Abstract
Epigenetics is the branch of genetics that studies the different mechanisms that influence gene expression without direct modification of the DNA sequence. An ever-increasing amount of evidence suggests that such regulatory processes may play a pivotal role both in the initiation of pregnancy and in the later processes of embryonic and fetal development, thus determining long-term effects even in adult life. In this narrative review, we summarize the current knowledge on the role of epigenetics in pregnancy, from its most studied and well-known mechanisms to the new frontiers of epigenetic regulation, such as the role of ncRNAs and the effects of the gestational environment on fetal brain development. Epigenetic mechanisms in pregnancy are a dynamic phenomenon that responds both to maternal–fetal and environmental factors, which can influence and modify the embryo-fetal development during the various gestational phases. Therefore, we also recapitulate the effects of the most notable environmental factors that can affect pregnancy and prenatal development, such as maternal nutrition, stress hormones, microbiome, and teratogens, focusing on their ability to cause epigenetic modifications in the gestational environment and ultimately in the fetus. Despite the promising advancements in the knowledge of epigenetics in pregnancy, more experience and data on this topic are still needed. A better understanding of epigenetic regulation in pregnancy could in fact prove valuable towards a better management of both physiological pregnancies and assisted reproduction treatments, other than allowing to better comprehend the origin of multifactorial pathological conditions such as neurodevelopmental disorders.
Keywords: Epigenetics, miRNA, Pregnancy, Review, Embryo and fetus development, Brain development
Introduction
The British epidemiologist David Barker first introduced the concept that “the womb may be more important than the home”, emphasizing the role of the gestational environment as a regulatory staple in the development of the embryo, of the fetus and, ultimately, of the adult [1].
After fertilization, oviductal and endometrial fluids nurture the embryo and regulate its development, before being replaced in this role by the placenta in the further stages of the pregnancy, while the embryo, and later the fetus, interact with the maternal environment in order to further its own implantation and development. This embryo-maternal and fetus-maternal crosstalk [2–6] is finely regulated by numerous cellular pathways, including epigenetic mechanisms.
Epigenetics is defined as the study of heritable changes in gene expression that do not involve modifications to the underlying DNA sequence. Knowledge of the phenomena related to such regulatory mechanisms carries a huge relevance in both physiology and pathophysiology. Both parental and environmental factors (such as nutrition, stress, socioeconomic status and exposure to teratogens and other environmental drivers) have been demonstrated to modulate prenatal development both in the preconception phase and afterwards during the pregnancy, via changes to the DNA methylation patterns (also known as methylome), histone modifications, and/or non-coding RNA (ncRNA) system.
Extensive evidence has progressively emerged that the gestational environment determines a remarkable impact on an epigenetic level, at least with two different mechanisms: by directly regulating the stages of implantation and placentation; and widely remodeling epigenetic patterns during prenatal development, thus determining long-term outcomes in the offspring [4, 5, 7–10].
This narrative review aims to recapitulate both the already established and the newly emerging environmental and parental factors that regulate pregnancy through epigenetic mechanisms.
Main epigenetics mechanisms
The epigenetic modulation of gene expression begins in the gametes and zygote through genomic imprinting and then continues throughout the whole gestation with changes to the fetal and maternal methyloma and the specific action of the ncRNA system. The main processes involved in the epigenetic regulation of pregnancy, namely the DNA methylation, the histone post-translational modifications, the non-coding RNA and the genomic imprinting, are summarized in Table 1.
Table 1.
Main epigenetic processes involved in the regulation of pregnancy
| Processes | Mechanisms | Enzymes/molecules | Effect |
|---|---|---|---|
| DNA methylation | Methylation of CpG and CGIs |
-DNMT1 -DNMT3A -DNMT3B -TET family -MBD4 -TDG |
-Transcription silencing (sporadically transcription permissive) -Chromatin remodeling |
| Histone post-translational modification |
Histones’: Acetylation (A) Methylation (M) Phosphorylation (P) Ubiquitination (U) |
-HATs (6 groups) -HDACs (4 classes, 18 enzymes) -HMTs -HDMs -DUBs (2 classes) |
-A: ↑ gene expression -M: 1. regulation of imprinting 2. ↓or ↑ gene expression -P: 1. DNA damage repair 2. Chromatin compaction 3. Transcription modulation -U: 1. DNA damage signalling 2. Protein translocation 3. Transcription regulation 4. Cellular signaling modulation |
| ncRNAs system |
-RNA–DNA interaction -RNA-RNA interaction -RNA–protein interaction -Protein recruitment (scaffolding &/or sponging) |
1. sncRNAs: -siRNAs -miRNAs -piRNAs 2. lncRNAs: -lincRNAs -ilncRNAs -eRNAs (including promoter/UTR/ telomere-associated lncRNAs) |
-Gene expression regulation -Promoter silencing/enhancing -RNA interference -mRNA splicing regulation -Histone-modifying complexes driver -Methylation processes involving -Post-transcriptional regulation |
| Imprinting |
Methylation of: -X-Chromosome -DMRs/ICRs -Histones |
DNMT3A DNMT3B DNMT3L |
-X- Inactivation -Parental/genomic imprinting control -Imprinting maintenance |
DNMT, DNA methyltransferase; TET, ten-eleven translocation methylcytosine dioxygenases; MBD4, methyl-CpG binding protein 4; TDG, thymine DNAglycosylase; (A), acetylation; (M), methylation; (P), phosphorylation; (U), ubiquitination; HATs, histone acetyltransferases; HDACs, histone deacetylases; HMTs, histone methyltransferases; HDMs, histone demethylases; DUBs, deubiquitinating enzymes; ncRNAs, non-coding RNAs; sncRNAs, short ncRNAs; siRNAs, short interfering RNAs; miRNAs, microRNAs; piRNAs, piwi interacting RNAs; lncRNAs, long ncRNAs; lincRNAs, long intergenic ncRNAs; ilncRNAs, intronic long ncRNAs; eRNAs, enhancer long ncRNAs; DMRs, differentially methylated regions; ICRs, imprinting control regions
DNA methylation
DNA methylation is the most widespread epigenetic modification of the human genome. The addition of a methyl group to the fifth carbon of the pyrimidine cytosine ring determines long-term silencing in processes such as genomic imprinting, tissue-specific regulation of gene expression, X-chromosome inactivation, and silencing of repetitive DNA elements. Correct patterns of DNA methylation, both prenatally and postnatally, are required for normal human development [11–14].
In multicellular eukaryotes, DNA methylation mostly occurs at cytosines of CpG (C–phosphate–G) dinucleotides; highly methylated sequences can also be found in satellite DNAs, repetitive element and non-repetitive intergenic DNA. When established on promoters or enhancers, methylation can repress transcription either directly, by inhibition of transcription factor binding, or indirectly, through recruitment of methyl-binding proteins and chromatin modifiers [11–15]. Even though the most widespread and well-known effect of DNA methylation is long-term silencing of transcription, it should be noted that recent works suggested that in some instances, hypermethylated promoters and enhancers could be permissive to transcription instead [16].
While most of the CpG sites in human DNA are methylated, the CpG-rich regions named CpG islands, that are located in approximately 60% of the promoters of human genes, display a baseline unmethylated pattern. Methylation of CpG islands located in the promoter region of a gene usually inhibits the transcription of that gene due to binding of methyl-CpG binding proteins, as they recruit chromatin remodelers, histone deacetylases, and methylases to the promoter of the gene, thereby blocking transcription [17, 18].
Histone post-translational modifications
Gene expression is also influenced by a variety of post-translational modifications to histones. Acetylating, methylating, phosphorylating, and ubiquitinating enzymes act alone or in combination to control chromatin compaction [19–22]. Histone acetylation generally results in higher gene expression, with rare exceptions [23–25]. Histone methylation has both permissive or repressive effect on transcription, depending on the location of target amino acid residues in the histone tail or on the number of methyl groups attached [9, 26, 27]. Histone phosphorylation is regulated by kinases (that bind phosphate groups) and phosphatases (which detach the phosphates) and has at least three activities: DNA damage repair, control of chromatin compaction connected with mitosis and meiosis, and modulation of transcriptional function [28, 29]. Histone mono-ubiquitination has a crucial role in protein translocation, DNA damage signaling, and regulation of transcription. On the other hand, poly-ubiquitination flags proteins as a mark for degradation or activation in some signaling pathways [22, 30].
Several studies and evidences report that precise chromatin modification patterns occur in the fetal membrane and decidua cells, changing dynamically during normal and pathological pregnancies [31].
Non-coding RNAs system
While it is known that most human transcripts are not translated, many of them actively participate in essential cellular functions nonetheless. Specifically, ncRNAs are a family of RNAs that do not encode functional proteins but are involved in pivotal regulatory and housekeeping mechanisms [32, 33].
The regulatory ncRNAs are divided into two main categories, based on size: short-chain non-coding RNAs (sncRNAs, including siRNAs, miRNAs, and piRNAs) when shorter than 200 nucleotides; long-chain non-coding RNA (lncRNAs, including lincRNAs, ilncRNAs, and eRNAs) when longer than 200 nucleotides. ncRNAs were originally thought to be only involved in gene expression at the post-transcriptional level, while nowadays it has been largely established that non-coding RNAs function as the most common regulatory RNAs, both at pre and post transcriptional level (e.g., by promoter silencing or RNA interference mechanisms), and are also involved in epigenetic control. Many of these transcripts are in fact necessary for proper targeting of histone modifying complexes or participate in the DNA methylation process and genomic imprinting. For such reasons, it has been proposed that a contemporary definition of epigenetics should also include the gene silencing or upregulation mediated by ncRNAs [34–36].
In particular, the study of miRNAs expression and transport appears to be the next frontier in terms of improving the understanding of epigenetic mechanisms in fetal development and fetal-maternal crosstalk. Their role in broader effects on fetal and long-term human development is yet to be fully investigated and could represent a promising research field for future studies, which could even expand to the optimization of in vitro fertilization [37, 38].
Imprinting
During gametogenesis and then again in the early moments after fertilization, mammals undergo a two-step DNA methylation reprogramming that affects more than 80% of the genome. Two events of extensive erasure occur in the primordial germ cells and pre-implantation embryo, followed by de novo DNA methylation, with differential kinetics and patterns during male and female gametogenesis and within cell lineage specification in post-implantation development. [37–41]. Additionally, the process of X-chromosome inactivation will occur in female embryos, leaving only one copy of almost all of the X-linked genes to be expressed [39].
However, specific differentially methylated regions (DMRs), in which DNA is methylated on one specific parental allele, escape the reprogramming process that happens in the pre-implantation embryo [37]. This phenomenon, that allows parent-of–origin specific gene expression, is defined as genomic imprinting. Most imprinted genes are gathered in distinct clusters of the size of about 1 Mb and contain both maternally and paternally expressed genes. In addition to protein-coding genes, these clusters typically contain lncRNA, which can regulate the imprinting of the nearby genes. Regulation of the clustered genes is coordinated through short DNA sequences called imprinting control regions (ICRs). Also, recent evidence is starting to highlight the contribution of post-translational histone modifications to the regulation of imprinting [42, 43].
The disruption of genomic imprinting leads to largely established human diseases with recognizable clinical features such as Beckwith-Wiedemann syndrome (MIM 130,650), Silver-Russell syndrome (MIM 180,860), Prader-Willi syndrome (MIM 176,270), and Angelman syndrome (MIM 105,830), but less dramatic and more subtle changes in the imprinted patterns, especially concerning multi-locus imprinting disturbances (MLID), can instead modulate fetal growth, resource acquisition, and organogenesis [44–46].
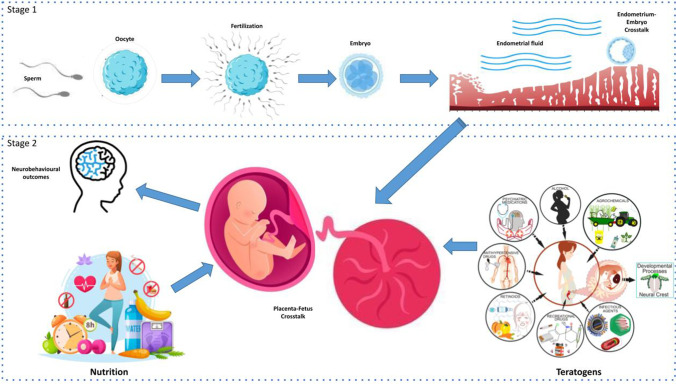
There are many epigenetic action’s hot spots along the entire pregnancy process, and it would have been too long and purely didactic to analyze them all in detail. We have therefore chosen the most significant steps and those for which there is already literature data that allow a reasoned review of the causative mechanism. Figure 1 details the main hot spots, divided into stage 1 (from gametes to embryo-endometrium cross-talk) and stage 2 (from placenta-fetus cross-talk to brain development, with an overview on environmental factors as well).
Fig. 1.
Hot spots of epigenetic action. Stage 1: from gametes to embryo-endometrium cross-talk; Stage 2: from placenta-fetus cross-talk to brain development, with an overview on environmental factors as well (nutrition and teratogens). Modified from Cerrizuela S et al, Birth Defects Res. 2020; other pictures obtained by Vecteezy.com
Hot spots of epigenetic action in gametes and embryo stage
Oocytes and spermatozoa
In addition to determining parent-of-origin specific gene expression by genomic imprinting, the epigenetic reprogramming that happens during gametogenesis regulates gametes development, and its disruption can also reverberate in the zygote, embryo, and postnatal life.
Fertile sperm production requires effective epigenetic regulation. In male gametes, epigenetic patterns are established soon after the demethylation event has occurred in the primordial germ cells. The sperm epigenome is well specialized because of its particular requirements in terms of motility and protection against the female reproductive tract’s hostile environment, exhibiting a unique pattern of histone modifications that includes both activating and silencing marks in the promoters of genes associated with development [47, 48]; sperm chromatin is highly compacted and organized, and spermatogenesis has been associated with fluctuating level of ncRNAs, particularly endo-siRNAs [49].
Sperm epigenetic abnormalities are associated with infertility, reduced embryogenesis capability, and alterations in early embryo development [50, 51], and the study of sperm epigenome is being evaluated as a potential diagnostic tool for idiopathic recurrent pregnancy loss and infertility [47, 50–55].
In the oocytes, de novo DNA methylation does not start as immediately as in their male counterparts and appears to be largely dispensable for the early oocyte development [56]. DNA methylation establishment in the oocyte is dynamically controlled during oocyte growth and it is proximally regulated by transcription events. As a result, DNA methylation is mainly restricted to the transcribed gene bodies, while intergenic regions are hypomethylated [57–59], as non-CpG hypermethylation plays an active role in gene expression regulation during oocyte maturation, potentially through the incorporation with transcription factors [57]. Any adverse factor altering the normal transcription program could therefore perturb the correct methylation patterns [60, 61]. Numerous studies also managed to identify maternal effect genes (MEGs) which encode factors that are present in the oocyte and are required for embryonic development. Pathogenic variants in MEGs or the absence of MEG factors are correlated to adverse outcomes, like zygotic cleavage failure, imprinting disorders, and structural birth defects [62–64]. There is also growing evidence that variants in the maternal genome affecting the imprinting status of the oocyte could cause MLID [44].
Fertilization and early embryo development
Fertilization is defined as the event when terminally differentiated gametes combine into a totipotent zygote, ultimately giving rise to the embryo. The earliest stages of embryonic development are characterized by major epigenetic remodeling events that include parental DNA methylation erasure and reprogramming (with the exception of DMRs), chromatin folding establishment, and spatial reorganization of the genome [65–67]. The precision and the timing of such processes are pivotal for avoiding developmental defects or embryonic lethality, as an incorrect establishment of the embryonic epigenome would cause a defective zygotic genome activation (ZGA). Such a brief but crucial phase represents a window of vulnerability towards interfering maternal and environmental factors (such as chronic metabolic disorders, polycystic ovary syndrome, diet, and teratogen exposure), as highlighted by an ever-increasing number of studies [68–71]. In particular, it has been suggested that the association between assisted reproductive treatment (ART) procedures and slightly higher frequences of imprinting syndromes may be due to ART causing an abnormal epigenome in the offspring [71]. Also, animal models suggested that chromatin changes occurring in the pluripotent embryo could determine regulatory information whose effects would reverberate in the adult life in terms of growth and brain development [72]. A better comprehension of the underlying mechanisms and their consequences could therefore improve diagnosis and treatment of human infertility and diseases.
Pre-receptive and receptive endometrium
Embryonal implantation is a crucial step of pregnancy establishment in mammals. The sensitivity of the endometrium to implanting embryos is commonly categorized into pre-receptive, receptive, and refractory phases. This period of receptivity, called “window of implantation” [73], is short and results from the programmed sequence of the action of estrogen and progesterone on the endometrium via multiple paracrine, juxtacrine, and autocrine signaling [26, 74].
High levels of progesterone in the post-ovulatory phase modulate the endometrial gene expression required for implantation by regulating DNA methylation [75–77], and alterations in the DNA methylation of 448 sites have been associated with defective endometrial receptivity and consequently in recurrent pregnancy failure [75, 78]. Moreover, different non-coding transcripts like lncRNAs and miRNAs are involved in endometrial receptivity regulation [79].
Several association studies have evaluated miRNA expression levels in the receptive and pre-receptive endometrium in fertile women [80–83]. The miR-30 family members (primarily miR-30b and miR-30d) were demonstrated to be significantly upregulated [84], while miR-494 and miR-923 were downregulated during the receptive phase. miR-30b and miR-30d were also consistently found to be elevated in the mid-secretory as compared with the proliferative phase. Expression arrays demonstrated that embryos treated with miR-30d exhibited increased expression of ten genes, including those encoding adhesion molecules such as ITGB3 (Integrin Subunit Beta 3), ITGA7 (Integrin Subunit Alpha 7), and CDH5 (Cadherin 5) [85].
A significant inverse association has been found between miR-31 and both FOXP3, a transcription factor for T regulatory cells, and CXCL12, a chemoattractant for uterine natural killer cells, which participates in the creation of an immune-tolerant environment in the secretory phase [86]. The modulation of immune response is considered an important target for miRNAs, because major histocompatibility complexes such as HLA-G appear to be extensively regulated by noncoding RNAs such as miR-133a [87, 88].
Also, members of the miR-17–92 cluster (including miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1) are upregulated at the implantation sites of the receptive uterus in humans. The functions of endometrial miR-17–92 during implantation are unclear. The cluster is known to target TGF-b signalling, which modulates events in decidualization and implantation [89].
Moreover, the miRNA-200 family (miR-200a, miR-200b, miR200c, miR-141, and miR-429) showed a differential expression in the receptive phase as well, when they appear to be downregulated [89].
Finally, by suppressing Dicer, miRNAs from the Lethal-7 family can regulate the expression of other miRNAs. It has been suggested that high levels of let-7 in the receptive uterus could be important for differentiation of a receptive endometrium [87].
Interestingly, histone acetylation is also involved in the early endometrial processes, being implicated in the vascular endothelial growth factor pathway during angiogenesis. Studies using histone deacetylase inhibitors suggest an involvement in endometrial proliferation and differentiation as well [77].
Decidualization
The term decidualization defines the functional and morphological transition of the endometrial cells to form the cellular environment into which the blastocyst is able to implant itself. Defects in decidualization can lead to recurrent implantation failure and recurrent spontaneous abortion [90, 91]. Human endometrial stem cells (hESC) have been decidualized in vitro to explore the effects of decidualization on miRNA expression. Studies found that miRNA expression profiles of decidualized hESC and control hESC departed from each other. According to Tochigi et al. [92], hESC transfection of miR-542-3p suppressed IGFBP-1 (insulin-like growth factor-binding protein) expression, leading to PRL (prolactin) and WNT4 suppression and thus to the inhibition of decidualization in human endometrial stroma cells. This suggests an important role of miR-542-3p in regulation of endometrial decidualization. Estrella et al. [93] found a total of 26 upregulated and 17 downregulated miRNAs following in vitro decidualization of hESC. Interestingly, only miR-155 was commonly downregulated in both studies, and no change was found in the expression of miR-542-3p in the study of Estrella et al. Despite several differences and significant variability, probably attributable to decidualization in vitro, these studies have nonetheless shown that the miRNA profiles of the proliferative versus secretory endometrium are significantly different, and therefore deserving of further research.
Endometrial fluid
Endometrial fluid is the medium where the most part of the exchanges between the embryo and the endometrium occur, allowing for a tight bidirectional regulation [94–96]. Among the substances conveyed in the endometrial fluid, extracellular vesicles (EVs) are emerging as one of the most important mediators, particularly in relation to their capability of transporting ncRNAs [2, 97–99]. The secretion of exosomes from the apical surface of endometrial glands, which was confirmed by electron microscopy, is consistent with the existence of endosomes in epithelial cells and of embryo-derived extracellular vesicles [100]. Furthermore, primary human endometrial endothelial cells (hEECs) were found to actively secrete large quantities of exosomes into the conditioned media, and labelling experiments with miR30-d revealed that miRNAs are internalized in vesicles and secreted in exosomes. Several experiments showed that exosomes loaded with miR-30d were secreted from hEECs and could be internalized by trophoblastic cells of murine embryos adhered to hEECs [96]. Also, hEECs have been demonstrated to be able to uptake embryo-derived miRNAs, such as the embryo–endometrial adhesion inhibitor miR-661 [101]. Recent literature revealed the presence, in the endometrial fluid of both fertile and infertile women, of 12 sncRNAs strongly associated with biological functions related to immune response, extracellular matrix and cell junction, highlighting the different expression patterns in the two subpopulations and suggesting that sncRNA could be used as biomarkers of endometrial receptivity and implantation success [102].
Even though the capability of numerous noncoding RNAs (both maternal and embryo-derived) of being transmitted through endometrial fluid has been clearly established, the role of such molecules in the regulation of pivotal phases of implantation and early embryonic development still requires much needed investigation [98].
Hot spots of epigenetic action at the fetal stage
Further processes of vascular remodeling, trophoblastic cellular migration, and immune regulation allow the formation of the placenta, a transient organ that connects the fetus to the mother [103]. The placenta supports fetus development supplying oxygen and nutrition, protects the semi-allogeneic fetus from immune rejection, and secretes hormones that affect maternal organs in order to promote the maintenance of pregnancy [104–107]. Many maternal influences, such as over and under-nutrition, drug and alcohol intake, smoking, infection, stress, and hormones activity (for example glucocorticoids [GCs] or thyroid hormones), can induce transformations in placental physiology. These placental modifications can range from alterations in aspects of macroscopic placental morphology to more subtle changes in placental gene expression which may have long-term effect on offspring health. However, many mechanisms of transitory prenatal insults that result in postnatal dysregulation remain unclear [108].
A pivotal role in the fetal stage of pregnancy is played by the amniochorionic membranes, which act as the feto-maternal interface, as they exhibit characteristic chromatin modification patterns, DNA (CpG) methylation, histone modifications, and non-coding RNA transcriptomes, whose dynamic changes during normal and pathological pregnancies are an important contributor to gene regulation throughout pregnancy [31]. Alterations in DNA methylation patterns and ncRNAs in amniochorionic membranes have also been detected in association with preterm birth and pathological conditions such as acute chorioamnionitis [109].
Numerous miRNAs are predominantly or exclusively expressed by the placenta and can be found clustered in specific chromosomal regions; they may be also controlled by the same promoters, have similar seed regions and targets, and work synergistically. Placental miRNome studies have also demonstrated its importance towards the coordination and modulation of the placental transcriptome [110, 111]. The three most relevant clusters are the chromosome 19 miRNA cluster (C19MC), the chromosome 14 miRNA cluster (C14MC), and the miR-371–3 cluster, which is localized on chromosome 19 as well. These miRNAs primarily regulate placental processes such as the migration of the trophoblast and the placental morphogenesis, but evidence has emerged that they are transported to the fetal and maternal compartments as well, even though their role in such districts is still unclear [80, 82, 83].
Epigenetic processes within the placenta are powerful mediators of maternal and environmental signals to the developing fetus. Furthermore, alterations of placental gene expression and signaling during fetal development can dramatically change the developmental program, particularly concerning brain development [110].
Placental metabolism and brain development
Among the organogenetic processes that occur during fetal development, one of the most critically affected by maternal, environmental, and epigenetic factors is brain development. The initial steps of nervous system development start as early as 2–3 weeks post fertilization in humans; however, neuronal proliferation begins later in the first trimester and synaptogenesis and neural migration mainly occur in the later stages of pregnancy, during the second and third trimesters. In the early pregnancy, the brain is extraordinarily plastic, but also exposed to environmental fluctuations which can influence long-term programming. The developing brain tissue requires considerable nutritional intake and it is particularly susceptible to overabundance or insufficiency of specific nutrients and growth factors [108, 110, 111].
Both maternal and fetal factors contribute to neurodevelopment during gestation [112], and the placenta is specifically believed to play a pivotal role [108, 113, 114].
The brain is an organ that consumes a lot of energy and absorbs a massive quantity of maternal resources during its growth and evolution. It is also specifically vulnerable to modifications in placental activity. Many signals regulate chromatin activation or repression; among these, those related to nutrient availability are relevant for epigenetic programming in the placenta.
In this scenario, the X-linked enzyme O-linked-N-acetylglucosamine transferase (OGT) seems to have a fundamental role in neurodevelopmental organization [108, 110, 115–118]. In fact, OGT represents a link between nutritional signals and chromatin regulation. It works as a nutrient sensor which modifies various proteins in order to change numerous cellular processes, like major epigenetic changes (such as methylation) [110].
The signaling of insulin, IGF-1 (insulin-like growth factor-1), and leptin receptors located at the maternal–fetal interface promotes amino acid transporter activity in trophoblast cells, combining maternal nutritional state to placenta function and, consequently, invalidating the accessibility of nutrients diffusing into the fetal circulation [119].
Some authors have reported that increased methylation of the leptin receptor gene in the human placenta is associated with increased lethargy and hypotonicity in male but not in female newborns [120]. This hypothesis supports both a connection between epigenetic regulation of the leptin receptor and the communication of fundamental information to the fetal brain and also a certain sex-specificity in these results.
To date, the relationship between placental metabolism and epigenetic programming has been especially studied in animals (and only marginally in humans) with particular attention to sex development, as sex differences in epigenetic mechanisms such as DNA methylation have been described across placental development. In mice, female placental tissue has higher amounts of global DNA methylation compared to male placentas, which may confer females additional protection from constant alterations in gene expression due to environmental insults [108, 120–122].
Therefore, the recognition of relevant placental biomarkers, such as OGT and leptin, that, in response to maternal nutrition and stress hormones levels, produce epigenetic modifications which sensibly affect the neurodevelopmental programming, may allow in the future for a better understanding and prevention of neurodevelopmental disorders in the offspring [123].
Placental neurotransmitters and neurobehavioral outcomes
The placenta is known to be involved in the transfer of nutrients, hormones, and metabolites, but it is important to remember that it also produces neurotransmitters, including serotonin (5-HT), dopamine (DA), norepinephrine/epinephrine, which enter the maternal–fetal circulation and interact with the development of the fetal brain. Therefore, it is reasonable to assume that some neurobehavioral disorders (such as autism spectrum disorders, ASD) may originate from placental changes in the production of these metabolites. For instance, 5-HT promotes various processes during fetal brain growth: neuronal migration, cell division, and differentiation and synaptogenesis. While placental hyperserotonaemia can disrupt early neural development, hyposerotonaemia too appears to impair cognitive, motor, or sensory capacities [118, 124].
Recently, DNA methylation and miRNA expression patterns within the placenta have been also studied for potential associations with later neurobehavioral disruptions in the offspring [118]. Although the exact mechanisms are still unclear, here, we summarize the literature about the connection between epigenetic modifications and neurobehavioral disorders.
Some authors reported, in a prospective study of high-risk pregnancies, that 400 DMRs discriminate placentas from offspring later diagnosed with ASD compared to those not diagnosed with such disorders [124].
Besides placental 5-HT affecting neurobehavioral development, methylation of placental HTR2A (the receptor which mediates the effects of 5-HT) may also be implicated. HTR2A methylation correlates inversely with infant quality of movement, but positively with infant attention [125].
Altered methylation patterns of 10 imprinted genes (DLX5, DHCR24, VTRNA2-1, PHLDA2, NPA1, FAM50B, GNAS-AS1, PAX8-AS1, SHANK2, and COPG2IT1) have been also associated with reduced quality of movement, elevated indices of asymmetrical and non-optimal reflexes, and increased likelihood of physiological stress [126].
Another type of epigenetic modification that can occur in the placenta and may regulate infant neurobehavioral patterns is represented by the alterations in miRNA expression profiles. Increased placental miR-16 expression is negatively associated with attention score. In contrast, miR-146a and miR-182 are positively related to quality of movement score [118]. In conclusion, even if the placenta is a temporary organ, there is no doubt that its alterations can strongly influence the development of the fetus, and in particular of the fetal brain. Biomedical research is currently focused on identifying placental biomarkers useful for identifying fetuses at risk of developing neurodevelopmental disorders, for starting an early clinical management [118, 125–129].
Role of maternal and environmental factors during pregnancy
Stress hormones
Several data suggest a direct association between prenatal early-life stress (i.e., maternal depression, chronic stress, abuse) and the incidence of psychopathology and cognitive impairments in the baby. Early life stress acts in different ways in different tissues, altering gene expression and inducing epigenetic modifications (e.g., increased or reduced DNA methylation or histone acetylation in many brain regions at the same time) [96].
In animals, stress during pregnancy and increased glucocorticoids (GCs) levels cause alteration in stress hormones receptors and impair the feedback regulation of the hypothalamic-pituitary adrenal axis in infancy and adulthood. Stress acts also on the amygdala and increases the incidence of anxiogenic and depressive-like behaviors [130].
In humans, excess amounts of corticotropin releasing hormones (CRH) and cortisol reaching the fetal brain can alter personality and predispose to attention deficits and depressive illness through changes in neurotransmitter activity [9, 73]. In fact, neurobehavioral alterations have been related to pre-gestational stress. In vitro studies showed that stress hormones can increase excitability hippocampal cells [131].
However, a recent review concluded that most of the published papers on this topic did not reveal a significant association between maternal cortisol in pregnancy and poorer offspring birth outcomes, lower cognitive outcomes, lower cognitive/motor development, or more behavioral problems in infancy and childhood. This result probably reflects the numerous confounding factors that act synergistically during pregnancy and in the postnatal period [132].
Nutrition and environment
The first 1000 days, which start from pre-conception until approximately two years of age, are considered the period during which early nutrition, thanks to its influence on epigenetic changes [133], can play a key role in developmental programming and can influence the individual susceptibility to diseases later in the life (cardiovascular diseases, obesity, diabetes, and other chronic condition). Patients with eating disorders show adverse pregnancy outcomes such as miscarriage, preterm delivery, or fetal anomalies as poor fetal growth or malformations. Altered levels of many nutrients (i.e., vitamin B, folic acid, zinc) can increase the risk of developing fetal disorders [134, 135].
Early life nutrition can modulate the epigenome through different mechanisms: the change in the structure of chromatin through histone modifications and the supply of methyl donors (i.e., methionine, choline, folate), the activity of DNA methyltransferases, and of specific transcription factors. In this way, altered maternal nutrition may induce epigenetic changes in the global expression pattern of the fetus, which will trigger biological and psychological alterations in offspring’s lifelong outcomes [136–139].
Moreover, some nutrients, such as vitamin B and polyphenols, can modulate the function of the methylation enzymes [140]. In particular, Vitamin B12 is associated with one carbon metabolic pathway and to substrate metabolism, as well as to the synthesis and stability of nucleic acids and methylation of DNA [137, 141]. High levels of vitamin B12 in the maternal blood was correlated with the reduction in the total level of DNA methylation of the neonate, while the elevated concentration of serum vitamin B12 in the new-born correlated with decrease methylation levels of the insulin-like growth factor-binding protein3 (IGFBP-3) gene, which is one of the candidate genes for intrauterine growth [142].
The lack of nutrients during pregnancy has been associated with various later-life consequences. Famine during fetal life, for example, can increase the risk of developing glucose intolerance or coronary artery disease in adulthood, creating persistent changes in DNA methylation that depend on the sex of the exposed individual and the gestational timing of the exposure [143, 144]. On the other hand, maternal obesity and a high fat diet can create not only metabolic disorders but also neurodevelopmental morbidity in the offspring. Maternal overeating and low protein consumption during pregnancy is associated with significant miRNA dysregulation in the offspring tissue, which may be associated with chronic inflammation status and metabolic health in offspring as early as the weaning age [145, 146]. As suggested in the recent literature, modifications to the offspring’s epigenome could even be influenced by paternal nutrition, mediated by epigenetic modifications in sperm [147].
One of the most common pregnancy complications worldwide correlated to nutrition is gestational diabetes (GDM). Unhealthy nutritional habits and excessive gestational weight gain during pregnancy can predispose to maternal GDM [148, 149].
GDM increases the incidence of short-term (i.e., large for gestational age, shoulder dystocia, higher body fat) and long-term complications (i.e., obesity, metabolic syndrome, diabetes mellitus type 2, attention problems, and depression) observed in the offspring [150, 151]. These effects are regulated by alteration in the epigenetic programming. Maternal GDM can create different effects like global DNA hypermethylation or alteration of miRNA expressions. A recent study shows that the effect of diabetic environment on miRNA regulated endothelial dysfunction is sex dependent, as females are more susceptible to metabolic derangements of GDM than males [151, 152].
GDM changes also placental microbiota: for example, in women with GDM, there are less bacteria belonging to the Pseudomonadales order and Acinetobacter genus associated with a more adverse metabolic and inflammatory phenotype. In this way, GDM could represent a state of placental microbiota-driven altered immunologic tolerance that can be the target of a new therapy [153].
Interestingly, epigenetic markers of the GDM offspring could become a tool for early detection and prognosis of adverse phenotypic outcomes [154].
Beyond food, there are many substances which influence epigenetic programming. Endocrine disruptors (EDs), for example, are environmental pollutants that mimic endogenous hormonal signals. The EDs most commonly associated with reproductive abnormalities are the xenoestrogens such as Bisphenol-A (BPA), polychlorinated biphenyls (PCBs), and antiandrogens such as phthalates. These compounds exhibit weak steroid-like activity and therefore can affect reproductive development along multiple points including the hypothalamus and the gonad. Exposure to EDs starts during fetal life and continues after birth: the link between prenatal exposures and latent health outcomes suggests that these exposures may result in long-term epigenetic reprogramming.
The epigenomic changes, due to prenatal exposure to ED, include altered global DNA methylation, gene specific CpG methylation, and microRNA expression [155]. In human cell lines, PCB exposure has been shown to modulate the activity of histone demethylases via androgen receptor binding [156]. Moreover, other EDs have been shown to interact with DNMTs and TETs enzymes, altering their activity in in vitro experiments [157]. Additionally, some EDs are known to interact with one-carbon metabolism, which produces methyl donors used for DNA methylation [158]. Finally, it is probable that EDs alter the transcription factors’ activity, changing the availability of DNA to epigenetic machinery and giving rise to gene-specific patterning of epigenomic markers [159]. Therefore, these epigenetic changes represent a critical mechanism essential to understand the changes of fetal epigenome causing adverse outcomes at birth and later in life, warranting further study.
Teratogens
In addition to stress, nutrients, and endocrine disruptors, other environmental factors, such as medications, alcohol or substances of abuse, produce changes in the global pattern of gene expression as well as hormonal imbalance, and ultimately adverse effects on embryonic development.
One of the best known and most studied teratogens is ethanol, for it can cause a wide range of developmental abnormalities. Alcohol-related neonatal abnormalities are commonly referred to as fetal alcohol spectrum disorder (FASD) [160]. The most severe form of FASD is called fetal alcohol syndrome (FAS) and manifests as growth retardation, facial abnormalities, and central nervous system deficiencies. Even if there is no specific time point during gestation when alcohol exposure is not accompanied by harmful consequences, the exposure during early embryonic stage is correlated with most severe birth defects [161]. Many researches have also found that alcohol interferes with gene expression levels and epigenetic processes by altering DNA methylation (particularly the expression of two methyltransferase enzymes, DNMT1 and DNMT3A), histone regulation, and non-coding RNAs [162]. Alcohol also induces alterations in the DNA methylome of the hypothalamus, including several differentially methylated regions (DMRs) that could underlie some of the deficits observed in FASD. DNA methylation profiles may not persist into adulthood but could alter developmental trajectories and induce lasting alterations in brain structure, connectivity, and function [163].
Children prenatally exposed to moderate-high levels of alcohol showed increased DNA methylation of stress regulatory genes proopiomelanocortin (POMC) and period 2 (PER2) resulting in increased levels of stress hormone cortisol and adrenocorticotropic hormone (ACTH), compared to controls. These results suggest the possibility that measuring DNA methylation levels of PER2 and POMC in biological samples from pregnant women or from children can serve as a biomarker of alcohol-related disorders [164].
Further studies are definitely needed to understand the correlation in detail between fetal alcohol spectrum disorder and epigenetic changes [165].
Unfortunately, smoking during pregnancy is still very widespread, especially in some populations where the level of attention to this teratogen is not so high. Maternal tobacco smoking is associated with smaller birth weight and head circumference, as well as reduced length for gestational age [166, 167]. Moreover, maternal tobacco smoking is associated with an increased risk of obesity [168], respiratory infection [169], and cardiovascular [170], psychiatric, and behavioral disorders [171–173].
In addition to maternal smoking, exposure to air particulate matter, polycyclic aromatic hydrocarbons, arsenic, heavy metals, cannabinoids, and persistent organic pollutants during pregnancy has been associated with epigenetic changes [174, 175]. Cigarette smoke creates alterations in DNA methylation of cord blood and placenta, histone modifications, and miRNA expression. In utero tobacco exposure, even in the absence of fetal growth restriction, may alter the epigenome, contributing to global DNA hypomethylation. Investigation towards miRNA and downstream transcriptional regulation are growing, while studies investigating histone modifications are still sparse [176].
Even second-hand smoke exposure among non-smoking women may alter DNA methylation in regions involved in development, carcinogenesis, and neuronal functioning. These novel findings suggest that even low levels of smoke exposure during pregnancy may be sufficient to alter DNA methylation of the fetus [177]. Hopefully, in the future, DNA methylation status could be used as a biomarker of prenatal insults for tobacco exposure as well.
Regarding substances universally recognized as teratogens, one of the most widespread is valproic acid (VPA). VPA is a well-tolerated antiepileptic drug and mood stabilizer that is used for the treatment of epilepsy and other non-psychiatric diseases like Alzheimer, HIV, and cancer [178]. VPA interacts with the inhibitory neurotransmitter gamma-Aminobutyric acid (GABA) and blocks voltage-gated ion channels. Several authors demonstrated on murine models that early increases in histone acetylation occur within the embryo and in murine decidua following gestational VPA exposure [179, 180]. This mechanism of action is known to result in teratogenicity and cell toxicity, causing a wide range of fetal alterations, both malformative and neurological.
Some other commonly used drugs have specific fetal body effects: a recent study, for example, has demonstrated that cyclophosphamide induces limb dysmorphogenesis by alterations in some miRNA expression (i.e., miRNA-34, miRNA-125b, and miRNA-155). These miRNAs, in fact, act in different ways: some miRNAs act to protect embryos, whereas other miRNAs boost a teratogen-induced process of maldevelopment to induce embryonic death. The analysis of correlations between the expression pattern of the miRNAs tested in the study and cyclophosphamide induced limb phenotypes implies that miRNAs regulating apoptosis may differ from each other with respect to their functional role in teratogenesis [181].
Finally, regarding another known teratogen inducing limb reduction anomalies (5-aza-2’-deoxycytidin or 5-aza), recent animal study focused the attention on miR-34 family, concluding that 5-aza and cyclophosphamide are able to activate miR-34 family in p53-independent fashion. This observation implies a scenario in which miR-34 family plays a regulatory role in the response to embryopathic stresses not activating the p53 pathway, but more studies are needed to explore these mechanisms. It is also undoubtedly important to reveal whether other teratogens can affect offspring in this way [182].
Conclusions
The knowledge regarding the feto-maternal cross talk is rapidly expanding and an ever-increasing amount of evidence has been highlighting the importance of epigenetic regulation both in determining the effectiveness of early impregnation processes and in influencing fetal development. While many of the epigenetic mechanisms affecting early and late embryo development have long been known and studied, a promising new frontier comprising ncRNAs and neurodevelopmental factors has yet to be fully explored and understood, especially regarding the human species. Determining the precise mechanisms underlying such phenomena could prove extremely valuable towards a better management of both physiological pregnancies and assisted reproduction treatments. A better understanding of the role of epigenetic agents in embryonic development may ultimately improve counselling in infertile couples, provide new pharmacological treatments to favor the early phases of the pregnancy, and allow a better recognition and management of potentially harmful or even beneficial environmental factors with regards to fetal development.
Funding
Open access funding provided by Università degli Studi dell’Aquila within the CRUI-CARE Agreement.
Declarations
Competing interests
Antonio Capalbo is full-time employee at Igenomix Italy. Other authors declare no conflicts of interest.
Footnotes
Daniela Zuccarello on behalf of SIERR
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Daniela Zuccarello and Ugo Sorrentino contributed equally to this work.
References
- 1.Barker DJ. The fetal and infant origins of adult disease. BMJ. 1990;301:1111. doi: 10.1136/bmj.301.6761.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurian NK, Modi D. Extracellular vesicle mediated embryo-endometrial cross talk during implantation and in pregnancy. J Assist Reprod Genet. 2019;36:189–198. doi: 10.1007/s10815-018-1343-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luddi A, Pavone V, Semplici B, Governini L, Criscuoli M, Paccagnini E, Gentile M, Morgante G, Leo VD, Belmonte G, Zarovni N, Piomboni P. Organoids of human endometrium: a powerful in vitro model for the endometrium-embryo cross-talk at the implantation site. Cells. 2020;9:E1121. doi: 10.3390/cells9051121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Argyraki M, Damdimopoulou P, Chatzimeletiou K, Grimbizis GF, Tarlatzis BC, Syrrou M, Lambropoulos A. In-utero stress and mode of conception: impact on regulation of imprinted genes, fetal development and future health. Hum Reprod Update. 2019;25:777–801. doi: 10.1093/humupd/dmz025. [DOI] [PubMed] [Google Scholar]
- 5.Legoff L, D’Cruz SC, Tevosian S, Primig M, Smagulova F. Transgenerational inheritance of environmentally induced epigenetic alterations during mammalian development. Cells. 2019;8:E1559. doi: 10.3390/cells8121559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rivera RM, Ross JW. Epigenetics in fertilization and preimplantation embryo development. Prog Biophys Mol Biol. 2013;113:423–432. doi: 10.1016/j.pbiomolbio.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 8.Tammen SA, Friso S, Choi S-W. Epigenetics: the link between nature and nurture. Mol Aspects Med. 2013;34:753–764. doi: 10.1016/j.mam.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Best JD, Carey N. The epigenetics of normal pregnancy. Obstet Med. 2013;6:3–7. doi: 10.1258/om.2011.110070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goyal D, Limesand SW, Goyal R. Epigenetic responses and the developmental origins of health and disease. J Endocrinol. 2019;242:T105–T119. doi: 10.1530/JOE-19-0009. [DOI] [PubMed] [Google Scholar]
- 11.Skvortsova K, Iovino N, Bogdanović O. Functions and mechanisms of epigenetic inheritance in animals. Nat Rev Mol Cell Biol. 2018;19:774–790. doi: 10.1038/s41580-018-0074-2. [DOI] [PubMed] [Google Scholar]
- 12.Borchiellini M, Ummarino S, Di Ruscio A. The bright and dark side of DNA methylation: a matter of balance. Cells. 2019;8:E1243. doi: 10.3390/cells8101243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aristizabal MJ, Anreiter I, Halldorsdottir T, Odgers CL, McDade TW, Goldenberg A, Mostafavi S, Kobor MS, Binder EB, Sokolowski MB, O’Donnell KJ. Biological embedding of experience: a primer on epigenetics. Proc Natl Acad Sci U S A. 2020;117:23261–23269. doi: 10.1073/pnas.1820838116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Lu Q, Chang C. Epigenetics in health and disease. Adv Exp Med Biol. 2020;1253:3–55. doi: 10.1007/978-981-15-3449-2_1. [DOI] [PubMed] [Google Scholar]
- 15.Liberman N, Wang SY, Greer EL. Transgenerational epigenetic inheritance: from phenomena to molecular mechanisms. Curr Opin Neurobiol. 2019;59:189–206. doi: 10.1016/j.conb.2019.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Angeloni A, Bogdanovic O. Enhancer DNA methylation: implications for gene regulation. Essays Biochem. 2019;63:707–715. doi: 10.1042/EBC20190030. [DOI] [PubMed] [Google Scholar]
- 17.Teschendorff AE, Relton CL. Statistical and integrative system-level analysis of DNA methylation data. Nat Rev Genet. 2018;19:129–147. doi: 10.1038/nrg.2017.86. [DOI] [PubMed] [Google Scholar]
- 18.Zhao L-Y, Song J, Liu Y, Song C-X, Yi C. Mapping the epigenetic modifications of DNA and RNA. Protein Cell. 2020;11:792–808. doi: 10.1007/s13238-020-00733-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suganuma T, Workman JL. Crosstalk among histone modifications. Cell. 2008;135:604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 20.Bellanti JA. Epigenetic studies and pediatric research. Pediatr Res. 2020;87:378–384. doi: 10.1038/s41390-019-0644-9. [DOI] [PubMed] [Google Scholar]
- 21.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 22.Alaskhar Alhamwe B, Khalaila R, Wolf J, von Bülow V, Harb H, Alhamdan F, Hii CS, Prescott SL, Ferrante A, Renz H, Garn H, Potaczek DP. Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy Asthma Clin Immunol. 2018;14:39. doi: 10.1186/s13223-018-0259-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gansen A, Tóth K, Schwarz N, Langowski J. Opposing roles of H3- and H4-acetylation in the regulation of nucleosome structure––a FRET study. Nucleic Acids Res. 2015;43:1433–1443. doi: 10.1093/nar/gku1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 25.Agricola E, Verdone L, Di Mauro E, Caserta M. H4 acetylation does not replace H3 acetylation in chromatin remodelling and transcription activation of Adr1-dependent genes. Mol Microbiol. 2006;62:1433–1446. doi: 10.1111/j.1365-2958.2006.05451.x. [DOI] [PubMed] [Google Scholar]
- 26.Cha J, Sun X, Dey SK. Mechanisms of implantation: strategies for successful pregnancy. Nat Med. 2012;18:1754–1767. doi: 10.1038/nm.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashary N, Tiwari A, Modi D. Embryo implantation: war in times of love. Endocrinology. 2018;159:1188–1198. doi: 10.1210/en.2017-03082. [DOI] [PubMed] [Google Scholar]
- 28.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossetto D, Avvakumov N, Côté J. Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics. 2012;7:1098–1108. doi: 10.4161/epi.21975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 31.Zakar T, Paul JW. Fetal membrane epigenetics. Front Physiol. 2020;11:588539. doi: 10.3389/fphys.2020.588539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattick JS, Makunin IV (2006) Non-coding RNA. Hum Mol Genet 15 Spec No 1:R17–29. 10.1093/hmg/ddl046 [DOI] [PubMed]
- 33.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 34.Wei J-W, Huang K, Yang C, Kang C-S. Non-coding RNAs as regulators in epigenetics (Review) Oncol Rep. 2017;37:3–9. doi: 10.3892/or.2016.5236. [DOI] [PubMed] [Google Scholar]
- 35.Chen J, Xue Y. Emerging roles of non-coding RNAs in epigenetic regulation. Sci China Life Sci. 2016;59:227–235. doi: 10.1007/s11427-016-5010-0. [DOI] [PubMed] [Google Scholar]
- 36.Peschansky VJ, Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9:3–12. doi: 10.4161/epi.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalish JM, Jiang C, Bartolomei MS. Epigenetics and imprinting in human disease. Int J Dev Biol. 2014;58:291–298. doi: 10.1387/ijdb.140077mb. [DOI] [PubMed] [Google Scholar]
- 38.Messerschmidt DM, Knowles BB, Solter D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014;28:812–828. doi: 10.1101/gad.234294.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 40.Zeng Y, Chen T. DNA methylation reprogramming during mammalian development. Genes. 2019;10:257. doi: 10.3390/genes10040257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meng H, Cao Y, Qin J, Song X, Zhang Q, Shi Y, Cao L. DNA methylation, its mediators and genome integrity. Int J Biol Sci. 2015;11:604–617. doi: 10.7150/ijbs.11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanna CW, Kelsey G. Genomic imprinting beyond DNA methylation: a role for maternal histones. Genome Biol. 2017;18:177. doi: 10.1186/s13059-017-1317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lozano-Ureña A, Montalbán-Loro R, Ferguson-Smith AC, Ferrón SR. Genomic imprinting and the regulation of postnatal neurogenesis. Brain Plast. 2017;3:89–98. doi: 10.3233/BPL-160041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elbracht M, Mackay D, Begemann M, Kagan KO, Eggermann T. Disturbed genomic imprinting and its relevance for human reproduction: causes and clinical consequences. Hum Reprod Update. 2020;26:197–213. doi: 10.1093/humupd/dmz045. [DOI] [PubMed] [Google Scholar]
- 45.Cassidy FC, Charalambous M. Genomic imprinting, growth and maternal-fetal interactions. J Exp Biol. 2018;221:jeb164517. doi: 10.1242/jeb.164517. [DOI] [PubMed] [Google Scholar]
- 46.Monk D, Mackay DJG, Eggermann T, Maher ER, Riccio A. Genomic imprinting disorders: lessons on how genome, epigenome and environment interact. Nat Rev Genet. 2019;20:235–248. doi: 10.1038/s41576-018-0092-0. [DOI] [PubMed] [Google Scholar]
- 47.Gannon JR, Emery BR, Jenkins TG, Carrell DT. The sperm epigenome: implications for the embryo. Adv Exp Med Biol. 2014;791:53–66. doi: 10.1007/978-1-4614-7783-9_4. [DOI] [PubMed] [Google Scholar]
- 48.Gold HB, Jung YH, Corces VG. Not just heads and tails: the complexity of the sperm epigenome. J Biol Chem. 2018;293:13815–13820. doi: 10.1074/jbc.R117.001561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheuquemán C, Maldonado R. Non-coding RNAs and chromatin: key epigenetic factors from spermatogenesis to transgenerational inheritance. Biol Res. 2021;54:41. doi: 10.1186/s40659-021-00364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schagdarsurengin U, Paradowska A, Steger K. Analysing the sperm epigenome: roles in early embryogenesis and assisted reproduction. Nat Rev Urol. 2012;9:609–619. doi: 10.1038/nrurol.2012.183. [DOI] [PubMed] [Google Scholar]
- 51.Jenkins TG, Carrell DT. The sperm epigenome and potential implications for the developing embryo. Reproduction. 2012;143:727–734. doi: 10.1530/REP-11-0450. [DOI] [PubMed] [Google Scholar]
- 52.Carrell DT. The sperm epigenome: implications for assisted reproductive technologies. Adv Exp Med Biol. 2019;1166:47–56. doi: 10.1007/978-3-030-21664-1_3. [DOI] [PubMed] [Google Scholar]
- 53.Khambata K, Raut S, Deshpande S, Mohan S, Sonawane S, Gaonkar R, Ansari Z, Datar M, Bansal V, Patil A, Warke H, Balasinor NH. DNA methylation defects in spermatozoa of male partners from couples experiencing recurrent pregnancy loss. Hum Reprod. 2021;36:48–60. doi: 10.1093/humrep/deaa278. [DOI] [PubMed] [Google Scholar]
- 54.Carrell DT, Hammoud SS. The human sperm epigenome and its potential role in embryonic development. Mol Hum Reprod. 2010;16:37–47. doi: 10.1093/molehr/gap090. [DOI] [PubMed] [Google Scholar]
- 55.Cannarella R, Crafa A, Condorelli RA, Mongioì LM, La Vignera S, Calogero AE. Relevance of sperm imprinted gene methylation on assisted reproductive technique outcomes and pregnancy loss: a systematic review. Syst Biol Reprod Med. 2021;67:251–259. doi: 10.1080/19396368.2021.1909667. [DOI] [PubMed] [Google Scholar]
- 56.Tomizawa S-I, Nowacka-Woszuk J, Kelsey G. DNA methylation establishment during oocyte growth: mechanisms and significance. Int J Dev Biol. 2012;56:867–875. doi: 10.1387/ijdb.120152gk. [DOI] [PubMed] [Google Scholar]
- 57.Yu B, Doni Jayavelu N, Battle SL, Mar JC, Schimmel T, Cohen J, Hawkins RD. Single-cell analysis of transcriptome and DNA methylome in human oocyte maturation. PLoS ONE. 2020;15:e0241698. doi: 10.1371/journal.pone.0241698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stewart KR, Veselovska L, Kelsey G. Establishment and functions of DNA methylation in the germline. Epigenomics. 2016;8:1399–1413. doi: 10.2217/epi-2016-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sendžikaitė G, Kelsey G. The role and mechanisms of DNA methylation in the oocyte. Essays Biochem. 2019;63:691–705. doi: 10.1042/EBC20190043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anvar Z, Chakchouk I, Demond H, Sharif M, Kelsey G, Van den Veyver IB. DNA methylation dynamics in the female germline and maternal-effect mutations that disrupt genomic imprinting. Genes (Basel) 2021;12:1214. doi: 10.3390/genes12081214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Osman E, Franasiak J, Scott R. Oocyte and embryo manipulation and epigenetics. Semin Reprod Med. 2018;36:e1–e9. doi: 10.1055/s-0039-1688801. [DOI] [PubMed] [Google Scholar]
- 62.Mitchell LE. Maternal effect genes: update and review of evidence for a link with birth defects. HGG Adv. 2022;3:100067. doi: 10.1016/j.xhgg.2021.100067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rong Y, Ji S-Y, Zhu Y-Z, Wu Y-W, Shen L, Fan H-Y. ZAR1 and ZAR2 are required for oocyte meiotic maturation by regulating the maternal transcriptome and mRNA translational activation. Nucleic Acids Res. 2019;47:11387–11402. doi: 10.1093/nar/gkz863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang J-J, Liu X, Chen L, Zhang S, Zhang X, Hao C, Miao Y-L (2020) Advanced maternal age alters expression of maternal effect genes that are essential for human oocyte quality. Aging (Albany NY) 12:3950–3961. 10.18632/aging.102864 [DOI] [PMC free article] [PubMed]
- 65.Xia W, Xie W. Rebooting the epigenomes during mammalian early embryogenesis. Stem Cell Reports. 2020;15:1158–1175. doi: 10.1016/j.stemcr.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu R, Li C, Liu X, Gao S. Insights into epigenetic patterns in mammalian early embryos. Protein Cell. 2021;12:7–28. doi: 10.1007/s13238-020-00757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xia W, Xu J, Yu G, Yao G, Xu K, Ma X, Zhang N, Liu B, Li T, Lin Z, Chen X, Li L, Wang Q, Shi D, Shi S, Zhang Y, Song W, Jin H, Hu L, Bu Z, Wang Y, Na J, Xie W, Sun Y-P. Resetting histone modifications during human parental-to-zygotic transition. Science. 2019;365:353–360. doi: 10.1126/science.aaw5118. [DOI] [PubMed] [Google Scholar]
- 68.Silver MJ, Saffari A, Kessler NJ, Chandak GR, Fall CHD, Issarapu P, Dedaniya A, Betts M, Moore SE, Routledge MN, Herceg Z, Cuenin C, Derakhshan M, James PT, Monk D, Prentice AM. Environmentally sensitive hotspots in the methylome of the early human embryo. Elife. 2022;11:e72031. doi: 10.7554/eLife.72031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Glaser J, Mundlos S (2021) 3D or not 3D: shaping the genome during development. Cold Spring Harb Perspect Biol a040188. 10.1101/cshperspect.a040188 [DOI] [PMC free article] [PubMed]
- 70.Lee TW, Katz DJ. Hansel, Gretel, and the consequences of failing to remove histone methylation breadcrumbs. Trends Genet. 2020;36:160–176. doi: 10.1016/j.tig.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y, Liu Q, Tang F, Yan L, Qiao J. Epigenetic regulation and risk factors during the development of human gametes and early embryos. Annu Rev Genomics Hum Genet. 2019;20:21–40. doi: 10.1146/annurev-genom-083118-015143. [DOI] [PubMed] [Google Scholar]
- 72.Greenberg MVC, Glaser J, Borsos M, Marjou FE, Walter M, Teissandier A, Bourc’his D, Transient transcription in the early embryo sets an epigenetic state that programs postnatal growth. Nat Genet. 2017;49:110–118. doi: 10.1038/ng.3718. [DOI] [PubMed] [Google Scholar]
- 73.Zhang S, Lin H, Kong S, Wang S, Wang H, Wang H, Armant DR. Physiological and molecular determinants of embryo implantation. Mol Aspects Med. 2013;34:939–980. doi: 10.1016/j.mam.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo S-W. The endometrial epigenome and its response to steroid hormones. Mol Cell Endocrinol. 2012;358:185–196. doi: 10.1016/j.mce.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 75.Retis-Resendiz AM, González-García IN, León-Juárez M, Camacho-Arroyo I, Cerbón M, Vázquez-Martínez ER. The role of epigenetic mechanisms in the regulation of gene expression in the cyclical endometrium. Clin Epigenetics. 2021;13:116. doi: 10.1186/s13148-021-01103-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Houshdaran S, Zelenko Z, Irwin JC, Giudice LC. Human endometrial DNA methylome is cycle-dependent and is associated with gene expression regulation. Mol Endocrinol. 2014;28:1118–1135. doi: 10.1210/me.2013-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Munro SK, Farquhar CM, Mitchell MD, Ponnampalam AP. Epigenetic regulation of endometrium during the menstrual cycle. Mol Hum Reprod. 2010;16:297–310. doi: 10.1093/molehr/gaq010. [DOI] [PubMed] [Google Scholar]
- 78.Pathare ADS, Hinduja I. Aberrant DNA methylation profiling affecting the endometrial receptivity in recurrent implantation failure patients undergoing in vitro fertilization. Am J Reprod Immunol. 2020;83:e13196. doi: 10.1111/aji.13196. [DOI] [PubMed] [Google Scholar]
- 79.Yu S-L, Kim T-H, Han Y-H, Kang Y, Jeong D-U, Lee DC, Kang J, Park S-R. Transcriptomic analysis and competing endogenous RNA network in the human endometrium between proliferative and mid-secretory phases. Exp Ther Med. 2021;21:660. doi: 10.3892/etm.2021.10092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morales-Prieto DM, Ospina-Prieto S, Chaiwangyen W, Schoenleben M, Markert UR. Pregnancy-associated miRNA-clusters. J Reprod Immunol. 2013;97:51–61. doi: 10.1016/j.jri.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 81.Chang G, Mouillet J, Mishima T, Chu T, Sadovsky E, Coyne CB, Parks WT, Surti U, Sadovsky Y. Expression and trafficking of placental microRNAs at the feto-maternal interface. FASEB j. 2017;31:2760–2770. doi: 10.1096/fj.201601146R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Paquette AG, Chu T, Wu X, Wang K, Price ND, Sadovsky Y. Distinct communication patterns of trophoblastic miRNA among the maternal-placental-fetal compartments. Placenta. 2018;72–73:28–35. doi: 10.1016/j.placenta.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paul ABM, Sadek ST, Mahesan AM. The role of microRNAs in human embryo implantation: a review. J Assist Reprod Genet. 2019;36:179–187. doi: 10.1007/s10815-018-1326-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Altmäe S, Martinez-Conejero JA, Esteban FJ, Ruiz-Alonso M, Stavreus-Evers A, Horcajadas JA, Salumets A. MicroRNAs miR-30b, miR-30d, and miR-494 regulate human endometrial receptivity. Reprod Sci. 2013;20:308–317. doi: 10.1177/1933719112453507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vilella F, Moreno-Moya JM, Balaguer N, Grasso A, Herrero M, Martínez S, Marcilla A, Simón C. Hsa-miR-30d, secreted by the human endometrium, is taken up by the pre-implantation embryo and might modify its transcriptome. Development. 2015;142:3210–3221. doi: 10.1242/dev.124289. [DOI] [PubMed] [Google Scholar]
- 86.Kresowik JDK, Devor EJ, Van Voorhis BJ, Leslie KK. MicroRNA-31 is significantly elevated in both human endometrium and serum during the window of implantation: a potential biomarker for optimum receptivity. Biol Reprod. 2014;91:17. doi: 10.1095/biolreprod.113.116590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang X, Li B, Wang J, Lei J, Liu C, Ma Y, Zhao H. Evidence that miR-133a causes recurrent spontaneous abortion by reducing HLA-G expression. Reprod Biomed Online. 2012;25:415–424. doi: 10.1016/j.rbmo.2012.06.022. [DOI] [PubMed] [Google Scholar]
- 88.Porto IOP, Mendes-Junior CT, Felício LP, Georg RC, Moreau P, Donadi EA, Chies JAB, Castelli EC. MicroRNAs targeting the immunomodulatory HLA-G gene: a new survey searching for microRNAs with potential to regulate HLA-G. Mol Immunol. 2015;65:230–241. doi: 10.1016/j.molimm.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 89.Liu W, Niu Z, Li Q, Pang RTK, Chiu PCN, Yeung WS-B. MicroRNA and embryo implantation. Am J Reprod Immunol. 2016;75:263–271. doi: 10.1111/aji.12470. [DOI] [PubMed] [Google Scholar]
- 90.Kim S-M, Kim J-S. A review of mechanisms of implantation. Dev Reprod. 2017;21:351–359. doi: 10.12717/DR.2017.21.4.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aplin JD, Ruane PT. Embryo–epithelium interactions during implantation at a glance. J Cell Sci. 2017;130:15–22. doi: 10.1242/jcs.175943. [DOI] [PubMed] [Google Scholar]
- 92.Tochigi H, Kajihara T, Mizuno Y, Mizuno Y, Tamaru S, Kamei Y, Okazaki Y, Brosens JJ, Ishihara O. Loss of miR-542-3p enhances IGFBP-1 expression in decidualizing human endometrial stromal cells. Sci Rep. 2017;7:40001. doi: 10.1038/srep40001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Estella C, Herrer I, Moreno-Moya JM, Quiñonero A, Martínez S, Pellicer A, Simón C. miRNA signature and Dicer requirement during human endometrial stromal decidualization in vitro. PLoS ONE. 2012;7:e41080. doi: 10.1371/journal.pone.0041080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Macklon NS, Brosens JJ. The human endometrium as a sensor of embryo quality. Biol Reprod. 2014;91:98. doi: 10.1095/biolreprod.114.122846. [DOI] [PubMed] [Google Scholar]
- 95.Bridi A, Perecin F, da Silveira JC. Extracellular vesicles mediated early embryo-maternal interactions. Int J Mol Sci. 2020;21:E1163. doi: 10.3390/ijms21031163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Homer H, Rice GE, Salomon C. Review: Embryo- and endometrium-derived exosomes and their potential role in assisted reproductive treatments-liquid biopsies for endometrial receptivity. Placenta. 2017;54:89–94. doi: 10.1016/j.placenta.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 97.Chen K, Liang J, Qin T, Zhang Y, Chen X, Wang Z. The role of extracellular vesicles in embryo implantation. Front Endocrinol (Lausanne) 2022;13:809596. doi: 10.3389/fendo.2022.809596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.O’Neil EV, Burns GW, Spencer TE. Extracellular vesicles: novel regulators of conceptus-uterine interactions? Theriogenology. 2020;150:106–112. doi: 10.1016/j.theriogenology.2020.01.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mishra A, Ashary N, Sharma R, Modi D. Extracellular vesicles in embryo implantation and disorders of the endometrium. Am J Reprod Immunol. 2021;85:e13360. doi: 10.1111/aji.13360. [DOI] [PubMed] [Google Scholar]
- 100.Vaiman D. Genes, epigenetics and miRNA regulation in the placenta. Placenta. 2017;52:127–133. doi: 10.1016/j.placenta.2016.12.026. [DOI] [PubMed] [Google Scholar]
- 101.Cuman C, Van Sinderen M, Gantier MP, Rainczuk K, Sorby K, Rombauts L, Osianlis T, Dimitriadis E. Human blastocyst secreted microRNA regulate endometrial epithelial cell adhesion. EBioMedicine. 2015;2:1528–1535. doi: 10.1016/j.ebiom.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li T, Greenblatt EM, Shin ME, Brown TJ, Chan C. Cargo small non-coding RNAs of extracellular vesicles isolated from uterine fluid associate with endometrial receptivity and implantation success. Fertil Steril. 2021;115:1327–1336. doi: 10.1016/j.fertnstert.2020.10.046. [DOI] [PubMed] [Google Scholar]
- 103.Knöfler M, Haider S, Saleh L, Pollheimer J, Gamage TKJB, James J. Human placenta and trophoblast development: key molecular mechanisms and model systems. Cell Mol Life Sci. 2019;76:3479–3496. doi: 10.1007/s00018-019-03104-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Okada H, Tsuzuki T, Murata H. Decidualization of the human endometrium. Reprod Med Biol. 2018;17:220–227. doi: 10.1002/rmb2.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu S, Diao L, Huang C, Li Y, Zeng Y, Kwak-Kim JYH. The role of decidual immune cells on human pregnancy. J Reprod Immunol. 2017;124:44–53. doi: 10.1016/j.jri.2017.10.045. [DOI] [PubMed] [Google Scholar]
- 106.Wallace AE, Fraser R, Cartwright JE. Extravillous trophoblast and decidual natural killer cells: a remodelling partnership. Hum Reprod Update. 2012;18:458–471. doi: 10.1093/humupd/dms015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Napso T, Yong HEJ, Lopez-Tello J, Sferruzzi-Perri AN. The role of placental hormones in mediating maternal adaptations to support pregnancy and lactation. Front Physiol. 2018;9:1091. doi: 10.3389/fphys.2018.01091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shallie PD, Naicker T. The placenta as a window to the brain: a review on the role of placental markers in prenatal programming of neurodevelopment. Int J Dev Neurosci. 2019;73:41–49. doi: 10.1016/j.ijdevneu.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 109.Konwar C, Price EM, Wang LQ, Wilson SL, Terry J, Robinson WP. DNA methylation profiling of acute chorioamnionitis-associated placentas and fetal membranes: insights into epigenetic variation in spontaneous preterm births. Epigenetics Chromatin. 2018;11:63. doi: 10.1186/s13072-018-0234-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bale TL. Epigenetic and transgenerational reprogramming of brain development. Nat Rev Neurosci. 2015;16:332–344. doi: 10.1038/nrn3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tau GZ, Peterson BS. Normal development of brain circuits. Neuropsychopharmacology. 2010;35:147–168. doi: 10.1038/npp.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Faa G, Manchia M, Pintus R, Gerosa C, Marcialis MA, Fanos V. Fetal programming of neuropsychiatric disorders. Birth Defects Res C Embryo Today. 2016;108:207–223. doi: 10.1002/bdrc.21139. [DOI] [PubMed] [Google Scholar]
- 113.Barker DJ, Bull AR, Osmond C, Simmonds SJ. Fetal and placental size and risk of hypertension in adult life. BMJ. 1990;301:259–262. doi: 10.1136/bmj.301.6746.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Warning JC, McCracken SA, Morris JM. A balancing act: mechanisms by which the fetus avoids rejection by the maternal immune system. Reproduction. 2011;141:715–724. doi: 10.1530/REP-10-0360. [DOI] [PubMed] [Google Scholar]
- 115.Bowers ME, Yehuda R. Intergenerational transmission of stress in humans. Neuropsychopharmacology. 2016;41:232–244. doi: 10.1038/npp.2015.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gil-Ibáñez P, Bernal J, Morte B. Thyroid hormone regulation of gene expression in primary cerebrocortical cells: role of thyroid hormone receptor subtypes and interactions with retinoic acid and glucocorticoids. PLoS ONE. 2014;9:e91692. doi: 10.1371/journal.pone.0091692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bonnin A, Goeden N, Chen K, Wilson ML, King J, Shih JC, Blakely RD, Deneris ES, Levitt P. A transient placental source of serotonin for the fetal forebrain. Nature. 2011;472:347–350. doi: 10.1038/nature09972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rosenfeld CS. The placenta-brain-axis. J Neurosci Res. 2021;99:271–283. doi: 10.1002/jnr.24603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lager S, Powell TL. Regulation of nutrient transport across the placenta. J Pregnancy. 2012;2012:179827. doi: 10.1155/2012/179827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lesseur C, Armstrong DA, Murphy MA, Appleton AA, Koestler DC, Paquette AG, Lester BM, Marsit CJ. Sex-specific associations between placental leptin promoter DNA methylation and infant neurobehavior. Psychoneuroendocrinology. 2014;40:1–9. doi: 10.1016/j.psyneuen.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Howerton CL, Bale TL. Prenatal programing: at the intersection of maternal stress and immune activation. Horm Behav. 2012;62:237–242. doi: 10.1016/j.yhbeh.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gallou-Kabani C, Gabory A, Tost J, Karimi M, Mayeur S, Lesage J, Boudadi E, Gross M-S, Taurelle J, Vigé A, Breton C, Reusens B, Remacle C, Vieau D, Ekström TJ, Jais J-P, Junien C. Sex- and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet. PLoS ONE. 2010;5:e14398. doi: 10.1371/journal.pone.0014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Howerton CL, Bale TL. Targeted placental deletion of OGT recapitulates the prenatal stress phenotype including hypothalamic mitochondrial dysfunction. Proc Natl Acad Sci U S A. 2014;111:9639–9644. doi: 10.1073/pnas.1401203111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rosenfeld CS. Placental serotonin signaling, pregnancy outcomes, and regulation of fetal brain development†. Biol Reprod. 2020;102:532–538. doi: 10.1093/biolre/ioz204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Paquette AG, Lesseur C, Armstrong DA, Koestler DC, Appleton AA, Lester BM, Marsit CJ. Placental HTR2A methylation is associated with infant neurobehavioral outcomes. Epigenetics. 2013;8:796–801. doi: 10.4161/epi.25358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Green BB, Kappil M, Lambertini L, Armstrong DA, Guerin DJ, Sharp AJ, Lester BM, Chen J, Marsit CJ. Expression of imprinted genes in placenta is associated with infant neurobehavioral development. Epigenetics. 2015;10:834–841. doi: 10.1080/15592294.2015.1073880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schmidt RJ, Schroeder DI, Crary-Dooley FK, Barkoski JM, Tancredi DJ, Walker CK, Ozonoff S, Hertz-Picciotto I, LaSalle JM. Self-reported pregnancy exposures and placental DNA methylation in the MARBLES prospective autism sibling study. Environ Epigenet. 2016;2:dvw024. doi: 10.1093/eep/dvw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhu Y, Mordaunt CE, Yasui DH, Marathe R, Coulson RL, Dunaway KW, Jianu JM, Walker CK, Ozonoff S, Hertz-Picciotto I, Schmidt RJ, LaSalle JM. Placental DNA methylation levels at CYP2E1 and IRS2 are associated with child outcome in a prospective autism study. Hum Mol Genet. 2019;28:2659–2674. doi: 10.1093/hmg/ddz084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Maccani MA, Padbury JF, Lester BM, Knopik VS, Marsit CJ. Placental miRNA expression profiles are associated with measures of infant neurobehavioral outcomes. Pediatr Res. 2013;74:272–278. doi: 10.1038/pr.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cratty MS, Ward HE, Johnson EA, Azzaro AJ, Birkle DL. Prenatal stress increases corticotropin-releasing factor (CRF) content and release in rat amygdala minces. Brain Res. 1995;675:297–302. doi: 10.1016/0006-8993(95)00087-7. [DOI] [PubMed] [Google Scholar]
- 131.Bögi E, Belovičová K, Moravčíková L, Csatlósová K, Dremencov E, Lacinova L, Dubovicky M. Pre-gestational stress impacts excitability of hippocampal cells in vitro and is associated with neurobehavioral alterations during adulthood. Behav Brain Res. 2019;375:112131. doi: 10.1016/j.bbr.2019.112131. [DOI] [PubMed] [Google Scholar]
- 132.Zijlmans MAC, Riksen-Walraven JM, de Weerth C. Associations between maternal prenatal cortisol concentrations and child outcomes: a systematic review. Neurosci Biobehav Rev. 2015;53:1–24. doi: 10.1016/j.neubiorev.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 133.Cai S, Quan S, Yang G, Chen M, Ye Q, Wang G, Yu H, Wang Y, Qiao S, Zeng X. Nutritional status impacts epigenetic regulation in early embryo development: a scoping review. Adv Nutr. 2021;12:1877–1892. doi: 10.1093/advances/nmab038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu H-Y, Liu S-M, Zhang Y-Z. Maternal folic acid supplementation mediates offspring health via DNA methylation. Reprod Sci. 2020;27:963–976. doi: 10.1007/s43032-020-00161-2. [DOI] [PubMed] [Google Scholar]
- 135.Keen CL, Uriu-Adams JY, Skalny A, Grabeklis A, Grabeklis S, Green K, Yevtushok L, Wertelecki WW, Chambers CD. The plausibility of maternal nutritional status being a contributing factor to the risk for fetal alcohol spectrum disorders: the potential influence of zinc status as an example. BioFactors. 2010;36:125–135. doi: 10.1002/biof.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sebastiani G, Andreu-Fernández V, Herranz Barbero A, Aldecoa-Bilbao V, Miracle X, Meler Barrabes E, Balada Ibañez A, Astals-Vizcaino M, Ferrero-Martínez S, Gómez-Roig MD, García-Algar O. Eating disorders during gestation: implications for mother’s health, fetal outcomes, and epigenetic changes. Front Pediatr. 2020;8:587. doi: 10.3389/fped.2020.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Oster M, Nuchchanart W, Trakooljul N, Muráni E, Zeyner A, Wirthgen E, Hoeflich A, Ponsuksili S, Wimmers K. Methylating micronutrient supplementation during pregnancy influences foetal hepatic gene expression and IGF signalling and increases foetal weight. Eur J Nutr. 2016;55:1717–1727. doi: 10.1007/s00394-015-0990-2. [DOI] [PubMed] [Google Scholar]
- 138.Bekdash RA. Early life nutrition and mental health: the role of DNA methylation. Nutrients. 2021;13:3111. doi: 10.3390/nu13093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Altundag Ö, Çelebi-Saltik B. From embryo to adult: one carbon metabolism in stem cells. Curr Stem Cell Res Ther. 2021;16:175–188. doi: 10.2174/1574888X15666200712191308. [DOI] [PubMed] [Google Scholar]
- 140.Alabduljabbar S, Zaidan SA, Lakshmanan AP, Terranegra A. Personalized nutrition approach in pregnancy and early life to tackle childhood and adult non-communicable diseases. Life (Basel) 2021;11:467. doi: 10.3390/life11060467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rush EC, Katre P, Yajnik CS. Vitamin B12: one carbon metabolism, fetal growth and programming for chronic disease. Eur J Clin Nutr. 2014;68:2–7. doi: 10.1038/ejcn.2013.232. [DOI] [PubMed] [Google Scholar]
- 142.McKay JA, Groom A, Potter C, Coneyworth LJ, Ford D, Mathers JC, Relton CL. Genetic and non-genetic influences during pregnancy on infant global and site specific DNA methylation: role for folate gene variants and vitamin B12. PLoS ONE. 2012;7:e33290. doi: 10.1371/journal.pone.0033290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, Slagboom PE, Heijmans BT. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18:4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Roseboom TJ, van der Meulen JH, Osmond C, Barker DJ, Ravelli AC, Schroeder-Tanka JM, van Montfrans GA, Michels RP, Bleker OP. Coronary heart disease after prenatal exposure to the Dutch famine, 1944–45. Heart. 2000;84:595–598. doi: 10.1136/heart.84.6.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zheng J, Xiao X, Zhang Q, Wang T, Yu M, Xu J. Maternal low-protein diet modulates glucose metabolism and hepatic microRNAs expression in the early life of offspring †. Nutrients. 2017;9:E205. doi: 10.3390/nu9030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hjort L, Martino D, Grunnet LG, Naeem H, Maksimovic J, Olsson AH, Zhang C, Ling C, Olsen SF, Saffery R, Vaag AA. Gestational diabetes and maternal obesity are associated with epigenome-wide methylation changes in children. JCI Insight. 2018;3:122572. doi: 10.1172/jci.insight.122572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Silver MJ. Intergenerational influences on child development: an epigenetic perspective. Nestle Nutr Inst Workshop Ser. 2020;93:145–152. doi: 10.1159/000503351. [DOI] [PubMed] [Google Scholar]
- 148.Yong HY, Mohd Shariff Z, Mohd Yusof BN, Rejali Z, Tee YYS, Bindels J, van der Beek EM. Independent and combined effects of age, body mass index and gestational weight gain on the risk of gestational diabetes mellitus. Sci Rep. 2020;10:8486. doi: 10.1038/s41598-020-65251-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mijatovic-Vukas J, Capling L, Cheng S, Stamatakis E, Louie J, Cheung NW, Markovic T, Ross G, Senior A, Brand-Miller JC, Flood VM. Associations of diet and physical activity with risk for gestational diabetes mellitus: a systematic review and meta-analysis. Nutrients. 2018;10:E698. doi: 10.3390/nu10060698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cai S, Qiu A, Broekman BFP, Wong EQ, Gluckman PD, Godfrey KM, Saw SM, Soh S-E, Kwek K, Chong Y-S, Meaney MJ, Kramer MS, Rifkin-Graboi A, GUSTO study group The influence of gestational diabetes on neurodevelopment of children in the first two years of life: a prospective study. PLoS One. 2016;11:e0162113. doi: 10.1371/journal.pone.0162113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fraser A, Nelson SM, Macdonald-Wallis C, Lawlor DA. Associations of existing diabetes, gestational diabetes, and glycosuria with offspring IQ and educational attainment: the Avon Longitudinal Study of Parents and Children. Exp Diabetes Res. 2012;2012:963735. doi: 10.1155/2012/963735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Strutz J, Cvitic S, Hackl H, Kashofer K, Appel HM, Thüringer A, Desoye G, Koolwijk P, Hiden U. Gestational diabetes alters microRNA signatures in human feto-placental endothelial cells depending on fetal sex. Clin Sci (Lond) 2018;132:2437–2449. doi: 10.1042/CS20180825. [DOI] [PubMed] [Google Scholar]
- 153.Bassols J, Serino M, Carreras-Badosa G, Burcelin R, Blasco-Baque V, Lopez-Bermejo A, Fernandez-Real J-M. Gestational diabetes is associated with changes in placental microbiota and microbiome. Pediatr Res. 2016;80:777–784. doi: 10.1038/pr.2016.155. [DOI] [PubMed] [Google Scholar]
- 154.Słupecka-Ziemilska M, Wychowański P, Puzianowska-Kuznicka M. Gestational diabetes mellitus affects offspring’s epigenome. Is there a way to reduce the negative consequences? Nutrients. 2020;12:E2792. doi: 10.3390/nu12092792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bommarito PA, Martin E, Fry RC. Effects of prenatal exposure to endocrine disruptors and toxic metals on the fetal epigenome. Epigenomics. 2017;9:333–350. doi: 10.2217/epi-2016-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Casati L, Sendra R, Poletti A, Negri-Cesi P, Celotti F. Androgen receptor activation by polychlorinated biphenyls: epigenetic effects mediated by the histone demethylase Jarid1b. Epigenetics. 2013;8:1061–1068. doi: 10.4161/epi.25811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Casati L, Sendra R, Sibilia V, Celotti F. Endocrine disrupters: the new players able to affect the epigenome. Front Cell Dev Biol. 2015;3:37. doi: 10.3389/fcell.2015.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pilsner JR, Liu X, Ahsan H, Ilievski V, Slavkovich V, Levy D, Factor-Litvak P, Graziano JH, Gamble MV. Genomic methylation of peripheral blood leukocyte DNA: influences of arsenic and folate in Bangladeshi adults. Am J Clin Nutr. 2007;86:1179–1186. doi: 10.1093/ajcn/86.4.1179. [DOI] [PubMed] [Google Scholar]
- 159.Martin EM, Fry RC. A cross-study analysis of prenatal exposures to environmental contaminants and the epigenome: support for stress-responsive transcription factor occupancy as a mediator of gene-specific CpG methylation patterning. Environ Epigenet. 2016;2:dvv011. doi: 10.1093/eep/dvv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mandal C, Kim SH, Chai JC, Oh SM, Lee YS, Jung KH, Chai YG. RNA Sequencing reveals the alteration of the expression of novel genes in ethanol-treated embryoid bodies. PLoS ONE. 2016;11:e0149976. doi: 10.1371/journal.pone.0149976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Grewal J, Carmichael SL, Ma C, Lammer EJ, Shaw GM. Maternal periconceptional smoking and alcohol consumption and risk for select congenital anomalies. Birth Defects Res A Clin Mol Teratol. 2008;82:519–526. doi: 10.1002/bdra.20461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wallén E, Auvinen P, Kaminen-Ahola N. The effects of early prenatal alcohol exposure on epigenome and embryonic development. Genes (Basel) 2021;12:1095. doi: 10.3390/genes12071095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lussier AA, Bodnar TS, Mingay M, Morin AM, Hirst M, Kobor MS, Weinberg J. Prenatal alcohol exposure: profiling developmental DNA methylation patterns in central and peripheral tissues. Front Genet. 2018;9:610. doi: 10.3389/fgene.2018.00610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sarkar DK, Gangisetty O, Wozniak JR, Eckerle JK, Georgieff MK, Foroud TM, Wetherill L, Wertelecki W, Chambers CD, Riley E, Zymak-Zakutnya N, Yevtushok L. Persistent changes in stress-regulatory genes in pregnant women or children exposed prenatally to alcohol. Alcohol Clin Exp Res. 2019;43:1887–1897. doi: 10.1111/acer.14148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mandal C, Halder D, Jung KH, Chai YG. Gestational alcohol exposure altered DNA methylation status in the developing fetus. Int J Mol Sci. 2017;18:E1386. doi: 10.3390/ijms18071386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Abraham M, Alramadhan S, Iniguez C, Duijts L, Jaddoe VWV, Den Dekker HT, Crozier S, Godfrey KM, Hindmarsh P, Vik T, Jacobsen GW, Hanke W, Sobala W, Devereux G, Turner S. A systematic review of maternal smoking during pregnancy and fetal measurements with meta-analysis. PLoS ONE. 2017;12:e0170946. doi: 10.1371/journal.pone.0170946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Quelhas D, Kompala C, Wittenbrink B, Han Z, Parker M, Shapiro M, Downs S, Kraemer K, Fanzo J, Morris S, Kreis K. The association between active tobacco use during pregnancy and growth outcomes of children under five years of age: a systematic review and meta-analysis. BMC Public Health. 2018;18:1372. doi: 10.1186/s12889-018-6137-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ino T. Maternal smoking during pregnancy and offspring obesity: meta-analysis. Pediatr Int. 2010;52:94–99. doi: 10.1111/j.1442-200X.2009.02883.x. [DOI] [PubMed] [Google Scholar]
- 169.McEvoy CT, Spindel ER. Pulmonary effects of maternal smoking on the fetus and child: effects on lung development, respiratory morbidities, and life long lung health. Paediatr Respir Rev. 2017;21:27–33. doi: 10.1016/j.prrv.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rauschert S, Melton PE, Burdge G, Craig JM, Godfrey KM, Holbrook JD, Lillycrop K, Mori TA, Beilin LJ, Oddy WH, Pennell C, Huang R-C. Maternal smoking during pregnancy induces persistent epigenetic changes into adolescence, independent of postnatal smoke exposure and is associated with cardiometabolic risk. Front Genet. 2019;10:770. doi: 10.3389/fgene.2019.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Dong T, Hu W, Zhou X, Lin H, Lan L, Hang B, Lv W, Geng Q, Xia Y. Prenatal exposure to maternal smoking during pregnancy and attention-deficit/hyperactivity disorder in offspring: a meta-analysis. Reprod Toxicol. 2018;76:63–70. doi: 10.1016/j.reprotox.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 172.He Y, Chen J, Zhu L-H, Hua L-L, Ke F-F. Maternal smoking during pregnancy and ADHD: results from a systematic review and meta-analysis of prospective cohort studies. J Atten Disord. 2020;24:1637–1647. doi: 10.1177/1087054717696766. [DOI] [PubMed] [Google Scholar]
- 173.Froggatt S, Covey J, Reissland N. Infant neurobehavioural consequences of prenatal cigarette exposure: a systematic review and meta-analysis. Acta Paediatr. 2020;109:1112–1124. doi: 10.1111/apa.15132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Knopik VS, Marceau K, Bidwell LC, Rolan E. Prenatal substance exposure and offspring development: does DNA methylation play a role? Neurotoxicol Teratol. 2019;71:50–63. doi: 10.1016/j.ntt.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cecconi S, Rapino C, Di Nisio V, Rossi G, Maccarrone M. The (endo)cannabinoid signaling in female reproduction: what are the latest advances? Prog Lipid Res. 2020;77:101019. doi: 10.1016/j.plipres.2019.101019. [DOI] [PubMed] [Google Scholar]
- 176.Ivorra C, Fraga MF, Bayón GF, Fernández AF, Garcia-Vicent C, Chaves FJ, Redon J, Lurbe E. DNA methylation patterns in newborns exposed to tobacco in utero. J Transl Med. 2015;13:25. doi: 10.1186/s12967-015-0384-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fuemmeler BF, Dozmorov MG, Do EK, Zhang JJ, Grenier C, Huang Z, Maguire RL, Kollins SH, Hoyo C, Murphy SK. DNA methylation in babies born to nonsmoking mothers exposed to secondhand smoke during pregnancy: an epigenome-wide association study. Environ Health Perspect. 2021;129:57010. doi: 10.1289/EHP8099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ornoy A, Becker M, Weinstein-Fudim L, Ergaz Z. S-Adenosine methionine (SAMe) and valproic acid (VPA) as epigenetic modulators: special emphasis on their interactions affecting nervous tissue during pregnancy. Int J Mol Sci. 2020;21:E3721. doi: 10.3390/ijms21103721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tung EWY, Winn LM. Epigenetic modifications in valproic acid-induced teratogenesis. Toxicol Appl Pharmacol. 2010;248:201–209. doi: 10.1016/j.taap.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 180.Shafique S, Winn LM. Gestational valproic acid exposure induces epigenetic modifications in murine decidua. Placenta. 2021;107:31–40. doi: 10.1016/j.placenta.2021.03.004. [DOI] [PubMed] [Google Scholar]
- 181.Gueta K, Molotski N, Gerchikov N, Mor E, Savion S, Fein A, Toder V, Shomron N, Torchinsky A. Teratogen-induced alterations in microRNA-34, microRNA-125b and microRNA-155 expression: correlation with embryonic p53 genotype and limb phenotype. BMC Dev Biol. 2010;10:20. doi: 10.1186/1471-213X-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mor E, He L, Torchinsky A, Shomron N. MicroRNA-34a is dispensable for p53 function as teratogenesis inducer. Arch Toxicol. 2014;88:1749–1763. doi: 10.1007/s00204-014-1223-9. [DOI] [PMC free article] [PubMed] [Google Scholar]