A critical evaluation of the construct of schizophrenia must consider the three primary purposes of diagnostic classifications: To facilitate research into the causes and treatments of illnesses, guide clinical decision making, and enable efficient and accurate communication between care providers and patients. A great deal of research has been facilitated by our current diagnostic systems but progress toward discovery of causal pathophysiology and novel therapeutics has been disappointing. Similarly, our diagnostic categories are only modestly helpful for clinical decision making, given high rates of phenotypic discontinuity, diagnostic heterogeneity, and co-occurrence among disorders. Diagnostic systems are, however, heavily relied upon for clinical communications and embedded into healthcare systems, reinforcing the need to proceed conservatively when considering changes to how disorders are defined while simultaneously exploring novel approaches to the first two purposes. As a diagnostic entity characterized by heterogeneity in phenotypic presentation, illness trajectory, and treatment response, schizophrenia challenges the field to confront these complexities.
Here we describe an integrative approach to refining the diagnosis of schizophrenia with the goal of enhancing clinical decision-making and facilitating research. This approach recognizes that Diagnostic and Statistical Manual (DSM)-based clinical diagnostics, conducted by expert clinicians, are a useful and important description of individuals with schizophrenia, but that they are insufficient to fully capture the biological and pathophysiological nature of the patient’s condition. Additional data that can inform our understanding of each patient are needed, but even more necessary is a framework for quantitatively evaluating the significance of these additional data for clinical prediction and/or biological research. Our proposed framework utilizes the NIMH Research Domain Criteria (RDoC) and a data-driven Bayesian inference approach to test the utility of deeper functional characterization in patients.
From a research perspective, we have now a decade’s worth of evidence that the RDoC initiative (Fig. 1) can provide an effective bridge between behavior and the brain. RDoC was launched with the purpose of stimulating research that has as its starting point the corpus of knowledge emerging from basic behavioral neuroscience research and brings that knowledge to bear on clinical problems, while not constraining clinical research questions to current diagnostic categories (Cuthbert and Insel, 2013). RDoC is one of several systems that seeks to break down behavior into its component domains. Importantly, function within a domain is evaluated along a dimension, rather than being categorically described as intact or deficient. Function can then be quantitatively mapped on to specific brain circuits and inferences made to dysfunction seen in mental illnesses.
Figure 1:
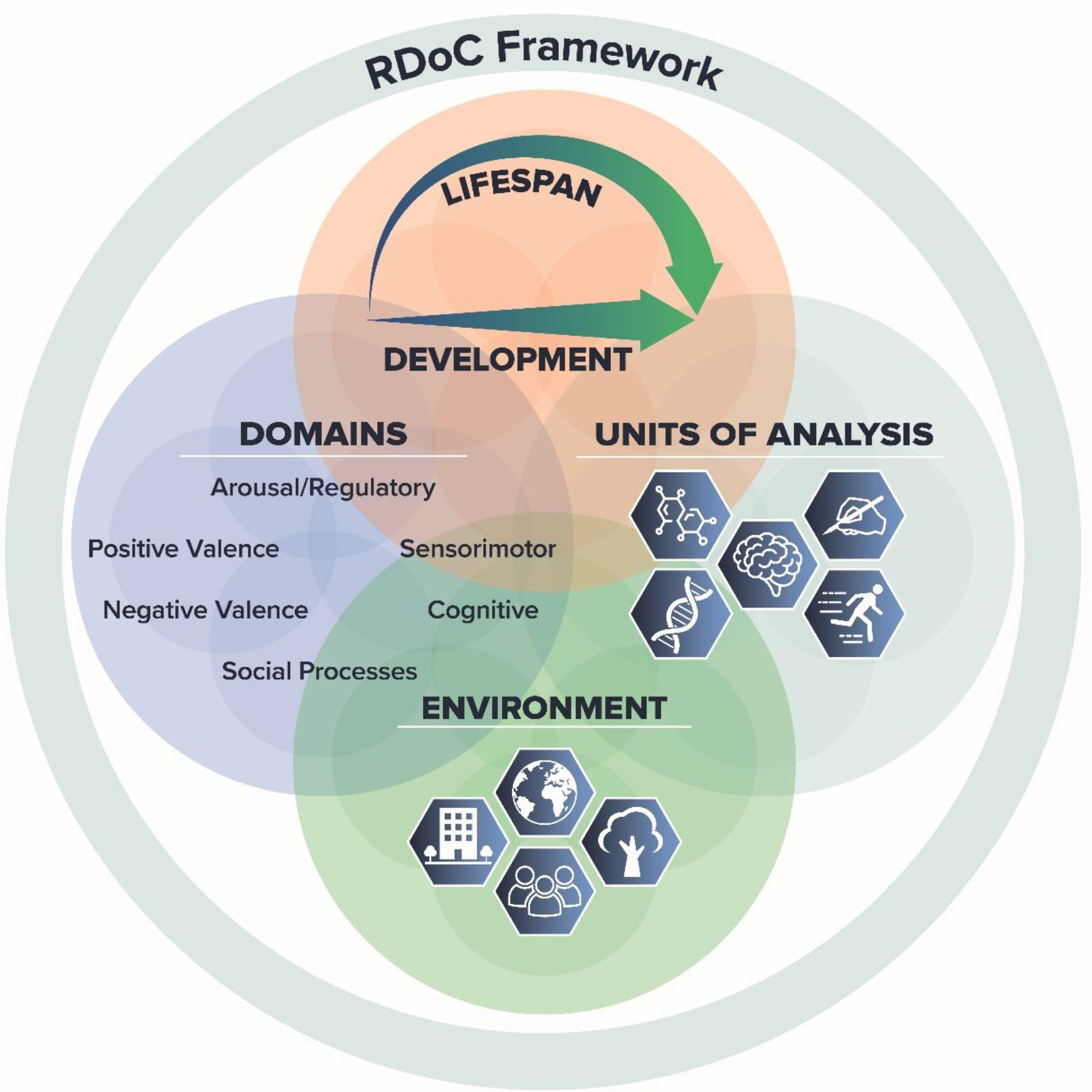
The Research Domain Criteria framework. The RDoC framework provides an organizational structure for research that considers mental health and illness in the context of major domains of human neurobehavioral functioning, taking into consideration environmental and developmental context.
Such work has the potential to improve our understanding of the different pathways via which one can develop psychosis and to point to new treatment targets. For example, the revolution in understanding the role of the midbrain dopamine system in learning that came about via basic behavioral neuroscience research has been used to create formal explanatory and predictive models relevant to the understanding of aberrant perceptions and beliefs seen in schizophrenia (Corlett and Schoenbaum, 2021). Resting-state functional connectivity has been shown to predict clinical response to anti-psychotic medication in people with schizophrenia (Doucet et al., 2020; Mehta et al., 2021) and may be a useful tool in the future for dissecting mechanistic heterogeneity within this disorder. Additional examples of new discoveries informed by cross-walking diagnoses and dimensional functions can be found in the mood and anxiety disorders literature. Classifying individuals according to reward learning ability rather than diagnosis yielded greater homogeneity in aspects of brain function (specifically medial prefrontal cortex glutamatergic function), and brain function provided stronger prediction of changes in (hypo)manic symptoms longitudinally than diagnosis alone, but the directionality of the association differed for individuals with unipolar versus bipolar depression (Whitton et al., 2021). In a study comparing threat conditioning and extinction across four anxiety disorders (Marin et al., 2020), investigators found that ventromedial prefrontal cortex hypoactivation and reduced skin conductance were common dysfunctions across disorders compared to healthy controls. Furthermore, when patients were grouped according to threat-induced arousal, between-group differences in psychophysiological reactivity and extinction-related brain activity were detected.
These and other studies suggest that diagnostic and functional approaches provide distinct information, raising the possibility of complementarity. Achieving this complementarity is a challenge. One potential pathway forward is through the recognition that the relationship between brain illness and its behavioral manifestations – whether the behavior is dimensional like an RDoC functional domain or categorical like a clinical diagnosis – is a probabilistic one. That is, a given disease state, caused by some set of etiologies and marked by some set of pathophysiological processes, results in a variable set of behavioral outcomes determined by chance and described by a given set of probabilities. Consider for example one of the several rare, large effect-size genetic alterations that raises the risk for schizophrenia: the 22q11.2 microdeletion (McDonald-McGinn et al., 2015). Arguably, the microdeletion represents a single disease state. Yet, if one has this microdeletion, there is an approximately 30% chance that one will develop a psychosis otherwise indistinguishable from idiopathic schizophrenia. There is also an approximately 60% chance of an anxiety disorder diagnosis, and a 15–25% chance of an autism diagnosis. [This probabilistic relationship is not limited to the CNS; approximately one third of individuals with the microdeletion will have a congenital heart defect]. Likewise, for functional outcomes like RDoC, working memory and other executive function deficits seen in 22q11.2 microdeletion carriers and individuals with schizophrenia are variable in severity. Thus, the microdeletion “disease state” causes a heterogeneous syndrome of potential behavioral outcomes.
The recognition that outcomes are probabilistically determined provides an opportunity to reconceptualize the diagnosis of schizophrenia. Rather than thinking of the diagnosis as a disease state – or as a collection of disease states, lumped together for lack of ability to differentiate them – we can think of schizophrenia as an observation about our patients, born of clinical experience, that tells researchers something useful (but not completely so) about what is going on in the brain, and helps guide clinicians (but not completely so) towards a range of prognoses and treatment options. Our task as researchers or clinicians is to use any additional data (for example, RDoC functional characterizations) that we can gather beyond this diagnosis to refine our understanding or clinical decision making.
Bayesian inference is one potential tool that can aid in the integration of data from probabilistic outcomes. Briefly, Bayesian inference describes a method to quantify the likelihood of a given outcome from an underlying state – and importantly, to quantify the likelihood of a given state based on the presence of a given outcome (see Flagel et al., 2016, for a more complete description of Bayesian inference in psychiatric diagnoses). One can also use this process to ask how much more information about these probabilistic relationships one gets by adding more data. Thus, Bayesian inference provides a framework to link disease states – including causes and pathophysiological mechanisms – with multiple types of outcomes – including RDoC functions and DSM diagnoses.
For example, knowing the base rate of schizophrenia in the population, the base rate of 22q11.2 microdeletions, and the likelihood of a schizophrenia diagnosis, one can calculate, in a straightforward way, the likelihood of a 22q11.2 microdeletion in a given patient with schizophrenia (see Box 1). This approach is routinely used to aid clinical decision making (Gill et al., 2005); prior knowledge about the base rate of a diagnosis, coupled with the accuracy of a test for that diagnosis, can be combined using Bayesian inference to help interpret the test and arrive at the likelihood of that diagnosis for a specific patient. For example, Bayesian inference has played a key role in interpreting results from ventilation/perfusion scans and other tests for diagnosing pulmonary embolisms (Luciani et al., 2007). For Schizophrenia and other mental illnesses, however, the pathway to clinical utility is not yet established. Suppose we wanted to use Bayesian inference to make specific predictions about longitudinal course or likelihood of treatment response with second generation antipsychotics. Here, making inferences about the probabilities of a given outcome requires data that currently do not exist, as well as a computational framework to evaluate the utility of acquiring these data for a given patient.
Box 1. Bayesian calculation of 22q11.2 frequency in sample of individuals with schizophrenia.
Bayesian Inference allows one to infer underlying states based upon their relationship to observable data. We can, for example, estimate the likelihood that an individual with schizophrenia has the 22q11.2 microdeletion using Bayes’ Theorem, which states:
Where p(SchZ|22q11.2) is the likelihood of having schizophrenia if an individual has the microdeletion; p(22q11.2) is the likelihood of the microdeletion in the general population; p(SchZ|non-22q11.2) is the likelihood of schizophrenia without the microdeletion, and p(not-22q11.2 is the probability of not having the microdeletion in the general population. Since all the parameters on the left side of the equation are known, the equation can be easily solved:
in rough agreement with the results from genetic studies, which suggest that 1–2% of individuals with schizophrenia have the microdeletion (Bassett and Chow, 2008).
One such framework (Fig. 2) is specifically constructed to permit a Bayesian inference approach to linking causes and outcomes and to evaluating and integrating diagnostic and functional data. Conceptually, the framework links causes to outcomes via hidden brain states and behavioral functions (technically considered “latent constructs”). By characterizing large samples of patients with both functional and diagnostic observations, one can create a data framework to ask how these observations relate to the underlying disease processes (causes, brain states and behavioral function), to each other, and to prognostic and treatment response data. One can also then quantitatively ask how much integration of RDoC measures and clinical diagnostics improve clinical prediction and neurobiological modeling of underlying disease states.
Figure 2:
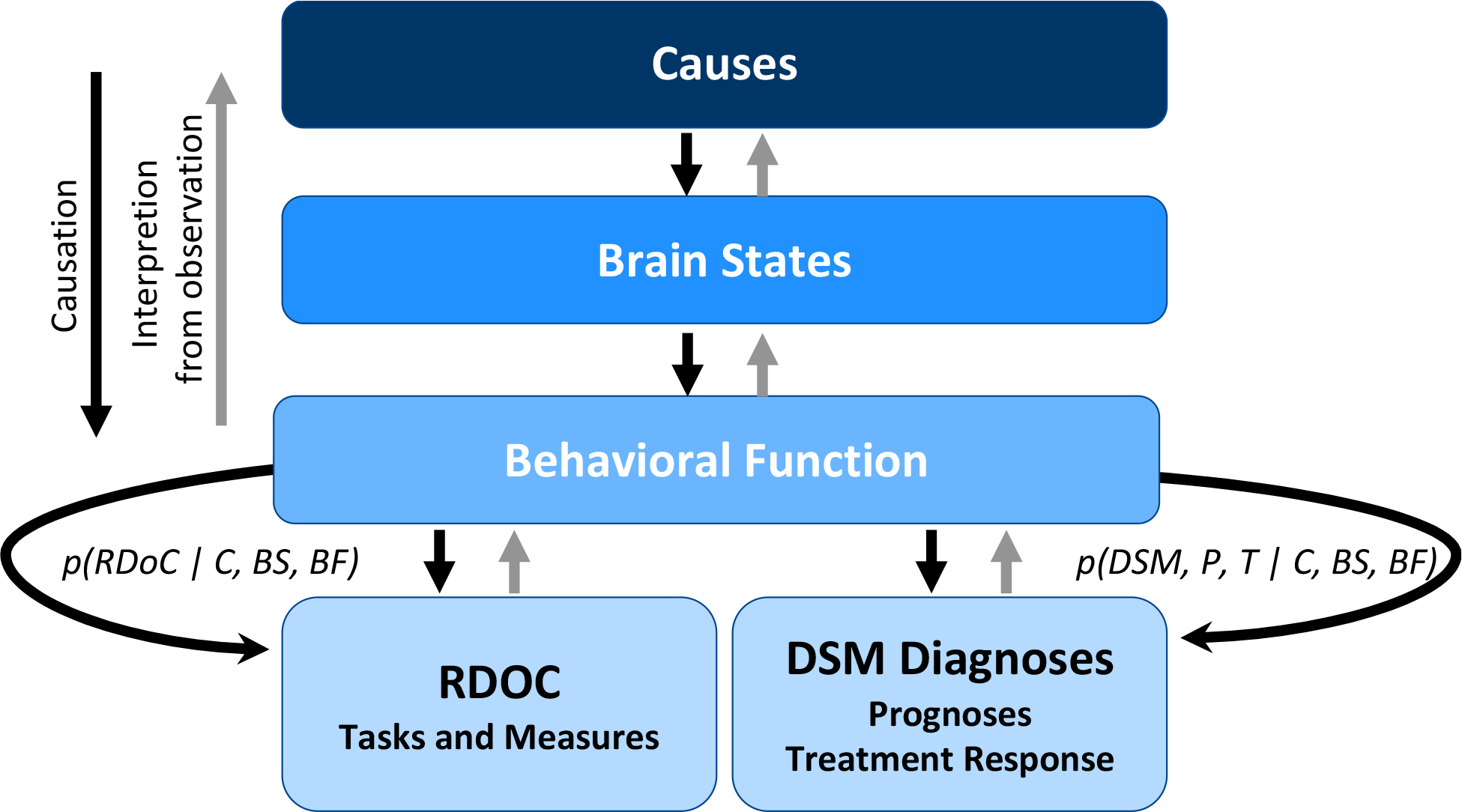
Approach to Integrating RDoC and DSM. Causes of mental illness result in changes in dimensional functions measured by RDoC behaviors as well as clinician-observed diagnoses, via altered brain states and behavioral functions. Bayesian approaches operating on data sets that combine both RDoC measures and clinical records have the potential to facilitate interpretation of underlying brain states and causes as well as quantification of the utility of integrating functional and diagnostic information. Adapted from Flagel et al. 2016
The data needed to initiate such a framework are significant but achievable. What is needed is a large cohort of individuals who consent to participating in remote behavioral assessments and allowing these to be combined with clinical information such as their electronic medical records for research. Biomarker data, genetics, environmental histories, and other data can help make mechanistic links. Several such cohorts exist or are being constructed at the NIH and elsewhere, such as the All of Us Program Research study (Denny et al., 2019) and the Accelerating Medicines Partnership® Schizophrenia initiative (National Institute of Mental Health). Together with a framework that facilitates progress, these new ways of thinking about schizophrenia and novel methods for studying mental illness make this an exciting time for the field.
Footnotes
Conflict of interest: The authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bassett AS, Chow EWC, 2008. Schizophrenia and 22q11.2 deletion syndrome. Curr Psychiatry Rep 10(2), 148–157. 10.1007/s11920-008-0026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corlett PR, Schoenbaum G, 2021. Leveraging basic science for the clinic—from bench to bedside. JAMA Psychiatry 78(3), 331–334. 10.1001/jamapsychiatry.2020.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbert BN, Insel TR, 2013. Toward the future of psychiatric diagnosis: The seven pillars of RDoC. BMC Medicine 11(1), 126. 10.1186/1741-7015-11-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Rutter JL, Goldstein DB, Philippakis A, Smoller JW, Jenkins G, Dishman E, 2019. The "All of Us" research program. New England Journal of Medicine 381(7), 668–676. 10.1056/NEJMsr1809937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet GE, Moser DA, Luber MJ, Leibu E, & Frangou S (2020). Baseline brain structural and functional predictors of clinical outcome in the early course of schizophrenia. Molecular Psychiatry, 25(4), 863–872. 10.1038/s41380-018-0269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel S, Pine D, Ahmari S, First M, Friston K, Mathys C, Redish A, Schmack K, Smoller J, Thapar A, 2016. A novel framework for improving psychiatric diagnostic nosology, in: Redish AD, Gordon JA (Eds.), Computational Psychiatry: New Perspectives on Mental Illness. MIT Press, Cambridge, MA. 10.7551/mitpress/9780262035422.003.0010. [DOI] [Google Scholar]
- Gill CJ, Sabin L, Schmid CH, 2005. Why clinicians are natural bayesians. BMJ 330:1080–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciani D, Cavuto S, Antiga L, Miniati M, Monti S, Pistolesi M, Bertolini G, 2007. Bayes pulmonary embolism assisted diagnosis: a new expert system for clinical use. Emerg Med J 24(3):157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin M-F, Hammoud MZ, Klumpp H, Simon NM, Milad MR, 2020. Multimodal categorical and dimensional approaches to understanding threat conditioning and its extinction in individuals with anxiety disorders. JAMA Psychiatry 77(6), 618–627. 10.1001/jamapsychiatry.2019.4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald-McGinn DM, Sullivan KE, Marino B, Philip N, Swillen A, Vorstman JAS, Zackai EH, Emanuel BS, Vermeesch JR, Morrow BE, Scambler PJ, Bassett AS, 2015. 22q11.2 deletion syndrome. Nature Reviews Disease Primers 1(1), 15071. 10.1038/nrdp.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta UM, Ibrahim FA, Sharma MS, Venkatasubramanian G, Thirthalli J, Bharath RD, Bolo NR, Gangadhar BN, & Keshavan MS (2021). Resting-state functional connectivity predictors of treatment response in schizophrenia - A systematic review and meta-analysis. Schizophrenia Research, 237, 153–165. 10.1016/j.schres.2021.09.004. [DOI] [PubMed] [Google Scholar]
- National Institute of Mental Health. Accelerating Medicines Partnership® Program - Schizophrenia (AMP SCZ). https://www.nimh.nih.gov/research/research-funded-by-nimh/research-initiatives/accelerating-medicines-partnershipr-program-schizophrenia-ampr-scz (accessed 26 October 2021).
- Whitton AE, Kumar P, Treadway MT, Rutherford AV, Ironside ML, Foti D, Fitzmaurice G, Du F, Pizzagalli DA, 2021. Mapping disease course across the mood disorder spectrum through a Research Domain Criteria framework. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging 6(7), 706–715. 10.1016/j.bpsc.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


