Abstract
In silico studies performed on the metabolites of four Cameroonian medicinal plants with a view to propose potential molecules to fight against COVID-19 were carried out. At first, molecular docking was performed for a set of 84 selected phytochemicals with SARS-CoV-2 main protease (PDB ID: 6lu7) protein. It was further followed by assessing the pharmacokinetics and pharmacological abilities of 15 compounds, which showed low binding energy values. As the screening criteria for their ADMET properties were performed, only two compounds have shown suitable pharmacological properties for human administration which were shortlisted. Furthermore, the stability of binding of these compounds was assessed by performing molecular dynamics (MD) simulations. Based on further analysis through molecular dynamics simulations and reactivity studies, it was concluded that only the Pycnanthuquinone C (17) and the Pycnanthuquinone A (18) extracted from the Pycnanthus angolensis could be considered as candidate inhibitors for targeted protein. Indeed, we expect that these compounds could show excellent in vitro and in vivo activity against SARS-CoV-2.
Supplementary information
The online version contains supplementary material available at 10.1007/s11224-022-01939-7.
Keywords: SARS-CoV-2, Cameroonian medicinal plants, Pycnanthuquinone, Pycnanthus angolensis, Molecular docking, Molecular dynamics, ADMET
Introduction
The respiratory system can be affected by illnesses ranging from acute infections such as pneumonia, bronchitis, influenza, and colds [1]; chronic conditions such as asthma and sinusitis; and chronic obstructive pulmonary disease or symptoms such as cough [2]. The control of these diseases in developing countries depends closely on traditional medicine and more specifically on herbal remedies made from local medicinal plants [3, 4]. With this, tremendous expansion in the use of traditional medicine, in particular, herbal medicines has led to a plethora of ethnobotanical investigations conducted. These, later, have proven to be one of the most reliable approaches for the discovery of new drugs [5]. Although the Cameroonian flora is rich in medicinal plant species, only a few ethnobotanical studies have been identified, some plants for their use to treat respiratory diseases [6]. However, respiratory diseases present a high cost for both patients and for society [1]. In addition, secondary metabolites have shown great and rapid action than generic drugs in the treatment of many microbial and viral infections. Moreover, secondary metabolites of some medicinal plant species have been isolated, purified, and characterized. It appears that these plants are rich with secondary metabolites, which are known for their potential biological activities [7, 8]. The Cameroonian flora could therefore be a source of interesting secondary metabolites that could be used to fight against emerging infection including the SARS-CoV-2 and its novel variants.
According to the World Health Organization, the worldwide situation update shows that from 31 December 2019 and as to the first week of October 2021, a total of 234 M cases (compare to 88.8 M cases in January) of COVID-19 have been reported worldwide, including 4.8 M deaths (compare 1.9 M death cases in January), which show an increase of infected case (145.2 M) and number of death (2.9 M) during 9 months. In Africa, the situation is also alarming with 6 M cases and 0.15 M deaths; the five countries reporting the most high number of cases are South Africa (2.9 M), Morocco (0.9 M), Tunisia (0.7 M), Ethiopia (0.35 M), and Egypt (0.31) [9]. These statistics demonstrate that developing countries are not preserved and are inundated by this viral infectious disease, which now occupies an important place in the global incidence of transmissible diseases. Several attempts to manage this pandemic, despite all the advances made in modern and orthodox medicine, have posed mankind a lot of health consequences. Traditional medicine has consequently gained renewed interest in health care services. In addition, it is estimated that at least 25% of all modern medicines are derived directly or indirectly from medicinal plants, mainly through the application of modern technologies to traditional knowledge [10].
As a part of our ongoing bioprospection for anti-COVID-19 agents, we observed significant antiviral activity of some secondary metabolites isolated from common medicinal plants of the Asteraceae family [11]. Things that prompted us to carry out an in silico study on secondary metabolites isolated from others medicinal plants, based on ethnobotanical survey made in some Cameroonian villages. The study aims to promote the use of traditional pharmacopeia in a possible treatment of COVID-19 infection. The secondary metabolites collected from the identified species were used to generate ligands, which will be used for docking studies towards main target protein of SARS-CoV-2.
Indeed, the interaction modes of 84 potential antiviral candidates were studied regarding the SARS-CoV-2 main protease (Mpro) protein using virtual screening, molecular docking, and molecular dynamics methods.
Materials and methods
Construction of phytochemical database
The dataset of this study was constructed by gathering 84 phytochemicals from plants belonging to Cameroonian flora commonly used to treat respiratory infections. This set was collected as follows: 28 from Pycnanthus angolensis (Myristicaceae) [12–16], 25 from Paullinia pinnata Linn (Sapindaceae) [17–26], 18 from Allanblackia monticola Staner L.C. (Guttiferae) [27–29], and 13 from Alchornea cordifolia (Euphorbiaceae) [30–33] (Table S1). The three-dimensional structures of these phytochemicals were obtained from the PubChem database.
In order to perform molecular docking analysis, the structures of the 84 phytochemicals were optimized and converted into a single database format (SDF) using Discovery Studio 2020 software. Molecular docking study was performed for each compound of the set with main protease (Mpro) SARS-CoV-2 protein.
Pharmacological and pharmacokinetic as well as ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) proprieties of the studied compounds were also assessed by using SwissADME webserver (http://www.swissadme.ch) [34]. The SwissADME server was used to determine the ADME characteristics of the studied compounds and involving the assessment of the drug-like characteristics as well as their possible toxicity.
Ligand–protein targets virtual screening
Molecular docking is considered as one of the best computational methods used to highlight the bonding modes of molecules (ligands) with the targets [35–38], because of its ability to predict the conformation and mode of binding of the ligand to the receptor binding site.
The three-dimensional structure of Mpro protein bounded with N3 inhibitor was downloaded from Protein Data Bank (PDB ID: 6lu7). Firstly, before performing the molecular docking, the two proteins are prepared. The N3 inhibitor, water molecules, and all non-protein elements were removed, and hydrogen atoms were added to the structure.
Next, the preparation of ligands that will be docked to proteins consists of adding hydrogen atoms to these ligands and optimizing their structure. The site of binding is defined as the volume occupied by the co-crystallized ligands in each of the receptors pocket.
The protein and ligands as well as the ligand entry site into the protein pockets are prepared in the present work by using Discovery Studio 2016 software. In addition, AutoDock software (ADT) MGLTools 1.5.6 packages are used in the re-docking of the co-ligands with the receptor and in the docking of molecules for the 84 molecules with Mpro SARS-CoV-2 protein.
The 3D grid box dimensions were (20 × 20 × 20) Angstrom, with coordinates of x = −10.641, y = 11.847, and z = 68.346 to suit the binding site of removed N3 inhibitor.
According to the results of recent molecular mechanics studies, Nelfinavir antiviral drug has been chosen to serve as a reference ligand with SARS-CoV-2 Mpro (6lu7) protein in this study [39–43].
This work explores molecular docking in the identification of the most important active sites and in the analyzing of the interactions that occur between the docked molecules and the identified active site.
The obtained results were evaluated according to the binding energy value, and the molecule with the lowest binding energy (best score) was considered as the best one that interacts with the target.
ADMET analysis of top drug candidates
Drug candidates should match many criteria as a basic step of drug discovery process in order to be considered as orally consumed drugs. Physicochemical properties (molecular weight, H bonding, heavy atoms, etc.), drug likeness properties (Lipinski et al. rule [44], bioavailability), water solubility, pharmacokinetics, stability, and low cost of production are evaluated to make a point of view about adopting this drug or not. In the present study, SwissADME webserver was used to evaluate the ADMET properties of the top selected molecules.
Determining the toxicity of potential drugs
Examining the toxicological characteristics of the drug candidates and ensuring their safety are one of the most important stages to be followed to approve any novel drug. In this study, ProTox-II16 online web server (https://tox-new.charite.de/protox_II) was utilized to evaluate the toxicological properties of the selected molecules; this server predicts the values of oral toxicity, cytotoxicity, carcinogenicity, immunotoxicity, and mutagenicity. Some toxicity-related evaluations have also been added, such as minnow toxicity (MT), hepatotoxicity (HP), and skin sensitization (SS). All previous properties are fundamental for the access to toxicological risk.
Molecular dynamics simulations
Compounds that have showed highest binding affinity were selected for further studies using molecular dynamics (MD) simulation. The MD simulation was performed using Gromacs-2018.1 packages with amber99sb-ILDN force field [45, 46]. The topology of both ligands was prepared using Antechamber packages in AmberTools19 [47]. The stability of the protein–ligand complexes for Mpro protein was studied using MD simulation. The protein and their complexes were solvated using TIP3P water model in triclinic boxes separately. All structures were neutralized by adding sodium or chlorine counter ions. The steepest minimization of 5000 steps was performed to remove weak Van der Waals contacts. All systems were then equilibrated for NVT and NPT for 1 ns each. The NVT equilibration was done at 300 K using V-rescale thermostat and the NPT equilibration was performed at 1.0 bar using Parrinello-Rahman barostat [48, 49]. The 100-ns MD simulation was carried out and a total of 10,000 frames of each system were saved at 10 ps intervals. All trajectories were subjected to PBC corrections before analysis using standard Gromacs utilities. One hundred frames from the trajectory of each complex were extracted from 60 to 100 ns for the MM-PBSA calculations [50].
Results and discussion
Molecular docking simulations
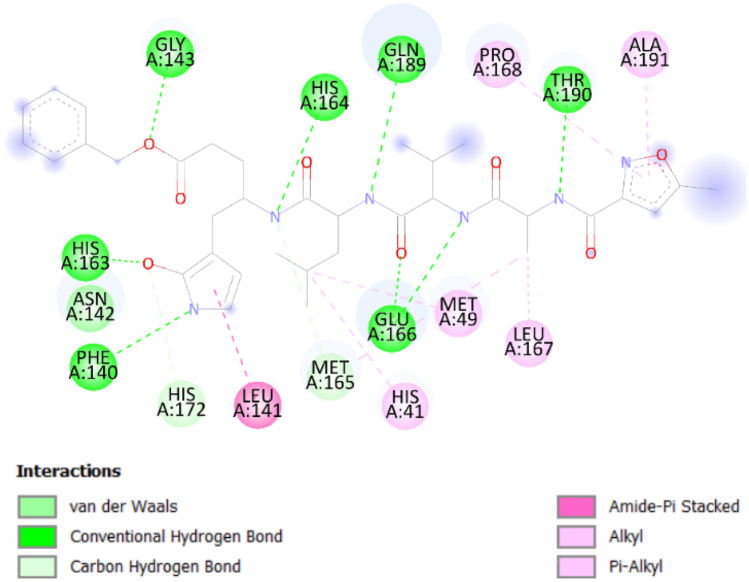
Firstly, the co-crystallized ligand is re-docked into the active site to validate the accuracy of molecular docking. Based on the inhibitor N3 with 6lu7, the binding site was found to be mostly located in the hydrophobic cleft lined by the following amino acids: Glu166, Thr190, Gln189, Phe140, His41, His163, His164, His172, Gly143, Leu141, Leu167, Asn142, Met49, Met167, Met165, Pro168, and Ala191.
It can be seen from Fig. 1 that there are eight hydrogen bond interactions with eight different amino acids, two with Glu166 and six with Phe140, Gly143, His163, His164, Gln189, and Thr190. There are two C-H interactions with Met165 and His172, and three hydrophobic Pi-alky interactions with Pro168, Ala191, and His41. Further inquiry indicates the presence of an Amide-Pi stacked interaction with Leu141 [51].
Fig. 1.
Different interactions for the inhibitor binding of 6lu7 with N3
Then, molecular docking was performed for the 84 ligands with the Mpro SARS-CoV-2 protein using AutoDock1.5.6 software. As it can be noticed from Table 1, molecules 40, 68, and 70 show the best binding affinity to the virus protein with binding energy of −8.5 kcal/mol.
Table 1.
The binding energies BE (kcal/mol) of the 84 studied ligands with the Mpro (PDB ID: 6lu7) SARS-CoV-2 protein using docking modeling
| N° | BE | N° | BE | N° | BE | N° | BE |
|---|---|---|---|---|---|---|---|
| 01 | − 7.2 | 22 | − 6.9 | 43 | − 4.8 | 64 | − 7.2 |
| 02 | − 7.2 | 23 | − 6.8 | 44 | − 5.0 | 65 | − 7.3 |
| 03 | − 7.1 | 24 | − 7.0 | 45 | − 5.5 | 66 | − 7.8 |
| 04 | − 7.0 | 25 | − 7.2 | 46 | − 7.5 | 67 | − 6.2 |
| 05 | − 7.7 | 26 | − 7.0 | 47 | − 6.2 | 68 | − 8.5 |
| 06 | − 7.1 | 27 | − 7.7 | 48 | − 7.9 | 69 | − 7.4 |
| 07 | − 7.2 | 28 | − 7.3 | 49 | − 7.4 | 70 | − 8.5 |
| 08 | − 7.5 | 29 | − 7.9 | 50 | − 8.4 | 71 | − 7.3 |
| 09 | − 5.4 | 30 | − 7.7 | 51 | − 6.3 | 72 | − 6.9 |
| 10 | − 7.2 | 31 | − 5.0 | 52 | − 7.6 | 73 | − 4.5 |
| 11 | − 7.0 | 32 | − 7.7 | 53 | − 7.4 | 74 | − 7.5 |
| 12 | − 8.0 | 33 | − 6.5 | 54 | − 7.4 | 75 | − 7.4 |
| 13 | − 7.7 | 34 | − 7.0 | 55 | − 7.6 | 76 | − 7.5 |
| 14 | − 6.9 | 35 | − 7.6 | 56 | − 7.3 | 77 | − 5.1 |
| 15 | − 7.8 | 36 | − 7.9 | 57 | − 7.6 | 78 | − 5.1 |
| 16 | − 7.2 | 37 | − 7.9 | 58 | − 7.1 | 79 | − 5.5 |
| 17 | − 7.8 | 38 | − 6.5 | 59 | − 7.0 | 80 | − 5.9 |
| 18 | − 8.3 | 39 | − 7.4 | 60 | − 6.9 | 81 | − 5.6 |
| 19 | − 7.5 | 40 | − 8.5 | 61 | − 7.1 | 82 | − 7.5 |
| 20 | − 7.0 | 41 | − 8.3 | 62 | − 8.0 | 83 | − 6.8 |
| 21 | − 6.8 | 42 | − 4.8 | 63 | − 7.2 | 84 | − 5.8 |
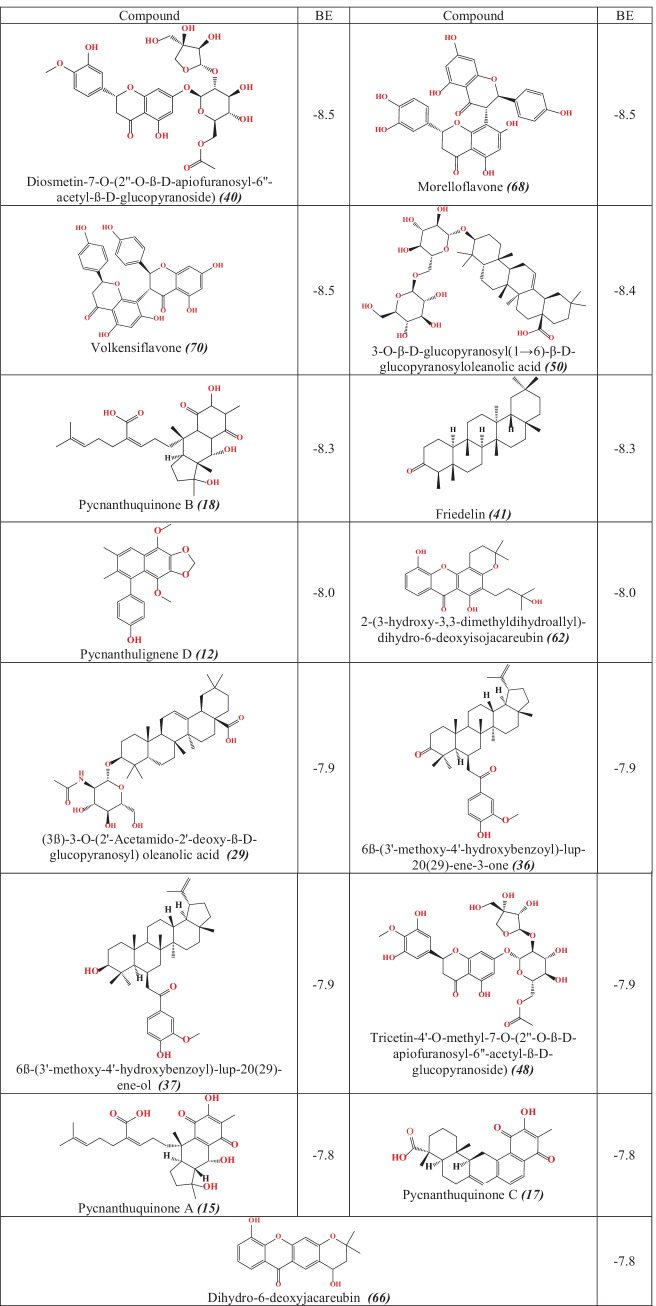
Structural conformation analysis of the complexes (ligand–protein) was carried out to unpick the drug surface hotspot of the target. Molecular interaction types between protein and ligand were identified also; ligand-bounded amino acid residues were determined. Table 2 gather studied compounds showed had the best molecular docking scores with Mpro SARS-CoV-2 protein.
Table 2.
2D structures and binding energy values (in kcal/mol) of the top 15 compounds
From the obtained outcomes of the molecular docking study, fifteen compounds have exhibited the best scores of binding energy values closely to the Nelfinavir (−8.2 kcal/mol). These results allowed us support to suggest the 15 studied compounds as promising SARS-CoV-2 inhibitors.
As depicted in Fig. 2C, Nelfinavir has shown the following interaction types with the Mpro protein of SARS-CoV-2: two H-bonds with Gly339 residue and four hydrophobic interactions with Phen374, Trp436, Leu335, and Val367 residues.
Fig. 2.
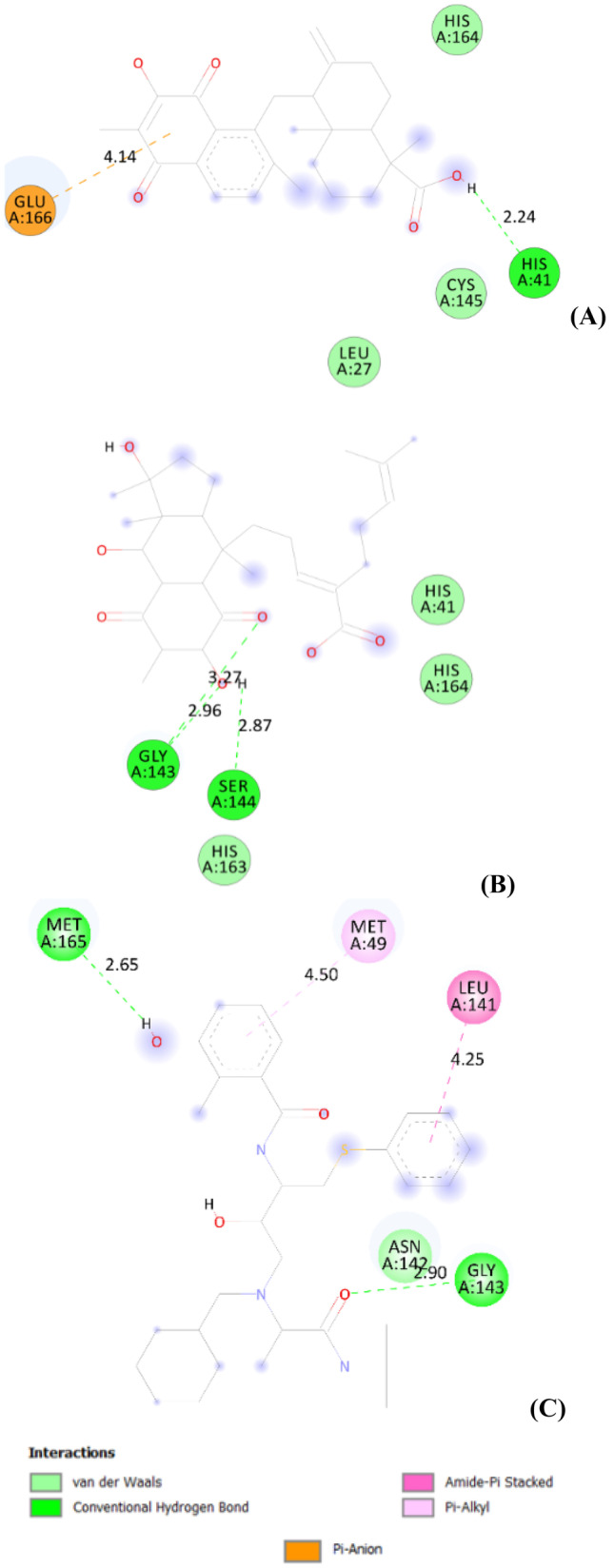
Different interactions and key residues for the inhibitor binding between 6lu7 and (A): compound 17, (B): compound 18 and (C): Nelfinavir (used as reference)
Although compound 18, obtained from Pycnanthus angolensis, was reported that it binds to 6lu7 protein with energy value of −8.3 kcal/mol; herein, it shows three H-bond interactions which maintain the stability of the complex. Two of them was formed with Gly143 amino acid residue and the third one with Ser144 amino acid residue of the Mpro protein, at respective distances of 2.96 Å, 3.27 Å, and 2.87 Å (Fig. 2B).
Compound 17, isolated from Pycnanthus angolensis, exhibits a binding energy of −7.8 kcal/mol with the SARS-CoV-2 Mpro protein. This compound interacts with the Mpro protein through H-bond with His41 amino acid residue at distance of 2.24 Å and pi-anion interaction with Glu166 residue at distance of 4.14 Å (Table 3). The interaction modes are illustrated in Fig. 2A.
Table 3.
Different interactions and key residues for the inhibitor binding between 6lu7 and compounds 17 and 18
| Amino acid | Distance (Å) | Interaction type | Type of HB interaction [57] | |
|---|---|---|---|---|
| 17 | HIS41 | 2.24 | H-Bond | Strong |
| GLU166 | 4.14 | Pi-Anion | ––- | |
| 18 | GLY143 | 2.96 | H-Bond | Strong |
| GLY143 | 3.27 | H-Bond | Average | |
| SER144 | 2.87 | H-Bond | Strong |
2.5 Å < d < 3.10 Å = > strong interaction; 3.1 Å < d < 3.55 Å = > average interaction
We can conclude that the following amino acid residues His41, Glu166, and Ser144 of the Mpro protein contribute significantly in the stability of compounds 17 and 18.
Drug-likeness and pharmacokinetic studies
Drug candidates should possess favorable ADME properties and ideally non-toxic. Therefore, the designed compounds were evaluated of their ADME profile, including drug-likeness, partition coefficient, solubility, human intestinal absorption, penetration of the blood brain barrier (BBB), cytochrome P450 inhibition, and several other parameters using SwissADME. The results show among the 15 studied molecules, two molecules were capable to pass ADME profiling to the toxicity aspect study used ProTox-II webserver with reference of Globally Harmonized System (UN, 2013).
Drug-likeness is a key criterion in screening drug candidates at the earlier phase of drug discovery and development. This parameter can be described as a mean to correlate physicochemical aspect of a compound with its biopharmaceutical aspect in the human body, especially its influence in bioavailability of per oral route [52].
Two different rule-based filters, Lipinski et al. [44, 53], were used to calculate the drug and lead likeness for the 15 selected compounds. The results exhibited that the compounds 29, 36, 37, 40, 48, 50, 68, and 70 have violated the understudy drug-likeness rules (Lipinski’s criteria: less than 5 HBD, less than 10 HBA, a MW less than 500 g/mol and log P not greater than 5 and Veber’s criteria: TPSA should be less than 140 Å2 and the NRB should be less than 10). These results suggest that these compounds have a low theoretical oral bioavailability according to the Lipinski’s rule-of-five and bioavailability score value.
Another important property of oral drugs is solubility in intestinal fluid, because insufficient solubility can limit intestinal absorption through the portal vein system. All of the compounds having low aqueous solubility level are shown in Table 4, except for compounds 12, 15, 17, 18, 40, 48, 62, and 66 that have shown moderate solubility. The aqueous solubility (log S) of a compound significantly affects its absorption and distribution characteristics.
Table 4.
Molecular properties and ADME prediction of ligand compounds
| N | MW g/mol | logP | HBA | HBD | N.rot | TPSA (Å2) | log S mol/l | Lipinski | Veber | Bioavail ability score | PAINS alert | GI | BBB | Cyp1A2 | Cyp2C19 | Cyp2C9 | Cyp2D6 | Cyp3A4 | Log kp |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 352.38 | 2.68 | 5 | 1 | 3 | 57.15 | −5.39 | Yes | Yes | 0.55 | 0 | High | Yes | Yes | Yes | Yes | No | No | −4.95 |
| 15 | 472.57 | 1.51 | 7 | 4 | 7 | 132.13 | −4.46 | Yes | Yes | 0.56 | 1 | Low | No | No | No | No | No | Yes | −6.75 |
| 17 | 436.54 | 3.04 | 5 | 2 | 3 | 91.67 | −5.90 | Yes | Yes | 0.56 | 1 | High | No | No | No | Yes | No | Yes | −5.11 |
| 18 | 490.63 | 1.87 | 7 | 4 | 7 | 132.13 | −4.49 | Yes | Yes | 0.56 | 0 | Low | No | No | No | No | No | Yes | −6.96 |
| 29 | 659.89 | 3.14 | 8 | 5 | 6 | 145.55 | −7.25 | Yes | No | 0.56 | 0 | Low | No | No | No | No | No | Yes | −6.14 |
| 36 | 588.86 | 5.97 | 4 | 1 | 5 | 63.60 | −9.69 | No | Yes | 0.17 | 0 | Low | No | No | No | No | No | No | −2.65 |
| 37 | 590.88 | 6.05 | 4 | 2 | 5 | 66.76 | −9.90 | No | Yes | 0.17 | 0 | Low | No | No | No | No | No | No | −2.44 |
| 40 | 638.57 | −2.84 | 16 | 7 | 10 | 240.36 | −2.97 | No | No | 0.17 | 0 | Low | No | No | No | No | No | No | −10.61 |
| 41 | 426.72 | 6.92 | 1 | 0 | 0 | 17.07 | −8.66 | Yes | Yes | 0.55 | 0 | Low | No | No | No | No | No | No | −1.94 |
| 48 | 654.57 | −3.30 | 17 | 8 | 10 | 260.59 | −2.85 | No | No | 0.17 | 0 | Low | No | No | No | No | No | No | −10.95 |
| 50 | 780.98 | 0.88 | 13 | 8 | 7 | 215.83 | −6.58 | No | No | 0.11 | 0 | Low | No | No | No | No | No | No | −8.40 |
| 62 | 398.45 | 1.86 | 6 | 3 | 3 | 100.13 | −5.20 | Yes | Yes | 0.55 | 0 | High | No | No | No | Yes | No | No | −5.66 |
| 66 | 312.32 | 1.29 | 5 | 2 | 0 | 79.90 | −3.79 | Yes | Yes | 0.55 | 0 | High | No | Yes | No | No | Yes | Yes | −6.44 |
| 68 | 558.49 | 0.09 | 11 | 7 | 3 | 194.21 | −6.18 | No | No | 0.17 | 1 | Low | No | No | No | Yes | No | Yes | −6.72 |
| 70 | 542.49 | 0.58 | 10 | 6 | 3 | 173.98 | −6.32 | No | No | 0.17 | 0 | Low | No | No | No | Yes | No | Yes | −6.38 |
The majority of the compounds identified have a low probability of being able to cross the BBB. In fact, only three compounds 12 have a good probability of being BBB permeates.
Pan-Assay Interference Compounds (PAINS) are very familiar to medicinal chemists who have spent time fruitlessly trying to optimize these nonprogressible compounds [54]. Table 4 shows that three of the compounds tested return a PAINS alert (15, 17, and 68).
The cytochrome P450 (CYP) superfamily is important in drug elimination through metabolic biotransformation. Inhibition of these isoenzymes is certainly a major cause of pharmacokinetic-related drug interactions leading to toxic or unwanted adverse effects due to the lower clearance and accumulation of the drug or its metabolites. Table 4 shows that all of compounds are found to be non-inhibitors of isoenzyme CYP1A2, except 12 and 66, and therefore the side effect (i.e., liver dysfunction) may be absent. CYP1A2 is expressed in the liver (10% of the total CYP content) and is responsible for the activation of aromatic and heterocyclic amines, PAHs, and many therapeutic drugs [55]. However, the compounds are predicted to be metabolized by CYP1A2, CYP2C19, CYP2C9, and CYP3A4 with more exceptions.
Skin permeability (log kp) is an important parameter for the evaluation of drugs, which may require transdermal administration [56]. The more negative the log Kp, the less skin permeant is the molecule. Except for compounds 40 and 48, the log Kp measurements of all the tested compounds are found to be within the limits (−10.0 to −1.0).
The toxicity prediction for the 7 compounds that did not violated Lipinski and Veber rules was based on different targets related to adverse drug reactions. The hepatotoxicity, carcinogenicity, mutagenicity, and cytotoxicity of the compounds have been determined [58]. Based on the results of ProTox II, it can be seen that the compound 66 is found to have mutagenicity. Compounds 12 and 62 are reported to be carcinogenicity. All compounds are not hepatotoxic and not cytotoxic (Table 5).
Table 5.
Toxicity prediction for ligands
| Compound | Hepatotoxicity | Carcinogenicity | Mutagenicity | Cytotoxicity | LD50 (mg kg−1) |
|---|---|---|---|---|---|
| 12 | Inactive | Active | Inactive | Inactive | 1000 |
| 15 | Inactive | Inactive | Inactive | Inactive | 340 |
| 17 | Inactive | Inactive | Inactive | Inactive | 2000 |
| 18 | Inactive | Inactive | Inactive | Inactive | 9000 |
| 41 | Inactive | Inactive | Inactive | Inactive | 500 |
| 62 | Inactive | Active | Inactive | Inactive | 2000 |
| 66 | Inactive | Inactive | Active | Inactive | 3850 |
Prediction of LD50 using ProTox II showed that compounds 12, 17, 18, 62, and 66 are predicted to have oral LD50 value ranging from 800 to 9000 mg kg−1 in rat model and have non-toxic effect predicted.
The results obtained from ADMET analysis reveal that compounds 17 and 18 are in good agreement with Lipinski’s and Veber’s rules with no violations; these two compounds have also acceptable ADMET properties. Compounds 17 and 18 could be orally administrated drug candidates. Moreover, compounds 17 and 18 were chosen to perform MD simulation study in order to investigate their stability on Mpro (6lu7) protein. The binding energy values observed for compounds 17 and 18 show that they possess good orientation shape with the active site of Mpro (6lu7) protein, through different interactions compared to chrysoeriol-7-O-β-D-glucuronopyranoside (BE = −8.0 kcal/mol) reported in our previous work [11].
Molecular dynamics simulations
The dynamic nature of the Mpro protein and their complexes were studied by simulating them in aqueous environment. The conformation exhibiting lowest biding energies was used in MD simulation studies.
Analysis of RMSD and RMSF
The initial analysis of trajectories for their stability was performed by calculating the root-mean square deviations (RMSD). The RMSD of each system was calculated with respect to their initial coordinates. The RMSD is presented in Fig. 3.
Fig. 3.

Root mean square deviation (RMSD) of Mpro, Mpro-compound_17 complex, and Mpro-compound_18 complex
The RMSD of Mpro alone shows some instability during initial duration but became stable after 60 ns. A similar observation was found for Mpro-compound_17 and Mpro-compound_18 complexes. The average RMSD of Mpro, Mpro-compound_17 complex, and Mpro-compound_18 complex was found to be 0.234, 0.249, and 0.255 nm, respectively. The RMSD of all systems remained below 0.30 nm indicating a good stability of all systems. The stability of the protein in the absence of ligands was also assessed by calculating the root mean square fluctuations (RMSF). The RMSF of Cα of all residues the protein is shown in Fig. 4.
Fig. 4.
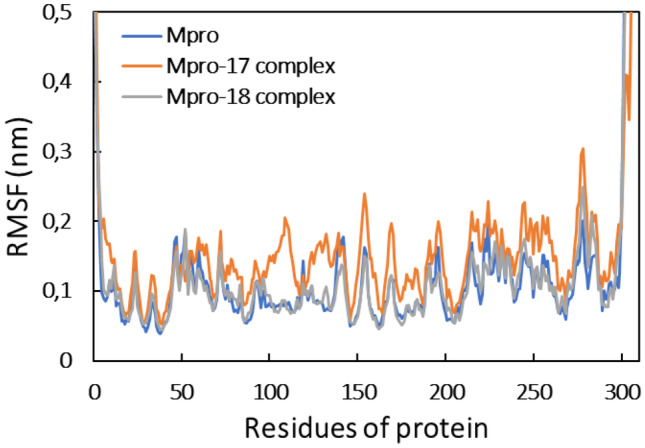
Root mean square fluctuation (RMSF) of Mpro, Mpro-compound_17 complex, and Mpro-compound_18 complex
The RMSF of almost all residues of Mpro was obtained below 0.2 nm, showing the good stability of the protein. The presence of both ligands exhibited a negligible effect on the RMSF of Mpro. The analysis of RMSD and RMSF that confirmed the protein and the complexes was well stable under aqueous environment. The RMSF of all atoms of the ligands was also calculated as shown in Fig. 5. Compound 18 showed relatively more fluctuation, which means that compound 18 underwent more dynamical shift at the binding from than compound 17.
Fig. 5.
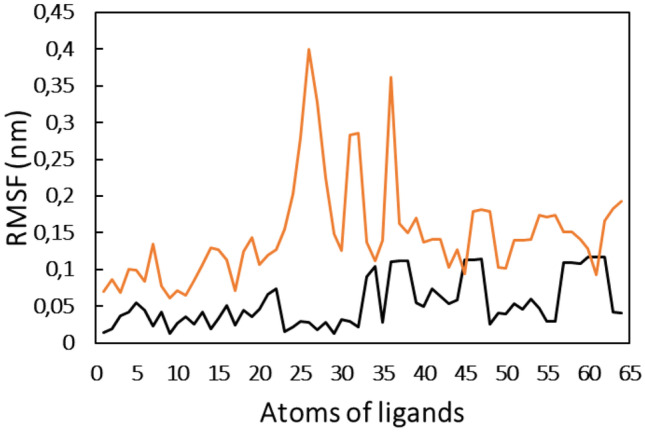
Root mean square fluctuation (RMSF) of each atom of compound 17 and compound 18 when complexed with Mpro
Assessment of Rg and SASA and energies
The mass-weighted root mean square distance of a collection of atoms from their common center of mass is defined as radius of gyration (Rg). This is also an important parameter to be considered while studying the stability of proteins in aqueous systems [59]. Generally, the compact and globular proteins exhibit less variation in their radius of gyration compared to the expanded form of protein [60]. The Rg of protein alone and their complexes are shown in Fig. 6.
Fig. 6.
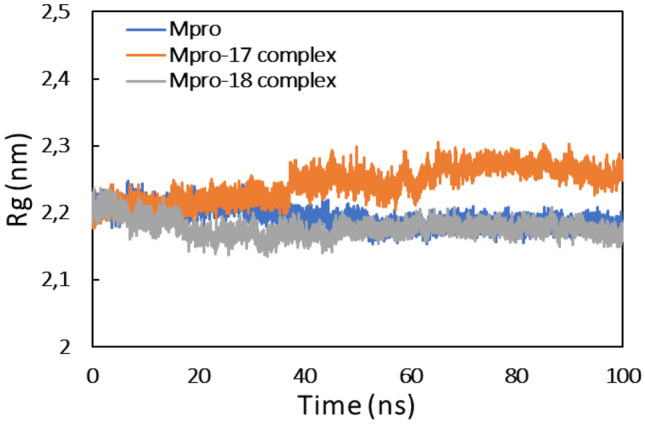
Radius of gyration (Rg) of Mpro, Mpro-compound_17 complex, and Mpro-compound_18 complex
As evident from the data, the Rg of Mpro remained constant during the entire simulation time. A similar observation was found for Mpro-compound_18 complex, while there was a slight variation in Rg of Mpro-compound_17 complex. These mean that all systems were stable during MD simulation. Moreover, the analysis of Rg also inducted that Mpro protein did not undergo major conformational changes [61].
Solvent accessible surface is another important parameter for staying the stability of proteins through MD simulation. The SASA of Mpro protein and their complexes are shown in Fig. 7.
Fig. 7.
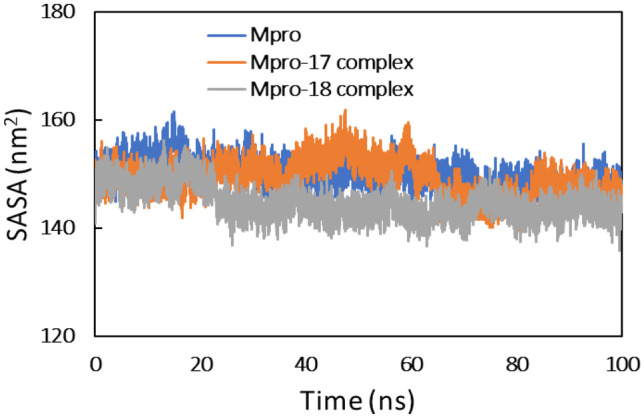
Solvent accessible surface area (SASA) of Mpro, Mpro-compound_17 complex, and Mpro-compound_18 complex
SASA of all systems was found to be persistent over the entire trajectory of simulation. This further validates the stability of complexes.
Finally, the physicochemical parameters of all systems were also calculated. The total and potential energies of all systems is presented in Fig. 8. Both these parameters were also found to be contact during simulation. The data finally validate the stability of all systems during MD simulation.
Fig. 8.
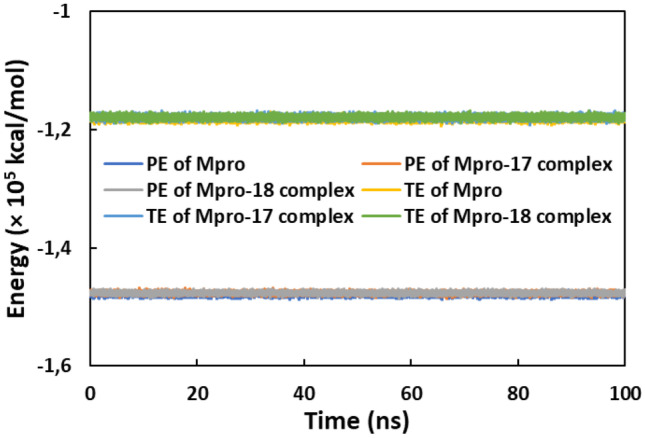
The potential energy and total energy of Mpro, Mpro-compound_17 complex, and Mpro-compound_18 complex as a function of simulation time
Assessment of the structural stability and hydrogen bonds
The effect of binding of both ligands in the structure stability of protein was studied by determining the secondary structural motifs of the proteins. Each secondary structure component of Mpro protein is shown in Fig. 9. The coils, β-sheet, β-bridge, bends, turns, and α-helix in Mpro were found to be 26.39, 25.99, 2.15, 7.20, 16.57, and 17.78%, respectively. All these secondary structures of Mpro change insignificantly when were complexed with compound 17 or compound 18. The data showed that the presence of ligands did not affect the structural stability of both the proteins.
Fig. 9.
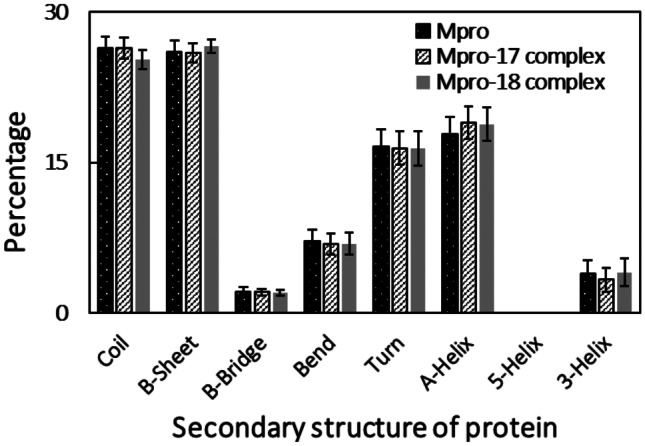
Percentage of secondary structure in Mpro, Mpro-compound_17 complex, and Mpro-compound_18 complex
The interaction between ligands and Mpro protein was studied by assessing the hydrogen bonds (Fig. 10). There was existence of H-bonds both in Mpro-compound_17 complex and Mpro-compound_18 complex throughout the entire simulation period. The average number of hydrogen bonds forming between Mpro and compound 18 was slightly more than that of compound 17. In all complexes, there was existence of H-bonds throughout the simulation period. However, there were slight variations in the H-bond profiles, which may be due to the dynamical shift of the ligands at the binding site.
Fig. 10.
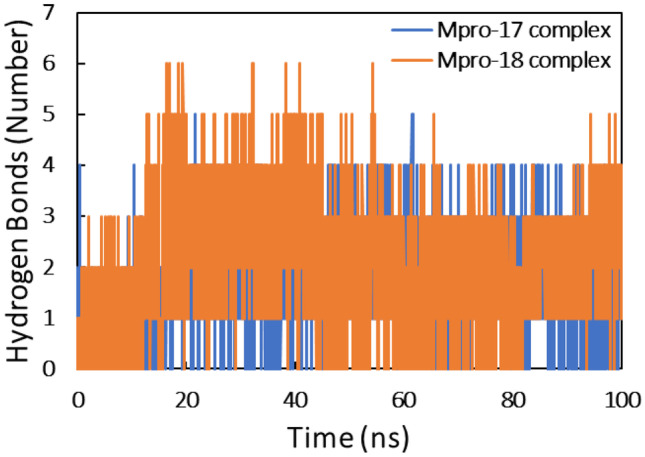
Number of hydrogen bonds for the interaction of compound 17 and compound 18 with Mpro
Principal component analysis
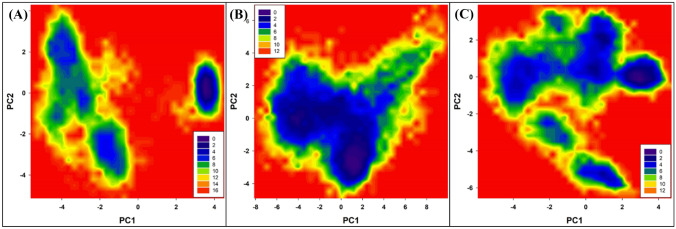
Principal component analysis (PCA) is a standard statistical procedure to analyze the large-scale motion in biological macromolecules such as proteins and DNA. PCA is performed by reducing the dimensionality of data set without losing important information, which is characterized by eigenvectors [62]. PCA was done to explore the flexibility of Mpro protein and their complexes. The projection of eigenvectors in uncomplexed and complexed forms is presented in Fig. 11.
Fig. 11.
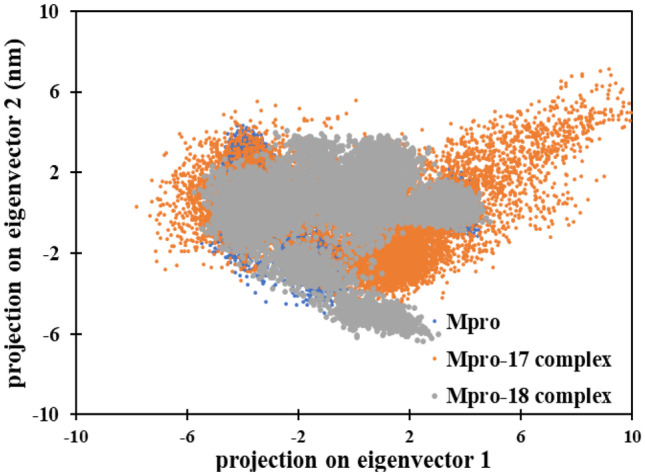
The 2D eigenvector projection plot for Mpro, Mpro-compound 17 complex, and Mpro-compound 19 complex
The data shows that Mpro alone and Mpro-compound_18 complex occupied a similar conformational space. However, Mpro-compound_17 complex occupied slightly more conformational space. This shows that the Mpro-compound_17 exhibited slightly more structural flexibility compared to Mpro alone and Mpro-compound_18 complex.
The free energy landscapes (FEL) of Mpro protein and complexes were also plotted to explore the variations in the patterns of protein folding (Fig. 12). The FEL shows that all systems reached energy minima in their respective landscapes. The lowest energy minima structures of the proteins were extracted to plot the respective Ramachandran plots. The Ramachandran plots of the protein and their complexes showed that almost all amino acids were found the favorable region (Fig. 13).
Fig. 12.
Free energy landscape plot of (A) Mpro, (B) Mpro-compound_17 complex, and (C) Mpro-compound_18 complex
Fig. 13.
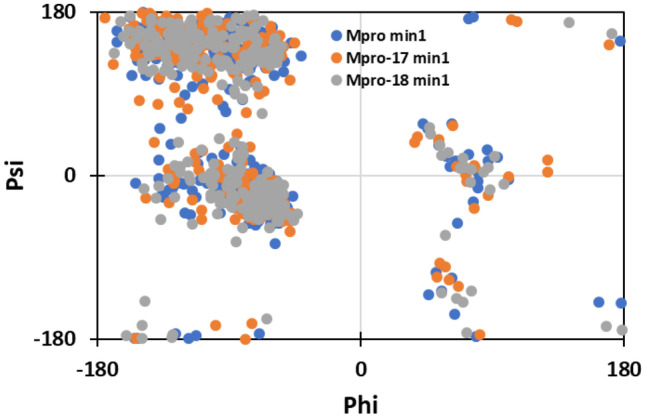
Ramachandran plot of the energy minima of BSA in the absence and presence of BR
MM-PBSA calculation
The detailed insight regarding various binding energies involved for the interaction of both ligands with Mpro protein as done using MM-PBSA calculations. Non-covalent forces mainly stabilize the protein ligand interactions mostly, which include electrostatic, hydrophobic interactions, hydrogen bonds, and Van Der Waals force. These forces may either contribute positively or negatively to the overall binding [63].
For MM-PBSA calculations, 100 frames were extracted from 60 to 100 ns from the trajectory of each complex. The MM-PBSA binding energies are enlisted in Table 6.
Table 6.
Binding free energy (kcal mol-1) for the interaction of compound 17 and compound 18 with Mpro using MMBSA analysis
| Type of energy | Comp. 17 | Comp. 18 | N3 |
|---|---|---|---|
| ΔEvdW | −30.46 ± 0.32 | −42.53 ± 0.38 | −173.639 ± 2.083 |
| ΔEele | −10.13 ± 0.23 | −7.27 ± 0.34 | −41.902 ± 1.424 |
| ΔEPSE | 29.61 ± 0.36 | 29.67 ± 0.36 | 170.300 ± 3.244 |
| ΔESSASA | −3.86 ± 0.03 | −4.57 ± 0.02 | −21.493 ± 0.234 |
| ΔEBE | −14.85 ± 0.27 | −24.71 ± 0.39 | −66.745 ± 2.640 |
ΔEvdW van der Waal energy, ΔEele electrostatic energy, ΔEPSE polar solvation energy, ΔESASA solvent accessible surface area energy, ΔEBE binding energy
The van Der Waals energy was found to be the major contributor in overall binding energy followed by the electrostatic energy. Moreover, there was also small contribution of SASA energy. However, polar solvation energy impaired the binding of both ligands to Mpro protein. The overall binding energy for interaction of compound 17 and compound 18 to Mpro was found to be −14.85 and −24.71 kcal mol−1, respectively. It is interesting to note that the MM-PBSA binding energy of compounds 17 and 18 with the Mpro protein (6lu7) was more compared to the respective controls. This shows the stronger binding affinity of compounds 17 and 18 compared to the control N3 for 6lu7.
The MM-PBSA data was further used to identify the key residues, which gave maximum contribution in the binding (Table 7). For interaction of compound 17 with Mpro, Thr25, Leu27, Val42, Ile43, Thr45, Ser46, Met49, Arg60, Lys61, Leu141, Gly143, Cys145, His164, and Glu166 were the major energy contributors. Thr25, Leu27, Val42, Met49, Leu141, Cys145, Met165, Glu166, Asp187, and Gln189 were the key energy contributors for compound 18 Mpro interaction.
Table 7.
The average polar, apolar, and total binding energies (kcal mol-1) of the key residues of Mpro
| Residues | Compound 17 | Compound 18 | |||||
|---|---|---|---|---|---|---|---|
| Epolar | EApolar | Etotal | Residues | Epolar | EApolar | Etotal | |
| Thr-25 | 2.786 | −0.274 | −1.240 | Thr-25 | 0.154 | −0.092 | −0.643 |
| Leu-27 | 0.150 | −0.036 | −0.700 | Leu-27 | 0.087 | −0.067 | −0.718 |
| Val-42 | 0.289 | −0.001 | −0.110 | Val-42 | 0.064 | −0.000 | −0.200 |
| Ile-43 | −0.185 | −0.000 | −0.176 | Met-49 | 0.520 | −0.251 | −1.717 |
| Thr-45 | 2.120 | −0.086 | −0.341 | Leu-141 | 0.106 | −0.014 | −0.324 |
| Ser-46 | 1.827 | −0.203 | −0.159 | Cys-145 | 0.786 | −0.123 | −0.834 |
| Met-49 | 0.349 | −0.123 | −0.658 | Met-165 | 1.093 | −0.202 | −1.689 |
| Arg-60 | 0.030 | −0.000 | −0.179 | Glu-166 | 1.166 | −0.093 | −0.257 |
| Lys-61 | −0.030 | −0.000 | −0.445 | Asp-187 | 0.946 | −0.045 | −0.476 |
| Leu-141 | −0.042 | 0.000 | −0.106 | Gln-189 | 1.114 | −0.159 | −0.534 |
| Gly-143 | 0.609 | −0.104 | −0.338 | ||||
| Cys-145 | 0.216 | −0.044 | −0.345 | ||||
| His-164 | 0.090 | −0.000 | −0.1201 | ||||
| Glu-166 | −0.158 | −0.001 | −0.422 | ||||
Epolar polar energy, EApolar apolar energy, Etotal total energy
Conclusion
Despite the huge effort made to control the current pandemic, the COVID-19 disease continues spreading throughout the globe. Nowadays, many vaccines are developed and are approved for emergency use even though most people of the world are not vaccinated yet, in particular, people belonging to developing country. It seems that the development of antiviral drugs could be a good strategy to fight against this continued pandemic. For this context, we have performed this in silico study, which aims to develop an inhibitor of both Mpro (6lu7) SARS-CoV-2 protein. The study was conducted on 84 molecules collected from four medicinal plants belonging to Cameroonian flora. Among the 84 compounds, only 15 compounds have shown good binding affinity in the molecular docking analysis compared to the reference (Nelfinavir). The total binding energies of the 15 compounds were in the range of −7.8 kcal/mol and −8.5 kcal/mol, which are near to that obtained for Nelfinavir (−8.2 kcal/mol). The selected 15 compounds were subjected to ADMET analysis to evaluate their absorption, distribution, metabolism, and toxicity properties. The results have shown that compounds 17 and 18 obtained from Pycnanthus angolensis are strongly recommended to be potential drugs against COVID-19. Further analysis was performed using molecular dynamics (MD) simulation to ensure the stability of compounds 17 and 18 on the 6lu7 protein. The obtained results have shown good stability for both compounds on the active sites of the protein during the simulation period. Relying on the result obtained from this study and in the light of previously described works, Pycnanthuquinone C (17) and Pycnanthuquinone B (18) could be considered as in silico inhibitors against Mpro SARS-CoV-2 protein. Experimental investigations should be carried out to assess their in vitro and in vivo efficiency against SARS-CoV-2.
Supplementary information
Below is the link to the electronic supplementary material.
Author contribution
Samir Chtita: writing, original draft preparation, conceptualization, methodology, software, and supervision; Romuald Tematio Fouedjou: data curation, writing, and original draft preparation; Loris Alvine Djoumbissie, Mohamed Bakhouch, and Mohammed Efendi: visualization and original draft preparation; Mebarka Ouassaf and Faizan Abul Qais: conceptualization, writing, and original draft preparation; Salah Belaidi, Tugba Taskin Tok, Mohammed Bouachrine, and Tahar Lakhlifi: visualization and validation.
Data availability
All data used in this work are private.
Code availability
The codes used in this work are not available.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fatima E, Ben AF, Dahmani J, Belahbib N, Zidane L. Étude ethnobotanique des plantes médicinales utilisées dans le traitement des infections du système respiratoire dans le plateau central marocain. Journal of Animal & Plant Sciences. 2015;25:3886–3897. [Google Scholar]
- 2.Fuhrman C, Delmas M-C. Épidémiologie descriptive de la bronchopneumopathie chronique obstructive (BPCO) en France. Rev Mal Respir. 2010;27(2):160–168. doi: 10.1016/j.rmr.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Adjanohoun E (2000) La biodiversité face au développement des industries pharmaceutiques africaines. In Réseau des espèces ligneuses médicinales, Eyog Matig O, Adjanohoun E, de Souza S et Sinsin B (eds). Compte rendu de la première réunion du réseau tenue 15–17 décembre 1999 à la station IITA Cotonou, Bénin, 88–103
- 4.Dibong SD, Mpondo ME, Ngoye A, Kwin NF. Plantes médicinales utilisées par les populations bassa de la région de Douala au Cameroun. International Journal of Biological and Chemical Sciences. 2011;5:1105–1117. doi: 10.4314/ijbcs.v5i3.72227. [DOI] [Google Scholar]
- 5.Koné D. Enquête ethnobotanique de six plantes médicinales maliennes - extraction, identification d’alcaloïdes - caractérisation, quantification de polyphénols: étude de leur activité antioxydante. Mali: Thèse de Doctorat Université de Bamako; 2009. p. 157p. [Google Scholar]
- 6.Benlamdini N, Elhafian M, Rochdi A, Zidane L. Étude floristique et ethnobotanique de la flore médicinale du Haut Atlas oriental (Haute Moulouya) Journal of Applied Biosciences. 2014;78:6771–6787. doi: 10.4314/jab.v78i1.17. [DOI] [Google Scholar]
- 7.Fouedjou TR, Teponno RB, Quassinti L, Bramucci M, Petrelli D, Vitali AL, Fiorini D, Tapondjou AL, Barboni L. Steroidal saponins from the leaves of Cordyline fruticosa (L.) A. Chev. and their cytotoxic and antimicrobial activity. Phytochemistry Letter. 2014;7:62–68. doi: 10.1016/j.phytol.2013.10.001. [DOI] [Google Scholar]
- 8.Fouedjou TR, Nguelefack-Mbuyo PE, Ponou KB, Nguelefack BT, Barboni L, Tapondjou AL. Antioxidant activities and chemical constituents of extracts from Cordyline fruticosa (Agavaceae) and Eriobotrya japonica (Rosaceae) Pharmacologia. 2016;7(103):113. [Google Scholar]
- 9.WHO World Health Organization (2021). https://covid19.who.int/
- 10.Biyiti LF, Meko'o DJL, Tamzc V, Amvam Zollo PH. Recherche de l’activité antibactérienne de quatre plantes médicinales camerounaises. Pharm Med Trad Afr. 2004;13:11–20. [Google Scholar]
- 11.Fouedjou RT, Chtita S, Bakhouch M, Belaidi S, Ouassaf M, Djoumbissie LA, Tapondjou LA, Abul Qais F (2021) Cameroonian medicinal plants as potential candidates of SARS-CoV-2 inhibitors. J Biomol Struct Dyn 1–15. 10.1080/07391102.2021.1914170 [DOI] [PubMed]
- 12.Fort DM, Ubillas RP, Mendez CD, Jolad SD, Inman WD, Carney JR, Chen JL, Ianiro TT, Hasbun C, Bruening RC, Luo J, Reed M, J., Iwu, M., Carlson, T.J., King, S.R., Bierer, D.E., Cooper, R. Novel antihyperglycemic terpenoid-quinones from Pycnanthus angolensis. J Org Chem. 2000;65:6534–6539. doi: 10.1021/jo000568q. [DOI] [PubMed] [Google Scholar]
- 13.Wabo H, Tatsimo S, Tane P, Connolly J. Pycnanthuquinone C: a new terpenoid-quinone from Pycnanthus angolensis. Planta Med. 2007;73:187–189. doi: 10.1055/s-2007-967103. [DOI] [PubMed] [Google Scholar]
- 14.Abrantes M, Mil-Homens T, Duarte N, Lopes D, Cravo P, Madureira M, Ferreira M-J. Antiplasmodial activity of lignans and extracts from Pycnanthus angolensis. Planta Med. 2008;74:1408–1412. doi: 10.1055/s-2008-1081317. [DOI] [PubMed] [Google Scholar]
- 15.Nono ECN, Mkounga P, Kuete V, Marat K, Hultin PG, Nkengfack AE. Pycnanthulignenes A−D, antimicrobial cyclolignene derivatives from the roots of Pycnanthus angolensis. J Nat Prod. 2010;73:213–216. doi: 10.1021/np9007393. [DOI] [PubMed] [Google Scholar]
- 16.Tsaassi VB, Hussain H, Tamboue H, Dongo E, Kouam SF, Krohn K (2010) Pycnangloside: a new cerebroside from bark of Pycnanthus Angolensis. Nat Prod Commun 5(11):1934578X1000501. 10.1177/1934578X1000501121 [PubMed]
- 17.Abourashed EA, Toyang NJ, Choinski J, Khan IA. Two new flavone glycosides from Paullinia pinnata. J Nat Prod. 1999;62:1179–1181. doi: 10.1021/np990063z. [DOI] [PubMed] [Google Scholar]
- 18.Miemanang RS, Krohn K, Hussain H, Dongo E (2007) Paullinoside A and paullinomide A: a new cerebroside and a new ceramide from leaves of Paullinia pinnata ChemInform 38. 10.1002/chin.200703188
- 19.Annan K, Houghton PJ. Two novel lupane triterpenoids from Paullinia pinnata L. with fibroblast stimulatory activity. J Pharm Pharmacol. 2010;62:663–668. doi: 10.1211/jpp.62.05.0016. [DOI] [PubMed] [Google Scholar]
- 20.Lasisi AA, Ayinde BW, Adeleye AO, Onocha PA, Oladosu IA, Idowu PA. New triterpene isovanniloyl and antibacterial activity of constituents from the roots of Paullinia pinnata Linn (Sapindaceae) J Saudi Chem Soc. 2015;19:117–122. doi: 10.1016/j.jscs.2011.12.012. [DOI] [Google Scholar]
- 21.Jackson N, Annan K, Mensah AY, Ekuadzi E, Mensah MLK, Habtemariam S (2015) A novel triterpene from the roots of Paullinia pinnata: 6α-(3′-methoxy-4′-hydroxybenzoyl)-lup-20(29)-ene-3-one. Nat Prod Commun 10:1934578X1501000. 10.1177/1934578X1501000405 [PubMed]
- 22.Lunga P-K, Qin X-J, Yang X-W, Kuiate J-R, Du Z-Z, Gatsing D. A new antimicrobial and radical-scavenging glycoside from Paullinia pinnata var. cameroonensis. Nat Prod Res. 2015;29:1688–1694. doi: 10.1080/14786419.2014.996756. [DOI] [PubMed] [Google Scholar]
- 23.Lunga PK, Qin X-J, Yang XW, Kuiate J-R, Du ZZ, Gatsing D. Antimicrobial steroidal saponin and oleanane-type triterpenoid saponins from Paullinia pinnata. BMC Complement Altern Med. 2014;14:369. doi: 10.1186/1472-6882-14-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miemanang RS, Krohn K, Hussain H, Dongo E. Paullinoside A and paullinomide A: a new cerebroside and a new ceramide from leaves of Paullinia pinnata. Zeitschrift für Naturforschung B. 2014;61(9):1123–1127. doi: 10.1515/znb-2006-0910. [DOI] [Google Scholar]
- 25.Awouafack MD, Ito T, Tane P, Kodama T, Tanaka M, Asakawa Y, Morita H. A new cycloartane-type triterpene and a new eicosanoic acid ester from fruits of Paullinia pinnata L. Phytochem Lett. 2016;15:220–224. doi: 10.1016/j.phytol.2016.02.002. [DOI] [Google Scholar]
- 26.Nkengfack AE, Azebaze GA, Vardamides JC, Fomum ZT, van Heerden FR. A prenylated xanthone from Allanblackia floribunda. Phytochemistry. 2002;60:381–384. doi: 10.1016/S0031-9422(02)00036-5. [DOI] [PubMed] [Google Scholar]
- 27.Kuete V, Azebaze AGB, Mbaveng ArmelleT, Nguemfo EL, Tshikalange ET, Chalard P, Nkengfack AE. Antioxidant, antitumor and antimicrobial activities of the crude extract and compounds of the root bark of Allanblackia floribunda. Pharm Biol. 2011;49:57–65. doi: 10.3109/13880209.2010.494673. [DOI] [PubMed] [Google Scholar]
- 28.Akpanika GA, Winters A, Wilson T, Ayoola GA, Adepoju-Bello AA, Hauck B. Polyphenols from Allanblackia floribunda seeds: identification, quantification and antioxidant activity. Food Chem. 2017;222:35–42. doi: 10.1016/j.foodchem.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Mountessou BYG, Tchamgoue J, Paul Dzoyem J, Tchuenguem RT, Surup F, Choudhary MI, Green IR, Kouam SF. Two xanthones and two rotameric (3⟶8) biflavonoids from the Cameroonian medicinal plant Allanblackia floribunda Oliv. (Guttiferae) Tetrahedron Lett. 2018;59:4545–4550. doi: 10.1016/j.tetlet.2018.11.035. [DOI] [Google Scholar]
- 30.Banzouzi J-T, Prado R, Menan H, Valentin A, Roumestan C, Mallie M, Pelissier Y, Blache Y. In vitro antiplasmodial activity of extracts of Alchornea cordifolia and identification of an active constituent: ellagic acid. J Ethnopharmacol. 2002;81:399–401. doi: 10.1016/S0378-8741(02)00121-6. [DOI] [PubMed] [Google Scholar]
- 31.Mavar-Manga H, Haddad M, Pieters L, Baccelli C, Penge A, Quetin-Leclercq J (2008) Anti-inflammatory compounds from leaves and root bark of Alchornea cordifolia (Schumach. & Thonn.) Mull Arg J Ethnopharmacol 115(1):25–9. 10.1016/j.jep.2007.08.043 [DOI] [PubMed]
- 32.Okwu DE, Ukanwa N. Isolation, characterization and antibacterial activity screening of anthocyanidine glycosides from Alchornea Cordifolia (Schumach. and Thonn.) Mull. Arg Leaves E-J Chem. 2010;7:41–48. doi: 10.1155/2010/586179. [DOI] [Google Scholar]
- 33.Kouakou K, Schepetkin IA, Yapi A, Kirpotina LN, Jutila MA, Quinn MT. Immunomodulatory activity of polysaccharides isolated from Alchornea cordifolia. J Ethnopharmacol. 2013;146:232–242. doi: 10.1016/j.jep.2012.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belhassan A, Zaki H, Chtita S, Alaqarbeh M, Alsakhen N, Benlyas M, Lakhlifi T, Bouachrine M (2021) Camphor, Artemisinin and sumac phytochemicals as inhibitors against COVID-19: computational approach, computers in biology and medicine 136:104758. 10.1016/j.compbiomed.2021.104758 [DOI] [PMC free article] [PubMed]
- 36.Aouidate A, Ghaleb A, Chtita S, Aarjane M, Ousaa A, Maghat H, Sbai A, Choukrad M, Bouachrine M, Lakhlifi T. Identification of a novel dual-target scaffold for 3CLpro and RdRp proteins of SARS-CoV-2 using 3D-similarity search, molecular docking, molecular dynamics and ADMET evaluation. J Biomol Struct Dyn. 2021;39(12):4522–4535. doi: 10.1080/07391102.2020.1779130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ouassaf M, Belaidi S, Chtita S, Aouidate A, Lanez T, Abul Qais F. Combined molecular docking and dynamics simulations studies of natural compounds as potent inhibitors against SARS-CoV-2 main protease. J Biomol Struct Dyn. 2021 doi: 10.1080/07391102.2021.1957712. [DOI] [PubMed] [Google Scholar]
- 38.Ouassaf M, Belaidi S, Al Mogren MM, Chtita S, Khan SU, Htar TT (2021) A docking-scoring approach to identify effective antiviral 2,5-diaminoBenzophenone derivatives against the main protease of SARS-CoV-2. J King Saud Univ Sci 33(2):101352. 10.1016/j.jksus.2021.101352 [DOI] [PMC free article] [PubMed]
- 39.Daoui O, Elkhattabi S, Chtita S, Zgou H, Benjelloun AT. QSAR, molecular docking, and ADMET properties in silico studies of novel 4,5,6,7-Tetrahydrobenzo[D]-Thiazol-2-Yl derivatives derived from Dimedone as potent antitumor, C-Met Tyrosine Kinase. Heliyon. 2021;7(7):e07463. doi: 10.1016/j.heliyon.2021.e07463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sargolzaei M. Effect of nelfinavir stereoisomers on coronavirus main protease: molecular docking, molecular dynamics simulation and MM/GBSA study. J Mol Graph Model. 2021;103:107803. doi: 10.1016/j.jmgm.2020.107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsieh LE, Lin CN, Su BL, Jan TR, Chen CM, Wang CH, Lin DS, Lin CT, Chueh LL. Synergistic antiviral effect of Galanthus nivalis agglutinin and nelfinavir against feline coronavirus. Antiviral Res. 2010;88:25–30. doi: 10.1016/j.antiviral.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto N, Yang R, Yoshinaka Y, Amari S, Nakano T, Cinatl J, Rabenau H, Doerr HW, Hunsmann G, Otaka A, Tamamura H. HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus. Biochem Biophys Res Commun. 2004;318:719–725. doi: 10.1016/j.bbrc.2004.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mothay D, Ramesh KV (2020) Binding site analysis of potential protease inhibitors of COVID-19 using Auto Dock Virus disease. 31(2):194–199. 10.1007/2Fs13337-020-00585-z [DOI] [PMC free article] [PubMed]
- 44.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 45.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718 . doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 46.Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C (2015) ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput 11(8):3696–3713. 10.1021/acs.jctc.5b00255 [DOI] [PMC free article] [PubMed]
- 47.Sousa Da Silva AW, Vranken WF. ACPYPE - AnteChamber PYthon Parser interface. BMC Res Notes. 2012;5:367. doi: 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126(1):014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 49.Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52(12):7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 50.Kumari R, Kumar R, Lynn A. G_mmpbsa- a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 51.Chtita S, Belhassan A, Aouidate A, Belaidi S, Bouachrine M, Lakhlifi T (2021) Discovery of potent SARS-CoV-2 inhibitors from approved antiviral drugs via docking screening. combinatorial chemistry & high throughput screening 24(3):441– 454. 10.2174/1386207323999200730205447. 10.1093/glycob/1.6.631 [DOI] [PubMed]
- 52.Bickerton GR, Paolini GV, Besnard J, Muresan S, Hopkins AL. Quantifying the chemical beauty of drugs. Nat Chem. 2012;4(2):90–98. doi: 10.1038/nchem.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 54.Baell JB, Nissink WM. Seven year itch: pan-assay interference compounds (PAINS) in 2017—utility and limitations. ACS Chem Biol. 2018;13(1):36–44. doi: 10.1021/acschembio.7b00903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Di L. The role of drug metabolizing enzymes in clearance. Expert Opin Drug Metab Toxicol. 2014;10:379–393. doi: 10.1517/17425255.2014.876006. [DOI] [PubMed] [Google Scholar]
- 56.Singh S, Singh J. Transdermal drug delivery by passive diffusion and iontophoresis: a review. Med Res Rev. 1993;13(5):569–621. doi: 10.1002/med.2610130504. [DOI] [PubMed] [Google Scholar]
- 57.Imberty A, Hardman KD, Carver JP, Perez S (1991) Molecular modelling of protein-carbohydrate interactions. docking of monosaccharides in the binding site of concanavalin A. Glycobiology 1(6):631–642 [DOI] [PubMed]
- 58.Lounkine E, Keiser MJ, Whitebread S, Mikhailov D, Hamon J, Jenkins JL, Lavan P, Weber E, Doak AK, Côté S, Shoichet BK, Urban L. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486(7403):361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qais FA, Sarwar T, Ahmad I, Khan RA, Shahzad SA, Husain FM. Glyburide inhibits non-enzymatic glycation of HSA: an approach for the management of AGEs associated diabetic complications. Int J Biol Macromol. 2021;169:143–152. doi: 10.1016/j.ijbiomac.2020.12.096. [DOI] [PubMed] [Google Scholar]
- 60.Fouedjou TR, Chtita S, Bakhouch M, Belaidi S, Ouassaf M, Djoumbissie AL, Tapondjou AL. Cameroonian medicinal plants as potential candidates of SARS CoV-2 inhibitors. J Biomol Struct Dyn. 2021;39:1–16. doi: 10.1080/07391102.2021.1914170. [DOI] [PubMed] [Google Scholar]
- 61.Rath B, Qais FA, Patro R, Mohapatra S, Sharma T (2021) Design, synthesis and molecular modeling studies of novel mesalamine linked coumarin for treatment of inflammatory bowel disease. Bioorg Med Chem Lett 128029. 10.1016/j.bmcl.2021.128029 [DOI] [PubMed]
- 62.Siddiqui S, Ameen F, Kausar T, Nayeem SM, Ur Rehman S, Tabish M. Biophysical insight into the binding mechanism of doxofylline to bovine serum albumin: an in vitro and in silico approach. Spectrochim Acta Part A Mol Biomol Spectrosc. 2021;249:119296. doi: 10.1016/j.saa.2020.119296. [DOI] [PubMed] [Google Scholar]
- 63.Siddiqui S, Ameen F, Jahan I, Nayeem SM, Tabish M. A comprehensive spectroscopic and computational investigation on the binding of the anti-asthmatic drug triamcinolone with serum albumin. New J Chem. 2019;43(10):4137–4151. doi: 10.1039/C8NJ05486J. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this work are private.
The codes used in this work are not available.