The expression of the predominant macrolide resistance genes in staphylococci, erm(A) and erm(C) (2, 7), is either inducible by 14- and 15-membered macrolides via translational attenuation or constitutive. Constitutively expressed erm(A) and erm(C) genes are of particular clinical relevance since they also confer resistance to 16-membered macrolides, lincosamides, streptogramin B antibiotics, and the new ketolide drugs. To date, very few data on the molecular basis of constitutive erm(A) gene expression in naturally occurring staphylococci are available (8).
In this study, we examined 64 clinical Staphylococcus aureus isolates from humans, collected at 24 European university hospitals within the SENTRY program (5, 6). All 64 isolates carried constitutively expressed erm(A) genes and exhibited the respective resistance phenotype as confirmed by MIC determination (5, 6). A previously described PCR assay (8) served for the detection of structural alterations in the erm(A) regulatory region. All 64 PCR amplicons were sequenced and compared to the sequence of the inducibly expressed erm(A) gene of Tn554 (3).
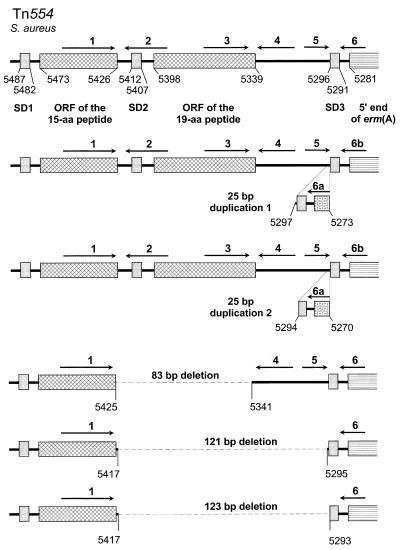
Five different types of structural alterations were seen: deletions of 83, 121, 123 bp and two closely related tandem duplications of 25 bp each (Fig. 1). The two tandem duplications of 25 bp (5′-TAAGGAGAAGGTTATAATGAACCAG-3′ and 5′-GGAGAAGGTTATAATGAACCAGAAA-3′) were detected in 1 and 51 isolates, respectively, and comprised the erm(A)-associated ribosome binding site (AGAAGG) as well as the inverted repeat 6 (IR6) sequence (GGTTATAATGAAC). The 83-bp deletion found in two of the isolates comprised the entire open reading frame (ORF) of the 19-amino-acid (aa) peptide including IR3. The 121-bp deletion detected in nine isolates and the 123-bp deletion observed in a single isolate are closely related. Both types of deletions comprise the ORF of the 19-aa peptide as well as the part immediately downstream of it which includes the IR4 and IR5 sequences. These deletions and tandem duplications favored the formation of mRNA secondary structures which allowed translation of the erm(A) transcripts in the absence of inducers.
FIG. 1.
Regulatory regions of the inducibly expressed erm(A) gene of Tn554 (2) and the constitutively expressed erm(A) genes analyzed in this study. SD1 to -3, Shine-Dalgarno sequences of the ORFs of the 15-aa peptide, the 19-aa peptide and the erm(A) gene, respectively; arrows, IR1 to IR6; stippled box, truncated erm(A) gene present in isolates which exhibit the 25-bp tandem duplications. Positions numbering corresponds to the Tn554 sequence (database accession no. K02987).
The data show that tandem duplications and deletions of different sizes account for constitutive erm(A) gene expression in naturally occurring S. aureus isolates. So far, deletions of 26 to 141 bp, a tandem duplication of 12 bp, and a point mutation, all of which cause constitutive erm(A) gene expression, have been derived under in vitro selection in the presence of noninducers (3, 4). Thus, these data on in vivo-occurring mutations in the erm(A) translational attenuator in human isolates complement both the in vitro-derived mutations (3, 4) and the mutation seen in a naturally occurring S. intermedius isolate of avian origin (8), thus confirming that similar mutations can arise in vivo and in vitro (1). The development of constitutive erm(C) and erm(A) mutants is a fast and irreversible process. Under in vitro conditions constitutive mutants can be obtained after overnight cultivation in the presence of noninducers (1, 3, 7, 9), and reversion to the inducible type has not been observed in any such mutants. Thus, noninducers, such as lincosamides or streptogramins, should not be recommended for the control of staphylococci which exhibit an inducible macrolide-lincosamide-streptogramin B resistance phenotype.
REFERENCES
- 1.Lampson B C, Parisi J T. Naturally occurring Staphylococcus epidermidis plasmid expressing constitutive macrolide-lincosamide-streptogramin B resistance contains a deleted attenuator. J Bacteriol. 1986;166:479–483. doi: 10.1128/jb.166.2.479-483.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leclercq R, Courvalin P. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob Agents Chemother. 1991;35:1267–1272. doi: 10.1128/aac.35.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy E. Nucleotide sequence of ermA, a macrolide-lincosamide-streptogramin B determinant in Staphylococcus aureus. J Bacteriol. 1985;162:633–640. doi: 10.1128/jb.162.2.633-640.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandler P, Weisblum B. Erythromycin-induced stabilization of ermA messenger RNA in Staphylococcus aureus and Bacillus subtilis. J Mol Biol. 1988;203:905–915. doi: 10.1016/0022-2836(88)90116-7. [DOI] [PubMed] [Google Scholar]
- 5.Schmitz F-J, Sadurski R, Kray A, Boos M, Geisel R, Köhrer K, Verhoef J, Fluit A C. Prevalence of macrolide-resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J Antimicrob Chemother. 2000;45:891–894. doi: 10.1093/jac/45.6.891. [DOI] [PubMed] [Google Scholar]
- 6.Schmitz F-J, Verhoef J, Fluit A C the SENTRY Participants Group. Prevalence of resistance to MLS antibiotics in 20 European university hospitals participating in the European SENTRY surveillance program. J Antimicrob Chemother. 1999;43:783–792. doi: 10.1093/jac/43.6.783. [DOI] [PubMed] [Google Scholar]
- 7.Weisblum B. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob Agents Chemother. 1995;39:797–805. doi: 10.1128/aac.39.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Werckenthin C, Schwarz S. Molecular analysis of the translational attenuator of a constitutively expressed erm(A) gene from Staphylococcusintermedius. J Antimicrob Chemother. 2000;46:785–788. doi: 10.1093/jac/46.5.785. [DOI] [PubMed] [Google Scholar]
- 9.Werckenthin C, Schwarz S, Westh H. Structural alterations in the translational attenuator of constitutively expressed ermC genes. Antimicrob Agents Chemother. 1999;43:1681–1685. doi: 10.1128/aac.43.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]