Abstract
Objectives
Solid organ transplant recipients (SOTR) receiving post‐transplant immunosuppression show increased COVID‐19‐related mortality. It is unclear whether an additional dose of COVID‐19 vaccines can overcome the reduced immune responsiveness against severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) variants.
Methods
We analysed humoral immune responses against SARS‐CoV‐2 and its variants in 53 SOTR receiving SARS‐CoV‐2 vaccination.
Results
Following the initial vaccination series, 60.3% of SOTR showed no measurable neutralisation and only 18.9% demonstrated neutralising activity of > 90%. More intensive immunosuppression, antimetabolites in particular, negatively impacted antiviral immunity. While absolute IgG levels were lower in SOTR than controls, antibody titres against microbial recall antigens were higher. By contrast, SOTR showed reduced vaccine‐induced IgG/IgA antibody titres against SARS‐CoV‐2 and its delta variants and fewer linear B‐cell epitopes, indicating reduced B‐cell diversity. Importantly, a third vaccine dose led to an increase in anti‐SARS‐CoV‐2 antibody titres and neutralising activity across alpha, beta and delta variants and to the induction of anti‐SARS‐CoV‐2 CD4+ T cells in a subgroup of patients analysed. By contrast, we observed significantly lower antibody titres after the third dose with the omicron variant compared to the ancestral SARS‐CoV‐2 and the improvement in neutralising activity was much less pronounced than for all the other variants.
Conclusion
Only a small subgroup of solid organ transplant recipients is able to generate functional antibodies after an initial vaccine series; however, an additional vaccine dose resulted in dramatically improved antibody responses against all SARS‐CoV‐2 variants except omicron where antibody responses and neutralising activity remained suboptimal.
Keywords: antibody responses, COVID‐19, omicron variant, SARS‐CoV‐2, solid organ transplant, vaccine
In this study, we found that only a small subgroup of solid organ transplant recipients is able to generate functional antibodies after an initial COVID‐19 vaccine series consisting of 1–2 doses. However, an additional vaccine dose resulted in dramatically improved antibody responses against all SARS‐CoV‐2 variants except omicron where antibody responses and their neutralizing activity remained suboptimal.

Introduction
COVID‐19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), 1 which contains several structural proteins including the surface‐exposed spike (S) and the internal nucleocapsid (N) proteins. 1 , 2 The S fusion protein consists of the S1/S2 components and the virus enters cells, such as pneumocytes in the lung, 3 through binding of the receptor‐binding domain (RBD) within the S1 protein, 4 to the angiotensin‐converting enzyme‐2 (ACE‐2) receptor. 2 , 5 Older patients and patients with pre‐existing medical conditions, including different types of cancer, 6 show a more complicated course of COVID‐19 and have a worse prognosis. 7 , 8 , 9 , 10 Solid organ transplant recipients (SOTR) with COVID‐19 also show an increased mortality of > 20%. 11 , 12 , 13 , 14 , 15 , 16 , 17 In these patients, advanced age and comorbidities such as cardiovascular and pulmonary disease seem to contribute to the reduced survival. 18 , 19 , 20
The development of anti‐SARS‐CoV‐2 antibodies following an active infection and/or vaccination, especially those directed against the S/RBD proteins of the virus, is crucial for the (1) protection from future COVID‐19 infections, (2) limiting disease severity and (3) controlling viral transmission. 21 , 22 Unfortunately, SOTR receive long‐term immunosuppressive treatment and are, therefore, less likely to build a protective immune response against SARS‐CoV‐2. 23 , 24 , 25 , 26 Indeed, it was shown that following an active COVID‐19 infection only ~50% of SOTR will mount an antibody response 27 with kidney transplants vs. other types of transplants, shorter time from transplant to diagnosis and more intensive immunosuppression being associated with a reduced humoral immune response. 27
The US Food and Drug Administration (FDA) has issued emergency use authorisations or full approval for 3 vaccines for the prevention of COVID‐19. 28 , 29 All three vaccines have been shown to elicit antibody‐ and T cell–mediated antiviral immune responses which confer almost complete protection against infection with the COVID‐19 virus in healthy individuals. 30 , 31 , 32 , 33 , 34 , 35 , 36 Unfortunately, recent studies have indicated that SOTR, in agreement with observations made after an active COVID‐19 infection, show reduced antibody responses following COVID‐19 vaccination 37 , 38 , 39 , 40 against the original SARS‐CoV‐2 virus.
Until very recently, the more infectious B.1.617.2 (delta) variant of SARS‐CoV‐2 contributed to a surge in cases across the globe. 41 Only modest differences in vaccine effectiveness were noted with the delta variant compared to the original viral strain following two doses of COVID‐19 mRNA vaccines; however, this may be very different in immunocompromised individuals, such as SOTR. 42 Additionally, the recently spreading omicron variant may even further promote viral immune escape 43 in these patients. Unfortunately, a detailed picture of vaccine‐induced antibody responses in SOTR, including a description of immunodominant antibody epitopes, has never been obtained. Furthermore, while recent data indicate that additional doses of COVID‐19 vaccines may help to overcome the reduced immune responsiveness in SOT patients, 44 , 45 , 46 , 47 it has remained unclear whether this also applies to different variants of SARS‐CoV‐2 such as the delta and the omicron mutants. Here, we present the results of our prospective study investigating in detail humoral immune responses against SARS‐CoV‐2 and its recent variants in fully vaccinated SOTR and in response to an additional dose of SARS‐CoV‐2 vaccine.
Results
Solid organ transplant recipients show a markedly reduced antibody‐mediated neutralising activity in response to mRNA COVID‐19 vaccines
As a first step, we screened a total of 53 SOTR recipients and 5 healthy controls for neutralising antibodies inhibiting RBD‐ACE2 interactions at 8 weeks after receiving the second dose of an mRNA COVID‐19 vaccine or 90 days after receiving the J&J vaccine. We found that all healthy controls (Figure 1) showed a viral neutralisation > 90% and were therefore classified as ‘Good Responders’ (GR). In marked contrast, most of the SOTR (32/53; 60.3%) showed a viral neutralisation of < 30% and were therefore classified as ‘Non‐Responders’ (NR). Of the 53 SOTR, 11 (20.8%) showed a neutralising activity between 30% and 90% and were classified as ‘Reduced Responders’ (RR). Only 10 (18.9%) showed a neutralising activity of above 90% and were classified as GR (Figure 1). Overall, the vast majority of SOTR (81.1%) showed a neutralising activity < 90% after completing the initial vaccine series.
Figure 1.
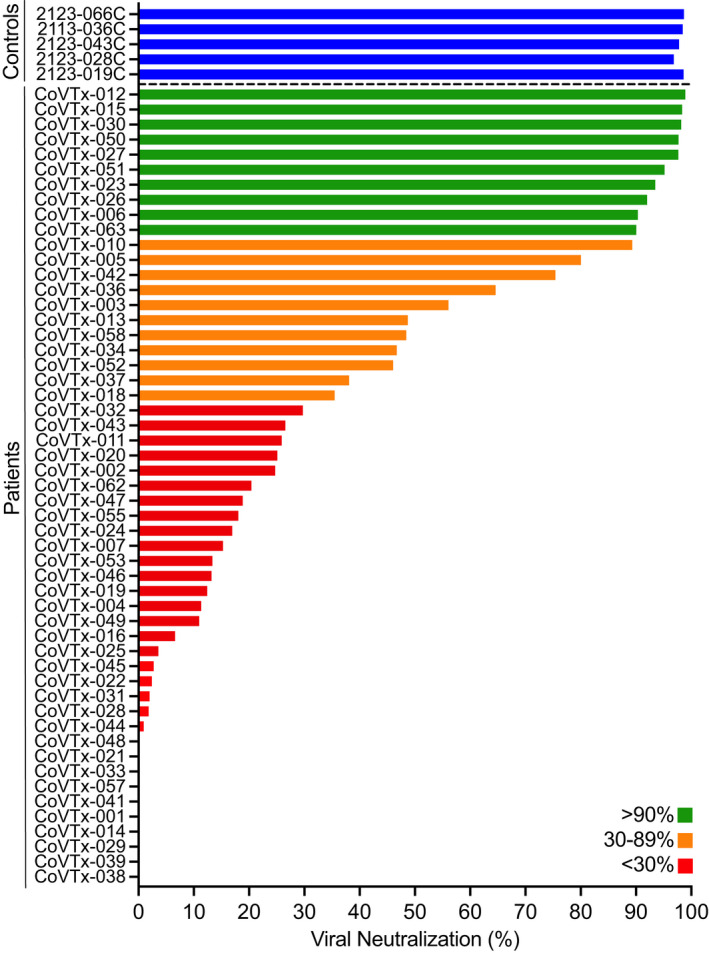
Neutralising activity in the peripheral blood of SOT recipients after two doses of a COVID‐19 mRNA vaccine. Neutralising activity of vaccine‐induced anti‐RBD antibodies in the peripheral blood of SOT recipients (N = 53) and healthy controls (N = 5; blue bars) after the second dose of the vaccine was measured as the degree of inhibition of RBD‐ACE2 interactions. Green, orange and red bars indicate different degrees of inhibition as indicated in the legend.
We did not find any significant associations (Table 1) of the patients’ demographic characteristics and their past medical history with the type of humoral immune response (GR vs. RR/NR). However, when we analysed the patients’ clinical characteristics at the time of the first dose of the vaccine (Table 2), we found that a higher overall number of immunosuppressants was associated with a reduced vaccine‐induced SARS‐CoV‐2 neutralising activity. Importantly, among immunosuppressants, only treatment with antimetabolites, which prevent lymphocyte proliferation, had a significant negative impact on the patients’ antiviral humoral immunity (Table 2), indicating that specifically antimetabolites interfere with the development of COVID‐19‐specific antibodies.
Table 1.
Demographic characteristics and medical history
| Total | GR | RR/NR | Sig. | |
|---|---|---|---|---|
| Patients | 53 | 10 (18.9) | 43 (81.2) | |
| Age | 64 (43–79) | 65 (45–78) | 64 (36–79) | 0.829 |
| Gender | ||||
| Male | 38 (71.7) | 5 (50.0) | 33 (76.7) | 0.124 |
| Female | 15 (28.3) | 5 (50.0) | 10 (23.3) | |
| Race | ||||
| Caucasian | 41 (77.4) | 9 (90.0) | 32 (74.4) | 0.800 |
| African American | 9 (17.0) | 1 (10.0) | 8 (18.6) | |
| Asian | 3 (5.7) | 0 (0.0) | 3 (7.0) | |
| Diabetes | ||||
| Yes | 14 (26.4) | 3 (30.0) | 11 (25.6) | 0.999 |
| No | 39 (73.6) | 7 (70.0) | 32 (74.4) | |
| Obesity | ||||
| Yes | 29 (54.7) | 7 (70.0) | 22 (51.2) | 0.318 |
| No | 24 (45.3) | 3 (30.0) | 21 (48.8) | |
| Heart failure | ||||
| Yes | 6 (11.3) | 0 (0.0) | 6 (14.0) | 0.581 |
| No | 47 (88.7) | 10 (100.0) | 37 (86.0) | |
| Myocardial infarction | ||||
| Yes | 3 (5.7) | 0 (0.0) | 3 (14.0) | 0.999 |
| No | 50 (94.3) | 10 (100.0) | 40 (86.0) | |
| Chronic kidney disease | ||||
| Yes | 30 (56.6) | 8 (80.0) | 22 (51.2) | 0.158 |
| No | 23 (43.4) | 2 (20.0) | 21 (48.8) | |
| Pulmonary disease | ||||
| Yes | 10 (18.9) | 1 (10.0) | 9 (20.9) | 0.665 |
| No | 43 (81.1) | 9 (90.0) | 34 (79.1) | |
Table 2.
Clinical characteristics at the time of vaccination (first dose)
| Total | GR | RR/NR | Sig. | |
|---|---|---|---|---|
| Patients | 53 | 10 (18.9) | 43 (81.1) | |
| Time from transplant (weeks) | 322 (30–1260) | 380 (84–698) | 308 (30–1260) | 0.630 |
| Type of transplant | ||||
| Kidney | 24 (45.3) | 3 (30.0) | 21 (48.8) | 0.175 |
| Liver | 12 (22.6) | 7 (70.0) | 5 (11.6) | |
| Lung | 6 (11.3) | 0 (0.0) | 6 (14.0) | |
| Heart | 5 (9.4) | 0 (0.0) | 5 (11.6) | |
| Kidney/Pancreas | 3 (5.7) | 0 (0.0) | 3 (7.0) | |
| Heart/Lung | 1 (1.9) | 0 (0.0) | 1 (2.3) | |
| Liver/Kidney | 1 (1.9) | 0 (0.0) | 1 (2.3) | |
| Pancreas | 1 (1.9) | 0 (0.0) | 1 (2.3) | |
| Induction | ||||
| Unknown | 3 (5.7) | 0 (0.0) | 3 (7.0) | 0.347 |
| Methylprednisolone | 18 (34.0) | 6 (60.0) | 12 (27.9) | |
| Alemtuzumab | 13 (4.5) | 1 (10.0) | 12 (27.9) | |
| ATG | 5 (9.4) | 0 (0.0) | 5 (11.6) | |
| Basiliximab | 6 (11.3) | 1 (10.0) | 5 (11.6) | |
| Unknown | 8 (15.1) | 2 (20.0) | 6 (14.0) | |
| Immunosuppressive Agents (total #) | 2.0 (1–3) | 1.5 (1–3) | 2.00 (1–3) | 0.018 |
| Antimetabolite | ||||
| Yes | 38 (71.7) | 3 (30.0) | 35 (81.4) | 0.007 |
| No | 15 (28.3) | 7 (70.0) | 8 (18.6) | |
| Calcineurin inhibitor | ||||
| Yes | 48 (90.6) | 10 (100.0) | 38 (88.4) | 0.647 |
| No | 5 (9.4) | 0 (0.0) | 5 (11.6) | |
| mTOR inhibitor | ||||
| Yes | 9 (17.0) | 2 (20) | 7 (16.3) | 0.448 |
| No | 44 (83.0) | 8 (80) | 36 (83.7) | |
| Steroids | ||||
| Yes | 21 (39.6) | 1 (10) | 20 (46.5) | 0.069 |
| No | 32 (60.4) | 9 (90) | 23 (53.3) | |
| Type of vaccine | ||||
| Pfizer | 32 (60.4) | 4 (40.0) | 28 (65.1) | 0.149 |
| Moderna | 18 (34.0) | 6 (60.0) | 12 (27.9) | |
| J&J | 3 (5.7) | 0 (0.0) | 3 (7.0) | |
Solid organ transplant recipients maintain their humoral immunity against microbial antigens other than SARS‐CoV‐2 proteins
To explore whether reduced neutralisation was the result of diminished total immunoglobulin levels, we next determined absolute levels of total IgG, IgM and IgA in the peripheral blood of our patients and controls. We found that the three patient groups (GR, RR and NR) indeed showed significantly lower levels of IgG than healthy controls (Figure 2). By contrast, total IgM levels were comparable in patients and controls and total levels of IgA were lower only in the SOTR who had shown a relatively good neutralising antibody response (Figure 2).
Figure 2.
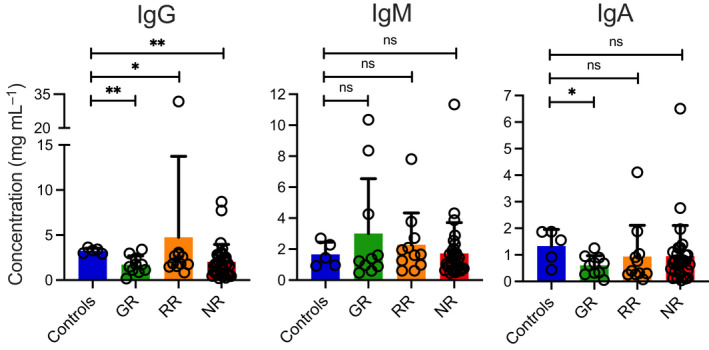
Absolute concentrations of immunoglobulins in the peripheral blood of SOT recipients. Absolute levels of IgG, IgM and IgA antibodies in our study subjects were measured after the second dose of the vaccine using a commercially available ELISA. Concentrations of total IgG, IgM and IgA are shown in ng mL–1 for healthy vaccinated controls and the three different groups of vaccinated SOT patients (Good Responders [GR], Reduced Responders [RR] and Non‐Responders [NR]) according to the degree of viral neutralisation after the second dose of the vaccine. Bars indicate means + SD. Differences between groups were analysed for statistical significance (*P < 0.05, **P < 0.01) using the Mann–Whitney U‐test.
Interestingly, when we determined titres of IgG antibodies against a variety of microbial antigens such as influenza A H1N1 nucleoprotein (Flu), tetanus toxoid (TT), cytomegalovirus glycoprotein B protein (CMV), Epstein–Barr virus glycoprotein gp350 protein (EBV) and herpes simplex virus type 1 gD protein (HSV), we found that NR patients showed even higher antibody titres against Flu, CMV and EBV. GR patients showed higher antibody levels against CMV than healthy controls (Figure 3).
Figure 3.
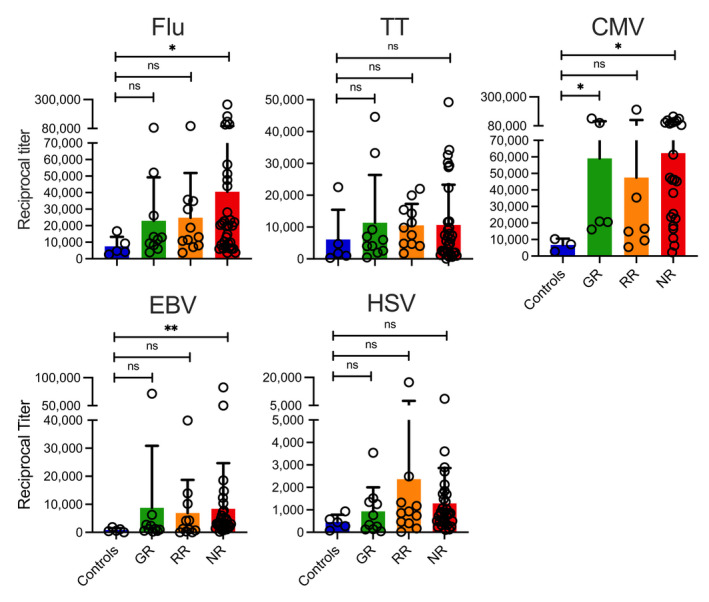
Titres of antibodies against different microbial antigens in the peripheral blood of SOT recipients. Titres of IgG antibodies against full‐length recombinant Influenza A nucleoprotein (Flu), tetanus toxoid (TT), cytomegalovirus (CMV), Epstein–Barr virus (EBV) and herpes simplex virus type 1 (HSV) were measured in an ELISA. Antibody titres are shown for healthy vaccinated controls and the three different groups of vaccinated SOT patients (Good Responders [GR], Reduced Responders [RR] and Non‐Responders [NR]) according to the degree of viral neutralisation after the second dose of the vaccine. Bars indicate means + SD. Differences between groups were analysed for statistical significance (*P < 0.05, **P < 0.01) using the Mann–Whitney U‐test.
Vaccine‐induced antibody titres against different SARS‐CoV‐2 proteins are suppressed in solid organ transplant recipients
Next, we measured IgG antibody titres against different SARS‐CoV‐2 proteins in SOTR post vaccination. We found that, compared to healthy controls, RR and NR patients, and in the case of the delta variant even GR patients, showed significantly lower vaccine‐induced antibody titres against the S1 and the RBD proteins as well as their ‘delta’ variants (Figure 4a). In addition, all SOTR showed lower vaccine‐induced IgA antibody titres against SARS‐CoV‐2 proteins S1, RBD and their respective delta variants except for the S2 protein where the GR patients showed antibody levels comparable to healthy donors (Figure 4b). None of the patients or controls showed IgG or IgA antibodies against the SARS‐CoV‐2 N protein (Figure 4a, b), indicating that all the antibodies detected were indeed vaccine‐induced and not based on a prior COVID‐19 infection.
Figure 4.
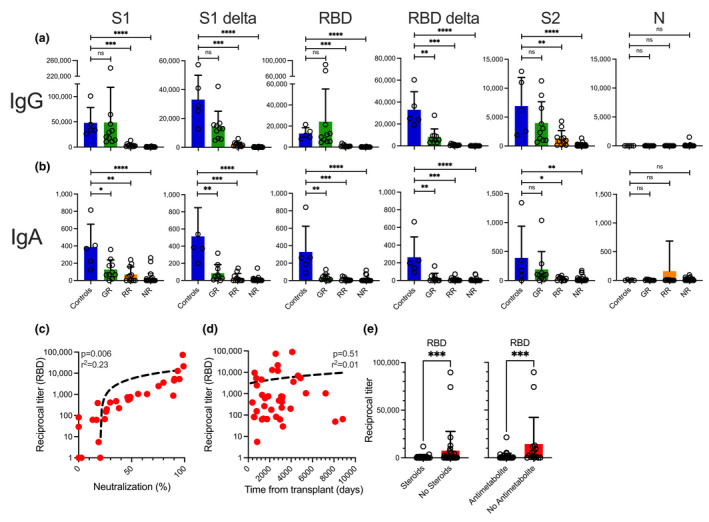
Titres and neutralising activity anti‐SARS‐CoV‐2 antibodies in SOT recipients after two doses of a COVID‐19 mRNA vaccine. Titres of (a) IgG and (b) IgA antibodies against different full‐length recombinant SARS‐CoV‐2 proteins and their delta variants were measured in an ELISA after two doses of a COVID‐19 mRNA vaccine. Antibody titres are shown for healthy vaccinated controls and the three different groups of vaccinated SOT patients (Good Responders [GR], Reduced Responders [RR] and Non‐Responders [NR]) according to the degree of viral neutralisation after the second dose of the vaccine. Correlation between anti‐RBD IgG antibody titres and (c) neutralising activity in the same sample and (d) time from SOT at the time of the first dose of the vaccine. (e) Impact of steroid or antimetabolite intake on anti‐RBD IgG antibody titres after two doses of the vaccine. Bars indicate means + SD. Differences between groups were analysed for statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001) using the Mann–Whitney U‐test.
Overall, these combined data already indicated an association between antibody titres and the neutralising activity of the patients’ serum. Accordingly, we were able to confirm a highly significant correlation between anti‐RBD antibody titres and the neutralising activity of the patients’ serum after two doses of the vaccine (Figure 4c). All patients with an anti‐RBD antibody > 4500 showed a viral neutralisation of > 90%. Analysing the impact of patient‐related characteristics on antibody titres, time from transplant did not have an effect (Figure 4d) titres but intake of steroids or antimetabolites did (Figure 4e).
Solid organ transplant recipients show a reduced breadth of SARS‐CoV‐2 vaccine‐induced antibody epitopes
Next, we aimed at identifying the most relevant, immunodominant target epitopes of S1 and RBD‐specific polyclonal IgG antibodies in vaccinated SOTR when compared to patients with active COVID‐19, vaccinated healthy controls (HC), HC before administration of the first dose of the vaccine and non‐vaccinated HC whose sera were collected before the COVID‐19 pandemic. When we used individual pools of peptides consisting of five 20mer peptides overlapping by 10 amino acids (aa) in an ELISA, we were able to identify regions in the complete S1 protein including its RBD domain that were preferentially targeted by the antiviral antibodies in patients with active COVID‐19. As described previously, 48 for anti‐S1 antibodies there were regions within the receptor‐binding motif (RBM) corresponding to peptide pool 46–50 (aa 451–510), the C‐terminal region of the RBD and the C‐terminal region of the S1 protein adjacent to the RBD that were preferentially targeted (Figure 5). Importantly, HC before administration of the first dose and non‐vaccinated HC did not show any measurable responses against any of the S1 protein peptide pools. Although we specifically selected SOTR with the comparably highest antibody titres against the full‐length S1 protein, vaccinated SOTR showed a dramatically reduced number of linear B‐cell epitopes with responses being restricted to a small region adjacent to the C‐terminal region of the RBD (Figure 5).
Figure 5.
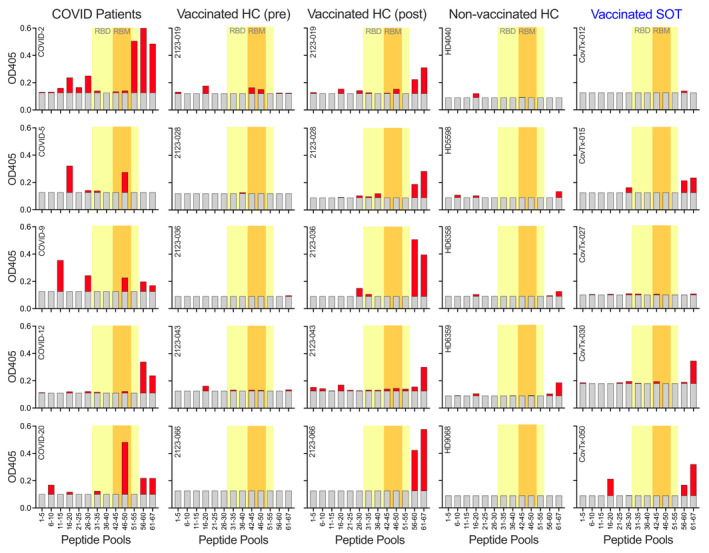
Peptide epitopes within the S1 protein targeted by vaccine‐induced antibodies. Plasma samples from five COVID‐19 patients, five vaccinated healthy controls after two doses of the vaccine, the same controls before receiving the first dose of the vaccine, five non‐vaccinated healthy controls and five SOT recipients with known anti‐SARS‐CoV‐2 reactivity after the second dose of the vaccine were analysed for immunodominant peptide epitopes. Peptide pools of 5 20mer peptides each overlapping by 10aa were used in an ELISA. Grey bars indicate background levels. RBD and RBM regions within the S1 protein are highlighted in yellow and orange, respectively.
In solid organ transplant recipients, an additional vaccine dose results in a dramatically improved antibody responses against all SARS‐CoV‐2 variants except omicron
Next, we analysed antibody responses induced by an additional vaccine dose in our SOTR. We found that an additional dose led to a highly significant increase in antibody titres against SARS‐CoV‐2 proteins RBD, S1 and S2 (Figure 6a). Importantly, the additional dose did not only lead to an increase in IgG antibody titres but also an enhanced neutralising activity of the polyclonal sera. Among the 32 SOTR who received an additional vaccine dose, numbers of GR (neutralising activity > 90%) increased from 6/32 (18.8%) after the second vaccine dose to 18/32 (56.3%) after the additional vaccine dose. The number of NR patients with no (< 30%) neutralising activity decreased from 15/32 (46.9%) to as few as 7/32 (21.9%) when the wild‐type RBD protein was used (Figure 6b). The improvement in neutralising activity was even more pronounced when we used the delta and ‘UK’ (alpha) variants of the RBD protein (Figure 6b). Numbers of GR for both variants increased from as few as 2/32 (0.6%) to 19/32 (59.4%) and numbers of NR patients decreased from 18/32 (56.3%) to 8/32 (25%) and from 21/32 (65.6%) to 7/32 (21.9%), respectively (Figure 6b). For the beta variant, there was not a single SOTR showing a neutralisation > 90% after just two vaccine doses. While the rate increased to 13/32 (40.6%) after the third dose, the proportion of ‘post‐third‐dose’ GRs was still substantially lower for the beta variant than for the aforementioned variants and the wild‐type RBD (Figure 6b). When we analysed patients not converting vs. converting from NR to RR/GR following an additional vaccine dose, we found no significant differences with regard to antimetabolite intake (5/7 vs. 7/8), or time between booster vaccination and blood sampling (37 [14–66] vs. 62 [18–186] days). Importantly, a preliminary analysis of anti‐S T‐cell responses in both patient groups indicated that most patients in both groups, SOTR with seroconversion vs. no seroconversion, evidenced anti‐SARS‐CoV‐2 CD4+ T cells after an additional dose of the vaccine (Supplementary figure 2).
Figure 6.

Effect of a third dose of an mRNA vaccine on anti‐SARS‐CoV‐2 antibodies and neutralising activity. (a) Titres of IgG antibodies against different full‐length recombinant SARS‐CoV‐2 proteins and their delta variants were measured in 32 SOT recipients using an ELISA before and after a third ‘booster’ dose of a COVID‐19 mRNA vaccine. (b, d) Neutralising activity before and after a third ‘booster’ dose of a COVID‐19 mRNA vaccine in the peripheral blood of the same SOT recipients. Green, orange and red areas indicate different degrees of inhibition (green, > 90%; orange, 30–89%; red, < 30%). (c) Titres of post‐booster IgG antibodies against the original anti‐SARS‐CoV‐2 RBD and S1 proteins vs. their omicron variants. Bars indicate median levels. Differences between groups were analysed for statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001) using the Mann–Whitney U‐test.
Finally, we compared binding of our patients’ post‐third‐dose anti‐RBD and anti‐S1 IgG antibodies to the respective wild‐type SARS‐CoV‐2 proteins vs. their omicron variants. We observed a highly significant decrease by 30% and 58% in median anti‐RBD and anti‐S1 antibody titres, respectively, when these were exposed to the omicron variants vs. the wild‐type proteins (Figure 6c). As a result, the third dose of the vaccine led to an improvement in the neutralising activity of our patients’ polyclonal sera; however, the degree of improvement was much less pronounced for the omicron variant than for all the other SARS‐CoV‐2 variants analysed (Figure 6d). For the omicron variant, there was not a single SOTR showing a neutralisation ≥ 30% after just two vaccine doses. While that rate increased to 8/32 (25%) after the third dose, the proportion of ‘post‐third‐dose’ RRs and GRs combined was still substantially lower than for all the aforementioned variants and the wild‐type RBD and only one single SOTR showed a neutralising activity of > 90% (Figure 6d).
Discussion
Here, we present the results of our prospective study investigating humoral immune responses to multiple SARS‐CoV‐2 viral variants following initial vaccination with COVID‐19 vaccines and following an additional vaccine dose in SOTR. Our analyses show that most SOTR do not respond adequately to the initial vaccine series. The majority of our vaccinated SOTR were ‘Non‐Responders’ with a viral neutralisation of < 30%. Less than 20% of SOTR showed a neutralising activity comparable to our healthy vaccinated controls. Few studies have assessed neutralising activity in SOTR after mRNA vaccination with one study demonstrating, in line with our results, a neutralising activity of > 30% in only 27% of all patients. 49 In conclusion, only a relatively small subgroup of SOTR is able to generate functionally relevant antibodies after two doses of a COVID‐19 mRNA vaccine.
When we asked whether the reduced immunoreactivity of our SOTR was an expression of a broadly suppressed humoral immunity, we found that absolute IgG levels in the patients’ peripheral blood at the time of the first dose of the vaccine were indeed lower than in healthy controls. However, titres of IgG antibodies against a variety of microbial antigens such as Influenza, CMV and EBV were even higher in the SOTR, especially the ones with a reduced anti‐SARS‐CoV‐2 response, than in healthy controls. These data indicate that humoral responses against recall antigens are not substantially impaired in SOTR but that the problem is a dysfunctional priming of antibody‐mediated immune response against novel antigens such as the S protein of SARS‐CoV‐2 that the patient has never encountered before, possibly based on impaired T cell help during priming and/or limited B cell expansion.
Analysing IgG antibody titres against different SARS‐CoV‐2 proteins in SOTR, we found that especially patients with a reduced anti‐SARS‐CoV‐2 neutralising activity showed significantly reduced vaccine‐induced antibody titres against the S1 and the RBD proteins of the virus including their delta variants. Importantly, anti‐SARS‐CoV‐2 antibody titres correlated significantly with the neutralising activity of the patients’ serum after two doses of the vaccine and all patients with an anti‐RBD antibody > 4500 showed a viral neutralisation of > 90%, supporting the functional relevance of measuring antibody titres in vaccinated patients, especially those with an impaired humoral immune response.
Out of a large number of clinical variables, we only found the overall intensity of the immunosuppressive treatment to be associated with a reduced vaccine‐induced SARS‐CoV‐2 neutralising activity and reduced levels of anti‐RBD antibodies. Specifically, treatment with antimetabolites had the most significant negative impact on the patients’ antiviral humoral immunity. This finding is in agreement with recent studies by other groups showing that SOTR on treatment with antimetabolites exhibit a reduced response to two doses of mRNA‐based SARS‐CoV‐2 vaccines. 48 , 50 , 51 , 52 , 53 Interestingly, it has previously been shown that SOTR show suboptimal responses to seasonal influenza vaccination with seroconversion rates as low as 34% and lower seroconversion rates were associated with high doses of MMF and lower numbers of postvaccine H1N1‐specific IL‐4+CD4+ T cells. 54
Mycophenolic acid (MPA) is a selective, non‐competitive and reversible inhibitor of inosine‐50‐monophosphate dehydrogenase (IMPDH). MPA inhibits the production of guanosine and deoxyguanosine nucleotides leading to a reduced proliferation of T and B lymphocytes and impaired production of immunoglobulins through the guanosine and deoxyguanosine nucleotides in depleted lymphocytes. 55 Mycophenolate mofetil (MMF) is a prodrug of MPA and suppresses T‐cell responses to allogeneic cells and other antigens. Importantly, in vivo the drug also suppresses primary, but not secondary, antibody responses. 56 In one study, patients with systemic lupus erythematosus (SLE) taking MMF showed lower numbers of plasmablasts, 57 a finding that was later confirmed by two different groups. 58 , 59 Interestingly, another study investigating B‐cell responses to the SARS‐CoV‐2 BNT162b2 vaccine in kidney transplant recipients also observed diminished seroconversion rates correlating with a reduced generation of plasmablasts and memory B cells. 40 Therefore, future studies should evaluate in detail the effect of MMF intake on B‐cell and T‐cell phenotype and function after COVID‐19 vaccination to help improve immune responses in SOTR.
Even though we specifically selected SOTR with the comparably highest anti‐S1 antibody titres, vaccinated SOTR showed a dramatically reduced number of linear B‐cell epitopes, indicating an overall reduced functional B‐cell diversity in these patients. Vaccine‐induced antibody responses against linear peptides in SOTR were primarily directed against a small region adjacent to the C‐terminal region of the RBD, a region that we and others have previously described as immunodominant. 48 , 60 Future studies should investigate whether the reduced breadth of the humoral immune response in SOTR further contributes to the immune escape of the SARS‐CoV‐2 virus in addition to lower overall antibody titres.
Next, we analysed whether an additional dose of SARS‐CoV‐2 vaccine in our SOTR could improve humoral responses observed after completion of the initial vaccine series. Importantly, we found that an additional dose not only led to a marked increase in antibody titres against the different SARS‐CoV‐2 proteins but also enhanced neutralising activity of the polyclonal sera. Remarkably, the increase in neutralising activity after the additional vaccine dose was even more pronounced for the delta and the alpha/UK variants of the RBD protein. By contrast, the beta variant, which shares immunorelevant mutations with the omicron variant, showed less improvement when compared to the aforementioned variants. Importantly, our preliminary analysis of antiviral T‐cell responses indicated that even patients without seroconversion after the additional dose of the vaccine are capable of mounting T‐cell responses against SARS‐CoV‐2.
The omicron (B.1.1.529) variant of SARS‐CoV‐2 was only very recently detected in South Africa, and due to enhanced transmissibility, it has rapidly spread and has become the dominant variant in many countries, including the United States. 61 A hallmark of the omicron variant is the large number of S protein mutations potentially causing immune escape even in fully vaccinated healthy individuals. 43 Here, we compared binding of our patients’ anti‐RBD and anti‐S1 IgG antibodies following an additional vaccine dose to the respective wild‐type SARS‐CoV‐2 proteins vs. their omicron variants. We observed a highly significant decrease in median anti‐RBD and anti‐S1 antibody titres, respectively, when these were exposed to the omicron variants. In addition, the post‐third‐dose improvement in neutralising activity was much less pronounced than for all the other variants and antibody responses as well as their neutralising activity remained suboptimal.
Limitations of our study include the limited number of patients enrolled, a certain degree of heterogeneity with regard to types of transplants and other patient characteristics, as well as the lack of data on serum levels of immunosuppressive drugs. Nevertheless, we think that findings presented herein have the potential to improve currently available prophylactic approaches for SOTR, for example, the use of additional vaccine doses or other types of pre‐ or post‐exposure prophylaxis, all of which may be particularly important given the recent occurrence of immune escape variants of SARS‐CoV‐2. In this context, we would like to stress that multi‐mutational variants of the virus are much more likely to arise in immunocompromised patients with a prolonged course of the COVID‐19 infection. 62 This is another important reason to find ways to prevent these individuals from becoming infected or at least limit the duration of viral persistence in case of an active COVID‐19 infection.
Methods
Study population and design
We performed a prospective cohort study of SOTR 18 years of age or older who received post‐transplant care at the University of Maryland Medical Center (UMMC) and had either received or were scheduled to receive any of the three SARS‐CoV‐2 vaccines approved by the FDA under emergency use authorisation. Those who were already vaccinated must have received their initial vaccine dose within approximately 90 days of study enrolment. SARS‐CoV‐2 vaccination was not provided as part of this study protocol. Patients were excluded if they were HIV positive, were receiving chemotherapy or radiation therapy, or previously had SARS‐CoV‐2 infection. Demographic and clinical data were collected from study participants and through medical chart abstractions. Up to 50 mL of heparinised blood was collected at the following time points (see Supplementary figure 1): visit 1 (within 7 days prior, up to 48 h after first vaccine dose), visit 2 (± 7 days), visit 3 (at 3 months [± 10 days] after 1st vaccine dose), visit 4 (at 6 months [± 14 days] after 1st vaccine dose), visit 5 (at 9 months [± 14 days] after 1st vaccine dose) and visit 6 (at 12 months [± 14 days] after 1st vaccine dose). All patients who received an mRNA COVID‐19 vaccine received the second dose 3–4 weeks after the first dose. For the post‐third‐dose analysis, samples were chosen from the available samples at least two weeks after the third dose was administered. Plasma was generated from peripheral blood samples after centrifugation at 400 g for 10 min and frozen immediately at −70°C. Peripheral blood mononuclear cells (PBMCs) were isolated using lymphocyte separation density gradient and immediately frozen in liquid nitrogen. This study was reviewed and approved by the University of Maryland (Baltimore) institutional review board (HP‐00095043).
For the epitope screening, samples from patients with an active COVID‐19 infection were used. These samples were collected as part of our prospective observational study enrolling COVID‐19 patients who were admitted to the University of Maryland Medical Center between June and August of 2020. For that study, informed consent was obtained, and blood samples were collected under IRB HP‐00091425. Samples from vaccinated healthy controls were collected as part of our prospective clinical study on immune responses to two doses of a COVID‐19 mRNA vaccine in cancer patients and healthy controls (IRB HP‐00095016).
Measurement of absolute immunoglobulin levels
Absolute serum concentrations of the different immunoglobulins were measured using Human IgG, IgM and IgA Enzyme‐linked Immunosorbent Assay (ELISA) Kits (Invitrogen, Cat. No. BMS2091, BMS2098, BMS2096) as per the manufacturer’s instructions. Absorbance was read at 450 nm with a reference wavelength of 620 nm in a microtitre plate reader (Tecan, Morrisville, NC).
Analysis of SARS‐CoV‐2‐specific antibodies
Serum antibody responses against recombinant, full‐length SARS‐CoV‐2 proteins (Supplementary table 2), viral control proteins (Supplementary table 1) or overlapping peptides covering the complete amino acid sequence of the SARS‐CoV‐2 S1 protein were determined by ELISA as previously described. 48 , 63 , 64 Briefly, high‐binding ELISA plates (Thermo Fisher, Cat. No. 44‐2404‐21) were coated with 5 µg mL–1 of the respective proteins in PBS (Gibco, Cat. No. 10010‐023) overnight at 4°C. The next day, plates were washed twice with PBS and twice with 0.1% PBS‐T (VWR, Cat. No. M147‐1L). Plates were then blocked with 5% non‐fat dry milk (Santa Cruz, Cat. No. sc2325) in PBS (MPBS) for 1 h at room temperature (RT) and then washed again as described above. Serum was diluted 1:40 for screening assays and for titration 1:100/1:400/1:1600/1:6400 and if necessary 1:25 000 and 1:100 000 in MPBS. Diluted sera were added to plates and incubated for 3H at RT. Plates were washed as described above before incubation with secondary antibodies against pan‐human IgG (Southern Biotech, Cat. No. 2040‐04) or IgA (Southern Biotech, Cat. No. 2050‐04). Secondary antibodies were diluted according to the manufacturers’ instructions and plates incubated for 1 h at RT. Plates were then washed as described above, PNPP tablets (Southern Biotech, Cat. No. 0201‐01) dissolved in diethanolamine (Thermo, Cat. No. 34064) and PNPP substrate solution added to each well for 10 min in the dark. 15 µL of 3N NaOH (VWR, Cat. No. BDH7472‐1) stop solution was added to each well, and absorbance was read at 405 nm with a reference wavelength of 620 nm in a microtitre plate reader (Tecan). Endpoint titres were calculated using serum titration curves for positive samples and pooled sera of 5 healthy donors. For non‐SARS‐CoV‐2 antigens, serum dilutions for anti‐GST (glutathione‐S‐transferase) antibodies (Supplementary table 1) were used as a negative control.
For peptide ELISAs, plates were first coated with 5 µg mL–1 neutravidin (Thermo, Cat. No. 31000) overnight at 4°C and then blocked with 2% bovine serum albumin (BSA; Thermo, Cat. No. 9048‐46‐8) in PBS for 1 h at RT. Plates were then incubated for 1 h at RT with either 1 µg mL–1 of the individual peptides or 5 µg mL–1 equimolar peptide pools in PBS as indicated. Plates were washed and then developed with serum at a dilution of 1:40 and with secondary reagents as described above using 2% BSA instead of M‐PBS.
SARS‐CoV‐2 neutralisation assay
Neutralising activity of patient sera was assessed using the cPass Neutralization Antibody Detection Kit (GenScript, Cat. No. L00847‐A), which is a surrogate test detecting circulating neutralising antibodies against SARS‐CoV‐2 that block the interaction between the receptor‐binding domain (RBD) of the viral spike glycoprotein with the ACE2 cell surface receptor. Briefly, samples and controls were diluted with sample dilution buffer and pre‐incubated with the Horseradish peroxidase (HRP) conjugated recombinant SARS‐CoV‐2 RBD fragment (HRP‐RBD) or one of its variants listed in Supplementary table 2 to allow the binding of the circulating neutralisation antibodies to HRP‐RBD. The mixture was then added to the capture plate, which was pre‐coated with the hACE2 protein. The unbound HRP‐RBD as well as any HRP‐RBD bound to non‐neutralising antibody was captured on the plate, while the circulating neutralisation antibodies HRP‐RBD complexes remained in the supernatant and were removed during washing. Following a wash cycle, TMB substrate solution was added followed by the Stop Solution. The absorbance of the final solution was read at 450 nm in a microtitre plate reader (Tecan).
Analysis of SARS‐CoV‐2‐specific T cells
T cells specific for SARS‐CoV‐2 were identified using different versions of the SARS‐CoV‐2 T Cell Analysis Kit (Miltenyi Biotec) and pools of lyophilised peptides, consisting mainly of 15‐mer sequences with 11 amino acids (aa) overlap, covering the complete sequence of the S protein (‘S complete’) or sequence domains 1‐692 (‘S1’) or aa 689‐895 (‘S plus’). For the analysis of T‐cell responses against the omicron variant of the SARS‐CoV‐2 Spike Glycoprotein, a peptide pool consisting of 315 peptides (15mers with 11 aa overlap) covering the entire sequence of the S protein was used (GenScript, cat. no. RP30121). For the analysis of T‐cell responses against microbial antigens other than SARS‐CoV‐2, a pool of 32 MHC class I‐specific peptides of 8–12 aa in length derived from cytomegalovirus (HCMV), Epstein–Barr virus (EBV) and influenza virus was used (CEF MHC Class I Plus, Miltenyi Biotec, Cat no. 130‐098‐426). Briefly, PBMC were thawed, plated cells at a density of 5 × 106 mL–1 in fresh cell culture medium in a 24‐well cell culture plate at 37°C and 5% CO₂ overnight. The next morning, cells were resuspended in culture medium at a density of 1 × 107 viable cells per mL and 100 µL of cell suspension per well was plated in a flat‐bottom 96‐well plate resulting in a total number of 1 × 106 cells per well. For antigen stimulation, 2 µL of peptide stock solution was added to the respective wells and mixed by pipetting up and down. As a positive control, 2 µL of the CytoStim crosslinking agent was used, and as a negative control, 2 µL of sterile water/10% DMSO solution was added to the respective wells. Cells were incubated at 37°C and 5% CO₂ for 2 h, 2 µL of Brefeldin A was added to each well, and cells were incubated at 37°C and 5% CO₂ for an additional 4 h. Cells were then resuspended in 100 µL reconstituted Viobility 405/452 Fixable Dye master mix and incubated for 10 min at room temperature. Fixation was performed by adding 100 µL Inside Fix to each well followed by incubation for 20 min at room temperature. The supernatant was removed, and permeabilisation was performed by adding 100 µL Inside Perm to each well. The supernatant was removed, and 100 µL of antibody staining mix was added to each well followed by incubation for 10 min at room temperature. Cells were washed and analysed by flow cytometry using a MACSQuant® Analyzer 10 (Miltenyi Biotec).
Statistical analyses
Statistical analyses for serological analyses were performed using GraphPad Prism 9 software (GraphPad Software, San Diego, CA). Groups were compared using the Mann–Whitney U‐test and paired analyses were performed using the Wilcoxon signed‐rank test. Correlations were calculated using the Pearson correlation coefficient. For the analysis of clinical characteristics, groups were compared using a Student’s t‐test.
Conflict of interest
The authors declare no conflict of interest.
Authors’ contributions
KKS, JSH, SVN, SD and APR designed the study, collected patient samples, analysed the data and wrote the manuscript. KS collected patient samples, analysed the data and wrote the manuscript. YJY, PL, HDM, AC, WYX, AC and JA collected patient samples. MM and AA collected patient data. TI, XF, DO and SVA performed experiments and wrote the manuscript. TL, NMH, EVM, KH, JB and OG analysed the data and wrote the manuscript. DA designed the study, performed experiments, analysed the data, made figures and wrote the manuscript.
Supporting information
Supplementary figure 1
Supplementary figure 2
Supplementary table 1
Supplementary table 2
Acknowledgments
We thank Natalie McNally for coordinating patient enrolment and Marion Williams for sample collection. This study was funded by two grants from the Kahlert Foundation. In addition, the study was supported in part by funds from the Division of Infectious Diseases and the Transplantation Program.
REFERENCES
- 1. Zhu NA, Zhang D, Wang W et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382: 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou P, Yang X‐L, Wang X‐G et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020; 579: 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chu H, Chan J‐W, Wang Y et al. Comparative replication and immune activation profiles of SARS‐CoV‐2 and SARS‐CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID‐19. Clin Infect Dis 2020; 71: 1400–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ou X, Liu Y, Lei X et al. Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nature Commun 2020; 11: 1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ge X‐Y, Li J‐L, Yang X‐L et al. Isolation and characterization of a bat SARS‐like coronavirus that uses the ACE2 receptor. Nature 2013; 503: 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Izcovich A, Ragusa MA, Tortosa F et al. Prognostic factors for severity and mortality in patients infected with COVID‐19: a systematic review. PLoS One 2020; 15: e0241955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Q, Guan X, Wu P et al. Early transmission dynamics in Wuhan, China, of novel coronavirus‐infected pneumonia. N Engl J Med 2020; 382: 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang C, Wang Y, Li X et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen R, Liang W, Jiang M et al. Risk factors of fatal outcome in hospitalized subjects with coronavirus disease 2019 from a nationwide analysis in China. Chest 2020; 158: 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guan W‐J, Liang W‐H, Zhao YI et al. Comorbidity and its impact on 1590 patients with Covid‐19 in China: a nationwide analysis. Eur Respir J 2020; 55: 2000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nair V, Jandovitz N, Hirsch JS et al. An early experience on the effect of solid organ transplant status on hospitalized COVID‐19 patients. Am J Transplant 2021; 21: 2522–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pereira MR, Mohan S, Cohen DJ et al. COVID‐19 in solid organ transplant recipients: initial report from the US epicenter. Am J Transplant 2020; 20: 1800–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roberts MB, Izzy S, Tahir Z, Al Jarrah A, Fishman JA, El Khoury J. COVID‐19 in solid organ transplant recipients: dynamics of disease progression and inflammatory markers in ICU and non‐ICU admitted patients. Transpl Infect Dis 2020; 22: e13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun J, Patel RC, Zheng Q et al. COVID‐19 disease severity among people with HIV infection or solid organ transplant in the United States: a nationally‐representative, multicenter observational cohort study. Medrxiv 2021. 10.1101/2021.07.26.21261028 [DOI] [Google Scholar]
- 15. Trapani S, Masiero L, Puoti F et al. Incidence and outcome of SARS‐CoV‐2 infection on solid organ transplantation recipients: a nationwide population‐based study. Am J Transplant 2021; 21: 2509–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ravanan R, Callaghan CJ, Mumford L et al. SARS‐CoV‐2 infection and early mortality of waitlisted and solid organ transplant recipients in England: a national cohort study. Am J Transplant 2020; 20: 3008–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Caillard S, Chavarot N, Francois H et al. Is COVID‐19 infection more severe in kidney transplant recipients? Am J Transplant 2021; 21: 1295–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caillard S, Anglicheau D, Matignon M et al. An initial report from the French SOT COVID Registry suggests high mortality due to COVID‐19 in recipients of kidney transplants. Kidney Int 2020; 98: 1549–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kates OS, Haydel BM, Florman SS et al. Coronavirus disease 2019 in solid organ transplant: a multicenter cohort study. Clin Infect Dis 2021; 73: e4090–e4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salto‐Alejandre S, Jiménez‐Jorge S, Sabé N et al. Risk factors for unfavorable outcome and impact of early post‐transplant infection in solid organ recipients with COVID‐19: a prospective multicenter cohort study. PLoS One 2021; 16: e0250796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bergwerk M, Gonen T, Lustig Y et al. Covid‐19 breakthrough infections in vaccinated health care workers. N Engl J Med 2021; 385: 1474–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lucas C, Klein J, Sundaram ME et al. Delayed production of neutralizing antibodies correlates with fatal COVID‐19. Nature Med 2021; 27: 1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aries JA, Davies JK, Auer RL et al. Clinical outcome of coronavirus disease 2019 in haemato‐oncology patients. Br J Haematol 2020; 190: e64–e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cattaneo C, Daffini R, Pagani C et al. Clinical characteristics and risk factors for mortality in hematologic patients affected by COVID‐19. Cancer 2020; 126: 5069–5076. [DOI] [PubMed] [Google Scholar]
- 25. Chari A, Samur MK, Martinez‐Lopez J et al. Clinical features associated with COVID‐19 outcome in multiple myeloma: first results from the International Myeloma Society data set. Blood 2020; 136: 3033–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Caballero‐Marcos A, Salcedo M, Alonso‐Fernández R et al. Changes in humoral immune response after SARS‐CoV‐2 infection in liver transplant recipients compared to immunocompetent patients. Am J Transplant 2021; 21: 2876–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burack D, Pereira MR, Tsapepas DS et al. Prevalence and predictors of SARS‐CoV‐2 antibodies among solid organ transplant recipients with confirmed infection. Am J Transplant 2021; 21: 2254–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. FDA . 12/11/2020. Available from: https://www.fda.gov/emergency‐preparedness‐and‐response/coronavirus‐disease‐2019‐covid‐19/pfizer‐biontech‐covid‐19‐vaccine
- 29. FDA . 12/18/2020. Available from: https://www.fda.gov/news‐events/press‐announcements/fda‐takes‐additional‐actionfight‐against‐covid‐19‐issuing‐emergency‐use‐authorization‐second‐covid
- 30. Walsh EE, Frenck RW, Falsey AR et al. Safety and immunogenicity of two RNA‐based Covid‐19 vaccine candidates. N Engl J Med 2020; 383: 2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Polack FP, Thomas SJ, Kitchin N et al. Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine. N Engl J Med 2020; 383: 2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anderson EJ, Rouphael NG, Widge AT et al. Safety and immunogenicity of SARS‐CoV‐2 mRNA‐1273 vaccine in older adults. N Engl J Med 2020; 383: 2427–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jackson LA, Anderson EJ, Rouphael NG et al. An mRNA vaccine against SARS‐CoV‐2 ‐ preliminary report. N Engl J Med 2020; 383: 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Widge AT, Rouphael NG, Jackson LA et al. Durability of responses after SARS‐CoV‐2 mRNA‐1273 vaccination. N Engl J Med 2021; 384: 80–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stankov MV, Cossmann A, Bonifacius A et al. Humoral and cellular immune responses against SARS‐CoV‐2 variants and human coronaviruses after single BNT162b2 vaccination. Clin Infect Dis 2021; 73: 2000–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wisnewski AV, Campillo Luna J, Redlich CA. Human IgG and IgA responses to COVID‐19 mRNA vaccines. PLoS One 2021; 16: e0249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marion O, Del Bello A, Abravanel F et al. Safety and immunogenicity of anti‐SARS‐CoV‐2 messenger RNA vaccines in recipients of solid organ transplants. Ann Intern Med 2021; 174: 1336–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mazzola A, Todesco E, Drouin S et al. Poor antibody response after two doses of SARS‐CoV‐2 vaccine in transplant recipients. Clin Infect Dis 2022; 74: 1093–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Korth J, Jahn M, Dorsch O et al. Impaired humoral response in renal transplant recipients to SARS‐CoV‐2 vaccination with BNT162b2 (Pfizer‐BioNTech). Viruses 2021; 13: 756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rincon‐Arevalo H, Choi M, Stefanski A‐L et al. Impaired humoral immunity to SARS‐CoV‐2 BNT162b2 vaccine in kidney transplant recipients and dialysis patients. Sci Immunol 2021; 6: eabj1031. [DOI] [PubMed] [Google Scholar]
- 41. Twohig KA, Nyberg T, Zaidi A et al. Hospital admission and emergency care attendance risk for SARS‐CoV‐2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect Dis 2022; 22: 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lopez Bernal J, Andrews N, Gower C et al. Effectiveness of Covid‐19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med 2021; 385: 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu L, Iketani S, Guo Y et al. Striking Antibody Evasion Manifested by the Omicron Variant of SARS‐CoV‐2. bioRxiv 2021. 2021.2012.2014.472719. [DOI] [PubMed] [Google Scholar]
- 44. Abedon AT, Teles MS, Alejo JL et al. Improved antibody response after a fifth dose of a SARS‐CoV‐2 vaccine in solid organ transplant recipients. Transplantation 2022. 10.1097/TP.0000000000004092. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kamar N, Abravanel F, Marion O, Couat C, Izopet J, Del Bello A. Three doses of an mRNA Covid‐19 vaccine in solid‐organ transplant recipients. N Engl J Med 2021; 385: 661–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hall VG, Ferreira VH, Ku T et al. Randomized trial of a third dose of mRNA‐1273 vaccine in transplant recipients. N Engl J Med 2021; 385: 1244–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Williams WW, Ingelfinger JR. Third time's a Charm ‐ Covid‐19 vaccine hope for solid‐organ transplant recipients. N Engl J Med 2021; 385: 1233–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Atanackovic D, Avila SV, Lutfi F et al. Deep dissection of the antiviral immune profile of patients with COVID‐19. Commun Biol 2021; 4: 1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hall VG, Ferreira VH, Ierullo M et al. Humoral and cellular immune response and safety of two‐dose SARS‐CoV‐2 mRNA‐1273 vaccine in solid organ transplant recipients. Am J Transplant 2021; 21: 3980–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rashidi‐Alavijeh J, Frey A, Passenberg M et al. Humoral response to SARS‐Cov‐2 vaccination in liver transplant recipients‐A single‐center experience. Vaccines 2021; 9: 738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Russo G, Lai Q, Poli L et al. SARS‐COV‐2 vaccination with BNT162B2 in renal transplant patients: Risk factors for impaired response and immunological implications. Clin Transplant 2021; 36: e14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Del Bello A, Abravanel F, Marion O et al. Efficiency of a boost with a third dose of anti‐SARS‐CoV‐2 messenger RNA‐based vaccines in solid organ transplant recipients. Am J Transplant 2022; 22: 322–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marinaki S, Adamopoulos S, Degiannis D et al. Immunogenicity of SARS‐CoV‐2 BNT162b2 vaccine in solid organ transplant recipients. Am J Transplant 2021; 21: 2913–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Egli A, Humar A, Widmer LA et al. Effect of immunosuppression on T‐helper 2 and B‐cell responses to influenza vaccination. J Infect Dis 2015; 212: 137–146. [DOI] [PubMed] [Google Scholar]
- 55. Tseng HT, Wu XC, Huang CY, Shih CM, Lin YW, Lin FY. The impact of SARS‐CoV‐2 infection, and application of immunosuppressive agents in kidney transplant recipients suffering from COVID‐19. Pharmaceuticals 2021; 14: 1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Allison AC, Eugui EM. Mycophenolate mofetil and its mechanisms of action. Immunopharmacology 2000; 47: 85–118. [DOI] [PubMed] [Google Scholar]
- 57. Eickenberg S, Mickholz E, Jung E, Nofer JR, Pavenstadt HJ, Jacobi AM. Mycophenolic acid counteracts B cell proliferation and plasmablast formation in patients with systemic lupus erythematosus. Arthritis Res Ther 2012; 14: R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Slight‐Webb S, Guthridge JM, Chakravarty EF et al. Mycophenolate mofetil reduces STAT3 phosphorylation in systemic lupus erythematosus patients. JCI Insight 2019; 4: e124575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fassbinder T, Saunders U, Mickholz E et al. Differential effects of cyclophosphamide and mycophenolate mofetil on cellular and serological parameters in patients with systemic lupus erythematosus. Arthritis Res Ther 2015; 17: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hansen CB, Jarlhelt I, Pérez‐Alós L et al. SARS‐CoV‐2 antibody responses are correlated to disease severity in COVID‐19 convalescent individuals. J Immunol 2021; 206: 109–117. [DOI] [PubMed] [Google Scholar]
- 61. Del Rio C, Omer SB, Malani PN. Winter of omicron‐the evolving COVID‐19 pandemic. JAMA 2022; 327: 319–320. [DOI] [PubMed] [Google Scholar]
- 62. Corey L, Beyrer C, Cohen MS, Michael NL, Bedford T, Rolland M. SARS‐CoV‐2 variants in patients with immunosuppression. N Engl J Med 2021; 385: 562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Atanackovic D, Luetkens T, Avila SV et al. Anti‐SARS‐CoV‐2 immune responses in patients receiving an allogeneic stem cell or organ transplant. Vaccines 2021; 9: 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Luetkens T, Metcalf R, Planelles V et al. Successful transfer of anti‐SARS‐CoV‐2 immunity using convalescent plasma in an MM patient with hypogammaglobulinemia and COVID‐19. Blood Adv 2020; 4: 4864–4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1
Supplementary figure 2
Supplementary table 1
Supplementary table 2


