Abstract
Gnathostomiasis is a food-borne zoonotic disease that can affect humans who eat improperly cooked meat containg infective third-stage larvae. Definitive diagnosis is through larval recovery. However, this is an invasive technique and is impractical if the larvae have encysted in inaccessible areas of the body. Antigen or antibody detection might be more interesting techniques for diagnosis. Proteomic could elucidate diagnostic markers and improve our understanding of parasite biology. However, proteomic studies on Gnathostoma spinigerum are hampered by the lack of a comprehensive database for protein identification. This study aimed to explore the protein and antigen profiles of advanced third-stage G. spinigerum larvae (aL3Gs) using interrogation of mass spectrometry data and an in-house transcriptomic database for protein identification. Immunoproteomic analysis found 74 proteins in 24-kDa SDS-PAGE bands, which is size-specific for the immunodiagnosis of gnathostomiasis. Moreover, 13 proteins were found in 2-DE 24-kDa bands. The data suggest that collagenase 3, cathepsin B, glutathione S-transferase 1, cuticle collagen 14, major antigen, zinc metalloproteinase nas-4, major egg antigen, peroxiredoxin, and superoxide dismutase [Cu–Zn] may be good candidates for novel human gnathostomiasis diagnostic assays. These findings improve our understanding of the parasite’s biology and provide additional potential targets for novel therapeutics, diagnostics, and vaccines.
Subject terms: Biomarkers, Proteomics
Introduction
Human gnathostomiasis is most commonly caused by Gnathostoma spinigerum. Infection occurs when humans ingest the infective third-stage larvae through consumption of raw or undercooked meat from second intermediate or paratenic hosts1. Gnathostomiasis is an emerging infectious disease with increasing reports of infection, particularly in travelers returning from endemic areas or from importation of infected fresh produce like eels2.
Definitive diagnosis of G. spinigerum is through identification of the nematode isolated from skin lesions or histopathological sections but this is invasive and is difficult when internal organs are affected. A presumptive diagnosis can be made in patients with eosinophilia coupled with a reported history of eating raw or undercooked fish or meat3,4. Another reliable diagnostic method for gnathostomiasis is immunoblotting to detect the 24-kDa crude worm antigen (CWA) from advanced third-stage G. spinigerum larvae (aL3Gs)5. This G. spinigerum-specific antigen has been found in gnathostomiasis and presumptive gnathostomiasis patients, but not in healthy individuals or patients infected with other parasites6.
Typically, aL3Gs are harvested from eel livers. The process of CWA production is complex and complicated by the seasonal prevalence of the aL3Gs in eels7,8. With advances in proteomics and mass spectrometry, the 24-kDa antigen has been further characterized and found to have high homology to peptide sequence regions of cyclophilin, actin, matrix metalloproteinase-like protein, and intermediate filament protein B. Related studies have shown the applicability of some of these peptides as antigens9,10. However, no commercial diagnostic tests are currently available for gnathostomiasis.
For many years, there was no effective treatment for gnathostomiasis and currently the only successful treatment option is surgical excision of the larvae. Various medications, including thiabendazole, praziquantel, metronidazole, diethylcarbamazine, and quinine, have been explored in animal models and humans without success11. Proteomic technology has provided crucial information about cellular and molecular processes, leading to an improved understanding of the biology of many different parasites. Aside from identifying antigens for diagnostic purposes, proteomic studies have made invaluable contributions in drug target identification and understanding host-parasite relationships. This work has culminated in the identification of possible drug and vaccine targets for a range of parasites. In contrast, recent proteomic analyses on G. spinigerum aL3Gs have only focused on identifying immunoreactive proteins10,12. To date, immunoproteomics is the only proteomic analysis that has been used to study G. spinigerum. Other proteomic studies have been limited by the lack of a database for G. spinigerum protein identification. Therefore, protein identification has mostly relied on using nucleotide sequences from other nematode species. Recently, next-generation sequencing (NGS) was performed to provide a transcriptomic dataset from G. spinigerum aL3Gs4. This G. spinigerum database was applied in this study to facilitate proteomic and immunoproteomic identification of aL3G proteins and antigens. The findings of this study should improve current knowledge of the aL3G proteome and may help to elucidate the molecular functions and biological processes occurring in the parasite to ultimately determine novel drug targets. In addition, these results have identified immunogenic proteins which could be useful for the improvement of vaccines and diagnostic assays for gnathostomiasis.
Results
Protein profile of G. spinigerum third-stage larvae
In this research, protein and antigen profiles of aL3G were explored (Fig. 1). Proteins from aL3Gs were separated by 12% gel electrophoresis. A Coomassie blue-stained gel is shown in Fig. 2. A total of 14 gel pieces were cut and in-gel digestion and mass spectrometry analysis were performed. Proteins were identified by searching against the in-house G. spinigerum transcriptomic database4. With a 95% confidence interval cut-off, 687 proteins were identified (Supplementary Dataset 1). These G. spinigerum proteins were semi-quantified based on the exponentially modified protein abundance index (emPAI). The 20 most abundant aL3G proteins are shown in Table 1. The highly abundant proteins had an emPAI value that ranged from 0.99 to 289.02 and molecular weight (MW) that ranged from 7.5 to 250.9 kDa. Among the proteins identified, actin-2 expression increased by 43-fold compared with actin-5c, the second most highly expressed protein. Similarly, other structure-related proteins such as myophilin and actin-1 were also highly expressed in aL3Gs. In addition, several proteases such as zinc metalloprotease nas-14 and matrix metalloproteinase-like protein were also abundant in aL3Gs. Metabolic enzymes, including glyceraldehyde-3-phosphate dehydrogenase and nucleoside diphosphate kinase, were also highly expressed. Furthermore, enzymes responsible for protein folding, such as peptidyl-prolyl-cis–trans-isomerase 3, were also present at high levels in aL3Gs.
Figure 1.
A diagrammatic flowchart summarizing the methods performed in the study.
Figure 2.
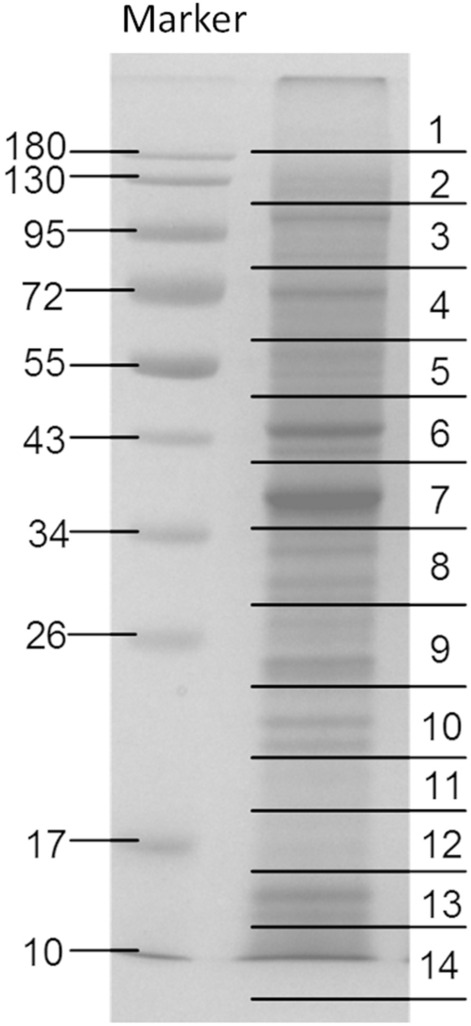
The Coomassie blue-stained gel of aL3Gs proteins. The proteins were separated using one-dimensional gel electrophoresis.
Table 1.
List of the top-20 most abundant proteins in aL3Gs determined based on emPAI values.
| No | Accession | Protein description | Score | M.W | No. of peptide | %coverage | emPAI |
|---|---|---|---|---|---|---|---|
| 1 | CL1711.Contig5_GsL3-Total-RNA | Actin-2 | 696.5 | 7582 | 9 | 87.1 | 289.02 |
| 2 | Unigene19421_GsL3-Total-RNA | Actin-5C | 719 | 10,846 | 10 | 71.6 | 6.7 |
| 3 | Unigene18798_GsL3-Total-RNA | Myoglobin | 648 | 24,935 | 13 | 48.2 | 5.48 |
| 4 | CL1262.Contig3_GsL3-Total-RNA | Ancylostoma secreted protein | 643 | 31,485 | 11 | 54.1 | 2.6 |
| 5 | CL7245.Contig1_GsL3-Total-RNA | Myophilin | 374 | 25,023 | 7 | 30.2 | 2.33 |
| 6 | Unigene9624_GsL3-Total-RNA | Glyceraldehyde-3-phosphate dehydrogenase | 849 | 44,630 | 17 | 47.5 | 2.13 |
| 7 | CL3958.Contig3_GsL3-Total-RNA | Small heat shock protein OV25-1 | 307 | 30,262 | 12 | 50.2 | 2.03 |
| 8 | Unigene14002_GsL3-Total-RNA | Peptidyl-prolyl cis–trans isomerase 3 | 228 | 31,937 | 8 | 33.2 | 1.58 |
| 9 | CL3958.Contig1_GsL3-Total-RNA | Small heat shock protein OV25-1 | 263 | 30,805 | 9 | 36.6 | 1.39 |
| 10 | CL7161.Contig1_GsL3-Total-RNA | Zinc metalloproteinase nas-14 | 253 | 15,103 | 6 | 43.3 | 1.37 |
| 11 | CL1711.Contig2_GsL3-Total-RNA | Actin, cytoplasmic 1 | 731 | 51,490 | 14 | 26.8 | 1.36 |
| 12 | Unigene6082_GsL3-Total-RNA | Nucleoside diphosphate kinase | 358 | 26,046 | 8 | 30.9 | 1.16 |
| 13 | CL3580.Contig2_GsL3-Total-RNA | Collagenase 3 | 236 | 31,095 | 10 | 32.6 | 1.13 |
| 14 | Unigene20056_GsL3-Total-RNA | Zinc metalloproteinase nas-14 | 209 | 17,429 | 9 | 67.1 | 1.12 |
| 15 | Unigene18966_GsL3-Total-RNA | Small heat shock protein OV25-2 | 91 | 32,250 | 12 | 47.6 | 1.07 |
| 16 | Unigene9664_GsL3-Total-RNA | Transthyretin-like protein 16 | 158 | 18,249 | 4 | 25.9 | 1.06 |
| 17 | Unigene18725_GsL3-Total-RNA | Calponin homolog OV9M | 643 | 72,439 | 23 | 38.5 | 1.03 |
| 18 | Unigene21925_GsL3-Total-RNA | Actin-1 | 897 | 73,407 | 23 | 32.1 | 1.01 |
| 19 | Unigene16420_GsL3-Total-RNA | Extracellular globin | 360 | 49,428 | 16 | 41.9 | 0.99 |
| 20 | Unigene21322_GsL3-Total-RNA | Major antigen | 4688 | 250,951 | 80 | 34 | 0.99 |
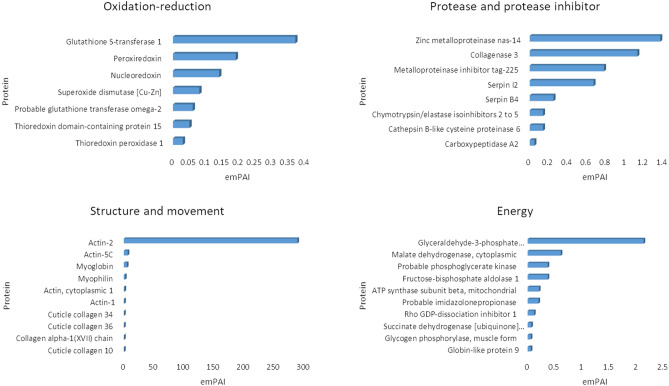
To gain more understanding into the biological processes occurring in aL3Gs, gene ontology (GO) was used to classify all identified G. spinigerum proteins (Supplementary Table 1). A total of 123 protein classes were found in aL3Gs. The top 20 protein classes are listed in Table 2. Proteins with unknown GO accounted for approximately 50% of the total identified G. spinigerum proteins. The major annotated protein classes observed in aL3Gs included embryo development ending in birth or egg hatching (GO:0009792), determination of adult lifespan (GO:0008340), and oviposition (GO:0018991). Other protein classes were involved in biological processes related to energy, movement, morphological development, and enzymes for protein structure conformational changes. In addition to GO classification by biological process terms, proteins essential for parasite survival and host immunity evasion were also considered. Proteins relating to oxidation–reduction, proteinase-protease inhibitors, structure-movement, and energy were identified in the aL3G protein profile. The top 10 most abundant G. spinigerum proteins involved in these four categories are shown in Fig. 3. Glutathione S-transferase (emPAI of 0.37), peroxiredoxin (emPAI of 0.19), and nucleoredoxin (emPAI of 0.14) were the major antioxidant proteins expressed in aL3Gs. The most prevalent proteases were zinc metalloproteinase nas-14 (emPAI of 1.37), matrix metalloproteinase-like protein (emPAI of 1.13), and cathepsin B-like cysteine proteinase 6 (emPAI of 0.14), while the most abundant protease inhibitors were metalloproteinase inhibitor tag-225 (emPAI of 0.78), serpin I2 (emPAI 0.67), and serpin B4 (emPAI of 0.25). Meanwhile, actin-2 (emPAI of 289.02), actin-5C (emPAI of 6.7), and myoglobin (emPAI of 5.48) were the most highly expressed proteins in the structure-movement group. Additionally, glyceraldehyde-3-phosphate dehydrogenase (emPAI of 2.13), malate dehydrogenase, cytoplasmic (emPAI of 0.61), and phosphoglycerate kinase (emPAI of 0.37) were the main energy-related proteins expressed in aL3Gs.
Table 2.
List of top-20 proteins classified by Gene Ontology (GO) according to the biological processes involved in aL3Gs.
| GO-Biological Process | No. of proteins |
|---|---|
| Unknown | 401 |
| GO:0,009,792//embryo development ending in birth or egg hatching | 33 |
| GO:0,008,340//determination of adult lifespan | 24 |
| GO:0,018,991//oviposition | 11 |
| GO:0,006,096//glycolytic process | 7 |
| GO:0,008,152//metabolic process | 7 |
| GO:0,019,915//lipid storage | 7 |
| GO:0,055,085//transmembrane transport | 6 |
| GO:0,000,003//reproduction | 5 |
| GO:0,010,171//body morphogenesis | 5 |
| GO:0,040,011//locomotion | 5 |
| GO:0,040,035//hermaphrodite genitalia development | 5 |
| GO:0,006,094//gluconeogenesis | 4 |
| GO:0,006,457//protein folding | 4 |
| GO:0,009,987//cellular process;GO:0,044,238 | 4 |
| GO:0,016,310//phosphorylation | 4 |
| GO:0,030,968//endoplasmic reticulum unfolded protein response | 4 |
| GO:0,044,763;GO:0,050,794//regulation of cellular process | 4 |
| GO:0,048,856//anatomical structure development | 4 |
| GO:0,071,688//striated muscle myosin thick filament assembly | 4 |
Figure 3.
The top-10 most abundant proteins classified by molecular function relating to oxidation–reduction (redox), protease-protease inhibitor activity, structure-movenent, and energy metabolism. Their abundance was estimated semi-quantitatively by emPAI. In each category, the x-axis indicates the emPAI value and the y-axis specifies the protein identification.
Comparison of aL3G and human protein sequences
To identify potential G. spinigerum drug target candidates, the nucleotide sequences of the 10 most abundant proteins relating to oxidation–reduction, proteinase-protease inhibitors, and structure-movement were retrieved from the aL3G transcriptome. The blastx algorithm was used to search the lowest E-value for human proteins in the GenBank database compared with G. spinigerum sequences. The percent homology between G. spinigerum and human sequences is shown in Table 3.
Table 3.
List of Abundant proteins of aL3Gs classified by Gene Ontology (GO) into four groups under the molecular function category that are aligned with human proteins to identify unique proteins of aL3Gs.
| Protein | Accession no. of human proteins | %identity | E-value | Query cover |
|---|---|---|---|---|
| Oxidation–reduction | ||||
| Thioredoxin peroxidase 1 | NP_006784.1 | 62.23% | 3.00E-79 | 17% |
| Thioredoxin domain-containing protein 15 | NP_001337664.1 | 30.34% | 2.00E-16 | 22% |
| Probable glutathione transferase omega-2 | 4YQM_A | 34.85% | 9.00E-36 | 42% |
| Superoxide dismutase [Cu–Zn] | 3GTV_A | 58.39% | 1.00E-51 | 37% |
| Nucleoredoxin | NP_001155097.1 | 36.36% | 8.00E-28 | 61% |
| Peroxiredoxin | NP_005800.3 | 74.61% | 8.00E-102 | 34% |
| Glutathione S-transferase 1 | 4EDY_A | 40.00% | 6.00E-08 | 68% |
| Protease-protease inhibitor | ||||
| Carboxypeptidase A2 | AAH14571.1 | 37.98% | 8.00E-88 | 63% |
| Cathepsin B-like cysteine proteinase 6 | NP_001304166.1 | 38.36% | 6.00E-29 | 69% |
| Chymotrypsin/elastase isoinhibitors 2 to 5* | – | – | – | – |
| Serpin B4* | – | – | – | – |
| Serpin I2* | – | – | – | – |
| Metalloproteinase inhibitor tag-225* | – | – | – | – |
| Collagenase 3 | BAD96700.1 | 36.99% | 6.00E-29 | 61% |
| Zinc metalloproteinase nas-14* | – | – | – | – |
| Structure-movement | ||||
| Actin-2 | BAG51757.1 | 98.55% | 9.00E-45 | 98% |
| Actin-5C | BAG62762.1 | 97.40% | 5.00E-50 | 75% |
| Myoglobin* | – | – | – | – |
| Myophilin | EAW52759.1 | 38.52% | 4.00E-24 | 55% |
| Actin, cytoplasmic 1 | AAH10417.2 | 98.16% | 9.00E-115 | 34% |
| Actin-1 | NP_001186883.1 | 92.51% | 0.00E + 00 | 57% |
| Cuticle collagen 34* | – | – | – | – |
| Cuticle collagen 36* | – | – | – | – |
| Collagen alpha–1(XVII) chain* | – | – | – | – |
| Cuticle collagen 10* | – | – | – | – |
| Energy metabolism | ||||
| Glycogen phosphorylase, muscle form | 1XOI_A | 67.45% | 0.00E + 00 | 74% |
| Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | XP_016865174.1 | 80.62% | 0.00E + 00 | 85% |
| Rho GDP-dissociation inhibitor 1 | BAG35268.1 | 48.08% | 9.00E-52 | 34% |
| Probable imidazolonepropionase | NP_689648.2 | 49.42% | 3.00E-136 | 81% |
| ATP synthase subunit beta, mitochondrial | BAA00016.1 | 84.43% | 0.00E + 00 | 73% |
| Fructose-bisphosphate aldolase 1 | NP_001121089.1 | 67.88% | 1.00E-171 | 72% |
| Probable phosphoglycerate kinase | 4AXX_A | 73.32% | 0.00E + 00 | 81% |
| Malate dehydrogenase, cytoplasmic | NP_005908.1 | 62.69% | 5.00E-142 | 84% |
| Glyceraldehyde-3-phosphate dehydrogenase | 6M61_O | 73.53% | 3.00E-168 | 81% |
| Globin-like protein 9* | – | – | – | – |
According to the alignment results, 10 G. spinigerum proteins including thioredoxin domain-containing protein 15, glutathione transferase omega-2, nucleoredoxin, glutathione S-transferase 1, carboxypeptidase A2, cathepsin B-like cysteinase 6, collagenase 3, myophilin, rho GDP-dissociation inhibitor 1, and imidazolonepropionase showed less than 50% homology to human sequences. Furthermore, no homology to human proteins was found in the following G. spinigerum proteins: chymotrypsin/elastase isoinhibitors 2 and 5; serpin B4; serpin I2; metalloproteinase inhibitor tag-225; zinc metalloproteinase nas-14; myoglobin; cuticle collagen 34; cuticle collagen 36; collagen alpha-1 (XVII) chain; and globin-like protein 9. Therefore, these G. spinigerum proteins might be potential candidates for drug development.
Protein identification in 24-kDa gel bands
Because identification of the 24-kDa aL3G crude worm antigen is an accepted diagnostic assay for gnathostomiasis, the aL3G proteins were separated by 12% gel electrophoresis then electro-transferred onto a membrane. Western blot analysis was performed using the sera of five individual patients with a confirmed diagnosis of gnathostomiasis as primary antibodies (Fig. 4). The aL3G proteins were transferred onto a membrane. The membrane was sliced into five 3 mm-strips. Each membrane strip (1–5) was individually incubated with sera from 5 different individuals diagnosed with gnathostomiasis As expected, the patient sera reacted with gel bands of approximately 24 kDa, a result reported to have high sensitivity and specificity for G. spinigerum diagnosis. Gel band numbers 9, 10, and 11 were excised for protein identification by mass spectrometry. The mass spectrometry data from three of the gel pieces were separately searched against NCBI and transcriptomic databases to identify their protein components (Table 4). Using the NCBI database, proteins from gel section numbers 9, 10, and 11 were identified as matrix metalloproteinase-like protein, unknown, and cyclophilin, respectively. Using the G. spinigerum transcriptomic database, matrix metalloproteinase-like protein and cyclophilin were identified from gels 9 and 11, respectively. Furthermore, 37, 26, and 23 proteins were also identified on gels 9, 10, and 11 using our database (Table 4). These data indicate the presence of other intriguing candidates, including antioxidative enzymes, cuticle collagens, and major antigens, which warrant further investigation as potential diagnostic targets.
Figure 4.
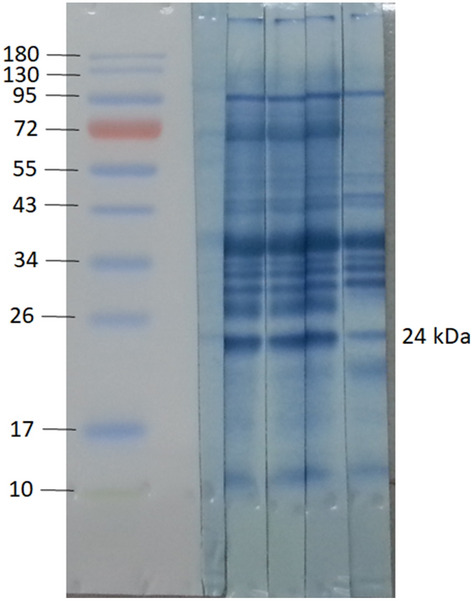
Western blot analyses of aL3Gs with the molecular weight (MW) markers indicated in kDa on the left. Each membrane strip (1–5) is incubated with sera from 5 different individuals diagnosed with gnathostomiasis. The 24-kDa gel regions were excised, and protein components explored against transcriptomic database individually.
Table 4.
The proteins identified from gel numbers 9, 10, and 11 using NCBI and transcriptomic data.
| Gel | NCBI | Protein description | Transcriptome | Protein description |
|---|---|---|---|---|
| 9 | AAF82802.1 | Matrix metalloproteinase-like protein [Gnathostoma spinigerum] | CL3580.Contig2_GsL3-Total-RNA | Collagenase 3 |
| Unigene18966_GsL3-Total-RNA | Small heat shock protein OV25-2 | |||
| CL1711.Contig1_GsL3-Total-RNA | Actin-2 | |||
| Unigene9422_GsL3-Total-RNA | Triosephosphate isomerase | |||
| Unigene21055_GsL3-Total-RNA | Glutamate dehydrogenase, mitochondrial | |||
| CL6152.Contig2_GsL3-Total-RNA | Glutathione S-transferase 1 | |||
| CL3958.Contig1_GsL3-Total-RNA | Small heat shock protein OV25-1 | |||
| Unigene18018_GsL3-Total-RNA | Succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial | |||
| Unigene16324_GsL3-Total-RNA | Cuticle collagen 14 | |||
| Unigene6075_GsL3-Total-RNA | Protein lethal(2)essential for life | |||
| Unigene17956_GsL3-Total-RNA | Adenylate kinase isoenzyme 1 | |||
| CL7292.Contig2_GsL3-Total-RNA | Stromelysin-2 | |||
| Unigene9624_GsL3-Total-RNA | Glyceraldehyde-3-phosphate dehydrogenase | |||
| Unigene6072_GsL3-Total-RNA | Twitchin | |||
| CL3283.Contig1_GsL3-Total-RNA | Loricrin | |||
| Unigene21925_GsL3-Total-RNA | Actin-1 | |||
| CL4911.Contig1_GsL3-Total-RNA | Myb-like protein D | |||
| Unigene21322_GsL3-Total-RNA | Major antigen | |||
| Unigene21850_GsL3-Total-RNA | Probable maleylacetoacetate isomerase | |||
| Unigene21057_GsL3-Total-RNA | Zinc metalloproteinase nas-4 | |||
| Unigene1222_GsL3-Total-RNA | Phosphoenolpyruvate carboxykinase [GTP] | |||
| CL2394.Contig1_GsL3-Total-RNA | Probable glutathione transferase omega-2 | |||
| CL845.Contig1_GsL3-Total-RNA | Uncharacterized protein ZK643.6 | |||
| CL437.Contig1_GsL3-Total-RNA | Disorganized muscle protein 1 | |||
| Unigene22528_GsL3-Total-RNA | Elongation factor Ts, mitochondrial | |||
| Unigene18459_GsL3-Total-RNA | Phosphoenolpyruvate carboxykinase [GTP] | |||
| Unigene21888_GsL3-Total-RNA | Propionyl-CoA carboxylase beta chain, mitochondrial | |||
| Unigene22645_GsL3-Total-RNA | E3 ubiquitin-protein ligase pellino homolog 2 | |||
| Unigene85_GsL3-Total-RNA | Eukaryotic translation initiation factor 4B | |||
| Unigene24479_GsL3-Total-RNA | Hypoxia up-regulated protein 1 | |||
| CL3117.Contig10_GsL3-Total-RNA | Nucleolar protein 10 | |||
| Unigene17966_GsL3-Total-RNA | Phosphoenolpyruvate carboxykinase [GTP] | |||
| CL1888.Contig1_GsL3-Total-RNA | Uncharacterized protein ZK688.3 | |||
| CL756.Contig10_GsL3-Total-RNA | GRIP1-associated protein 1 | |||
| CL607.Contig12_GsL3-Total-RNA | Probable splicing factor, arginine/serine-rich 7 | |||
| CL3810.Contig4_GsL3-Total-RNA | Troponin T | |||
| CL3881.Contig1_GsL3-Total-RNA | cathepsin B [EC:3.4.22.1] | |||
| CL2383.Contig1_GsL3-Total-RNA | Transformation/transcription domain-associated protein | |||
| 10 | unknown | Unknown | CL3958.Contig1_GsL3-Total-RNA | Small heat shock protein OV25-1 |
| CL6067.Contig1_GsL3-Total-RNA | Small heat shock protein OV25-2 | |||
| Unigene19421_GsL3-Total-RNA | Actin-5C | |||
| Unigene17198_GsL3-Total-RNA | Major egg antigen | |||
| CL187.Contig1_GsL3-Total-RNA | Phosphatidylethanolamine-binding protein homolog F40A3.3 | |||
| Unigene18966_GsL3-Total-RNA | Small heat shock protein OV25-2 | |||
| CL2984.Contig1_GsL3-Total-RNA | Beta-ureidopropionase | |||
| CL221.Contig3_GsL3-Total-RNA | Peroxiredoxin | |||
| CL1711.Contig1_GsL3-Total-RNA | Actin-2 | |||
| CL5074.Contig2_GsL3-Total-RNA | Elongation of very long-chain fatty acids protein 5 | |||
| Unigene20106_GsL3-Total-RNA | Methyltransferase-like protein 17, mitochondrial | |||
| CL6024.Contig2_GsL3-Total-RNA | Ferric-chelate reductase 1 | |||
| Unigene1383_GsL3-Total-RNA | E3 ubiquitin-protein ligase MYLIP-B | |||
| CL2656.Contig4_GsL3-Total-RNA | CDKN2AIP N-terminal-like protein | |||
| CL5237.Contig1_GsL3-Total-RNA | DNA polymerase delta subunit 2 | |||
| Unigene19315_GsL3-Total-RNA | Rho GDP-dissociation inhibitor 1 | |||
| Unigene21055_GsL3-Total-RNA | Glutamate dehydrogenase, mitochondrial | |||
| Unigene2602_GsL3-Total-RNA | Carboxypeptidase A2 | |||
| CL5122.Contig1_GsL3-Total-RNA | Barrier-to-autointegration factor 1 | |||
| Unigene21322_GsL3-Total-RNA | Major antigen | |||
| Unigene25087_GsL3-Total-RNA | Bromodomain and WD repeat-containing protein 3 | |||
| CL2434.Contig1_GsL3-Total-RNA | Thioredoxin peroxidase 1 | |||
| CL68.Contig11_GsL3-Total-RNA | Coiled-coil domain-containing protein 18 | |||
| CL7147.Contig1_GsL3-Total-RNA | Lipoma-preferred partner homolog | |||
| CL498.Contig10_GsL3-Total-RNA | Stromal interaction molecule 1 | |||
| CL1701.Contig10_GsL3-Total-RNA | DmX-like protein 2 | |||
| 11 | ACX47902.1 | Cyclophilin [Gnathostoma spinigerum] | Unigene14002_GsL3-Total-RNA | Peptidyl-prolyl cis–trans isomerase 3 (cyclophilin) |
| Unigene9664_GsL3-Total-RNA | Transthyretin-like protein 16 | |||
| Unigene16365_GsL3-Total-RNA | OV-16 antigen | |||
| Unigene6082_GsL3-Total-RNA | Nucleoside diphosphate kinase | |||
| CL1711.Contig1_GsL3-Total-RNA | Actin-2 | |||
| CL7161.Contig1_GsL3-Total-RNA | Zinc metalloproteinase nas-14 | |||
| CL6541.Contig1_GsL3-Total-RNA | Transthyretin-like protein 46 | |||
| Unigene26369_GsL3-Total-RNA | Putative uncharacterized transposon-derived protein F52C9.6 | |||
| CL221.Contig3_GsL3-Total-RNA | Peroxiredoxin | |||
| Unigene14764_GsL3-Total-RNA | 60S ribosomal protein L12 | |||
| CL3958.Contig1_GsL3-Total-RNA | Small heat shock protein OV25-1 | |||
| CL187.Contig1_GsL3-Total-RNA | Phosphatidylethanolamine-binding protein homolog F40A3.3 | |||
| Unigene20506_GsL3-Total-RNA | Superoxide dismutase [Cu–Zn] | |||
| Unigene20053_GsL3-Total-RNA | Myosin, essential light chain | |||
| Unigene9624_GsL3-Total-RNA | Glyceraldehyde-3-phosphate dehydrogenase | |||
| Unigene21322_GsL3-Total-RNA | Major antigen | |||
| CL1294.Contig1_GsL3-Total-RNA | Intermediate filament protein B | |||
| CL1614.Contig1_GsL3-Total-RNA | Endoplasmic reticulum-Golgi intermediate compartment protein 3 | |||
| CL198.Contig1_GsL3-Total-RNA | Mitochondrial Rho GTPase | |||
| Unigene25389_GsL3-Total-RNA | Beta-lactamase domain-containing protein 2 | |||
| CL7147.Contig1_GsL3-Total-RNA | Lipoma-preferred partner homolog | |||
| Unigene22939_GsL3-Total-RNA | DDB1- and CUL4-associated factor 5 | |||
| CL756.Contig10_GsL3-Total-RNA | GRIP1-associated protein 1 |
2D-immunoblot aL3G antigen profile
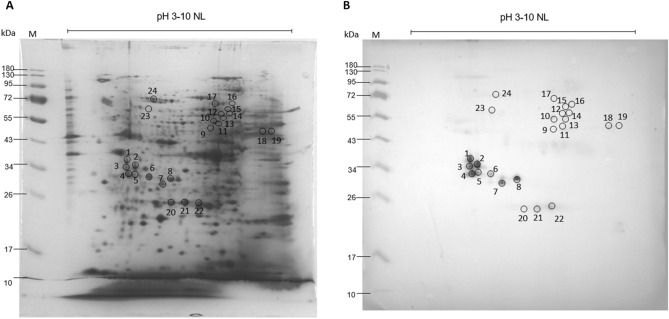
To explore the aL3G immunome, aL3G proteins were separated by 2-DE and transferred onto a nitrocellulose membrane and later analyzed by western blotting (Fig. 5). Pooled patient sera served as the primary antibody. A total of 24 immunoreactive spots was observed. These spots were identified by comparing mass spectrometry data with the G. spinigerum transcriptomic database. A total of 115 proteins were identified as G. spinigerum antigens (Table 5). Spots 8, 17, and 22 demonstrated a lot number of protein identification at 21, 17 and 12, respectively. The proteins 32-kDa beta-galactoside-binding lectin, alkylated DNA repair protein alkB homolog 8, and mitogen-activated protein kinase were identified with the highest score in spot 8. Polyphosphoinositide phosphatase, UPF0378 protein, and heat shock factor binding protein 1 were observed with the highest confidence in gel 17. Matrix metalloproteinase-like protein, e3 ubiquitin-protein ligase trim13, and isocitrate isopropylmalate dehydrogenase domain containing protein were found in gel 22.
Figure 5.
Images of protein from crude extract of aL3Gs subjected to two-dimensional electrophoresis (2-DE). Using IEF, the proteins are separated on a linear pH range of 3–10 in the first dimension and followed by 12% SDS-PAGE for the second dimension. The gel on the left is stained with Coomassie blue (A). The right side shows the nitrocellulose membrane with proteins electro-transferred from the gel and then probed with pooled sera from patients diagnosed with gnathostomiasis (B). Numbers at the left of each image indicate protein molecular weight (MW) markers.
Table 5.
List of proteins identified from aL3Gs after 2D-immunoblot analysis to identify proteins of importance related to diagnostic development of gnathostomiasis infection.
| Gel | No | Protein description | Score | M.W | No. of sequence | %cov |
|---|---|---|---|---|---|---|
| 1 | 1 | Unknown | 63 | 28,771 | 4 | 16.2 |
| 2 | Unknown | 42 | 54,746 | 1 | 1.6 | |
| 2 | 1 | Unknown | 70 | 28,771 | 4 | 19.7 |
| 2 | Cytochrome c oxidase assembly protein cox15-like protein | 30 | 64,100 | 3 | 7.5 | |
| 3 | Grip and coiled-coil domain-containing protein 2 | 30 | 80,345 | 2 | 3.4 | |
| 4 | f-box wd repeat-containing protein 5 | 29 | 21,484 | 1 | 5 | |
| 5 | Hemicentin-1 | 25 | 9709 | 1 | 8.5 | |
| 6 | unknown | 24 | 71,901 | 1 | 3.1 | |
| 3 | 1 | unknown | 37 | 28,771 | 2 | 7.7 |
| 2 | unknown | 35 | 9753 | 1 | 9.2 | |
| 3 | unknown | 26 | 8063 | 1 | 21.9 | |
| 4 | 1 | unknown | 130 | 28,771 | 4 | 16.2 |
| 2 | Translational activator GCN1 | 55 | 293,891 | 4 | 2.3 | |
| 3 | Adenosine monophosphate-protein transferase FICD | 52 | 92,486 | 3 | 2.9 | |
| 4 | Otopetrin-1 | 48 | 67,029 | 2 | 2.8 | |
| 5 | 1 | Otopetrin-1 | 74 | 131,621 | 4 | 4.8 |
| 2 | Nucleoside-diphosphatase mig- | 55 | 58,592 | 2 | 3.6 | |
| 3 | Anaphase-promoting complex subunit 5 | 52 | 119,152 | 3 | 2.7 | |
| 4 | Trafficking protein particle complex subunit 8 | 51 | 154,261 | 3 | 2.8 | |
| 5 | zinc metallopeptidase 2 MEP2 | 48 | 74,140 | 3 | 5 | |
| 6 | 1 | Steroidogenic acute regulatory-like protein 1 | 132 | 43,117 | 11 | 19.7 |
| 2 | Ankyrin-2 | 36 | 74,803 | 1 | 2.4 | |
| 3 | Methionine synthase reductase | 30 | 90,007 | 1 | 1.1 | |
| 4 | Cytochrome c oxidase assembly protein cox15-like protein | 30 | 63,522 | 2 | 3.3 | |
| 5 | Grip and coiled-coil domain-containing protein 2 | 29 | 80,285 | 2 | 2.1 | |
| 6 | Transcription factor Dp- | 23 | 110,765 | 2 | 2.6 | |
| 7 | sn1-specific diacylglycerol lipase beta | 19 | 80,114 | 2 | 2.5 | |
| 7 | 1 | Unknown | 78 | 28,771 | 2 | 6.6 |
| 2 | Adenosine monophosphate-protein transferase FICD homolog | 55 | 92,486 | 2 | 1.8 | |
| 8 | 1 | 32 kDa beta-galactoside-binding lectin | 892 | 43,181 | 17 | 34.7 |
| 2 | Unknown | 52 | 13,040 | 1 | 7.4 | |
| 3 | Alkylated DNA repair protein alkB homolog 8 | 48 | 39,451 | 2 | 2.5 | |
| 4 | Unknown | 48 | 25,280 | 1 | 3.5 | |
| 5 | Mitogen-activated protein kinase 15 | 48 | 117,237 | 1 | 0.7 | |
| 6 | Unknown | 35 | 54,746 | 1 | 1.6 | |
| 7 | sh2 domain-containing protein 3c | 34 | 86,816 | 2 | 2.3 | |
| 8 | Cytochrome c oxidase assembly protein cox15-like protein | 29 | 63,522 | 1 | 1.4 | |
| 9 | Protein HID1 | 29 | 194,912 | 3 | 1.6 | |
| 10 | Ubiquitin carboxyl-terminal hydrolase 31 | 29 | 174,633 | 1 | 0.5 | |
| 11 | Grip and coiled-coil domain-containing protein 2 | 29 | 77,955 | 1 | 1.2 | |
| 12 | e3 ubiquitin-protein ligase trim13 | 27 | 92,227 | 1 | 1.1 | |
| 13 | putative ubiquitin carboxyl-terminal hydrolase 46 | 23 | 83,972 | 1 | 1.2 | |
| 14 | Sestrin-1 | 23 | 45,954 | 1 | 2.1 | |
| 15 | Metallophosphoesterase | 23 | 111,504 | 3 | 3.2 | |
| 16 | Ras GTPase-activating protein gap-2 | 23 | 195,165 | 1 | 0.6 | |
| 17 | 1-acyl-sn-glycerol-3-phosphate acyltransferase alpha | 23 | 77,675 | 1 | 1.3 | |
| 18 | 2-oxoglutarate | 23 | 115,162 | 1 | 0.9 | |
| 19 | Magnesium transporter 1 | 23 | 108,173 | 1 | 0.9 | |
| 20 | Homeobox protein cut-like ceh- | 23 | 120,530 | 1 | 0.9 | |
| 21 | Putative leucine carboxyl methyltransferase 1 | 22 | 69,138 | 1 | 1.1 | |
| 9 | 1 | Fructose-bisphosphate aldolase 1 | 79 | 53,610 | 4 | 12.6 |
| 2 | Unknown | 64 | 151,052 | 4 | 2.7 | |
| 3 | Spectrin beta chain | 63 | 337,895 | 5 | 1.9 | |
| 4 | Proteasome activator complex subunit 4 | 52 | 168,147 | 4 | 3.5 | |
| 5 | Nucleoside-diphosphatase mig-23 | 51 | 58,592 | 2 | 3.6 | |
| 6 | Uncharacterized protein F54D1.6 | 51 | 181,401 | 3 | 2.3 | |
| 7 | Uncharacterized protein DDB_G0271670 | 50 | 170,273 | 4 | 3.3 | |
| 10 | 1 | Unknown | 33 | 54,746 | 1 | 1.6 |
| 2 | Protogenin B (Fragment) | 27 | 239,030 | 2 | 1.3 | |
| 3 | Probable phosphoserine aminotransferase | 16 | 71,802 | 1 | 2.8 | |
| 4 | 2-methoxy-6-polyprenyl- -benzoquinol | 16 | 74,095 | 1 | 2.6 | |
| 5 | von Willebrand factor and Proteinase inhibitor I15 domain-containing protein | 16 | 197,901 | 1 | 0.9 | |
| 6 | Unknown | 16 | 27,113 | 1 | 7 | |
| 7 | Decaprenyl-diphosphate synthase subunit 1 | 16 | 70,245 | 1 | 3 | |
| 11 | 1 | Fructose-bisphosphate aldolase 1 | 344 | 56,589 | 12 | 23.4 |
| 2 | Fructose-bisphosphate aldolase 1 | 198 | 53,610 | 10 | 23.5 | |
| 3 | Unknown | 41 | 54,746 | 1 | 1.6 | |
| 4 | Myosin-2 | 33 | 21,125 | 1 | 7.1 | |
| 5 | Transcriptional regulator ATRX | 21 | 100,673 | 1 | 1.2 | |
| 12 | 1 | Neuroserpin | 68 | 274,509 | 2 | 0.6 |
| 3 | DNA polymerase kappa | 35 | 53,850 | 1 | 1.8 | |
| 2 | Unknown | 35 | 9753 | 1 | 9.2 | |
| 13 | 1 | DNA polymerase kappa | 28 | 53,850 | 1 | 1.8 |
| 14 | - | - | - | - | - | - |
| 15 | 1 | Putative helicase mot1 | 74 | 265,846 | 5 | 2.5 |
| 2 | Protein split ends | 60 | 310,778 | 5 | 2.2 | |
| 3 | Lon protease homolog, mitochondrial | 52 | 277,509 | 3 | 1.1 | |
| 4 | NAD(P) transhydrogenase, mitochondrial | 51 | 128,218 | 4 | 5.1 | |
| 16 | 1 | Unknown | 39 | 54,746 | 1 | 1.6 |
| 17 | 1 | Unknown | 40 | 30,189 | 1 | 3.7 |
| 2 | Polyphosphoinositide phosphatase | 34 | 134,822 | 1 | 0.7 | |
| 3 | Unknown | 25 | 33,690 | 1 | 4.3 | |
| 4 | UPF0378 protein | 25 | 295,143 | 2 | 1 | |
| 5 | Heat shock factor binding protein 1 | 25 | 44,624 | 1 | 4.9 | |
| 6 | Unknown | 22 | 7327 | 1 | 9.1 | |
| 7 | Nuclear pore complex protein nup160 | 22 | 182,675 | 1 | 0.7 | |
| 8 | Unknown | 17 | 51,481 | 1 | 2.8 | |
| 9 | Unknown | 17 | 15,910 | 1 | 9.5 | |
| 10 | Transgelin-2 | 17 | 38,743 | 1 | 4.2 | |
| 11 | Unknown | 17 | 27,190 | 1 | 4.4 | |
| 12 | Multiple C2 and transmembrane domain-containing protein 1 | 17 | 117,365 | 2 | 2.5 | |
| 13 | Tyrosine-protein phosphatase non-receptor type 13 | 17 | 212,990 | 1 | 0.7 | |
| 14 | Reverse transcriptase | 17 | 69,975 | 1 | 2.2 | |
| 15 | Unknown | 17 | 36,926 | 1 | 4.2 | |
| 16 | Unknown | 17 | 12,421 | 1 | 12.1 | |
| 17 | Unknown | 17 | 8180 | 1 | 18.7 | |
| 18 | 1 | Major antigen | 74 | 250,951 | 3 | 1.9 |
| 2 | Hypothetical protein LOAG_11590 | 20 | 11,291 | 1 | 21.3 | |
| 19 | 1 | Calponin homolog OV9M | 41 | 72,439 | 4 | 8 |
| 2 | Unknown | 36 | 8848 | 1 | 19.8 | |
| 3 | Unknown | 34 | 54,746 | 1 | 1.6 | |
| 4 | Major antigen | 33 | 250,951 | 1 | 0.5 | |
| 5 | Inositol polyphosphate multikinase | 29 | 55,418 | 1 | 1.4 | |
| 6 | Unknown | 29 | 7351 | 1 | 10 | |
| 7 | Unknown | 29 | 12,755 | 1 | 6.3 | |
| 8 | Acetyl-CoA acetyltransferase, mitochondrial | 27 | 72,535 | 1 | 1.8 | |
| 20 | 1 | Collagenase 3 | 144 | 31,095 | 6 | 21.5 |
| 2 | Small heat shock protein ov25-1 | 54 | 31,353 | 1 | 3.1 | |
| 3 | unknown | 42 | 30,189 | 2 | 11.5 | |
| 21 | 1 | Collagenase 3 | 323 | 31,095 | 7 | 28 |
| 2 | Hypothetical protein LOAG_06805 | 40 | 25,230 | 1 | 4.2 | |
| 3 | Unknown | 37 | 13,565 | 1 | 17.4 | |
| 4 | Unknown | 35 | 54,746 | 1 | 1.6 | |
| 5 | sh2 domain-containing protein 3c | 34 | 86,816 | 1 | 1 | |
| 22 | 1 | Collagenase 3 | 64 | 31,095 | 4 | 13.8 |
| 2 | e3 ubiquitin-protein ligase trim13 | 29 | 92,227 | 2 | 3.1 | |
| 3 | Unknown | 22 | 42,073 | 1 | 3.4 | |
| 4 | Isocitrate isopropylmalate dehydrogenase domain-containing protein | 17 | 67,366 | 1 | 2.3 | |
| 5 | PREDICTED: 5-oxoprolinase | 17 | 102,576 | 2 | 3.7 | |
| 6 | putative f-box lrr-repeat protein | 17 | 45,640 | 1 | 3.1 | |
| 7 | N-alpha-acetyltransferase 60 | 17 | 19,499 | 1 | 7.9 | |
| 8 | Unknown | 17 | 17,177 | 1 | 7.8 | |
| 9 | Hypothetical protein WUBG_00429 | 17 | 55,682 | 1 | 2.4 | |
| 10 | Hypothetical protein DICPUDRAFT_160192 | 17 | 7537 | 1 | 18.3 | |
| 11 | Unknown | 17 | 13,878 | 1 | 11.1 | |
| 12 | Cathepsin B | 17 | 73,569 | 1 | 2.1 | |
| 23 | 1 | Unknown | 36 | 22,949 | 1 | 5.6 |
| 2 | WD-repeat protein 22 | 33 | 113,846 | 1 | 0.8 | |
| 3 | sh2 domain-containing protein 3c | 29 | 86,816 | 2 | 2.3 | |
| 4 | Unknown | 29 | 54,746 | 1 | 1.6 | |
| 5 | Glutamate-gated chloride channel subunit beta | 29 | 67,212 | 1 | 1.3 | |
| 6 | Unconventional myosin-IXb | 23 | 231,645 | 1 | 0.7 | |
| 7 | Na-dependent Cl/HCO3 exchanger | 17 | 134,665 | 1 | 1.2 | |
| 24 | 1 | Intermediate filament protein B | 95 | 83,781 | 4 | 7 |
| 2 | Unknown | 32 | 32,006 | 1 | 5.6 | |
| 3 | Mucin-5AC | 19 | 141,479 | 1 | 0.6 | |
| 4 | Hypothetical protein Y032_0138g2053 | 19 | 114,441 | 1 | 0.7 | |
| 5 | Kinesin-like protein KIF3B | 19 | 8000 | 1 | 10.3 | |
| 6 | Cytochrome b5 reductase 4 | 19 | 36,260 | 1 | 2.6 | |
| 7 | Histone-lysine N-methyltransferase SETMAR | 19 | 22,618 | 1 | 3.8 |
Potentially immunogenic proteins are known to be found in the 24-kDa region. A total of three, five, and 12 24-kDa proteins were identified in gels 20, 21, and 22, respectively. Interestingly, matrix metalloproteinase-like protein) was located in spots 20, 21, and 22, while cathepsin B was observed in spot 22 only. These two proteins are considered to be antigens in several parasites. Therefore, we predicted their protein properties, including molecular weight, isoelectric point, secretory and transmembrane domains, n-glycosylation, and o-glycosylation (Table 6). The results showed that matrix metalloproteinase-like protein and cathepsin B were secreted proteins through signaling peptides and both of them contained n-glycosylation and o-glycosylation sites. As some clinical manifestations of G. spinigerum infection similar to Angiostrongylus spp., Strongyloides spp., and Sparganum spp. infection for example eosinophilic meningitis, larval migration and migration to the brain, distinguishing between G. spinigerum infection and the relating parasites might be helpful for diagnosis. Comparison of G. spinigerum matrix metalloproteinase-like protein and cathepsin B protein sequences with the highest % homology with protein sequences from humans and from related helminths, including Angiostrongylus spp., Strongyloides spp., and Sparganum spp., was performed. Matrix metalloproteinase-like protein and cathepsin B protein demonstrated 28%–31% and 34%–64% homology to related helminths and humans, respectively. Therefore, these proteins could be potential candidates for novel diagnostic assays to detect gnathostomiasis.
Table 6.
The protein properties of G. spinigerum matrix metalloproteinase-like protein and cathepsin B.
| Properties | Software | Matrix metalloproteinase-like protein | Cathepsin B |
|---|---|---|---|
| Molecular weight | Expasy Compute pI/Mw tool | 27,661.1 Da | 52,709.06 Da |
| Isolectric point | Expasy Compute pI/Mw tool | 7.09 | 8.54 |
| Signal peptide | SignalP 5.1 | Cleavage site between pos. 23 and 24 | Cleavage site between pos. 31 and 32 |
| Non-classical secretory | SecretomeP-2.0 Server | Containing signal peptide | Containing signal peptide |
| Transmembrane protein | TMHMM—2.0 | Inside ( pos 1–6), Tmhelix (pos 7–26), outside (27–245) | Outside (1–462) |
| N-glycosylation | NetNGlyc—1.0 | position 41, 59 | Position 156, 243 |
| O-glycosylation | NetOGlyc—4.0 | position 220, 226, 230, 234, 236 | Position 175, 301, 303, 304, 315, 456 |
| % similarity | Clustal omega | 28.74% with VDM58434.1 of Angiostrongylus spp. | 62.21% with VDM57237.1 of Angiostrongylusspp. |
| 30.29% with XP_024501568.1 of Strongyloides spp. | 64.16% with XP_024507370.1 of Strongyloides spp. | ||
| 30.40% with VZI42166.1 of Sparganum spp. | 34.38% with VZI02086.1 of Sparganum spp. | ||
| 31.9% with NP_002414.1 of H. sapiens | 46.91% with NP_071447.1 of H. sapiens | ||
| Gene ontology | UniProt | Hydrolase, Protease and Metal-binding | Cysteine-type peptidase activity |
| Molecular weight | Expasy Compute pI/Mw tool | 27,661.1 Da | 52,709.06 Da |
| Isolectric point | Expasy Compute pI/Mw tool | 7.09 | 8.54 |
| Signal peptide | SignalP 5.1 | Cleavage site between pos. 23 and 24 | Cleavage site between pos. 31 and 32 |
| Non-classical secretory | SecretomeP-2.0 Server | Containing signal peptide | Containing signal peptide |
| Transmembrane protein | TMHMM—2.0 | Inside ( pos 1–6), Tmhelix (pos 7–26), outside (27–245) | Outside (1–462) |
| N-glycosylation | NetNGlyc—1.0 | Position 41, 59 | Position 156, 243 |
| O-glycosylation | NetOGlyc—4.0 | Position 220, 226, 230, 234, 236 | Position 175, 301, 303, 304, 315, 456 |
Discussion
Our data showed that actin-2 is highly expressed in aL3Gs. In addition, high levels of other G. spinigerum structure-related proteins, such as myophilin and actin-1, were also found. Actin is a cytoskeletal protein found in myofibrils in muscle cells and microfilaments in a variety of other cell types13. In trematodes, actin has been widely researched. It appears to be a crucial component of the platyhelminth tegument and is engaged in a variety of critical functions, including movements of secretory vesicles, muscle contraction, cytokinesis, and maintenance of cell shape14. In parasitic nematodes, cytoskeletal proteins play roles similar to those in trematodes and also have a function in nutrient uptake through transcuticular absorption15. The possibility of anthelmintic medications that attack helminth cytoskeletal proteins has piqued researchers' interest. In recent years, research on the microtubular network and its interaction with benzimidazole anthelmintics has been published16,17. Therefore, G. spinigerum structure-related proteins could be anthelmintic drug targets. In our study, GO classification of all identified aL3Gs proteins highlighted embryo development ending in birth or egg hatching (GO:0009792) as the major protein class expressed in aL3Gs. This protein class controls how an embryo develops over time, from zygote formation to the end of the embryonic life stage18. Similarly, transcriptomic analysis of the murine parasite Heligmosomoides polygyrus also found this protein class to be the most abundant GO term19. In G. spinigerum, the life cycle begins when feces containing parasite eggs reach fresh water. Second-stage larvae hatch from the eggs and early third-stage larvae develop after being consumed by cyclopoid copepods. When infected copepods are eaten by various intermediate hosts, such as fish, amphibians and reptiles, the larvae develop into aL3Gs. When the aL3Gs are consumed by definitive hosts, they migrate into the stomach wall and eventually mature into the adult stage, thus completing their life cycle20. Because aL3Gs are an intermediate stage of development, proteins related to embryo development ending in birth or egg hatching could contribute to and facilitate the maturation process.
As well as GO classification by biological process terms, various protein classes required for parasite survival and host immunity evasion were also considered. The host immune system's oxidative burst can prevent intracellular parasite invasion and proliferation. Therefore, an antioxidant response promotes parasite survival, reduces inflammation, and changes the metabolism of the host cell21. In aL3Gs, the most abundant proteins relating to oxidation–reduction were glutathione S-transferase and peroxiredoxin. Moreover, these two proteins were also identified as G. spinigerum antigens (gels 9 and 10 in the 24-kDa region). Helminth glutathione S-transferase represents the main mechanism of detoxifying reactive oxygen intermediates produced by the parasite's endogenous metabolism or by the host immune system22. A 28-kDa glutathione-S-transferase was identified as an anti-schistosome vaccine candidate and recently evaluated in human clinical trials23. The glutathione-S-transferases have also been identified as potential immunotherapy or chemotherapy targets to treat infection with several other parasites, including Heligmosomoides polygyrus, Onchocerca gutturosa, and Dirofilaria immitis24. Peroxiredoxins are cysteine-dependent peroxidases that also play an important role in antioxidant, regulatory, and signaling systems. They are involved in defense against both endogenous and host-derived reactive oxygen species25. Treatment of mouse peritoneal macrophages with recombinant Toxoplasma gondii peroxiredoxin resulted in the production of IL-12p40 and IL-626. This result suggests that peroxiredoxin induces both humoral and cellular immunological responses. Therefore, it could be used as a toxoplasmosis vaccine antigen. Peroxiredoxins have also been found in several parasitic helminths that affect humans such as Brugia malayi27, Onchocerca volvulus28, Taenia solium29, Opisthorchis viverrini30, and Schistosoma japonicum31. Furthermore, these proteins have been reported to be attractive therapeutic targets and vaccine candidates to treat and prevent helminth infections32. Nucleoredoxin was one of most abundantly expressed G. spinigerum proteins relating to oxidation–reduction. This protein is a newly discovered member of the thioredoxin family and plays a role in cell proliferation and differentiation33. Nucleoredoxin also regulates phosphofructokinase activity to balance between glycolysis and the pentose phosphate pathway34. However, there is limited information about this protein’s role in parasites. Therefore, the function of nucleoredoxin in G. spinigerum needs to be further explored.
Proteases and protease inhibitors are another important protein class expressed in G. spinigerum. The most prevalent proteases in aL3Gs were zinc metalloproteinase nas-14, matrix metalloproteinase-like protein, and cathepsin B-like cysteine proteinase 6. These three proteases had limited homology with human proteins. Therefore, they might be potential candidates for anthelmintic drug development. Moreover, they were also identified as G. spinigerum antigens (gels 9 and 11 in the 24-kDa region).
The ability of nematodes to molt is critical for their survival and development. Zinc metalloproteases, leucine aminopeptidases, and cysteine proteases are implicated in molting35,36. In the parasitic nematode Haemonchus contortus, the zinc metalloprotease nas-33 is required for molting and survival37. In Strongyloides ratti, collagenase plays a role in adult females at the time of migration through the host intestinal mucosa during oviposition38. In Radopholus similis, an important plant parasitic nematode, the cysteine proteinase cathepsin B-like plays an important role in worm development and hatching39.
Owing to the various protease functions in parasitic worms, nematode proteases have been proposed as new anthelmintic and vaccine targets. The most abundant protease inhibitors in aL3Gs were metalloproteinase inhibitor tag-225, serpin I2, and serpin B4. There was no homology between these three protease inhibitors and human proteins. G. spinigerum serpins have been identified in excretory–secretory products of aL3Gs and have been shown to react with sera from gnathostomiasis patients4. Therefore, they may be good candidates for therapeutic and diagnostic development. Parasite protease inhibitors create a safer environment in the host by suppressing host protease activity and modulating host immunity40 . Serpins from the parasitic worms Brugia malayi41 , Ancylostoma caninum42, and Trichuris suis43 have been identified. Brugia malayi serpin functions by neutralizing the immunostimulatory properties of the host cathepsin G44. Onchocerca volvulus serpin reduces the enzymatic activity of a panel of serine proteases including host elastase, chymotrypsin, trypsin, and cathepsin G45. Helminth protease inhibitors protect the worms from the hydrolytic action of host proteases and allow them to break through protective barriers and evade immunological responses. Protease inhibitors are, therefore, beneficial to parasite survival and colonization in the host46. Consequently, G. spinigerum proteases and protease inhibitors might have similar protective roles and may be good anthelmintic candidates.
Several G. spinigerum cuticle collagens—cuticle collagen 34, cuticle collagen 36, and collagen alpha-1 (XVII) chain—were identified in aL3Gs. These cuticle collagens had no homology with human proteins. However, these three cuticle collagens were conserved among other nematodes (data not shown). As a result, they may be suitable universal vaccine or drug targets for novel pan-nematicide therapeutics. Moreover, cuticle collagen 14 was identified as a 24-kDa G. spinigerum antigen and it showed less homology with protein sequences from other nematodes (data not shown). Therefore, this protein might be a potential target for the development of novel gnathostomiasis diagnostics and vaccines.
The exoskeleton of Caenorhabditis elegans, a free-living nematode, comprises a complex collagen matrix. Individual cuticle collagen gene mutations can result in exoskeletal abnormalities that change the shape of a nematode47. RNA interference targeted to the cuticle collagen genes of the root-knot nematode Meloidogyne incognita caused a 30.80%–35.00% reduction in the number of adult females and a 76.47%–82.59% reduction in the number of eggs. This study demonstrates the role of cuticle collagen genes in the structure and development of the nematode cuticle48. Furthermore, external enzymes that induce structural damage to the cuticle, such as papain, bromelain, collagenase, chitinase, and lipase, cause parasitic worms to die. After incubating Heligmosomoides polygyrus, Trichuris muris, and Protospirula muricola with plant cysteine proteases, the nematodes died as a result of cuticle damage49. These data suggest that cuticle collagens could be fascinating targets for nematicide development.
Western blot analysis has recently identified diagnostic targets for gnathostomiasis by detecting specific total IgG against a 24-kDa protein in aL3G extract5 . This study detected specific IgG subclasses as well as total IgG against a 24-kDa antigen. When compared with other subclasses, IgG4 had the best sensitivity and specificity (91.6% and 87.8%, respectively)6. Although the detection of specific IgG against crude worm antigen (CWA) has a high sensitivity and specificity, the antigen preparation procedure is complex, time-consuming and laborious, and batch-to-batch quality is inconsistent. Furthermore, natural sources of G. spinigerum are limited and depend on the season and climatic conditions. Therefore, identification of G. spinigerum candidate antigens is crucial for recombinant protein production to improve gnathostomiasis diagnosis. The 24-kDa diagnostic protein was first identified as matrix metalloprotease9. In another report, matrix metalloproteinase-like protein and cyclophilin with approximate molecular weights of 23–24 kDa were identified as G. spinigerum antigens50.
However, identification of the protein composition of aL3Gs has been hampered by the lack of genomic sequence information required for proteomic analysis. In our study, the global aL3G antigen profile and specific 24-kDa aL3G antigens were identified using next-generation sequencing transcriptomic information4. Matrix metalloproteinase-like protein and cyclophilin were identified in the 24-kDa region, corresponding with previous reports. However, additional G. spinigerum antigen candidates were also identified. A total of 74 proteins were observed in 24-kDa bands on our SDS-PAGE gels. Our data suggest that glutathione S-transferase 1, cuticle collagen 14, major antigen, zinc metalloproteinase nas-4, major egg antigen, peroxiredoxin, and superoxide dismutase [Cu–Zn] might also be good candidates to explore for human gnathostomiasis diagnostic assays. On 2-DE, 13 proteins were also observed in 24-kDa bands.
A recombinant G. spinigerum matrix metalloproteinase protein has been produced. This protein was shown to have a sensitivity and specificity of 100% in a study comparing 40 patients with 30 healthy controls51. In another study, recombinant G. spinigerum matrix metalloproteinase protein was analyzed by immunoblot of serum samples from proven and clinically suspected cases of gnathostomiasis, patients with other parasitic diseases, and healthy volunteers. The sensitivity and specificity in this study were 100% and 94.7%, respectively52. Although matrix metalloproteinase protein is a potential candidate for G. spinigerum diagnostics, it may be important to assess the different isoforms of this protein to improve its diagnostic capability. Our data also show that cathepsin B may be another good diagnostic candidate for human gnathostomiasis.
In this study, SDS-PAGE identified more proteins than 2-DE, possibly because 2-DE cannot resolve proteins that are too basic or too acidic, too large or too small. In addition, highly hydrophobic proteins are hard to dissolve in the 2-DE buffer system53. Therefore, integrating SDS-PAGE and 2-DE data from the 24-kDa region might be a useful way to explore novel diagnostic candidates for G. spinigerum. This study identified both matrix metalloproteinase like protein and cathepsin B from SDS-PAGE and 2-DE. Furthermore, these proteins were predicted to be secreted proteins and, therefore, might be useful for circulating antigen detection in patient serum. Moreover, both antigens were predicted to have glycosylation. Because glycosylation contributes to protein antigenic properties54, this post translational modification might need to be taken into account during any recombinant antigen production. the limitation of proteomics study using other G. spinigerum stages is to obtain adequate amount of parasite samples. In conclusion, the data from this proteomic and immunoproteomic analysis could help researchers better understand not only the parasite's biology but also possible targets for future treatments, diagnostics, and vaccines.
Methods
Preparation of third-stage G. spinigerum larvae
The aL3Gs were obtained from the livers of naturally-infected eels using an acid-pepsin digestion technique. Eel livers were brought from markets in Bangkok. They were chopped and subjected to 1% acid-pepsin at 37 °C for 2 h in a water bath stirred frequently, then were washed with tap water several times. Worms were then collected via a simple sedimentation technique. The collected worms were further identified using a dissecting microscope then washed again with tap water followed by normal saline solution (0.85% NaCl). Specie of G. spinigerum was confirmed by morphology. Approximately 20 collected worms were pooled in a microfuge tube and kept in -80 °C prior to analysis.
Sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE)
The pooled aL3Gs were ground using a mortar and pestle after short snap freezing. Lysis buffer was then added and further homogenization was performed using an ultrasonicator. After sonication, the mixture was centrifuged at 12 000 rpm for 15 min at 4 °C. The supernatant was collected and the protein concentration was determined by the Bradford method. Proteins from the lysate were separated by 12% SDS-PAGE gel and later stained with Coomassie Brilliant Blue G250 solution (Bio-Rad, Hercules, CA, USA). After running the SDS-PAGE, the gel was cut into 14 rectangles followed by in-gel digestion.
Two dimensional gel electrophoresis (2-DE)
For the first dimension, a non-linear immobilized pH gradient (IPG) strip (pH 3–10; Amersham Bioscience, USA) was rehydrated overnight in immobilized pH gradient (IPG) sample buffer (8 M urea, 2% (w/v) 3-[(3-cholamidopropyl)dimethlyammonio]-1 propanesulfonate (CHAPS), 15 mM dithiothreitol (DTT), and 0.5% IPG sample buffer) and crude aL3G extract. Isoelectric focusing (IEF) was then performed using the following parameters: 30 V for 14 h; 200 V for 1 h; 500 V for 1 h; 1000 Vfor 1 h; 3500 V for 1 h; and 8000 V for 18 h. After focusing, the strips were equilibrated with DTT for 15 min and with iodoacetamide (C2H4INO) for 15 min. The strips were then subjected to 12% SDS-PAGE at 120 V until the bromophenol blue dye front reached the bottom of the gel. All three 2-DE gels were stained with silver and the immunoreactive spots in these gels were excised and pooled for mass spectrometry analysis. A different 2-DE gel was used for immunoblotting.
Immunoblotting
All methods were carried out in accordance with relevant guidelines and regulations. All experimental protocols were approved by the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University (MUTM 2020–058-02). Informed consent was obtained from all subjects and/or their legal guardian(s). Proteins from SDS-PAGE were transferred onto a nitrocellulose membrane. The membrane was cut into five strips and immunoblotting was performed using sera from five different patients as the primary antibody. The proteins separated by 2-DE were transferred onto a nitrocellulose membrane and pooled patient sera was used as the primary antibody. Membranes were blocked using 5% (w/v) non-fat milk in phosphate buffered saline (PBS) for 2 h at room temperature, then rinsed with PBS containing 0.05% (v/v) Tween-20. Serum samples diluted 1:200 in PBS containing 1% non-fat milk were added to the membranes and incubated overnight at 4 °C. After incubation, the membranes were washed three times with PBS containing 0.05% (v/v) Tween-200. Then horseradish peroxidase-conjugated goat anti-human IgG secondary antibodies were added and incubated for 1 h. Immunogen spots were visualized by detection of peroxidase activity using the Ultra TMB-Blotting Solution (ThermoFisher Scientific, UK). Immunoreactive protein spots were excised from silver-stained 2-DE gels and subjected to in-gel digestion.
In-gel tryptic digestion
Gel slices from SDS-PAGE and gels from 2-DE were destained until colorless. The former was destained with 50% acetonitrile (ACN, Sigma-Aldrich) in 50 mM ammonium bicarbonate (ABC, Sigma-Aldrich), while 30 mM potassium ferricyanide (K3Fe(CN)6, Sigma-Aldrich) and 100 nM sodium thiosulfate (Na2S2O3, Sigma-Aldrich) was used for the latter. After destaining, gel pieces were incubated in 4 mM DL-Dithiothreitol (DTT, Sigma-Aldrich) at 60 °C for 15 min. The DTT was subsequently removed and proteins were alkylated by adding 250 mM iodoacetamide (ICH2CONH2, Sigma-Aldrich) and incubated at room temperature in the dark for 30 min. The reaction was quenched with 4 mM DTT and dehydrated in 100% ACN. To digest the proteins, the gel pieces were again rehydrated with 10 ng/µL trypsin in 50 mM ABC and incubated at 37 °C overnight. The peptides were recovered by adding ACN. The supernatant was collected and dried using a vacuum centrifuge (TOMY, Japan). Dried peptides were resuspended in 0.1% formic acid for LC–MS/MS analysis.
Mass spectrometry
A MicroTOF Q II mass spectrometer interfaced with an Ultimate™ 3000 nano-LC system was used for LC–MS/MS. An Acclaim PepMap RSLC 75 µm × 15 cm nanoviper C18 column with 2 µm particle size and 100 Å pore size (Thermo Scientific, Waltham, MA) was used. Data were acquired using a MicroTOF Q II mass spectrometer set with a scan range of 500–3500 m/z. Mass spectrometry data were analyzed using the MASCOT search engine 2.3 (Matrix Science, Ltd) for peptide identification and a previously reported in-house transcriptomic database1 as the reference proteome. Search parameters were set as follows: one miss cleavage; trypsin digestion; 0.8 Da peptide tolerance; ± 0.8 fragment mass tolerance; carbamidomethyl (C) and oxidation (M) variable modifications. Significance threshold was 0.05. The protein abundance was also determined based on the LC-MS/MS output55.
Bioinformatic analysis
After protein identification using the in-house transcriptomic database based on the recently published ES proteome of infective stage G. spinigerum larvae4, the identified proteins were further classified using the GO database (http://www.geneontology.org) to determine and predict the biological processes affected by these parasitic proteins. Furthermore, the sequences of proteins identified by the transcriptomic data were also subjected to the Basic Local Alignment Search Tool (BLAST) translated (BLAST:blastx) and searched against the human non-redundant protein database to identify possible drug and vaccine targets. The % identity, E-value, and query coverage were reported.
For matrix metalloproteinase-like protein and cathepsin B, property predictions and molecular weight and isoelectric point calculations were performed by the Expasy Compute pI/Mw tool (https://web.expasy.org/compute_pi/). Prediction of signaling peptides, non-classical secretory domains, transmembrane protein domains, n-glycosylation, and o-glycosylation were performed using SignalP version 5.1 (https://services.healthtech.dtu.dk/service.php?SignalP-5.0), SecretomeP version 2.0 Server (https://services.healthtech.dtu.dk/service.php?SecretomeP-2.0), TMHMM version 2.0 (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0), NetNGlyc version 1.0 (https://services.healthtech.dtu.dk/service.php?NetNGlyc-1.0), and NetOGlyc version 4.0 (https://services.healthtech.dtu.dk/service.php?NetOGlyc-4.0), respectively. For sequence alignment, all sequences were retrieved from the non-redundant protein sequence NCBI database. The alignments and identity calculations were performed using the Clustal Omega software.
Supplementary Information
Acknowledgements
This research project was supported by international postdoctoral fellowships awarded by Mahidol university to K.N. and O.R., Research Grant from the Faculty of Tropical Medicine, Mahidol University, Fiscal Year 2018 to T.T. and Partnership Research Award for Clinical and Pre-Clinical Departments FY2019 awarded by Faculty of Tropical Medicine, Mahidol university to P.D. and S.M.
Author contributions
All authors participated in the design and interpretation of the study. P.A., P.D. and O.R. analyzed the results. K.N., T.T., O.R. conducted the proteomics and immunomics experiments. P.A. prepared parasites. S.M. and C.T. prepared patient sera. K.N. and T.T. performed bioinformatic analysis. All authors wrote, revised, and approved the final manuscript.
Dat availability
The dataset used in this study might be shared upon reasonable request to Onrapak Reamtong, PhD.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kathyleen Nogrado and Tipparat Thiangtrongjit.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-10826-4.
References
- 1.Maleewong W, et al. Gnathostoma spinigerum: Growth and development of third-stage larvae in vitro. J. Parasitol. 1995;81:800–803. doi: 10.2307/3283983. [DOI] [PubMed] [Google Scholar]
- 2.Diaz J. Gnathostomiasis: An emerging infection of raw fish consumers in Gnathostoma nematode-endemic and nonendemic countries. J. Travel Med. 2015;22:318–324. doi: 10.1111/jtm.12212. [DOI] [PubMed] [Google Scholar]
- 3.Sivakorn C, et al. Case Report: The first direct evidence of Gnathostoma spinigerum migration through human lung. Am. J. Trop. M. Hyg. 2020;103:1129–1134. doi: 10.4269/ajtmh.20-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nuamtanong, S. et al. Transcriptome and excretory–secretory proteome of infective-stage larvae of the nematode Gnathostoma spinigerum reveal potential immunodiagnostic targets for development. Parasite26 (2019). [DOI] [PMC free article] [PubMed]
- 5.Tapchaisri P, Nopparatana C, Chaicumpa W, Setasuban P. Specific antigen of Gnathostoma spinigerum for immunodiagnosis of human gnathostomiasis. Int. J. Parasitol. 1991;21:315–319. doi: 10.1016/0020-7519(91)90033-4. [DOI] [PubMed] [Google Scholar]
- 6.Laummaunwai P, et al. Evaluation of human IgG class and subclass antibodies to a 24 kDa antigenic component of Gnathostoma spinigerum for the serodiagnosis of gnathostomiasis. Parasitol. Res. 2007;101:703–708. doi: 10.1007/s00436-007-0538-3. [DOI] [PubMed] [Google Scholar]
- 7.Nopparatana C, Setasuban P, Chaicumpa W, Tapchaisri P. Purification of Gnathostoma spinigerum specific antigen and immunodiagnosis of human gnathostomiasis. Int. J. Parasitol. 1991;21:677–687. doi: 10.1016/0020-7519(91)90079-M. [DOI] [PubMed] [Google Scholar]
- 8.Saksirisampant W, Kulkaew K, Nuchprayoon S, Yentakham S, Wiwanitkit V. A survey of the infective larvae of Gnathostoma spinigerum in swamp eels bought in a local market in Bangkok, Thailand. Ann. Trop. Med. Parasitol. 2002;96:191–195. doi: 10.1179/000349802125000295. [DOI] [PubMed] [Google Scholar]
- 9.Uparanukraw P, Morakote N, Harnnoi T, Dantrakool A. Molecular cloning of a gene encoding matrix metalloproteinase-like protein from Gnathostoma spinigerum. Parasitol. Res. 2001;87:751–757. doi: 10.1007/s004360100440. [DOI] [PubMed] [Google Scholar]
- 10.Janwan P, et al. Proteomic analysis identification of antigenic proteins in Gnathostoma spinigerum larvae. Exp. Parasitol. 2015;159:53–58. doi: 10.1016/j.exppara.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Kraivichian P, Kulkumthorn M, Yingyourd P, Akarabovorn P, Paireepai C-C. Albendazole for the treatment of human gnathostomiasis. Trans. R. Soc. Trop. Med. Hyg. 1992;86:418–421. doi: 10.1016/0035-9203(92)90248-B. [DOI] [PubMed] [Google Scholar]
- 12.Kongkerd N, Uparanukraw P, Morakote N, Sajid M, McKerrow JH. Identification and characterization of a cathepsin L-like cysteine protease from Gnathostoma spinigerum. Mol. Biochem. Parasitol. 2008;160:129–137. doi: 10.1016/j.molbiopara.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Dominguez R, Holmes KC. Actin structure and function. Annu. Rev. of Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stitt AW, Fairweather I, Trudgett AG, Johnston CF, Anderson SM. Localisation of actin in the liver fluke. Fasciola hepatica. Parasitol. Res. 2004;78:96–102. doi: 10.1007/BF00931648. [DOI] [PubMed] [Google Scholar]
- 15.Geary TG, et al. Haemonchus contortus: Ivermectin-induced paralysis of the pharynx. Exp. Parasitol. 1993;77:88–96. doi: 10.1006/expr.1993.1064. [DOI] [PubMed] [Google Scholar]
- 16.Criado-Fornelio A, Armas-Serra CD, Jimenez-Gonzlez A, Casado-Escribano N, Rodrìguez-Caabeiro F. Biochemical effects of luxabendazole on Trichinella spiralis. Parasitol. Res. 2004;76:518–520. doi: 10.1007/BF00931057. [DOI] [PubMed] [Google Scholar]
- 17.Lacey E. The role of the cytoskeletal protein, tubulin, in the mode of action and mechanism of drug resistance to benzimidazoles. Int. J. Parasitol. 1988;18:885–936. doi: 10.1016/0020-7519(88)90175-0. [DOI] [PubMed] [Google Scholar]
- 18.Hall, D. H., Herndon, L. A. & Altun, Z. in WormAtlas (ed Herndon L.A.); 10.3908/wormatlas.4.1 (2002–2021).
- 19.Moreno Y, et al. Proteomic analysis of excretory-secretory products of Heligmosomoides polygyrus assessed with Next-Generation Sequencing transcriptomic information. PLoS Negl. Trop. Dis. 2011;5:e1370. doi: 10.1371/journal.pntd.0001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janwan P, et al. Growth and development of Gnathostoma spinigerum (Nematoda: Gnathostomatidae) larvae in Mesocyclops aspericornis (Cyclopoida: Cyclopidae) Parasit Vectors. 2011;4:93. doi: 10.1186/1756-3305-4-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reverte M, et al. The antioxidant response favors Leishmania parasites survival, limits inflammation and reprograms the host cell metabolism. PLoS Pathog. 2021;17:e1009422. doi: 10.1371/journal.ppat.1009422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brophy PM, Barrett J. Glutathione transferase in helminths. Parasitology. 1990;100:345–349. doi: 10.1017/S0031182000061369. [DOI] [PubMed] [Google Scholar]
- 23.Capron A, Riveau G, Capron M, Trottein F. Schistosomes: the road from host-parasite interactions to vaccines in clinical trials. Trends Parasitol. 2005;21:143–149. doi: 10.1016/j.pt.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Torres-Rivera A, Landa A. Glutathione transferases from parasites: A biochemical view. Acta Trop. 2008;105:99–112. doi: 10.1016/j.actatropica.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Hall A, Nelson K, Poole LA, Andrew Karplus P. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox Signal. 2011;15:795–815. doi: 10.1089/ars.2010.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fereig RM, Kuroda Y, Terkawi MA, Mahmoud ME, Nishikawa Y. Immunization with Toxoplasma gondii peroxiredoxin 1 induces protective immunity against toxoplasmosis in mice. PLoS ONE. 2017;12:e0176324. doi: 10.1371/journal.pone.0176324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh I, Eisinger SW, Raghavan N, Scott AL. Thioredoxin peroxidases from Brugia malayi. Mol. Biochem. Parasitol. 1998;91:207–220. doi: 10.1016/S0166-6851(97)00213-2. [DOI] [PubMed] [Google Scholar]
- 28.Lu W, Egerton GL, Bianco AE, Williams SA. Thioredoxin peroxidase from Onchocerca volvulus: A major hydrogen peroxide detoxifying enzyme in filarial parasites. Mol. Biochem. Parasitol. 1998;91:221–235. doi: 10.1016/S0166-6851(97)00230-2. [DOI] [PubMed] [Google Scholar]
- 29.Molina-López J, Jiménez L, Ochoa-Sánchez A, Landa A. Molecular cloning and characterization of a 2-cys peroxiredoxin from Taenia solium. J. Parasitol. 2006;92:796–802. doi: 10.1645/GE-754R.1. [DOI] [PubMed] [Google Scholar]
- 30.Suttiprapa S, et al. Characterization of the antioxidant enzyme, thioredoxin peroxidase, from the carcinogenic human liver fluke, Opisthorchis viverrini. Mol. Biochem. Parasitol. 2008;160:116–122. doi: 10.1016/j.molbiopara.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumagai T, Osada Y, Kanazawa T. 2-Cys peroxiredoxins from Schistosoma japonicum: The expression profile and localization in the life cycle. Mol. Biochem. Parasitol. 2006;149:135–143. doi: 10.1016/j.molbiopara.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 32.Gretes MC, Poole LBA, Andrew Karplus P. Peroxiredoxins in parasites. Antioxid. Redox Signal. 2012;17:608–633. doi: 10.1089/ars.2011.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Funato Y, Miki H. Nucleoredoxin, a novel thioredoxin family member involved in cell growth and differentiation. Antioxid. Redox Signal. 2007;9:1035–1058. doi: 10.1089/ars.2007.1550. [DOI] [PubMed] [Google Scholar]
- 34.Funato Y, Hayashi T, Irino Y, Takenawa T, Miki H. Nucleoredoxin regulates glucose metabolism via phosphofructokinase 1. Biochem. Biophys. Res. Commun. 2013;440:737–742. doi: 10.1016/j.bbrc.2013.09.138. [DOI] [PubMed] [Google Scholar]
- 35.Rogers WP. Enzymes in the exsheathing fluid of nematodes and their biological significance. Int. J. Parasitol. 1982;12:495–502. doi: 10.1016/0020-7519(82)90043-1. [DOI] [PubMed] [Google Scholar]
- 36.Lustigman S. Molting, enzymes and new targets for chemotherapy of Onchocerca volvulus. Parasitol. Today. 1993;9:294–297. doi: 10.1016/0169-4758(93)90128-3. [DOI] [PubMed] [Google Scholar]
- 37.Huang Y, et al. A Zinc metalloprotease nas-33 is required for molting and survival in parasitic nematode Haemonchus contortus. Front. Cell Dev. Biol. 2021;9:1828. doi: 10.3389/fcell.2021.695003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wertheim G, Lustigman S, Silberman H, Shoshan S. Demonstration of collagenase activity in adult Strongyloides ratti (Nematoda: Strongyloididae) and its absence in the infective larvae. J. Helminthol. 1983;57:241–246. doi: 10.1017/S0022149X0000955X. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, et al. Cathepsin B cysteine proteinase is essential for the development and pathogenesis of the plant parasitic nematode Radopholus similis. Int. J. Biol. Sci. 2015;11:1073–1087. doi: 10.7150/ijbs.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ranasinghe SL, McManus DP. Protease inhibitors of parasitic flukes: Emerging roles in parasite survival and immune defence. Trends Parasitol. 2017;33:400–413. doi: 10.1016/j.pt.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 41.Zang X, et al. The serpin secreted by Brugia malayi microfilariae, Bm-SPN-2, elicits strong, but short-lived, immune responses in mice and humans. J. Immun. 2000;165:5161–5169. doi: 10.4049/jimmunol.165.9.5161. [DOI] [PubMed] [Google Scholar]
- 42.Duggan B, Dyson J, Wright P. Inherent flexibility in a potent inhibitor of blood coagulation, recombinant nematode anticoagulant protein c2. Europ. J. Biochem. 1999;265:539–548. doi: 10.1046/j.1432-1327.1999.00781.x. [DOI] [PubMed] [Google Scholar]
- 43.Rhoads M, Fetterer R, Hill D, Urban J. Trichuris suis: A secretory chymotrypsin/elastase inhibitor with potential as an immunomodulator. Exp. Parasitol. 2000;95:36–44. doi: 10.1006/expr.2000.4502. [DOI] [PubMed] [Google Scholar]
- 44.Chertov O, et al. Identification of human neutrophil-derived cathepsin G and zzurocidin/CAP37 as chemoattractants for mononuclear cells and neutrophils. J. Exp. Med. 1997;186:739–747. doi: 10.1084/jem.186.5.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poole CB, Jin J, McReynolds LA. Cloning and biochemical characterization of blisterase, a subtilisin-like convertase from the filarial parasite, Onchocerca volvulus*. J. Biol. Chem. 2003;278:36183–36190. doi: 10.1074/jbc.M302601200. [DOI] [PubMed] [Google Scholar]
- 46.Gamble HR, et al. International commission on Trichinellosis: Recommendations on methods for the control of Trichinella in domestic and wild animals intended for human consumption. Vet. Parasitol. 2000;93:393–408. doi: 10.1016/S0304-4017(00)00354-X. [DOI] [PubMed] [Google Scholar]
- 47.Johnstone IL. Cuticle collagen genes: Expression in Caenorhabditis elegans. Trends Genet. 2000;16:21–27. doi: 10.1016/S0168-9525(99)01857-0. [DOI] [PubMed] [Google Scholar]
- 48.Banerjee S, et al. Host delivered RNAi of two cuticle collagen genes, Mi-col-1 and Lemmi-5 hampers structure and fecundity in Meloidogyne incognita. Front. Plant Sci. 2018;8:2266. doi: 10.3389/fpls.2017.02266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Page AP, Stepek G, Winter AD, Pertab D. Enzymology of the nematode cuticle: A potential drug target? Int. J. Parasitol. Drugs Drug Resist. 2014;4:133–141. doi: 10.1016/j.ijpddr.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laummaunwai P, Intapan PM, Wongkham C, Lulitanond VA, Maleewong W. Identification of antigenic components of Gnathostoma spinigerum advanced third stage larvae by two-dimensional gel electrophoresis and mass spectrometry. Southeast Asian J. Trop. Med. Public Health. 2008;39:19–25. [Google Scholar]
- 51.Janwan P, et al. A Recombinant matrix metalloproteinase protein from Gnathostoma spinigerum for serodiagnosis of neurognathostomiasis. Korean J. Parasitol. 2013;51:751–754. doi: 10.3347/kjp.2013.51.6.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janwan P, et al. Application of Recombinant Gnathostoma spinigerum matrix metalloproteinase-like protein for serodiagnosis of human gnathostomiasis by immunoblotting. Am. J. Trop. Med. Hyg. 2013;89:63–67. doi: 10.4269/ajtmh.12-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Issaq HJ, Veenstra TD. Two-dimensional polyacrylamide gel electrophoresis (2D-PAGE): advances and perspectives. Biotechniques. 2008;44:697–700. doi: 10.2144/000112823. [DOI] [PubMed] [Google Scholar]
- 54.Lisowska E. The role of glycosylation in protein antigenic properties. Cell. Mol. Life Sci. 2002;59:445–455. doi: 10.1007/s00018-002-8437-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ishihama Y, et al. Exponentially Modified Protein Abundance Index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per Protein*s. Mol. Cell. Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset used in this study might be shared upon reasonable request to Onrapak Reamtong, PhD.