc-104 and unc-31. The consensus binding motifs for CEH-48 and CEH-38, extracted from the ChIP-seq datasets using MEME-ChIP 45 are shown on the right. See Data S1A–C for a full list of genes with CEH-48 and CEH-38 ChIP peaks. See Figure S2 for how CUT ChIP binding correlates with the cis-regulatory elements that we previously defined in pan-neuronally expressed genes 3.
(B-D) Schematic representation of rab-3 (B), ric-4 (C) and unc-10 (D) gene loci (left) showing the location of CEH-48/CEH-38 ChIP peaks, CUT homeodomain binding sites, endogenous GFP tags for CRISPR reporters rab-3(syb3072[rab-3::T2A::3xNLS::GFP]), ric-4(syb2878[ric-4::T2A::3xNLS::GFP]), unc-10(syb2898 syb3252[unc-10::T2A::3xNLS::GFP])), and small promoters tested (rab-3prom10::2xNLS-GFP[otEx7814], ric-4prom30::2xNLS-GFP[otEx7645], unc-10prom12::2xNLS-GFP[otEx7646]). Blue ovals indicate binding based on ChIP-seq peak data, red ovals indicate binding site based on sequence. Worm head GFP images showing a reduction in pan-neuronal gene expression when the CUT homeodomain binding site is mutated compared to WT (middle, left). Mutation of the same CUT homeodomain binding sites endogenously in the context of CRISPR reporters affects pan-neuronal expression (middle, right). ric-4 gfp-tagged allele expression is only affected upon mutation of additional CUT homeodomain binding sites (site 1 and 2). unc-10 gfp-tagged allele expression is very dim and expression is not visible in all neurons. All images correspond to worms at the L4 larval stage.
(E) Quantification of small promoters and CRISPR reporters (shown in B-D) head neurons fluorescence intensity in wild-type and upon CUT homeodomain binding site mutations in the regulatory control regions of rab-3 (left), ric-4 (center) and unc-10 (right). The data are presented as individual values with each dot representing the expression level of one worm with the mean ± SEM indicated. Unpaired t-test, **P < 0.01, ***P < 0.001. For ric-4(syb2878[ric-4::T2A::3xNLS::GFP]), one-way ANOVA followed by Tukey’s multiple comparisons test; ***P < 0.001. n ≥ 10 for all genotypes.
(F) Aldicarb-sensitivity defects in wild-type animals, ric-4 and rab-3 CRISPR reporter alleles (rab-3(syb3072), ric-4(syb287)), ric-4 and rab-3 cis-regulatory alleles (ric-4(ot1123 syb2878), rab-3(ot1178 syb3072)), and ric-4 and rab-3 null alleles (ric-4(md1088), rab-3(js49)). Wild-type data is represented with black dots, the CRISPR reporter alleles with purple dots, the cis-regulatory alleles with green dots, and null alleles with orange dots. Two-way ANOVA followed by Tukey’s multiple comparisons test, comparisons for ric-4 and rab-3 cis-regulatory alleles vs wild-type indicated; *P < 0.05, **P < 0.01, ***P < 0.001. n ≥ 3 independent experiments (25 animals per independent experiment). Mean and SEM values are provided in Data S5A.
TSS, transcription start site; WT, wild-type; a.u., arbitrary units. Scale bars 15 μm for all panels except for CRISPR reporters in (B-D), where scale bars equal 10 μm.
We assessed the functional relevance of these CUT homeodomain binding sites in two different ways: First, we deleted these sites in the context of enhancer fragments, isolated from pan-neuronal gene loci, that drive broad neuronal if not pan-neuronal expression in transgenic, multicopy reporter arrays. We observed a loss of expression upon deletion of CUT homeodomain binding sites from isolated cis-regulatory enhancer elements derived from the rab-3/RAB3, ric-4/SNAP25 and unc-10/RIM genes (Figure 2B–E). Second, we used CRISPR/Cas9 genome engineering to first tag several pan-neuronal genes (rab-3/RAB3, ric-4/SNAP25, unc-10/RIM, ehs-1/EPS15) with a gfp reporter tag, and to subsequently delete their respective CUT homeodomain binding site from the respective endogenous locus. Deletion of CUT homeodomain binding sites affected expression of all four pan-neuronal genes that we tested (Figure 2B–E and S5D).
We tested the functional significance of the CUT homeodomain binding site mutations by asking whether these potential cis-regulatory alleles displayed behavioral defects expected from the loss of function of these pan-neuronal genes. rab-3/RAB3 and ric-4/SNAP25 null alleles show defects in synaptic transmission that can be measured via the sensitivity of animals to a drug that affects synaptic transmission at the neuromuscular junction, aldicarb 18,19. We found that rab-3/RAB3 and ric-4/SNAP25 alleles carrying CUT homeodomain binding site mutations show resistance to aldicarb (Figure 2F), which correlates with the reduction in ric-4/SNAP25 and rab-3/RAB3 expression observed in these alleles, and indicate impairment on synaptic transmission. Taken together, the functional relevance of presumptive CUT homeodomain binding sites hints toward a function of the CUT family of transcription factors as potential regulators of pan-neuronal gene expression.
Dosage-dependent requirement of CUT homeobox genes for pan-neuronal gene expression and neuronal behavior.
We next analyzed the consequences of genetic removal of the two pan-neuronally expressed ceh-44 and ceh-48 genes. We used a ceh-48 null allele from a C. elegans knockout consortium 20 and engineered a ceh-44 null allele using the CRISPR/Cas9 system (Fig. 1B,C). As a first step to assess gene function, we analyzed the expression of a rab-3 reporter construct in single and double ceh-44 and ceh-48 null mutant backgrounds. Given the functional importance of the CUT homeodomain binding site in the rab-3 locus described above, we were surprised to observe no rab-3/RAB3 expression defects in either single or ceh-44; ceh-48 double null mutant animals (Figure 3A).
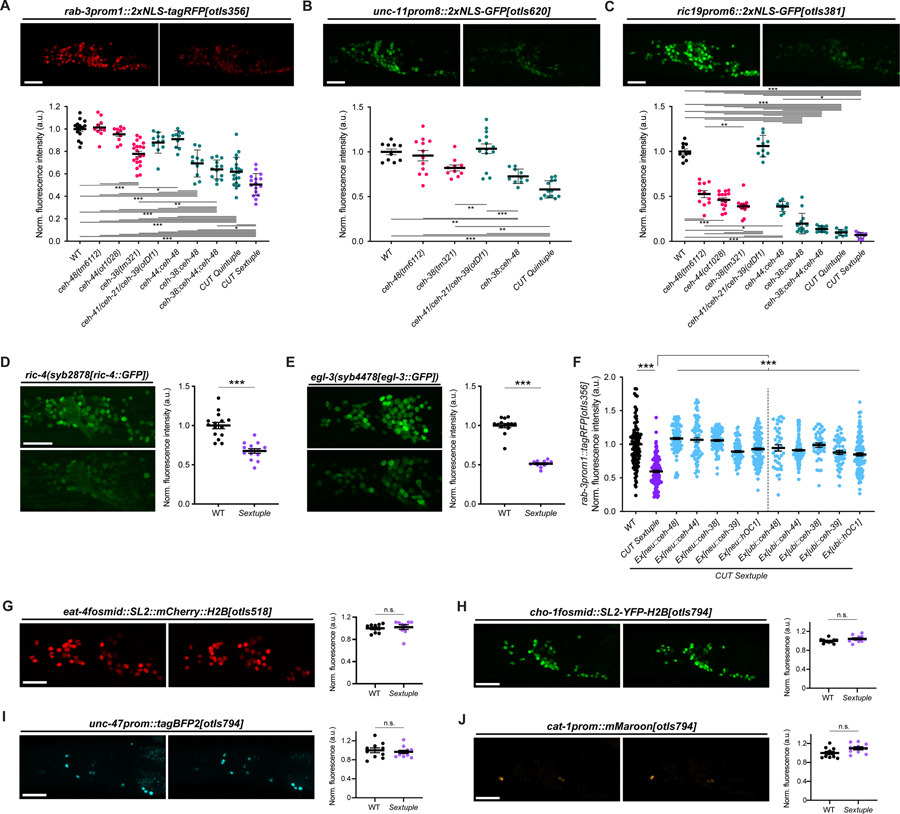
Figure 3. CUT genes act in a dosage-dependent manner to control pan-neuronal gene expression.
(A-C) Expression of rab-3prom1::2xNLS-tagRFP[otIs356] (A), unc-11prom8::2xGFP[otIs620] (B) and ric19prom6::2xNLS-GFP[otIs381] (C) in wild-type (left) and CUT sextuple mutant (right). Lateral views of the worm head at the L4 stage are shown. Quantification of fluorescence intensity in head neurons (bottom) in wild-type, individual CUT mutants (ceh-48(tm6112), ceh-44(ot1028) and ceh-38(tm321)) and compound CUT mutants (otDf1, which deletes ceh-41, ceh-21 and ceh-39; double ceh-44;ceh-48, double ceh-38;ceh-48, triple ceh-38;ceh-44;ceh-48, quintuple ceh-38;ceh-48;otDf1, and sextuple ceh-38;ceh-44;ceh-48;otDf1). unc-11prom::2xGFP[otIs620] and ceh-44 are located in the same chromosome (chr. III) and cannot be recombined together. The data are presented as individual values with each dot representing the expression level of one worm with the mean ± SEM indicated. Wild-type data is represented with black dots, individual CUT mutants with pink dots, the sextuple CUT mutant with purple dots, and other compound CUT mutants with green dots. One-way ANOVA followed by Tukey’s multiple comparisons test; *P < 0.05, **P < 0.01, ***P < 0.001. n ≥ 10 for all genotypes. All genotypes were compared, but only those comparisons that show statistically significant differences are indicated with lines.
(D-E) Expression of ric-4(syb2878[ric-4::GFP]) (ric-4(syb2878[ric-4::T2A-3xNLS-GFP])) (D), egl-3(syb4478[egl-3::GFP]) (egl-3(syb4478[egl-3::SL2-GFP-H2B])) (E) in wild-type (top) and CUT sextuple mutant (bottom). Lateral views of the worm head at the L4 stage are shown. Quantification of CRISPR alleles fluorescence intensity in head neurons. The data are presented as individual values with each dot representing the expression level of one worm with the mean ± SEM indicated. Unpaired t-test, ***P < 0.001. n ≥ 12 for all genotypes.
(F) Expression of rab-3prom::2xNLS-tagRFP[otIs356] was compared between wild-type, CUT sextuple mutant, and CUT sextuple mutant rescue (pan-neuronal, ceh-48 promoter (“neu”, see Figure S4), or ubiquitous, eft-3 promoter (“ubi”), expression of ceh-48, ceh-44, ceh-38, ceh-39 or hOC1). Quantification of fluorescence intensity analyzed by COPAS system (“worm sorter”). The data are presented as individual values with each dot representing the expression level of one worm with the mean ± SEM indicated. Wild-type data is represented with black dots, the sextuple CUT mutant with purple dots, and rescue lines with blue dots. One-way ANOVA followed by Tukey’s multiple comparisons test; ***P < 0.001. n ≥ 40 for all genotypes.
(G-J) Neurotransmitter reporter transgenes in CUT gene mutants. Transgenes are otIs518 (eat-4fosmid::SL2::mCherry::H2B) (G) and otIs794 which contains cho-1fosmid::NLS-SL2-YFP-H2B (H), unc-47prom::tagBFP2 (I), and cat-1prom::mMaroon (J), analyzed in a wild-type (left) or CUT sextuple mutant background (right). Lateral views of the worm head at the L4 stage are shown. Quantification of fluorescence intensity in head neurons. The data are presented as individual values with each dot representing the expression level of one worm with the mean ± SEM indicated. n ≥ 10 for all genotypes.
WT, wild-type; a.u., arbitrary units; n.s., not significant. Scale bars 15 μm.
ChIP analysis from the modENCODE project shows that the conserved and ubiquitously expressed CEH-38 ONECUT protein displays the same binding profile to pan-neuronal genes as the CEH-48 protein 17 (Figures 2A and S2; Data S1A–C). Moreover, motif extraction from the ChIP-seq data reveals that CEH-48 and CEH-38 consensus binding motifs are identical (Figure 2A). To test the possibility that CEH-38 could compensate for loss of ceh-44 and ceh-48, we generated a triple ceh-44; ceh-48; ceh-38 null mutant strain and indeed now found a reduction of rab-3/RAB3 expression (Figure 3A). Since rab-3/RAB3 expression was reduced but not eliminated, and since the ceh-38 result indicates that even a ubiquitously expressed CUT gene contributes to the regulation of pan-neuronal gene expression, we also considered a role of the three remaining, ubiquitously expressed CUT genes, ceh-41, ceh-21 and ceh-39. We used CRISPR/Cas9 to generate a precise deletion of those three genes, all located in an operon on the X chromosome, and found that this deletion (otDf1; Figure 1D) alone has no significant effect on rab-3 reporter expression (Figure 3A). However, adding this triple gene deletion to a ceh-44; ceh-48; ceh-38 triple mutant revealed that the sextuple CUT mutant strain displayed the strongest effect on rab-3 expression throughout the nervous system (Figure 3A). Sextuple CUT mutants further displayed a significant reduction in the expression of four other pan-neuronal genes, unc-11/SNAP91, ric-19/ICA1, ric-4/SNAP25 and egl-3/PCSK2 (Figure 3B–3E; for unc-11, due to linkage issues, we only generated a quintuple mutant). We tested two of these additional pan-neuronal genes, unc-11/SNAP91 and ric-19/ICA1, for whether they show cumulative expression defects upon removal of individual and multiple CUT genes in combination and found this to be the case (Figure 3B and 3C). The joint involvement of multiple CUT genes provides an explanation for why previous screens for mutants affecting pan-neuronal gene expression were unsuccessful 11 and are a testament to the robustness of pan-neuronal gene expression control.
Defects observed in the compound CUT mutants appear complementary to the gene expression defects observed in neuron type-specific terminal selector mutants. Specifically, several exemplary genes that are more selectively expressed in the nervous system, including cho-1/ChT (a marker that is exclusive to cholinergic neurons), eat-4/VGLUT (a marker specific to glutamatergic neurons), unc-47/VGAT (a marker specific for GABAergic neurons) and cat-1/VMAT (monoaminergic neuron marker), were not affected in sextuple CUT mutant animals (Figure 3G–3J). This result is consistent with these genes lacking ChIP peaks of CUT protein binding (Data S1A–C). Thus, the sextuple CUT mutant phenotype appears to be a mirror image of the phenotype of terminal selector transcription factors, whose removal results in loss of neuron type-specific identity features (such as the tested cho-1/ChT, eat-4/VGLUT, unc-47/VGAT, cat-1/VMAT), but not pan-neuronal identity features 4.
As expected from a loss of pan-neuronal gene expression, sextuple CUT mutant animals are severely deficient in nervous system function (Figures 4A–B and E). Animals display an almost complete paralysis in swimming assays, a very sensitive and well quantifiable read-out of animal locomotion (Figure 4A)21–23. Crawling behavior on an agar surface, quantified using a semi-automated WormTracker system, is also severely affected in sextuple CUT mutant animals (Figure 4B). Synaptic transmission defects, scored via responsiveness to aldicarb, are also very obvious, CUT sextuple mutants display a strong resistance to aldicarb (Figure 4E). We have found that these crawling and synaptic transmission defects are again cumulative, i.e. worsen the more CUT genes are removed (Figure 4B and E). Overall nervous system anatomy is unaffected in CUT sextuple mutants, including general cell body and fascicle position, (Figure S3). However, a visualization of synaptic punctae with the active zone marker CLA-1 24 or with a neuroligin-based GRASP strain 25 reveal defects in synapse abundance in compound CUT gene mutants (Figure 4H–I).
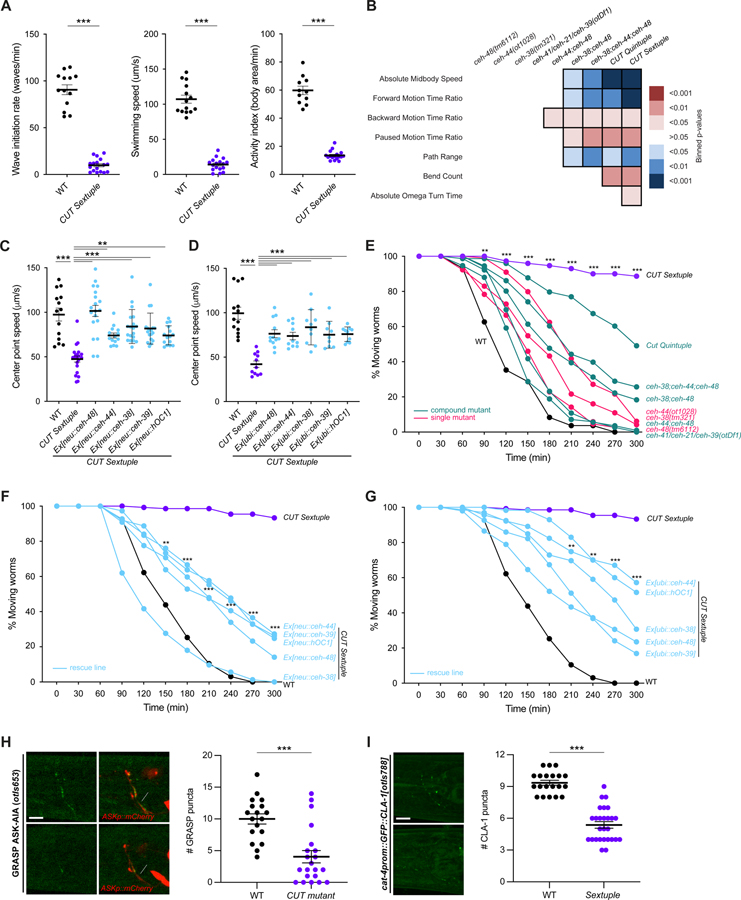
Figure 4. CUT genes are required for proper neuronal function.
(A) Swimming behavior: wave initiation rate (left), swimming speed (center), and activity index (right) were compared between wild-type and CUT sextuple mutant using a multi-worm tracker system 46. The data are presented as individual values with each dot representing the value of one worm with the mean ± SEM indicated. Unpaired t-test, ***P < 0.001. n ≥ 11 for all genotypes.
(B) Behavioral phenotypic summaries of representative locomotion features for individual and compound CUT mutants, analyzed using an automated worm tracker system 47. Heat map colors indicate the p-value for each feature for the comparison between each of the mutant strains and the wild-type strain. Red indicates a significant increase for the tested feature, while blue indicates a significant decrease. One-way ANOVA followed by Tukey’s multiple comparisons test. n ≥ 10 for all genotypes. Time ratio = (total time spent performing behavior)/(total assay time).
(C-D) Worm speed was compared between wild-type, CUT sextuple mutant, and CUT sextuple mutant rescue (panneuronal, ceh-48 promoter (“neu”, see Figure S4) (C), or ubiquitous, eft-3 promoter (“ubi”) (D), expression of ceh-48, ceh-44, ceh-38, ceh-39 or hOC1) using a multi-worm tracker system 46. The data are presented as individual values with each dot representing the value of one worm with the mean ± SEM indicated. Wild-type data is represented with black dots, the sextuple CUT mutant with purple dots, and rescue lines with blue dots. One-way ANOVA followed by Tukey’s multiple comparisons test, comparisons with CUT sextuple mutant indicated; **P < 0.01, ***P < 0.001. n ≥ 10 for all genotypes. See Figure S4 for additional locomotion features.
(E) Aldicarb-sensitivity defects in individual CUT mutants (ceh-48(tm6112), ceh-44(ot1028), ceh-38(tm321)) and compound CUT mutants (otDf1, which deletes ceh-41, ceh-21 and ceh-39; double ceh-44;ceh-48, double ceh-38;ceh-48, triple ceh-38;ceh-44;ceh-48, quintuple ceh-38;ceh-48;otDf1, and sextuple ceh-38;ceh-44;ceh-48;otDf1) compared to wild-type animals. Aldicarb is an acetylcholinesterase inhibitor that paralyzes worms. Decreased sensitivity to aldicarb correlates with a reduction in synaptic transmission 48. Worms were tested every 30 min for paralysis by touching the head and tail three times each. The data are presented as the percentage of moving worms at the indicated time point, dots represent the mean of independent experiments for each genotype. Wild-type data is represented with black dots, individual CUT mutants with pink dots, the sextuple CUT mutant with purple dots, and other compound CUT mutants with green dots. Two-way ANOVA followed by Tukey’s multiple comparisons test, comparisons for wild-type vs CUT sextuple mutant indicated; **P < 0.01, ***P < 0.001. n ≥ 3 independent experiments (25 animals per independent experiment). Mean and SEM values are provided in Data S5B.
(F-G) Aldicarb-sensitivity defects in wild-type animals, CUT sextuple mutant, and CUT sextuple mutant rescue lines (pan-neuronal (F), or ubiquitous (G) rescue lines). Wild-type data is represented with black dots, the sextuple CUT mutant with purple dots, and rescue lines with blue dots. Two-way ANOVA followed by Tukey’s multiple comparisons test, comparisons for CUT sextuple mutant vs Ex[neu::ceh-44] (F), and CUT sextuple mutant vs Ex[ubi::ceh-44] (G) indicated; **P < 0.01, ***P < 0.001. n ≥ 3 independent experiments (25 animals per independent experiment). Mean and SEM values are provided in Data S5B.
(H) ASK-AIA GRASP signal for the ASK>AIA (otIs653) in wild-type (top) and CUT compound mutant (ceh-38(tm321); ceh-44(ot1028); otDf1) (bottom). Lateral views of L1 worm heads at the nerve ring level are shown. ASK axon is labelled with cytoplasmic mCherry. Arrowheads indicate GRASP GFP synaptic puncta. otIs653 and ceh-48 are located in the same chromosome (chr. IV) and cannot be recombined together. Quantification of puncta along the ASK axon in the nerve ring. The data are presented as individual values with each dot representing the number of puncta in one worm with the mean ± SEM indicated. Unpaired t-test, ***P < 0.001. n ≥ 18 for all genotypes.
(I) HSN presynaptic specializations labeled by GFP-CLA-1 (cat-4prom::GFP::CLA-1[otIs788]) in wild-type (top) and CUT sextuple mutant (bottom). Lateral views of young adult worm heads at the nerve ring level are shown. Arrowheads indicate CLA-1 presynaptic specializations. Quantification of CLA-1 puncta along the HSN axon in the nerve ring. The data are presented as individual values with each dot representing the number of puncta in one worm with the mean ± SEM indicated. Unpaired t-test, ***P < 0.001. n ≥ 20 for all genotypes. See Figure S3 for overall nervous system anatomy.
WT, wild-type; Scale bars 5 μm.
The cumulative effects of CUT homeobox gene removal suggest a scenario in which it is primarily the overall dosage of CUT genes, rather than specific features of each individual CUT gene that is important to specify pan-neuronal gene expression. To further test this notion, we re-introduced individual CUT genes into the sextuple CUT mutant background. We used two separate drivers – a ubiquitous driver (eft-3prom) or a pan-neuronal driver (a fragment from the ceh-48 locus, ceh-48prom4, Figure S4A) – to generate multicopy transgenic arrays for overexpression. We found that each individually tested, overexpressed C. elegans CUT gene is alone able to rescue (a) the pan-neuronal gene expression defects (Figure 3F) and (b) the crawling and synaptic transmission defects of sextuple mutant animals (Figures 4C–D, 4F–G and S4B–E).
To assess potential phylogenetic conservation of CUT gene function, we also over-expressed a human ONECUT homolog, hOC1, and found that it is also capable of rescuing the C. elegans CUT sextuple mutant phenotype (Figures 3F, 4C–D, 4F–G and S4B–E).
Taken together, these results allow us to draw four conclusions: First, the usage of the postmitotic, pan-neuronal ceh-48 promoter indicates that CUT genes indeed act cell-autonomously in postmitotic neurons; second, CUT genes are functionally interchangeable; and, third, CUT gene dosage in the nervous system appears to be the main determinant of CUT gene function as regulators of pan-neuronal gene expression. Fourth CUT gene function may be phylogenetically conserved.
Genome-wide analysis of CUT homeobox gene targets.
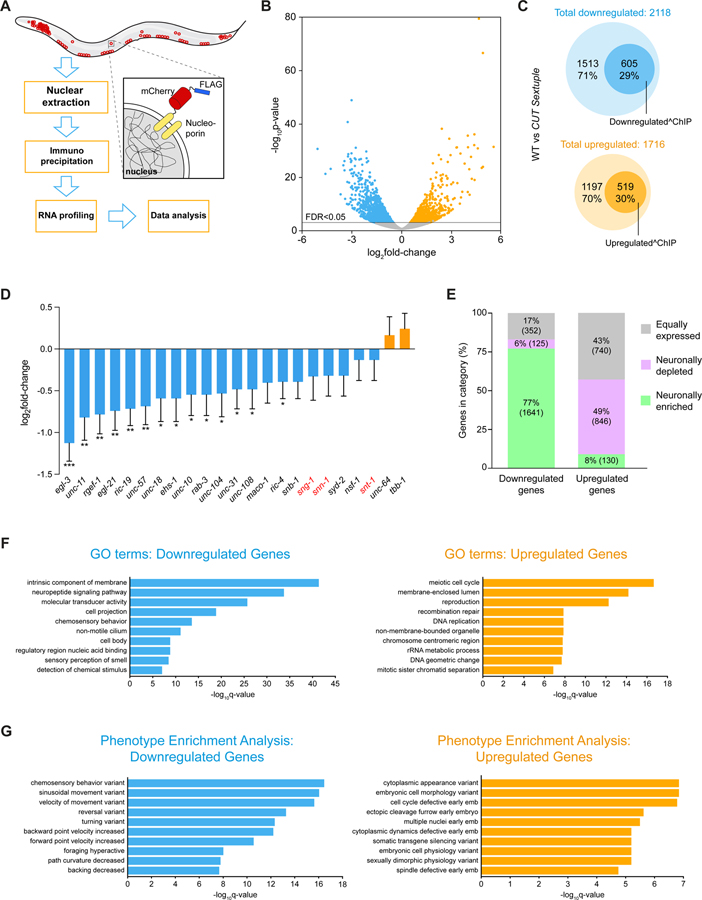
We further expanded our characterization of CUT gene function by RNA transcriptome profiling of CUT gene mutant animals. To this end, we used Isolation of Nuclei TAgged in specific Cell Types (INTACT) technology 26,27 to isolate all neuronal nuclei and compared neuronal transcriptomes of wild-type animals with those of sextuple CUT mutant animals (Figure 5A). Apart from upregulated genes, we found >2,000 genes to be downregulated (FDR < 0.05) and about 605 (29%) of those have CUT homeodomain binding ChIP peaks (Figure 5B–C, Data S2A–D). Downregulated genes with CUT homeodomain binding peak include known pan-neuronally expressed genes involved in the synaptic vesicle cycle (e.g. unc-57/SH3GL3, ric-19/ICA1, unc-11/SNAP91), synaptic activity zone assembly (e.g. cla-1/PCLO), neuronal transport (e.g. unc-116/JIP3), axon pathfinding (e.g. unc-14/RUSC1), neuronal cytoskeleton (e.g. unc-119/UNC119, unc-69/SCOC), neuropeptide processing (e.g. egl-3/PCSK2, egl-21/CPE, pamn-1/PAM) and other previously known pan-neuronal genes (e.g. rgef-1/RASGRP3, a commonly used pan-neuronal marker). Focusing on the battery of 23 pan-neuronal genes whose expression patterns we had defined in a previous analysis 3, we found that most of them show reduced transcript levels in the CUT sextuple mutant (Figure 5D). As described above, we have validated these changes in expression for rab-3, unc-11, ric-19, ric-4 and egl-3 (Figure 3A–E).
Figure 5. Transcriptional profiling of CUT sextuple mutants.
(A) Schematic and experimental design for INTACT sample collection, protocol, and data analysis for neuronal transcriptome profiling.
(B) Volcano plot of differentially expressed genes in CUT sextuple mutant neurons showing significantly (FDR < 0.05) downregulated (blue) or upregulated (orange) genes (RNA-seq, n = 3). See Data S2A for full list of differentially expressed genes.
(C) Diagrams showing overlap between differentially expressed genes in CUT sextuple mutant and genes bound by CEH-48 or CEH-38 in a wild-type ChIP-seq 17. Downregulated genes are marked in blue, upregulated genes are marked in orange, and the genes that contain CUT peaks are marked with dark circles within both clusters. See Figure S5 for effect on ubiquitously expressed genes containing CUT peaks.
(D) Changes of previously described pan-neuronal gene battery 3 in CUT sextuple mutant animals. The data are presented as the log2FoldChange ± standard error calculated by DESeq2, comparing neuronal samples from wild-type and CUT sextuple mutant. The two-stage step-up method of Benjamini, Krieger and Yekutieli (FDR 10%) was used to calculate the q-values for this subset of genes, analyzing the individual p-values obtained from the DESeq2 comparison. *Q < 0.05, **Q < 0.01, ***Q < 0.001 (RNA-seq, n = 3).
(E) Vertical slices representation of the distribution (in percentage) of the downregulated and the upregulated gene sets between the neuronally enriched (green), neuronally depleted (purple) and equally expressed (gray) gene sets. See Data S3A–C for full list of neuronally enriched and depleted genes. See Figure S6 for the validation of pan-neuronal expression of a neuronally-enriched CUT gene target.
(F-G) GO enrichment analysis (F) and phenotype enrichment analysis (G) using gene sets of significantly downregulated (blue) or upregulated (orange) transcripts. Graphs illustrate the 10 most significant terms. Analysis performed using the Gene Set Enrichment Analysis Tool from Wormbase. See Data S4A–D for full list of enriched terms.
WT, wild-type.
The use of INTACT technology to isolate the entire nervous system from wild-type animals identifies 6372 neuronally enriched genes through comparison of neuronal-nuclei to total nuclei samples (Figure 5E, Data S3A–B). Among the differentially expressed genes in CUT sextuple mutants, a large proportion (77%) of the downregulated gene set corresponds to this neuronally enriched gene set, while only 8% of the upregulated genes belong to the neuronally enriched gene set. Around half of the upregulated genes are actually neuronally depleted genes, whereas the other half corresponds to genes equally distributed between the nervous system and the whole animal (Figure 5E, Data S3A–C). Moreover, the downregulated gene set, but not the upregulated set, display significantly GO term enrichment for several neuronal processes (e.g. neuropeptide signaling pathway, chemosensory behavior)(Figure 5F; Data S4A–B). Similarly, phenotype enrichment analysis for the downregulated, but not upregulated gene set shows a large amount of locomotion phenotypes (Figure 5G; Data S4C–D). These findings are consistent with our reporter gene analysis, as well as our behavioral analysis, confirming that CUT homeodomain proteins are critical activators of pan-neuronal genes essential for proper neuronal function.
We find that the expression of some ubiquitously expressed genes, with potential selective functions in the nervous system, can also be CUT gene dependent. For example, we find that the C. elegans orthologs of the vertebrate neuronal splicing regulator NOVA1 28, the C. elegans ortholog of the alternative splicing factor RBM25, and the C. elegans homolog of a regulator of endocytosis, EPS15 29, show diminished transcript levels in the transcriptome analysis of CUT sextuple mutants. All three loci show binding of CUT proteins by ChIP analysis in the modENCODE dataset (Data S1A–C). gfp reporter alleles that we generated using CRISPR/Cas9-genome engineering revealed ubiquitous expression of nova-1/NOVA1, rbm-25/RBM25 and ehs-1/EPS15 throughout all tissue types (Figure S5A–S5C). We confirmed the CUT dependence of these genes in number of different manners. First, we crossed the nova-1 reporter allele into a CUT sextuple mutant background and found diminished expression in the nervous system. Second, we deleted the CUT homeodomain binding site from nova-1 gene locus and also observed diminished expression in the nervous system (Figure S5D). Similarly, a deletion of the CUT homeodomain binding site from the ubiquitously expressed ehs-1 gene locus also resulted in diminished neuronal expression (Figure S5E). In the case of ehs-1 this downregulation was specific to the nervous system since non-neuronal cells did not show downregulation (Figure S5E). Taken together, these results demonstrate the critical role of CUT-dependent gene expression of even ubiquitously expressed genes.
Lastly, we used the CUT-dependent transcriptome dataset to identify novel pan-neuronally expressed genes. Due to its uncommon primary sequence, we honed in on a small, 76 amino acid long protein, Y44A6D.2, with no predicted signal sequence, which is (a) downregulated in CUT sextuple mutants and (b) displays binding of CUT proteins in the modENCODE ChIP dataset (Data S1A–C). We used CRISPR/Cas9 to engineer gfp coding sequences at the 3’ end of the gene, and found that the resulting fusion protein is cytoplasmically expressed in all neurons throughout the nervous system, but no other tissue types (Figure S6). We named this locus tpan-1 for “tiny panneuronal protein”. Hence, the CUT-dependent transcriptome indeed identifies, as expected, novel pan-neuronal genes.
Collaboration of CUT homeobox genes with terminal selectors.
One notable feature of our CUT gene mutant analysis is that even in the sextuple CUT mutant, pan-neuronal gene expression is not uniformly eliminated. Nor do sextuple mutants display the larval lethality observed upon genetic removal of synaptic transmission machinery 30. To address the apparently incomplete nature of these phenotypes, we considered our previous functional analysis of neuron-type specific terminal selectors, which are required for the initiation of neuron-type specific gene expression profiles 1,3,4,6. While terminal selector removal alone does not generally affect pan-neuronal gene expression, we had found that pan-neuronal genes do contain terminal selector binding sites, and we had shown that these binding sites are functionally relevant, but only in the context of isolated cis-regulatory elements 3. Based on these findings, we had suggested that terminal selectors may provide redundant regulatory input into pan-neuronal gene expression (Figure 6A)3. Hence, an explanation for the lack of a complete loss of pan-neuronal gene expression in CUT sextuple mutants would be that terminal selectors are responsible for residual pan-neuronal gene expression.
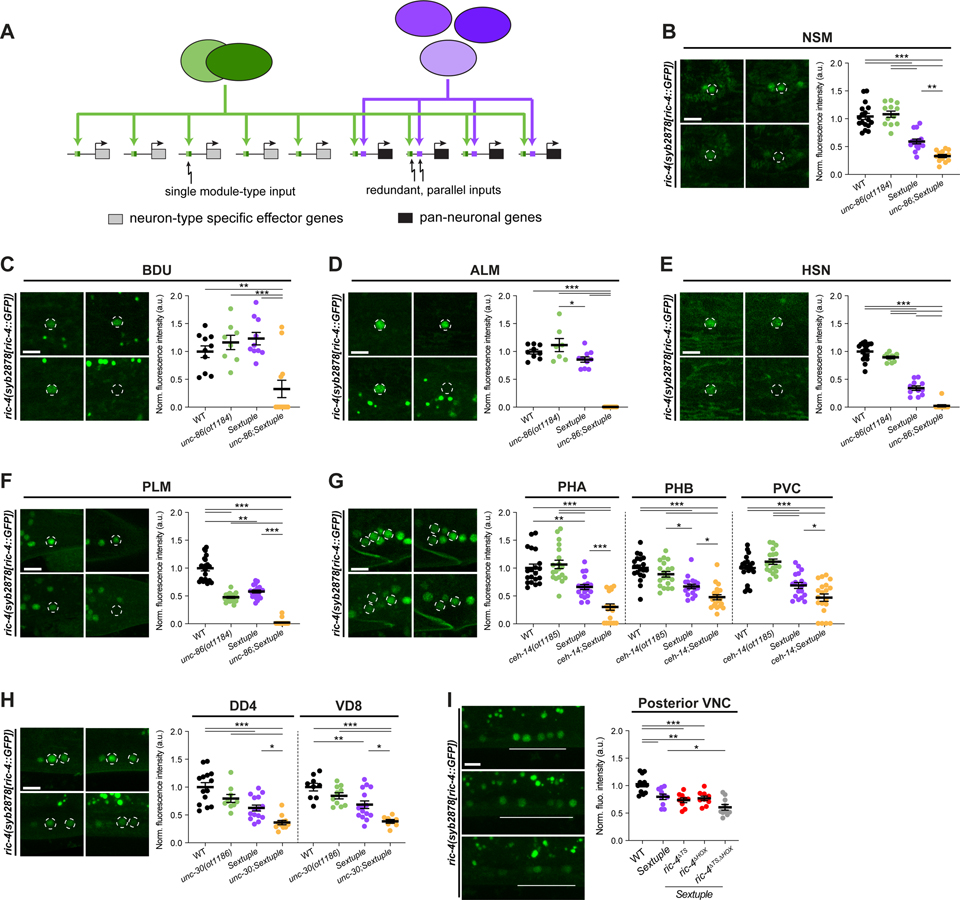
Figure 6. CUT genes cooperate with terminal selectors to control pan-neuronal gene expression.
(A) Illustration for how terminal selectors contribute to the regulation of pan-neuronal gene expression.
(B-H) Expression of ric-4(syb2878[ric-4::GFP]) (ric-4(syb2878[ric-4::T2A-3xNLS-GFP])) in wild-type (top left), terminal selector mutant (unc-86(ot1184) (B-F), ceh-14(ot1185) (G) or unc-30(ot1186) (H); top right), CUT sextuple mutant (bottom left), and compound terminal selector and CUT sextuple mutant (bottom right). Lateral views of the head (B), midbody (C-E, H) and tail (F and G) are shown. All images correspond to worms at the L4 larval stage, except for HSN (E) where young adults are shown. Quantification of ric-4(syb2878[ric-4::T2A-3xNLS-GFP]) fluorescence intensity in individual neurons. The data are presented as individual values with each dot representing the expression level of NSM (B), BDU (C), ALM (D), HSN (E), PLM (F), PHA, PHB, PVC (G), DD4, or VD8 (H) neuron, with the mean ± SEM indicated. Wild-type data is represented with black dots, terminal selector mutants with green dots, the sextuple CUT mutant with purple dots, and compound terminal selector and CUT sextuple mutant with yellow dots. One-way ANOVA followed by Tukey’s multiple comparisons test; *P < 0.05, **P < 0.01, ***P < 0.001. n ≥ 8 for all genotypes.
(I) Expression of ric-4(syb2878[ric-4::T2A-3xNLS-GFP]) in wild-type (top), CUT sextuple mutant (middle), and upon mutation of HOX and terminal selector binding sites on the ric-4 endogenous locus in a CUT sextuple mutant background (bottom). Individual mutation of the HOX (ric-4(ot1182 syb2878)) or terminal selector binding sites (ric-4(ot1181 syb2878)) has no effect on ric-4(syb2878[ric-4::T2A-3xNLS-GFP]) expression, but the expression is reduced in posterior ventral nerve cord (VNC) neurons when binding site mutations are combined (ric-4(ot1183 ot1181 syb2878)). Lateral views of the posterior VNC in L4 worms are shown. Quantification of ric-4(syb2878[ric-4::T2A-3xNLS-GFP]) fluorescence intensity in posterior VNC neurons. The data are presented as individual values with each dot representing the expression level of one worm with the mean ± SEM indicated. Wild-type data is represented with black dots, the sextuple CUT mutant with purple dots, the sextuple mutant with individual binding sites mutated with red dots, and the sextuple mutant with both binding sites mutated with gray dots. One-way ANOVA followed by Tukey’s multiple comparisons test; *P < 0.05, **P < 0.01, ***P < 0.001. n ≥ 9 for all genotypes.
WT, wild-type; a.u., arbitrary units. Scale bars 5 μm.
We addressed this possibility by generating different septuple null mutant strains in which we jointly removed all six CUT genes together with different terminal selectors that were previously found to regulate distinct neuron type-specific gene batteries. We indeed found that joint removal of terminal selectors and CUT genes strongly enhanced the reduction of pan-neuronal gene expression. For example, pan-neuronal gene expression in CUT sextuple mutants in ALM/PLM, HSN, BDU and NSM is further reduced, if not completely eliminated, upon CRISPR/Cas9-mediated deletion of the POU homeobox gene unc-86, which is a terminal selector of these neuron classes 31 (Figure 6B–6F). Similarly, the CUT sextuple effect in the PVC, PHA and PHB tail neurons is enhanced upon CRISPR/Cas9-mediated deletion of the LIM homeobox gene ceh-14, the terminal selector of PVC, PHA and PHB (Figure 6G)32–34. Likewise, the DD and VD GABAergic motor neurons of the ventral nerve cord, which lose neuron-type specific identity features, but not pan-neuronal identity features upon removal of the unc-30 Pitx homeobox gene 35,36, show a further reduction of pan-neuronal gene expression in a septuple CUT; unc-30 mutant background, compared to the CUT sextuple or unc-30 single mutant background alone (Figure 6H).
As an independent approach to removal of a terminal selector-encoding locus, we also mutated terminal selector binding sites in a pan-neuronal gene locus and asked whether this would enhance the effect of removal of CUT genes. Indeed, mutating binding sites for terminal selectors for ventral nerve cord motor neurons into a gfp-tagged ric-4/SNAP25 locus, further decreased ric-4/SNAP25 expression in a CUT sextuple null mutant background. (Figure 6I). These results indicate that CUT factors act in concert with terminal selectors to control pan-neuronal gene batteries.
DISCUSSION
We have shown here how a critical, but previously little understood component of neuronal gene expression programs – the expression of pan-neuronal gene batteries - is controlled. We identified an entire family of transcription factors, the CUT homeodomain transcription factors, as key regulators of pan-neuronal gene expression. CUT homeobox genes are also candidate regulators of pan-neuronal gene expression in other organisms. Drosophila, sea urchin and the simple chordate Ciona contain a single ONECUT gene with strikingly restricted, pan-neuronal gene expression 37–39. In vertebrates, CUX and ONECUT gene numbers have expanded and display complex expression patterns within and outside the nervous system 40,41. Encouragingly, a recent analysis of Ciona ONECUT revealed changes in gene expression of synaptic transmission molecules upon manipulation of ONECUT function in photoreceptor differentiation 42. Another recent study revealed that ONECUT proteins can indeed induce neuronal features in a fibroblast-to-neuronal reprogramming approach in vitro 43. Vertebrate ONECUT and CUX homologs are expressed in the nervous system 40,41, but a systematic, comparative, side-by-side analysis of all family members remains to be conducted to assess how broadly all family members cover the entire nervous system. Our finding that a vertebrate ONECUT protein, human OC1, can rescue the CUT sextuple mutant phenotype provides an encouraging hint that vertebrate CUT proteins may similarly be involved in pan-neuronal gene regulation. Our genetic loss of function analysis predicts that compound mutants may need to be generated in mice to assess CUT family function in vertebrate pan-neuronal gene expression.
The identification of CUT genes as regulators of pan-neuronal genes in C. elegans provides a complement to the much better understood regulation of neuron type-specific gene batteries. Pan-neuronal genes require at least two distinct sets of direct regulatory inputs to initiate (and presumably also maintain) their expression: a proper dosage of broadly expressed CUT homeobox genes and neuron type-specific terminal selector transcription factors (Figure 6A). Only the cumulative removal of all these regulatory inputs results in strong disruptions of pan-neuronal gene expression, illustrating a striking robustness of pan-neuronal gene regulation. The multitude of regulatory inputs into pan-neuronal gene loci that we define here by a genetic analysis of trans-acting factors predicts that the cis-regulatory control regions of pan-neuronal gene loci are very complex, combining inputs from CUT genes plus whatever type of terminal selector transcription factor a given neuron type employs. Our previous dissection of cis-regulatory regions of pan-neuronal gene corroborates this notion by describing a striking complexity of regulatory inputs 3, and therefore providing a satisfying complement to our present analysis of trans-acting factors.
The robustness of pan-neuronal gene regulatory architecture contrasts with the regulation of neuron-type specific gene batteries, where removal of individual cis-regulatory elements, or individual terminal selector transcription factors that act through such cis-regulatory elements, completely eliminates expression of neuron type-specific genes 3,4. These dichotomous regulatory strategies may speak to (a) the evolvability of neuron type-specific gene expression programs and (b) the evolutionary stability of pan-neuronal gene batteries. Brain evolution involves an increase in neuronal cell type diversity, and is essentially a “variation on a theme” process, characterized by an increase in neuronal cell type diversity, in which certain parameters remain stable (pan-neuronal identity), while others rapidly evolve. The two distinct regulatory strategies for neuron type-specific and pan-neuronal gene expression may lie at the basis of such evolutionary plasticity and stability.
Our studies underscore the centrality of homeobox genes in controlling multiple aspects of neuronal identity, not just in terms of conferring neuron-type specific features as has been shown before 12,44, but also in broadly defining what distinguishes non-neuronal from neuronal cells, a cell type that has gained the ability to communicate with others via a shared synaptic machinery and neuropeptides. These points indicate that the homeobox gene family may have been recruited into the control of neuronal gene expression very early in the evolution of nervous systems.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Oliver Hobert (or38@columbia.edu).
Materials Availability
All newly generated strains will be available at the Caenorhabditis Genetics Center (CGC).
Data and Code Availability
Raw and processed RNA-seq data will be available at GEO accession #GSE188489.
No original code has been generated for this paper.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Caenorhabditis elegans strains and handling
Worms were grown at 20°C on nematode growth media (NGM) plates seeded with E. coli (OP50) bacteria as a food source unless otherwise mentioned. Worms were maintained according to standard protocol 49. Wild-type strain used is Bristol variety, strain N2. A complete list of strains and transgenes generated and used in this study is listed in the Key Resources Table. A few of the strains were previously published, and/or obtained from the CGC, the National BioResource Project (NBRP, Japan) or the Transgeneome project 50, as detailed in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-FLAG M2 | Sigma-Aldrich | Cat# F1804 |
| Bacterial and virus strains | ||
| E. coli | Caenorhabditis Genetics Center (CGC) | WormBase: OP50; WormBase: WBStrain00041969 |
| Chemicals, peptides, and recombinant proteins | ||
| Aldicarb | ChemService | Cat# N-11044-100MG |
| Sodium Azide | Sigma-Aldrich | Cat# 71289 |
| RNase-free DMF | Acros Organics | Cat# AC327175000 |
| OptiPrep | Cosmo Bio USA | Cat # AXS-1114542 |
| Dynabeads Protein G | Thermo Fisher | Cat# 10003D |
| Dynabeads M-270 Carboxylic Acid | Thermo Fisher | Cat# 14305D |
| Alt-R S.p. Cas9 Nuclease V3 | IDT | Cat# 1081059 |
| Alt-R CRISPR-Cas9 tracrRNA | IDT | Cat# 1072533 |
| Critical commercial assays | ||
| NucleoSpin Tissue XS | Takara | Cat# 740901.250 |
| Universal RNA-seq with NuQauant | Tecan | Cat# 0533-32 |
| Deposited data | ||
| Raw and analyzed RNA-seq data | This paper | GEO: #GSE188489 |
| CEH-48 ChIP-seq dataset | 17 | https://www.encodeproject.org/; Experiment: ENCSR844VCY |
| CEH-38 ChIP-seq dataset | 17 | http://www.modencode.org/; Accession # modEncode_4800 |
| Experimental models: Organisms/strains | ||
| C. elegans: Strain N2 | Caenorhabditis Genetics Center (CGC) | WormBase: N2; WormBase: WBStrain00000001 |
| ceh-38(tm321) II | 20 | FX00321 |
| ceh-48(tm6112) IV | 20 | FX06112 |
| rab-3(js49) | 19 | NM791 |
| otIs356(rab-3prom1::2xNLS-tagRFP) V | 3 | OH10690 |
| otIs381(ric-19prom6::2xNLS-GFP) V | 3 | OH11062 |
| otIs620(unc-11prom8::2xNLS-GFP) III | 11 | OH13606 |
| otIs518(eat-4fosmid::SL2::mCherry::H2B, pha-1(+)) V; pha-1(e2123) III | 32 | OH13645 |
| otIs653(srg-8prom::mCherry, cho-1prom::mCherry, srg-8prom::NLG-1::spGFP1-10, cho-1prom::NLG-1::spGFP11) | This study | OH15034 |
| otIs748(rab-3prom1::GFP, ttx-3prom::mCherry) X | This study | OH16085 |
| otDf1 X | This study | OH16102 |
| ceh-44(ot1015[ceh-44::GFP]) III | 12 | OH16219 |
| ceh-49(ot1016[ceh-49::GFP]) V | 12 | OH16224 |
| otEx7463(ceh-48prom4:: 2xNLS-GFP, pha-1(+)); pha-1(e2123) III | This study | OH16284 |
| otEx7481(mir-228prom::ceh-48::GFP); otIs356(rab-3prom1::2xNLS-tagRFP) V | This study | OH16356 |
| ceh-44(ot1028) III | This study | OH16376 |
| ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otIs356 V; otDf1 X | This study | OH16377 |
| ceh-38(tm321) II; ceh-48(tm6112) IV; otIs356 V; otDf1 X | This study | OH16397 |
| ceh-38(tm321) II; ceh-48(tm6112) IV | This study | OH16583 |
| ceh-44(ot1028) III; ceh-48(tm6112) IV | This study | OH16584 |
| ceh-38(tm321) II; otIs356 V | This study | OH16586 |
| ceh-38(tm321) II; ceh-48(tm6112) IV; otIs356 V | This study | OH16587 |
| ceh-48(tm6112) IV; otIs356 V | This study | OH16590 |
| ceh-38(tm321) II;ceh-44(ot1028) III; ceh-48(tm6112) IV | This study | OH16593 |
| nsIs198(mir-228prom::GFP); otIs356(rab-3prom1::2xNLS-tagRFP) V | This study | OH16602 |
| otEx7617(unc-10prom12(ΔCUT)::2xNLS-GFP, pha-1(+)); pha-1(e2123) III | This study | OH16654 |
| otEx7619(rab-3prom10(ΔCUT)::2xNLS-GFP, pha-1(+)); pha-1(e2123) III | This study | OH16656 |
| otEx7644(ric-4prom30(ΔCUT)::2xNLS-GFP, pha-1(+)); pha-1(e2123) III | This study | OH16707 |
| otEx7645(ric-4prom30:: 2xNLS-GFP, pha-1(+)); pha-1(e2123) III | This study | OH16708 |
| otEx7646(unc-10prom12::2xNLS-GFP, pha-1(+)); pha-1(e2123) III | This study | OH16709 |
| otIs788(cat-4prom::GFP::CLA-1, cat-4prom::mCherry, inx-16prom::tagRFP) | This study | OH16737 |
| otIs790(UPN::npp-9::mCherry::blrp::3xflag) | 27 | OH16748 |
| otIs794(cho-1fosmid::NLS-SL2-YFP-H2B, eat-4fosmid::SL2:: LSSmOrange-H2B, unc-47prom::tagBFP2, cat-1prom::mMaroon, rab3prom1::2xNLS-tagRFP) | This study | OH16765 |
| ric-4(ot1123 syb2878) V | This study | OH17045 |
| ceh-48(ot1125[ceh-48::GFP]) IV | This study | OH17051 |
| ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otDf1 X; otIs790(UPN::npp-9::mCherry::blrp::3xflag) | This study | OH17055 |
| rab-3(ot1178 syb3072) II | This study | OH17504 |
| ric-4(ot1179 ot1123 syb2878) V | This study | OH17505 |
| unc-10(ot1180 syb2898 syb3252) X | This study | OH17506 |
| ric-4(ot1181 syb2878) V | This study | OH17507 |
| ric-4(ot1182 syb2878) V | This study | OH17508 |
| ric-4(ot1183 ot1181 syb2878) V | This study | OH17509 |
| ceh-38(tm321) II; ceh-44(ot1028) III; unc-86(ot1184) III; ceh-48(tm6112) IV; otDf1 X | This study | OH17510 |
| ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otDf1 X; ceh-14(ot1185) X | This study | OH17511 |
| ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; unc-30(ot1186) IV; otDf1 X | This study | OH17512 |
| unc-86(ot1184) III | This study | OH17513 |
| ceh-14(ot1185) X | This study | OH17514 |
| unc-30(ot1186) IV | This study | OH17515 |
| otEx7814(rab-3prom10::2xNLS-GFP, pha-1(+)); pha-1(e2123) III | This study | OH17517 |
| ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; ric-4(syb2878) V; otDf1 X | This study | OH17518 |
| ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; egl-3(syb4478) V; otDf1 X | This study | OH17519 |
| ceh-38(tm321) II; ceh-48(tm6112) IV; otDf1 X | This study | OH17520 |
| ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otDf1 X | This study | OH17521 |
| ceh-38(tm321) II; ceh-44(ot1028) III; otDf1 X; otIs653 | This study | OH17522 |
| ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otDf1 X; otIs788 | This study | OH17523 |
| ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otIs518 V; otDf1 X | This study | OH17524 |
| ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otDf1 X; otIs794 | This study | OH17525 |
| ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otDf1, otIs748 X | This study | OH17526 |
| otEx7815(ceh-48prom4::ceh-48, ttx-3prom::GFP); ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otIs356 V; otDf1 X | This study | OH17527 |
| otEx7816(ceh-48prom4::ceh-44, ttx-3prom::GFP); ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otIs356 V; otDf1 X | This study | OH17528 |
| otEx7817(ceh-48prom4::ceh-38, ttx-3prom::GFP); ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otIs356 V; otDf1 X | This study | OH17529 |
| otEx7818(ceh-48prom4::ceh-39, ttx-3prom::GFP); ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otIs356 V; otDf1 X | This study | OH17530 |
| otEx7819(ceh-48prom4::hOC1, ttx-3prom::GFP); ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otIs356 V; otDf1 X | This study | OH17531 |
| otEx7820(eft-3prom::ceh-48, ttx-3prom::GFP); ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otIs356 V; otDf1 X | This study | OH17532 |
| otEx7821(eft-3prom::ceh-44, ttx-3prom::GFP); ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otIs356 V; otDf1 X | This study | OH17533 |
| otEx7822(eft-3prom::ceh-38, ttx-3prom::GFP); ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otIs356 V; otDf1 X | This study | OH17534 |
| otEx7823(eft-3prom::ceh-39, ttx-3prom::GFP); ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otIs356 V; otDf1 X | This study | OH17535 |
| otEx7824(eft-3prom::hOC1, ttx-3prom::GFP); ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otIs356 V; otDf1 X | This study | OH17536 |
| ceh-44(ot1028) III; otIs356 V | This study | OH17537 |
| otIs356 V; otDf1 X | This study | OH17538 |
| ceh-44(ot1028) III; ceh-48(tm6112) IV; otIs356 V | This study | OH17539 |
| ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otIs356 V | This study | OH17540 |
| otIs620 III; ceh-48(tm6112) IV | This study | OH17541 |
| ceh-38(tm321) II; otIs620 III | This study | OH17542 |
| otIs620 III; otDf1 X | This study | OH17543 |
| ceh-38(tm321) II; otIs620 III; ceh-48(tm6112) IV | This study | OH17544 |
| ceh-38(tm321) II; otIs620 III; ceh-48(tm6112) IV; otDf1 X | This study | OH17545 |
| ceh-48(tm6112) IV; otIs381 V | This study | OH17546 |
| ceh-44(ot1028) III; otIs381 V | This study | OH17547 |
| ceh-38(tm321) II; otIs381 V | This study | OH17548 |
| otIs381 V; otDf1 X | This study | OH17549 |
| ceh-44(ot1028) III; ceh-48(tm6112) IV; otIs381 V | This study | OH17550 |
| ceh-38(tm321) II; ceh-48(tm6112) IV; otIs381 V | This study | OH17551 |
| ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otIs381 V | This study | OH17552 |
| ceh-38(tm321) II; ceh-48(tm6112) IV; otIs381 V; otDf1 X | This study | OH17553 |
| ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; otIs381 V; otDf1 X | This study | OH17554 |
| ceh-38(tm321) II; ceh-44(ot1028) III; ceh-48(tm6112) IV; nova-1(syb4373) V; otDf1 X | This study | OH17584 |
| wgIs241(ceh-38fosmid::TY1::EGFP::3xFLAG + unc-119(+)) | 50 | OP241 |
| wgIs631(ceh-48fosmid::TY1::EGFP::3xFLAG + unc-119(+)) | 50 | OP631 |
| wgIs759(ceh-41fosmid::TY1::EGFP::3xFLAG + unc-119(+)) | 50 | OP759 |
| ric-4(syb2878[ric-4::T2A::3xNLS::GFP]) V | This study | PHX2878 |
| rab-3(syb3072[rab-3::T2A::3xNLS::GFP]) II | This study | PHX3072 |
| unc-10(syb2898 syb3252[unc-10::T2A::3xNLS::GFP]) X | This study | PHX3252 |
| nova-1(syb4373[nova-1::GFP]) V | This study | PHX4373 |
| rbm-25(syb4376[rbm-25::GFP]) V | This study | PHX4376 |
| ehs-1(syb4426[ehs-1::SL2-GFP-H2B]) II | This study | PHX4426 |
| egl-3(syb4478[egl-3::SL2-GFP-H2B]) V | This study | PHX4478 |
| ehs-1(syb4426 syb4716) II | This study | PHX4716 |
| ceh-38(syb4799[ceh-38::GFP]) II | This study | PHX4799 |
| ceh-41(syb4901[ceh-41::GFP]) X | This study | PHX4901 |
| tpan-1(syb5349[tpan-1::GFP]) V | This study | PHX5349 |
| nova-1(syb4373 syb5446) V | This study | PHX5446 |
| ric-4(md1088) V | 18 | RM956 |
| Software and algorithms | ||
| ImageJ | 56 | https://imagej.nih.gov/ij/ |
| Worm Tracker v2.0 | 47 | https://www.mrc-lmb.cam.ac.uk/wormtracker/ |
| Wormlab | 46 | MBF Bioscience |
| STAR | 57 | https://code.google.com/archive/p/rna-star/ |
| featurecounts | 58 | http://subread.sourceforge.net/ |
| DeSeq2 | 59 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Gene Set Enrichment Analysis tool | 60 | https://wormbase.org/tools/enrichment/tea/tea.cgi |
| MEME-ChIP | 45 | https://meme-suite.org/meme/tools/meme-chip |
| Tomtom Motif Comparison Tool | 61 | https://meme-suite.org/meme/tools/tomtom |
| Other | ||
| Confocal Laser Scanning Microscope | Zeiss | LSM 880 |
| Sequencing Platform | Illumina | NextSeq 500 |
| Sorting Platform | Union Biometrica | COPAS FP-250 |
| Disposable Tissue Grinder | Fisher Scientific | Cat# 02-542-09 |
METHOD DETAILS
CRISPR/Cas9-based genome engineering
ceh-48(ot1125[ceh-48::GFP]), ceh-44(ot1028), otDf1, rab-3(ot1178 syb3072), unc-10(ot1180 syb2898 syb3252), ric-4(ot1123 syb2878), ric-4(ot1179 ot1123 syb2878), ric-4(ot1181 syb2878), ric-4(ot1182 syb2878), ric-4(ot1183 ot1181 syb2878), unc-86(ot1184), ceh-14(ot1185), unc-30(ot1186) were generated using Cas9 protein, tracrRNA, and crRNAs from IDT, as previously described 51. For ceh-48(ot1125[ceh-48::GFP]), one crRNA (atatgattattaggtgatta) and an assymetric double stranded GFP-loxP-3xFLAG cassette, amplified from a plasmid, were used to insert the fluorescent tag at the C-terminal. For ceh-44(ot1028), two crRNAs (ttaaggcgacgaagttatga and ccgaggaggcgaacagctat) and a ssODN donor (ataatatgatttctataattaaggcgacgaagttatatcggcagaagaatacggattctgaacttattga) were used to delete 80 bp of ceh-44 exon 8, introducing a frameshift in the CUT isoform of the Y54F10AM.4 locus (isoform a; the b isoform of this locus generates a different, non-homeodomain containing isoform, homologous to CASP protein; 15). For otDf1, two crRNAs (ggcatacatcttttcgaaag and atgaagaaaattatcaggat) and a ssODN donor (gaaaagggaattcggaaatgaagaaaattatcagtcgaaaagatgtatgcccgaaatgttccgagaaac) were used to generate a 8968 bp deletion (from position −159 upstream ceh-39 ATG, to 89 bp downstream ceh-41 stop codon) affecting 4 genes (deficiency, Df). The genes deleted in otDf1 are ceh-41, ceh-21, T26C11.9 and ceh-39. For rab-3(ot1178 syb3072), one crRNA (gctcacaaaaatggatcgat) and a ssODN donor (ctatctctctccgtgagcaacgagctagtcaacccaaaaaaccatttttgtgagcacacacagagagagactcaaa) were used to mutate a CUT homeodomain binding site on rab-3(syb3072[rab-3::T2A::3xNLS::GFP]) CRISPR reporter (details on binding site mutations on section below). For unc-10(ot1180 syb2898 syb3252), one crRNA (tcgtgcttcacggaattgtg) and a ssODN donor (gcagagagagaaaagtagtcgtgcttcacggaattgtggagagaaaaaaagagatctcaagtcagagagcgcgagc ttcgtttct) were used to mutate a CUT homeodomain binding site on unc-10(syb2898 syb3252[unc-10::T2A::3xNLS::GFP]) CRISPR reporter. For ric-4(ot1123 syb2878), one crRNA (atgagagccaatcgatacgt) and a ssODN donor (acgaagtgagccagaaagggaagcccgcacccacgtaaaaaaaaactctcatagagagaaagagagtctctgttttc tct) were used to mutate a CUT homeodomain binding site (“site 1”) on ric-4(syb2878[ric-4::T2A::3xNLS::GFP]) CRISPR reporter. For ric-4(ot1179 ot1123 syb2878), two crRNA (gaaaaatggaagtcacttgg and gggaaacagagaaaagacta) and a ssODN donor (aaatttcatataatttcccatccttcccacccccactaaggcttcatagtgcaaccttataactattagt) were used to delete a 431 bp section containing 9 CUT homeodomain binding sites (“site 2”) within ric-4 intron 1, on top of ric-4(ot1123 syb2878). For ric-4(ot1181 syb2878), one crRNA (ttgacgataacagagaccca) and a ssODN donor (ttgttcagtctttcccaaatttttgtgcccaatctAAAAAAAAAAAAAActctgttatcgtcaaaagtgacatcttttctttc g) were used to mutate COE (UNC-3) and UNC-30 binding sites on ric-4(syb2878[ric-4::T2A::3xNLS::GFP]) CRISPR reporter. For ric-4(ot1182 syb2878) and ric-4(ot1183 ot1181 syb2878), one crRNA (cgaaaagagctcagcgaaaa) and a ssODN donor (tcttcgtgccatccattcaaacaacgcttattttaaaaaaaaaaacatttttcgctgagctcttttcgtttcgtctttcttgtttc) were used to mutate a HOX binding site on ric-4(syb2878[ric-4::T2A::3xNLS::GFP]) or ric-4(ot1183 ot1181 syb2878). For unc-86(ot1184), two crRNAs (caaggtccccctcttttcca and acaacatacaatgggctacc) and a ssODN donor (tctgtctcctcccagcttcaaggtccccctcttttaccttgattctttgattagtttcgttttcgtgaac) were used to delete the entire unc-86 locus. For ceh-14(ot1185), two crRNAs (tcttggcgagtgcgatgagc and tgtactgtggagtcatgtgt) and a ssODN donor (gggacacaacattttgactcttggcgagtgcgatgcatgactccacagtacatttgaactggagaaaaac) were used to delete the entire ceh-14 locus. For unc-30(ot1186), two crRNAs (taagacggtaataatccttg and gtagtaaagttgaaaaggcg) and a ssODN donor (ccgatcactgactttgcgtaagacggtaataatcccttttcaactttactactgttcaataaacaattaa) were used to delete the entire unc-30 locus.
rab-3(syb3072), ric-4(syb2878), unc-10(syb2878), egl-3(syb4478), ceh-38(syb4799), ceh-41(syb4901), nova-1(syb4373), rbm-25(syb4376), ehs-1(syb4426), ehs-1(syb4426 syb4716), nova-1(syb4373 syb5446) and tpan-1(syb5349) were generated by SUNY Biotech. ceh-38(syb4799) and ceh-41(syb4901) were generated with the exact same GFP-loxP-3xFLAG cassette as in ceh-48(ot1125[ceh-48::GFP]) for direct comparison of CUT gfp-tagged CRISPR alleles.
For CUT homeodomain binding site mutations, we looked for CEH-48 sites centered within the region covered by CEH-48 and/or CEH-38 ChIP peaks in rab-3, ric-4, unc-10 and ehs-1 regulatory regions. The CEH-48 binding motif (consensus ATCGA), is cataloged in the CIS-BP (Catalog of Inferred Sequence Binding Preferences) database (http://cisbp.ccbr.utoronto.ca/)52. The CEH-48 motif matches known motifs for other ONECUT and CUX proteins (see ChIP-seq section below) (Data S6A–B). Deletions of CEH-48 binding sites were done by replacement of the binding site by adenines.
In rab-3(syb3072[rab-3::T2A::3xNLS::GFP]), ATCGAT (+2399, +2404) was mutated to AAAAAA. This site was centered within CEH-48 (+2326, +2452) and CEH-38 (+2211, +2719) ChIP peaks.
In unc-10(syb2898 syb3252[unc-10::T2A::3xNLS::GFP]), ATCGAT (−4558, −4553) was mutated to AAAAAA. This site was centered within CEH-48 (−4784, −4415) and CEH-38 (−4811, −4366) ChIP peaks.
In ric-4(syb2878[ric-4::T2A::3xNLS::GFP]), ATCGATTGG (−3683, −3675; “site 1”) was mutated to AAAAAAAAA. This site was centered within CEH-48 (−3832, −3598) and CEH-38 (−4062, −3521) ChIP peaks.
In ehs-1(syb4426[ehs-1::SL2-GFP-H2B]), ATCGAT (−220, −215) was mutated to AAAAAA. This site was centered within CEH-48 (−311, −106) and CEH-38 (−373, −168) ChIP peaks.
In nova-1(syb4373[nova-1::GFP]), ATCGATTTTCGAT (−1976, −1964) was mutated to AAAAAATTAAAAA. This site was centered within CEH-48 (−2196, −1826) and CEH-38 (−2223, −1709) ChIP peaks.
For ric-4, a second set of CUT homeodomain binding sites (“site 2”) was mutated within ric-4prom25 (cis-regulatory element found to be broadly expressed in head neurons)3. A 431 bp section (+4947, +5378) in ric-4 intron 1, containing 9 CUT homeodomain binding sites, was deleted.
The HOX/EXD motif, COE (UNC-3) motif, and UNC-30 motifs on ric-4 were mutated following prior experiments in small cis-regulatory elements 3, but here these mutations were done on the ric-4 CRISPR reporter allele, ric-4(syb2878[ric-4::T2A::3xNLS::GFP]). The HOX motif TGAATAATTG (−1064, −1055) was mutated to AAAAAAAAAA. The COE, TCCCTTGGGT (−1349, −1340), and UNC-30, TAATCC (−1352, −1347), motifs partially overlap and were mutated together: CTAATCCCTTGGGT was mutated to AAAAAAAAAAAAAA.
In the small cis-regulatory element reporters (see below) mutations in the same CUT homeodomain binding sites described here for rab-3, ric-4 and unc-10 were introduced in rab-3prom10, ric-4prom30 (site 1) and unc-10prom12.
Reporter transgenes
The rab-3, ric-4 and unc-10 cis-regulatory element reporters were generated using a PCR fusion approach 53. The rab-3prom10 (+2326, +2452) (promoter fragment number continues the series generated for cis-regulatory analysis in 3), ric-4prom30 (−3832, −3598) and unc-10prom12 (−4784, −4415) promoter fragments were amplified from N2 genomic DNA and fused to 2xNLS-GFP. These promoter fragment coordinates match those of the CEH-48 ChIP peaks in the regulatory regions of these genes. The resulting PCR fusion DNA fragments were injected as simple extrachromosomal arrays (50 ng/mL) into pha-1(e2123) animals, using a pha-1 rescuing plasmid (pBX at 50 ng/μL) as co-injection marker. Extrachromosomal array lines were selected according to standard protocol. For rab-3prom10, ric-4prom30 and unc-10prom12 harboring the CUT homeodomain binding site mutations, promoters were obtained as gBlocks (IDT) and fused to 2xNLS-GFP.
To assess neurotransmitter identity, we generated a transgene that expresses multiple reporters that assess neurotransmitter usage, including: a cho-1 fosmid reporter construct (cho-1fosmid::NLS-SL2-YFP-H2B; 3), to label cholinergic neurons; an eat-4 fosmid reporter construct (eat-4fosmid::SL2::mCherry::H2B 32, where mCherry was replaced with LSSmOrange) to label glutamatergic neurons; unc-47prom (coordinates −2778, −1) fused with TagBFP2 to label GABAergic neurons, cat-1prom (−1599, −1) fused with mMaroon to label monoaminergic neurons, and rab-3prom1 (−1462, +2921) fused with tagRFP to label all neurons (pan-neuronal marker). The cho-1fosmid::NLS-SL2-YFP-H2B (20 ng/μL), eat-4fosmid::SL2:: LSSmOrange::H2B (20 ng/μL), unc-47prom::tagBFP2 (5 ng/μL), cat-1prom::mMaroon (5 ng/μL) and rab3prom1::2xNLStagRFP (10 ng/ μL) constructs were injected together, and the resulting extrachromosomal array strain was integrated into the genome using standard UV irradiation methods. This was followed by 3 rounds of backcrossing to N2 to generate otIs794.
To generate cat-4prom::GFP::CLA-1(S) (pMM13), cat-4prom8 (−629, −299; expressed in HSN; 54) was amplified from N2 genomic DNA. The PCR fragment was cloned into PK065 (kindly shared by Peri Kurshan). cat-4prom::mCherry (pMM11) was generated similarly and cloned into pPD95.75. The constructs pMM13 and pMM11 were injected at 5 and 30 ng/μL, respectively, with an inx-16prom::tagRFP co-injection marker (10 ng/μL). The resulting extrachromosomal array strain was integrated into the genome using standard UV irradiation methods.
To label the ASK-AIA synapse with GRASP 25, we generated otIs653(srg-8prom::mCherry, cho-1prom::mCherry, srg-8prom::NLG-1::spGFP1–10, cho-1prom::NLG-1::spGFP11). For this transgene, a 2kb srg-8prom (coordinates −2000, −1; expressed in ASK) was cloned into MVC2 (pSM::NLG-1::spGFP1–10) using RF cloning to generate srg-8prom::NLG-1::spGFP1–10 (pMM14). srg-8prom::mCherry (pMM02) was generated by subcloning srg-8prom into pPD95.75. A 364bp cho-1prom (3006, −2642; expressed strongly in AIA, AIY, AIN; 55) PCR fragment amplified from genomic DNA was cloned into MVC3 (pSM::NLG-1::spGFP11) and pPD95.75 to generate cho-1prom::NLG-1::spGFP11 (pMM08) and cho-1prom::mCherry (pMM07), respectively. The constructs were injected at a total of 90 ng/μL, transgenic lines were picked based on the mCherry cytoplasmic expression, and the resulting extrachromosomal array strain was integrated into the genome using standard UV irradiation methods.
To generate the rab-3 cytoplasmic reporter (rab-3prom1::GFP), rab-3 promoter (“prom1” 3) was cloned into pPD95.67 (plasmid containing 2xNLS-GFP), where the 2xNLS was removed. The resulting plasmid was injected injected as simple extrachromosomal array (50 ng/μL) into N2 animals, using ttx-3prom::mCherry as a co-injection marker (25 ng/μL). The resulting extrachromosomal array strain was integrated into the genome using standard UV irradiation methods. This was followed by 6 rounds of backcrossing to N2 to generate otIs748.
Automated worm tracking
Automated single worm tracking was performed using the Wormtracker 2.0 system at room temperature 47. Young adult animals were recorded for 5 min and tracked on NGM plates with a small patch of food in the center (5 μL OP50 bacteria). Analysis of the tracking videos was performed as previously described 47. For the tracking of the CUT rescue lines and controls, tracking was performed using the WormLab automated multi-worm tracking system (MBF Bio-science)46 at room temperature. In each plate, 5 young adult animals were recorded for 5 min and tracked on NGM plates with a small patch of food in the center (5 μL OP50 bacteria). Videos were segmented to extract the worm contour and skeleton for phenotypic analysis. Raw WormLab data was exported to Prism (GraphPad) for further statistical analysis. Statistical significance between each group was calculated using One-way ANOVA followed by Tukey’s multiple comparisons test.
Swimming analysis
The swimming assay was performed as previously described 23 using the WormLab automated multi-worm tracking system (MBF Bio-science)46 at room temperature. In brief, 5 young adult animals were transferred into 50 μl M9 buffer and recorded for 1 min. Multiple features of the swim behavior were then analyzed using the WormLab software. Swimming metrics are based on the metrics described in 23. WormLab data was exported to Prism (GraphPad) for further statistical analysis.
Aldicarb assays
Aldicarb assays were performed as previously described 48. Briefly, 25 young adult animals (24 h after L4 stage, blinded for genotype) were picked into freshly seeded NGM plates containing 1 mM aldicarb (ChemService). Worms were assayed for paralysis every 30 min by prodding with a platinum wire. A worm was considered paralyzed if it did not respond to prodding to the head and tail three times each at a given time point. Strains were grown and assayed at room temperature. Statistical significance between each group was calculated in Prism (GraphPad) using Two-way ANOVA followed by Tukey’s multiple comparisons test.
Microscopy
Worms were anesthetized using 100mM of sodium azide and mounted on 5% agarose on glass slides. All images were acquired using a Zeiss confocal microscope (LSM 880). Image reconstructions were performed using Zen software tools. Maximum intensity projections of representative images were shown. Fluorescence intensity was quantified using the ImageJ software 56. Figures were prepared using Adobe Illustrator.
INTACT for purification of affinity-tagged neuronal nuclei
UPN::INTACT control worms (otIs790) as well as CUT sextuple mutant were grown on large plates (150mm) with enriched peptone media coated with NA22 bacteria to allow for the growth of large quantities of worms: 100,000 worms can grow from synchronized L1 stage to gravid adults on a single plate. ~600,000 animals were collected for each replicate at the L1 larval stage after egg preparation according to standard protocol. Animals were washed off the plate with M9, washed 3x with M9, lightly fixed with cold RNAse-free DMF for 2 minutes before washing with 1xPBS 3x. We followed the modified INTACT protocol 27 to optimize pull-down of neuronal nuclei. All steps following were done in cold rooms (4 °C) to minimize RNA and protein tag degradation. The animals were homogenized mechanically using disposable tissue grinders (Fisher) in 1x hypotonic buffer (1x HB: 10 mM Tris pH 7.5, 10 mM NaCl, 10 mM KCl, 2 mM EDTA, 0.5 mM EGTA, 0.5 mM Spermidine, 0.2 mM Spermine, 0.2 mM DTT, 0.1% Triton X-100, 1x protease inhibitor). After each round of mechanical grinding (60 turns of the grinder), the grinder was washed with 1 mL 1x HB and the entire homogenate was centrifuged at 100xg for 3 min. The supernatant was collected for later nuclei extraction and the pellet was put under mechanical grinding and centrifugation for 4 additional rounds. The supernatant collected from each round were pooled, dounced in a glass dounce, and gently passed through an 18-gauge needle 20x to further break down small clumps of cells. The supernatant was then centrifuged at 100xg for 10 min to further remove debris and large clumps of cells. Nuclei was isolated from the supernatant using Optiprep (Sigma): supernatant after centrifugation was collected in a 50mL tube, added with nuclei purification buffer (1x NPB: 10 mM Tris pH 7.5, 40 mM NaCl, 90 mM KCl, 2mM EDTA, 0.5 mM EGTA, 0.5 mM Spermidine, 0.2 mM Spermine, 0.2 mM DTT, 0.1% Triton X-100, 1x protease inhibitor) to 20 mL, and layered on top of 5 mL of 100% Optiprep and 10 mL of 40% Optiprep. The layered solution was centrifuged at 5000xg for 10 min in a swinging bucket centrifuge at 4 °C. The nuclei fraction was collected at the 40/100% Optiprep interface. After removal of the top and bottom layers, leaving a small volume containing the nuclei, the process was repeated 2 additional times. After final collection of the crude nuclei fraction, the volume was added to 4 mL with 1xNPB and precleared with 10 μL of Protein-G Dynabeads and 10 μL of M270 Carboxylated beads for 30 min to 1 h (Invitrogen). The precleared nuclei extract was then removed, and 50 μL was taken out as input samples (total nuclei). The rest was incubated with 30 μL of Protein G Dynabeads and 3 μL of anti-FLAG M2 antibody (Sigma) overnight to immunoprecipitate (IP) the neuronal nuclei. The following day, the IPed neuronal nuclei/beads was washed 6–8 times with 1xNPB for 10–15 min each time. The resulting IPed neuronal nuclei/beads were resuspended in 50 μL 1xNPB and a small aliquot was used to check with DAPI staining to quality-check the procedure for the following: 1) sufficient quantities of nuclei was immunoprecipitated; 2) nuclei are intact and not broken; 3) the majority of bound nuclei are single, mCherry-labelled neuronal nuclei and minimal nuclei clumps and large tissue chunks were immunoprecipitated. Anything not satisfying these quality checks were not used for downstream processing. The resulting input and neuronal IP samples were used for isolation of total RNA using Nucleospin RNA XS kit according to manufacturer’s protocol (Takara).
RNA-seq and data analysis
RNA-seq libraries were prepared using the Universal RNA-seq kit (Tecan) according to manufacturer’s protocol. The libraries were sequenced on Illumina NextSeq 500 machines with 75bp single-end reads. After initial quality check, the reads were mapped to WS220 using STAR 57 and assigned to genes using featurecounts 58. Differential gene expression analysis was conducted using DESeq2 59. 3834 genes were found to be differentially expressed in CUT sextuple mutants compared to wild-type animals (FDR < 0.05) (Data S2A). Gene Ontology and Phenotype Enrichment Analysis were performed using the Gene Set Enrichment Analysis tool from Wormbase (https://wormbase.org)60(Data S4A–D).
ChIP-seq datasets analysis
The CEH-48 ChIP-seq dataset (Experiment: ENCSR844VCY, bigBed file containing peak information: ENCFF784CKU) was obtained from the ENCODE portal (https://www.encodeproject.org/). The CEH-38 ChIP-seq dataset (Accession # modEncode_4800, gff3 interpreted data file containing peak information for combined replicates) was obtained from the modENCODE portal (http://www.modencode.org/). To identify the genes associated with these peak regions, peak coordinates were intersected with gene promoter regions (defined as from 5kb upstream of the transcription start site to 1kb downstream), and overlapping genes were identified (Data S1A–C). The consensus binding motif for CEH-48 and CEH-38 was obtained using MEME-ChIP 45, which returned similar motifs for both factors (consensus AATCGATA). Comparison of these motifs, and of the CEH-48 motif defined in 52, to known motifs using the Tomtom Motif Comparison Tool in MEME Suite 61 returned matches to known motifs for other ONECUT and CUX proteins (Data S6A–C).
QUANTIFICATION AND STATISTICAL ANALYSIS
All microscopy fluorescence quantifications were done in the ImageJ software 56. For all images used for fluorescence intensity quantification, the acquisition parameters were maintained constant among all samples (same pixel size, laser intensity), with control and experimental conditions imaged in the same imaging session. For quantification of head neurons (Figure 2, Figure 3), nerve ring neurons (Figure S1, Figure S6) and ventral nerve cord neurons (Figure 6I), fluorescence intensity was measured in maximum intensity projections using a single rectangular region of interest. A common standard threshold was assigned to all the control and experimental conditions being compared. For quantification of individual neurons (Figure 6), fluorescence intensity was measured in the focal plane with the strongest neuronal nucleus signal within the z-stack (circular region of interest around the nucleus). For each worm, a single circular region of interest was also used to measure the background intensity in an adjacent area, and this value was then subtracted from the reporter fluorescence intensity value. For quantification of GFP::CLA-1 and GRASP puncta (Figure 4), manual counting was performed using the ImageJ software. For quantification of hypodermal cells (Figure S6), fluorescence intensity was measured as described above for individual neurons. For each worm five hypodermal cells were measured, and the fluorescence intensity averaged. The same hypodermal cells were measured in all animals compared. For fluorescence quantification of CUT rescue lines (Figure 3), synchronized day 1 adult worms were grown on NGM plates seeded with OP50 and incubated at 20°C. The COPAS FP-250 system (Union Biometrica; “worm sorter”) was used to measure the fluorescence of 40–150 worms for each strain.
For all behavioral assay, randomization and blinding was done wherever possible. All statistical tests for fluorescence quantifications and behavior assays were conducted using Prism (Graphpad) as described in figure legends.
Supplementary Material
Data S1. Genes associated to CEH-48 and CEH-38 ChIP datasets. Related to Figure 2.
CEH-48 (Experiment: ENCSR844VCY, https://www.encodeproject.org/) and CEH-38 (Accession # modEncode_4800, http://www.modencode.org/) ChIP-seq datasets were used to identify genes associated to the peak regions. Those genes which promoter region (defined as from 5kb upstream of the transcription start site to 1kb downstream) overlapped with the ChIP-seq peaks were identified. A list of genes associated to CEH-48 peaks (4709 genes, Data S1A), genes associated to CEH-38 peaks (7795 genes, Data S1B), and genes associated to CEH-48 or CEH-38 peaks (9578 genes, Data S1C) are provided in separate tabs. The genes are sorted by WormBase Gene ID. Brief gene descriptions obtained from WormMine.
Data S2. Differentially expressed genes in CUT sextuple mutant. Related to Figure 5.
Comparison of neuronal-nuclei immunoprecipitated samples from wild-type and CUT sextuple mutant is conducted using DESeq2 59 to determine differentially expressed genes (DEG, 3834 genes have a padj < 0.05, Data S2A). The genes are sorted by padj from smallest to largest. baseMean is the average normalized read counts of all samples. log2FoldChange is calculated using the formula log2 (average read counts of mutant samples)/(average read counts of wild-type samples). A list of downregulated genes (genes with a log2FoldChange < 0; 2118 genes, Data S2B), upregulated genes (genes with a log2FoldChange > 0; 1716 genes, Data S2C), and differentially expressed genes that are found associated to CEH-48 or CEH-38 ChIP-seq peaks (1124 genes, Data S2D) are provided in separate tabs, with the genes sorted by WormBase Gene ID. Brief gene descriptions obtained from WormMine.
Data S3: Neuronally-enriched genes. Related to Figure 5.
Comparison of wild-type neuronal-nuclei immunoprecipitated (IP) samples to wild-type input (total nuclei) samples is conducted using DESeq2 to determine enrichment (12639 genes have a padj < 0.05, Data S3A). The genes are sorted by padj from smallest to largest. baseMean is the average normalized read counts of all IP and input samples. log2FoldChange is calculated using the formula log2 (average read counts of mutant samples)/(average read counts of wild-type samples). A list of neuronally-enriched genes (genes with a log2FoldChange > 0; 6372 genes, Data S3B), and neuronally-depleted genes (genes with a log2FoldChange < 0; 6267 genes, Data S3C) are provided in separate tabs, with the genes sorted by WormBase Gene ID. Brief gene descriptions obtained from WormMine.
Data S4. Full list of enrichment analysis GO and phenotype terms. Related to Figure 5.
Full list of enriched terms for GO enrichment analysis (Data S4A–B) and phenotype enrichment analysis (Data S4C–D) using gene sets of significantly downregulated or upregulated transcripts. The terms are sorted by q-value from smallest to largest. Analysis performed using the Gene Set Enrichment Analysis Tool from Wormbase.
Data S5. Aldicarb assay measurements mean and standard error. Related to Figure 2 and Figure 4.
Mean and standard error of the mean (SEM) values corresponding to the aldicarb data shown in Figure 2F (Data S5A) and Figure 4E–G (Data S5B).
Data S6. Known transcription factor motifs matching CEH-48 and CEH-38 motifs. Related to the STAR Methods.
Tomtom Motif Comparison Tool 61 output list of known motifs matching the CEH-48 motif defined in 52 (Data S6A), the CEH-48 motif extracted from the ChIP-seq dataset (Experiment: ENCSR844VCY, https://www.encodeproject.org/) using MEME-ChIP 45 (Data S6B), and the CEH-38 motif extracted from the ChIP-seq dataset (Accession # modEncode_4800, http://www.modencode.org/) using MEME-ChIP (Data S6C). These three motifs were compared with an eukaryote DNA motif database (vertebrates, in vivo and in silico) provided in MEME Suite 61 (JASPAR2018_CORE_vertebrates_non-redundant, uniprobe_mouse, and jolma2013; total 1808 known motifs). The motifs are sorted by q-value from smallest to largest.
Highlights:
C. elegans CUT family homeobox genes directly control pan-neuronal gene expression
Proper CUT gene dosage ensures robust pan-neuronal gene expression
CUT genes are required for proper neuronal function and synapse density
CUT genes collaborate with terminal selectors of neuron type-specific identities
ACKNOWLEDGEMENTS
We thank Chi Chen for generating transgenic lines, HaoSheng Sun for help with the INTACT protocol, Peri Kushan for CLA-1 constructs, Maryam Majeed for providing the HSN CLA-1 and GRASP ASK-AIA strains, and for help with the analysis of GRASP data, Eviatar Yemini for help with worm tracking, Cyril Cros and Molly Reilly for providing the neurotransmitter strain, Berta Vidal for providing the rab-3 cytoplasmic strain, Waleed Ali, Kevin Gonzalez and Mayeesa Rahman for genotyping and strain maintenance, Hynek Wichterle, Wes Gruber, Esteban Mazzoni and members of the Hobert lab for comments on this manuscript, and Wormbase and CGC (funded by NIH Office of Research Infrastructure Programs, P40 OD010440) for providing resources and reagents. This work was funded by the Howard Hughes Medical Institute. E.L.-D. was supported by an EMBO long-term fellowship (ALTF 962-2014).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hobert O, Carrera I, and Stefanakis N (2010). The molecular and gene regulatory signature of a neuron. Trends in neurosciences 33, 435–445. S0166–2236(10)00082–2 [pii] 10.1016/j.tins.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor SR, Santpere G, Weinreb A, Barrett A, Reilly MB, Xu C, Varol E, Oikonomou P, Glenwinkel L, McWhirter R, et al. (2021). Molecular topography of an entire nervous system. Cell 184, 4329–4347 e4323. 10.1016/j.cell.2021.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stefanakis N, Carrera I, and Hobert O (2015). Regulatory Logic of Pan-Neuronal Gene Expression in C. elegans. Neuron 87, 733–750. 10.1016/j.neuron.2015.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hobert O (2016). Terminal Selectors of Neuronal Identity. Curr Top Dev Biol 116, 455–475. 10.1016/bs.ctdb.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Qiu Z, and Ghosh A (2008). A brief history of neuronal gene expression: regulatory mechanisms and cellular consequences. Neuron 60, 449–455. 10.1016/j.neuron.2008.10.039. [DOI] [PubMed] [Google Scholar]
- 6.Hobert O, and Kratsios P (2019). Neuronal identity control by terminal selectors in worms, flies, and chordates. Curr Opin Neurobiol 56, 97–105. 10.1016/j.conb.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Bertrand N, Castro DS, and Guillemot F (2002). Proneural genes and the specification of neural cell types. Nat Rev Neurosci 3, 517–530. [DOI] [PubMed] [Google Scholar]
- 8.Kratsios P, Stolfi A, Levine M, and Hobert O (2011). Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat Neurosci 15, 205–214. 10.1038/nn.2989 nn.2989 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altun-Gultekin Z, Andachi Y, Tsalik EL, Pilgrim D, Kohara Y, and Hobert O (2001). A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development 128, 1951–1969. [DOI] [PubMed] [Google Scholar]
- 10.Dykes IM, Tempest L, Lee SI, and Turner EE (2011). Brn3a and Islet1 act epistatically to regulate the gene expression program of sensory differentiation. J Neurosci 31, 9789–9799. 10.1523/JNEUROSCI.0901-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leyva-Diaz E, Stefanakis N, Carrera I, Glenwinkel L, Wang G, Driscoll M, and Hobert O (2017). Silencing of Repetitive DNA Is Controlled by a Member of an Unusual Caenorhabditis elegans Gene Family. Genetics 207, 529–545. 10.1534/genetics.117.300134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reilly MB, Cros C, Varol E, Yemini E, and Hobert O (2020). Unique homeobox codes delineate all the neuron classes of C. elegans. Nature 584, 595–601. 10.1038/s41586-020-2618-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alkema MJ, Hunter-Ensor M, Ringstad N, and Horvitz HR (2005). Tyramine Functions independently of octopamine in the Caenorhabditis elegans nervous system. Neuron 46, 247–260. [DOI] [PubMed] [Google Scholar]
- 14.Burglin TR, and Affolter M (2016). Homeodomain proteins: an update. Chromosoma 125, 497–521. 10.1007/s00412-015-0543-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burglin TR, and Cassata G (2002). Loss and gain of domains during evolution of cut superclass homeobox genes. Int J Dev Biol 46, 115–123. [PubMed] [Google Scholar]
- 16.Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. (2013). DNA-binding specificities of human transcription factors. Cell 152, 327–339. 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 17.Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK, et al. (2018). The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res 46, D794–D801. 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen M, Alfonso A, Johnson CD, and Rand JB (1995). Caenorhabditis elegans mutants resistant to inhibitors of acetylcholinesterase. Genetics 140, 527–535. 10.1093/genetics/140.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nonet ML, Staunton JE, Kilgard MP, Fergestad T, Hartwieg E, Horvitz HR, Jorgensen EM, and Meyer BJ (1997). Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J Neurosci 17, 8061–8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Consortium, C.e.D.M. (2012). large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda) 2, 1415–1425. 10.1534/g3.112.003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Croll NA (1975). Behavioural analysis of nematode movement. Adv Parasitol 13, 71–122. [DOI] [PubMed] [Google Scholar]
- 22.Pierce-Shimomura JT, Chen BL, Mun JJ, Ho R, Sarkis R, and McIntire SL (2008). Genetic analysis of crawling and swimming locomotory patterns in C. elegans. Proc Natl Acad Sci U S A 105, 20982–20987. 0810359105 [pii] 10.1073/pnas.0810359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Restif C, Ibanez-Ventoso C, Vora MM, Guo S, Metaxas D, and Driscoll M (2014). CeleST: computer vision software for quantitative analysis of C. elegans swim behavior reveals novel features of locomotion. PLoS Comput Biol 10, e1003702. 10.1371/journal.pcbi.1003702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xuan Z, Manning L, Nelson J, Richmond JE, Colon-Ramos DA, Shen K, and Kurshan PT (2017). Clarinet (CLA-1), a novel active zone protein required for synaptic vesicle clustering and release. eLife 6. 10.7554/eLife.29276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, and Bargmann CI (2008). GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57, 353–363. 10.1016/j.neuron.2007.11.030 S0896–6273(07)01020–3 [pii]. [DOI] [PubMed] [Google Scholar]
- 26.Steiner FA, Talbert PB, Kasinathan S, Deal RB, and Henikoff S (2012). Cell-type-specific nuclei purification from whole animals for genome-wide expression and chromatin profiling. Genome Res 22, 766–777. 10.1101/gr.131748.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun H, and Hobert O (2021). Temporal transitions in the post-mitotic nervous system of Caenorhabditis elegans. Nature 600, 93–99. 10.1038/s41586-021-04071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, and Darnell RB (2000). Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 25, 359–371. 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 29.Ioannou MS, and Marat AL (2012). The role of EHD proteins at the neuronal synapse. Sci Signal 5, jc1. 10.1126/scisignal.2002989. [DOI] [PubMed] [Google Scholar]
- 30.Kohn RE, Duerr JS, McManus JR, Duke A, Rakow TL, Maruyama H, Moulder G, Maruyama IN, Barstead RJ, and Rand JB (2000). Expression of multiple UNC-13 proteins in the Caenorhabditis elegans nervous system. Mol Biol Cell 11, 3441–3452. 10.1091/mbc.11.10.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leyva-Diaz E, Masoudi N, Serrano-Saiz E, Glenwinkel L, and Hobert O (2020). Brn3/POU-IV-type POU homeobox genes-Paradigmatic regulators of neuronal identity across phylogeny. Wiley interdisciplinary reviews. Developmental biology, e374. 10.1002/wdev.374. [DOI] [PubMed] [Google Scholar]
- 32.Serrano-Saiz E, Poole Richard J., Felton T, Zhang F, De La Cruz Estanisla D., and Hobert O (2013). Modular Control of Glutamatergic Neuronal Identity in C. elegans by Distinct Homeodomain Proteins. Cell 155, 659–673. 10.1016/j.cell.2013.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pereira L, Kratsios P, Serrano-Saiz E, Sheftel H, Mayo AE, Hall DH, White JG, LeBoeuf B, Garcia LR, Alon U, and Hobert O (2015). A cellular and regulatory map of the cholinergic nervous system of C. elegans. eLife 4. 10.7554/eLife.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kagoshima H, Cassata G, Tong YG, Pujol N, Niklaus G, and Burglin TR (2013). The LIM homeobox gene ceh-14 is required for phasmid function and neurite outgrowth. Dev Biol 380, 314–323. 10.1016/j.ydbio.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Cinar H, Keles S, and Jin Y (2005). Expression profiling of GABAergic motor neurons in Caenorhabditis elegans. Curr Biol 15, 340–346. [DOI] [PubMed] [Google Scholar]
- 36.Yu B, Wang X, Wei S, Fu T, Dzakah EE, Waqas A, Walthall WW, and Shan G (2017). Convergent Transcriptional Programs Regulate cAMP Levels in C. elegans GABAergic Motor Neurons. Dev Cell 43, 212–226 e217. 10.1016/j.devcel.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 37.Lowe EK, and Stolfi A (2018). Developmental system drift in motor ganglion patterning between distantly related tunicates. Evodevo 9, 18. 10.1186/s13227-018-0107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen DN, Rohrbaugh M, and Lai Z (2000). The Drosophila homolog of Onecut homeodomain proteins is a neural-specific transcriptional activator with a potential role in regulating neural differentiation. Mech Dev 97, 57–72. 10.1016/s0925-4773(00)00431-7. [DOI] [PubMed] [Google Scholar]
- 39.Poustka AJ, Kuhn A, Radosavljevic V, Wellenreuther R, Lehrach H, and Panopoulou G (2004). On the origin of the chordate central nervous system: expression of onecut in the sea urchin embryo. Evol Dev 6, 227–236. 10.1111/j.1525-142X.2004.04028.x. [DOI] [PubMed] [Google Scholar]
- 40.Kropp PA, and Gannon M (2016). Onecut transcription factors in development and disease. Trends Dev Biol 9, 43–57. [PMC free article] [PubMed] [Google Scholar]
- 41.Weiss LA, and Nieto M (2019). The crux of Cux genes in neuronal function and plasticity. Brain Res 1705, 32–42. 10.1016/j.brainres.2018.02.044. [DOI] [PubMed] [Google Scholar]
- 42.Vassalli QA, Colantuono C, Nittoli V, Ferraioli A, Fasano G, Berruto F, Chiusano ML, Kelsh RN, Sordino P, and Locascio A (2021). Onecut Regulates Core Components of the Molecular Machinery for Neurotransmission in Photoreceptor Differentiation. Front Cell Dev Biol 9, 602450. 10.3389/fcell.2021.602450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Raadt J, van Gestel SHC, Nadif Kasri N, and Albers CA (2019). ONECUT transcription factors induce neuronal characteristics and remodel chromatin accessibility. Nucleic Acids Res 47, 5587–5602. 10.1093/nar/gkz273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hobert O (2021). Homeobox genes and the specification of neuronal identity. Nat Rev Neurosci 22, 627–636. 10.1038/s41583-021-00497-x. [DOI] [PubMed] [Google Scholar]
- 45.Machanick P, and Bailey TL (2011). MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697. 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roussel N, Sprenger J, Tappan SJ, and Glaser JR (2014). Robust tracking and quantification of C. elegans body shape and locomotion through coiling, entanglement, and omega bends. Worm 3, e982437. 10.4161/21624054.2014.982437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yemini E, Jucikas T, Grundy LJ, Brown AE, and Schafer WR (2013). A database of Caenorhabditis elegans behavioral phenotypes. Nat Methods 10, 877–879. 10.1038/nmeth.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahoney TR, Luo S, and Nonet ML (2006). Analysis of synaptic transmission in Caenorhabditis elegans using an aldicarb-sensitivity assay. Nat Protoc 1, 1772–1777. 10.1038/nprot.2006.281. [DOI] [PubMed] [Google Scholar]
- 49.Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarov M, Murray JI, Schanze K, Pozniakovski A, Niu W, Angermann K, Hasse S, Rupprecht M, Vinis E, Tinney M, et al. (2012). A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell 150, 855–866. 10.1016/j.cell.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dokshin GA, Ghanta KS, Piscopo KM, and Mello CC (2018). Robust Genome Editing with Short Single-Stranded and Long, Partially Single-Stranded DNA Donors in Caenorhabditis elegans. Genetics 210, 781–787. 10.1534/genetics.118.301532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. (2014). Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443. 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hobert O (2002). PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. BioTechniques 32, 728–730. [DOI] [PubMed] [Google Scholar]
- 54.Lloret-Fernandez C, Maicas M, Mora-Martinez C, Artacho A, Jimeno-Martin A, Chirivella L, Weinberg P, and Flames N (2018). A transcription factor collective defines the HSN serotonergic neuron regulatory landscape. eLife 7. 10.7554/eLife.32785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Serrano-Saiz E, Gulez B, Pereira L, Gendrel M, Kerk SY, Vidal B, Feng W, Wang C, Kratsios P, Rand JB, and Hobert O (2020). Modular Organization of Cis-regulatory Control Information of Neurotransmitter Pathway Genes in Caenorhabditis elegans. Genetics 215, 665–681. 10.1534/genetics.120.303206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 59.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Angeles-Albores D, RY NL, Chan J, and Sternberg PW (2016). Tissue enrichment analysis for C. elegans genomics. BMC Bioinformatics 17, 366. 10.1186/s12859-016-1229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gupta S, Stamatoyannopoulos JA, Bailey TL, and Noble WS (2007). Quantifying similarity between motifs. Genome Biol 8, R24. 10.1186/gb-2007-82-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Genes associated to CEH-48 and CEH-38 ChIP datasets. Related to Figure 2.
CEH-48 (Experiment: ENCSR844VCY, https://www.encodeproject.org/) and CEH-38 (Accession # modEncode_4800, http://www.modencode.org/) ChIP-seq datasets were used to identify genes associated to the peak regions. Those genes which promoter region (defined as from 5kb upstream of the transcription start site to 1kb downstream) overlapped with the ChIP-seq peaks were identified. A list of genes associated to CEH-48 peaks (4709 genes, Data S1A), genes associated to CEH-38 peaks (7795 genes, Data S1B), and genes associated to CEH-48 or CEH-38 peaks (9578 genes, Data S1C) are provided in separate tabs. The genes are sorted by WormBase Gene ID. Brief gene descriptions obtained from WormMine.
Data S2. Differentially expressed genes in CUT sextuple mutant. Related to Figure 5.
Comparison of neuronal-nuclei immunoprecipitated samples from wild-type and CUT sextuple mutant is conducted using DESeq2 59 to determine differentially expressed genes (DEG, 3834 genes have a padj < 0.05, Data S2A). The genes are sorted by padj from smallest to largest. baseMean is the average normalized read counts of all samples. log2FoldChange is calculated using the formula log2 (average read counts of mutant samples)/(average read counts of wild-type samples). A list of downregulated genes (genes with a log2FoldChange < 0; 2118 genes, Data S2B), upregulated genes (genes with a log2FoldChange > 0; 1716 genes, Data S2C), and differentially expressed genes that are found associated to CEH-48 or CEH-38 ChIP-seq peaks (1124 genes, Data S2D) are provided in separate tabs, with the genes sorted by WormBase Gene ID. Brief gene descriptions obtained from WormMine.
Data S3: Neuronally-enriched genes. Related to Figure 5.
Comparison of wild-type neuronal-nuclei immunoprecipitated (IP) samples to wild-type input (total nuclei) samples is conducted using DESeq2 to determine enrichment (12639 genes have a padj < 0.05, Data S3A). The genes are sorted by padj from smallest to largest. baseMean is the average normalized read counts of all IP and input samples. log2FoldChange is calculated using the formula log2 (average read counts of mutant samples)/(average read counts of wild-type samples). A list of neuronally-enriched genes (genes with a log2FoldChange > 0; 6372 genes, Data S3B), and neuronally-depleted genes (genes with a log2FoldChange < 0; 6267 genes, Data S3C) are provided in separate tabs, with the genes sorted by WormBase Gene ID. Brief gene descriptions obtained from WormMine.
Data S4. Full list of enrichment analysis GO and phenotype terms. Related to Figure 5.
Full list of enriched terms for GO enrichment analysis (Data S4A–B) and phenotype enrichment analysis (Data S4C–D) using gene sets of significantly downregulated or upregulated transcripts. The terms are sorted by q-value from smallest to largest. Analysis performed using the Gene Set Enrichment Analysis Tool from Wormbase.
Data S5. Aldicarb assay measurements mean and standard error. Related to Figure 2 and Figure 4.
Mean and standard error of the mean (SEM) values corresponding to the aldicarb data shown in Figure 2F (Data S5A) and Figure 4E–G (Data S5B).
Data S6. Known transcription factor motifs matching CEH-48 and CEH-38 motifs. Related to the STAR Methods.
Tomtom Motif Comparison Tool 61 output list of known motifs matching the CEH-48 motif defined in 52 (Data S6A), the CEH-48 motif extracted from the ChIP-seq dataset (Experiment: ENCSR844VCY, https://www.encodeproject.org/) using MEME-ChIP 45 (Data S6B), and the CEH-38 motif extracted from the ChIP-seq dataset (Accession # modEncode_4800, http://www.modencode.org/) using MEME-ChIP (Data S6C). These three motifs were compared with an eukaryote DNA motif database (vertebrates, in vivo and in silico) provided in MEME Suite 61 (JASPAR2018_CORE_vertebrates_non-redundant, uniprobe_mouse, and jolma2013; total 1808 known motifs). The motifs are sorted by q-value from smallest to largest.
Data Availability Statement
Raw and processed RNA-seq data will be available at GEO accession #GSE188489.
No original code has been generated for this paper.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.