Abstract
Membrane contact sites are specialized areas where the membranes of two distinct organelles are physically connected and allow for the exchange of molecules and for signaling processes. Understanding the mechanisms whereby proteins localize to and function in these structures is of special interest; however, methods allowing for reconstitution of these contact sites are few and only based on synthetic membranes and recombinant proteins. Here, we devised a strategy to create in situ artificial contact sites between synthetic and endogenous organelle membranes. Liposomes functionalized with a peptide containing a two phenylalanines in an acidic tract (FFAT) motif were added to adherent cells whose plasma membrane was perforated. Confocal and super-resolution microscopy revealed that these liposomes associated with the endoplasmic reticulum via the specific interaction of the FFAT motif with endoplasmic reticulum–resident vesicle-associated membrane protein–associated proteins. This approach allowed for quantification of the attachment properties of peptides corresponding to FFAT motifs derived from distinct proteins and of a protein construct derived from steroidogenic acute regulatory protein–related lipid transfer domain-3. Collectively, these data indicate that the creation of in situ artificial contact sites represents an efficient approach for studying the membrane-tethering activity of proteins and for designing membrane contact site reconstitution assays in cellular contexts.
Keywords: cell biology, liposome, confocal microscopy, endoplasmic reticulum, lipid transport, membrane contact sites, FFAT motif, VAP, STARD3, OSBP
Abbreviations: ATTO647N-PE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-ATTO647N; convFFAT, conventional FFAT; DIC, differential interference contrast; DLS, dynamic light scattering; DMEM, Dulbecco's modified Eagle’s medium; DOGS–NTA–Ni2+, 1,2-di-(9Z-octadecenoyl)-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl]–nitrilotriacetic acid (nickel salt); DOPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; ER, endoplasmic reticulum; FFAT, two phenylalanines in an acidic tract; GST, glutathione-S-transferase; ISACS, in situ artificial contact sites; L/P, lipid-to-protein; MCS, membrane contact sites; MPB–PE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[4-(p-maleimidophenyl) butyramide]; MSP, major sperm protein; OSBP, oxysterol-binding protein; PD, polydispersity; PM, plasma membrane; Rhod–PE, rhodamine–1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; RH, hydrodynamic radius; StAR, steroidogenic acute regulatory protein; STARD, steroidogenic acute regulatory protein–related lipid transfer domain; STED, stimulated emission depletion; TN, Tris and NaCl; VAP, vesicle-associated membrane protein–associated protein
Membrane contact sites (MCS) correspond to subcellular sites in which two organelle membranes are in close apposition, only separated by a distance of 10 to 80 nm (1, 2, 3). These MCS are active regions of lipid and ion exchange, signal transmission, as well as anchoring points where membrane remodeling activities, such as organelle biogenesis, positioning, fission, or autophagy, are controlled. A variety of tether proteins, such as vesicle-associated membrane protein–associated proteins (VAPs) (4), nonfusogenic SNARE-like syntaxin1 (Stx1) and Sec22b (5), junctophilins (6), or annexin A1 (7), have been identified to be associated with MCS. They usually partner with other factors to connect two organelles. Amongst these tether proteins, VAPs are certainly the most prominent ones and contribute to the fact that the endoplasmic reticulum (ER), which constitutes a membrane network spreading throughout the cell, is highly engaged in contact sites with all organelles as well as the plasma membrane (PM). These proteins are evolutionary conserved (with VAP-A and VAP-B expressed in human (8) and Scs2p and Scs22p expressed in yeast (9, 10)) and consist of an N-terminal major sperm protein (MSP) domain, a central coiled-coil region, and a C-terminal transmembrane region that is anchored in the ER membrane (4, 8, 11). The MSP domain is poised with the ability to recruit different protein partners by recognizing a stretch of amino acids referred to as two phenylalanines in an acidic tract (FFAT) motif, with a core sequence EFFDAxE (using the single-letter amino acid code, where x is any amino acid) and flanking regions rich in acidic residues D and E. This motif, later called conventional FFAT (convFFAT) motif, is notably present in proteins such as oxysterol-binding protein (OSBP) and many related homologs in human and yeast (10, 12, 13), steroidogenic acute regulatory protein (StAR)–related lipid transfer domain 11 (STARD11)/ceramide transfer protein (CERT) (14), and Nir2/3 (15)). Recently, we established that the sterol transporter STARD3, which is anchored to the limiting membrane of late endosomes (16), binds to VAP owing to an unconventional FFAT motif named phospho-FFAT (17, 18). The phosphorylation of this motif controls the association of STARD3 with VAP and its sterol-transfer activity. In addition to their critical function in organizing lipid fluxes through MCS (18, 19, 20), VAPs contribute to other cellular functions, such as endosome positioning, Ca2+ homeostasis, autophagosome biogenesis, or ion channel clustering (21, 22), by recruiting diverse types of proteins with FFAT motifs. In total, convFFAT and nonconventional FFAT motifs have been identified or predicted in more than 100 proteins (17, 23, 24); for many of these, whether they interact with VAPs and play a functional role in MCS remain to be addressed.
In vitro approaches using minimal reconstitution systems have been instrumental to dissect the function of proteins at MCS, and in particular, how VAPs and lipid transfer proteins work together. Notably, spectroscopy approaches combined with the use of liposomes and recombinant proteins made possible to establish how OSBP associates not only with VAP but also with Golgi determinants to create ER–Golgi contacts and exchange lipids between these two organelles (25). Using similar approaches, we found how STARD3 forms ER–endosome contact sites in a VAP-dependent manner and can therefore transfer cholesterol between these two compartments (18). Likewise, the mode of action of other lipid-transfer proteins, which do not rely on VAPs, such as E-synaptotagmins/tricalbins or autophagy-related protein 2, has been established in vitro (26, 27, 28, 29, 30). Although these approaches are instructive as they are based on perfectly controlled components, they have limits as they cannot faithfully mimic what occurs in cellular contexts. Here, we designed a new strategy to generate in situ artificial contact sites (ISACS) between synthetic and organelle membranes under more physiological conditions. This approach relies on a perforation protocol, which gives access to the cytoplasmic content of cells in culture (31, 32, 33), and on the use of functionalized liposomes. As a proof of concept, we examined whether chemically defined liposomes decorated with peptides containing either FFAT motifs of STARD3 and OSBP, or the cytosolic part of STARD3, formed contact sites with the ER. Using confocal and stimulated emission depletion (STED) microscopy combined with semi-automatized image analyses, we showed that these liposomes were recruited to the ER through specific interactions with VAPs. Moreover, this approach allowed quantitative measurement of tethering activities at contact sites. Thus, our approach to create ISACS constitutes a relevant strategy to measure tethering activities between synthetic and cellular membranes and might be useful to devise novel functional assays.
Results
Liposomes bearing the FFAT motif of STARD3 associate with the ER of opened cells
Our aim was to examine whether artificial contact sites could be generated in the context of a whole cell by exposing the ER to functionalized liposomes mimicking a second organelle. The strategy was based on a perforation method that gives access to the cytoplasmic content of the cells (31). We selected the STARD3–VAP complex as a model tethering machinery to study contacts in this hybrid setting. STARD3 comprises a transmembrane domain composed of four transmembrane helices, anchored to the limiting membrane of endosomes followed by a cytosolic part constituted by a linker and a sterol-transfer domain. The first part of the linker (20 amino acids) contains a nonconventional FFAT motif, which represents the minimal interacting region of this protein with the MSP domain of VAPs. Our recent investigations have defined this motif as a new functional FFAT motif called phospho-FFAT that requires phosphorylation at the fourth position of the core sequence (S209 in STARD3) to allow association with VAPs (17). Moreover, we found that phosphorylation of serine residues flanking this core motif (S203 and S213) maximizes the affinity of STARD3 for VAP. Therefore, we used a peptide (CLFSGALpSEGQFYpSPPEpSFAG, hereafter referred to as 3P peptide) corresponding to the phospho-FFAT motif of STARD3 with three phosphorylated serines. This peptide also contained an extra cysteine residue at its N terminus to be anchored to liposomes composed of phosphatidylcholine and doped with thiol-reactive 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine–N-[4-(p-maleimidophenyl) butyramide] (MPB–PE) lipids (3 mol%). With this strategy, the peptide was positioned on liposome surface like the corresponding cytosolic segment of the full-length STARD3 at the surface of endosomes (Fig. 1A).
Figure 1.
Liposomes decorated with an FFAT-containing 3P peptide associate with the ER of perforated cells.A, LA liposomes (total lipid concentration of 50 μM) bearing 3P peptide (380 nM) were mixed with LB liposomes (50 μM), covered by wildtype or K94D–M96D mutant VAPHis6 (300 nM), attached to DOGS–NTA–Ni2+. The mean radius (dots) and polydispersity (shaded area) are represented over time. B, RH distribution determined before (gray bars) and after a representative aggregation experiment (blue or black bars, n = 3). The mean RH and polydispersity of the liposome population are given. C, schematic representation of the liposomes applied onto the opened cells. D, principle of our approach to create ISACS. HeLa cells were transfected with GFP-VAP-A or GFP-Sec61, in order to visualize the ER, and were opened with a nitrocellulose filter, which gave access to their internal content. Then, the cells were incubated with Rhod–PE–containing liposomes covered by 3P peptide, washed, fixed, and observed by confocal microscopy. E, DOPC liposomes (total lipid of 125 μM), doped with 1 mol% Rhod–PE, covered by 3P peptide (at lipid/peptide molar ratio = 250) via covalent attachment to MPB–PE lipid (3 mol%), were incubated at room temperature for 5 min with perforated cells prior to washing and fixation. DIC microscopy allows detecting perforated cells (indicated by asterisks) in which the nuclear envelope and the nucleolus were brighter than in nonperforated cells. Dots (red channel) were observed by confocal microscopy at the top of perforated cells, colocalizing with GFP-VAP-A (green channel). Objective 63×. The scale bar represents 20 μm. F, quantification of the Rhod–PE fluorescence in intact and opened cells incubated with Rhod–PE–containing liposomes covered with 3P peptide (each dot represents one cell; intact cells, n = 95; opened cells; n = 125, from three independent experiments, mean ± SD are represented). G, colocalization analysis. Pictures corresponding to one plane in which Rhod–PE–containing liposomes covered with 3P peptide colocalize with the ER labeled with GFP-VAP-A. The scale bar represents 5 μm. Inset magnification panel, a line-scan analysis shows that liposomes (magenta line) colocalized with the ER membrane (green line). The scale bar represents 1 μm. DIC, differential interference contrast; DOGS–NTA–Ni2+, 1,2-di-(9Z-octadecenoyl)-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl]–nitrilotriacetic acid (nickel salt); DOPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; ER, endoplasmic reticulum; FFAT, two phenylalanines in an acidic tract; ISACS, in situ artificial contact sites; MPB–PE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[4-(p-maleimidophenyl) butyramide]; Rhod–PE, rhodamine–1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; VAP, vesicle-associated membrane protein–associated protein.
Prior to experiments with cells, we ascertained in vitro whether the peptide can act with VAP as an efficient membrane tether. MPB–PE–containing LA liposomes, extruded through a filter with pore size of 200 nm, were functionalized with 3P peptide at a lipid-to-peptide molar ratio of 250 and diluted in a quartz cuvette to perform dynamic light scattering (DLS) experiments (total lipids of 50 μM). As estimated by size distribution analysis (see Experimental procedures section), the mean hydrodynamic radius (RH) of these liposomes was about 100 nm with a small polydispersity (PD <50 nm) (Fig. 1B). Then, LB liposomes including 2 mol% 1,2-di-(9Z-octadecenoyl)-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl]–nitrilotriacetic acid (nickel salt) (DOGS–NTA–Ni2+), with a mean radius similar to that of LA liposomes, were added to the sample. After 1 min, VAPHis6, a recombinant protein in which the C-terminal transmembrane region of VAP is substituted by a short His tag (25), was injected; the association of VAPHis6 with LB liposomes generated ER-like liposomes (Fig. 1A). An immediate increase in the mean radius was observed, indicating that the 3P peptide interacted with VAP to attach the two liposome populations (Fig. 1A). Size distribution analysis indicated that aggregates were formed at the expense of free liposomes (RH = 872 nm, PD = 607 nm; Fig. 1B). The mean RH of the liposome population estimated at the end of independent kinetics with a cumulant algorithm was equal to 641.9 ± 73.9 nm (SD, n = 3). No aggregation was seen when this experiment was repeated with LB liposomes, exposing a VAPHis6 construct unable to recognize an FFAT motif because of a K94D–M96D double mutation (11). These data indicated that 3P peptide–functionalized liposomes were efficiently recruited at the surface of a second membrane covered by functional VAP and were thus suitable to devise our assay in cells.
As a next step, we prepared LA liposomes bearing 3P peptide and doped with 1 mol% rhodamine–1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (Rhod–PE) to have endosome-like fluorescent liposomes that could be observed by microscopy. In parallel, HeLa cells were transfected with a plasmid encoding GFP-VAP-A to visualize the ER. The day of the experiment, the PM at the top of cells was peeled off using a nitrocellulose filter according to the procedure described by Kobayashi and Pagano (31). The cells were incubated for 5 min with the functionalized Rhod–PE–containing liposomes, washed, and fixed prior to imaging by microscopy (Fig. 1, C and D). Using differential interference contrast (DIC) microscopy, we were able to recognize opened cells, as their nuclear envelope and nucleolus were brighter than those of unperforated cells (Fig. 1E). From one experiment to the other, the percentage of perforated cells ranged from 30 to 60%. Remarkably, under epifluorescence, we observed that many fluorescent dots corresponding to liposomes were associated with opened cells but almost none with intact cells. A quantification of Rhod–PE fluorescence associated with intact and opened cells confirmed that liposomes were specifically attached to opened cells (Fig. 1F). Then, by examining the localization of the fluorescent dots relative to that of GFP-VAP-A, we noticed that the liposomes were clustered in the upper region of the perforated cells, whose thickness was 11.1 ± 1.4 μm (SD, Fig. S1A) and associated with the VAP signal (Figs. 1G and S1, B–D). Besides, we noted that the tubular structure of the ER was slightly altered as we observed a predominance of rounded structures labeled with GFP-VAP-A, probably a consequence of the perforation process and the loss of cytosolic content. The average diameter of these structures was 0.72 ± 0.27 μm (SD) (Fig. S2, A–D). Line-scan analyses of confocal Z-stack images revealed a partial colocalization between the ER and liposomes with a juxtaposition as well as a superimposition of rhodamine and GFP signals (Figs. 1G and S2E). Collectively, these data suggest that liposomes bearing an FFAT peptide were recruited onto the ER membrane of opened cells.
Attachment of individual liposomes with the ER was observed by STED microscopy
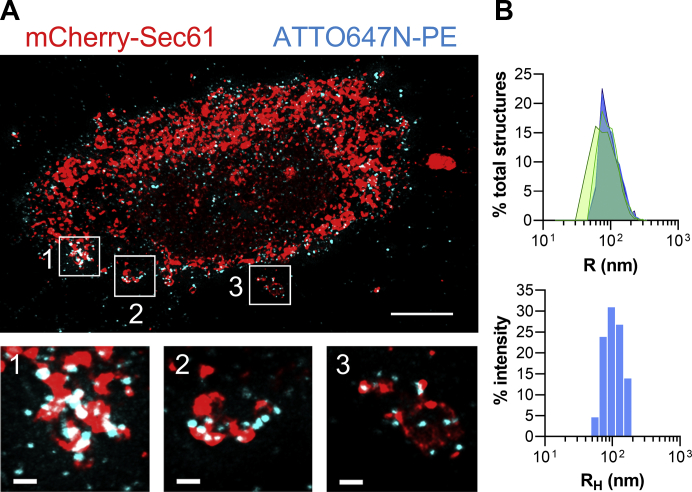
Next, we adapted our protocol for STED microscopy to visualize the association of liposomes with the ER at higher resolution. To this end, liposomes were doped with 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-ATTO647N (ATTO647N-PE) (at 0.5 mol%) instead of Rhod–PE as ATTO647N is a dye suitable for the depletion procedure. These liposomes were functionalized with the 3P peptide and added to opened cells expressing the ER marker mCherry-Sec61, which is suitable for STED microscopy. Cells were then washed, fixed, and mounted in a medium adapted for STED microscopy prior to imaging. We built 3D-STED stacks of images followed by deconvolution to obtain tridimensional views with a pixel size of 30 nm. As shown in Figure 2A, the super-resolution mode clearly revealed that individual liposomes were in contact with the ER (the organelle being observed in a standard confocal mode). An image analysis procedure was used to determine the volume and thus the radius of liposomes associated with the cell. The radius distribution of this liposome population, measured in five different opened cells, was centered around a mean radius of 102.2 ± 7.1 nm (SD) and remarkably close to the distribution measured for the same liposome population in vitro by DLS, characterized by a mean RH of 107 nm (Fig. 2B). Collectively, these data confirm that liposomes functionalized with 3P peptides and added to opened cells individually associated with the ER, and thus that our procedure could generate ISACS between synthetic and organelle membranes.
Figure 2.
Association of individual liposomes with the ER observed by STED microscopy.A, top view of a tridimensional reconstruction of an opened cell decorated with liposomes bearing 3P peptide and doped with 0.5 mol% ATTO647N-PE (cyan labeling). mCherry-Sec61 was expressed to visualize the ER. Objective 63×. The scale bar represents 10 μm. The three bottom images correspond to higher magnification (3.85×) images of the areas outlined in white. The scale bar represents 1 μm. B, top panel, size distribution of ATTO647N-PE–containing liposomes associated with the opened cells (one profile per cell; number of cells, n = 5; average number of liposomes per cell, n = 1781 ± 444 [SD]). The average radius of the liposome population associated with the ER was equal to 102.2 ± 7.1 nm (SD). Bottom panel, size distribution of ATTO647N-PE–containing liposomes measured by DLS. Mean RH = 107 nm. ATTO647N-PE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-ATTO647N; DLS, dynamic light scattering; ER, endoplasmic reticulum; STED, stimulated emission depletion.
The association of functionalized liposomes with the ER depends on the 3P peptide–VAP interaction
Since LA liposomes were functionalized with an FFAT motif that is recognized by the MSP domains of VAPs, we then studied the specificity of the observed attachment by measuring how liposomes that do not bear the 3P peptide associated with opened cells compared with liposomes functionalized with this peptide. We imaged large fields of view (40× objective) in 3D and quantified the fluorescent dots, which corresponded to liposomes associated with opened cells, using a novel image analysis procedure (Fig. S3 and Experimental procedures section). Faint fluorescent dots, outside the cells, likely corresponding to liposomes bound in a nonspecific manner to the dish surface and resistant to washing, were automatically excluded from the counting. Based on their appearance in DIC, opened cells were manually counted, and the mean number of fluorescent dots per opened cell was determined (Fig. S3). We determined that liposomes covered with the 3P peptide were significantly associated with opened cells compared with naked liposomes, which were barely detectable (Fig. 3, A and B). This suggests that liposomes functionalized with 3P associate with cells owing to a specific interaction with VAP.
Figure 3.
Attachment of liposomes to the ER depends on their functionalization with 3P peptide and on the ER-resident VAP protein.A, representative images of perforated cells incubated with liposomes functionalized or not with 3P peptide. Objective 63×. The scale bar represents 30 μm. B, quantification of dots per opened cell indicating that the association of liposomes with the ER depends on their functionalization with 3P peptide. Each data point corresponds to a unique microscopy field (number of fields; 3P, n = 25 [488 cells in total]; no peptide, n = 25 [752 cells in total], from three independent experiments). Means and error bars (SD) are shown. (Mann–Whitney test: ∗∗∗∗p ≤ 0.0001). C, quantification of VAP-A and VAP-B expression levels in HeLa cells transfected with control siRNA control or siRNA targeting VAP-A and VAP-B by immunoblotting. ∗aspecific band. A representative experiment is shown. D, HeLa cells were transfected with a pool of control siRNA (siControl) or a pool of siRNA targeting VAP-A and VAP-B (siVAP-A/VAP-B) 2 days prior to the perforation of cells and their incubation with liposomes covered by 3P peptide. Representative images of perforated cells incubated with liposomes in the two siRNA conditions. Objective 63×. The scale bar represents 30 μm. E, quantification of dots per opened cell indicating that the association of liposomes with the ER was strongly inhibited when VAP-A and VAP-B were silenced. The ER was visualized using the GFP-Sec61 marker (not shown). Each data point corresponds to a unique microscopy field (number of fields; siControl, n = 22 [146 cells in total]; siVAP-A/VAP-B, n = 19 [204 cells in total], from three independent experiments). Means and error bars (SD) are shown. (Mann–Whitney test: ∗∗∗∗p ≤ 0.0001). F, tridimensional view of perforated and nonperforated cells overexpressing GFP-VAP-A and of Rhod–PE–containing liposomes, exposing 3P peptide at their surface, attached to perforated cells. The overexpression of GFP-VAP-A in the two cells present in the center of the image (white asterisks) did not promote any extra recruitment of liposomes on the ER compared with a nontransfected cell (white arrows) (images scale: each square of the grid is a 13.45 × 13.45 μm2 square). G, fluorescence intensity in green and red channels (GFP-VAP-A and Rhod–PE liposomes, respectively) associated with transfected cells perforated and incubated with liposomes covered by 3P peptide. Each dot corresponds to a single cell (n = 91 cells, from three independent experiments). ER, endoplasmic reticulum; Rhod–PE, rhodamine–1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; VAP, vesicle-associated membrane protein–associated protein.
To further support this conclusion, we conducted experiments in which liposomes functionalized with the 3P peptide were added to VAP-depleted cells. The expression of VAP-A and VAP-B was reduced by 60% 2 days after transfection of anti-VAP-A and anti-VAP-B siRNAs (Fig. 3C). Using our quantification procedure, we determined that the number of attached liposomes was reduced by 64% in VAP-silenced perforated cells compared with perforated cells treated with control siRNAs (Fig. 3, D and E). This further indicated that the localization of FFAT-functionalized liposomes at the ER was dependent on the presence of VAPs.
We then performed the reciprocal experiment by determining the amount of endosome-like liposomes recruited to the ER under conditions of VAP overexpression. To do so, we quantified the Rhod–PE and GFP-VAP-A fluorescence in opened cells expressing different levels of GFP-VAP-A (Fig. 3F) and represented the Rhod–PE signal as a function of the GFP signal (Fig. 3G). As we found no correlation between the two signals, we concluded that the overexpression of the GFP-VAP-A protein did not promote extra attachment of liposomes to the ER. Of note, VAP-A and to a lesser extent VAP-B are quite abundant proteins in HeLa cell (34). Therefore, it is quite likely that the level of endogenous VAP was not limiting for the formation of ISACS. Thus, we conclude that the labeling of liposomes functionalized with FFAT-mimicking peptide is dependent on the presence of their receptors on the ER, that is, the VAP-A and VAP-B proteins. Moreover, we conclude that the endogenous amounts of VAPs are not limiting for the creation of ISACS.
Finally, we examined the intracellular localization of functionalized liposomes using costaining with a series of organelle markers to ask whether they were specifically associated with the ER. Line-scan analyses indicated that the liposome signal overlapped predominantly with the ER using GFP-VAP and GFP-Sec61 signals as readout (Fig. 4A). In contrast, almost no overlap was seen between liposomes and signals from markers of major organelles, such as the Golgi apparatus (GalNAcT2, cis-Golgi; Rab6A, trans-Golgi) and mitochondrion (Mit). Signal quantifications showed that liposomes were significantly and predominantly associated with VAP-A (Fig. 4B). A significant yet lower association was measured between liposomes and the second ER marker Sec61. In contrast, a much lower or almost null degree of colocalization was found between liposomes and markers of mitochondria and Golgi apparatus. Overall, these data indicated that liposomes are predominantly engaged with ER regions where VAP is present, in agreement with the idea that liposomes specifically associate with this protein. Thus, our strategy seemed relevant to measure a specific protein–protein interaction mediating the formation of MCS.
Figure 4.
Colocalization of liposomes bearing 3P peptide with VAP and other organelle markers. HeLa cells expressing GFP-VAP-A (ER), GFP-Sec61 (ER), GalNAcT2-GFP (cis-Golgi), Rab6A-GFP (trans-Golgi), or Mit-GFP (mitochondrion) were perforated, incubated with liposomes covered by 3P peptide, washed, and fixed. Image stacks of opened cells labeled with liposomes were acquired by confocal microscopy. A, for each condition, a single Z-section of a representative image stack as well as a representative profile from line-scan analyses are shown (liposome, magenta line, organelle marker, green line). The scale bar represents 10 μm. B, Manders’s correlation coefficient M2, expressing the fraction of Rhod–PE signal overlapping with GFP signal, was determined from image stacks for each marker. Each dot represents a single cell (number of cells: GFP-VAP-A/liposome: 71; GFP-Sec61/liposome: 41; GalNAcT2-GFP/liposome: 36; Rab6A-GFP/liposome: 25; GFP-Mit/liposome: 36, from at least three independent experiments). Means and error bars (SD) are shown. (Kruskal–Wallis test with Dunn’s multiple comparison: ∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001). ER, endoplasmic reticulum; Rhod–PE, rhodamine–1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; VAP, vesicle-associated membrane protein–associated protein.
In situ quantification of the membrane-tethering ability of FFAT-containing peptides
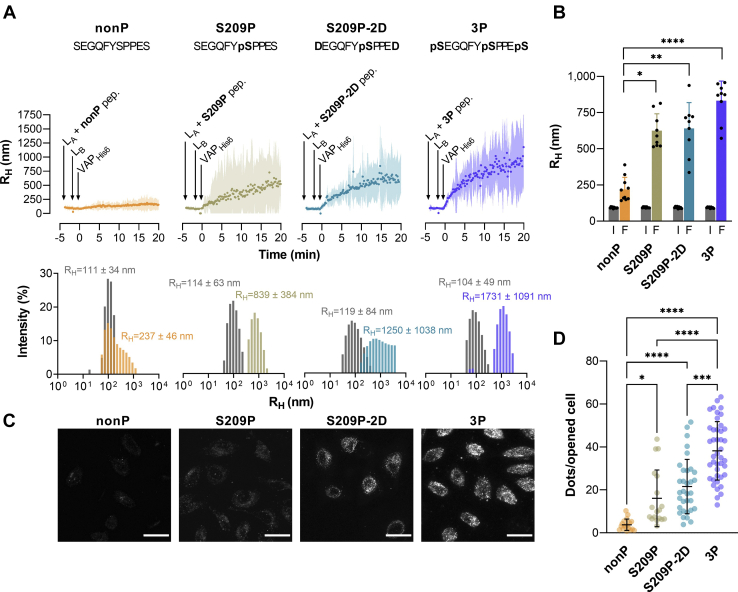
We then examined whether the ISACS strategy could provide evidence that structural determinants can fine-tune contact site formation. To this end, we studied the phospho-FFAT motif of STARD3 as a model for a regulatable attachment motif as its association with VAP is conditioned to the presence of a phosphorylation on serine S209 in the core sequence and modulated by the phosphorylation of flanking serine residues S203 and S213 (17). Therefore, we compared the ability of peptides corresponding to the phospho-FFAT motif of STARD3 in its unphosphorylated form (CLFSGALSEGQFYSPPESFAG, nonP), with a single phosphorylation on S209 residue (CLFSGALSEGQFYpSPPESFAG, pS209), and having three phosphorylations on S203, S209, and S213 residues (3P peptide) to create ISACS in opened cells. We also tested a peptide in which the S209 residue was phosphorylated and S203 and S213 were replaced by aspartate residues to mimic the presence of phosphorylated serines (CLFSGALDEGQFYpSPPEDFAG, pS209-2D).
First, we measured using DLS the tethering activity of each peptide in a complete reconstituted system (Fig. 5, A and B). LA liposomes, functionalized with a given peptide, were mixed with LB liposomes and VAPHis6. As previously observed (Fig. 1A), a rapid increase in the initial mean RH was observed with liposomes bearing the 3P peptide. Its values, estimated by a cumulant analysis, shifted from 92.7 ± 4.5 nm (SD, n = 9) to 831 ± 135 nm within 20 min, indicative of a massive aggregation process. Size distribution analysis at the end of the kinetics indicated that large liposome aggregates were formed at the expense of free liposomes (Fig. 5A, lower panel). Aggregation also occurred with liposomes decorated by pS209 and pS209-2D peptide, but at a slightly lesser extent (final mean: RH = 623.9 ± 117.6 nm and 639.3 ± 178.9 nm, respectively; Fig. 5, A and B). Finally, in those conditions, almost no aggregation occurred with liposomes covered by the nonP peptide. These data confirmed that additional phosphorylation in the STARD3 FFAT region modulated its capacity to bind to VAP and thus to drive membrane tethering.
Figure 5.
In situ determination of the tethering ability of FFAT peptides with different phosphorylation status.A, DLS experiments. Representative aggregation kinetics measured with LA liposomes bearing nonP, pS209, pS209-2D, or 3P peptide (380 nM) mixed with LB liposomes containing DOGS–NTA–Ni2+ lipids and VAPHis6 (300 nM). Upper panels, mean radius (dots) and polydispersity (shaded area) over time. Lower panels, RH distribution before (gray bars) and after the reaction (colored bars). B, average RH measured at the beginning (I: initial RH) and the end (F: final RH) of independent kinetic measurements (nonP, n = 10; pS209, pS209-2D, and 3P, n = 9). Means and error bars (SD) are shown. (Kruskal–Wallis test with Dunn’s multiple comparison: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗∗p ≤ 0.0001). C, representative images (Z-projection of image stacks, red channel) of perforated cells incubated with liposomes decorated with nonP, pS209, pS209-2D, or 3P peptide. Images were acquired by confocal microscopy with a 40× objective. The scale bar represents 30 μm. D, quantification of liposomes attached to the ER by different peptides. Each dot corresponds to a unique microscopy field (number of fields; 3P, n = 42 [499 cells in total], from seven independent experiments; nonP, n= 17 [531 cells]; pS209, n = 17 [415 cells]; pS209-2D, n = 33 [302 cells], from four independent experiments). Means and error bars (SD) are shown (Kruskal–Wallis test with Dunn’s multiple comparison: ∗p ≤ 0.05, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001). DLS, dynamic light scattering; DOGS–NTA–Ni2+, 1,2-di-(9Z-octadecenoyl)-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl]–nitrilotriacetic acid (nickel salt); ER, endoplasmic reticulum; FFAT, two phenylalanines in an acidic tract; VAP, vesicle-associated membrane protein–associated protein.
Second, we quantified the ability of each peptide to connect liposomes to the ER of opened cells (Fig. 5, C and D). When covered by nonP peptide, liposomes were not recruited onto the ER, whereas a slight recruitment was seen with liposomes functionalized with pS209 peptide. In contrast, as observed before, a strong recruitment of liposomes was observed if they were decorated with 3P peptide (Fig. 5, C and D). Quantification indicated that the binding to the ER was 10.2-fold higher than with liposomes covered by nonP peptide. An intermediate degree of association was observed with the pS209-2D peptide (5.7-fold higher compared with the nonP peptide). Finally, a 4.3-fold increase in association was seen with liposomes bearing pS209 peptide compared with the nonP peptide. We concluded that our in situ approach and quantification procedure were robust enough to provide quantitative data on a given interaction leading to the formation of contact sites.
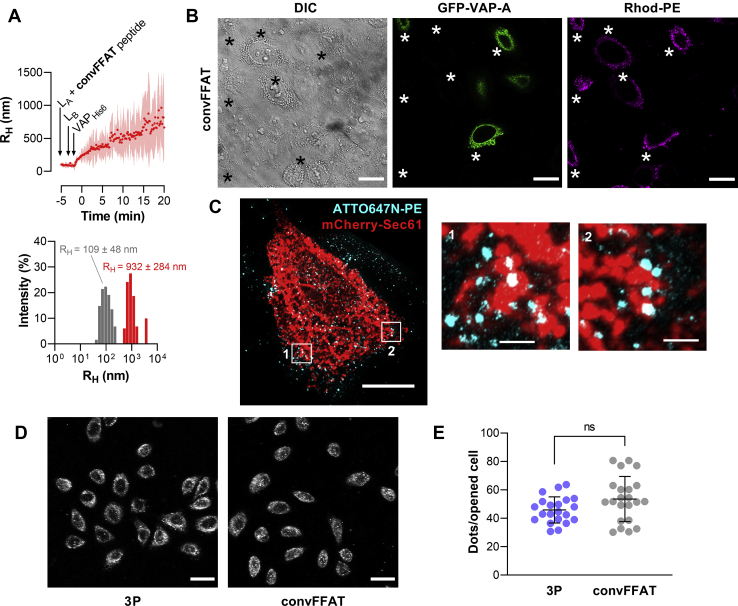
Third, we tested whether ISACS could be generated with liposomes bearing a convFFAT motif. MPB–PE–containing liposomes were functionalized with a peptide named convFFAT corresponding to the FFAT motif of OSBP (WCSG KGDMSDEDDENEFFDAPEIITMPENLGH with an extra N-terminal WC motif for covalent binding to MPB–PE and determination of the peptide concentration by spectrophotometry). As before, the attachment was done at a lipid-to-peptide molar ratio of 250. We observed that the incubation of peptide-functionalized liposomes with liposomes decorated with VAPs induced a strong aggregation at a degree similar to aggregations observed with the 3P peptide (Fig. 6A). Next, we observed in confocal microscopy that numerous Rhod–PE–containing liposomes bearing the convFFAT peptide were attached to the ER, labeled with GFP-VAP-A, of opened cells (Fig. 6B). We confirmed this observation using 3D-STED microscopy, which allows distinguishing individual liposomes associated with the ER membrane (Fig. 6C). Our quantification procedure indicated that the peptide with a convFFAT motif guaranteed, like the 3P peptide, the association of a large number of liposomes with the ER (Fig. 6, D and E). All these data indicated that our approach could be used to generate ISACS with the ER with peptide encompassing nonconventional FFAT or convFFAT motif. Moreover, our strategy was sensitive enough to highlight subtle attachment characteristics as observed in vitro in a pure controlled system.
Figure 6.
Efficient attachment of liposomes bearing a conventional FFAT-containing peptide with the ER.A, DLS experiments. LA liposomes bearing convFFAT peptide were mixed with LB liposomes containing DOGS–NTA–Ni2+ lipids to which VAPHis6 was anchored. Upper panels, mean radius (dots) and polydispersity (shaded area) over time. Lower panels, size distribution before (gray bars) and after the reaction (red bars) of a representative kinetic measurement (n = 3). RH and polydispersity are given. B, perforated cells were incubated with liposomes functionalized with convFFAT peptide and doped with Rhod–PE. DIC microscopy allows detecting perforated cells (indicated by black asterisks). Dots (red channel) were observed by confocal microscopy colocalizing with GFP-VAP-A (green channel). The scale bar represents 20 μm. C, STED microscopy. Top view of a tridimensional reconstruction of an opened cell decorated with liposomes bearing convFFAT peptide and doped with 0.5 mol% ATTO647N-PE (cyan labeling). mCherry-Sec61 was expressed to visualize the ER (red). The scale bars represent 10 μm. The right images correspond to higher magnification (5.3×) image of the areas outlined in white. The scale bar represents 1 μm. D, representative images (Z-projection of stacks) of perforated cells incubated with liposomes decorated with 3P or convFFAT peptide. Images were acquired in the red channel by confocal microscopy. Objective 40×. The scale bar represents 30 μm. E, quantification of liposomes attached to the ER by 3P and convFFAT peptide. Each dot corresponds to a unique microscopy field (3P, n = 38 fields [387 cells in total], from six independent experiments; convFFAT, n = 13 fields [336 cells], from three independent experiments). Means and error bars (SD) are shown (Mann–Whitney test: ns p = 0.0615). ATTO647N-PE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-ATTO647N; convFFAT, conventional FFAT; DIC, differential interference contrast; DLS, dynamic light scattering; DOGS–NTA–Ni2+, 1,2-di-(9Z-octadecenoyl)-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl]–nitrilotriacetic acid (nickel salt); ER, endoplasmic reticulum; FFAT, two phenylalanines in an acidic tract; ns, not significant; Rhod–PE, rhodamine–1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; STED, stimulated emission depletion; VAP, vesicle-associated membrane protein–associated protein.
STARD3 tethers liposomes to the ER via its cytosolic FFAT-containing region
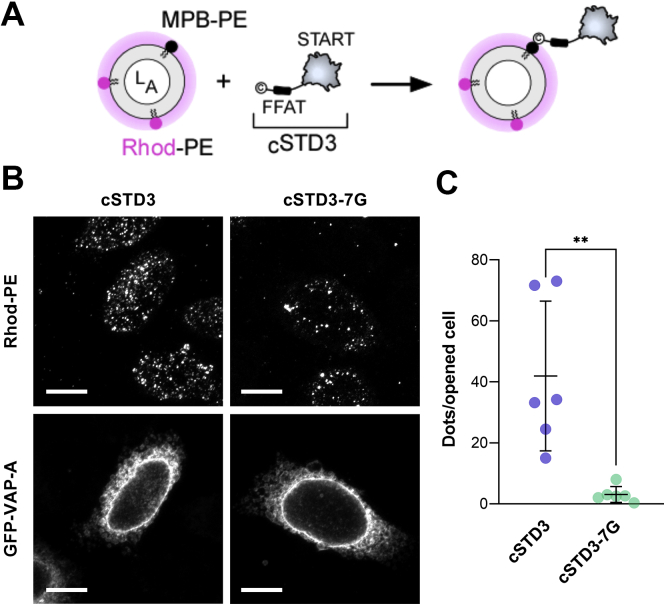
Once we demonstrated the feasibility of our approach to create ISACS using liposomes functionalized with peptides, we examined whether it could be used using liposomes bearing an FFAT-containing protein construct. Again, as a proof of principle, we analyzed whether STARD3 might connect liposomes to the ER. Because STARD3 is a transmembrane protein, we used its cytosolic part thereafter named cSTD3, which contains the lipid transfer StAR-related lipid transfer domain and a phospho-FFAT motif. Because it is not possible to obtain high yields of the recombinant cSTD3 in a phosphorylated form, we replaced its FFAT sequence by the convFFAT of OSBP. In parallel, we used a mutant protein devoid of the FFAT motif called cSTD3-7G where the FFAT motif was replaced by a stretch of seven glycine residues (18). A cysteine was present at the N-terminal end of these proteins to covalently attach them to MPB–PE–containing liposomes (Fig. 7A).
Figure 7.
Liposomes decorated by the cytosolic part of STARD3 are recruited onto the ER.A, experimental strategy. LA liposomes containing Rhod–PE and MPB–PE lipids, to which was attached cSTD3 construct, were incubated with perforated cells. B, representative images of perforated HeLa cells incubated with liposomes covered with cSTD3 or cSTD3-7G at L/P = 1000 (Rhod–PE: Z-projection of rhodamine labeling of a representative image stack, GFP-VAP-A: corresponding labeling of the ER by GFP-VAP-A [one plane]). Objective 63×. The scale bar represents 10 μm. C, quantification of the association of liposomes, covered at L/P = 1000 with cSTD3 protein, with opened HeLa cells. Each dot corresponds to a unique microscopy field (number of fields: cSTD3: 6 [22 cells], cSTD3-7G: 6 [25 cells], from two independent experiments). Means and error bars (SD) are shown. (Mann–Whitney test: ∗∗p = 0.0022). ER, endoplasmic reticulum; L/P, lipid-to-protein; MPB–PE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[4-(p-maleimidophenyl) butyramide]; Rhod–PE, rhodamine–1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; STARD3, steroidogenic acute regulatory protein–related lipid transfer domain 3.
We prepared liposomes covered with cSTD3 or cSTD3-7G by incubating each construct with MPB–PE–containing liposomes at different lipid-to-protein (L/P) ratios (250, 500, 750, and 1000). We measured the size of these liposomes and found that their average radius was higher (RH from 111 ± 49 nm to 131 ± 70 nm) for L/P ratio that decreased from 1000 to 500. At L/P = 250, two populations were detected (106 ± 31 nm and 1176 ± 318 nm; Fig. S4A), the one with the largest radius likely corresponding to liposomes that were self-aggregated. Similar data were obtained with cSTD3-7G (Fig. S4A). This suggested that liposomes covered by cSTD3 construct were prone to self-aggregate when the protein density at the membrane surface was too high. To perform in situ experiments and unambiguously analyze the obtained results, we incubated perforated cells with liposomes onto which cSTD3 or cSTD3-7G was anchored at L/P = 1000, a ratio preventing self-aggregation. We observed that liposomes bearing cSTD3 were bound to opened cells, whereas little binding was observed with the FFAT-less mutant cSTD3-7G (Fig. 7B). Our quantification procedure indicated that the binding of liposomes bearing cSTD3 to cells was on average 14.3-fold higher than that of liposomes decorated with cSTD3-7G (Fig. 7C). When similar experiments were performed with liposomes decorated with a higher amount of cSTD3 (L/P = 500 or 750), the labeling differed from the one observed at L/P = 1000, with red liposomes forming a thicker layer around the ER, likely because of their ability to self-aggregate in addition to being attached to the organelle surface (Fig. S4B). A similar labeling was seen with cSTD3-7G, yet at a lesser degree. These observations were more difficult to interpret but still suggested that liposomes covered by cSTD3 were more prone to bind to the ER than liposomes bearing cSTD3-7G. This showed that an FFAT-containing protein can tether liposomes to the ER and that the presence of a functional FFAT motif is mandatory for this process to occur.
Discussion
Here, we devised an approach to create ISACS between synthetic and organelle membranes. It differs from existing methods that are either completely in vivo or in vitro. To establish a proof of principle, we based our strategy on a characterized endosome–ER tethering complex formed by FFAT-containing proteins and VAPs. We adapted our protocol from previous studies showing that small liposomes incubated with perforated cells associate with the Golgi surface (31), likely as they are recruited by Golgi microtubule–associated protein 210, a peripheral protein whose function is to trap transport vesicles with similar radius (32). We modified this methodology by functionalizing the liposome surface with purified peptides or proteins containing an FFAT motif. Thus, instead of using naked liposomes as done previously, we engineered organelle-like liposomes mimicking the membrane composition of a given organelle and exposing at its surface a functional tether. Alongside, we developed a sensitive and semiquantitative analytical method to quantify the association of these liposomes to endomembranes, as a readout for the formation of artificial contact sites. This approach offers a way to examine in situ key protein–protein interactions responsible for the building of MCS and opens the way for further experimental setup, as the composition of liposome and the nature of lipid-anchored proteins can be easily modified.
We observed by confocal microscopy that fluorescent red liposomes covered by peptide corresponding to a triphosphorylated form of the phospho-FFAT motif of STARD3, and able to firmly bind to VAP (17), associated with the ER network of opened cells. The labeling was granular as the ER adopted a more rounded morphology and formed digitations that were surrounded by a layer of fluorescent liposomes. Quite powerfully, STED microscopy allowed to visualize the association of individual liposomes with the ER. When liposomes were not functionalized or when the expression of endogenous VAP-A and VAP-B was silenced, this association was abrogated. We also observed that liposomes colocalized predominantly with VAP and the ER; little association was seen with other organelles. Moreover, we showed that a peptide with the convFFAT motif of OSBP promoted a robust recruitment of liposomes to the ER, showing that our protocol can be used to test other FFAT motifs. Finally, we showed that protein domains too could be used to functionalize liposomes to be tested for their ability to create contact sites. Importantly, the observations and measurements made with ISACS were consistent with in vitro results showing that the recruitment of the FFAT motif by VAP was modulated by subtle structural determinants (phosphorylation). Jointly, these data show that our strategy allows detecting quantitatively and in situ a recruitment of liposomes with the ER that relies on a specific interaction with VAP.
Liposomes were found to be attached to the ER only in the upper region of opened HeLa cells, which was ∼11 μm thick and represented approximately half of the average height of these cells. In contrast, the ER region that is underneath the nucleus and near the bottom of the cell was never labeled. That liposomes were absent from the lowest half of cells might be due to a number of factors. First, there are likely enough anchoring points at the surface of the ER network present in the top region of the cell (i.e., VAP copies) to retain liposomes and thereby act as a barrier. As a confirmation, we found that overexpressing GFP-VAP did not increase the number of liposomes that could be recruited. Moreover, the presence of other organelles and the viscosity of the cytoplasm presumably limit the diffusion of liposomes inside opened cells. As mentioned before, the ER underwent slight morphological changes, shifting from a tubular to a rounded aspect, and formed digitations, as a result of the perforation process, presumably because of the loss of soluble factors and proteins along with the rupture of contacts with other organelles (e.g., between the ER and the PM). Nevertheless, this morphological change did not impede the recognition of FFAT-containing peptides and proteins by membrane-embedded VAPs.
The procedure that we developed to quantify the liposome attachment to the opened cells is based on the Z-projection of all the slices of confocal image stacks. We noted that the dynamic range of signal was between 1 and 14 between two extreme conditions, one obtained with a cSTD3 construct with no FFAT motif, and one obtained with a cSTD3 construct that interacts with VAP (Fig. 7), meaning that our quantification procedure should allow the ranking of series of proteins or peptides on the basis of their ability to form ISACS. This procedure cannot be entirely automatized because of the low contrast of DIC images that are acquired to identify and count opened cells. We also noted that the fraction of the cells that were opened during the peeling step cannot exceed more than 60% because of the uneven geometry of these cells in culture. Flat cells, which are more abundant when cells are just at confluence, seem more prone to be opened. Moreover, the degree of opening can vary from cell to cell, and sometimes, only a small part of the PM is peeled out. The variability in cell geometry can thus constitute an inherent limit to our strategy that can, however, be overcome by the analysis of a large number of cells.
A main asset of our strategy is its versatility; on the one hand, liposomes of distinct composition can be engineered and, on the other hand, the cell context can be modified. Indeed, it is easy to change the lipid composition, diameter, color, and brightness of liposomes as well as the nature of proteins or peptides that they display, in a very controlled manner. The features and quality of liposome sample can be verified, and the surface density of peptides or proteins can be adjusted. Of note, using STED microscopy, we can double-check in situ the size distribution of liposomes that have been added to the opened cells. This capacity to prepare well-defined liposomes with pure peptide allowed us to precisely quantify how the phosphorylation degree of the FFAT motif of STARD3 governed its interaction with endogenous VAP and thereby its membrane-tethering ability. In the future, our approach could be useful, using series of peptides, to define precisely whether different proteins with putative FFAT, phospho-FFAT (17, 23) or the recently described two phenylalanines in a neutral tract motifs (35) interact with not only ER-resident VAPs but also other MSP-containing proteins, such as motile sperm domain–containing protein 2 (34, 35) to form MCS. Furthermore, because of its versatility, the ISACS approach might serve to examine the role of other transmembrane and peripheral proteins, which do not necessarily interact with VAPs, in the formation of MCS. Both the nature and specificity of the interaction between liposomes and endomembrane components could be assessed by silencing the cellular receptors by siRNAs, as done here to silence VAPs, or by other approaches (CRISPR–Cas9, shRNA viruses). One can also envision incubating several populations of liposomes, each with a specific color and functionalized with a given peptide or protein, with opened cells to determine how proteins compete for binding a common organelle adaptor. This might allow defining the respective ability of diverse proteins to populate a particular type of MCS, which is a piece of information that is otherwise difficult to obtain.
Importantly, several cell types in culture have been successfully opened with the perforation protocols (31, 32). This means that the ISACS approach should be tractable to compare how functionalized liposomes associate with the endomembrane of diverse cell types, either primary cell lines or transformed cell lines. This might serve to quantitatively define whether cells, depending on their type, harbor a particular number of MCS or different subsets of MCS that are linked to specific requirements in terms of interorganelle communication. It is well appreciated that a strong interplay exists between the function of MCS, notably lipid transfer, and lipid metabolism and that the lipid composition of organelle membrane regulates in turn the formation of MCS (36). This aspect might also be interrogated by ISACS by precisely changing the composition of liposomes and/or by modifying the lipid content of organelles via specific diet or the silencing of given lipid-synthesizing or lipid-modifying enzymes. Furthermore, our approach might be also useful to gain more insights into the role of MCS not only in the response to different cellular stress including nutrient, oxidative, and ER stress (2, 37) but also in aging and disease (38, 39) by working with cell lines subjected to particular conditions or with cells engaged in disease-related dysfunctions. Finally, the ISACS approach might serve as the basis for the design of more complex functional assays aiming to examine processes such as lipid and Ca2+ transfer at MCS. We conclude that the creation of artificial contact sites between liposomes and cellular membranes in situ might represent a potent strategy to study the molecular basis of membrane-tethering events in different cellular contexts and might be useful to devise novel functional assays.
Experimental procedures
Cell culture
HeLa cells were maintained in Dulbecco's modified Eagle’s medium (DMEM) (Gibco Thermo Fisher Scientific) supplemented with 10% fetal bovine serum and 1% ZellShield (Minerva Biolabs) in a 5% humidified CO2 incubator at 37 °C in T-75 cell culture plastic flasks (Gibco Thermo Fisher Scientific).
Lipids
1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), DOGS–NTA–Ni2+, Rhod–PE, and MPB–PE were purchased from Avanti Polar Lipids. ATTO647N-PE was purchased from ATTO-TEC.
Peptides
The synthesis of peptides was done using an Applied Biosystems 433A peptide synthesizer using standard Fmoc chemistry. Peptides were subsequently purified by reverse-phase HPLC using a preparative scale column (Phenomenex; Kinetex EVO C18, 100 Å, 5 μm, 250 × 21.2 mm). Molecular weight and purity of the peptides were confirmed by mass spectrometry.
Protein expression and purification
Glutathione-S-transferase (GST)-cSTD3 and GST-cSTD3-7G were expressed in Escherichia coli at 37 °C for 4 h upon induction with 1 mM IPTG at an absorbance equal to 0.6 at λ = 600 nm. All purification steps were carried out using a 50 mM Tris, pH 7.4, 150 mM NaCl (TN) buffer containing 2 mM DTT. The buffer was supplemented during the first purification steps with 1 mM PMSF, 10 μM bestatin, 1 μM pepstatin, and EDTA-free protease inhibitor tablets (Roche). Bacteria were lysed using a Cell Disrupter (TS Series; Constant Systems Ltd), and the lysate was centrifuged at 40,000g for 90 min. Then, the supernatant was applied to Glutathione Sepharose 4B beads (Cytiva Sweden AB). After three washing steps with TN buffer containing 2 mM DTT, the beads were incubated with thrombin at 4 °C overnight to cleave off cSTD3 from the GST tag. The protein was recovered in the supernatant by three successive washing of the beads with TN buffer followed by centrifugation. The fractions were pooled, concentrated, flash-frozen in liquid nitrogen, and stored at −80 °C with 10% (v/v) glycerol. Concentration of protein stocks was estimated using a bicinchoninic acid assay (Thermo Fisher Scientific). VAP-A[8–212]His6 (called VAPHis6) and its K94D–M96D version were expressed in E. coli at 37 °C for 3 h upon induction with 1 mM IPTG (at an absorbance at 600 nm = 0.6). Bacteria cells were harvested and resuspended in buffer containing 50 mM Tris, pH 7.5, 300 mM NaCl, 5 mM imidazole supplemented with 1 mM PMSF, 10 μM bestatin, 1 μM pepstatin, 10 μM phosphoramidon, and protease inhibitor tablets (Roche). Bacteria cells were lysed using a French press, and the lysate was centrifuged at 200,000g for 1 h. Then, the supernatant was mixed with NTA–Ni2+ agarose beads (QIAGEN). After three washing steps, the proteins were eluted from the beads with buffer containing first 250 mM and then 500 mM imidazole.
Liposome preparation
Lipids, stored in stock solutions in CHCl3, were mixed at the desired molar ratio in a glass flask. The flask was connected to a rotary evaporator, and the solvent was removed under vacuum. The lipid mixtures containing DOGS–NTA–Ni2+ or MPB–PE were prewarmed to 33 °C for 5 min before creating the vacuum. Once dry, the lipids were resuspended in TN buffer to obtain a suspension of multilamellar liposomes. These liposomes were subsequently extruded through polycarbonate filters of 0.2 μm pore size using a mini-extruder (Avanti Polar Lipids). Liposomes were stored at 4 °C and in the dark when containing light-sensitive lipids (Rhod–PE and ATTO647N-PE) and used within 2 days.
Preparation of functionalized liposomes for cell experiments
Peptides corresponding to the FFAT motif of STARD3 and OSBP, with an N-terminal cysteine, stored in powder, were resuspended in distilled water or TN buffer, freshly degassed, and their concentration was determined by UV spectroscopy at λ = 280 nm. Each peptide was covalently attached to liposomes containing 96 mol% DOPC, 3 mol% MPB–PE, and 1 mol% Rhod–PE, extruded through polycarbonate filters of 0.2 μm pore size, with a lipid-to-peptide molar ratio of 250. The attachment was performed by mixing liposomes (total lipids of 500 μM) and peptide (2 μM) in 100 μl of freshly degassed TN buffer during 5 min at 850 rpm at 37 °C in a thermomixer. Afterward, a volume of 300 μl of perforation buffer (25 mM Hepes–KOH, pH 7.2, 115 mM KCl, and 2.5 mM MgCl2) supplemented with 1 mM DTT was added to the mix to block any potential reaction between MPB–PE and free cysteines during experiments with perforated cells. In order to covalently attach cSTD3 to MPB–PE-containing liposomes, 100 μl of a stock solution of protein was applied onto an illustra NAP-5 column (Cytiva Sweden AB) and eluted with freshly degassed TN buffer according to manufacturer’s indications to remove DTT. The concentration of the proteins was then determined by UV spectroscopy at λ = 280 nm using an extinction coefficient of 28,420 M−1 cm−1. The attachment of the proteins was performed by extemporaneously mixing cSTD3 or cSTD3-7G (0.5, 0.66, 1, or 2 μM for L/P ratio equal to 1000, 750, 500, and 250, respectively) with liposomes (total lipids of 500 μM) in 100 μl of freshly degassed TN buffer during 5 min at 850 rpm at 37 °C in a thermomixer. Then, a volume of 300 μl of perforation buffer supplemented with 1 mM DTT was added to the mix.
Cell perforation and incubation with liposomes
The attachment of liposomes to endomembranes in situ was observed by harnessing a method to perforate the cells in culture (31, 33). Briefly, HeLa cells were seeded (1.1 × 105 cells per condition) in Ibidi-treated μ-Dish 35 mm (Ibidi) precoated with collagen I (Corning) and used at day 4 once confluence was reached. To visualize the ER, the cells were transfected at day 3 with a plasmid that encodes GFP-VAP-A or at day 2 with a plasmid that encodes GFP-Sec61, using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. To visualize other compartments, the cells were transfected at day 3 with plasmid that either encodes GalNAcT2-GFP (cis-Golgi marker, gift from F. Perez), Rab6A-GFP (trans-Golgi marker, gift from B. Goud), or Mit-GFP (mitochondrion marker, gift from M. Chami). The day of the experiment, the cells were washed once with 1 ml of DMEM devoid of phenol red (Gibco Thermo Fisher Scientific). Then, the excess of moisture was aspirated with a Pasteur pipette. A filter of 25 mm diameter (HATF; 0.45 μm pore size; Millipore) was recut to reduce its diameter down to 20 mm and soaked in perforation buffer (25 mM Hepes–KOH, pH 7.2, 115 mM KCl, and 2.5 mM MgCl2), blotted against a filter paper, and positioned on the top of cells in the μ-Dish 35 mm. After 1 min in contact with the cells at room temperature, the filter was slowly peeled off from them. Next, the cells were incubated for 5 min at room temperature with 400 μl of a suspension of functionalized liposomes covered by peptide or protein in perforation buffer supplemented with 1 mM DTT. Thereafter, cells were washed twice with 1 ml of phenol red–free DMEM and fixed with 3% paraformaldehyde and 0.5% glutaraldehyde in PBS for 15 min. The fixation solution was removed and replaced by PBS. The samples were stored at 4 °C before imaging.
Visualization and quantification of liposomes associated with perforated cells
The attachment of liposomes to cells was visualized using a confocal microscope LSM780 Zeiss driven by the Zen software (Zeiss). First, an image in the DIC mode was recorded to identify the cells that were perforated. Then, image stacks were acquired to visualize the Rhod–PE–containing liposomes in the red channel and to visualize GFP-Sec61 or GFP-VAP-A and thereby see the limit of the ER network, in the green channel, using a 63× or a 40× objective. Each image of the stack corresponds to an optical slice of 280 nm thickness; a sufficient number of slices were recorded to acquire the totality of the Rhod–PE signal per cell. Tridimensional views were built from these image stacks using the Volocity software (PerkinElmer). The thickness (along z-axis) of the cellular region in which red granules were observed was measured using the Volocity software. To quantify the degree of association of liposomes with endomembranes, the images were analyzed using a home-made ImageJ (NIH, 40) macro to determine the total number of dots per cell in a microscopy field. First, for each field, the number of perforated cells per image was manually counted in DIC images based on the specific visual appearance of nuclei and nucleoli. Then, an image stack was acquired in the red channel. The images of the stack were projected onto a unique XY plan to sum the signal, followed by a filtering procedure (“bandpass” type) in the frequency domain to bring out the granules from the background. Next, these granules were detected and counted by applying a recursive “Find maxima” function with user-defined tolerance intervals (commonly 10 intervals from 50 to 500) in order to optimize the detection. A tolerance interval value of 250 was set for the detection of dots in all the images as, for this value, the number of dots that were detected outside the cells was minimal, and the background noise was suppressed. The results were expressed in term of dots per opened cell by dividing the total number of dots present in the field by the number of opened cells.
STED microscopy
All the samples were mounted in Abberior MOUNT SOLID Antifade medium (Abberior GmbH). STED images were acquired using a Leica SP8 STED 3× (Leica Microsystems) equipped with a pulsed white light laser as an excitation source and a 775 nm pulsed laser as depletion light source. All images were acquired at 400 Hz through a 100×/1.4 numerical aperture oil objective using the LAS X software (Leica Microsystems). mCherry-Sec61 was imaged in a confocal mode using an excitation wavelength at 561 nm. Liposomes doped with 0.5 mol% ATTO647N-PE were imaged using an excitation wavelength at 633 nm. The pixel resolution is 17 and 30 nm for dimensional and tridimensional STED images, respectively. All images were subjected to deconvolution using the Huygens Core, version 21.04/Huygens Remote Manager, version 3.7 (Scientific Volume Imaging; http://svi.nl/), using the classic maximum likelihood estimation algorithm, with 40 iterations to achieve a resolution with a signal-to-noise ratio equal to 20 for the confocal mode and five iterations to reach a resolution with a signal-to-noise ratio equal to 100 for the STED mode. Imaris Cell Imaging software (Oxford Instruments) was used to create tridimensional views from STED images and to measure liposome volume.
Colocalization of liposomes with organelle markers in perforated cells
To quantify the colocalization between liposomes and diverse organelle markers in opened cells, image stacks in green and red channels were acquired with an SP8 STED 3X TCS SP8 confocal using a 63× objective. The fraction of liposome colocalizing with organelle markers in opened cells was estimated with the Manders’s correlation coefficient M2, which corresponds to the fraction of red fluorescence having a counterpart in the green channel, using the Volocity software with an autothresholding performed using the Costes method (41).
Anti-VAP-A and VAP-B siRNA
HeLa cells were seeded and transfected 2 days after with ON-TARGETplus human VAP-A (9218) and human VAP-B (9217) siRNA SMARTpools (Dharmacon) to conjointly silence VAP-A and VAP-B expression. Each siRNA pool (20 pmol) was transfected with RNAiMax Lipofectamine (Invitrogen). A pEGFP or pEGFP-Sec61 plasmid was cotransfected in order to verify the efficiency of the transfection. As a negative control, cells were transfected by ON-TARGETplus nontargeting pool. Cells were used for experiments 48 h after transfection.
Quantification of endogenous VAP proteins by Western blot
In silencing experiments, the VAP-A and VAP-B expression levels were individually analyzed by immunodetection using an anti-VAP-A antibody (rabbit polyclonal H-40; sc-98890; Santa Cruz) and an anti-VAP-B antibody (rabbit polyclonal antibody, gift from L. Dupuis (42)). GAPDH level, detected by an anti-GAPDH polyclonal antibody (1:2500 dilution, rabbit; ab9485 from Abcam) was measured for each lane to ascertain that an equivalent amount of sample was loaded into each well. Immobilon Western Chemiluminescent horseradish peroxidase substrate (Millipore) was used for the revelation. The band intensities were quantified with the AIDA software (Elysia-raytest, version 4.27.039).
DLS
DLS experiments were performed at 25 °C in a Dynapro apparatus (Protein Solutions). Peptides corresponding to the FFAT motif of STARD3 or OSBP were resuspended in TN buffer, and their concentration was determined by UV spectroscopy at λ = 280 nm. Each peptide was covalently attached to LA liposomes containing 96 mol% DOPC, 3 mol% MPB–PE, extruded through polycarbonate filters of 0.2 μm pore size. The attachment was performed by mixing liposomes (total lipids of 50 μM) with peptide (420 nM) in 200 μl of freshly degassed buffer during 30 min at 900 rpm at 25 °C. Then, a final concentration of 1 mM DTT was added to each mix to block any potential reaction between MPB–PE and free cysteines. A volume of functionalized liposomes (18 μl) was added to the quartz cuvette. A first set of 12 autocorrelation curves was acquired to measure the initial size distribution of liposomes in the suspension. Then, 1 μl of a LB liposome suspension (90 mol% DOPC and 10 mol% DOGS–NTA–Ni2+, 50 μM total lipid final concentration) was added and mixed thoroughly. After an incubation of 1 min, 1 μl of a stock solution of VAPHis6 or VAP(K94D–M96D)His6 was subsequently added to the sample (final concentration of 300 nM). The aggregation kinetics was measured by acquiring one autocorrelation curve every 10 s. At the end of each kinetics, a set of 12 autocorrelation functions was also acquired. The data were analyzed using two distinct algorithms implemented in the Dynamics, version 6.1, software (Protein Solutions). During the kinetics, the autocorrelation functions were fitted assuming that the size distribution is a simple Gaussian function. This mode, referred to as the cumulant analysis, provides the mean RH and the PD of the liposome suspension. The PD can reach very large values because of the concomitant presence of free liposomes and liposome aggregates of various sizes. The set of autocorrelation curves acquired before and after the aggregation process was also fitted using a more refined algorithm, referred as a regularization algorithm, which gives size distribution. This algorithm is able to detect the existence of several particle populations, whose mean size is different, such as free liposomes and liposome aggregates. For the experiments with cSTD3 constructs, each protein was first mixed with MPB–PE–containing liposomes at L/P = 250, 500, 750, or 1000 in 100 μl of freshly TN buffer during 10 min at 850 rpm at 37 °C in a thermomixer. Afterward, 50 μl of this liposome suspension was mixed with 150 μl of perforation buffer (25 mM Hepes–KOH, pH 7.2, 115 mM KCl, and 2.5 mM MgCl2) supplemented with 1 mM DTT. Next, 8 μl of this mixture was added to 12 μl of perforation buffer in the cuvette for the DLS measurement. A set of 12 autocorrelation functions was also acquired and analyzed using the regularization algorithm.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supporting information
This article contains supporting information (Figs. S1–S4).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We acknowledge the flow cytometry and microscopy facility from the Institut de Pharmacologie Moléculaire et Cellulaire, which is part of the GIS IBiSA platform MICA (Microscopie Imagerie Côte d’Azur). We thank P. Eberling (IGBMC [Institut de Génétique et de Biologie Moléculaire et Cellulaire] peptide synthesis facility) for the peptide syntheses. This work was supported by the CNRS, France; Université Côte d’Azur, France; and by a grant from the Agence Nationale de la Recherche, France (grant no.: ANR-19-CE44-0003).
Author contributions
J. M. and G. D. conceptualization; J. M. and G. D. methodology; F. B. software; J. M. and M. M. validation; J. M., M. M., and N. F. formal analysis; J. M., M. M., N. F., and S. A. investigation; F. A. resources; J. M. writing–original draft; F. A., C. T., and G. D. writing–review & editing; G. D. visualization; G. D. supervision.
Edited by Karen Fleming
Supporting information
References
- 1.Li C., Qian T., He R., Wan C., Liu Y., Yu H. Endoplasmic reticulum-plasma membrane contact sites: Regulators, mechanisms, and physiological functions. Front. Cell Dev. Biol. 2021;9:627700. doi: 10.3389/fcell.2021.627700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prinz W.A., Toulmay A., Balla T. The functional universe of membrane contact sites. Nat. Rev. Mol. Cell Biol. 2020;21:7–24. doi: 10.1038/s41580-019-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scorrano L., De Matteis M.A., Emr S., Giordano F., Hajnóczky G., Kornmann B., Lackner L.L., Levine T.P., Pellegrini L., Reinisch K., Rizzuto R., Simmen T., Stenmark H., Ungermann C., Schuldiner M. Coming together to define membrane contact sites. Nat. Commun. 2019;10:1287. doi: 10.1038/s41467-019-09253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skehel P.A., Martin K.C., Kandel E.R., Bartsch D. A VAMP-binding protein from Aplysia required for neurotransmitter release. Science. 1995;269:1580–1583. doi: 10.1126/science.7667638. [DOI] [PubMed] [Google Scholar]
- 5.Petkovic M., Jemaiel A., Daste F., Specht C.G., Izeddin I., Vorkel D., Verbavatz J.M., Darzacq X., Triller A., Pfenninger K.H., Tareste D., Jackson C.L., Galli T. The SNARE Sec22b has a non-fusogenic function in plasma membrane expansion. Nat. Cell Biol. 2014;16:434–444. doi: 10.1038/ncb2937. [DOI] [PubMed] [Google Scholar]
- 6.Takeshima H., Komazaki S., Nishi M., Iino M., Kangawa K. Junctophilins: A novel family of junctional membrane complex proteins. Mol. Cell. 2000;6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 7.Eden E.R., Sanchez-Heras E., Tsapara A., Sobota A., Levine T.P., Futter C.E. Annexin A1 tethers membrane contact sites that mediate ER to endosome cholesterol transport. Dev. Cell. 2016;37:473–483. doi: 10.1016/j.devcel.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishimura Y., Hayashi M., Inada H., Tanaka T. Molecular cloning and characterization of mammalian homologues of vesicle-associated membrane protein-associated (VAMP-associated) proteins. Biochem. Biophys. Res. Commun. 1999;254:21–26. doi: 10.1006/bbrc.1998.9876. [DOI] [PubMed] [Google Scholar]
- 9.Loewen C.J., Levine T.P. A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J. Biol. Chem. 2005;280:14097–14104. doi: 10.1074/jbc.M500147200. [DOI] [PubMed] [Google Scholar]
- 10.Loewen C.J., Roy A., Levine T.P. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaiser S.E., Brickner J.H., Reilein A.R., Fenn T.D., Walter P., Brunger A.T. Structural basis of FFAT motif-mediated ER targeting. Structure. 2005;13:1035–1045. doi: 10.1016/j.str.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Wyles J.P., McMaster C.R., Ridgway N.D. Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J. Biol. Chem. 2002;277:29908–29918. doi: 10.1074/jbc.M201191200. [DOI] [PubMed] [Google Scholar]
- 13.Delfosse V., Bourguet W., Drin G. Structural and functional specialization of OSBP-related proteins. Contact. 2020;3 doi: 10.1177/2515256420946627. [DOI] [Google Scholar]
- 14.Kawano M., Kumagai K., Nishijima M., Hanada K. Efficient trafficking of ceramide from the endoplasmic reticulum to the Golgi apparatus requires a VAMP-associated protein-interacting FFAT motif of CERT. J. Biol. Chem. 2006;281:30279–30288. doi: 10.1074/jbc.M605032200. [DOI] [PubMed] [Google Scholar]
- 15.Amarilio R., Ramachandran S., Sabanay H., Lev S. Differential regulation of endoplasmic reticulum structure through VAP-nir protein interaction. J. Biol. Chem. 2005;280:5934–5944. doi: 10.1074/jbc.M409566200. [DOI] [PubMed] [Google Scholar]
- 16.Alpy F., Rousseau A., Schwab Y., Legueux F., Stoll I., Wendling C., Spiegelhalter C., Kessler P., Mathelin C., Rio M.C., Levine T.P., Tomasetto C. STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER. J. Cell Sci. 2013;126:5500–5512. doi: 10.1242/jcs.139295. [DOI] [PubMed] [Google Scholar]
- 17.Di Mattia T., Martinet A., Ikhlef S., McEwen A.G., Nomine Y., Wendling C., Poussin-Courmontagne P., Voilquin L., Eberling P., Ruffenach F., Cavarelli J., Slee J., Levine T.P., Drin G., Tomasetto C., et al. FFAT motif phosphorylation controls formation and lipid transfer function of inter-organelle contacts. EMBO J. 2020;39 doi: 10.15252/embj.2019104369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilhelm L.P., Wendling C., Vedie B., Kobayashi T., Chenard M.P., Tomasetto C., Drin G., Alpy F. STARD3 mediates endoplasmic reticulum-to-endosome cholesterol transport at membrane contact sites. EMBO J. 2017;36:1412–1433. doi: 10.15252/embj.201695917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peretti D., Dahan N., Shimoni E., Hirschberg K., Lev S. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol. Biol. Cell. 2008;19:3871–3884. doi: 10.1091/mbc.E08-05-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yadav S., Thakur R., Georgiev P., Deivasigamani S., Krishnan H., Ratnaparkhi G., Raghu P. RDGBα localization and function at membrane contact sites is regulated by FFAT–VAP interactions. J. Cell Sci. 2018;131 doi: 10.1242/jcs.207985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamemura K., Chihara T. Multiple functions of the ER-resident VAP and its extracellular role in neural development and disease. J. Biochem. 2019;165:391–400. doi: 10.1093/jb/mvz011. [DOI] [PubMed] [Google Scholar]
- 22.Johnson B., Leek A.N., Tamkun M.M. Kv2 channels create endoplasmic reticulum/plasma membrane junctions: A brief history of Kv2 channel subcellular localization. Channels (Austin) 2019;13:88–101. doi: 10.1080/19336950.2019.1568824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy S.E., Levine T.P. VAP, a versatile access point for the endoplasmic reticulum: Review and analysis of FFAT-like motifs in the VAPome. Biochim. Biophys. Acta. 2016;1861:952–961. doi: 10.1016/j.bbalip.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 24.James C., Kehlenbach R.H. The interactome of the VAP family of proteins: An overview. Cells. 2021;10:1780. doi: 10.3390/cells10071780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mesmin B., Bigay J., Moser von Filseck J., Lacas-Gervais S., Drin G., Antonny B. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 2013;155:830–843. doi: 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- 26.Osawa T., Kotani T., Kawaoka T., Hirata E., Suzuki K., Nakatogawa H., Ohsumi Y., Noda N.N. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 2019;26:281–288. doi: 10.1038/s41594-019-0203-4. [DOI] [PubMed] [Google Scholar]
- 27.Saheki Y., Bian X., Schauder C.M., Sawaki Y., Surma M.A., Klose C., Pincet F., Reinisch K.M., De Camilli P. Control of plasma membrane lipid homeostasis by the extended synaptotagmins. Nat. Cell Biol. 2016;18:504–515. doi: 10.1038/ncb3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bian X., Zhang Z., Xiong Q., De Camilli P., Lin C. A programmable DNA-origami platform for studying lipid transfer between bilayers. Nat. Chem. Biol. 2019;15:830–837. doi: 10.1038/s41589-019-0325-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valverde D.P., Yu S., Boggavarapu V., Kumar N., Lees J.A., Walz T., Reinisch K.M., Melia T.J. ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 2019;218:1787–1798. doi: 10.1083/jcb.201811139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maeda S., Otomo C., Otomo T. The autophagic membrane tether ATG2A transfers lipids between membranes. Elife. 2019;8 doi: 10.7554/eLife.45777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobayashi T., Pagano R.E. ATP-dependent fusion of liposomes with the Golgi apparatus of perforated cells. Cell. 1988;55:797–805. doi: 10.1016/0092-8674(88)90135-3. [DOI] [PubMed] [Google Scholar]
- 32.Magdeleine M., Gautier R., Gounon P., Barelli H., Vanni S., Antonny B. A filter at the entrance of the Golgi that selects vesicles according to size and bulk lipid composition. Elife. 2016;5 doi: 10.7554/eLife.16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simons K., Virta H. Perforated MDCK cells support intracellular transport. EMBO J. 1987;6:2241–2247. doi: 10.1002/j.1460-2075.1987.tb02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Mattia T., Wilhelm L.P., Ikhlef S., Wendling C., Spehner D., Nomine Y., Giordano F., Mathelin C., Drin G., Tomasetto C., Alpy F. Identification of MOSPD2, a novel scaffold for endoplasmic reticulum membrane contact sites. EMBO Rep. 2018;19 doi: 10.15252/embr.201745453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cabukusta B., Berlin I., van Elsland D.M., Forkink I., Spits M., de Jong A.W.M., Akkermans J., Wijdeven R.H.M., Janssen G.M.C., van Veelen P.A., Neefjes J. Human VAPome analysis reveals MOSPD1 and MOSPD3 as membrane contact site proteins interacting with FFAT-related FFNT motifs. Cell Rep. 2020;33:108475. doi: 10.1016/j.celrep.2020.108475. [DOI] [PubMed] [Google Scholar]
- 36.Lipp N.F., Ikhlef S., Milanini J., Drin G. Lipid exchangers: Cellular functions and mechanistic links with phosphoinositide metabolism. Front. Cell Dev. Biol. 2020;8:663. doi: 10.3389/fcell.2020.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senft D., Ronai Z.A. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem. Sci. 2015;40:141–148. doi: 10.1016/j.tibs.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janikiewicz J., Szymanski J., Malinska D., Patalas-Krawczyk P., Michalska B., Duszynski J., Giorgi C., Bonora M., Dobrzyn A., Wieckowski M.R. Mitochondria-associated membranes in aging and senescence: Structure, function, and dynamics. Cell Death Dis. 2018;9:332. doi: 10.1038/s41419-017-0105-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petkovic M., O'Brien C.E., Jan Y.N. Interorganelle communication, aging, and neurodegeneration. Genes Dev. 2021;35:449–469. doi: 10.1101/gad.346759.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costes S.V., Daelemans D., Cho E.H., Dobbin Z., Pavlakis G., Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kabashi E., El Oussini H., Bercier V., Gros-Louis F., Valdmanis P.N., McDearmid J., Mejier I.A., Dion P.A., Dupre N., Hollinger D., Sinniger J., Dirrig-Grosch S., Camu W., Meininger V., Loeffler J.-P., et al. Investigating the contribution of VAPB/ALS8 loss of function in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2013;22:2350–2360. doi: 10.1093/hmg/ddt080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.