Summary
Salt taste is one of the most ancient of all sensory modalities. However, the molecular basis of salt taste remains unclear in invertebrates. Here we show that the response to low, appetitive salt concentrations in Drosophila depends on Ir56b, an atypical member of the Ionotropic receptor (Ir) family. Ir56b acts in concert with two co-receptors, Ir25a and Ir76b. Mutation of Ir56b virtually eliminates an appetitive behavioral response to salt. Ir56b is expressed in neurons that also sense sugars via members of the Gr (Gustatory receptor) family. Misexpression of Ir56b in bitter-sensing neurons confers physiological responses to appetitive doses of salt. Ir56b is unique among tuning Irs in containing virtually no N-terminal region, a feature that is evolutionarily conserved. Moreover, Ir56b is a "pseudo-pseudogene": its coding sequence contains a premature stop codon that can be replaced with a sense codon without loss of function. This stop codon is conserved among many Drosophila species, but is absent in a number of species associated with cactus in arid regions. Thus, Ir56b serves the evolutionarily ancient function of salt detection, in neurons that underlie both salt and sweet taste modalities.
eTOC Blurb
Dweck et al. find that Ir56b is a salt tuning receptor that mediates an appetitive salt behavior. Ir56b is expressed in sugar-sensing neurons and acts together with the co-receptors Ir25a and Ir76b. Ir56b lacks a typical N-terminal region. Ir56b harbors a conserved premature stop codon, which is absent in cactophilic flies that live in the desert.
INTRODUCTION
Salt taste is the most widespread taste modality of the animal world. While cats have lost sweet sensation, pandas have lost a receptor for umami taste, and whales are believed to have lost four of the five basic tastes, salt taste appears indispensable across the animal kingdom 1-6. Sodium chloride is critical to many aspects of animal physiology, ranging from neuronal firing to the control of blood volume, and regulation of its intake is critical 2. Animals have evolved taste cells that detect NaCl, allow evaluation of its concentration, and drive feeding decisions 7-12.
In flies, as in many other animals, low salt concentrations are appetitive 10,12, but the receptor that detects low NaCl levels and underlies its ingestion has not been identified. Two Ionotropic receptors (IRs), Ir25a and Ir76b, have been found to be required for NaCl response 10,12,13, but both are widely expressed co-receptors that operate in concert with various individual tuning receptors in the response to many diverse sensory stimuli 10,12-17. A tuning receptor that confers sensitivity to low NaCl levels has not been identified.
The principal taste organ of the fly head, the labellum, contains 31 stereotyped taste sensilla in Drosophila melanogaster: these sensilla are large (L), intermediate (I), or small (S) 18. Each sensillum contains up to four taste neurons 19. Different subsets of taste neurons respond to sugars, bitter compounds or water (osmolarity) 5,20. Many taste neurons respond to high concentrations of NaCl, e.g. 1 M 10, although it is not clear how often flies encounter such high concentrations in nature. Many if not all of the fruits on which flies feed and breed contain 10 mM concentrations or less 21. To identify taste neurons on the labellum that respond to such low concentrations of NaCl we systematically tested all 31 taste sensilla via electrophysiology.
RESULTS AND DISCUSSION
Ir56b underlies salt-sensing
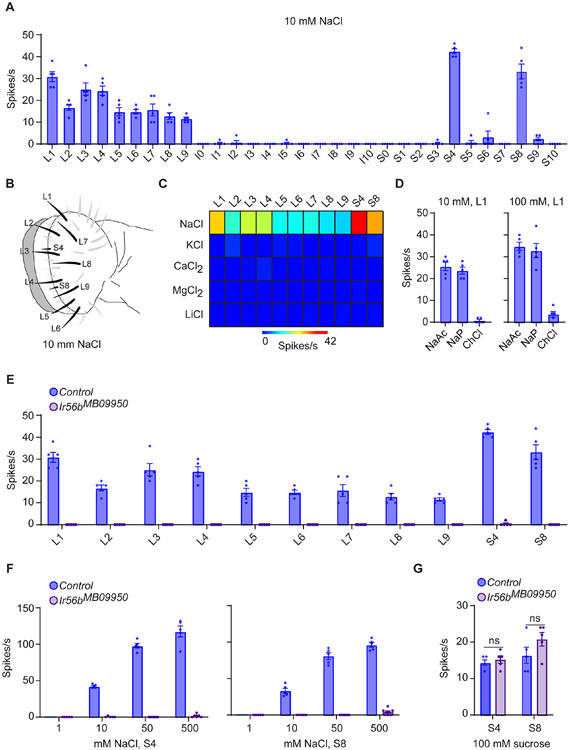
We identified 11 sensilla that gave robust responses to 10 mM NaCl: all nine of the L sensilla and two of the S sensilla, S4 and S8 (Figures 1A-1C; Data S1A and S1C). To determine the specificity of these salt responses, we tested KCl, CaCl2, MgCl2, and LiCl, again at 10 mM concentrations, and found no responses (Figure 1C; Data S1C). These results suggested that the NaCl responses are elicited by Na+ and not by Cl−. Consistent with this suggestion, two additional Na+ salts elicited strong responses from L1, and an additional chloride salt did not, even at a 100mM concentration (Figure 1D; Data S1D).
Figure 1. Response of labellar taste sensilla to salt.
(A) Responses of labellar sensilla to 10 mM NaCl, n = 5. Error bars are S.E.M. and are too small to be seen in some cases.
(B) Map of labellum; the labeled sensilla respond to 10 mM NaCl.
(C) Heatmap of responses of sensilla to salts, each tested at 10 mM. n = 5.
(D) Responses of L1 to other sodium and chloride salts. n = 5.
(E) Responses to 10 mM NaCl in control and Ir56bMB09950 flies. Mann-Whitney test; n =5. Values for control and mutant flies were measured in parallel; the control values are also shown in Figure 1A.
(F) Responses of S4 and S8 to NaCl. Mann-Whitney test; n = 5. The values for control flies to 10 mM NaCl are from Figure 1E.
(G) Responses of S4 and S8 to 100 mM sucrose. Mann-Whitney test; n = 5.
We noticed that exactly two Ir genes have been mapped to all 11 of these salt-sensitive sensilla among the Ir20a clade of ionotropic receptor genes: Ir47a and Ir56b 16. In an initial test of an existing Ir56b mutant, Ir56bMB09950, no responses to 10 mM NaCl were found in any of the 11 sensilla (Figure 1E; Data S1E). Thus the mutation eliminated response in all of the sensilla that respond to low salt concentrations. Further testing of two sensilla, S4 and S8, showed that responses were essentially eliminated across a broad range of concentrations (Figure 1F; Data S1F), although responses to sucrose were normal (Figure 1G; Data S1G).
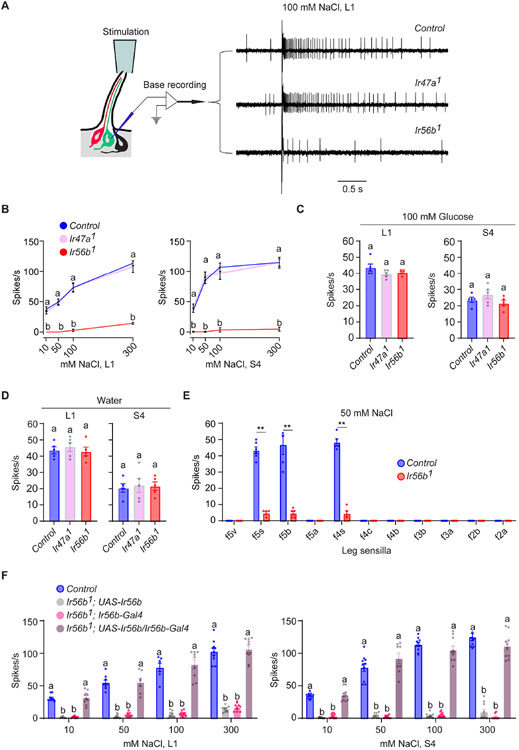
To further investigate the roles of Ir47a and Ir56b in salt reception, we created new alleles of both genes using CRISPR/Cas9 genome editing. We backcrossed each new allele six times to our control genetic background to minimize genetic background effects, and then tested them with a different recording technique. Conventional single-unit electrophysiology uses a single electrode both to deliver taste stimuli to the tip of the sensillum and to record the response. We used an alternative method of recording in which the stimulus is delivered to the tip of the sensillum and a tungsten recording electrode is inserted in the base of the sensillum (Figure 2A) 22. Because of the technical challenges of this preparation we focused on two sensilla, L1 and S4, whose positions offer convenient access.
Figure 2. Ir56b is required for salt sensing.
(A) Left, "base recording" electrophysiological method. Right, sample traces of recordings from L1. Control is w Canton-S (w CS).
(B) Responses to NaCl. One-way ANOVA followed by Tukey’s multiple comparison test; n = 5. Values indicated with different letters are significantly different. "a" applies to both Ir47a1 and Ir56b1. Control is w CS.
(C) Responses of L1 and S4 in control, Ir47a1, and Ir56b1 to 100 mM glucose. One-way ANOVA followed by Tukey’s multiple comparison test; n = 5. Error bars = S.E.M.
(D) Responses to water. One-way ANOVA followed by followed by Tukey’s multiple comparison test; n = 5.
(E) Responses of tarsal sensilla on leg to 50 mM NaCl. Mann-Whitney test; n = 5-6. **p<0.01.
(F) Responses of L1 and S4 in the indicated genotypes to NaCl. One-way ANOVA followed by Tukey’s multiple comparison test; n = 5. Values indicated with different letters are significantly different.
The Ir47a1 mutant gave a normal response, but the Ir56b1 mutant gave a severely reduced response across a broad range of concentrations in both L1 and S4 (Figures 2A and 2B; Data S2B). Both mutants gave normal responses to glucose (Figure 2C; Data S2C). Normal responses were also observed for a pure water stimulus (Figure 2D; Data S2D); water response can be measured directly with this base-recording technique but not with conventional tiprecording, because in tip-recording an electrolyte is required in the single electrode that is used to deliver the stimulus 22.
Ir56b has also been mapped to neurons in taste sensilla of the legs 16,17. We tested the four most distal tarsal segments of the female foreleg and found that three tarsal sensilla that express Ir56b, f5s, f5b, and f4s, responded to NaCl in control flies, consistent with previous observations 23,24. These responses were severely reduced in Ir56b1, indicating that Ir56b is also required for the response of tarsal sensilla to NaCl (Figure 2E; Data S2E).
To confirm that the physiological phenotype observed in Ir56b1 is indeed due to the loss of Ir56b, we carried out a rescue experiment. When two different Ir56b-GAL4 constructs were used to drive UAS-Ir56b, in two different Ir56b mutants, responses to NaCl were fully rescued across a broad range of concentrations, in both L1 and S4 sensilla (Figures 2F and S1; Data S2F). The responses of the parental lines were dramatically lower. Taken together, our results demonstrate that Ir56b is required for salt sensing.
Ir56b acts in concert with the co-receptors Ir25a and Ir76b
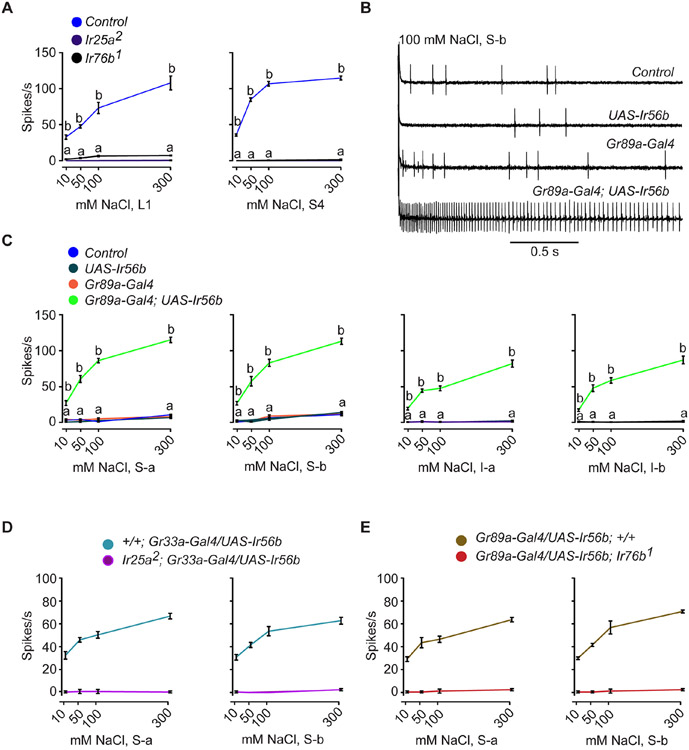
The broadly expressed Ir co-receptors Ir25a and Ir76b have previously been implicated in salt reception 10,12,13. We confirmed that both are required for physiological responses to NaCl across a broad range of concentrations in both L1 and S4 sensilla (Figure 3A; Data S3A). The phenotypes of both Ir25a and Ir76b mutants are similar to those of Ir56bMB09950 and Ir56b1, (Figures 1E and 2B), suggesting the possibility that Ir25a, Ir76b, and Ir56b act together to confer taste response to Na+.
Figure 3. Misexpression of Ir56b confers salt sensitivity to bitter neurons.
(A) Responses of L1 and S4 in control w Canton-S (w CS), Ir25a2, and Ir76b1 to NaCl. One-way ANOVA followed by Tukey’s multiple comparison test; n = 5. Values indicated with different letters are significantly different. Measurements were taken for all concentrations; values equal to zero are not visible as points. Error bars are S.E.M. and are too small to be seen in some cases. The values for control were from Figure 2B.
(B) Sample traces from L1 in the indicated genotypes presented with 100 mM NaCl. Control = w CS.
(C) Responses of the indicated classes of bitter neurons in the indicated genotypes to NaCl. Control = w CS. One-way ANOVA followed by Tukey’s multiple comparison test; n = 5. Values indicated with different letters are significantly different. "a" applies to all three of the control genotypes.
(D) Salt responses of S-a and S-b bitter neurons that ectopically express Ir56b in an Ir25a mutant. Gr33a-Gal4, rather than the Gr89a-Gal4 driver, was used to drive expression in the bitter neuron because the Gr89a-Gal4 insertion is located on the same chromosome as Ir25a2.
(E) Salt responses of S-a and S-b bitter neurons that ectopically express Ir56b in an Ir76b mutant.
To test the hypothesis that Ir56b functions together with the two Ir co-receptors, we expressed Ir56b in all four classes of bitter-sensing neurons of the labellum: S-a, S-b, I-a, and I-b 18. In wild type these bitter neurons express Ir25a and Ir76b, but not Ir56b 16,17. All of these bitter neurons also express the bitter taste receptor Gr89a (Gustatory receptor 89a) 18, and we used a Gr89a-GAL4 construct to drive Ir56b expression in them.
All four classes of bitter neurons responded to NaCl in a dose-dependent manner in flies expressing Ir56b, but not in control flies that do not express Ir56b (Figures 3B and 3C; Data S3C). In a control experiment, the bitter compound coumarin (COU) elicited comparable responses among all four genotypes in all four classes (Figure S2). These results demonstrate that ectopic expression of Ir56b confers NaCl response to the four classes of bitter neurons.
To determine whether the NaCl response conferred upon bitter neurons by Ir56b depends on Ir25a and Ir76b, we expressed Ir56b in the same four classes of bitter neurons, but in mutants lacking either Ir25a or Ir76b. We found that expression of Ir56b did not confer response to any concentration of NaCl in an Ir25a or Ir76b mutant background (Figures 3D and 3E; Data S3D and S3E). The simplest interpretation of these results is that Ir56b, Ir25a and Ir76b act together to allow response to NaCl.
Ir56b senses salt in sugar-sensing neurons
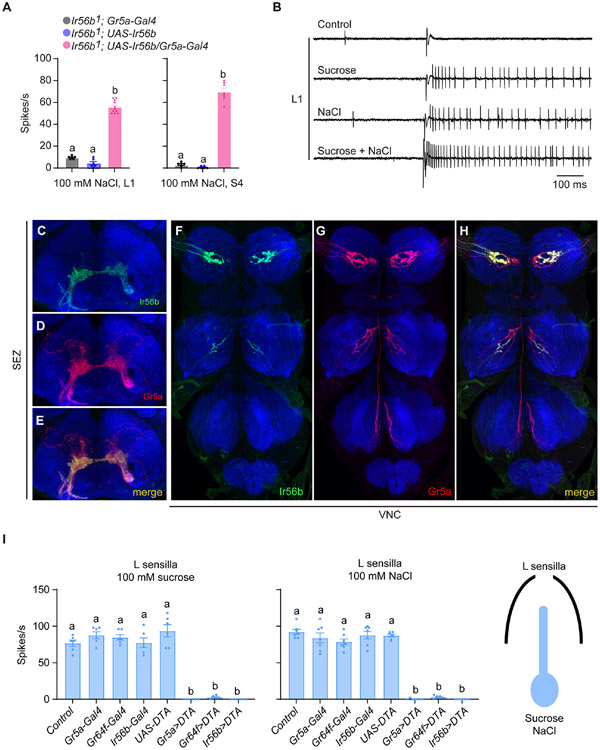
In what neuron does Ir56b operate in wild type? Labellar taste sensilla contain a neuron that responds to sugars via members of the Gustatory receptor (Gr) family 25,26. We first asked whether Ir56b can function in sugar-sensing neurons by driving expression of a UAS-Ir56b construct in the sugar-sensing neuron of mutant Ir56b1 sensilla. In both L1 and S4 sensilla, expression driven in sugar-sensitive neurons by either of two GAL4 constructs conferred NaCl response to the sensilla (Figure 4A and S3A; Data S4A).
Figure 4. Ir56b is expressed in a subset of sugar-sensitive neurons.
(A) Responses of L1 and S4 in the indicated genotypes to 100 mM NaCl. One-way ANOVA followed by Tukey’s multiple comparison test; n = 6-11. Values indicated with different letters are significantly different.
(B) Sample traces of electrophysiological recordings from L1 in control flies presented with diluent control (30 mM TCC), 50 mM sucrose, 50 mM NaCl, and mixture of 50 mM NaCl and 50 mM sucrose. Sucrose, NaCl, and the mixture were all dissolved in 30 mM TCC.
(C-E) Projection patterns of Ir56a-GAL4- and Gr5a-LexA-expressing neurons in the suboesophageal ganglion (SEZ).
(F-H) Projection patterns of Ir56a-GAL4- and Gr5a-LexA-expressing neurons in the ventral nerve cord (VNC).
(I) Expression of diphtheria toxin under the control of Gr5a-, Gr64f-, or IR56b-Gal4 drivers in L sensilla severely reduced response to both sucrose and NaCl. One-way ANOVA followed by Tukey's multiple comparison test; n=6-7. Values indicated by different letters are different. p<0.05.
If the sugar-sensing neuron of these sensilla is in fact the same neuron as the salt-sensing neuron, then we would expect the action potentials produced by both sugar and salt to be of the same amplitude. We first tested mixtures of sucrose and salt on L sensilla, which give robust sucrose responses and account for most of the labellar sensilla that respond to low salt concentrations (Figure 1). We found that stimulation of L sensilla with a mixture of sucrose and NaCl produced a train of action potentials of uniform amplitude (Figures 4B and S3B). Similar results were found in salt-sensitive sensilla of the leg (Figure S3C). The response of L1 to a mixture (62 spikes/s ± 4 spikes/s for a mixture of 50 mM sucrose and 50 mM NaCl) was greater than the response to either stimulus alone (48 spikes/s ± 2 spikes/s for 50 mM NaCl; 43 spikes/s ± 2 spikes/s for 50 mM sucrose; p<0.001, one-way ANOVA, n=5), but less than the sum of the two individual responses (91 spikes/s ± 2 spikes/s; p<0.01, Mann-Whitney), supporting the conclusion that the two stimuli activate the same neuron. Moreover, if two distinct neurons were producing spikes of the same amplitude, we would expect to observe some spikes of larger amplitude due to summation of coincident spikes; these were not observed. Nor did we observe closely spaced "doublet" spikes, which would also be observed if two distinct neurons were firing.
We have found that all or almost all cells labeled by Ir56b-GAL4 are also labeled in the labellum by Gr5a-LexA, a marker of sugar neurons; co-labeling was also observed in the leg (Koh et al., 2014; Figures S3D-S3I). We confirmed and extended these results by examining the CNS projections of neurons labeled by Ir56b and Gr5a drivers. We again found overlap, in both the subesophageal zone (Figures 4C-4E) and the ventral nerve cord (Figures 4F-4H).
Finally, we found that ablation of sugar cells in the L sensilla with drivers of either of two sugar receptor genes eliminated response to both sugar and salt; likewise, ablation of salt-sensing cells with the Ir56b driver eliminated response to both sugar and salt (Figure 4I; Data S4I). The simplest interpretation of all these results is that Ir56b confers salt response primarily if not exclusively in neurons that respond to sugars via Gr receptors.
Appetitive response to salt depends on Ir56b
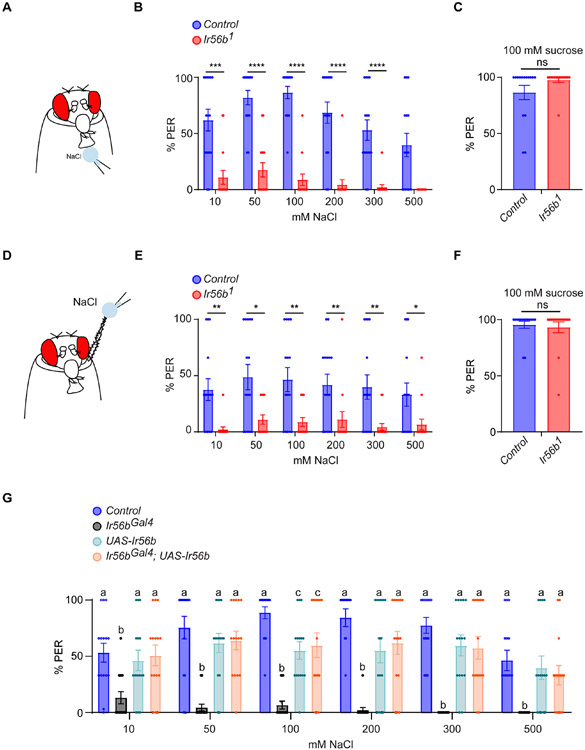
Next we tested the role of Ir56b in the appetitive response to salt using the proboscis extension response (PER) paradigm, in which a fly extends its proboscis in response to contact with taste stimuli (Figure 5A). Control flies displayed strong PER responses across a broad range of salt concentrations applied to the labellum, consistent with a previous report 27. By contrast, Ir56b1 mutants showed severely diminished responses (Figure 5B; Data S5B). PER responses to a sugar stimulus were normal in the mutant (Figure 5C; Data S5C). The same results were obtained when salt stimuli were applied to the legs (Figures 5D-5F; Data S5F).
Figure 5. Ir56b is required for appetitive behavioral responses to NaCl.
(A) The labellar PER assay. A NaCl stimulus is presented to the labellum; the percentage of stimulus presentations that produce a proboscis extension is indicated.
(B) Labellar PER responses in control and Ir56b1 to NaCl. Mann-Whitney test; n =15. Error bars = S.E.M.
(C) Labellar PER responses to 100 mM sucrose. Mann-Whitney test; n =15.
(D) Leg PER assay. The stimulus is presented to the tarsal segments of the foreleg.
(E) Leg PER responses to NaCl. Mann-Whitney test; n =15.
(F) Leg PER responses in control and Ir56b1 to 100 mM sucrose. Mann-Whitney test; n =15.
(G) Labellar PER responses to NaCl in the indicated genotypes. One-way ANOVA followed by Tukey’s multiple comparison test; n = 15. Values indicated with different letters are significantly different. See also Table S2.
To confirm that the PER defect in fact arose from the loss of Ir56b, we repeated the experiment using an independent allele, Ir56bGAL4, which we constructed by inserting a GAL4 transcription factor gene within Ir56b by CRISPR/Cas9 genome editing, and then backcrossing this allele five generations to a control strain. The Ir56bGAL4 mutant again showed a reduced response to NaCl (Figure 5G; Data S5G). The phenotype was rescued when the Ir56bGAL4 construct was allowed to drive expression of a UAS-Ir56b transgene.
Ir56b has a conserved function
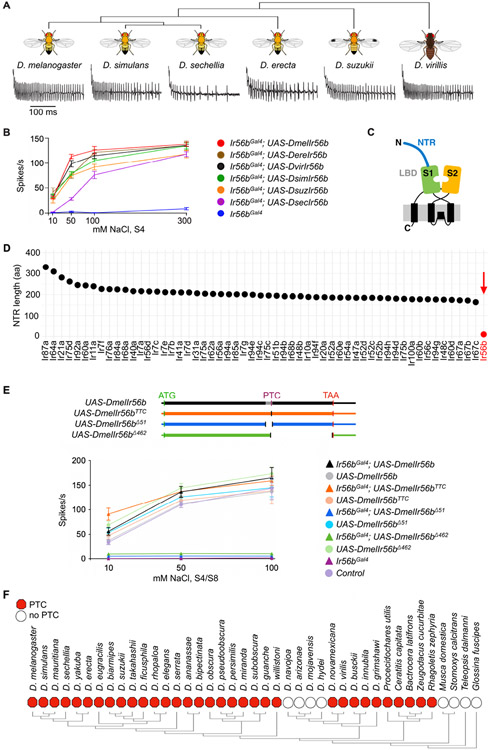
We then asked whether a functional role for Ir56b in salt reception has been conserved through evolution. We generated UAS-Ir56b constructs from five other species that diverged from D. melanogaster at times ranging from 2-3 million years ago (D. simulans) to 40 million years ago (D. virilis) (Figure 6A) 28-33. All constructs were capable of restoring salt response to the Ir56bGAL4 mutant of D. melanogaster (Figures 6A and 6B; Data S6B). Interestingly, however, the D. sechellia transgene conferred a less sensitive response than the other alleles; it responded only at concentrations higher than 10 mM. D. sechellia has adapted to feed and breed on a single fruit, the noni fruit of Morinda citrifolia 34, and its need to detect and evaluate salt concentration may differ from that of other Drosophila species.
Figure 6. Ir56b has a conserved function and an atypical structure.
(A) Electrophysiological traces from D. melanogaster IR56bGal4; UAS-DxIr56b, where x represents each of the indicated species. Recordings were from S4 sensilla tested with 50 mM NaCl.
(B) Responses of S4 sensilla to NaCl in each of the indicated genotypes. n= 5-10. Error bars = S.E.M.
(C) Diagram of tuning IRs, showing N-terminal region (NTR), and the S1 and S2 half-domains of the LBD. Adapted from Abuin et al. (2019).
(D) Length of the N-terminal regions of all tuning IRs (all Irs except Ir8a, Ir25a, Ir76b, and Ir93a, which are considered co-receptors17).
(E) The variant D. melanogaster Ir56b UAS constructs tested (top). All constructs include untranslated regions (thin boxes), the start codon (ATG), and the normal termination codon (TAA). The UAS-DmelIr56bTTC construct lacks the premature termination codon (PTC) and replaces it with a TTC codon, the UAS-DmelIr56bΔ51 construct lacks the annotated intron (grey), and the UAS-DmelIr56bΔ462 construct lacks the coding sequence from the PTC until the TAA, which it includes. Responses of S4/S8 sensilla to NaCl in each of the indicated genotypes (bottom). n= 5-10. Error bars = S.E.M.
(F) Presence (red hexagon) or absence (empty circle) of the PTC in the 41 IR56b orthologs identified through BLAST searches.
See also Figures S4-S6, Table S1 and Table S2.
Ir56b has an atypical structure and is encoded by a pseudo-pseudogene
Tuning Irs are predicted to include an N-terminal region (NTR) and an extracellular ligand-binding domain (LBD) 35 (Figure 6C). The LBD contains two half-domains, S1 and S2, that together form a "Venus flytrap". Ligand binding leads to currents that are carried by Na+ and other cations 36,37. Given that Ir56b has an atypical function, i.e. signaling the presence of a cation that it uses for conduction, we wondered whether its structure might also be atypical. Interestingly, Ir56b has virtually no N-terminal region (NTR) (Figure 6D and Table S1). Among 56 Irs considered as tuning IRs in D. melanogaster, 55 have an NTR ranging from 164 to 331 amino acids in length. Ir56b, by contrast, has an NTR of only 11 amino acids. Moreover, the severely shortened NTR of Ir56b is conserved among all of 40 Ir56b orthologs examined (Figure S4).
The genome annotation of Ir56b in D. melanogaster indicates a 51 bp intron (Figure S5A, asterisk) that is unusual in three ways: i) its degree of sequence conservation is similar to that of Ir56b coding sequences (CDS) (Figures S5A and S5B); ii) the putative splice sites show extremely low prediction scores: 0.01 and 0 on a scale from 0 to 1 (Figure S5C); iii) its GC content is similar to that of the coding sequences (Figure S5D). We found that the annotated intronic sequences are in fact retained in Ir56b transcripts, as determined by RNAseq analysis (Fig. S5E), by RT-PCR analysis of six species (Figure S5F), and by sequence analysis of the UAS-Ir56b constructs derived from these species, all of which encoded a single-exon transcript (Figure S5G).
It was surprising that this annotated 51 bp sequence is in fact included in the Ir56b transcripts of all six species because it contains a premature termination codon (PTC) that would be predicted to truncate the receptor in the S2 domain and render the receptor non-functional (Figure S6A). However, all six of the constructs, despite containing the PTC, encode functional Ir56b (Figure 6B). By contrast, we engineered a UAS construct in which the 51 bp sequence was removed (UAS-Ir56bΔ51) and found that it does not express a functional Ir56b protein (Figure 6E; Data S6E), consistent with our hypothesis that the 51 bp annotated intron in fact contains essential coding sequences.
Translational readthrough of a PTC has been reported in an olfactory receptor gene of D. sechellia, referred to as a "pseudo-pseudogene"; readthrough of a PTC has also been found in a few olfactory receptor genes of individual strains of D. melanogaster 38 . Unlike those cases, however, the Ir56b PTC is conserved across 28 Drosophila species of 32 analyzed, as well as 5 other dipteran species among 9 analyzed (Figures 6F and S6B). In all of the Ir56b genes with PTCs, the stop codon is TGA followed by a C nucleotide, which is considered to be the "leakiest" termination codon 39. The exceptional Drosophila species without PTCs are D. mojavensis, D. arizonae, D. navojoa, and D. hydei, members of the D. repleta group that is associated with cactus 40.
To test the possibility of translational readthrough in Ir56b, we generated a UAS-Ir56b construct in which the PTC is replaced by a TTC codon (UAS-Ir56bTTC), which is found in place of the PTC in most of the cactophilic Drosophila species, and which encodes phenylalanine (Figure S6B). We also generated a UAS-Ir56b construct in which the sequence from the PTC until the last codon was deleted (UAS-Ir56bΔ462). The UAS-Ir56bTTC construct rescued the salt response of the Ir56bGal4 mutant to a similar extent as the wild type UAS-Ir56b construct (Figure 6E). Conversely, the UAS-Ir56bΔ462 construct did not rescue the salt response of the Ir56bGal4 mutant, consistent with our hypothesis that the Ir56b PTC is read through.
Readthrough may occur when a near-cognate tRNA inserts an amino acid, and its efficiency varies across cell type and conditions 41,42. We speculate that the conserved Ir56b PTC could provide a mechanism that allows the level of Ir56b to be modulated by environmental conditions such as salinity. Such a mechanism could have been lost in the cactophilic species that evolved in arid regions and that may have experienced a narrower range of salt concentrations or may have special needs to maintain salt balance 40.
Here we have demonstrated that Ir56b underlies the response to ecologically relevant concentrations of salt, which may be a limiting resource in many environments. Ir56b depends on the essential co-receptors Ir25a and Ir76b and is expressed in neurons that also sense sugars, consistent with a role in appetitive taste. Thus, receptors of two distinct families, Irs and Grs, are co-expressed in a subset of taste neurons, where they underlie two distinct taste modalities.
This coding logic is fundamentally distinct from that in mammals. In mice, the epithelial sodium channel (ENaC) acts as the receptor for low salt concentrations, and knocking out its α subunit eliminates both physiological responses and attractive behavioral responses to low concentrations 6,7,43. These ENaC-expressing cells are distinct from those that sense sugar and other taste modalities 6. Thus the salt and sugar in food sources are encoded by different circuits in mice but a common circuit in flies 5,44. This fly circuit also mediates responses to other appetitive taste stimuli, including fatty acids, glycerol, and acetic acid 45-48. These fly neurons thus signal the presence of appetitive stimuli representing a wide variety of chemical identities.
Other neurons and other receptors also respond to salt at high concentrations in both flies and mammals 5. A recent study found that Ir56b was not required for an aversive response to high concentrations of salt, consistent with its expression in sugar neurons; however, it was reported to act in an aversive response to zinc, which invites further investigation into the role of this gene in fly taste 49.
Here we have found that mutation of Ir56b virtually eliminates an appetitive response to the low concentrations of NaCl that the fly encounters in food sources. Thus Ir56b, a member of the ionotropic receptor family, serves in Drosophila one of the most evolutionary ancient of all sensory functions.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, John R. Carlson (john.carlson@yale.edu).
Materials availability
All reagents generated in this study are available from the Lead Contact without restriction.
Data and code availability
This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila stocks
Flies were reared on corn syrup and soy flour culture medium (Archon Scientific) at 25°C and 60% relative humidity in a 12:12-hour light–dark cycle. Ir56bMB09950 (LB27818) was obtained from the Bloomington Drosophila Stock Center, as was Ir56b-GAL4 (#60707). D. biarmipes (14023-0361.04), D. simulans (14021-0251.001), D. sechellia (14021-0248.27) and D. erecta (14021-0224.01) were obtained from the Drosophila Species Stock Center. The D. suzukii stock was collected in Connecticut.
METHOD DETAILS
Transgenic flies
Ir56b and Ir47a deletions were generated using CRISPR/Cas9 homologous recombination. Guide sequences were cloned into pCFD5 (Addgene 73914) using Gibson assembly (New England BioLabs) to create Ir56b-gRNA-pCFD5. Homology-driven repair template cloning was constructed by incorporation of homology arms and Gal4 into multiple cloning sites of the pHD-DsRed-attP vector (Addgene 51019). (Restriction enzymes used were as follows: left arm, EcoRI/XbaI; Gal4, XbaI/NdeI; right arm, SapI; all were purchased from New England BioLabs) to generate Ir56b-HomologyArms-pHD-DsRed-attP. Homology arms and Gal4 sequences were amplified by PCR using Q5 (New England BioLabs); all primers are provided in Table S2. The guide RNA and donor plasmids were injected into embryos by Bestgene, Inc. (Chino Hills, CA). DsRed positive alleles were then backcrossed to our control w1118 line for five generations.
Ir56b− Gal4 line, referred to as Ir56bGAL4: the Gal4 core promoter fragment was amplified by PCR with Q5 and inserted in the homology-driven repair template targeting the Ir56b locus using Gibson assembly (New England BioLabs) to generate Ir56b-HomologyArms-Gal4-pHD-DsRed-attP. Primers are provided in Table S2. The Ir56b-gRNA-pCFD5 and Ir56b-HomologyArms-pHD-DsRed-attP plasmids were injected into embryos by Bestgene, Inc. (Chino Hills, CA). DsRed positive alleles were then backcrossed to our control w1118 line.
UAS lines: The Ir56b gene was amplified from genomic DNA from different species by PCR using Q5 and incorporated in pUAST-attB-QS (Addgene 24366) plasmid using Gibson assembly to create DmelIr56b-gene-pUAST-attB-QS, DsimIr56b-gene-pUAST-attB-QS, DsecIr56b-gene-pUAST-attB-QS, DereIr56b-gene-pUAST-attB-QS, DsuzIr56b-gene-pUAST-attB-QS, and DvirIr56b-gene-pUAST-attB-QS. Finally, DmelIr56b-gene-TTC-pUAST-attB-QS, DmelIr56b-gene-Δ51-pUAST-attB-QS, and DmelIr56b-gene-Δ462-pUAST-attB-QS constructs were generated by mutating DmelIr56b-gene-pUAST-attB-QS using Q5-mutagenesis (New England BioLabs). Primers are provided in Table S2. The plasmids were injected into embryos by Bestgene, Inc. (Chino Hills, CA).
Tastants
All tastants were obtained at the highest available purity. All tastants were dissolved in 30 mM tricholine citrate (TCC), an electrolyte that inhibits the water neuron, until otherwise indicated. All tastants were prepared fresh and used for no more than one day.
Tip-recording technique
The tip recording technique was used in Figures. 1, 2E, 3B-E, 6A,B, S1, S2, and S3B. 5–7d old mated female flies were used. Flies were immobilized in pipette tips, and the labellum or the female foreleg was placed in a stable position on a glass coverslip. A reference tungsten electrode was inserted into the eye of the fly. The recording electrode consisted of a fine glass pipette (10–15μm tip diameter) and connected to an amplifier with a silver wire. This pipette performed the dual function of recording electrode and container of the stimulus. Recording started the moment the glass capillary electrode was brought into contact with the tip of the sensillum. Signals were amplified (10x; Syntech Universal AC/DC Probe; www.syntech.nl), sampled (10,667 samples/s), and filtered (100–3000 Hz with 50/60-Hz suppression) via a USB-IDAC connection to a computer (Syntech). Action potentials were extracted using Syntech Auto Spike 32 software. Responses were quantified by counting the number of spikes generated over a 500 ms period after contact. Response to the TCC diluent was not subtracted.
No more than one dose of a given tastant was tested on an individual sensillum of a given fly, with 2–3 minutes between presentations. Sensilla on both sides of the labellum were tested.
Base-recording technique
The base-recording technique was used in Figures. 2A-D, 2F, 3A, and 4A,B. Female flies, 5–7d old, were immobilized in pipette tips, and the labellum was placed in a stable position on a glass coverslip. A reference tungsten electrode was inserted into the eye of the fly. A recording tungsten electrode was inserted at the base of a taste sensillum. Sensilla on the left half of the labellum were tested. Stimuli were dissolved in 30 mM tricholine citrate (TCC), an electrolyte that inhibits the water neuron, and delivered in a glass capillary to the tip of a sensillum using a motorized micromanipulator (EC1 60-0571 standard motorized control micromanipulator, Harvard Apparatus). Signals were amplified (10x; Syntech Universal AC/DC Probe; http://www.syntech.nl), sampled (10,667 samples/s), and filtered (100–3000 Hz with 50/60-Hz suppression) via a USB-IDAC connection to a computer (Syntech). Action potentials were extracted using Syntech Auto Spike 32 software. Responses were quantified by counting the number of spikes generated over a 500 ms period after contacting the tip of a sensillum with the stimulus-containing glass capillary.
Immunohistochemistry and confocal imaging
CNS and labellar dissections were performed as described previously 50 with minor modifications. Briefly, flies incubated at 25°C that were ~7 days old were cold-anaesthetized on ice, then dipped into 100% ethanol in an effort to make their cuticles less hydrophobic. Flies were then dissected in cold PBS. Fixation was in 2% PFA in PBS for 55 minutes. After fixation, samples were washed 4 times, 15 minutes each in 0.3% PBST (0.3% Triton-X in PBS) at room temperature, then blocked in 5% Western Blocking Solution (Roche, #11921673001) in 0.3% PBST for at least 1.5 hours. Samples were then incubated with primary antibodies diluted in 0.3% PBST at 4°C for 2 days, washed 4 times, 15 minutes each in 0.3% PBST at room temperature, and incubated with secondary antibodies for another 2 days in darkness.
Before mounting, samples were balanced in SlowFade Gold antifade reagent (ThermoFisher, S36937) for 1 hour. Then samples were mounted on a slide (ThermoFisher Superfrost Plus, 4951PLUS4) using SlowFade Gold antifade reagent.
Antibodies used in this study were: chicken anti-GFP (Abcam, ab13970, 1:1000), rabbit anti-RFP (TakaraBio, 632496, 1:500), mouse anti-Bruchpilot (DSHB, 1:20), donkey anti-mouse AF647 (Invitrogen, 1900251, 1:1,000), donkey anti-rabbit AF568 (Invitrogen, A-11011,1:1,000), and goat anti-chicken AF488 (Abcam, ab150169, 1:1,000). The labeling was done on w, Gr5a-LexA; +; UAS-mCD8GFP, LexAop-mtdTomato/Ir56b-GAL4 flies. Leg samples were not stained; raw fluorescence images of GFP and mtdTomato were taken directly by mounting legs in the same antifade mountant.
A series of overlapping tiled Z-stack images were taken with a 40X oil objective using a Zeiss LSM880 confocal microscope, at 1μm intervals. Images were then stitched using ZEN software.
Proboscis extension reflex (PER) assay
PER assays were carried out as described in Sloane et al. 51 and Ahn et al. 45 with some modifications. Briefly, flies were collected on the day of eclosion and kept on standard corn meal food for 3–5 days at 25°C. Before performing PER assays, mated female flies were starved for 24 hr at 25°C in vials with water-saturated kimwipes. Flies were then mounted inside pipette tips and allowed to recover for 30 min at room temperature. Before the PER assay, flies were allowed to drink water until satiation to ensure that PER responses were derived from nutrients. Flies that did not respond to water were excluded (~5-10%). Taste solutions were delivered with a 10 μl pipette to the labellum or the tarsal segments of the female foreleg for up to ~4 s. Each fly was tested three times with one individual taste solution, and flies were allowed to drink water between each new application. A PER response was recorded as positive (1) if the proboscis was fully extended, otherwise it was recorded as negative (0). PER response scores (%) from a single fly were 0% (0/3 responses in the three applications), 33% (1/3), 66% (2/3) or 100% (3/3). The scoring of the different genotypes was performed blind to genotype.
Bioinformatics
Ir56b mRNA and protein sequences were identified using BLASTN within the NCBI nucleotide collection which includes GenBank, EMBL, DDBJ, PDB, and RefSeq sequences 52. Sequence alignment was performed using Clustal Omega 53 and visualized with Mview 54.
The N-terminus region length was determined by generating a homology-based model for each IR using SWISS-MODEL55 and identifying the region outside of the predicted S1 lobe. Splice site prediction scores were estimated using NNSPLICE v0.9 (https://www.fruitfly.org/seq_tools/splice.html).
RNA purification and RT-PCR
Labella were hand-dissected on ice from about 100 animals and immediately dropped in lysis buffer (RTL buffer, Qiagen). After tissue disruption, RNA was extracted using hot acid phenol. DNA was removed using DNase-Zero (Lucigen), cDNA was generated with Episcript (Lucigen), and PCR was carried out with Apex Taq (Genesee Science). Primers used are provided in Table S2.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical tests were performed in GraphPad Prism (version 6.01). All error bars are SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Material
Data S1. Response of labellar taste sensilla to salt. Related to STAR Methods and Figure 1.
A) Responses of labellar sensilla to 10 mM NaCl.
C) Responses of sensilla to salts, each tested at 10 mM.
D) Responses of L1 to other sodium and chloride salts.
E) Responses to 10 mM NaCl in control and Ir56bMB09950 flies.
F) Responses of S4 and S8 to NaCl.
G) Responses of S4 and S8 to 100 mM sucrose.
Data S2. Response of Ir47a1, and Ir56b1 to NaCl. Related to STAR Methods and Figure 2.
B) Responses of L1 and S4 to NaCl.
C) Responses of L1 and S4 in control, Ir47a1, and Ir56b1 to 100 mM glucose.
D) Responses of L1 and S4 to water.
E) Responses of tarsal sensilla on leg to 50 mM NaCl.
F) Responses of L1 and S4 in the indicated genotypes to NaCl.
Data S3. Response of the indicated genotypes to NaCl. Related to STAR Methods and Figure 3.
A) Responses of L1 and S4 in control w Canton-S (w CS), Ir25a2, and Ir76b1 to NaCl.
C) Responses of the indicated classes of bitter neurons in the indicated genotypes to NaCl.
D) Salt responses of S-a and S-b bitter neurons that ectopically express Ir56b in an Ir25a mutant.
E) Salt responses of S-a and S-b bitter neurons that ectopically express Ir56b in an Ir76b mutant.
Data S4. Response of the indicated genotypes to sucrose and NaCl. Related to STAR Methods and Figure 4.
A) Responses of L1 and S4 in the indicated genotypes to 100 mM NaCl.
I) Expression of diphtheria toxin under the control of Gr5a-, Gr64f-, or IR56b-Gal4 drivers in L sensilla severely reduced response to both sucrose and NaCl.
Data S5. Behavioral responses to NaCl. Related to STAR Methods and Figure 5.
B) Labellar PER responses in control and Ir56b1 to NaCl.
C) Labellar PER responses to 100 mM sucrose.
E) Leg PER responses to NaCl.
F) Leg PER responses in control and Ir56b1 to 100 mM sucrose.
G) Labellar PER responses to NaCl in the indicated genotypes.
Data S6. Response of the indicated genotypes to NaCl. Related to STAR Methods and Figure 6.
B) Responses of S4 sensilla to NaCl in each of the indicated genotypes.
E) Responses of S4/S8 sensilla to NaCl in each of the indicated genotypes.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant proteins | ||
| Sodium chloride (NaCl) | Millipore Sigma | Cat# S7653 |
| Potassium chloride (KCl) | Millipore Sigma | Cat# P9333 |
| Calcium chloride (CaCl2) | Millipore Sigma | Cat# 499609 |
| Magnesium chloride (MgCl2) | Millipore Sigma | Cat# M8266 |
| Lithium chloride (LiCl) | Millipore Sigma | Cat# 203637 |
| Sodium acetate (NaAc) | Millipore Sigma | Cat# 241245 |
| Sodium phosphate monobasic dihydrate (NaP) | Millipore Sigma | Cat# 71505 |
| Choline chloride (ChCl) | Millipore Sigma | Cat# C7017 |
| Sucrose | Millipore Sigma | Cat# S7903 |
| D-Glucose | Millipore Sigma | Cat# NIST917C |
| Coumarin (COU) | Millipore Sigma | Cat# C4261 |
| EcoRI | New England BioLabs | Cat# R0101S |
| XbaI | New England BioLabs | Cat# R0145S |
| NdeI | New England BioLabs | Cat# R0111S |
| SapI | New England BioLabs | Cat# R0569S |
| Gibson assembly | New England BioLabs | Cat# E5510S |
| Q5 | New England BioLabs | Cat# M0543S |
| Q5 site-directed mutagenesis kit | New England BioLabs | Cat# E0554S |
| RLT buffer | Qiagen | Cat # 79216 |
| Apex Taq | Genesee Science | Cat #: 42-138 |
| Episcript | Lucigen | ERT12925K |
| Baseline-ZERO DNase | Lucigen | DB0715K |
| Blocking Solution | Roche | Cat# 11921673001 |
| SlowFade Gold antifade | Thermo Fisher | Cat# S36937 |
| Antibodies | ||
| chicken anti-GFP | Abcam | Cat # ab13970 |
| Rabbit anti-RFP | TakaraBio | Cat #632496 |
| mouse anti-Bruchpilot | DSHB | Cat# nc82 |
| donkey anti-mouse AF647 | Invitrogen | Cat #1900251 |
| donkey anti-rabbit AF568 | Invitrogen | Cat # A-11011 |
| goat anti-chicken AF488 | Abcam | Cat # ab150169 |
| Experimental models: Organisms/strains | ||
| D. melanogaster: Ir56bMB09950 | Bloomington Drosophila Stock Center | Stock# 27818 |
| D. melanogaster: wCS; Ir47a1 | Figure 2 | N/A |
| D. melanogaster: wCS; Ir56b1 | Figure 2 | N/A |
| D. melanogaster: wCS; UAS-Ir56b | Figures 2 and 3 | N/A |
| D. melanogaster: wCS; Ir56b-Gal4 | Dr. John Carlson’s lab | Koh et al., 2014 |
| D. melanogaster: wCS; Ir25a2 | Bloomington Drosophila Stock Center | Stock# 41737 |
| D. melanogaster: Ir76b1 | Bloomington Drosophila Stock Center | Stock# 51309 |
| D. melanogaster: wCS; Gr89a-Gal4 | Dr. John Carlson’s lab | Weiss et al., 2011 |
| D. melanogaster: Gr5a-Gal4 | Bloomington Drosophila Stock Center | Stock# 57592 |
| D. melanogaster: w; Gr5a-LexA; UAS-mCD8GFP LexAop-mtdTomato | Dr. John Carlson’s lab | Koh et al., 2014 |
| D. melanogaster: wCS; Gr64f-Gal4 | Dr. John Carlson’s lab | Dahanukar et al., 2007 |
| D. melanogaster: UAS-DTA | Bloomington Drosophila Stock Center | Stock# 25039 |
| D. melanogaster: wCS; Ir56bGal4 | Figure 5 | N/A |
| D. melanogaster: UAS-DmelIr56b | Figure 6 | N/A |
| D. melanogaster: UAS-DereIr56b | Figure 6 | N/A |
| D. melanogaster: UAS-DvirIr56b | Figure 6 | N/A |
| D. melanogaster: UAS-DsimIr56b | Figure 6 | N/A |
| D. melanogaster: UAS-DsecIr56b | Figure 6 | N/A |
| D. melanogaster: UAS-DsuzIr56b | Figure 6 | N/A |
| D. melanogaster: UAS-DmelIr56bTCC | Figure 6 | N/A |
| D. melanogaster: UAS-DmelIr56bΔ51 | Figure 6 | N/A |
| D. melanogaster: UAS-DmelIr56bΔ462 | Figure 6 | N/A |
| Software and algorithms | ||
| AutoSpike 32 software | Syntech | http://www.ockenfels-syntech.com/ |
| BLASTN | NCBI | https://blast.ncbi.nlm.nih.gov/ |
| Clustal Omega | EMBL-EBI | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| Mview | EMBL-EBI | https://www.ebi.ac.uk/seqdb/confluence/display/THD/Mview |
| SWISS-MODEL | SIB Swiss Institute | https://swissmodel.expasy.org/ |
| NNSPLICE | Berkeley Drosophila Genome Project | https://www.fruitfly.org/seq_tools/splice.html |
| Oligonucleotides | ||
| See Table S3 | This paper | N/A |
| Recombinant DNA | ||
| pCFD5 | Addgene | Plasmid #73914 |
| pHD-DsRed-attP | Addgene | Plasmid #51019 |
| pUAST-attB-QS | Addgene | Plasmid #24366 |
| Ir56b-gRNA-pCFD5 | This paper | N/A |
| Ir56b-HomologyArms-pHD-DsRed-attP | This paper | N/A |
| Ir56b-HomologyArms-Gal4-pHD-DsRed-attP | This paper | N/A |
| DmelIr56b-gene-pUAST-attB-QS | This paper | N/A |
| DsimIr56b-gene-pUAST-attB-QS | This paper | N/A |
| DsecIr56b-gene-pUAST-attB-QS | This paper | N/A |
| DereIr56b-gene-pUAST-attB-QS | This paper | N/A |
| DsuzIr56b-gene-pUAST-attB-QS | This paper | N/A |
| DvirIr56b-gene-pUAST-attB-QS | This paper | N/A |
| DmelIr56b-gene-TTC-pUAST-attB-QS | This paper | N/A |
| DmelIr56b-gene-Δ51-pUAST-attB-QS | This paper | N/A |
| DmelIr56b-gene-Δ462-pUAST-attB-QS | This paper | N/A |
| DmelIr56b-gene-pUAST-attB-QS | This paper | N/A |
Highlights.
Ir56b is required for the appetitive behavioral response to salt.
Ir56b is expressed in neurons that also sense sugars.
Ir56b acts together with Ir25a and Ir76b to confer salt responses.
Ir56b is a "pseudo-pseudogene", harboring a conserved premature stop codon.
ACKNOWLEDGMENTS
We thank Zina Berman for support and the other members of the Carlson laboratory for discussion. Supported by a Merck fellowship from the Life Sciences Research Foundation to H.K.M.D, 1F32DC018445 to S.A.M.E., 1F32DC019302-01A1 to G.J.S.T., and NIH R01 DC11697, NIH R01 DC02147, and NIH R01 DC04729 to J.R.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Beauchamp G, and Jiang P (2014). Comparative biology of taste: Insights into mechanism and function. Flavour 4, 1–3. [Google Scholar]
- 2.Dethier VG (1977). The taste of salt. Am Sci 65, 744–751. [PubMed] [Google Scholar]
- 3.Feng P, Zheng J, Rossiter SJ, Wang D, and Zhao H (2014). Massive losses of taste receptor genes in toothed and baleen whales. Genome Biol Evol 6, 1254–1265. 10.1093/gbe/evu095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, et al. (2010). The sequence and de novo assembly of the giant panda genome. Nature 463, 311–317. 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montell C (2021). Drosophila sensory receptors-a set of molecular Swiss Army Knives. Genetics 217 1, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yarmolinsky DA, Zuker C, and Ryba N (2009). Common Sense about Taste: From Mammals to Insects. Cell 139, 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandrashekar J, Kuhn C, Oka Y, Yarmolinsky DA, Hummler E, Ryba N, and Zuker C (2010). The cells and peripheral representation of sodium taste in mice. Nature 464, 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dethier VG, and Hanson FE (1968). Electrophysiological responses of the chemoreceptors of the blowfly to sodium salts of fatty acids. Proc Natl Acad Sci U S A 60, 1296–1303. 10.1073/pnas.60.4.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiroi M, Meunier N, Marion-Poll F, and Tanimura T (2004). Two antagonistic gustatory receptor neurons responding to sweet-salty and bitter taste in Drosophila. J Neurobiol 61, 333–342. 10.1002/neu.20063. [DOI] [PubMed] [Google Scholar]
- 10.Jaeger AH, Stanley M, Weiss ZF, Musso PY, Chan RC, Zhang H, Feldman-Kiss D, and Gordon MD (2018). A complex peripheral code for salt taste in Drosophila. Elife 7. 10.7554/eLife.37167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oka Y, Butnaru M, Buchholtz L.V.v., Ryba N, and Zuker C (2013). High salt recruits aversive taste pathways. Nature 494, 472–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang YV, Ni J, and Montell C (2013). The molecular basis for attractive salt-taste coding in Drosophila. Science 340, 1334–1338. 10.1126/science.1234133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee MJ, Sung HY, Jo H, Kim HW, Choi MS, Kwon JY, and Kang K (2017). Ionotropic Receptor 76b Is Required for Gustatory Aversion to Excessive Na+ in Drosophila. Mol Cells 40, 787–795. 10.14348/molcells.2017.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganguly A, Pang L, Duong VK, Lee A, Schoniger H, Varady E, and Dahanukar A (2017). A Molecular and Cellular Context-Dependent Role for Ir76b in Detection of Amino Acid Taste. Cell Rep 18, 737–750. 10.1016/j.celrep.2016.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hussain A, Zhang M, Üçpunar HK, Svensson T, Quillery E, Gompel N, Ignell R, and Grunwald Kadow IC (2016). Ionotropic Chemosensory Receptors Mediate the Taste and Smell of Polyamines. PLoS Biol 14, e1002454. 10.1371/journal.pbio.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koh TW, He Z, Gorur-Shandilya S, Menuz K, Larter NK, Stewart S, and Carlson JR (2014). The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron 83, 850–865. 10.1016/j.neuron.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez-Alcaniz JA, Silbering AF, Croset V, Zappia G, Sivasubramaniam AK, Abuin L, Sahai SY, Munch D, Steck K, Auer TO, et al. (2018). An expression atlas of variant ionotropic glutamate receptors identifies a molecular basis of carbonation sensing. Nature communications 9, 4252. 10.1038/s41467-018-06453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiss LA, Dahanukar A, Kwon JY, Banerjee D, and Carlson JR (2011). The molecular and cellular basis of bitter taste in Drosophila. Neuron 69, 258–272. S0896-6273(11)00002-X [pii] 10.1016/j.neuron.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shanbhag SR, Park SK, Pikielny CW, and Steinbrecht RA (2001). Gustatory organs of Drosophila melanogaster: fine structure and expression of the putative odorant-binding protein PBPRP2. Cell Tissue Res 304, 423–437. [DOI] [PubMed] [Google Scholar]
- 20.Joseph RM, and Carlson JR (2015). Drosophila Chemoreceptors: A Molecular Interface Between the Chemical World and the Brain. Trends Genet 31, 683–695. 10.1016/j.tig.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.USDA. https://fdc.nal.usda.gov.
- 22.French A, Ali Agha M, Mitra A, Yanagawa A, Sellier MJ, and Marion-Poll F (2015). Drosophila Bitter Taste(s). Front Integr Neurosci 9, 58. 10.3389/fnint.2015.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meunier N, Ferveur JF, and Marion-Poll F (2000). Sex-specific non-pheromonal taste receptors in Drosophila. Curr Biol 10, 1583–1586. 10.1016/s0960-9822(00)00860-5. [DOI] [PubMed] [Google Scholar]
- 24.Meunier N, Marion-Poll F, Rospars JP, and Tanimura T (2003). Peripheral coding of bitter taste in Drosophila. J Neurobiol 56, 139–152. [DOI] [PubMed] [Google Scholar]
- 25.Clyne PJ, Warr CG, and Carlson JR (2000). Candidate taste receptors in Drosophila. Science 287, 1830–1834. [DOI] [PubMed] [Google Scholar]
- 26.Dahanukar A, Lei YT, Kwon JY, and Carlson JR (2007). Two Gr genes underlie sugar reception in Drosophila. Neuron 56, 503–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker SJ, Corrales-Carvajal VM, and Ribeiro C (2015). Postmating Circuitry Modulates Salt Taste Processing to Increase Reproductive Output in Drosophila. Curr Biol 25, 2621–2630. 10.1016/j.cub.2015.08.043. [DOI] [PubMed] [Google Scholar]
- 28.Campesan S, Dubrova Y, Hall JC, and Kyriacou CP (2001). The nonA gene in Drosophila conveys species-specific behavioral characteristics. Genetics 158, 1535–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garrigan D, Kingan SB, Geneva AJ, Andolfatto P, Clark AG, Thornton KR, and Presgraves DC (2012). Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res 22, 1499–1511. 10.1101/gr.130922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hey J, and Kliman RM (1993). Population genetics and phylogenetics of DNA sequence variation at multiple loci within the Drosophila melanogaster species complex. Mol Biol Evol 10, 804–822. 10.1093/oxfordjournals.molbev.a040044. [DOI] [PubMed] [Google Scholar]
- 31.Lachaise D, and Silvain JF (2004). How two Afrotropical endemics made two cosmopolitan human commensals: the Drosophila melanogaster-D. simulans palaeogeographic riddle. Genetica 120, 17–39. 10.1023/b:gene.0000017627.27537.ef. [DOI] [PubMed] [Google Scholar]
- 32.Petrov DA, and Hartl DL (1998). High rate of DNA loss in the Drosophila melanogaster and Drosophila virilis species groups. Mol Biol Evol 15, 293–302. 10.1093/oxfordjournals.molbev.a025926. [DOI] [PubMed] [Google Scholar]
- 33.Russo CA, Takezaki N, and Nei M (1995). Molecular phylogeny and divergence times of drosophilid species. Mol Biol Evol 12, 391–404. 10.1093/oxfordjournals.molbev.a040214. [DOI] [PubMed] [Google Scholar]
- 34.Lachaise D, David JR, Lemeunier F, Tsacas L, and Ashburner M (1986). The reproductive relationships of Drosophila sechellia with D. mauritiana, D. simulans and D. melanogaster from the afrotropical region. Evolution 40, 262–271. 10.1111/j.1558-5646.1986.tb00468.x. [DOI] [PubMed] [Google Scholar]
- 35.Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, and Benton R (2010). Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet 6, e1001064. 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, and Benton R (2011). Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69, 44–60. 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, and Dingledine R (2010). Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 62, 405–496. 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prieto-Godino LL, Rytz R, Bargeton B, Abuin L, Arguello JR, Peraro MD, and Benton R (2016). Olfactory receptor pseudo-pseudogenes. Nature 539, 93–97. 10.1038/nature19824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jungreis I, Lin MF, Spokony R, Chan CS, Negre N, Victorsen A, White KP, and Kellis M (2011). Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res 21, 2096–2113. 10.1101/gr.119974.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliveira DC, Almeida FC, O'Grady PM, Armella MA, DeSalle R, and Etges WJ (2012). Monophyly, divergence times, and evolution of host plant use inferred from a revised phylogeny of the Drosophila repleta species group. Mol Phylogenet Evol 64, 533–544. 10.1016/j.ympev.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 41.Gerashchenko MV, Lobanov AV, and Gladyshev VN (2012). Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc Natl Acad Sci U S A 109, 17394–17399. 10.1073/pnas.1120799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hudson AM, Szabo NL, Loughran G, Wills NM, Atkins JF, and Cooley L (2021). Tissue-specific dynamic codon redefinition in Drosophila. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2012793118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Augustine V, Gokce SK, and Oka Y (2018). Peripheral and Central Nutrient Sensing Underlying Appetite Regulation. Trends Neurosci 41, 526–539. 10.1016/j.tins.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liman ER, Zhang YV, and Montell C (2014). Peripheral coding of taste. Neuron 81, 984–1000. 10.1016/j.neuron.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahn JE, Chen Y, and Amrein H (2017). Molecular basis of fatty acid taste in Drosophila. Elife 6. 10.7554/eLife.30115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devineni AV, Sun B, Zhukovskaya A, and Axel R (2019). Acetic acid activates distinct taste pathways in Drosophila to elicit opposing, state-dependent feeding responses. Elife 8. 10.7554/eLife.47677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tauber JM, Brown EB, Li Y, Yurgel ME, Masek P, and Keene AC (2017). A subset of sweet-sensing neurons identified by IR56d are necessary and sufficient for fatty acid taste. PLoS Genet 13, e1007059. 10.1371/journal.pgen.1007059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wisotsky Z, Medina A, Freeman E, and Dahanukar A (2011). Evolutionary differences in food preference rely on Gr64e, a receptor for glycerol. Nature Neuroscience 14, 1534–1541. [DOI] [PubMed] [Google Scholar]
- 49.Luo R, Zhang Y, Jia Y, Zhang Y, Li Z, Zhao J, Liu T, and Zhang W (2021). Molecular basis and homeostatic regulation of Zinc taste. Protein Cell. 10.1007/s13238-021-00845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He Z, Luo Y, Shang X, Sun JS, and Carlson JR (2019). Chemosensory sensilla of the Drosophila wing express a candidate ionotropic pheromone receptor. PLoS Biol 17, e2006619. 10.1371/journal.pbio.2006619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slone J, Daniels J, and Amrein H (2007). Sugar receptors in Drosophila. Curr Biol 17, 1809–1816. 10.1016/j.cub.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Z, Schwartz S, Wagner L, and Miller W (2000). A greedy algorithm for aligning DNA sequences. J Comput Biol 7, 203–214. 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 53.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown NP, Leroy C, and Sander C (1998). MView: a web-compatible database search or multiple alignment viewer. Bioinformatics 14 4, 380–381. [DOI] [PubMed] [Google Scholar]
- 55.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Research 46, W296–W303. 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Response of labellar taste sensilla to salt. Related to STAR Methods and Figure 1.
A) Responses of labellar sensilla to 10 mM NaCl.
C) Responses of sensilla to salts, each tested at 10 mM.
D) Responses of L1 to other sodium and chloride salts.
E) Responses to 10 mM NaCl in control and Ir56bMB09950 flies.
F) Responses of S4 and S8 to NaCl.
G) Responses of S4 and S8 to 100 mM sucrose.
Data S2. Response of Ir47a1, and Ir56b1 to NaCl. Related to STAR Methods and Figure 2.
B) Responses of L1 and S4 to NaCl.
C) Responses of L1 and S4 in control, Ir47a1, and Ir56b1 to 100 mM glucose.
D) Responses of L1 and S4 to water.
E) Responses of tarsal sensilla on leg to 50 mM NaCl.
F) Responses of L1 and S4 in the indicated genotypes to NaCl.
Data S3. Response of the indicated genotypes to NaCl. Related to STAR Methods and Figure 3.
A) Responses of L1 and S4 in control w Canton-S (w CS), Ir25a2, and Ir76b1 to NaCl.
C) Responses of the indicated classes of bitter neurons in the indicated genotypes to NaCl.
D) Salt responses of S-a and S-b bitter neurons that ectopically express Ir56b in an Ir25a mutant.
E) Salt responses of S-a and S-b bitter neurons that ectopically express Ir56b in an Ir76b mutant.
Data S4. Response of the indicated genotypes to sucrose and NaCl. Related to STAR Methods and Figure 4.
A) Responses of L1 and S4 in the indicated genotypes to 100 mM NaCl.
I) Expression of diphtheria toxin under the control of Gr5a-, Gr64f-, or IR56b-Gal4 drivers in L sensilla severely reduced response to both sucrose and NaCl.
Data S5. Behavioral responses to NaCl. Related to STAR Methods and Figure 5.
B) Labellar PER responses in control and Ir56b1 to NaCl.
C) Labellar PER responses to 100 mM sucrose.
E) Leg PER responses to NaCl.
F) Leg PER responses in control and Ir56b1 to 100 mM sucrose.
G) Labellar PER responses to NaCl in the indicated genotypes.
Data S6. Response of the indicated genotypes to NaCl. Related to STAR Methods and Figure 6.
B) Responses of S4 sensilla to NaCl in each of the indicated genotypes.
E) Responses of S4/S8 sensilla to NaCl in each of the indicated genotypes.
Data Availability Statement
This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.