Abstract
Candida species have a major role in nosocomial infections leading to high morbidity and mortality. Increased resistance to various antifungals, especially azoles is a significant problem. One of the main mechanisms for azole resistance is the up-regulation of efflux pump genes including CDR1 and MDR1. In the current study, clinical Candida isolates were identified to the species level and the antifungal susceptibility (AFS) of different Candida species was determined by disk diffusion method. Furthermore, the main mechanisms of azole resistance were investigated. Finally, haloperidol and pantoprazole were tested for their potential synergistic effect against fluconazole-resistant isolates.
One hundred and twenty-two Candida clinical isolates were used in this study. 70 isolates were Candida albicans (57.4%), the non-albicans Candida species include: C. krusei (20.5%), C. tropicalis (6.6%), C. parapsilosis (5.7%), C. dubliniensis (4.9%) and C. glabrata (4.9%). The AFS testing showed that resistance to fluconazole and voriconazole were 13.1% (n = 16) and 9.8% (n = 12), respectively. Among the 16 resistant isolates, eight isolates (50%) were strong biofilm producers, seven (43.8 %) formed intermediate biofilm and one had no biofilm. All resistant strains overexpressed efflux pumps. Using RT-PCR, the efflux genes CDR1, MDR1 and ABC2 were over-expressed in azole resistant isolates. Haloperidol-fluconazole and pantoprazole-fluconazole combinations reduced the MIC of fluconazole in resistant isolates. The current study showed an increase in azole resistance of Candida species. The majority of resistant isolates form biofilm, and overexpress efflux pumps. Pantoprazole and Haloperidol showed a noteworthy effect as efflux pump inhibitors which oppose the fluconazole resistance in different Candida species.
Keywords: Candida species, Azole resistance, Efflux pump, Pantoprazole, Haloperidol
1. Introduction
Candida species have emerged as significant cause of nosocomial infection in recent decades that leading to high morbidity and mortality (Jahagirdar et al., 2018). Candida are responsible for both superficial (mucosal and cutaneous) and life threatening invasive infection. The most common form of invasive infections is candidemia, at least 8% of nosocomial bloodstream infections are caused by Candida species (Papon et al., 2013). The increased incidence of invasive fungal infections can be correlated with an expansion in the number of immune-compromised patients, which include AIDS patients and cancer chemotherapy patients (Cornet et al., 2011).
The five species most commonly associated with candidiasis are Candida albicans, C. glabrata, C. krusei, C. tropicalis and C. parapsilosis (Turner and Butler, 2014). C. albicans remains the most commonly isolated species, but the proportion of non-albicans Candida (NAC) species has increased over the past few years in Egypt and worldwide (El-Ganiny et al., 2021, Ricotta et al., 2021).
The number of therapeutic options for treatment of fungal infections is quite limited, only three main structural classes of antifungal drugs are available, namely polyenes, azoles, and echinocandins (Sanguinetti et al., 2015). The azoles are the most widely used class of antifungal drugs, as they have low toxicity and more bioavailability (Fisher et al., 2018). Azole drugs inhibit ergosterol biosynthesis by binding to cytochrome P450-dependent enzyme 14α demethylase, (encoded by ERG11 gene). Inhibition of this enzyme results in unstable fungal cell membrane and membrane leakage (Francois et al., 2006). Resistance to azoles was associated with mutations or overexpression of the ERG11 or up-regulation of several drug efflux pumps (Whaley et al., 2017).
One of the main mechanisms of azole resistance in Candida is through active efflux of drugs mediated by ATP-binding cassette (ABC) transporters and major facilitators superfamily (MFS) pumps; encoded by CDR and MDR genes, respectively (Bhattacharya et al., 2020, Lee et al., 2020). The ABC transporter CDR1 is reported to be the major contributor to azole resistance in clinical isolates of C. albicans and C. glabrata. While, in the emerging pathogen, C. krusei, the efflux pump Abc1 is crucial in azole resistance. Furthermore, it is believed that ABC2-encoded efflux pump was important in C. krusei's innate fluconazole resistance (Holmes et al., 2012). In C. tropicalis, ERG11 mutation was the main mechanism responsible for azole resistance, however, overexpression of CDR1 and MDR1 also contributed to azole resistance (Fan et al., 2019). It has been reported that activation of MDR1 gene in clinical C. albicans strains induced fluconazole resistant during therapy (Wirsching et al., 2000).
Phenotypic evaluation of efflux pump activity usually uses efflux pump substrates such as ethidium bromide (Martins et al., 2011). And acridine Orange (AO) which is a non-toxic fluorescent dye that is used for staining eukaryotic cells. AO has been shown to be a substrate of bacterial efflux pumps, hence it is used for screening of efflux activity (Martins and Amaral, 2012). Other fluorescent dyes that is used for detection of the efflux pump include: Rhodamine 6G, Alanine B-napthylamide and Nile Red (Maesaki, 1999, Bhattacharya et al., 2016). Furthermore, Nile Red is a dye that can be used to investigate intracellular lipids and protein hydrophobic regions. When Nile Red is present in a highly hydrophobic environment, it fluoresces brightly (Ivnitski-Steele et al., 2009).
Another significant aspect of Candida resistance and pathogenesis is biofilm formation. This phenomenon enables Candida to bind to mucosal cells and medical devices leading to device associated infections (Bitar et al., 2014). Biofilms formation in Candida species is mostly associated with increased resistance to antifungal drugs (Rautemaa and Ramage, 2011, Marak and Dhanashree, 2018). It is assumed that formation of polysaccharide matrix in biofilm diminishes the diffusion of drugs to fungal cells through the formation of transmission barrier (Sardi et al., 2013).
The high mortality rate of invasive candidiasis and increased resistance to conventional antifungals make it necessary to develop new antifungal therapies (Faustino and Pinheiro, 2020). Researchers have explored natural and synthetic agents that can sensitize antifungal agents. Several drug classes have been investigated for their effect on virulence and resistance (Lu et al., 2020, El-Baz et al., 2021, Peyclit et al., 2021). Repurposing of USA food and drug administration (FDA)-approved drugs is a new approach that can reduce the time and economic costs of developing de novo drugs (Miró-canturri et al., 2019). Screening for efflux pump inhibitors (EPIs) is an attractive way of increasing intracellular concentration of antifungal drugs and hence reducing resistance (Vega-Chacón et al., 2021, Tong et al., 2021). Few studies used in silico methods for identification and validation of EPI (Lamping et al., 2017). Among the in silico methods, molecular docking has been recently used to explore compounds that would be capable of binding to efflux pump proteins (Zárate et al., 2019).
The aim of the current study was to investigate the phenotypic mechanisms of azole drugs resistance in clinical Candida isolates. Furthermore, the expressions of efflux pump genes in resistant Candida isolates from different species was explored using RT-PCR. In addition the effect of two FDA approved drugs was evaluated on fluconazole resistant isolates using phenotypic and molecular docking approaches.
2. Materials and methods
2.1. Isolation and identification of Candida species
All the clinical specimens were collected from different sources (blood, urine, and sputum) in clinical laboratories at Mansoura and Zagazig University Hospitals, Egypt. First, the specimens were inoculated on Sabouraud dextrose agar (SDA) plates and incubated at 37 °C then the Candida identification started with microscopical examination after Gram staining followed by biochemical identification and PCR as described previously (El-Ganiny et al., 2021).
2.2. Investigating antifungal susceptibility by disk diffusion method
Candida isolates were tested for their susceptibility to different antifungal agents by disk diffusion method according to Clinical and Laboratory Standard Institute guidelines (CLSI, 2009). The tested disks include; amphotericin B (AMB, 10 μg) as a polyene drug, fluconazole (FLU, 25 μg) as representative to the 1st generation azole, and voriconazole (VOR, 1 μg) as representative to 2nd generation azole. The antifungal disks were obtained from Bioanalyse, (Ankara, Turkey).
Briefly, 3–5 Candida colonies were inoculated into 4–5 mL Sabouraud dextrose broth (Oxoid, Hampshire, England) and incubated for 24 h at 37 °C. The turbidity of the suspension was adjusted to 0.5 McFarland turbidity standard. A sterile cotton swab was immediately dipped into the prepared suspension, and then used to streak over Muller Hinton agar (Oxoid, Hampshire, England) plate (containing 2% glucose and 0.5 µg/mL methylene blue). Antifungal disks were placed on MHA plates using sterile forceps and pressed firmly against the agar surface. The plates were inverted and incubated at 37 °C for 24–48 h. The diameter of inhibition zones around each antifungal disk were measured in millimeter, and interpreted as susceptible, intermediate or resistant according to interpretative criteria of CLSI (2009).
2.3. Biofilm formation assay
Biofilm formation assay was performed on the azole-resistant strains as described previously (Stepanović et al., 2007) with some modifications. Overnight cultures of candida isolates were prepared, diluted with Sabouraud dextrose broth (SDB), and adjusted to a cell density of 1 × 106 CFU/mL. Aliquots of 100 μL of adjusted candida suspension were inoculated to the wells of sterile 96-well polystyrene microtiter plates. After incubation for 48 h at 37 °C, the contents of the wells were gently aspirated, and the wells were washed with sterile phosphate-buffered saline (PBS, pH 7.2). The adherent cells were fixed with 100 μL of 99% methanol for 20 min, then stained with 100 μL crystal violet (1%) for 20 min. The excess dye was then removed under running distilled water, then the plates were left to air-dry. The bound dye was dissolved by addition of 80 μL of 33% glacial acetic acid, and the optical densities were read at 540 nm using a microtiter plate reader. The experiment was performed in triplicate, the mean optical densities were calculated, and wells using media only were used as a negative control. The cut-off OD (ODc) was defined as three times standard deviations above the mean OD of the negative control. The test isolates were categorized into four groups: non-biofilm forming (OD ≤ ODc), weak biofilm (OD > ODc, but ≤ 2 × ODc), moderate biofilm (OD > 2 × ODc, but ≤ 4 × ODc), and strong biofilm-forming (OD > 4 × ODc).
2.4. Acridine orange (AO) assay for phenotypic detection of efflux pump activity
Candida isolates were grown on SDA then a 0.5 McFarland suspension was prepared. Single sterile swabs were dipped into suspension. Plates of SDA containing increasing concentrations (1, 5, 10, and 20 mg/mL) of AO (Sigma-Aldrich, USA) were swabbed from the center to the periphery of the plate, the cartwheel design of swabbing was used. The swabbed plates were incubated for 16 h and fluorescence of the overlying fungal streak was determined with Cole-Parmer UV trans-illuminator (Vernon Hills, USA). The concentration of AO that first produced evidence of green fluorescence of the fungal streak was recorded for each strain (Martins and Amaral, 2012).
2.5. Synthesis of cDNA and quantitative real time PCR
Total RNA was extracted from fungal colony lysate with Direct-zol RNA Miniprep Plus (Zymo Research Corp. USA), then quantity and quality of RNA were assessed by Beckman dual spectrophotometer (Fullerton, USA). The primers used in this study (listed in Table 1) were purchased from Operon Biotechnologies (GmbH Biocompus Cologne, Germany).
Table 1.
Primers used for qRT-PCR in this study.
| Primer name | Primer sequence | Tm (°C) | Size | Reference |
|---|---|---|---|---|
| Ct-MDR1F | GGGTGCATCATTCCAGCCTA | 57.3 | 189 | Jin et al. (2018) |
| Ct-MDR1-R | GGGATGGCAATCATCACGAG | 56 | ||
| Ct-ACT1F | TTTACGCTGGTTTCTCCTTGCC | 57.9 | 322 | |
| Ct-ACT1R | GCAGCTTCCAAACCTAAATCGG | 56.6 | ||
| Cg-CDR1F | TAGCACATCAACTACACGAACGT | 56.3 | 170 | Szweda et al. (2015) |
| Cg-CDR1R | AGAGTGAACATTAAGGATGCCATG | 55.2 | ||
| Cg-URAF | GAAAACCAATCTTTGTGCTTCTCT | 54 | 125 | |
| Cg-URAR | CATGAGTCTTAAGCAAGCAAATGT | 54.1 | ||
| Ca-ACT1F | GCTTTTGGTGTTTGACGAGTTTCT | 56.4 | 72 | Watamoto et al. (2011) |
| Ca-ACT1R | GTGAGCCGGGAAATCTGTATAGTC | 56.9 | ||
| Ca-CDR1F | GTACTATCCATCAACCATCAGCACTT | 56.6 | 79 | |
| Ca-CDR1R | GCCGTTCTTCCACCTTTTTGTA | 55.7 | ||
| Ca-MDR1F | TCAGTCCGATGTCAGAAAATGC | 55.3 | 91 | |
| Ca-MDR1R | GCAGTGGGAATTTGTAGTATGACAA | 55.4 | ||
| Ck-ABC2-F | CCTTTTGTTCAGTGCCAGATTG | 57.4 | 301 | He et al. (2015) |
| Ck-ABC2-R | GTAACCAGGGACACCAGCAA | 57.3 | ||
| Ck-ACT1-F | TGGGCCAAAAGGATTCTTATG | 53 | 300 | |
| Ck-ACT1-R | AGATCTTTTCCATATCATCCCAG | 52.1 |
The qRT-PCR master mix was prepared in 20 μL final volume containing: 10 μL of 2 × SensiFAST™ SYBR (Bioline, London, UK), 0.8 μL of each primer (10 μM), 0.2 μL reverse transcriptase, 0.4 μL RiboSafe-RNase Inhibitor, water up to 16 μL and 4 μL of RNA (0.2 µg/µL) template was added. The qRT-PCR conditions were: an initial denaturation at 95 °C for 2 min, followed by 40 cycles of denaturation (95 °C for 10 s), annealing temperature for 10 s, and extension (72 °C for 30 s); then final extension step at 72 °C for 5 min. The test was performed in triplicates. Samples were normalized to house-keeping gene expression, then the transcript levels of azole-resistant isolates were compared with the average expression level of the azole susceptible isolates. Fold changes in gene expression were calculated using the comparative Ct method (2−ΔΔCT) as described previously (Livak and Schmittgen, 2001).
2.6. Evaluation of potential efflux pump inhibitors phenotypically
The Minimum inhibitory concentrations (MICs) of fluconazole (Diflucan® 50 mg vials, Pfizer, USA), haloperidol (haloperidol® 50 mg/mL ampules, Marcyrl, Egypt) and pantoprazole (Controloc® 40 mg vials, Takeda, USA) were determined using the broth microdilution method according to the (CLSI-document M27-A3) with some modifications (CLSI, 2008). Fluconazole, pantoprazole and haloperidol stock solutions were prepared using Muller-Hinton broth (MHB, Oxoid, Hampshire, UK), then serial dilutions were prepared in microtiter plate using MHB so the range of concentration was from 2048 to 2 µg/mL. Overnight culture of well isolated colonies of each isolate in MHB was prepared and its turbidity was adjusted to 0.5 McFarland. The prepared suspension was diluted 1:100 with MHB to have an approximate cell density of 104 CFU/mL. Then 50 µL of the prepared suspensions were transferred to the wells of a microtiter plate containing 50 µL of serial dilutions of drug in MHB. The plates were incubated at 37 °C for 24 h and the MIC was determined as the lowest concentration of the drug that inhibited any visible growth. To evaluate the potential inhibitory effect of haloperidol and pantoprazole, the MIC of fluconazole was evaluated again in presence of ¼ MIC of haloperidol and pantoprazole.
2.7. Evaluation of potential efflux pump inhibitors genotypically by qRTPCR
The qRTPCR was carried out as described previously to evaluate the difference in the expression of efflux genes (MDR1 and CDR1) in presence of the tested efflux pump inhibitors using ¼ MIC of haloperidol and pantoprazole in comparison to control stains grown in absence of the tested efflux pump inhibitors.
2.8. Molecular docking study
Dock module of MOE (Molecular Operating Environment) version MOE 2019.0102,2 (Chemical Computing Group Inc., 2019) on a computer having Pentium 1.6 GHz workstation, 512 MB memory using windows operating system, was utilized in docking studies. Haloperidol and Pantoprazole were drawn into Marvin Sketch of Marvin suite (http://www.chemaxon.com) to generate the lowest energy conformer to investigate their binding mode and affinity to UniProtKB – Q5ABU7 (MDR1_CANAL) Multidrug resistance protein 1 of efflux pump encoded by MDR1 gene in C. albicans, UniProtKB-Q5ANA3 (CDR1_CANAL) pleiotropic ABC efflux transporter of multiple drugs CDR1 expressed by CDR1 gene in both C. albicans and C. glabrata, and UniProtKB-K7WIP0 (K7WIP0_PICKU) ATP-binding cassette transporter protein encoded by ABC2 gene in C. krusei The expressed amino acids sequences of aforementioned genes were obtained as a primary structure in FASTA form UniProt Knowledgebase (https://www.uniprot.org/help/uniprotkb). The predicted 3D structures of all protein were obtained from the IntFOLD Server (Version 5.0) as described previously (Mcguffin et al., 2019).
2.9. Statistics
Statistical analysis was performed using GraphPad Prism software version 6 (GraphPad Software Inc., La Jolla, CA, USA). Differences between groups were analyzed by one-way analysis of variance (ANOVA) followed by Tukey's Kramer multiple comparison test. Data are presented as mean ± standard error (SEM). A value of P ≤ 0.0001 was considered to indicate statistical significance.
3. Results:
3.1. Isolation and identification of Candida species
One hundred twenty-two non-duplicate Candida clinical isolates were identified in the present study. The isolates were identified to the species level using PCR, 70 isolates (57.4%) were identified as C. albicans, while 52 isolates (42.6%) were non-albicans candida (NAC. The NAC species include: C. krusei (25 strains, 20.5%), C. tropicalis (8 isolates, 6.6%), C. parapsilosis (7 isolates, 5.7%), and 6 isolates for both C. dubliniensis and C. glabrata (4.9% each).
3.2. Susceptibility to antifungals
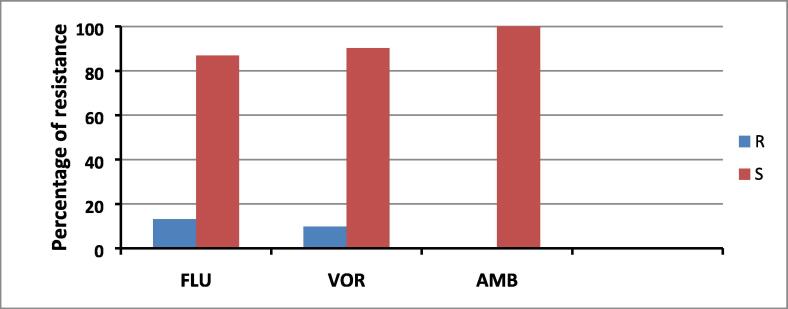
The antifungal susceptibility testing of the 122 Candida isolates against 3 antifungal drugs (fluconazole, voriconazole and amphotericin B) revealed resistance for both fluconazole and voriconazole The data presented in Fig. 1 showed that the percentages of resistance to fluconazole and voriconazole were 13.1% (16 isolates) and 9.8% (12 isolates), respectively. On the other hand, there was no resistance to amphotericin B.
Fig. 1.
Antifungal susceptibility testing of candida isolates. FLU: fluconazole, VOR: voriconazole, AMB: Amphotericin B. R resistant, S susceptible.
Sixteen isolates out of 122 tested isolates were resistant to antifungals (Table 2). Seven isolates of C. albicans showed resistance to azoles (6 to both fluconazole and voriconazole and 1 to fluconazole only), 4 of C. krusei isolates showed resistance to azoles, (3 to both fluconazole and voriconazole and 1 to fluconazole only), one C. tropicalis isolate was resistant to both fluconazole and voriconazole. On the other hand, 4 isolates of C. glabrata were resistant to azoles (2 for both fluconazole and voriconazole and 2 to fluconazole alone).
Table 2.
Distribution of clinical Candida species and their resistance to azoles.
| Candida species | No. of isolates | No. of resistant isolates in the species (%) | Fluconazole | Voriconazole |
|---|---|---|---|---|
| C. albicans | 70 | 7 (10%) | 7 | 6 |
| C. krusei | 25 | 4 (16%) | 4 | 3 |
| C. tropicalis | 8 | 1 (12.5%) | 1 | 1 |
| C. glabrata | 6 | 4 (66.7%) | 4 | 2 |
| C. parapsilosis | 7 | – | – | – |
| C. dubliniensis | 6 | – | – | – |
| Total | 122 | 16 (13.1%) | 16 (13.1%) | 12 (9.8%) |
3.3. Biofilm formation in Candida isolates
All the azole resistant strains are tested for the biofilm formation. Fifteen isolates have biofilm producing activity and one strain has no biofilm activity. The data revealed that eight isolates form strong biofilm. While 7 isolates were intermediate biofilm producer, and only one isolate has no biofilm activity (Table 3).
Table 3.
Biofilm and efflux pump activities in azole-resistant Candida isolates.
| Isolates no | Candida species | VOR | FLU | Biofilm activity | Efflux activity |
|---|---|---|---|---|---|
| 4 | C. tropicalis | R | R | Strong | Strong |
| 8 | C. albicans | S | R | Intermediate | Strong |
| 11 | C. krusei | S | R | Intermediate | Strong |
| 22 | C. glabrata | S | R | Intermediate | Strong |
| 26 | C. krusei | R | R | Strong | Strong |
| 27 | C. glabrata | R | R | Intermediate | Strong |
| 28 | C. glabrata | S | R | Intermediate | Strong |
| 30 | C. krusei | R | R | Intermediate | Intermediate |
| 32 | C. krusei | R | R | Strong | Strong |
| 33 | C. albicans | R | R | Strong | Strong |
| 38 | C. glabrata | R | R | Intermediate | Strong |
| 39 | C. albicans | R | R | Strong | Strong |
| 40 | C. albicans | R | R | None | Strong |
| 41 | C. albicans | R | R | Strong | Strong |
| 55 | C. albicans | R | R | Strong | Strong |
| 73 | C. albicans | R | R | Strong | Strong |
3.4. Phenotypic detection of efflux pump activity by AO
All resistant isolates were tested for detecting the presence of the efflux pump. It was observed that all strains have efflux pump activity. Fifteen isolate showed strong activity because they did not give any fluorescence at all tested AO concentrations. While one strain of C. krusei showed intermediate efflux activity as it began to fluoresce at the highest concentration (20 mg/mL) of AO (Table 3).
3.5. Expression levels of CDR1, ABC2 and MDR1 genes
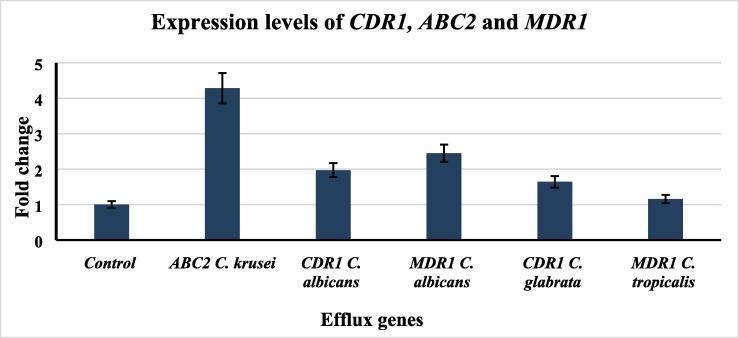
The expression of efflux genes was measured in the azole resistant isolates. Relative expression levels of the efflux gene were calculated using the 2−ΔΔCt method. The highest over-expression level (4.29-fold in relative to the control strain) was observed in ABC2 gene in C. krusei. Furthermore, gene up-regulation of CDR1and MDR1 was observed in C. albicans (1.97 and 2.45 fold increase, respectively). Up-regulation of the CDR1 gene was observed in C. glabrata to (1.64 -fold increase relative to control strain) and the lowest up-regulation level was (1.16 -fold increase) was observed in MDR1 gene of C. tropicalis (Fig. 2).
Fig. 2.
Expression levels of efflux pump genes in resistant Candida species.
3.6. Evaluation of potential efflux pump inhibitors phenotypically
First, the MIC of EPIs was determined, it was 256 µg/mL for pantoprazole and 64 µg /mL for haloperidol. Then, the MIC of fluconazole was measured for the 16 resistant isolates in presence and absence of ¼ MIC of haloperidol and pantoprazole. Pantoprazole was able to cause reduction (3–5 fold decrease) in fluconazole MIC in all tested isolates. While, haloperidol was able to cause reduction in the MIC in 15 isolates with a reduction ranging from 1 to 4 fold change. (Table 4).
Table 4.
The MIC (µg/mL) of Fluconazole alone and in combination with ¼ MIC of haloperidol and pantoprazole.
| Isolates no. | Candida Species | FLU alone | FLU + haloperidol | Fold decrease in MIC | FLU + pantoprazole | Fold decrease in MIC |
|---|---|---|---|---|---|---|
| 4 | C. tropicalis | 1024 | 64 | 4 fold | 32 | 5 fold |
| 8 | C. albicans | 512 | 64 | 3 fold | 32 | 4 fold |
| 11 | C. krusei | 512 | 32 | 4 fold | 32 | 4 fold |
| 22 | C. glabrata | 256 | 64 | 2 fold | 32 | 3 fold |
| 26 | C. krusei | 1024 | 1024 | None | 32 | 5 fold |
| 27 | C. glabrata | 512 | 64 | 3 fold | 64 | 3 fold |
| 28 | C. glabrata | 512 | 64 | 3 fold | 32 | 4 fold |
| 30 | C. krusei | 512 | 64 | 3 fold | 32 | 4 fold |
| 32 | C. krusei | 1024 | 128 | 3 fold | 32 | 5 fold |
| 33 | C. albicans | 1024 | 64 | 4 fold | 32 | 5 fold |
| 38 | C. albicans | 1024 | 512 | 1 fold | 32 | 5 fold |
| 39 | C. albicans | 512 | 32 | 4 fold | 32 | 4 fold |
| 40 | C. albicans | 512 | 64 | 3 fold | 16 | 5 fold |
| 41 | C. albicans | 256 | 32 | 3 fold | 32 | 3 fold |
| 55 | C. albicans | 1024 | 64 | 4 fold | 32 | 5 fold |
| 73 | C. albicans | 512 | 64 | 3 fold | 64 | 3 fold |
3.7. Evaluation of potential efflux pump inhibitors genotypically
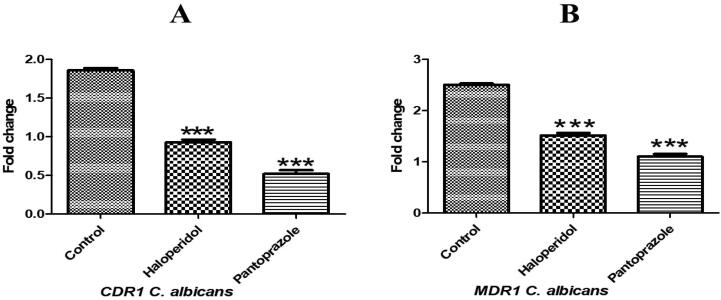
The expression levels of efflux pump genes CDR1 and MDR1 in resistant Candida albicans isolates in presence and absence of tested efflux pump inhibitors (haloperidol and pantoprazole) were evaluated using qRT-PCR. Haloperidol reduced the expression level of CDR1 from 1.97 to be quite similar to sensitive standard strain (1) and reduced the expression level of MDR1 from 2.45 to approximately its half (1.3). Pantoprazole showed stronger inhibitory effects, as it reduced the expression of CDR1 to 0.55 and MDR1 to 1 (see Fig. 3.).
Fig. 3.
Expression levels of efflux pump genes in resistant Candida albicans isolates in presence and absence of tested efflux pump inhibitors (Haloperidol and pantoprazole). A: expression of CDR1B: expression of MDR1. Data are expressed as means ± SEM. Results are considered significant when P < 0.0001. The control is Candida albicans in absence of inhibitors. *** Significance vs control group.
3.8. Molecular docking of haloperidol and pantoprazole on Candida efflux pump proteins CDR1, MDR1 and ABC2
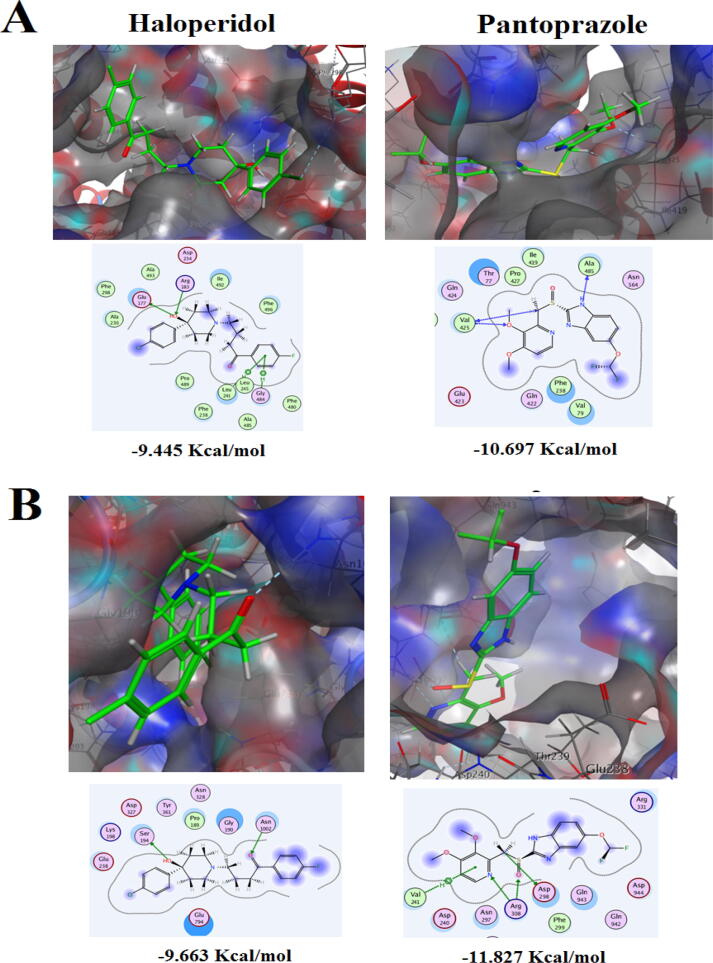
The docking results of the putative EPIs Haloperidol and Pantoprazole into Candida albicans and NAC species efflux pump proteins are shown in Fig. 4 and 5. For C. albicans MDR1 protein, the docking results revealed that Haloperidol hydroxyl group at position-4 of the piperidine ring constructed a distinguished bifurcated H-bond with the pocket conserved H-donor amino acid Arg183, and with the H-acceptor amino acid Glu177 in the core of the active site. Moreover, the p-fluorinated phenyl ring formed a bifurcated arene-H bond with Leu241 and Gly484. Eventually, Haloperidol scored a free binding energy − 9.445 Kcal/mol. Concerning Pantoprazole, Val425, as a pocket conserved amino acid, acted as H-acceptor/donor and formed a bifurcated H-bond with the oxygen atom of the 3-methoxy group of ligand pyridine ring and methyl group which acted as a linker between sulfinyl group and 3,4-dimethoxylated pyridine. In addition, the hydrogen atom at position-1 of benzimidazole moiety formed H-bond with the conserved amino acid Ala485, giving rise to score free binding energy − 10.697 Kcal/mol (Fig. 4A).
Fig. 4.
The Putative binding modes (2D & 3D) of Haloperidol and Pantoprazole and their free binding energies expressed in Kcal/mol in the active site of the predicted 3D structure of (A) Multidrug resistance protein 1 expressed by MDR1 gene in C. albicans. (B) Pleiotropic ABC efflux transporter expressed by CDR1 gene in C. albicans. The blue and cyan shadow of the ligand and active site amino acids respectively, indicated strong hydrophobic/hydrophilic interactions.
Fig. 5.
The Putative binding modes (2D & 3D) of Haloperidol and Pantoprazole and their free binding energies expressed in Kcal/mol in the active site of the predicted 3D structure of (A) CDR1 protein in C. glabrata. (B) ABC efflux transporter expressed by ABC2 gene in C krusei. The blue and cyan shadow of the ligand and active site amino acids respectively, indicated strong hydrophobic/hydrophilic interactions.
In case of C. albicans CDR1 protein, the docking results showed again the crucial role of the hydroxyl group of Haloperidol that formed a H-bond with Ser194, whereas another H-bond was constructed between Asn1002 and its carbonyl group, leading to high free binding energy − 9.663 Kcal/mol. For Pantoprazole, the methyl linker once more played a fundamental role in forming H-bond with Asp298. Additionally, Arg308 formed a bifurcated H-bond with nitrogen atom of the pyridine ring and oxygen atom of the sulfinyl group, fixing the ligand inside the active site. Furthermore, Val241 constructed a H-arene bond with the pyridine ring. Hence, Pantoprazole scored the highest free binding energy − 11.827 Kcal/mol (Fig. 4B).
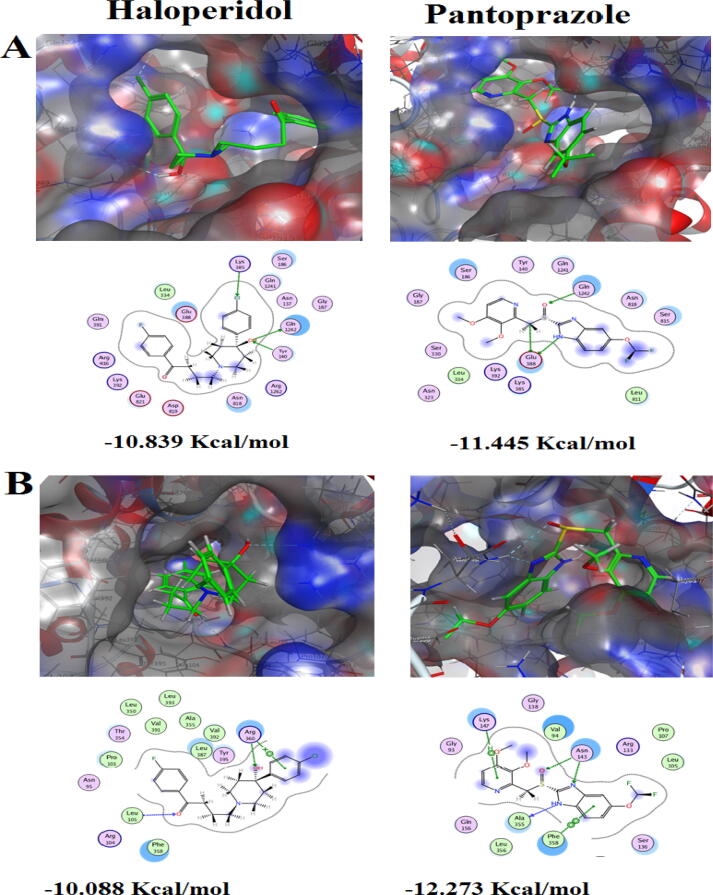
Docking into the Pleiotropic ABC efflux transporter CDR1 protein in C. glabrata revealed that Haloperidol scored high binding energy − 10.839 Kcal/mol. Its hydroxyl group proceeded as H-donor/acceptor and formed bifurcated H-bond with H-acceptor Gln1242 and H-donor Tyr140. Interestingly, Haloperidol as a halogenated compound, its chlorine atom formed a unique intermolecular interaction- halogen-bond- with Lys385. Furthermore, Pantoprazole was seen to be stabilized via fixation to the core of the active site of the receptor by bifurcated H-bond between hydrogen atom attached to N − 1 of the imidazole moiety and methyl linker in ligand side, with Glu388 that performed as tent wedge, from receptor side. From the other side of the ligand, another H-bond wad discerned between sulfinyl oxygen and the H-donor Gln1242 that augmented the stability of ligand /receptor complex to score − 11.445 Kcal/mol (Fig. 5A).
Upon docking against ABC2 protein in C. krusei, we found that Haloperidol was tied to the efflux pump protein receptor by three bonds; two H-bonds established between the two oxygen atoms of hydroxyl and carbonyl groups with Arg360 and Leu105 respectively, added with a characteristic cation-arene bond between Arg360 and p-chlorinated phenyl ring. Hence, the total stability of ligand/receptor complex has achieved − 10.088 Kcal/mol. Otherwise, Pantoprazole scored higher binding free energy of − 12.273 Kcal/mol through the virtuoso role of its benzimidazole nucleus whose fused benzene ring and two nitrogen atoms of parent heterocyclic ring at positions-1,3 constructed an arene-arene bond and two H-bonds with Phe358, Ala355 and Asn143 respectively. Moreover, an arene-H bond was constructed between the terminal 3,4-dimethoxylated pyridine ring while at the middle of the ligand a H-bond was conspicuously observed between sulfinyl oxygen and Asn143 (Fig. 5B).
4. Discussion
Fungal infections affect over a billion people worldwide, leading to high morbidity and mortality as well as expensive medical costs (Bongomin et al., 2017). Despite the availability of antifungal medications, the fatality rates of candidiasis are estimated to be as high as 45% (Cheng et al., 2005). The usage of new antifungal drugs hasn't improved the prognosis for infected patients. Candida resistance has increased especially to azoles as a result of widespread and long-term use (Marak and Dhanashree, 2018). In this sense, monitoring Candida's resistance mechanisms is critical for predicting clinical response (Pristov and Ghannoum, 2019). The current study tried to clarify the main mechanism by which different Candida species resist the azoles through phenotypic and genotypic approaches and searched for solution to this elevated resistance.
Regarding antifungal susceptibility, the highest resistance in the current study was detected for FLU (13.1%) followed by VOR (9.8%). Prolonged and extensive use of fluconazole in treating candida infections led to emergence of resistance in all Candida species (Bassetti et al., 2016, Whaley et al., 2017). Furthermore, the species that showed fluconazole resistance were C. glabrata (66.6%), C. krusei (16%), C. tropicalis (12.5%) and C. albicans (10%). Our results are in consistent with other Egyptian study by Khairat et al. (2019) who reported azole resistance is more in NAC species (44%) than C. albicans (38.9%). Furthermore, Kadry et al. (2018) reported that 9.4% of C. albicans isolates were resistant to fluconazole. C. glabrata has innate low susceptibility to azoles and acquired azole resistance has been reported during therapy (Vermitsky and Edlind, 2004). C. krusei has intrinsic resistance to fluconazole (Whaley et al., 2017). While C. tropicalis high level rate of azole resistance (up to 83%) was reported in South Korea (Yoo et al., 2009) and low rates (4–9%) in USA (Berkow and Lockhart, 2017).
Amphotericin B is a fungicidal drug that has been used for >60 years for treatment of fungal infections (Mesa-Arango et al., 2012). The renal toxicity of AmB limits its use, however, several lipid formulations are developed to overcome this problem (Faustino and Pinheiro, 2020). Fortunately, none of isolates under investigation showed resistance to AmB. Similarly, previous studies in Egypt reported either no resistance to AmB (Kadry et al., 2018), or very low (3%) resistance (Khairat et al., 2019). It is well known that Candida rapidly evolves resistance to azoles, however, resistance to AmB remains extremely limited (Vincent et al., 2013).
Biofilm formation is an important virulence factor in pathogenic fungi that also contributes to the development of drug resistance (Wuyts et al., 2018). The present study showed that the majority of Candida resistant isolates (15 out of 16) were able to form biofilm, among which 8 isolates form strong biofilm and 7 isolates were intermediate producer. Among the C. albicans resistant isolates, 5 were strong producer, one is intermediate producer and one was none biofilm producer. This result is nearly similar to previous study showed that 80% of resistant isolates (4 out of 5) were able to form strong biofilm. However, C. albicans was the only species under investigation in their study (Kadry et al., 2018). Moreover, a study carried by Shi et al (2019) revealed positive correlation between biofilm-producer and azole-resistance in C. albicans clinical isolates. These results were on the same line with results reported in our study where almost all azole resistant isolates were biofilm producer.
Efflux pumps overexpression is one of the most common mechanisms of Candida resistance to azoles (Brilhante et al., 2016). The efflux pump genes usually associated with azole resistance are CDR1, CDR2 and MDR1 in C. albicans (Watamoto et al., 2011), CDR1 in C. glabrata (Szweda et al., 2015) and MDR1in C. tropicalis (Jin et al., 2018). While two kinds of efflux gene are associated with C. krusei resistance, ABC1 and ABC2 (He et al., 2015).
Phenotypically, all resistant strains showed overexpression of efflux pumps. Genotypically, qRT-PCR was utilized for the quantifying the expression of common efflux-pump genes in Candida species, efflux pump genes were up-regulated in azole-resistant isolates compared with the susceptible strain. The highest over-expression level was observed in ABC2 gene in C. krusei. In addition, CDR1 and MDR1 overexpression in C. albicans were observed to be 1.97 and 2.45 fold, respectively. While, 1.64 -fold increase of CDR1 was observed in C. glabrata and 1.16 -fold increase was observed in C. tropicalis MDR1.
Similarly, it was reported that CDR1, CDR2 and MDR1 were up-regulated in all azole-resistant C. albicans isolates compared with control strain (Chen et al., 2010). In addition, it was reported that azole-resistant clinical isolates of C. albicans display overexpression of CDR1 (Prasad and Rawal, 2014). Furthermore, a previous study reported that 86.6% of azole-resistant isolates showed a significant increase in the level of expression of CDR1 in C. glabrata (Szweda et al., 2015). Regarding C. tropicalis, Pandey et al. (2020) revealed that fluconazole resistance is correlated to overexpression of efflux pump transporter genes CDR1 and MDR1. For C. krusei, ABC2 gene levels was significantly higher in itraconazole and voriconazole resistant isolates than susceptible isolates (Ricardo et al., 2014, He et al., 2015). All of the above are in consistent with the results of the current study.
Multidrug efflux pump represents one major mechanism of resistance to azole antifungals. Recently, the strategy of using EPIs to combat such resistance is explored (Holmes et al., 2012). In the current study, the EPIs pantoprazole and haloperidol were able to reduce fluconazole resistance in all Candida species under investigation. Pantoprazole showed stronger chemo-sensitization of fluconazole than haloperidol. Similarly, a recent study showed a synergistic effect between fluconazole and proton pump inhibitors (PPIs) including pantoprazole but the study investigates the effect on C. albicans only (Lu et al., 2020). Another study showed that the PPI verapamil inhibits efflux pumps in C. albicans, and exhibits synergism with fluconazole (Vega-chacón et al., 2021). Consistent with our results, the MIC of fluconazole against Malassezia spp. decreased in presence of sub-MIC of haloperidol (Iatta et al., 2017). In the current study, both Haloperidol and pantoprazole significantly reduced the expression of efflux pump genes CDR1 and MDR1 in C. albicans, Similarly, Ji and coworkers stated that the haloperidol derivative B11 down-regulated the overexpression of ERG11 and MDR1 genes when used in combination with fluconazole (Ji et al., 2020). It is worth mentioning that this study is the first study to assess the effect of pantoprazole and haloperidol, not only in C. albicans but also in non-albicans Candida species resistant to azoles and our investigation showed a promising results on all tested strains.
For validation of our results, Molecular docking was used to evaluate the interaction between potential EPIs and CDR1 and MDR1 efflux pump proteins. The results showed that Pantoprazole binds into the active site cavity of C. albicans CDR1 and MDR1 which was indicated by excellent minimum binding energy of − 11.827 Kcal/mol and − 10.697 Kcal/mol, respectively. Slightly higher minimum binding energy was observed for haloperidol with C. albicans CDR1 (−9.663 Kcal/mol) and MDR1 (−9.445 Kcal/mol). Similarly, a recent study reported that the natural monoterpenoid geraniol actively bind to C. albicans CDR1 (−8.8 Kcal/mol) and showed synergism with fluconazole (Singh et al., 2018). Furthermore, Beauvericin compounds show good binding affinity to CDR1 and CDR2 and block the efflux function of ABC transporters in C. albicans (Tong et al., 2016).
It was proven that introducing halogen atoms into a lead compounds, have several processes in drug-target complexes rather than steric effect (Hernandes et al., 2010). Inspecting the results of this docking from structure activity relationship (SAR) point of view, both ligands are halogenated, comprising three rings; two terminal substituted aromatic rings with middle heterocycle, and flexible with rotatable bonds whether in side chains or spacers. However, the middle imidazole-ring (five-membered heterocycle) in Pantoprazole contributed effectively with all types of intermolecular interactions in enhancing the stability and improving the recognition of ligand/receptor complex rather than its six-membered heterocycle (piperidine) counterpart in Haloperidol.
Of note, the interaction energy expressed in Kcal/mol, reflects how stable is the ligand–protein complex, the lower the interaction energy the more stable is the formed ligand/receptor complex and the higher the affinity of investigated compound into the target receptor. Consequently, Pantoprazole showed higher blockade activity in both efflux pump protein receptors, which explains the highest activity of the Pantoprazole rather than Haloperidol in the reduction of fluconazole MIC.
5. Conclusion
Candida albicans remains the most isolated Candida species, however an increase of NAC species was observed. The NAC species showed a higher resistance to azole drugs. Efflux pump was detected in all resistant isolates indicated that it is a major mechanism of resistance in all Candida species. Pantoprazole and Haloperidol were able to chemo-sensitize fluconazole in all resistant species indicating their possible usage to strengthen azole drugs for better therapy outcome.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
References
- Bhattacharya S., Sobel J.D., White T.C. A Combination fluorescent assay demonstrates increased in efflux pump activity as a resistance mechanism in Azole-resistant vaginal Candida albicans isolates. Antimicrob. Agents Chemother. 2016;60(10):5858–5866. doi: 10.1128/aac.01252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S., Sae-Tia S., Fries B.C. Candidiasis and mechanisms of antifungal resistance. Antibiotics (Basel) 2020;9(6):312. doi: 10.3390/antibiotics9060312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassetti, M., Peghin, M., Timsit, J.F., 2016. The Current Treatment Landscape: Candidiasis, J. Antimicrob. Chemother. 71, ii13–22. [DOI] [PubMed]
- Berkow E.L., Lockhart S.R. Infection and Drug Resistance Dovepress Fluconazole resistance in Candida species: a current perspective. Infect. Drug Res. 2017;10:237–245. doi: 10.2147/IDR.S118892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitar, I., Khalaf, R.A., Harastani, H., Tokajian. S., 2014. Identification, typing, antifungal resistance profile, and biofilm formation of Candida albicans isolates from Lebanese hospital patients. BioMed. Res. Int. 2014 (2014). Article ID 931372. [DOI] [PMC free article] [PubMed]
- Bongomin F., Gago S., Oladele R.O., Denning D.W. Global and multi-national prevalence of fungal diseases-estimate precision. J. Fungi. 2017;3:57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilhante R.S., Paiva M.A., Sampaio C.M., Castelo-Branco D.S., Teixeira C.E., de Alencar L.P., Bandeira T.J., Monteiro A.J., Cordeiro R.A., Pereira-Neto W.A., Sidrim J.J., Moreira J.L., Rocha M.F. Azole resistance in Candida spp. isolated from Catú Lake, Ceará, Brazil: an efflux-pump-mediated mechanism. Braz. J. Microbiol. 2016;47(1):33–38. doi: 10.1016/j.bjm.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemical Computing Group Inc. Molecular Operating Environment (MOE), 2019.01.: Montreal, QC, Canada, 2019.
- Chen L.M., Xu Y.H., Zhou C.L., Zhao J., Li C.Y., Wang R. Overexpression of CDR1 and CDR2 genes plays an important role in fluconazole resistance in Candida albicans with G487T and T916C Mutations. J. Int. Med. Res. 2010;38:536–545. doi: 10.1177/147323001003800216. [DOI] [PubMed] [Google Scholar]
- Cheng, M.F., Yang, Y.L., Yao, T.J., Lin, C.Y., Liu, J.S., Tang, R.B., Yu, K.W., Fan, Y.H., Hsieh, K.S., Ho, M., Lo, H.J, 2005. Risk factors for fatal candidemia caused by Candida albicans and non-albicans Candida species. BMC Infect Dis. 5, Article number: 22. [DOI] [PMC free article] [PubMed]
- Clinical and Laboratory Standards Institute, 2008. M27–A3: Reference Method for Broth Dilution Antifungal susceptibility testing of yeasts; Approved Standard-3rd ed., Wayne, PA, USA CLSI.
- Clinical and Laboratory Standards Institute, 2009. M44-A2: Method for Antifungal Disk Diffusion Susceptibility Testing of Yeasts; Approved Guideline—second ed. CLSI Doc, 29(17).
- Cornet M., Sendid B., Fradin C., Gaillardin C., Poulain D., Nguyen H.V. Molecular identification of closely related Candida species using two ribosomal intergenic spacer fingerprinting methods. J. Mol. Diagnost. 2011;13:12–22. doi: 10.1016/j.jmoldx.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Baz A.M., Mosbah R.A., Goda R.M., Mansour B., Sultana T., Dahms T., El-Ganiny A.M. Back to nature : combating Candida albicans biofilm, phospholipase and hemolysin using plant essential oils. Antibiotics. 2021;10:81. doi: 10.3390/antibiotics10010081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Ganiny A.M., Yossef N.E., Kamel H.A. Prevalence and antifungal drug resistance of nosocomial Candida species isolated from two university hospitals in Egypt. Curr. Med. Mycol. 2021;7(1):31–37. doi: 10.18502/cmm.7.1.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Xiao M., Zhang D., Huang J.J., Wang H., Hou X., Zhang L., Kong F., Chen S.A., Tong Z.H., Xu Y.C. Molecular mechanisms of azole resistance in Candida tropicalis isolates causing invasive candidiasis in China. Clin. Microbiol. Infect. 2019;25(7):885–891. doi: 10.1016/j.cmi.2018.11.007. [DOI] [PubMed] [Google Scholar]
- Faustino C., Pinheiro L. Lipid systems for the delivery of amphotericin B in antifungal therapy. Pharmaceutics. 2020;12:29. doi: 10.3390/pharmaceutics12010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher M.C., Hawkins N.J., Sanglard D., Gurr S.J. Worldwide emergence of resistance to antifungal drugs challenges health and food security. Science. 2018;360:739–742. doi: 10.1126/science.aap7999. [DOI] [PubMed] [Google Scholar]
- Francois I.E., Cammue B., Borgers M., Ausma J., Dispersyn G.D., Thevissen K. Azoles: mode of antifungal action and resistance development. Effect of miconazole on endogenous reactive oxygen species production in Candida albicans. Anti-infect. Agents Med. Chem. 2006;5(1):3–13. [Google Scholar]
- He X., Zhao M., Chen J., Wu R., Zhang J., Cui R., Jiang Y., Chen J., Cao X., Xing Y., Zhang Y., Meng J., Deng Q., Sui T. Overexpression of both ERG11 and ABC2 genes might be responsible for itraconazole resistance in clinical isolates of Candida krusei. PLoS One. 2015;10:e0136185. doi: 10.1371/journal.pone.0136185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A.R., Keniya M.V., Ivnitski-Steele I., Monk B.C., Lamping E., Sklar L.A., Cannon R.D. The monoamine oxidase A inhibitor clorgyline is a broad-spectrum inhibitor of fungal ABC and MFS transporter efflux pump activities which reverses the azole resistance of Candida albicans and Candida glabrata clinical isolates. Antimicrob. Agents Chemother. 2012;56(3):1508–1515. doi: 10.1128/AAC.05706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandes M.Z., Cavalcanti S.M.T., Moreira D.R.M., de Azevedo Junior W.F., Leite A.C.L. Halogen atoms in the modern medicinal chemistry: hints for the drug design. Curr. Drug Targets. 2010;11(3):303–314. doi: 10.2174/138945010790711996. [DOI] [PubMed] [Google Scholar]
- Iatta R., Puttilli M.R., Immediato D., Otranto D., Cafarchia C. The role of drug efflux pumps in Malassezia pachydermatis and Malassezia furfur defence against azoles. Mycoses. 2017;60(3):178–182. doi: 10.1111/myc.12577. [DOI] [PubMed] [Google Scholar]
- Ivnitski-Steele I., Holmes A.R., Lamping E., Monk B.C., Cannon R.D., Sklar L.A. Identification of Nile red as a fluorescent substrate of the Candida albicans ATP-binding cassette transporters Cdr1p and Cdr2p and the major facilitator superfamily transporter Mdr1p. Anal. Biochem. 2009;394(1):87–91. doi: 10.1016/j.ab.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahagirdar V.L., Davane M.S., Aradye S.C., Nagoba B.S. Candida species as potential nosocomial pathogens–a review. Electron. J Gen. Med. 2018;15(2):em05. [Google Scholar]
- Ji C., Liu N., Tu J., Li Z., Han G., Li J., Sheng C. Drug rrepurposing of haloperidol: discovery of new benzocyclane derivatives as potent antifungal agents against Cryptococcosis and Candidiasis. ACS Infect. Dis. 2020;6(5):768–786. doi: 10.1021/acsinfecdis.9b00197. [DOI] [PubMed] [Google Scholar]
- Jin L., Cao Z., Wang Q., Wang Y., Wang X., Chen H., Wang H. MDR1 overexpression combined with ERG11 mutations induce high-level fluconazole resistance in Candida tropicalis clinical isolates. BMC Infect Dis. 2018;18(1):162. doi: 10.1186/s12879-018-3082-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadry A.A., El-Ganiny A.M., El-Baz A.M. Relationship between Sap prevalence and biofilm formation among resistant clinical isolates of Candida albicans. Afr. Health Sci. 2018;18:1166–1174. doi: 10.4314/ahs.v18i4.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khairat S.M., Sayed A.M., Nabih M., Soliman N.S., Hassan Y.M. Prevalence of Candida blood stream infections among children in tertiary care hospital: detection of species and antifungal susceptibility. Infect. Drug Resist. 2019;12:2409–2416. doi: 10.2147/IDR.S196972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamping E., Madani G., Lee H.J., Niimi M., Cannon R.D. In: Candida albicans: Cellular and Molecular Biology. Prasad R., editor. Springer; Cham: 2017. Structure-function analyses of multidrug transporters; pp. 379–406. [Google Scholar]
- Lee Y., Puumala E., Robbins N., Cowen L.E. Antifungal drug resistance: molecular mechanisms in Candida albicans and beyond. Chem. Rev. 2020;121(6):3390–3411. doi: 10.1021/acs.chemrev.0c00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real- time quantitative PCR and the 2ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu M., Yan H., Yu C., Yuan L., Sun S. Proton pump inhibitors act synergistically with fluconazole against resistant Candida albicans. Sci. Rep. 2020;10(1):498. doi: 10.1038/s41598-019-57174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maesaki S. Rhodamine 6G efflux for the detection of CDR1-overexpressing azole-resistant Candida albicans strains. J. Antimicrob. Chemother. 1999;44(1):27–31. doi: 10.1093/jac/44.1.27. [DOI] [PubMed] [Google Scholar]
- Marak, M., Dhanashree B., 2018. Antifungal susceptibility and biofilm production of Candida spp . isolated from clinical samples, Int. J. Microbiol. 2018, Article ID 7495218. [DOI] [PMC free article] [PubMed]
- Martins M., Viveiros M., Couto I., Costa S.S., Pacheco T., Fanning S., Pagès J.M., Amaral L. Identification of efflux pump-mediated multidrug-resistant bacteria by the Ethidium Bromide-agar cartwheel method. In Vivo. 2011;25(2):171–178. [PubMed] [Google Scholar]
- Martins A.N.A., Amaral L. Screening for efflux pump systems of bacteria by the new acridine orange agar method. In Vivo. 2012;26:203–206. [PubMed] [Google Scholar]
- McGuffin L.J., Adiyaman R., Maghrabi A.H., Shuid A.N., Brackenridge D.A., Nealon J.O., Philomina L.S. IntFOLD: An integrated web resource for high performance protein structure and function prediction. Nucl. Acids Res. 2019;47:W408–W413. doi: 10.1093/nar/gkz322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa-Arango A.C., Scorzoni L., Zaragoza O. It only takes one to do many jobs: amphotericin B as antifungal and immunomodulatory drug. Front. Microbiol. 2012;3:286. doi: 10.3389/fmicb.2012.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miró-Canturri A., Ayerbe-Algaba R., Smani Y. Drug repurposing for the treatment of bacterial and fungal infections. Front Microbiol. 2019;10:41. doi: 10.3389/fmicb.2019.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey N., Tripathi M., Gupta M.K., Tilak R. Overexpression of efflux pump transporter genes and mutations in ERG11 pave the way to fluconazole resistance in Candida tropicalis: a study from a North India region. J Glob Antimicrob Resist. 2020;22:374–378. doi: 10.1016/j.jgar.2020.02.010. [DOI] [PubMed] [Google Scholar]
- Papon N., Courdavault V., Clastre M., Bennett R.J. Emerging and emerged pathogenic Candida species: Beyond the Candida albicans paradigm. PLoS Pathog. 2013;9(9):e1003550. doi: 10.1371/journal.ppat.1003550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyclit L., Yousfi H., Rolain J.M., Bittar F. Drug repurposing in medical mycology: Identification of compounds as potential antifungals to overcome the emergence of multidrug-resistant fungi. Pharmaceuticals. 2021;14(5):488. doi: 10.3390/ph14050488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R., Rawal M.K. Efflux pump proteins in antifungal resistance. Front. Pharmacol. 2014;5:202. doi: 10.3389/fphar.2014.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pristov K.E., Ghannoum M.A. Resistance of Candida to azoles and echinocandins worldwide. Clin. Microbiol. Infect. 2019;25(7):792–798. doi: 10.1016/j.cmi.2019.03.028. [DOI] [PubMed] [Google Scholar]
- Rautemaa R., Ramage G. Oral candidosis clinical challenges of a biofilm disease. Crit. Rev. Microbiol. 2011;37(4):328–336. doi: 10.3109/1040841X.2011.585606. [DOI] [PubMed] [Google Scholar]
- Ricardo E., Miranda I.M., Faria-Ramos I., Silva R.M., Rodrigues A.G., Pina-Vaz C. In vivo and in vitro acquisition of resistance to voriconazole by Candida krusei. Antimicrob. Agents Chemother. 2014;58(8):4604–4611. doi: 10.1128/AAC.02603-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricotta E.E., Lai Y.L., Babiker A., Strich J.R., Kadri S.S., Lionakis M.S., Prevots D.R., Adjemian J. Invasive candidiasis species distribution and trends, United States, 2009–2017. J. Infect. Dis. 2021;223(7):1295–1302. doi: 10.1093/infdis/jiaa502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti M., Posteraro B., Lass-Flörl C. Antifungal drug resistance among Candida species: mechanisms and clinical impact. Mycoses. 2015;58(S2):2–13. doi: 10.1111/myc.12330. [DOI] [PubMed] [Google Scholar]
- Sardi J., Scorzoni L., Bernardi T., Fusco-Almeida A.M., Mendes Giannini M. Candida species: Current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J. Med. Microbiol. 2013;62:10–24. doi: 10.1099/jmm.0.045054-0. [DOI] [PubMed] [Google Scholar]
- Shi C., Liu J., Li W., Zhao Y., Meng L., Xiang M. Expression of fluconazole resistance-associated genes in biofilm from 23 clinical isolates of Candida albicans. Braz. J. Microbiol. 2019;50(1):157–163. doi: 10.1007/s42770-018-0009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Fatima Z., Ahmad K., Hameed S. Fungicidal action of geraniol against Candida albicans is potentiated by abrogated CaCdr1p drug efflux and fluconazole synergism. PLoS One. 2018;13(8):e0203079. doi: 10.1371/journal.pone.0203079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanović S., Vuković D., Hola V., Di Bonaventura G., Djukić S., Cirković I., Ruzicka F. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115(8):891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- Szweda P., Gucwa K., Romanowska E., Dzierz K., Naumiuk Ł., Brillowska-Da A., Wojciechowska-Koszko I., Milewski S. Mechanisms of azole resistance among clinical isolates of Candida glabrata in Poland. J Med Microbiol. 2015;64(6):610–619. doi: 10.1099/jmm.0.000062. [DOI] [PubMed] [Google Scholar]
- Tong Y., Liu M., Zhang Y., Liu X., Huang R., Song F., Dai H., Ren B., Sun N., Pei G., Bian J. Beauvericin counteracted multi-drug resistant Candida albicans by blocking ABC transporters. Synth. Syst. Biotechnol. 2016;1(3):158–168. doi: 10.1016/j.synbio.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y., Zhang J., Sun N., Wang X., Wei Q., Zhang Y., Huang R., Pu Y., Dai H., Ren B., Pei G., Song F., Zhu G., Wang X., Xia X., Chen X., Jiang L., Wang S., Ouyang L., Xie N., Zhang B., Jiang Y., Liu X., Calderone R., Bai F., Zhang L., Alterovitz G. Berberine reverses multidrug resistance in Candida albicans by hijacking the drug efflux pump Mdr1p. Chin. Sci. Bull. 2021;66(18):1895–1905. doi: 10.1016/j.scib.2020.12.035. [DOI] [PubMed] [Google Scholar]
- Turner S.A., Butler G. The Candida pathogenic species complex. Cold Spring Harb. Perspect. Med. 2014;4(9):a019778. doi: 10.1101/cshperspect.a019778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Chacón Y., de Albuquerque M.C., Pavarina A.C., Goldman G.H., Mima E. Verapamil inhibits efflux pumps in Candida albicans, exhibits synergism with fluconazole, and increases survival of Galleria mellonella. Virulence. 2021;12(1):231–243. doi: 10.1080/21505594.2020.1868814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermitsky J.P., Edlind T.D. Azole resistance in Candida glabrata: Coordinate upregulation of multidrug transporters and evidence for a Pdr1-like transcription factor. Antimicrob. Agents Chemother. 2004;48(10):3773–3781. doi: 10.1128/AAC.48.10.3773-3781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent B.M., Lancaster A.K., Scherz-Shouval R., Whitesell L., Lindquist S. Fitness trade-offs restrict the evolution of resistance to amphotericin B. PLoS Biol. 2013;11(10):e1001692. doi: 10.1371/journal.pbio.1001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watamoto T., Samaranayake L.P., Egusa H., Yatani H., Seneviratne C.J. Transcriptional regulation of drug-resistance genes in Candida albicans biofilms in response to antifungals. J. Med. Microbiol. 2011;60:1241–1247. doi: 10.1099/jmm.0.030692-0. [DOI] [PubMed] [Google Scholar]
- Whaley S.G., Berkow E.L., Rybak J.M., Nishimoto A.T., Barker K.S., Rogers P.D. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front. Microbiol. 2017;7:2173. doi: 10.3389/fmicb.2016.02173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirsching S., Michel S., Köhler G., Morschhäuser J. Activation of the multiple drug resistance gene MDR1 in fluconazole- resistant, clinical Candida albicans strains is caused by mutations in a trans-regulatory factor. J. Bacteriol. 2000;182(2):400–404. doi: 10.1128/jb.182.2.400-404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts J., Van Dijck P., Holtappels M. Holtappels, Fungal persister cells: The basis for recalcitrant infections? PLoS Pathog. 2018;14(10):e1007301. doi: 10.1371/journal.ppat.1007301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J.I., Choi C.W., Lee K.M., Kim Y.K., Kim T.U., Kim E.C., Joo S.I., Yun S.H., Lee Y.S., Kim B.S. National surveillance of antifungal susceptibility of Candida species in South Korean hospitals. Med. Mycol. 2009;47(5):554–558. doi: 10.1080/13693780802354037. [DOI] [PubMed] [Google Scholar]
- Zárate S.G., Morales P., Świderek K., Bolanos-Garcia V.M., Bastida A. A Molecular modeling approach to identify novel inhibitors of the major facilitator superfamily of efflux pump transporters. Antibiotics. 2019;8(1):25. doi: 10.3390/antibiotics8010025. [DOI] [PMC free article] [PubMed] [Google Scholar]