Abstract
Background
HER2-low breast cancers were reported to have distinct clinicopathological characteristics from HER2-zero; however, the difference in their genetic features remains unclear. This study investigated the clinical and molecular features of breast tumors according to HER2 status.
Methods
We analyzed the clinicopathological and genomic data of 523 Chinese women with breast cancer. Genomic data was generated by targeted next-generation sequencing (NGS) of breast tumor samples using a commercial 520 gene panel. The cohort was stratified according to HER2 status as HER2-zero (n = 90), HER2-low (n = 231), and HER2-positive (n = 202) according to their immunohistochemistry and fluorescence in situ hybridization results.
Results
HER2-low breast tumors were enriched with hormone receptor-positive tumors, and who had lower Ki67 expression levels. Genes were differentially mutated across HER2 subgroups. HER2-low tumors had significantly more mutations involved in PI3K-Akt signaling than HER2-positive (p < 0.001) and HER2-zero breast tumors (p < 0.01). HER2-zero tumors had more mutations in checkpoint factors (p < 0.01), Fanconi anemia (p < 0.05), and p53 signaling and cell cycle pathway (p < 0.05) compared to HER2-low breast tumors. Compared with HER2-zero tumors, HER2-low tumors had significantly lower pathological complete response rates after neoadjuvant therapy (15.9% vs. 37.5%, p = 0.042) and proportion of relapsed/progressed patients across follow-up time points (p = 0.031), but had comparable disease-free survival (p = 0.271).
Conclusion
Our results demonstrate the distinct clinical and molecular features and clinical outcomes of HER2-low breast tumors.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12916-022-02346-9.
Keywords: Breast cancer, Human epidermal growth factor receptor 2 (HER2), Genomic alteration, HER2-low, Next-generation sequencing (NGS), Targeted therapy
Background
The overexpression of the human epidermal growth factor receptor 2 (HER2) is associated with shorter overall survival and higher risk of disease recurrence. HER2 overexpression is detected in 15–40% of unselected patients with breast cancer [1–4]. Genetic alterations in HER2, also known as Erb-B2 receptor tyrosine kinase 2 (ERBB2), which bring about protein overexpression, have become important biomarkers of response to HER2-targeting agents, including trastuzumab, ado-trastuzumab emtansine (T-DM1), and lapatinib [5–11]. These HER2-targeted therapies have substantially improved the clinical outcomes and survival rates of patients with HER2-positive breast cancer [5–11].
Currently, HER2 status is assayed using standard molecular methods based on the assessment of HER2 protein overexpression by immunohistochemistry (IHC) and estimation of HER2 gene amplification by fluorescence in situ hybridization (FISH). The results from these molecular methods were assessed based on the algorithm recommended by an expert panel from the American Society of Clinical Oncology (ASCO) and the College of American Pathologists (CAP) [12–14]. The most recent update (2018) of this algorithm refined the IHC and FISH criteria into a binary classification system as either HER2-positive or HER2-negative, which eliminated the definition of HER2 equivocal status. This in turn, led to a considerable increase in HER2 FISH-negative cases and a slight decrease in HER2 FISH-positive cases [15, 16].
However, the clinical implications of classifying HER2 equivocal breast tumors as HER2-negative remain unclear. Some studies have demonstrated similar clinical and survival outcomes between HER2 equivocal and HER2-negative/zero breast tumors [15, 16], whereas other studies have revealed distinct clinical, molecular, and survival outcomes between HER2-low and HER2-zero breast tumors [17–20]. A recent report that analyzed a cohort of 2310 patients pooled from four clinical trials demonstrated the distinct biology, clinicopathological features, therapeutic response, and clinical outcome of HER2-low breast cancers (defined by IHC 1+/2+ with FISH-negative result) as compared with HER2-zero, suggesting that HER2-low should be recognized as a new subtype of breast cancer [21].
At present, novel HER2-targeting therapies with mechanisms independent of overexpressed HER2 protein have been developed. These include neratinib, a pan-EGFR tyrosine kinase inhibitor (TKI) [22], and trastuzumab deruxtecan (T-DXd, formerly DS8201a), a novel antibody-drug conjugate (ADC), which elicited 40% objective response for patients with HER2-low metastatic breast cancer [23–25]. Other novel treatments targeting HER2 protein include HER2 vaccines and bispecific antibodies [26–28]. These therapeutic advances raise an important question about the need to reclassify HER2-low as a separate entity. They also highlight the need for alternative methods to comprehensively evaluate HER2 expression and its alteration status to identify patients who might benefit from these novel targeted therapies. Therefore, it is essential to elucidate the underlying genetic background of breast tumors with HER2-zero, HER2-low, or HER2-positive expression status to gain a better understanding of their distinct features. Contemporary diagnostic methods, such as next-generation sequencing (NGS), can be explored to simultaneously evaluate HER2 gene amplification status and other concomitant gene alterations.
We conducted this study to investigate the genomic characteristics of breast tumors with HER2-zero (IHC 0), HER2-low (IHC 1+ or IHC 2+/FISH-negative), and HER2-positive (IHC 3+ or IHC 2+/FISH-positive) status, by retrospectively analyzing the molecular, clinical, and survival data from 523 Chinese women with breast cancer. All breast tumor tissues were subjected to targeted NGS using a large gene panel to comprehensively elucidate their somatic mutational landscape. We then compared the breast cancer subtypes, molecular features, treatment outcome, and disease-free survival data according to HER2 expression status.
Methods
Patient selection
Women diagnosed with breast cancer at Guangdong Provincial People’s Hospital (GDPH) were prospectively recruited for genomic studies and voluntarily submitted samples for NGS as previously described [29]. The study cohort was unselected for clinical or pathological features; however, clinical and molecular data were only retrieved for the patients who met the following study inclusion criteria: (1) patients who were treatment naïve and have not received any systemic therapy; (2) patients with NGS results generated using a 520-gene targeted panel from DNA extracted from formalin-fixed paraffin-embedded (FFPE) tissue sample of the primary breast cancer obtained either by excisional biopsy or core-needle biopsy; (3) available data for HER2 status determined by IHC and/or FISH; (4) complete clinical and pathological results, including estrogen receptor (ER), progesterone receptor (PR), and Ki-67 status. IHC-based surrogate molecular subtypes were classified according to the St. Gallen 2013 guideline [30]. Patients who did not meet all the inclusion criteria were excluded. All the patients provided written informed consent for NGS-based genomic testing. This study was conducted according to the ethical principles of the Declaration of Helsinki, with the study protocol reviewed and approved by the GDPH Ethics Committee (approval no. GDREC2014122H).
Standard HER2 testing
Thin FFPE sections of breast tumor samples from all patients were subjected to initial HER2 testing with IHC using HercepTest (Dako/Agilent, Santa Clara, CA, USA). HER2-positivity was determined as an IHC result of 3+, defined as having >10% intact membrane staining. With an IHC result of 2+ or upon patient request, the slides containing a thin section from the same breast tumor sample of the patient were submitted for subsequent FISH-based HER2 testing using HER2 IQFISH pharmDx (Dako/Agilent, Santa Clara, CA, USA). FISH-based HER2 amplification positivity was defined as HER2 to chromosome 17 centromere (CEP17) signal ratio of ≥2.0 with HER2 signals exceeding 6.0 per cell. The slides were scored by experienced pathologists and/or technicians according to the 2018 ASCO/CAP guidelines [14].
Tissue DNA extraction and capture-based targeted DNA sequencing
Genomic DNA was extracted from FFPE breast tumor tissue with at least 10% tumor content using QIAamp DNA FFPE tissue kit as per the manufacturer’s instructions (Qiagen, Valencia, CA, USA). DNA concentration was determined using Qubit dsDNA high-sensitivity quantification assay on the Qubit fluorometer according to the manufacturer’s instructions (Life Technologies, Carlsbad, CA, USA). Target capture of the tissue DNA samples was performed using a 520-gene panel (OncoScreen Plus, Burning Rock Biotech, Guangzhou, China) following previously published protocols [29, 31, 32]. The 520-gene panel covered 1.64 megabases (Mb) of the human genome and consisted of whole exons of 312 genes and critical exons, introns, and promoter regions of the remaining 208 genes. Briefly, tissue DNA was acoustically fragmented, with DNA fragments of 200–400 bp purified, amplified, and hybridized with capture probes to construct the NGS library. After assessing the quality and fragment size, the indexed samples were sequenced on a Nextseq500 sequencer (Illumina, Inc., San Diego, CA, USA) with paired-end reads at a target average sequencing depth of 1000×. Bioinformatics pipeline developed by Burning Rock Biotech was used to analyze the sequencing data. First, the sequencing reads were aligned to the human reference genome (version hg19) using Burrows-Wheeler aligner (version 0.7.10). Local alignment optimization, variant calling, and annotation of single-nucleotide variations (SNVs) and small insertion-deletions (Indels) were performed using GATK (version 3.2; Broad Institute, Cambridge, MA, USA) and VarScan (version 2.4.3; Genome Institute, Washington University, USA). Analysis of structural variations (SVs) was performed using Factera version 1.4.3. Copy number variations (CNVs) were analyzed based on the depth of coverage. Sequencing bias arising from guanine-cytosine (G-C) content and probe design was corrected against the coverage data. The sequencing coverage of different samples was normalized to comparable scales using the average coverage of all captured regions. CNV was called when the coverage data of the gene region was statistically different from its reference control and with a p-value of <0.005 and a ratio of >0.5. The normal copy number is set at 2 as per the normal diploid genome, copy number ≤1.5 is considered copy number deletion, and copy number ≥2.75 is considered copy number amplification.
Tumor mutation burden (TMB) calculation
TMB per patient was computed as a ratio between the total number of non-synonymous mutations detected and the total coding region size of the panel using the equation below. The mutation count included non-synonymous SNVs and Indels detected within the coding region and ±2 bp upstream or downstream region and does not include hot mutation events, CNVs, SVs, and germline single-nucleotide polymorphisms (SNPs). Only mutations with allelic fractions (AF) of ≥ 2% for tissue samples and ≥ 0.2% for plasma samples were included in the mutation count. For accurate TMB calculation, maximum AF (maxAF) was calculated as ≥ 5% for tissue samples and ≥ 1% for plasma samples. The total size of the coding region for estimating TMB was 1.003 Mb for the 520-gene OncoScreen Plus panel.
Neoadjuvant therapy and treatment outcomes
Neoadjuvant regimens were administered as various combinations of systemic chemotherapy/targeted inhibitors according to the patient’s HER2 and HR status and the physician’s decision. Regimens included were taxanes (T), epirubicin (E), doxorubicin (A), cyclophosphamide (C), 5-fluorouracil (F), trastuzumab (T), pertuzumab (P), carboplatin (Cb), and capecitabine (X). All combination regimens were administered at standard dosing schedules. Pathological complete response (pCR) was assessed using the completely resected breast specimen and all regional lymph nodes sampled after neoadjuvant therapy. pCR was defined as the absence of residual invasive cancer cells on hematoxylin and eosin evaluation of all specimens.
Survival analysis
Disease-free survival (DFS) was defined as the time in months from the date of diagnosis until the date of either disease relapse/disease progression or last follow-up. The proportion of patients who had radiologically confirmed disease relapse (PD ratio) was calculated as the ratio of patients with relapse to the total number of patients who remained disease-free at that particular year of follow-up, excluding the number of patients who were lost to follow-up. The data cut-off date was October 15, 2021.
Statistical analysis
The R statistics package (R version 4.0.5; R: The R-Project for Statistical Computing, Vienna, Austria) was used to analyze the data. Patient characteristics and sequencing results were summarized with descriptive statistics. Mutation profiles of HER2-positive, HER2-low, and HER2-zero groups in the GDPH cohort were compared using Fisher’s exact test and Wilcoxon signed-rank test, as deemed appropriate. Data clustering was performed using non-negative matrix factorization (NMF) based on Euclidean distance. R package NMF was implemented to estimate the best rank using Lee’s algorithm with the following initial parameters: 2:8 for rank, and numeric random seed (seed = 123456). The most ideal values for cophenetic consensus were observed at ranks 3–4; hence, we selected rank 3 to cluster our data into three subgroups. Kaplan-Meier survival curves with log-rank statistics were plotted to compare DFS among subgroups. All statistical tests were performed as two-sided test and significance level was set at P < 0.05.
Results
Baseline characteristics of the cohort
Figure 1 illustrates our study design. This study included 523 women with breast cancer with a median age of 48 years (range 25–85 years). The majority of the cohort was comprised of patients with early-stage breast cancer (72.3%, n = 378), with invasive carcinoma of no special type (NST) (92.4%, n = 483), and HR-positive breast cancer (72.7%, n = 380). The cohort was further stratified according to HER2 status based on the 2018 ASCO/CAP guidelines. A total of 321 patients were HER2-negative as per the guidelines, with 90 patients (17.2%) having IHC 0, defined as HER2-zero, and 231 patients (44.2%) having either IHC 1+ or IHC 2+ with negative FISH results, defined as HER2-low. The remaining 202 patients (38.6%) had either IHC 3+ or IHC 2+ with positive FISH results, defined as HER2-positive. Table 1 summarizes the clinicopathological characteristics of the cohort and the patient subgroups. The patients stratified according to HER2 status had similar age, menopausal status, and pathological stage distribution, but had statistically different histological types, histological grades, hormone receptor (HR) status, Ki67 expression levels, and IHC-based molecular subgroup status.
Fig. 1.
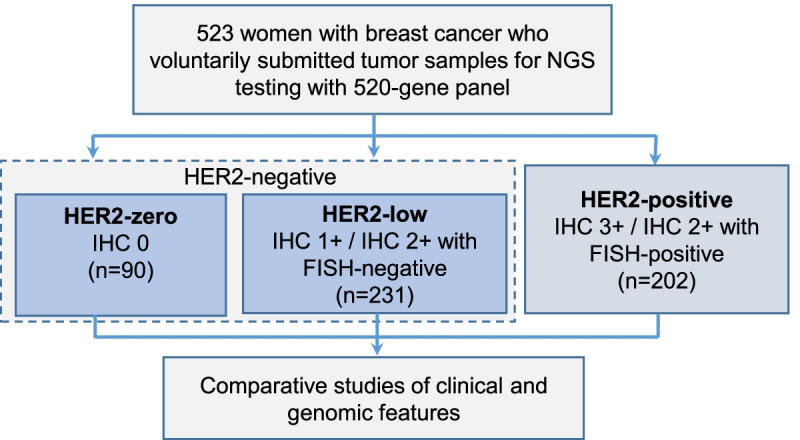
Diagram of the study design
Table 1.
Baseline clinicopathological characteristics of the cohort
| Clinicopathological characteristics | Total (n=523); n(%) |
IHC/FISH-based HER2 status subgroup; n(%) | |||
|---|---|---|---|---|---|
| HER2-zero (n=90) |
HER2-low (n=231) |
HER2-positive (n=202) |
p-value | ||
| Age (median [range], years) | 48 [25-85] | 46[25-85] | 49[27-76] | 49.5[27-74] | ns |
| Menopausal status | |||||
| Pre-menopause | 297(56.8%) | 56(62.2%) | 131(56.7%) | 110(54.5%) | ns |
| Post-menopause | 226(43.2%) | 34(37.8%) | 100(43.3%) | 92(45.5%) | |
| Tumor (T) stage | |||||
| T1-T2 | 469(89.7%) | 80(88.9%) | 208(90.0%) | 181(89.6%) | ns |
| T3-T4 | 51(9.7%) | 8(8.9%) | 22(9.5%) | 21(10.4%) | |
| Unknown | 3(0.6%) | 2(2.2%) | 1(0.5%) | 0 | |
| Lymph node (N) stage | |||||
| N0-N1 | 400(76.5%) | 68(75.6%) | 176(76.2%) | 156(77.2%) | ns |
| N2-N3 | 121(23.1%) | 20(22.2%) | 55(23.8%) | 46(22.8%) | |
| Unknown | 2(0.4%) | 2(2.2%) | 0 | 0 | |
| Metastasis (M) stage | |||||
| M0 | 495(94.6%) | 84(93.3%) | 226(97.8%) | 185(91.6%) | Zero vs Low, p =0.036 |
| M1 | 26(5.0%) | 4(4.4%) | 5(2.2%) | 17(8.4%) | Zero vs Positive, ns |
| Mx | 2(0.4%) | 2(2.2%) | 0 | 0 | Low vs Positive, p =0.004 |
| Pathological stage | |||||
| I-II | 378(72.3%) | 65(72.2%) | 168(72.7%) | 145(71.8%) | ns |
| III-IV | 142(27.1%) | 23(25.6%) | 62(26.8%) | 57(28.2%) | |
| Unknown | 3(0.6%) | 2(2.2%) | 1(0.5%) | 0 | |
| Histological grade | |||||
| 1 | 13(2.5%) | 3(3.3%) | 8(3.5%) | 2(1.0%) | Zero vs Low, p <0.001 |
| 2 | 248(47.4%) | 31(34.4%) | 137(59.3%) | 80(39.6%) | Zero vs Positive, ns |
| 3 | 248(47.4%) | 50(55.6%) | 82(35.5%) | 116(57.4%) | Low vs Positive, p <0.001 |
| Unknown | 14(2.7%) | 6(6.7%) | 4(1.7%) | 4(2.0%) | |
| Histological type | |||||
| Invasive carcinoma of no special type (NST) | 483(92.4%) | 77(85.6%) | 214(92.6%) | 192(95.0%) | Zero vs Low, p =0.043 |
| Lobular, invasive | 15(2.9%) | 4(4.4%) | 10(4.3%) | 1(0.5%) | Zero vs Positive, p =0.008 |
| Other invasive histology | 25(4.7%) | 9(10.0%) | 7(3.1%) | 9(4.5%) | Low vs Positive, p =0.023 |
| Estrogen Receptor (ER) status | Zero vs Low, p <0.001 | ||||
| Positive | 360(68.8%) | 59(65.6%) | 198(85.7%) | 103(51.0%) | Zero vs Positive, p =0.022 |
| Negative | 163(31.2%) | 31(34.4%) | 33(14.3%) | 99(49.0%) | Low vs Positive, p <0.001 |
| Progesterone Receptor (PR) status | Zero vs Low, p <0.001 | ||||
| Positive | 335(64.1%) | 52(57.8%) | 191(82.7%) | 92(45.5%) | Zero vs Positive, ns |
| Negative | 188(35.9%) | 38(42.2%) | 40(17.3%) | 110(54.5%) | Low vs Positive, p <0.001 |
| Hormone Receptor (HR) status | Zero vs Low, p <0.001 | ||||
| Positive | 380(72.7%) | 60(66.7%) | 202(87.4%) | 118(58.4%) | Zero vs Positive, ns |
| Negative | 143(27.3%) | 30(33.3%) | 29(12.6%) | 84(41.6%) | Low vs Positive, p <0.001 |
| Ki67 expression level (median, IQR) | 30.0%[15.0%, 50.0%] | ||||
| Ki67 expression status (cut-off 30%) | Zero vs Low, p =0.013 | ||||
| Low (<30%) | 225(43.4%) | 38(42.2%) | 132(57.9%) | 55(27.5%) | Zero vs Positive, p =0.015 |
| High (≥30%) | 293(56.6%) | 52(57.8%) | 96(42.1%) | 145(72.5%) | Low vs Positive, p <0.001 |
| Immunohistochemistry-based surrogate molecular subgrouping | |||||
| Luminal A (ER±/PR±/HER2-/Ki67<14%) | 91(17.4%) | 25(27.8%) | 66(28.6%) | 0 | Zero vs Low, p <0.001 |
| Luminal B HER2- (ER±/PR±/HER2-/Ki67≥14%) | 171(32.7%) | 35(38.9%) | 136(58.9%) | 0 | Zero vs Positive, p <0.001 |
| Luminal B HER2+ (ER±/PR±/HER2+/Ki67≥14%) | 118(22.6%) | 0 | 0 | 118(58.4%) | Low vs Positive, p <0.001 |
| HER2-enriched (ER-/PR-/HER2+) | 84(16.1%) | 0 | 0 | 84(41.6%) | |
| Triple-negative (ER-/PR-/HER2-) | 59(11.3%) | 30(33.3%) | 29(12.6%) | 0 | |
Abbreviations: ns not statistically significant
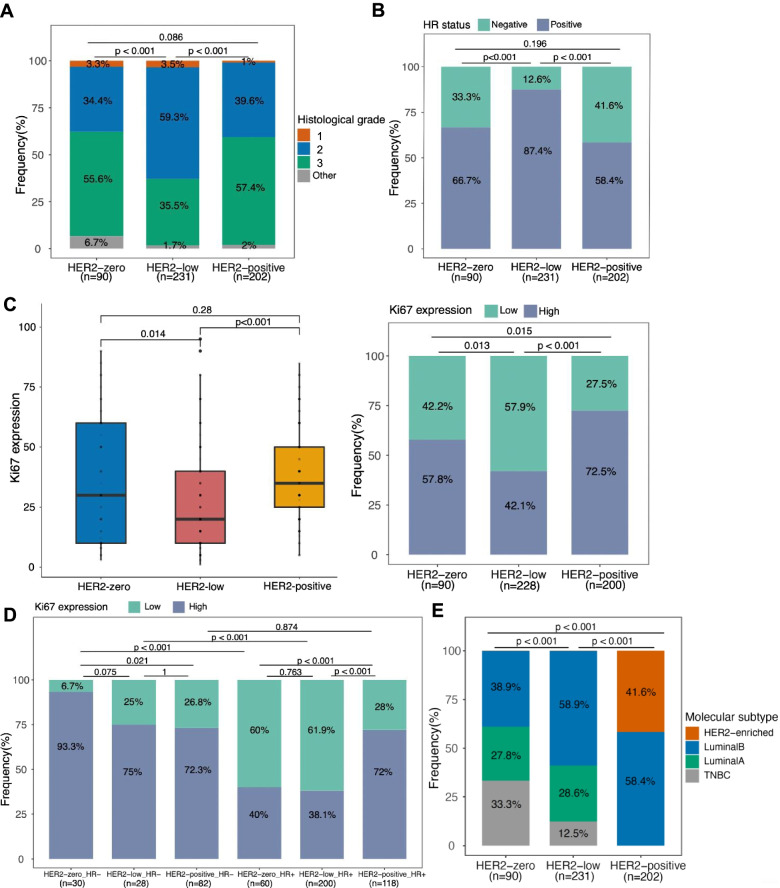
Distinct clinical features of HER2-low breast tumors
The HER2-low subgroup were distinctive based on three criteria: (1) the subgroup exhibited more histological grade 2 than HER2-zero (59.3% vs. 34.4%, p < 0.001) and more than HER2-positive (59.3% vs. 39.6%, p < 0.001) (Fig. 2A); (2) The HER2-low subgroup had significantly more patients with HR-positive status than HER2-zero patients (87.4% vs. 66.7%, p < 0.001) and HER2-positive patients (87.4% vs. 41.6%, p < 0.001) subgroups (Fig. 2B); and (3) the HER2-low subgroup had significantly lower Ki67 expression levels than the HER2-zero patients (p = 0.014) and the HER2-positive patients (p < 0.001)(Fig. 2C). The HER2-zero and HER2-positive subgroups had no statistical difference in histological grade (p = 0.086), HR status (p = 0.196), and Ki67 expression level (p = 0.28). We also used a median cut-off of 30% to stratify the Ki67 as low (<30%) and high (≥30%). Among the HR-negative tumors, the HER2-zero subgroup had a significantly higher proportion of tumors with high Ki67 expression than the HER2-positive subgroup (99.3% vs. 72.3%, p = 0.021). Among the HR-positive tumors, the HER2-positive subgroup had a significantly higher proportion of tumors with high Ki67 expression than HER2-low (72.0% vs. 38.1%, p < 0.001) and HER2-zero subgroup (72.0% vs. 40.0%, p < 0.001) (Fig. 2D). Moreover, among the HER2-low subgroup, those with HR-positive status had a significantly higher proportion of low Ki67 expression than those with HR-negative status (61.9% vs. 25.0%, p < 0.001) (Fig. 2D). According to IHC-based molecular subtype distribution, the HER2-low subgroup was comprised predominantly of luminal B tumors (58.9%) and was 28.6% of luminal A tumors, and 12.5% triple-negative breast cancer (TNBC). This distribution was significantly different from HER2-zero (p < 0.001) and HER2-positive (p < 0.001) subgroups (Fig. 2E). As compared with the HER2-zero subgroup, the HER2-low subgroup had a significantly higher proportion of luminal B tumors (58.9% vs. 38.9%, p = 0.002), and a significantly lower proportion of TNBC tumors (12.5% vs. 33.3%, p < 0.001). It should be noted that all luminal B tumors in HER2-low and HER2-zero subgroups were HER2-negative. Taken together, these findings indicate the difference in clinical features among HER2-zero, HER2-low, and HER2-positive breast tumors.
Fig. 2.
HER2 subgroups had different clinical features, including A histological grade, B hormone receptor (HR) expression status, C,D Ki-67 expression levels, and E IHC-based molecular subtype distribution
Mutation landscape of the cohort
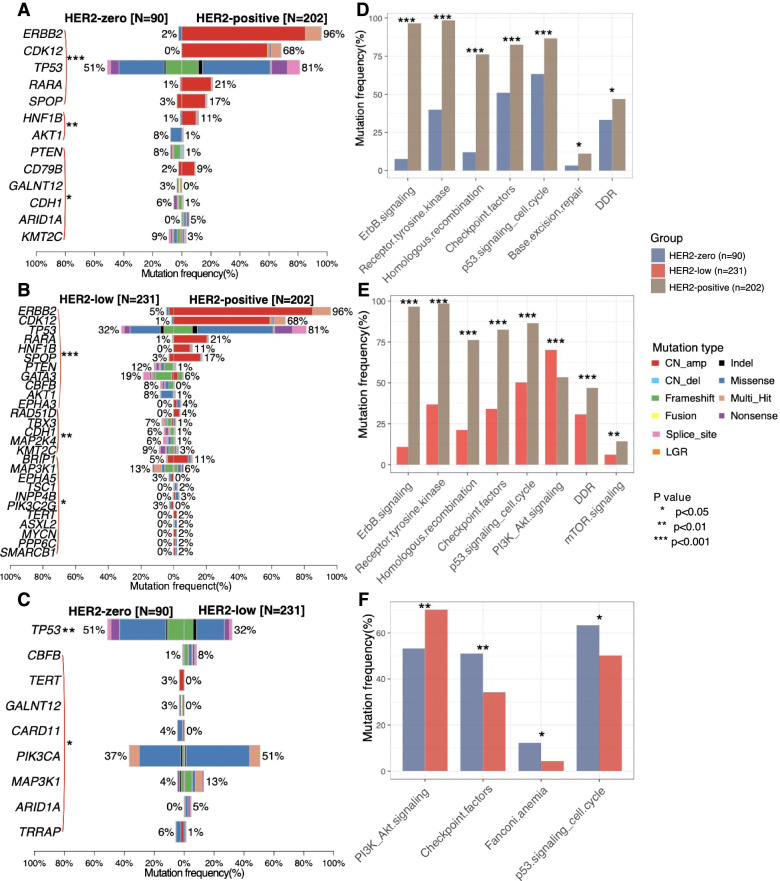
Genomic results from targeted sequencing of breast tumors using a large 520-gene panel were analyzed and are summarized in supplementary Additional file 1: Fig. S1. Tumor protein 53 (TP53) and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), respectively, were detected in 54% and 46% of the cohort and were the two most frequently mutated genes. Copy number amplifications in cyclin-dependent kinase 12 (CDK12) were only exclusively detected together with copy number amplifications in HER2 (n = 113; p < 0.001; Additional file 1: Fig. S1).
Distinct somatic mutation landscape and mutated pathways across HER2 subgroup
We then compared the somatic mutation landscape according to the HER2 subgroups. Besides HER2 gene amplifications, HER2-positive breast tumors were detected with more copy number amplification in genes including CDK12 (p < 0.001), retinoic acid receptor alpha (RARA; p < 0.001), and speckle-type POZ protein (SPOP; p < 0.001) and significantly higher TP53 mutation rates (p < 0.001) as compared with HER2-zero (Fig. 3A, Additional file 1: Fig. S2A) and HER2-low breast tumors (Fig. 3B, Additional file 1: Fig. S2B). HER2-low breast tumors had significantly more frequent mutations in phosphatase and tensin homolog (PTEN; p < 0.001), GATA binding protein 3 (GATA3; p < 0.001), core-binding factor subunit beta (CBFB; p < 0.001), and AKT serine/threonine kinase 1 (AKT1; p < 0.001) than HER2-positive breast tumors (Fig. 3B, Additional file 1: Fig. S2B). HER2-low breast tumors also had significantly higher mutations rates in CBFB (p < 0.05), PIK3CA (p < 0.05), mitogen-activated protein kinase kinase kinase 1 (MAP3K1; p < 0.05), and AT-rich interaction domain 1A (ARID1A; p < 0.05) as compared with HER2-zero breast tumors (Fig. 3C, Additional file 1: Fig. S2C). Moreover, HER2-zero breast tumors had significantly higher mutation rates in TP53 (p < 0.01), telomerase reverse transcriptase (TERT; p < 0.05), polypeptide N-acetylgalactosaminyltransferase 12 (GALNT12; p < 0.05), caspase recruitment domain family member 11 (CARD11; p < 0.05), and transformation/transcription domain-associated protein (TRRAP; p < 0.05) than HER2-low tumors (Fig. 3C, Additional file 1: Fig. S2C).
Fig. 3.
HER2 subgroups had differentially mutated genes (A–C) and molecular signaling pathways (D–F)
Of the TP53 mutations detected, approximately 80% were located across exons 5–8 for all HER2 subgroups (Additional file 1: Fig. S3). Overall, the TP53 exon 5–8 mutation rate was 40.0% (36/90) for HER2-zero, 26.0% (60/231) for HER2-low, and 67.8% (137/202) for HER2-positive breast tumors. PIK3CA mutations were distributed across the functional domains (Additional file 1: Fig. S4). PIK3CA E545K (n = 11) and H1047R/L (n = 11) were the most common PIK3CA mutations in HER2-zero. PIK3CA H1047R/L was the most common in HER2-low (n = 71) and HER2-positive (n = 46) breast tumors.
We also explored the genes and signaling pathways commonly mutated in HER2 subgroups and analyzed them according to HR status and IHC-based molecular subtypes. The two commonly mutated genes across HER2 subgroups and HR status were TP53 and PIK3CA (Additional file 1: Fig. S5A). No genes or signaling pathways were differentially mutated between HER2-low and HER2-zero tumors with HR-negative status (Additional file 1: Fig. S5B). Only the mutation rate of RPTOR-independent companion of mammalian target of rapamycin (RICTOR) was found to be significantly higher in HER2-zero as compared to HER2-low tumors with HR-positive status (7% [4/60] vs. 0% [0/220], p = 0.011; Additional file 1: Fig. S5C). Despite being numerically higher in HER2-zero subgroup, TP53 mutation rate was not statistically different between HER2-zero and HER2-low breast tumors regardless of HR status (HR-positive [HER2-zero vs. HER2-low], 33.3% vs. 26.2%, p = 0.325; HR-negative 86.7% vs. 72.4%, p = 0.209; Additional file 1: Fig. S6A). Likewise, PIK3CA mutations were numerically higher in HER2-low subgroup; however, no statistical difference was observed between HER2-low and HER2-zero subgroups with HR-negative (23.3% vs. 37.9%, p = 0.267) and HR-positive status (43.3% vs. 52.5%, p = 0.241) (Additional file 1: Fig. S6B). However, as compared with HR-negative breast tumors, those with HR-positive status had significantly fewer TP53 mutations among HER2-zero (33.3% [20/60] vs. 86.7% [26/30], p < 0.001) and HER2-low (26.2% [53/202] vs. 72.4% [21/29], p < 0.001), but not among HER2-positive breast tumors (79.7% [94/118] vs. 85.7% [72/84], p = 0.351) (Additional file 1: Fig. S6A). PIK3CA and TP53 were the two commonly mutated genes in HER2-low and HER2-zero tumors across IHC-based molecular subgrouping (Additional file 1: Fig. S7). TP53 mutation rates were numerically higher in HER2-zero tumors but not statistically different between HER2-zero and HER2-low tumors with TNBC (86.7% vs. 72.4%, p = 0.209), luminal A (16.0% vs. 12.1%, p = 0.730), and luminal B (45.7% vs. 33.1%, p = 0.172; Additional file 1: Fig. S8A). According to IHC-based molecular subgrouping, TP53 mutations were more common in TNBC and less frequent in luminal A for both HER2-zero and HER2-low tumors (Additional file 1: Fig. S8A). PIK3CA mutation rates were numerically higher in HER2-low tumors but not statistically different between HER2-low and HER2-zero tumors with TNBC (37.9% vs. 23.3%, p = 0.267), luminal A (63.3% vs. 56.0%, p = 0.630), and luminal B (47.1% vs. 34.3%, p = 0.118) (Additional file 1: Fig. S8A). PIK3CA mutations were more frequent in luminal A and least common in TNBC (Additional file 1: Fig S8B). No genes or signaling pathways were differentially mutated between TNBC tumors from HER2-low and HER2-zero subgroups (Additional file 1: Fig. S9A). The lack of statistical significance in these subgroup analyses might be due to the smaller sample size in each subgroup after stratification. In luminal A tumors, the mutation rate of spliceosome factor 3b (SF3B1) was found to be significantly higher for HER2-zero than HER2-low subgroup (12% [3/25] vs. 0% [0/66], p < 0.05; Additional file 1: Fig. S9B). In luminal B tumors, the mutations rates of TERT (6% [2/35] vs. 0% [0/136], p < 0.05), GALNT12 (6% [2/35] vs. 0% [0/136], p < 0.05), and RICTOR (11% [3/35] vs. 0% [0/136], p < 0.05) were significantly higher for HER2-zero than HER2-low subgroup (Additional file 1: Fig. S9C).
We further investigated the signaling pathways commonly mutated in each HER2 subgroup. Aside from the ErbB/HER pathway, HER2-positive breast tumors had frequent mutations in genes involved in receptor tyrosine kinase signaling (p < 0.001), homologous recombination (p < 0.001), checkpoint factors (p < 0.01), and p53 signaling and cell cycle (p < 0.01) as compared with HER2-zero (Fig. 3D) and HER2-low breast tumors (Fig. 3E). In contrast, HER2-low tumors had significantly more mutations involved in PI3K-Akt signaling pathways as compared with HER2-positive (p < 0.001) (Fig. 3E) and HER2-zero breast tumors (p < 0.01) (Fig. 3F). HER2-zero tumors had significantly more mutations in checkpoint factors (p < 0.01), Fanconi anemia (p < 0.05), and p53 signaling and cell cycle pathway (p < 0.05) than HER2-low breast tumors (Fig. 3F).
Although mutation detection rates were varied across the HER2 subgroups, the comparison of their TMB demonstrated no statistical difference (Additional file 1: Fig. S10). The median TMB was 3.67 (range 0–32.11) mutations/Mb for HER2-positive, 3.67 (range 0–19.27) mutations/Mb for HER2-low, and 2.75 (range 0–32.11) mutations/Mb for HER2-zero breast tumor.
Taken together, these data suggest some significant differences in the genomic features for breast tumors with HER2-positive, HER2-low, and HER2-zero status.
Mutations in HER family among the HER2 subgroups
We also explored the mutation frequency of other members of the ErbB/HER family. The HER family consists of four receptor tyrosine kinases, epidermal growth factor receptor (EGFR or ERBB1), ERBB2 (or HER2), ERBB3, and ERBB4. Additional file 1: Table S1 summarizes the distribution of HER family mutations of the cohort. HER2-positive breast tumors had the highest mutation rate in genes in the HER family (11.9%) but were not statistically different from the mutation rate in HER2-low (8.7%) and HER2-zero breast tumors (7.8%) (Additional file 1: Fig. S11A). SNVs affecting the ERBB2 gene, such as missense mutations and splice-site mutations, were detected in two HER2-zero patients (Additional file 1: Fig. S11B), four HER2-low patients (Additional file 1: Fig. S11C), and 13 HER2-positive patients (Additional file 1: Fig. S11D). Concurrent ERBB2 amplification and ERBB2 P122A and D277E missense mutations were detected in two patients with IHC 3+. EGFR mutations were more frequently detected in HER2-positive tumors (4.9%). ERBB3 mutations were detected in four patients (4.4%) with HER2-zero tumors. ERBB4 mutations were detected in four patients each with HER2-low (1.7%) and HER2-positive tumors (2.0%).
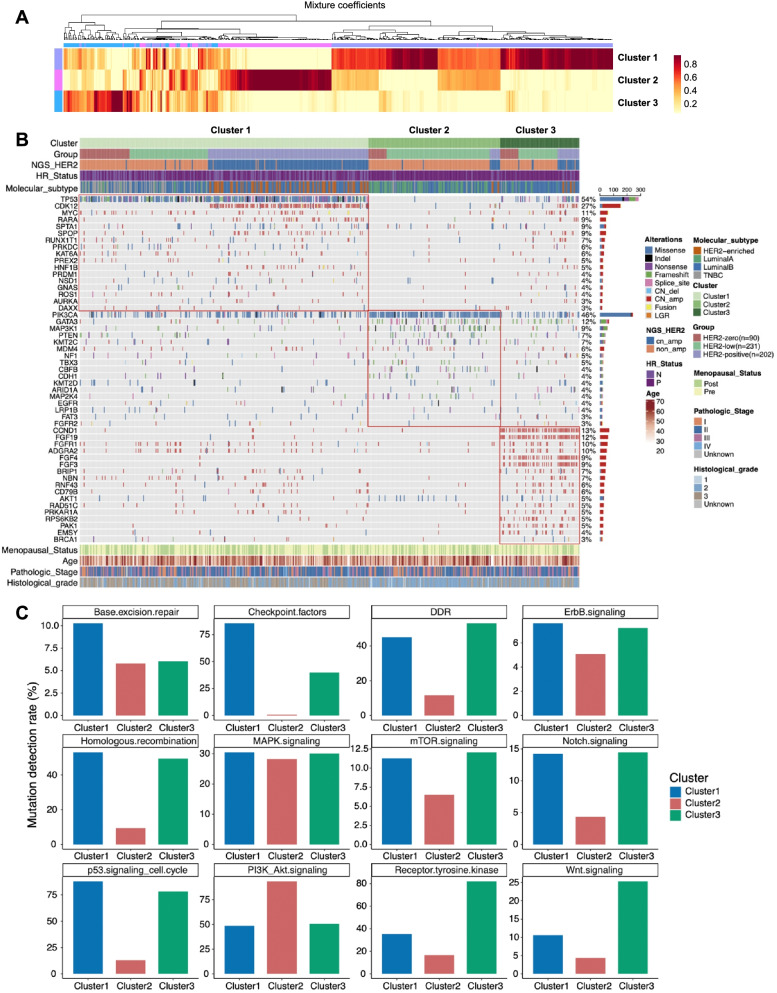
Clustering analysis reveals distinct mutational features and pathways
We then performed data clustering using the NMF approach to identify common clusters of mutations from the mutational profile of the cohort. HER2 gene amplification was excluded for this analysis to eliminate the confounding effects of the HER2 subgrouping.
Unsupervised clustering of the mutation profile of the cohort revealed three molecularly distinct clusters as shown by the heat map in Fig. 4A and the oncoprint in Fig. 4B. Cluster 1 was enriched in TP53 mutations, CDK12, MYC, and RARA amplifications. Cluster 2 was enriched in PIK3CA, GATA3, and MAP3K1 mutations, with a lesser number of gene amplification and the absence of TP53 mutation. Cluster 3 was enriched in copy number amplifications of various genes, including cyclin D1 (CCND1), fibroblast growth factor (FGF) 3 (FGF3), FGF4, FGF19, ribosomal protein S6 kinase B2 (RPS6KB2), P21 activated kinase 1 (PAK1), and EMSY BRCA2-interacting transcriptional repressor (EMSY), which are associated with chromosome 11q13 loci amplification, as well as FGF receptor 1 (FGFR1) and adhesion G protein-coupled receptor A2 (ADGRA2), which are both located in chromosome 8p12.
Fig. 4.
Distinct mutational features as demonstrated by clustering of mutation profiles of the cohort. A Heat map illustrating the unsupervised hierarchical clustering of mutation profile of the cohort. B Oncoprint summarizing the mutational features of each cluster. C Comparison of the molecular signaling pathways that are differentially mutated in each cluster
Pathway analysis demonstrated that genes in Cluster 1 were involved in base excision repair, homologous recombination, p53 signaling and cell cycle, checkpoint factor, ErbB signaling, and Notch signaling. Cluster 2 was comprised of genes involved in PI3K/Akt pathway. Cluster 3 was comprised of genes involved in DNA damage response, receptor tyrosine kinase, and Wnt signaling pathway (Fig. 4C).
According to HER2 subgrouping, Cluster 1 comprised of 55.6% (168/302) HER2-positive, 27.2%% (82/302) HER2-low, and 17.2% (52/302) HER2-zero breast tumors. Of them, 178 patients (58.9%) were HR-positive. Cluster 2 comprised of 78.3% (108/138) HER2-low, 13.7% (19/139) HER2-zero, and 8.0% (11/138) HER2-positive breast tumors. Cluster 3 comprised of 49.4% (41/83) HER2-low, 27.7% (23/83) HER2-positive, and 22.9% (19/83) HER2-zero breast tumors. A majority of the patients in Cluster 2 (92.8%, 128/138) and Cluster 3 (89.2%, 74/83) were HR-positive (Additional file 1: Fig. S12A). According to the IHC-based molecular subgrouping, Cluster 1 was comprised of 54.0% (163/302) luminal B, 24.5% (74/302) HER2-enriched, 16.6% (50/302) TNBC, and 5% (15/302) luminal A. Cluster 2 was comprised of 47.1% (65/138) luminal A, 45.7% (63/138) of luminal B, 4.3% (6/138) TNBC, and 2.9% (4/138) HER2-enriched. Cluster 3 was comprised of a majority of luminal B (75.9%, 63/83), 13.3% (11/83) luminal A, 7.2% (6/83) HER2-enriched, and 3.6% (3/83) TNBC (Additional file 1: Fig. S12B).
These findings suggest the existence of molecular subsets within HER2-positive, HER2-low, and HER2-zero subgroups harboring certain mutational features that could contribute to the molecular heterogeneity of these tumors.
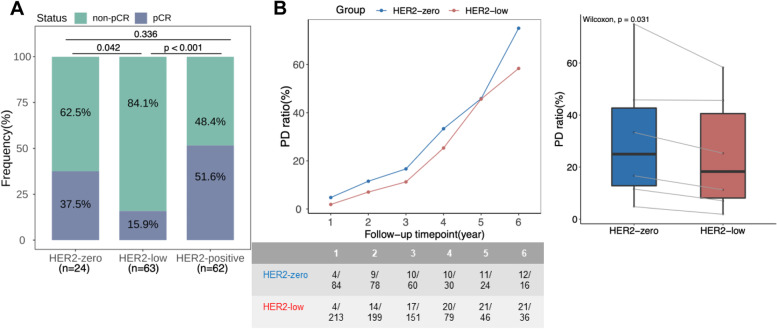
Clinical outcomes
We further analyzed the treatment outcomes of 149 patients who received neoadjuvant therapy with available pathological response data. HER2-low subgroup had significantly lower pCR rates than HER2-zero (15.9% [10/63] vs. 37.5% [9/24], p = 0.042) and HER2-positive subgroups (15.9% [10/63] vs. 51.6% [32/62], p < 0.001) (Fig. 5A). pCR rates were numerically lower in HER2-low subgroup with HR-positive (9.3% vs. 20.0%, p = 0.358), HR-negative (55.6% vs. 66.7%, p = 1), TNBC (55.6% vs. 66.7%, p = 1), and luminal B (10.8% vs. 25.0%, p = 0.340), but was numerically higher in luminal A tumors (5.9% vs. 0.0%, p = 1); however, statistical analysis showed no significant difference between HER2-low and HER2-zero subgroups across HR status and IHC-based molecular subtypes possibly due to lower sample size in some subgroups (Additional file 1: Fig. S13).
Fig. 5.
HER2-low breast tumor had different clinical outcomes than HER2-zero. A HER2-low subgroup had lower pathological complete response (pCR) rates. B Proportion of relapsed/progressed patients were fewer in HER2-low subgroup across follow-up time points. The table below summarizes the number of relapsed patients over the total number of included patients per time point (in year) per subgroup. The gray connecting lines in the box plot denote the follow-up data per year corresponding to the data point in the dot plot on the left
We also analyzed the DFS for all patients. DFS were comparable between HER2-low and HER2-zero subgroups (p = 0.271; Additional file 1: Fig. S14A), regardless of pCR status (Additional file 1: Fig. S15), HR status (Additional file 1: Fig. S16), and IHC-based molecular subtypes (Additional file 1: Fig. S17). It should be noted that the DFS data was only 10.1% (53/523) mature and results might be inconclusive. Hence, we performed further analysis to compare the DFS of patients who experienced disease relapse (n = 53) according to HER2 subgroups. As expected, women with HER2-zero (n = 12; p = 0.025) and HER2-low (n = 21; p = 0.031) tumors had significantly shorter DFS than HER2-positive tumors (n = 20); however, DFS was comparable between HER2-zero and HER2-low subgroups (Additional file 1: Fig. S14B). Interestingly, the proportion of relapsed/progressed patients was significantly lower in HER2-low subgroup than HER2-zero subgroup across follow-up time points (p = 0.031; Fig. 5B). Despite comparable DFS, these findings suggest that HER2-low tumors had different clinical outcomes than HER2-zero tumors as shown by the significantly lower pCR rate but a higher proportion of disease-free patients across follow-up time points.
Discussion
In this study, we included a cohort of 523 Chinese women with breast cancer stratified into three subgroups according to their HER2-expression status as HER2-positive, HER2-low, and HER2-zero to compare their clinicopathological characteristics and NGS-based genetic alteration landscape. Our results describe the differences among the three HER2 categories at the clinicopathological and molecular level. From a clinical perspective, HER2-low patients were significantly more likely to be HR positive, with significantly lower Ki-67 expression level, and had significantly lower pCR rate with neoadjuvant chemotherapy than HER2-zero patients. From a molecular perspective, HER2-low tumors displayed a different somatic mutation landscape and mutated pathways, such as the higher prevalence of PIK3CA mutations and lower prevalence of TP53 mutations than HER2-zero tumors. However, whether these differences in terms of molecular features are at least partly driven by the distinct clinicopathological features remains to be established.
In recent years, anti-HER2 treatment has successfully expanded its clinical application to target HER2-low expressing tumors using novel ADCs such as T-DXd and SYD 985. These novel ADCs were designed to effectively be anchored and delivered into the cell even those with low HER2 expression level to exert their cytotoxic effects via by-stander effect with significantly enhanced potency compared to the previous generation of ADCs [25, 33]. Since HER2-low expressing breast tumors constitute nearly half of all breast tumors, the recent availability of an established therapeutic strategy with novel ADCs for this subgroup have triggered new investigations to comprehensively characterize the clinical and genetic features of this new patient subgroup. Recently, Denkert et al. conducted a study by pooling individual patient data out of 2310 patients with HER2-negative primary breast cancer defined by the ASCO/CAP algorithm from four prospective neoadjuvant clinical trials [21]. Their findings, in the setting of clinical trial cohorts, revealed the distinct clinical features of HER2-low-positive breast cancer, such as significantly higher HR-positive expression, lower Ki-67 expression, and lower complete pathological response to neoadjuvant chemotherapy but longer overall survival, as compared with HER2-zero breast cancer. Moreover, they also found that HER2-low-positive breast tumors had a higher prevalence of PIK3CA mutations and a lower prevalence of TP53 mutations than HER2-zero breast tumors. Their study provided evidence to consider HER2-low tumors as a new subtype of breast cancer with distinct biology, clinical characteristics, and prognosis from HER2-zero tumors. Our findings, while in the real-world patient setting, are consistent with the findings reported by Denkert et al [21]. Findings from both cohorts similarly revealed that HER2-low breast tumors had higher frequency of PIK3CA mutations, lower frequency of TP53 mutations, and lower pCR rates with neoadjuvant therapy as compared with HER2-zero breast tumors.
In our study, the lower pCR rates of HER2-low subgroup had a similar trend in HR-positive, HR-negative/TNBC, and luminal B tumors; however, these comparisons across HR status and IHC-based molecular subtypes were inconclusive due to the small sample size of some of the subgroups (i.e., HR-negative and TNBC only had nine patients each per subgroup, luminal A only had three patients in HER2-zero and 17 in HER2-low, and luminal B only had 12 and 37 patients). Despite the inconclusive DFS due to the largely immature survival data from our cohort, our findings still showed that a significantly lesser proportion of women with HER2-low breast tumors experienced disease relapse up to 6 years of follow-up. This finding suggest that women with HER2-low are expected to have longer DFS and better survival outcomes than HER2-zero breast tumors.
The observed differences in clinical outcomes regarding pCR rates and survival trends could be attributed to differences in IHC-based molecular subtype distribution and molecular features, such as more luminal B and fewer TNBC subtypes, less TP53 mutations but more PIK3CA mutation in the HER2-low tumors than in the HER2-zero tumors. The enriched mutations in genes involved in the PI3K-Akt pathway suggest a potential therapeutic strategy of combining chemotherapy and PI3K-related targeted therapy to improve the pCR rate in women with HER2-low tumors, which have been successfully demonstrated in various clinical trials [34–36]. It might also be possible to add these inhibitors to ADC when treating the HER2-low patients with metastatic disease to enhance the overall response rates, which is 40% with T-DXd and 28% with SYD985 [23, 24, 33, 37]. Nonetheless, these speculations are only hypothesis-generating and needs further confirmation by rigidly designed clinical trials.
Consistent with our previous study and other existing reports [32, 38, 39], somatic mutational profiling using a 520-gene panel NGS showed that HER2-positive breast tumors had more frequent TP53 mutations, CDK12 amplifications, and RARA amplifications. It should be noted that in our cohort, 81% of HER2-positive tumors were detected with TP53 mutations while all CDK12 amplifications were concurrently detected with HER2 amplification. TP53 mutations, particularly in the DNA-binding domain spanning exons 5–8, results in the malfunctioning of DNA damage repair pathways and are the most frequently reported mutation in solid tumors, including breast cancer [29, 40]. CDK12 is located in chromosome 17q12, which is a neighboring gene of HER2, and hence is frequently amplified with HER2 [41, 42]. CDK12 regulates the alternative splicing of the genes involved in DNA damage response pathway and the pathogenesis of breast cancer [42, 43]. Consistently, our results demonstrate that the DNA damage response pathway and homologous recombination repair pathway were significantly altered with a higher frequency among HER2-positive breast tumors. In addition, our study showed that HER2-positive tumors also harbored the most frequent HER family genes mutations than the other two subgroups.
Our findings from the molecular data clustering showed that our breast cancer cohort can be classified into three clusters of differential gene mutation. Cluster 1 was enriched in TP53 mutations. Cluster 2 was enriched in PIK3CA mutation but lack TP53 mutation. Cluster 3 was enriched in copy number amplifications of various genes associated with the chromosome 11q13 loci, wherein coamplification of CCND1 and FGF3/4/19 was the most common. Chromosome 11q13 locus is considered gene-rich and is one of the most commonly aberrant chromosome loci in breast cancer [44]. Amplification of chromosome 11q13 has been reported in up to 15% of patients with breast cancer and is associated with endocrine therapy resistance and poor prognosis, particularly for ER-positive and invasive lobular carcinomas [44–46]. CCND1 amplification has been associated with immunosuppression of the tumor microenvironment and poor outcomes with immune checkpoint inhibitors [47]. Furthermore, the amplification of chromosome 11q13 locus is commonly observed to be associated with amplifications at chromosome 8p12, particularly FGFR1, and appear to be mutually exclusive from HER2 amplification [48].
Our study has some inherent limitations. Our study was carried out in a single institution, which could introduce patient selection bias. Our study only included tumor samples sequenced using a 520-gene panel, which might introduce patient sampling bias, such as the higher HER2 positive rate of our cohort despite being unselected for clinical or pathological features (~38.6%). However, a study with a larger cohort of unselected invasive breast cancer (n = 12,467) have reported an average HER2-positive rate of 24.7%, ranging 13.7 to 35.7% across 19 different institutions from China [4]. Hence, we consider that the HER2-positive rate of our cohort is within the acceptable range, albeit being at the higher end of the limit. Since both the HER2-low and HER2-zero subgroups were classified as HER2-negative, our analysis was unaffected by the HER2-positive rate except for a smaller subgroup when analyzing HR status and IHC-based molecular subtypes. Since the study was not selected for clinicopathological features, our cohort did not include an adequate sample size to perform subgroup matching based on HR or IHC-based molecular subtype, which limited our findings to determine whether the difference in molecular features observed for HER2-low tumors were driven by differences in clinicopathological features. Moreover, only the genomic profiles of the breast tumors were analyzed and lacked other omics data such as gene expression profiling or protein expression, which limits our conclusions on the signaling pathways involved in various HER2 subgroups. Our cohort lacked treatment outcomes for HER2-antibody-drug conjugates, PI3K inhibitors, and immune checkpoint inhibitors. Due to the retrospective nature of our study, the treatment and survival data of our cohort were incomplete, with some patients lost to follow-up. The survival data of this cohort remains immature (approximately 10% maturity); hence, a longer follow-up period is necessary for definitive analysis. Nevertheless, our observations suggest that HER2 low breast tumors are clinically and molecularly different from HER2-zero tumors. Our findings are hypothesis-generating. Further investigations are warranted to elucidate and establish the distinct features of HER2 low breast tumors.
Conclusion
Our study demonstrates the complex genetic heterogeneity across breast tumors with varied HER2 status and identifies potential oncogenic mechanism that drives HER2-low breast tumor. Our findings indicate that patients with either IHC 1+ or IHC 2+ with FISH-negative results have distinct clinical and mutational features as compared with HER2-zero and HER2-positive breast tumors. Moreover, the accumulating evidence that HER2-low is clinically and biologically distinct from HER2-zero raises the need to explore diagnostic techniques and personalized therapeutic approaches that could identify and potentially benefit the subset of patients with HER2-low breast cancer.
Supplementary Information
Additional file 1: Table S1. Mutations in other members of the HER family detected from our cohort. Figure S1. Somatic mutation landscape of the cohort. Figure S2. Comparison of mutational profile of the HER2 subgroups. Figure S3. Distribution of TP53 mutations among the HER2 subgroups. Figure S4. Distribution of PIK3CA mutations among the HER2 subgroups. Figure S5. Genes frequently mutated across HER2 subgroups according to hormone receptor (HR) status. Figure S6. Comparison of TP53 (A) and PIK3CA (B) mutation rates across HER2 subgroups according to hormone receptor (HR) status. Figure S7. Genes frequently mutated across immunohistochemistry-based molecular subgrouping between HER2-low and HER2-zero subgroups. TNBC, triple negative breast cancer. Figure S8. Comparison of TP53 (A) and PIK3CA (B) mutation rates between HER2-zero and HER2-low subgroups according to immunohistochemistry-based molecular subgrouping. Figure S9. Comparison of genes (top 11) and signaling pathways (top 5) differentially mutated between HER2-low and HER2-zero subgroups according to immunohistochemistry-based molecular subgrouping: A. triple negative breast cancer (TNBC), B. luminal A, and C. luminal B. Figure S10. Tumor mutational burden (TMB) were comparable across HER2 subgroups. TMB were not statistically different among patients with HER2-zero (IHC 0), HER2-low (IHC1+ or IHC 2+/FISH-negative), and HER2-positive (IHC 3+ or IHC 2+/FISH-positive) breast tumors. Figure S11. Distribution of ERBB/HER family mutations according to HER2 status. Figure S12. Molecular clustering across hormone receptor (HR) status (A) and immunohistochemistry-based molecular subgrouping (B). Figure S13. Pathological complete response (pCR) rate is comparable between HER2-low and HER2-zero subgroups across hormone receptor (HR) status (A-B) and immunohistochemistry-based molecular subgrouping (C-E). Figure S14. Disease-free survival (DFS) was comparable between women with HER2-low and HER2-zero tumors as shown by Kaplan-Meier survival plot of the whole cohort (A) and when considering only the relapsed patients per group (B). Figure S15. Disease-free survival (DFS) was comparable between HER2-zero and HER2-low subgroups regardless of pathological complete response (pCR) status. Figure S16. Disease-free survival (DFS) was comparable between HER2-zero and HER2-low subgroups regardless of hormone receptor (HR) status. Figure S17. Disease-free survival (DFS) was comparable between HER2-zero and HER2-low subgroups across immunohistochemistry-based molecular subgrouping.
Acknowledgements
We thank all the patients and their families for participation in our study. We would also like to appreciate our colleagues, nurses, and other health workers at the Guangdong Provincial People’s Hospital. We would also like to acknowledge the staff of Burning Rock Biotech, in particular Drs. Analyn Lizaso, Xinze Lv, Ting Hou, Jing Liu, Min Li, Jianxing Xiang, Zhou Zhang, and Junjun Li of Burning Rock Biotech for their active support.
Abbreviations
- ADC
Antibody drug conjugate
- ASCO
American Society of Clinical Oncology
- CAP
College of American Pathologists
- CEP17
Chromosome 17 centromere
- CNVs
Copy number variations
- DFS
Disease-free survival
- ER
Estrogen receptor
- ERBB2
Erb-B2 receptor tyrosine kinase 2
- FFPE
Formalin-fixed paraffin-embedded
- FISH
Fluorescence in situ hybridization
- GDPH
Guangdong Provincial People’s Hospital
- HER2
Human epidermal growth factor receptor 2
- HR
Hormone receptor
- IHC
Immunohistochemistry
- Indels
Insertion-deletions
- NGS
Next-generation sequencing
- NMF
Non-negative matrix factorization
- pCR
Pathological complete response
- PD
Progressive disease
- PR
Progesterone receptor
- SNP
Single-nucleotide polymorphism
- SNVs
Single-nucleotide variations
- SVs
Structural variations
- TKI
Tyrosine kinase inhibitor
- TMB
Tumor mutation burden
Authors’ contributions
GZ, CR, CL, and NL conceived and designed the study. GZ drafted the manuscript. GZ, YW, and BC acquired funding. GZ, CR, CL, YW, BC, LW, MJ, KL, HM, LC, XQC, JL, GW, YL, and YZ participated in the data collection, data analysis and interpretation, manuscript writing, manuscript editing, and review. CB provided critical comments and suggestions. GZ and NL supervised the study. All authors reviewed the manuscript and approved the final version of the manuscript.
Funding
This work was supported by funds from the Natural Science Foundation of Guangdong Province (grant number: 2018A030313292 to GZ), Research Funds from Guangzhou Municipal Science and Technology Project (grant number: 201804010430 to GZ), National Natural Science Foundation of China (grant number: 81602645 to YW), and High-level Hospital Construction Project (grant number: DFJH201921 to BC). The funding agency had no role in the conceptualization, design, patient recruitment, data collection, analysis, interpretation, decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets analyzed for this study are available in the National Omics Data Encyclopedia (NODE), hosted by the Bio-Med Big Data Center (BMDC) available as accession number OEP001295.
Declarations
Ethics approval and consent to participate
The study protocol was reviewed and approved by the ethics committee of the Guangdong Provincial People’s Hospital (approval no. GDREC2014122H). The study was performed in accordance with the ethics standards of the participating institutions and the Declaration of Helsinki. All the patients provided written informed consent for NGS-based genomic testing.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Guochun Zhang, Chongyang Ren and Cheukfai Li contributed equally to this work.
References
- 1.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 2.Seshadri R, Firgaira FA, Horsfall DJ, McCaul K, Setlur V, Kitchen P. Clinical significance of HER-2/neu oncogene amplification in primary breast cancer. The south Australian breast cancer study group. J Clin Oncol. 1993;11(10):1936–1942. doi: 10.1200/JCO.1993.11.10.1936. [DOI] [PubMed] [Google Scholar]
- 3.Press MF, Bernstein L, Thomas PA, Meisner LF, Zhou JY, Ma Y, et al. HER-2/neu gene amplification characterized by fluorescence in situ hybridization: poor prognosis in node-negative breast carcinomas. J Clin Oncol. 1997;15(8):2894–2904. doi: 10.1200/JCO.1997.15.8.2894. [DOI] [PubMed] [Google Scholar]
- 4.Shui R, Liang X, Li X, Liu Y, Li H, Xu E, et al. Hormone receptor and human epidermal growth factor receptor 2 detection in invasive breast carcinoma: a retrospective study of 12,467 patients from 19 Chinese representative clinical centers. Clin Breast Cancer. 2020;20(1):e65–e74. doi: 10.1016/j.clbc.2019.07.013. [DOI] [PubMed] [Google Scholar]
- 5.Slamon D, Eiermann W, Robert N, Pienkowski T, Martin M, Press M, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365(14):1273–1283. doi: 10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geyer CE, Forster J, Lindquist D, Chan S, Romieu CG, Pienkowski T, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med. 2006;355(26):2733–2743. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- 7.Verma S, Miles D, Gianni L, Krop IE, Welslau M, Baselga J, et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N Engl J Med. 2012;367(19):1783–1791. doi: 10.1056/NEJMoa1209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fehrenbacher L, Cecchini RS, Geyer CE, Jr, Rastogi P, Costantino JP, Atkins JN, et al. NSABP B-47/NRG oncology phase III randomized trial comparing adjuvant chemotherapy with or without Trastuzumab in high-risk invasive breast cancer negative for HER2 by FISH and with IHC 1+ or 2. J Clin Oncol. 2020;38(5):444–453. doi: 10.1200/JCO.19.01455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Press MF, Finn RS, Cameron D, Di Leo A, Geyer CE, Villalobos IE, et al. HER-2 gene amplification, HER-2 and epidermal growth factor receptor mRNA and protein expression, and lapatinib efficacy in women with metastatic breast cancer. Clin Cancer Res. 2008;14(23):7861–7870. doi: 10.1158/1078-0432.CCR-08-1056. [DOI] [PubMed] [Google Scholar]
- 10.Burris HA, 3rd, Rugo HS, Vukelja SJ, Vogel CL, Borson RA, Limentani S, et al. Phase II study of the antibody drug conjugate trastuzumab-DM1 for the treatment of human epidermal growth factor receptor 2 (HER2)-positive breast cancer after prior HER2-directed therapy. J Clin Oncol. 2011;29(4):398–405. doi: 10.1200/JCO.2010.29.5865. [DOI] [PubMed] [Google Scholar]
- 11.Krop IE, LoRusso P, Miller KD, Modi S, Yardley D, Rodriguez G, et al. A phase II study of trastuzumab emtansine in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer who were previously treated with trastuzumab, lapatinib, an anthracycline, a taxane, and capecitabine. J Clin Oncol. 2012;30(26):3234–3241. doi: 10.1200/JCO.2011.40.5902. [DOI] [PubMed] [Google Scholar]
- 12.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014;138(2):241–256. doi: 10.5858/arpa.2013-0953-SA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25(1):118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 14.Wolff AC, Hammond MEH, Allison KH, Harvey BE, Mangu PB, Bartlett JMS, et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J Clin Oncol. 2018;36(20):2105–2122. doi: 10.1200/JCO.2018.77.8738. [DOI] [PubMed] [Google Scholar]
- 15.Hoda RS, Brogi E, Xu J, Ventura K, Ross DS, Dang C, et al. Impact of the 2018 American Society of Clinical Oncology/College of American Pathologists HER2 guideline updates on HER2 assessment in breast cancer with equivocal HER2 immunohistochemistry results with focus on cases with HER2/CEP17 ratio <2.0 and average HER2 copy number >/=4.0 and <6.0. Arch Pathol Lab Med. 2020;144(5):597–601. doi: 10.5858/arpa.2019-0307-OA. [DOI] [PubMed] [Google Scholar]
- 16.Crespo J, Sun H, Wu J, Ding QQ, Tang G, Robinson MK, et al. Rate of reclassification of HER2-equivocal breast cancer cases to HER2-negative per the 2018 ASCO/CAP guidelines and response of HER2-equivocal cases to anti-HER2 therapy. PLoS One. 2020;15(11):e0241775. doi: 10.1371/journal.pone.0241775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarantino P, Hamilton E, Tolaney SM, Cortes J, Morganti S, Ferraro E, et al. HER2-low breast cancer: pathological and clinical landscape. J Clin Oncol. 2020;38(17):1951–1962. doi: 10.1200/JCO.19.02488. [DOI] [PubMed] [Google Scholar]
- 18.Schettini F, Chic N, Braso-Maristany F, Pare L, Pascual T, Conte B, et al. Clinical, pathological, and PAM50 gene expression features of HER2-low breast cancer. NPJ Breast Cancer. 2021;7(1):1. doi: 10.1038/s41523-020-00208-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agostinetto E, Rediti M, Fimereli D, Debien V, Piccart M, Aftimos P, et al. HER2-low breast cancer: molecular characteristics and prognosis. Cancers (Basel) 2021;13(11):2824. doi: 10.3390/cancers13112824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dieci MV, Miglietta F. HER2: a never ending story. Lancet Oncol. 2021;22(8):1051–1052. doi: 10.1016/S1470-2045(21)00349-1. [DOI] [PubMed] [Google Scholar]
- 21.Denkert C, Seither F, Schneeweiss A, Link T, Blohmer JU, Just M, et al. Clinical and molecular characteristics of HER2-low-positive breast cancer: pooled analysis of individual patient data from four prospective, neoadjuvant clinical trials. Lancet Oncol. 2021;22(8):1151–1161. doi: 10.1016/S1470-2045(21)00301-6. [DOI] [PubMed] [Google Scholar]
- 22.Hyman DM, Piha-Paul SA, Won H, Rodon J, Saura C, Shapiro GI, et al. HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature. 2018;554(7691):189–194. doi: 10.1038/nature25475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modi S, Saura C, Yamashita T, Park YH, Kim SB, Tamura K, et al. Trastuzumab Deruxtecan in previously treated HER2-positive breast cancer. N Engl J Med. 2020;382(7):610–621. doi: 10.1056/NEJMoa1914510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Tsurutani J, Takahashi S, Iwata H, Krop IE, Redfern C, et al. Trastuzumab deruxtecan (DS-8201a) in patients with advanced HER2-positive breast cancer previously treated with trastuzumab emtansine: a dose-expansion, phase 1 study. Lancet Oncol. 2019;20(6):816–826. doi: 10.1016/S1470-2045(19)30097-X. [DOI] [PubMed] [Google Scholar]
- 25.Modi S, Park H, Murthy RK, Iwata H, Tamura K, Tsurutani J, et al. Antitumor activity and safety of Trastuzumab Deruxtecan in patients with HER2-low-expressing advanced breast cancer: results from a phase Ib study. J Clin Oncol. 2020:JCO1902318. [DOI] [PMC free article] [PubMed]
- 26.Mittendorf EA, Lu B, Melisko M, Price Hiller J, Bondarenko I, Brunt AM, et al. Efficacy and safety analysis of Nelipepimut-S vaccine to prevent breast cancer recurrence: a randomized, multicenter, phase III clinical trial. Clin Cancer Res. 2019;25(14):4248–4254. doi: 10.1158/1078-0432.CCR-18-2867. [DOI] [PubMed] [Google Scholar]
- 27.Meric-Bernstam F, Beeram M, Mayordomo JI, Hanna DL, Ajani JA, Blum Murphy MA, et al. Single agent activity of ZW25, a HER2-targeted bispecific antibody, in heavily pretreated HER2-expressing cancers. J Clin Oncol. 2018;36(15_suppl):2500. [Google Scholar]
- 28.Weisser N, Wickman G, Davies R, Rowse G. Preclinical development of a novel biparatopic HER2 antibody with activity in low to high HER2 expressing cancers. Cancer Res. 2017;77(13):Abstract nr 31. [Google Scholar]
- 29.Zhang G, Wang Y, Chen B, Guo L, Cao L, Ren C, et al. Characterization of frequently mutated cancer genes in Chinese breast tumors: a comparison of Chinese and TCGA cohorts. Ann Transl Med. 2019;7(8):179. doi: 10.21037/atm.2019.04.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang P, Tse GM. Immunohistochemical surrogates for molecular classification of breast carcinoma: a 2015 update. Arch Pathol Lab Med. 2016;140(8):806–814. doi: 10.5858/arpa.2015-0133-RA. [DOI] [PubMed] [Google Scholar]
- 31.Chen B, Zhang G, Li X, Ren C, Wang Y, Li K, et al. Comparison of BRCA versus non-BRCA germline mutations and associated somatic mutation profiles in patients with unselected breast cancer. Aging (Albany NY) 2020;12(4):3140–3155. doi: 10.18632/aging.102783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen B, Zhang G, Wei G, Wang Y, Guo L, Lin J, et al. Heterogeneity of genomic profile in patients with HER2-positive breast cancer. Endocr Relat Cancer. 2020;27(3):153–162. doi: 10.1530/ERC-19-0414. [DOI] [PubMed] [Google Scholar]
- 33.Banerji U, van Herpen CML, Saura C, Thistlethwaite F, Lord S, Moreno V, et al. Trastuzumab duocarmazine in locally advanced and metastatic solid tumours and HER2-expressing breast cancer: a phase 1 dose-escalation and dose-expansion study. Lancet Oncol. 2019;20(8):1124–1135. doi: 10.1016/S1470-2045(19)30328-6. [DOI] [PubMed] [Google Scholar]
- 34.Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019;380(20):1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 35.Xing Y, Lin NU, Maurer MA, Chen H, Mahvash A, Sahin A, et al. Phase II trial of AKT inhibitor MK-2206 in patients with advanced breast cancer who have tumors with PIK3CA or AKT mutations, and/or PTEN loss/PTEN mutation. Breast Cancer Res. 2019;21(1):78. doi: 10.1186/s13058-019-1154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J, Nie J, Ma X, Wei Y, Peng Y, Wei X. Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol Cancer. 2019;18(1):26. doi: 10.1186/s12943-019-0954-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Modi S, Park H, Murthy RK, Iwata H, Tamura K, Tsurutani J, et al. Antitumor activity and safety of Trastuzumab Deruxtecan in patients with HER2-low-expressing advanced breast cancer: results from a phase Ib study. J Clin Oncol. 2020;38(17):1887–1896. doi: 10.1200/JCO.19.02318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross DS, Zehir A, Cheng DT, Benayed R, Nafa K, Hechtman JF, et al. Next-generation assessment of human epidermal growth factor receptor 2 (ERBB2) amplification status: clinical validation in the context of a hybrid capture-based, comprehensive solid tumor genomic profiling assay. J Mol Diagn. 2017;19(2):244–254. doi: 10.1016/j.jmoldx.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Press MF, Seoane JA, Curtis C, Quinaux E, Guzman R, Sauter G, et al. Assessment of ERBB2/HER2 status in HER2-equivocal breast cancers by FISH and 2013/2014 ASCO-CAP guidelines. JAMA Oncol. 2019;5(3):366–375. doi: 10.1001/jamaoncol.2018.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang S, Hu L, Wu Z, Chen Z, Liu S, Xu X, et al. CDK12: a potent target and biomarker for human cancer therapy. Cells. 2020;9(6):1483. doi: 10.3390/cells9061483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naidoo K, Wai PT, Maguire SL, Daley F, Haider S, Kriplani D, et al. Evaluation of CDK12 protein expression as a potential novel biomarker for DNA damage response-targeted therapies in breast cancer. Mol Cancer Ther. 2018;17(1):306–315. doi: 10.1158/1535-7163.MCT-17-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tien JF, Mazloomian A, Cheng SG, Hughes CS, Chow CCT, Canapi LT, et al. CDK12 regulates alternative last exon mRNA splicing and promotes breast cancer cell invasion. Nucleic Acids Res. 2017;45(11):6698–6716. doi: 10.1093/nar/gkx187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ormandy CJ, Musgrove EA, Hui R, Daly RJ, Sutherland RL. Cyclin D1, EMS1 and 11q13 amplification in breast cancer. Breast Cancer Res Treat. 2003;78(3):323–335. doi: 10.1023/a:1023033708204. [DOI] [PubMed] [Google Scholar]
- 45.Borg A, Sigurdsson H, Clark GM, Ferno M, Fuqua SA, Olsson H, et al. Association of INT2/HST1 coamplification in primary breast cancer with hormone-dependent phenotype and poor prognosis. Br J Cancer. 1991;63(1):136–142. doi: 10.1038/bjc.1991.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Courjal F, Cuny M, Simony-Lafontaine J, Louason G, Speiser P, Zeillinger R, et al. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups. Cancer Res. 1997;57(19):4360–4367. [PubMed] [Google Scholar]
- 47.Chen Y, Huang Y, Gao X, Li Y, Lin J, Chen L, et al. CCND1 amplification contributes to immunosuppression and is associated with a poor prognosis to immune checkpoint inhibitors in solid tumors. Front Immunol. 2020;11(1620):1620. doi: 10.3389/fimmu.2020.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karlseder J, Zeillinger R, Schneeberger C, Czerwenka K, Speiser P, Kubista E, et al. Patterns of dna amplification at band q13 of chromosome 11 in human breast cancer. Genes Chromosom Cancer. 1994;9(1):42–48. doi: 10.1002/gcc.2870090108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Mutations in other members of the HER family detected from our cohort. Figure S1. Somatic mutation landscape of the cohort. Figure S2. Comparison of mutational profile of the HER2 subgroups. Figure S3. Distribution of TP53 mutations among the HER2 subgroups. Figure S4. Distribution of PIK3CA mutations among the HER2 subgroups. Figure S5. Genes frequently mutated across HER2 subgroups according to hormone receptor (HR) status. Figure S6. Comparison of TP53 (A) and PIK3CA (B) mutation rates across HER2 subgroups according to hormone receptor (HR) status. Figure S7. Genes frequently mutated across immunohistochemistry-based molecular subgrouping between HER2-low and HER2-zero subgroups. TNBC, triple negative breast cancer. Figure S8. Comparison of TP53 (A) and PIK3CA (B) mutation rates between HER2-zero and HER2-low subgroups according to immunohistochemistry-based molecular subgrouping. Figure S9. Comparison of genes (top 11) and signaling pathways (top 5) differentially mutated between HER2-low and HER2-zero subgroups according to immunohistochemistry-based molecular subgrouping: A. triple negative breast cancer (TNBC), B. luminal A, and C. luminal B. Figure S10. Tumor mutational burden (TMB) were comparable across HER2 subgroups. TMB were not statistically different among patients with HER2-zero (IHC 0), HER2-low (IHC1+ or IHC 2+/FISH-negative), and HER2-positive (IHC 3+ or IHC 2+/FISH-positive) breast tumors. Figure S11. Distribution of ERBB/HER family mutations according to HER2 status. Figure S12. Molecular clustering across hormone receptor (HR) status (A) and immunohistochemistry-based molecular subgrouping (B). Figure S13. Pathological complete response (pCR) rate is comparable between HER2-low and HER2-zero subgroups across hormone receptor (HR) status (A-B) and immunohistochemistry-based molecular subgrouping (C-E). Figure S14. Disease-free survival (DFS) was comparable between women with HER2-low and HER2-zero tumors as shown by Kaplan-Meier survival plot of the whole cohort (A) and when considering only the relapsed patients per group (B). Figure S15. Disease-free survival (DFS) was comparable between HER2-zero and HER2-low subgroups regardless of pathological complete response (pCR) status. Figure S16. Disease-free survival (DFS) was comparable between HER2-zero and HER2-low subgroups regardless of hormone receptor (HR) status. Figure S17. Disease-free survival (DFS) was comparable between HER2-zero and HER2-low subgroups across immunohistochemistry-based molecular subgrouping.
Data Availability Statement
The datasets analyzed for this study are available in the National Omics Data Encyclopedia (NODE), hosted by the Bio-Med Big Data Center (BMDC) available as accession number OEP001295.